 Open Access
Open Access
ARTICLE
MED4 gene positively affects preadipocyte differentiation in Chinese red steppe cattle
1 Institute of Animal Biotechnology, Jilin Academy of Agricultural Sciences, Gongzhuling, 136100, China
2 Institute of Muscle Biology and Growth, Research Institute for Farm Animal Biology (FBN), Dummerstorf, 18196, Germany
3 Key Laboratory of Beef Cattle Genetics and Breeding, Ministry of Agriculture and Rural Affairs, Gongzhuling, 136100, China
* Corresponding Authors: YANG CAO. Email: ; YUMIN ZHAO. Email:
# These authors contributed equally to this work
(This article belongs to the Special Issue: Cellular Signal Transduction in Biological Activities)
BIOCELL 2023, 47(9), 2115-2123. https://doi.org/10.32604/biocell.2023.030364
Received 02 April 2023; Accepted 01 June 2023; Issue published 28 September 2023
Abstract
Background: The regulatory genes related to lipid metabolism affect the intramuscular fat (IMF) content and improve meat quality traits. Mediator Complex Subunit 4 (MED4), a vitamin D receptor protein, affects vitamin D, livestock growth, carcass traits, and triglyceride deposition. However, the physiological function of the MED4 gene on bovine adipocyte differentiation remains unknown. Methods: This study explored the function of the MED4 gene in preadipocyte differentiation in Chinese Red Steppe cattle. The overexpression plasmid and the interference sequences of the MED4 gene were constructed to detect the effects of the MED4 gene on adipogenesis and biomakers using quantitative polymerase chain reaction and western blotting. Results: The MED4 gene had significantly high expression during preadipocyte differentiation (p < 0.05). Overexpression of the MED4 gene increased the expression of the PPARγ gene, a preadipocyte differentiation biomarker, improved cellular lipid droplets and triglycerides accumulation, and positively accelerated adipocyte maturation (p < 0.05). Interference of the MED4 gene can negatively regulate preadipocyte differentiation. Conclusion: This study showed that the MED4 gene may affect the preadipocyte differentiation and adipogenesis in Chinese Red Steppe cattle by regulating the PPARγ gene.Keywords
Supplementary Material
Supplementary Material FileChinese Red Steppe cattle, a special breed with roughage tolerance, strong stress resistance, and high meat quality in China, is widely raised in northern China (Fang et al., 2021). As the economy and livestock industry continues to develop, higher meat quality is becoming more popular with consumers. However, the growth rate and meat quality of the Red Steppe cattle are not able to meet the rising demand, making it crucial to improve the growth rate and meat quality trait under keeping the original characteristics of Red Steppe cattle. The intramuscular fat (IMF) content significantly affects meat quality traits, such as taste, flavor, tenderness, juiciness, and nutrients (Hocquette et al., 2010; Ahmed et al., 2022). High IMF content stored in the muscle is also called marbling, which provides more fatty acids composition and significantly improves the meat trait, and is highly appealing to consumers (Wen et al., 2020; Yao et al., 2022). Genetics plays a vital role in IMF formation and accumulation under the same feeding condition; therefore, it is important to explore genes affecting adipocyte differentiation and lipid metabolism (Estany et al., 2017; Miao et al., 2021).
IMF comprises white adipose tissue, an endocrine organ that stores energy in the form of triglycerides (TG), regulates the endocrine balance, and affects insulin sensitivity (Wang et al., 2013; Kawai et al., 2021). Preadipocyte derived from mesenchymal stem cells near the stromal vascular fraction (SVF) differentiate into mature adipocytes; this process is regulated by various biomarkers, for example, peroxisome proliferator-activated receptorγ (PPARγ) (Mota de Sá et al., 2017; Wu et al., 2022). PPARγ, a member of the nuclear hormone receptor superfamily, is a transcription factor that plays a vital and indispensable role in initiating and inducing adipocyte differentiation, which fails if PPARγ is not expressed (Yang et al., 2018).
A mediator complex, comprising mediator complex subunit 4 (MED4) and another subunit, regulates the transcriptional function and influences the proliferation and differentiation of the cells (Verger et al., 2019). Some studies have shown that the MED4 gene affects poultry growth and carcass traits, and regulates IMF deposition in livestock (Zhang et al., 2021). However, the function of the MED4 gene and its regulatory mechanism in bovine preadipocytes are not fully elucidated. This study explored the consequences of the MED4 gene on adipogenesis, biomarker gene, lipid droplets, and TG accumulation in Chinese Red Steppe cattle. The study may provide a candidate gene or reference for improving the meat quality in cattle.
Male healthy Chinese Red Steppe calves at 2 months of age were obtained from the Jilin Academy of Agricultural Sciences and slaughtered for preadipocyte isolation. The animal experiment strictly complied with ethical requirements and operating rules. All animal procedures, such as welfare and ethical issues, were approved by the Committee for the Ethics of Animal Experiments by AWEC 2019A05, 16 May 2019.
Preadipocyte isolation, proliferation, and maturation
Subcutaneous white adipose tissue of calve was isolated and washed with PBS (Sigma-Aldrich, St. Louis, MO, US) containing 6% penicillin/streptomycin (Sigma). The samples were sliced into 1 mm2 piece with sterile scissors and were digested with collagenase Type II (Sigma) and 0.25% trypsin (Sigma) for 1 h. The solution containing blocks and cells was filtered with a 40 μm filter to remove the undigested tissue pieces and obtain the preadipocytes. The cells were placed in a 37°C incubator (Thermo Fisher Scientific, Waltham, MA, United States) with 5% CO2 to culture in a complete culture medium consisting of 10% fetal bovine serum (Gemini Bio-Products, Woodland, CA, United States), 1% penicillin/streptomycin (Sigma), and Dulbecco’s modified Eagle medium-basic medium (Sigma), the fresh culture medium was replaced once 48 h. After culturing for 6 h, the preadipocyte proliferation and differentiation at the end of proliferation was induced into the mature adipocyte with the exogenous supply of inducer containing inducer I and II. Inducer I solution consisted of 10 mg/mL insulin (Sigma), 1.0 mM dexamethasone (Sigma), 0.5 mM IBMX (Sigma) and was added to the culture medium to treat the cells for 2 days. Inducer II solution consisted of 10 mg/mL insulin (Sigma) and was added to the culture medium to treat the cells for 2 days. The cells were cultured in a fresh culture medium until preadipocyte maturation.
The lipid droplets in the mature adipocytes can be stained by Oil Red O dye to red color, and it is a general way to verify adipocyte maturation. The adipocytes were washed with cold PBS three times, treated with 4% paraformaldehyde for 30 min in a 37°C incubator, and then washed again. Oil Red O solution containing Oil Red O dye and ddH2O in a volume ratio of 3:2 was used to treat cells for 30 min in an incubator. The mature adipocytes were photographed and analyzed under a microscope after being washed with PBS. Finally, an isopropanol solution was used to absorb the lipid droplets, then the solution was mixed with the lipid to determine the lipid droplet content at an OD value of 490 nm.
Construction of overexpression plasmid and synthesis of interference sequences
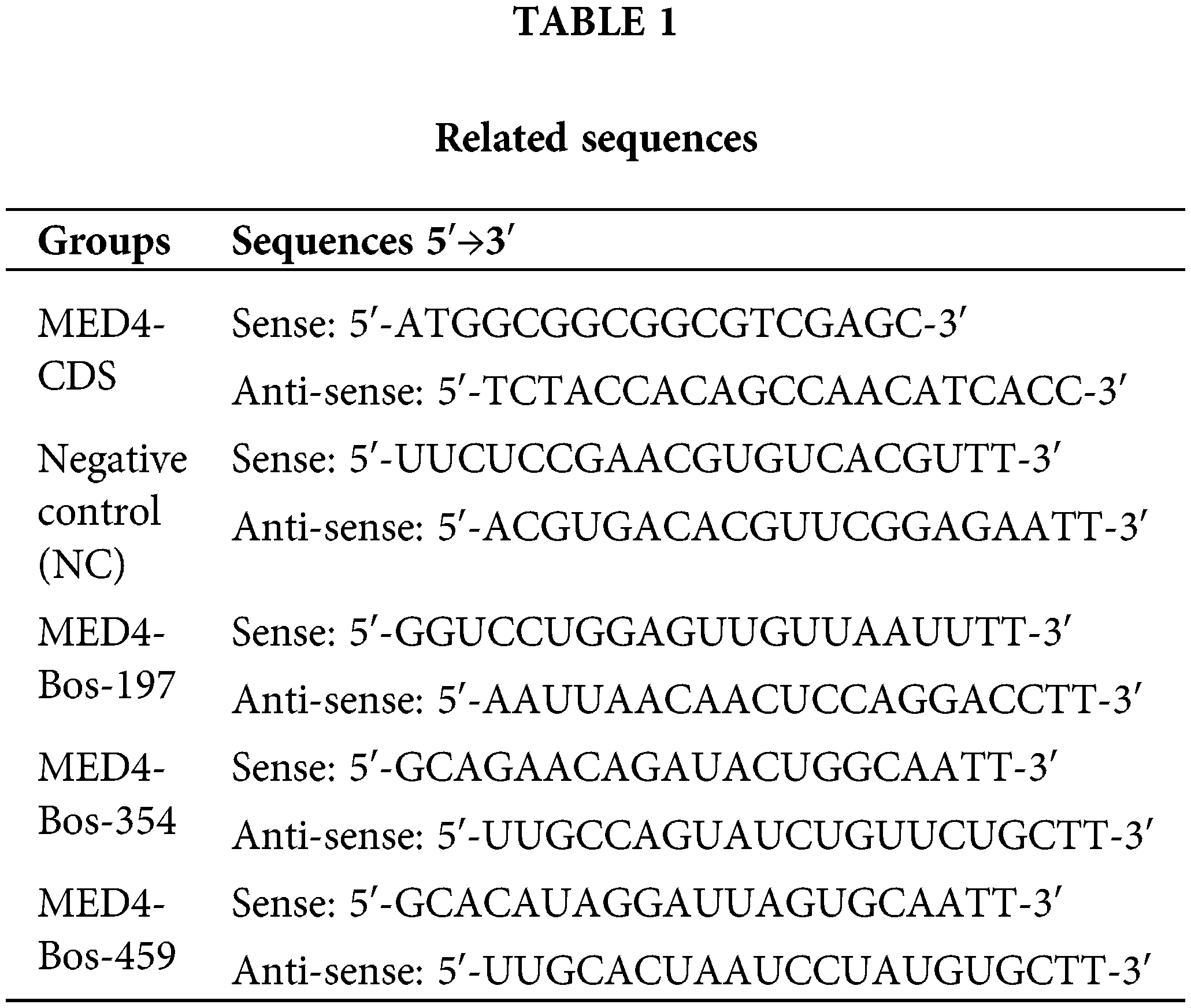
The primers to amplify the coding-sequence region (CDs) region of the MED4 gene were designed according to the bovine sequences (NM_001034486.1) available on the NCBI website. Overexpression plasmid pEX4 vector was used to construct the recombination vector containing MED4 CDs sequence at XhoI and EcoRI restriction endonuclease, and pEX4 empty plasmid was the negative control. Based on the sequence of MED4 mRNA, three small interference RNA sequences and negative control were designed and synthesized by GenePharma Company, Shanghai (Table 1). The gel image mapping the CDs region of MED4 gene and sequencing comparison results are showed in the Suppl. Fig. S1.
Transfection and verification of transformants
The plasmid or interference sequences were transfected into the preadipocytes when the cells fused 70% culture dishes. Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA, USA) reagent gets the room temperature before use. One hundred picomoles of interference sequences, 5 μL lip2 000, and 200 μL Opti-MEM (Invitrogen, Carlsbad, CA, USA) were mixed well for 10 min. The mixture was added into the cells with 1.8 mL fresh medium for 6 h and replaced with a fresh complete culture medium. Transfection efficiency was determined by quantitative polymerase chain reaction (qPCR) test after 48 h. FuGENE reagent (Roche Applied Science, Indianapolis, IN, USA) was used to mix with 400 μL Opti-MEM, 3 ng plasmid; it was placed at room temperature for 20 min and added into the cells for 48 h, the transfection efficiency was determined by qPCR test.
RNA extraction and verification through the quantitative polymerase chain reaction
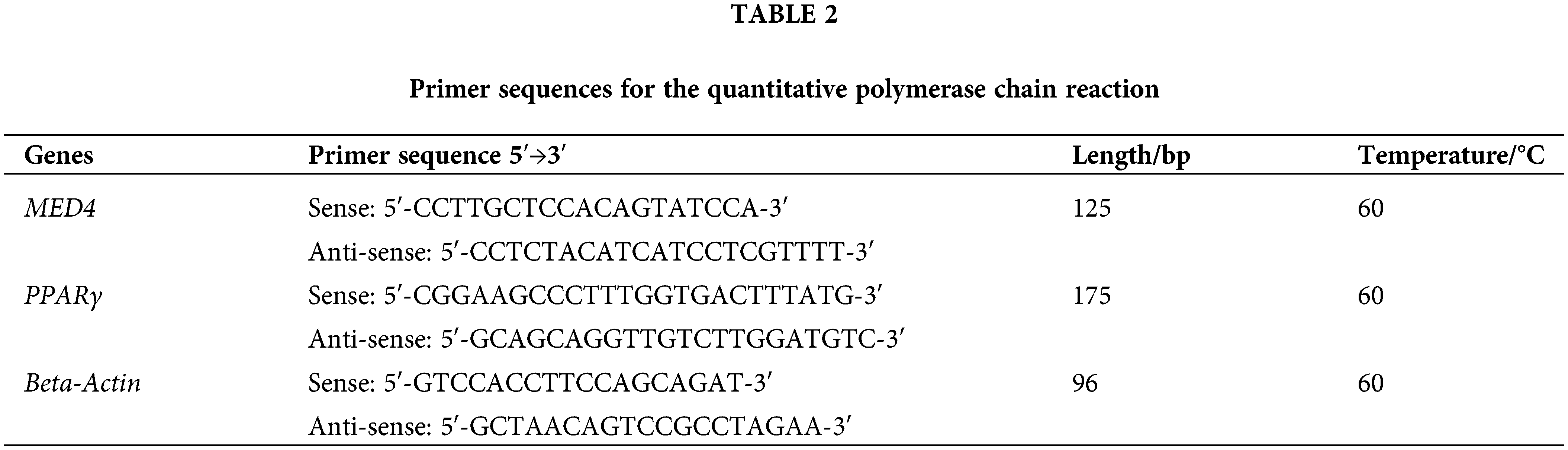
TRIzol reagent (Thermo Fisher Scientific) was used to extract total cellular RNA following the procedure. RNA contamination and degradation were validated via 1% agarose gels, and concentration was detected via a Quawell Q5000 spectrophotometer (Quawell Technology, San Jose, CA, USA). The reverse transcription kit (Takara, Japan) reversed the total RNA into cDNA. The reaction volume includes 2 μL of 5 × PrimeScript RT Master Mix, 500 ng of total RNA, and RNase Free ddH2O up to 10 μL. Reaction condition: 37°C for 15 min, 85°C for 5 s, and the PCR products were normalized with ddH2O and stored in a 4°C fridge. Roche LightCycler® 480 (Roche Applied Science), a quantitative fluorescent PCR device, detected the relative expression of the mRNA. Beta-actin was an internal control. The reaction volume: 10 μL of 2 × Light Cycler 480 SYBR Green I Master, 8 μL of ddH2O, 1 μL of cDNA, and 0.5 μL of sense and anti-sense primers. The reaction condition: 40 cycles of 95°C for 5 min, 95°C for 10 s, 60°C for 15 s, and 72°C for 20 s, followed by 95°C for 5 s, 65°C for 1 min, and 40°C for 10 s. The detailed information on primers was showed in Table 2.
Triglyceride content determination
Cell TG content was determined using a Triglyceride kit (Prilax, Beijing, China) following the manufacturer’s procedure. Clean mature adipocytes without medium were digested with 0.25% trypsin (Sigma-Aldrich), and the mixture was centrifuged at 1500 rpm for 20 min. The sediments were digested for 10 min with lysis solution, heated at 70°C for 10 min, then centrifuged at 2000 rpm for 5 min. The OD of the enzyme-labeled supernatant was detected using an instrument at 550 nm wavelength and TG content was calculated.
Total protein from the cells was extracted using a Protein extraction kit (Solarbio, Shanghai, China). Adipocytes without medium were lysed for 25 min on ice, and the mixture was transferred into a tube and centrifuged at 12000 rpm at 4°C for 30 min to collect the protein. The BCA assay kit was used to determine the protein concentration. All proteins of samples were normalized with those in the lysate solution. The proteins were heated at 99°C for 8 min and mixed with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) buffer (Beyotime, Jiangsu, China). The upper and lower gels were made with the PAGE Gel Fast Preparation kit (EpiZyme, Shanghai, China). Thirty micrograms of protein in each sample and 6 μL standard marker (Thermo Fisher Scientific) were loaded to the gel wells and separated by electrophoresis at 80 V for 20 min. Then 120 V for 40 min, the gel with samples was transferred into a 0.45 mm Immobilon poly-vinylidene difluoride (PVDF) membrane (Millipore, Bedford, MA, USA) at 200 mA for 1 h. PVDF membrane was washed with TBST (Tris-HCl buffered saline with tween 20; Beyotime, Jiangsu, China), blocked with 5% skimmed milk for 2 h, then washed again, and incubated with antibody (dilution ratio at 1:1 500; Bioss Beijing, China) and placed overnight on a shaker at 4°C. The clean membrane was incubated with a secondary antibody (anti-rabbit or anti-mouse IgG-HRP, 1:5000) at room temperature for 2 h and washed again. ECL hypersensitive photoluminescence solution (Pripril, Beijing, China) was used for a chromogenic reaction with the proteins. Then, protein bands were determined using the ChemiScope 6000 Touch imaging system (Clinx Science Instruments, Shanghai, China), and quantified using the ImageJ software. β-actin was an internal reference protein.
SPSS 17.0 software was used for statistical analyses. Each experiment was repeated three times. All data was described as means ± SD. The results compared two groups by t-test, and multiple group comparisons by one-way ANOVA. The statistical significance levels were set at p < 0.05. The raw data and images are presented in the Suppl. Table S1 and Fig. S1.
Preadipocyte culture and differentiation
The preadipocytes were isolated and cultured in a normal incubator, and they started proliferating until full fusion in the plates. Then, the inducer solution was added to promote the preadipocyte differentiation into mature adipocytes. The whole process of differentiation took around 10 days. The mature adipocytes were shown in Fig. 1.
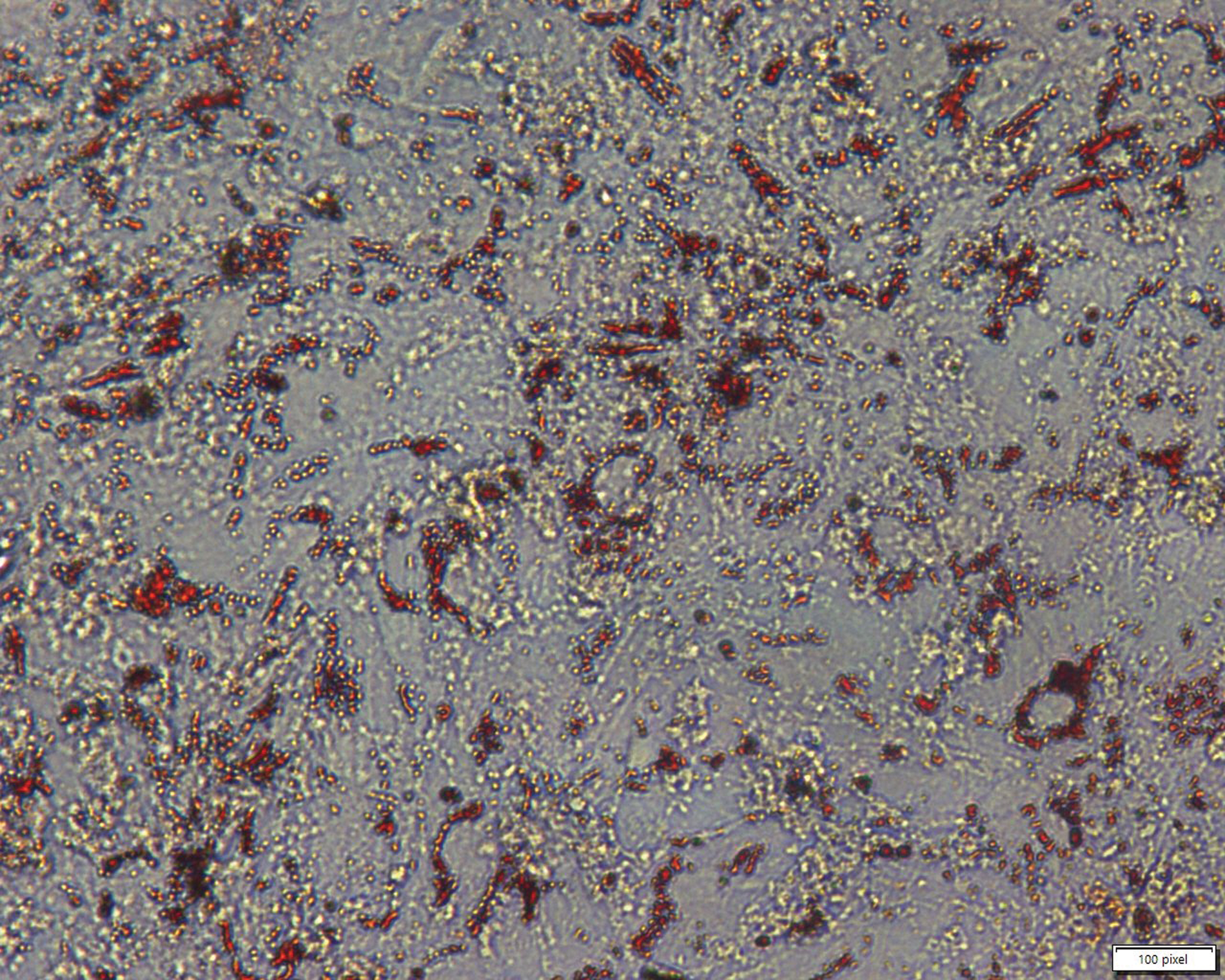
Figure 1: Morphology of mature adipocytes. Red dots represent lipid droplets in the adipocytes. Scales bar: 100 pixel.
Expression of the MED4 gene increased during preadipocyte differentiation
To investigate the changing pattern of the MED4 gene in preadipocyte differentiation. RNA in different periods (days 0, 2, 4, 6, 8) was extracted to verify the level of MED4 gene expression by qPCR. The inducer I solution was added to the preadipocytes on day 0 and inducer solution II was added on day 2. On days 4 and 6, fresh medium was added into the preadipocytes. Some small lipid droplets we noticed in the preadipocytes on day 8 and the majority of preadipocytes became mature adipocytes on day 10. These results showed that the expression of the MED4 gene was constant from day 0 to day 6 and significantly increased on day 8 (Fig. 2).
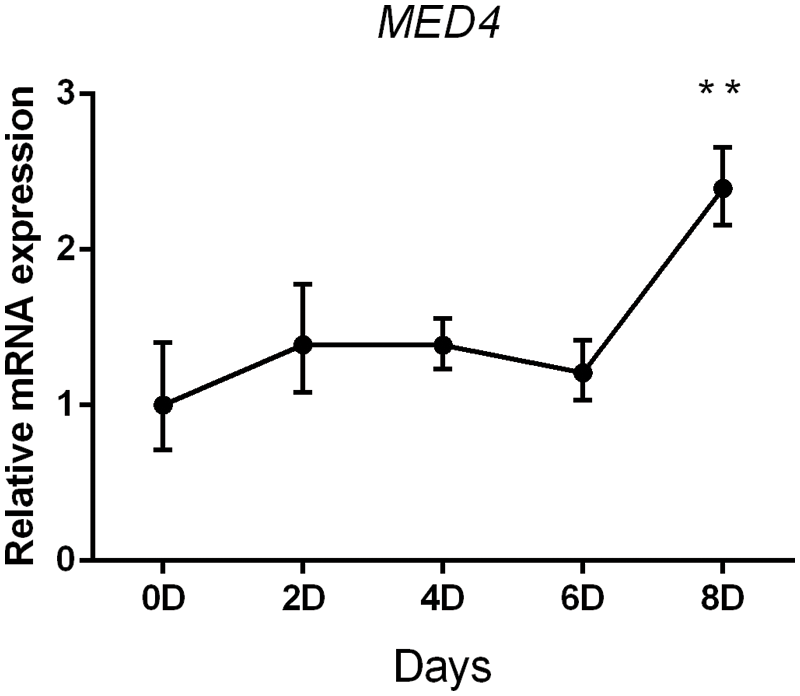
Figure 2: The change in the pattern of the MED4 gene expression during preadipocyte differentiation **p < 0.05.
Verification of transfection efficiency
After 48 h of transfection, the efficiency was determined by qPCR test. The recombination plasmid exhibited significantly increased expression of the MED4 gene compared to the empty plasmid (p < 0.05) (Fig. 3A). Subsequent assessment showed that three interference sequences had different effects, of which siR354 significantly decreased the MED4 gene expression compared to the negative control (p < 0.05) (Fig. 3B). The results of western blotting had a similar trend as the qPCR results (Figs. 3C–3F).
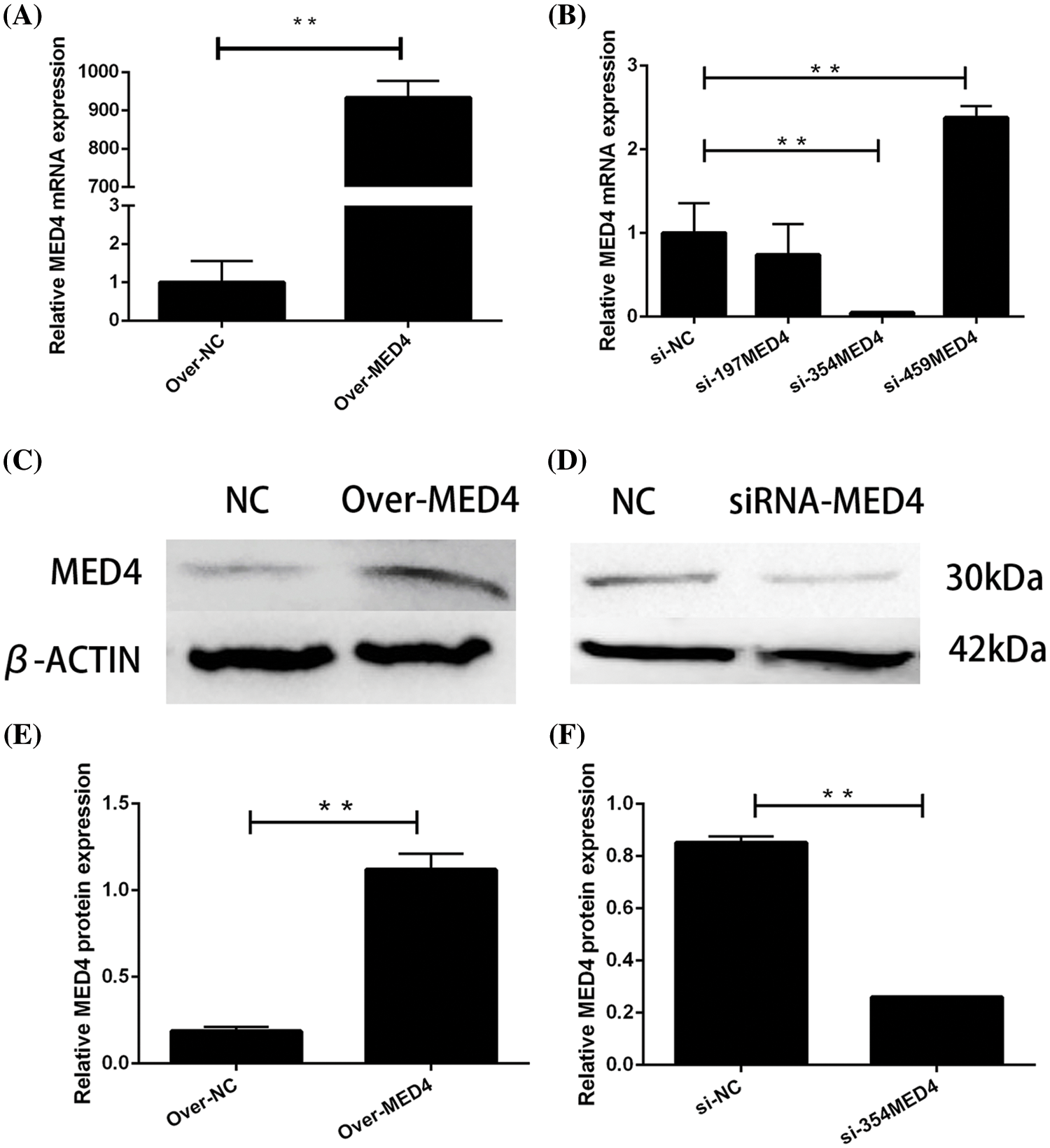
Figure 3: Verification of transfection efficiency. (A) Overexpression plasmid verification by quantitative polymerase chain reaction (qPCR); (B) verification of the interference sequence by qPCR; (C) overexpression plasmid verified by western blotting; (D) verification of the interference sequence by western blotting; (E and F) gray value analysis **p < 0.05.
MED4 overexpression promotes adipocyte differentiation
To assess the effect of the MED4 gene on adipocyte differentiation, the expression of differentiation-related biomarker, was detected by qPCR test in different periods (days 0, 4, 8), and the TG content and lipid droplets were measured on day 10. With the increase in the MED4 gene, the PPARγ gene expression significantly increased (p < 0.05). Meanwhile, the MED4 gene also improved the accumulation of the TG content and lipid droplets (p < 0.05) (Fig. 4) and may improve adipocyte differentiation by affecting the PPARγ gene expression.
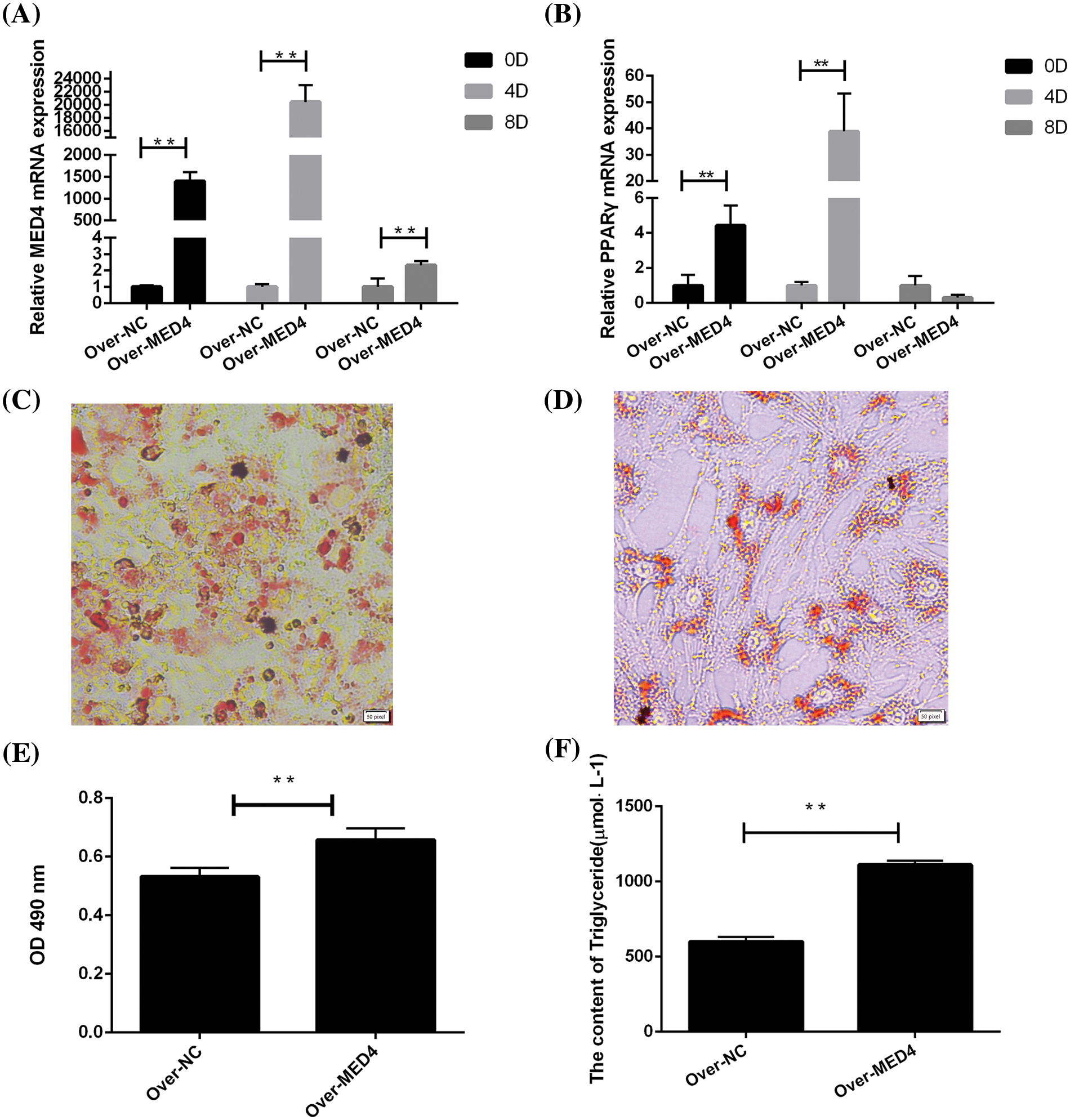
Figure 4: MED4 overexpression affects the differentiation of adipocytes. (A) MED4 gene expression for different periods; (B) PPARγ gene expression in the different periods; (C) oil red O staining in the MED4 overexpression group; (D) oil red O staining in the control group; scale bar: 50 pixel; (E) lipid droplets in the adipocytes test; (F) the triglyceride content test; **p < 0.05.
MED4 interference inhibits adipocyte differentiation
To investigate the effect of the MED4 gene on adipocyte differentiation. The differentiation-related biomarker expression in different periods (days 0, 4, 8) was detected by qPCR, and the TG content and lipid droplets were measured on day 10. Inhibition of the MED4 gene significantly reduced the PPARγ gene expression (p < 0.05) and decreased the OD value of TG content and lipid droplets (p < 0.05) (Fig. 5). Thus, the MED4 gene may inhibit adipocyte differentiation by affecting the expression of the PPARγ gene.
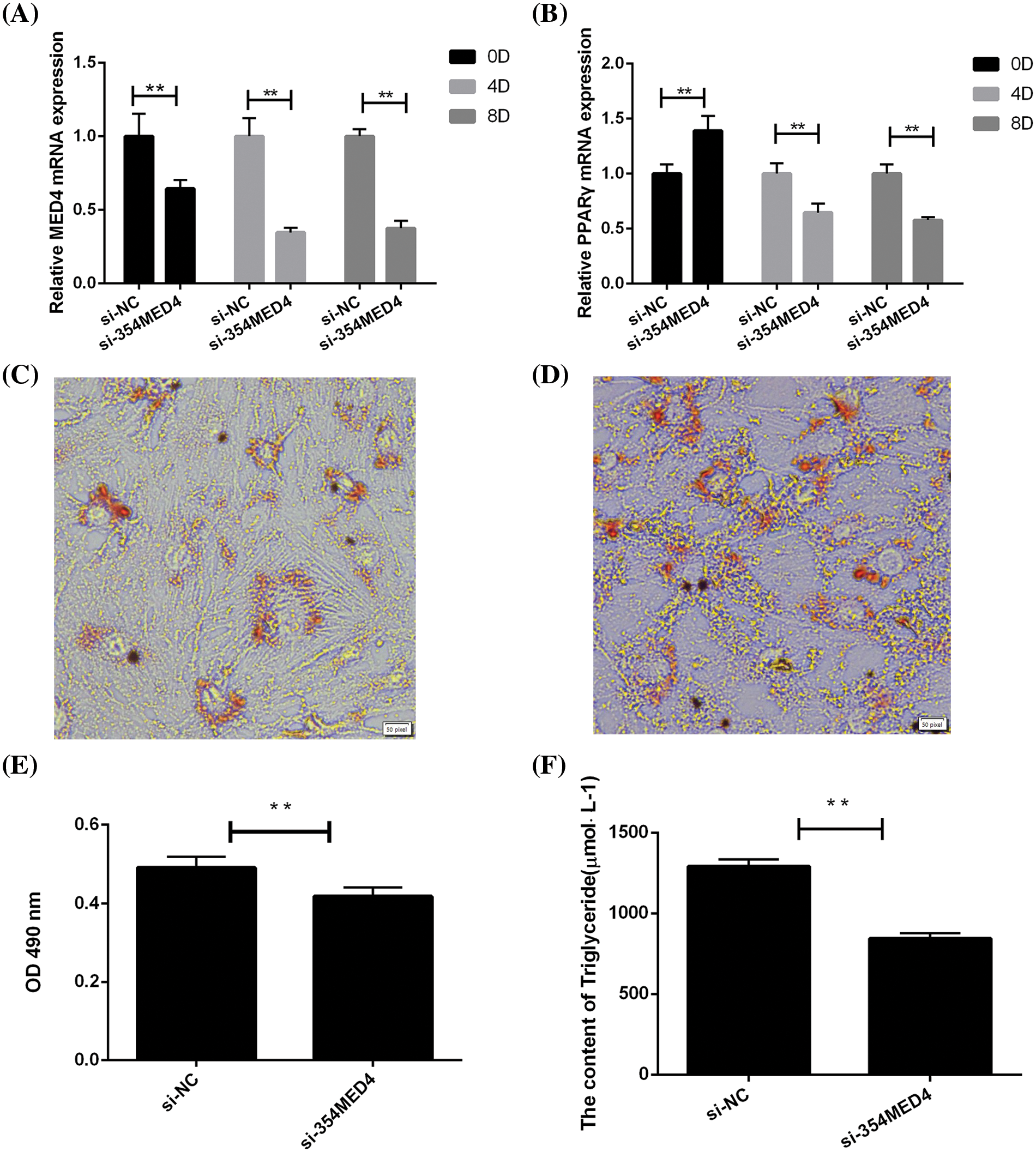
Figure 5: MED4 inhibition affects adipocyte differentiation. (A) MED4 gene expression in the different periods; (B) PPARγ gene expression in the different periods; (C) oil red O staining in the MED4 inhibited group; (D) oil red O staining in the control group; scale bar: 50 pixel; (E) lipid droplets in the adipocytes test; (F) the triglyceride content estimation test; **p < 0.05.
Chinese Red Steppe cattle, a unique breed resource in China, mainly provides meat products in the northeast of China, but its meat quality needs to be improved to meet the consumption demand (Li et al., 2022). Visible adipose tissue in the muscle forms marbling, and is positively correlated with beef juiciness, tenderness, and palatability, and significantly improves the meat quality (Shahrai et al., 2021). Albrecht reported that compared to Holstein, the Japanese black cattle have a stranger ability to accumulate the IMF content (marbling), even under the same feeding condition (Albrecht et al., 2011). Thus, the exploration mechanism of adipogenesis and lipid metabolism is crucial for increasing the IMF content and promoting meat quality (Nematbakhsh et al., 2021). The process of adipocyte differentiation and IMF accumulation involves several genes, molecules, transcription factors, and so on (Tong and Sun, 2015; Xiao et al., 2021), and this study aimed to find new candidate genes regulating the adipogenesis and provide a valuable reference for improving the meat quality by molecular breeding.
We investigated the physiological function of MED4 in the process of adipocyte differentiation. MED4, one of the subunits in a mediator complex protein, supports the cellular structure, affects the transcription and activity of RNA polymerase II, and also encodes vitamin D receptor protein to interact with nuclear receptors (Rachez et al., 1999; Koschubs et al., 2010; Balykova et al., 2022). Only a few studies are reported on the MED4 gene for adipogenesis; in this study, changing pattern of MED4 in the differentiation process was detected. MED4 gene expression first increased (day 2) and became constant (day 2–6), and then increased in the later stage of differentiation (day 8). Therefore, the MED4 gene may be involved in adipogenesis and lipid formation. PPARγ, an important transcription factor for adipocyte differentiation, can activate several genes that play vital roles in adipogenesis (Wafer et al., 2017), such as those coding for CCAAT/enhancer-binding protein alpha (C/EBPA), lipoprotein lipase (LPL), fatty acid-binding protein 4 (FABP4), Krüppel-like factor (KLF) family members to initiate the preadipocyte differentiation (Moseti et al., 2016; Salmerón, 2018). The activated PPARγ gene is necessary for preadipocyte differentiation, even in cattle, sheep, pigs, poultry, and rodents (Soret et al., 2016; Martínez Del Pino et al., 2017; Stachecka et al., 2019; Sun et al., 2019; Guru et al., 2021). Knockout or interference of the PPARγ gene leads to differentiation failure. Nonetheless, exogenous PPARγ recovers the ability of preadipocyte differentiation (Ren et al., 2002), and is thus used as a biomarker for differentiation in general. The trend of MED4 expression is similar to that of PPARγ and C/EBPα in preadipocytes. The change in MED4 can influence the expression of the PPARγ gene, and may have a positive correlation (Wallberg et al., 2003). With the increase in MED4, the PPARγ expression constantly increased; consequently, the accumulation of the TG content and lipid droplets significantly improved. However, MED4 interference assay, Oil Red O staining, and TG detection showed a negative effect. In one study, the MED1 gene was found to bind to the PPARγ gene and regulate adipogenesis (Ge et al., 2002; Grøntved et al., 2010). MED1 and MED4 can form a mediator complex and have a close relationship, suggesting that MED4 may affect the expression of PPARγ and lipid metabolism; one study described higher expression of the MED4 gene in high TG content samples than in low TG samples in Jinghuang chicken (Li et al., 2020). Another study a conducted genome-wide association study in Gushi chicken and reported the association of MED4 with growth and carcass traits (Zhang et al., 2021). MED4 gene may regulate the adipocytes differentiation by affecting PPARγ expression. Unfortunately, in this study, we did not detect the PPARγ protein expression due to technical limitations; however, the results provide useful proof of MED4-mediated regulation of adipogenesis.
MED4 gene significantly increases in the late-stage duration of adipocyte differentiation and positively regulates lipid droplets and TG accumulation.
Acknowledgement: We would like to thank the researchers who contributed to this work but are not included in the author list.
Funding Statement: Supported by the China Agriculture Research System of MOF and MARA (CARS37). Agricultural Science and Technology Innovation Program of Jilin Province CXGC2022DX007.
Author Contributions: Yumin, Zhao: fund support. Yang, Cao: experiment design. Minghong, Wei, and Lihong, Qin: performed experiments and analyzed and interpreted results: Jian, Wu, and Hongliang Liu: data collection. Cheng, Xiao: draft manuscript preparation.
Availability of Data and Materials: All data generated or analyzed during this study are included in this published article (and its supplementary information files).
Ethics Approval: All procedures involving animals, such as welfare and ethical issues, were approved by the Committee for the Ethics of Animal Experiments (AWEC2017A01, 9 March 2017).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Supplementary Materials: The supplementary materials are available at https://doi.org/10.32604/biocell.2023.030364.
References
Ahmed I, Ijaz A, Ahmad I, Linsen Z (2022). Transcriptional regulation of adipogenic marker genes for the improvement of intramuscular fat in Qinchuan beef cattle. Animal Biotechnology 33: 776–795. [Google Scholar]
Albrecht E, Gotoh T, Ebara F, Xu JX, Viergutz T, Nürnberg G, Maak S, Wegner J (2011). Cellular conditions for intramuscular fat deposition in Japanese Black and Holstein steers. Meat Science 89: 13–20. [Google Scholar]
Balykova AN, Kukleva LM, Naryshkina EA, Eroshenko GA, Kutyrev VV (2022). Five draft genome sequences of historical yersinia pestis strains of phylogroups 2.MED4 and 2.MED1 of the medieval biovar. Microbiology Resource Announcements 11: e0004422. [Google Scholar]
Estany J, Ros-Freixedes R, Tor M, Pena RN (2017). Triennial growth and development symposium: Genetics and breeding for intramuscular fat and oleic acid content in pigs. Journal of Animal Science 95: 2261–2271. [Google Scholar]
Fang X, Qin L, Yu H, Jiang P, Xia L, Gao Z, Yang R, Zhao Y, Yu X, Zhao Z (2021). Comprehensive analysis of miRNAs and target mRNAs between immature and mature testis tissue in Chinese red steppes cattle. Animals 11: 3024. [Google Scholar]
Ge K, Guermah M, Yuan CX, Ito M, Wallberg AE, Spiegelman BM, Roeder RG (2002). Transcription coactivator TRAP220 is required for PPARγ2-stimulated adipogenesis. Nature 417: 563–567. [Google Scholar]
Grøntved L, Madsen MS, Boergesen M, Roeder RG, Mandrup S (2010). MED14 tethers mediator to the N-terminal domain of peroxisome proliferator-activated receptor gamma and is required for full transcriptional activity and adipogenesis. Molecular and Cellular Biology 30: 2155–2169. [Google Scholar]
Guru A, Issac PK, Velayutham M, Saraswathi NT, Arshad A, Arockiaraj J (2021). Molecular mechanism of down-regulating adipogenic transcription factors in 3T3-L1 adipocyte cells by bioactive anti-adipogenic compounds. Molecular Biology Reports 48: 743–761. [Google Scholar]
Hocquette JF, Gondret F, Baéza E, Médale F, Jurie C, Pethick DW (2010). Intramuscular fat content in meat-producing animals: Development, genetic and nutritional control, and identification of putative markers. Animal: An International Journal of Animal Bioscience 4: 303–319. [Google Scholar]
Kawai T, Autieri MV, Scalia R (2021). Adipose tissue inflammation and metabolic dysfunction in obesity. American Journal of Physiology. Cell Physiology 320: C375–C391. [Google Scholar]
Koschubs T, Lorenzen K, Baumli S, Sandström S, Heck A J, Cramer P (2010). Preparation and topology of the mediator middle module. Nucleic Acids Research 38: 3186–3195. [Google Scholar]
Li J, Xing S, Wang X (2020). Screening out candidate genes related to intramuscular fat deposition in chicken (in Chinese). China Animal Husbandry and Veterinary Medicine 47: 1828–1836. [Google Scholar]
Li G, Yang R, Lu X, Liu Y, He W et al. (2022). RNA-seq analysis identifies differentially expressed genes in the longissimus dorsi of wagyu and Chinese red steppe cattle. International Journal of Molecular Sciences 24: 387. [Google Scholar]
Martínez Del Pino L, Arana A, Alfonso L, Mendizábal JA, Soret B (2017). Adiposity and adipogenic gene expression in four different muscles in beef cattle. PLoS One 12: e0179604. [Google Scholar]
Miao Y, Qin H, Zhong Y, Huang K, Rao C (2021). Novel adipokine asprosin modulates browning and adipogenesis in white adipose tissue. The Journal of Endocrinology 249: 83–93. [Google Scholar]
Moseti D, Regassa A, Kim WK (2016). Molecular regulation of adipogenesis and potential anti-adipogenic bioactive molecules. International Journal of Molecular Sciences 17: 124. [Google Scholar]
Mota de Sá P, Richard AJ, Hang H, Stephens JM (2017). Transcriptional regulation of adipogenesis. Comprehensive Physiology 7: 635–674. [Google Scholar]
Nematbakhsh S, Chong PP, Selamat J, Nordin N, Idris LH, Abdull Razis AF (2021). Molecular regulation of lipogenesis, adipogenesis and fat deposition in chicken. Genes 12: 414. [Google Scholar]
Rachez C, Lemon BD, Suldan Z, Bromleigh V, Gamble M, Näär AM, Erdjument-Bromage H, Tempst P, Freedman LP (1999). Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature 398: 824–828. [Google Scholar]
Ren D, Collingwood TN, Rebar EJ, Wolffe AP, Camp HS (2002). PPARγ knockdown by engineered transcription factors: Exogenous PPARγ2 but not PPARγ1 reactivates adipogenesis. Genes & Development 16: 27–32. [Google Scholar]
Salmerón C (2018). Adipogenesis in fish. The Journal of Experimental Biology 221, Suppl 1: jeb161588. [Google Scholar]
Shahrai NN, Babji AS, Maskat MY, Razali AF, Yusop SM (2021). Effects of marbling on physical and sensory characteristics of ribeye steaks from four different cattle breeds. Animal Bioscience 34: 904–913. [Google Scholar]
Soret B, Mendizabal JA, Arana A, Alfonso L (2016). Expression of genes involved in adipogenesis and lipid metabolism in subcutaneous adipose tissue and longissimus muscle in low-marbled pirenaica beef cattle. Animal 10: 2018–2026. [Google Scholar]
Stachecka J, Nowacka-Woszuk J, Kolodziejski PA, Szczerbal I (2019). The importance of the nuclear positioning of the PPARG gene for its expression during porcine in vitro adipogenesis. Chromosome Research 27: 271–284. [Google Scholar]
Sun GR, Zhang M, Sun JW, Li F, Ma XF et al. (2019). Krüppel-like factor KLF9 inhibits chicken intramuscular preadipocyte differentiation. British Poultry Science 60: 790–797. [Google Scholar]
Tong J, Sun X (2015). Genetic and genomic analyses for economically important traits and their applications in molecular breeding of cultured fish. Science China Life Sciences 58: 178–186. [Google Scholar]
Verger A, Monté D, Villeret V (2019). Twenty years of mediator complex structural studies. Biochemical Society Transactions 47: 399–410. [Google Scholar]
Wafer R, Tandon P, Minchin JEN (2017). The role of peroxisome proliferator-activated receptor gamma (PPARG) in adipogenesis: Applying knowledge from the fish aquaculture industry to biomedical research. Frontiers in Endocrinology 8: 102. [Google Scholar]
Wallberg AE, Yamamura S, Malik S, Spiegelman BM, Roeder RG (2003). Coordination of p300-mediated chromatin remodeling and TRAP/mediator function through coactivator PGC-1α. Molecular Cell 12: 1137–1149. [Google Scholar]
Wang QA, Tao C, Gupta RK, Scherer PE (2013). Tracking adipogenesis during white adipose tissue development, expansion and regeneration. Nature Medicine 19: 1338–1344. [Google Scholar]
Wen Y, Liu H, Liu K, Cao H, Mao H, Dong X, Yin Z (2020). Analysis of the physical meat quality in partridge (Alectoris chukar) and its relationship with intramuscular fat. Poultry Science 99: 1225–1231. [Google Scholar]
Wu SC, Lo YM, Lee JH, Chen CY, Chen TW et al. (2022). Stomatin modulates adipogenesis through the ERK pathway and regulates fatty acid uptake and lipid droplet growth. Nature Communications 13: 4174. [Google Scholar]
Xiao C, Wei T, Liu LX, Liu JQ, Wang CX, Yuan ZY, Ma HH, Jin HG, Zhang LC, Cao Y (2021). Whole-transcriptome analysis of preadipocyte and adipocyte and construction of regulatory networks to investigate lipid metabolism in sheep. Frontiers in Genetics 12: 662143. [Google Scholar]
Yang W, Yang C, Luo J, Wei Y, Wang W, Zhong Y (2018). Adiponectin promotes preadipocyte differentiation via the PPARγ pathway. Molecular Medicine Reports 17: 428–435. [Google Scholar]
Yao D, Su R, Zhang Y, Wang B, Hou Y, Luo Y, Sun L, Guo Y, Jin Y (2022). Impact of dietary Lactobacillus supplementation on intramuscular fat deposition and meat quality of sunit sheep. Journal of Food Biochemistry 46: e14207. [Google Scholar]
Zhang Y, Wang Y, Li Y, Wu J, Wang X et al. (2021). Genome-wide association study reveals the genetic determinism of growth traits in a Gushi-Anka F2 chicken population. Heredity 126: 293–307. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools