 Open Access
Open Access
ARTICLE
Light-controlled phosphorylation in the TrkA-Y785 site by photosensitive UAAs activates the MAPK/ERK signaling pathway
1 School of Life Sciences, Nantong University, Nantong, 226019, China
2 Institute National de la Sante et de la Recherche Med
icale (INSERM) U1195, Bicetre Hospital, Paris-Saclay University, Le Kremlin-Bicêtre, 94276, France
* Corresponding Author: Shu Zhao,
BIOCELL 2023, 47(6), 1377-1388. https://doi.org/10.32604/biocell.2023.023874
Received 19 May 2022; Accepted 06 September 2022; Issue published 19 May 2023
Abstract
Background: This paper aims to establish a light-controlled phosphorylation detection method at the Y785 site of tropomyosin receptor kinase A (TrkA) receptor in mammalian cells by using genetic code expansion technology and detecting the effects of optical activation of this site on the downstream MAPK/ERK pathway. The study is based on the current situation that the regulatory mechanism of TrkA phosphorylation has not been fully elucidated. Methods: Two photosensitive unnatural amino acids, p-azido-L-phenylalanine (AzF) and photo-caged tyrosine (ONB) were introduced into the TrkA-Y785 site by genetic code expansion technology and site-directed mutagenesis. Western blotting and laser confocal imaging were conducted to analyze the effects of this site on activating the MAPK/ERK pathway and nerve cell differentiation before and after photostimulation. Results: Our results supplemented the light-controlled results of the TrkA-Y785 site based on our previous research and verified that Y785 also makes important contributions in regulating the MAPK/ERK pathway. Conclusion: This study demonstrated the significant contributions of the TrkAY785 site in regulating the ERK pathway by precisely controlling the phosphorylation state of a single tyrosine site.Keywords
Tropomyosin receptor kinase A (TrkA) is a membrane receptor widely involved in the physiological processes of the cells, such as cell differentiation, growth, and synaptic plasticity (Huang and Reichardt, 2003). The general concept of TrkA activation is that when nerve growth factor (NGF) stimulates TrkA, it causes the autophosphorylation of five tyrosine residues (Tyr, Y) in the intracellular domain and then recruit the corresponding effector proteins to activate downstream signaling pathways. Among these sites, Y490 and Y785 are the two main sites for activating the mitogen-activated protein kinase/extracellular signal-regulated kinase 1/2 (MAPK/ERK) signaling pathway and the PLCγ pathway (Adriaenssens et al., 2008; Marlin and Li, 2015); Y670, Y674, and Y675 in the kinase domain region are closely related to the activation of the enzyme (Grewal et al., 1999; Biarc et al., 2013). The activation effects of these sites and their combinations are associated with the development of many diseases, but the mechanism has not been fully elucidated.
Our strategy for a better analysis of the TrkA signaling pathway was to regulate the phosphorylation state of the tyrosine side chain by introducing photosensitive unnatural amino acids (UAAs), thus making TrkA phosphorylation sites artificially controllable (Chen et al., 2017; Chin, 2017; Lateef et al., 2022). We chose two well-developed photosensitive tyrosine analogs: 1) photo-caged tyrosine (ONB), which can revert to tyrosine upon release of the photolabile moiety from the side chain after exposure to light (Arbely et al., 2012; Zhou and Deiters, 2021); 2) azido-L-phenylalanine (AzF), which can crosslink with nearby molecules within a distance of 3–4 Å after light stimulation (Grunbeck and Sakmar, 2013). To encode UAAs into protein, we used the genetic code expansion (GCE) technology to suppress the expression of the stop codon (usually an amber codon) in the mRNA of the target gene (here, TrkA) with the help of two pairs of engineered orthogonal aminoacyl tRNA synthetase (aaRS)/suppressor tRNA. The amber codon was used to encode specific UAAs (here, ONB or AzF), thereby enabling the read-through of the site-specific UAAs in the protein of interest (Davis and Chin, 2012; Xiao and Schultz, 2016; Brown et al., 2018; Gautier et al., 2014). In our previous study, we successfully used mammalian cell models (HEK293T and SH-SY5Y cells) to introduce AzF and ONB at the four phosphorylation sites (Y490, Y670, Y674, and Y675) of TrkA, respectively (Zhao et al., 2020). When these two photosensitive UAAs were introduced into these sites, all showed sensitivity to light and could specifically activate the downstream MAPK/ERK signaling pathway under the drive of UV stimulation.
As one of the main docking sites for TrkA, Y785 has been reported in previous studies to act on the ERK pathway depending on PLCγ and PKC protein, and it can regulate the differentiation of PC12 neural cells together with TrkA-Y490 (Obermeier et al., 1993; Khamo et al., 2019). But our light-controlled method established before has not been tested in the Y785 site (Zhao et al., 2020). We speculate that after the introduction of ONB at the TrkA-Y785 site, the downstream MAPK/ERK signaling pathway can also be activated by the light-controlled test according to the previous study, while the effects of the introduction of AzF at Y785 are unknown. Applying this light-controlled strategy to the TrkA-Y785 site is expected to help us further study the regulatory mechanism of TrkA phosphorylation. Therefore, based on our previous research, this paper intends to introduce two photosensitive UAAs (ONB and AzF) into the TrkA-Y785 site and combine the light-controlled method to detect its roles in regulating the MAPK/ERK signaling pathway. Our research will expand the application of the genetic code expansion (GCE) system in mammalian cells and provide a basis for understanding the contribution of the five phosphorylation sites in TrkA to specific signaling pathways (MAPK/ERK pathway) (Qin and Liu, 2022; Han et al., 2017).
HEK293T, SH-SY5Y cell lines required for this experiment were purchased from ATCC; the plasmid for TrkA-Y785 amber mutation was constructed according to the site-directed mutagenesis kit, and the wild-type TrkA plasmid (PLNCX-TrkA-ΔO-GFP) was from UPMC for Prof Brain. For AzF, ONB, two orthogonal RS/tRNA pairs that specifically recognize and fit them, were required. The plasmid AzFRS-(tRNA)4X (Ye et al., 2008), which recognizes AzF, carries the genes AzFRS (the mutant Escherichia coli Tyr tRNA-synthetase for AzF) and amber suppressor tRNA (derived from Bacillus stearothermophilus Tyr-tRNACUA). The plasmid was obtained from East China Normal University. The plasmid pONBYRS/U6-PyltRNA, which recognizes “photo-caged” tyrosine ONB, carries the plasmid pONBYRS (derived from the mutant Methanosarcina barkeri pyrrolysine tRNA synthetase for ONB) and the amber suppressor tRNA PylRS/tRNACUA pair, which has also been reported previously (Arbely et al., 2012). This plasmid was obtained from Peking University.
The expression of TrkA-785AzF and 785ONB mutants in HEK293T and SH-SY5Y cells
The TrkA-Y785amb plasmid was co-transfected with RS-tRNA (RS/tRNA pairs corresponding to AzF or ONB) into cells. The experimental procedures are as follows: (1) Preparation of cell culture before transfection: cells were plated the day before transfection to ensure that the cell density of the experimental group reached about 70% confluency the next day. (2) The medium was changed 1 h before transfection to DMEM/high glucose medium (serum-free). (3) Preparation of polyethylenimine (PEI)-DNA mixture: In a 24-well plate, for example, 0.5 µg of AzFRS-(tRNA)4X plasmid was mixed with 0.5 µg of PLNCX-TrkA (Y785amber)-ΔO-GFP plasmid and the mixture was co-transfected in 50 μL empty DMEM medium, marked as “A”; the ratio of DNA (µg): PEI (μL) = 1:3, and 3 μL PEI was added to another centrifuge tube containing 50 μL empty DMEM medium and marked as “B”. After allowing it to stand for 5 min, “B” was added to “A”. (4) After 20 min, the “AB” mixture was added to the corresponding well plate. (5) After 4–6 h, the medium was changed to the corresponding DMEM medium containing 1 mM AzF (for expressing ONB, the medium was replaced with a fresh one containing 100 µM ONB). Any exposure to light was carefully avoided. (6) Cells were collected 24–48 h after transfection for fluorescence imaging or western blotting. Suppression of the stop codon in TrkA-Y785amb produces full-length TrkA whose EGFP tag reports the binding of AzF or ONB at the stop codon TAG. We added AzF (1 mM) or ONB (100 μM) to the cell growth medium, and observed the fluorescence expression at 24–48 h. If the fluorescence intensity of EGFP in the cells was significantly enhanced compared with that without AzF or ONB, it indicated a successful addition of AzF or ONB to TrkA.
Analysis of photo-caged tyrosine and azido-L-phenylalanine incorporation efficiency at Y785 site of TrkA in HEK293T cells by fluorescence microscopy
After 24–48 h of transfection, photos were taken using a fluorescence microscope, and six different cell fields were taken for each mutant for statistics. The experiment was repeated no less than three times for each mutant. The pictures taken were processed by the ImageJ software, and the average fluorescence intensity was counted. The rates of ONB and AzF incorporation at the Y785 site were expressed as the mean fluorescence intensity of cells incorporating ONB or AzF divided by the mean fluorescence intensity of cells expressing WT TrkA. The R language was used to draw statistical graphs.
Nerve growth factor and ultraviolet treatment for protein expression and cell phenotype analysis
After 48 h of transfection, NGF stimulation and light treatment were performed. Before this step, the medium supplemented with 1 mM AzF or 100 uM ONB was replaced with fresh phosphate buffer saline (PBS). Then cell plates that needed to be exposed to UV were placed on ice and irradiated with light for 5–10 min in a 365 nm UV cross-linker (40 W). After UV treatment, the PBS was replaced with a serum-free medium, and cells were cultured for 1 h in a carbon dioxide (5%) incubator at 37°C. Cells were then stimulated by adding human NGF to the medium at a final concentration of 50 ng/mL. As a control, an equal volume of NGF-free medium-well plates was used. HEK293T cells needed to be incubated at 37°C for 20 min for western blot analysis, while neuronal cells needed to be treated in an NGF-containing medium for 24–48 h (dark) for cell growth and differentiation ratio analysis (The control group (without NGF treatment) also needed to ensure the same treatment).
Detecting the expression levels of TrkA and its activated downstream signaling proteins in HEK293T cells by western blot
HEK293T cells were collected 48 h after transfection, washed with PBS, and protein samples were prepared and quantified according to the instructions of the BCA protein quantitative detection kit. Then the protein was resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto the membrane. The membrane was then blocked and incubated with primary and secondary antibodies. The membranes were probed with primary antibodies against the TrkA epitope (Sigma-Aldrich, United States; 1:2000), p-ERK1/2 (Millipore, United States; 1:1000), actin (Sigma-Aldrich, United States; 1:1000), and a horse-radish peroxidase-conjugated secondary antibody (Bio-Rad, United States; 1:5,000–1:10,000; anti-mouse or anti-rabbit IgG). Signals were visualized by ECL treatment (Perkin Elmer Life Sciences, USA) and exposure to ImageQuant. Equal amounts of total protein were loaded per lane. ERK activation levels were expressed by the relative expression level of p-ERK/total ERK. All detailed statistical datasets were included in the corresponding legends. The data in the statistical chart was expressed as mean ± SD, and two groups was compared by student’s t-test (unpaired, two-tailed). p < 0.05 was considered statistically significant. All experiments were performed at least three times, and the results were consistent.
Neuronal cells climbing for confocal laser imaging analysis
The coverslips were treated in advance; they were soaked overnight in 1% dilute hydrochloric acid, rinsed with water the next day, then autoclaved and dried, and soaked in 75% ethanol for later use. The slides were treated with 0.01 mg/mL polylysine for 10 min before inoculation, then repeatedly washed with ddH2O and dried for later transfection. The treated slides were seeded in 24-well plates one day before transfection. After 48 h of transfection, the slides were washed with PBS, fixed with ice methanol, and then stained with DAPI. Finally, the covered slides were stored in a wet and dark box for confocal laser imaging analysis.
SH-SY5Y cell differentiation assay
Cells were harvested 48 h post-transfection and treated with NGF and with or without UV, as mentioned above. Differentiation ratios for each mutant at various conditions were calculated at various conditions: Differentiation ratios = (Nm/Nmt)/(NWT/NWTt) (Zhao et al., 2020).
Nm = Number of green fluorescing cells expressing a specific mutant with neurite length longer than the cell body diameter.
Nmt = Number of total green fluorescing cells expressing a specific mutant.
NWT = Number of green fluorescing cells expressing WT TrkA with neurite length longer than the cell body diameter.
NWTt = Number of total green fluorescing cells expressing WT TrkA.
Quantification and statistical analysis
Quantification of western blots was carried out using ImageJ. Details of all statistical datasets are included in the corresponding figure legends.
Introducing azido-L-phenylalanine and photo-caged tyrosine at the TrkA-Y785 site
To introduce ONB and AzF at the TrkA-Y785 site, we constructed the TrkA-Y785amb plasmid based on the obtained WT-TrkA plasmid. TrkA-Y785amb is structurally the same as WT-TrkA; that is, the TrkA intracellular domain is flagged with an EGFP tag at the C-terminus. The only difference between them is that the Y785 site has an amber-stop codon mutation. So only after a successful introduction of AzF or ONB at the Y785 site the expression of TrkA-EGFP fusion protein can be detected; That is, the expression of TrkA can be visualized by EGFP tag (Fig. 1a). We introduced ONB or AzF (Fig. 1b) at the Y785 site in HEK293T cells by co-transfecting the TrkA-Y785amb plasmid with a single-plasmid system pONBYRS/U6-PyltRNA (recognizing ONB) or the TrkA-Y785amb plasmid with a single-plasmid system AzFRS-tRNA4X (identifying AzF) in HEK293T cells. We detected the green fluorescence signals when 100 µM ONB or 1 mM AzF was added to the medium, but the fluorescence was weak and undetectable without the addition of UAAs (Figs. 1c and 1e).
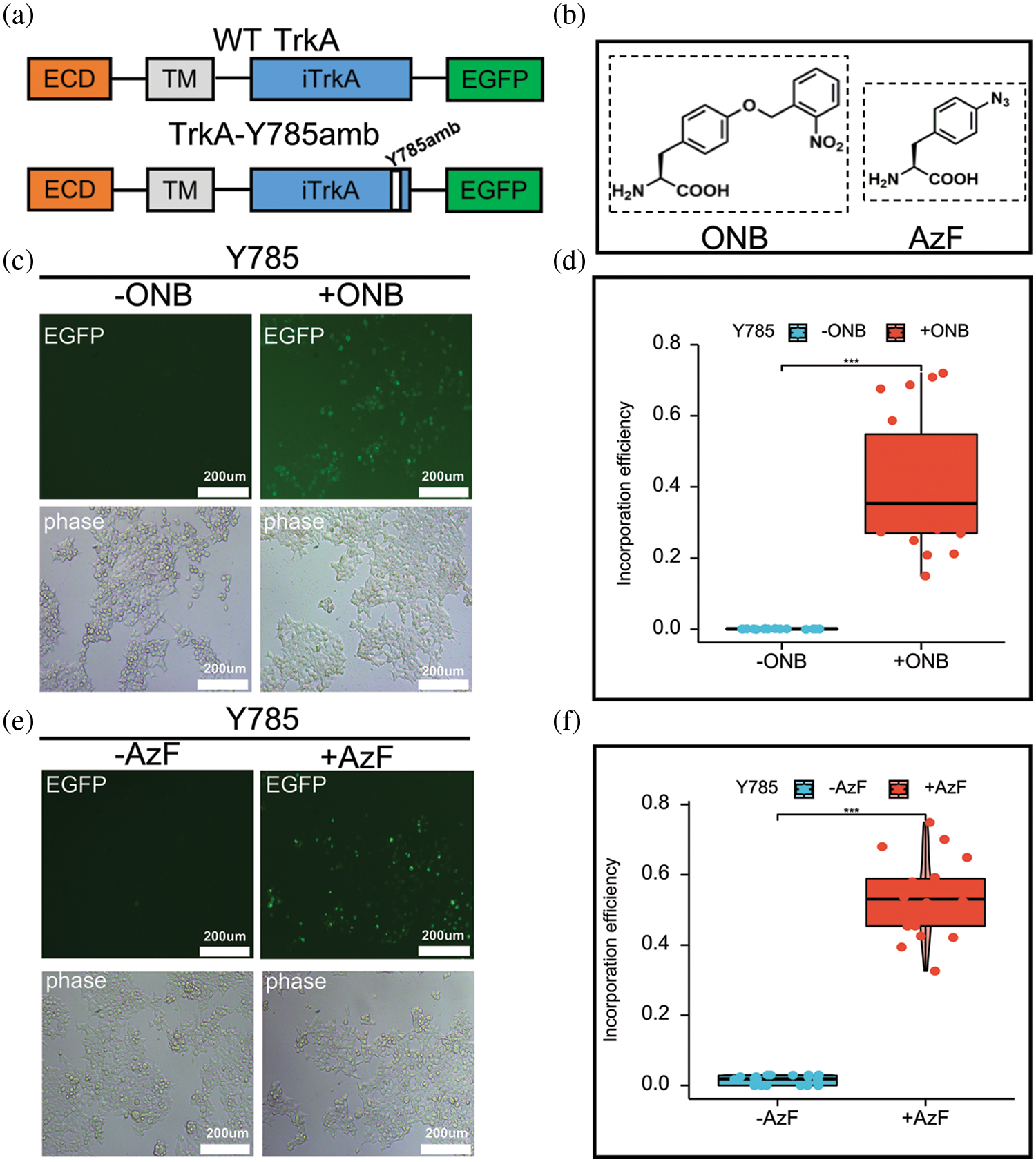
Figure 1: Incorporation of AzF and ONB at TrkA-Y785 site in HEK293T cells. (a) Schematic diagram of the constructed TrkA receptor tagged with EGFP at the C-terminal. The full-length WT-TrkA receptor includes three domains: an extracellular domain (ECD), a transmembrane domain (TM), an intracellular domain (iTrkA), and a fusion protein (EGFP). TrkA-Y785amb has an amber mutation TAG introduced into the Y785 position in the iTrkA domain. (b) Chemical structures of ONB and AzF. Fluorescent images of HEK293T cells co-transfected with AzFRS-tRNA4x (AzF) or pONBYRS/U6-PyltRNA (ONB) at one site out of tyrosine kinase domain Y785 in the presence (+) or absence (−) of 100 µM ONB (c) and 1 mM AzF (e). 24–48 h post-transfection, cells were imaged using fluorescence microscopy for GFP expression (Scale bar: 200 µm). (d) The incorporation efficiency of TrkA-ONB mutants in the absence (grey) or presence (dark) of ONB. (f) The incorporation efficiency expression of TrkA-AzF mutants in the absence (grey) or presence (dark) of AzF. The incorporation efficiency was described as average fluorescence intensity (%) in cells expressing UAA-incorporating receptors divided by the moderate fluorescence intensity into cells expressing the wt TrkA receptors. Mean fluorescence intensity = Intensity/area; a sample of six fields was taken. Sample numbers are three. Error bars show SD.
The above fluorescence images were statistically analyzed. The ratio of the average fluorescence intensity of Y785 to the ratio of WT-TrkA was calculated to consider the difference in counts and the intensity of the fluorescence expression. The two mutants were compared with WT-TrkA (data from our previous study) to assess fluorescent expression levels and UAAs’ introduction efficiency. Analysis of fluorescence imaging results showed that the yield of the TrkA-AzF mutant was about 53% of that of WT-TrkA (Fig. 1f). In contrast, the expression level of the TrkA-ONB mutant was slightly lower than that of TrkA-AzF, accounting for about 40.0% of WT TrkA (Fig. 1d). The expression levels of two UAAs at the same site were slightly different. At the same time, in the absence of AzF and ONB, both groups showed quite low fluorescence expression levels (Figs. 1d and 1f).
Thus, the difference in mean fluorescence intensity of the two mutants, as counted by fluorescence imaging, generally indicates the efficient amber-stop codon suppression at the TrkA-Y785 target, resulting in the specific synthesis of AzF or ONB in TrkA. Both tRNA/RS pairs exhibited good orthogonality at the Y785 site.
Light-controlled extracellular signal-regulated kinase 1/2 activation by TrkA-Y785 dependent on NGF treatment
In addition to the fluorescence analysis, we used the expression characteristics of the TrkA-EGFP fusion protein to reflect the read-through of TrkA. Through laser confocal microscopy, we observed that TrkA-785AzF and TrkA-785ONB mutants were mainly expressed on the cell membrane, where the blue fluorescence represented the nuclear localization, and the green fluorescence represented the expression of full-length TrkA-EGFP. This experiment further demonstrated that the TrkA-Y785 site could efficiently and specifically introduce AzF and ONB in HEK293T cells, thereby expressing the photosensitive TrkA-UAA mutants on the cell membrane (Fig. 2a).
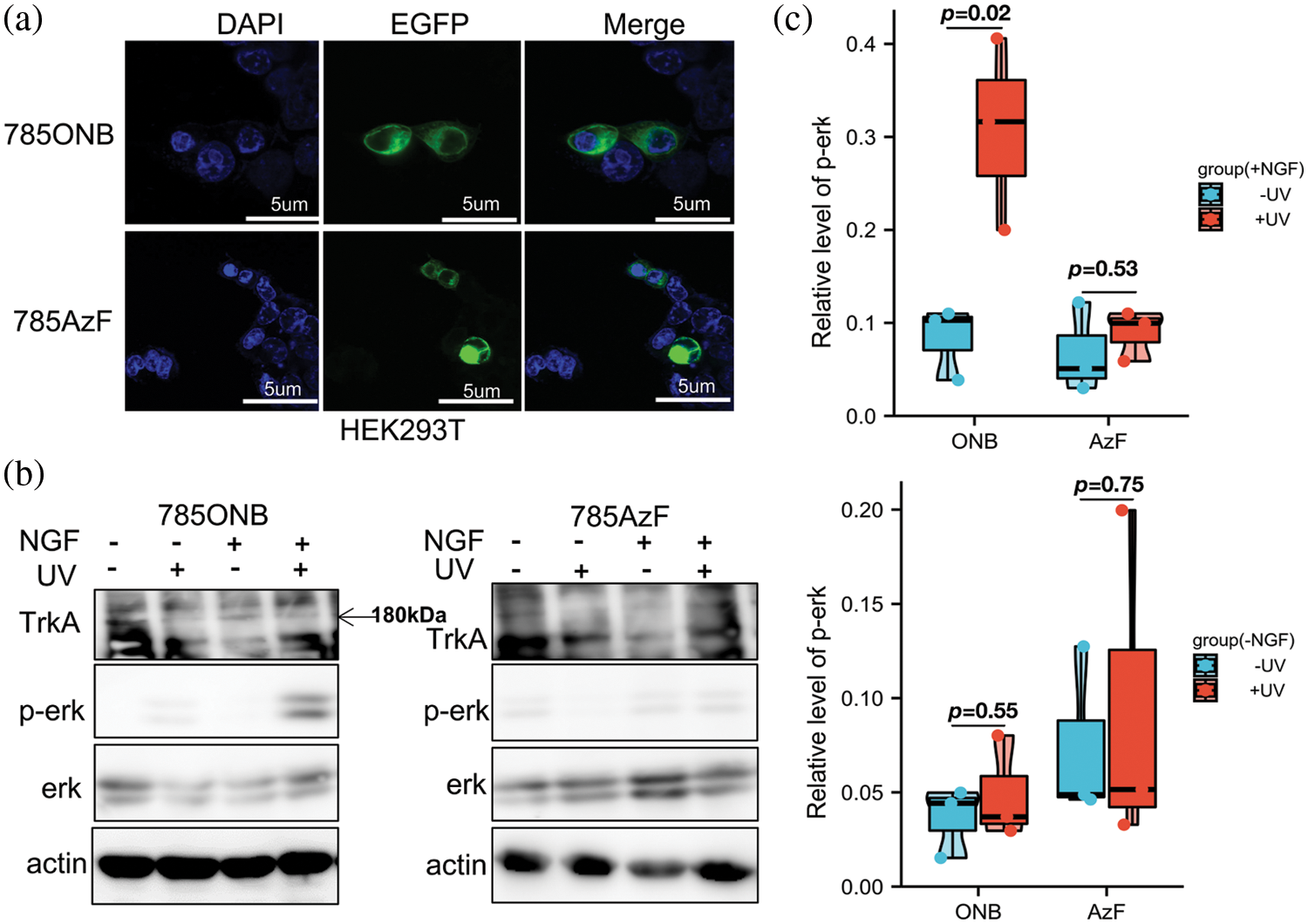
Figure 2: MAPK/ERK pathway activated by photosensitive TrkA-Y785ONB. (a) Fluorescence images of TrkA-785AzF or TrkA-785ONB mutants expressed in HEK293T cells. Blue, nuclei stained with DAPI (4′, 6-diamidino-2-phenylindole); green, enhanced green fluorescent protein (EGFP) tagged TrkA (Scale bar: 5 µm). (b) Western blotting analysis of HEK293T cells expressing TrkA-Y785ONB and 785AzF mutants for phosphorylated ERK (Thr202 and Tyr204) and ERK, without (−) or with (+) NGF and without (−) or with (+) UV light. TrkA and β-actin were used as a loading control. (c) The expression of phosphorylated ERK for TrkA-785ONB and 785AzF mutants in the absence (blue) or presence (red) of ultraviolet (UV) Light. Cells were transfected with the amber mutant at the Y785 site of TrkA and the RS/tRNA pair for each unnatural amino acid (UAA), expressing TrkA-ONB and TrkA-AzF at Y785 in the presence and absence of NGF. Error bars show SD p < 0.05 represents statistically significant.
Next, to test whether this light-controlled strategy testing at the Y785 site could activate the MAPK/ERK pathway, we examined the activation of p-ERK by TrkA-Y785ONB and TrkA-Y785AzF mutants after NGF and UV treatment. Under UV irradiation, ONB released the “photocage” group from the introduction site and recovered to tyrosine, followed by autophosphorylation of the hydroxyl group on the tyrosine site. According to our previous study, we tested whether UV light stimulation in TrkA could directly activate ERK without NGF. Wt-TrkA was expressed and assayed in the absence and presence of NGF and served as a control with non-detectable p-ERK neither without nor with UV treatment, confirming that the UV treatment does not cause significant functional changes. To assess whether the photoactivation of TrkA-Y785ONB leads to functional changes in ERK signaling, we performed immunoblot analysis of p-ERK activity following UV treatment in live cells which express photosensitive ONB. The TrkA-Y785ONB mutant was expressed in HEK293T cells by co-transfecting the Y785amb plasmid and the pONBYRS/U6-PyltRNA pair. Then cells were treated with NGF and UV stimulation. After extracting protein from cells, we continued to detect TrkA, p-ERK, ERK, and actin levels. It is found that p-ERK was not detected in TrkA-Y785ONB in the absence of UV due to the presence of photolabile groups in ONB, while UV stimulation induced apparent expression of p-ERK in the presence of NGF (Fig. 2b and Suppl. Fig. S1c). To quantify p-ERK activation levels, we calculated the average ratio of p-ERK to total ERK in each mutant before and after UV treatment. Quantification of p-ERK levels revealed about 30% TrkA-Y785ONB expression level under UV and NGF treatment, which showed a stronger expression level than the untreated UV group (8.3%) (Fig. 2c). We also examined the photo-induced effect of AzF at the Y785 site. Compared with ONB, AzF undergoes a different photochemical process. Upon UV treatment, AzF typically generates a nitrogen radical, which may subsequently be reduced to an amine or form a covalent bond with an adjacent atom within a 3–4 Å distance. Unlike the previous results in the kinase domain region (Zhao et al., 2020), the Y785 site did not show an apparent photo-controllable p-ERK activation after the introduction of AzF under NGF and UV treatment (Fig. 2b). However, in the absence of NGF stimulation, there was no significant difference in p-ERK expression levels with or without light stimulation in both ONB and AzF mutants (Figs. 2b, 2c, Suppl. Figs. S1a, S1b and S1d).
Taken together, our results demonstrate that TrkA-Y785ONB could induce p-ERK activation by light control only in the presence of NGF, while theTrkA-Y785AzF could not. Among them, ONB acts as a powerful light-controlled “switch” to control the phosphorylation state of this tyrosine site.
The photosensitive TrkA-Y785ONB mutant regulates SH-SY5Y cell differentiation dependent on nerve growth factor
Compared to HEK293T cells, which were able to efficiently transfect and successfully express TrkA-ONB and TrkA-AzF mutants, the successful application of this technology to neurons required extensive experimental optimization. So, in our previous study, we tested light-sensitive TrkA mutants (Y490, Y670, Y674, and Y675) in the SH-SY5Y cell model due to its lesser expression of endogenous TrkA, aiming to establish a light-controlled method for neurite outgrowth (Zhao et al., 2020). Therefore, using the same strategy, we introduced AzF and ONB into the Y785 site and carried out fluorescence expression detection in SH-SY5Y cells. In the case of adding AzF or ONB in the medium, only weaker EGFP expression (about 7%) could be detected in two groups of cells. In the absence of AzF or ONB, almost no fluorescence was detected. The two tRNA/RS still showed good orthogonality in SH-SY5Y cells (Suppl. Figs. S2 and S3).
In addition, immunoblotting experiments showed that in WT SH-SY5Y cells with or without NGF treatment, the p-ERK signal was almost undetectable (Fig. 3a). After transfecting exogenous WT TrkA in cells, the ERK activation could only be detected after NGF treatment (Fig. 3a). We tested SH-SY5Y cells expressing WT TrkA and found that after treatment with 50 ng/mL NGF for 24–48 h, cells differentiated significantly with or without UV light treatment. No obvious cell differentiation effect was seen when cells were treated without NGF stimulation (Fig. 3b), which indicated that the exogenous TrkA retained ligand sensitivity and signaling activity, and UV light had no obvious effect on cell differentiation.
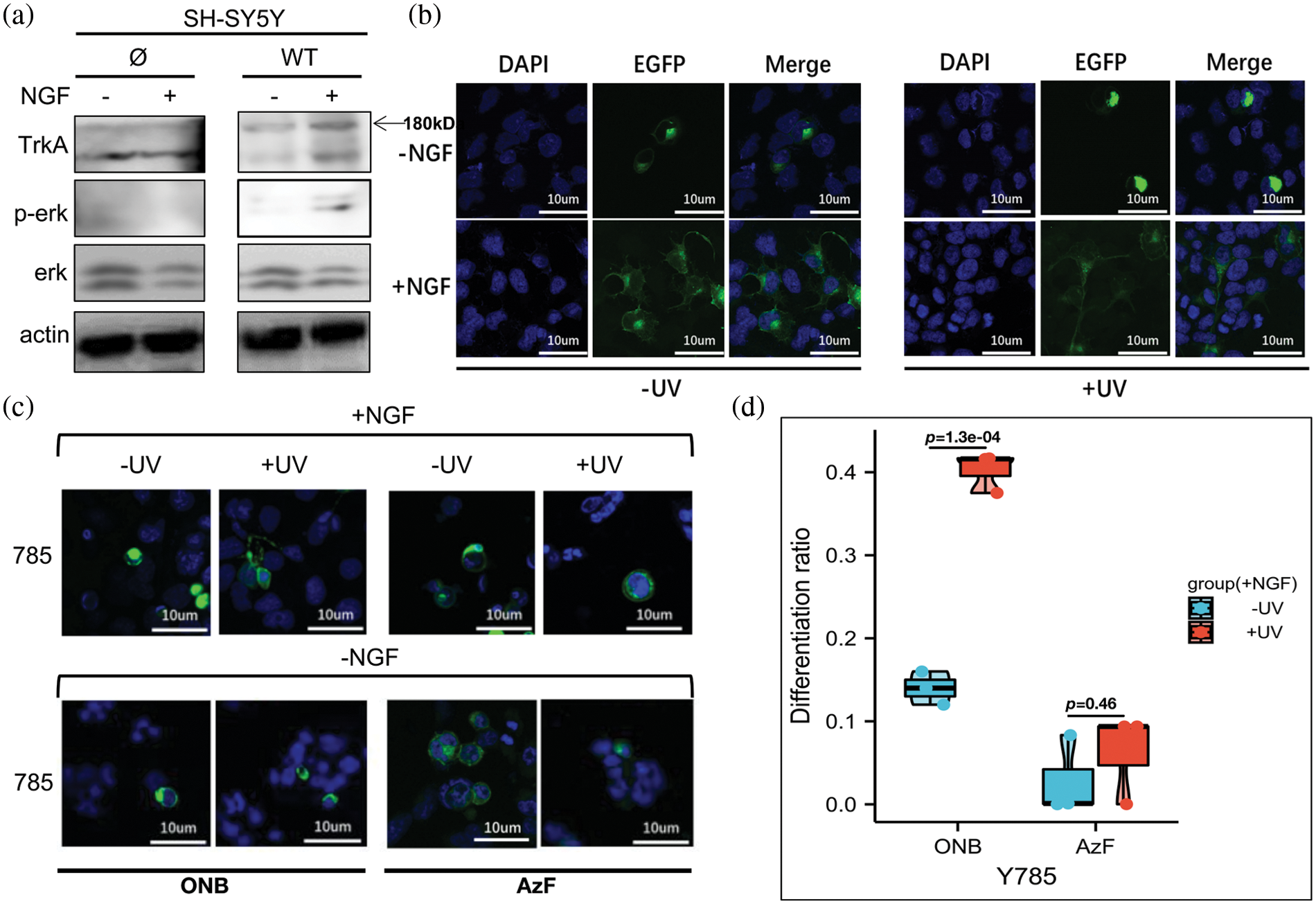
Figure 3: TrkA-785ONB mutant promotes SH-SY5Y cell differentiation in response to UV stimulation with nerve growth factor (NGF). (a) Western blot analysis of SH-SY5Y cells expressing endogenous TrkA (Ø) or WT TrkA for phosphorylated ERK (Thr202 and Tyr204) and ERK without (−) or with (+) NGF treatment. TrkA and β-actin were used as a loading control. (b) Representative fluorescence images depicting SH-SY5Y cells expressing wt TrkA before (−) and after (+) UV treatment, with medium supplied without (−) or with (+) 50 ng/mL NGF kept in the dark for 24–48 h before imaging. (c) Representative fluorescence images depicting SH-SY5Y cells expressing TrkA-785ONB (two columns on the left) or 785AzF (two columns on the right) mutants with (+) and without (−) UV treatment, with medium supplied with NGF (50 ng/mL). Cells were cultured in the cell incubator for 24–48 h (dark) before imaging. (d) Differentiation ratios calculated for various mutants expressed in SH-SY5Y cells in the presence of NGF with or without UV light. Differentiation ratios were calculated using the ratios (No. of green fluorescing cells with neurite longer than cell body/No. of green fluorescing cells) for Y785 and wt TrkA at each condition. Then the ratios of each mutant/wt TrkA were treated in the same condition. Values above represent the mean ± SD of three biological replicates (n = 3) with >200 cells counted per replicate. Scale bar: 10 µm.
Hence, we continued to test the TrkA-Y785 site in SH-SY5Y cells. We successfully expressed light-sensitive UAA-mutants at the TrkA-Y785 site in SH-SY5Y cells. The next step was to analyze neurite outgrowth and cell differentiation with and without NGF treatment through the light-controlled Y785 site. TrkA-785ONB and TrkA-785AzF mutants were expressed by separately co-transfecting two sets of genetic code systems into SH-SY5Y cells, which were consistent with the previous western blotting treatment. Neuronal cells were cultured in the dark for 24–48 h in the serum-free medium containing 50 ng/mL NGF, and their neuronal growth were compared using WT TrkA. In the presence of NGF, cells expressing the Y785ONB mutant showed differences in UV-induced differentiation relative to the untreated UV group (differentiation rates: UV-treated group: 40.6%; UV-untreated group: 13.9%) (Figs. 3c and 3d). However, the mutant Y785AzF without obvious p-ERK activation effects before and after UV irradiation also showed no obvious difference in cell differentiation in neural cells (differentiation rates: UV-treated group: 0.6%; UV-untreated group: 0.3%). In the absence of NGF treatment, there was no apparent cell differentiation phenotype for either UAAs (Fig. 3c and Suppl. Fig. S4), which showed consistent results with p-ERK expression levels in HEK293T cells.
Thus, our results suggest that activation of p-ERK in the TrkA-Y785 site exhibits substantial consistency with neuronal cell differentiation regardless of NGF stimulation.
The general concept of TrkA signaling activation is that the ligand NGF from the extracellular domain promotes the homodimerization of TrkA at the cell membrane, which in turn activates downstream signaling pathways. However, because the TrkA signaling pathway is intricate and related to five intracellular phosphory-lation sites (Y490, Y670, Y674, Y675, and Y785) and their combinations, the mechanism has not yet been fully elucidated (Duan et al., 2018).
We were interested in developing systems for light-controlled protein function through site-specific control of the kinase receptor TrkA and using genetic code expansion (GCE) technology to combine photosensitive UAAs with site-directed mutagenesis (phosphorylation sites). This method is used for precise, light-directed activation of the membrane kinase receptor TrkA, as it provides a novel concept for studying NGF/TrkA signaling pathways. At present, we have tried at the five phosphorylation sites of TrkA (two docking sites, Y490 and Y785; three sites, Y670, Y674, and Y675, in the kinase domain) and found that the downstream MAPK/ERK pathway can be specifically activated by light-controlled means through the introduction of photosensitive UAAs into these five sites. The two introduced molecules, ONB and AzF, are chemically structurally distinct from tyrosine, and both can prevent phosphorylation before photoactivation. However, different photochemical reactions occur after UV illumination, which provides a light-controlled method for analyzing the contribution of each tyrosine to the activation of specific pathways during the TrkA signal transduction (Pless and Ahern, 2013; Coin et al., 2013; Huber and Sakmar, 2014). This advantage cannot be provided by conventional mutagenesis methods (Arbely et al., 2012; Baker and Deiters, 2014).
Our light control study of the TrkA-Y785 site indicated two points of interest: 1) For the difference between ONB and AzF in terms of photoactivation at the Y785 position. Our interpretation is that Y490 and Y785 are the main sites to activate the MAPK/ERK pathway, and inhibiting the Y785 site results in a dramatic decrease in the level of ERK (Thr-202/Tyr-204, Thr-183/Tyr-185) signaling. When the Y785 site was mutated to ONB or AzF, the ERK activity was inhibited; for ONB, the ERK pathway could not be fully activated by UV irradiation unless the “caged” compound was released. When Y785 is mutated to AzF, it results in the inactivation of MAPK/ERK signaling similar to the conventional site-directed mutagenesis, such as phenylalanine (F), prior to the UV treatment. We observed that ERK still could not be activated after UV treatment, which proves that the photoproduct of AzF at the Y785 site cannot be phosphorylated. Our experiments were performed under the same conditions; the only difference was whether the Y785 site was treated with UV or not. So the obtained results are all reasonable and non-artificial. It is suggested that the ligand-dependent activation of Y785ONB by light-controlled p-ERK may reflect the ligand-induced site-state-dependent activation process. 2) The two docking sites, Y490 and Y785, are the main activation sites of the MAPK/ERK pathway. SH-SY5Y cells were significantly differentiated at the Y785 site compared with before and after light control only under the condition of introducing ONB and NGF treatment. Light-controlled activation of the Y785 site indicated that this site activates the MAPK/ERK pathway dependent on the NGF ligand (Figs. 4a and 4b). Using the SH-SY5Y cell model with less endogenous TrkA interference, we combined this (microscopic) detection of the ERK pathway in photosensitive TrkA mutants with neural cell differentiation (cellular phenotype), which all showed good consistency. This is reasonable because Y785 has been reported in previous studies to activate ERK in a PLCγ-PKC signaling-dependent manner, and cell differentiation was significantly reduced after simultaneous inhibition of both Y490 and Y785 phosphorylation, suggesting that Y785 makes important contributions in regulating the MAPK/ERK pathway. Our light-controlled approach offers advantages in explaining the phenotypic roles of the Y785 site and other single tyrosine sites (Zhao et al., 2020). We expect that separation and combination light-control studies of these critical sites will provide a powerful approach to resolving the polymorphism of the tyrosine kinase receptor phosphorylations and the diversity of its functions.
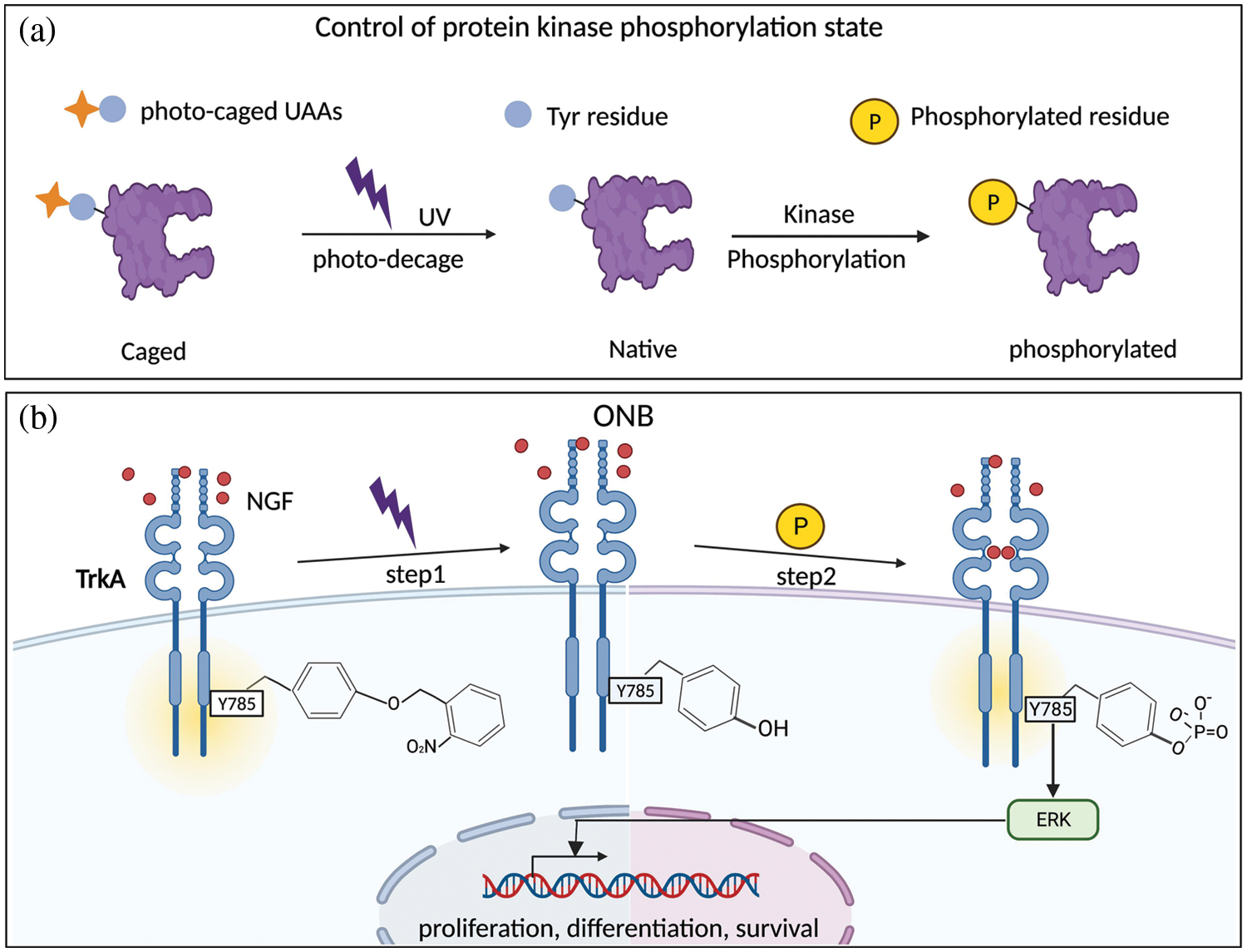
Figure 4: Reactions caused by light-controlled photosensitive unnatural amino acids (UAAs). (a) Control of protein phosphorylation state using photocaged UAAs. Introduction of photocaged UAAs into specific phosphorylation sites of proteins blocked the phosphorylation of the tyrosine sites. Then the sites were restored by light-induced deprotection and subsequently phosphorylated. (b) The introduction of ONB analog can switch “on” and “off” tyrosine phosphorylation at a possible site; here, it is the TrkA-Y785 site.
To sum up, we supplemented the light-controlled results of the TrkA-Y785 site based on our previous research and further expanded the application of genetic code expansion technology in kinase receptors. Combined with our previous results (Zhao et al., 2020), we propose that these five phosphorylation sites may have different contributions to the activation of the MAPK pathway, and site-dependent phosphorylations in TrkA may have polymorphic control over the signaling process. Through precise optical control, the activity of kinase receptors can be regulated at a single phosphorylation site to achieve the effect of site-directed light-controlled phosphorylation (Ye et al., 2010; Broichhagen et al., 2015; Zhou and Deiters, 2021). This approach lays the foundation for a comprehensive analysis of kinase-related pathways and screening of compounds that intervene in site-specific phosphorylation pathways in targeted therapy.
Acknowledgement: We thank the biorender website for providing the convenience of drawing.
Funding Statement: The authors received no specific funding for this study.
Author Contributions: Study conception and design: Shu Zhao. Data collection: Shu Zhao. Analysis and interpretation of results: Shu Zhao. Draft manuscript preparation: Shu Zhao and Shixin Ye. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
Adriaenssens E, Vanhecke E, Saule P, Mougel A, Page A, Romon R, Nurcombe V, Le Bourhis X, Hondermarck H (2008). Nerve growth factor is a potential therapeutic target in breast cancer. Cancer Research 68: 346–351. https://doi.org/10.1158/0008-5472.CAN-07-1183 [Google Scholar] [PubMed] [CrossRef]
Arbely E, Torres-Kolbus J, Deiters A, Chin JW (2012). Photocontrol of tyrosine phosphorylation in mammalian cells via genetic encoding of photocaged tyrosine. Journal of the American Chemical Society 134: 11912–11915. https://doi.org/10.1021/ja3046958 [Google Scholar] [PubMed] [CrossRef]
Baker AS, Deiters A (2014). Optical control of protein function through unnatural amino acid mutagenesis and other optogenetic approaches. ACS Chemical Biology 9: 1398–1407. https://doi.org/10.1021/cb500176x [Google Scholar] [PubMed] [CrossRef]
Biarc J, Chalkley RJ, Burlingame AL, Bradshaw RA (2013). Dissecting the roles of tyrosines 490 and 785 of TrkA protein in the induction of downstream protein phosphorylation using chimeric receptors. Journal of Biological Chemistry 288: 16606–16618. https://doi.org/10.1074/jbc.M113.475285 [Google Scholar] [PubMed] [CrossRef]
Broichhagen J, Damijonaitis A, Levitz J, Sokol KR, Leippe P, Konrad D, Isacoff EY, Trauner D (2015). Orthogonal optical control of a G protein-coupled receptor with a SNAP-tethered photochromic ligand. ACS Central Science 1: 383–393. https://doi.org/10.1021/acscentsci.5b00260 [Google Scholar] [PubMed] [CrossRef]
Brown W, Liu J, Deiters A (2018). Genetic code expansion in animals. ACS Chemical Biology 13: 2375–2386. https://doi.org/10.1021/acschembio.8b00520 [Google Scholar] [PubMed] [CrossRef]
Chen Y, Lu L, Ye S (2017). Genetic code expansion and optoproteomics. Yale Journal of Biology and Medicine 90: 599–610. https://www.ncbi.nlm.nih.gov/pubmed/29259524 [Google Scholar] [PubMed]
Chin JW (2017). Expanding and reprogramming the genetic code. Nature 550: 53–60. https://doi.org/10.1038/nature24031 [Google Scholar] [PubMed] [CrossRef]
Coin I, Katritch V, Sun T, Xiang Z, Siu FY, Beyermann M, Stevens RC, Wang L (2013). Genetically encoded chemical probes in cells reveal the binding path of urocortin-I to CRF class B GPCR. Cell 155: 1258–1269. https://doi.org/10.1016/j.cell.2013.11.008 [Google Scholar] [PubMed] [CrossRef]
Davis L, Chin JW (2012). Designer proteins: Applications of genetic code expansion in cell biology. Nature Reviews Molecular Cell Biology 13: 168–182. https://doi.org/10.1038/nrm3286 [Google Scholar] [PubMed] [CrossRef]
Duan L, Hope JM, Guo S, Ong Q, Francois A, Kaplan L, Scherrer G, Cui B (2018). Optical activation of TrkA signaling. ACS Synthetic Biology 7: 1685–1693. https://doi.org/10.1021/acssynbio.8b00126 [Google Scholar] [PubMed] [CrossRef]
Gautier A, Gauron C, Volovitch M, Bensimon D, Jullien L, Vriz S (2014). How to control proteins with light in living systems. Nat Chemical Biology 10: 533–541. https://doi.org/10.1038/nchembio.1534 [Google Scholar] [PubMed] [CrossRef]
Grewal SS, York RD, Stork PJ (1999). Extracellular-signal-regulated kinase signalling in neurons. Current Opinion in Neurobiology 9: 544–553. https://doi.org/10.1016/S0959-4388(99)00010-0 [Google Scholar] [PubMed] [CrossRef]
Grunbeck A, Sakmar TP (2013). Probing G protein-coupled receptor-ligand interactions with targeted photoactivatable cross-linkers. Biochemistry 52: 8625–8632. https://doi.org/10.1021/bi401300y [Google Scholar] [PubMed] [CrossRef]
Han S, Yang A, Lee S, Lee HW, Park CB, Park HS (2017). Expanding the genetic code of Mus musculus. Nature Communications 8: 14568. https://doi.org/10.1038/ncomms14568 [Google Scholar] [PubMed] [CrossRef]
Huang EJ, Reichardt LF (2003). Trk receptors: Roles in neuronal signal transduction. Annual Review of Biochemistry 72: 609–642. https://doi.org/10.1146/annurev.biochem.72.121801.161629 [Google Scholar] [PubMed] [CrossRef]
Huber T, Sakmar TP (2014). Chemical biology methods for investigating G protein-coupled receptor signaling. Chemistry & Biology 21: 1224–1237. https://doi.org/10.1016/j.chembiol.2014.08.009 [Google Scholar] [PubMed] [CrossRef]
Khamo JS, Krishnamurthy VV, Chen Q, Diao J, Zhang K (2019). Optogenetic delineation of receptor tyrosine kinase subcircuits in PC12 cell differentiation. Cell Chemical Biology 26: 400–410.e3. https://doi.org/10.1016/j.chembiol.2018.11.004 [Google Scholar] [PubMed] [CrossRef]
Lateef OM, Akintubosun MO, Olaoba OT, Samson SO, Adamczyk M (2022). Making sense of “nonsense” and more: Challenges and opportunities in the genetic code expansion, in the world of tRNA modifications. International Journal of Molecular Sciences 23: 938. https://doi.org/10.3390/ijms23020938 [Google Scholar] [PubMed] [CrossRef]
Marlin MC, Li G (2015). Biogenesis and function of the NGF/TrkA signaling endosome. International Review of Cell and Molecular Biology 314: 239–257. https://doi.org/10.1016/bs.ircmb.2014.10.002 [Google Scholar] [PubMed] [CrossRef]
Obermeier A, Halfter H, Wiesmuller KH, Jung G, Schlessinger J, Ullrich A (1993). Tyrosine 785 is a major determinant of Trk--substrate interaction. EMBO Journal 12: 933–941. https://doi.org/10.1002/j.1460-2075.1993.tb05734.x [Google Scholar] [PubMed] [CrossRef]
Pless SA, Ahern CA (2013). Unnatural amino acids as probes of ligand-receptor interactions and their conformational consequences. Annual Review of Pharmacoloy and Toxicology 53: 211–229. https://doi.org/10.1146/annurev-pharmtox-011112-140343 [Google Scholar] [PubMed] [CrossRef]
Qin X, Liu T (2022). Recent advances in genetic code expansion techniques for protein phosphorylation studies. Journal of Molecular Biology 434: 167406. https://doi.org/10.1016/j.jmb.2021.167406 [Google Scholar] [PubMed] [CrossRef]
Xiao H, Schultz PG (2016). At the interface of chemical and biological synthesis: An expanded genetic code. Cold Spring Harbor Perspect Biology 8: a023945. https://doi.org/10.1101/cshperspect.a023945 [Google Scholar] [PubMed] [CrossRef]
Ye S, Kohrer C, Huber T, Kazmi M, Sachdev P, Yan ECY, Bhagat A, RajBhandary UL, Sakmar TP (2008). Site-specific incorporation of keto amino acids into functional G protein-coupled receptors using unnatural amino acid mutagenesis. Journal of Biological Chemistry 283: 1525–1533. https://doi.org/10.1074/jbc.M707355200 [Google Scholar] [PubMed] [CrossRef]
Ye S, Zaitseva E, Caltabiano G, Schertler GF, Sakmar TP, Deupi X, Vogel R (2010). Tracking G-protein-coupled receptor activation using genetically encoded infrared probes. Nature 464: 1386–1389. https://doi.org/10.1038/nature08948 [Google Scholar] [PubMed] [CrossRef]
Zhao S, Shi J, Yu G, Li D, Wang M et al. (2020). Photosensitive tyrosine analogues unravel site-dependent phosphorylation in TrkA initiated MAPK/ERK signaling. Communications Biology 3: 706. https://doi.org/10.1038/s42003-020-01396-0 [Google Scholar] [PubMed] [CrossRef]
Zhou W, Deiters A (2021). Chemogenetic and optogenetic control of post-translational modifications through genetic code expansion. Current Opinion in Chemical Biology 63: 123–131. https://doi.org/10.1016/j.cbpa.2021.02.016 [Google Scholar] [PubMed] [CrossRef]
Supplementary Materials
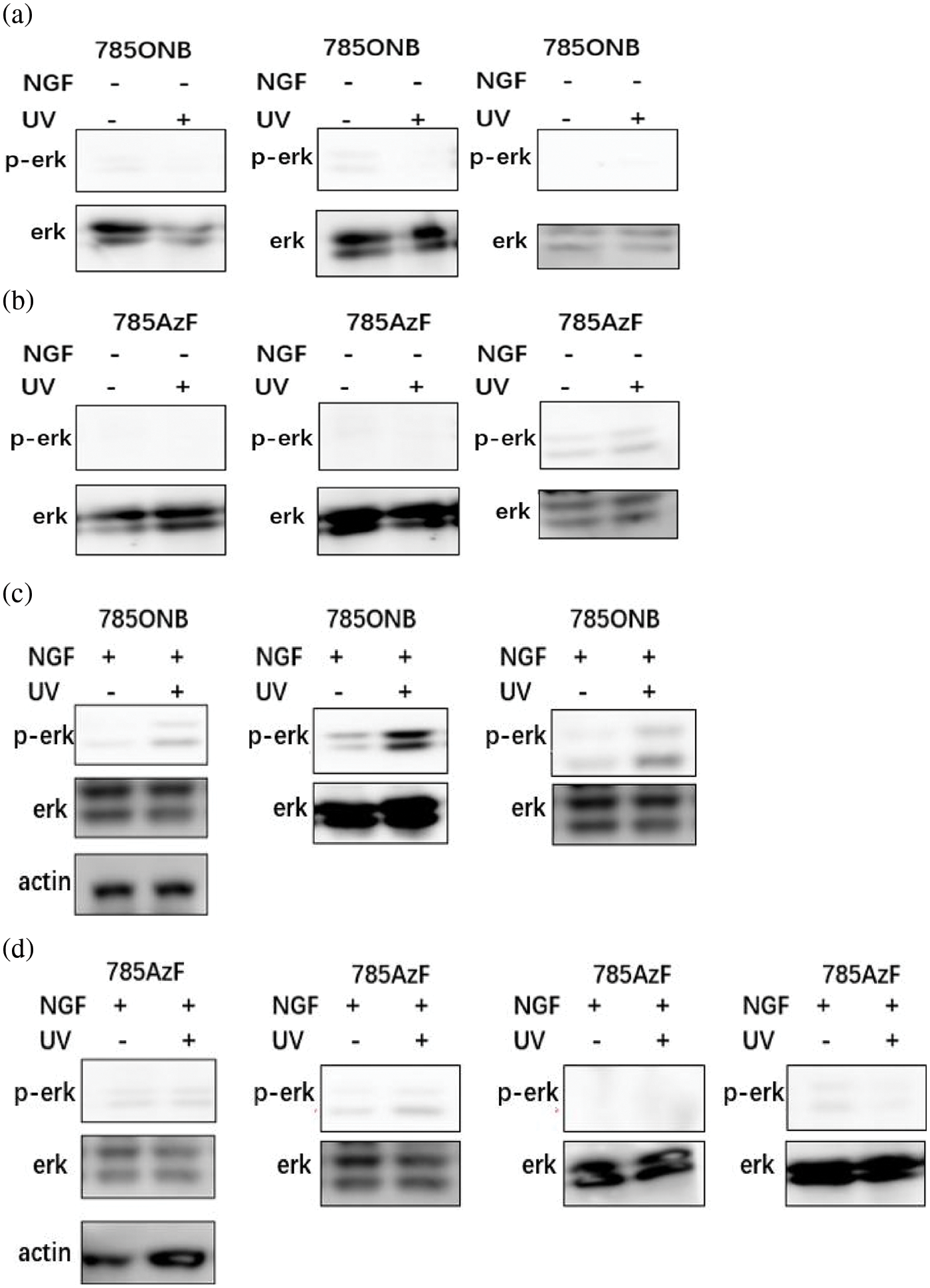
Figure S1: Repeats for gel experiments Western blotting analysis of HEK293T cells expressing TrkA-785ONB and TrkA-785AzF mutants for phosphorylated erk in the presence or absence of ligand NGF without or with UV.
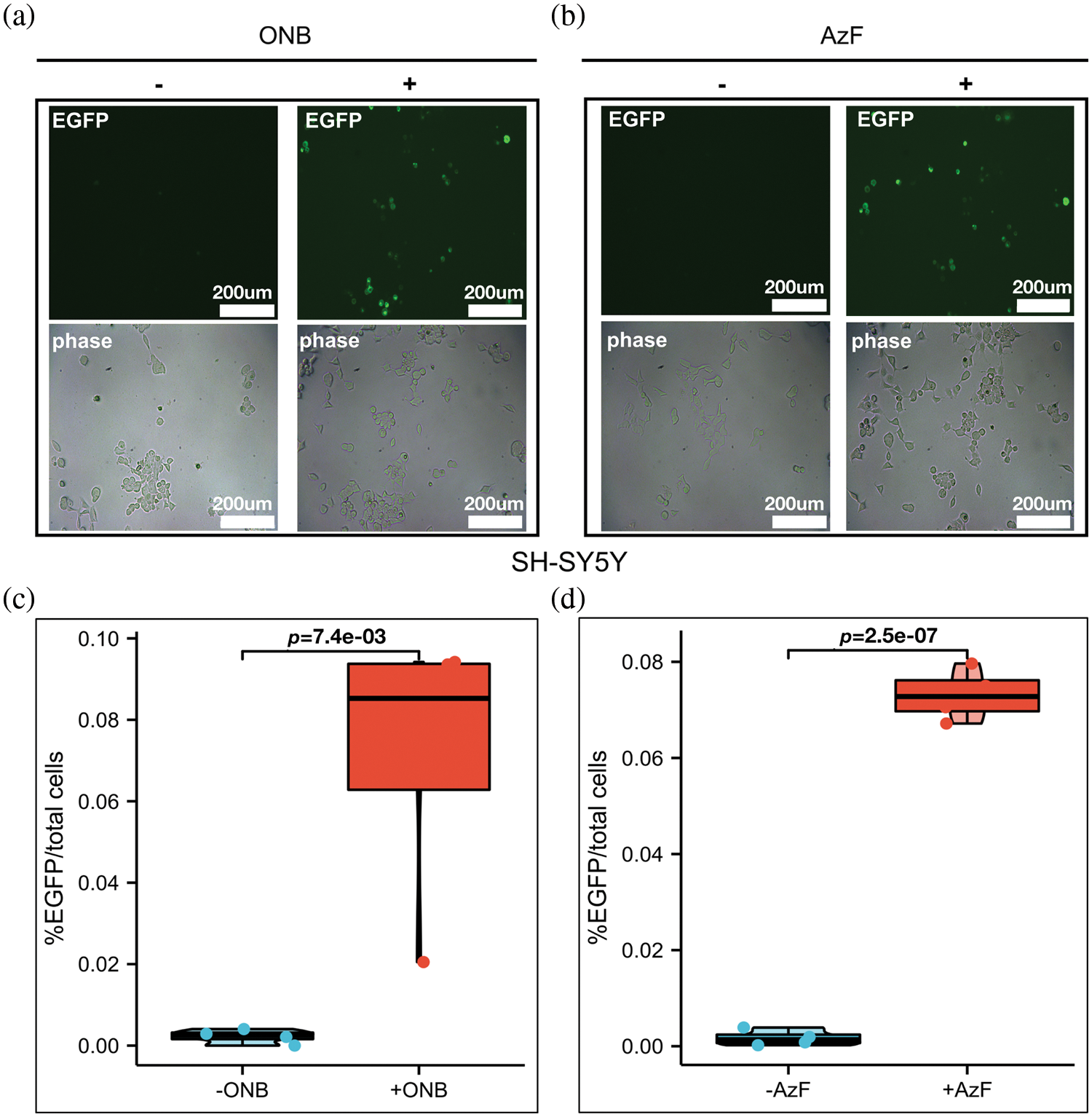
Figure S2: The incorporation efficiency of ONB or AzF at Y785 sites. (a and b) Fluorescent images of HEK293T cells and SH-SY5Y cells co-transfected with AzFRS-tRNA4x (AzF) or pONBYRS/U6-PyltRNA (ONB) at the Y785 site in the presence (+) or absence (−) of 100 uM ONB (left) and 1 mM AzF (right). 24–48 h post-transfection, cells were imaged using fluorescence microscopy for GFP expression (Scale bar: 200 um). (c and d) TrkA-Y785 mutant measured for indicated conditions in the absence (grey) or presence (dark) of 1 mM AzF or 100 um ONB, quantified by the percentage of fluorescent cells (EGFP) to total cells (phase). Error bars show SD
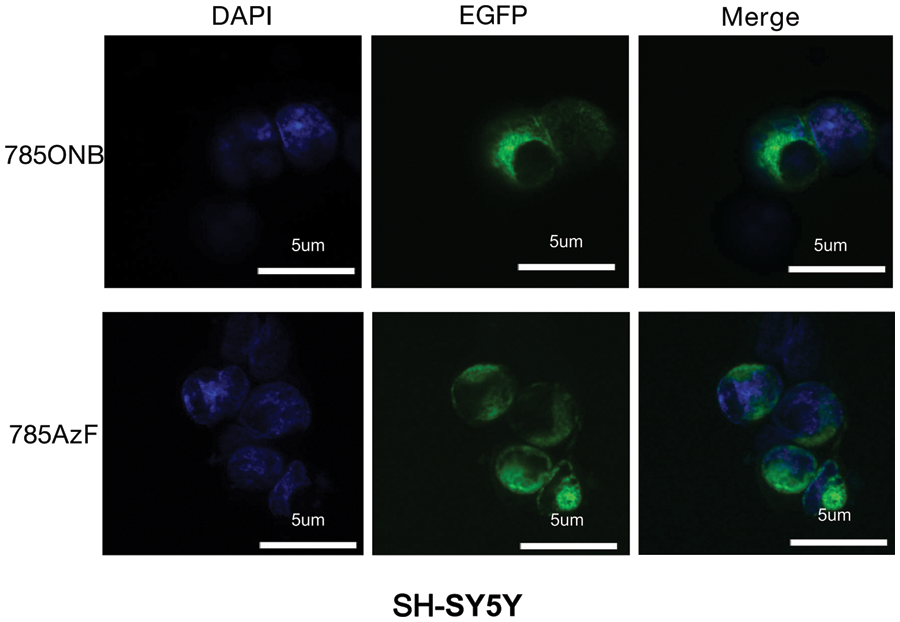
Figure S3: Fluorescence images of TrkA-785AzF or TrkA-785ONB mutants expressed in SH-SY5Y cells. Blue, nuclei stained with DAPI (4′, 6-diamidino-2-phenylindole); green, EGFP tagged TrkA (Scale bar: 5 um).
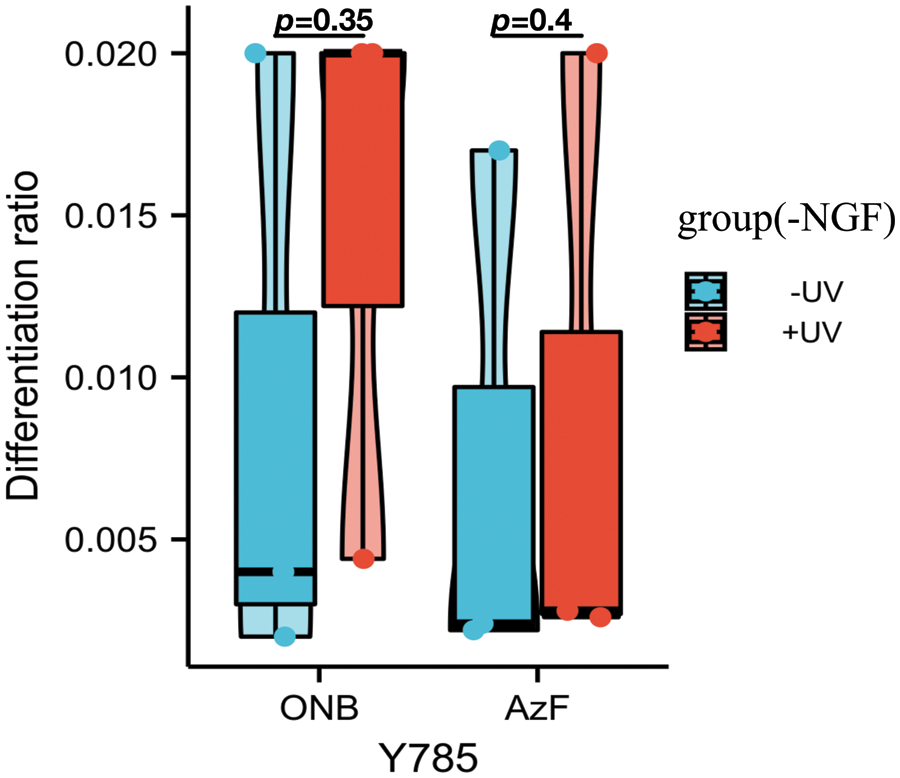
Figure S4: Differentiation ratios calculated for various mutants expressed in SH-SY5Y cells in the absence of NGF with or without UV light. Differentiation ratios were calculated as the following: taking the ratios (No. of green fluorescing cells with neurite longer than cell body/No. of green fluorescing cells) for Y785 and wt TrkA at each condition. Then take the ratios of each mutant/wt TrkA treated in the same condition. Values above represent the mean ± SD of three biological replicates (n = 3) with >200 cells counted per replicate. Scale bar: 10 um.
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools