 Open Access
Open Access
ARTICLE
Initial steps on the analysis of the underlying pharmacological mechanisms of Wendan decoction on sudden deafness using network pharmacology and molecular docking
1 Department of Otolaryngology, Yizheng People’s Hospital, Yangzhou, 211400, China
2 Department of Otolaryngology-Head and Neck Surgery, Nanjing Drum Tower Hospital Clinical College of Traditional Chinese and Western Medicine,
Nanjing University of Chinese Medicine, Nanjing, 210008, China
3 Department of Pulmonary and Critical Care Medicine, Jiangyin Hospital of Traditional Chinese Medicine; Jiangyin Hospital Affiliated to Nanjing University of
Chinese Medicine, Jiangyin, 214400, China
4 Department of Gastroenterology, Jiangyin Hospital of Traditional Chinese Medicine; Jiangyin Hospital Affiliated to Nanjing University of Chinese Medicine,
Jiangyin, 214400, China
5 Department of Internal Medicine, Jiangyin Hospital of Traditional Chinese Medicine; Jiangyin Hospital Affiliated to Nanjing University of Chinese Medicine,
Jiangyin, 214400, China
* Corresponding Authors: Wandong She, ; Haibing Hua,
# Co-first authors
(This article belongs to the Special Issue: Natural Products for Chronic Inflammatory Diseases: Pharmacology and Toxicology)
BIOCELL 2023, 47(9), 1947-1964. https://doi.org/10.32604/biocell.2023.029333
Received 13 February 2023; Accepted 19 April 2023; Issue published 28 September 2023
Abstract
Background: Despite its widespread therapeutic use and effectiveness, the underlying pharmacologic mechanisms of Wendan decoction (WDD) and how it works to treat sudden deafness (SD) remain unclear. In this study, the pharmacological mechanisms of WDD underlying SD were analyzed using network pharmacology and molecular docking. Methods: The Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP) was employed to identify the active compounds and target genes of WDD, and genes associated with SD were screened on five databases. RGUI conducted Gene Ontology (GO) functional and the Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses. A compound-target network was established using Cytoscape, and the STRING database created a protein-protein interaction (PPI) network to identify the key compounds and targets. Subsequently, a network of crucial compound-target was generated for further molecular docking analysis. For molecular docking simulations of the macromolecular target proteins and their matching ligand molecules, AutoDock Vina and AutoDockTool were utilized. Results: TCMSP identified 162 active target genes and 36 active compounds for WDD. The active target genes were compared with the 2271 genes associated with SD to identify 70 intersecting active target genes linked to 34 active compounds. The GO functional enrichment and KEGG pathway enrichment analyses were undertaken, and compound–target, and PPI networks were built. The key compounds and protein targets were identified and integrated to form a key compound–target network. Eventually, molecular docking was performed to investigate the interactions of the protein targets with their respective compounds. Conclusion: This study highlights the mechanisms of multi-compounds, targets, and pathways of WDD acting on SD and provides further evidence of crucial compounds and their matching target proteins of WDD acting on SD.Keywords
Supplementary Material
Supplementary Material FileSudden deafness (SD), also known as sudden sensorineural hearing loss, is a prevalent ear disease with unexplained etiology occurring suddenly within 72 h, affecting at least two adjacent frequencies and a minimum of 30 dB (Chandrasekhar et al., 2019). SD affects 160 individuals per 100,000 individuals every year. While SD can occur at any age, it is more common in individuals aged 40 to 60 years, with no gender difference (Yoon et al., 2022). However, the incidence of SD in China has increased significantly in recent years, particularly among younger individuals (Meng et al., 2022; Tai et al., 2021).
Although there is no specific treatment for SD, corticosteroid hormone therapy is the most commonly used treatment option. However, due to the unclear underlying pathogenesis, the response to treatment and prognosis can vary significantly among patients (Kordis et al., 2020). According to traditional Chinese medicine (TCM), SD is associated with several factors, such as phlegm and fire depression, qi and blood deficiencies, qi stagnation, blood stasis, kidney essence loss, wind and heat invasion, and liver fire disturbance (Wang et al., 2022). Given the limitations of Western medicine in addressing the unknown mechanism of SD, TCM is increasingly being used in the clinical management of SD (Castaneda et al., 2019; Zhu et al., 2022).
Wendan decoction (WDD), a conventional TCM formulation first described in Ji Yan Fang during the Southern and Northern Dynasties, was adopted by the National Administration of TCM in 2018 and included in the “Ancient Classical Chinese Medicine Formula Catalogue (First Edition)” (Wang et al., 2021; Zhang et al., 2022a). WDD primarily consists of banxia (Rhizoma Pinelliae (RP)), chenpi (Pericarpium Citri Reticulate (PCR)), zhishi (Fructus Aurantii Immaturus (FAI)), zhuru (Caulis Bambusae in Taeniam (CBT)), fuling (Poria (PA)), and gancao (Radix Glycyrrhizae (RG)) (Che et al., 2016).
WDD is commonly used to invigorate the spleen (Pi), harmonize the stomach (Wei), regulate qi, and dispel phlegm when treating phlegm syndrome, which is intimately linked to several diseases, including digestive reflux disorders, dyslipidemia, cardiovascular disease, primary insomnia, and others diseases (Feng et al., 2019; Ling et al., 2015; Yan et al., 2017; Zhang et al., 2022b). Recent studies have demonstrated that WDD has a remarkable therapeutic impact in treating SD (Xia and Ji, 2021). For the quantification of compounds in WDD, a validated and reliable ultra-high performance liquid chromatography combined with triple quadrupole mass spectrometry (UHPLC-QQQ-MS/MS) method was developed (Zhang et al., 2019). Despite its widespread therapeutic use and effectiveness, the underlying pharmacologic mechanisms of WDD and the mode of action to treat SD remains unclear.
In this study, we aimed to investigate the pharmacologic mechanisms of WDD in the treatment of SD using network pharmacology and molecular docking. We first identified the active compounds in WDD and its target genes and compared these target genes with SD-associated genes to determine the active target genes of WDD in treating SD. Next, we performed functional enrichment analyses using Gene Ontology (GO) and pathway enrichment analyses using the Kyoto Encyclopedia of Genes and Genomes (KEGG). We then constructed the protein-protein interaction (PPI) network, compound–target, and key compound–target networks and selected key compounds and protein targets. Finally, molecular docking analysis was performed to examine the binding of the protein targets to the related compounds.
Identification of active compounds in Wendan decoction
The Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP) (http://tcmspw.com/tcmsp.php) (Ru et al., 2014) was utilized to identify the compounds present within WDD. TCMSP provides valuable information regarding the interrelation between Chinese herbal medicines, their targets, and the applicable disorders. The widely employed criteria to identify active compounds in network pharmacology are oral bioavailability (OB) ≥30% and drug-likeness (DL) ≥0.18 (Chen et al., 2022; Gao et al., 2022). The OB value represents the absorption rate of the compound in the body, while the DL parameter indicates the similarity of the functional groups of the compound or physical properties with those of existing medications (Xia et al., 2020; Xing et al., 2021).
Active compound target genes screening
The TCMSP was also employed to obtain the matching target genes for the active compounds. The gene targets were uploaded to the UniProt Knowledgebase, a rich repository for protein sequence and annotation data (http://www.uniprot.org/) (UniProt, 2018), with the retrieval format designated as the phrase “homo sapiens”. Subsequently, the official gene symbols for humans were determined and identified as the active target genes of WDD.
Acquisition of sudden deafness-associated genes and determination of active target genes of Wendan decoction acting on sudden deafness
Multiple databases were explored, including DrugBank (https://www.drugbank.ca/), GeneCards (https://www.genecards.org/), Therapeutic Target Database (TTD; http://db.idrblab.net/ttd/), PharmGkb platform (https://www.pharmgkb.org/), and Online Mendelian Inheritance in Man (OMIM; https://omim.org/) databases, to obtain a comprehensive collection of genes associated with SD.
The GeneCards database is a retrievable, integrated repository of annotated and predicted human genes (Stelzer et al., 2016). At the same time, OMIM contains a detailed and up-to-date collection of genes and genetic traits in humans (Amberger and Hamosh, 2017). PharmGkb is a pharmacogenetic informational platform that includes clinical data such as clinical instructions and medication labels, possibly clinically actionable gene-drug connections, and genotype-phenotype interactions (Whirl-Carrillo et al., 2021). On the contrary, TTD contains data on discovered and investigated nucleic acid targets and therapeutic proteins, as well as data concerning the target illness, pathway details, and the related pharmaceuticals directed at each target (Zhou et al., 2022). DrugBank Online is a detailed, freely accessible online resource containing information about medications and drug candidates (Wishart et al., 2018).
These genes were acquired from a pooled collection, and SD-associated genes were obtained. Subsequently, the active target genes of WDD were matched with the SD-associated genes, where the genes that overlapped were identified as the active target genes of WDD acting on SD.
Analyses of Gene Ontology functional enrichment and Kyoto Encyclopedia of Genes and Genomes pathway enrichment
The RGUI 3.6.1 was used in combination with the org.Hs.eg.db package to obtain the entrezIDs of the active target genes identified in the previous step (Chen et al., 2021b). Subsequently, RGUI, clusterProfiler package, and GOplot package were employed to perform functional enrichment analyses on GO terms, and pathway enrichment analysis on KEGG terms (Walter et al., 2015; Wu et al., 2021). The GO functional enrichment analysis included molecular function, cellular component, and biological process.
Compound-target network establishment
The compound–target network was created and analyzed using the Cytoscape 3.6.0 software and its NetworkAnalyzer module. Nodes denoted compounds and gene targets, while edges denoted their links. A subsequent examination of the network was carried out under the degree of connectivity between the compound and the target gene (the greater the extent of connections, the greater the degree value) (Assenov et al., 2008; Shannon et al., 2003).
The Establishment of the protein-protein interaction network
To create a PPI network, the active target genes were incorporated into the STRING database (https://string-db.org/), a repository that facilitates the identification of functional relationships in large-scale gene experimental datasets (Szklarczyk et al., 2019). A PPI network was created by designating the study species as “homo sapiens” and using the least interaction value of 0.4. Subsequently, the PPI network data were employed to undertake a topology assessment using Cytoscape 3.6.0 software and its NetworkAnalyzer module. Finally, the primary target proteins of WDD acting on SD were identified based on the degree values of each protein target (the greater the extent of connections, the greater the degree value) (Assenov et al., 2008; Shannon et al., 2003).
Validation of molecular docking
The molecular docking technique was used to investigate the interaction of the protein target with its matching compound. The Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB; http://www.rcsb.org/) was employed to acquire the target protein structures (Kouranov et al., 2006), while the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) was utilized to retrieve the relative compounds (Kim et al., 2021). AutoDockTool 1.5.6 and AutoDock Vina software were used to conduct molecular docking simulations of protein targets with their matching compounds (Eberhardt et al., 2021).
For molecular docking and network pharmacology, automatically generated statistical analyses were carried out using the bioinformatic tools provided by the software and platforms mentioned above. A q-value < 0.05 indicated statistical significance in the GO functional and KEGG pathway enrichment.
Screening of active compounds in Wendan decoction
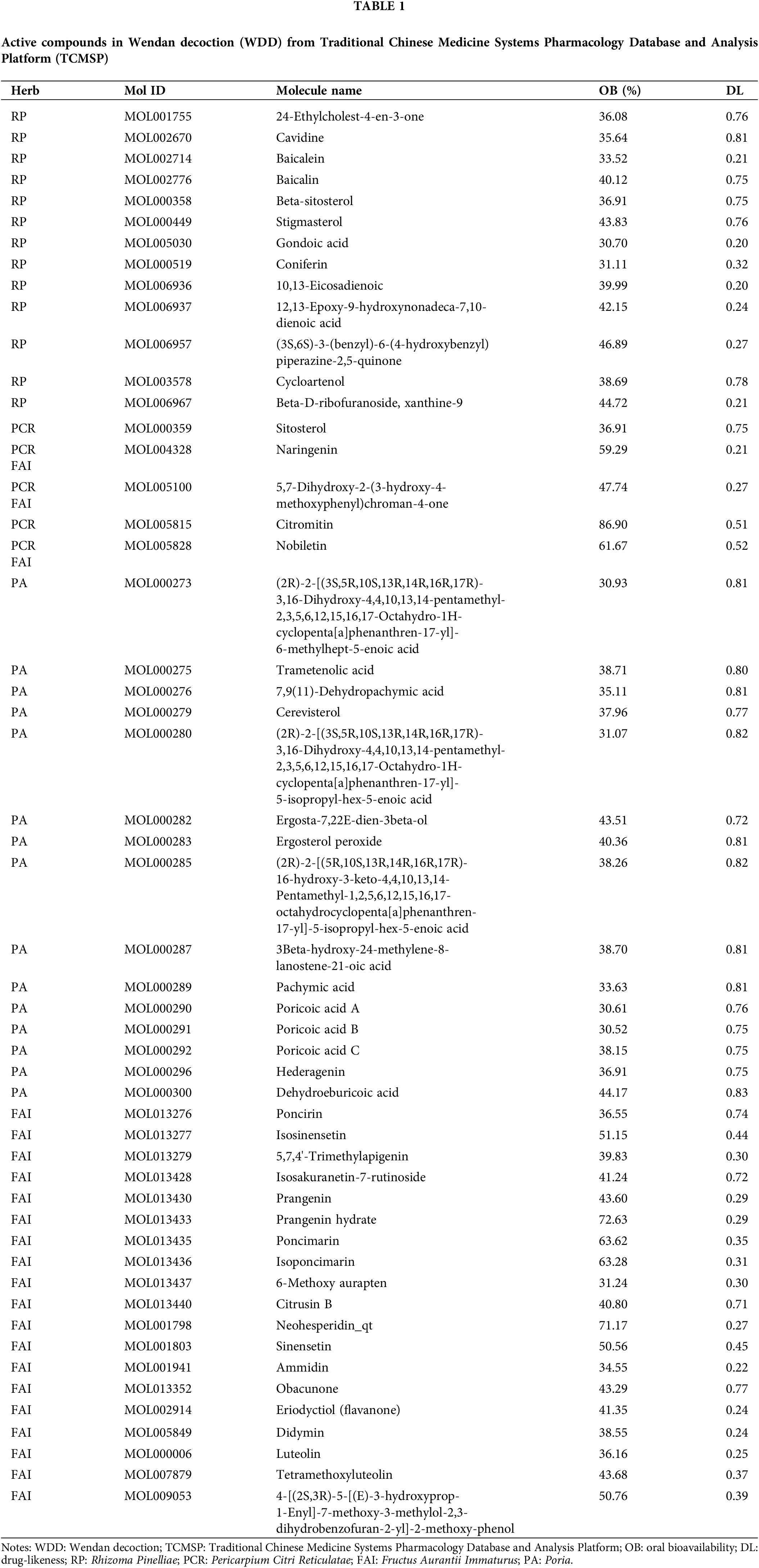
We retrieved 116 compounds from RP, 63 from PCR, 34 from PA, and 65 from FAI using the TCMSP database (The database did not have relevant information for CBT; RG as a guide herb was not included, Suppl. Table S1). We identified 13 active compounds from RP, 5 from PCR, 15 from PA, and 22 from FAI after the screening conditions of DL ≥ 0.18 and OB ≥ 30%. Finally, after removing duplicates from the samples, we found 52 active compounds in WDD. Table 1 summarizes the basic details of the active compounds in WDD.
Identification of active target genes of Wendan decoction
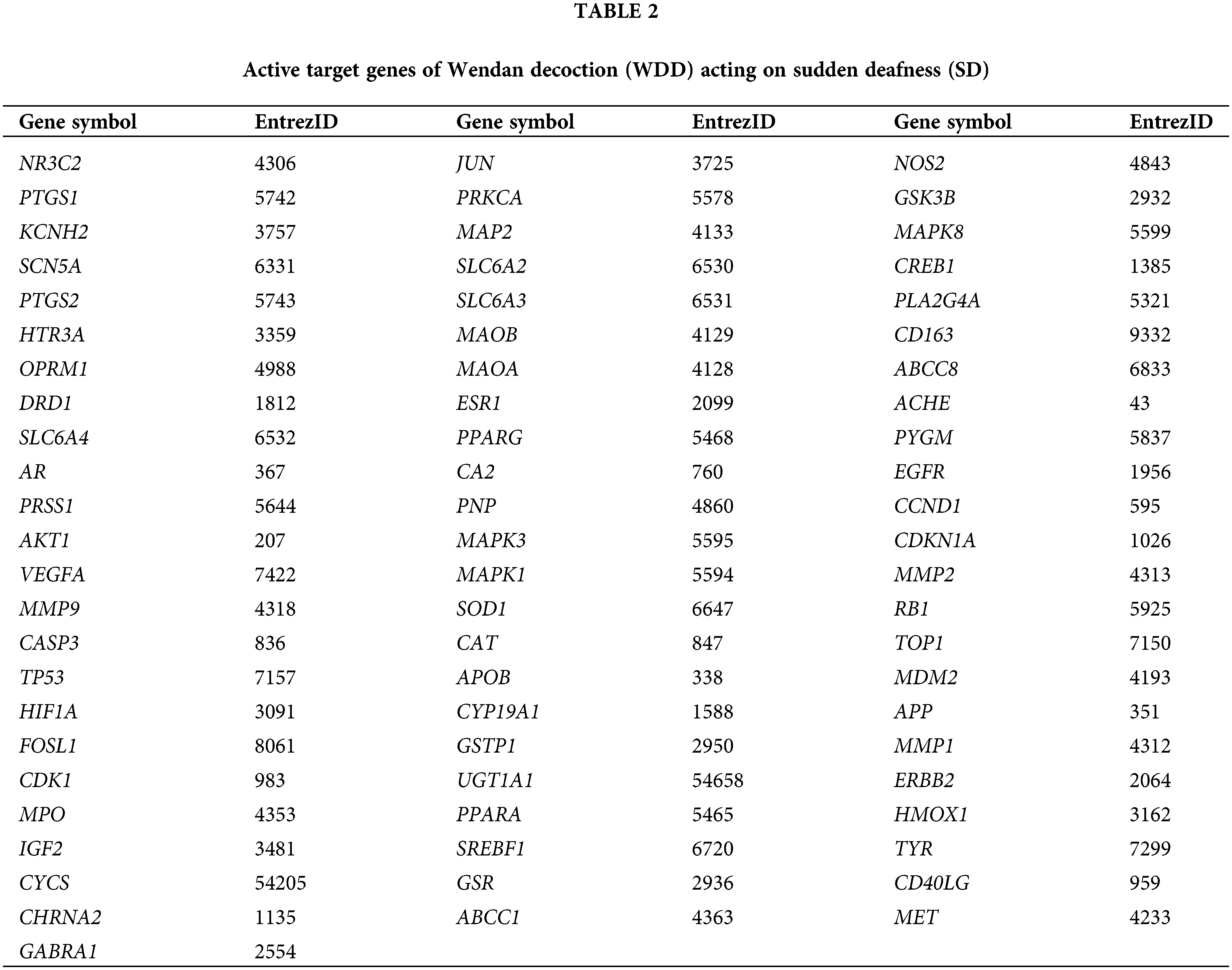
The TCMSP was also used to identify the target genes for the 52 active compounds in WDD, of which 13 had no corresponding target genes (Suppl. Table S2). The matching gene symbols were further filtered using the “homo sapiens” format of UniProt Knowledgebase, resulting in three compounds that did not have matching gene symbols (Suppl. Table S3). Ultimately, we identified 162 active target genes for the 36 active compounds in WDD (Suppl. Table S4).
Acquisition of sudden deafness-related genes
We retrieved 2189 associated genes in the GeneCards database, 152 in the OMIM database, 7 in the PharmGkb platform, one in the TTD, and one in the DrugBank database using “sudden deafness” as the keyword (Suppl. Table S5). After removing duplicates, we considered a pooled set of these genes and obtained 2271 SD-related genes (Fig. 1).
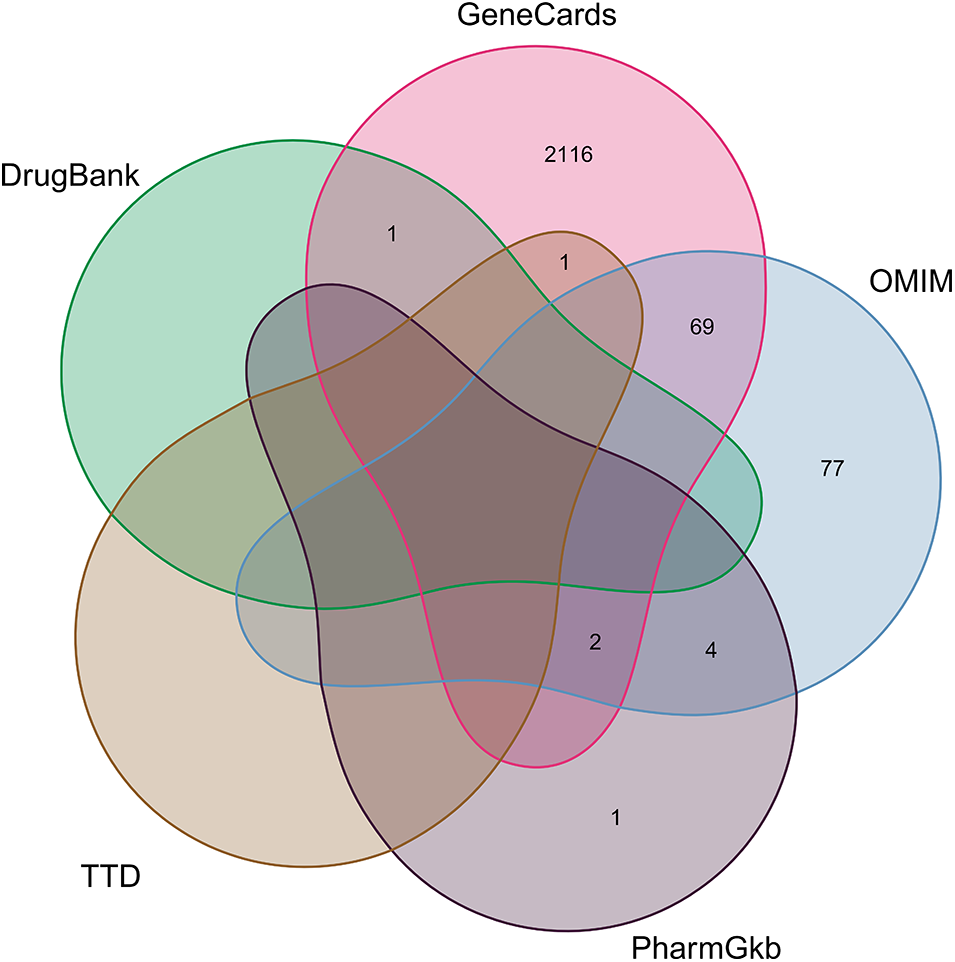
Figure 1: Genes related to sudden deafness (SD). A total of 2189 SD-related genes were retrieved in the GeneCards database, 152 in the Online Mendelian Inheritance in Man (OMIM) database, 7 in the PharmGkb platform, 1 in the Therapeutic Target Database (TTD), and 1 in the DrugBank database.
Identification of active genes targets of Wendan decoction acting on sudden deafness
We uncovered 70 active WDD target genes that acted on SD after comparing the 162 active WDD target genes to the 2271 genes associated with SD (Fig. 2, Table 2).
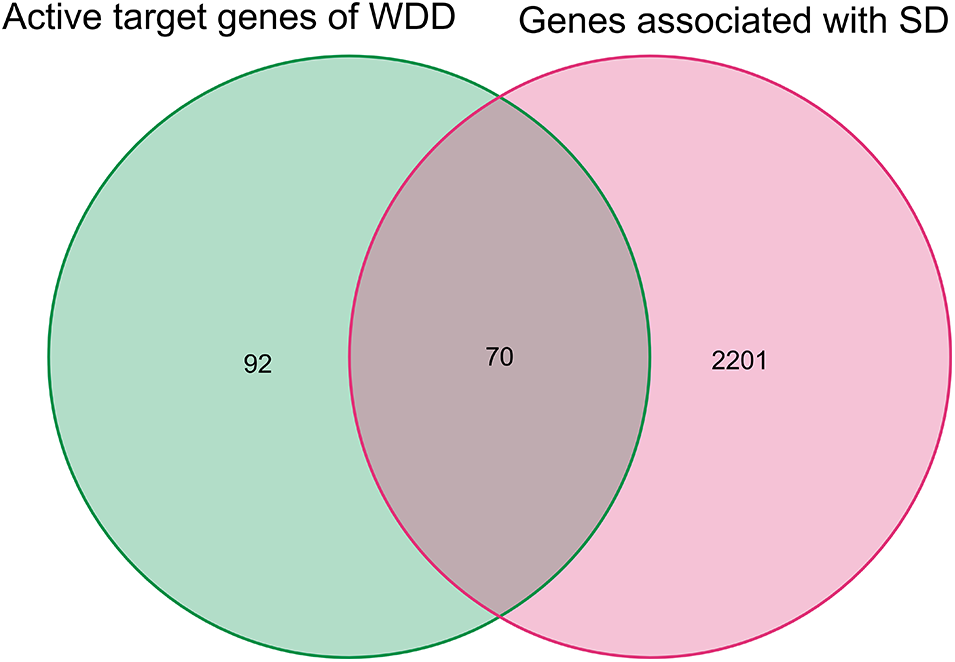
Figure 2: Active target genes of Wendan decoction (WDD) that influence sudden deafness (SD). The 162 WDD active target genes were matched to the 2271 genes associated with SD, identifying 70 WDD active target genes that acted on SD.
Analyses of Gene Ontology functional enrichment and Kyoto Encyclopedia of Genes and Genomes pathway enrichment
We employed RGUI and org.Hs.eg.db to retrieve the entrezIDs of the WDD active target genes that acted on SD (Table 2). Then, we conducted GO functional and KEGG pathway enrichment studies utilizing RGUI and clusterProfiler. Finally, RGUI and GOplot displayed the terms with their enriched genes on GO and KEGG chord plots.
The GO biological process enrichment analysis revealed that the WDD active target genes acting on SD were strongly enriched in processes such as sensitivity to toxic substances, responsiveness to oxidative stress, reaction to extracellular stimuli, response to metal ions, and response to nutrient levels. The analytical findings from GO cellular component enrichment illustrated a substantial enrichment in the membrane region, caveola, membrane microdomain, vesicle lumen, membrane raft, and other components. Further, the GO molecular function enrichment analysis illustrated a significant enrichment in several functions, including ubiquitin-protein ligase binding, tetrapyrrole binding, heme binding, protein phosphatase binding, and phosphatase binding (Suppl. Table S6). Fig. 3 depicts the top 10 GO functional enrichment terms sorted by q-value. The top 10 GO biological function enrichments with enriched genes are displayed as GO chord plots (Fig. 4).
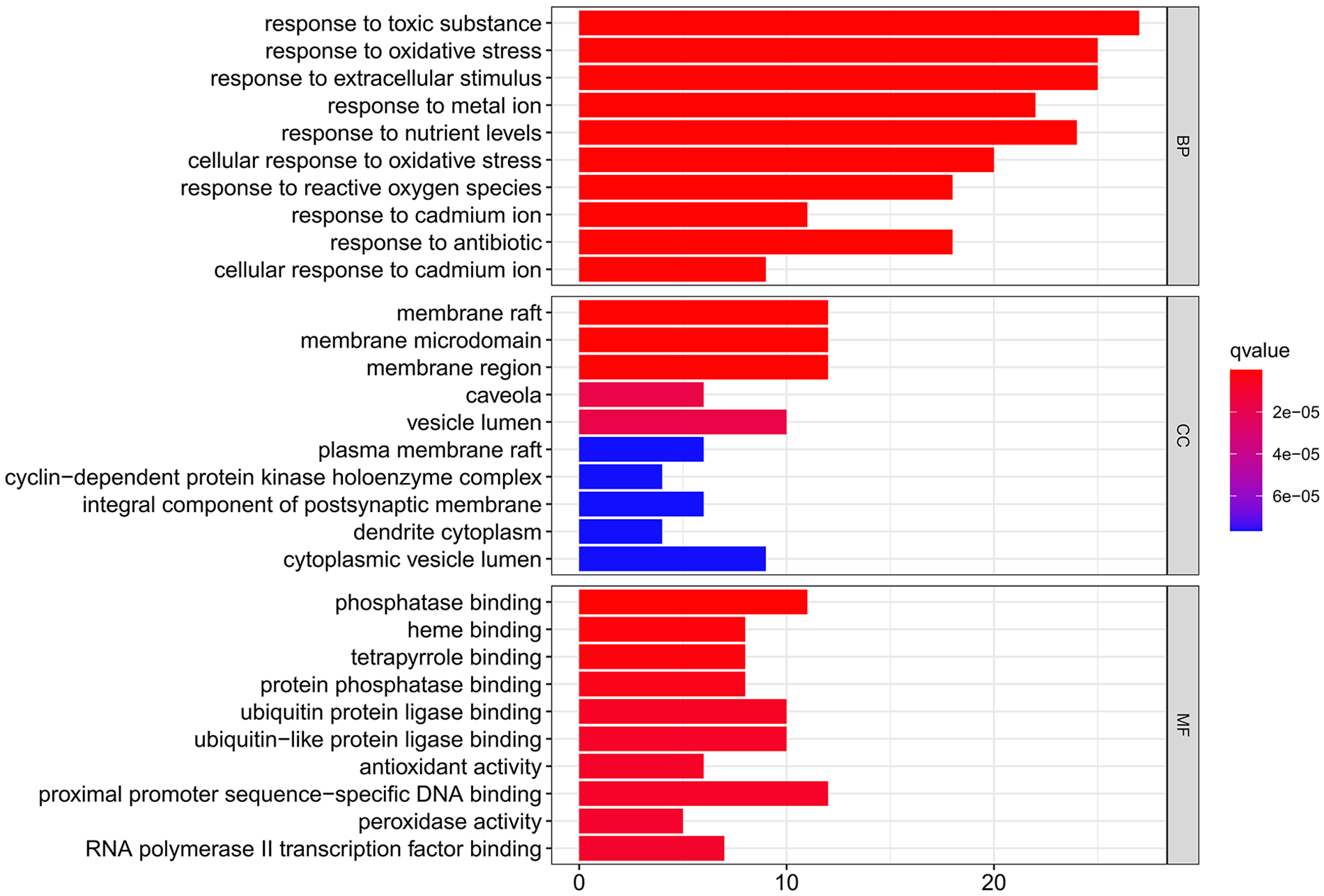
Figure 3: Analysis of Gene Ontology (GO) functional enrichments. The top 10 GO functional enrichments included biological process (BP), cellular component (CC), and molecular function (MF). The smaller the q-value, the greater the significance of the enrichment.
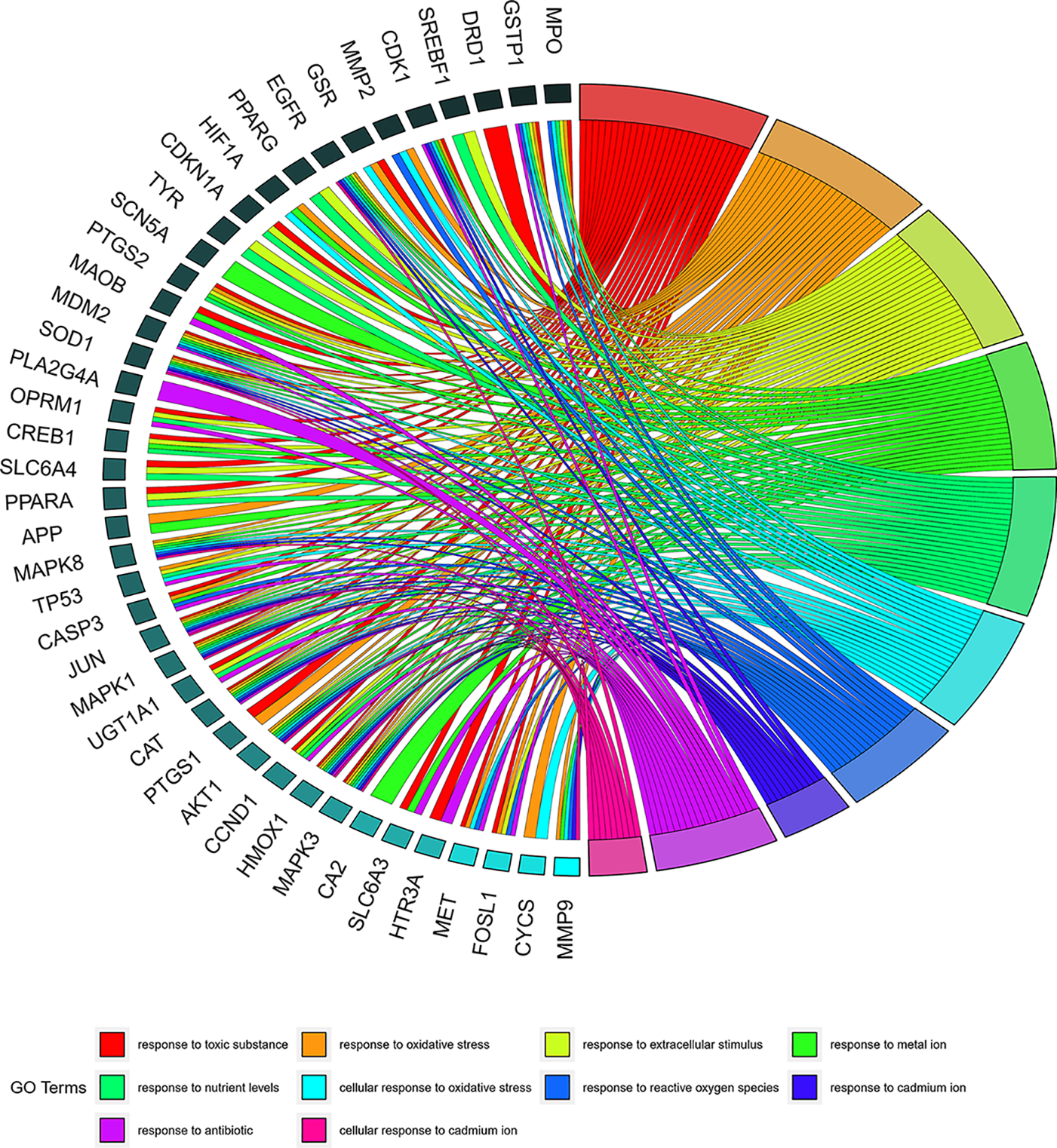
Figure 4: Chord plot of Gene Ontology (GO) biological process (BP). The GO terms represent the top 10 GO BP functional enrichments, and gene names with the connection represent their enriched genes.
The analytical findings recorded from the KEGG pathway enrichment illustrated a significant enrichment of the WDD active target genes acting on SD in proteoglycans in cancer, Kaposi sarcoma-associated herpesvirus infection, prostate cancer, endocrine resistance, and bladder cancer (Suppl. Table S7). Fig. 5 depicts the top 20 enriched KEGG pathways sorted by q-value. The top 10 KEGG pathway enrichments with their enriched genes are represented as a KEGG chord plot (Fig. 6).
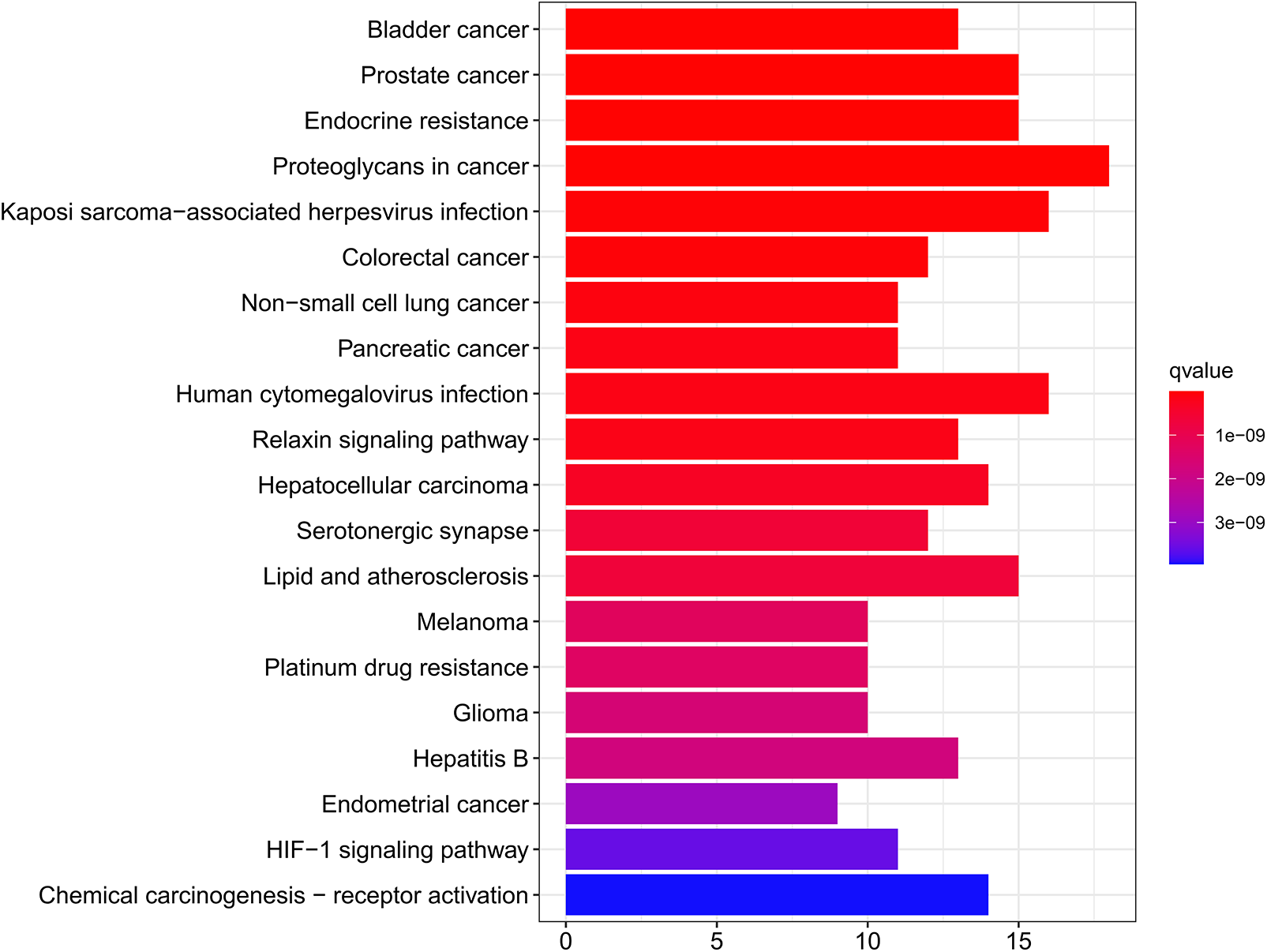
Figure 5: Analysis of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichments. The top 20 KEGG pathway enrichments. The smaller the q-value, the greater the significance of the enrichment.
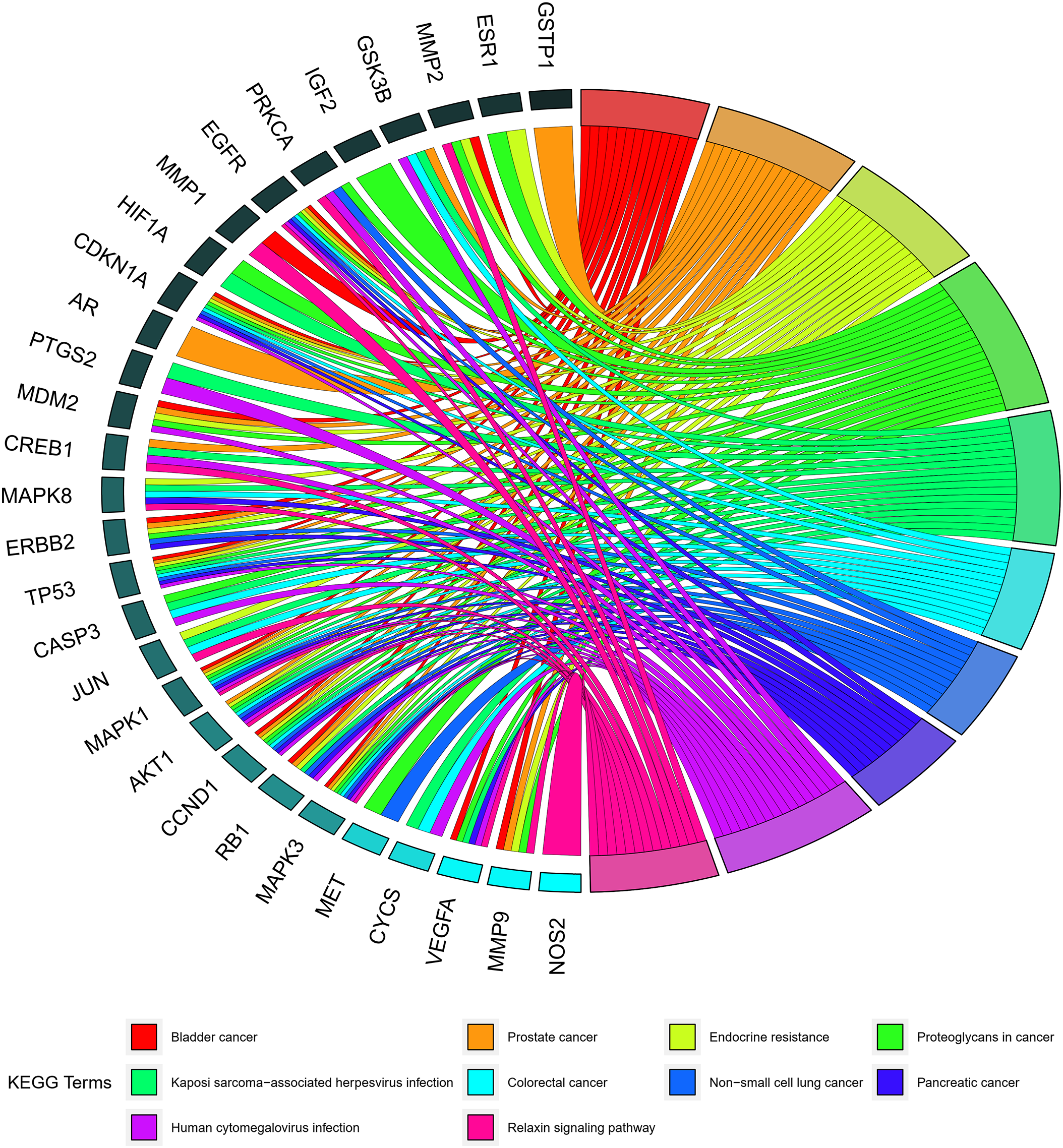
Figure 6: Chord plot of Kyoto Encyclopedia of Genes and Genomes (KEGG). The KEGG terms represent the top 10 KEGG pathway enrichments and gene names, with the connection representing their enriched genes.
Establishment of a compound–target network
We next employed NetworkAnalyzer to evaluate the network of compounds and targets constructed using the Cytoscape software. However, two compounds (MOL000282 and MOL000283) did not exhibit matching overlapped target genes; therefore, 70 intersecting active target genes were linked to 34 active compounds. The resulting network contained 198 edges and 104 nodes (34 compounds and 70 target genes) (Suppl. Table S8, Fig. 7).
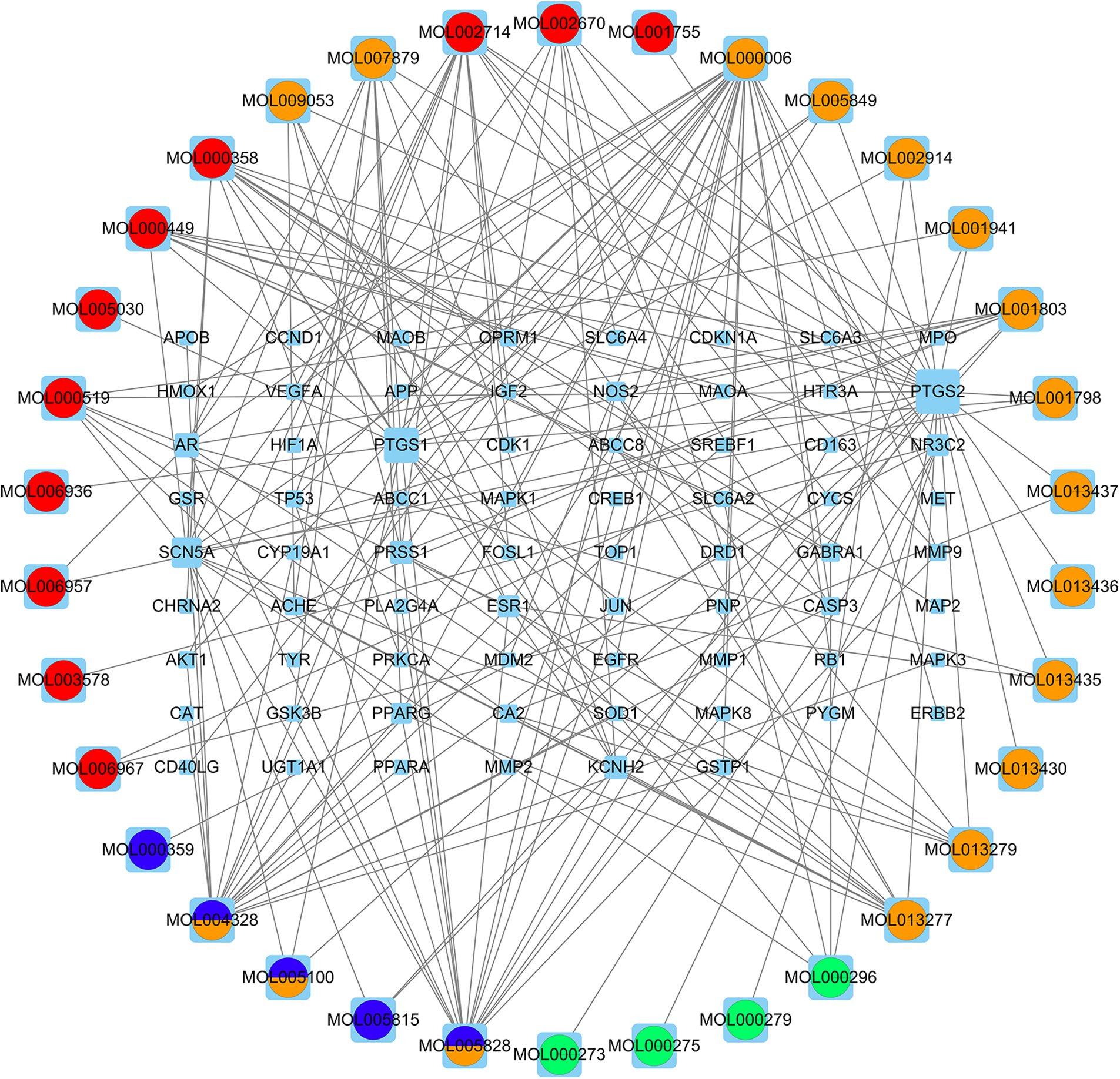
Figure 7: The compound-target network. The compound–target network containing 198 edges and 104 nodes (70 target genes and 34 compounds). Active compounds are shown by circles (different colors depict distinct compound types), the rectangles indicate active target genes, and the edges denote the links between the nodes. The increase in degree value is directly related to the extent of interconnections between the compounds and target genes.
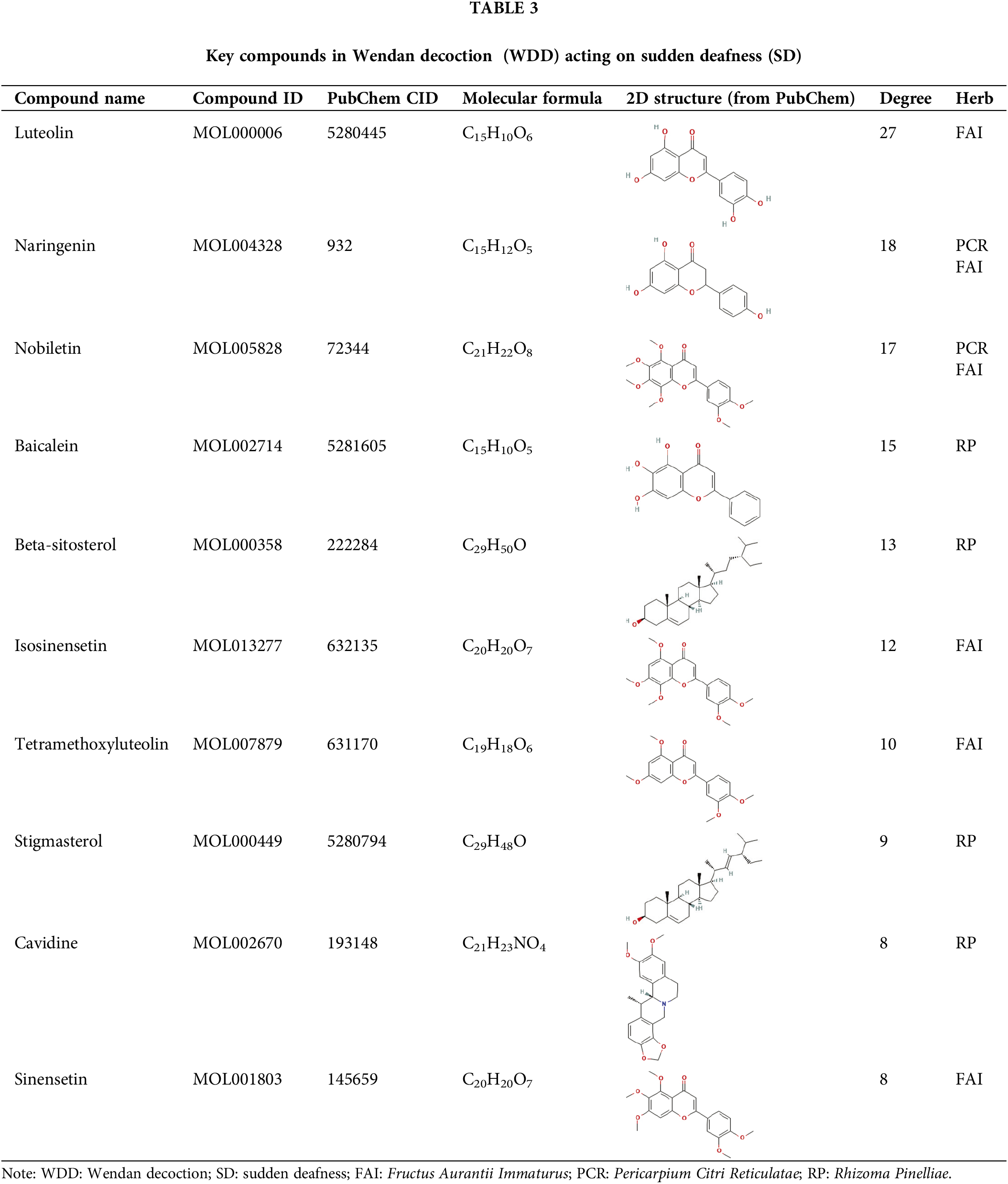
Using NetworkAnalyzer, the top 10 compounds in the network were identified based on their degree value within the network. These key compounds may be important for the action of WDD on SD. Table 3 presents the basic details of these compounds, including their two-dimensional structure derived from the PubChem database, sorted by the degree value.
Generation of the protein-protein interaction network
A PPI network was constructed by mapping the 70 matching target genes into the STRING database. When the minimum interaction value was adjusted to 0.40, the target proteins were observed to interact within the network, while 697 edges denoted the interconnections among the proteins (Suppl. Table S9, Fig. 8).
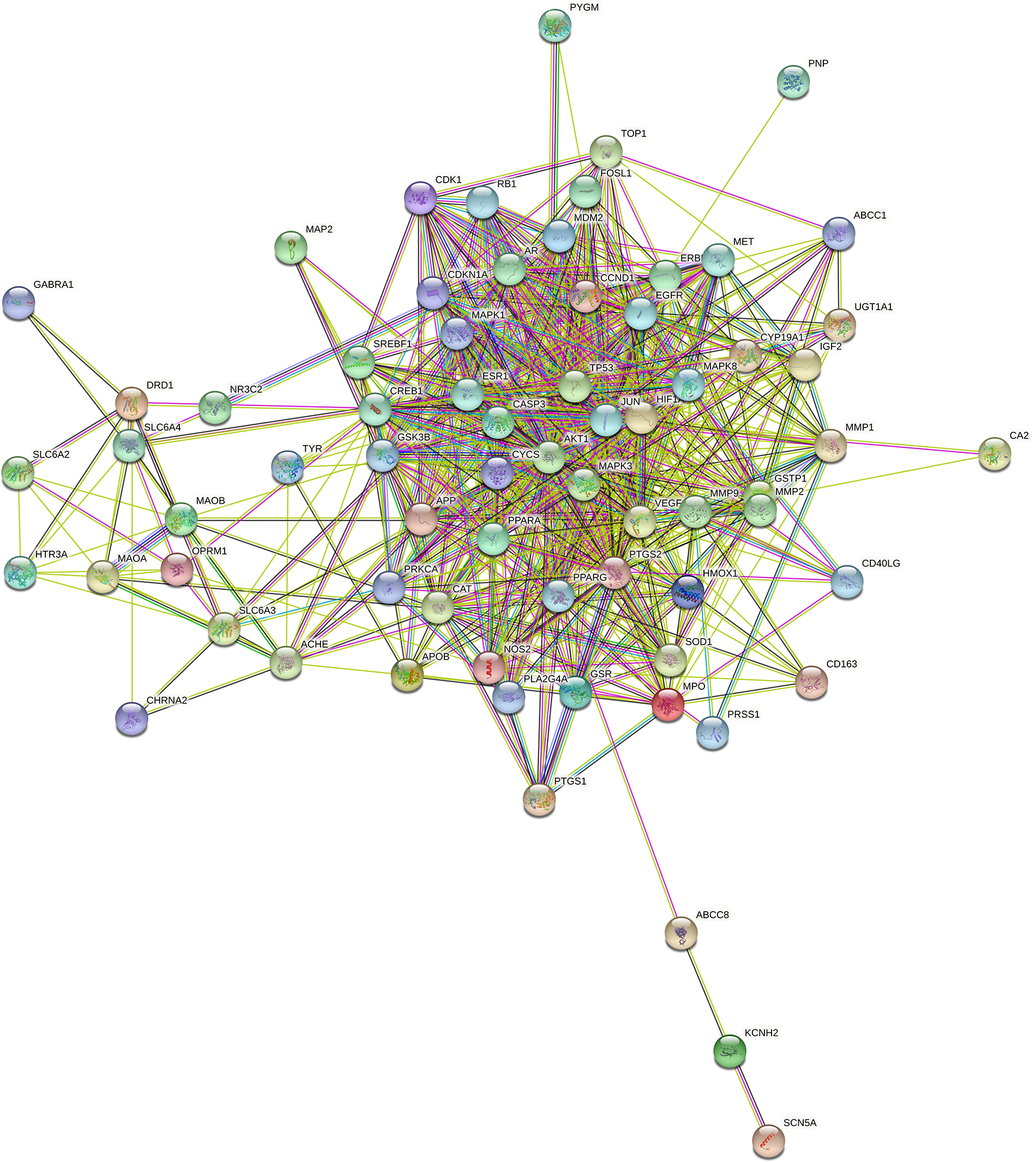
Figure 8: The protein-protein interaction (PPI) network. When the minimum interaction value was adjusted to 0.40, the network exhibited 70 interacting target proteins, resulting in 697 edges, which represents the interconnections among the proteins. The increase in the degree value is directly related to the extent of connections.
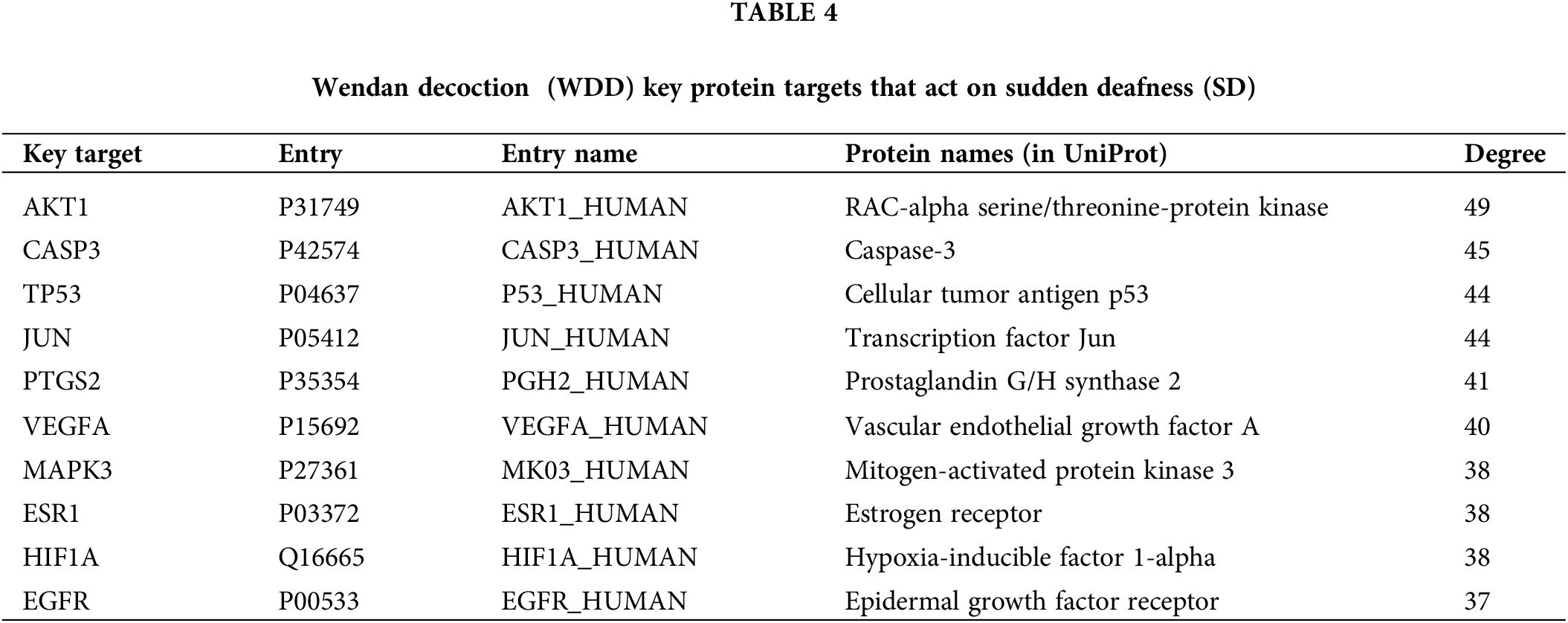
The top 10 target proteins sorted using Cytoscape software and the NetworkAnalyzer module based on their degree value in the PPI network are displayed in Table 4; these might be considered the key target proteins of WDD acting on SD.
Establishment of key compound-target network
The key compound-target network was established by uploading the key compounds (small-molecular compounds) and targets (macromolecular target protein receptors) along with their associations into the Cytoscape software, which was further used for molecular docking analysis (Suppl. Table S10, Fig. 9).
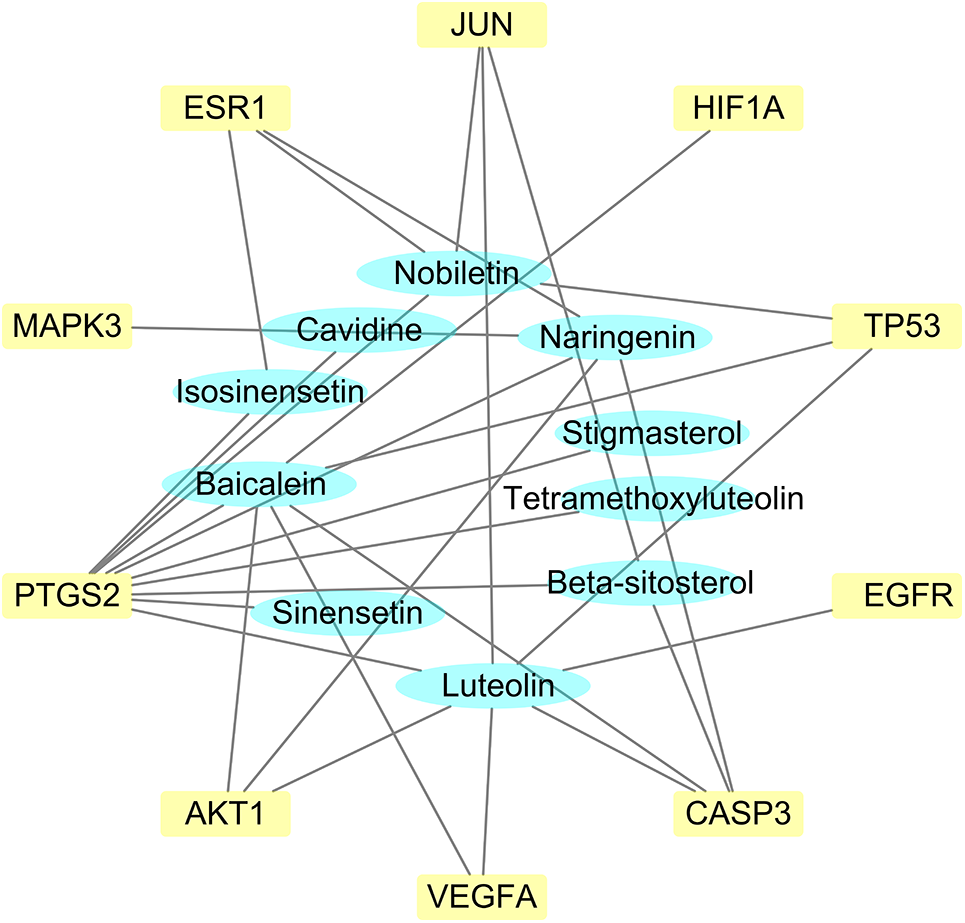
Figure 9: The key compound-target network. The network comprised 31 edges and 120 nodes (110 target genes and 10 compounds). The degree value increases directly to the extent of interconnections between the compounds and target genes.
The PubChem and RCSB PDB databases were employed to obtain the three-dimensional structures of the compounds and the target proteins, respectively. AutoDock Vina and AutoDockTool software performed docking simulations of the target proteins and their associated compounds. Using the molecular docking technique, we then investigated the interaction between the target proteins with their respective compounds. Finally, we selected the primary proteins and compounds as examples based on the key compound–target network.
The molecular docking simulations of prostaglandin-endoperoxide synthase 2 (PTGS2)-luteolin revealed grid center values of 22.594, 40.999, and 39.56, a minimum affinity of −9.8 kcal/mol, and a distance from the best mode of 0.000 rmsd u. b and 0.000 rmsd l. b. The results of molecular docking simulations of PTGS2-luteolin are displayed in Fig. 10.
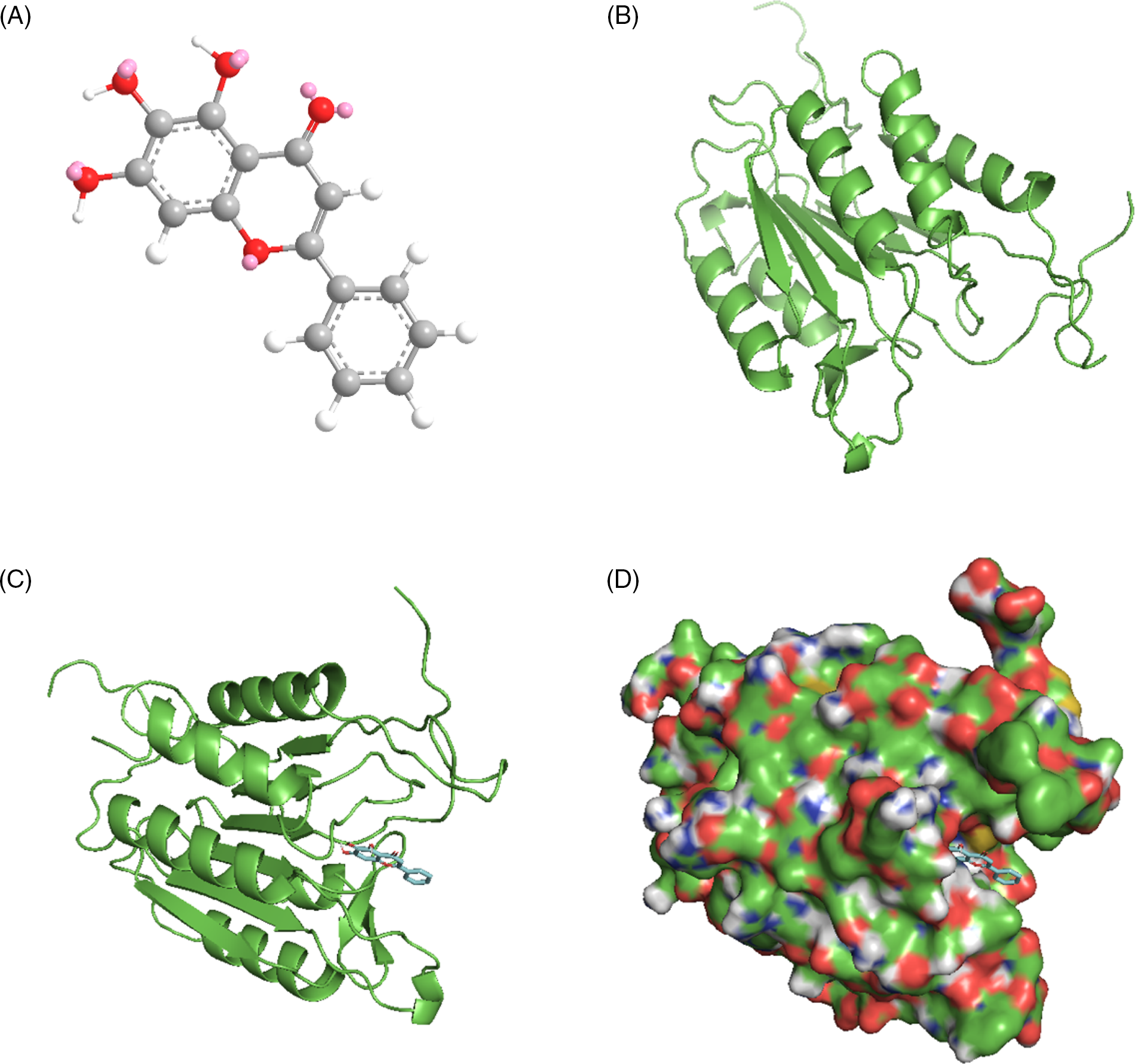
Figure 10: Molecular docking of prostaglandin-endoperoxide synthase 2 (PTGS2) and luteolin. (A) Three-dimensional structures of luteolin were obtained from the PubChem database. (B) Three-dimensional structures of PTGS2 were obtained from the Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB) database. (C) Molecular docking simulation. (D) The surface of the protein is shown in a molecular docking simulation.
The molecular docking simulations of caspase 3 (CASP3)-baicalein demonstrated grid center values of 27.82, 102.814, and 10.952, a minimum affinity of −8.0 kcal/mol, and a distance from the best mode of 0.000 rmsd u. b and 0.000 rmsd l. b. The results of molecular docking simulations of CASP3-baicalein are summarized in Fig. 11.
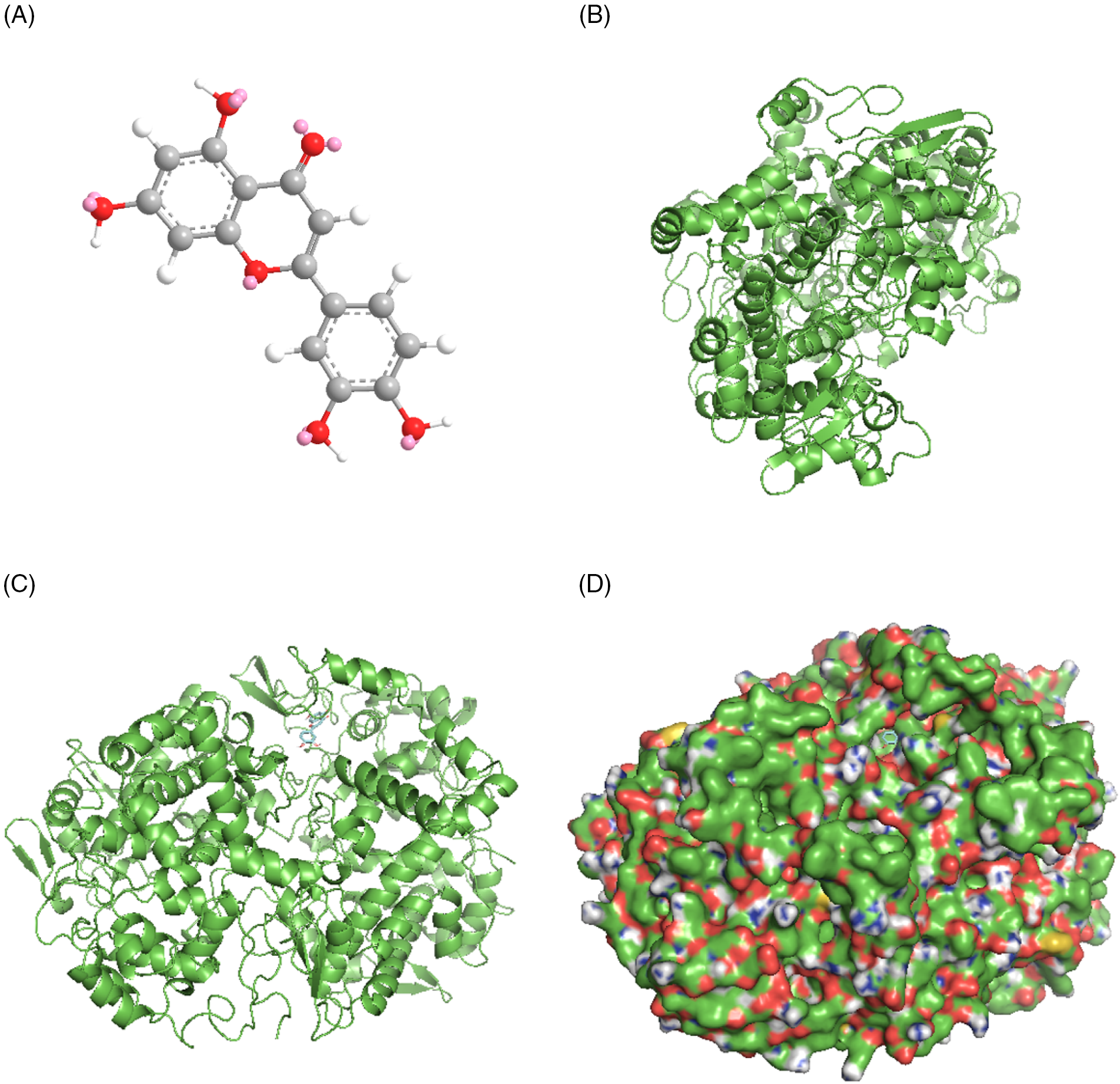
Figure 11: Molecular docking of caspase3 (CASP3) and baicalein. (A) Three-dimensional structures of baicalein from the PubChem database. (B) Three-dimensional structures of CASP3 from the Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB) database. (C) Molecular docking simulation. (D) The surface of the protein is shown in a molecular docking simulation.
The TCM theory suggests that the etiology of SD is associated with various factors, including the lack of qi and blood, kidney essence deficit, disturbance in the liver, phlegm fire, and blood stasis. Thus, TCM theory implies that treating patients with SD relies on modulating qi and dispelling phlegm (Wang et al., 2022; Zhu et al., 2022).
Among the components of WDD (RP, PCR, FAI, CBT, PA, and RG), RP is a monarch (Jun) herb considered the primary constituent of WDD, PCR, FAI, and CBT are minister (Chen) herbs, PA is an assistant (Zuo) herb, and RG is a guide (Shi) herb (Wang et al., 2021; Zhang et al., 2022a). Studies indicate that WDD can regulate qi and dispel phlegm and is extensively used in treating phlegm syndrome, strongly linked to digestive reflux disorders, dyslipidemia, cardiovascular disease, primary insomnia, and other diseases (Feng et al., 2019; Ling et al., 2015; Yan et al., 2017; Zhang et al., 2022b).
Clinical investigations have demonstrated that the use of WDD in treating SD can enhance clinical outcomes (Xia and Ji, 2021). However, a more comprehensive understanding of the pharmacological mechanisms underlying WDD’s therapeutic effect on SD is warranted. Network pharmacology is a useful, innovative tool in TCM research that could offer an method for transforming TCM from an experience-based medicine to an evidence-based medical field for systematic investigation of medicinal herbs and the discovery of pharmacodynamic compounds (Gu and Pei, 2017; Li and Zhang, 2013). Network pharmacology proposes a multi-component treatment strategy consistent with the multi-pathway, multi-target, and multi-component aspects of TCM (Huang et al., 2018; Luo et al., 2020). Thus, we adopted a network pharmacological method to examine the pharmacologic molecular mechanisms of WDD in the treatment of SD.
We could identify 70 active target genes of WDD that act on SD. Based on the functional enrichment of the GO biological process, the WDD active target genes that act on SD exhibited a significant enrichment in processes such as sensitivity to toxic substances, responsiveness to oxidative stress, reaction to extracellular stimuli, sensitivity to metal ions, and responsiveness to the levels of the nutrient. The functional enrichment of the GO cellular component illustrated a significant enrichment in the membrane region, caveola, membrane microdomain, vesicle lumen, and membrane raft, among other components. Moreover, functional enrichment of GO molecular function illustrated a significant enrichment in ubiquitin-protein ligase binding, tetrapyrrole binding, heme binding, phosphatase binding, protein phosphatase binding, and so on. These functions are strongly linked to the etiopathogenesis of SD, primarily characterized by inflammation and immunoregulation (Chen et al., 2019, 2021a; Kum et al., 2015; Li et al., 2021).
The KEGG pathway enrichment analysis revealed a strong association between the pathophysiology of SD and several pathways, with the predominant pathways being the relaxin signaling pathway, non-small cell lung cancer, human cytomegalovirus (HCMV) infection, pancreatic cancer, Kaposi sarcoma-associated herpesvirus infection, colorectal cancer, proteoglycans in cancer, endocrine resistance, prostate cancer, and bladder cancer. Several of these pathways have been shown to be intimately associated with SD, indicating multifaceted mechanisms of WDD in treating SD. For instance, the occurrence of SD is associated with cancer-related genes (Nam et al., 2011; Rakusic et al., 2015), the extracellular matrix performs an integral function in the adaptive responses to SD on multiple levels along the central auditory pathway (Heusinger et al., 2019), and HCMV infection has been associated with SD and acute labyrinthitis (Strauss, 1990). These findings suggest that WDD could effectively alleviate SD via various mechanisms involving multiple pathways and target genes. Future research should further investigate these pathways and target genes for a better understanding of the pharmacological mechanisms of WDD in treating SD.
In this study, a comprehensive network of compound-target interaction was established, and the compounds of WDD that act on SD were identified. Our results revealed the association of 70 active target genes with 34 active compounds, corresponding to different herbs in the WDD. Therefore, we hypothesized that every herb in the WDD has a function in responding to SD. Interestingly, active compounds in RP were found to perform a critical function in the compound-target network, consistent with the TCM theory of RP being the predominant active herb in WDD. The key compounds identified in the study were luteolin, naringenin, nobiletin, baicalein, beta-sitosterol, isosinensetin, tetramethoxyluteolin, stigmasterol, cavidine, and sinensetin. Luteolin has been shown to have clinical significance in the treatment of age-associated hearing loss caused by oxidative stress (Zhu et al., 2021), while baicalein may protect auditory function in noise-induced hearing loss (NIHL) (Kang et al., 2010). Thus, the impact of WDD on SD might be the consequence of the synergistic interactions among multiple compounds. However, further studies are needed to investigate the individual and combined effects of these compounds on SD. Nonetheless, only a few studies have been conducted on the effects of these compounds on SD, which means that further research is needed in this field.
The PPI network highlighted that the impact of WDD on SD was linked to several different targets, including AKT1, CASP3, TP53, JUN, PTGS2, VEGFA, MAPK3, ESR1, HIF1A, and EGFR. Studies have shown that the activities of the phosphoinositide-3 kinase/protein kinase B (PI3K/Akt) and mitogen-activated protein kinase (MAPK) signaling pathways are crucial in the onset and survival of auditory hair cells in sudden sensorineural hearing loss (Liu et al., 2021); Akt1, Akt2, and Akt3 are all implicated in the normal function of the ear, with Akt2 and Akt3 regulating the hair cell survival within the inner ear of mammals (Brand et al., 2015). Polymorphisms in CASP3 have been linked to the risk of developing NIHL (Wu et al., 2017) and mice lacking caspase-3 display hearing loss due to the loss of spiral ganglion and cochlear hair cells (Makishima et al., 2011). In the zebrafish lateral line, p53, Bcl2, and Bax modulate neomycin- and gentamicin-mediated hair cell apoptosis (Coffin et al., 2013). Hearing loss and auditory hair cell death caused by aminoglycosides and acoustic stress are prevented by a peptide antagonist of c-Jun N-terminal kinase (Wang et al., 2003). PTGS2 performs a function in the etiology of NIHL, and pharmacological suppression of PTGS2 has been shown to have therapeutic significance in the treatment of this condition (Sun et al., 2016). Vascular endothelial growth factor A signaling can generate new vessels in the ear (Wang et al., 2019). WBP2 codes for the WW domain-binding protein 2 (Wbp2), which functions as a transcriptional coactivator for estrogen receptor alpha (ESR1), and Wbp2 deficiency causes a gradual loss of high-frequency hearing in mice (Buniello et al., 2016). Hypoxia-inducible factor 1-alpha is involved in the protection against noise-induced inner-ear damage (Chung et al., 2011). Our results reveal an association between these genes and SD, paving the way for additional investigation into the pharmacologic processes and underlying mechanisms.
We developed a key compound–target network and performed molecular docking to investigate specific interconnections between key compounds and their target proteins, which may potentially increase the reliability of the network model. The results of molecular docking studies revealed that the key active compounds in WDD exhibited strong binding abilities with their respective target proteins. By targeting similar signaling pathways, these active compounds might be able to enhance the therapeutic role of WDD in SD. Future research may explore compounds associated with the matched target proteins.
In this study, we utilized a network pharmacological method to explore the impact of WDD on SD for its pharmacological mechanisms and examined the interaction of the target with the relevant compound using molecular docking. Nonetheless, the research still has some notable constraints. First, the screening parameters and definitions employed to identify active compounds and gene targets associated with WDD in the TCMSP were fixed. The genes associated with SD were examined using five databases. Even though these databases are rather comprehensive, certain compounds and target genes might have been missed. Additionally, relevant data for CBT are currently lacking in the TCMSP database. But our main purpose was to find the primary compounds and targets for preliminary research, as our results showed that the main compounds and main targets, which could provide the basis for further mechanistic studies. Second, the fact that WDD is a mixture makes it challenging for other researchers to replicate experiments. However, HPLC studies were performed to facilitate the quantification of compounds in WDD, and HPLC studies on key compounds can be carried out to further validate the findings (Wang et al., 2021; Zhang et al., 2019). We can also start by studying the key compounds first. Third, further empirical analysis of the candidate target genes and pathways is necessary although GO functional enrichment and KEGG pathway enrichment analyses were conducted, and a PPI network was established to examine the gene targets and pathways of WDD that act on SD. Finally, although exploratory docking analyses were performed in this study, additional experimental investigations of small-molecular compounds and macromolecular target proteins are required to validate these findings in the future.
In this study, a network pharmacological approach was employed to investigate the multi-compounds, targets, and pathway mechanisms underlying the therapeutic effect of WDD on SD. Furthermore, molecular docking analysis of key compounds and their matching protein targets indicated that these compounds could serve as major modulators of the therapeutic action of WDD on SD. Overall, our study provides valuable insights into the underlying mechanisms of WDD in treating SD and may contribute to the development of novel therapeutic strategies for this condition.
Acknowledgement: We express our gratitude to the developers who developed online database platforms and the programs utilized for data analysis.
Funding Statement: This research was partly funded by the Traditional Chinese Medicine Science and Technology Development Plan Project of Jiangsu Province (ZT202113 to Haibing Hua), the 510 Talent Training Project of Yizheng City (2022 to Shiming Ye), the Scientific Research Project of Jiangyin Association of Chinese Medicine (Y202205 to Yufeng Zhang), and the ChengXing Talent Training Plan of Jiangyin Hospital of Traditional Chinese Medicine (2022 to Yufeng Zhang).
Author Contributions: Shiming Ye, Yufeng Zhang, Zheng Yan, Haibing Hua, and Wandong She conceptualized and designed the study and drafted and edited the manuscript. Shiming Ye, Yufeng Zhang, Ting Liu, Cong Wang, and Haibing Hua collected and extracted data from various sources and performed data analysis. Zheng Yan, Haibing Hua, and Wandong She were in charge of project administration and supervision. The final version was reviewed and approved by all authors.
Availability of Data and Materials: All data generated or analyzed during this study are included in this published article (and its supplementary files).
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare they have no conflicts of interest to report regarding the present study.
Supplementary Materials: The supplementary materials are available online at DOI: https://doi.org/10.32604/biocell.2023.029333.
References
Amberger JS, Hamosh A (2017). Searching online mendelian inheritance in man (OMIMA knowledgebase of human genes and genetic phenotypes. Current Protocols in Bioinformatics 58: 1–2. https://doi.org/10.1002/cpbi.27 [Google Scholar] [PubMed] [CrossRef]
Assenov Y, Ramirez F, Schelhorn SE, Lengauer T, Albrecht M (2008). Computing topological parameters of biological networks. Bioinformatics 24: 282–284. https://doi.org/10.1093/bioinformatics/btm554 [Google Scholar] [PubMed] [CrossRef]
Brand Y, Levano S, Radojevic V, Naldi AM, Setz C, Ryan AF, Pak K, Hemmings BA, Bodmer D (2015). All Akt isoforms (Akt1, Akt2, Akt3) are involved in normal hearing, but only Akt2 and Akt3 are involved in auditory hair cell survival in the mammalian inner ear. PLoS One 10: e121599. https://doi.org/10.1371/journal.pone.0121599 [Google Scholar] [PubMed] [CrossRef]
Buniello A, Ingham NJ, Lewis MA, Huma AC, Martinez-Vega R et al. (2016). Wbp2 is required for normal glutamatergic synapses in the cochlea and is crucial for hearing. EMBO Molecular Medicine 8: 191–207. https://doi.org/10.15252/emmm.201505523 [Google Scholar] [PubMed] [CrossRef]
Castaneda R, Natarajan S, Jeong SY, Hong BN, Kang TH (2019). Traditional oriental medicine for sensorineural hearing loss: Can ethnopharmacology contribute to potential drug discovery? Journal of Ethnopharmacology 231: 409–428. https://doi.org/10.1016/j.jep.2018.11.016 [Google Scholar] [PubMed] [CrossRef]
Chandrasekhar SS, Tsai DB, Schwartz SR, Bontempo LJ, Faucett EA et al. (2019). Clinical practice guideline: Sudden hearing loss (Update). Otolaryngology-Head and Neck Surgery 161: S1–S45. https://doi.org/10.1177/0194599819859885 [Google Scholar] [PubMed] [CrossRef]
Che YW, Yao KY, Xi YP, Chen ZJ, Li YL, Yu N, Zhai SQ (2016). Wendan decoction (温胆汤) for treatment of schizophrenia: A systematic review of randomized controlled trials. Chinese Journal of Integrative Medicine 22: 302–310. https://doi.org/10.1007/s11655-015-2047-z [Google Scholar] [PubMed] [CrossRef]
Chen X, Fu YY, Zhang TY (2019). Role of viral infection in sudden hearing loss. Journal of International Medical Research 47: 2865–2872. https://doi.org/10.1177/0300060519847860 [Google Scholar] [PubMed] [CrossRef]
Chen Y, Sun J, Zhang Z, Liu X, Wang Q, Yu Y (2022). The potential effects and mechanisms of hispidulin in the treatment of diabetic retinopathy based on network pharmacology. BMC Complementary Medicine and Therapies 22: 141. https://doi.org/10.1186/s12906-022-03593-2 [Google Scholar] [PubMed] [CrossRef]
Chen X, Zhang Q, Yang C, Liu Y, Li L (2021a). GRβ regulates glucocorticoid resistance in sudden sensorineural hearing loss. Current Pharmaceutical Biotechnology 22: 1206–1215. https://doi.org/10.2174/1389201021666201008163534 [Google Scholar] [PubMed] [CrossRef]
Chen L, Zhu T, Qi J, Zhang Y, Zhang Z, Liu H (2021b). Pharmacological mechanism of JiaWeiSiWu granule in the treatment of hypertension based on network pharmacology. Annals of Palliative Medicine 10: 7486–7513. https://doi.org/10.21037/apm-21-1140 [Google Scholar] [PubMed] [CrossRef]
Chung JW, Shin JE, Han KW, Ahn JH, Kim YJ, Park JW, So HS (2011). Up-regulation of hypoxia-inducible factor-1 alpha by cobalt chloride prevents hearing loss in noise-exposed mice. Environmental Toxicology and Pharmacology 31: 153–159. https://doi.org/10.1016/j.etap.2010.10.002 [Google Scholar] [PubMed] [CrossRef]
Coffin AB, Rubel EW, Raible DW (2013). Bax, Bcl2, and p53 differentially regulate neomycin- and gentamicin-induced hair cell death in the zebrafish lateral line. Journal of the Association for Research in Otolaryngology 14: 645–659. https://doi.org/10.1007/s10162-013-0404-1 [Google Scholar] [PubMed] [CrossRef]
Eberhardt J, Santos-Martins D, Tillack AF, Forli S (2021). AutoDock vina 1.2.0: New docking methods, expanded force field, and python bindings. Journal of Chemical Information and Modeling 61: 3891–3898. https://doi.org/10.1021/acs.jcim.1c00203 [Google Scholar] [PubMed] [CrossRef]
Feng W, Ye X, Lv H, Hou C, Chen Y (2019). Wendan decoction for dyslipidemia: Protocol for a systematic review and meta-analysis. Medicine 98: e14159. https://doi.org/10.1097/MD.0000000000014159 [Google Scholar] [PubMed] [CrossRef]
Gao Y, Ji W, Lu M, Wang Z, Jia X, Wang D, Cao P, Hu C, Sun X, Wang Z (2022). Systemic pharmacological verification of Guizhi Fuling decoction in treating endometriosis-associated pain. Journal of Ethnopharmacology 297: 115540. https://doi.org/10.1016/j.jep.2022.115540 [Google Scholar] [PubMed] [CrossRef]
Gu S, Pei J (2017). Chinese herbal medicine meets biological networks of complex diseases: A computational perspective. Evidence-Based Complementary and Alternative Medicine 2017: 7198645. https://doi.org/10.1155/2017/7198645 [Google Scholar] [PubMed] [CrossRef]
Heusinger J, Hildebrandt H, Illing RB (2019). Sensory deafferentation modulates and redistributes neurocan in the rat auditory brainstem. Brain and Behavior 9: e1353. https://doi.org/10.1002/brb3.1353 [Google Scholar] [PubMed] [CrossRef]
Huang T, Zhong L, Lin CY, Zhao L, Ning ZW, Hu DD, Zhang M, Tian K, Cheng CW, Bian ZX (2018). Approaches in studying the pharmacology of Chinese medicine formulas: Bottom-up, top-down-and meeting in the middle. Chinese Medicine 13: 15. https://doi.org/10.1186/s13020-018-0170-4 [Google Scholar] [PubMed] [CrossRef]
Kang TH, Hong BN, Park C, Kim SY, Park R (2010). Effect of baicalein from Scutellaria baicalensis on prevention of noise-induced hearing loss. Neuroscience Letters 469: 298–302. https://doi.org/10.1016/j.neulet.2009.12.009 [Google Scholar] [PubMed] [CrossRef]
Kim S, Chen J, Cheng T, Gindulyte A, He J et al. (2021). PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Research 49: D1388–D1395. https://doi.org/10.1093/nar/gkaa971 [Google Scholar] [PubMed] [CrossRef]
Kordis S, Vozel D, Hribar M, Urbancic NB, Battelino S (2020). The outcome of prompt concomitant Single-Dose High-Concentration intratympanic and tapered Low-Dose oral systemic corticosteroid treatment for sudden deafness. The Journal of International Advanced Otology 16: 201–206. https://doi.org/10.5152/iao.2020.8341 [Google Scholar] [PubMed] [CrossRef]
Kouranov A, Xie L, de la Cruz J, Chen L, Westbrook J, Bourne PE, Berman HM (2006). The RCSB PDB information portal for structural genomics. Nucleic Acids Research 34: D302–D305. https://doi.org/10.1093/nar/gkj120 [Google Scholar] [PubMed] [CrossRef]
Kum RO, Ozcan M, Baklaci D, Yurtsever KN, Yilmaz YF, Unal A, Avci Y (2015). Investigation of neutrophil-to-lymphocyte ratio and mean platelet volume in sudden hearing loss. Brazilian Journal of Otorhinolaryngology 81: 636–641. https://doi.org/10.1016/j.bjorl.2015.08.009 [Google Scholar] [PubMed] [CrossRef]
Li X, Chen W, Yi H, Xu J, Lu M, Guo F, Li X, Ye J (2021). The association between the levels of autoimmune related indicators and clinical condition in sudden deafness patients. Annals of Palliative Medicine 10: 10535–10541. https://doi.org/10.21037/apm-21-2388 [Google Scholar] [PubMed] [CrossRef]
Li S, Zhang B (2013). Traditional Chinese medicine network pharmacology: Theory, methodology and application. Chinese Journal of Natural Medicines 11: 110–120. https://doi.org/10.1016/S1875-5364(13)60037-0 [Google Scholar] [PubMed] [CrossRef]
Ling W, Huang Y, Xu JH, Li Y, Huang YM, Ling HB, Sui Y, Zhao HL (2015). Consistent efficacy of wendan decoction for the treatment of digestive reflux disorders. The American Journal of Chinese Medicine 43: 893–913. https://doi.org/10.1142/S0192415X15500524 [Google Scholar] [PubMed] [CrossRef]
Liu Y, Wei M, Mao X, Chen T, Lin P, Wang W (2021). Key signaling pathways regulate the development and survival of auditory hair cells. Neural Plasticity 2021: 5522717. https://doi.org/10.1155/2021/5522717 [Google Scholar] [PubMed] [CrossRef]
Luo TT, Lu Y, Yan SK, Xiao X, Rong XL, Guo J (2020). Network pharmacology in research of Chinese medicine formula: Methodology, application and prospective. Chinese Journal of Integrative Medicine 26: 72–80. https://doi.org/10.1007/s11655-019-3064-0 [Google Scholar] [PubMed] [CrossRef]
Makishima T, Hochman L, Armstrong P, Rosenberger E, Ridley R, Woo M, Perachio A, Wood S (2011). Inner ear dysfunction in caspase-3 deficient mice. BMC Neuroscience 12: 102. https://doi.org/10.1186/1471-2202-12-102 [Google Scholar] [PubMed] [CrossRef]
Meng X, Wang J, Sun J, Zhu K (2022). COVID-19 and sudden sensorineural hearing loss: A systematic review. Frontiers in Neurology 13: 883749. https://doi.org/10.3389/fneur.2022.883749 [Google Scholar] [PubMed] [CrossRef]
Nam SI, Yu GI, Kim HJ, Park KO, Chung JH, Ha E, Shin DH (2011). A polymorphism at -1607 2G in the matrix metalloproteinase-1 (MMP-1) increased risk of sudden deafness in Korean population but not at -519A/G in MMP-1. The Laryngoscope 121: 171–175. https://doi.org/10.1002/lary.21334 [Google Scholar] [PubMed] [CrossRef]
Rakusic Z, Misir KA, Stupin PD, Jakovcevic A, Bisof V (2015). Sudden bilateral hearing loss in gastric cancer as the only symptom of disease. OncoTargets and Therapy 8: 1285–1289. https://doi.org/10.2147/OTT [Google Scholar] [CrossRef]
Ru J, Li P, Wang J, Zhou W, Li B et al. (2014). TCMSP: A database of systems pharmacology for drug discovery from herbal medicines. Journal of Cheminformatics 6: 13. https://doi.org/10.1186/1758-2946-6-13 [Google Scholar] [PubMed] [CrossRef]
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Research 13: 2498–2504. https://doi.org/10.1101/gr.1239303 [Google Scholar] [PubMed] [CrossRef]
Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M et al. (2016). The GeneCards suite: From gene data mining to disease genome sequence analyses. Current Protocols in Bioinformatics 54: 1–30. https://doi.org/10.1002/cpbi.5 [Google Scholar] [PubMed] [CrossRef]
Strauss M (1990). Human cytomegalovirus labyrinthitis. American Journal of Otolaryngology 11: 292–298. https://doi.org/10.1016/0196-0709(90)90057-3 [Google Scholar] [PubMed] [CrossRef]
Sun Y, Yu J, Lin X, Tang W (2016). Inhibition of cyclooxygenase-2 by NS398 attenuates noise-induced hearing loss in mice. Scientific Reports 6: 22573. https://doi.org/10.1038/srep22573 [Google Scholar] [PubMed] [CrossRef]
Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S et al. (2019). STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Research 47: D607–D613. https://doi.org/10.1093/nar/gky1131 [Google Scholar] [PubMed] [CrossRef]
Tai SY, Shen CT, Wang LF, Chien CY (2021). Association of sudden sensorineural hearing loss with dementia: A nationwide cohort study. BMC Neurology 21: 88. https://doi.org/10.1186/s12883-021-02106-x [Google Scholar] [PubMed] [CrossRef]
UniProt CT (2018). UniProt: The universal protein knowledgebase. Nucleic Acids Research 46: 2699. https://doi.org/10.1093/nar/gky092 [Google Scholar] [PubMed] [CrossRef]
Walter W, Sanchez-Cabo F, Ricote M (2015). GOplot: An R package for visually combining expression data with functional analysis. Bioinformatics 31: 2912–2914. https://doi.org/10.1093/bioinformatics/btv300 [Google Scholar] [PubMed] [CrossRef]
Wang J, van de Water TR, Bonny C, de Ribaupierre F, Puel JL, Zine A (2003). A peptide inhibitor of c-Jun N-terminal kinase protects against both aminoglycoside and acoustic trauma-induced auditory hair cell death and hearing loss. The Journal of Neuroscience 23: 8596–8607. https://doi.org/10.1523/JNEUROSCI.23-24-08596.2003 [Google Scholar] [PubMed] [CrossRef]
Wang Y, Wang J, Chen X (2022). Clinical efficacy of Xueshuantong plus urokinase in the treatment of sudden deafness. Evidence-Based Complementary and Alternative Medicine 2022: 7775556. https://doi.org/10.1155/2022/7775556 [Google Scholar] [PubMed] [CrossRef]
Wang X, Zhang J, Li G, Sai N, Han J, Hou Z, Kachelmeier A, Shi X (2019). Vascular regeneration in adult mouse cochlea stimulated by VEGF-A165 and driven by NG2-derived cells ex vivo. Hearing Research 377: 179–188. https://doi.org/10.1016/j.heares.2019.03.010 [Google Scholar] [PubMed] [CrossRef]
Wang Q, Zou Z, Zhang Y, Lin P, Lan T, Qin Z, Xu D, Wu H, Yao Z (2021). Characterization of chemical profile and quantification of major representative components of Wendan decoction, a classical traditional Chinese medicine formula. Journal of Separation Science 44: 1036–1061. https://doi.org/10.1002/jssc.202000952 [Google Scholar] [PubMed] [CrossRef]
Whirl-Carrillo M, Huddart R, Gong L, Sangkuhl K, Thorn CF, Whaley R, Klein TE (2021). An evidence-based framework for evaluating pharmacogenomics knowledge for personalized medicine. Clinical Pharmacology & Therapeutics 110: 563–572. https://doi.org/10.1002/cpt.2350 [Google Scholar] [PubMed] [CrossRef]
Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A et al. (2018). DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Research 46: D1074–D1082. https://doi.org/10.1093/nar/gkx1037 [Google Scholar] [PubMed] [CrossRef]
Wu T, Hu E, Xu S, Chen M, Guo P et al. (2021). ClusterProfiler 4.0: A universal enrichment tool for interpreting omics data. The Innovation 2: 100141. https://doi.org/10.1016/j.xinn.2021.100141 [Google Scholar] [PubMed] [CrossRef]
Wu Y, Ni J, Qi M, Cao C, Shao Y, Xu L, Ma H, Yang L (2017). Associations of genetic variation in CASP3 gene with noise-induced hearing loss in a Chinese population: A case-control study. Environmental Health 16: 78. https://doi.org/10.1186/s12940-017-0280-y [Google Scholar] [PubMed] [CrossRef]
Xia JJ, Ji ZN (2021). Clinical effect on Wendan decoction in the treatment of phlegm fire syndrome of sudden deafness with low frequency decrease. Guangming Journal of Chinese Medicine 36: 214–216. https://doi.org/10.3969/j.issn.1003-8914.2021.02.020 [Google Scholar] [CrossRef]
Xia Q, Liu M, Li H, Tian L, Qi J, Zhang Y (2020). Network pharmacology strategy to investigate the pharmacological mechanism of HuangQiXiXin decoction on cough variant asthma and evidence-based medicine approach validation. Evidence-Based Complementary and Alternative Medicine 2020: 3829092. https://doi.org/10.1155/2020/3829092 [Google Scholar] [PubMed] [CrossRef]
Xing G, Zhang Y, Wu X, Wang H, Liu Y, Zhang Z, Hou M, Hua H (2021). Analysis of the efficacy and pharmacological mechanisms of action of Zhenren Yangzang decoction on ulcerative colitis using meta-analysis and network pharmacology. Evidence-Based Complementary and Alternative Medicine 2021: 4512755. https://doi.org/10.1155/2021/4512755 [Google Scholar] [PubMed] [CrossRef]
Yan X, Wang Y, Li X, Li Z, Zhang Y, Cai X, Wang D (2017). Wendan decoction for primary insomnia: Protocol for a systematic review and meta-analysis. Medicine 96: e8906. https://doi.org/10.1097/MD.0000000000008906 [Google Scholar] [PubMed] [CrossRef]
Yoon CY, Kong TH, Lee J, Seo YJ, Ahn JJ (2022). Epidemiology of idiopathic sudden sensorineural hearing loss in the era of big data. European Archives of Oto-Rhino-Laryngology 280: 2181–2190. https://doi.org/10.1007/s00405-022-07693-0 [Google Scholar] [PubMed] [CrossRef]
Zhang Y, Liu T, Zhang L, Pu Z, Yan Z, Hua H (2022a). Wendan decoction in the treatment of nonalcoholic fatty liver disease: A systematic review and meta-analysis. Frontiers in Pharmacology 13: 1039611. https://doi.org/10.3389/fphar.2022.1039611 [Google Scholar] [PubMed] [CrossRef]
Zhang B, Qi D, Deng X, Ma Z, Wu Y et al. (2019). Quantification of nineteen bioactive components in the ancient classical Chinese medicine formula of Wen-Dan decoction and its commercial preparations by UHPLC-QQQ-MS/MS. Molecules 24: 2031. https://doi.org/10.3390/molecules24112031 [Google Scholar] [PubMed] [CrossRef]
Zhang X, Wang Y, Liu L, Jiang H, Wang J, Xiao Y, Wang J (2022b). Efficacy of Wen-Dan Decoction in the treatment of patients with coronary heart disease: A protocol for systematic review and meta-analysis. Medicine 101: e28041. https://doi.org/10.1097/MD.0000000000028041 [Google Scholar] [PubMed] [CrossRef]
Zhou Y, Zhang Y, Lian X, Li F, Wang C, Zhu F, Qiu Y, Chen Y (2022). Therapeutic target database update 2022: Facilitating drug discovery with enriched comparative data of targeted agents. Nucleic Acids Research 50: D1398–D1407. https://doi.org/10.1093/nar/gkab953 [Google Scholar] [PubMed] [CrossRef]
Zhu RZ, Li BS, Gao SS, Seo JH, Choi BM (2021). Luteolin inhibits H2O2-induced cellular senescence via modulation of SIRT1 and p53. The Korean Journal of Physiology & Pharmacology 25: 297–305. https://doi.org/10.4196/kjpp.2021.25.4.297 [Google Scholar] [PubMed] [CrossRef]
Zhu Z, Wu Q, Hu G, Wang X, Chang W, Bin J, Yang W (2022). Efficacy of ginkgo biloba extract combined with hormones in the treatment of sudden deafness and its effect on the reactivity of peripheral blood T cell subsets. Computational and Mathematical Methods in Medicine 2022: 2903808. https://doi.org/10.1155/2022/2903808 [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools