 Open Access
Open Access
ARTICLE
LncRNA ZFAS1 regulates cardiomyocyte differentiation of human embryonic stem cells
1 Department of Pharmacy, The Second Affiliated Hospital of Harbin Medical University (Institute of Clinical Pharmacy, The University Key Laboratory of Drug Research, Heilongjiang Higher Education Institutions), Harbin, 150086, China
2 Department of Pharmacology (State-Province Key Laboratories of Biomedicine-Pharmaceutics of China, Key Laboratory of Cardiovascular Medicine Research, Ministry of Education), College of Pharmacy, Harbin Medical University, Harbin, 150081, China
* Corresponding Author: BENZHI CAI. Email:
# The first two authors contributed equally to this work
BIOCELL 2023, 47(6), 1407-1416. https://doi.org/10.32604/biocell.2023.029080
Received 31 January 2023; Accepted 20 March 2023; Issue published 19 May 2023
Abstract
Background: Cardiomyocytes derived from human embryonic stem cells (hESCs) are regulated by complex and stringent gene networks during differentiation. Long non-coding RNAs (lncRNAs) exert critical epigenetic regulatory functions in multiple differentiation processes. However, the involvement of lncRNAs in the differentiation of hESCs into cardiomyocytes has not yet been fully elucidated. Here, we identified the key roles of ZFAS1 (lncRNA zinc finger antisense 1) in the differentiation of cardiomyocytes from hESCs. Methods: A model of cardiomyocyte differentiation from stem cells was established using the monolayer differentiation method, and the number of beating hESCs-derived cardiomyocytes was calculated. Gene expression was analyzed by quantitative real-time PCR (qRT-PCR). Immunofluorescence assays were performed to assess the expression of cardiac troponin T (cTnT) and α-actinin protein in cardiomyocytes. Results: qRT-PCR showed that ZFAS1 expression in the mesoderm was significantly higher than that in embryonic stem cells, cardiac progenitor cells, and cardiomyocytes. Knockdown of ZFAS1 inhibited cardiomyocyte differentiation from hESCs, which was characterized by reduced expression of the cardiac-specific markers cTnT, α-actinin, myosin heavy chain 6 (MYH6), and myosin heavy chain 7 (MYH7). In contrast, ZFAS1 overexpression remarkably increased the percentage of spontaneously beating cardiomyocytes. In terms of the mechanism, we found that ZFAS1 is an antisense lncRNA at the 5′ end of the protein-coding gene ZNFX1. Knockdown of ZFAS1 could increase the mRNA expression level of ZNFX1. Furthermore, qRT-PCR demonstrated that the silencing of ZNFX1 led to an increase in cardiac-specific markers that predicted the promotion of cardiomyocyte differentiation. Conclusion: Altogether, these data suggest that lncRNA-ZFAS1 is required for cardiac differentiation by functionally inhibiting the expression of ZNFX1, which may provide a reference for the treatment of heart disease to a certain extent.Keywords
Cardiovascular disease, particularly myocardial infarction (MI), remains the leading cause of death worldwide (Andersson and Vasan, 2018; Benjamin et al., 2019). Mammalian cardiomyocytes (CMs) exit the cell cycle shortly after birth, which limits the regenerative potential of the adult heart and results in poor replacement of lost tissue after acute ischemic injury, ultimately leading to heart failure (Laflamme and Murry, 2011; Chen et al., 2021). Therefore, how to deal with the inactivated myocardium has become an urgent medical problem. Currently, several avenues to promote cardiac regeneration after acute ischemic injury have been evaluated (Li et al., 2020; Bae et al., 2021; Han et al., 2021). In addition to promoting the endogenous cardiomyocyte proliferation and reprogramming of non-cardiomyocytes to cardiomyocytes, stem cell therapy for the treatment of inactivated myocardium in fatal cardiovascular disease has been extensively studied (Song et al., 2012; Protze et al., 2019; Dhahri et al., 2022).
Human embryonic stem cells (hESCs) have the potential to proliferate indefinitely, self-renew, and differentiate into a variety of cell lineages including CMs in vitro (Wang, 2006; Mummery et al., 2012; Frame et al., 2020). Ideally, mesoderm-derived CMs are highly similar to the primitive cardiomyocyte in terms of cell morphology, electrophysiology, and energy metabolism, which not only provide an adequate source of cardiomyocytes for cell replacement therapy but also serve as an important model for exploring human heart development (Abu-Issa and Kirby, 2007; Caspi et al., 2007; Chong et al., 2014). Cardiomyocytes derived from hESCs (hESC-CMs) are regulated by complex gene networks and despite substantial progress in inducing cardiomyocyte differentiation in vitro, the potential application of hESC-CMs is limited due to the low purity and yield. Therefore, it is of great significance to uncover the underlying mechanisms of cardiomyocyte differentiation to address this issue.
Long non-coding RNAs (LncRNAs) are defined as transcripts longer than 200 nucleotides and do not code for any transcripts. They have emerged as key regulators of diverse biological and pathophysiology processes, including cell metabolism, epigenetic regulation, cell proliferation, and apoptosis (Ni et al., 2019; Palazzo and Koonin, 2020; Zhao et al., 2020; Park et al., 2021). Functionally, lncRNAs can regulate gene expression through a variety of mechanisms such as histone modification, chromatin remodeling, interaction with transcription factors, and competition with endogenous mRNA or microRNAs (miRNAs) (Zhu et al., 2013; Beermann et al., 2016; Qian et al., 2019; Tan et al., 2021). Notably, accumulated evidence has suggested that lncRNAs play critical roles in the differentiation fate of hESCs. For instance, cardiac mesoderm enhancer-associated noncoding RNA (CARMEN) and Braveheart (Bvht) are lncRNAs located on the opposite strands adjacent to each other, which are essential for cardiac mesoderm specification and cardiomyocyte differentiation (Klattenhoff et al., 2013; Ounzain et al., 2015). Furthermore, downregulation of cardiomyocyte maturation-associated lncRNA (CARMA) levels in differentiating ESCs promote cardiogenic commitment and cardiomyocyte differentiation in mice (Kay et al., 2022). The identification of such lncRNAs that play a significant role in regulating cardiomyocyte terminal differentiation will facilitate the development of therapeutic strategies for cardiac repair.
LncRNA zinc finger antisense 1 (ZFAS1) is the antisense strand of the encoded gene ZNFX1 and has emerged as a key molecule in governing multiple biological processes. In breast cancer, ZFAS1 knockout significantly increased the proliferation and differentiation ability of breast cancer cells, suggesting that ZFAS1 may be a new target for breast cancer therapy as a tumor suppressor gene (Fan et al., 2018). Moreover, recent studies found that ZFAS1 is involved in the regulation of cardiomyocyte apoptosis, intracellular calcium overload, and cardiac dysfunction after MI (Zhang et al., 2018; Jiao et al., 2019). However, the specific function of ZFAS1 in cardiomyocyte differentiation is still lacking in adequate research evidence. In this study, we showed that ZFAS1 is required for cardiac differentiation by functionally inhibiting the expression of ZNFX1. This finding may provide a reference for the treatment of heart disease to a certain extent.
The hESCs line H9 was routinely maintained in PSCeasy® II medium (Cellapy, Beijing, China) on Matrix-coated plates (Cellapy, Beijing, China) and passaged every 3 days using 0.5 mM ethylenediaminetetraacetic acid (EDTA) (Cellapy, Beijing, China). The cells were cultured under the condition of 37°C with 5% CO2, and the medium was changed every day. In addition, MycoBlue™ mycoplasma kits (Vazyme, Nanjing, China) were used to periodically detect mycoplasma infection.
The small interfering RNA siRNA-ZFAS1, siRNA-ZNFX1, and negative control (siNC) were synthesized from GenePharma Co., Ltd. (Shanghai, China), which were transfected into hESCs using RNAiMAX reagent (Invitrogen, Carlsbad, USA) according to the manufacturer’s instructions. 100 μL of Opti-MEM containing 6 μL RNAiMAX reagent and 100 μL of Opti-MEM containing 2 μL siRNA were mixed thoroughly for 5 min and then incubated into a 12-well plate with 80% stem cell fusion. The target sequences are listed in Table 1.
The plasmid pcDNA3.1(+)-ZFAS1 was purchased from OBiO Technology Co., Ltd. (Shanghai, China), which were transfected into hESCs using ViaFect Transfection Reagent (Promega, Madison, Wisconsin, USA). DNA was diluted to 2 μg per 200 μL of Opti-MEM, and 8 μL ViaFect Transfection Reagent was added to achieve the proper ratio of reagent to DNA. Transfection was performed when hESCs fusion reached about 80%, and samples were collected after 48 h for subsequent experimental studies.
Cardiac differentiation from human embryonic stem cells
Cardiac differentiation was induced from hESCs according to a previous report (Han et al., 2019). Briefly, at the onset of differentiation, hESCs were cultured in RPMI 1640 medium containing B27 supplement minus insulin (Thermo Fisher, Waltham, USA). On days 0–1, 6 μm CHIR (MCE, New Jersey, USA) was added to the medium. On days 3–5, 2 μm wnt-C59 (MCE, New Jersey, USA) was added into the medium. On day 7 of differentiation, the medium was replaced with RPMI 1640 plus B27 supplement (Thermo Fisher, Waltham, USA), and the medium was changed every 48 h until pulsing cardiomyocytes were observed.
RNA extraction and quantitative real-time PCR
Total RNA extractions from hESCs and CMs were performed using TRIzol reagent (Invitrogen, Carlsbad, USA) and reverse-transcribed into single-strand cDNA using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Carlsbad, USA). SYBR Green Master mix (Roche, Basel, Switzerland) and 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) were used for the relative quantification of RNA. The reaction conditions were: 95°C for 10 min, 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s, a total of 40 cycles. The relative expression levels of mRNA were calculated and analyzed by the 2–ΔΔCT method (Livak and Schmittgen, 2001). GAPDH was used as the internal control. The corresponding primer sequences are listed in Table 2.
HESC-CMs of each group were fixed with 4% paraformaldehyde (Solarbio, Beijing, China) for 20 min before being permeabilized with 0.4% Triton X-100 (Beyotime, Shanghai, China) for 10 min at room temperature. After blocking with goat serum (BOSTER, Wuhan, China) for 30 min at 37°C to reduce nonspecific binding, the samples were incubated with primary antibodies against α-actinin (GeneTex, San Antonio, USA) and cTnT (Invitrogen, Carlsbad, USA) overnight at 4°C. Then, the cells were incubated with Alexa Fluor-488 (Abcam, Cambridge, UK) and Alexa Fluor-594 (Abcam, Cambridge, UK) secondary antibodies for 1 h at room temperature in darkness. Images were acquired by a live cell imaging system (Olympus Fluoview 10i) and analyzed by Image-Pro Plus (Media Cybernetics, USA).
The Two-tailed Student’s t-test was performed for comparisons between two groups, and ANOVA was used to compare the differences among multiple groups. Statistical significance was defined as a p value less than 0.05. The data are presented as mean ± SEM (standard error of mean). All statistical analyses were carried out using GraphPad Prism 8.2.1 (Graph Pad Software, Inc., USA).
LncRNA ZFAS1 was significantly upregulated during cardiomyocyte differentiation
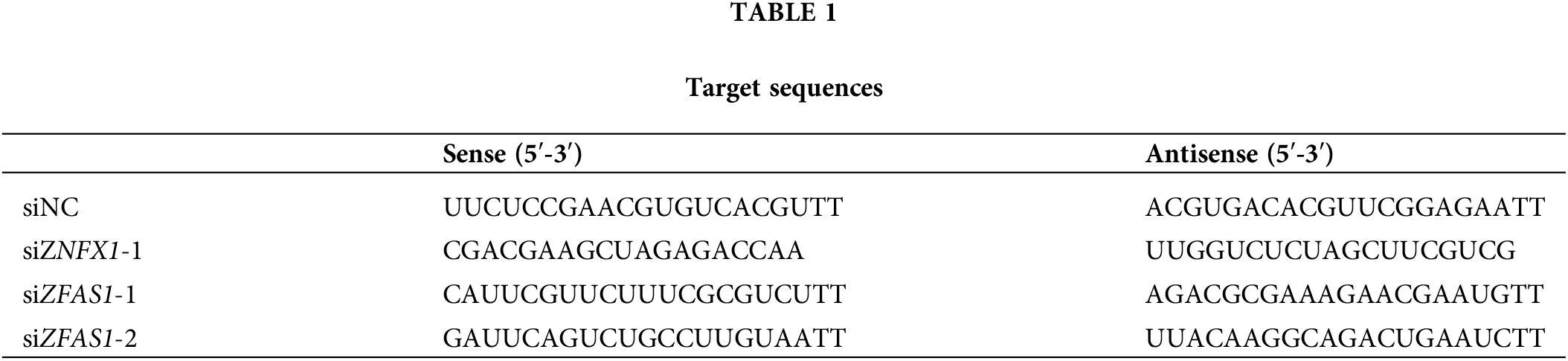
In order to define the implication of ZFAS1 in the process of cardiomyocyte differentiation, we induced the differentiation of hESCs into cardiomyocytes in vitro, which was consistent with the previous procedure (Fig. 1A). The differentiation of hESCs into cardiomyocytes was divided into the following stages based on the gene expression pattern: hESCs (human embryonic cells, day 0), MES (mesoderm, day 2), CPCs (cardiac progenitor cells, day 5) and CMs (cardiomyocytes, day 8). Firstly, we performed the qRT-PCR analysis for ZFAS1 at different stages of differentiation and found that the expression level of ZFAS1 increased significantly during the hESCs to MES phase (Fig. 1B). This elevated expression of ZFAS1 suggests that it may be involved in the differentiation of hESCs into CMs.
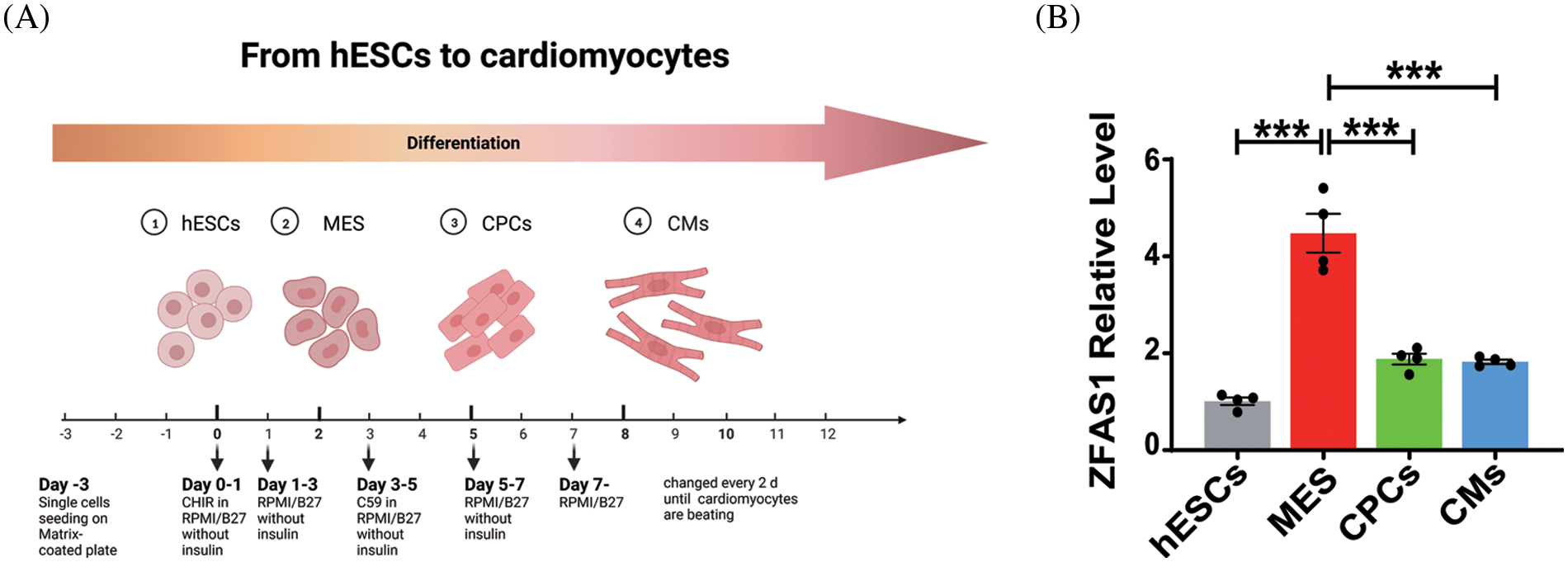
Figure 1: LncRNA ZFAS1 was upregulated during hESCs to MES. (A) Schematic diagram of the differentiation system of hESC-CMs. hESCs: human embryonic stem cells (day 0). MES: mesoderm (day 2). CPCs: cardiac progenitor cells (day 5). CMs: cardiomyocytes (day 8). (B) The mRNA expressions of ZFAS1 in hESCs (day 0), MES (day 2), CPCs (day 5) and CMs (day 8) were analyzed by quantitative real-time PCR (qRT-PCR). Two-tailed Student’s t-test was used to compare the differences between the two experimental groups. Error bars represent mean ± SEM. ***p < 0.001. n = 4.
Knockdown of ZFAS1 inhibited cardiac differentiation from hESCs
Based on the above findings, we further explored whether the alteration of ZFAS1 is related to the differentiation of CMs. The hESCs were differentiated into CMs in the presence or absence of siRNA targeting lncRNA ZFAS1. A significant decrease in ZFAS1 expression was observed following siRNA administration (Fig. 2A). Silencing of ZFAS1 did not affect the pluripotency of hESCs, as no significant change was detected in the stem cell marker OCT4 from day 0 to day 8 after differentiation compared with the control group (Fig. 2B). Notably, the levels of BRACHYURY as the mesodermal marker and T-box transcription factor 5 (TBX-5) as the cardiac progenitor cells marker, were significantly reduced from day 2 to day 5 as compared to the control cells treated with scrambled siRNAs (Figs. 2C and 2D). As expected, after the transfection of siZFAS1 in hESCs, we found an inhibition of cardiomyocyte differentiation, which was characterized by reduced mRNA expression of cardiac-specific markers cTnT, α-actinin, MYH6, and MYH7 (Figs. 2E–2H). Similarly, the cTnT and α-actinin protein expression in CMs of the siZFAS1 group was decreased compared with the control group observed by immunofluorescence technique (Figs. 2I and 2J). In addition, the percentage of spontaneously beating CMs decreased with the knockdown of ZFAS1 as observed under the microscope (Fig. 3C).
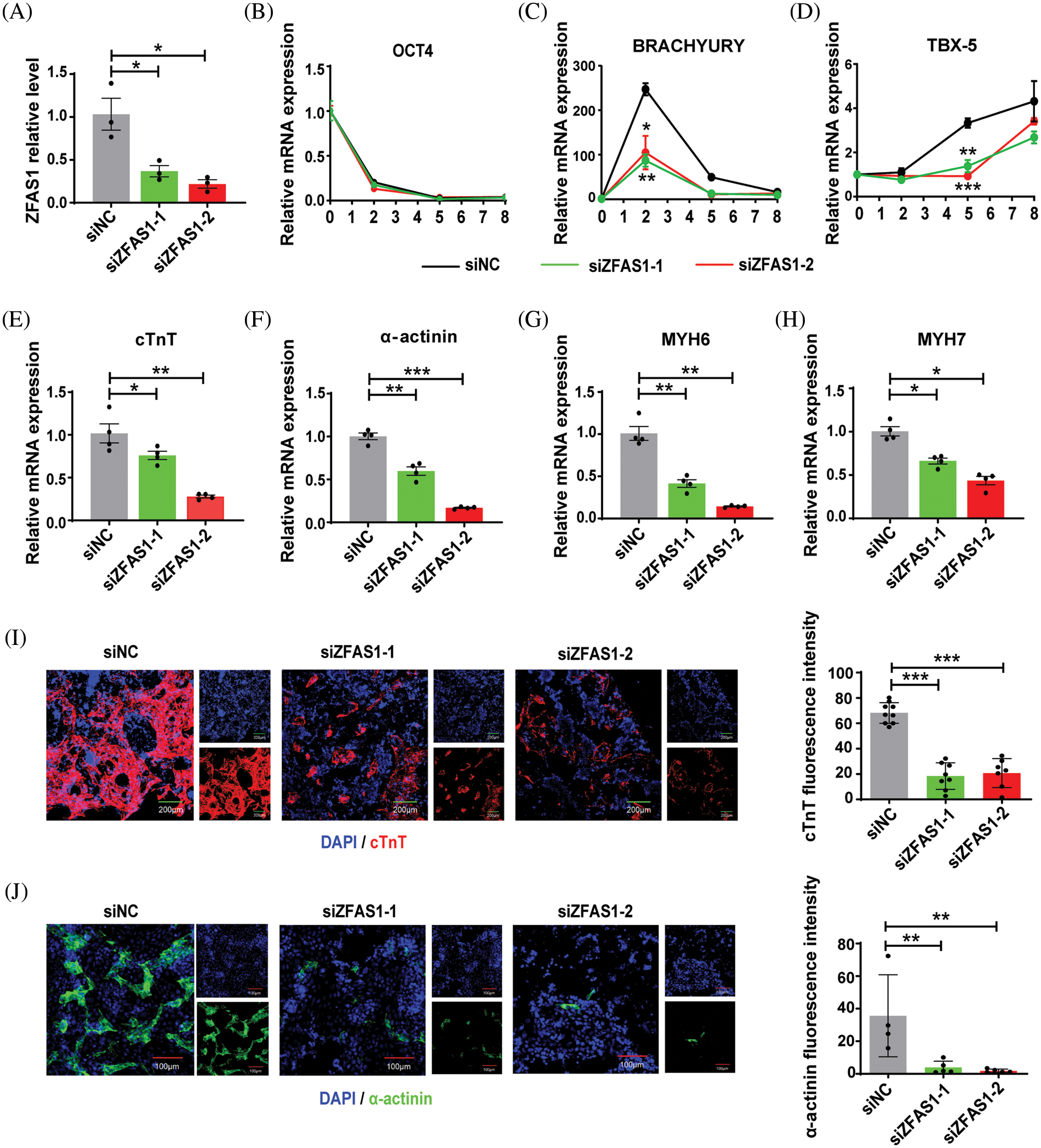
Figure 2: Knockdown of ZFAS1 inhibited cardiac differentiation from human embryonic stem cells (hESCs). (A) qRT-qPCR was performed to evaluate the expression of ZFAS1 in siNC and siZFAS1 cells at day 3 of cardiomyocyte (CM) differentiation. At least three independent batches of cells for each group were done. *p < 0.05 vs. siNC. (B–D) The mRNA expression of OCT4, BRACHYURY and T-box transcription factor 5 (TBX-5) at day 0 to day 8 of CM differentiation. n = 3. (E–H) The mRNA expression of cardiac troponin T (cTnT), α-actinin, myosin heavy chain 6 (MYH6), and myosin heavy chain 7 (MYH7) at day 8 of CM differentiation. *p < 0.05, **p < 0.01, ***p < 0.001. n = 4. (I–J) Immunofluorescence staining for cardiac troponin T (cTnT) (red) and α-actinin (green) in hESCs-induced CMs at day 8. Nuclei were stained in blue with 4′,6-diamidino-2-phenylindole (DAPI). The scale bar indicates 100 μm/200 μm. Two-tailed Student’s t-test was used to compare the differences between the two experimental groups. Error bars represent mean ± SEM. **p < 0.01, ***p < 0.001.
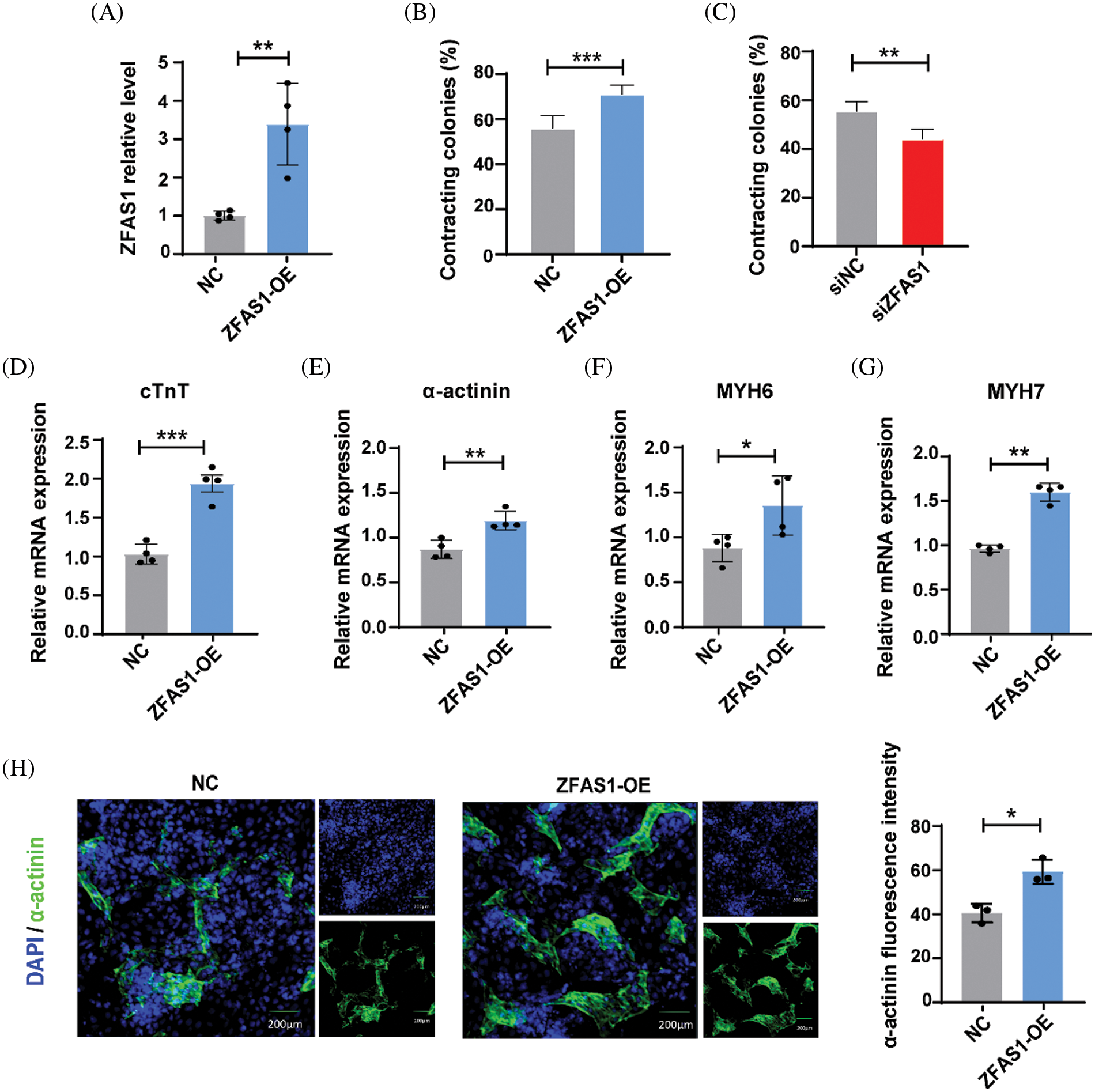
Figure 3: Overexpression of ZFAS1 promoted cardiac differentiation in human embryonic stem cells (hESCs). (A) Quantitative real-time PCR (qRT-qPCR) was performed to evaluate the mRNA expression of ZFAS1 in NC and ZFAS1-OE cells at day 3 of CM differentiation. **p < 0.01. n = 4. (B and C) Percentage of spontaneously contracting CMs on the days of differentiation following ZFAS1 overexpression and knockdown. **p < 0.01, ***p < 0.001. n = 4. (D–G) The mRNA level of cardiac troponin T (cTnT), α-actinin, myosin heavy chain 6 (MYH6), and myosin heavy chain 7 (MYH7) in NC and ZFAS1-OE CMs at day 8 of CM differentiation. (H) Immunofluorescence staining for α-actinin (green) in hESC-induced CMs on day 8. Nuclei were stained in blue with 4′,6-diamidino-2-phenylindole (DAPI). The scale bar indicates 200 μm. Error bars represent mean ± SEM. *p < 0.05. n = 3.
Overexpression of ZFAS1 promoted cardiac differentiation in hESCs
We next characterized the role of ZFAS1 in cardiac differentiation by overexpressing this lncRNA in hESCs. As shown in Figs. 3A and 3B, with the effective overexpression of ZFAS1 by plasmid transfection at the initiation of differentiation, the percentage of spontaneously beating CMs was obviously increased. Consistently, overexpression of ZFAS1 markedly elevated the mRNA expression levels of cTnT, α-actinin, MYH6, and MYH7 compared with the control group (Figs. 3D–3G). The immunofluorescence analysis further revealed that the α-actinin protein expression was also increased in CMs of ZFAS1 overexpression group (Fig. 3H). Together, these results indicated that lncRNA-ZFAS1 could promote cardiac differentiation in hESCs.
ZNFX1 was the target of ZFAS1 that regulates the cardiac differentiation in hESCs
Increasing evidence had shown that antisense lncRNAs could form sense-antisense pairs by pairing with protein-coding gene mRNAs on the opposite strand to regulate transcription, variable splicing, and mRNA stability. Given that ZFAS1 is the antisense lncRNA at the 5′ end of protein-coding gene ZNFX1 (Fig. 4A), we hypothesized that the regulatory role of antisense ZFAS1 in hESC differentiation may be linked to the regulation of ZNFX1. Firstly, we examined the expression of ZNFX1 in hESCs and found that the knockdown of ZFAS1 could increase the mRNA expression level of ZNFX1 (Fig. 4B). To further investigate whether ZNFX1 is directly involved in regulating cardiomyocyte differentiation, we then utilized siRNAs to knockdown the expression of ZNFX1 in differentiating hESCs, and the efficiency was verified by qRT-PCR (Fig. 4C). As shown in Figs. 4D–4H, compared with the control group, the expression of cardiac cell markers cTnT, α-actinin, MYH6, and MYH7 were upregulated upon ZNFX1 knockdown. Notably, in the presence of siZNFX1, the knockdown of ZFAS1 failed to inhibit the cardiac differentiation in hESCs (Figs. 4I–4L). Overall, these results suggest that ZFAS1 is required for cardiac differentiation by functionally inhibiting the expression of ZNFX1.
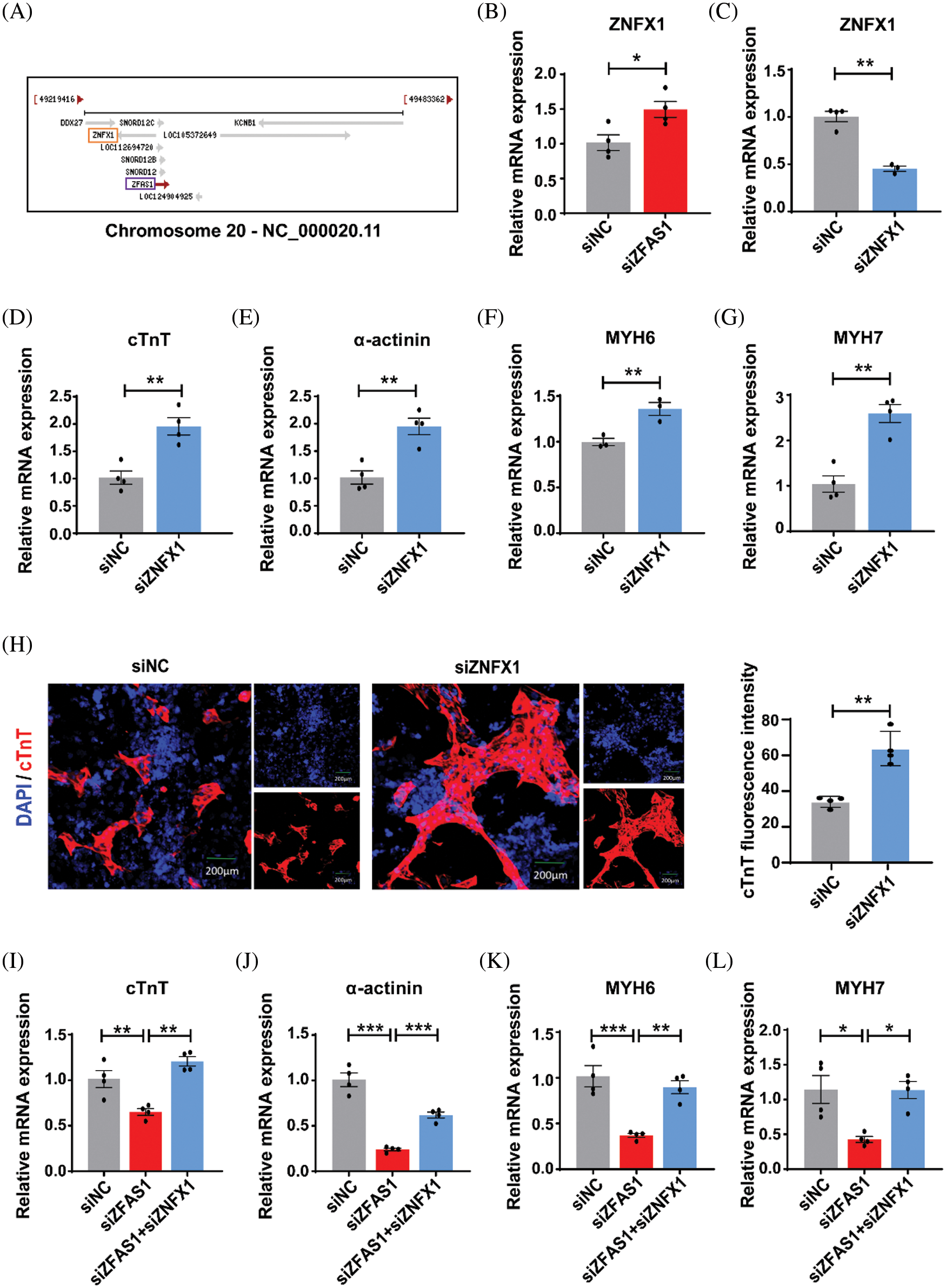
Figure 4: ZNFX1 was the target of ZFAS1 that regulates cardiac differentiation in human embryonic stem cells (hESCs). (A) Genomic organization of the human ZNFX1 gene cluster. (B) The mRNA expression level of ZNFX1 after knockdown of ZFAS1 at day 0 of cardiomyocyte (CM) differentiation. *p < 0.05. n = 4. (C) Quantitative real-time PCR (qRT-PCR) analysis of ZNFX1 in siNC and siZNFX1-transfected cells at the beginning of differentiation. **p < 0.05. n = 4. (D–G) The expression levels of cardiac troponin T (cTnT), α-actinin, myosin heavy chain (MYH6), and myosin heavy chain (MYH7) mRNA in siNC and siZNFX1-transfected cells on day 8 of differentiation. **p < 0.01. n = 3–4. (H) Immunofluorescence staining for cTnT (red) in hESCs-induced CMs at day 8. Nuclei were stained in blue with 4′,6-diamidino-2-phenylindole (DAPI). The scale bar indicates 200 μm. **p < 0.01. n = 3–4. (I–L) The mRNA expression levels of cTnT, α-actinin, MYH6, and MYH7 in siNC, siZFAS1 and siZFAS1+siZNFX1-transfected cells on day 8 of differentiation. *p < 0.05, **p < 0.01, ***p < 0.001. n = 4.
In recent years, it has been found that the combined transplantation of pluripotent stem cell-derived cardiomyocytes and stem cells can significantly increase the retention of transplanted cardiomyocytes, improve the survival rate, promote the formation of new blood vessels and anti-inflammatory effects through the paracrine pathway (Khan et al., 2015; Liu et al., 2018; Lima Correa et al., 2021), thus repairing the damaged heart and improving the heart function. Accumulated evidence has suggested that multiple signaling pathways like Wnt, Notch, Fibroblast growth factor 2 (FGF2), and the Bone morphogenetic protein-4 (BMP4) pathway can be involved in regulating the efficiency of inducing differentiation of pluripotent stem cells to cardiomyocytes (Willems et al., 2011; Tung et al., 2014; Pallotta et al., 2017; Rajabi et al., 2018; Wu et al., 2021). Notably, lncRNAs also play a critical role in the differentiation fate of hESCs. Improving the differentiation efficiency and maturation of cardiomyocytes by lncRNAs is expected to be a new therapeutic strategy for myocardial repair after myocardial infarction (Fico et al., 2019; Hunkler et al., 2022).
ZFAS1 (zinc finger antisense 1) is a splicing and polyadenylated lncRNA transcribed from the 5′ end of ZNFX1. It is derived from chromosome 20q13.13 and is involved in different types of malignancies including gastric cancer, colorectal cancer, and hepatocellular carcinoma (Zhou et al., 2019; Lin et al., 2021; Zhu et al., 2021; Zhuo et al., 2022). Previous studies have reported that ZFAS1 was an endogenous sarcoplasmic/endoplasmic reticulum Ca2+ ATPase 2a (SERCA2a) inhibitor. It was involved in the regulation of the Wnt/β-Catenin signal pathway, which was linked to the pathogenesis of acute myocardial infarction (Ying et al., 2018). However, the specific function of ZFAS1 in cardiomyocyte differentiation is still lacking in adequate research evidence.
In this study, we investigated the regulatory role of lncRNA-ZFAS1 in the differentiation of hESCs into CMs. We found for the first time that ZFAS1 is highly expressed in the mesoderm during cardiomyocyte differentiation. The downregulation or overexpression of ZFAS1 during differentiation revealed that ZFAS1 regulated the process of cardiac differentiation from hESCs. Mechanistically, there was a negative feedback relationship between lncRNA-ZFAS1 and its host gene ZNFX1, and ZNFX1 could be directly involved in the cardiac differentiation process. Previous studies reported that ZNFX1 localizes to germ granules (P granules) of C. elegans in early germline blastomeres. Furthermore, the biological function of ZNFX1 was to interact with silencing RNAs in the C. elegans germline to direct transgenerational epigenetic inheritance (TEI) (Wan et al., 2018). In addition, human ZNFX1 was also associated with stress granules and was essential for both monocyte homeostasis and protective immunity against mycobacteria (Le Voyer et al., 2021). In this study, we found that inhibition of ZNFX1 not only promoted cardiac differentiation efficiency but also reversed the altered differentiation caused by ZFAS1 silencing. However, the underlying mechanism by which ZFAS1 and ZNFX1 mediate cardiac differentiation is still not well understood and needs to be defined in our future studies.
To summarize, in this study, we found that lncRNA-ZFAS1 is required for cardiac differentiation by functionally inhibiting the expression of ZNFX1, which will expand our understanding of the role of lncRNAs in cardiac lineage commitment.
Funding Statement: This work was supported by the National Natural Science Foundation of China [81573434 to BZC]. Heilongjiang Touyan Innovation Team Program [BZC], HMU Marshal Initiative Funding (HMUMIF-21018 to BZC). National Nature Science Youth Foudation of China [82000226 to XFG].
Author Contributions: The authors confirm their contribution to the paper as follows: study conception and design: Benzhi Cai, Yang Cao, and Yining Liu; data collection: Yang Yu, Xiaofei Guo, Xiuxiu Wang, Wenya Ma; analysis and interpretation of results: Hanjing Li, Zhongyu Ren, Sijia Li, Haoyu Ji, Hongyang Chen; draft manuscript preparation: Hong Yan, Xinlu Gao, Yanan Tian, Xin Wang. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The datasets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Ethics Approval: The study fully complies with the guidelines of the ethical criteria. We adhered to the principles of informed consent, safety and efficacy, and prevention of commercialization. We strictly abided by the Ethical Guidelines for Human Embryonic Stem Cell Research in China, and did not conduct any unethical experimental operations. The whole experimental process was carried out according to the procedures approved by the Institutional Animal Care and Use Committee of Harbin Medical University.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
Abu-Issa R, Kirby ML (2007). Heart field: From mesoderm to heart tube. Annual Review of Cell and Developmental Biology 23: 45–68. https://doi.org/10.1146/annurev.cellbio.23.090506.123331 [Google Scholar] [PubMed] [CrossRef]
Andersson C, Vasan RS (2018). Epidemiology of cardiovascular disease in young individuals. Nature Reviews Cardiology 15: 230–240. https://doi.org/10.1038/nrcardio.2017.154 [Google Scholar] [PubMed] [CrossRef]
Bae J, Salamon RJ, Brandt EB, Paltzer WG, Zhang Z, Britt EC, Hacker TA, Fan J, Mahmoud AI (2021). Malonate promotes adult cardiomyocyte proliferation and heart regeneration. Circulation 143: 1973–1986. https://doi.org/10.1161/CIRCULATIONAHA.120.049952 [Google Scholar] [PubMed] [CrossRef]
Beermann J, Piccoli MT, Viereck J, Thum T (2016). Non-coding RNAs in development and disease: Background, mechanisms, and therapeutic approaches. Physiological Reviews 96: 1297–1325. https://doi.org/10.1152/physrev.00041.2015 [Google Scholar] [PubMed] [CrossRef]
Benjamin EJ, Muntner P, Alonso A, Bittencourt MS, Callaway CW et al. (2019). Heart disease and stroke statistics-2019 update: A report from the american heart association. Circulation 139: e56–e528. https://doi.org/10.1161/CIR.0000000000000659 [Google Scholar] [PubMed] [CrossRef]
Caspi O, Huber I, Kehat I, Habib M, Arbel G, Gepstein A, Yankelson L, Aronson D, Beyar R, Gepstein L (2007). Transplantation of human embryonic stem cell-derived cardiomyocytes improves myocardial performance in infarcted rat hearts. Journal of the American College of Cardiology 50: 1884–1893. https://doi.org/10.1016/j.jacc.2007.07.054 [Google Scholar] [PubMed] [CrossRef]
Chen Y, Lüttmann FF, Schoger E, Schöler HR, Zelarayán LC, Kim KP, Haigh JJ, Kim J, Braun T (2021). Reversible reprogramming of cardiomyocytes to a fetal state drives heart regeneration in mice. Science 373: 1537–1540. https://doi.org/10.1126/science.abg5159 [Google Scholar] [PubMed] [CrossRef]
Chong JJ, Yang X, Don CW, Minami E, Liu YW et al. (2014). Human embryonic-stem-cell-derived cardiomyocytes regenerate non-human primate hearts. Nature 510: 273–277. https://doi.org/10.1038/nature13233 [Google Scholar] [PubMed] [CrossRef]
Dhahri W, Sadikov Valdman T, Wilkinson D, Pereira E, Ceylan E et al. (2022). In vitro matured human pluripotent stem cell-derived cardiomyocytes form grafts with enhanced structure and function in injured hearts. Circulation 145: 1412–1426. https://doi.org/10.1161/circulationaha.121.053563 [Google Scholar] [PubMed] [CrossRef]
Fan S, Fan C, Liu N, Huang K, Fang X, Wang K (2018). Downregulation of the long non-coding RNA ZFAS1 is associated with cell proliferation, migration and invasion in breast cancer. Molecular Medicine Reports 17: 6405–6412. https://doi.org/10.3892/mmr.2018.8707 [Google Scholar] [PubMed] [CrossRef]
Fico A, Fiorenzano A, Pascale E, Patriarca EJ, Minchiotti G (2019). Long non-coding RNA in stem cell pluripotency and lineage commitment: Functions and evolutionary conservation. Cellular and Molecular Life Sciences 76: 1459–1471. https://doi.org/10.1007/s00018-018-3000-z [Google Scholar] [PubMed] [CrossRef]
Frame JM, Kubaczka C, Long TL, Esain V, Soto RA et al. (2020). Metabolic regulation of inflammasome activity controls embryonic hematopoietic stem and progenitor cell production. Developmental Cell 55: 133–149.e136. https://doi.org/10.1016/j.devcel.2020.07.015 [Google Scholar] [PubMed] [CrossRef]
Han Z, Wang X, Xu Z, Cao Y, Gong R et al. (2021). ALKBH5 regulates cardiomyocyte proliferation and heart regeneration by demethylating the mRNA of YTHDF1. Theranostics 11: 3000–3016. https://doi.org/10.7150/thno.47354 [Google Scholar] [PubMed] [CrossRef]
Han Z, Yu Y, Xu J, Bao Z, Xu Z et al. (2019). Iron homeostasis determines fate of human pluripotent stem cells via glycerophospholipids-epigenetic circuit. Stem Cells 37: 489–503. https://doi.org/10.1002/stem.2967 [Google Scholar] [PubMed] [CrossRef]
Hunkler HJ, Gross S, Thum T, Bar C (2022). Non-coding RNAs: Key regulators of reprogramming, pluripotency, and cardiac cell specification with therapeutic perspective for heart regeneration. Cardiovascular Research 118: 3071–3084. https://doi.org/10.1093/cvr/cvab335 [Google Scholar] [PubMed] [CrossRef]
Jiao L, Li M, Shao Y, Zhang Y, Gong M et al. (2019). lncRNA-ZFAS1 induces mitochondria-mediated apoptosis by causing cytosolic Ca overload in myocardial infarction mice model. Cell Death & Disease 10: 942. https://doi.org/10.1038/s41419-019-2136-6 [Google Scholar] [PubMed] [CrossRef]
Kay M, Soltani BM, Nemir M, Aghagolzadeh P, Pezzuto I et al. (2022). The conserved long non-coding RNA CARMA regulates cardiomyocyte differentiation. Cardiovascular Research 118: 2339–2353. https://doi.org/10.1093/cvr/cvab281 [Google Scholar] [PubMed] [CrossRef]
Khan M, Nickoloff E, Abramova T, Johnson J, Verma SK et al. (2015). Embryonic stem cell-derived exosomes promote endogenous repair mechanisms and enhance cardiac function following myocardial infarction. Circulation Research 117: 52–64. https://doi.org/10.1161/CIRCRESAHA.117.305990 [Google Scholar] [PubMed] [CrossRef]
Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA et al. (2013). Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell 152: 570–583. https://doi.org/10.1016/j.cell.2013.01.003 [Google Scholar] [PubMed] [CrossRef]
Laflamme MA, Murry CE (2011). Heart regeneration. Nature 473: 326–335. https://doi.org/10.1038/nature10147 [Google Scholar] [PubMed] [CrossRef]
Le Voyer T, Neehus AL, Yang R, Ogishi M, Rosain J et al. (2021). Inherited deficiency of stress granule ZNFX1 in patients with monocytosis and mycobacterial disease. Proceedings of the National Academy of Sciences of the United States of America 118: e2102804118. https://doi.org/10.1073/pnas.2102804118 [Google Scholar] [PubMed] [CrossRef]
Li Y, Feng J, Song S, Li H, Yang H et al. (2020). gp130 controls cardiomyocyte proliferation and heart regeneration. Circulation 142: 967–982. https://doi.org/10.1161/CIRCULATIONAHA.119.044484 [Google Scholar] [PubMed] [CrossRef]
Lima Correa B, El Harane N, Gomez I, Rachid Hocine H, Vilar J et al. (2021). Extracellular vesicles from human cardiovascular progenitors trigger a reparative immune response in infarcted hearts. Cardiovascular Research 117: 292–307. https://doi.org/10.1093/cvr/cvaa028 [Google Scholar] [PubMed] [CrossRef]
Lin JC, Yang PM, Liu TP (2021). PERK/ATF4-dependent ZFAS1 upregulation is associated with sorafenib resistance in hepatocellular carcinoma cells. International Journal of Molecular Sciences 22: 5848. https://doi.org/10.3390/ijms22115848 [Google Scholar] [PubMed] [CrossRef]
Liu YW, Chen B, Yang X, Fugate JA, Kalucki FA et al. (2018). Human embryonic stem cell-derived cardiomyocytes restore function in infarcted hearts of non-human primates. Nature Biotechnology 36: 597–605. https://doi.org/10.1038/nbt.4162 [Google Scholar] [PubMed] [CrossRef]
Livak KJ, Schmittgen TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 25: 402–408. https://doi.org/10.1006/meth.2001.1262 [Google Scholar] [PubMed] [CrossRef]
Mummery CL, Zhang J, Ng ES, Elliott DA, Elefanty AG, Kamp TJ (2012). Differentiation of human embryonic stem cells and induced pluripotent stem cells to cardiomyocytes: A methods overview. Circulation Research 111: 344–358. https://doi.org/10.1161/CIRCRESAHA.110.227512 [Google Scholar] [PubMed] [CrossRef]
Ni W, Yao S, Zhou Y, Liu Y, Huang P, Zhou A, Liu J, Che L, Li J (2019). Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m6A reader YTHDF3. Molecular Cancer 18: 143. https://doi.org/10.1186/s12943-019-1079-y [Google Scholar] [PubMed] [CrossRef]
Ounzain S, Micheletti R, Arnan C, Plaisance I, Cecchi D et al. (2015). CARMEN, a human super enhancer-associated long noncoding RNA controlling cardiac specification, differentiation and homeostasis. Journal of Molecular and Cellular Cardiology 89: 98–112. https://doi.org/10.1016/j.yjmcc.2015.09.016 [Google Scholar] [PubMed] [CrossRef]
Palazzo AF, Koonin EV (2020). Functional long non-coding RNAs evolve from junk transcripts. Cell 183: 1151–1161. https://doi.org/10.1016/j.cell.2020.09.047 [Google Scholar] [PubMed] [CrossRef]
Pallotta I, Sun B, Wrona EA, Freytes DO (2017). BMP protein-mediated crosstalk between inflammatory cells and human pluripotent stem cell-derived cardiomyocytes. Journal of Tissue Engineering and Regenerative Medicine 11: 1466–1478. https://doi.org/10.1002/term.2045 [Google Scholar] [PubMed] [CrossRef]
Park MK, Zhang L, Min KW, Cho JH, Yeh CC et al. (2021). NEAT1 is essential for metabolic changes that promote breast cancer growth and metastasis. Cell Metabolism 33: 2380–2397.e2389. https://doi.org/10.1016/j.cmet.2021.11.011 [Google Scholar] [PubMed] [CrossRef]
Protze SI, Lee JH, Keller GM (2019). Human pluripotent stem cell-derived cardiovascular cells: From developmental biology to therapeutic applications. Cell Stem Cell 25: 311–327. https://doi.org/10.1016/j.stem.2019.07.010 [Google Scholar] [PubMed] [CrossRef]
Qian X, Zhao J, Yeung PY, Zhang QC, Kwok CK (2019). Revealing lncRNA structures and interactions by sequencing-based approaches. Trends in Biochemical Sciences 44: 33–52. https://doi.org/10.1016/j.tibs.2018.09.012 [Google Scholar] [PubMed] [CrossRef]
Rajabi S, Pahlavan S, Ashtiani MK, Ansari H, Abbasalizadeh S et al. (2018). Human embryonic stem cell-derived cardiovascular progenitor cells efficiently colonize in bFGF-tethered natural matrix to construct contracting humanized rat hearts. Biomaterials 154: 99–112. https://doi.org/10.1016/j.biomaterials.2017.10.054 [Google Scholar] [PubMed] [CrossRef]
Song K, Nam YJ, Luo X, Qi X, Tan W et al. (2012). Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature 485: 599–604. https://doi.org/10.1038/nature11139 [Google Scholar] [PubMed] [CrossRef]
Tan YT, Lin JF, Li T, Li JJ, Xu RH, Ju HQ (2021). LncRNA-mediated posttranslational modifications and reprogramming of energy metabolism in cancer. Cancer Communications 41: 109–120. https://doi.org/10.1002/cac2.12108 [Google Scholar] [PubMed] [CrossRef]
Tung JC, Paige SL, Ratner BD, Murry CE, Giachelli CM (2014). Engineered biomaterials control differentiation and proliferation of human-embryonic-stem-cell-derived cardiomyocytes via timed Notch activation. Stem Cell Reports 2: 271–281. https://doi.org/10.1016/j.stemcr.2014.01.011 [Google Scholar] [PubMed] [CrossRef]
Wan G, Fields BD, Spracklin G, Shukla A, Phillips CM, Kennedy S (2018). Spatiotemporal regulation of liquid-like condensates in epigenetic inheritance. Nature 557: 679–683. https://doi.org/10.1038/s41586-018-0132-0 [Google Scholar] [PubMed] [CrossRef]
Wang L (2006). Endothelial and hematopoietic cell fate of human embryonic stem cells. Trends in Cardiovascular Medicine 16: 89–94. https://doi.org/10.1016/j.tcm.2006.01.001 [Google Scholar] [PubMed] [CrossRef]
Willems E, Spiering S, Davidovics H, Lanier M, Xia Z, Dawson M, Cashman J, Mercola M (2011). Small-molecule inhibitors of the Wnt pathway potently promote cardiomyocytes from human embryonic stem cell-derived mesoderm. Circulation Research 109: 360–364. https://doi.org/10.1161/CIRCRESAHA.111.249540 [Google Scholar] [PubMed] [CrossRef]
Wu Y, Zhou L, Liu H, Duan R, Zhou H et al. (2021). LRP6 downregulation promotes cardiomyocyte proliferation and heart regeneration. Cell Research 31: 450–462. https://doi.org/10.1038/s41422-020-00411-7 [Google Scholar] [PubMed] [CrossRef]
Ying Z, Lei J, Sun LH, Li Y, Yang B (2018). LncRNA ZFAS1 as a SERCA2a inhibitor to cause intracellular Ca2+ overload and contractile dysfunction in a mouse model of myocardial infarction. Circulation Research 122: 1354–1368. https://doi.org/10.1161/CIRCRESAHA.117.312117 [Google Scholar] [PubMed] [CrossRef]
Zhang Y, Jiao L, Sun L, Li Y, Gao Y et al. (2018). ZFAS1LncRNA as a SERCA2a inhibitor to cause intracellular ca overload and contractile dysfunction in a mouse model of myocardial infarction. Circulation Research 122: 1354–1368. https://doi.org/10.1161/CIRCRESAHA.117.312117 [Google Scholar] [PubMed] [CrossRef]
Zhao X, Su L, He X, Zhao B, Miao J (2020). CA7-4Long noncoding RNA promotes autophagy and apoptosis via sponging and in high glucose-induced vascular endothelial cells. Autophagy 16: 70–85. https://doi.org/10.1080/15548627.2019.1598750 [Google Scholar] [PubMed] [CrossRef]
Zhou HL, Zhou YF, Feng ZT (2019). Long noncoding RNA ZFAS1 promotes hepatocellular carcinoma proliferation by epigenetically repressing miR-193a-3p. European Review for Medical and Pharmacological Sciences 23: 9840–9847. https://doi.org/10.26355/eurrev_201911_19547 [Google Scholar] [PubMed] [CrossRef]
Zhu J, Fu H, Wu Y, Zheng X (2013). Function of lncRNAs and approaches to lncRNA-protein interactions. Science China Life Sciences 56: 876–885. https://doi.org/10.1007/s11427-013-4553-6 [Google Scholar] [PubMed] [CrossRef]
Zhu T, Wang Z, Wang G, Hu Z, Ding H, Li R, Sun J (2021). Long non-coding RNA ZFAS1 promotes the expression of EPAS1 in gastric cardia adenocarcinoma. Journal of Advanced Research 28: 7–15. https://doi.org/10.1016/j.jare.2020.06.006 [Google Scholar] [PubMed] [CrossRef]
Zhuo ZL, Xian HP, Sun YJ, Long Y, Liu C, Liang B, Zhao XT (2022). Long noncoding RNA ZNFX1-AS1 promotes the invasion and proliferation of gastric cancer cells by regulating LIN28 and CAPR1N1. World Journal of Gastroenterology 28: 4973–4992. https://doi.org/10.3748/wjg.v28.i34.4973 [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools