 Open Access
Open Access
ARTICLE
Label-free quantitative proteomics analysis models in vivo and in vitro reveal key proteins and potential roles in sciatic nerve injury
1 Department of Trauma Orthopedics Surgery, Ningbo No. 6 Hospital, Ningbo University School of Medicine, Ningbo, 315000, China
2 Department of Orthopedics, Ningbo Medical Center, Lihuili Hospital, Ningbo University School of Medicine, Ningbo, 315000, China
3 Department of Orthopedic Surgery, Fujian Medical University Union Hospital, Fuzhou, 350001, China
4 School of Medicine, Ningbo University, Ningbo, 315000, China
* Corresponding Authors: YANG GU. Email: ; JIANMING CHEN. Email:
# Yang Gu and Mingguang Bi made the equal contribution to this study and should be listed co-first authors
BIOCELL 2023, 47(9), 2069-2080. https://doi.org/10.32604/biocell.2023.029989
Received 17 March 2023; Accepted 27 April 2023; Issue published 28 September 2023
Abstract
Background: The underlying mechanism of sciatic nerve injury (SNI) is a common motor functional disorder, necessitates further research. Methods: A rat model of SNI was established, with the injury group subjected to compressive injury of the right sciatic nerve exposed at the midpoint of the thigh and the sham surgery group undergoing the same surgical procedure. An oxygen-glucose deprivation model was employed to simulate in vitro SNI in PC12 cells. Following data acquisition and quality control, differentially expressed proteins (DEPs) in each model were identified through differential analysis, and enrichment analysis was used to explore the potential functions and pathways of the DEPs. Venn diagrams were drawn, and DEPs from both in vivo and in vitro SNI models were imported into the STRING database to construct a protein-protein interaction network and screen for hub proteins. Results: After the peptide segments obtained from rat nerve blockade and PC12 cells met quality requirements, 258 DEPs were identified in rat nerve samples, and 119 DEPs were screened in PC12 cells. Enrichment analysis revealed that DEPs in the rat model were predominantly concentrated in biological functions such as myogenic cell proliferation and signaling related to lipid and energy metabolism. DEPs in the in vitro model were mainly enriched in biological processes such as phagocytosis and were associated with lipid transport and metabolism. Two hub proteins, amyloid precursor protein (APP) and fibronectin 1 (FN1), were identified through MCC, MCODE, and Degree scoring. Both PC12 cells and external validation sets showed relatively higher expression of APP and FN1 in injured samples. Results of gene set enrichment analysis indicated that these two proteins were associated with metabolic pathways, such as biosynthesis of glycosaminoglycan chondroitin sulfate and biosynthesis of unsaturated fatty acids. Conclusion: APP and FN1 are potential key molecules involved in SNI and are associated with various metabolic pathways in nerve repair. These findings provide a theoretical basis for the development of therapeutic targets for SNI.Keywords
Authors are encouraged to use the Microsoft Word template when preparing the final version of their manuscripts. In introduction, authors should provide a context or background for the study (that is, the nature of the problem and its significance). State the specific purpose or research objective of, or hypothesis tested by, the study or observation. Cite only directly pertinent references, and do not include data or conclusions from the work being reported.
Peripheral nerve injury (PNI) is a common clinical condition (Kamble et al., 2019) that occurs due to the fragility of peripheral nerves, which are susceptible to sensory and motor nerve abnormalities resulting from acute trauma or chronic disease; in severe cases, it can lead to severe pain, severe disruption to life, and even permanent disability (Bray and Huggett, 2016; Hussain et al., 2020). Due to the complexity of the extent and type of injury and the interaction between the microenvironment of the injury, nerve damage is often difficult to treat and the long healing period is often accompanied by complications that erode the physical and mental health of the patient; besides, there is still a lack of perfect treatment to allow full recovery of nerve function (Arthur-Farraj and Coleman, 2021; Menorca et al., 2013).
Among these, sciatic nerve injury (SNI) is a common type of PNI, and in the past, PNI models were often constructed by exposing and stimulating the sciatic nerve (Lee et al., 2020; Yu et al., 2023). SNI is usually accompanied by extensive soft tissue and skeletal injury, presenting significant nerve damage with slow recovery (Craig et al., 2023; Ferdinand and Raduma, 2019; Jones et al., 2018). Despite great progress in nerve repair research over the past few years, clinical outcomes are still unsatisfactory (Garg et al., 2021; Stavrakakis et al., 2022). If the damaged nerve is not repaired in time, it may even involve the muscle, leading to irreversible loss of muscle function (Sayad-Fathi et al., 2019; Yue et al., 2019). Therefore, uncovering the molecular mechanism of SNI is of crucial importance to facilitate the research and development of new drugs for nerve repair.
With the advancement of biotechnology, researchers have gained a deeper understanding of neural injuries. For example, microarray and RNA sequencing technologies have emerged in elucidating transcriptional changes in neural injury (Brosius Lutz et al., 2022; Sun et al., 2020a; Yao et al., 2020). In this study, we used both in vivo and in vitro models to simulate SNI, constructing a rat SNI model through compressive injury to investigate the repair and functional recovery after neural injury and to explore the possibility that neural injury may deprive certain functions of neural innervation areas, leading to motor and sensory dysfunction. PC12 is a rat adrenal chromaffin cell line that is commonly used to study neural cell differentiation, regeneration, and neuron growth (Wiatrak et al., 2020). One of the common neural cell injury models is the oxygen-glucose deprivation (OGD) model of PC12 cells (Bork et al., 2015; Wang et al., 2022b). Both in vivo and in vitro model samples were used. In this study, we used unlabeled quantitative proteomics to explore key proteins in sciatic nerve injury at the protein level. Through this approach, we hope to identify potential molecular targets and signaling pathways to promote neural injury repair and provide a basis for novel treatment strategies.
Establishment of nerve injury models of rats
All rats involved in experiments were acquired from the Institute of Zoology, Chinese Academy of Sciences (Shanghai, China), and all assays were done with permission from the Laboratory Animal Ethics Committee of Ningbo University with approval No. 11837. These adopted rats were eight-week-old male SD rats (200–220 g). Before the assay, the rats were raised at 23°C–25°C for at least 7 days under alternative day and night (12 h/12 h) and were allowed to have water and food freely. After a week of adaptive feeding, the 6 rats were assigned to an injury group (n = 3) or a sham operation group (n = 3). The rats were anesthetized through intraperitoneal injection of 10% chloral hydrate (3.5 mL/kg, aladdin, CellMark Biotech Ltd., Shenzhen, China). After skin preparation, the right sciatic nerve was horizontally exposed at the middle thigh through blunt dissection of the biceps femoris (Fig. 1A). At the proximal end (about 7 mm) of the trigeminal orifice of the sciatic nerve, the hemostatic forceps of the two blood vessels were placed about 2 mm apart to form moderate compression injury (Fig. 1B). After the suture of muscle and skin, cefazolin sodium (50 mg/kg, Meryer, Meryer Biochemical Technology, Ltd., Shanghai, China) was injected intravenously for seven days. The sham operation group received the same operation procedure, but the sciatic nerve was not compressed. After 7 days, the compressed nerve was acquired (Fig. 1C) and its pathological index was evaluated (Bennett and Xie, 1988).
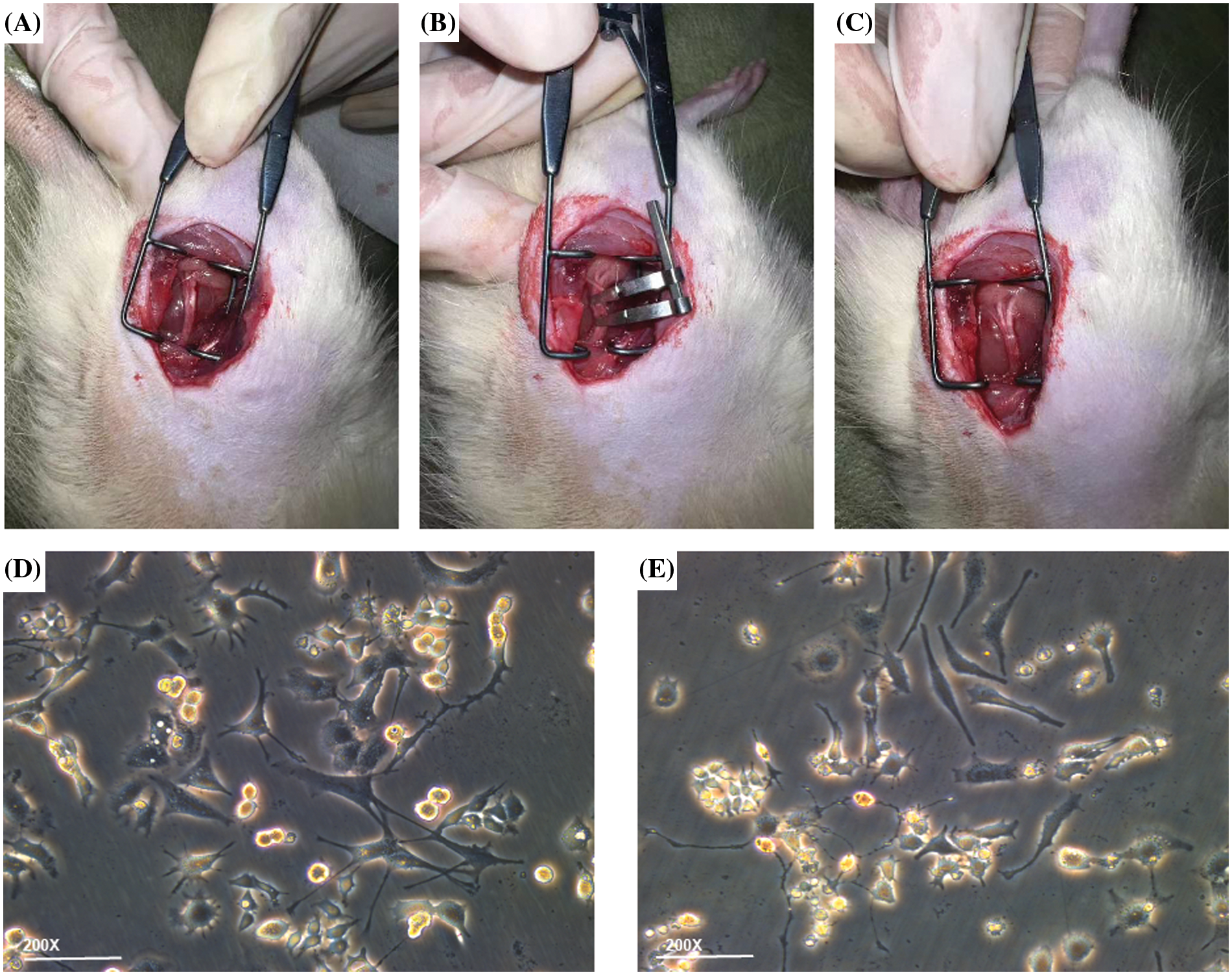
Figure 1: Establishment of in vitro and in vivo experimental models. (A) Sciatic nerve exposure, (B) sciatic nerve injury modeling, (C) edema of the sciatic nerve after being compressed. Compared to (D) untreated PC12 cell group, the (E) PC12 group after oxygen-glucose deprivation injury showed more swollen cells in the visual field; some cell protrusions appeared discontinuous and shortened in length, and there was less intertwining of intercellular protrusions and more cellular debris floating in the medium.
Cell culture and establishment of oxygen-glucose deprivation models
PC12 cells were subjected to incubation (37°C, 5% CO2) in RPMI-1640 complete culture medium (Solarbio, Solarbio Technology, Ltd., Beijing, China) with 5% fetal bovine serum (FBS), 10% equine serum (ES), and 1% double-antibody. The medium was refreshed every 2 days, and the passage was made every 3–4 days. Before the establishment of OGD models, first, 0.01% sterile poly-L-lysine (PLL) solution (Macklin, Trinity Biotechnology, Ltd., Guangzhou, China) was dripped onto the surface of the Petri dish until it completely covered the surface and then let to stand still at indoor temperature for 1 h. PLL solution was removed and recycled, and the Petri dish was placed in an incubator at 37°C for drying overnight. The plate was washed with sterile deionized water three times before planting. Then, PC12 cells was digested and removed, followed by counting. Then the PC12 cells were seeded in a 6-cm culture dish coated with PLL (1 × 106 cells per dish). After 24 h, the cells adhered to the wall. The medium was replaced with a complete culture medium having nerve growth factor (NGF) (Procell, Procell Life Sciences, Ltd., Wuhan, China) with a final concentration of 50 ng/mL, 10%FBS and 1% double-antibody. The medium was replaced every 2 days until the length of the nerve axon greatly increased on the 5th day, and the subsequent treatment was carried out at the end of induced differentiation. To build an OGD model, the air Inlet of the three-gas incubator (anaerobic workstation) was connected with the mixed gas containing 95% N2, 4% CO2, and 1% O2, respectively, and the mixed gas was continuously ventilated to drain the air in the box for use. The culture medium of differentiated PC12 cells was absorbed, and the cells were cleaned gently through PBS 3 times, followed by the addition of a proper amount of glucose-free Hank’s Balanced Salt Solutions (HBSS) medium (Gibco. Visions Biotechnology, Ltd., Guangzhou, China), and then the cells were transferred to the preset hypoxia three-gas incubator (Kang Heng, R&B Technology Development, Ltd., Beijing, China). The control group was subjected to incubation in the complete medium to induce differentiation under normal conditions. Cells were collected after 2 h of continuous hypoxia and sugar deficiency (Fig. 1E), respectively, for follow-up experiments. The control group consisted of the same batch of differentiated PC12 cells there were not treated for OGD modeling (Fig. 1D).
Label-free quantitative proteomics analysis
The samples were taken out from −80°C refrigerators (AS-ONE, Myriad Biochemical Technology, Ltd., Shanghai, China), placed in one mortar precooled by liquid nitrogen, and ground to powder. Each group was added with lysis buffer with 4 times the volume of power (8 M urea, 1% protease inhibitor) and cracked ultrasonically, followed by 10-min centrifugation (4°C, 12000 g) to eliminate cell debris. The supernatant was then placed in one new centrifuge tube, followed by the determination of protein concentration by BCA kit (Beyotime, Beyotime Biotechnology, Shanghai, China). An equal amount of every sample protein was adopted for enzymolysis, and the volume was changed to be consistent with the lysate. The protein was added with TCA with a final concentration of 20%, followed by vortex mixing and 2-h precipitation (4°C), and then by 5-min centrifugation (4500 g) to remove the supernatant. The precipitate was cleaned through precooled acetone 2--3 times. After drying, the precipitate was added with TEAB (ThermoFisher, Sui-Ho Biotechnology Co., Ltd., Guangzhou, China) with the final concentration of 200 mM and then broken by ultrasonic waves (Ningbo Xinzhi,Gert Biotechnology Co., Zhuhai, China), followed by the addition of trypsin (Beyotime, Beyotime Biotechnology, Shanghai, China) at 1:50 (protease: protein, m/m) and enzymolysis all night long. Subsequently, it was put with dithiothreitol (DTT, Merck millipore, Sigma Aldrich (Shanghai) Trading Co., Shanghai, China) to change its final concentration to 5 mM, followed by a 30-min reduction (56°C). After that, iodoacetamide (IAA, Macklin, Trinity Biotechnology, Ltd., Guangzhou, China) was added to change its final concentration to 11 mM, followed by 15-min incubation (room temperature) with dark surroundings.
Liquid chromatography-mass spectrometry analysis
The peptide was dissolved by mobile phase A in liquid chromatography and then isolated by mobile phase system in EASY-nLC 1200 liquid chromatographer (ThermoFisher, Sui-Ho Biotechnology Co., Ltd., Guangzhou, China). The mobile phase A was an aqueous solution with 0.1% formic acid as well as 2% acetonitrile, and the mobile phase B was a solution with 0.1% formic acid as well as 100% acetonitrile. Liquid phase gradient setting: 0–68 min, 6%–23%B; 68–82 min, 23%–35%B; 82–86 min, 35%–80%B; 86–90 min, 80%B. The flow rate was kept at 550 nL/min. The peptides were isolated via ultra-high-performance liquid chromatography, followed by injection into the Capillary ion source for ionization and analysis through None mass spectrometry. The voltage of the ion source was set at 1.6 kV, and the parent ion of the peptide and its secondary fragments were determined and evaluated through high-resolution time-of-flight mass spectrometry. The scanning range of secondary mass spectrometry was set to 400–1200 m/z. The data acquisition mode was the parallel accumulation-serial fragmentation (PASEF) mode. One secondary spectrogram with the charge number of the parent ion within 0–5 was acquired through 10 times of PASEF mode after the first-order mass spectrometry was collected, and the dynamic exclusion time of tandem mass spectrum scanning was set to 20 s to avoid repeated scanning.
Selection of differentially expressed proteins (DEPs)
The reproducibility of protein quantification was assessed using principal component analysis (PCA) and relative standard deviation (RSD) methods. Fold Change (FC) was calculated between groups. After log2 transformation, the protein differences were calculated using the following formula:
The screening criteria were set as p ≤ 0.05, with FC > 1.5.
The R package “clusterProfiler” was used to perform enrichment analysis of DEPs at three levels: Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and protein domain, to identify the enrichment trends of these proteins in terms of function, pathway, or structure. The R package ggplot2 was used for visualization. GSEA v4.3.2 software was used to perform single sample gene set enrichment analysis (ss GSEA) on hub molecules according to c2.cp.kegg.v2022.1.Hs.symbols.gmt.
Construction of protein-protein interaction (PPI) network
Venn diagrams were drawn based on DEPs, and the PPI network of DEPs was constructed using the STRING database. Cytoscape and Metascape were employed to analyze and visualize the PPI network. Utilizing the CytoHubba plugin, assign values to each node and calculate their attributes.
Dataset download and statistical analysis
Expression profiles from the SNI rat model (GSE24982, GSE63442) and the GSE150408 dataset of human SNI were selected and downloaded via Gene Expression Omnibus (GEO). GraphPad Prism 9.0 was used for data calculation and visualization. Quantitative data were analyzed using an independent samples t-test with a significance threshold set at p < 0.05.
Sample collection and analysis of differentially expressed proteins
Peptide segments retrieved from the samples in the database (Figs. 2A and 2F) were analyzed for their length (Figs. 2B and 2G), quantity (Figs. 2C and 2H), protein coverage (Figs. 2D and 2I), and distribution of protein molecular weight (Figs. 2E and 2J) to ensure that the peptide segments obtained from both rat and PC12 cell samples met the quality standards.
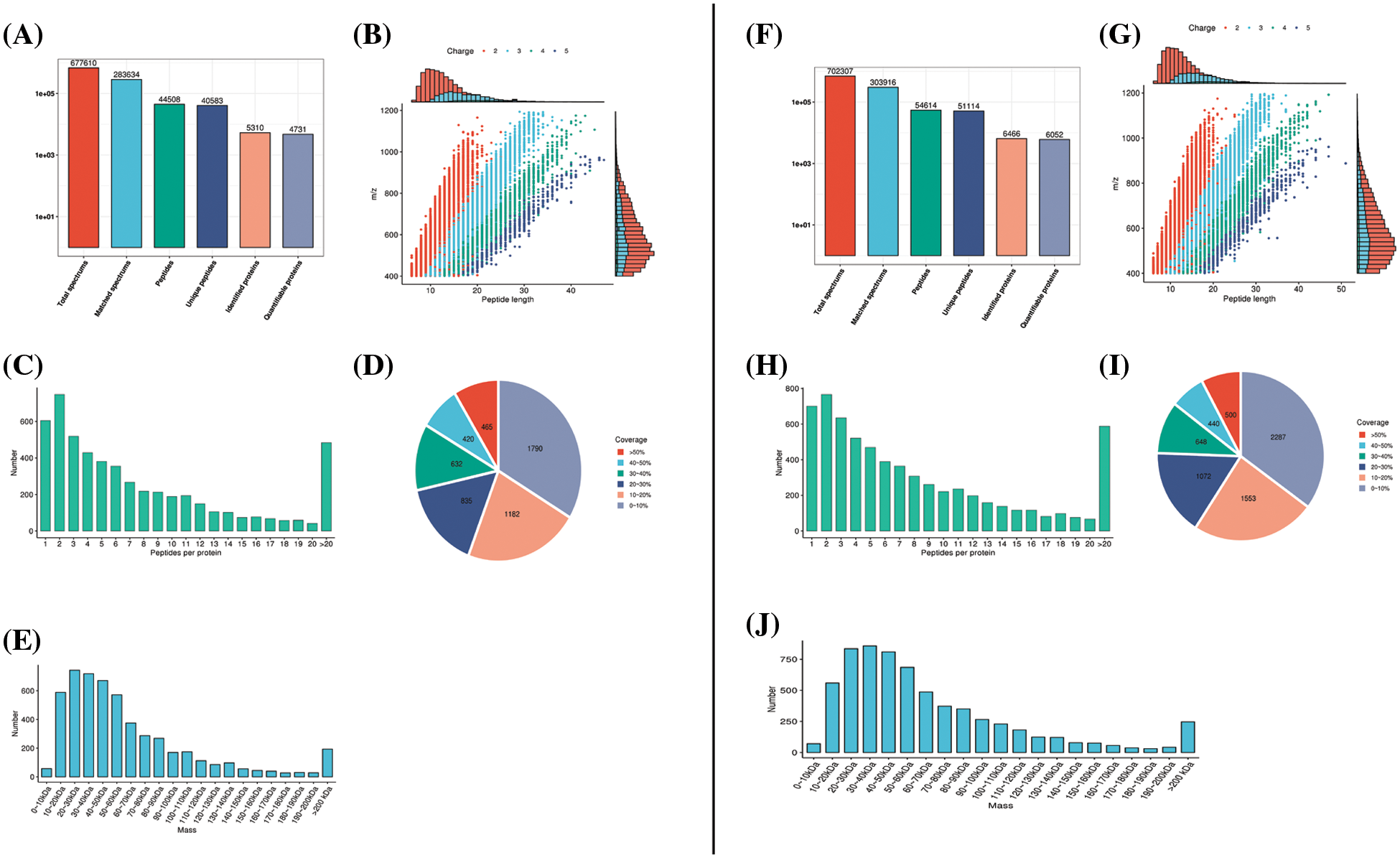
Figure 2: Mass spectrometry data were subjected to database retrieval and quality control analysis. In the rat sciatic nerve samples, (A) protein identification, (B) peptide length, (C) quantity analysis, (D) protein coverage, and (E) molecular weight distribution analysis was conducted. The right panels (F–J) represent the corresponding database and quality control analyses in PC12 cells.
Sciatic nerve samples of rats were collected from the injury group and the Sham group. PCA was performed to evaluate whether the biological repeatability within the group was consistent with the statistical consistency. The results revealed good repeatability of samples (Figs. 3A and 3B). Similarly, the PCA results of PC12 cells in the control and OGD groups also revealed good repeatability of samples (Figs. 2D and 2F).
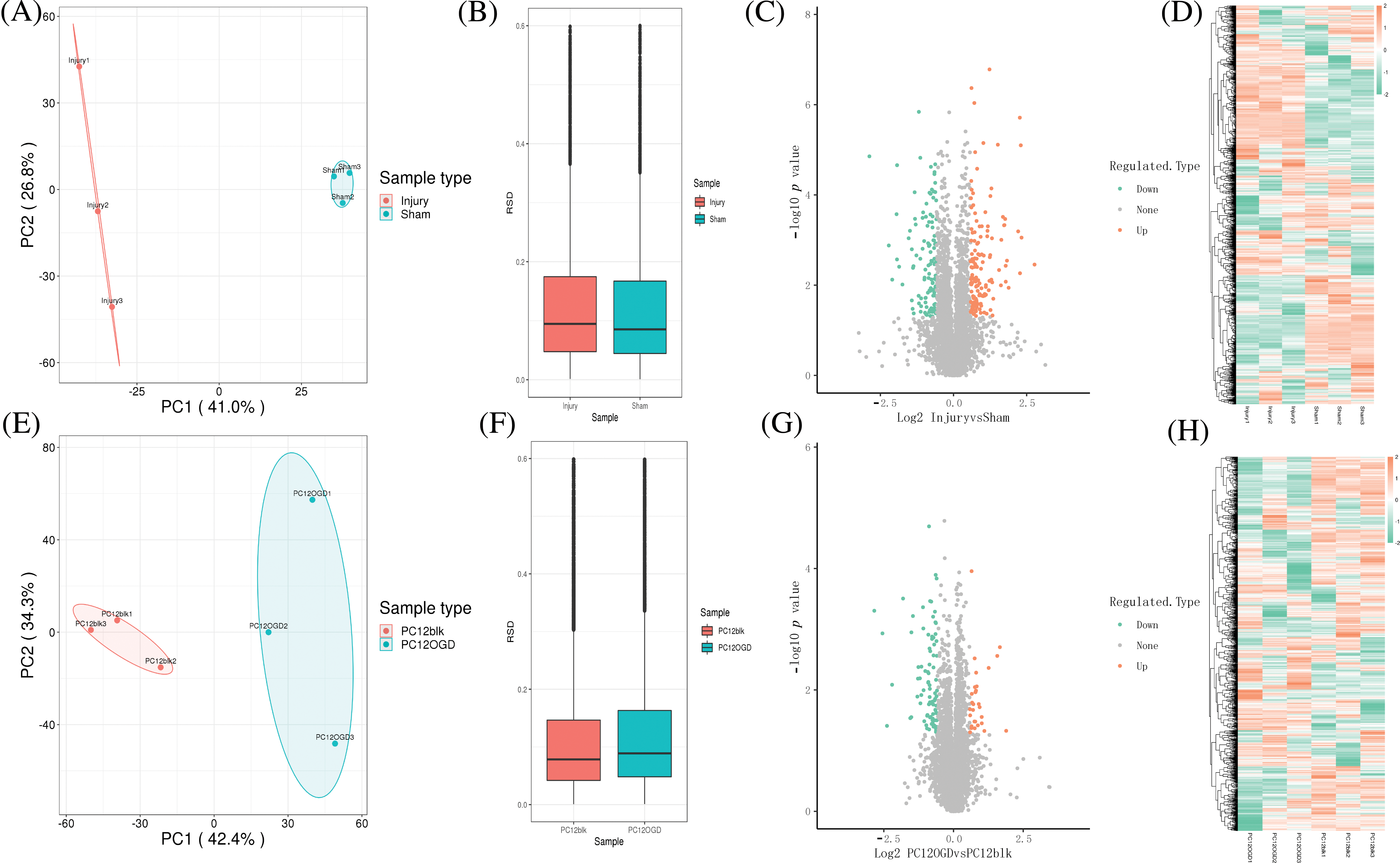
Figure 3: Sample collection and analysis of differentially expressed proteins (DEPs). (A) Principal component analysis (PCA) and (B) relative standard deviation (RSD) of rat sciatic nerve samples, volcano plot (C), and heat map (D) for differential expression analysis; (E) PCA, (F) RSD of PC12 cell samples, volcano plot (G), heat map, and (H) for differential expression analysis.
Then, the DEPs of sciatic nerve samples of rats and PC12 cell samples were analyzed, and volcano plots and heat maps (orange represents proteins whose expression was up-regulated in the injury group/OGD group, and green represents proteins with down-regulated expression in the injury group/OGD group). Totally 258 DEPs were identified in sciatic nerve samples of rats, including 141 up-regulated proteins and 117 down-regulated ones (Figs. 3C and 3D). Totally 119 DEPs were identified in PC12 cells, including 29 up-regulated proteins and 90 down-regulated ones (Figs. 3G and 3H).
Function of differentially expressed proteins and pathway enrichment
The GO enrichment analysis showed the enrichment of DEPs in biological processes (BP), molecular functions (MF), and cellular components (CC) for both rat sciatic nerve samples and PC12 cells. In the edematous rat nerve samples after compression, DEPs were widely involved in BPs such as assembly of the spine apparatus, regulation of myogenic cell proliferation, and production of interleukin-8 (Fig. 4A), associated with CCs like endoplasmic reticulum lumen and MHC class I (Fig. 4B), and significantly enriched in MFs such as titin Z domain binding and retinol oxidase activity (Fig. 4C). In OGD-induced PC12 cells, DEPs repeatedly appeared in BPs like vitamin D metabolic processes, recognition, and response to BMP (Fig. 4F), CCs like layer-adhering protein-11 complex (Fig. 4G), and MFs like vitamin D metabolic processes and phagocytosis (Fig. 4H).
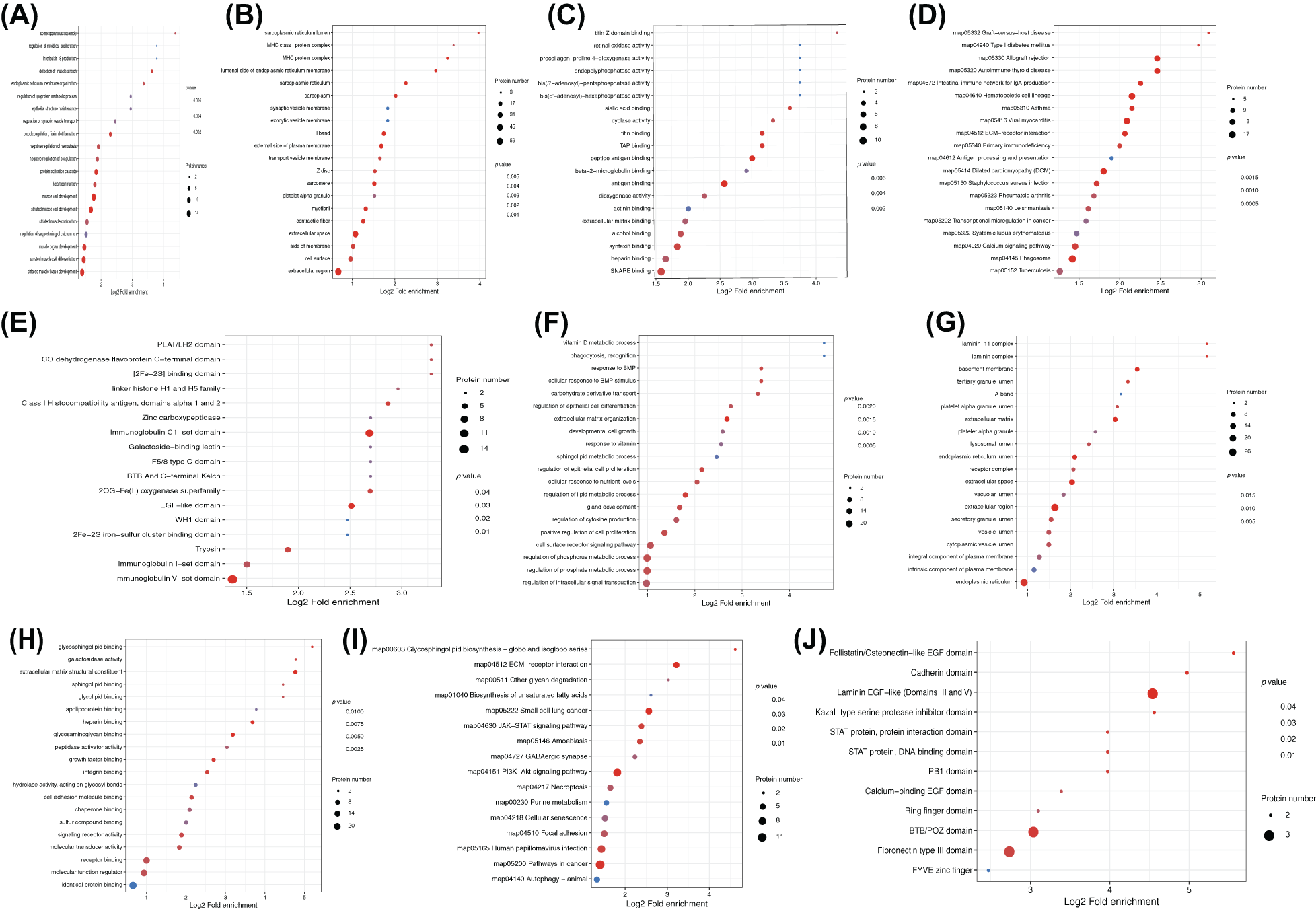
Figure 4: Functional and pathway enrichment of differentially expressed proteins (DEPs). Enrichment analysis of DEPs in rat sciatic nerve samples for (A) biological processes (BP), (B) cellular components (CC), (C) molecular functions (MF), as well as (D) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis, and (E) protein-protein domain analysis. Enrichment analysis results for PC12 cells are shown in (F) BP, (G) CC, (H) MF, (I) KEGG, and (J) domain analysis.
KEGG analysis revealed that DEPs in rat sciatic nerve samples were mainly enriched in pathways including graft-vs.-host disease, type I diabetes, autoimmune thyroid disease, allograft rejection, and IgA production in the intestinal immune network (Fig. 4D). DEPs in PC12 cells were mainly enriched in pathways like the biosynthesis of glycosphingolipids - globo and isoglobo series, extracellular matrix (ECM)-receptor interaction, other glycolysis, and biosynthesis of unsaturated fatty acids (Fig. 4I).
DEPs in rat sciatic nerve samples were repeatedly enriched in domains such as PLAT/LH2, CO dehydrogenase flavoprotein C-terminal domain, and [2F--2S] binding domain (Fig. 4E). In PC12 cells, DEPs were mainly significantly enriched in domains like PLAT/LH2, CO dehydrogenase flavoprotein C-terminal domain, and [2F–2S] binding domain (Fig. 4J).
Prediction of hub proteins based on protein-protein interaction network
With the Venn diagram, the DEPs in sciatic nerve samples of rats and PC12 cells were combined. After the removal of three duplicate proteins, 374 DEPs were obtained (Fig. 5A). Based on the STRING database, a PPI network consisting of 374 proteins is established and is visually displayed through Metascape (Fig. 5B). A total of 13 sub-modules are identified (Fig. 5C). The functions and pathways of the individual modules are detailed in Table 1 and are highly correlated with ECM receptor interactions in MCODE 1. Genes in MCODE1 are screened by calculating the attributes of each node, with the top ten hub proteins based on MCC and Degree rankings, leaving only two proteins: amyloid precursor protein (APP) and fibronectin 1 (FN1) (Fig. 5D). Further, the expression of APP and FN1 in external datasets was determined by comparing the expression of APP and FN1 in the online database of sciatic nerve-related datasets (Fig. 6).
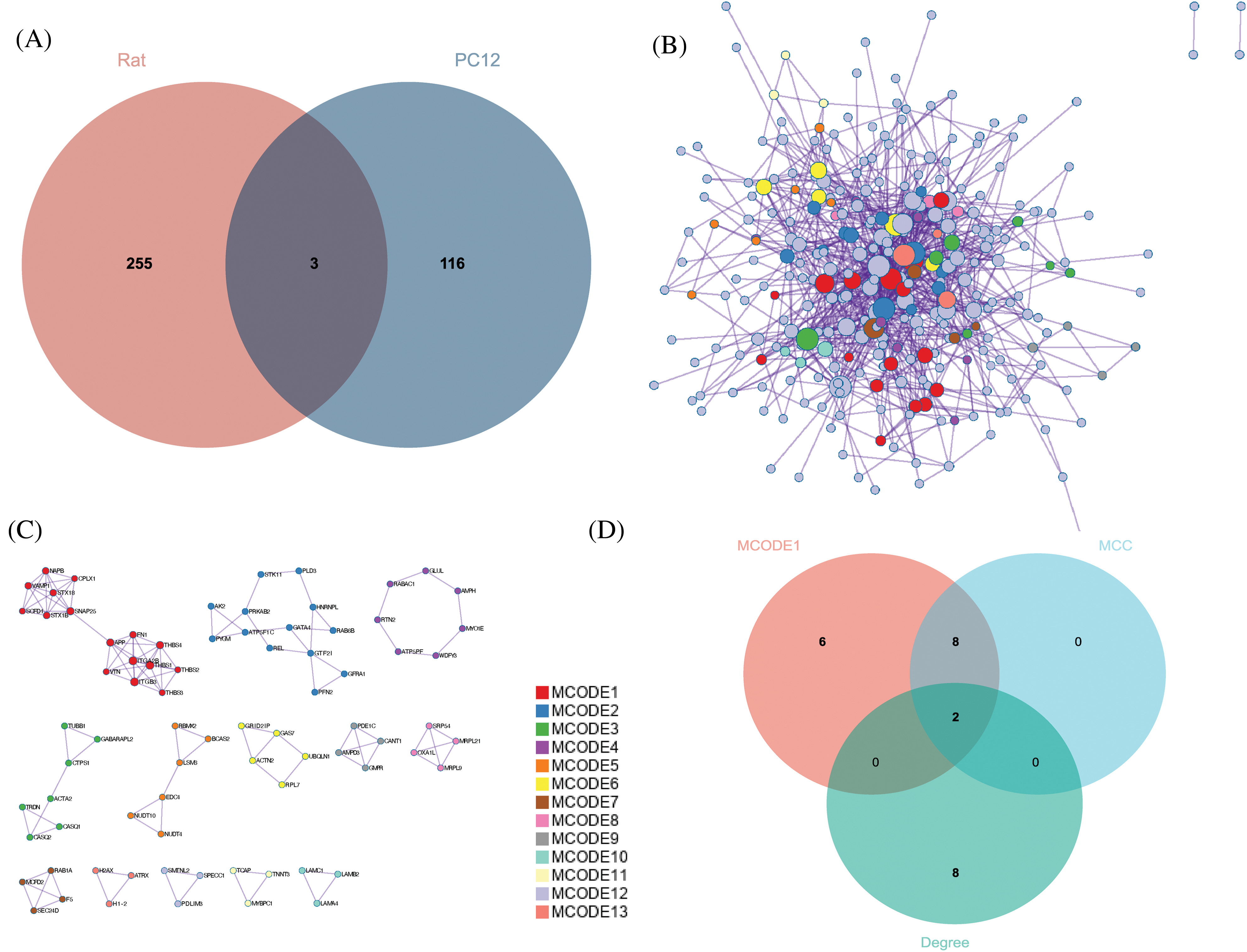
Figure 5: Identification of hub proteins. (A) Overlap of the two model differentially expressed proteins (DEPs), (B) Protein-protein interaction network (PPI) consisting of 347 DEPs, (C) 13 identified MCODE components, and (D) screening of hub proteins.
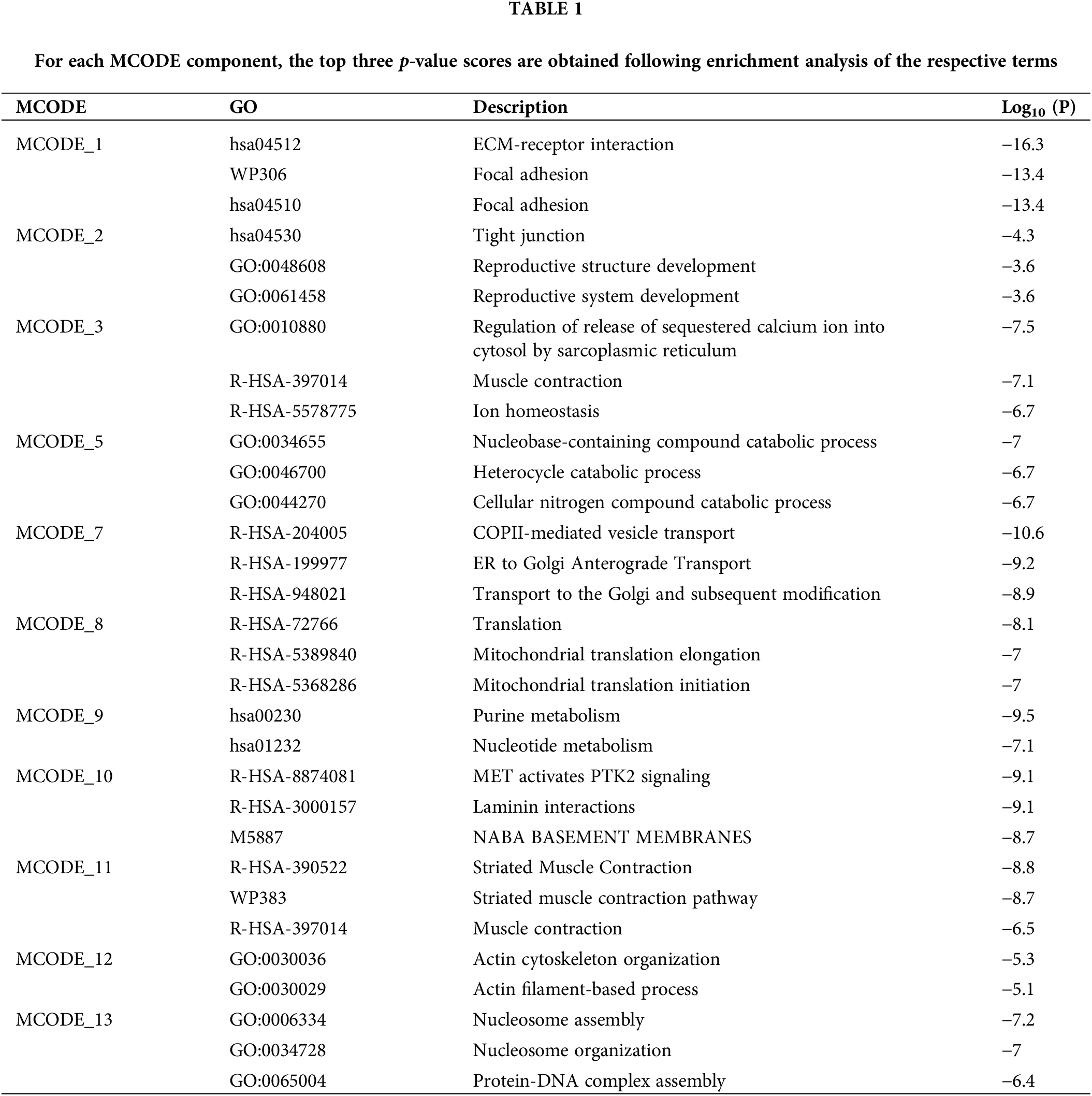
Figure 6: Amyloid precursor protein (APP) APP and fibronectin 1 (FN1) expression levels. (A–B) Relative protein expression of APP and FN1 in oxygen-glucose deprivation-induced and untreated PC12 cells, and APP and FN1 mRNA relative expression in (C–D) GSE63442 dataset and (E–F) GSE24982 dataset for the Injury group and the Sham group, respectively. Note: *p < 0.05; **p < 0.01; ***p < 0.001.
The potential functions of hub proteins
To further explore the pathways involved with hub proteins, we conducted GSEA enrichment analysis on two hub proteins in the GSE150408 dataset. The results revealed that in the low expression group of APP, biosynthesis of glycosaminoglycan chondroitin sulfate, lysosomes, and Fc Gamma R mediated phagocytosis were significantly enriched (p < 0.05). In contrast, FN1 was associated with the biosynthesis of unsaturated fatty acids, glycosaminoglycan biosynthesis of keratan sulfate, and the pentose phosphate pathway (p < 0.05). For more details, see Fig. 7.
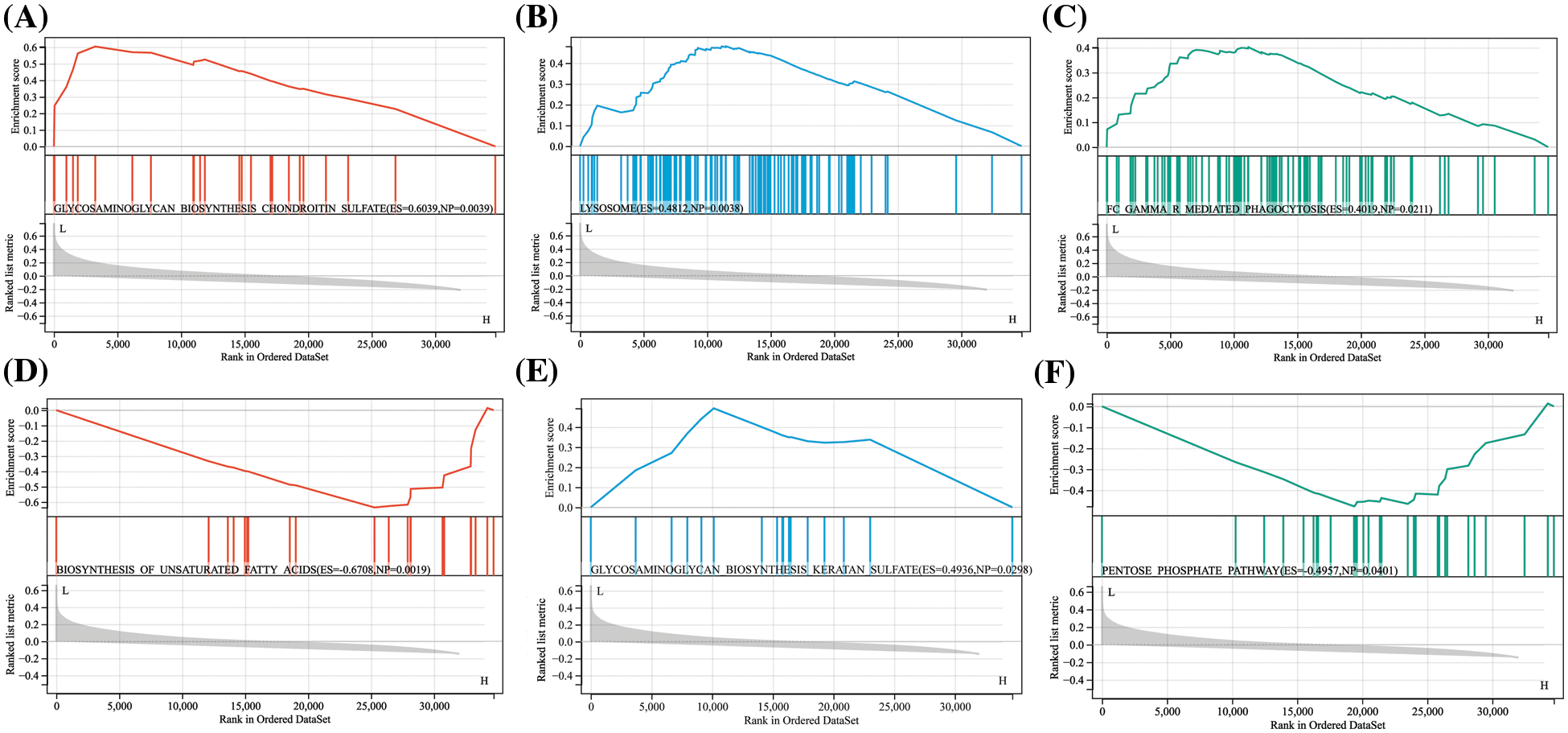
Figure 7: Single gene set enrichment analysis (GSEA) results for APP and FN1 APP was enriched in the GSE150408 dataset in (A) biosynthesis of glycosaminoglycan chondroitin sulfate, (B) lysosomes, and (C) Fc gamma R mediated Phagocytosis. fn1 was enriched in the GSE150408 dataset in (D) unsaturated fatty acid biosynthesis, (E) glycosaminoglycan biosynthesis keratin sulfate, and (F) pentose phosphate pathway.
Acute mechanical and physical injury, chronic nerve compression, or nerve degeneration can all result in injury to the peripheral nerve, leading to functional impairment or neuropathic pain (Wang et al., 2022a; Wang et al., 2023). In contrast to the central nerve, the axons of injured peripheral nerves already have a remarkable capacity for regeneration (Gaudet et al., 2011), although nerve regeneration involves a large pathophysiological network that requires the coordination of damaged parts and distal neurons, and the interaction between various growth factors, cytokines, inflammatory factors, etc. (Vijayavenkataraman, 2020). The complex inflammatory response to injury and the proliferation of microglia often impede nerve regeneration in SNIs (Molnár et al., 2022), and the different trajectories followed by various cytokines after injury (Yuan et al., 2021), make the outcome of nerve repair but the results are not encouraging. Repair of peripheral nerves takes months to years, and in some cases, cannot even be achieved (DeLeonibus et al., 2021; Sulaiman and Gordon, 2013), especially in patients with SNI or shoulder brachial plexus injuries, where the prognosis is often poor. This is due to the long distances required for growth and repair, during which time the muscle atrophies and degenerates due to a prolonged lack of innervation, leading to irreversible loss of function. Accelerating nerve repair is an effective way to improve and prevent peripheral muscle atrophy (Gaudet et al., 2011; Jiang et al., 2020). However, the molecular mechanisms underlying nerve injury repair have not yet been explained, so uncovering the molecular mechanisms of SNI has become an important prerequisite for translational research and the development of new drugs to promote nerve recovery.
Analysis of the differences in translational and post-translational modifications in the nervous system that occur after injury by SNI can help identify key biological markers of SNI that can be used as major molecular targets for drug therapy and improve the efficacy of SNI treatment (Niederberger and Geisslinger, 2008). A number of studies have been conducted using different animal models to simulate the onset of SNI and for the triggers of SNI (e.g., viruses (Wang et al., 2022c)), therapeutic agents (Chen et al., 2022b; Xiong et al., 2022), temporal changes (Ujcikova et al., 2022; Vergara et al., 2018). In contrast, the current study constructed ex vivo models of SNI using compressive injury and OGD induction and explored DEPs in SNI that suffered an injury based on label-free quantitative proteomics extraction and identification of proteins.
Enrichment analysis revealed the involvement of the DEPs from these rat models in the regulation of myogenic cell proliferation and interleukin-8 production, suggesting differences in muscle regeneration and inflammatory responses in rats after injury (Dort et al., 2021; Jungen et al., 2019) and that the search for molecular markers targeting myogenic cells and inflammatory factors may be able to promote muscle regeneration after SNI injury and ameliorate inflammation-induced sciatica. Vitamin D is known to have significant positive effects on nerve regeneration (El Soury et al., 2021) and can significantly reduce neuropathic pain caused by sciatic nerve constriction in rats (Poisbeau et al., 2019) and that DEPs in PC12 cells are significantly enriched in vitamin D metabolic processes and ECM-receptor interaction-related terms, suggesting that these DEPs may be key molecules in repairing nerve damage and reducing neuroinflammation (Hart and Karimi-Abdolrezaee, 2020; Kalinski et al., 2020).
After further screening, APP and FN1 were identified as hub proteins, and both showed high expression in models of sciatic nerve injury. APP is a common transmembrane protein localized to chromosome 21 and is widely expressed in axons and dendrites, contributing to neurite growth and synaptogenesis (Hunter et al., 2003; Kotulska et al., 2010; Reinhard et al., 2005). Also, APP is an important regulator in myelin formation and has now been identified as a key protein in neural development and neural repair processes after injury (Truong et al., 2019). After PNI, APP expression was found to increase in the dorsal root ganglion and does not return to baseline levels until the nerve is reinnervated (Scott et al., 1991); therefore, overexpressed APP is also considered one of the markers to identify nerve injury (Dzreyan et al., 2021). This is consistent with our findings that APP can be used as a biological marker of SNI injury. In addition, low expression of APP seems to be associated with lysosomal activation. Lysosomal cytosolic action in Schwann cells contributes to axonal myelin regeneration and that regenerating sciatic nerve myelin regeneration is blocked in the presence of lysosomal dysfunction (Chen et al., 2012). Most studies have also shown that lysosomal activation is an important target for the treatment of many inflammatory and neurological pathologies (Bonam et al., 2019; Plotegher and Duchen, 2017). We speculate that APP inhibits lysosomal function and that targeted inhibition of APP may be able to mediate lysosomal action and promote myelin regeneration.
FN1 is a member of the integrin receptor family of receptors that primarily mediates ECM interactions and is associated with cytoskeleton, adhesion, and growth (Li et al., 2019; Pankov and Yamada, 2002; Sun et al., 2020b). FN is also key to repairing nerve injury (Tsuda et al., 2008), where fibronectin deposited in the sciatic nerve after injury alters the composition of the ECM and inhibits Schwann cell migration (Akassoglou et al., 2003). In recent years, it has been shown that Sox2 and FN are co-expressed in a time-dependent manner in pro-regenerative Chevron cells in vivo (Torres-Mejía et al., 2020). Even in vitro, FN1 expression is shown to increase through increased FN1 expression and neural recovery through fibrillogenic fibril formation (Chen et al., 2022a). Besides, FN1 has been indicated as a key molecular target for axonal regeneration in DRG neurons after SNI (Mao et al., 2019). These studies also corroborate our findings that FN1 may be an important target in neural repair after SNI injury.
We acknowledge our study’s limitations, which include the absence of western blotting validation for key protein expression and sciatic functional index (SFI) behavioral tests to confirm model construction success. In future research, we plan to address these limitations by conducting western blotting, SFI tests, and additional wet lab experiments to provide a more comprehensive validation of our results, ultimately strengthening the reliability of our findings.
In these studies, we have identified two hub proteins of SNI, namely, APP and FN1, through quantitative proteomics and bioinformatics analysis. Many studies have revealed their critical role in nerve-associated diseases, but the role they play in SNI and whether there is a regulatory relationship between them still need further experimental research.
Acknowledgement: None.
Funding Statement: This research was funded by the Project of 2022 Health Commission Technology Plan of Zhejiang Province (Project Number: 2022KY1169), and the 2022 Ningbo Natural Science Foundation Young Doctoral Project (Project Number: 2022J027).
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Yang Gu, Mingguang Bi; data collection: Dehui Chen; analysis and interpretation of results: Ning Ni; draft manuscript preparation: Yang Gu, Mingguang Bi, Jianming Chen. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics Approval: All assays were done with permission from the Laboratory Animal Ethics Committee of Ningbo University with approval No. 11837.
Conflicts of Interest: The authors declare that they have no conflict of interest.
References
Akassoglou K, Akpinar P, Murray S, Strickland S (2003). Fibrin is a regulator of Schwann cell migration after sciatic nerve injury in mice. Neuroscience Letters 338: 185–188. [Google Scholar] [PubMed]
Arthur-Farraj P, Coleman MP (2021). Lessons from injury: How nerve injury studies reveal basic biological mechanisms and therapeutic opportunities for peripheral nerve diseases. Neurotherapeutics 18: 2200–2221. [Google Scholar] [PubMed]
Bennett GJ, Xie YK (1988). A peripheral mononeuropathy in rat that produces disorders of pain sensation like those seen in man. Pain 33: 87–107. https://doi.org/10.1016/0304-3959(88)90209-6 [Google Scholar] [PubMed] [CrossRef]
Bonam SR, Wang F, Muller S (2019). Lysosomes as a therapeutic target. Nature Reviews Drug Discovery 18: 923–948. https://doi.org/10.1038/s41573-019-0036-1 [Google Scholar] [PubMed] [CrossRef]
Bork K, Wurm F, Haller H, Strauss C, Scheller C, Gnanapragassam VS, Horstkorte R (2015). Neuroprotective and neuroregenerative effects of nimodipine in a model system of neuronal differentiation and neurite outgrowth. Molecules 20: 1003–1013. [Google Scholar] [PubMed]
Bray GM, Huggett DL (2016). Neurological diseases, disorders and injuries in Canada: Highlights of a national study. Canadian Journal of Neurological Sciences 43: 5–14. https://doi.org/10.1017/cjn.2015.312 [Google Scholar] [PubMed] [CrossRef]
Brosius Lutz A, Lucas TA, Carson GA, Caneda C, Zhou L, Barres BA, Buckwalter MS, Sloan SA (2022). An RNA-sequencing transcriptome of the rodent Schwann cell response to peripheral nerve injury. Journal of Neuroinflammation 19: 105. [Google Scholar] [PubMed]
Chen W, Chang S, Yang C, Zhou J, Zhang H, Nie K, Wei Z (2022a). Schwann cell‐like cells derived from human amniotic mesenchymal stem cells promote sciatic nerve repair through an exosome‐induced SOX2/FN1 pathway in vitro. International Journal of Molecular Medicine 49: 1–12. https://doi.org/10.3892/ijmm.2022.5136 [Google Scholar] [PubMed] [CrossRef]
Chen G, Zhang Z, Wei Z, Cheng Q, Li X, Li W, Duan S, Gu X (2012). Lysosomal exocytosis in Schwann cells contributes to axon remyelination. Glia 60: 295–305. [Google Scholar] [PubMed]
Chen Y, Zhou Y, Dong Q (2022b). Proteomics reveals the key molecules involved in curcumin-induced protection against sciatic nerve injury in rats. Neuroscience 501: 11–24. [Google Scholar] [PubMed]
Craig J, Linnecor BG, Neil M (2023). Soft tissue, peripheral nerve and brachial plexus injury. In: Orthopaedic and Trauma Nursing: An Evidence-Based Approach to Musculoskeletal Care, pp. 304–315. [Google Scholar]
DeLeonibus A, Rezaei M, Fahradyan V, Silver J, Rampazzo A, Bassiri Gharb B (2021). A meta-analysis of functional outcomes in rat sciatic nerve injury models. Microsurgery 41: 286–295. [Google Scholar] [PubMed]
Dort J, Orfi Z, Fabre P, Molina T, Conte TC, Greffard K, Pellerito O, Bilodeau JF, Dumont NA (2021). Resolvin-D2 targets myogenic cells and improves muscle regeneration in Duchenne muscular dystrophy. Nature Communications 12: 6264. [Google Scholar] [PubMed]
Dzreyan V, Rodkin S, Nikul V, Pitinova M, Uzdensky A (2021). The expression of E2F1, p53, and Caspase 3 in the rat dorsal root ganglia after sciatic nerve transection. Journal of Molecular Neuroscience 71: 826–835. https://doi.org/10.1007/s12031-020-01705-6 [Google Scholar] [PubMed] [CrossRef]
El Soury M, Fornasari BE, Carta G, Zen F, Haastert-Talini K, Ronchi G (2021). The role of dietary nutrients in peripheral nerve regeneration. International Journal of Molecular Sciences 22: 7417. [Google Scholar] [PubMed]
Ferdinand NW, Raduma OS (2019). Post thermal sciatic nerve injury successfully repaired with a sural nerve graft: Case report. Journal of Surgery 7: 119–122. [Google Scholar]
Garg SP, Hassan AM, Patel AA, Perez MM, Stoehr JR, Ketheeswaran S, Chappell AG, Galiano RD, Ko JH (2021). Outcomes of tibial nerve repair and transfer: A structured evidence-based systematic review and meta-analysis. The Journal of Foot and Ankle Surgery 60: 1280–1289. [Google Scholar] [PubMed]
Gaudet AD, Popovich PG, Ramer MS (2011). Wallerian degeneration: Gaining perspective on inflammatory events after peripheral nerve injury. Journal of Neuroinflammation 8: 1–13. [Google Scholar]
Hart CG, Karimi-Abdolrezaee S (2020). Bone morphogenetic proteins: New insights into their roles and mechanisms in CNS development, pathology and repair. Experimental Neurology 334: 113455. [Google Scholar] [PubMed]
Hunter CL, Isacson O, Nelson M, Bimonte-Nelson H, Seo H, Lin L, Ford K, Kindy MS, Granholm AC (2003). Regional alterations in amyloid precursor protein and nerve growth factor across age in a mouse model of Down’s syndrome. Neuroscience Research 45: 437–445. [Google Scholar] [PubMed]
Hussain G, Wang J, Rasul A, Anwar H, Qasim M et al. (2020). Current status of therapeutic approaches against peripheral nerve injuries: A detailed story from injury to recovery. International Journal of Biological Sciences 16: 116–134. https://doi.org/10.7150/ijbs.35653 [Google Scholar] [PubMed] [CrossRef]
Jiang JP, Liu XY, Zhao F, Zhu X, Li XY, Niu XG, Yao ZT, Dai C, Xu HY, Ma K (2020). Three-dimensional bioprinting collagen/silk fibroin scaffold combined with neural stem cells promotes nerve regeneration after spinal cord injury. Neural Regeneration Research 15: 959–968. [Google Scholar] [PubMed]
Jones PE, Meyer RM, Faillace WJ, Landau ME, Smith JK, McKay PL, Nesti LJ (2018). Combat injury of the sciatic nerve--an institutional experience. Military Medicine 183: e434–e441. [Google Scholar] [PubMed]
Jungen MJ, Ter Meulen BC, van Osch T, Weinstein HC, Ostelo RW (2019). Inflammatory biomarkers in patients with sciatica: A systematic review. BMC Musculoskeletal Disorders 20: 1–9. [Google Scholar]
Kalinski AL, Yoon C, Huffman LD, Duncker PC, Kohen R, Passino R, Hafner H, Johnson C, Kawaguchi R, Carbajal KS (2020). Analysis of the immune response to sciatic nerve injury identifies efferocytosis as a key mechanism of nerve debridement. eLife 9: e60223. [Google Scholar] [PubMed]
Kamble N, Shukla D, Bhat D (2019). Peripheral nerve injuries: Electrophysiology for the neurosurgeon. Neurology India 67: 1419. https://doi.org/10.4103/0028-3886.273626 [Google Scholar] [PubMed] [CrossRef]
Kotulska K, Larysz-Brysz M, LePecheur M, Marcol W, Lewin-Kowalik J, Paly E, London J (2010). APP overexpression prevents neuropathic pain and motoneuron death after peripheral nerve injury in mice. Brain Research Bulletin 81: 378–384. [Google Scholar] [PubMed]
Lee JI, Hur JM, You J, Lee DH (2020). Functional recovery with histomorphometric analysis of nerves and muscles after combination treatment with erythropoietin and dexamethasone in acute peripheral nerve injury. PLoS One 15: e0238208. [Google Scholar] [PubMed]
Li B, Shen W, Peng H, Li Y, Chen F, Zheng L, Xu J, Jia L (2019). Fibronectin 1 promotes melanoma proliferation and metastasis by inhibiting apoptosis and regulating EMT. OncoTargets and Therapy 12: 3207–3221. [Google Scholar] [PubMed]
Mao S, Huang T, Chen Y, Shen L, Zhou S, Zhang S, Yu B (2019). Circ-Spidr enhances axon regeneration after peripheral nerve injury. Cell Death & Disease 10: 787. https://doi.org/10.1038/s41419-019-2027-x [Google Scholar] [PubMed] [CrossRef]
Menorca RM, Fussell TS, Elfar JC (2013). Nerve physiology: Mechanisms of injury and recovery. Hand Clinics 29: 317–330. https://doi.org/10.1016/j.hcl.2013.04.002 [Google Scholar] [PubMed] [CrossRef]
Molnár K, Nógrádi B, Kristóf R, Mészáros Á, Pajer K, Siklós L, Nógrádi A, Wilhelm I, Krizbai IA (2022). Motoneuronal inflammasome activation triggers excessive neuroinflammation and impedes regeneration after sciatic nerve injury. Journal of neuroinflammation 19: 68. [Google Scholar]
Niederberger E, Geisslinger G (2008). Proteomics in neuropathic pain research. Anesthesiology 108: 314–323. https://doi.org/10.1097/01.anes.0000299838.13368.6e [Google Scholar] [PubMed] [CrossRef]
Pankov R, Yamada KM (2002). Fibronectin at a glance. Journal of Cell Science 115: 3861–3863. [Google Scholar] [PubMed]
Plotegher N, Duchen MR (2017). Mitochondrial dysfunction and neurodegeneration in lysosomal storage disorders. Trends in Molecular Medicine 23: 116–134. https://doi.org/10.1016/j.molmed.2016.12.003 [Google Scholar] [PubMed] [CrossRef]
Poisbeau P, Aouad M, Gazzo G, Lacaud A, Kemmel V, Landel V, Lelièvre V, Feron F (2019). Cholecalciferol (vitamin D3) reduces rat neuropathic pain by modulating opioid signaling. Molecular Neurobiology 56: 7208–7221. [Google Scholar] [PubMed]
Reinhard C, Hébert SS, de Strooper B (2005). The amyloid-beta precursor protein: Integrating structure with biological function. The EMBO Journal 24: 3996–4006. https://doi.org/10.1038/sj.emboj.7600860 [Google Scholar] [PubMed] [CrossRef]
Sayad-Fathi S, Nasiri E, Zaminy A (2019). Advances in stem cell treatment for sciatic nerve injury. Expert Opinion on Biological Therapy 19: 301–311. [Google Scholar] [PubMed]
Scott JN, Parhad IM, Clark AW (1991). β-Amyloid precursor protein gene is differentially expressed in axotomized sensory and motor systems. Molecular Brain Research 10: 315–325. [Google Scholar] [PubMed]
Stavrakakis IM, Kritsotakis EI, Giannoudis PV, Kapsetakis P, Dimitriou R, Bastian JD, Tosounidis TH (2022). Sciatic nerve injury after acetabular fractures: A meta-analysis of incidence and outcomes. European Journal of Trauma and Emergency Surgery 48: 2639–2654. [Google Scholar] [PubMed]
Sulaiman W, Gordon T (2013). Neurobiology of peripheral nerve injury, regeneration, and functional recovery: From bench top research to bedside application. Ochsner Journal 13: 100–108. [Google Scholar] [PubMed]
Sun W, Kou D, Yu Z, Yang S, Jiang C, Xiong D, Xiao L, Deng Q, Xie H, Hao Y (2020a). A transcriptomic analysis of neuropathic pain in rat dorsal root ganglia following peripheral nerve injury. NeuroMolecular Medicine 22: 250–263. [Google Scholar] [PubMed]
Sun Y, Zhao C, Ye Y, Wang Z, He Y, Li Y, Mao H (2020b). High expression of fibronectin 1 indicates poor prognosis in gastric cancer. Oncology Letters 19: 93–102. [Google Scholar] [PubMed]
Torres-Mejía E, Trümbach D, Kleeberger C, Dornseifer U, Orschmann T, Bäcker T et al. (2020). Sox2 controls Schwann cell self-organization through fibronectin fibrillogenesis. Scientific Reports 10: 1984. https://doi.org/10.1038/s41598-019-56877-y [Google Scholar] [PubMed] [CrossRef]
Truong PH, Ciccotosto GD, Merson TD, Spoerri L, Chuei MJ, Ayers M, Xing YL, Emery B, Cappai R (2019). Amyloid precursor protein and amyloid precursor-like protein 2 have distinct roles in modulating myelination, demyelination, and remyelination of axons. Glia 67: 525–538. [Google Scholar] [PubMed]
Tsuda M, Toyomitsu E, Komatsu T, Masuda T, Kunifusa E, Nasu-Tada K, Koizumi S, Yamamoto K, Ando J, Inoue K (2008). Fibronectin/integrin system is involved in P2X4 receptor upregulation in the spinal cord and neuropathic pain after nerve injury. Glia 56: 579–585. [Google Scholar] [PubMed]
Ujcikova H, Robles D, Yue X, Svoboda P, Lee YS, Navratilova E (2022). Time-dependent changes in protein composition of medial prefrontal cortex in rats with neuropathic pain. International Journal of Molecular Sciences 23: 955. [Google Scholar] [PubMed]
Vergara D, Romano A, Stanca E, La Pesa V, Aloisi L, de Domenico S, Franck J, Cicalini I, Giudetti A, Storelli E (2018). Proteomic expression profile of injured rat peripheral nerves revealed biological networks and processes associated with nerve regeneration. Journal of Cellular Physiology 233: 6207–6223. [Google Scholar] [PubMed]
Vijayavenkataraman S (2020). Nerve guide conduits for peripheral nerve injury repair: A review on design, materials and fabrication methods. Acta Biomaterialia 106: 54–69. https://doi.org/10.1016/j.actbio.2020.02.003 [Google Scholar] [PubMed] [CrossRef]
Wang SL, Liu XL, Kang ZC, Wang YS (2023). Platelet-rich plasma promotes peripheral nerve regeneration after sciatic nerve injury. Neural Regeneration Research 18: 375–381. [Google Scholar] [PubMed]
Wang Y, Xiong ZL, Ma XL, Zhou C, Huo MH, Jiang XW, Yu WH (2022a). Acetyl-11-keto-beta-boswellic acid promotes sciatic nerve repair after injury: Molecular mechanism. Neural Regeneration Research 17: 2778–2784. https://doi.org/10.4103/1673-5374.339494 [Google Scholar] [PubMed] [CrossRef]
Wang Y, Xiong Z, Zhou C, Zhang Q, Liu S, Dong S, Jiang X, Yu W (2022b). AKBA promotes axonal regeneration via RhoA/rictor to repair damaged sciatic nerve. International Journal of Molecular Sciences 23: 15903. [Google Scholar] [PubMed]
Wang X, Yang H, Wang Z, Wang G, Yang L, Yuan Y, Li X, Zhang D, Shen K, Wang Z (2022c). Label-free proteomics-based analysis of peripheral nerve injury induced by Japanese encephalitis virus. Journal of Proteomics 264: 104619. https://doi.org/10.1016/j.jprot.2022.104619 [Google Scholar] [PubMed] [CrossRef]
Wiatrak B, Kubis-Kubiak A, Piwowar A, Barg E (2020). PC12 cell line: Cell types, coating of culture vessels, differentiation and other culture conditions. Cells 9: 958. [Google Scholar] [PubMed]
Xiong ZL, Wang Y, Ma XL, Zhou C, Jiang XW, Yu WH (2022). Based on proteomics to explore the mechanism of mecobalamin promoting the repair of injured peripheral nerves. Canadian Journal of Physiology and Pharmacology 100: 562–572. [Google Scholar] [PubMed]
Yao Y, Wang J, He T, Li H, Hu J, Zheng M, Ding Y, Chen YY, Shen Y, Wang LL (2020). Microarray assay of circular RNAs reveals cicRNA. 7079 as a new anti-apoptotic molecule in spinal cord injury in mice. Brain Research Bulletin 164: 157–171. [Google Scholar] [PubMed]
Yu C, Wang X, Qin J (2023). Effect of necrostatin-1 on sciatic nerve crush injury in rat models. Journal of Orthopaedic Surgery and Research 18: 1–9. https://doi.org/10.1186/s13018-023-03565-3 [Google Scholar] [PubMed] [CrossRef]
Yuan YS, Yu F, Zhang YJ, Niu SP, Xu HL, Kou YH (2021). Changes in proteins related to early nerve repair in a rat model of sciatic nerve injury. Neural Regeneration Research 16: 1622–1627. https://doi.org/10.4103/1673-5374.301025 [Google Scholar] [PubMed] [CrossRef]
Yue L, Talukder M, Gurjar A, Lee JI, Noble M, Dirksen RT, Chakkalakal J, Elfar JC (2019). 4-Aminopyridine attenuates muscle atrophy after sciatic nerve crush injury in mice. Muscle & Nerve 60: 192–201. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools