 Open Access
Open Access
ARTICLE
Changes in intracellular and extracellular proteins after ERGIC3 knockdown in lung cancer: Proteins interacting with ERGIC3, HORN, and FLNA
1 Special Key Laboratory of Oral Disease Research and High Education Institute in Guizhou Province, School of Stomatology, Zunyi Medical University, Zunyi, 563006, China
2 Department of Genetics, Zunyi Medical University, Zunyi, 563006, China
3 Department of Pathology, Affiliated Hospital of Zunyi Medical University, Zunyi, 563008, China
* Corresponding Authors: XUEYING LI. Email: ; MINGSONG WU. Email:
(This article belongs to the Special Issue: Extracellular Vesicles and Cancer)
BIOCELL 2023, 47(8), 1821-1833. https://doi.org/10.32604/biocell.2023.027175
Received 17 October 2022; Accepted 20 March 2023; Issue published 28 August 2023
Abstract
Objective: Endoplasmic reticulum-Golgi intermediate compartment 3 (ERGIC3) promotes cell proliferation and metastasis in lung cancer, but its molecular mechanism is unclear. Methods: The GLC-82 cells were randomly divided into the ERGIC3i group and the negative control group. The cells were transfected with ERGIC3 siRNA or control siRNA in the groups, respectively. The ERGIC3-interacting proteins expressed in cells or extracellularly were isolated by the immunoprecipitation method and detected by isobaric tags for relative and absolute quantitation and liquid chromatography-tandem mass spectrometry. The differentially expressed proteins were determined by bioinformatic methods. Results: After ERGIC3 knockdown, 88 extracellular differentially expressed proteins, 41 up-regulated and 47 down-regulated, were detected in the supernatant of cultured cells. Among 52 intracellular differentially expressed proteins, 33 were up-regulated and 19 down-regulated. Cluster analysis showed that the extracellular differential proteins are mainly involved in Ca2+ binding and transport and I-kappa B kinase/NF-kappa B signal transduction. The upregulated proteins are mainly involved in the biological process of H3-K27 and H3-K4 methylation in cells. Co-immunoprecipitation assay showed that proteins interacting with ERGIC3 were rich in cytoskeleton construction and RHO GTPases activated p21-activated kinases. The intersection of these two research methods shows that ERGIC3 interacts with HORN and filamin A (FLNA). Conclusion: Proteomic analyses reveal that ERGIC3 acts as a vesicle transmembrane protein on the distribution of various extracellular and intracellular proteins and regulates the extracellular and intracellular biological processes by specifically binding hornin (HORN) and FLNA proteins. These findings maybe provide new methods and ideas for ERGIC3 as a therapeutic target for lung cancer.Keywords
Lung cancer is a malignant tumor originating from the trachea, bronchial mucosa, and glands. The incidence and mortality of lung cancer are extremely high and on the rise around the world. Lung adenocarcinoma accounts for about 40% of lung malignancies (Chen et al., 2014). Although a diagnosis of lung cancer might not be the definitive death sentence it used to be, it is still very bad news: only about 20% of people survive for five years with the disease (Brody, 2020; Herbst et al., 2018). Targeted therapy has changed the treatment landscape for non-small cell lung cancer (NSCLC) (Imyanitov et al., 2021; Shokoohi et al., 2022). But these checkpoints alone are insufficient. Therefore, identifying novel predictive biomarkers has become key to detecting and treating this deadly cancer.
Endoplasmic reticulum-Golgi intermediate compartment 3 (ERGIC3), also known as Erv46 and ERp43, an ERGIC membrane protein family member, is a type II transmembrane protein located in the endoplasmic reticulum, and Golgi body and ERGIC (Otte and Barlowe, 2002). It is involved in forward vesicle transport between the ER and Golgi bodies (Liu et al., 2022; Yoo et al., 2019). ERGIC3 plays an important role in the tumorigenesis and development of lung cancer and is a recently identified marker for NSCLC (Wu et al., 2013).
ERGIC3 is also abnormally expressed in liver cancer, breast cancer, and other tumors. However, the specific biological function of ERGIC3 in lung cancer remains yet to be elucidated. In previous studies, ERGIC3 was found to regulate tumor development as a direct binding site for multiple miRNAs and proteins. In hepatic cell carcinoma (HCC), as a direct target of miRNA-490-3p, up-regulation of ERGIC3 not only enhances the viability and colony-forming ability of HCC cells but also facilitates the cell migration and invasion ability (Zhang et al., 2013). Sharma et al. (2017) found that the palmitoylation of ERGIC3 protein and DHHC3 is one of the key mechanisms through which DHHC3 promotes breast tumor growth. DHHC3 ablation significantly reduces ERGIC3 palmitoylation, facilitates endoplasmic reticulum (ER) stress, and alters ERGIC3 subcellular distribution (Sharma et al., 2017). Moreover, ERGEC3 is not expressed in normal lung tissues, but it is overexpressed in lung cancer tissues (Nishikawa et al., 2007; Zhao et al., 2020), which results in tumor cell proliferation, high vitality, and anti-apoptosis. Cell growth is inhibited after the knockdown of ERGEC3 expression; for instance, silencing ERGIC3 leads to growth inhibition and persistent autophagy in A549 cells. However, the specific mechanism of ERGIC3, especially the proteins that are involved in the biological function of ERGIC3, needs more research.
Quantitative proteomics has been widely used in oncology to identify biomarkers with diagnostic and therapeutic potential (Wang et al., 2018; Wang et al., 2022). Isobaric tags for relative and absolute quantitation (iTRAQ), a new protein quantitation technique, has the advantages of a simpler labeling process, higher sensitivity, and a wider application range compared with traditional quantitative methods. In the study, lung cancer cells were selected to explore the intracellular and extracellular differentially expressed proteins after ERGIC3 knockdown to study the specific mechanism of ERGIC3 and its influence of development in lung cancer at a large scale through iTRAQ-coupled with liquid chromatography-tandem mass spectrometry (LC-MS/MS) comparative proteomic analysis.
GLC-82 cell line was a gift from Dr. Cao Yi at Kunming Institute of Zoology, Chinese Academy of Sciences. GLC-82 cells were maintained in Roswell Park Memorial Institute-1640 medium (Gibco, California, USA) containing 1% penicillin/streptomycin and incubated at 37°C in a humidified incubator with 5% CO2.
Approximately 7.0 × 105 GLC-82 were placed in each well of a 6-well plate and incubated overnight at 37°C in the presence of 5% CO2. The cells were divided into two groups: the ERGIC3i group and the negative control (NC) group. Meanwhile, in these two groups, supernatant and cell mass were collected separately, and finally, four groups of samples were obtained (each group contained two biological replicates): the intracellular (RNAi_intra) and extracellular (RNAi_sec) proteins in the ERGIC3i group, and the intracellular (NC_intra) and extracellular (NC_sec) of the NC group. Proteins in the NC group were analyzed to predict the effects of ERGIC3 on intracellular and extracellular protein levels and cell physiological function (Fig. 1).
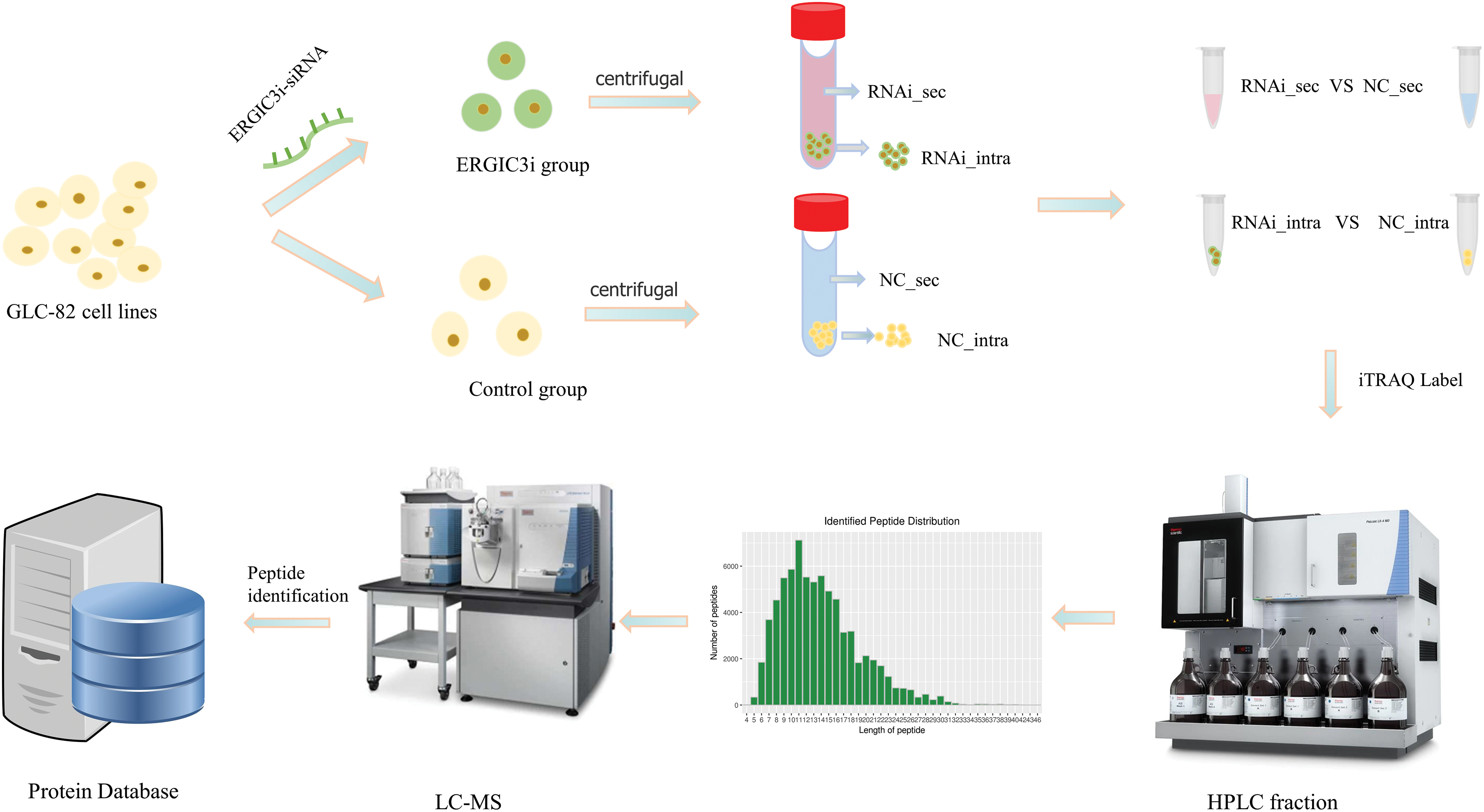
Figure 1: The experimental grouping and process.
Transfection of endoplasmic reticulum-Golgi intermediate compartment 3 siRNA
For the ERGIC3i group, 70%–80% of fused cells were transfected with ERGIC3 siRNA (5´-CCUGUUCAAGCAA CGACUATT-3´) in the ERGIC3i group or control siRNA in the NC group (Thermo Fisher Scientific, Massachusetts, USA) using lipofectamine 3000 reagent (Invitrogen, California, USA). In short, 8 μL of lipofectamine 3000 was diluted in 192 μL OPti-MEM medium and mixed, and 8 μL ERGIC3 siRNA (20 μm) was diluted in 192 μL OPti-MEM medium and mixed. Diluted Lipofectamine 3000 and ERGIC3 siRNA were mixed, and 400 μL mixture was added to 1600 μL of the medium. Cells were harvested for protein extraction after 48 h of siRNA treatment.
Total RNA extraction and reverse transcription‑quantitative polymerase chain reaction (RT‑qPCR) analysis
Total RNA from cells was extracted using RNAisoTM Plus (TaKaRa Biotechnology, Beijing, China) reagent and reverse transcribed into cDNA. Total RNA concentration was measured using a Nanodrop 2000 Ultra-Micro spectrophotometer (Thermo Fisher Scientific, Massachusetts, USA). The cDNA synthesis was performed according to provided specifications and used at a 1:10 dilution for qPCR. Quantitative real-time polymerase chain reaction (qRT-PCR) was performed with CFX96 TouchTM fluorescent quantitative PCR instrument (Bio-Rad, California, USA). The program was set as follows: 40 cycles of 94°C for 60 s, 95°C for 20 s, and 60°C for 30 s. The primers for qPCR were as follows: GAPDH, forward: 5'-GGCCTCCAAGGAGTAAGACC-3', reverse: 5'-AGGGGAGATTCAGTGTGGTG-3'; ERGIC3, forward: 5'-GGAGAGGTACTGAGGACAAATCA-3', reverse: 5'-AGCTCATAGAGGACGAAGACTC-3'. The mRNA levels were quantified using the 2−ΔΔCt method and normalized to the internal reference gene GAPDH. qPCR experiments were performed in triplicate.
Cells were collected and cleaved on ice for 10 min using RIPA cleavage buffer (Solarbio Lifesciences, Beijing, China) and protease inhibitor (BBI, Shanghai, China). Total protein was obtained after centrifugation, and the protein concentration was determined by the BCA method. The protein was denatured by boiling and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) electrophoresis. The primary antibody, GAPDH (ProteinTech Group, Wuhan, China), 1:10000 dilution, ERGIC3 (Abcam, Cambridge, UK), 1:1000 dilution, was incubated at 4°C overnight, while the secondary antibody was incubated at 37°C for 2 h. ECL illuminant was added and recorded by a machine (Bio-Rad, California, USA).
Isobaric tags for relative and absolute quantitation
Protein extraction and quantification
The proteins of four groups of experimental samples were extracted and quantified. Briefly, lysis Buffer (including 8M urea, 2 mM ethylenediaminetetraacetate, 10 mM dithiothreitol, 1% protease phenylmethyl sulfonyl fluoride (Beyotime, Shanghai, China)) was added to lysis cell samples. The supernatants (12,000 rpm, centrifugation at 4°C for 5 min) were again added with 3 to 5 times the volume of pre-cooled acetone and precipitated at −20°C for more than 3 h. The acetone was discarded after centrifugation (16000 g, 4°C, 5 min). Protein precipitates were dissolved in an appropriate amount of resolution buffer (including 8M urea, 100 mM triethylammonium bicarbonate buffer, pH 8.0). Bradford (Beyotime, Shanghai, China) method was used to determine the protein concentration.
Protein digestion and isobaric tags for relative and absolute quantitation labeling
The four groups of protein samples were quantitatively analyzed by Monitor Helix Company (http://www.mhelix.cn/index.html, Shanghai, China). The procedure used for the iTRAQ analyses was as follows.
First, 100 μg samples were taken respectively, and 8M urea buffer (Sigma, Saint Louis, USA) was used to adjust the equal volume. The final concentration of 10 mM dithiothreitol (Sigma, Saint Louis, USA) was added and incubated at 37°C for 45 min to reduce the disulfide bonds in the proteins. Then 25 mM iodoacetamide (IAM, Sigma, Saint Louis, USA) was added and incubated at room temperature for 55 min. Alkylation was selected to protect the sulfhydryl groups.
Enzymolysis was performed after dilution with 100 mM triethylammonium bicarbonate buffer (Thermo Fisher Scientific, Massachusetts, USA) to urea concentration below 2M. According to the mass ratio of 1:50, trypsin was added, and the process of enzymolysis was performed overnight at 37°C. Subsequently, trypsin was added at 1:100 and the enzymolysis was repeated at 37°C for 4 h. The NC_intra group was labeled 113 and 115; the NC_sec group was labeled 116 and 117. Likewise, the RNAi_intra group was labeled 117, 121, and the RNAi_sec group was labeled 119, 121.
The mixed labeled peptide was pre-separated into 12 components by high-performance liquid chromatography using a C18 column (Phenomenex, Strata X C18, Waters, Massachusetts, USA). The samples were separated by a high pH C18RP column (Bridge Peptide BEH C18, 130A 3.5 µm 4.6 * 250 mm, Waters, Massachusetts, USA) using Shimadzu LC-15 chromatograph. The samples were dissolved in liquid A (2% acetonitrile, pH 10). The peptides were eluted using a linear gradient of liquid B (98% acetonitrile, pH 10); the liquid phase flow rate was 0.7 mL/min, and the liquid phase gradient: 0–5 min, 0% B; 5–55 min, 0%–30% B; 55–67 min, 30%–100% B; 67–75 min, 100% B; 75–78 min, 100%–0% B; 78-88 min, 100% A. The detection wavelength was 214 nm.
Liquid chromatography-tandem mass spectrometry analysis for protein samples
The 12 components were dissolved in 20 μL nanoLC A solution, and 2 µL the samples were injected. After the samples were loaded onto the column, the analysis column was gradient eluted into the Q-exactiveHF combined mass spectrometer for identification. The sample was upgraded with the UltiMate3000 NA HPLC system; Acclaim PepMap® 100 C18, 3 μm, 100 A (75 μm × 2 cm) column and Acclaim PepMap® RSLC C18, 2 μm, 100 A (50 μm × 15 cm) column were used. The separation flow rate was 300 nL/min with a linear (3%–100%) gradient of liquid B (98% acetonitrile and 0.1% formic acid) over 120 min. The parameter of MS1 was Orbitrap, with a resolution of 120,000 and an activation type of higher-energy collisional dissociation. MS2 had an Orbitrap resolution of 30000 and a dynamic exclusion duration of 10 s. The scanning range was 400–2000 m/z, and the data collection mode was high-speed DDA.
Protein identification and bioinformatics analysis
The project used Proteome Discoverer 1.3 (Thermo Fisher Scientific, Massachusetts, USA) software to submit the original atlas file (.raw file) of peptide identification by Q ExactiveHF to MASCOT software for database search. Mass spectrometry proteomics data already through iProX partners were saved the repository to ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org).
Gene Ontology (GO) annotation of the proteome was derived from the UniProt-GOA database. Then proteins were classified by GO annotation based on three categories: biological process, cellular component, and molecular function. Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.genome.jp/kegg/) annotation was performed using KOBAS 2.1.1 software to predict the signaling pathways.
Co-immunoprecipitation (Co-IP) assay and proteome analysis
Identification of the ERGIC3 interacting proteins by co-immunoprecipitation
Proteins interacting with ERGIC3 were detected by Co-IP using Pierce Classic IP Kit (Thermo Fisher Scientific, Massachusetts, USA) and Clean-Blot IP Detection Kit (HRP) (Thermo Fisher Scientific, Massachusetts, USA). Approximately 1 × 107 cells were seeded in a 6 cm dish and then collected. The cell lysate was added to a resinous centrifuge column and incubated at 4°C for 1 h while the column was gently turned to blend. The protein was then incubated overnight with an anti-ERGIC3 antibody (Abcam, Cambridge, UK) or the same IgG (1 μg antibody :1 mg cell protein) at 4°C. Then, protein A/G agarose (20–80 μL) was added to shake and incubated for 2 h, and the resin was washed three times with 200 µL IP cracking/washing buffer and the 100 μL 1× conditioned buffer. Finally, the precipitated proteins were eluted by boiling in the sample buffer and subjecting to SDS-PAGE. The immunoprecipitated protein samples were cooled to room temperature and electrophoresed at 40 mA on a 15% acrylamide gel. The gel was then stained with Coomassie blue R-250 (Sangon, Shanghai, China).
Protein identification and bioinformatics analysis
Protein bands were excised from the gel and then analyzed by LC-MS. The obtained MS data were searched against the UniProt Homo sapiens database downloaded from the Institute website (Homo sapiens (Human) (uniprot.org)). Proteome Discoverer 1.3 (Thermo Fisher Scientific, Massachusetts, USA) software was used to match and score the original map file (.raw file) of peptide identification by Q Exactive by Sequest software. The data were screened according to the standard of FDR < 0.01 to obtain highly reliable qualitative results. GO clustering analysis was performed on the obtained proteins by ImageGP to predict cellular components, biological processes, and biological functions.
Protein-protein interaction enrichment analysis
We analyzed the proteins by mass spectrometry using Metascape. Protein-protein interaction enrichment analysis was carried out with the following databases: STRING, BioGrid, OmniPath, and InWeb_IM. Only physical interactions in STRING (physical score > 0.132) and BioGrid were used. The resultant network contains the subset of proteins that form physical interactions with at least one other member in the list. If the network contains between 3 and 500 proteins, the Molecular Complex Detection (MCODE) algorithm has been applied to identify densely connected network components. Pathway and process enrichment analysis has been applied to each MCODE component independently, and the three best-scoring terms by p-value have been retained as the functional description of the corresponding components.
The MS proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository with the dataset identifier PXD035616. All other data that support the conclusions of this study are available from the corresponding author upon request.
Western blotting and qRT-PCR data were analyzed by GraphPad Prism 9.0 software. The t-test was used to compare the mean values of two independent samples for intra-group analysis. p < 0.05 was considered statistically significant. t-test function in R language was used to calculate the difference significance p-value among samples. In this project, the number of different proteins under different screening smell values was counted. Enrichment analysis was performed using the software DAVID (https://david-d.ncifcrf.gov/), Fisher’s exact test. In general, when the p-value < 0.05, the GO function is considered significantly enriche.
Successful knockdown of endoplasmic reticulum-Golgi intermediate compartment 3 in human hepatoma GLC-82cell
The molecular mechanism by which ERGIC3 promotes the growth of lung cancer cells, RNAi technology was used to knock down ERGIC3 expression in GLC-82 cells. The results of Q-RT-PCR and western blotting revealed that ERGIC3 expression was significantly reduced 48 h after ERIGC3 siRNA treatment (Fig. 2). The obtained control and knockdown groups were used in subsequent experiments.

Figure 2: The expression of ERGIC3 at mRNA and protein was significantly reduced with ERGIC3 siRNA. Notes: ** indicating extremely significant difference (p < 0.01). ERGIC3, endoplasmic reticulum-Golgi intermediate compartment 3; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Analysis of the differentially accumulated proteins
iTRAQ and LC–MS were used to explore differential
Proteins after the ERGIC3 knockdown. Through this approach, 4600 proteins were detected, and 3711 proteins were quantified in the knockdown and control groups. Proteins with a quantitative ratio of more than 1.2 (fold change > 1.2) and a p-value < 0.05 were considered differentially expressed proteins. After ERGIC3 knockdown, 52 differentially expressed proteins were detected in secreted proteins, of which, 33 were up-regulated and 19 down-regulated (Fig. 3A). In the intracellular protein, 88 differentially expressed proteins were detected, of which 41 were up-regulated and 47 down-regulated (Fig. 3B).
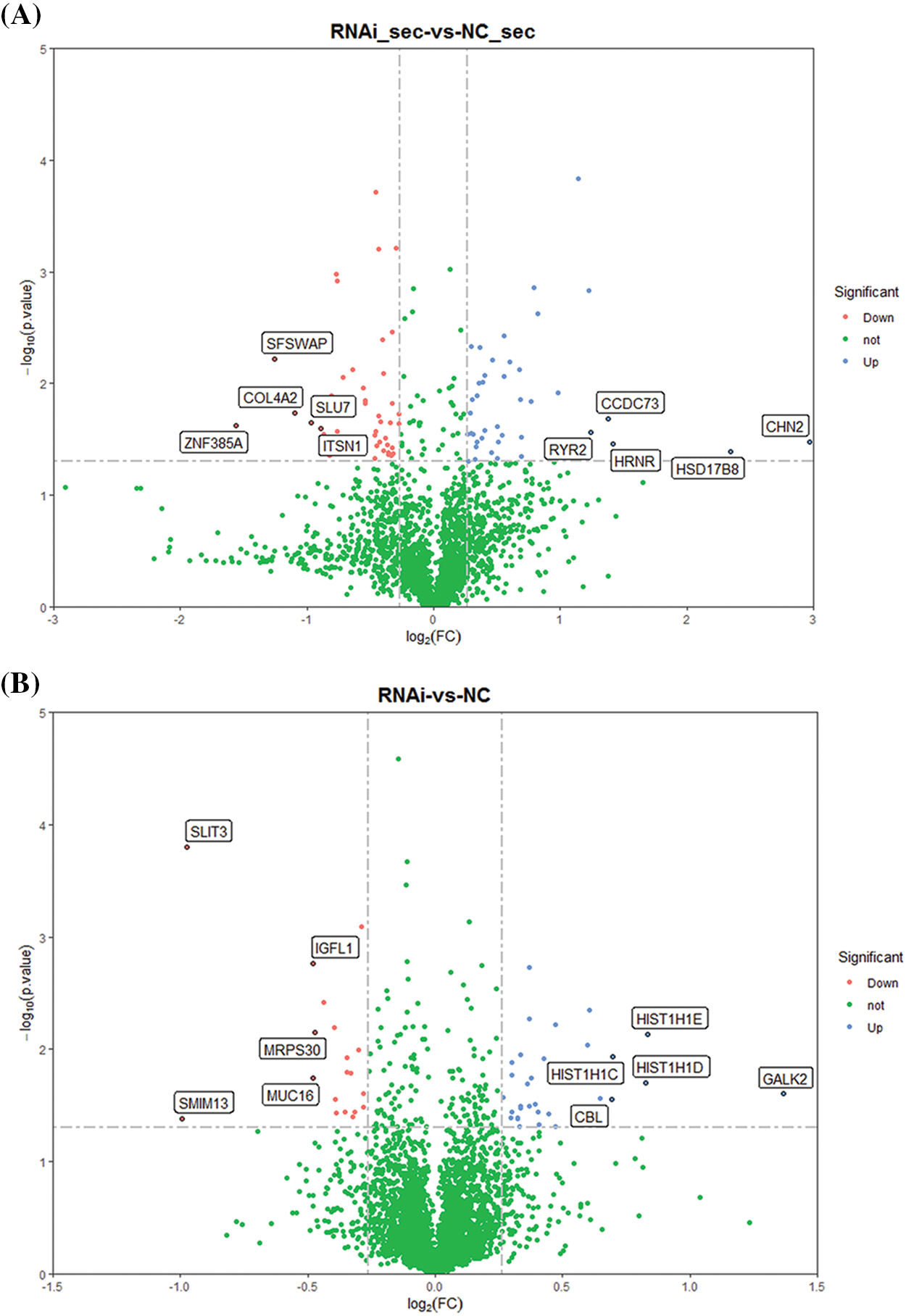
Figure 3: The volcanic map of differentially expressed proteins after ERGIC3 knockdown. Blue dots represented up-regulated genes, red dots represented down-regulated genes, and green dots represented genes with no significant difference. Y-axis was -log10 (p-value), and the larger quantity meant less difference (Only the data of fold change >1.2, p-value < 0.05 are shown). ERGIC3, endoplasmic reticulum-Golgi intermediate compartment 3.
Functional classification of extracellular differential proteins subcellular localization of extracellular differentially accumulated proteins (DPS) was mostly distributed in the nuclear and cytoplasm (Fig. 4A). The distribution of cell components showed up-regulation of mainly structural proteins like a ciliary membrane (GO:0060170), transmembrane transporter complex (GO:1902495), cation channel complex (GO:0034703), membrane region (GO:0098589), and plasma membrane region (GO:0098590). The observed down-regulated proteins were associated with cell anchoring and adhesion like actin filament (GO:0005884), focal adhesion (GO:0005925), and cell-substrate adherens junction cell-substrate junction (GO:0005924). Molecular functions of the differentially expressed proteins were enriched in NAD+ADP-ribosyltransferase activity (GO:0003950), transferase activity, transferring pentosyl groups (GO:0016763), calcium ion binding (GO:0005509), and various GTPase binding (GO:0017016, GO:0031267, GO:0051020, GO:0008536). Concurrently, ERGIC3 played an essential role in biological processes, such as the regulation of I-kappaB kinase/NF-κB signaling (GO:0007249, GO:0043122, GO:0043123) and mucosal innate immune response (GO:0002227), etc. (Figs. 4B and 4D). From the clusters of orthologous groups of proteins/eukaryotic orthologous groups (COG/KOG) classification, it was predicted that these proteins may be involved in signal transduction, post-translational modification, protein turnover, and chaperones (Fig. 4E). Importantly, these proteins were enriched in poly (ADP-ribose) polymerase, catalytic domain, EF-hand domain, WWE domain, and calcium-binding site, etc. (Fig. 4C). These results showed that ERGIC3 is linked to many of Ca2+ related molecular and biological functions, suggesting that ERGIC3 knockdown significantly alters exocrine protein expression level associated with Ca2+ activity. This finding is consistent with the hypothesis proposed by Nishikawa et al. (2007) that ERGIC3 may be one of the Ca2+ regulators in the cytoplasmic or endoplasmic network lumen (Nishikawa et al., 2007).
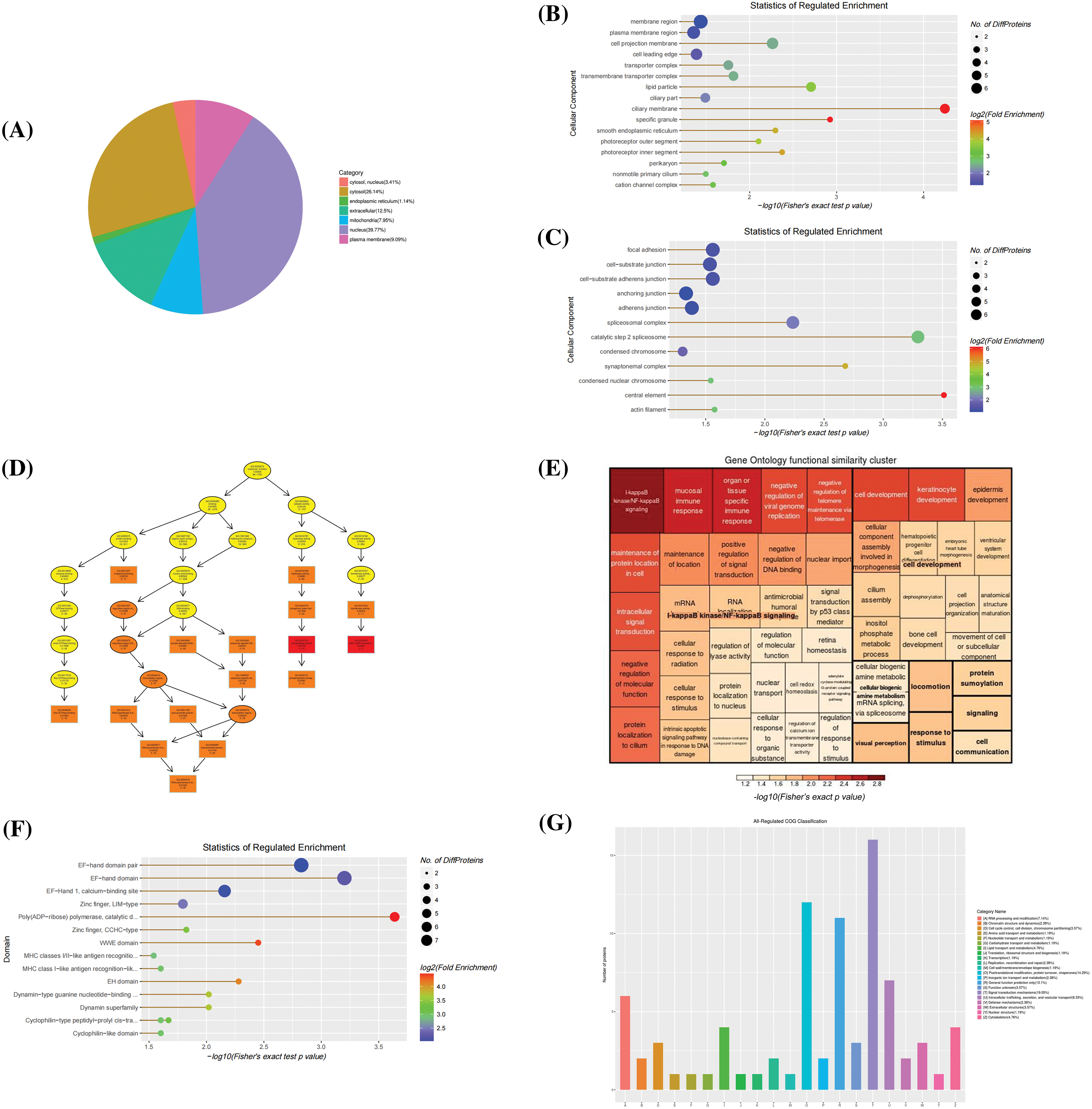
Figure 4: Bioinformatics analysis of extracellular differentially expressed proteins. (A) The Sub-cellular location terms distribution of proteins, (B, C) Analysis of cellular components of up-regulated and down-regulated proteins, (D, E) The visualization of significantly enriched Gene Ontology terms of molecular function and biological process enrichment similarity cluster, (F) Domain enrichment analysis, and (G) Clusters of orthologous groups of proteins/ eukaryotic orthologous groups (COG/KOG) classification.
Functional classification of intracellular differential proteins
The subcellular localization of intracellular differentially expressed proteins was mainly in the nucleus, cytoplasm, and plasma membrane (Fig. 5A), as well as on protein-DNA complex and cytosolic large ribosomal subunit (Fig. 5B). Most of these proteins perform molecular functions such as chromatin DNA binding, ATP-dependent 3′-5′ DNA helicase activity, transmembrane receptor protein serine/threonine kinase activity, calcium-dependent protein binding, etc. (Fig. 5D). The results of biological processes demonstrated that DPS are closely correlated with H3 like histone H3-K27 and H3-K4 trimethylation (Fig. 5C). COG enrichment analysis revealed that these proteins participate in signal transduction, chromatin structure and dynamics, intracellular transport, secretion, vesicle transport and translation, ribosomal structure, and biogenesis (Fig. 5E).
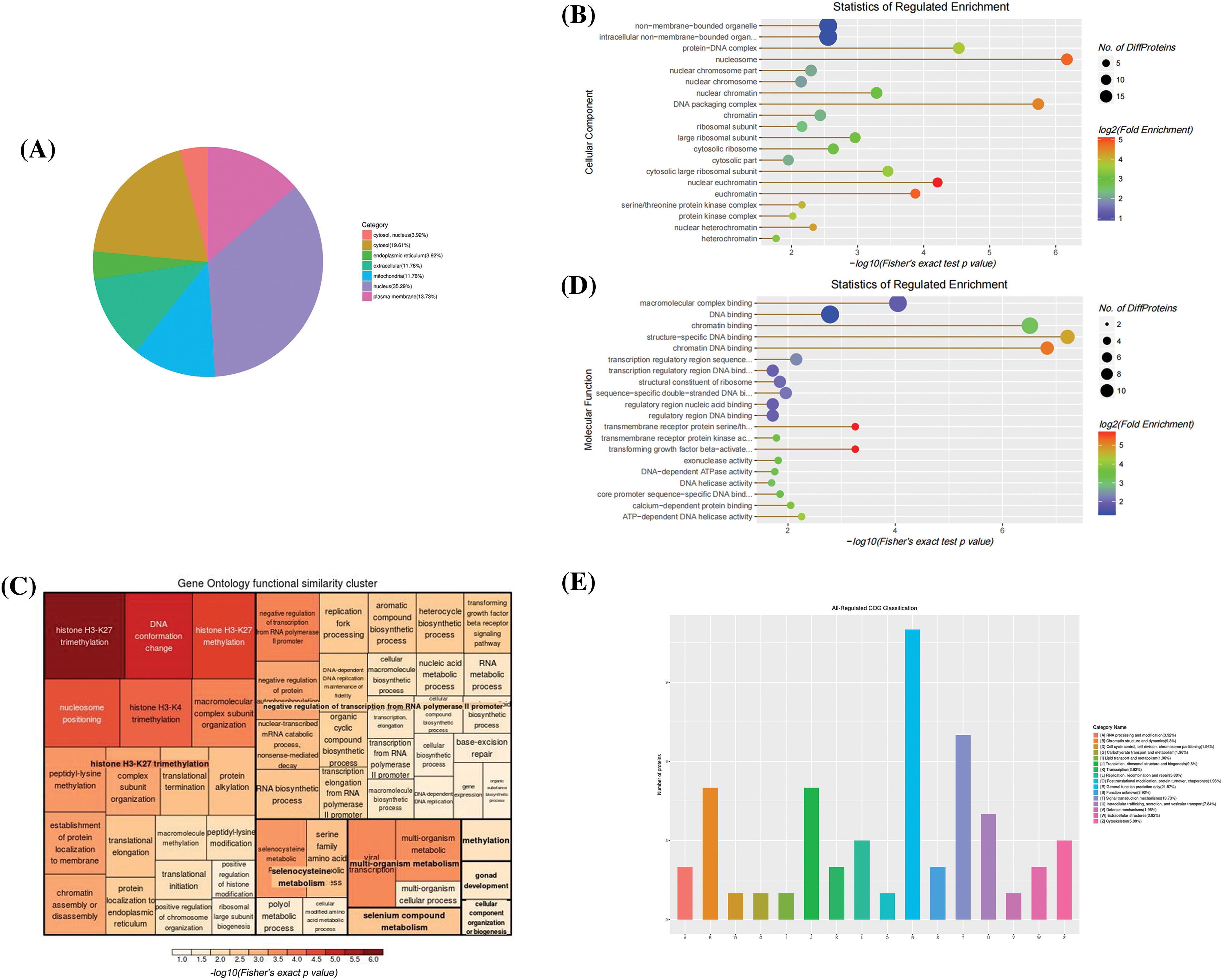
Figure 5: Bioinformatics analysis of intracellular differential proteins. (A) The Sub-cellular location terms distribution of proteins, (B) Analysis of cellular components of proteins, (C, D) The visualization of significantly enriched Gene Ontology (GO) terms of molecular function and biological process enrichment similarity cluster, (E) Clusters of orthologous groups of proteins/ eukaryotic orthologous groups (COG/KOG) classification.
Identification of proteins interacting with endoplasmic reticulum-Golgi intermediate compartment 3
The ERGIC3 interacting proteins were efficiently and specially screened via SDS-PAGE (Fig. 6A). In this figure, the protein lanes stained by Coomassie staining showed obvious differentiated protein bands near 245, 100, 75, and 63–48 KDa. Bands larger than 245 KDa were selected for MS analysis; in total, 42 proteins closely related to ERGIC3 were obtained (Table 1). GO analysis of proteins was performed by ImageGP (Fig. 6B). Cluster analysis of cell components showed that 32 proteins belong to extracellular exosome, among which 26 proteins belong to both cytoplasm and nuclear structure, such as HNRPU, K1C16, HS90A, FLNB, DESP, etc. Many proteins are involved in intermediate filament organization and keratinization biological process. According to the three best-scoring terms by MCODE (Fig. 6C), these proteins interact with ERGIC3 to participate in RHO GTPases-mediated activation of p21-activated kinases (PAKs; GO:R-HAS-5627123) and participate in forming intermediate filament organization (GO:0045109), keratinization (GO:0031424), and intermediate filament cytoskeleton organization (GO:0045104).
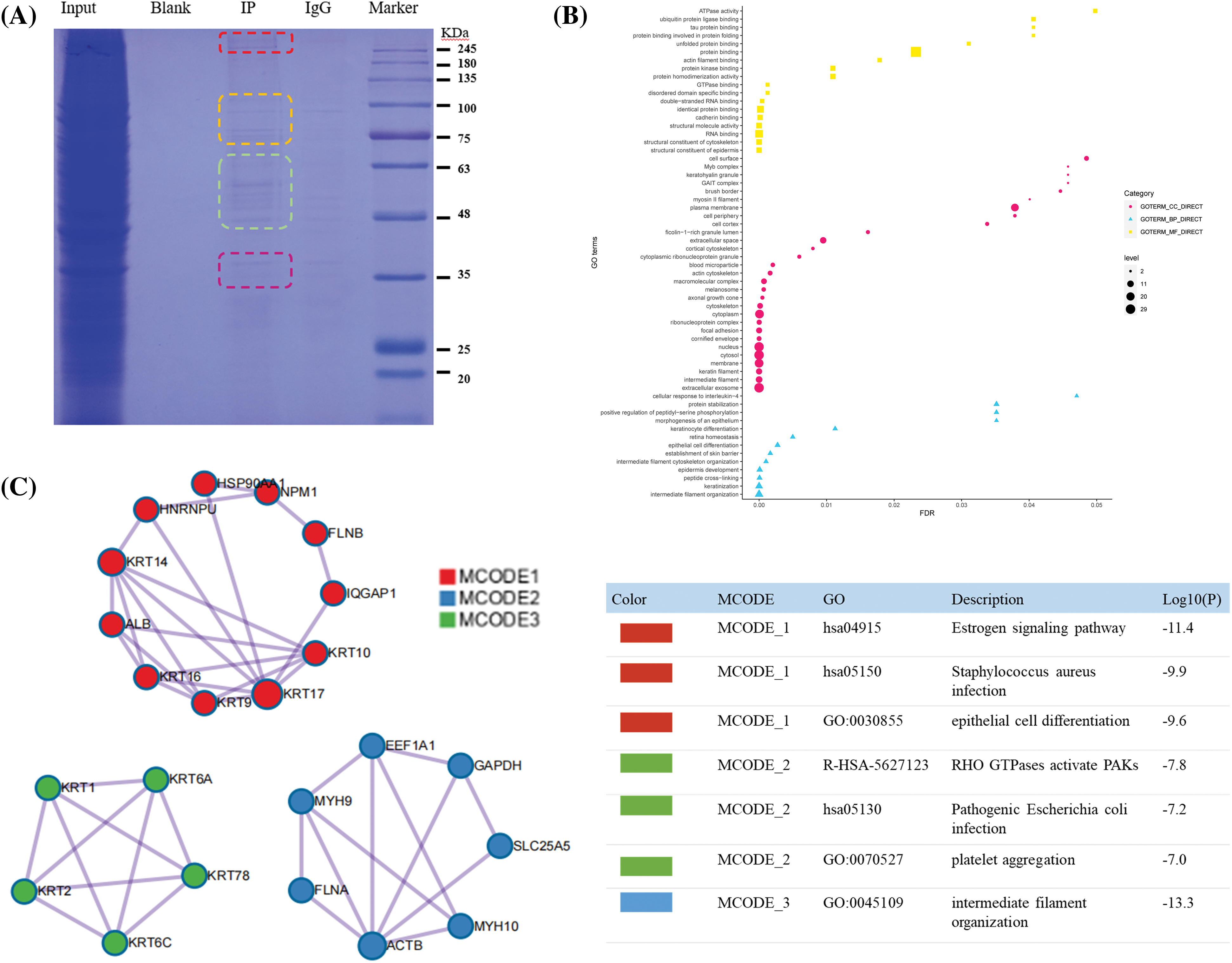
Figure 6: Co-IP results and proteome analysis. (A) From left to right, Input was sodium dodecyl sulfate-polyacrylamide gel electrophoresis of total protein, blank, IP sample, IgG, Maker, (B) The Y-axis represents the protein enrichment term, and the X-axis represents the FDR value. According to the number of protein enrichment items, the size of different shapes changes. The more protein-enriched terms, the larger the shape was. (C) Protein-protein interaction networks of the endoplasmic reticulum-Golgi intermediate compartment 3 (ERGIC3) interacting proteins.
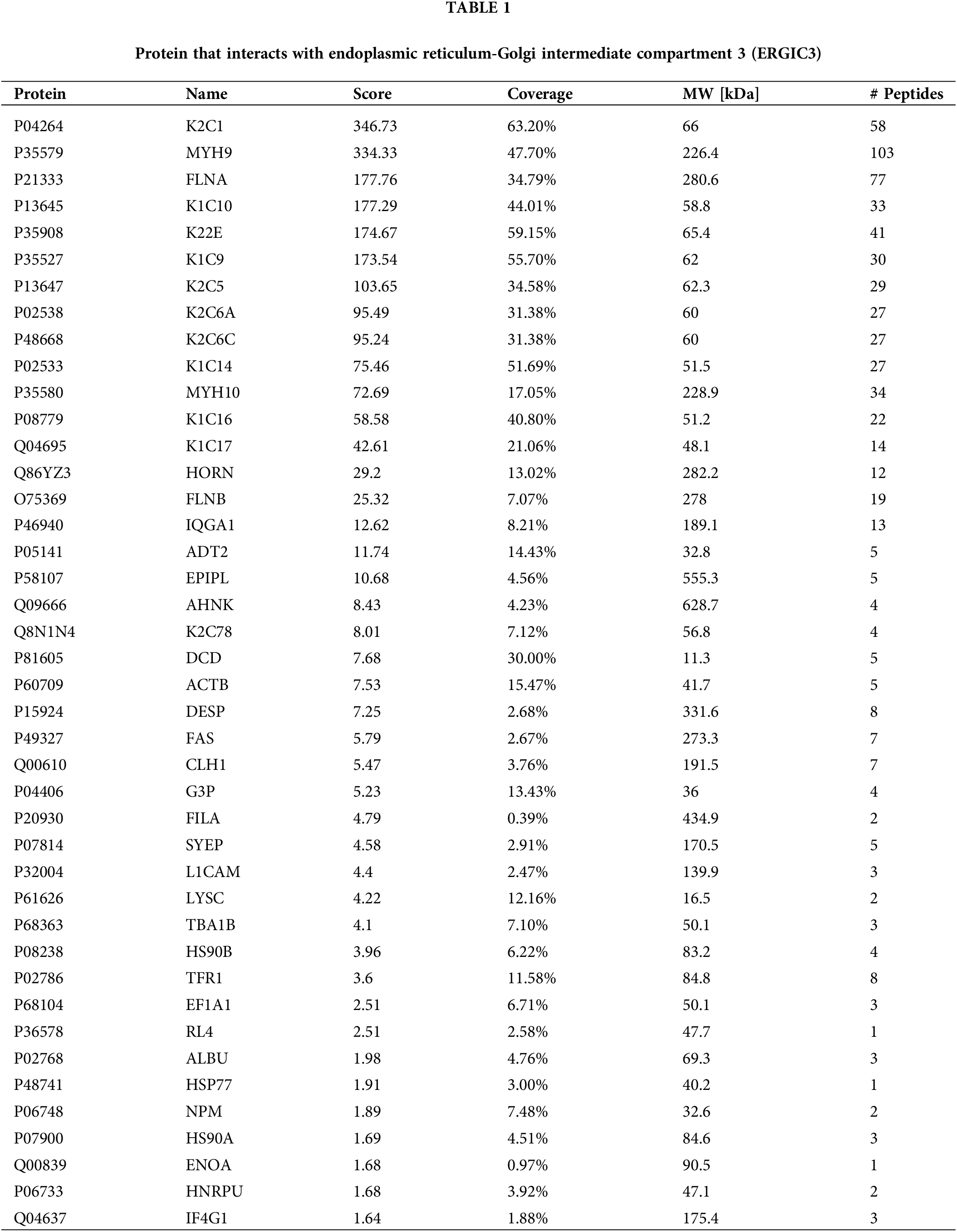
Differentially expressed proteins that interact with endoplasmic reticulum-Golgi intermediate compartment 3
Immunoprecipitation and differentially expressed proteins were treated with R4.1.0, and the intersection of proteins between groups (Fig. 7) indicated interaction with ERGIC3 and their differential expression after ERGIC3 was knocked down (Table 2). HORN and FLNA were the common proteins between coprecipitated proteins and extracellular or intracellular differentially expressed proteins. However, the differentially expressed proteins during intracellular proteins did not appear in the coprecipitate proteins.
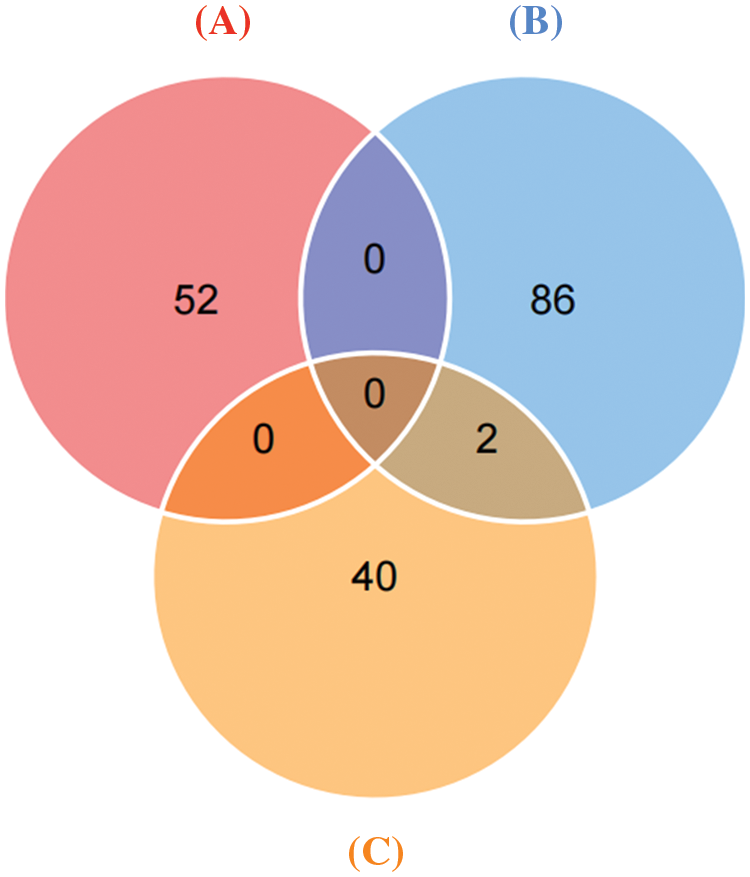
Figure 7: Differentially expressed proteins that interact with ERGIC3. Venn diagram of co-precipitation and differentially expressed proteins. (A) Intracellular differential proteins, (B) extracellular differential proteins, (C) Co-immunoprecipitated proteins.
Proteins are the executor of cell functions. Finding proteins interacting with ERGIC3 is the most direct evidence to explain the regulation of ERGIC3 on the proliferation and migration of lung cancer. Therefore, we applied proteomics to study the function of ERGIC3. First, the changes in intracellular and extracellular secreted proteins in ERGIC3-knockdown lung cancer cells were observed. The extracellular differentially expressed protein showed that the abundance of many binding- and transport-related calcium proteins changed after ERGIC3 knockdown. Some differential proteins, such as CPNE1, FLNA, TRIP6, and ZCCHY, are involved in important biological processes such as I-KappaB kinase/NF-KappaB signal transduction and are closely associated with cancer development (Hayden and Ghosh, 2008; Pflug and Sitcheran, 2020; Rasmi et al., 2020). These results suggest that ERGIC3 may play an important role in the development of cancer. The upregulated intracellular proteins are involved in biological processes such as histone H3-K27 and H3-K4 methylation, suggesting that ERGIC3 may affect biological processes in the nucleus. Moreover, proteins interacting with ERGIC3 were studied by CO-IP. These proteins are mainly involved in cytoskeleton construction and some proteins regulate RHO GTPases-activated PAKs. Furthermore, only extracellular proteins were obtained with CO-IP. Thus, the ERGIC3 protein complex may affect tumor cells by acting on proteins in the extracellular matrix.
The primary function of ERGIC3 is its participation in vesicle transport (Yoo et al., 2019). To our surprise, none of these differential proteins were significantly enriched in signaling pathways; this may be due to the complexity of vesicle transport, which has a broad spectrum (Adolf et al., 2019) and that the selection of proteins is wide from pathway analysis, while CO-IP has very narrow selectivity. Fortunately, two common proteins, HORN and FLNA, were observed in exocrine proteins and immunoprecipitation assay in ERGIC3-knocked-down lung cancer cells. HORN was up-regulated, while FLNA was down-regulated in the extracellular matrix. These proteins may be closely related to the function of ERGIC3 and form protein complexes with ERGIC3 to participate in the occurrence and development of lung cancer. In the experiment, to screen for differentially expressed proteins outside lung cancer cells, we collected cells treated with a serum-free medium for 24 h. Hence, the proteins in the serum would not interfere with the secreted proteins. FLNA is an actin-binding protein and cytoskeletal protein involved in cell diffusion and migration (Cheng and Tong, 2021; Zhou et al., 2010). In the early stage, it is mainly expressed in the cytoplasm (Adolf et al., 2019) and limited to biological processes in the cytoplasm. FLNA-knockout cells had worsened diffusion ability (Kim et al., 2008) and significantly reduced the number of cell extensions. D’Addario et al. (2001) found that binding of FLNA protein to β1 integrin mediates cell proliferation and prevents apoptosis (D’Addario et al., 2001). When integrin binds to extracellular matrix ligands, small GTPases are activated, leading to actin polymerization, forming lamellar lipid membranes and filamentous basal membranes. FLNA also interacts with small GTPases and their upstream and downstream factors, including Rac, Rho, Cdc42, and Rala (Ohta et al., 1999). After ERGIC3 knockdown, FLNA protein and numerous small GTP-binding proteins, PHP14, RBP17, and RBP2, were found in the extracellular down-regulated proteins of lung cancer. Immunoprecipitation demonstrated the association of FLNA protein with ERGIC3 vesicle transport, suggesting that FLNA protein is likely transported to the extracellular matrix through vesicle transport interacting with ERGIC3 to participate in extracellular biological processes. FLNA indeed has complex biological effects and is not expressed only in the cytoplasm. But it is now known to interact with a variety of cytoplasmic membranes and nuclear proteins (Berry et al., 2005; Feng and Walsh, 2004; Stossel et al., 2001). With the development of FLNA research, Bachmann et al. (2006) demonstrated for the first time that FLNA is not only expressed in cells but also on the cell surface, and its C-terminal part is exposed to the extracellular matrix (Bachmann et al., 2006). Combined with our results, FLNA may be the transporter of ERGIC3, and the high expression of ERGIC3 in tumor cells accelerates the transport of FLNA, thus accelerating the regulation of related signaling pathways and cell migration. After ERGIC3 knockdown, the transport of FLNA out of the vesicle decreases, thus reducing the amount of extracellular FLNA protein. As described above, FLNA is of great significance in cell movement, apoptosis, and even angiogenesis (Nallapalli et al., 2012). Due to ERGIC3 knockdown, the decrease in FLNA protein in extracellular matrix of lung cancer cells may affect the migration of lung cancer cells and promote tumor cell apoptosis. ERGIC3 has been found to promote the growth of liver cancer cells by affecting the collagen-integrin receptor-actin axis (Liu et al., 2022). In our study, we further confirmed the feasibility of FLNA as a potential lung cancer target and identified the pathway that may interact with ERCIC3 to promote lung cancer development.
HORN, whose extracellular level is significantly increased after ERGIC3 knockdown, is a member of the S100 fusion protein family and has a Ca2+ binding EF-hand domain at the N-terminal (Sun et al., 2018). After ERGIC3 knockdown, extracellular proteins with EF binding domain and ion transport domain and proteins with Ca2+ binding biological function were enriched and upregulated. The S100 protein family comprises more than 20 members and is the largest subgroup of calcium-binding proteins. These proteins are distributed in multiple structures of cells and have complex biological functions. Differentially expressed S100 family proteins have been found in many tumors, promoting tumor-genesis processes such as cell proliferation, metastasis, angiogenesis, and immune evasion (Babini et al., 2011; Fleming et al., 2012; Marenholz et al., 2004). HORN protein is easily detected in the cytoplasm of breast cancer cells and is focally positive in the nucleus. Moreover, as an exosome protein, HORN may play an important role in the invasion process of breast cancer progression, suggesting potential utility as a marker for predicting recurrence or potential metastasis (Choi et al., 2016). HORN is up-regulated as a key protein in breast cancer, pancreatic cancer, and liver cancer and plays an important role in maintaining the tumor vascular system in pancreatic cancer (Gutknecht et al., 2017). Inhibition of AKT (protein kinase B) expression in liver cancer significantly inhibited the proliferation, colony formation, migration, and invasion of liver cancer cells and led to a decrease in AKT phosphorylation (Fu et al., 2018). Nonetheless, the role of the protein in lung cancer is still poorly studied. Our data indicate that the number of HORN outside lung cancer cells is up-regulated after ERGIC3 knockdown. Consequently, ERGIC3 may have an inhibitory effect on HORN. In previous studies, ERGIC3 overexpression inhibited ER stress (ERS)-induced cell death (Nishikawa et al., 2007). ERS occurs after calcium metabolism disorder, oxidative stress, protein unfolding or misfolding, etc. ERGIC3 knockdown results in an increase in the expression of HORN. Due to its Ca2+-binding biological characteristics, HORN is likely to cause intracellular calcium metabolism disorder, resulting in ERS and cell death.
These proteomic results are preliminary data and need further experimental validation. In particular, multiple cellular components should be isolated for analysis. In addition to secreted proteins, cell membrane proteins, cytoplasmic proteins, nuclear proteins, and even sub-organelle proteins, such as mitochondrial proteins, can be isolated for correlation analysis with ERGIC3, to fully understand the protein regulation function of ERGIC3. Based on our current results, we propose a hypothesis that has not been reported in previous studies: ERGIC3, as a transmembrane protein transported through vesicles, also appears to affect many intranuclear proteins. This also suggests that ERGIC3 may be involved in the forward transport of ER vesicles to the Golgi apparatus and in the reverse transport of Golgi apparatus vesicles to ER or even the nucleus (Adolf et al., 2019). HORN and FLNA may play a core regulatory role in vesicular transport. These results suggest that ERGIC3 may not only participate in the biological processes in the cytoplasm but also affect some functions in the nucleus to regulate the occurrence and development of lung cancer. Therefore, more evidence is needed to elucidate the pathophysiological function of ERGIC3 in the development and progression of lung cancer.
To conclude, ERGIC3, as a transmembrane protein, binds to a large number of cargo proteins, and the molecular mechanisms are more complex. As one of the markers of lung cancer, the above-mentioned results can demonstrate the reliability of ERGIC3 as a marker of lung cancer, reveal some specific molecular mechanisms, and provide new methods and ideas for the future treatment of lung cancer.
Acknowledgement: We thank our tutor Mingsong Wu for his guidance on this article, the reviewers who reviewed this article, and the editors who helped us edit it.
Funding Statement: This work was supported by the National Natural Science Foundation of China (Grant No. 81760508); Zunyi City “15851 Talent Elite” Project (Grant No. 81760508); Natural Science and Technology Foundation of Guizhou Province (QiankeheZhicheng [2022] YiBan182); Young Scientific and Technological Talents Growth Project of Guizhou Provincial Department of Education [Grant No. QianJiaoHe KY Zi [2022] 280 Hao].
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: MINGSONG WU, WEI ZHAI, QIURONG ZHAO; data collection: XIANG ZHENG, SHANSHAN FENG, JINJING WANG; analysis and interpretation of results: WEI ZHAI, XUEYING LI; draft manuscript preparation: MENGYUAN LIU, YUSHU ZHANG, YUQING LUO. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
Adolf F, Rhiel M, Hessling B, Gao Q, Hellwig A, Béthune J, Wieland F T (2019). Proteomic profiling of mammalian COPII and COPI Vesicles. Cell Reports 26: 250–265. https://doi.org/10.1016/j.celrep.2018.12.041 [Google Scholar] [PubMed] [CrossRef]
Babini E, Bertini I, Borsi V, Calderone V, Hu X, Luchinat C, Parigi G (2011). Structural characterization of human S100A16, a low-affinity calcium binder. Journal of Biological Inorganic Chemistry 16: 243–256. https://doi.org/10.1007/s00775-010-0721-3 [Google Scholar] [PubMed] [CrossRef]
Bachmann AS, Howard JP, Vogel CW (2006). Actin-binding protein filamin A is displayed on the surface of human neuroblastoma cells. Cancer Science 97: 1359–1365. https://doi.org/10.1111/j.1349-7006.2006.00327.x [Google Scholar] [PubMed] [CrossRef]
Berry FB, O’Neill MA, Coca-Prados M, Walter MA (2005). FOXC1 transcriptional regulatory activity is impaired by PBX1 in a filamin A-mediated manner. Molecular and Cellular Biology 25: 1415–1424. https://doi.org/10.1128/MCB.25.4.1415-1424.2005 [Google Scholar] [PubMed] [CrossRef]
Brody H (2020). Lung cancer. Nature 587: S7. https://doi.org/10.1038/d41586-020-03152-0 [Google Scholar] [PubMed] [CrossRef]
Chen Z, Fillmore CM, Hammerman PS, Kim CF, Wong KK (2014). Non-small-cell lung cancers: A heterogeneous set of diseases. Nature Reviews Cancer 14: 535–546. https://doi.org/10.1038/nrc3775 [Google Scholar] [PubMed] [CrossRef]
Cheng L, Tong Q (2021). Interaction of FLNA and ANXA2 promotes gefitinib resistance by activating the Wnt pathway in non-small-cell lung cancer. Molecular and Cellular Biochemistry 476: 3563–3575. https://doi.org/10.1007/s11010-021-04179-1 [Google Scholar] [PubMed] [CrossRef]
Choi J, Kim DI, Kim J, Kim BH, Kim A (2016). Hornerin is involved in breast cancer progression. Journal of Breast Cancer 19: 142–147. https://doi.org/10.4048/jbc.2016.19.2.142 [Google Scholar] [PubMed] [CrossRef]
D’Addario M, Arora PD, Fan J, Ganss B, Ellen RP, McCulloch CA (2001). Cytoprotection against mechanical forces delivered through beta 1 integrins requires induction of filamin A. Journal of Biological Chemistry 276: 31969–31977. https://doi.org/10.1074/jbc.M102715200 [Google Scholar] [PubMed] [CrossRef]
Feng Y, Walsh CA (2004). The many faces of filamin: A versatile molecular scaffold for cell motility and signalling. Nature Cell Biology 6: 1034–1038. https://doi.org/10.1038/ncb1104-1034 [Google Scholar] [PubMed] [CrossRef]
Fleming JM, Ginsburg E, Oliver SD, Goldsmith P, Vonderhaar BK (2012). Hornerin, an S100 family protein, is functional in breast cells and aberrantly expressed in breast cancer. BMC Cancer 12: 266. https://doi.org/10.1186/1471-2407-12-266 [Google Scholar] [PubMed] [CrossRef]
Fu SJ, Shen SL, Li SQ, Hua YP, Hu WJ, Guo B, Peng BG (2018). Hornerin promotes tumor progression and is associated with poor prognosis in hepatocellular carcinoma. BMC Cancer 18: 815. https://doi.org/10.1186/s12885-018-4719-5 [Google Scholar] [PubMed] [CrossRef]
Gutknecht MF, Seaman ME, Ning B, Cornejo DA, Mugler E, Antkowiak PF, Moskaluk CA, Hu S, Epstein FH, Kelly KA (2017). Identification of the S100 fused-type protein hornerin as a regulator of tumor vascularity. Nature Communication 8: 552. https://doi.org/10.1038/s41467-017-00488-6 [Google Scholar] [PubMed] [CrossRef]
Hayden MS, Ghosh S (2008). Shared principles in NF-κB signaling. Cell 132: 344–362. https://doi.org/10.1016/j.cell.2008.01.020 [Google Scholar] [PubMed] [CrossRef]
Herbst R, Morgensztern D, Boshoff C (2018). The biology and management of non-small cell lung cancer. Nature 553: 446–454. https://doi.org/10.1038/nature25183 [Google Scholar] [PubMed] [CrossRef]
Imyanitov EN, Iyevleva AG, Levchenko EV (2021). Molecular testing and targeted therapy for non-small cell lung cancer: Current status and perspectives. Critical Reviews in Oncology/Hematology 157: 103194 [Google Scholar] [PubMed]
Kim H, Sengupta A, Glogauer M, McCulloch CA (2008). Filamin A regulates cell spreading and survival via β1 integrins. Experimental Cell Research 314: 834–846. https://doi.org/10.1016/j.yexcr.2007.11.022 [Google Scholar] [PubMed] [CrossRef]
Liu M, Zhao Q, Zheng X, Yang L, Zhao Y, Li X, Wu M (2022). Transcriptome changes in ERGIC3-knockdown hepatocellular carcinoma cells: ERGIC3 is a novel immune function related gene. PeerJ 10: e13369. https://doi.org/10.7717/peerj.13369 [Google Scholar] [PubMed] [CrossRef]
Marenholz I, Heizmann CW, Fritz G (2004). S100 proteins in mouse and man: From evolution to function and pathology (including an update of the nomenclature). Biochemical & Biophysical Research Communications 322: 1111–1122. https://doi.org/10.1016/j.bbrc.2004.07.096 [Google Scholar] [PubMed] [CrossRef]
Nallapalli RK, Ibrahim MX, Zhou AX, Bandaru S, Sunkara SN et al. (2012). Targeting filamin A reduces K-RAS-induced lung adenocarcinomas and endothelial response to tumor growth in mice. Molecular Cancer 11: 50. https://doi.org/10.1186/1476-4598-11-50 [Google Scholar] [PubMed] [CrossRef]
Nishikawa M, Kira Y, Yabunaka Y, Inoue M (2007). Identification and characterization of endoplasmic reticulum-associated protein, ERp43. Gene 386: 42–51. https://doi.org/10.1016/j.gene.2006.06.030 [Google Scholar] [PubMed] [CrossRef]
Ohta Y, Suzuki N, Nakamura S, Hartwig JH, Stossel TP (1999). The small GTPase RalA targets filamin to induce filopodia. Proceedings of the National Academy of Sciences of the United States of America 96: 2122–2128. https://doi.org/10.1073/pnas.96.5.2122 [Google Scholar] [PubMed] [CrossRef]
Otte S, Barlowe C (2002). The Erv41p-Erv46p complex: Multiple export signals are required in trans for COPII-dependent transport from the ER. The EMBO Journal 21: 6095–6104. https://doi.org/10.1093/emboj/cdf598 [Google Scholar] [PubMed] [CrossRef]
Pflug KM, Sitcheran R (2020). Targeting NF-κB-inducing kinase (NIK) in immunity, inflammation, and cancer. International Journal of Molecular Sciences 21: 8470. https://doi.org/10.3390/ijms21228470 [Google Scholar] [PubMed] [CrossRef]
Rasmi RR, Sakthivel KM, Guruvayoorappan C (2020). NF-κB inhibitors in treatment and prevention of lung cancer. Biomedicine & Pharmacotherapy 130: 110569. https://doi.org/10.1016/j.biopha.2020.110569 [Google Scholar] [PubMed] [CrossRef]
Sharma C, Wang HX, Li Q, Knoblich K, Reisenbichler ES, Richardson AL, Hemler ME (2017). Protein acyltransferase DHHC3 regulates breast tumor growth, oxidative stress, and senescence. Cancer Research 77: 6880–6890. https://doi.org/10.1158/0008-5472.CAN-17-1536 [Google Scholar] [PubMed] [CrossRef]
Shokoohi A, Al-Hashami Z, Moore S, et al. (2022). Effect of targeted therapy and immunotherapy on advanced nonsmall-cell lung cancer outcomes in the real world. Cancer Medicine 11: 86–93 [Google Scholar] [PubMed]
Stossel TP, Condeelis J, Cooley L, Hartwig JH, Noegel A, Schleicher M, Shapiro SS (2001). Filamins as integrators of cell mechanics and signalling. Nature Reviews Molecular Cell Biology 2: 138–145. https://doi.org/10.1038/35052082 [Google Scholar] [PubMed] [CrossRef]
Sun X, Wang T, Zhang C, Ning K, Guan ZR, Chen SX, Hong TT, Hua D (2018). S100A16 is a prognostic marker for colorectal cancer. Journal of Surgical Oncology 117: 275–283. https://doi.org/10.1002/jso.24822 [Google Scholar] [PubMed] [CrossRef]
Wang K, Chen Z, Long L, Tao Y, Wu Q et al. (2018). iTRAQ-based quantitative proteomic analysis of differentially expressed proteins in chemoresistant nasopharyngeal carcinoma. Cancer Biology & Therapy 19: 809–824. https://doi.org/10.1080/15384047.2018.1472192 [Google Scholar] [PubMed] [CrossRef]
Wang Z, Sheng B, Wei Z, Li Y, Liu Z (2022). Identification of a metastasis-related protein IFI16 in esophageal cancer using a proteomic approach. Journal of Cancer 13: 1630–1639. https://doi.org/10.7150/jca.59286 [Google Scholar] [PubMed] [CrossRef]
Wu M, Tu T, Huang Y, et al. (2013). Suppression subtractive hybridization identified differentially expressed genes in lung adenocarcinoma: ERGIC3 as a novel lung cancer-related gene. BMC Cancer 13: 44 [Google Scholar] [PubMed]
Yoo W, Cho EB, Kim S, Yoon JB (2019). The E3 ubiquitin ligase MARCH2 regulates ERGIC3-dependent trafficking of secretory proteins. Journal of Biological Chemistry 294: 10900–10912. https://doi.org/10.1074/jbc.RA119.007435 [Google Scholar] [PubMed] [CrossRef]
Zhang LY, Liu M, Li X, Tang H (2013). miR-490-3p modulates cell growth and epithelial to mesenchymal transition of hepatocellular carcinoma cells by targeting endoplasmic reticulum-Golgi intermediate compartment protein 3 (ERGIC3). Journal of Biological Chemistry 288: 4035–4047. https://doi.org/10.1074/jbc.M112.410506 [Google Scholar] [PubMed] [CrossRef]
Zhao Q, Wu M, Zheng X, Yang L, Zhang Z, Li X, Chen J (2020). ERGIC3 silencing additively enhances the growth inhibition of BFA on lung adenocarcinoma cells. Current Cancer Drug Targets 20: 67–75. https://doi.org/10.2174/1568009619666190917145906 [Google Scholar] [PubMed] [CrossRef]
Zhou AX, Hartwig JH, Akyürek LM (2010). Filamins in cell signaling, transcription and organ development. Trends in Cell Biology 20: 113–123. https://doi.org/10.1016/j.tcb.2009.12.001 [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF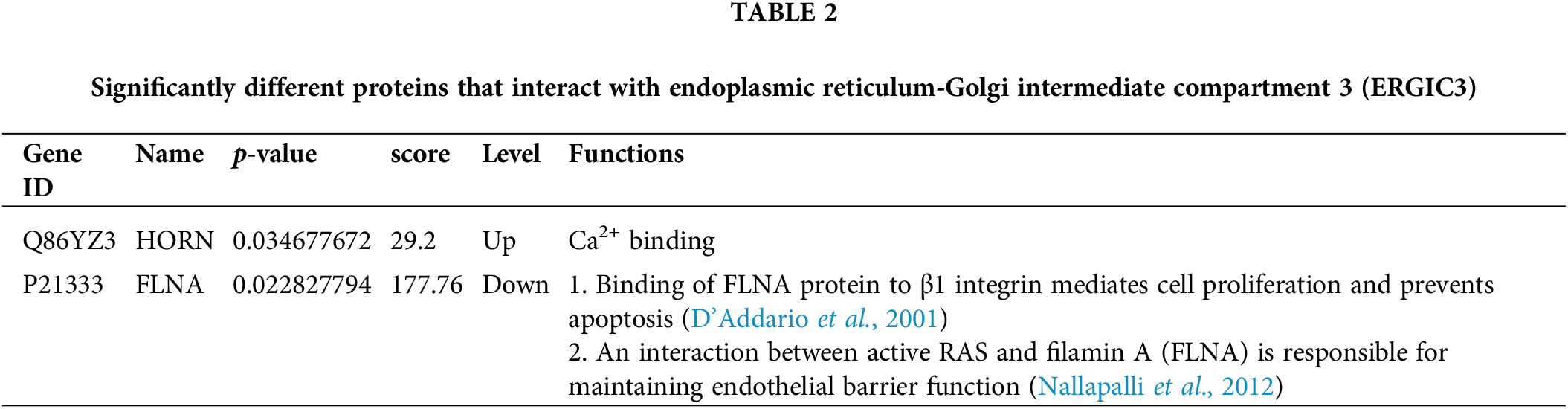
 Downloads
Downloads
 Citation Tools
Citation Tools