 Open Access
Open Access
ARTICLE
Heterogeneity beyond tumor heterogeneity—SULF2 involvement in Wnt/β-catenin signaling activation in a heterogeneous side population of liver cancer cells
1 Division of Gastroenterology & Hepatology, The University of Hong Kong-Shenzhen Hospital, Shenzhen, 518053, China
2 Shenzhen Key Laboratory of Gene and Antibody Therapy, Center for Biotechnology and Biomedicine, State Key Laboratory of Health Sciences and Technology, State Key Laboratory of Chemical Oncogenomics, Precision Medicine and Healthcare Research Center, Tsinghua-Berkeley Shenzhen Institute (TBSI), Shenzhen International Graduate School, Tsinghua University, Shenzhen, 518055, China
3 Department of Chemistry, Tsinghua University, Beijing, 100084, China
* Corresponding Authors: DONGYE YANG. Email: ; LAIQING HUANG. Email:
# These authors contributed equally to this work
(This article belongs to the Special Issue: Application of Deep Learning in Cancer)
BIOCELL 2023, 47(9), 2037-2049. https://doi.org/10.32604/biocell.2023.028863
Received 16 January 2023; Accepted 28 June 2023; Issue published 28 September 2023
Abstract
Introduction: Sulfatase 2 (SULF2), an endogenous extracellular sulfatase, can remove 6-O-sulfate groups of glucosamine residues from heparan sulfate (HS) chains to modulate the Wnt/β-catenin signaling pathway, which plays an important role in both liver carcinogenesis and embryogenesis. Side population (SP) cells are widely identified as stem-like cancer cells and are closely related to carcinoma metastasis, recurrence, and poor patient prognosis. However, the roles of SULF2 in SP cells of hepatomas are unclear, and the underlying mechanism is undefined. Objectives: This study aimed to compare the heterogeneity between SP cells and non-side population (NSP) cells derived from three different liver cancer cell lines and to elucidate the involvement of the SULF2-Wnt/β-catenin axis in liver cancer stem cells (CSCs) and its impact on the processes of carcinogenesis and invasiveness. Methods: In this work, three different liver cancer SP cells (HepG2, Huh7, and PRC/PRL/5) were sorted by flow cytometry. We also examined the migration and invasion behaviors of SP and NSP cells. To determine if this high tumorigenic potential of SP cells is correlated to SULF2, qPCR, western blotting, and immunofluorescence analysis were conducted. We also performed nude mouse xenograft experiments for in vivo analysis. Results: The results from the in vitro colony formation assay showed that SP cells exhibited a 2-fold higher colony formation efficiency compared to their NSP counterparts. The SP cells exhibited significantly higher potentials in terms of their migratory capacity and invasive ability compared to NSP cells. We found that higher expression of SULF2 in SP cells was associated with greater capabilities for clonogenicity, migration, and invasion. It was also linked to higher activation of the Wnt/β-catenin signaling pathway via stimulation of key downstream factors, particularly β-catenin, c-Myc, and cyclin D1. Further, a positive correlation between the upregulated SULF2 expression and tumorigenesis in the in vivo nude mouse xenograft models was demonstrated, highlighting that the potential underlying mechanism was Wnt/β-catenin signaling pathway activation. Conclusion: Our findings show that variable SULF2 expression was associated with differential activation of the Wnt/β-catenin signaling pathway, which could lead to behavioral differences between SP and NSP cells and also among the SP cells of the three liver cancer cell lines assessed. It was reasonably concluded that the SULF2-Wnt/β-catenin axis could play an important role in the tumorigenicity of liver cancer stem cells.Graphic Abstract
Keywords
Abbreviations
| qPCR | quantitative real-time polymerase chain reaction |
| SULF2 | sulfatase 2 |
| HS | heparan sulfate |
| SP | side population |
| CSCs | cancer stem cells |
| EpCAM | epithelial cell adhesion molecule |
| ABC | adenosine triphosphate (ATP)-binding cassette |
| HSPGs | heparan sulfate proteoglycans |
| GPC3 | glypican 3 |
| OD | optical density |
| PBS | phosphate-buffered saline |
| MTT | 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide |
| HSGAGs | heparan sulfate glycosaminoglycans. |
Hepatocellular carcinoma (HCC) is a common malignancy worldwide, and more than 53% of HCC patients are from China (Wu and Qin, 2013). Currently, the main curative treatment for HCC is surgical resection, which is also the first choice for early-stage HCC patients with tumor sizes less than 3 cm (Dahiya et al., 2010). However, the current clinical outcome is still unsatisfactory, partially due to postoperative tumor recurrence. It has been reported that approximately 50% of patients may experience tumor recurrence, which manifests as intra- or extra-hepatic metastasis within 2 years (Chun et al., 2011; Portolani et al., 2006; Zhou et al., 2014) after curative hepatectomy. Therefore, it is still of great importance to explore the molecular mechanism underlying liver cancer recurrence to improve clinical prognosis.
Cancer stem cells (CSCs), a relatively small population of cancer cells, have “stem cell-like” characteristics and have the ability to initiate and maintain primary tumorigenesis (Kucia et al., 2005; Nguyen et al., 2017; Wang et al., 2014). Accumulating experimental and clinical evidence shows that a low abundance of CSCs contributes to tumor recurrence and metastasis in multiple types of cancers, including liver cancer (Chang, 2016; Guo et al., 2016; Peitzsch et al., 2017). CSCs can detach from the tumor mass, invade the lymph nodes and peripheral blood, and then arrive at a unique “niche”, where they are protected from damage and initiate tumor recurrence or metastasis (Kreso and Dick, 2014; Lloyd et al., 2016; Vicente-Duenas et al., 2010). Therefore, CSCs are an ideal therapeutic target for suppressing tumor recurrence or metastasis to improve clinical prognosis.
It has been reported that CSC populations in liver cancer can express several cell surface markers, such as CD13, CD44, CD90+, CD133, or epithelial cell adhesion molecule (EpCAM) (Haraguchi et al., 2010; Yamashita et al., 2013; Yang et al., 2008; Zhu et al., 2010). However, isolation of liver CSCs is still a challenge due to the lack of a standard set of surface markers. One of the strategies for isolating liver CSCs is the sorting of SP cells. SP cells have shown functions different from those of NSP cells and have been considered a substitute for CSCs, to some extent, in several studies (Dubrovska et al., 2012; Feng et al., 2015; Guo et al., 2016; Liao et al., 2015; Summer et al., 2003; Teshima et al., 2014; Van den Broeck et al., 2013; Wang et al., 2017; Zhao et al., 2016). Isolation of SP cells is typically based on their ability to pump out Hoechst 33342 dye given the high expression of adenosine triphosphate (ATP)-binding cassette (ABC) membrane transporters on their membranes (27, 28 Goodell, 2005; Patrawala et al., 2005), a characteristic of stem cells (Ding et al., 2010; Hu et al., 2017; Tiezzi et al., 2013). Further, SP cells cannot pump out Hoechst 33342 dye when verapamil, a calcium channel antagonist, is used to inhibit ABC membrane transporters, whereas the influence of verapamil on NSP cells is negligible (Scharenberg et al., 2002).
Sulfatase 2 (SULF2), a heparin-degrading endosulfatase, can selectively remove 6-O-sulfate groups from heparan sulfate proteoglycans (HSPGs) and plays a role in organogenesis and tumorigenesis. SULF2 can also promote liver carcinogenesis by interacting with glypican 3 (GPC3) on the cytoplasmic membrane to activate downstream signaling pathways such as the Wnt/β-catenin pathway. In addition, SULF2 has been reported to have an oncogenic function in promoting tumorigenesis (Lai et al., 2010). Besides, elevated expression of SULF2 in HCC patients has been linked to a worse prognosis and a higher risk of recurrence after surgery (Lai et al., 2008).
The Wnt/β-catenin signaling pathway can not only stimulate stem/progenitor cell proliferation and differentiation (Masckauchan et al., 2005; Reya et al., 2003) but also play a crucial role in liver carcinogenesis due to its abnormal activation. Wnts, such as Wnt3a, can activate diverse cancer-associated processes of proliferation, survival, and metastasis (Wei et al., 2017) by promoting the translocation of cytoplasmic β-catenin into the nucleus to regulate the expression of downstream proto-oncogenes such as c-Myc (Lim et al., 2008; Wang et al., 2011).
This study aimed to investigate the role of the SULF2-Wnt/β-catenin axis in liver CSCs and its contribution to carcinogenesis and invasiveness. Specifically, SP cells derived from three human liver cancer cell lines (HepG2, Huh7, and PLC/PRF/5) were compared, as these cell lines originate from hepatitis B virus (HBV)-infected patients and exhibit varying expression levels of SULF2 and GPC3. Phenotypic properties of SP cells from each cell line (HepG2: SULF2+/GPC3+, Huh7: SULF2+/GPC3−, and PLC/PRF/5: SULF2-/GPC3−) were examined and compared with their respective NSP counterparts. Moreover, the activation of the SULF2-Wnt/β-catenin axis was evaluated by analyzing the localization and expression levels of key factors, including β-catenin, c-Myc, and cyclin D1. Notably, significantly higher levels of these factors were observed in SP cells derived from both the cell lines and tumor tissues. These findings contribute to our understanding of the SULF2-Wnt/β-catenin axis in liver cancer CSCs, shedding light on its potential significance in liver cancer development and progression.
Relative abundance of side population cells
The proportions of SP and NSP cells in liver cancer cells [HepG2 (SULF+/GPC3+), Huh7 (SULF2+/GPC3−) and PLC/PRF/5 (SULF2−/GPC3−)] were analyzed by flow cytometry based on the ability of intracellular retention of Hoechst 33342 dye. Dead cells and debris were easily removed because they emitted intense red fluorescence from labeling with propidium iodide, a red fluorescent dye. The percentages of SP cells in HepG2, Huh7, and PLC/PRF/5 cells were 0.442% ± 0.3%, 0.619% ± 0.5%, and 8.71% ± 0.8%, respectively. Notably, the percentage of SP cells within PLC/PRF/5 cells was 8.71% ± 0.8%, which was significantly higher than the percentages of SP cells within HepG2 and Huh7 cells (Fig. 1; p < 0.001).
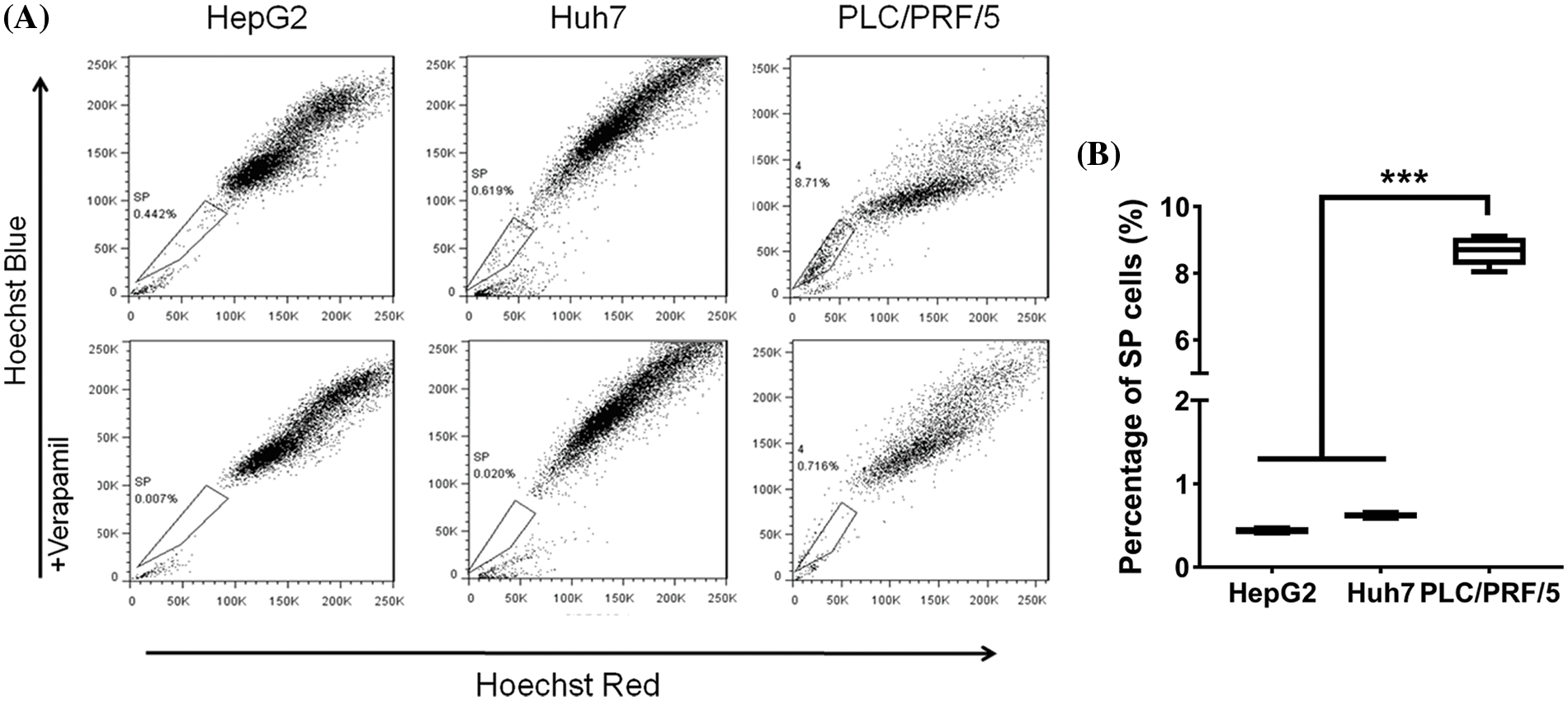
Figure 1: (A) Isolation of SP (boxed) and NSP cells from HepG2, Huh7, and PLC/PRF/5 using flow cytometry. Addition of 50 µM verapamil (bottom row) served to block SP cells from pumping out Hoechst Blue dye. (B) The percentage of SP cells differed among the three cell lines: PLC/PRF/5 (8.71%) > Huh7 (0.619%) > HepG2 (0.442%). The results are presented as mean ± s.d., n = 3, ***p < 0.001.
Comparison of sulf2 mRNA expression in side population and non-side population cell lines
The sulf2 mRNA level was measured in HepG2, Huh7, and PLC/PRF/5 cells. Among these three cell lines, sulf2 was expressed at the highest level in Huh7 cells, while it was barely detected in PLC/PRF/5 cells (Fig. 2). This was consistent with a previous report indicating that Huh7 cells are positive for SULF2 expression (Lai et al., 2010). As shown in Fig. 2, sulf2 mRNA expression was approximately 3-fold higher in PLC/PRF/5-SP cells than in PLC/PRF/5-NSP cells, this expression was also seen in HepG2 and Huh7 cell lines. Overall, the mRNA expression of sulf2 was higher in SP cells than in their NSP counterparts.
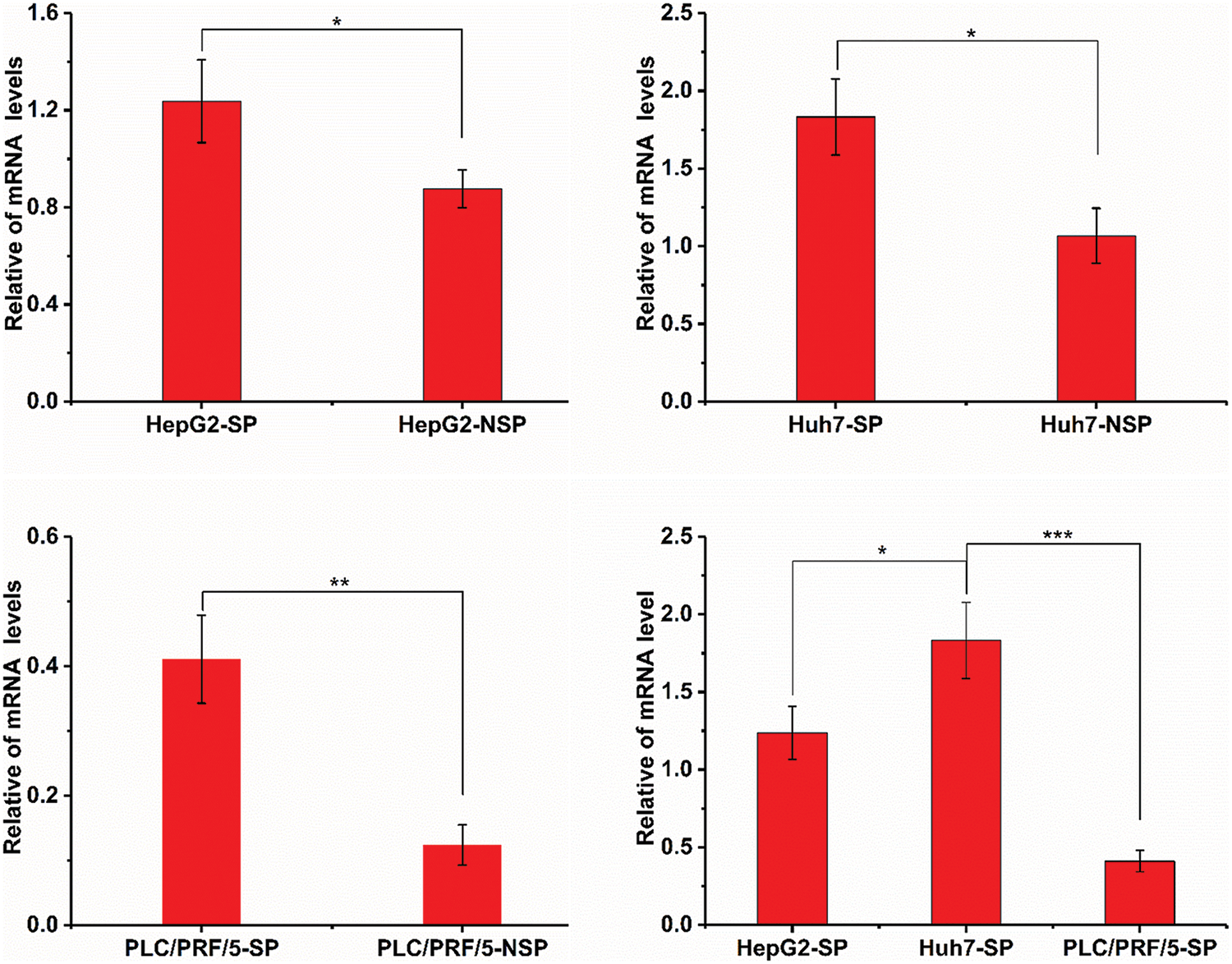
Figure 2: Detection of sulf2 mRNA levels in SP and NSP cells of HepG2, Huh7, and PLC/PRF/5 cell lines. The expression of sulf2 mRNA is relatively higher in SP cells than in NSP cells. Among the three SP cells, Huh7-SP shows the highest sulf2 mRNA level. The results are presented as mean ± s.d., n = 5, *p < 0.05, **p < 0.01, ***p < 0.001.
High viability of side population cells
MTT assays were performed to demonstrate the in vitro proliferation of SP and NSP cells. The number of living cells stained by MTT was determined by measurement of the optical density (OD) at 490 nm. In all evaluated cell lines, SP cells had higher proliferation rates than NSP cells, especially at 48 h (Fig. 3; p < 0.01). As each cell population was cultured for up to 4 days, appreciable differences were observed between SP and NSP cells, with the most significant differences observed in Huh7 cells. Specifically, the relative number of cells in the Huh7-NSP cell population was only 0.71 ± 0.3 on day 4, while it was 1.27 ± 0.7 in the Huh7-SP cell population, and this trend became more evident on the fifth day. In addition, the relative number of SP cells decreased in the following order: Huh7-SP > HepG2-SP > PLC/PRF/5-SP.
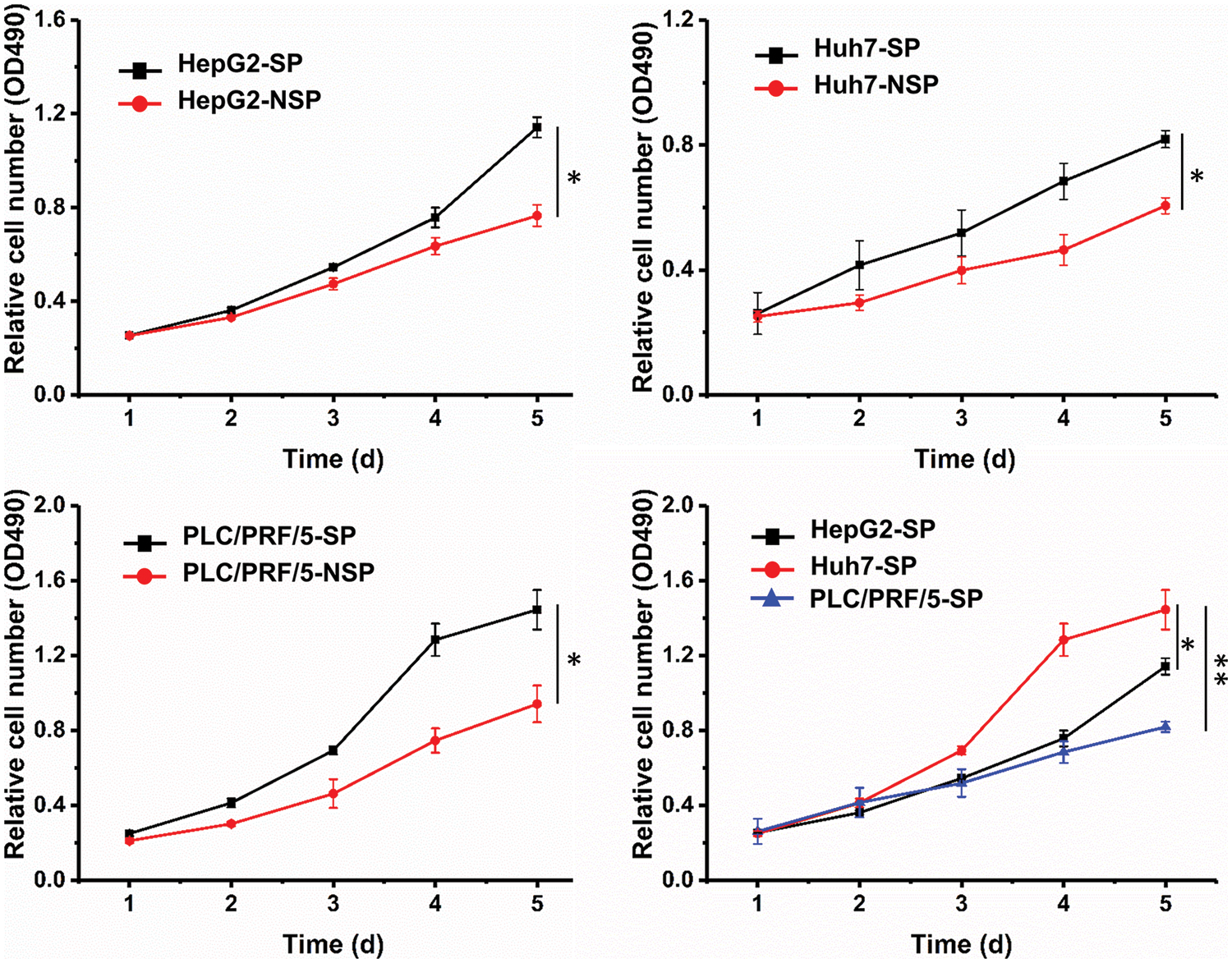
Figure 3: SP and NSP cells were seeded in 96-well plates at the indicated time points. The number of living cells stained by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) was determined by measurement of the optical density (OD) at 490 nm. The viability of SP cells was significantly higher. The results are presented as mean ± s.d., n = 5, *p < 0.05, **p < 0.01.
High clonogenicity of side population cells
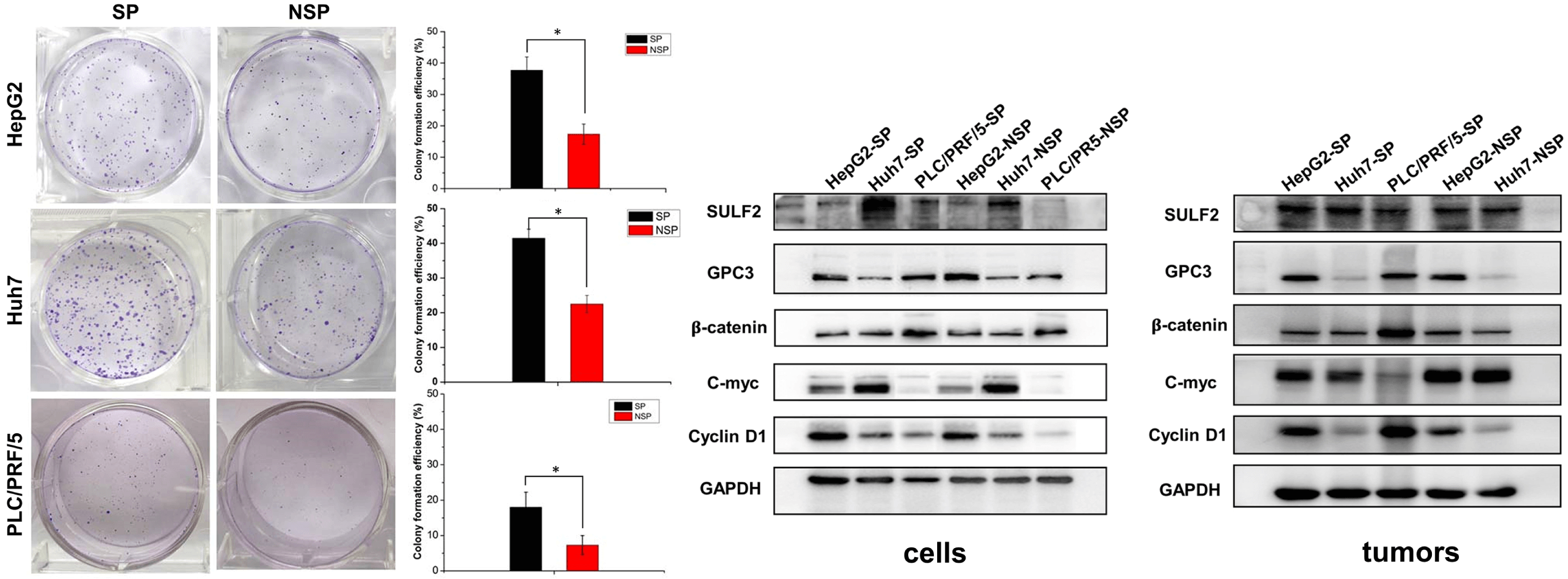
To investigate the difference in the clonogenic ability of SP and NSP cells, we carried out a colony formation assay (Fig. 4). Compared with that of NSP cells of each cell line, the colony-forming capacity of SP cells was significantly higher after 7 days of culture on agar. The colony formation rate was calculated with the following formula: clonogenic number/cell seed number. Semiquantitative analysis suggested that the colony formation rates of SP HepG2, Huh7, and PLC/PRF/5 cells were 38.1%, 40.9%, and 17.7%, respectively, approximately 2-fold higher than those of the counterpart NSP cells (Fig. 4). These findings were consistent with the in vitro proliferation rates of SP and NSP cells.
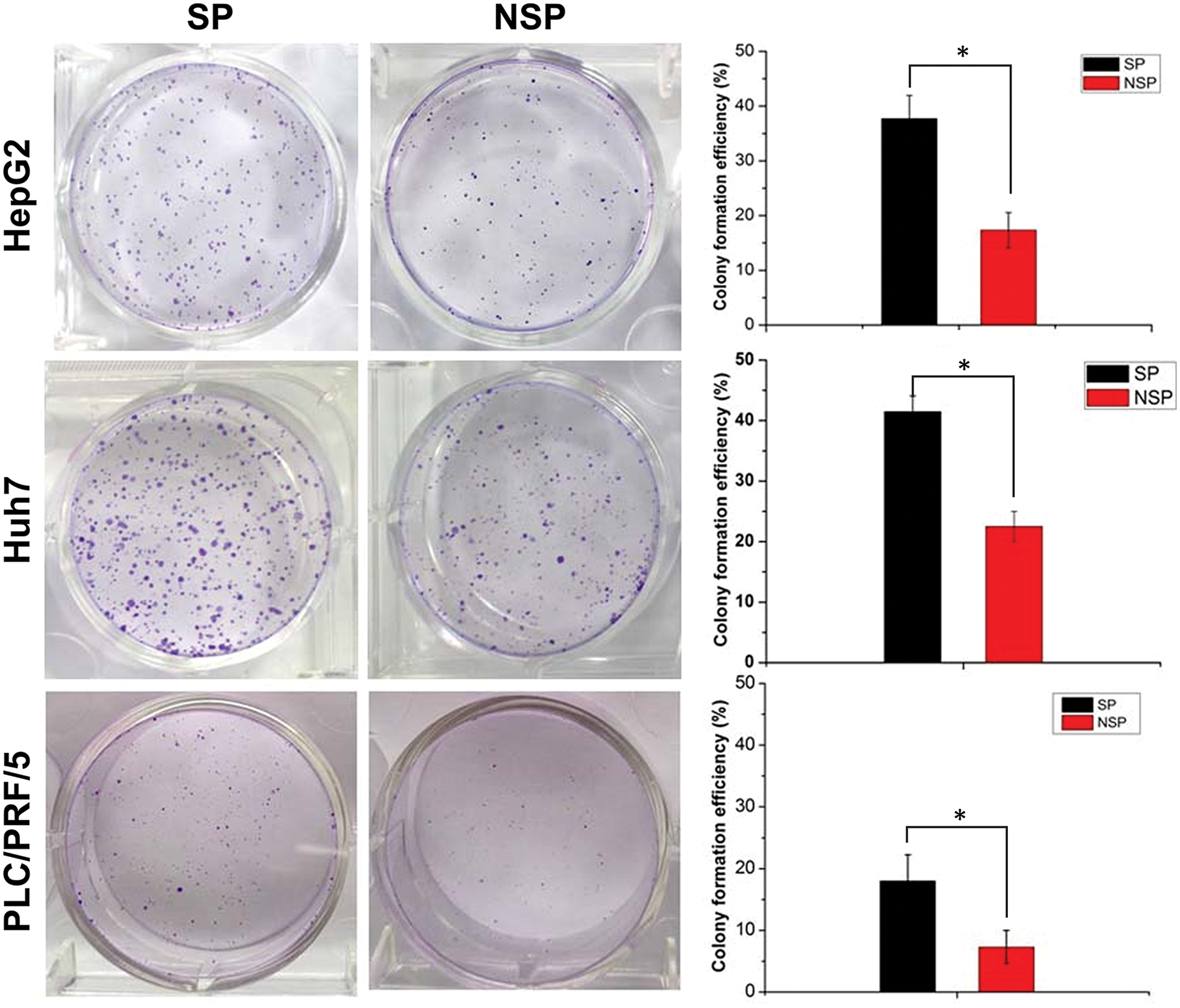
Figure 4: The colony formation (clonogenicity) assay where SP and NSP cells were seeded in six-well plates and cultured for 7 days, and then subjected to crystal violet staining. The histogram represents the statistical analysis of the efficiency of clone formation. The results are presented as mean ± s.d., n = 3, *p < 0.05.
Migration and invasion capabilities of different cell subpopulations
With HepG2-NSP cells as control cells, we performed Transwell-based migration and invasion assays to demonstrate the in vitro migration and invasion capabilities of SP cells. HepG2-SP cells had a higher migration ability than the corresponding NSP cells (Fig. 5A). The Transwell invasion assay also showed this result (Fig. 5A). Further, Huh7 and PLC/PRF/5 SP cells showed a similar pattern, which further confirmed this observation (Figs. 5B and 5C).
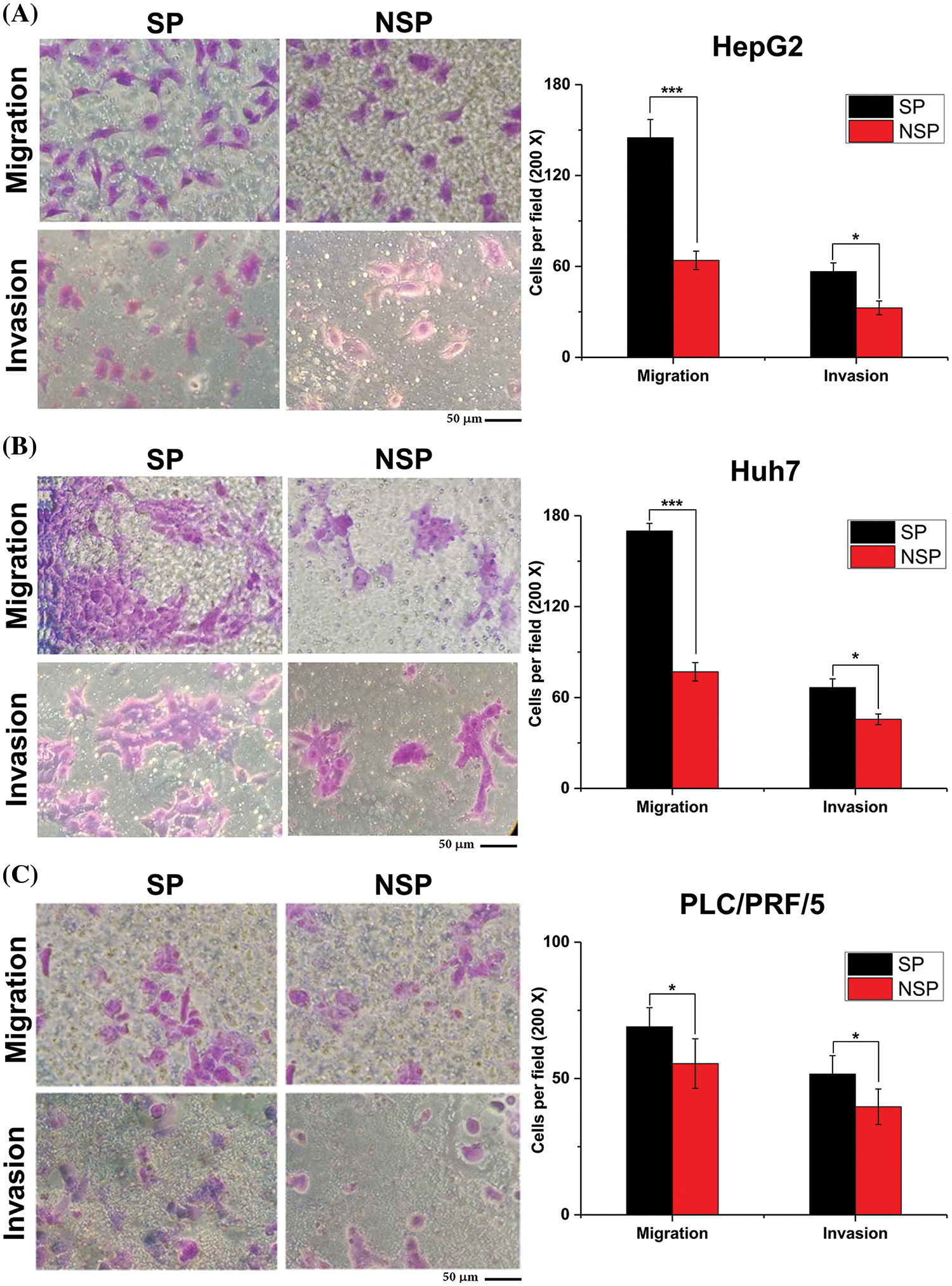
Figure 5: Transwell assays were performed to detect migration and invasion capabilities of SP and NSP cells from HepG2, Huh7, and PLC/PRF/5 cell lines. The histogram represents the statistical analysis of the number of cells in each visual field. Scale bars: 50 μm. Staining was done with crystal violet; magnification, 200×. The results are presented as mean ± s.d., n = 3, *p < 0.05, ***p < 0.001.
We performed immunofluorescence and immunoblot assays to examine the cellular localization and expression level of β-catenin in SP cells of the three cell lines. As shown in Fig. 6, β-catenin was expressed not only in the cytoplasm but also in the nucleus of HepG2-SP cells. However, β-catenin was expressed only in the cytoplasm of both Huh7-SP and PLC/PRF/5-SP cells. The expression level of β-catenin was higher in SP cells than in NSP cells. Next, we conducted a double immunofluorescence (IF) staining assay to further demonstrate whether the total expression of β-catenin was associated with the upstream SULF2 and downstream c-Myc molecules. It was shown high SULF2 expression was accompanied by high c-Myc expression. Further validation was conducted by immunoblotting.
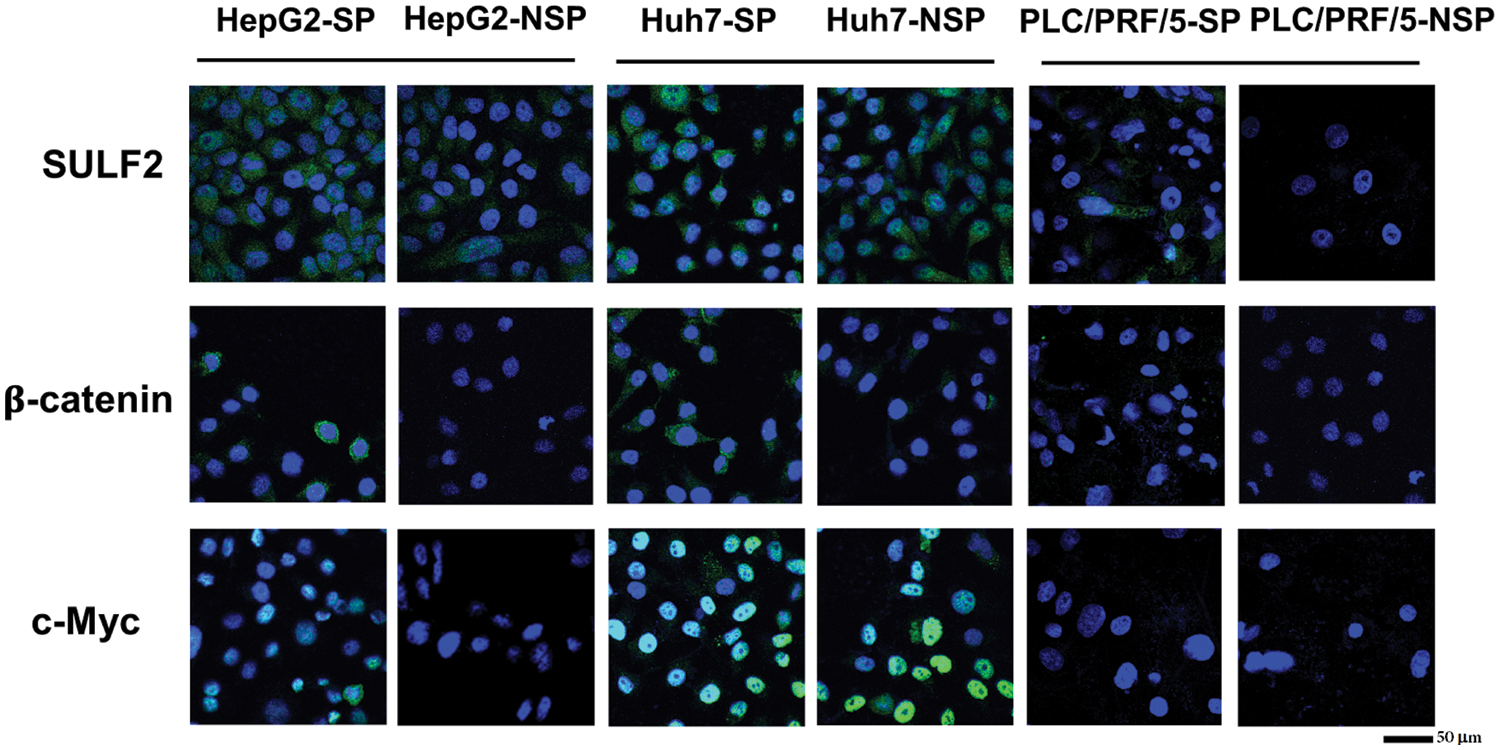
Figure 6: The expression levels of SULF2, β-catenin, and c-Myc in SP and the corresponding NSP cells of HepG2, Huh7, and PLC/PRF/5 cell lines were evaluated by immunofluorescence staining. Scale bars: 50 μm.
Evaluation of in vivo tumor formation
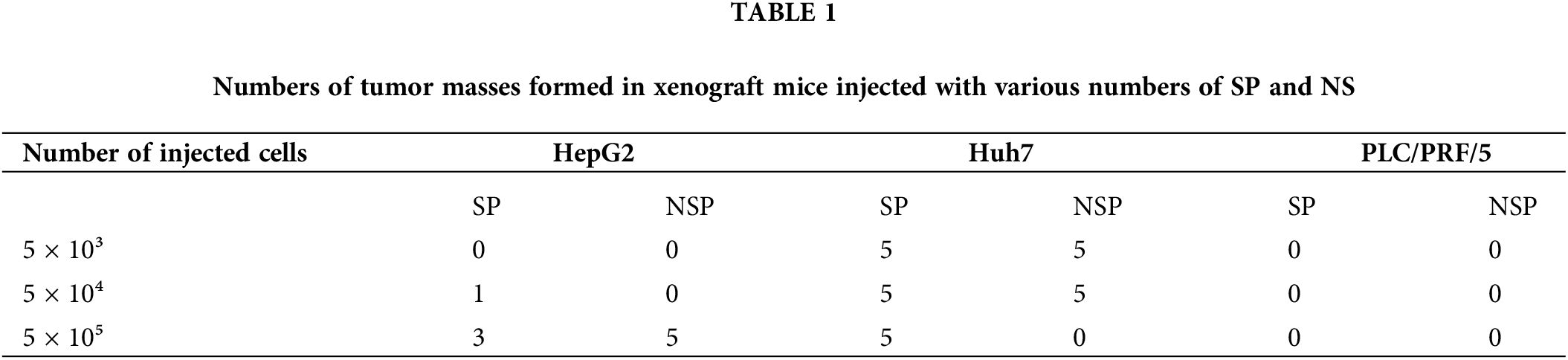
To investigate tumorigenesis in vivo, we established nude mouse xenograft models with both SP and NSP cells of different liver cancer cell lines. As shown in Table 1, both Huh7-SP and Huh7-NSP cells exhibited significant tumor formation ability, while PLC/PRF/5 cells could not form tumors even at the highest injected concentration. Overall, the tumourigenic ability of the cells in the three SP subpopulations decreased in the following order: Huh7-SP > HepG2-SP > PLC/PRF/5-SP. Compared with SP cells, NSP cells hardly formed tumors in our mouse models, consistent with the results of the in vitro assays.
Immunohistochemical assays were performed to examine the key factors involved in the SULF2-Wnt/β-catenin axis, including SULF2, GPC3, β-catenin, c-Myc, and cyclin D1. Since PLC/PRF/5-NSP cells engrafted into nude mice did not form tumors due to their poor proliferative ability, as indicated in Table 1, only 5 groups of mouse tissues (HepG2-SP/NSP, Huh7-SP/NSP, and PLC/PRF/5-SP) were analyzed by immunofluorescence and Western blotting. The expression of SULF2 was highest in tumor tissues that were harvested from the five groups of xenografted mice. Further, β-catenin had the highest expression level in PLC/PRF/5-SP mice (Fig. 7). The expression of c-Myc and cyclin D1 was also detected simultaneously where c-Myc expression was not only increased but also localized mainly in the nucleus in SP cells compared to NSP cells. However, cyclin D1 expression did not differ significantly among the indicated cells. In addition, while GPC3 expression was greatly increased in PLC/PRF/5-SP cells, it was only slightly increased in the other cells.
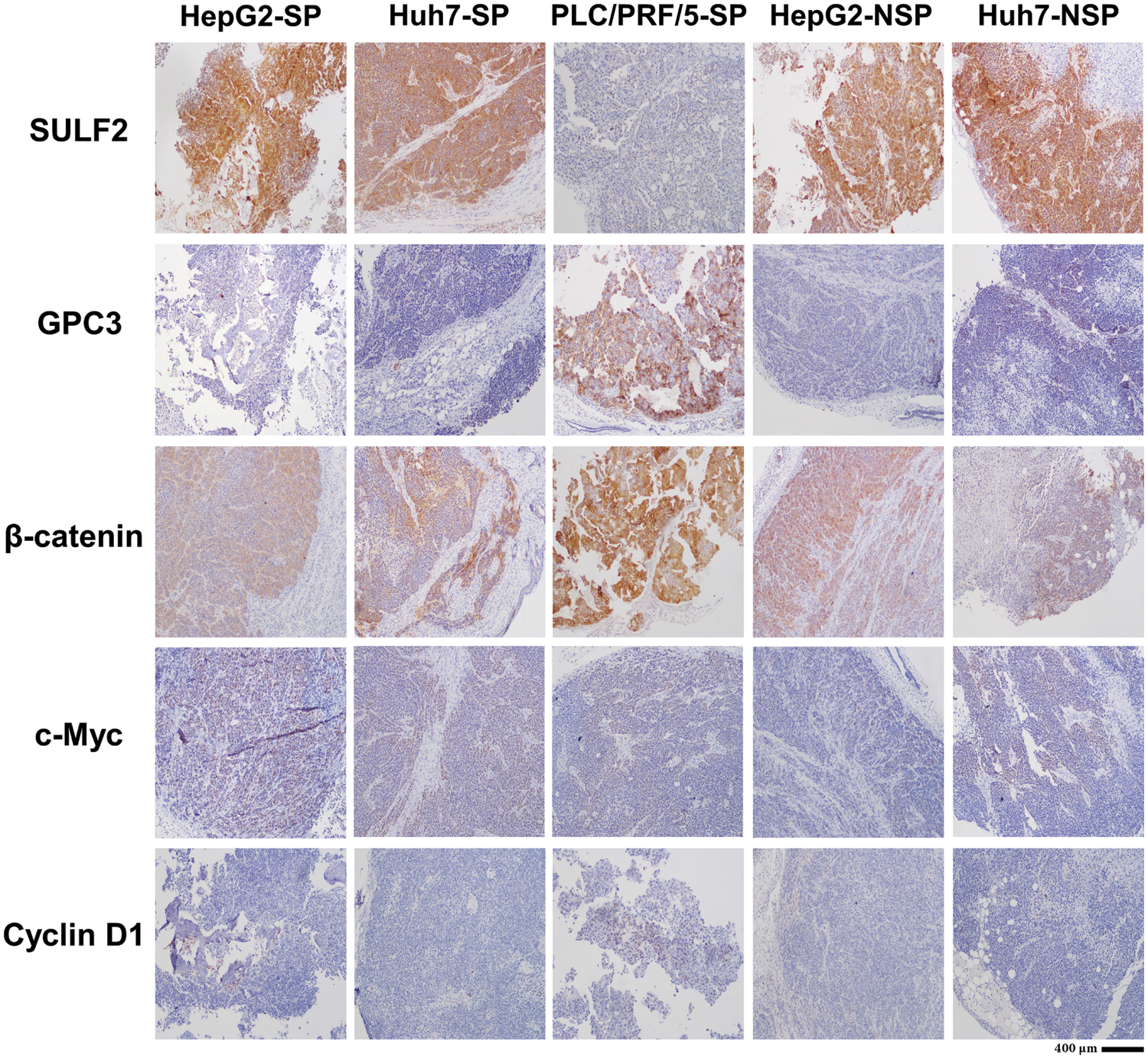
Figure 7: Immunohistochemical analysis of SULF2, GPC3, β-catenin, c-Myc, and Cyclin D1 expression in tumor tissues from the indicated groups of nude mice. Nuclei were stained by 4′,6-diamidino-2-phenylindole (DAPI). Scale bars: 400 µm.
Western blotting analysis results
The expression patterns of SULF2 and GPC3 proteins were similar to that of the mRNA expression patterns observed. However, SULF2 expression was higher in SP cells, especially Huh7-SP cells, than in the corresponding NSP cells. In parallel experiments, increased c-Myc expression was correlated with increased SULF2 expression in both tumor tissue and cell lines. However, the expression of the other downstream component, cyclin D1, was variable (Fig. 8).
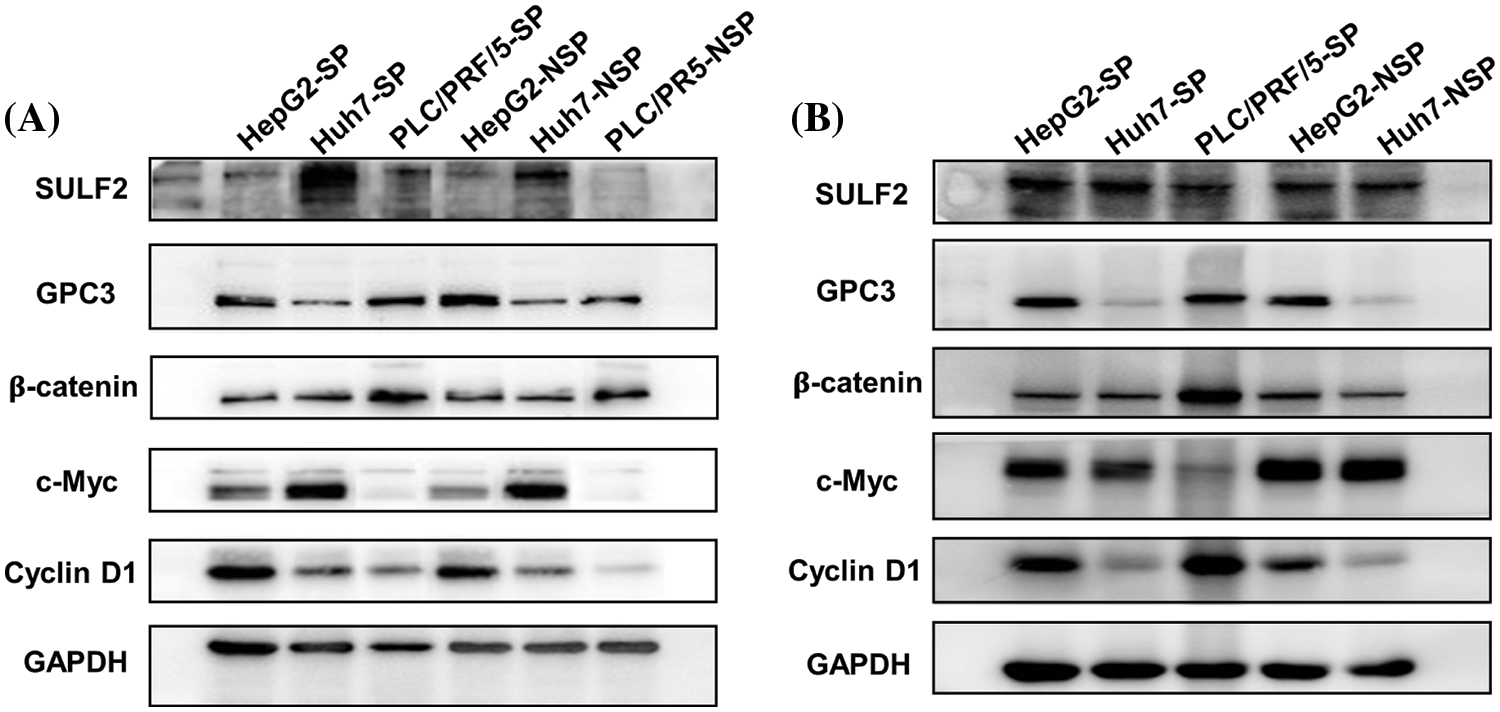
Figure 8: Western blot analysis of (A) different HepG2, Huh7, and PLC/PRF/5 cell lines including SP and the corresponding NSP cells, and (B) lysates of harvested tumor tissue from xenografted mice.
SULF2 has a highly conserved role in embryo development and has been documented to have a high expression in several cancers (Rosen and Lemjabbar-Alaoui, 2010). SULF2 can also promote tumorigenesis in the liver by activating the Wnt/β-catenin signaling pathway (Nakamura et al., 2013). Thus, we hypothesized that the SULF2-Wnt/β-catenin axis plays an important role in maintaining CSC-like properties associated with liver cancer recurrence and metastasis (Jang et al., 2015).
In this study, SP cells, a proxy for CSCs, and NSP cells were sorted by flow cytometry from two HCC cell lines with different SULF2 and GPC3 expression levels—Huh7 (SULF2+/GPC3−) and PLC/PRF/5 (SULF2−/GPC3−) and one hepatoblastoma cell line—HepG2 (SULF+/GPC3+) derived from patients with HBV-related liver cancer. Despite their lower proportions (i.e., PLC/PRF/5 (8.71%) > Huh7 (0.619%) > HepG2 (0.442%)), SP cells exhibited higher proliferation than their NSP counterparts, which implied that a minority of SP cells may be responsible for tumor initiation and progression. In addition, we initially verified that the SULF2 expression level not only dominantly determined the difference between SP and NSP cells of various liver cancer cell lines but also activated the Wnt/β-catenin signaling pathway. Both the proliferation and invasion capabilities of SP cells were much stronger than those of NSP cells, as validated by the in vitro MTT, colony formation, and transwell-based migration and invasion assays. Moreover, the SULF2-Wnt/β-catenin axis was further studied through Western blot and immunofluorescence analyses. Overall, SP cells had stronger tumor formation and metastasis abilities, consistent with previous studies (Britton et al., 2011; Fukuda et al., 2009; Szotek et al., 2006).
The relationships between SULF2 expression and SP cell behaviors were evaluated, and the probable molecular mechanism was explored. The main findings from this study are as follows:
1. SULF2 expression was higher in SP cells than in NSP cells, and the expression decreased in the following order: Huh7-SP > HepG2-SP > PLC/PRF/5-SP, which was also consistent with cell behaviors, such as migration, invasion, and tumorigenesis both in vitro and in vivo.
2. SULF2 activated the Wnt/β-catenin signaling pathway, as demonstrated by the elevated in vitro and in vivo expression of signaling components such as β-catenin, c-Myc, and cyclin D1 in SP cells.
3. In the xenograft mouse models, SP cells from HepG2, Huh7, and PLC/PRF/5 cell lines were more likely to contribute to tumor formation and xenograft tumor growth. Further, SP cells proliferated significantly faster than the corresponding NSP cells, which demonstrated the enhanced tumourigenicity of SP cells compared to NSP cells.
Wnt signaling is the key driver of cell growth and is constitutively activated in stem and stem-like cells (Katoh, 2017). Accumulating evidence suggests that abnormal Wnt signaling occurs in HCC and that such aberrant Wnt signaling mediates a multitude of signal transduction pathways in HCC cells (Khalaf et al., 2018; Perugorria et al., 2019; Wang et al., 2019). The higher the level of SULF2 expressed by liver cancer cells, the proliferation and invasion capacities were greater, while the tumourigenicity was weaker when the level of SULF2 was lower. This implied that the SULF2-Wnt/β-catenin axis may have been activated to varying degrees.
The immunofluorescence assay results indicated that the fluorescence intensity of β-catenin, which is indispensable in Wnt/β-catenin signaling, was fairly strong in SP cells, though the expression of β-catenin in NSP cells remained at a rather lower level. In addition, upregulated SULF2 expression correlated with increased expression of downstream transcription factors, including c-Myc and cyclin D1, as validated by immunofluorescence assays, which indicated that aberrant activation of the Wnt/β-catenin signaling pathway occurs in SP cells. Furthermore, fluorescence images of some cells (e.g., HepG2-SP, Huh7-SP) showed that β-catenin was present not only in the cytoplasm but also in the nucleus, which was consistent with the canonical Wnt/β-catenin signaling pattern. Based on our knowledge, c-Myc is involved in the Wnt signaling pathway and regulates the aberrant proliferation of cancer cells upon activation. Although SULF2 expression was positively correlated with c-Myc in most cells (HepG2-SP, Huh7-SP, Huh7-NSP), the relationship between the two factors remains unclear, as contrasting results have been observed in other cell types (HepG2-NSP, PLC/PRF/5-SP), which may be associated with low expression of β-catenin. We, therefore, hypothesize that SULF2-Wnt/β-catenin is a complete cellular axis comprising upstream, midstream, and downstream factors and that SULF2 may play a role in triggering and driving the Wnt/β-catenin signaling pathway. These results were confirmed by Western blot analysis, which was used to detect β-catenin along with other essential components in the Wnt/β-catenin pathway.
Since β-catenin is a downstream effector of the Wnt/β-catenin pathway, its expression is not only impacted by the SULF2-Wnt/β-catenin axis, but also may be influenced by crosstalk with other pathways, which probably is the reason why β-catenin expression commonly varies (Kim et al., 2017). Collectively, these observations indicate that SULF2 positively regulates tumorigenesis and progression. When SULF2 expression is elevated, the Wnt/β-catenin pathway can be activated to lead cells to exhibit a more malignant stem-like cell phenotype. However, when SULF2 expression is insufficient, the stem-like cell properties would not be maintained even though the downstream components of the pathway are upregulated.
We also experimentally examined the expression of GPC3, and our results are in agreement with previous results showing that GPC3 is not able to transmit signals to activate its downstream pathway in the absence of SULF2 (Gao et al., 2014; Lai et al., 2008). This is probably because SULF2 influences the sulfation of GPC3 heparan sulfate glycosaminoglycans (HSGAGs) and subsequently induces changes in Wnt/β-catenin signaling in liver cancer cells. In addition, our experiments show that the trends in the expression of c-Myc and cyclinD1 were not exactly the same. This can be explained by the observation that although β-catenin is an upstream mediator controlling both factors, the cyclinD1 gene is closely related to the cell cycle and its expression may be associated with the cell cycle phase, while c-Myc is an oncogene that is probably simultaneously regulated by other pathways.
Different behaviors of SP and NSP cells imply the heterogeneity of liver cancer cells. To further investigate the Wnt/β-catenin signaling pathway in vivo, xenograft experiments were conducted, and the results showed that tumorigenesis correlated with the SULF2 expression level. Additionally, immunohistochemical analysis of SP cell-derived tumor tissues showed greater Wnt/β-catenin signaling pathway activity in HepG2-SP and Huh7-SP cells than in PLC/PRF/5-SP cells. This was consistent with the immunofluorescence results but suggested that GPC3 expression could change the tumorigenesis of SP cells via an unknown mechanism, which needs further investigation.
In summary, we conducted a comprehensive comparison between SP and NSP cells derived from three HBV-related liver cancer cell lines by examining various aspects of their characteristics. Our findings demonstrated that SP cells exhibited enhanced abilities in proliferation, clonogenicity, migration, and invasion compared to NSP cells. These observations suggest that SP cells possess the hallmark features associated with CSCs, including heightened proliferative and clonogenic potentials, as well as increased migratory and invasive capabilities. Notably, our investigations revealed that the upregulation of SULF2 expression significantly contributed to the excessive activation of the Wnt/β-catenin signaling pathway, specifically in SP cells across the three liver cancer cell lines studied. Conversely, corresponding NSP cells did not exhibit this phenomenon. Importantly, the distinct subpopulations of cells sorted from the same parental cancer cell population displayed notable differences, such as the activation of diverse signaling pathways, variable degrees of pathway activation, or differential expression levels of genes and proteins. These findings underscore the intricate nature of tumor heterogeneity, necessitating in-depth exploration and elucidation of the underlying mechanisms. In addition, The SULF2 overexpression and knockdown experiments are a robust way to validate our claims, so further studies can be explored in more depth, and the follow-up experiments would consolidate the conclusions.
Many reports in the literature have reached a consensus regarding tumor heterogeneity, stating that tumors are constituted by multiple heterogeneous populations of cells, which is one manifestation of tumor heterogeneity. In this study, we also found heterogeneity in selected cell lines, which means that there are still large differences in the biological behavior of tumor cells even if they are the same species as we have always thought. Our study suggests that SULF2 could be a key regulator in liver cancer cells to trigger a range of abnormal downstream signaling activations, particularly Wnt/β-catenin signaling. Tumors have long been studied on the basis of bulk mass; however, the present study emphasizes that exploration at the single-cell level is urgently needed in cancer research. Collectively, these findings prompt us to further understand tumors at the single-cell level and to reveal the relationship between the behaviors of single cells and subpopulations of cancer cells. These studies lay a solid foundation for future studies and will help us to understand and prevent liver cancer recurrence and metastasis and right (1.31 cm). Acceptable font is Minion Pro, 10 pt., except for writing special symbols and mathematical equations. Use 0.7 cm intend on the first line of second paragraph.
Three human liver cancer cell lines, HepG2, Huh7, and PLC/PRF/5, were purchased from the American Type Culture Collection (ATCC, Rockville, MD, USA). All cells were cultured in high-glucose Dulbecco’s modified Eagle’s medium (DMEM, Gibco, Paisley, UK) containing 10% (v/v) heat-inactivated fetal bovine serum (FBS, Gibco, Paisley, UK) and 100 U/ml streptomycin/penicillin in an incubator at 37°C under 5% CO2.
Isolation of side population cells
All cultured cells were detached from the culture dishes by using Gibco TrypLE Express Enzyme (Thermo Fisher Scientific, CA, USA) and re-suspended in DMEM containing 5% FBS for adjustment of the final concentration to 1 × 106 cells/ml. Cells were incubated with 5 mg/l Hoechst 33342 dye (Sigma-Aldrich, St. Louis, MO, USA) at 37°C for 90 min in the presence or absence of 50 µM verapamil (Sigma-Aldrich, St. Louis, MO, USA). Cell suspensions were washed once with cold phosphate-buffered saline (PBS), centrifuged, and re-suspended in DMEM containing 5% FBS and 2 mg/l propidium iodide (PI; Invitrogen, Thermo Fisher Scientific, Oregon, USA) to label dead cells and filtered through a 40-µm cell strainer (BD Biosciences, Franklin Lakes, NJ, USA) to obtain single-cell suspensions. These suspensions were maintained at 4°C for further analysis.
SP and NSP cells were separated by using flow cytometry (FACSComp™; BD Biosciences, USA) based on the retention of Hoechst 33342 dye. Since verapamil can block Hoechst 33342 efflux, the SP gate was defined as the region from which cells disappeared after exposure to verapamil. All samples were evaluated in three independent experiments.
RNA isolation and quantitative real-time polymerase chain reaction
Total RNA from liver cancer cells was purified by utilizing an RNeasy mini-Kit (Takara Biomedical Technology, Beijing, China) after extraction with TRIzol according to the manufacturer’s instructions. The isolated RNA was stored at −80°C or immediately reverse transcribed to cDNA using a Takara Reverse Transcription Kit (Takara Biomedical Technology, Beijing, China) following the manufacturer’s protocols. The reverse transcription product was used to perform qPCR with a Takara SYBR Green Master Mix Kit (Takara Biomedical Technology, Beijing, China), and all steps were performed according to the protocols provided by the manufacturer. The primer sequences specific for sulf2 used in this study were as follows: forward, 5′-ggcaggtttcagagggacc-3′, reverse, 3′-gaaggcgttgatgaagtgcg-5′. Actin was used as the internal control for calculating relative expression levels using the 2−ΔΔCt method.
Cells (3 × 103 cells/well) were seeded and incubated in 96-well plates containing 100 μL of DMEM. We added 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), thiazolyl blue tetrazolium bromide (final concentration of 5 mg/ml; Aladdin, Shanghai, China) to the cells at designated time intervals (1, 2, 3, 4, or 5 day(s)) and cultured for another 4 h. After the MTT solution was removed by aspiration, 150 μL of dimethyl sulfoxide (DMSO; Sigma-Aldrich, St. Louis, MO, USA) was added to each well to dissolve the formazan crystals, and the absorbance of each well was measured in a microplate reader (Bio-Rad Model 680, UK) at 490 nm. All experiments were performed three times with five replicates per experiment.
SP and NSP cells isolated from HepG2, Huh7, and PLC/PRF/5 cells were seeded in 6-well plates at a density of 1 × 103 cells/well and incubated for 7 days. The culture medium was changed at designated time intervals (2, 4, or 6 day(s)). Methanol was added to fix the cells for 15 min after removal of the medium by aspiration on day 7. Subsequently, crystal violet solution (ZSGB-BIO, Beijing, China) was added to each well, and the cells were stained for 30 min. All cell colonies were photographed by native camera of iPhone8.
SP and NSP cells were re-suspended in FBS-free high-glucose DMEM. A total of 1 × 105 cells (in 200 μL) were sequentially added to the upper compartments (with Matrigel-precoated membranes) of a 24-well Transwell chamber plate (8-µm pore size; Corning Incorporated, Corning, USA) while 500 μL of DMEM supplemented with 20% FBS was added to the lower compartments of the Transwell chambers. After incubating for 48 h, the cells remaining in the upper chambers that did not pass through the filter membranes were wiped off with a cotton swab. Methanol was then added to the lower compartments for 15 min at room temperature to fix the cells remaining on the filter membranes. The cells on the filter membranes were stained with crystal violet solution (ZSGB-BIO, Beijing, China) for 15 min.
Cells were allowed to grow to the proper density (logarithmic phase) on coverslips and were then fixed with 4% glutaraldehyde at room temperature for 15 min. Cells were rinsed with PBS before and after fixation. Each coverslip was covered with 0.1% Triton-X 100 (Sangon Biotech, Shanghai, China) for 15 min and was then blocked with 5% BSA (Solarbio Science & Technology, Beijing, China) for 30 min. Cells were then incubated with primary antibodies, including anti-SULF2 (Abcam, Cambridge, UK), anti-GPC3 (Abcam, Cambridge, UK), and anti-β-catenin (Cell Signaling Technology, MA, USA) antibodies, at 4°C overnight. The next morning, after three washes with PBS, the coverslips containing cells were incubated with the following secondary antibodies for 1 h at room temperature in a darkroom: goat anti-rat Alexa Fluor 488 (ABclonal, Wuhan, China) and goat anti-rat Alexa Fluor 594 (ABclonal, Wuhan, China). All antibodies were diluted in accordance with the recommendations in the manufacturer’s manual. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) (Cell Signaling Technology, MA, USA) at a 1:1000 dilution and incubated in the dark for 5 min. The coverslips were then sealed and observed under a confocal laser scanning microscope (CLSM; FV1000, Olympus, Japan).
Cells were collected and were then sonicated in lysis buffer (20 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1 mM ethylenediaminetetraacetic acid or EDTA, 1 mM ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid or EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerol phosphate, 1 mM sodium orthovanadate, 2 μg/ml leupeptin, and 1 mM phenylmethylsulfonyl fluoride or PMSF). The samples of tumor tissues collected from the xenograft mice were crushed in the lysis buffer to extract protein for subsequent analysis. Western blotting was performed with cell and tissue lysates according to a standard protocol. In brief, protein fractions were separated by sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and were then transferred to polyvinylidene difluoride (PVDF) membranes. Membranes were blocked with 5% non-fat milk for 1 h at room temperature and were then incubated with the desired primary antibodies, including anti-SULF2 (1:100) (Abcam, Cambridge, UK, cat. No. ab232835), anti-GPC3 (1:1000) (Abcam, Cambridge, UK, cat. No. ab207080), anti-β-catenin (1:1000) (Cell Signaling Technology, MA, USA, cat. No. 8480), anti-c-Myc (1:1000) (Cell Signaling Technology, MA, USA, cat. No. 18583), and anti-cyclin D1 (1:1000) (Cell Signaling Technology, MA, USA, cat. No. 55506) antibodies overnight at 4°C. Following 1 h of incubation at room temperature with the appropriate HRP-conjugated secondary antibody, specific proteins were detected with the enhanced chemiluminescence (ECL) reagent. Finally, the protein bands were scanned with Quantity One software (Bio-Rad, USA).
Both SP and NSP cells separated from the different liver cancer cell lines were engrafted into nude mice for further study. Three- to five-week-old nude mice were purchased from Guangdong Medical Laboratory Animal Center and divided randomly into three groups (n = 5 mice per group). Cells were diluted to 5 × 103, 5 × 104 and 5 × 105 cells/μL in PBS. SP cells were injected subcutaneously into the right flanks of the mice, while NSP cells were injected into the left flanks. Tumor formation was monitored and sizes were recorded every two days for five weeks, and tumor volumes were calculated using the following equation: volume = width2 × length/2. After the mice were euthanized, the tumor tissues were excised for the subsequent analysis.
After 35 days of observations, nude mice were sacrificed following the instructions of the institutional ethics committee of Tsinghua University. Tumors were harvested, washed three times with PBS, and then fixed with formalin and embedded in paraffin to generate tissue slices following standard protocols. Immunohistochemical analysis was performed on 5-µm-thick sections of each tumor tissue after deparaffinization and rehydration through a graded alcohol series. After antigen retrieval and protein blocking, slices were incubated with the primary antibodies used for Western blot analysis, washed with PBS buffer three times, and incubated with the appropriate secondary antibodies. Finally, all tumor samples were stained with diaminobenzidine, counterstained with hematoxylin, and detected with a horseradish peroxidase (HRP)-based detection system.
All experimental values are presented as the mean ± standard deviation (SD) values. Paired two-tailed Student’s t-tests were used to calculate p-values and the significance was displayed as *p < 0.05, **p < 0.01, ***p < 0.001. A p-value < 0.05 was considered statistically significant.
Acknowledgement: None.
Funding Statement: This study was supported by the Shenzhen Science and Technology Innovation Commission Fundamental Research Key Projects (Nos. JCYJ20180508153013853, JCYJ20180508152130899), the Shenzhen Science and Technology Innovation Fund (No. JCYJ20150331142757381), and the National Natural Science Foundation of China (Nos. 81641051, 81872368).
Author Contributions: The authors confirm contribution to the paper as follows: Dongye Yang and Laiqiang Huang designed and supervised the research. Dongdong Guo, Yunmei Peng, and Dongye Yang performed the research, analyzed the data, and wrote the paper. Xuanang He performed the research and analyzed the data. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The datasets generated and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Ethics Approval: All animal experiments were approved and conducted under the regulation of the institutional ethics committee of Tsinghua University.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
Britton KM, Kirby JA, Lennard TW, Meeson AP (2011). Cancer stem cells and side population cells in breast cancer and metastasis. Cancers 3: 2106–2130. https://doi.org/10.3390/cancers3022106 [Google Scholar] [PubMed] [CrossRef]
Chang JC (2016). Cancer stem cells: Role in tumor growth, recurrence, metastasis, and treatment resistance. Medicine 95: S20–S25. https://doi.org/10.1097/MD.0000000000004766 [Google Scholar] [PubMed] [CrossRef]
Chun JM, Kwon HJ, Sohn J, Kim SG, Park JY, Bae HI, Yun YK, Hwang YJ (2011). Prognostic factors after early recurrence in patients who underwent curative resection for hepatocellular carcinoma. Journal of Surgical Oncology 103: 148–151. https://doi.org/10.1002/jso.21786 [Google Scholar] [PubMed] [CrossRef]
Dahiya D, Wu TJ, Lee CF, Chan KM, Lee WC, Chen MF (2010). Minor versus major hepatic resection for small hepatocellular carcinoma (HCC) in cirrhotic patients: A 20-year experience. Surgery 147: 676–685. https://doi.org/10.1016/j.surg.2009.10.043 [Google Scholar] [PubMed] [CrossRef]
Ding XW, Wu JH, Jiang CP (2010). ABCG2: A potential marker of stem cells and novel target in stern cell and cancer therapy. Life Sciences 86: 631–637. https://doi.org/10.1016/j.lfs.2010.02.012 [Google Scholar] [PubMed] [CrossRef]
Dubrovska A, Hartung A, Bouchez LC, Walker JR, Reddy VA, Cho CY, Schultz PG (2012). CXCR4 activation maintains a stem cell population in tamoxifen-resistant breast cancer cells through AhR signalling. British Journal of Cancer 107: 43–52. https://doi.org/10.1038/bjc.2012.105 [Google Scholar] [PubMed] [CrossRef]
Feng L, Wu JB, Yi FM (2015). Isolation and phenotypic characterization of cancer stem-like side population cells in colon cancer. Molecular Medicine Reports 12: 3531–3536. https://doi.org/10.3892/mmr.2015.3801 [Google Scholar] [PubMed] [CrossRef]
Fukuda K, Saikawa Y, Ohashi M, Kumagai K, Kitajima M, Okano H, Matsuzaki Y, Kitagawa Y (2009). Tumor initiating potential of side population cells in human gastric cancer. International Journal of Oncology 34: 1201–1207. [Google Scholar] [PubMed]
Gao W, Kim H, Feng M, Phung Y, Xavier CP, Rubin JS, Ho M (2014). Inactivation of Wnt signaling by a human antibody that recognizes the heparan sulfate chains of glypican-3 for liver cancer therapy. Hepatology 60: 576–587. https://doi.org/10.1002/hep.26996 [Google Scholar] [PubMed] [CrossRef]
Goodell MA (2005). Stem cell identification and sorting using the Hoechst 33342 side population (SP). Current Protocols in Cytometry 34: 9–18. https://doi.org/10.1002/0471142956.cy0918s34 [Google Scholar] [PubMed] [CrossRef]
Guo Z, Jiang JH, Zhang J, Yang HJ, Zhong YP, Su J, Yang RR, Li LQ, Xiang BD (2016). Side population in hepatocellular carcinoma HCCLM3 cells is enriched with stem-like cancer cells. Oncology Letters 11: 3145–3151. https://doi.org/10.3892/ol.2016.4343 [Google Scholar] [PubMed] [CrossRef]
Haraguchi N, Ishii H, Mimori K, Tanaka F, Ohkuma M et al. (2010). CD13 is a therapeutic target in human liver cancer stem cells. Journal of Clinical Investigation 120: 3326–3339. https://doi.org/10.1172/JCI42550 [Google Scholar] [PubMed] [CrossRef]
Hu J, Li J, Yue X, Wang JC, Liu JZ, Sun L, Kong DL (2017). Expression of the cancer stem cell markers ABCG2 and OCT-4 in right-sided colon cancer predicts recurrence and poor outcomes. Oncotarget 8: 28463–28470. https://doi.org/10.18632/oncotarget.15307 [Google Scholar] [PubMed] [CrossRef]
Jang GB, Kim JY, Cho SD, Park KS, Jung JY, Lee HY, Hong IS, Nam JS (2015). Blockade of Wnt/β-catenin signaling suppresses breast cancer metastasis by inhibiting CSC-like phenotype. Scientific Reports 5: 124651-15. [Google Scholar]
Katoh M (2017). Canonical and non-canonical WNT signaling in cancer stem cells and their niches: Cellular heterogeneity, omics reprogramming, targeted therapy and tumor plasticity. International Journal of Oncology 51: 1357–1369. https://doi.org/10.3892/ijo.2017.4129 [Google Scholar] [PubMed] [CrossRef]
Khalaf AM, Fuentes D, Morshid AI, Burke MR, Kaseb AO, Hassan M, Hazle JD, Elsayes KM (2018). Role of Wnt/beta-catenin signaling in hepatocellular carcinoma, pathogenesis, and clinical significance. Journal of Hepatocellular Carcinoma 5: 61–73. https://doi.org/10.2147/JHC [Google Scholar] [CrossRef]
Kim W, Khan SK, Gvozdenovic-Jeremic J, Kim Y, Dahlman J et al. (2017). Hippo signaling interactions with Wnt/β-catenin and Notch signaling repress liver tumorigenesis. Journal of Clinical Investigation 127: 137–152. https://doi.org/10.1172/JCI88486 [Google Scholar] [PubMed] [CrossRef]
Kreso A, Dick JE (2014). Evolution of the cancer stem cell model. Cell Stem Cell 14: 275–291. https://doi.org/10.1016/j.stem.2014.02.006 [Google Scholar] [PubMed] [CrossRef]
Kucia M, Reca R, Miekus K, Wanzeck J, Wojakowski W, Janowska-Wieczorek A, Ratajczak J, Ratajczak MZ (2005). Trafficking of normal stem cells and metastasis of cancer stem cells involve similar mechanisms: Pivotal role of the SDF-1-CXCR4 axis. Stem Cells 23: 879–894. https://doi.org/10.1634/stemcells.2004-0342 [Google Scholar] [PubMed] [CrossRef]
Lai JP, Oseini AM, Moser CD, Yu C, Elsawa SF et al. (2010). The oncogenic effect of sulfatase 2 in human hepatocellular carcinoma is mediated in part by glypican 3-dependent Wnt activation. Hepatology 52: 1680–1689. https://doi.org/10.1002/hep.23848 [Google Scholar] [PubMed] [CrossRef]
Lai JP, Sandhu DS, Yu C, Han T, Moser CD, Jackson KK et al. (2008). Sulfatase 2 up-regulates glypican 3, promotes fibroblast growth factor signaling, and decreases survival in hepatocellular carcinoma. Hepatology 47: 1211–1222. https://doi.org/10.1002/hep.22202 [Google Scholar] [PubMed] [CrossRef]
Liao K, Xia B, Zhuang QY, Hou MJ, Zhang YJ et al. (2015). Parthenolide inhibits cancer stem-like side population of nasopharyngeal carcinoma cells via suppression of the NF-kappa B/COX-2 pathway. Theranostics 5: 302–321. https://doi.org/10.7150/thno.8387 [Google Scholar] [PubMed] [CrossRef]
Lim JH, Chun YS, Park JW (2008). Hypoxia-inducible factor-1α obstructs a Wnt signaling pathway by inhibiting the hARD1-mediated activation of β-catenin. Cancer Research 68: 5177–5184. https://doi.org/10.1158/0008-5472.CAN-07-6234 [Google Scholar] [PubMed] [CrossRef]
Lloyd MC, Cunningham JJ, Bui MM, Gillies RJ, Brown JS, Gatenby RA (2016). Darwinian dynamics of intratumoral heterogeneity: Not solely random mutations but also variable environmental selection forces. Cancer Research 76: 3136–3144. https://doi.org/10.1158/0008-5472.CAN-15-2962 [Google Scholar] [PubMed] [CrossRef]
Masckauchan TN, Shawber CJ, Funahashi Y, Li CM, Kitajewski J (2005). Wnt/β-catenin signaling induces proliferation, survival and interleukin-8 in human endothelial cells. Angiogenesis 8: 43–51. https://doi.org/10.1007/s10456-005-5612-9 [Google Scholar] [PubMed] [CrossRef]
Nakamura I, Fernandez-Barrena MG, Ortiz-Ruiz MC, Almada LL, Hu C et al. (2013). Activation of the transcription factor GLI1 by WNT signaling underlies the role of SULFATASE 2 as a regulator of tissue regeneration. Journal of Biological Chemistry 288: 21389–21398. https://doi.org/10.1074/jbc.M112.443440 [Google Scholar] [PubMed] [CrossRef]
Nguyen CB, Houchen CW, Ali N (2017). Tumor/cancer stem cell marker doublecortin-like kinase 1 in liver diseases. Experimental Biology and Medicine 242: 242–249. https://doi.org/10.1177/1535370216672746 [Google Scholar] [PubMed] [CrossRef]
Patrawala L, Calhoun T, Schneider-Broussard R, Zhou JJ, Claypool K, Tang DG (2005). Side population is enriched in tumorigenic, stem-like cancer cells, whereas ABCG2+ and ABCG2− cancer cells are similarly tumorigenic. Cancer Research 65: 6207–6219. https://doi.org/10.1158/0008-5472.CAN-05-0592 [Google Scholar] [PubMed] [CrossRef]
Peitzsch C, Tyutyunnykova A, Pantel K, Dubrovska A (2017). Cancer stem cells: The root of tumor recurrence and metastases. Seminars in Cancer Biology 44: 10–24. https://doi.org/10.1016/j.semcancer.2017.02.011 [Google Scholar] [PubMed] [CrossRef]
Perugorria MJ, Olaizola P, Labiano I, Esparza-Baquer A, Marzioni M, Marin JJG, Bujanda L, Banales JM (2019). Wnt-β-catenin signalling in liver development, health and disease. Nature Reviews Gastroenterology and Hepatology 16: 121–136. https://doi.org/10.1038/s41575-018-0075-9 [Google Scholar] [PubMed] [CrossRef]
Portolani N, Coniglio A, Ghidoni S, Giovanelli M, Benetti A, Tiberio GAM, Giulini SM (2006). Early and late recurrence after liver resection for hepatocellular carcinoma—Prognostic and therapeutic implications. Annals of Surgery 243: 229–235. https://doi.org/10.1097/01.sla.0000197706.21803.a1 [Google Scholar] [PubMed] [CrossRef]
Reya T, Duncan AW, Ailles L, Domen J, Scherer DC, Willert K, Hintz L, Nusse R, Weissman IL (2003). A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature 423: 409–414. https://doi.org/10.1038/nature01593 [Google Scholar] [PubMed] [CrossRef]
Rosen SD, Lemjabbar-Alaoui H (2010). Sulf-2: An extracellular modulator of cell signaling and a cancer target candidate. Expert Opinion on Therapeutic Targets 14: 935–949. https://doi.org/10.1517/14728222.2010.504718 [Google Scholar] [PubMed] [CrossRef]
Scharenberg CW, Harkey MA, Torok-Storb B (2002). The ABCG2 transporter is an efficient Hoechst 33342 efflux pump and is preferentially expressed by immature human hematopoietic progenitors. Blood 99: 507–512. https://doi.org/10.1182/blood.V99.2.507 [Google Scholar] [PubMed] [CrossRef]
Summer R, Kotton DN, Sun X, Ma B, Fitzsimmons K, Fine A (2003). Side population cells and Bcrp1 expression in lung. American Journal of Physiology-Lung Cellular and Molecular Physiology 285: L97–L104. https://doi.org/10.1152/ajplung.00009.2003 [Google Scholar] [PubMed] [CrossRef]
Szotek PP, Pieretti-Vanmarcke R, Masiakos PT, Dinulescu DM, Connolly D, Foster R, Dombkowski D, Preffer F, Maclaughlin DT, Donahoe PK (2006). Ovarian cancer side population defines cells with stem cell-like characteristics and Mullerian Inhibiting Substance responsiveness. Proceedings of the National Academy of Sciences of the United States of America 103: 11154–11159. https://doi.org/10.1073/pnas.0603672103 [Google Scholar] [PubMed] [CrossRef]
Teshima K, Nara M, Watanabe A, Ito M, Ikeda S, Hatano Y, Oshima K, Seto M, Sawada K, Tagawa H (2014). Dysregulation of BMI1 and microRNA-16 collaborate to enhance an anti-apoptotic potential in the side population of refractory mantle cell lymphoma. Oncogene 33: 2191–2203. https://doi.org/10.1038/onc.2013.177 [Google Scholar] [PubMed] [CrossRef]
Tiezzi DG, Sicchieri RD, Mouro LR, Oliveira TMG, Silveira WA, Antonio HMR, Muglia VF, de Andrade JM (2013). ABCG2 as a potential cancer stem cell marker in breast cancer. Journal of Clinical Oncology 31: 12007. https://doi.org/10.1200/jco.2013.31.15_suppl.e12007 [Google Scholar] [CrossRef]
Van den Broeck A, Vankelecom H, van Delm W, Gremeaux L, Wouters J, Allemeersch J, Govaere O, Roskams T, Topal B (2013). Human pancreatic cancer contains a side population expressing cancer stem cell-associated and prognostic genes. PLoS One 8: e73968. https://doi.org/10.1371/journal.pone.0073968 [Google Scholar] [PubMed] [CrossRef]
Vicente-Duenas C, Cobaleda C, Perez-Losada J, Sanchez-Garcia I (2010). The evolution of cancer modeling: The shadow of stem cells. Disease Models & Mechanisms 3: 149–155. https://doi.org/10.1242/dmm.002774 [Google Scholar] [PubMed] [CrossRef]
Wang W, Smits R, Hao H, He C (2019). Wnt/β-catenin signaling in liver cancers. Cancers 11: 926–946. https://doi.org/10.3390/cancers11070926 [Google Scholar] [PubMed] [CrossRef]
Wang RH, Sun Q, Cheng B (2014). Joint detection in liver cancer stem cells’ surface markers plays a role in predicting the prognosis of hepatocellular carcinoma. Gastroenterology 146: S998. https://doi.org/10.1016/S0016-5085(14)63629-5 [Google Scholar] [CrossRef]
Wang WQ, Tabu K, Hagiya Y, Sugiyama Y, Kokubu Y, Murota Y, Ogura S, Taga T (2017). Enhancement of 5-aminolevulinic acid-based fluorescence detection of side population-defined glioma stem cells by iron chelation. Scientific Reports 7: 1–12. https://doi.org/10.1038/srep42070 [Google Scholar] [PubMed] [CrossRef]
Wang D, Wang Y, Kong T, Fan F, Jiang Y (2011). Hypoxia-induced beta-catenin downregulation involves p53-dependent activation of Siah-1. Cancer science 102: 1322–1328. https://doi.org/10.1111/j.1349-7006.2011.01950.x [Google Scholar] [PubMed] [CrossRef]
Wei ZZ, Zhang JY, Taylor TM, Gu X, Zhao Y, Wei L (2017). Neuroprotective and regenerative roles of intranasal Wnt-3a Administration after focal ischemic stroke in mice. Journal of Cerebral Blood Flow and Metabolism 38: 404–421. https://doi.org/10.1177/0271678X17702669 [Google Scholar] [PubMed] [CrossRef]
Wu Q, Qin SK (2013). Features and treatment options of Chinese hepatocellular carcinoma. Chinese Clinical Oncology 2: 38. [Google Scholar] [PubMed]
Yamashita T, Honda M, Nakamoto Y, Baba M, Nio K et al. (2013). Discrete nature of EpCAM+ and CD90+ cancer stem cells in human hepatocellular carcinoma. Hepatology 57: 1484–1497. https://doi.org/10.1002/hep.26168 [Google Scholar] [PubMed] [CrossRef]
Yang ZF, Ho DW, Ng MN, Lau CK, Yu WC, Ngai P, Chu PWK, Lam CT, Poon RTP, Fan ST (2008). Significance of CD90+ cancer stem cells in human liver cancer. Cancer Cell 13: 153–166. https://doi.org/10.1016/j.ccr.2008.01.013 [Google Scholar] [PubMed] [CrossRef]
Zhao R, Liu XX, Wang YS, Jie XX, Qin RH et al. (2016). Integrated glycomic analysis of ovarian cancer side population cells. Clinical Proteomics 13: 1–14. https://doi.org/10.1186/s12014-016-9131-z [Google Scholar] [PubMed] [CrossRef]
Zhou L, Rui JA, Wang SB, Chen SG, Qu Q (2014). Early recurrence in large hepatocellular carcinoma after curative hepatic resection: Prognostic significance and risk factors. Hepato-Gastroenterology 61: 2035–2041. [Google Scholar] [PubMed]
Zhu Z, Hao XF, Yan MX, Yao M, Ge C, Gu JR, Li JJ (2010). Cancer stem/progenitor cells are highly enriched in CD133+ CD44+ population in hepatocellular carcinoma. International Journal of Cancer 126: 2067–2078. [Google Scholar] [PubMed]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools