 Open Access
Open Access
ARTICLE
Bioinformatic analysis of lncRNA-associated competing endogenous RNA regulatory networks in synovial tissue of temporomandibular joint osteoarthritis
1 School of Stomatology, Qingdao University, Qingdao, 266000, China
2 Department of Oral Surgery, Ninth People’s Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai Key Laboratory of Stomatology & Shanghai
Research Institute of Stomatology, National Clinical Research Center of Stomatology, Shanghai, 200011, China
* Corresponding Author: Dongmei He,
# Co-first authors
(This article belongs to the Special Issue: Bioinformatics Study of Diseases)
BIOCELL 2023, 47(6), 1293-1306. https://doi.org/10.32604/biocell.2023.028199
Received 04 December 2022; Accepted 15 February 2023; Issue published 19 May 2023
Abstract
Background: Temporomandibular joint osteoarthritis (TMJOA) is an end-stage disease that seriously affects the patients’ quality of life. Molecular insights in advancing our understanding of TMJOA are the need of the hour. Methods: We performed RNA high-throughput sequencing and bioinformatics analysis of differentially expressed (DE) long non-coding RNA (lncRNAs), microRNAs (miRNAs), and messenger RNA (mRNAs) in human synovial TMJOA tissues. Firstly, synovium samples of TMJOA patients and non-TMJOA controls were collected for highthroughput sequencing of lncRNAs, miRNAs, and mRNAs. We then performed biological function analysis of the top 100 mRNAs with more than 2-fold differential expression, and their upstream regulated miRNAs and lncRNAs were predicted separately. Intersections between predicted miRNAs/lncRNAs and differentially expressed miRNAs/ lncRNAs were determined, respectively. Regulatory networks among the selected lncRNAs, miRNAs, and mRNAs were constructed. Finally, Western Blotting and reverse transcription-quantitative polymerase chain reaction (RTqPCR) were used to explore the function of selected lncRNA and mRNAs. Results: Our analysis showed that the only upregulated lncRNAxist, was involved in intracellular protein degradation, cartilage matrix degradation, and osteoclast differentiation through four regulatory axes: miR-1271-5p/ctsb, miR-365a-3p/mmp3, miR-199a-3p/fos, and miR-27b-3p/ miR-1271-5p/grb2. RT-qPCR results showed that xist expression increased in both TMJOA synovial tissue and inflammatory-stimulated synovial cells. Inhibition of xist could promote the proliferation of synovial cells upon inflammatory stimulation, and xist could positively regulate the expression of GRB2. Targeting GRB2 could inhibit the expression of matrix degrading enzymes in synovial cells, thereby attenuating the inflammatory environment. Conclusion: Given the current lack of reports of lncRNA regulation in TMJOA synovitis, our results revealed that xist was significantly differentially expressed in human synovial TMJOA and could regulate the expression of GRB2, which may play a role in the progression of TMJOA.Keywords
Temporomandibular joint osteoarthritis (TMJOA) is an end-stage disease of temporomandibular joint disorder (TMD) characterized pathologically by synovial inflammation, cartilage degeneration, and subchondral bone remodeling. The clinical manifestations include chronic pain and masticatory difficulties that seriously affect a patient’s quality of life (Bush and Beier, 2013; Li et al., 2019). Although there are many studies on the etiology of TMJOA, its exact pathogenesis remains unclear (Zheng et al., 2018).
In recent years, studies on RNA sequencing have found that a variety of noncoding RNAs, including miRNA, lncRNA, and circRNA play an important role through competing endogenous RNA (ceRNA) networks in the development of osteoarthritis (OA) (Kostopoulou et al., 2015; Yin et al., 2017). CircRNAs and lncRNAs can act as endogenous miRNA molecular sponges and competitively inhibit miRNA regulation of target mRNAs (Memczak et al., 2013; Wang et al., 2016; Zheng et al., 2016; Wang et al., 2017; Xiong et al., 2018). Our previous study found that the expression of circRNAgcn1l1 was elevated in synovial TMJOA tissues and was able to regulate Tumor Necrosis Factor-alpha (TNF-a) secretion by synoviocytes through miRNA-330-3p, aggravating cartilage degeneration (Zhu et al., 2020). LncRNAs are another important group of miRNA sponges in addition to circRNAs and may be an important therapeutic target of TMJOA as an indispensable part of ceRNA regulation. For example, Fu et al. (2021) found that the lncRNA-h19/miR-124a/cdk2/mcp-1 axis could promote the proliferation of human rheumatoid arthritis (RA) synoviocytes and synovial inflammation, which may represent a new RA therapeutic strategy. Bi et al. (2019) concluded that inhibition of lncRNA picsar/miRNA-4701-5p could effectively inhibit the proliferation and invasion of synoviocytes as well as secretion of inflammatory factors in human RA. Thus, lncRNA picsar could be used as a new biomarker for RA. In addition, the lncRNA neat1/miRNA‑ 204‑5p/NF-κB axis and the lncRNA pint/miRNA-155-5p/socs1 axis have also been reported in human RA synovitis symptoms (Wang and Zhao, 2020; Xiao et al., 2021).
As there is a current lack of reports on lncRNA regulation in TMJOA synovitis, in this study, we performed high-throughput sequencing of lncRNAs, miRNAs, and mRNAs in human TMJOA synovial tissues. By constructing lncRNA-related ceRNA networks using bioinformatics analysis, we aimed to probe their potential role in the development of TMJOA.
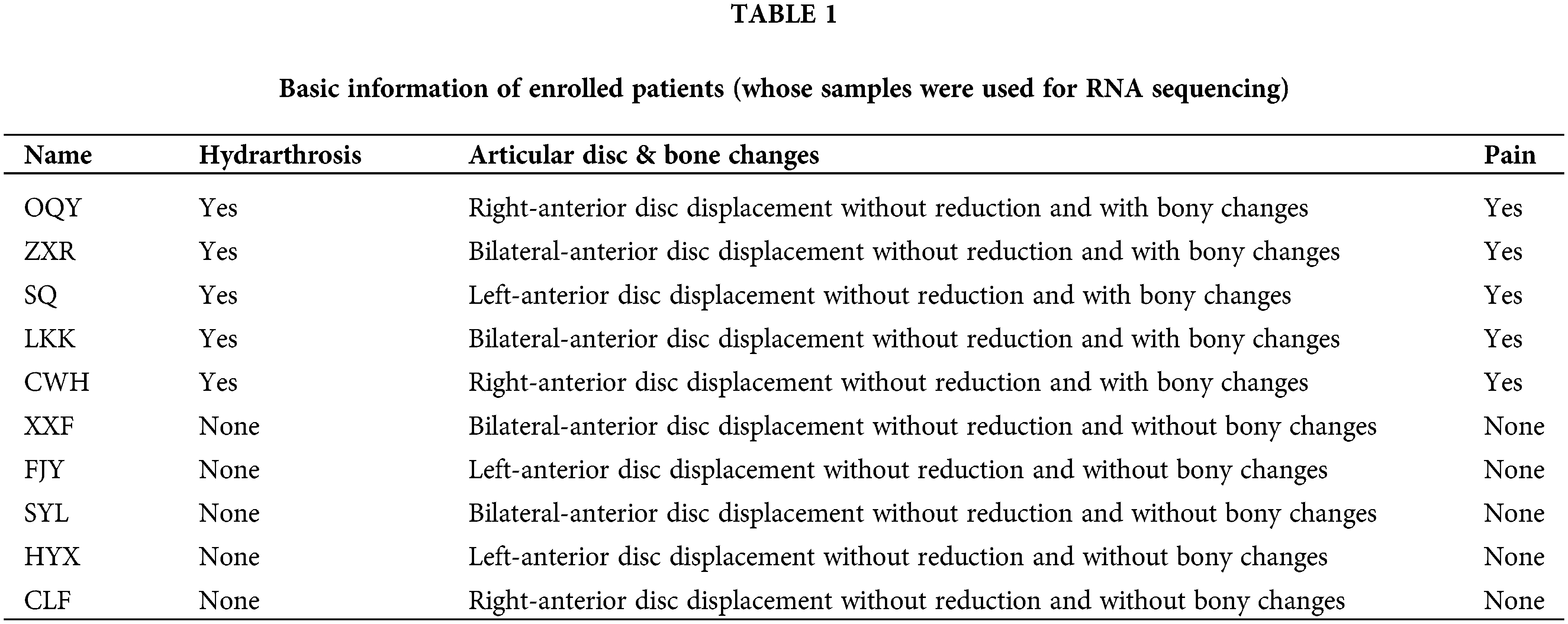
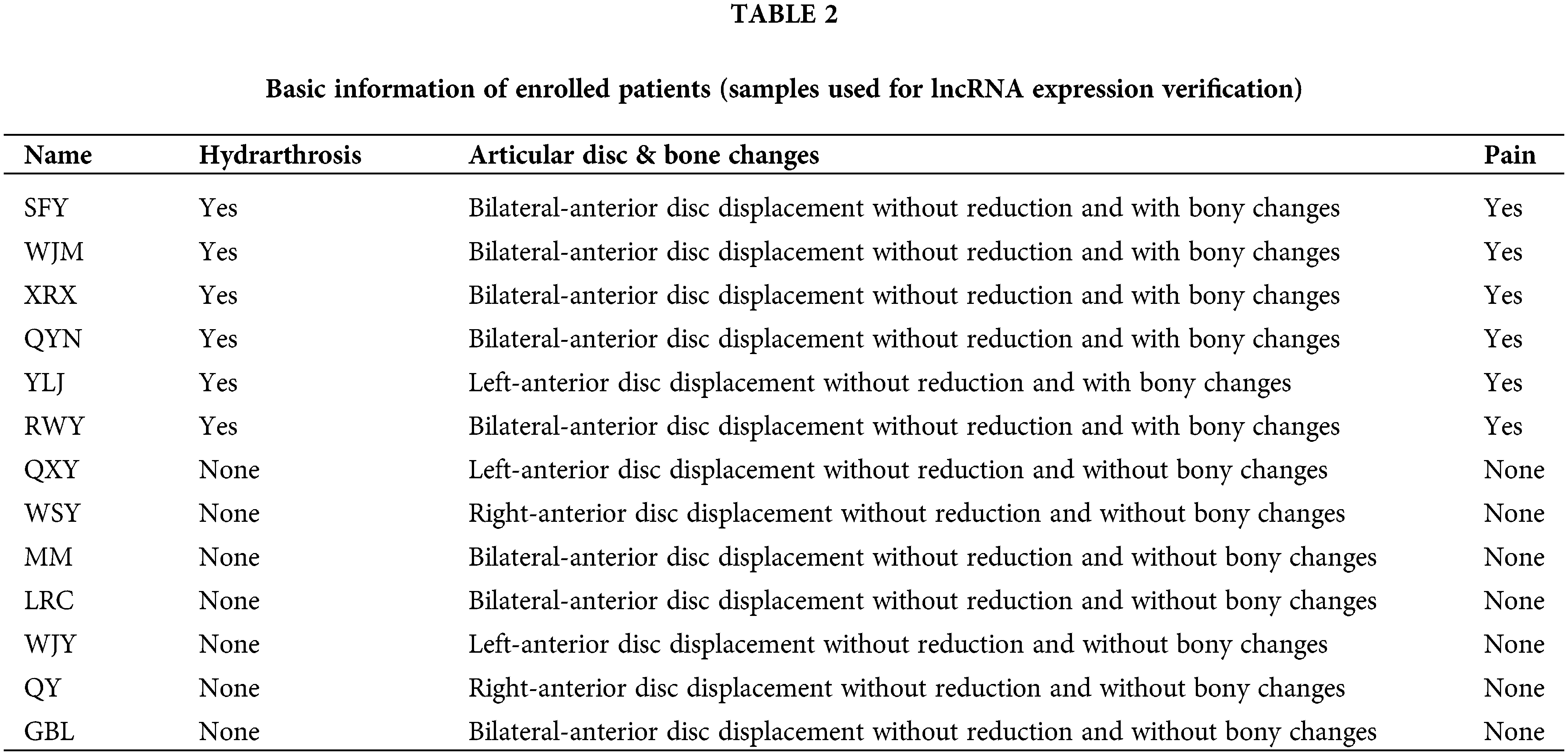
Patients with TMJOA were chosen as the experimental group. Since the synovium samples of healthy people cannot be collected due to ethical requirements, after patients were evaluated for clinical symptoms and imaging manifestations, we chose patients without any synovitis as controls. The inclusion criteria for the control group were as follows. (1) Patients with anterior disc displacement and in need of disc repositioning surgery; (2) Patients who had no clinical manifestations of pain; (3) Patients who had neither bony changes nor effusion. On the other hand, the inclusion criteria for the experimental group were as follows. (1) Patients with anterior disc displacement without reduction; (2) Patients who had clinical manifestations of pain; (3) Patients who had bony changes. Ten patients were enrolled for RNA seq (Table 1). Another 13 patients were enrolled for lncRNA expression verification (Table 2). During the collection operation, we cut the tissue in the posterior region of the articular disc, and carefully removed the surrounding muscle and fibrous layer. We then placed the sample in liquid nitrogen for preservation. The collection of all specimens was approved by the Ethics Committee of Shanghai Ninth People’s Hospital (Shanghai, China). The protocol number is SH9H-2021-T141-1.
Total RNA was extracted from five TMJOA tissues and five control tissues using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). All samples were tested for RNA concentration, purity, and integrity and qualified for RNA library construction. The RNA library was then sequenced by Cloud-seq Biotechnology (Shanghai, China).
Differential expression analysis and further screening of mRNAs
Differential expression (DE) analysis of lncRNAs, miRNAs, and mRNAs was performed by the DESeqDataSetFromMatrix function in the DESeq2 package (https://bioconductor.org/packages/release). DE RNAs with absolute log2FoldChange (FC) > 1 and p-value < 0.05 were selected using the dplyr: filter function in the tidyverse package (https://bioconductor.org/packages/release). Then all DE mRNAs were ranked and assigned according to log2FC, and the top 100 mRNAs were selected. TBtools (https://github.com/CJ-Chen/TBtools/releases) was used for differential gene heatmaps.
Pathway and functional analysis of differentially expressed mRNAs
Pathway and functional analysis of the top 100 ranked mRNAs was performed using the clusterProfiler package (https://bioconductor.org/packages/release). For the Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis with the help of the enrichKEGG function and the enrichGO function for Gene Ontology (GO) analysis, pathways with p-value < 0.05 and gene ratio > 0.05 were selected. The bubble diagram and histogram were drawn with the Rgraphviz package (https://bioconductor.org/packages/release). For a more comprehensive analysis of all DE mRNAs, this study also utilized a gene set enrichment analysis (GSEA) to analyze all DE mRNAs with a p-value < 0.05 (without setting a threshold for fold change). After screening the pathways of interest, KEGG mapping tools (https://www.kegg.jp/) were used to draw the pathway diagram. The cytoHubba function in Cytoscape software (https://cytoscape.org/) was used to analyze the top 10 hub genes.
miRNA and lncRNA screening and ceRNA regulatory network construction
DE lncRNAs with fragments per kilobase of exon model per million mapped fragments (FPKM) values greater than 0 in the control and experimental groups were selected. Among them, intergenic lncRNAs (lincRNAs) with sequences that did not overlap with any known gene exons were used for further analysis. Upstream regulatory miRNAs capable of regulating the top 100 mRNAs were predicted using the FunRich software (http://www.funrich.org/), and the prediction results were intersected with DE miRNAs. The starBase database was used to predict upstream lncRNAs of selected miRNAs. Then, the intersection between predicted lncRNAs and the previously screened DE lincRNAs was determined. In this way, we constructed the complete lncRNA-miRNA-mRNA network in the range of DE significant RNAs (Fig. 1).
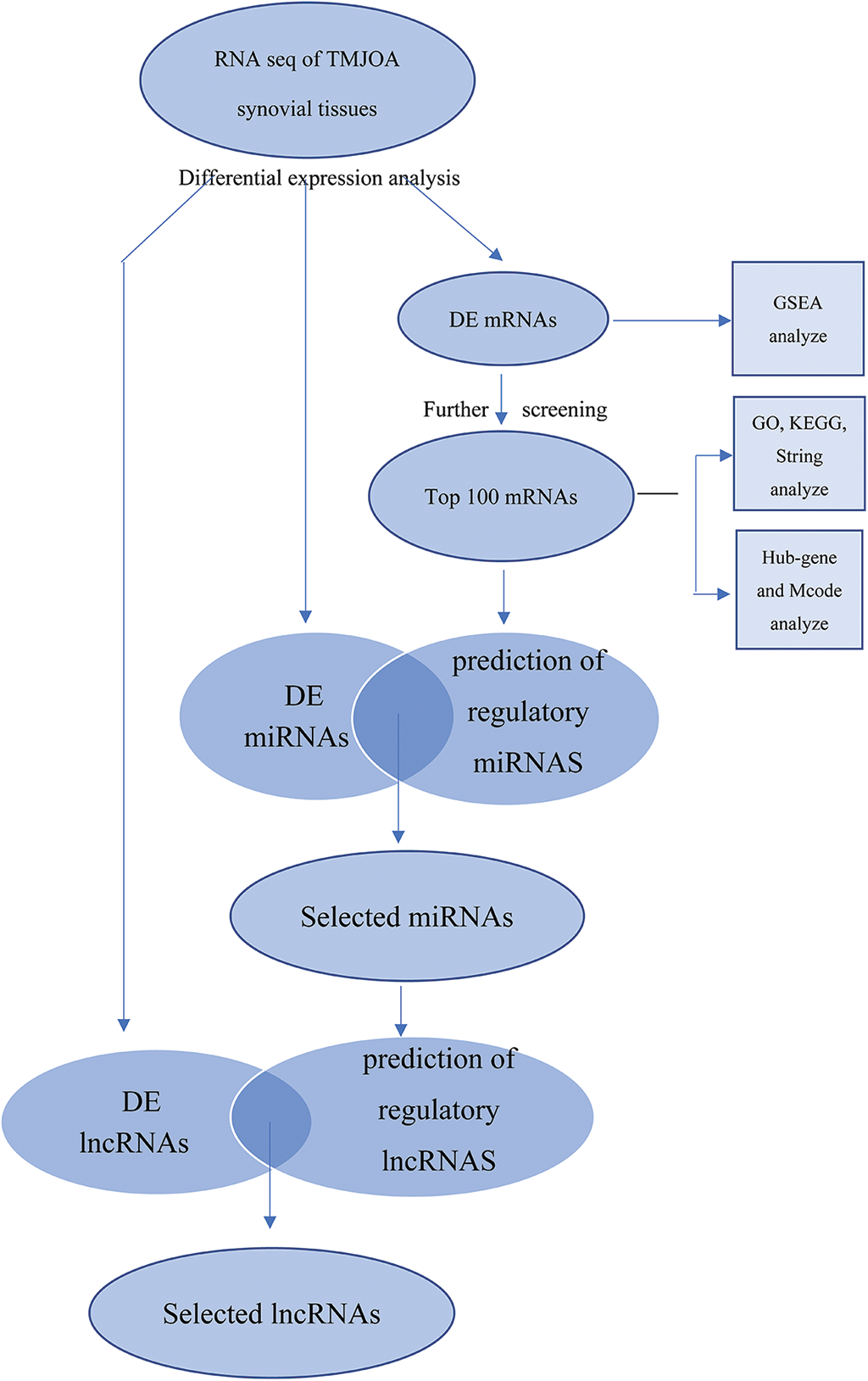
Figure 1: Schematic of this study.
Preliminary validation of significant lncRNAs
SW982 cells were cultured in Leibovitz’s L-15 medium supplemented with 10% fetal bovine serum (FBS). When the cell confluence reached 70%, TNF-a was added to the culture medium in a 5, 10, and 20 ng/ml concentration gradient. The cells were then cultured for another 48 h.
Specific xist antisense oligonucleotide (ASO) and grb2 small interfering RNA (si-RNA) was designed by Genomeditech (Shanghai, China) and transfected into SW982 cells using Lipofectamine 3000 transfection reagent (Thermo Fisher Scientific, Waltham, MA, USA). The primer sequences of xist were as follows:
Forward primer: ACCACACGTCAAGCTCTTCAT.
Reverse primer: CCTCAAGTGCTAGAGTGCCA.
RNA extraction and reverse transcription-quantitative polymerase chain reaction analysis
Total RNA was extracted from cells and tissues with Trizol reagent. Reverse transcription was performed using the Hifair® III 1st Strand cDNA Synthesis Kit (Yeasen, Shanghai, China). Then the reactions were subsequently measured on a Roche LightCycler®480II PCR instrument (Basel, Switzerland). All reactions were analyzed in triplicate. LncRNA expression was normalized to beta-actin (ACTB), and relative lncRNA expression levels were calculated using the 2−▵▵t method.
Cells were lysed in lysis buffer supplemented with protease inhibitors for 30 min on ice. Protein fractions were collected by centrifugation at 10,000×g for 10 min, separated on 10% sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gels, and then electrotransferred onto nitrocellulose membranes (Whatman, Piscataway, NJ, USA). Membranes were blocked with 5% bovine serum albumin (BSA) and then incubated with specific antibodies overnight at 4°C. The primary antibodies used were as follows: matrix metallopeptidase 13 (MMP13) (1:1000, 18165-1-AP, Proteintech), matrix metallopeptidase 3 (MMP3) (1:1000, abs130153, Absin), matrix metallopeptidase 2 (MMP2) (1:1000, ab86607, Abcam), AP-1 transcription factor subunit (c-FOS) (1:1000, A2444, Abclonal), cathepsin B (CTSB) (1:1000, A0967, Abclonal), growth factor receptor bound protein 2 (GRB2) (1:1000, A19059, Abclonal), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (1:2000, AF7021, Affinity).
LncRNA was inhibited in SW982 cells under inflammatory stimuli, and fluorescence staining of proliferating cells was performed using the BeyoClick™EdU Cell Proliferation Kit with Alexa Fluor 488 (Beyotime Biotechnology, Shanghai, China) after 48 h of culture. Image acquisition was performed using a fluorescence microscope (Nikon, Japan).
All data from the control and experimental groups were analyzed using the paired two-tailed Student’s t-test or two-way ANOVA. The statistical difference was set as p < 0.05.
A total of 10 female patients were enrolled, including 5 patients in the experimental group with a mean age of 35.0 ± 15.7 years and 5 patients in the control group with a mean age of 39.6 ± 15.8 years. A total of another 13 female patients were enrolled for lncRNA expression verification, including 6 patients (8 synovial tissues) in the experimental group with a mean age of 25.4 ± 17.0 years and 7 patients (8 synovial tissues) in the control group with a mean age of 23.1 ± 15.6 years. The basic information about the enrolled patients is provided in Tables 1 and 2.
Quality of RNA and the library
The A260/280 of RNA from all samples ranged from 1.8 to 2.0, indicating that the total RNA had high purity. The total amount of RNA extracted from each sample was ≥0.5 μg, which met the criteria and sequencing requirements. The bands of 28S and 18S rRNA in gel electrophoresis were clear, and the intensity of 28S rRNA was approximately twice that of 18S rRNA, suggesting that the extracted RNA had no genomic DNA (gDNA) contamination. The library quality control results met the sequencing requirements. Our source data have been uploaded to GSE205389 and GSE205684.
Differential expression analysis and further screening of differentially expressed mRNAs
In this study, we found a total of 384 DE lncRNAs (upregulated 200, downregulated 184), 115 DE miRNAs (upregulated 33, downregulated 82), and 1767 DE mRNAs (upregulated 1158, downregulated 609). The volcano plot documented the differential expression of the three outcomes in TMJOA (Figs. 2A–2C). The heatmap showed the difference in expression of the top 100 upregulated/downregulated DE mRNAs between the two groups (Fig. 2D and Suppl. Fig. S1A).
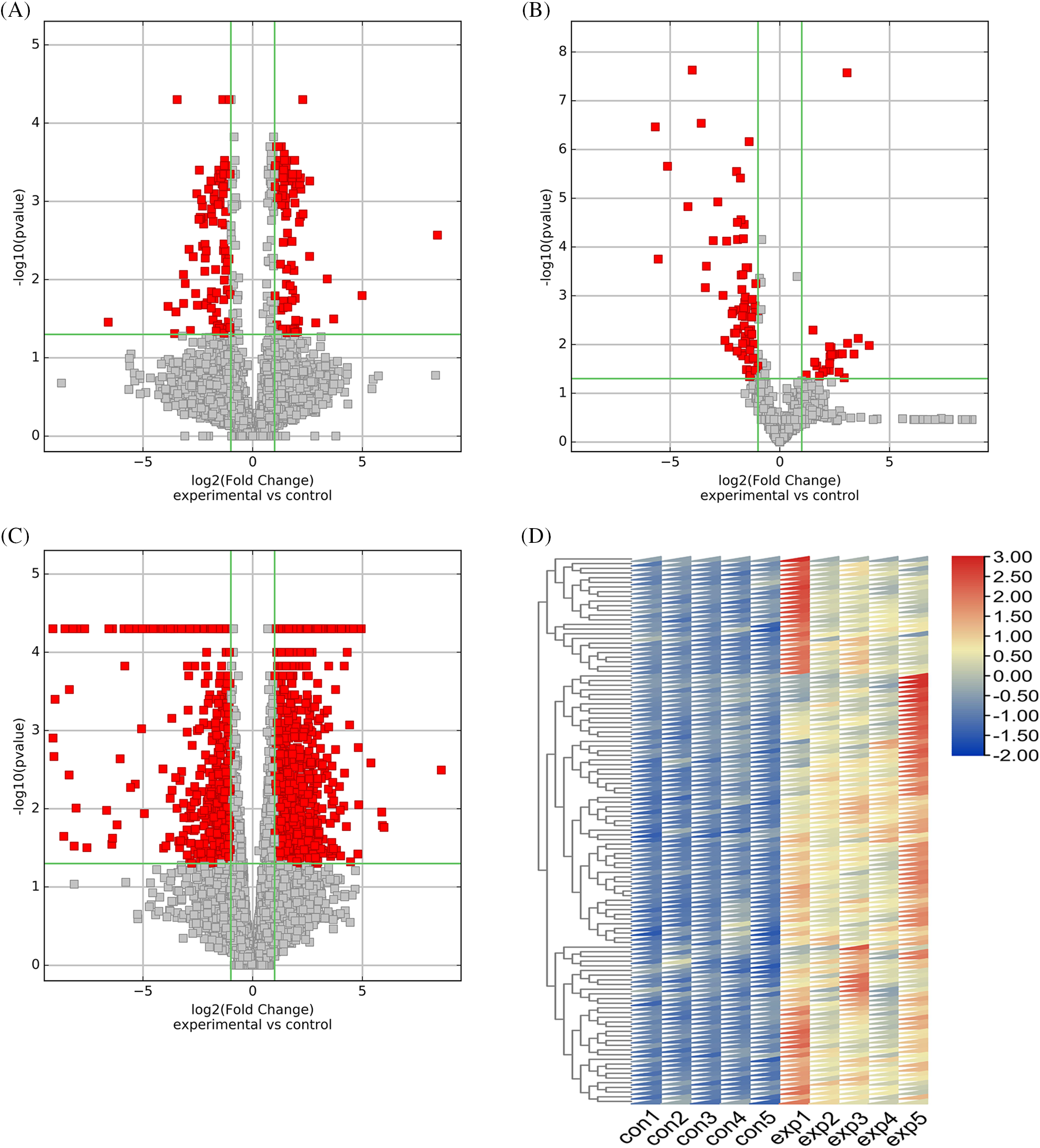
Figure 2: Differential expression of lncRNAs, miRNAs and mRNAs and further screening of differentially expressed (DE) mRNAs. Volcano plot of differentially expressed lncRNAs (A). Volcano plot of differentially expressed miRNAs (B). Volcano plot of differentially expressed mRNAs (C). Differential expression of top 100 upregulated mRNAs (D).
Pathway and functional analysis of selected differentially expressed mRNAs
KEGG analysis showed that upregulated genes were mainly enriched in immune-related pathways. In addition, the osteoclast differentiation pathway was also enriched (Fig. 3A). Downregulated genes were mainly enriched in the cytoskeleton-regulated pathway (Suppl. Fig. S1B). The top 5 pathways (p < 0.05) enriched in GSEA were shown in Suppl. Fig. S1C. The GSEA analysis also showed osteoclast differentiation pathway similar to the KEGG analysis (Fig. 3B). The schematic of the osteoclast differentiation pathway has been illustrated with the relevant mRNAs involved in this pathway (Suppl. Fig. S1D).
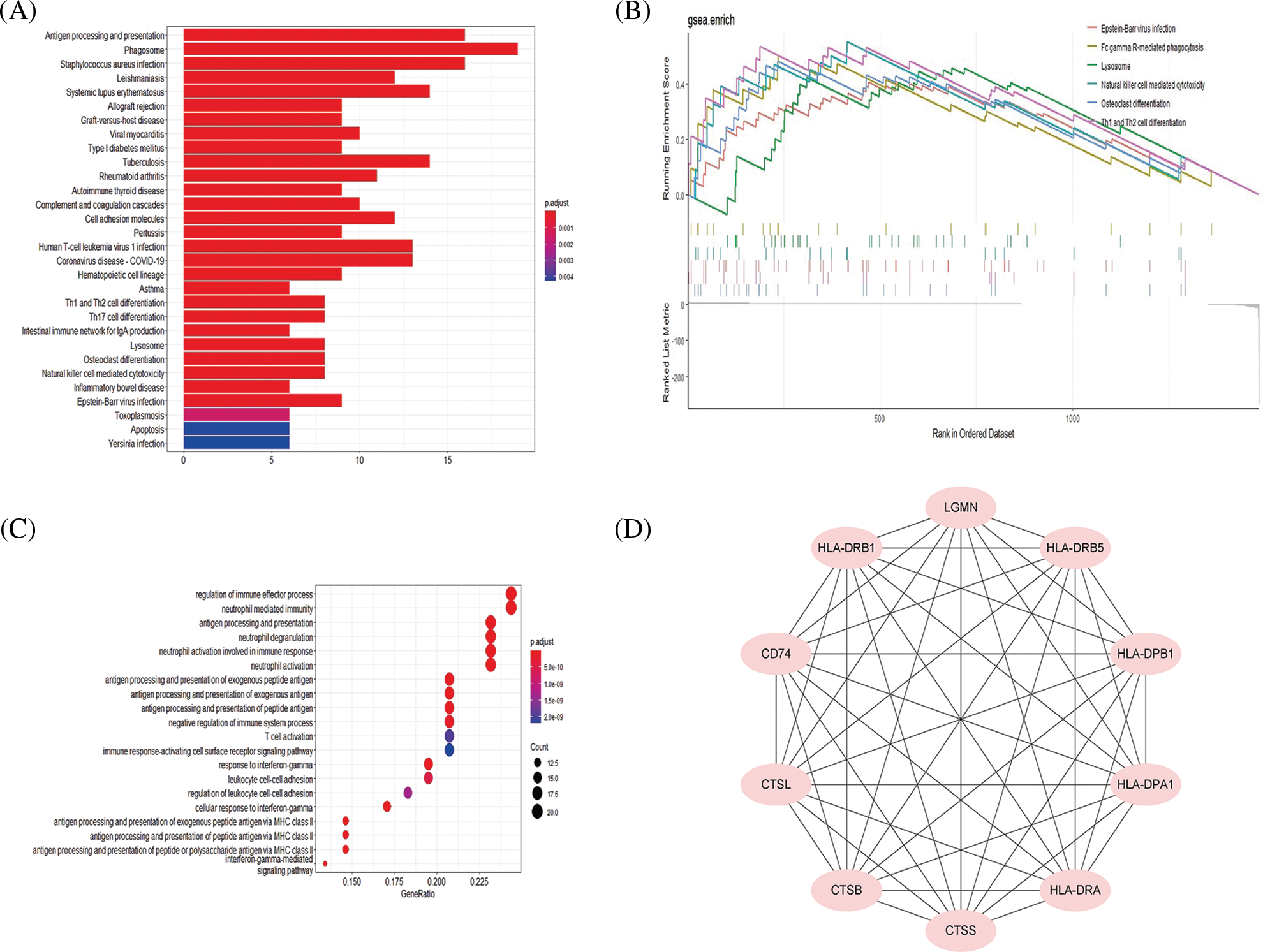
Figure 3: Pathway and functional analysis of top 100 upregulated mRNAs. Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of top 100 up-regulated mRNAs (A). The osteoclast differentiation pathway was also enriched in the gene set enrichment analysis (GSEA) analysis (B). Biological Process analysis of top 100 upregulated mRNAs (C). Hub genes of top 100 upregulated mRNAs (D).
By comparing GSEA with KEGG analysis results, it was shown that the main involved genes in each pathway were upregulated mRNAs, so we further analyzed the upregulated mRNAs and their upstream regulated miRNAs and lncRNAs. GO analysis showed that the upregulated genes mostly involved immune regulation-related biological processes such as antigen processing and presentation (Fig. 3C). Hub gene analysis further revealed the top 10 hub genes in upregulated mRNAs (Fig. 3D).
miRNA and lncRNA screening and ceRNA regulatory network construction
Among the 384 DE lncRNAs, 25 met the screening conditions (11 upregulated and 14 downregulated) (Table 3). A total of 15 downregulated DE miRNAs could constitute a regulatory relationship with DE mRNAs, of which 6 could simultaneously bind to DE lncRNAs (Fig. 4A). The results showed that among the upregulated DE lncRNAs, only one lncRNA, xist, could regulate 6 DE miRNAs: miR-125b-5p, miR-1271-5p, miR-195-5p, miR-365a-3p, miR-27b-3p, and miR-199a-3p (Fig. 4B). Further, these 6 downregulated miRNAs could form regulatory relationships with 9 upregulated mRNAs, including ctsb, lcp1, mt2a, mmp3, fos, grb2, cd163, vamp8, and wdr1 (Fig. 4C).
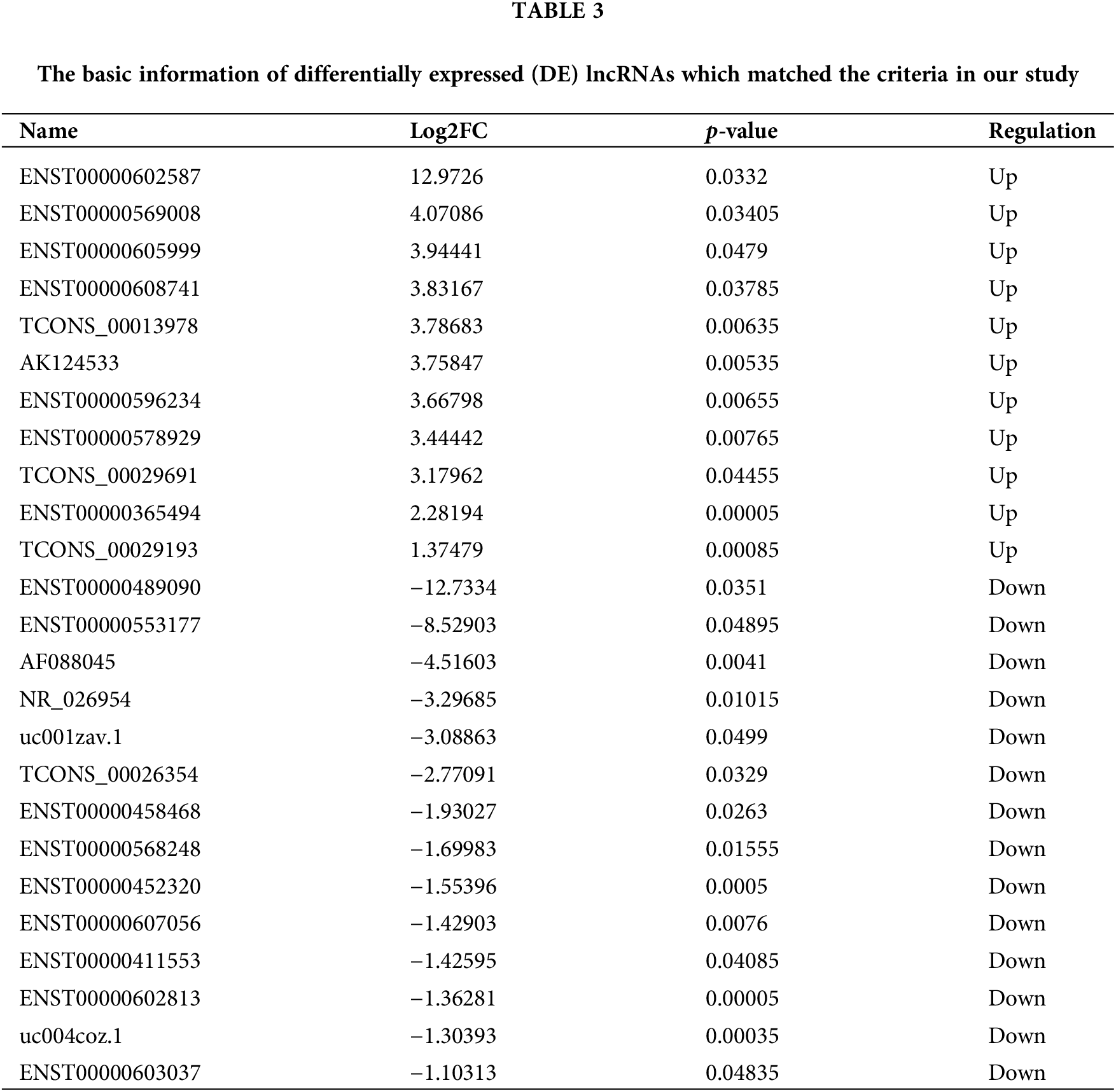
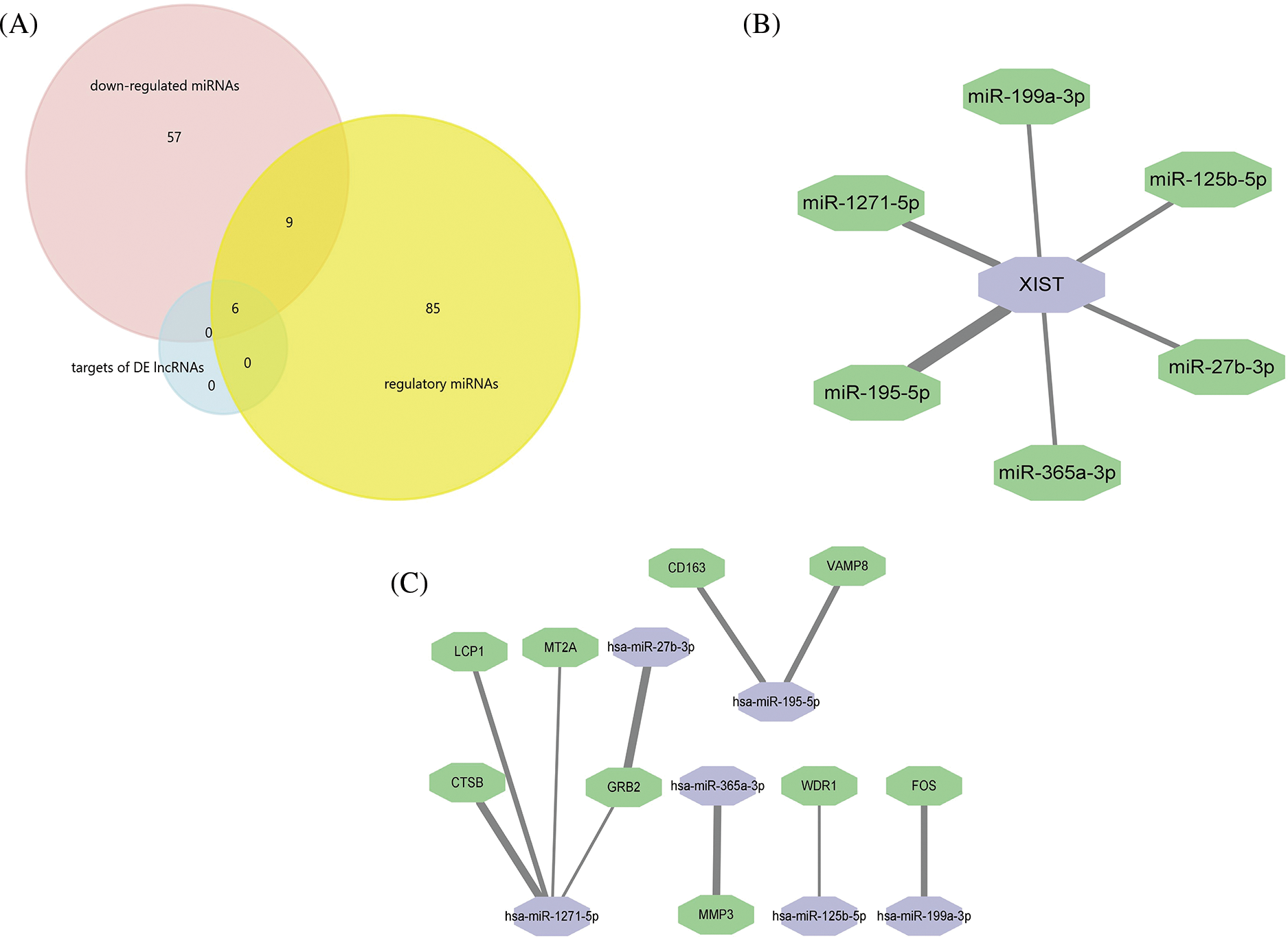
Figure 4: Professional website prediction showed upstream regulatory miRNAs and lncRNAs. Six down-regulated differentially expressed (DE) miRNAs were capable of forming regulatory relationships with DE mRNAs as well as DE lncRNAs (A). LncRNA xist could regulate six down-regulated DE miRNAs, the thicker the line, the greater the possibility of connection (B). LncRNA xist could regulate 9 DE mRNAs through 6 down-regulated DE miRNAs: the thicker the line, the greater the possibility of connection (C).
Expression and functional verification of xist
RT-qPCRresults showed that the xist expression in TMJOA synovial tissue was about 3-fold higher than in the control group (Fig. 5A). Under 20 ng/ml TNF-a stimulation, the expression of xist in SW982 cells increased by about 2.6 times (Fig. 5B). Western blotting results showed that under the same concentration of TNF-a stimulation, the expression of CTSB, c-FOS and GRB2 protein in synovial cells increased, and cells secreted more MMP3, promoting matrix degradation (Fig. 5C). We screened the effective concentration of ASO-xist, and RT-qPCR results showed that the minimal effective dose was 100 nM. The xist expression was reduced by about 50% (Fig. 5D). Western blotting results showed that, under 20 ng/ml TNF-a stimulation, the protein expression levels of target genes in the network, including CTSB, c-FOS, and GRB2 were lowered with diminished xist levels. In the meantime, synovial cells produced less MMP3 and MMP13 (Fig. 5E). EdU fluorescence staining showed that xist also affected cell proliferation. Upon TNF-a stimulation, the cell proliferation rate reduced significantly in the ASO-NC group while the cell proliferation rate of the ASO-xist group only reduced slightly (Fig. 5F). Since GRB2 levels dropped most significantly when xist was inhibited, we further tested the function of GRB2. Western blotting showed that under 20 ng/ml TNF-a stimulation, the synthesis of a series of matrix degrading enzymes was blocked after GRB2 was inhibited (Fig. 5G).
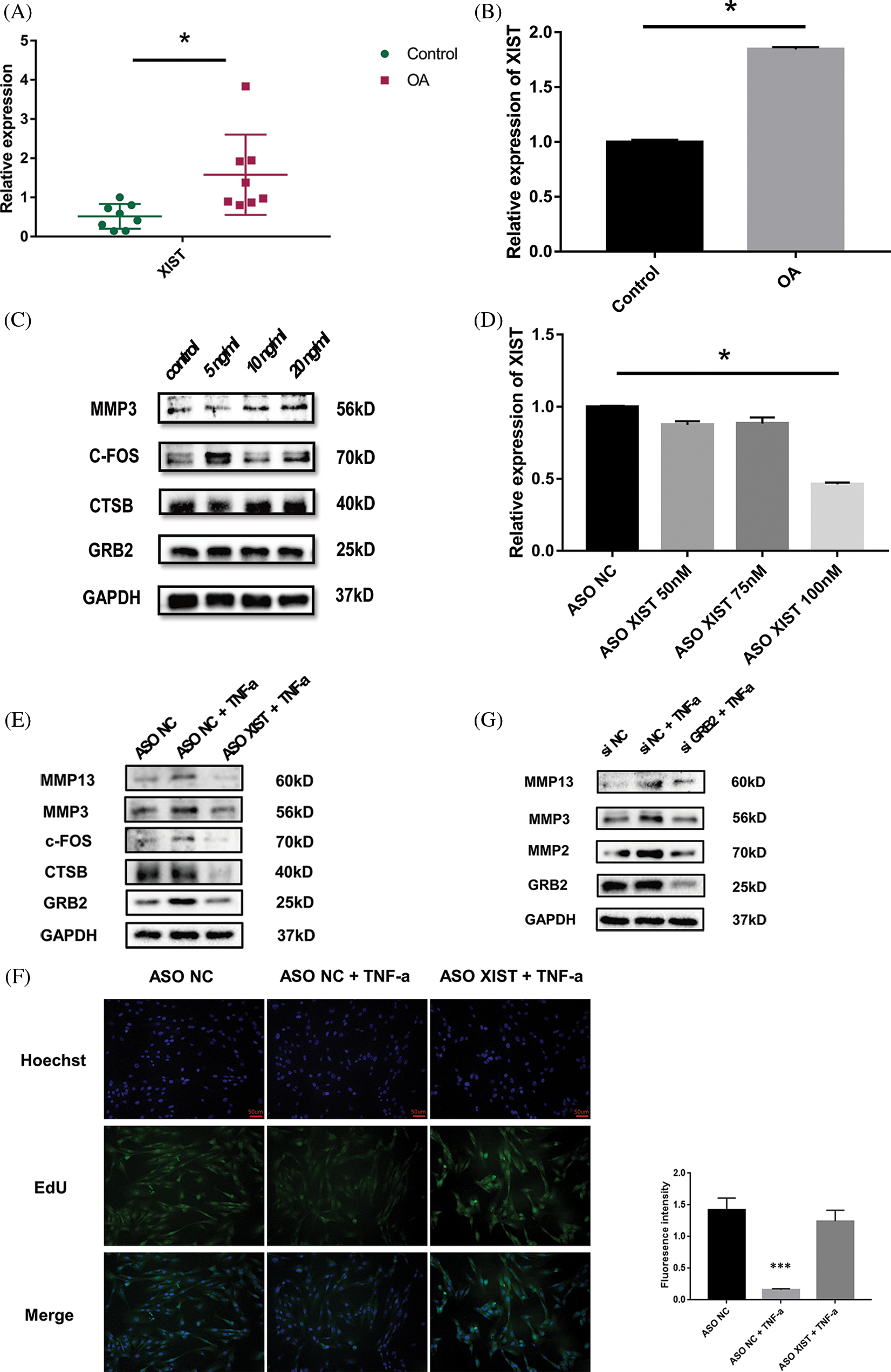
Figure 5: Expression profile and functional verification of xist. Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) results showed that the xist expression in TMJOA synovial tissue was about 3-fold higher than in the control group (A). RT-qPCR results showed that upon TNF-a stimulation, the expression of xist in SW982 increased about 2.6 times (B). Western blot results showed that upon TNF-a stimulation, the expression of CTSB, c-FOS, and GRB2 protein in synovial cells increased, and cells secreted more MMP3 (C). RT-qPCR results showed that the minimal effective dose of ASO-xist was 100 nM, xist expression was reduced by about 50% (D). Western blot results showed that upon TNF-a stimulation, CTSB, c-FOS, and GRB2 decreased upon xist inhibition. In the meantime, synovial cells produced less MMP3 and MMP13 (E). EdU fluorescence staining showed that under TNF-a stimulation, the cell proliferation rate of the ASO-xist group was much higher than in the ASO-NC group (F). Western blotting showed that under TNF-a stimulation, the synthesis of a series of matrix degrading enzymes was blocked after GRB2 was inhibited (G). *, p < 0.05; ***, p < 0.001.
LncRNAs are noncoding RNAs of more than 200 nucleotides in length (Xie et al., 2021). They can work as miRNA sponges to participate in regulating gene expression (Wang et al., 2019c; Wu et al., 2020). There is ample evidence that the relationship between lncRNAs and miRNAs plays an important role in OA progression (Xie et al., 2020). For example, Zhang et al. (2019) found that lncRNA malat1 could play a role in knee osteoarthritis by regulating chondrocyte proliferation, apoptosis, and extracellular matrix degradation through miR-150-5p/akt3. In another study, Liu et al. (2018) found that exosomes secreted by human stem cells contain lncRNA klf3-as1, which could regulate miR-206/git1, thereby affecting chondrocyte apoptosis. In addition, the lncRNA malat1/miR-146a-PI3K/Akt/mTOR axis and lncRNA foxd2-as1/miR-27a-3p/tlr4 axis have also been reported to regulate chondrocyte matrix degradation (Wang et al., 2019a; Li et al., 2020b). Some scholars have sequenced lncRNAs and analyzed differentially expressed lncRNAs in knee OA cartilage tissues, providing a basis for studying the regulatory role of lncRNAs in knee OA cartilage degeneration (Fu et al., 2015; Liu et al., 2016). As the initiating factor of OA, synovial inflammation can lead to the secretion of various inflammatory factors to promote cartilage degeneration, which is closely related to the severity and progression of OA (Kortekaas et al., 2016; Bhattaram and Chandrasekharan, 2017; van den Bosch, 2019). Many scholars have therefore also conducted many aspects of research on OA synovial inflammation. For example, Xiang et al. (2019) identified differentially expressed circRNAs in synovial tissues of knee OA by RNA sequencing, while further validation is however needed. This study previously identified differentially expressed circRNAs in TMJOA synovial tissues and found that circRNA_gcn1l1 was able to regulate TNF-a secretion in the synovium through miRNA-330-3p. Additionally, Xu et al. (2016) performed miRNA sequencing in TMJOA synovial tissues and found that miR-221-3p could regulate IL-1B and MMP1/9 secretion. However, none of the above studies involved the role of lncRNAs in OA and synovial inflammation symptoms. Studies on the relationship between lncRNAs and synovial inflammation have mostly come from RA. For example, Zou et al. (2018) sequenced lncRNAs in RA synovial tissue and found that lncRNA lerfs has a protective role in RA. Due to the current lack of reports of lncRNA regulation in TMJOA synovitis, we conducted lncRNA, miRNA, and mRNA sequencing in TMJOA synovial tissues in the hope of finding lncRNA/miRNA/mRNA axes that may play an important role in TMJOA.
The results of this study revealed that only one lncRNA molecule xist, was predicted to affect the expression of DE mRNAs through DE miRNAs.
LncRNA xist classically functions to silence the X chromosome to compensate for differences in the expression of X chromosome-associated genes between males and females (Czermiński and Lawrence, 2020). It can also play a role as an oncogene in some parenchymal tumors, including lung cancer and liver cancer (Yang et al., 2020). CeRNA network-related research is also mainly in the field of oncology. For instance, Li et al. (2020a) found that xist was significantly increased in both synovial and cartilage tissues of knee OA, which was consistent with the results of the analysis in this paper. They found that xist could help express osteopontin (OPN) in M1 macrophages in synovial tissues by binding miR-376c-5p, promoting macrophage cytotoxicity to chondrocytes, and then resulting in chondrocyte apoptosis. In another report, Liu et al. (2020) also confirmed that there was a significant upregulation of xist in knee OA cartilage and found that xist could facilitate DNA methyltransferase 3A (DNMT3A) expression and eventually exacerbate the destruction of chondrocytes by inhibiting miR-149-5p. Further, Wang et al. (2019b) reported that significant upregulation of xist was also confirmed in knee OA cartilage, and the xist/miR-1277-5p/mmp 13/adamts5 axis was found to have an important role in regulating OA disease development. Hence, xist can impact the course of OA through different targets and therefore may become an important target for the treatment of OA. Among lncRNAs, xist is a research hotspot with important functions. However, as most of the above studies were on cartilage tissue, the role of xist in synovitis is still unclear. Our in vitro experiments proved that blocking xist could protect synovial cells under inflammatory conditions.
Among the nine DE mRNAs predicted to be regulated by xist, ctsb was the hub gene selected by analysis, and mmp3 is considered an important cartilage matrix degradation marker. Besides, fos and grb2 products are molecules involved in the osteoclast differentiation pathway. The remaining five target genes are not involved in pathway enrichment or hub gene prediction. Thus ctsb, mmp3, fos, and grb2 were screened as subsequent research points in this work. The known functions of these four mRNAs in TMJOA have been summarized in Table 4. Our primary results showed that under inflammatory conditions, protein expression of CTSB, MMP3, c-FOS, and GRB2 decreases after inhibition of xist. This provided the evidence for our network construction.
Cathepsin B (CTSB), a lysosomal cysteine protease, is also a marker of the dedifferentiated phenotype of chondrocytes (Li et al., 2017). Research revealed that the DE miRNA hsa-miR-1271-5p could regulate CTSB. Zhang et al. (2021) documented that in chondrocytes, miR-140-5p could attenuate OA cartilage destruction by inhibiting CTSB/NLRP3. Ben-Aderet et al. (2015) found that CTSB was significantly higher in the synovial fluid in OA. However, the mechanism of CTSB in synovial tissue and synovitis is still unclear, and the analysis in this study yielded that the ceRNA network xist/miR-1271-5p/ctsb may function in synovitis.
Matrix metalloproteinase-3 (MMP3) is a classical molecule involved in cartilage destruction that is able to directly degrade the cartilage extracellular matrix and cause cartilage destruction by activating MMP9. MMP3 expression is directly related to the degree of inflammatory cell infiltration in the synovium. Further, increased metalloproteinase levels such as MMP3 are typical findings in OA (Berenbaum and Sellam, 2010). We found that the DE miRNA miR-365a-3p, a target of xist, was able to inhibit MMP3. It, therefore, constitutes the second ceRNA network, xist/miR-365a-3p/mmp3, in this study.
The results of GSEA and KEGG analysis both showed that the osteoclast differentiation pathway and osteoclast formation play an important role in the degeneration of subchondral bone (Zhu et al., 2019). This may be a bridge between synovitis and pathological remodeling of the subchondral bone. There are two core genes, fos and grb2 in the osteoclast differentiation pathway. The fos-encoded C-FOS protein is an important transcription factor in osteoclast formation, which is activated by the classical osteoclast formation regulator Receptor activator of nuclear factor kappa-B ligand (RANKL) and then activates the downstream NFATC1. Together with transcription factors such as MYC and NF-κB, C-FOS forms a complex network to function in promoting osteoclast formation. However, in this process, the regulation of C-FOS expression after activation by RANKL is not fully understood (Fujii et al., 2021). Therefore, whether C-FOS is also regulated by other molecules and thus affects its final expression needs to be confirmed. We found that the DE miRNA miR-199a-3p is able to bind fos, so it is worth exploring whether xist/miR-199a-3p can play a role in osteoclast formation through fos and whether it can be a therapeutic target for TMJOA.
GRB2 is a key factor in several tyrosine kinase signaling conversion pathways. Some studies have reported that GRB2 can promote osteoclast survival by activating Erk and can strengthen the adhesion of osteoclasts to the bone surface, leading to bone resorption (Levy-Apter et al., 2014). A recent study found that after the knockout of PIP5k1β kinase, GRB2 expression increased, and the ERK1/2 signaling pathway was more active, which promoted osteoclast formation (Zhao et al., 2020). Our preliminary in vitro experiments demonstrated that under inflammatory conditions, inhibition of GRB2 contributes to the decline of matrix degrading enzymes in synoviocytes. This provides a research basis for alleviating joint destruction by regulating GRB2.
Through the bioinformatics analysis above, four ceRNA regulatory axes were found in this study, including xist/miR-1271-5p/ctsb, xist/miR-365a-3p/mmp3, xist/miR-199a-3p/fos, and xist/miR-27b-3p/miR-1271-5p/grb2. These four networks may be involved in the development of synovial inflammation and are one of the important reasons why synovial inflammation can promote cartilage and subchondral bone degeneration. Their roles still need to be confirmed by further in vitro and in vivo studies. Thus, in our future work, we will focus on the validation of the aforementioned ceRNA networks.
Funding Statement: This study was supported by grants from the National Natural Science Foundation of China (32071313, 82270996), the Science and Technology Commission of Shanghai Municipality Science Research Project (20S31902500, 20Y11903900), and the Cross-Disciplinary Research Fund of Shanghai Ninth People’s Hospital, Shanghai Jiao Tong University School of Medicine (JYJC202203).
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Dongmei He; data collection: Chuyao Wang; analysis and interpretation of results: Chuyao Wang, Chuan Lu; draft manuscript preparation: Luxiang Zou. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: All data generated or analyzed during this study are included in this published article (and its supplementary information files).
Ethics Approval: The collection of all specimens was approved by the Ethics Committee of Shanghai Ninth People’s Hospital (Shanghai, China). The protocol number is SH9H-2021-T141-1 and the date of adoption was June 01, 2021. All patients signed an informed consent approved by the Institutional Review Board.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
Ben-Aderet L, Merquiol E, Fahham D, Kumar A, Reich E et al. (2015). Detecting cathepsin activity in human osteoarthritis via activity-based probes. Arthritis Research & Therapy 17: 1–14. https://doi.org/10.1186/s13075-015-0586-5 [Google Scholar] [PubMed] [CrossRef]
Berenbaum F, Sellam J (2010). The role of synovitis in pathophysiology and clinical symptoms of osteoarthritis. Nature Reviews Rheumatology 6: 625–635. https://doi.org/10.1038/nrrheum.2010.159 [Google Scholar] [PubMed] [CrossRef]
Bhattaram P, Chandrasekharan U (2017). The joint synovium: A critical determinant of articular cartilage fate in inflammatory joint diseases. Seminars in Cell & Developmental Biology 62: 86–89. https://doi.org/10.1016/j.semcdb.2016.05.009 [Google Scholar] [PubMed] [CrossRef]
Bi X, Guo XH, Mo BY, Wang ML, Luo XQ, Chen YX, Liu F, Olsen N, Pan YF, Zheng SG (2019). LncRNA PICSAR promotes cell proliferation, migration and invasion of fibroblast-like synoviocytes by sponging miRNA-4701-5p in rheumatoid arthritis. eBioMedicine 50: 408–420. https://doi.org/10.1016/j.ebiom.2019.11.024 [Google Scholar] [PubMed] [CrossRef]
Bush JR, Beier F (2013). TGF-β and osteoarthritis--The good and the bad. Nature Medicine 19: 667–669. https://doi.org/10.1038/nm.3228 [Google Scholar] [PubMed] [CrossRef]
Czermiński JT, Lawrence JB (2020). Silencing trisomy 21 with XIST in neural stem cells promotes neuronal differentiation. Developmental Cell 52: 294–308. https://doi.org/10.1016/j.devcel.2019.12.015 [Google Scholar] [PubMed] [CrossRef]
Du S, Liang C, Sun Y, Ma B, Gao W, Geng W(2021). The attenuating effect of low-intensity pulsed ultrasound on hypoxia-induced rat chondrocyte damage in TMJ osteoarthritis based on TMT labeling quantitative proteomic analysis. Frontiers in Pharmacology 12: 752734. https://doi.org/10.3389/fphar.2021.752734 [Google Scholar] [PubMed] [CrossRef]
Fu M, Huang G, Zhang Z, Liu J, Zhang Z, Huang Z, Yu B, Meng F (2015). Expression profile of long noncoding RNAs in cartilage from knee osteoarthritis patients. Osteoarthritis and Cartilage 23: 423–432. https://doi.org/10.1016/j.joca.2014.12.001 [Google Scholar] [PubMed] [CrossRef]
Fu X, Song G, Ni R, Liu H, Xu Z, Zhang D, He F, Huang G (2021). LncRNA-H19 silencing suppresses synoviocytes proliferation and attenuates collagen-induced arthritis progression by modulating miR-124a. Rheumatology 60: 430–440. https://doi.org/10.1093/rheumatology/keaa395 [Google Scholar] [PubMed] [CrossRef]
Fujii T, Murata K, Mun S, Bae S, Lee YJ, Pannellini T, Kang K, Oliver D, Park-Min KH, Ivashkiv LB (2021). MEF2C regulates osteoclastogenesis and pathologic bone resorption via c-FOS. Bone Research 9: 4. https://doi.org/10.1038/s41413-020-00120-2 [Google Scholar] [PubMed] [CrossRef]
Kortekaas MC, Kwok W, Reijnierse M, Stijnen T, Kloppenburg M (2016). Brief report: Association of inflammation with development of erosions in patients with hand osteoarthritis: A prospective ultrasonography study. Arthritis & Rheumatology 68: 392–397. https://doi.org/10.1002/art.39438 [Google Scholar] [PubMed] [CrossRef]
Kostopoulou F, Malizos KN, Papathanasiou I, Tsezou A (2015). MicroRNA-33a regulates cholesterol synthesis and cholesterol efflux-related genes in osteoarthritic chondrocytes. Arthritis Research & Therapy 17: 1–13. https://doi.org/10.1186/s13075-015-0556-y [Google Scholar] [PubMed] [CrossRef]
Levy-Apter E, Finkelshtein E, Vemulapalli V, Li SSC, Bedford MT, Elson A (2014). Adaptor protein GRB2 promotes src tyrosine kinase activation and podosomal organization by protein-tyrosine phosphatase ε in osteoclasts. Journal of Biological Chemistry 289: 36048–36058. https://doi.org/10.1074/jbc.M114.603548 [Google Scholar] [PubMed] [CrossRef]
Li Y, Fang J, Ao G (2017). Cathepsin B and L inhibitors: A patent review (2010-present). Expert Opinion on Therapeutic Patents 27: 643–656. https://doi.org/10.1080/13543776.2017.1272572 [Google Scholar] [PubMed] [CrossRef]
Li H, Guo H, Lei C, Liu L, Xu LQ et al. (2019). Nanotherapy in joints: Increasing endogenous hyaluronan production by delivering hyaluronan synthase 2. Advanced Materials 31: 1904535. https://doi.org/10.1002/adma.201904535 [Google Scholar] [PubMed] [CrossRef]
Li L, Lv G, Wang B, Kuang L (2020a). XIST/miR-376c-5p/OPN axis modulates the influence of proinflammatory M1 macrophages on osteoarthritis chondrocyte apoptosis. Journal of Cellular Physiology 235: 281–293. https://doi.org/10.1002/jcp.28968 [Google Scholar] [PubMed] [CrossRef]
Li H, Xie S, Li H, Zhang R, Zhang H (2020b). LncRNA MALAT1 mediates proliferation of LPS treated-articular chondrocytes by targeting the miR-146a-PI3K/Akt/mTOR axis. Life Sciences 254: 116801. https://doi.org/10.1016/j.lfs.2019.116801 [Google Scholar] [PubMed] [CrossRef]
Liu Q, Hu X, Zhang X, Dai L, Duan X, Zhou C, Ao Y (2016). The TMSB4 pseudogene LncRNA functions as a competing endogenous RNA to promote cartilage degradation in human osteoarthritis. Molecular Therapy 24: 1726–1733. https://doi.org/10.1038/mt.2016.151 [Google Scholar] [PubMed] [CrossRef]
Liu Y, Lin L, Zou R, Wen CY, Wang Z, Lin F (2018). MSC-derived exosomes promote proliferation and inhibit apoptosis of chondrocytes via lncRNA-KLF3-AS1/miR-206/GIT1 axis in osteoarthritis. Cell Cycle 17: 2411–2422. https://doi.org/10.1080/15384101.2018.1526603 [Google Scholar] [PubMed] [CrossRef]
Liu Y, Liu K, Tang C, Shi Z, Jing K, Zheng J (2020). Long non-coding RNA XIST contributes to osteoarthritis progression via miR-149-5p/DNMT3A axis. Biomedicine & Pharmacotherapy 128: 110349. https://doi.org/10.1016/j.biopha.2020.110349 [Google Scholar] [PubMed] [CrossRef]
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Rajewsky N (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495: 333–338. https://doi.org/10.1038/nature11928 [Google Scholar] [PubMed] [CrossRef]
van den Bosch MHJ (2019). Inflammation in osteoarthritis: Is it time to dampen the alarm(in) in this debilitating disease? Clinical and Experimental Immunology 195: 153–166. https://doi.org/10.1111/cei.13237 [Google Scholar] [PubMed] [CrossRef]
Wang Y, Cao L, Wang Q, Huang J, Xu S (2019a). LncRNA FOXD2-AS1 induces chondrocyte proliferation through sponging miR-27a-3p in osteoarthritis. Artificial Cells Nanomedicine Biotechnology 47: 1241–1247. https://doi.org/10.1080/21691401.2019.1596940 [Google Scholar] [PubMed] [CrossRef]
Wang P, Chen D, Ma H, Li Y (2017). LncRNA MEG3 enhances cisplatin sensitivity in non-small cell lung cancer by regulating miR-21-5p/SOX7 axis. OncoTargets and Therapy 10: 5137–5149. https://doi.org/10.2147/OTT [Google Scholar] [CrossRef]
Wang T, Liu Y, Wang Y, Huang X, Zhao W, Zhao Z (2019b). Long non-coding RNA XIST promotes extracellular matrix degradation by functioning as a competing endogenous RNA of miR-1277-5p in osteoarthritis. International Journal of Molecular Medicine 44: 630–642. https://doi.org/10.3892/ijmm.2019.4240 [Google Scholar] [PubMed] [CrossRef]
Wang K, Long B, Liu F, Wang J, Liu CY, Zhao B, Zhou LY, Sun T, Wang M, Yu T (2016). A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. European Heart Journal 37: 2602–2611. https://doi.org/10.1093/eurheartj/ehv713 [Google Scholar] [PubMed] [CrossRef]
Wang M, Mao C, Ouyang L, Liu Y, Lai W et al. (2019c). Long noncoding RNA LINC00336 inhibits ferroptosis in lung cancer by functioning as a competing endogenous RNA. Cell Death & Differentiation 26: 2329–2343. https://doi.org/10.1038/s41418-019-0304-y [Google Scholar] [PubMed] [CrossRef]
Wang J, Zhao Q (2020). LncRNA LINC-PINT increases SOCS1 expression by sponging miR-155-5p to inhibit the activation of ERK signaling pathway in rheumatoid arthritis synovial fibroblasts induced by TNF-α. International Immunopharmacology 84: 106497. https://doi.org/10.1016/j.intimp.2020.106497 [Google Scholar] [PubMed] [CrossRef]
Wu P, Mo Y, Peng M, Tang T, Zhong Y, Deng X, Xiong W (2020). Emerging role of tumor-related functional peptides encoded by lncRNA and circRNA. Molecular Cancer 19: 1–14. https://doi.org/10.1186/s12943-020-1147-3 [Google Scholar] [PubMed] [CrossRef]
Xiang S, Li Z, Bian Y, Wei X (2019). RNA sequencing reveals the circular RNA expression profiles of osteoarthritic synovium. Journal of Cellular Biochemistry 120: 18031–18040. https://doi.org/10.1002/jcb.29106 [Google Scholar] [PubMed] [CrossRef]
Xiao J, Wang R, Zhou W, Cai X, Ye Z (2021). LncRNA NEAT1 regulates the proliferation and production of the inflammatory cytokines in rheumatoid arthritis fibroblast-like synoviocytes by targeting miR-204-5p. Human Cell 34: 372–382. https://doi.org/10.1007/s13577-020-00461-4 [Google Scholar] [PubMed] [CrossRef]
Xie J, Li H, Chen L, Cao Y, Hu Y, Zhu Z, Wang M, Shi J (2021). A novel pyroptosis-related lncRNA signature for predicting the prognosis of skin cutaneous melanoma. International Journal of General Medicine 14: 6517–6527. https://doi.org/10.2147/IJGM.S335396 [Google Scholar] [PubMed] [CrossRef]
Xie F, Liu YL, Chen XY, Li Q, Zhong J, Dai BY, Shao XF, Wu GB (2020). Role of MicroRNA, LncRNA, and exosomes in the progression of osteoarthritis: A review of recent literature. Orthopaedic Surgery 12: 708–716. https://doi.org/10.1111/os.12690 [Google Scholar] [PubMed] [CrossRef]
Xiong D, Li Z, Liang L, He R, Ma F, Luo D, Hu X, Chen G (2018). The LncRNA NEAT1 accelerates lung adenocarcinoma deterioration and binds to Mir-193a-3p as a competitive endogenous RNA. Cellular Physiology and Biochemistry 48: 905–918. https://doi.org/10.1159/000491958 [Google Scholar] [PubMed] [CrossRef]
Xu J, Liu Y, Deng M, Li J, Cai H, Meng Q, Fang W, Long X, Ke J (2016). MicroRNA221-3p modulates Ets-1 expression in synovial fibroblasts from patients with osteoarthritis of temporomandibular joint. Osteoarthritis and Cartilage 24: 2003–2011. https://doi.org/10.1016/j.joca.2016.06.011 [Google Scholar] [PubMed] [CrossRef]
Yang X, Zhang S, He C, Xue P, Zhang L, He Z, Zang L, Feng B, Sun J, Zheng M (2020). METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Molecular Cancer 19: 1–16. https://doi.org/10.1186/s12943-020-1146-4 [Google Scholar] [PubMed] [CrossRef]
Yin C, Suen W, Lin S, Wu XM, Li G, Pan XH (2017). Dysregulation of both miR-140-3p and miR-140-5p in synovial fluid correlate with osteoarthritis severity. Bone Joint Research 6: 612–618. https://doi.org/10.1302/2046-3758.611.BJR-2017-0090.R1 [Google Scholar] [PubMed] [CrossRef]
Zhang L, Qiu J, Shi J, Liu S, Zou H (2021). MicroRNA-140-5p represses chondrocyte pyroptosis and relieves cartilage injury in osteoarthritis by inhibiting cathepsin B/Nod-like receptor protein 3. Bioengineered 12: 9933–9948. https://doi.org/10.1080/21655979.2021.1985342 [Google Scholar] [PubMed] [CrossRef]
Zhang Y, Wang F, Chen G, He R, Yang L (2019). LncRNA MALAT1 promotes osteoarthritis by modulating miR-150-5p/AKT3 axis. Cell & Bioscience 9: 1–12. https://doi.org/10.1186/s13578-019-0302-2 [Google Scholar] [PubMed] [CrossRef]
Zhao X, Cui P, Hu G, Wang C, Jiang L, Zhao J, Xu J, Zhang X (2020). PIP5k1β controls bone homeostasis through modulating both osteoclast and osteoblast differentiation. Journal of Molecular Cell Biology 12: 55–70. https://doi.org/10.1093/jmcb/mjz028 [Google Scholar] [PubMed] [CrossRef]
Zheng Q, Bao C, Guo W, Li S, Chen J et al. (2016). Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nature Communications 7: 1–13. https://doi.org/10.1038/ncomms11215 [Google Scholar] [PubMed] [CrossRef]
Zheng L, Pi C, Zhang J, Fan Y, Cui C et al. (2018). Aberrant activation of latent transforming growth factor-beta initiates the onset of temporomandibular joint osteoarthritis. Bone Research 6: 1–10. https://doi.org/10.1038/s41413-018-0027-6 [Google Scholar] [PubMed] [CrossRef]
Zhu H, Hu Y, Wang C, Zhang X, He D (2020). CircGCN1L1 promotes synoviocyte proliferation and chondrocyte apoptosis by targeting miR-330-3p and TNF-α in TMJ osteoarthritis. Cell Death & Disease 11: 1–16. https://doi.org/10.1038/s41419-020-2447-7 [Google Scholar] [PubMed] [CrossRef]
Zhu S, Zhu J, Zhen G, Hu Y, An S et al. (2019). Subchondral bone osteoclasts induce sensory innervation and osteoarthritis pain. Journal of Clinical Investigation 129: 1076–1093. https://doi.org/10.1172/JCI121561 [Google Scholar] [PubMed] [CrossRef]
Zou Y, Xu S, Xiao Y, Qiu Q, Shi M et al. (2018). Long noncoding RNA LERFS negatively regulates rheumatoid synovial aggression and proliferation. Journal of Clinical Investigation 128: 4510–4524. https://doi.org/10.1172/JCI97965 [Google Scholar] [PubMed] [CrossRef]
Supplementary Materials
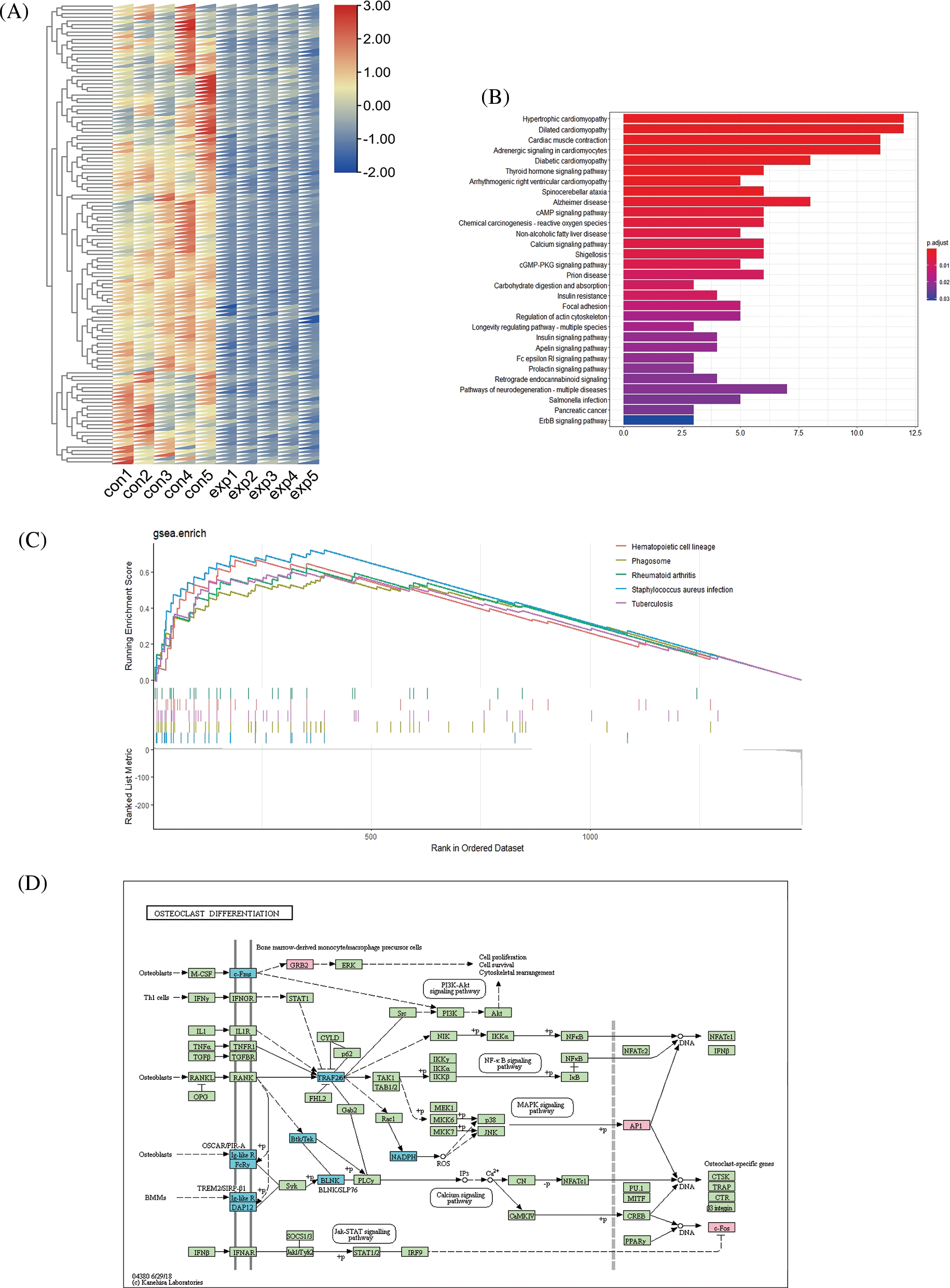
Figure S1: Details of pathway and functional analysis results of top 100 differentially expressed (DE) mRNAs. Differential expression of top 100 downregulated mRNAs between the experimental group and control group (A). Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of top 100 downregulated mRNAs (B). Top five pathways in the gene set enrichment analysis (GSEA) analysis of all DE mRNAs with p value < 0.05 (C). Schematic diagram of the osteoclast differentiation pathway (D).
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF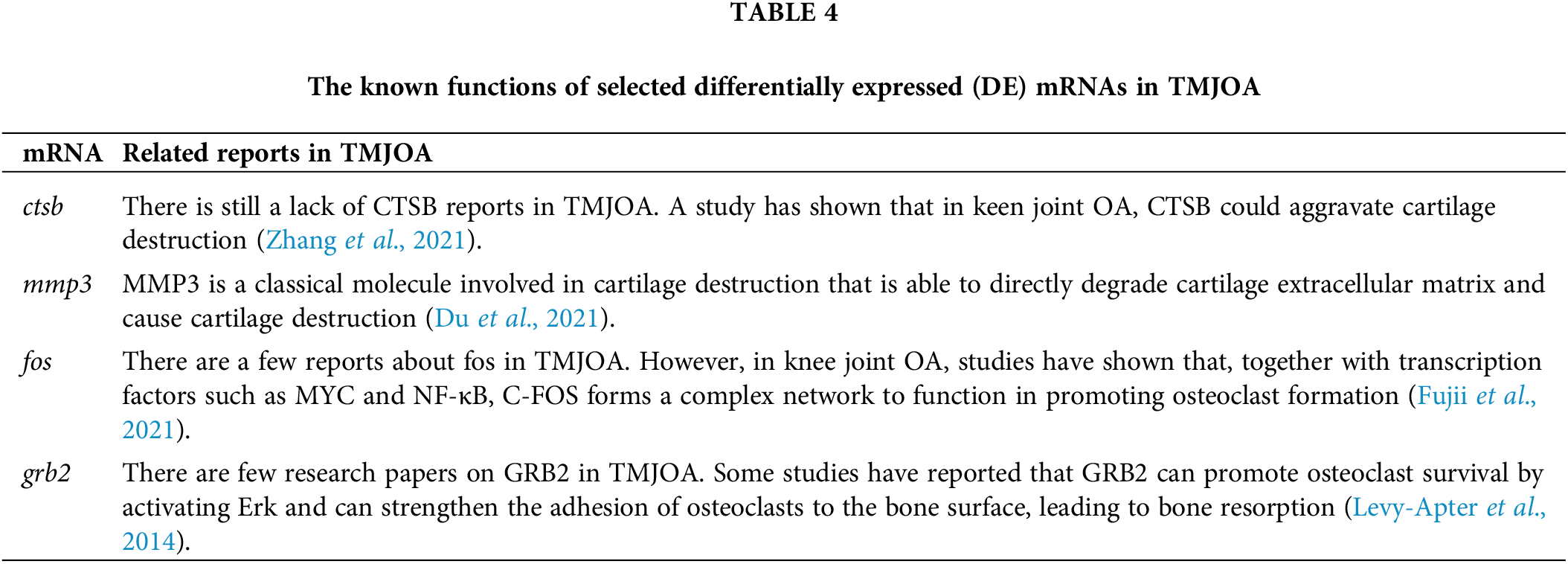
 Downloads
Downloads
 Citation Tools
Citation Tools