 Open Access
Open Access
ARTICLE
Comparative transcriptome analysis provides insights into the molecular mechanism of the anti-nematode role of Arachis hypogaea (Fabales: Fabaceae) against Meloidogyne incognita (Tylenchida: Heteroderidae)
1 College of Life Science, Institute of Life Science and Green Development, Hebei University, Baoding, 071002, China
2 Hebei Innovation Center for Bioengineering and Biotechnology, Hebei University, Baoding, 071002, China
* Corresponding Authors: Jianfeng Liu, ; Dandan Cao,
# These authors contributed equally to this work
BIOCELL 2023, 47(9), 2101-2113. https://doi.org/10.32604/biocell.2023.029581
Received 27 February 2023; Accepted 01 June 2023; Issue published 28 September 2023
Abstract
Background: Plant root-knot nematode (RKN) disease is a serious threat to agricultural production across the world. Meloidogyne incognita is the most prominent pathogen to the vegetables and cash crops cultivated. Arachis hypogaea can effectively inhibit M. incognita, but the underlying defense mechanism is still unclear. Methods: In our study, the chemotaxis and infestation of the second-stage juveniles (J2s) of M. incognita to A. hypogaea root tips were observed by the Pluronic F-127 system and stained with sodium hypochlorite acid fuchsin, respectively. The transcriptome data of A. hypogaea roots with non-infected or infected by J2s were analyzed. Results: The J2s could approach and infect inside of A. hypogaea root tips, and the chemotactic migration rate and infestation rate were 20.72% and 22.50%, respectively. Differential gene expression and pathway enrichment analyses revealed ubiquinone and other terpenoid-quinone biosynthesis pathway, plant hormone signal transduction pathway, and phenylpropanoid biosynthesis pathway in A. hypogaea roots responded to the infestation of M. incognita. Furthermore, the AhHPT gene, encoding homogentisate phytyltransferase, was considered to be an ideal candidate gene due to its higher expression based on the transcriptome data and quantitative real-time PCR analysis. Conclusion: Therefore, the key gene AhHPT might be involved in the A. hypogaea against M. incognita. These findings lay a foundation for revealing the molecular mechanism of A. hypogaea resistance to M. incognita and also provide a prerequisite for further gene function verification, aiming at RKN-resistant molecular breeding.Keywords
Root-knot nematode (RKN; Meloidogyne spp.) is an important sedentary plant endoparasitic pathogenic nematode with a wide range of hosts (Aioub et al., 2022; Nishat et al., 2022), which can infect more than 5,500 kinds of plants such as food crops and vegetables (Jaouannet et al., 2012). Global agricultural losses caused by RKN are estimated at $157 billion annually (Abad et al., 2008; Eldeeb et al., 2022). The second stage juveniles (J2s) of RKN search for the root system of the host plant by receiving chemical signals (Reynolds et al., 2015), further invade meristem from the plant root tips, and subsequently travel intracellularly along the vascular cylinder and remodel the parenchymal cells into giant cells as their feeding sites. The physiology and metabolism of plants will be seriously affected once they are infected by RKN (Davis et al., 2000). For example, the aboveground part of plants is short, and the leaf color is abnormally yellow with deciduous and withered, which has a significant inhibitory effect on the growth of plant seedlings (Oka et al., 2015; Elnahal et al., 2022). With global climate change, reform of planting systems, and the rapid development of large-scale, mechanized, and high-value agriculture, RKN have gradually become one of the most serious pathogenic nematodes affecting agriculture cultivation (Jones et al., 2013). Meloidogyne incognita, M. hapla, M. javanica, and M. arenaria are the main RKN species, among which M. incognita is recognized as the most prominent pathogen (Moens et al., 2009). Therefore, the safe and effective prevention to control M. incognita has become the bottleneck problem of high-quality and -yield for agricultural production and double reduction of chemical fertilizers and pesticides (Ali et al., 2022; EI-Ashry et al., 2023).
Arachis hypogaea L. is the main oil crop belonging to the family Leguminosae, subfamily Papilionatae, and is economically important as a source of proteins, minerals, and vitamins. It is widely grown in the semi-arid tropics and plays a role in the world agricultural economy. From the 1990s, the global planting area for A. hypogaea gradually increased, with a global production of about 48 million tons in 2019 (Çiftçi and Suna, 2022). However, in North America, South America, Africa, Asia, and Europe, A. hypogaea production areas have been reported to be mainly infested with three pathogens, namely, M. arenaria, M. javanica, and M. hapla, resulting in considerable yield loss (Dong et al., 2008). Previous studies showed that only M. arenaria was isolated from the root-knot of the A. hypogaea root system grown in soils containing M. incognita and M. arenaria (Johnson et al., 2000). To verify the reproduction of M. incognita on A. hypogaea plants, 2000 eggs of M. incognita were inoculated into A. hypogaea roots; however, no M. incognita could reproduce on A. hypogaea plants (Faske and Starr, 2009). In addition, the population density of M. incognita in soil was effectively reduced by continuous cropping of A. hypogaea (Johnson et al., 1998). Compared with planting tomatoes, planting A. hypogaea could reduce the infestation rate of M. incognita by 70% (Belcher and Hussey, 1977). Several studies have proved that A. hypogaea is a non-host plant of M. incognita and has a certain inhibitory (or killing) effect on M. incognita.
The approach of screening and identifying resistance genes in non-host and high-resistant plant varieties is the key to successful molecular breeding for anti-RKN properties. The resistance genes Mi-1, Mi-2, Mi-3, Mi-4, Mi-5, Mi-6, Mi-7, Mi-8, Mi-9, and Mi-HT against M. incognita have been identified in tomato (El-Sappah et al., 2019; Du et al., 2020). When Capsicum annuum L. was infected by M. incognita, the expression level of the CaRKNR gene significantly increased, and silencing this gene by the virus-induced gene silencing system significantly reduced the resistance to M. incognita of pepper (Mao et al., 2015). The MIC gene was also found to be closely related to the resistance of M. incognita in Gossypium hirsutum L. (Wubben et al., 2008). At present, transcriptome analysis has been used to study the mechanism of plant and RKN interactions. Gene expression of soybean (Glycine max) infested with M. incognita for 12 days and 10 weeks revealed changes in expressions of genes involved in carbohydrate and cell wall metabolism, cell cycle control, and plant defense (Ibrahim et al., 2011). Comparison of the transcriptome of Polianthes tuberosa uninfected and infected by M. incognita in the early, middle, and late periods revealed differential gene functions mainly in the pathways related to carbohydrate metabolism, signal transduction, and translation (Singh et al., 2022). The screening of resistance genes, especially the obtaining of more accurate and effective resistance genes based on transcriptomics, has become the main strategy for plants to breed resistant-nematode varieties.
At present, the study of the interaction between A. hypogaea and M. incognita only focused on the inhibition phenomenon manifested in reducing the activity and the density population of M. incognita. The internal mechanism of A. hypogaea-M. incognita interaction is still poorly understood. The main hypothesis of our study is that M. incognita J2s can collect and infect the A. hypogaea root tips. M. incognita J2s infestation can cause differential expression of genes, which maybe play a key role in the interaction between A. hypogaea and M. incognita. Especially, the key gene AhHPT might be involved in A. hypogaea against M. incognita. Our findings will provide a strong background for identifying possible gene targets in A. hypogaea to develop broad resistance of plants to M. incognita by gene silencing technology or gene super-expression technology.
The initial sample of M. incognita, which was provided by the Academy of Agriculture and Forestry Sciences of Beijing, China, was propagated in living tomato plants in the laboratory incubator. The tomato variety was Qinshu Shanghai 908, provided by Xi’an Qinshu Agriculture Company Limited (China). Xiaobaisha (Fuhuasheng as female parent and Shitouqi as male parent), a widely cultivated A. hypogaea variety, was used as the experiment material. The study was conducted at the Agricultural Environmental Ecology Research Laboratory of Hebei Innovation Center for Bioengineering and Biotechnology of Hebei University, China (38°87' N, 115°51' E).
Nematodes collection and Arachis hypogaea cultivation
Tomato roots infected with M. incognita were selected, and the surface soil was removed by washing. The mature egg masses on the roots were picked with sterilized forceps under the stereomicroscope. The selected egg masses were thoroughly washed with 2% sodium hypochlorite solution for 3 min, and the remaining sodium hypochlorite solution was washed 3–4 times with sterile water. The washed egg masses were placed in a 500-mesh screen, placed in a Petri dish with sterile water, and incubated for 3–4 d in the dark constant temperature incubator at 27°C. The hatched J2s were collected.
A. hypogaea seeds (Cultivar Xiaobaisha) of the same size were soaked in 5% sodium hypochlorite solution for 15 min, washed 3–4 times with sterile water, placed in sterile Petri dishes containing sterile water for germination promotion at 27°C in the dark. After 3 d, each A. hypogaea was transplanted into 250 g sterilized soil (nutritive soil: vermiculite = 1:1 v/v) in a plastic pot of 11 cm in height and diameter and cultured in an intelligence artificial climate chamber until the A. hypogaea grew into two true leaves. The intelligence artificial climate chamber was set to two stages; the first stage was 16 h, 26°C, relative humidity 60%, light intensity 350 μmol·m−2·s−1, and the second stage was 8 h, 20°C, relative humidity 75%, light intensity 0 μmol·m−2·s−1.
Nematode treatment of excised Arachis hypogaea root tips
Twenty-three grams of Pluronic F-127 was added to a beaker containing 80 mL cold, sterile water and a magnetic stirring rod, stirred slowly at 4°C until dissolved. In the 6-well cell culture plate, 3 mL of 23% Pluronic F-127 and 10 μL suspension containing 100 J2s were added into each well. After mixing, a 1 cm of A. hypogaea root tip with similar shape and size was added into each well. 27°C in dark conditions after 3, 6, 9, 12, 18, 24, 30, 36, 48, 54 and 60 h, the stereomicroscope was used to observe the behavioral responses. The number of J2s in the range of 1 mm near the root tip of excised A. hypogaea was counted. At 30, 36, 42, 48, 54, 60, 66 and 72 h, the protocol of Byrd et al. (1983) was used to stain the nematodes in A. hypogaea root tips. Six replicates were set in each period, and the experiment was repeated three times.
Nematode treatment of potted Arachis hypogaea
The incubation of the J2s and the cultivation of A. hypogaea were the same as mentioned in the previous experiment. The A. hypogaea with strong and uniform growth were randomly divided into two groups. One group was inoculated with sterile water as the control group, the other group was inoculated with a suspension containing 2000 J2s per A. hypogaea plant. The amount of sterile water inoculated was consistent with the amount of suspension. The glass rod was used to dig two holes of 2 cm symmetrically, 1 cm away from the root of A. hypogaea as the inoculation site. The A. hypogaea roots in the two groups of 3, 9, and 15 d after treatment were selected and dyed according to the above staining method. Each treatment was repeated three times, and there were 18 samples marked as C3d_1, C3d_2, C3d_3, C9d_1, C9d_2, C9d_3, C15d_1, C15d_2, C15d_3, T3d_1, T3d_2, T3d_3, T9d_1, T9d_2, T9d_3, T15d_1, T15d_2 and T15d_3. All the collected A. hypogaea root samples were frozen in liquid nitrogen, stored at −80°C, and sent to Shanghai Majorbio Bio-pharm Technology Co., Ltd. (China). For transcriptome sequencing.
RNA extraction, purification, and detection
Total RNA was extracted from the A. hypogaea roots using TRIzol® Reagent according to the manufacturer’s instructions, and genomic DNA was removed using DNase I (TaKara). RNA quality was determined by 2100 Bioanalyser (Agilent) and quantified using the ND-2000 (NanoDrop Technologies). The high-quality RNA samples (OD260/280 = 1.8 − 2.2, OD260/230 ≥ 2.0, RIN ≥ 6.5, 28S:18S ≥ 1.0, > 1 μg) were used to construct sequencing library.
Library preparation and Illumina Hiseq xten/Nova seq 6000 sequencing
RNA-seq transcriptome library was prepared following TruSeqTM RNA sample preparation Kit from Illumina (San Diego, CA) using 1 μg of total RNA. Messenger RNA was isolated following to polyA selection method by oligo (dT) beads and fragmented by fragmentation buffer. Double-stranded cDNA was synthesized using a SuperScript double-stranded cDNA synthesis Kit (Invitrogen, CA) and random hexamer primers (Illumina). The synthesized cDNA was subjected to end-repair, phosphorylation, and ‘A’ base addition according to Illumina’s library construction protocol. Libraries were size-selected for cDNA target fragments of 300 bp on 2% Low Range Ultra Agarose followed by PCR amplified using Phusion DNA polymerase (NEB) for 15 PCR cycles. After quantification by TBS380, paired-end RNA-seq sequencing library was sequenced with the Illumina HiSeq xten/NovaSeq 6000 sequencer (2 × 150 bp read length).
Sequencing data quality control, comparison, and assembly
The raw paired-end reads were trimmed and quality controlled by SeqPrep (https://github.com/jstjohn/SeqPrep) and Sickle (https://github.com/najoshi/sickle) with default parameters. The clean reads were separately aligned to the reference genome with orientation mode using HISAT2 (http://ccb.jhu.edu/software/hisat2) software (Kim et al., 2015). The mapped reads of each sample were assembled by StringTie (https://ccb.jhu.edu/software/stringtie) in a reference-based approach (Pertea et al., 2015).
Analysis of differentially expressed genes (DEGs) and functional enrichment
To identify DEGs in different treatment groups and corresponding control groups, the expression level of each transcript was calculated according to the transcripts per million reads (TPM) method. RSEM (http://deweylab.github.io/RSEM/) software (Li and Dewey, 2011) was used to quantify gene abundances. DEGs analysis was performed using the DESeq2 (Love et al., 2014) with p ≤ 0.05 and |log2FC| > 1. The DEGs were screened by comparing all the genes expressed in A. hypogaea roots at different periods under nematode treatment. Functional-enrichment analysis through Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) was performed to identify which DEGs were significantly enriched in GO terms and metabolic pathways at Bonferroni-corrected p ≤ 0.05 and compared with all transcriptome data. GO functional enrichment, and KEGG pathway analysis were performed by Goatools (https://github.com/tanghaibao/GOatools) software and KOBAS (http://kobas.cbi.pku.edu.cn) software (Xie et al., 2011).
Quantitative real-time PCR (qRT -PCR) analysis
Extraction of total RNA and removal of residual DNA from RNA were as described above. The first strand cDNA of RNA was synthesized by the UEIris II RT-PCR System for First-Strand cDNA Synthesis Kit (R2028, US Everbright®Inc., China). QRT-PCR was done using 2 × AugeGreen qPCR Master Mix (S2008S, US Everbright®Inc., China). The thermal cycling program was as follows: 95°C for 5 min, followed by 45 cycles at 95°C for 15 s and 53°C for 60 s. AhActin was used as a reference gene to normalize target gene expression levels in the samples. The 2−ΔΔCT method was used to calculate the relative expression levels of stress-response genes. Primers were designed using Oligo 7 software, and the primer sequences are listed in Table 1.
Origin 2022b software was used to analyze the chemotactic migration rate and infestation rate of J2s to excise A. hypogaea root tips. Kruskal-Wallis test and one-way ANOVA were used to analyze the significance of the differences among different time periods. False discovery rate correction with Benjamini/Hochberg (BH) was adopted to conduct multiple test and corrections for p-value of DEGs analysis. Images were visualized by XT-10C stereomicroscope and Photoshop 2020 software.
Aggregation of J2s near the excised Arachis hypogaea root tips
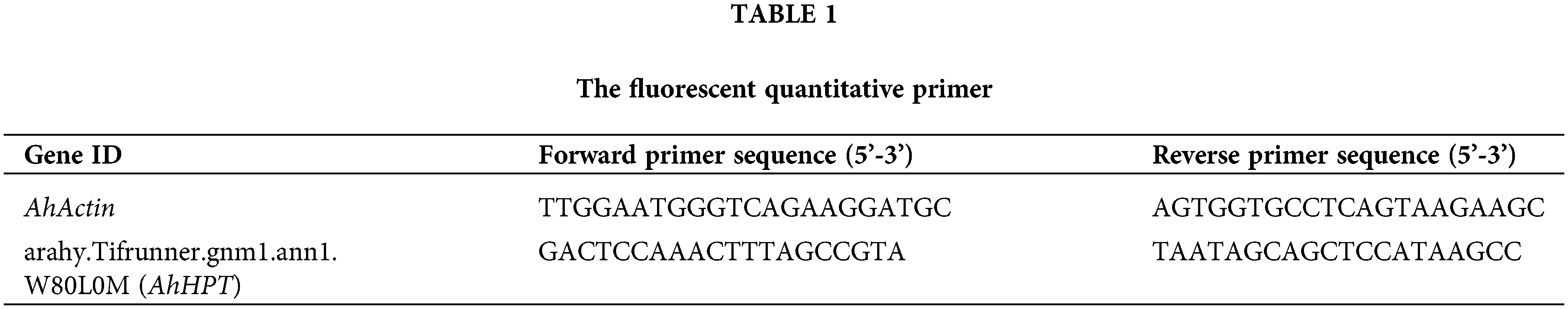
As shown in Fig. 1A, the J2s moved toward and aggregated near the excised A. hypogaea root tips. From 3 to 24 h, the number of the J2s near the root tip of A. hypogaea increased gradually. At 24 h, the chemotactic migration rate of J2s to the root tip of excised A. hypogaea was the highest, which was 20.72%. The chemotactic migration rate of the J2s to the excised A. hypogaea root tip decreased significantly to 10.22% at 30 h. The chemotactic migration rate of the J2s to the excised A. hypogaea root tip was 2.00% at 60 h (Fig. 1B). Staining of the root tips isolated from A. hypogaea showed red J2s (Fig. 1C), indicating the invasion of J2s into the root of excised A. hypogaea, with an infestation rate of 22.50% at 66 h (Fig. 1D).
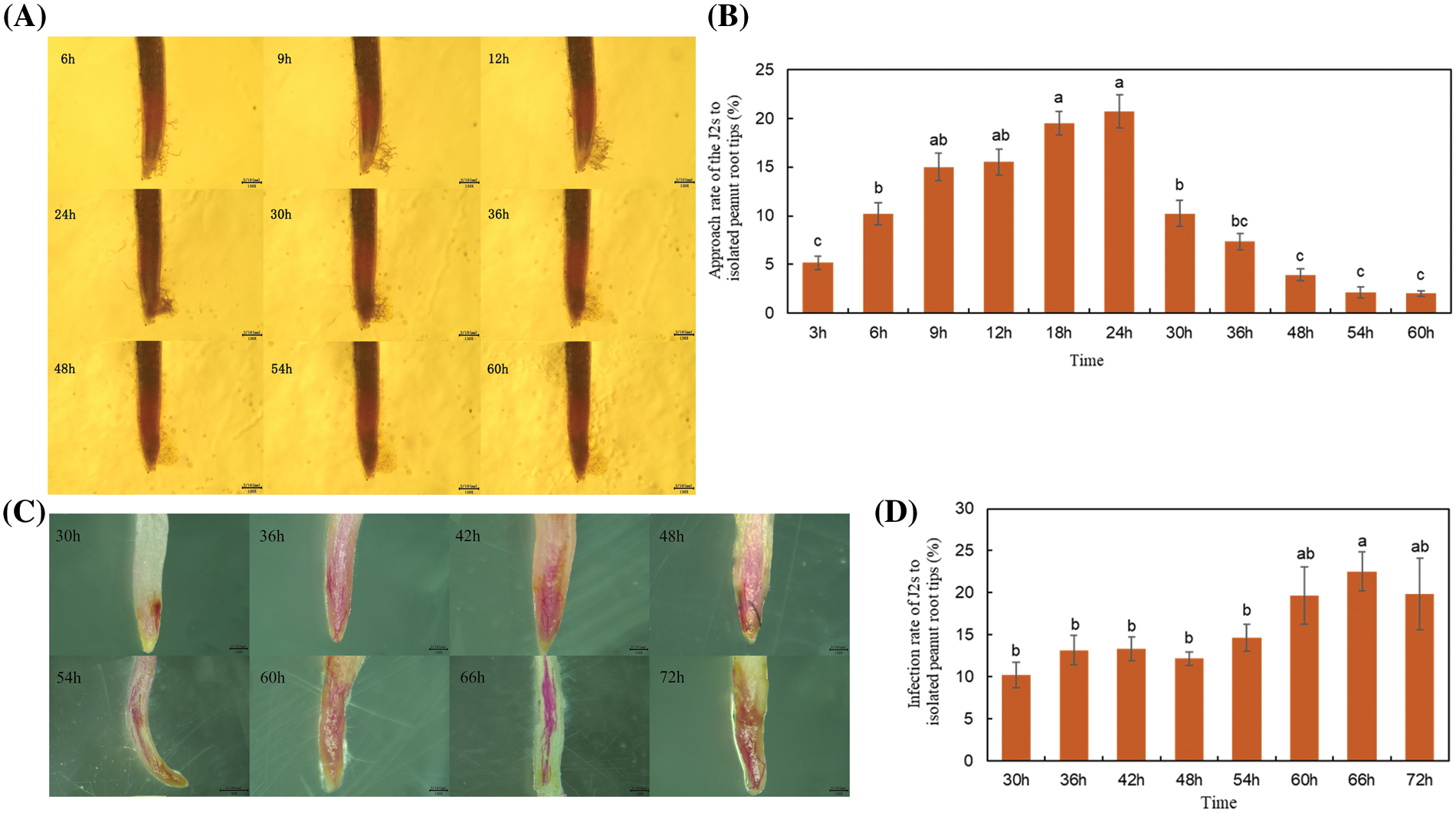
Figure 1: Assessment of chemotaxis and infestation of Meloidogyne incognita J2s to excised Arachis hypogaea root tips at different time points. (A) M. incognita J2s moved toward and aggregated near the excised A. hypogaea root tip at 6, 9, 12, 24, 30, 36, 48, 54, and 60 h. The scale is in the lower right corner. (B) Statistical representation of the chemotactic migration rate of the J2s to excised A. hypogaea root tip at different time points. (C) Staining of M. incognita J2s after entering the excised A. hypogaea root tip at different time points. The M. incognita that enters the peanut root is stained red. The scale is in the lower right corner. (D) The percentage of J2s invading the excised A. hypogaea root tip at different time points.
Note: Different bars in the bar charts are presented as means ± standard error of the mean. The different lowercase letters above each bar chart indicated statistical significance at 0.05 level.
The Arachis hypogaea roots infected by J2s in the soil environment
The pot experiment was carried out considering that the activity of excised A. hypogaea root tips would affect the chemotaxis of J2s. Sodium hypochlorite acid fuchsin staining of A. hypogaea roots at 3, 9, and 15 d after J2s inoculation showed that J2s were observed in the meristem and elongation regions of A. hypogaea root crowns (Fig. 2).
Figure 2: Pictures of J2s dyed after invading the Arachis hypogaea root tip at 3, 9, and 15 d in soil environment.
Note: The black arrow points to the J2s, which were stained red.
Statistical analysis and quality evaluation of RNA-Seq
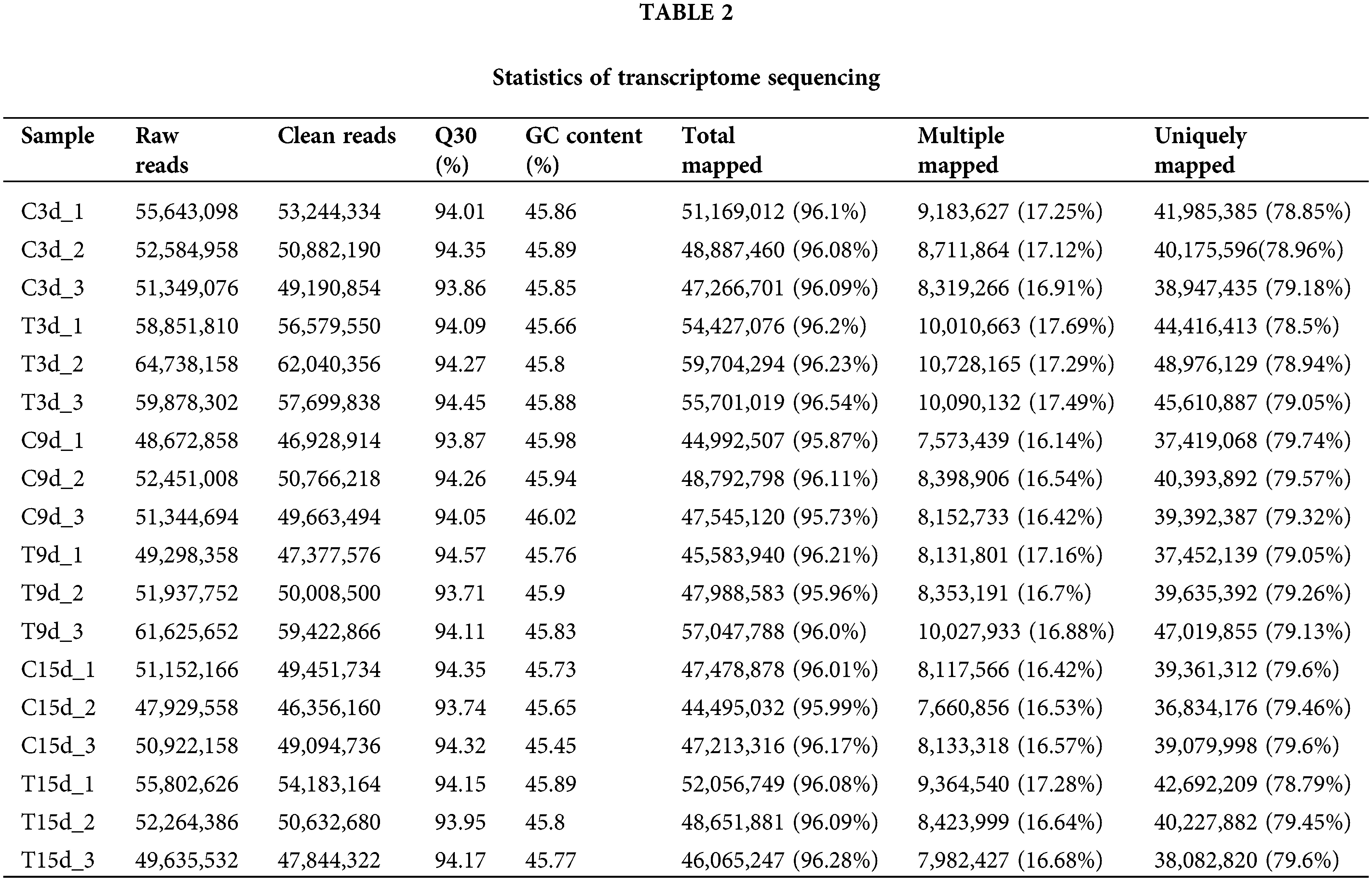
In this experiment, the genome of A. hypogaea (https://data.legumeinfo.org/Arachis/hypogaea/annotations/Tifrunner.gnm1.ann1.CCJH/) was used as a reference genome. The results of statistical analysis and comparison of sequencing data are shown in Table 2. The similarity analysis of sequences of 18 samples and the reference transcriptome database yielded a total of 135.6 G clean data. More than 6.73 G of clean data from each sample were obtained. The distribution of Q30 bases was 93.71%–94.57%, and the GC content was 45.45%–46.02%. The matching degree between the clean reads of each sample and the reference genome sequence was above 95%, with the highest matching rate of 96.54% and the lowest matching rate of 95.73%, indicating the accuracy and reliability of the sequencing.
Statistical analysis of differentially expressed genes
The results of principal component analysis (PCA) showed overlapping in the samples of C3d and T3d groups and the distance among the other groups was long (Fig. 3A), indicating that the gene expression difference between the C3d and T3d groups was not significant, while the difference between the other groups was significant. Three replicates within the same group were close to each other and indicated consistent and uniform gene expression in the samples within the group. Comparison and analysis of the DEGs among the C3d vs. T3d, C9d vs. T9d, and C15d vs. T15d groups, a total of 3072 DEGs were obtained. The heat map of Fig. 3B showed the aggregation of DEGs. The gene expression of C3d was similar to that of T3d, with highly-expressed genes were mainly distributed in the upper parts of the heat map and low-expressed genes mainly distributed in the lower parts of the heat map. The genes with high expression of C9d were mainly distributed in the middle and upper parts of the heat map, while the genes with low expression were mainly distributed in the lower parts of the heat map. The high-expression genes in T9d were mainly distributed in the upper middle parts and the lower middle parts of the heat map. The high expression genes of C15d were mainly distributed in the lower parts of the heat map, and the low expression genes were mainly distributed in the upper parts of the heat map, while the high expression genes of T15d were mainly distributed in the middle and the upper parts of the heat map, and the low expression genes were mainly distributed in the lower parts of the heat map.
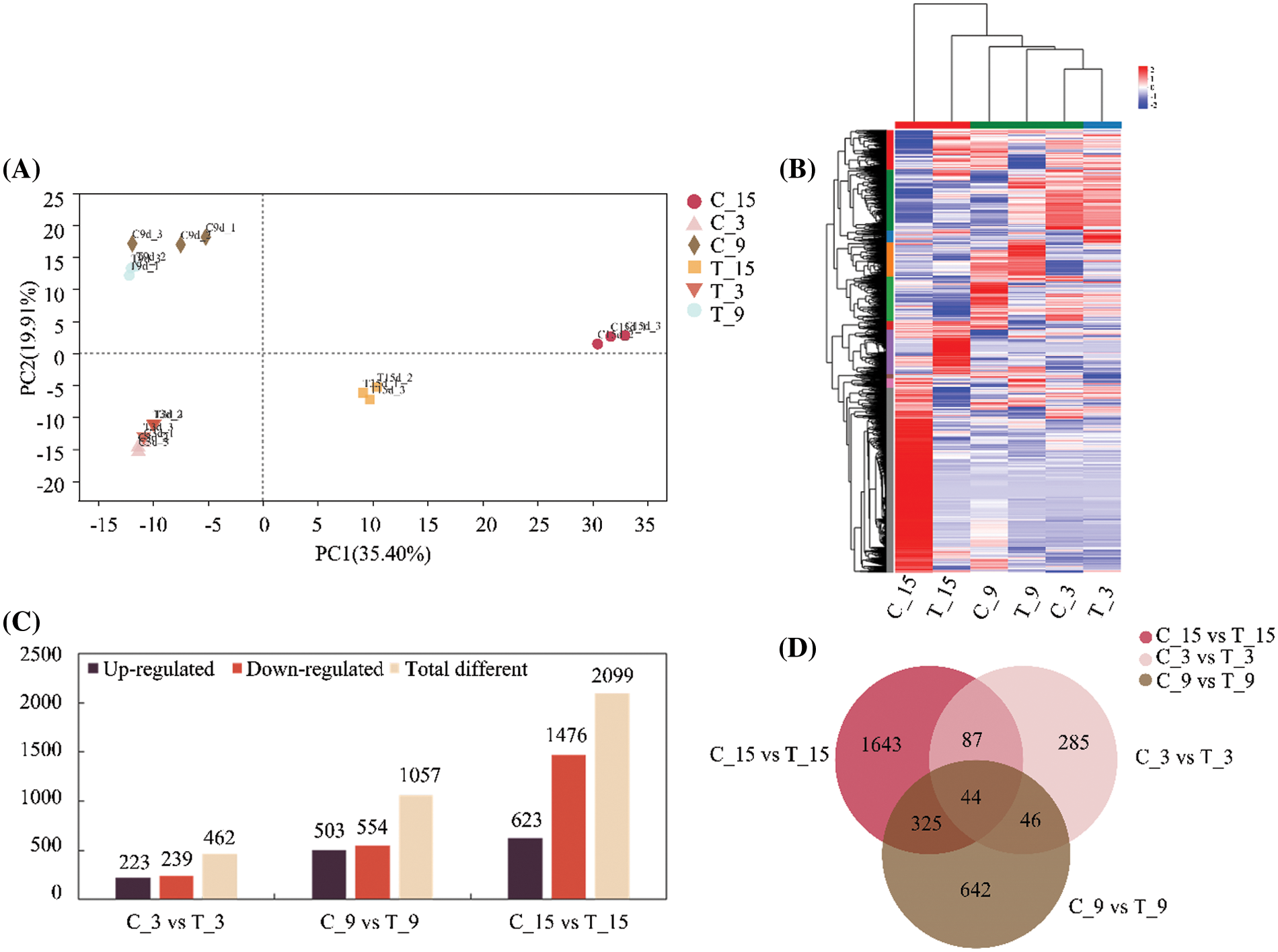
Figure 3: Analysis of differentially expressed genes (DEGs) in Arachis hypogaea roots responding to Meloidogyne incognita stress. (A) Principal component analysis of all samples. (B) Heatmap clustering of DEGs under different days of nematode treatment conducted using Hierarchical Clustering (HCL). The colors in the heatmap represent the normalized expression of the gene in each group. (C) Statistical presentation of the number of genes significantly regulated by nematode treatment on different days. (D) Venn diagram analysis of significant regulatory genes under different durations of nematode treatment.
The DEGs of the three groups of nematode treatment groups and their respective control groups were statistically analyzed, and the results are shown in Fig. 3C. Statistical analysis showed that 462 DEGs were detected between C3d and T3d groups, of which 223 were up-regulated and 239 were down-regulated. Among C9d and T9d groups, 1057 DEGs were detected, including 503 genes with up-regulated expression and 554 genes with downregulated expression. The number of DEGs between C15d and T15d groups was 2099, including 623 with up-regulated expression and 1476 with down-regulated expression. The number of DEGs increased with the extension of nematode treatment time. There were 44 identical DEGs in the three treatment groups. The C3d vs. T3d, C9d vs. T9d, and C15d vs. T15d groups contained 285, 642, and 1643 unique DEGs, respectively. The C9d vs. T9d, and C15d vs. T15d groups had the largest number of genes with common differential expression, which was 369 (Fig. 3D).
Gene ontology functional enrichment analysis of differentially expressed genes in response to nematode treatment
GO function enrichment analysis was performed on the DEGs of the treatment groups and the control groups with various durations after inoculation. Statistical analysis of GO Terms, which considered significantly enriched DEGs (p < 0.05) in each group and ranked them in the top 30 of the enrichments, showed enrichment in GO terms related to plant defense response (Figs. 4A–4C). Twenty-two DEGs of C3d vs. T3d were significantly enriched in GO Term of plant defense response, of which 15 were up-regulated and 7 were down-regulated (Fig. 4D). In C9d vs. T9d, 43 DEGs were significantly enriched in GO terms of plant defense response, of which 24 were up-regulated, and 19 were down-regulated (Fig. 4E). There were 81 DEGs in C15d vs. T15d that were significantly enriched in the plant defense response GO Term, of which 24 were up-regulated, and 57 were down-regulated (Fig. 4F). The two genes with their IDs arahy.Tifrunner.gnm1.ann1.NH8LJ7 and arahy.Tifrunner.gnm1.ann1.VFVG10, respectively, were present in the GO term of the plant defense response enriched by DEGs in the C3d vs. T3d, C9d vs. T9d, and C15d vs. T15d. The gene with ID arahy.Tifrunner.gnm1.ann1.NH8LJ7 was up-regulated in C3d vs. T3d and C9d vs. T9d and down-regulated in C15d vs. T15d, and is a gene encoding MLP-like protein 43. The gene with the gene ID arahy.Tifrunner.gnm1.ann1.VFVG10 was up-regulated in C3d vs. T3d, C9d vs. T9d, and C15d vs. T15d and is a gene encoding the disease resistance protein (TIR-NBS-LRR class).
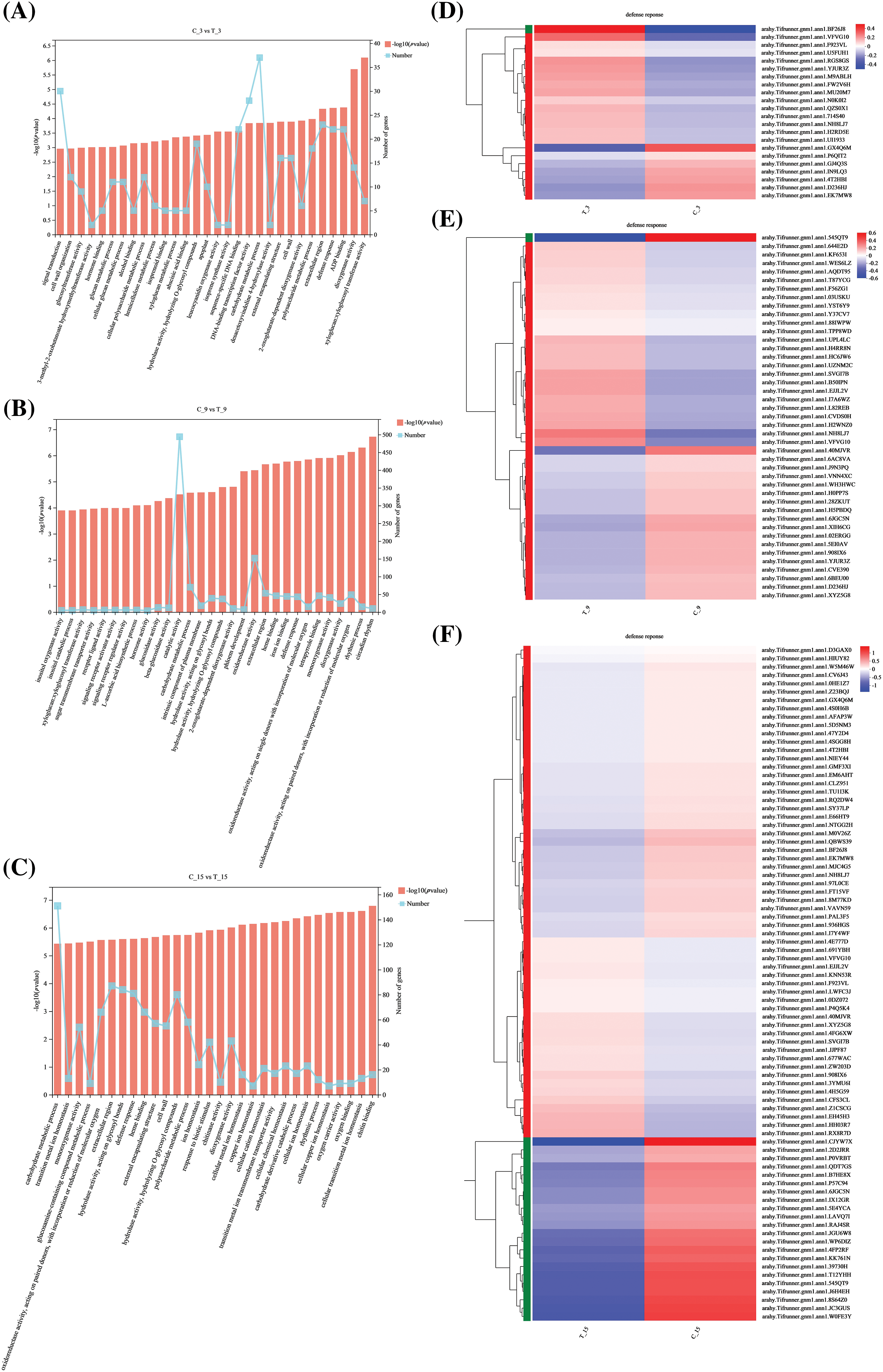
Figure 4: Gene Ontology (GO) enrichment results and heat map clustering analysis of C3d vs. T3d, C9d vs. T9d, and C15d vs. T15d (A–C) Statistical analysis of GO terms with significant enrichment and the top 30 GO terms in C3d vs. T3d, C9d vs. T9d, and C15d vs. T15d. The broken line graph shows the number of genes in the corresponding GO terms. (D–F) Heat map clustering analysis of plant defense response function entries differentially in C3d vs. T3d, C9d vs. T9d, and C15d vs. T15d.
Note: The colors in the heatmap represent the normalized expression of the gene in each group.
Analysis of the kyoto encyclopedia of genes and genomes enrichment pathway of differentially expressed genes in response to nematode treatment
Statistical analysis of the DEGs in C3d vs. T3d, C9d vs. T9d, and C15d vs. T15d with significant enrichment in the top 20 KEGG pathways revealed that ubiquinone and other terpenoid-quinone biosynthesis pathway, plant hormone signal transduction pathway and phenylpropanoid biosynthesis pathway were associated with nematode treatment and more genes enrichments in these three pathways (Figs. 5A–5C).
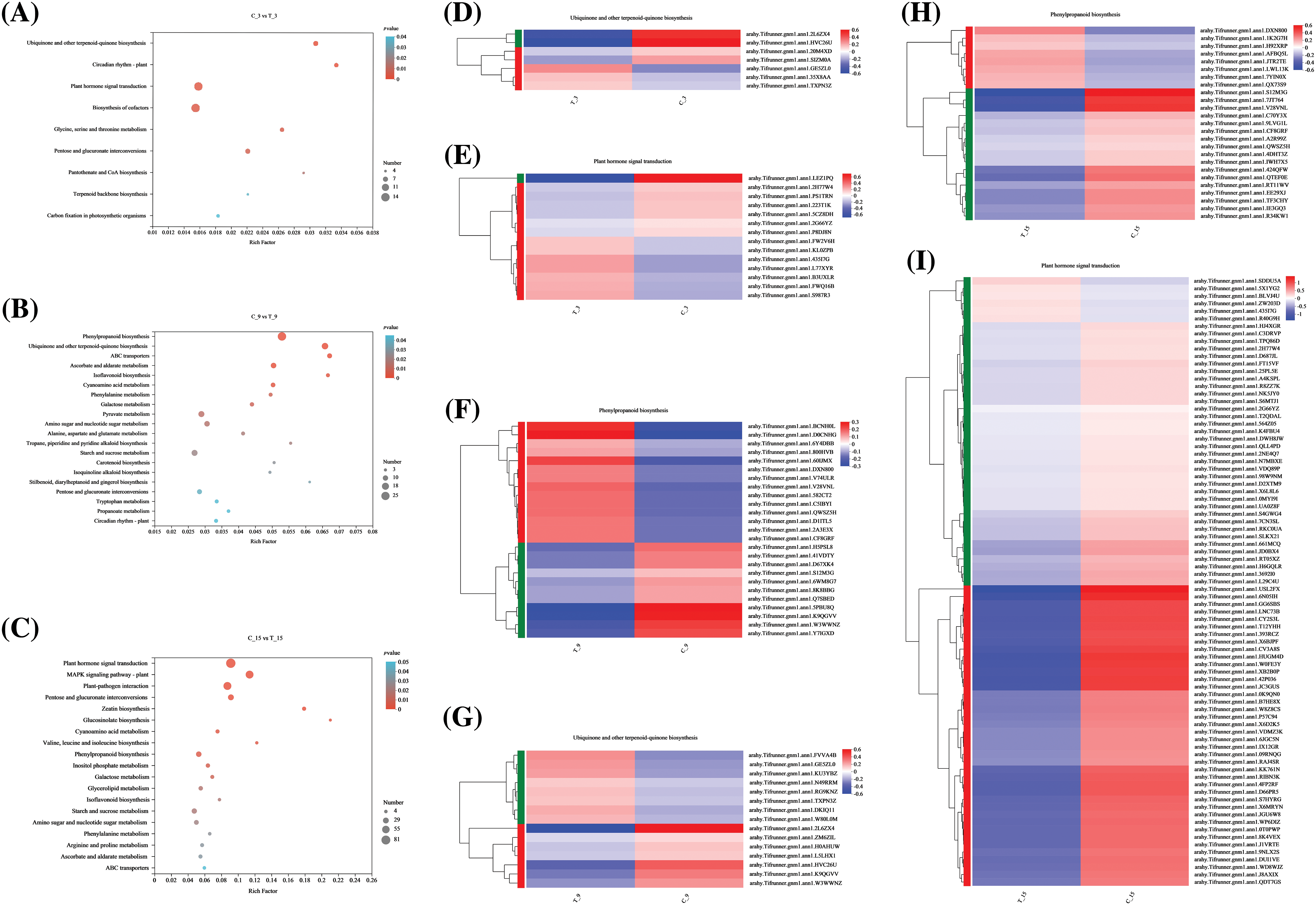
Figure 5: Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment results and heat map clustering analysis. (A–C) Statistical analysis of KEGG pathway with significant enrichment and enrichment in the top 30 KEGG pathway in C3d vs. T3d, C9d vs. T9d, and C15d vs. T15d. Bubble size represents the number of genes with the difference. The color of the bubble represents the degree of significant enrichment. (D–I) Heat map clustering analysis of differential genes in ubiquinone and other terpenoid-quinone biosynthesis pathway, plant hormone signal transduction pathway, and phenylpropanoid biosynthesis pathway.
Note: The colors in the heatmap represent the normalized expression of the gene in each group.
In C3d vs. T3d, 7 DEGs were enriched in ubiquinone and other terpenoid-quinone biosynthesis pathway, wherein 3 were up-regulated and 4 were down-regulated (Fig. 5D), and 14 DEGs were enriched in plant hormone signal transduction pathway, wherein 7 were up-regulated, and 7 were down-regulated (Fig. 5E). Twenty-five genes in C9d vs. T9d were enriched in the phenylpropanoid biosynthesis pathway, of which 14 were up-regulated and 11 were down-regulated (Fig. 5F). Moreover, 15 genes were enriched in the ubiquinone and other terpenoid-quinone biosynthesis pathway, of which 8 were up-regulated and 7 were down-regulated (Fig. 5G). Twenty-five genes in C15d vs. T15d were enriched in the phenylpropanoid biosynthesis pathway, in which 8 were up-regulated and 17 were down-regulated (Fig. 5H). In the KEGG enrichment plots of C15d vs. T15d, the largest number of genes was enriched in the plant hormone signaling metabolic pathway. Eighty-one genes were enriched in the plant hormone signal transduction pathway, of which 6 genes were up-regulated and 75 were down-regulated (Fig. 5I). The results showed that the above-mentioned DEGS regulate the ubiquinone and other terpenoid-quinone biosynthesis pathway, plant hormone signal transduction pathway, and phenylpropanoid biosynthesis pathway inside the A. hypogaea root by activating or inhibiting its own expression in response to the stress of nematodes.
Quantitative real-time PCR (qRT -PCR) analysis validation of differentially expressed genes
Statistical analysis of the changes of the genes number in ubiquinone and other terpenoid-quinone biosynthesis, plant hormone signal transduction, and phenylpropanoid biosynthesis KEGG pathway at 3, 9, and 15 d showed that the number of genes in ubiquinone and other terpenoid-quinone biosynthesis varied by 7, 15 and 5, respectively, with the prolongation of nematode treatment time. The number of genes annotated to the plant hormone signal transduction pathway with increased nematode treatment time was 14, 13, and 81, respectively. The number of genes annotated to the phenylpropanoid biosynthesis pathway with increased nematode treatment time was 5, 25, and 25, respectively (Fig. 6A). At the time limit set by this experiment, there were more DEGs in the early stage of nematode stress than in the later stage to regulate the ubiquinone and other terpenoid-quinone biosynthesis pathway. In the later stage of nematode stress, there were more DEGs that regulate plant hormone signal transduction pathway and phenylpropanoid biosynthesis pathway.
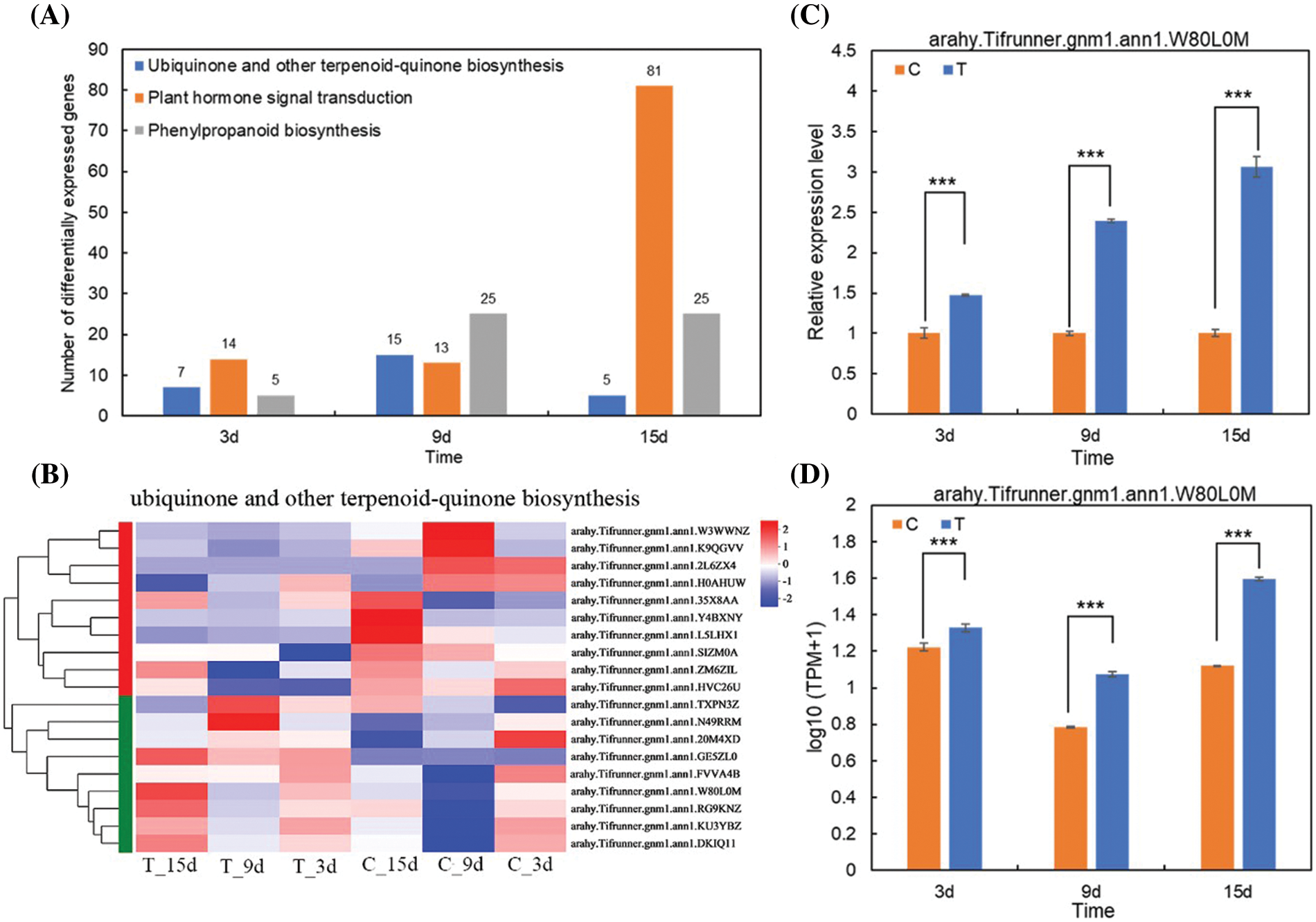
Figure 6: Analysis of target gene selection and validation. (A) Changes in the genes number in ubiquinone and other terpenoid-quinone biosynthesis, plant hormone signal transduction and phenylpropanoid biosynthesis Kyoto Encyclopedia of Genes and Genomes pathway at 3, 9, and 15 d. (B) Heat map clustering analysis of different genes of ubiquinone and other terpenoid-quinone biosynthesis pathway in each group. (C) Quantitative real-time PCR verification of target genes in 3, 9, and 15 d treatment groups and control groups. (D) Statistics of target gene RNA-Seq values in 3, 9, and 15 d treatment groups and control groups.
Note: *** indicates extremely significant difference at 0.001 level.
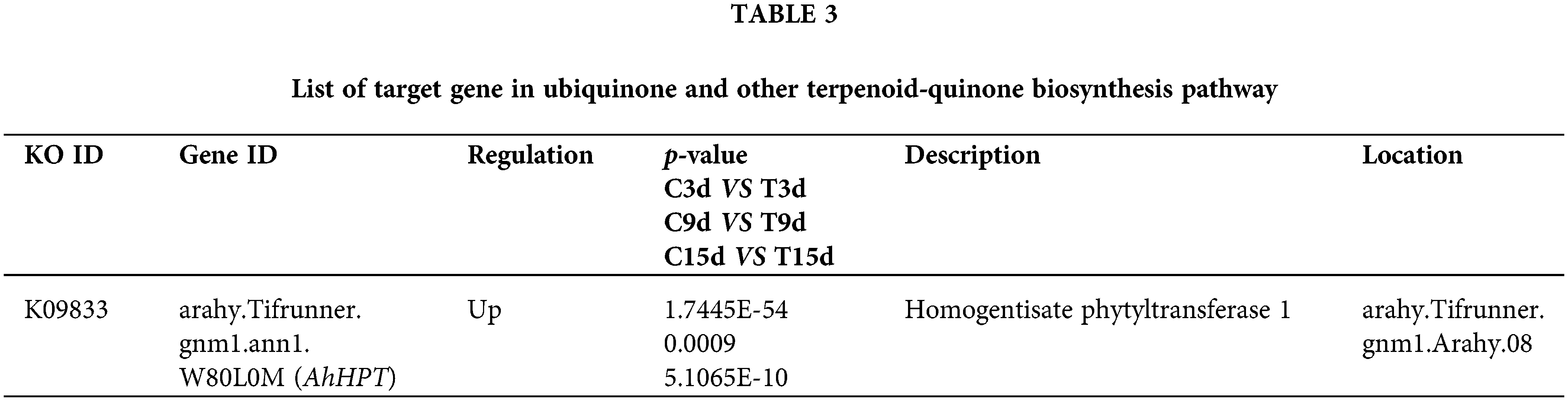
Nineteen DEGs were enriched in the ubiquinone and other terpenoid-quinone biosynthesis pathway in the C3d vs. T3d, C9d vs. T9d, and C15d vs. T15d groups (Fig. 6B). Among these 19 genes, a significantly up-regulated gene in C3d vs. T3d, C9d vs. T9d, and C15d vs. T15d groups was selected (Table 3) to verify the accuracy of transcriptome sequencing analysis and provide reference to find target genes with positive effects on resistance to nematode stress. The verified gene was located on chromosome 8 of A. hypogaea and encodes homogentisate phytyltransferase (HPT). QRT-PCR results showed that the expression levels of the verified gene in the 3, 9, and 15 d treatment groups were higher than that of the corresponding control groups (Fig. 6C). The expression trend of the gene verified by qRT-PCR was consistent with its transcriptome sequencing analysis (Fig. 6D), indicating that transcriptome sequencing data were accurate and reliable.
Many studies have shown that the relationship between A. hypogaea and M. incognita is incompatible and interactive, but the resistance mechanism was unclear. A full understanding of the inhibiting effects of A. hypogaea will provide more ways for the biological control of M. incognita. In this study, the Pluronic F-127 system was used to verify the tendency of the J2s to the excised root tips of A. hypogaea under visual conditions, similar to the phenomenon reported in studies (Wang et al., 2009; Williamson and Čepulytė, 2017). The number of nematodes near the plant root tip is also different due to the different factors such as nematode species, number of inoculations, selected plant species, root tip length, and other factors in the system. The chemotactic migration rate of M. incognita J2s to the excised A. hypogaea root tips increased initially and then decreased along with the extension of observation; the staining result showed that the J2s of M. incognita could infect the A. hypogaea root tips. However, there were no knots at the surface of A. hypogaea roots inoculated with M. incognita J2s in the pot experiment, consistent with an earlier report (Vijayalakshni and Goswami, 1987). These phenomena indicated that A. hypogaea may be a trap plant capable of attracting and inducing M. incognita J2s infestation, while further development and propagation of M. incognita were inhibited in A. hypogaea root system (Waisen et al., 2019; Sacchi et al., 2021). The environment of the A. hypogaea root system is not conducive to the survival and breeding of M. incognita (Bendezu and Starr, 2003).
High-throughput sequencing technology, with its advantages of high sensitivity, high accuracy and low cost, has been gradually applied by many scholars in the study of species and diseases (Sapkota and Nicolaisen, 2015; Yang et al., 2016). In this study, statistical analysis was conducted to find the DEGs after M. incognita J2s infestation.
Through transcriptome sequencing of 6 groups (18 samples), the Q30 (%) value of sequence data after quality control was more than 93.71%, and the matching rate of reference gene was more than 95.73%, indicating the accuracy and reliability of sequencing results. The number of significant DEGs in C3d vs. T3d, C9d vs. T9d, and C15d vs. T15d was counted by transcriptomic sequencing based on RNA-seq, and the number of genes with significant differential expression increased with the increase in the number of stress days. After 7, 14, and 28 days of infestation by M. incognita, the DEG numbers in the eggplant (Solanum melongena L.) also increased (Zhang et al., 2021), but the DEGs numbers in the rice (Oryza sativa L.) root system after 6 days of M. incognita infestation was higher than that after 18 days (Zhou et al., 2020). Dicotyledonous plants and monocotyledonous plants have some differences in the infection response to root-knot nematode (Przybylska and Obrępalska-Stęplowska, 2020).
GO functional enrichment analysis of the three groups of DEGs showed that the DEGs in each group were enriched in the GO term of plant defense response, and one gene encoding defense protein (TIR-NBS-LRR class) was found in all three groups. The invasion of plant pathogens triggers the immune defense system, in which the second defense layer (ETI) in plants is mediated by the plant resistance gene (R gene) (Meyers et al., 1999). Most R genes have been found in plants with nucleotide binding sites (NBS) and leucine-rich repeat (LRR) domains. A subfamily of NBS-LRR proteins and TIR-NBS-LRR proteins with a Toll/interleukin-1 receptor domain at the N-terminus have been found in Arabidopsis (Arabidopsis thaliana L.) (Botella et al., 1998), soybean (Glycine max L.) (Li et al., 2022), chickpea (Cicer arietinum L.) (Sagi et al., 2017) and other plants and are associated with plant disease resistance.
KEGG pathway enrichment analysis of the three groups of DEGs showed that ubiquinone and other terpenoid-quinone biosynthesis, plant hormone signal transduction, and phenylpropanoid biosynthesis were the main metabolic pathways in A. hypogaea responding to M. incognita stress. The candidate gene selected from the ubiquinone and other terpenoid-quinone biosynthesis pathway encodes HPT, the key rate-limiting enzyme for the synthesis of α-tocopherol (Hunter and Cahoon, 2007; Kurmaz et al., 2022). The HPT super-expressing plants had significantly increased content of α-tocopherol (Crowell et al., 2008). In addition, the silencing of the HPT gene in tobacco by dsRNAi technology reduced the content of tocopherol in tobacco and increased the sensitivity of plants to abiotic stress compared with wild-type plants (Abbasi et al., 2007). At present, many studies have found that α-tocopherol plays a positive role in plant growth under the stress of abiotic factors such as low temperature (Maeda et al., 2006), drought (Liu et al., 2008), high salt (Abbasi et al., 2007), and heavy metal ions (Collin et al., 2008). Tocopherol has antioxidant effects, which can remove oxygen free radicals to affect the growth and development of parasites (Apel and Hirt, 2004; Taylor et al., 1997; Ibrahim et al., 2012). In addition, α-tocopherol can slow down plant damage and aging (Austin et al., 2006; Sattler et al., 2006; Abbasi et al., 2009). α-Tocopherol is also involved in the transduction of cell signals in plants, and studies have shown that hormone levels in plants, such as jasmonic acid and ethylene, are related to the synthesis and regulation of tocopherol. In fact, plant hormones have been shown to play a key role in coordinating the signal response to nematode infection (Falk et al., 2002; Cela et al., 2009; Singh et al., 2011). The treatment of soybeans with methyl jasmonate and sodium nitroprusside could improve the resistance of soybeans to Spodoptera littoralis, and α-tocopherol detection was performed on treated soybeans, both of which were significantly increased (Mohamed et al., 2021). Therefore, we suggest that there is a correlation between the AhHPT gene and α-tocopherol expression, which further plays a key role in the resistance of A. hypogaea against M. incognita. The results indicated that the AhHPT gene may be an active regulatory gene that can be applied to plant molecular breeding with resistance against M. incognita.
The excised root tips of A. hypogaea had a certain attraction effect on M. incognita J2s, which could invade the excised and intravital A. hypogaea root. The DEGs of C3d vs. T3d, C9d vs. T9d, and C15d vs. T15d were 462,1057 and 2099, respectively. These DEGs were mainly related to plant defense response function and were enriched in ubiquinone and other terpenoid-quinone biosynthesis, plant hormone signal transduction, and phenylpropanoid biosynthesis pathways. The candidate gene AhHPT was identified in ubiquinone and other terpenoid-quinone biosynthesis pathway, which may be involved in the regulation of A. hypogaea resistance to M. incognita.
Acknowledgement: None.
Funding Statement: This work was supported by the Post-Doctoral Program of Hebei Province (2019003011) and Foundation of President of Hebei University (XZJJ201924).
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Jianfeng Liu and Dandan Cao; data collection: Yuanyuan Zhou and Jianqing Ma; analysis and interpretation of results: Xinyi Peng and Xiaohong Fu; draft manuscript preparation: Xuejin Yang. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The datasets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
Abad P, Gouzy J, Aury J, Castagnone-Sereno P, Danchin EG et al. (2008). Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nature Biotechnology 26: 909–915. https://doi.org/10.1038/nbt.1482 [Google Scholar] [PubMed] [CrossRef]
Abbasi AR, Hajirezaei M, Hofius D, Sonnewald U, Voll LM (2007). Specific roles of α- and γ-tocopherol in abiotic stress responses of transgenic tobacco. Plant Physiology 143: 1720–1738. https://doi.org/10.1104/pp.106.094771 [Google Scholar] [PubMed] [CrossRef]
Abbasi AR, Saur A, Hennig P, Tschiersch H, Hajirezaei M, Hofius D, Sonnewald U, Voll LM (2009). Tocopherol deficiency in transgenic tobacco (Nicotiana tabacum L.) plants leads to accelerated senescence. Plant Cell Environment 32: 144–157. https://doi.org/10.1111/j.1365-3040.2008.01907.x [Google Scholar] [PubMed] [CrossRef]
Aioub AA, Elesawy AE, Ammar EE (2022). Plant growth promoting rhizobacteria (PGPR) and their role in plant‐parasitic nematodes control: A fresh look at an old issue. Journal of Plant Diseases and Protection 129: 1305–1321. https://doi.org/10.1007/s41348-022-00642-3 [Google Scholar] [CrossRef]
Ali AA, EI-Ashry RM, Aioub AAA (2022). Animal manure rhizobacteria co‐fertilization suppresses phytonematodes and enhances plant production: Evidence from field and greenhouse. Journal of Plant Diseases and Protection 129: 155–169. https://doi.org/10.1007/s41348-021-00529-9 [Google Scholar] [CrossRef]
Apel K, Hirt H (2004). Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annual Review of Plant Biology 55: 373–399. https://doi.org/10.1146/annurev.arplant.55.031903.141701 [Google Scholar] [PubMed] [CrossRef]
Austin JN, Frost E, Vidi PA, Kessler F, Staehelin LA (2006). Plastoglobules are lipoprotein subcompartments of the chloroplast that are permanently coupled to thylakoid membranes and contain biosynthetic enzymes. Plant Cell 18: 169–1703. https://doi.org/10.1105/tpc.105.039859 [Google Scholar] [PubMed] [CrossRef]
Belcher JV, Hussey RS (1977). Influence of Tagetes patula and Arachis hypogaea on Meloidogyne incognita. Plant Disease Reporter 61: 525–528. [Google Scholar]
Bendezu IF, Starr JL (2003). Mechanism of resistance to Meloidogyne arenaria in the peanut cultivar COAN. Journal of Nematology 35: 115–118. [Google Scholar] [PubMed]
Botella MA, Parker JE, Frost LN, Bittner-Eddy PD, Beynon JL, Daniels MJ, Holub EB, Jones JD (1998). Three genes of the Arabidopsis RPP1 complex resistance locus recognize distinct Peronospora parasitica avirulence determinants. The Plant Cell 10: 1847–1860. https://doi.org/10.1105/tpc.10.11.1847 [Google Scholar] [PubMed] [CrossRef]
Byrd DW, Kirkpatrick T, Barker KR (1983). An improved technique for clearing and staining plant tissue for detection of nematodes. Journal of Nematology 15: 142–143. [Google Scholar]
Cela J, Falk J, Munné-Bosch S (2009). Ethylene signaling may be involved in the regulation of tocopherol biosynthesis in Arabidopsis thaliana. FEBS Letters 583: 992–996. https://doi.org/10.1016/j.febslet.2009.02.036 [Google Scholar] [PubMed] [CrossRef]
Çiftçi S, Suna G (2022). Functional components of peanuts (Arachis hypogaea L.) and health benefits: A review. Future Foods 5: 100140. [Google Scholar]
Collin VC, Eymery F, Genty B, Rey P, Havaux M (2008). Vitamin E is essential for the tolerance of Arabidopsis thaliana to metal-induced oxidative stress. Plant Cell and Environment 31: 244–257. https://doi.org/10.1111/j.1365-3040.2007.01755.x [Google Scholar] [PubMed] [CrossRef]
Crowell EF, McGrath JM, Douches DS (2008). Accumulation of vitamin E in potato (Solanum tuberosum) tubers. Transgenic Research 17: 205–217. https://doi.org/10.1007/s11248-007-9091-1 [Google Scholar] [PubMed] [CrossRef]
Davis EL, Baum TJ, Bakker J, Schots A, Hussey RS (2000). Nematode parasitism genes. Annual Review of Phytopathology 38: 365–396. [Google Scholar] [PubMed]
Dong WB, Holbrook CC, Timper P, Brenneman TB, Chu Y, Ozias-Akins P (2008). Resistance in peanut cultivars and breeding lines to three root-knot nematode species. Plant Disease 92: 631–638. https://doi.org/10.1094/PDIS-92-4-0631 [Google Scholar] [PubMed] [CrossRef]
Du C, Jiang J, Zhang H, Zhao T, Yang H, Zhang D, Zhao Z, Xu X, Li J (2020). Transcriptomic profiling of Solanum peruvianum LA3858 revealed a Mi-3-mediated hypersensitive response to Meloidogyne incognita. BMC Genomics 21: 250. https://doi.org/10.1186/s12864-020-6654-5 [Google Scholar] [PubMed] [CrossRef]
EI-Ashry RM, Aioub AAA, Awad SE (2023). Suppression of Meloiodogyne incognita (Tylenchida: Heteroderidae) and Tylenchulus semipenterans (Tylenchida: Tylenchulidae) using Tilapia fish powder and plant growth promoting rhizobacteria in vivo and in vitro. European Journal of Plant Pathology 165: 665–676. https://doi.org/10.1007/s10658-023-02637-8 [Google Scholar] [CrossRef]
El-Sappah AH, Islam MM, El-awady HH, Yan S, Qi S, Liu J, Cheng G, Liang Y (2019). Tomato natural resistance genes in controlling the root-knot nematode. Genes 10: 925. https://doi.org/10.3390/genes10110925 [Google Scholar] [PubMed] [CrossRef]
Eldeeb AM, Farag AAG, AI-Harbi MS, Kesba H, Sayed S, Elesawy AE, Hendawi MA, Mostafa EM, Aioub AAA (2022). Controlling of Meloidgyne incognita (Tylenchida: Heteroderidae) using nematicides, Linum usitatissimum extract and certain organic acids on four peppers cultivars under greenhouse conditions. Saudi Journal of Biological Sciences 29: 3107–3113. https://doi.org/10.1016/j.sjbs.2022.03.018 [Google Scholar] [PubMed] [CrossRef]
Elnahal ASM, El‐Saadony MT, Saad AM, Desoky EM, El‐Tahan AM, Rady MM, AbuQamar SF, EI-Tarabily KA (2022). The use of microbial inoculants for biological control, plant growth promotion, and sustainable agriculture: A review. European Journal of Plant Pathology 162: 759–792. https://doi.org/10.1007/s10658-021-02393-7 [Google Scholar] [CrossRef]
Falk J, Krauß N, Dähnhardt D, Krupinska K (2002). The senescence associated gene of barley encoding 4-hydroxyphenyl-pyruvate dioxygenase is expressed during oxidative stress. Journal of Plant Physiology 159: 1245–1253. https://doi.org/10.1078/0176-1617-00804 [Google Scholar] [CrossRef]
Faske TR, Starr JL (2009). Reproduction of Meloidogyne marylandi and M. incognita on several Poaceae. Journal of Nematology 41: 2–4. [Google Scholar] [PubMed]
Hunter SC, Cahoon EB (2007). Enhancing vitamin E in oilseeds: Unraveling tocopherol and tocotrienol biosynthesis. Lipids 42: 97–108. https://doi.org/10.1007/s11745-007-3028-6 [Google Scholar] [PubMed] [CrossRef]
Ibrahim HM, Hosseini P, Alkharouf NW, Hussein EH, El-Din AE, Aly MA, Matthews BF (2011). Analysis of gene expression in soybean (Glycine max) roots in response to the root knot nematode Meloidogyne incognita using microarrays and KEGG pathways. BMC Genomics 12: 220. https://doi.org/10.1186/1471-2164-12-220 [Google Scholar] [PubMed] [CrossRef]
Ibrahim MA, Zuwahu MM, Isah MB, Jatau ID, Aliyu AB, Umar IA (2012). Effects of vitamin E administration on Plasmodium berghei induced pathological changes and oxidative stress in mice. Tropical Biomedicine 29: 98–106. [Google Scholar] [PubMed]
Jaouannet M, Perfus-Barbeoch L, Deleury E, Magliano M, Engler G et al. (2012). A root-knot nematode-secreted protein is injected into giant cells and targeted to the nuclei. The New phytologist 194: 924–931. https://doi.org/10.1111/j.1469-8137.2012.04164.x [Google Scholar] [PubMed] [CrossRef]
Johnson AW, Dowler CC, Handoo ZA (2000). Population dynamics of Meloidogyne incognita, M. arenaria, and other nematodes and crop yields in rotations of cotton, peanut, and wheat under minimum tillage. Journal of Nematology 32: 52–61. [Google Scholar] [PubMed]
Johnson AW, Minton NA, Brenneman TB, Todd JW, Herzog GA, Gascho GJ, Baker SH, Bondari Y (1998). Peanut-cotton-rye rotations and soil chemical treatment for managing nematodes and thrips. Journal of Nematology 30: 211–225. [Google Scholar] [PubMed]
Jones JT, Haegeman A, Danchin EG, Gaur HS, Helder J et al. (2013). Top 10 plant-parasitic nematodes in molecular plant pathology. Molecular Plant Pathology 14: 946–961. https://doi.org/10.1111/mpp.12057 [Google Scholar] [PubMed] [CrossRef]
Kim D, Langmead B, Salzberg SL (2015). HISAT: A fast spliced aligner with low memory requirements. Nature methods 12: 357–360. https://doi.org/10.1038/nmeth.3317 [Google Scholar] [PubMed] [CrossRef]
Kurmaz SV, Fadeeva NV, Skripets JA, Komendant RI, Ignatiev VM, Emel’yanova NS, Soldatova YV, Faingold II, Poletaeva DA, Kotelnikova RA (2022). New water-soluble forms of a-tocopherol: Preparation and study of antioxidant activity in vitro. Mendeleev Communications 32: 117–119. https://doi.org/10.1016/j.mencom.2022.01.038 [Google Scholar] [CrossRef]
Li B, Dewey CN (2011). RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12: 323. https://doi.org/10.1186/1471-2105-12-323 [Google Scholar] [PubMed] [CrossRef]
Li Z, Deng S, Xuan H, Fan X, Sun R, Zhao J, Wang H, Guo N, Xing H (2022). A novel TIR-NBS-LRR gene regulates immune response to Phytophthora root rot in soybean. The Crops Journal 10: 1644–1653. https://doi.org/10.1016/j.cj.2022.03.003 [Google Scholar] [CrossRef]
Liu X, Hua X, Guo J, Qi D, Wang L, Liu Z, Jin Z, Chen S, Liu G (2008). Enhanced tolerance to drought stress in transgenic tobacco plants overexpressing VTE1 for increased tocopherol production from Arabidopsis thaliana. Biotechnology Letters 30: 1275–1280. [Google Scholar] [PubMed]
Love MI, Huber W, Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 15: 550. https://doi.org/10.1186/s13059-014-0550-8 [Google Scholar] [PubMed] [CrossRef]
Maeda H, Song W, Sage TL, DellaPenna D (2006). Tocopherols play a crucial role in low-temperature adaptation and phloem loading in Arabidopsis. Plant Cell 18: 2710–2732. https://doi.org/10.1105/tpc.105.039404 [Google Scholar] [PubMed] [CrossRef]
Mao Z, Zhu P, Liu F, Huang Y, Ling J, Chen G, Yang Y, Feng D, Xie B (2015). Cloning and functional analyses of pepper CaRKNR involved in Meloidogyne incognita resistance. Euphytica 205: 903–913. https://doi.org/10.1007/s10681-015-1438-8 [Google Scholar] [CrossRef]
Meyers BC, Dickerman AW, Michelmore RW, Sivaramakrishnan S, Sobral BW, Young ND (1999). Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant Journal 20: 317–332. https://doi.org/10.1046/j.1365-313x.1999.t01-1-00606.x [Google Scholar] [PubMed] [CrossRef]
Moens M, Perry RN, Starr JL (2009). Meloidogyne Species: A Diverse Group of Novel and Important Plant Parasites, pp. 1–17. Wallingford, UK: CABI Publishing. [Google Scholar]
Mohamed HI, Mohammed AH, Mohamed NM, Ashry NA, Zaky LM, Mogazy AM (2021). Comparative effectiveness of potential elicitors of soybean plant resistance against Spodoptera littoralis and their effects on secondary metabolites and antioxidant defense system. Gesunde Pflanzen 73: 273–285. https://doi.org/10.1007/s10343-021-00546-6 [Google Scholar] [CrossRef]
Nishat Y, Danish M, Mohamed HI, Shaikh H, Elhakem A (2022). Biological control of root-knot nematode Meloidogyne incognita in Psoralea corylifolia plant by enhancing the biocontrol efficacy of Trichoderma harzianum using Press Mud. Phyton-International Journal of Experimental Botany 91: 1757–1777. https://doi.org/10.32604/phyton.2022.021267 [Google Scholar] [CrossRef]
Oka YJ, Koltai H, Bar-Eyal M, Mor M, Sharon E, Chet I, Spiegel Y (2015). New strategies for the control of plant-parasitic nematodes. Pest Management Science 56: 983–988. https://doi.org/10.1002/1526-4998(200011)56:11<983::aid-ps233>3.0.co;2-x [Google Scholar] [CrossRef]
Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell J, Salzberg SL (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nature Biotechnology 33: 290–295. https://doi.org/10.1038/nbt.3122 [Google Scholar] [PubMed] [CrossRef]
Przybylska A, Obrępalska-Stęplowska A (2020). Plant defense responses in monocotyledonous and dicotyledonous host plants during root-knot nematode infection. Plant and Soil 451: 239–260. https://doi.org/10.1007/s11104-020-04533-0 [Google Scholar] [CrossRef]
Reynolds AM, Dutta TK, Curtis RH, Powers SJ, Gaur HS, Kerry BR (2015). Chemotaxis can take plant-parasitic nematodes to the source of a chemo-attractant via the shortest possible routes. Journal of the Royal Society Interface 8: 568–577. https://doi.org/10.1098/rsif.2010.0417 [Google Scholar] [PubMed] [CrossRef]
Sacchi S, Torrini G, Marianelli L, Mazza G, Fumagalli A, Cavagna B, Ciampitti M, Roversi PF (2021). Control of Meloidogyne graminicola a root-knot nematode using rice plants as trap crops: Preliminary results. Agriculture 11: 37. https://doi.org/10.3390/agriculture11010037 [Google Scholar] [CrossRef]
Sagi MS, Deokar AA, Tar’an B (2017). Genetic analysis of NBS-LRR gene family in chickpea and their expression profiles in response to Ascochyta blight infection. Frontiers in Plant Science 8: 838. https://doi.org/10.3389/fpls.2017.00838 [Google Scholar] [PubMed] [CrossRef]
Sapkota R, Nicolaisen M (2015). High-throughput sequencing of nematode communities from total soil DNA extractions. BMC Ecology 15: 3. https://doi.org/10.1186/s12898-014-0034-4 [Google Scholar] [PubMed] [CrossRef]
Sattler SE, Mène-Saffrané L, Farmer EE, Krischke M, Mueller MJ, DellaPenna D (2006). Nonenzymatic lipid peroxidation reprograms gene expression and activates defense markers in Arabidopsis tocopherol-deficient mutants. Plant Cell 18: 3706–3720. https://doi.org/10.1105/tpc.106.044065 [Google Scholar] [PubMed] [CrossRef]
Singh KB, Jayaswal P, Chandra S, Jayanthi M, Mandal PK (2022). Comparative transcriptome profiling of Polianthes tuberosa during a compatible interaction with root-knot nematode Meloidogyne incognita. Molecular Biology Reports 49: 4503–4516. https://doi.org/10.1007/s11033-022-07294-4 [Google Scholar] [PubMed] [CrossRef]
Singh RK, Ali SA, Nath P, Sane VA (2011). Activation of ethylene-responsive P-hydroxyphenylpyruvate dioxygenase leads to increased tocopherol levels during ripening in mango. Journal of Experimental Botany 62: 3375–3385. https://doi.org/10.1093/jxb/err006 [Google Scholar] [PubMed] [CrossRef]
Taylor DW, Levander OA, Krishna VR, Evans CB, Morris VC, Barta JR (1997). Vitamin E-eficient diets enriched with fish oil suppress lethal Plasmodium yoelii infections in athymic and scid/bg mice. Infection and Immunity 65: 197–202. https://doi.org/10.1128/iai.65.1.197-202.1997 [Google Scholar] [PubMed] [CrossRef]
Vijayalakshni K, Goswami BK (1987). Effect of root dip treatment of tomato seedlings of aqueous extracts of some oil-seed cakes on root-knot nematode infestation. Annals of Agricultural Research 8: 168–171. [Google Scholar]
Waisen P, Sipes BS, Wang KH (2019). Potential of biofumigant cover crops as open-end trap crops against root-knot and reniform nematodes. Nematropica 49: 254–264. [Google Scholar]
Wang C, Lower S, Williamson VM (2009). Application of Pluronic gel to the study of root-knot nematode behaviour. Nematology 11: 453–464. https://doi.org/10.1163/156854109x447024 [Google Scholar] [CrossRef]
Williamson VM, Čepulytė R (2017). Assessing attraction of nematodes to host roots using Pluronic gel medium. Methods in Molecular Biology 1573: 261–268. https://doi.org/10.1007/978-1-4939-6854-1_19 [Google Scholar] [PubMed] [CrossRef]
Wubben MJ, Callahan FE, Hayes RW, Jenkins JN (2008). Molecular characterization and temporal expression analyses indicate that the MIC (Meloidogyne Induced Cotton) gene family represents a novel group of root-specific defense-related genes in upland cotton (Gossypium hirsutum L.). Planta 228: 111–123. https://doi.org/10.1007/s00425-008-0723-3 [Google Scholar] [PubMed] [CrossRef]
Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li C, Wei L (2011). KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Research 39: W316–W322. https://doi.org/10.1093/nar/gkr483 [Google Scholar] [PubMed] [CrossRef]
Yang MX, Wang QX, Xiao H, Yang C (2016). Sequencing complete mitochondrial genome of Lanius sphenocercus sphenocercus (Passeriformes: Laniidae) using Illunima HiSeq 2500. Mitochondrial DNA Part B-Resources 1: 306–307. https://doi.org/10.1080/23802359.2016.1167640 [Google Scholar] [PubMed] [CrossRef]
Zhang M, Zhang H, Tan J, Huang S, Chen X, Jiang D, Xiao X (2021). Transcriptome analysis of eggplant root in response to root-knot nematode infection. Pathogens 10: 470. https://doi.org/10.3390/pathogens10040470 [Google Scholar] [PubMed] [CrossRef]
Zhou Y, Zhao D, Shuang L, Xiao D, Xuan Y et al. (2020). Transcriptome analysis of rice roots in response to root-knot nematode infection. International Journal of Molecular Sciences 21: 848. https://doi.org/10.3390/ijms21030848 [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools