 Open Access
Open Access
ARTICLE
A novel mutation in ROR2 led to the loss of function of ROR2 and inhibited the osteogenic differentiation capability of bone marrow mesenchymal stem cells (BMSCs)
1 Prenatal Diagnosis Center, Shijiazhuang Obstetrics and Gynecology Hospital, Key Laboratory of Maternal and Fetal Medicine of Hebei Province, Shijiazhuang, 050011, China
2 Department of Stomatology, Fifth Medical Center of Chinese PLA General Hospital, Beijing, 100039, China
3 Institute of Hematology, Fifth Medical Center of Chinese PLA General Hospital, Beijing, 100039, China
4 Prenatal Diagnosis Center, Beijing Obstetrics and Gynecology Hospital, Beijing Maternal and Child Health Care Hospital, Capital Medical University, Beijing, 100026, China
5 Department of Orthodontics, Beijing Stomatological Hospital, Capital Medical University School of Stomatology, Capital Medical University, Beijing, 100050, China
* Corresponding Authors: JING ZHANG. Email: ; KAI YANG. Email:
# These authors contributed equally to this work
BIOCELL 2023, 47(7), 1561-1569. https://doi.org/10.32604/biocell.2023.028851
Received 11 January 2023; Accepted 20 March 2023; Issue published 21 June 2023
Abstract
Background: Receptor tyrosine kinase-like orphan receptor 2 (ROR2) has a vital role in osteogenesis. However, the mechanism underlying the regulation of ROR2 in osteogenic differentiation is still poorly comprehended. A previous study by our research group showed that a novel compound heterozygous ROR2 variation accounted for the autosomal recessive Robinow syndrome (ARRS). This study attempted to explore the impact of the ROR2: c.904C>T variant specifically on the osteogenic differentiation of BMSCs. Methods: Coimmunoprecipitation (CoIP)-western blotting was carried out to identify the interaction between ROR2 and Wnt5a. Double-immunofluorescence staining was used for determining the expressions and co-localization of ROR2 and Wnt5a in bone marrow mesenchymal stem cells (BMSCs). Western blot (WB) analysis and quantitative reverse transcription polymerase chain reaction (RT-qPCR) were conducted to identify the expression levels of ROR2 in the BMSCs transfected with LV-shROR2 or LV-ROR2-c.904C>T. The alkaline phosphatase (ALP) activity was detected, and Alizarin Red S staining was done for evaluating the osteogenic differentiation of BMSCs. RT-qPCR was employed to identify the expression of the sphingomyelin synthase 1 (SMS1) mRNA in the BMSCs transfected with LV-shROR2 or LV-ROR2-c.904C>T and the mRNA expression levels of Runt-related transcription factor 2 (RUNX2), osteocalcin (OCN), and osteopontin (OPN). WB was performed to confirm the protein expressions of extracellular regulated protein kinases1 (ERK), P-ERK, Smad family member1/5/8 (Smad1/5/8), P-Smad1/5/8, P-P38, P38, RUNX2, OCN, and OPN in the BMSCs transfected with LV-shROR2/LV-ROR2-c.904C>T and sphingomyelin (SM). Results: The ROR2: c.904C>T mutant altered the subcellular localization of the ROR2 protein, which caused an impaired interaction between ROR2 and Wnt5a. The depletion of ROR2 restricted the osteogenic differentiation capability of BMSCs and downregulated the expression of SMS1. SM treatment could reverse the inhibition of osteoblastic differentiation in ROR2-depleted BMSCs. Conclusion: The findings of this work revealed that the ROR2: c.904C>T variant led to the loss of function of ROR2, which impaired the interaction between ROR2 and Wnt5a and also controlled the osteogenic differentiation capability of BMSCs. Furthermore, SM was revealed to be engaged in the osteoblastic differentiation of BMSCs regulated by ROR2, which renders SM a potential target in the therapy for ARRS.Graphic Abstract
Keywords
Receptor tyrosine kinase-like orphan receptor 2 (ROR2) refers to a 943-amino-acid transmembrane protein tyrosine kinase encoded by the ROR2 gene (van Bokhoven et al., 2000). ROR2 codes for a member of the ROR receptor tyrosine kinase family that is expressed widely in multiple tissues. This includes the heart, the lung, and the brain, and is involved in developing the nervous and skeletal systems (Huang et al., 2015). Researchers have verified the vital role of ROR2 in controlling cell polarity, migration, proliferation, differentiation, developmental morphogenesis, and tissue/organogenesis (Debebe and Rathmell, 2015). The biallelic gene mutations in ROR2 are responsible for the autosomal recessive Robinow syndrome (ARRS). Missense, nonsense, and frameshift mutations have been reported in both intracellular and extracellular domains of ROR2, which suggests that ARRS is associated with the loss of function of ROR2 (Chen et al., 2005). A previous study by our research group showed that a novel missense variant, ROR2: c.904C>T accounted for ARRS (Yang et al., 2020). Accumulating evidence from studies conducted using ROR2-knockout mice and ARRS patients indicates a significant function of ROR2 in bone formation and development (DeChiara et al., 2000; Schwabe et al., 2004; Weissenböck et al., 2019). However, till now, the mechanism underlying the regulation of ROR2 in osteogenesis is poorly comprehended.
ROR2 is the inherent receptor of Wnt5a and a component of the non-canonical Wnt pathway (Yu et al., 2016; Hasegawa et al., 2018). Mutations in any of the components of the non-canonical Wnt pathway, including Wnt5a, are reported to be associated with dysmorphic skeletal disorders, the manifestations of which include short limbs and facial anomalies (Michigami, 2019). Mutations in Wnt5a are responsible for the autosomal dominant Robinow syndrome (ADRS) (Bolzoni et al., 2013). The binding of ROR2 to Wnt5a has been documented to contribute to the cytoskeletal arrangement, cell migration, cell polarity, and cell proliferation (Ma et al., 2021). Evidence suggests that Wnt5a stimulates osteoblastogenesis in humans via its co-receptor ROR2 (Bolzoni et al., 2013) and that the loss of function of ROR2 could impair the Wnt5a/ROR2 signaling pathway to result in skeletal abnormalities (Kaissi et al., 2020). However, whether c.904C>T could impair the interaction between ROR2 and Wnt5a remains unknown so far.
Sphingolipids are important components of the mammalian cell membranes of which SM is the major subgroup (Wang et al., 2019). The biogenesis of SM involves two key synthetases belonging to the SM synthase (SMS) family, namely, SM synthase 1 (SMS1) and SM synthase 2 (SMS2), which are selectively localized in the Golgi apparatus and the plasma membrane of the cells (Villani et al., 2008; Yeang et al., 2008; Matsumoto et al., 2019). SM forms an SM-rich microdomain on the cell membrane to control signal transduction for various processes, such as cell proliferation/survival, migration, and inflammation (Taniguchi and Okazaki, 2021). It was reported that local SM catabolism is vital for the mineralization process in healthy bones (During et al., 2015). Nuclear magnetic resonance (NMR) analysis also revealed that the amount of SM is increased during osteogenic differentiation in human adipose tissue stem cells (Bispo et al., 2022). On the basis of a previous study performed by our research group, SM was involved in the senescence of dental pulp stem cells (DPSCs) regulated by ROR2 (Dong et al., 2021). BMSCs are reported to possess a high osteogenic capacity (Lee et al., 2019). The potential involvement of SM in the osteoblast differentiation of BMSCs mediated by ROR2 warrants further investigation.
Hence, this work aimed to explore the specific impact of the ROR2: c.904C>T variant on the osteogenic differentiation of BMSCs and the potential novel mechanism underlying ARRS.
Cell isolation, culture, and transfection
The approval of this study was acquired from the Ethics Committee of Shijiazhuang Obstetrics and Gynecology Hospital. BMSCs isolation has been described in a previous study (You et al., 2022). The SM transfection was performed as follows. A mixture of sphingomyelin (125 µM, Sigma, MO, USA, #85615) together with Lipofectamine (10 µL, Thermo, MA, USA, STEM00003) was prepared by thorough mixing, followed by 15–20-min incubation at room temperature for allowing the formation of Lipo-SM. Next, BMSCs were placed the into the freshly prepared medium and subsequently incubated using Lipo or Lipo-SM at 37°C for 72 h to allow SM transfection (Ishida et al., 2016).
The RIPA lysis buffer was used to isolate the protein from the cultured BMSCs, according to the protocol in previous studies (Yang et al., 2019; Yang et al., 2017). Next, the isolated proteins were subjected to separation through sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE: 8% or 10%) followed by electro-transfer onto the polyvinylidene difluoride (PVDF) membrane (Millipore, MA , USA,). Then, membranes were blocked for a 2-h period with 5% milk diluted in TBS (Mahmood and Yang, 2012). The membranes were later incubated overnight with primary antibodies at 4°C. The antibodies used were anti-Smad1/5/8 (Abcam, Cambridge, UK, 1:1000), anti-P-Smad1/5/8 (Cell Signaling Technology, Boston, USA, 1:1000), anti-ERK (Abcam, Cambridge, UK, 1:1000), anti-P-P38 (Abcam, Cambridge, UK, 1:1000), anti-RUNX2 (Abcam, Cambridge, UK, 1:1000), anti-P-ERK (Abcam, Cambridge, UK, 1:1000), anti-P38 (Abcam, Cambridge, UK, 1:1000), anti-ROR2 (Abcam, Cambridge, UK, 1:1000), anti-OPN (Abcam, Cambridge, UK, 1:1000), anti-OCN (Abcam, Cambridge, UK, 1:1000), and anti-GAPDH (Abcam, Cambridge, UK, 1:1000). Using FuazonFx (VilberLourmat), next day, blots were examined by ECL (enhanced chemiluminescence). After capturing blot images, FusionCapt Advance Fx5 software (VilberLourmat) was employed for processing. Moreover, each assay was carried out three times.
RNA isolation and quantitative reverse transcription polymerase chain reaction
BMSC cells were lysed using QIAzol Lysis Reagent (QIAGEN, Dusseldorf, GER) for subsequent RNA extraction. The quantity and quality of the extracted RNA were analyzed using NanoDrop 2000 (Thermo Fisher, MA, USA) (Sun et al., 2017). The mRNA was reverse transcribed by using the M-MLV First-Strand Kit (Life Technologies, Cal, USA). Later, qRT-PCR analysis of the mRNAs was carried out using the Platinum SYBR Green qPCR SuperMix-UDG Kit (Invitrogen, Cal, USA). In addition, RT-PCR assay was performed with the CFX96™ Real-Time System (Bio-Rad). Gene levels were calculated by the 2–ΔΔCt method, with GAPDH being the reference.
To evaluate the cell osteogenic differentiation potential in vitro, we inoculated BMSCs (1 × 105 cells) into 6-well plates. Then, the cells were subjected to the appropriate treatments in accordance with the experimental design. The Lv-Subconfluent cultures were subject to 14-day incubation in an osteogenic medium (Invitrogen, Cal, USA), with the medium being changed at 2-day intervals. Subsequently, 70% ethanol was added for cell fixation, followed by the identification of cell osteogenic differentiation potential based on their ALP activity and Alizarin Red S staining. In addition, the Image-Pro Plus 6.0 program (Media Cybernetics) was used for measuring mineralization.
4% formaldehyde was added to fix cells for 15 min, followed by a wash with PBS. Next, cell smears were prepared and subjected to 30-min pre-incubation using 10% normal goat serum (710027, KPL, MA, USA). Subsequently, we incubated cell smears using primary antibodies anti-Wnt5a and anti-ROR2 for 2 h, at 37°C, followed by incubation with the fluorescence-labeled rabbit IgG secondary antibody (021516, KPL, MA, USA). Finally, 4′,6-diamidino-2-phenylindole (DAPI) was added to the cell smears and incubated for 15 min to counterstain the nucleus. A confocal microscope (DM6000CFS, Leica) was employed for image acquisition, followed by digitization using the LAS AF software.
We incubated the cell lysates using a specific antibody at 4°C overnight, followed by another protein A-agarose incubation for 1 h at 4°C. The resultant protein A-agarose-antigen-antibody complex was harvested by centrifugation at 12000 g for 2 min at 4°C, and washed with the immunoprecipitation-HAT buffer (each wash was performed for 20 min using 1 mL of the buffer) five times under 4°C. Subsequently, SDS-PAGE separation was carried out to resolve bound proteins, and western blot analysis was conducted using specific antibodies.
Results were depicted as mean ± standard error of mean (SEM). All assays were conducted in triplicate. Student’s t-test was adopted to analyze the differences in the averages between the two groups. The threshold of statistical significance for the differences was p < 0.05.
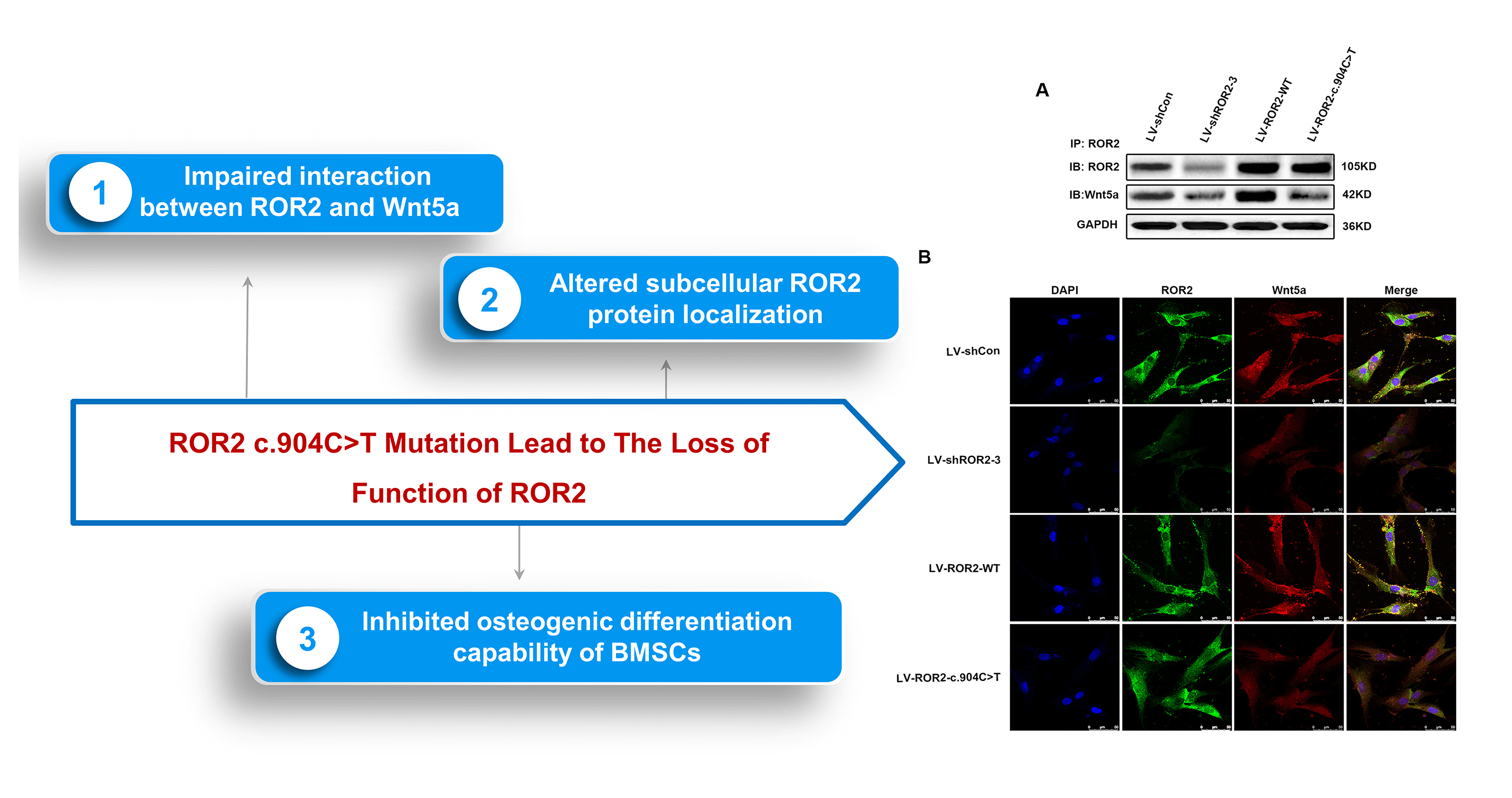
The ROR2: c.904C>T mutation led to the loss of function of ROR2 and subsequent impaired of ROR2-Wnt5a interaction
ROR2, the receptor of Wnt5a, has a critical effect on regulation via the activation of the non-canonical Wnt/β-catenin pathway (Dave et al., 2019; Yu et al., 2016).According to the results of the present study, mutant ROR2 c.904C>T was retained in the endoplasmic reticulum (ER), while wild-type ROR2 was exported to the plasma membrane (Fig. 1B). The co-immunoprecipitation analysis revealed that the ROR2: c.904C>T mutation impaired the interaction between ROR2 and Wnt5a (Fig. 1A). The in vitro findings indicated that the ROR2 c.904C>T mutation altered subcellular ROR2 protein localization which consequently induced the loss of function of ROR2, thereby affecting the interaction between ROR2 and Wnt5a.
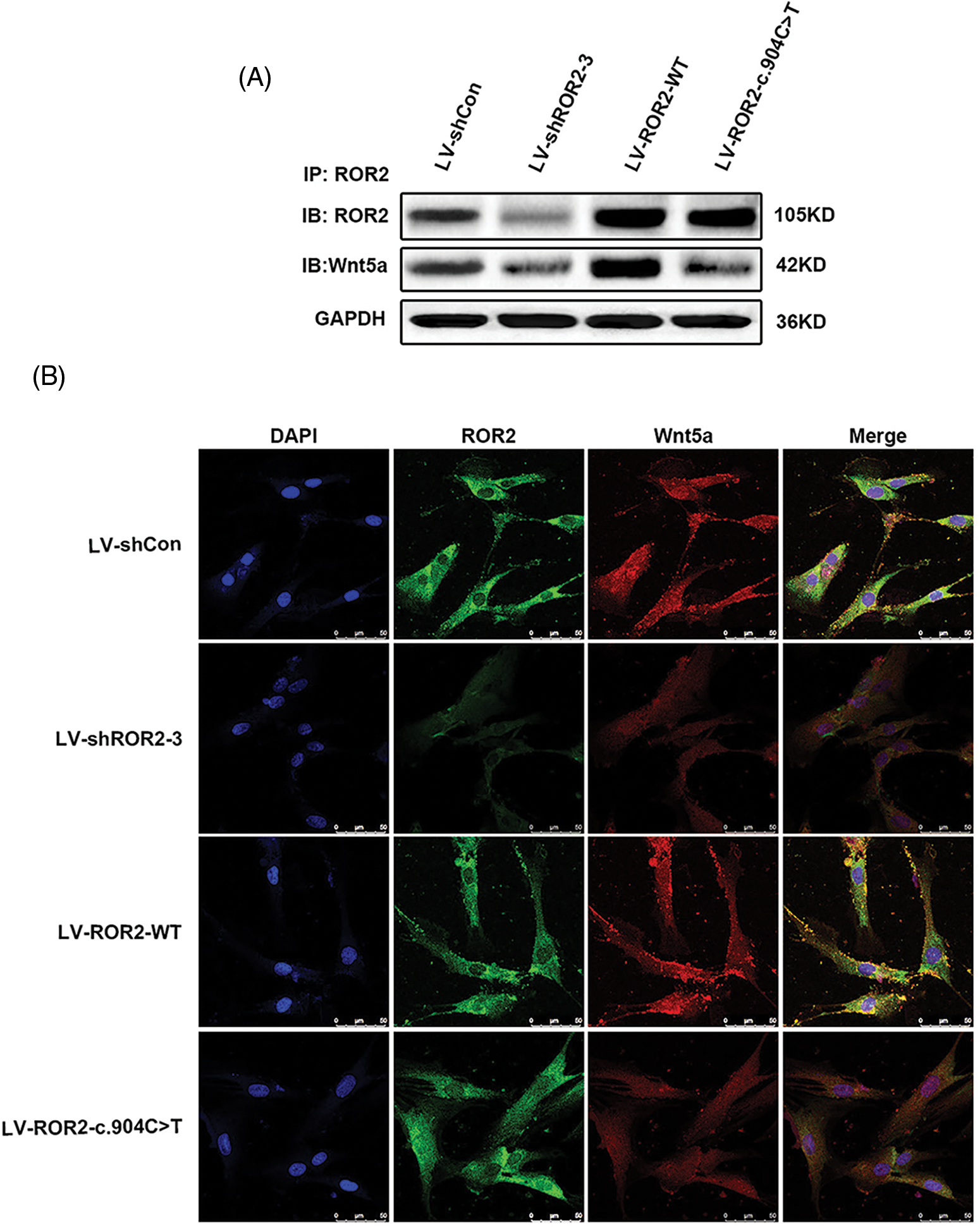
Figure 1: The ROR2: c.904C>T mutation led to a loss of function of receptor tyrosine kinase-like orphan receptor 2 (ROR2), which resulted in an impaired interaction between Wnt5a and ROR2. Coimmunoprecipitation (CoIP)–Western blotting was performed for detecting the interaction between ROR2 and Wnt5a (A). Double-Immunoflorescence (IF) staining was carried out for determining expressions and ROR2 and Wnt5a co-localization in BMSCs. Scale bar = 50 µm (B).
The loss of function of ROR2 led to an inhibited cell osteogenic differentiation capability
For exploring the effect of ROR2 on osteogenic differentiation, we evaluated the osteogenic capacity following osteogenic induction in ROR2-depleted BMSCs transfected with LV-shROR2 and LV-ROR2-c.904C>T. Based on quantitative RT-PCR as well as WB assays, ROR2 expression was downregulated in the ROR2-depleted BMSCs (Figs. 2A and 2B). Furthermore, , ROR2 depletion inhibited the Alizarin Red S accumulation in the Alizarin Red S staining assay and ALP staining in ALP activity assays (Figs. 2C and 2D). These results demonstrated that the loss of function of ROR2 led to an inhibited osteogenic differentiation capability of BMSCs.
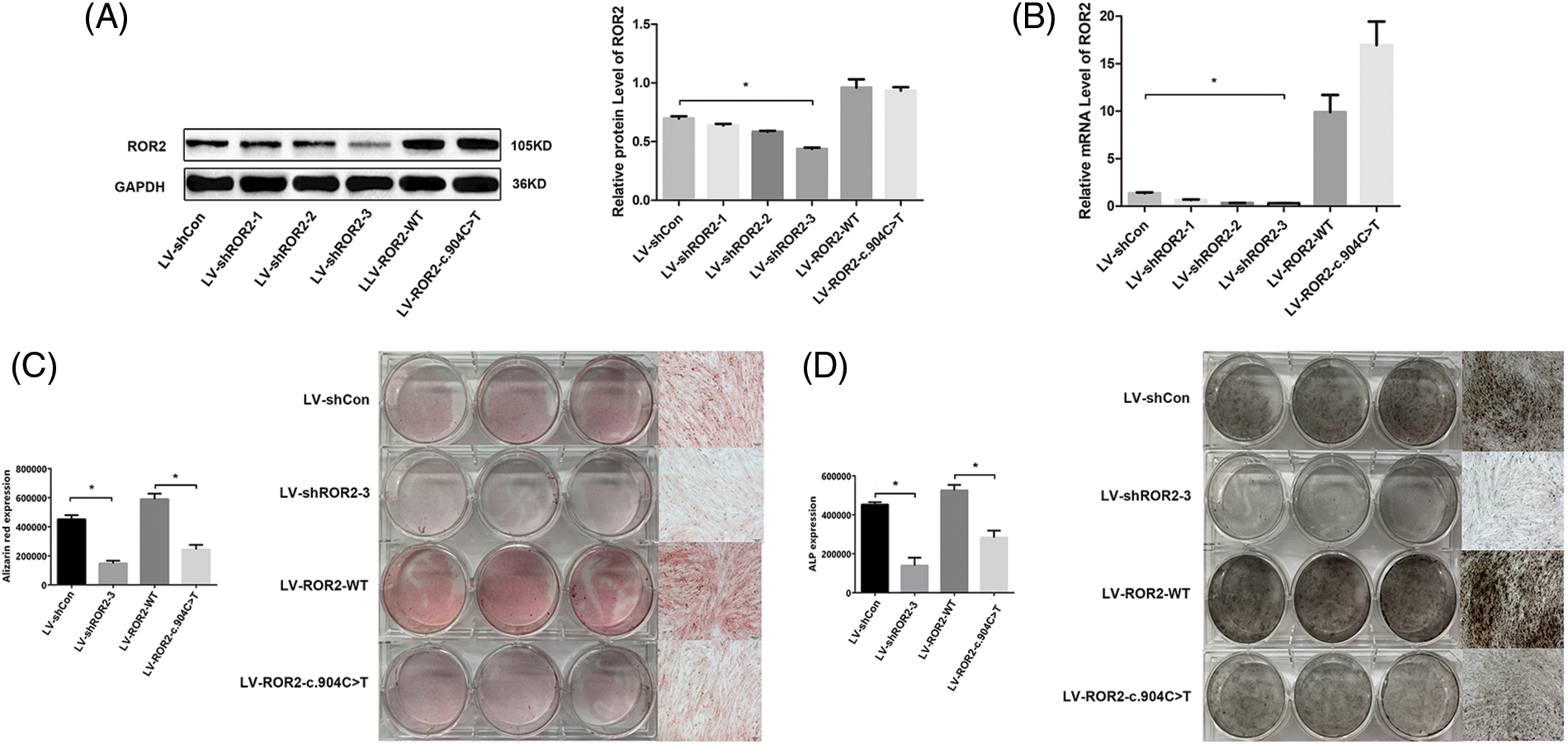
Figure 2: The loss of function of receptor tyrosine kinase-like orphan receptor 2 (ROR2) led to a lowered osteogenic differentiation capability of bone marrow mesenchymal stem cells (BMSCs). We conducted WB assays for detecting ROR2 protein levels in the BMSCs subjected to LV-shCon, LV-shROR2, LV-ROR2-WT, and LV-ROR2-c.904C>T transfections (A). We carried out RT-qPCR for identifying ROR2 mRNA levels in BMSCs subjected to LV-shCon, LV-shROR2, LV-ROR2-WT, and LV-ROR2-c.904C>T transfections (B). Alizarin Red S staining together with quantification of BMSC osteogenesis were performed after transfection of the cells with LV-shCon, LV-shROR2, LV-ROR2-WT, and LV-ROR2-c.904C>T (C). The alkaline phosphatase (ALP) activity determination and the quantitative measurements of the BMSC osteogenesis were performed after transfection with LV-shCon, LV-shROR2, LV-ROR2-WT, and LV-ROR2-c.904C>T (D). All data are shown as the Mean ± SEM (n = 3). In each analysis, *p < 0.05 relative to the corresponding controls was considered the threshold of significance.
Sphingomyelin was related to BMSCs osteoblastic differentiation mediated via ROR2
Previous studies have verified that SM belongs to the sphingolipid signaling pathway (Bienias et al., 2016). SM production and metabolism involve several reactions and enzymes (Brodowicz et al., 2018). Our prior work documented that ROR2 downregulation facilitated DPSC aging by promoting SM biosynthesis (Dong et al., 2021). However, whether SM is related to cell osteogenic differentiation remains unclear. SMS1 is a key enzyme that generates SM (Dong et al., 2021). According to our RT-PCR analysis, SMS1 expression was downregulated in ROR2-depleted BMSCs (Fig. 3A). In order to determine whether SM is related to ROR2-mediated osteogenic differentiation of BMSCs, SM was supplemented to the osteogenic medium, and the cells were evaluated for osteogenic markers. The canonical Smad-dependent pathway and the MAPK pathway have been shown to be inextricably linked with the osteogenesis process (Tang et al., 2018; Zhang et al., 2019). Smad1/5/8 phosphorylation levels together with associated proteins involved in the MAPK pathway were, therefore, examined. The depletion of ROR2 was observed to inhibit the phosphorylation of P38, Smad1/5/8, and ERK1/2. Moreover, SM treatment could reverse the downregulation of P-P38, P-Smad1/5/8, and P-ERK1/2 in the ROR2-depleted BMSCs (Figs. 4A and 4B). RUNX2 has a critical effect on stem cell osteogenic differentiation, which can directly stimulate the transcription of downstream targets that encode OCN and OPN (Huang et al., 2014). In the present study, RT-PCR and WB assays revealed that the expression of OPN, OCN, and RUNX2 was obviously lowered in ROR2-depleted BMSCs. The SM treatment could reverse the downregulation of OCN, OPN, and RUNX2 expression (Figs. 3B–3D; Figs. 4A and 4B). Consequently, it was inferred that SM positively regulated osteoblast differentiation. Based on these results, SM was related to ROR2-mediated BMSC osteoblastic differentiation. Hence, SM supplementation could maintain the osteoblastic differentiation of BMSCs.
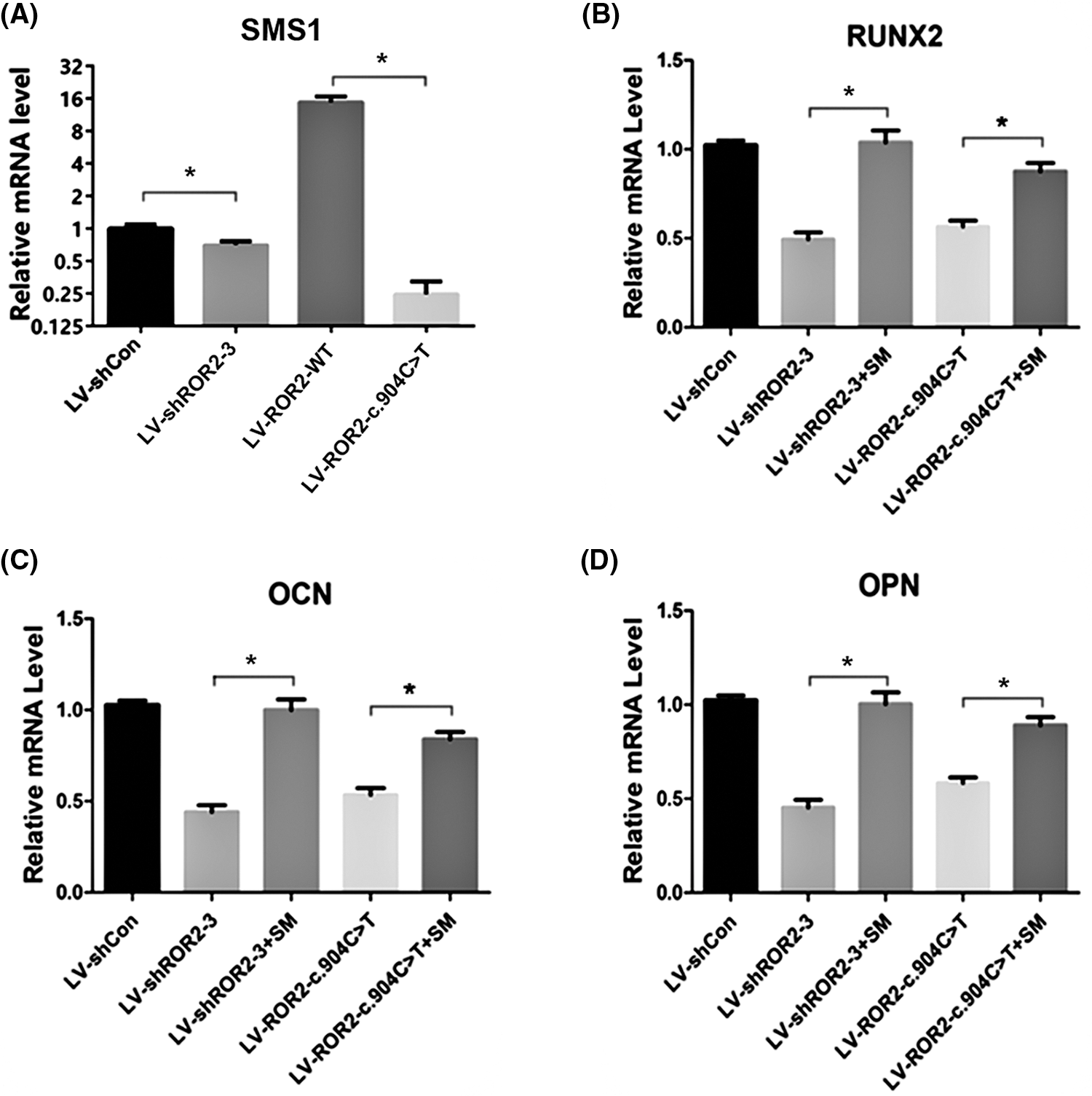
Figure 3: The quantitative reverse transcription polymerase chain reaction (RT-qPCR) confirmed that sphingomyelin (SM) was related to receptor tyrosine kinase-like orphan receptor 2 (ROR2)-mediated bone marrow mesenchymal stem cells (BMSC) osteoblastic differentiation. RT-qPCR was done for determining sphingomyelin synthase 1 (SMS1) mRNA levels in BMSCs subjected to LV-shCon, LV-shROR2, LV-ROR2-WT, and LV-ROR2-c.904C>T transfection (A). RUNX2, OCN, and OPN mRNA expression in BMSCs subjected to LV-shROR2, LV-ROR2-WT, LV-ROR2-c.904C>T, and lipo-SM (lipofectamine-SM) transfection was analyzed by RT-qPCR (B–D). Results are represented as the Mean ± SEM (n = 3). *p < 0.05 vs. controls was considered the threshold of significance.
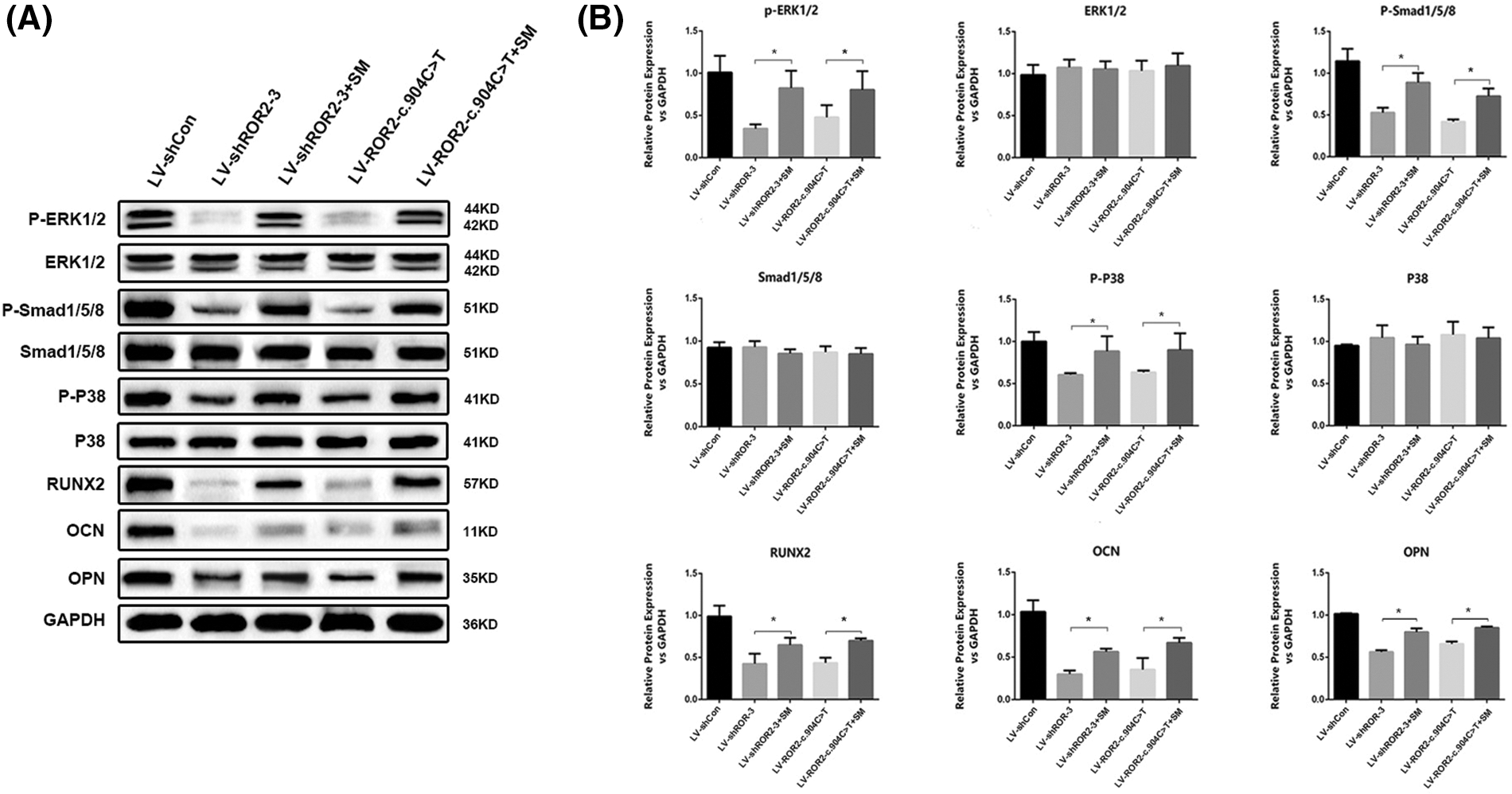
Figure 4: Western blotting confirmed that SM was related to receptor tyrosine kinase-like orphan receptor 2 (ROR2)-mediated bone marrow mesenchymal stem cells (BMSC) osteoblastic differentiation. WB assays were done for verifying ERK, P-ERK, Smad1/5/8, P-Smad1/5/8, P-P38, P38, RUNX2, OCN, and OPN protein expression in the BMSCs transfected with LV-shROR2, LV-ROR2-WT, LV-ROR2-c.904C>T, and lipo-SM (A–B). All data are shown as the Mean ± SEM (n = 3). In all analyses, *p < 0.05 vs. controls was considered to be the threshold of significance.
ROR2 is an essential gene for bone development. Nonetheless, the molecular mechanism related to the role of ROR2 in regulating osteogenic differentiation remains largely unknown so far. Intending to shed more light on cell mechanisms underlying ROR2-regulated, this study analyzed how decreased ROR2 levels affected in vitro osteogenesis. It was revealed that the ROR2 c.904C>T mutation resulted in the loss of function of ROR2, resulting in an impaired interaction between Wnt5a and ROR2. This consequently inhibited the osteogenic differentiation of BMSCs. Furthermore, SM participated in the ROR2-mediated osteoblastic differentiation of BMSCs. Indeed, SM supplementation could maintain the osteoblastic differentiation of BMSCs.
ROR2 was previously suggested to have a significant effect on cell polarity, migration, growth, aging, and differentiation (Debebe and Rathmell, 2015). ROR2 reportedly promotes osteoblast differentiation and enhances bone formation (Awais et al., 2020). Our prior work suggested that the novel variation c.904C>T of the ROR2 gene, which caused ARRS expanded the disease mutation spectrum (Dong et al., 2021). Nonetheless, its associated mechanism in regulating ROR2 during osteogenesis differentiation remained unknown. In the present study, according to functional assays conducted in vitro, the ROR2 c.904C>T mutation could alter ROR2 protein subcellular localization. This protein with the ROR2 c.904C>T mutation was retained in the ER and was not transported to the plasma membrane, which presumably led to its loss of function. Wnt5a, the non-canonical Wnt agonist, can promote human osteoblastogenesis via the co-receptor ROR2 (Bolzoni et al., 2013). Evidence suggests that the ROR2 loss-of-function would probably reduce limb deformity and Wnt signal transduction (Wang et al., 2011). The present study demonstrated that the ROR2 c.904C>T mutation resulted in an impaired interaction between ROR2 and Wnt5a, thereby restraining BMSC osteogenic differentiation capability. It is hence suggested that ROR2 c.904C>T mutation led to its loss of function, and the mutated ROR2 protein was still in the ER because of misfolding and, therefore, did not reach its site of action, i.e., the plasma membrane. Accordingly, the impact of the ROR2 c.904C>T mutation on osteogenic differentiation might be consistent with the above mechanism.
SM is an integral component in different lipid membranes and is a critical second messenger in several pathways that affects cell proliferation and differentiation (Lahiri and Futerman, 2007). Previous studies by our research group have demonstrated that decreased ROR2 expression enhances DPSCs aging by inhibiting the STK4-FOXO1/SMS1 axis during SM biosynthesis (He et al., 2022). According to our RT-qPCR analysis, the SMS1 level decreased in ROR2-depleted BMSCs. SM is recognized to be the growth-regulating molecule that regulates tissue homeostasis as well as skeletal development (Matsumoto et al., 2019). These findings raised a query whether SM could be engaged in ROR2-regulated BMSCs osteogenic differentiation. There are different transcriptional factors regulating osteogenic differentiation. Several studies have also reported the involvement of MAPK pathways, such as the P38 and ERK pathways during osteoblastogenesis (Kim et al., 2015; Zhu et al., 2018). The constitutively active form of ERK could reportedly activate in vivo and in vitro osteoblast differentiation (Ge et al., 2007; Lai et al., 2001), while P38 was reported to be involved in the Smad-1 phosphorylation, thereby activating osteoblasts (Nöth et al., 2003). RUNX2 was shown to be expressed in preosteoblasts, which has vital effects on preosteoblast differentiation into immature osteoblasts (Komori, 2020). OPN and OCN are formed in osteogenesis as well as the late stage of the mineralization process and are known to exert synergistic effects on detecting bone shape and size (Bailey et al., 2017). In the present study, the expressions of P-ERK, P-Smad1/5/8, P38, RUNX2, OCN, and OPN were observed to be significantly decreased in ROR2-depleted BMSCs. This suggested that ROR2 had a critical effect on BMSC osteogenic differentiation. Furthermore, the treatment with SM rescued impaired osteogenic differentiation in ROR2-depleted BMSCs, and SM supplementation could maintain BMSC osteogenic differentiation. Further, research also suggested that ROR2 might facilitate osteogenic differentiation through STAT3 (Lei et al., 2020). That means there is no single mechanism through which ROR2 works. There must be multiple pathways through which ROR2 facilitates osteogenic differentiation and SM is not a lone potential target for ARRS therapy. Therefore, whether ROR2 interacted directly with SM in BMSC osteogenesis warrants further research.
Collectively, the findings of the current work revealed that the ROR2: c.904C>T variant could cause the loss of function of ROR2, which resulted in an impaired interaction between ROR2 and Wnt5a, and inhibited the osteogenic differentiation capability of BMSCs. Moreover, SM was engaged in osteoblastic differentiation of BMSCs regulated by ROR2, which renders SM a potential target in the therapy for ARRS.
Funding Statement: The present study was funded by the Project Funded by China Postdoctoral Science Foundation (No. 2022T150445), the Beijing Hospitals Authority Youth Programme (No. QML20211401), the Young Talent Foundation of PLA General Hospital (2019-YQPY-002), and Beijing Nova Program (Z201100006820057).
Author Contributions: The authors confirm their contribution to the paper as follows: Wenqi Chen contributed to the primary manuscript drafting. Xiaoyang Chu and Yang Zeng were responsible for database organization. Yousheng Yan and Yipeng Wang performed data curation and visualization. Donglan Sun wrote sections of the manuscript. Dongliang Zhang, Jing Zhang, and Kai Yang were in charge of the study conception and design. The authors assisted in revising, reading the manuscript, and approving the version for submission.
Availability of Data and Materials: The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
Awais S, Balouch SS, Riaz N, Choudhery MS (2020). Human dental pulp stem cells exhibit osteogenic differentiation potential. Open Life Sciences 15: 229–236. https://doi.org/10.1515/biol-2020-0023 [Google Scholar] [PubMed] [CrossRef]
Bailey S, Karsenty G, Gundberg C, Vashishth D (2017). Osteocalcin and osteopontin influence bone morphology and mechanical properties. Annals of the New York Academy of Sciences 1409: 79–84. https://doi.org/10.1111/nyas.13470 [Google Scholar] [PubMed] [CrossRef]
Bienias K, Fiedorowicz A, Sadowska A, Prokopiuk S, Car H (2016). Regulation of sphingomyelin metabolism. Pharmacological Reports 68: 570–581. https://doi.org/10.1016/j.pharep.2015.12.008 [Google Scholar] [PubMed] [CrossRef]
Bispo DSC, Michálková L, Correia M, Jesus CSH, Duarte IF, Goodfellow BJ, Oliveira MB, Mano JF, Gil AM (2022). Endo- and exometabolome crosstalk in mesenchymal stem cells undergoing osteogenic differentiation. Cells 11: 1257. https://doi.org/10.3390/cells11081257 [Google Scholar] [PubMed] [CrossRef]
Bolzoni M, Donofrio G, Storti P, Guasco D, Toscani D et al. (2013). Myeloma cells inhibit non-canonical wnt co-receptor ror2 expression in human bone marrow osteoprogenitor cells: Effect of wnt5a/ror2 pathway activation on the osteogenic differentiation impairment induced by myeloma cells. Leukemia 27: 451–463. https://doi.org/10.1038/leu.2012.190 [Google Scholar] [PubMed] [CrossRef]
Brodowicz J, Przegaliński E, Müller CP, Filip M (2018). Ceramide and its related neurochemical networks as targets for some brain disorder therapies. Neurotoxicity Research 33: 474–484. https://doi.org/10.1007/s12640-017-9798-6 [Google Scholar] [PubMed] [CrossRef]
Chen Y, Bellamy WP, Seabra MC, Field MC, Ali BR (2005). ER-associated protein degradation is a common mechanism underpinning numerous monogenic diseases including Robinow syndrome. Human Molecular Genetics 14: 2559–2569. https://doi.org/10.1093/hmg/ddi259 [Google Scholar] [PubMed] [CrossRef]
Dave H, Butcher D, Anver M, Bollard CM (2019). ROR1 and ROR2-novel targets for neuroblastoma. Pediatric Hematology and Oncology 36: 352–364. https://doi.org/10.1080/08880018.2019.1646365 [Google Scholar] [PubMed] [CrossRef]
Debebe Z, Rathmell WK (2015). Ror2 as a therapeutic target in cancer. Pharmacology & Therapeutics 150: 143–148. https://doi.org/10.1016/j.pharmthera.2015.01.010 [Google Scholar] [PubMed] [CrossRef]
DeChiara TM, Kimble RB, Poueymirou WT, Rojas J, Masiakowski P, Valenzuela DM, Yancopoulos GD (2000). Ror2, encoding a receptor-like tyrosine kinase, is required for cartilage and growth plate development. Nature Genetics 24: 271–274. https://doi.org/10.1038/73488 [Google Scholar] [PubMed] [CrossRef]
Dong XY, Huang YX, Yang Z, Chu XY, Wu J et al. (2021). Downregulation of ROR2 promotes dental pulp stem cell senescence by inhibiting STK4-FOXO1/SMS1 axis in sphingomyelin biosynthesis. Aging Cell 20: e13430. https://doi.org/10.1111/acel.13430 [Google Scholar] [PubMed] [CrossRef]
During A, Penel G, Hardouin P (2015). Understanding the local actions of lipids in bone physiology. Progress in Lipid Research 59: 126–146. https://doi.org/10.1016/j.plipres.2015.06.002 [Google Scholar] [PubMed] [CrossRef]
Ge C, Xiao G, Jiang D, Franceschi RT (2007). Critical role of the extracellular signal-regulated kinase-MAPK pathway in osteoblast differentiation and skeletal development. The Journal of Cell Biology 176: 709–718. https://doi.org/10.1083/jcb.200610046 [Google Scholar] [PubMed] [CrossRef]
Hasegawa D, Wada N, Yoshida S, Mitarai H, Arima M, Tomokiyo A, Hamano S, Sugii H, Maeda H (2018). Wnt5a suppresses osteoblastic differentiation of human periodontal ligament stem cell-like cells via Ror2/JNK signaling. Journal of Cellular Physiology 233: 1752–1762. https://doi.org/10.1002/jcp.26086 [Google Scholar] [PubMed] [CrossRef]
He X, Yang Z, Chu XY, Li YX, Zhu B et al. (2022). ROR2 downregulation activates the MSX2/NSUN2/p21 regulatory axis and promotes dental pulp stem cell senescence. Stem Cells 40: 290–302. https://doi.org/10.1093/stmcls/sxab024 [Google Scholar] [PubMed] [CrossRef]
Huang J, Shi Y, Li H, Tan D, Yang M, Wu X (2015). Knockdown of receptor tyrosine kinase-like orphan receptor 2 inhibits cell proliferation and colony formation in osteosarcoma cells by inducing arrest in cell cycle progression. Oncology Letters 10: 3705–3711. https://doi.org/10.3892/ol.2015.3797 [Google Scholar] [PubMed] [CrossRef]
Huang RL, Yuan Y, Tu J, Zou GM, Li Q (2014). Opposing TNF-α/IL-1β- and BMP-2-activated MAPK signaling pathways converge on Runx2 to regulate BMP-2-induced osteoblastic differentiation. Cell Death & Disease 5: e1187. https://doi.org/10.1038/cddis.2014.101 [Google Scholar] [PubMed] [CrossRef]
Ishida Y, Kiyokawa Y, Asai T, Oku N (2016). Ameliorating effects of sphingomyelin-based liposomes on sarcopenia in senescence-accelerated mice. Biological & Pharmaceutical Bulletin 39: 786–793. https://doi.org/10.1248/bpb.b15-00915 [Google Scholar] [PubMed] [CrossRef]
Kaissi AA, Kenis V, Shboul M, Grill F, Ganger R, Kircher SG (2020). Tomographic study of the malformation complex in correlation with the genotype in patients with robinow syndrome: Review article. Journal of Investigative Medicine High Impact Case Reports 8: 2324709620911771. https://doi.org/10.1177/2324709620911771 [Google Scholar] [PubMed] [CrossRef]
Kim BS, Kang HJ, Park JY, Lee J (2015). Fucoidan promotes osteoblast differentiation via JNK- and ERK-dependent BMP2-Smad 1/5/8 signaling in human mesenchymal stem cells. Experimental & Molecular Medicine 47: e128. https://doi.org/10.1038/emm.2014.95 [Google Scholar] [PubMed] [CrossRef]
Komori T (2020). Molecular mechanism of Runx2-dependent bone development. Molecules and Cells 43: 168–175. https://doi.org/10.14348/molcells.2019.0244 [Google Scholar] [PubMed] [CrossRef]
Lahiri S, Futerman AH (2007). The metabolism and function of sphingolipids and glycosphingolipids. Cellular and Molecular Life Sciences 64: 2270–2284. https://doi.org/10.1007/s00018-007-7076-0 [Google Scholar] [PubMed] [CrossRef]
Lai CF, Chaudhary L, Fausto A, Halstead LR, Ory DS, Avioli LV, Cheng SL (2001). Erk is essential for growth, differentiation, integrin expression, and cell function in human osteoblastic cells. The Journal of Biological Chemistry 276: 14443–14450. https://doi.org/10.1074/jbc.M010021200 [Google Scholar] [PubMed] [CrossRef]
Lee YC, Chan YH, Hsieh SC, Lew WZ, Feng SW (2019). Comparing the osteogenic potentials and bone regeneration capacities of bone marrow and dental pulp mesenchymal stem cells in a rabbit calvarial bone defect model. International Journal of Molecular Sciences 20: 5015. https://doi.org/10.3390/ijms20205015 [Google Scholar] [PubMed] [CrossRef]
Lei L, Huang Z, Feng J, Huang Z, Tao Y, Hu X, Zhang X (2020). Loss of receptor tyrosine kinase-like orphan receptor 2 impairs the osteogenesis of mBMSCs by inhibiting signal transducer and activator of transcription 3. Stem Cell Research & Therapy 11: 137. https://doi.org/10.1186/s13287-020-01646-2 [Google Scholar] [PubMed] [CrossRef]
Ma Y, Jing J, Feng J, Yuan Y, Wen Q, et al. (2021). Ror2-mediated non-canonical Wnt signaling regulates Cdc42 and cell proliferation during tooth root development. Development 148: 1. https://doi.org/10.1242/dev.196360 [Google Scholar] [PubMed] [CrossRef]
Mahmood T, Yang PC (2012). Western blot: Technique, theory, and trouble shooting. North American Journal of Medical Sciences 4: 429–434. https://doi.org/10.4103/1947-2714.100998 [Google Scholar] [PubMed] [CrossRef]
Matsumoto G, Hashizume C, Watanabe K, Taniguchi M, Okazaki T (2019). Deficiency of sphingomyelin synthase 1 but not sphingomyelin synthase 2 reduces bone formation due to impaired osteoblast differentiation. Molecular Medicine 25: 56. https://doi.org/10.1186/s10020-019-0123-0 [Google Scholar] [PubMed] [CrossRef]
Michigami T (2019). Wnt signaling and skeletal dysplasias. Clinical Calcium 29: 323–328. https://doi.org/10.20837/4201903323 [Google Scholar] [PubMed] [CrossRef]
Nöth U, Tuli R, Seghatoleslami R, Howard M, Shah A, Hall DJ, Hickok NJ, Tuan RS (2003). Activation of p38 and Smads mediates BMP-2 effects on human trabecular bone-derived osteoblasts. Experimental Cell Research 291: 201–211. https://doi.org/10.1016/S0014-4827(03)00386-0 [Google Scholar] [PubMed] [CrossRef]
Schwabe GC, Trepczik B, Süring K, Brieske N, Tucker AS, Sharpe PT, Minami Y, Mundlos S (2004). Ror2 knockout mouse as a model for the developmental pathology of autosomal recessive Robinow syndrome. Developmental Dynamics 229: 400–410. https://doi.org/10.1002/dvdy.10466 [Google Scholar] [PubMed] [CrossRef]
Sun Y, Yang Z, Zheng B, Zhang XH, Zhang ML, Zhao XS, Zhao HY, Suzuki T, Wen JK (2017). A novel regulatory mechanism of smooth muscle α-Actin expression by NRG-1/circACTA2/miR-548f-5p axis. Circulation Research 121: 628–635. https://doi.org/10.1161/circresaha.117.311441 [Google Scholar] [PubMed] [CrossRef]
Tang Q, Tong M, Zheng G, Shen L, Shang P, Liu H (2018). Masquelet’s induced membrane promotes the osteogenic differentiation of bone marrow mesenchymal stem cells by activating the Smad and MAPK pathways. American Journal of Translational Research 10: 1211–1219. [Google Scholar] [PubMed]
Taniguchi M, Okazaki T (2021). Role of ceramide/sphingomyelin (SM) balance regulated through “SM cycle” in cancer. Cellular Signalling 87: 110119. https://doi.org/10.1016/j.cellsig.2021.110119 [Google Scholar] [PubMed] [CrossRef]
van Bokhoven H, Celli J, Kayserili H, van Beusekom E, Balci S et al. (2000). Mutation of the gene encoding the ROR2 tyrosine kinase causes autosomal recessive Robinow syndrome. Nature Genetics 25: 423–426. https://doi.org/10.1038/78113 [Google Scholar] [PubMed] [CrossRef]
Villani M, Subathra M, Im YB, Choi Y, Signorelli P, Del Poeta M, Luberto C (2008). Sphingomyelin synthases regulate production of diacylglycerol at the Golgi. The Biochemical Journal 414: 31–41. https://doi.org/10.1042/BJ20071240 [Google Scholar] [PubMed] [CrossRef]
Wang C, Ming B, Wu X, Wu T, Cai S et al. (2019). Sphingomyelin synthase 1 enhances BCR signaling to promote lupus-like autoimmune response. eBioMedicine 45: 578–587. https://doi.org/10.1016/j.ebiom.2019.06.038 [Google Scholar] [PubMed] [CrossRef]
Wang B, Sinha T, Jiao K, Serra R, Wang J (2011). Disruption of PCP signaling causes limb morphogenesis and skeletal defects and may underlie Robinow syndrome and brachydactyly type B. Human Molecular Genetics 20: 271–285. https://doi.org/10.1093/hmg/ddq462 [Google Scholar] [PubMed] [CrossRef]
Weissenböck M, Latham R, Nishita M, Wolff LI, Ho HH, Minami Y, Hartmann C (2019). Genetic interactions between Ror2 and Wnt9a, Ror1 and Wnt9a and Ror2 and Ror1: Phenotypic analysis of the limb skeleton and palate in compound mutants. Genes to Cells 24: 307–317. https://doi.org/10.1111/gtc.12676 [Google Scholar] [PubMed] [CrossRef]
Yang Z, Chen JS, Wen JK, Gao HT, Zheng B et al. (2017). Silencing of miR-193a-5p increases the chemosensitivity of prostate cancer cells to docetaxel. Journal of Experimental & Clinical Cancer Research 36: 178. https://doi.org/10.1186/s13046-017-0649-3 [Google Scholar] [PubMed] [CrossRef]
Yang Q, Lang C, Wu Z, Dai Y, He S et al. (2019). MAZ promotes prostate cancer bone metastasis through transcriptionally activating the KRas-dependent RalGEFs pathway. Journal of Experimental & Clinical Cancer Research 38: 391. https://doi.org/10.1186/s13046-019-1374-x [Google Scholar] [PubMed] [CrossRef]
Yang K, Zhu J, Tan Y, Sun X, Zhao H, Tang G, Zhang D, Qi H (2020). Whole-exome sequencing identified compound heterozygous variants in ROR2 gene in a fetus with Robinow syndrome. Journal of Clinical Laboratory Analysis 34: e23074. https://doi.org/10.1002/jcla.23074 [Google Scholar] [PubMed] [CrossRef]
Yeang C, Varshney S, Wang R, Zhang Y, Ye D, Jiang XC (2008). The domain responsible for sphingomyelin synthase (SMS) activity. Biochimica et Biophysica Acta 1781: 610–617. https://doi.org/10.1016/j.bbalip.2008.07.002 [Google Scholar] [PubMed] [CrossRef]
You M, Ai Z, Zeng J, Fu Y, Zhang L, Wu X (2022). Bone mesenchymal stem cells (BMSCs)-derived exosomal microRNA-21-5p regulates Kruppel-like factor 3 (KLF3) to promote osteoblast proliferation in vitro. Bioengineered 13: 11933–11944. https://doi.org/10.1080/21655979.2022.2067286 [Google Scholar] [PubMed] [CrossRef]
Yu J, Chen L, Cui B, Widhopf GFII, Shen Z et al. (2016). Wnt5a induces ROR1/ROR2 heterooligomerization to enhance leukemia chemotaxis and proliferation. The Journal of Clinical Investigation 126: 585–598. https://doi.org/10.1172/JCI83535 [Google Scholar] [PubMed] [CrossRef]
Zhang Y, Gu X, Li D, Cai L, Xu Q (2019). METTL3 regulates osteoblast differentiation and inflammatory response via Smad signaling and MAPK signaling. International Journal of Molecular Sciences 21: 199. https://doi.org/10.3390/ijms21010199 [Google Scholar] [PubMed] [CrossRef]
Zhu D, Deng X, Han XF, Sun XX, Pan TW, Zheng LP, Liu YQ (2018). Wedelolactone enhances osteoblastogenesis through ERK- and JNK-mediated BMP2 expression and Smad/1/5/8 phosphorylation. Molecules 23: 561. https://doi.org/10.3390/molecules23030561 [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools