 Open Access
Open Access
ARTICLE
Proteomics analysis provides novel biomarkers and therapeutic target candidates in the treatment of the Huang-Pu-Tong-Qiao formula in an AD rat model
1 School of Integrated Chinese and Western Medicine, Anhui University of Chinese Medicine, Hefei, 230012, China
2 Anhui Province Key Laboratory of Chinese Medicinal Formula, Anhui University of Chinese Medicine, Hefei, 230012, China
* Corresponding Authors: SHU YE. Email: ; BIAO CAI. Email:
# Qian Chen, Xin Lei and Guanhua Hu contributed equally to this work
(This article belongs to the Special Issue: Bioinformatics Study of Diseases)
BIOCELL 2023, 47(6), 1265-1277. https://doi.org/10.32604/biocell.2023.028811
Received 09 January 2023; Accepted 09 March 2023; Issue published 19 May 2023
Abstract
Background: Huang-Pu-Tong-Qiao formula (HPTQ), a traditional Chinese herbal formula, has a variety of pharmacological effects. It has been used to treat Alzheimer’s disease (AD) for decades. This study aimed to screen differentially expressed proteins in the hippocampus of AD model rats treated with HPTQ. Proteomic studies of the effects of HPTQ on AD are key to understanding the therapeutic mechanisms of HPTQ and identifying potential therapeutic targets. Methods: We hence used the isobaric tags for relative and absolute quantification (ITRAQ) approach to investigate the differentially expressed proteins in the hippocampus of AD model rats before and after HPTQ administration and to identify the potential therapeutic target proteins of HPTQ. In this study, the learning and memory abilities of AD rats were examined by the Morris water maze test. After HPTQ administration, the differentially expressed proteins in the hippocampus of AD rats were quantified and analyzed in silico. Furthermore, western blotting was used to verify the expression of related proteins. Results: The Morris water maze results showed that HPTQ could improve the learning and memory ability of AD model rats. The proteomics analysis results showed that 57 proteins were differentially expressed, of which 35 were up-regulated and 22 were down-regulated. Bioinformatics analysis indicated that proteins with altered expression after HPTQ treatment were involved in several biological processes that have the potential to exert neuroprotective effects. These included promoting the translation of ribosomes, improving the deposition of amyloid-beta (Aβ), regulating autophagy, regulating neuronal synaptic function and plasticity, and alleviating oxidative stress. Conclusion: In conclusion, we identified several potential therapeutic target proteins and related mechanistic pathways of HPTQ in the treatment of AD, laying the foundation for further investigation of the therapeutic effects of HPTQ.Keywords
Alzheimer’s disease (AD) is a common neurodegenerative disease in the elderly. In recent years, the number of AD patients worldwide has been on the rise (Fjell et al., 2014). The World Alzheimer’s Report of 2021 reported that over 55 million people live with dementia worldwide, and the number of AD patients is predicted to reach around 78 million by 2030. The cognitive impairment, memory loss, and behavioral disorders caused by AD inevitably reduce the quality of life in patients and increase the financial burden on families and the national healthcare system (Lane et al., 2018). Senile plaques formed by the deposition of amyloid-beta (Aβ) and neurofibrillary tangles formed by aggregation of hyperphosphorylated Tau are two pathological markers of AD. The presence of both these manifestations indicates the activation of multiple pathogenic mechanisms and neuronal damage in the brain (Sharma et al., 2019; Busche and Hyman, 2020). Most single-target drugs developed over the years to reduce Aβ or hyperphosphorylated Tau have failed in recent clinical trials. The reason can be attributed to the fact that AD is a multifactorial disease and therefore drugs targeting a single AD target cannot achieve substantial therapeutic effects (Huang and Mucke, 2012; Knopman et al., 2021). A growing number of studies have shown that a multi-target combination is an effective treatment for AD (Cummings et al., 2019; Veitch et al., 2019; Kabir et al., 2020). Therefore, determining the integrated protein changes in the hippocampal region before and after drug administration in AD patients is important to elucidate the potential molecular mechanisms of drug therapy. The hippocampus is an important area of the brain responsible for memory function and cognitive ability. The systematic study of protein expression in the hippocampus is key to discovering the actual pathogenic mechanisms of AD and developing effective therapeutic drugs (Poll et al., 2020). The hippocampal proteins show different expression levels at different stages of AD, and these changes undoubtedly include neurodegenerative processes triggered by pathophysiological abnormalities and adaptive responses. Although many researchers have performed extensive experimental research on hippocampal disease-related genes and proteins, studies on the differentially expressed proteins in the hippocampal tissue after drug administration are still lacking.
Huang-Pu-Tong-Qiao formula (HPTQ) has been widely used for decades in treating AD at the First Affiliated Hospital of Anhui University of Traditional Chinese Medicine. HPTQ is composed of 6 herbs: Rheum, Acorus tatarinowiischott, Alpiniaeoxyphyllae fructus, Polygoni Multiflori Radix Preparata, Ligusticum wallichii, and Panax ginseng. Previous studies have found that HPTQ could reduce neuronal loss and improve AD symptoms. For example, it could reduce the over phosphorylated tau in AD model rats and primary hippocampal neurons cultured in vitro by inhibiting the calcium/calmodulin-dependent protein kinase IV (CAM-CAMKIV) pathway (Ye et al., 2020). The administration also reduced oxidative stress and inhibited the mitochondrial apoptosis pathway in AD model rats (Cai et al., 2018). In another study, it modulated the epidermal growth factor receptor (EGFR)- phospholipase Cγ (PLCγ) signaling pathway and inhibited calcium overload in neurons (Wang et al., 2020). Further, inhibition of the abnormal activation of the c-Jun N-terminal kinases (JNK) signaling pathway and reducing the inflammatory response in animal models were also documented (Jiang et al., 2019). These results confirm that HPTQ has a variety of pharmacological effects. However, the total protein expression changes in the hippocampus before and after HPTQ administration have not been documented to this date.
Proteomics is a method for systematically studying the total protein expression changes and an important tool for elucidating disease mechanisms and discovering potential therapeutic targets (Bai et al., 2020). With rapid biotechnological development, isobaric tags for relative and absolute quantification (ITRAQ), which is a quantitative proteomics method can be used to perform quantitative studies of protein expression (Li et al., 2017). Currently, proteomics is a major tool being used to study the therapeutic mechanism of Chinese Traditional Medicine and to identify disease-related proteins (Xu et al., 2019, 2020).
In this study, we established an animal model of AD with an intraperitoneal injection of D-galactose and an intracranial hippocampal injection of Aβ25-35. Isobaric tag for relative and absolute quantitation (ITRAQ) method based on ITRAQ Labeling, HPLC Fractionation, and LC-MS/MS analysis was used to identify the full range of proteins in rat hippocampal tissue samples, and the differentially expressed proteins were obtained by bioinformatics analysis (Rhein et al., 2009; Yang et al., 2012). The proteomics results were then further validated by western blotting (WB) experiments. This study aimed to identify the potential therapeutic target proteins and related mechanistic pathways of HPTQ and to provide a basis for the therapeutic potential of HPTQ.
Preparation of Huang-Pu-Tong-Qiao formula
HPTQ was soaked for 30 min with tenfold water content and boiled three times every 30 min for a total of 90 min. The boiled liquid was then collected, filtered, concentrated to a crude drug with the amount of 1 g/ml, and stored at 4°C for use. A ten-fold amount of water was added and refluxed for 2 h for the first time, and for the second time, an 8-fold amount of water was added and refluxed for 1.5 h. The solution was then collected, concentrated to 50 mg/mL, and stored at 4°C.
A total of 30 male Sprague Dawley rats (250–300 g, 8–10 months old) were obtained from the Animal breeding base, Qinglongshan, Nanjing, China (Certificate no. SXCK (Su) 2017–0001). The model was established by the method previously described by Ye et al. (2020). A brief description is given below: during the week of acclimatization, the rats were housed in an environment with a temperature of 25°C–30°C and humidity of 40%–60%. The rats were then randomly divided into three groups: the control group (CTRL), the model group (MODEL), and the HPTQ group (1.41 g/kg, three times the clinical dose, calculated from the difference in the volumetric surface area between human and rat). Animal experiments were conducted for a total of 42 days, and rats in the MODEL and HPTQ groups were injected intraperitoneally with 100 mg/kg D-galactose once a day. After 14 days of animal experiments, rats in the HPTQ group were fed according to their body weight for 28 days. Intracerebral Aβ25-35 injection was administered to rats in the MODEL and HPTQ groups after 21 days of animal experiments. The cranial surface of rats was adjusted to a horizontal level and two positions: 2.2 mm on the left side and 2.2 mm to the right side were selected as injection sites based on 4.4 mm behind the fontanelle. A microinjector (Anting Scientific Instrument, Shanghai, China) was inserted 3.0 mm below the injection site, and 5 μL (10 μg) of Aβ25-35 was slowly injected into the left and right hippocampus of the rats, respectively. The detailed process has been shown in Fig. 1.
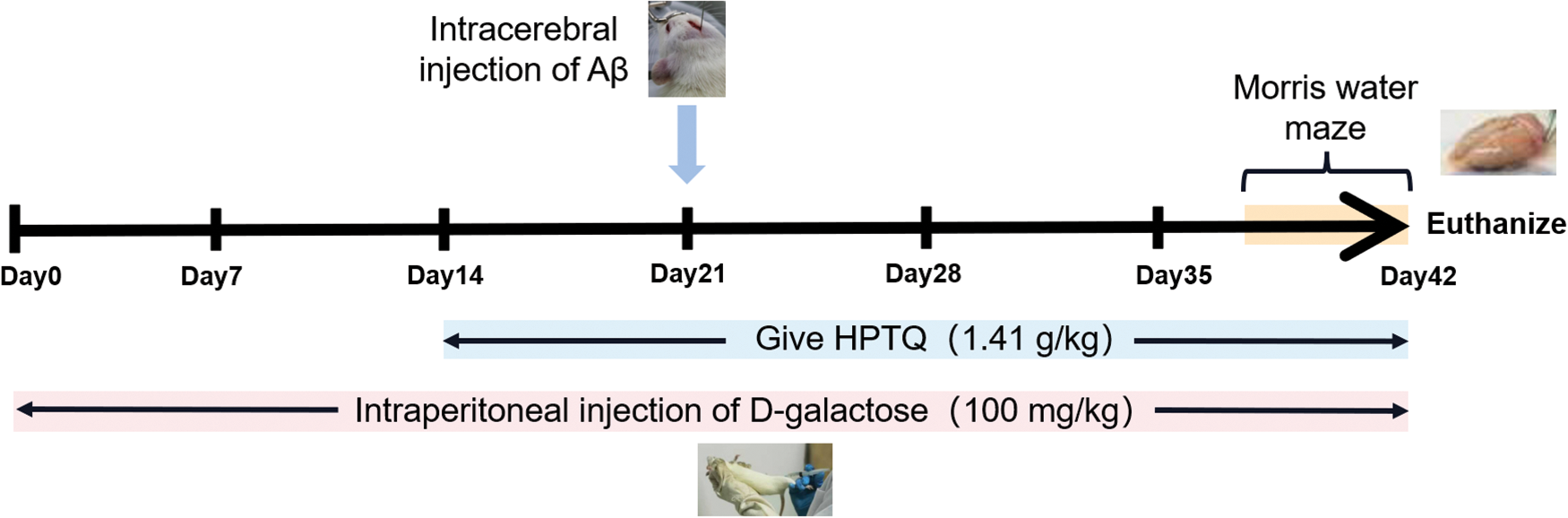
Figure 1: The scheme of Huang-Pu-Tong-Qiao formula (HPTQ) treatment for AD rats. The experiment was divided into three groups: CTRL group, MODEL group and HPTQ group. The MODEL group and HPTQ group were intraperitoneally injected with d-galactose once a day for 42 days. After intraperitoneal injection of d-galactose for 14 days, in the HPTQ group, HPTQ was administered by gavage continuously for 28 days. On the 21st day, the MODEL and HPTQ groups were injected with Aβ25-35 in the bilateral hippocampal region. Finally, a behavioral test was performed with the Morris Water Maze in the last week.
The Morris water maze test was done after 38 days of the animal experiment (Vorhees and Williams, 2006). The experiment lasted for 5 days, including the navigation training for the first 4 days and the escape latency test on day 5. To summarize, during the navigation training, the rats were placed into one of the four quadrants and observed if they could reach the platform within 90 s. If the rats failed to find the platform within the required time, they were moved to the platform to memorize this location. For each quadrant, one navigation experiment was conducted on rats. On day 5, the rats were placed into water in the first quadrant only, and the time required for the rats to reach the platform was recorded.
After the Morris water maze test, rats were anesthetized intraperitoneally with 1.5% sodium pentobarbital (60 mg/kg). Brain tissues were promptly obtained after the rats were sacrificed, and bilateral hippocampus tissues inside the brain were isolated.
The rat hippocampal tissues were ground into a powder in liquid nitrogen and then mixed with 4 volumes of protein lysis buffer (8 M urea, 1 mM phenylmethylsulfonyl fluoride) and placed on ice for 5 min followed by ultrasonication for 15 min. Gradient centrifugation was then performed at 13,000 g for 15 min at 4°C to remove tissue debris in the mixed isolate. The protein concentration of the separated supernatant was quantified using the Bicinchoninic Acid Kit (Beyotime Biotechnology, Shanghai, China).
The supernatant samples were subsequently reduced with 5mM dithiothreitol at 56°C for 30 min and treated with 11 mM iodoacetamide at 25°C for 15 min in the dark. Then, 200 mM of Triethylammonium bicarbonate was added for diluting the samples to reduce the urea concentration to less than 2 M. Trypsin was added to the samples for a mass ratio of trypsin-to-protein: 1:50, and overnight digestion was performed. On the next day, secondary digestion was performed with a 1:100 mass ratio of trypsin-to-protein for 4 h. After trypsin digestion, Strata X C18 SPE column (Phenomenex, USA) was used to desalt and dry the peptides of samples. ITRAQ labeling of the peptides was performed by mixing the peptides with the labeling reagent from the ITRAQ kit (AB Sciex, USA) and placing them at 25°C for 2 h. Subsequently, the peptides were subjected to vacuum desalination and dehydration. High pH reversed-phase HPLC was performed using an Agilent 300Extend C18 (5 μm particles, 4.6 mm inner diameter, 250 mm length) system to fractionate peptides into fractions. In the liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis, the peptides were mixed with the solvent I (0.1% formic acid and 2% acetonitrile) and combined with solution II (0.1% formic acid, 90% acetonitrile) on the EASY-nLC 1000 UPLC system. The gradient of peptide separation was set to increase the ratio of solution II from 9% to 80% within 40 min, and the flow rate was set at 400 nL/min. The separated peptides were ionized in the nanospray ionization (NSI) ion source and the tandem mass spectrometry analysis was performed in a Q ExactiveTM Plus system (Thermo, China). The Orbitrap was used to detect and analyze a full scan of the peptide and their secondary fragments. The automatic gain control was selected as 5E4. Scan options were set to a full scan range of 400–1500 m/z at 70,000 resolutions. The fragments of the peptides were identified by setting a scan range of 100 m/z at 17,500 resolutions. Data-dependent acquisition was used as the collection method and the maximum signal value was set to 63000 ions/s. The dynamic exclusion time was set to 30 s and the injection time was limited to 64 ms.
The data obtained in the LC-MS/MS analysis were processed using the Maxquant search engine (v.1.5.2.8). The analytical conditions were set according to the requirements of the proteomics. The database was Rattus_norvegicus_10116_PR_20191115 (29938 sequences). Protein quantification was performed using the iTRAQ-4plex method and the false discovery rate was adjusted to less than 1%.
Proteomics data were compared with the UniProt-GO database (http://www.ebi.ac.uk/GOA/) using gene ontology (GO) analysis to obtain protein information and classify the proteins according to their characteristics. The identified protein IDs from the experimental data were converted to GO IDs using the website tools. Protein sequences that did not annotate in the UniProt-GOA database were annotated using InterProScan software. Based on the obtained GO annotation data, proteins were classified into three categories: biological process, cellular component, and molecular function. GO enrichment analysis was performed afterward using a two-tailed Fisher’s exact test to determine the enrichment degree of various GO annotations in proteins and identify the number of proteins with the corresponding GO ID. The enrichment with a corrected p-value < 0.05 was considered to be statistically significantly different.
Kyoto encyclopedia of genes and genomes pathway analysis
The related protein pathways information from proteomics was annotated in the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (https://www.kegg.jp/). Pathway results of proteins were annotated using the KAAS tool on the website and the annotated pathways were then compared with the KEGG mapper database to obtain protein pathway data. A two-tailed Fisher’s exact test was used to identify pathways with significant enrichment and their degree of enrichment. The pathway with a corrected p-value < 0.05 was considered to be statistically significantly enriched.
Establishment of the protein-protein interaction network
The Protein-Protein Interaction (PPI) network was constructed using the STRING 11.0 database (https://string-db.org/), followed by network topology analysis and visualization of the obtained results using Cytoscape 3.9.1. This was done to understand the interactions between differentially expressed proteins and identify key genes linked to HPTQ in the treatment of AD.
The total protein of the hippocampus was extracted from the samples, and the protein concentration was measured using a protein quantification kit (Sparkjade Scientific, China). The total protein was electrophoresed on 8%–12% sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gels (Bio-Rad, USA) and subsequently transferred to a polyvinylidene fluoride membrane. The membrane was blocked with 5% skim milk for 2 h, followed by overnight incubation with relevant antibodies at 4°C. On the next day, the membrane was incubated with the HRP-conjugated goat anti-rabbit IgG for 2 h at 25°C. The protein bands in the membrane were visualized using electrochemiluminescence (ECL; Affinity, UK) and analyzed on the Tanon 5200 gel imager (Tanon, China). The rabbit anti-rat monoclonal antibodies against (Carboxylic ester hydrolase) ACHE, CaM kinase-like vesicle-associated (CAMKV), and Regulating synaptic membrane exocytosis protein 4 (Rims4) were obtained from Affinity (1:1000, Affinity, UK), Peroxisomal membrane protein 4 (Pxmp4) antibody was obtained from LSBio (1: 800, LSBio, China), and the GAPDH antibody was obtained from Zen bio (1:5000, Zen bio, China). The goat anti-rabbit IgG was obtained from Zen bio (1:20000, Zen bio, China). Fig. 2 demonstrates the entire experimental protocol.
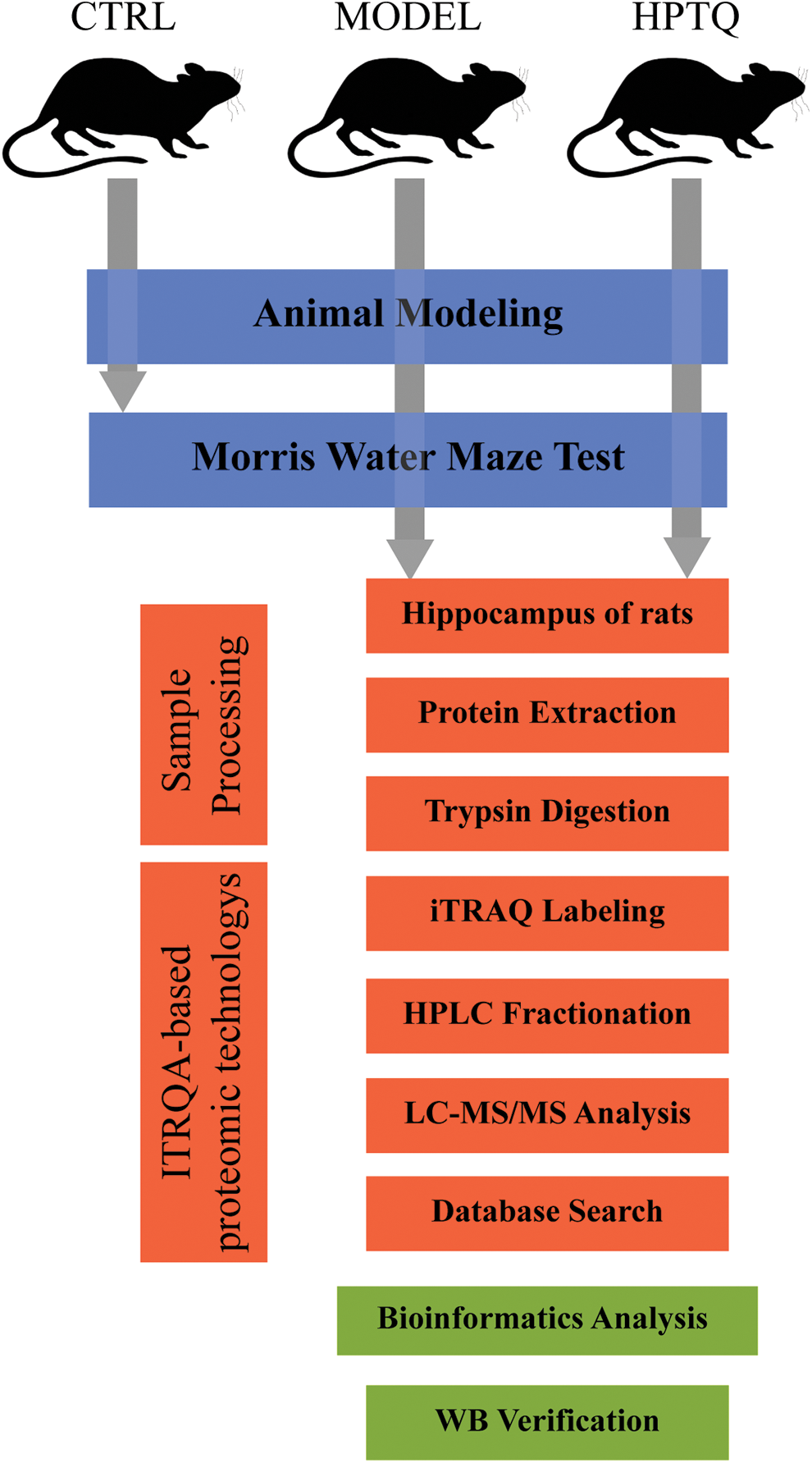
Figure 2: Schematic diagram of the present study. The animals in the MODEL and Huang-Pu-Tong-Qiao formula (HPTQ) groups were prepared by Alzheimer’s Disease (AD) rat models. After the behavioral test, hippocampus samples were extracted from the MODEL and HPTQ groups. After trypsin digestion, the samples were subjected to isobaric tags for relative and absolute quantification (ITRAQ)-based proteomics techniques to identify the full range of protein species. Subsequently, the differentially expressed proteins were obtained by database search and bioinformatics analysis was performed to elucidate the potential molecular mechanisms. Finally, four proteins were validated by Western blot analysis.
The SPSS version 23.0 was selected as the data analysis software for this experiment. Data results were expressed as mean ± standard deviation (SD). The Independent samples t-test was used to compare the means between two groups while one-way analysis of variance (ANOVA) and Least Significance Difference (LSD) were used to compare the means of multiple groups. The results with a p-value < 0.05 were considered to be statistically and significantly different while one-way analysis of variance (ANOVA) and Least Significance Difference (LSD) were used to compare the means of multiple groups. The results with a p-value < 0.05 were considered to be statistically and significantly different.
Learning and the memory ability of AD model rats
The Morris water maze was performed to evaluate the spatial reference memory of rats. The results showed that rats in the MODEL group took longer to reach the platform than those in the CTRL group (p < 0.01, Fig. 3), while rats in the HPTQ group took less time to reach the platform compared to the MODEL group (p < 0.01, Fig. 3).
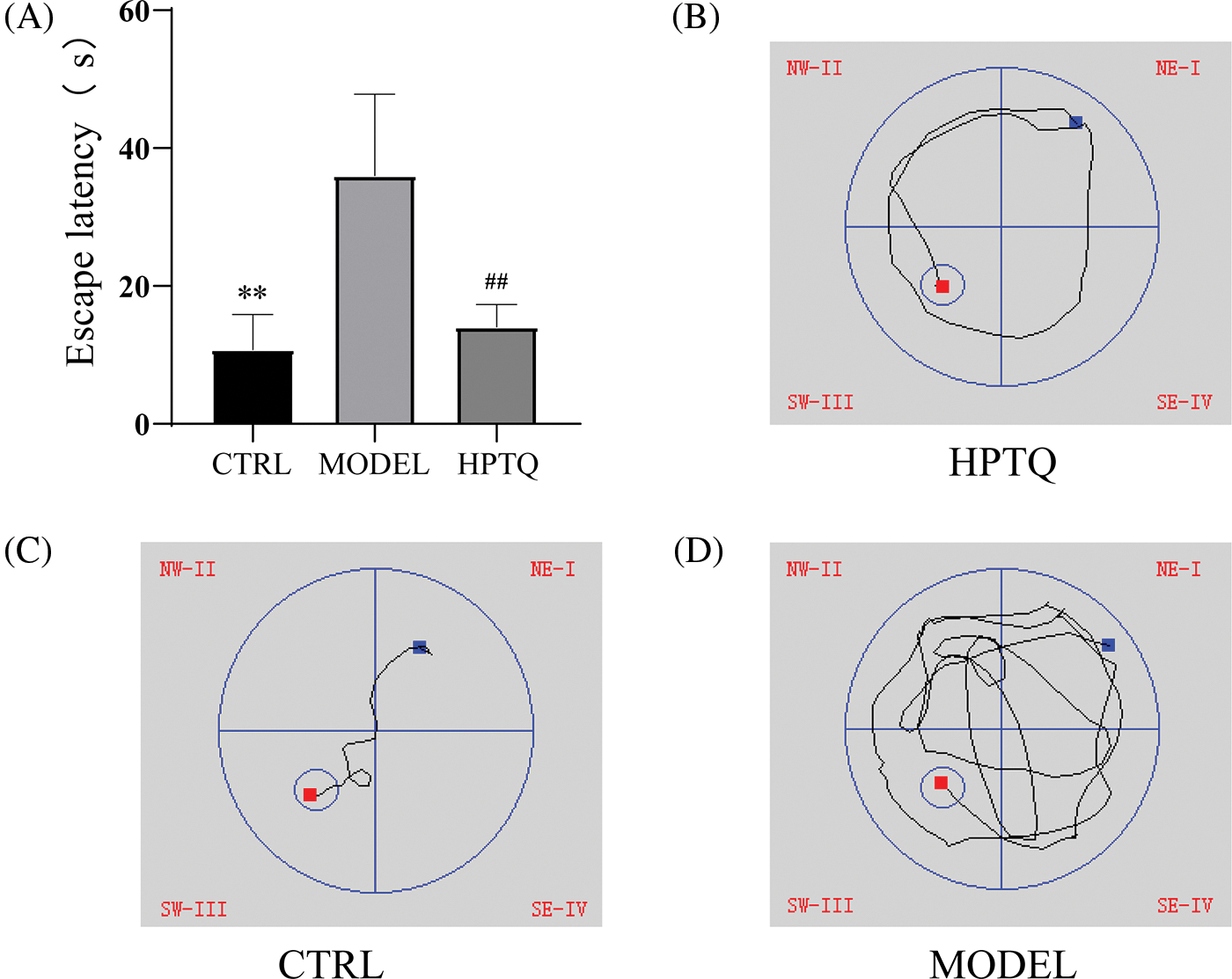
Figure 3: Effects of Huang-Pu-Tong-Qiao formula (HPTQ) on learning and the memory ability of Alzheimer’s Disease (AD) rats. (A) The escape latency of rats in CTRL, MODEL, and HPTQ groups; (B–D) Path diagram of the escape latency for CTRL, MODEL, and HPTQ groups. Values are expressed as the mean±SD. **p < 0.01, vs. CTRL group; ##p < 0.01, vs. MODEL group.
Differentially expressed proteins in the rat hippocampus
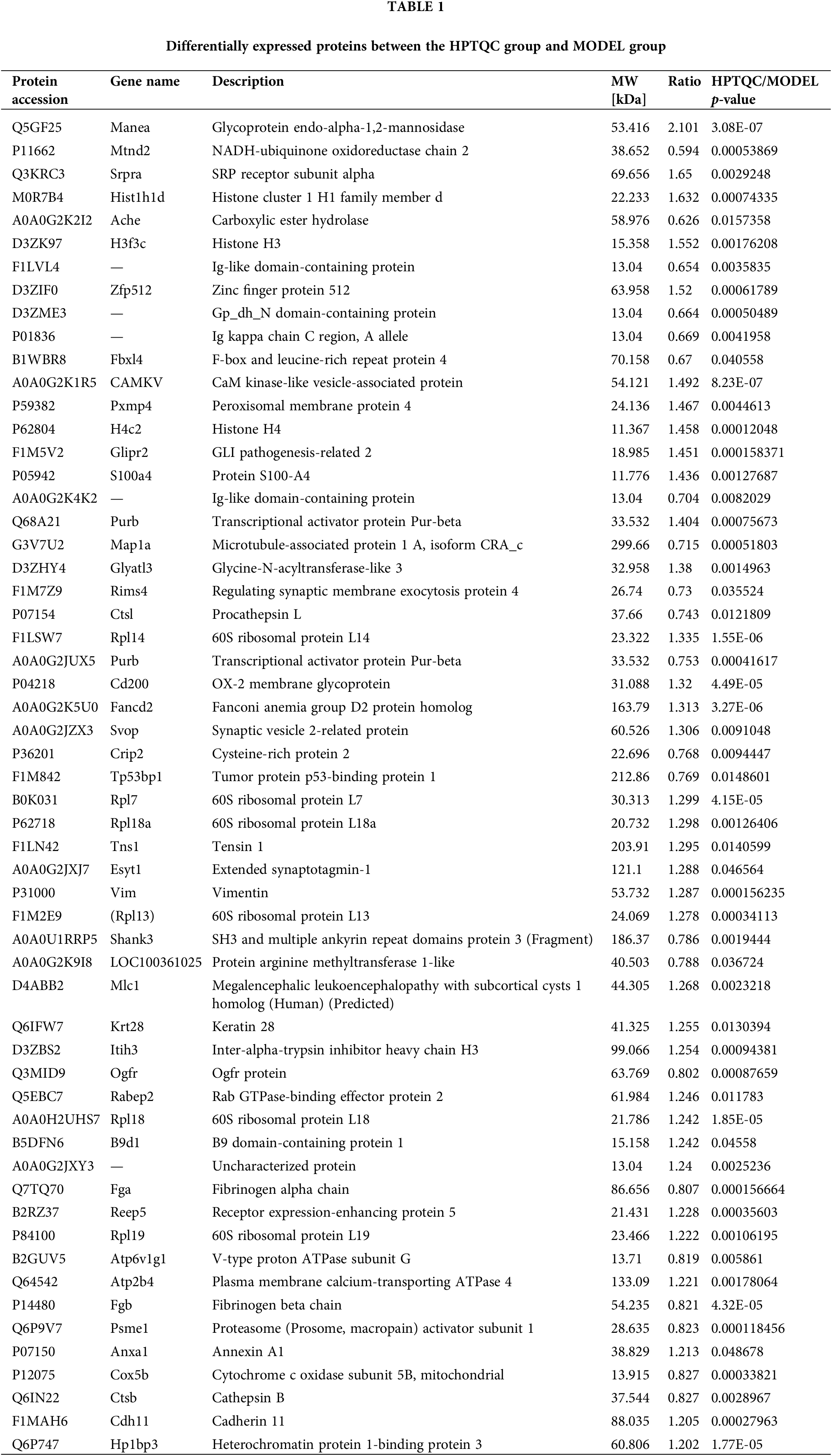
We used proteomics analysis based on ITRAQ quantification to successfully obtain protein expression levels in AD rat models before and after HPTQ administration. A 1.2-fold cutoff value was used as the threshold of the difference in the degree of protein expression levels, and a p-value < 0.05 was used as the threshold of significance. Comparative quantification of the hippocampus between the HPTQ and the MODEL groups revealed that 57 proteins were significantly differentially expressed. These, included 35 proteins with upregulated expression (61%) and 22 proteins with downregulated expression (39%, Fig. 4). All differentially expressed proteins in the HPTQ and Model groups have been shown in Table 1 ranked by the degree of expression differences.
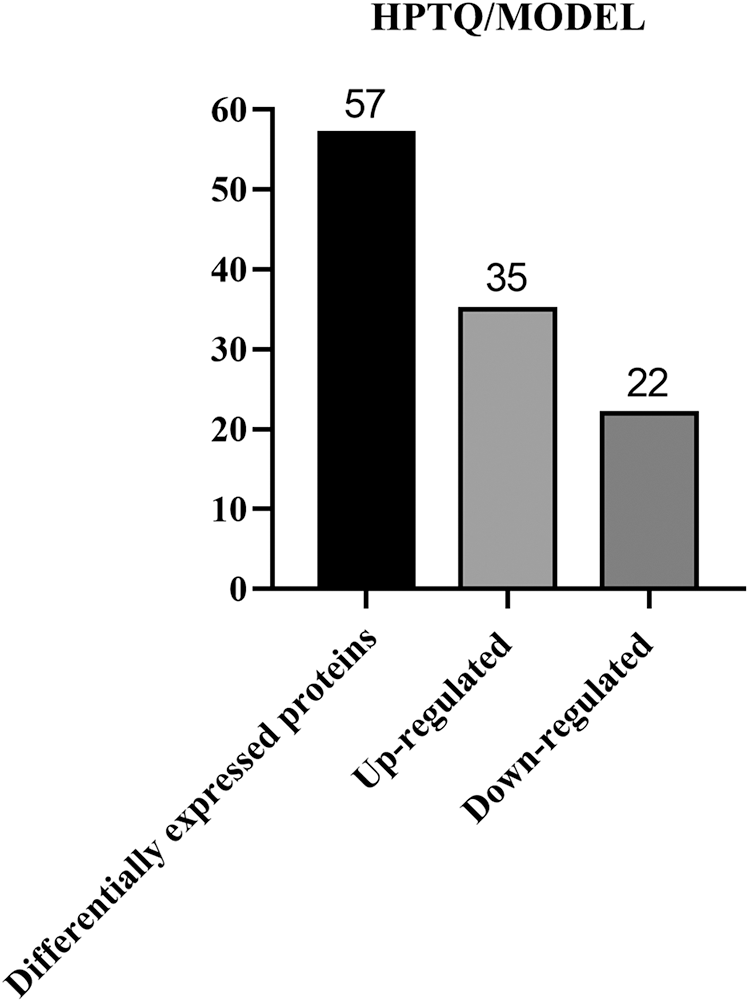
Figure 4: Total differentially expressed proteins in Huang-Pu-Tong-Qiao formula (HPTQ) and MODEL groups. The differential proteins detected included both up- and down-regulated proteins. The number of the upregulated proteins was larger than that of downregulated proteins, suggesting that HPTQ could promote protein synthesis.
Bioinformatics analysis of differentially expressed proteins
Based on GO analysis, the differentially expressed proteins in the HPTQ and MODEL groups were classified into three categories: biological process, cellular compartment, and molecular function, as shown in Figs. 5A–5C. In biological process, the GO analysis with the largest number of differentially expressed proteins included cellular processes (50.9%), single biological processes (40.4%), biological regulation (40.4%), metabolic processes (38.6%), and responses to stimuli (29.8%) were significantly enriched. In the molecular function, the GO terms with the largest number of differentially expressed proteins were binding (70.2%), catalytic activity (24.6%), structural molecule activity (17.5%), transporter activity (8.8%), and molecular function regulator (7.0%). The cellular components showed that proteins were mostly located in the cell (75.4%), organelle (68.4%), membrane (43.9%), macromolecular complex (36.8%), and extracellular region (29.8%). GO enrichment analysis was used to analyze the differentially expressed proteins in the HPTQ and MODEL groups, the enrichment results have been shown in Figs. 6A–6C. Biological processes were enriched in regulation of heterotypic cell-cell adhesion, blood coagulation, fibrin clot formation, antimicrobial humoral response, skeletal muscle tissue regeneration, response to X-ray, tissue regeneration, negative regulation of hemostasis, maternal process involved in female pregnancy, regulation of wound healing, protein activation cascade, protein-DNA complex assembly, DNA packaging, defense response to bacteria, DNA conformation change, nucleosome organization, chromatin assembly or disassembly, regulation of response to wounding, protein processing, reproductive structure development, and reproductive system development. The enrichment analysis of GO cellular components revealed that the identified proteins were mainly involved in fibrinogen complex, nucleosome, DNA packaging complex, replisome, nuclear replisome, DNA replication factor A complex, protein-DNA complex, platelet alpha granule, cytosolic large ribosomal subunit, cytosolic ribosome, external side of plasma membrane, large ribosomal subunit, ribosomal subunit, cytosolic part, chromosome, chromosomal part, ribosome, extracellular space, non-membrane-bounded organelle, and intracellular non-membrane-bounded organelle. In terms of molecular function, there was significant enrichment in single-stranded DNA binding, protein binding and bridging, histone binding, cysteine-type endopeptidase activity, binding and bridging, cation-transporting ATPase activity, calcium-dependent protein binding, structural constituent of ribosome, structural molecule activity, and DNA binding.
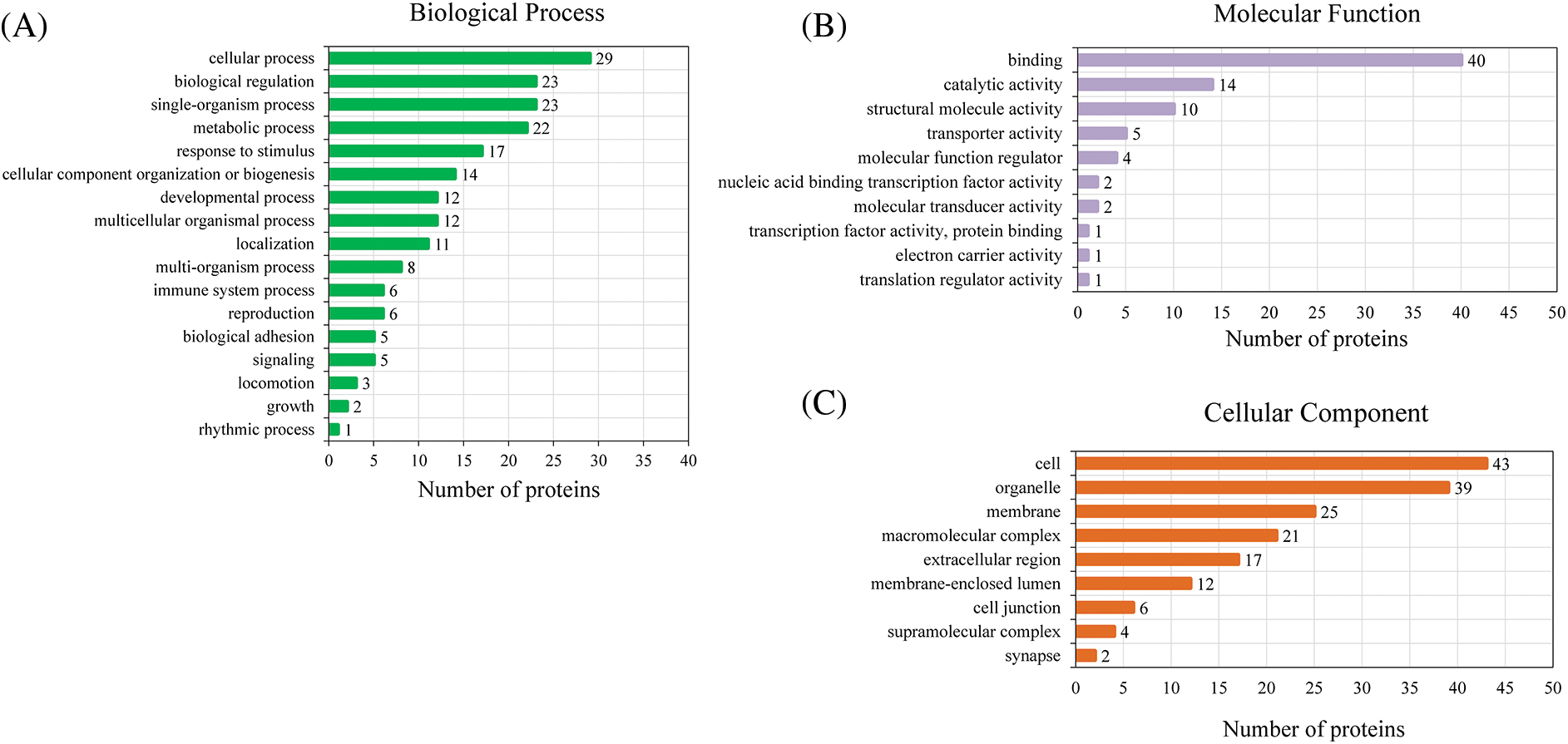
Figure 5: Three classes of differentially expressed proteins analyzed by Gene Ontology (GO). (A) Biological Process; (B) Molecular Function; (C) Cellular Component.
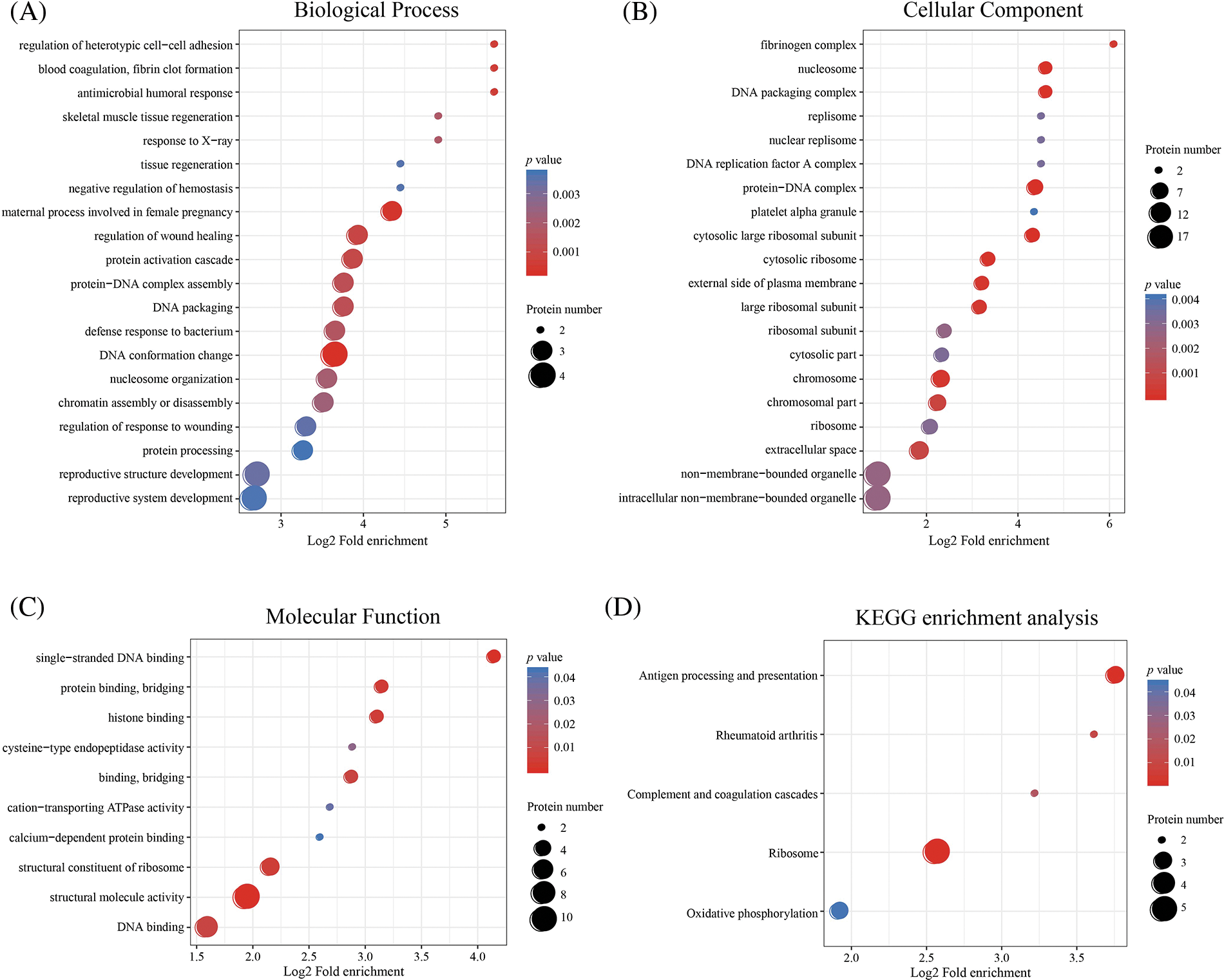
Figure 6: Enrichment analysis of Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG). (A) Biological Process; (B) Molecular Function; (C) Cellular Component; (D) KEGG enrichment analysis. GO and KEGG enrichment analysis using two-tailed Fisher’s exact test. The results with a p-value < 0.05 were considered to be statistically and significantly different.
Next, KEGG pathway analysis revealed that the proteins with a differential expression before and after HPTQ administration were involved in 51 kinds of signaling pathways. These included Alzheimer’s disease, Ribosome, Autophagy-animal, Apoptosis, Peroxisome, Parkinson’s disease, Complement and coagulation cascades, Oxidative phosphorylation, Glycolysis/Gluconeogenesis, Huntington’s disease, cGMP-PKG signaling pathway, cAMP signaling pathway, HIF-1 signaling pathway, FoxO signaling pathway, Calcium signaling pathway, NOD-like receptor signaling pathway, Proteasome, Glutamatergic synapse, mTOR signaling pathway, and Cholinergic synapse. KEGG enrichment analysis identified five pathways, of which Antigen processing and presentation, Complement and coagulation cascades, Ribosome, and Oxidative phosphorylation were associated with AD, as shown in Fig. 6D.
Subsequently, the Protein-Protein Interaction (PPI) network results showed the interconnection between differentially expressed proteins. The results obtained from the analysis of the STRING database indicated that a total of 46 differentially expressed proteins had interactions, among which 60S ribosomal-related proteins, Fga, Fgb, Ctsb, S100a4, CAMKV, and Rims4 were associated with AD pathogenesis, as shown in Fig. 7.
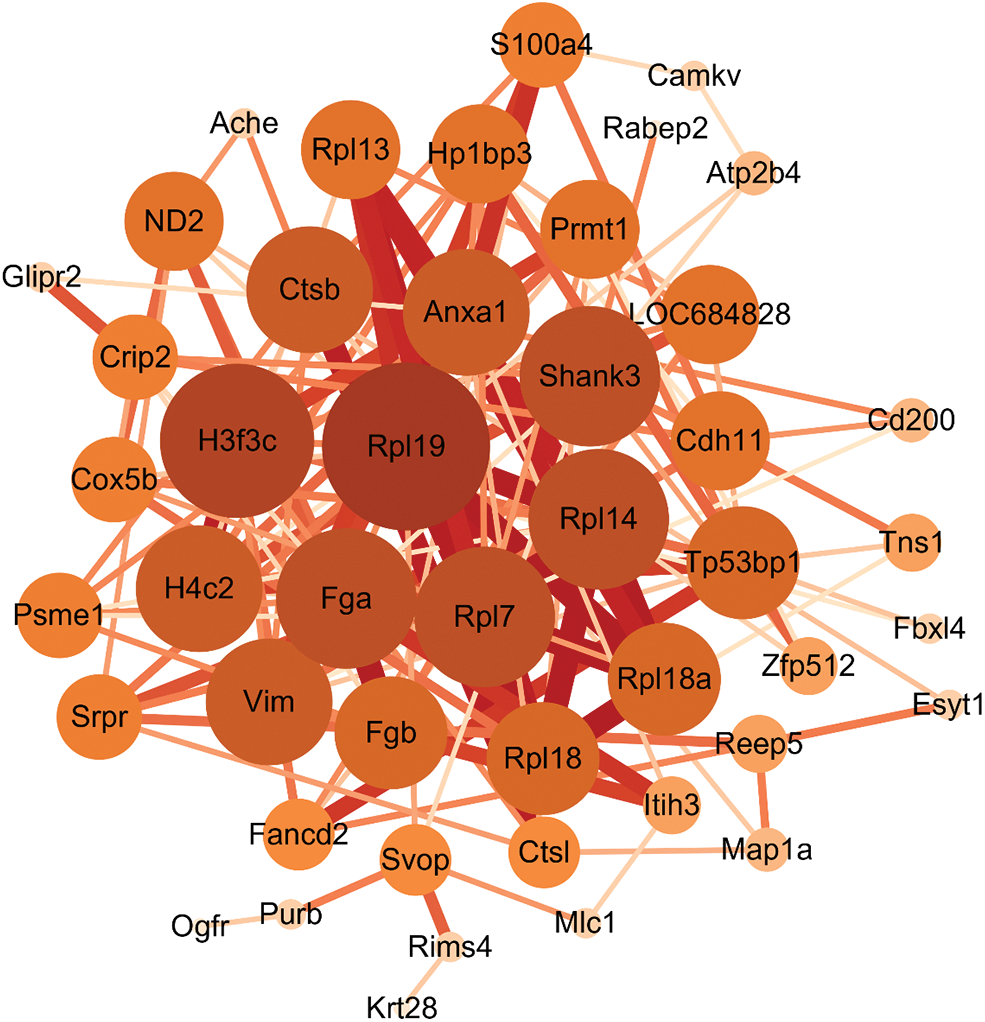
Figure 7: Protein Interaction (PPI) network construction. The larger the node, the more important it is in this network, the thicker the connection line between nodes, and the stronger the interaction between nodes.
Finally, the results of proteomic assays were further validated by Western blot analysis. The expression levels of four differential proteins CAMKV, ACHE, Rims4, and Pxmp4, were consistent with that obtained in the proteomics analysis. The protein bands and the expression ratios are shown in Fig. 8.
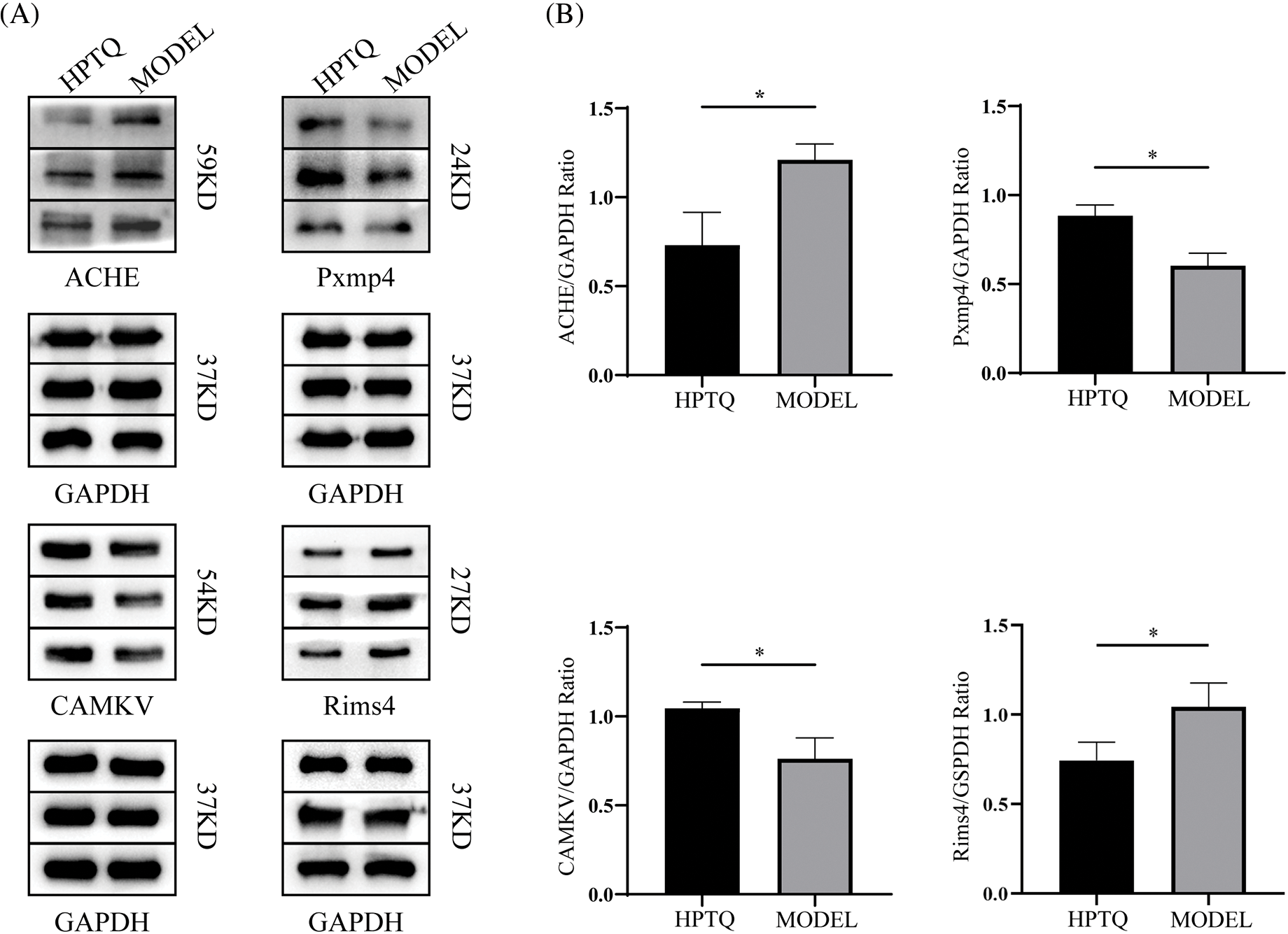
Figure 8: Effect of Huang-Pu-Tong-Qiao formula (HPTQ) on the protein expressions of ACHE, Pxmp4, CAMKV, and Rims4 in Alzheimer’s Disease (AD) rat models. (A) The protein bands of the indicated proteins; (B) These graphs show the relative expression of Carboxylic ester hydrolase (ACHE), Peroxisomal membrane protein 4 (Pxmp4), CaM kinase-like vesicle-associated protein (CAMKV), and membrane exocytosis protein 4 (Rims4). Values are expressed as the mean ± SD (n = 3). *p < 0.05, vs. MODEL group.
In this study, we applied the ITRAQ-based proteomics approach and performed a comparative proteomics analysis of the hippocampus in AD model rats after HPTQ treatment. The AD rat model was established using intraperitoneal injections of D-galactose combined with an intraventricular injection of Aβ25-35. The Morris water maze results pointed at the ability of HPTQ to improve learning memory in AD model rats. Subsequently, the ITRAQ results showed 57 differentially expressed proteins in hippocampal tissues of the HPTQ and MODEL groups, of which 35 proteins were upregulated (61%) and 22 proteins were downregulated (39%). This result indicated that the protein expression level in the hippocampal tissue of AD rats was enhanced after HPTQ treatment. To elucidate the potential molecular mechanisms of HPTQ for the treatment of AD, we analyzed the effect and mechanism of proteins by GO analysis and KEGG analysis and assessed the degree of association between proteins through the construction of PPI networks and KEGG enrichment analysis. AD-related biological functions of differentially expressed proteins obtained by bioinformatics analysis included DNA transcription and mRNA translation, coagulation, fibrin clot formation, regulation of autophagy, inhibition of cholinesterase, improvement of neuronal survival, anti-inflammation, and alleviation of oxidative stress. In addition, the PPI network showed significant associations between most of the differentially expressed proteins. Some differentially expressed proteins with potential neuroprotective effects have been selected for further analysis.
Ribosomal protein is an important constituent of the ribosome unit. The main function of the ribosomes is to participate in the assembly and translation of protein ribosomes. In neurodegenerative diseases, ribosomal dysfunction leads to impaired protein biosynthesis (Ding et al., 2005; Ding et al., 2006). For instance, pathological Tau protein formation in AD-induced ribosomal dysfunction led to reduced protein synthesis (Meier et al., 2016). It was found in another study that the levels of 60S ribosomal protein subunits were significantly decreased in Tau-induced frontotemporal dementia mice compared to the normal group (Evans et al., 2021). Moreover, after the Aβ1-42 intervention, the phosphorylation of Tau protein led to reduced protein transcription and translation levels by inducing ribosomal dysfunction in SHSY5Y cells (Maina et al., 2018). In this study, the KEGG enrichment analysis indicated that the ribosomal pathway was significantly enriched and the PPI network showed that several genes of the 60S ribosomal protein subunits occupied the center of the network and interacted with other differentially expressed proteins. The expression of L7, L14, L18, L18Aa, L19, and L13 of 60S ribosomal protein subunits was upregulated in the HPTQ group. Combined with the results of our previous study in which we found that HPTQ could reduce the level of pathologically phosphorylated Tau protein in AD rat models (Ye et al., 2020), we suggest that HPTQ could play a neuroprotective role by enhancing protein biosynthesis through restoring ribosomal dysfunction in AD. The complement and coagulation cascade pathways involved in Fga and Fgb proteins were found to be significantly enriched in the KEGG enrichment analysis. Fibrinogen alpha chain (Fga) and Fibrinogen beta chain (Fgb) are proteins that are closely associated with vascular diseases, and play an essential role in thrombosis, wound healing, and other biological functions (Simurda et al., 2020). The protein expression levels of Fga and Fgb were previously reported to be positively correlated with the degree of Aβ deposition in AD humans and mice brains (Bian et al., 2021). Interestingly, our proteomics results showed that the protein expression levels of Fga and Fgb were decreased after HPTQ treatment. Therefore, Fga and Fgb might be potential therapeutic targets for HPTQ in reducing Aβ deposition.
Autophagy is a clearance mechanism that degrades abnormal or redundant intracellular components to promote cell survival. The ability of autophagy activation in AD to eliminate abnormal Aβ deposition and phosphorylated Tau tangles makes autophagy a vital mechanism for neuronal cell protection (Nilsson et al., 2013; Liu et al., 2015). Among the differentially expressed proteins obtained by proteomics analysis, Histone cluster 1 H1 family member (Hist1h1d) and Cathepsin B (Ctsb) were closely associated with autophagy. Hist1h1d is a histone family member responsible for gene transcription and protein folding. The up-regulated expression of Hist1h1d in neurons promoted Aβ clearance through autophagy (Bastrup et al., 2021). As an important cysteine protease in lysosomes, Ctsb was shown to be involved in several pathways connected with neuroinflammation and injury, including the autophagy pathway, lysosomal pathway, apoptotic pathway, and nucleotide-binding oligomerization domain (NOD)-like receptor signaling pathway associated with the NLRP3 inflammasome. A previous study has shown that overexpression of Ctsb led to lysosomal damage, causing autophagy dysfunction and the activation of the NLRP3 signaling pathway, ultimately leading to neuronal inflammation (Wang et al., 2017). In addition, the antigen processing and presentation pathways involving Ctsb were significantly enriched in the KEGG enrichment analysis, where the MHC II pathway could eventually generate pro-inflammatory CD4+ T cells (Monsonego et al., 2013). A study showed that over-activated CD4+ T cells in AD lead to excessive immune responses and promote the formation of β-secretase, which cleaves APP and forms pathological Aβ (Dai et al., 2021), while Ctsb has been shown to positively affect the activity of the MHC II pathway (Ma et al., 2022). Therefore, the reduction of the Ctsb expression level can reduce the overexpression of CD4+ T cells in AD and alleviate the inflammatory response of neurons. Among the obtained proteomics data, we found that the Hist1h1d levels were significantly increased and the Ctsb levels were significantly decreased after HPTQ treatment. Taken together, the above results suggest that HPTQ can regulate the expression of Hist1h1d and Ctsb, both of which might be potential therapeutic targets for HPTQ to exert its neuroprotective effects.
To validate the results of ITRAQ quantitative proteomics, four proteins showing high variation in expression, ACHE, CAMKV, Pxmp4, and Rims4, were screened and validated by western blotting analysis. ACHE hydrolyzes acetylcholine, and a normal ACHE expression is necessary for the brain to maintain healthy neuron signal transmission. The cholinergic system of the AD brain is defective, and over-activation of cholinesterase leads to decreased synaptic acetylcholine levels and ultimately to various neurodegenerative symptoms (Schliebs and Arendt, 2011; Ferreira-Vieira et al., 2016; Ahmed et al., 2017). Currently, cholinesterase inhibitors are an important class of drugs for the treatment of AD and have shown efficacy in clinical trials for the treatment of mild and moderate AD. In this study, the expression level of ACHE was decreased in the HPTQ group, indicating that the administration of HPTQ in AD has the effect of inhibiting ACHE and restoring the normal function of the cholinergic system. CAM kinase-like vesicle-associated protein (CAMKV) is a synaptic protein that belongs to the calmodulin-dependent protein kinase family and does not possess kinase activity. CAMKV plays an essential role in learning and memory by regulating dendritic spine generation, synaptic transmission, and neural plasticity (Liang et al., 2016). Combined with the recovery of spatial memory ability shown in the water maze experiment and increased levels of CAMKV expression in the HPTQ group in the proteomics data, we surmised that CAMKV is a potential therapeutic target of HPTQ to improve spatial memory deficits in AD rats. Peroxisome membrane protein 4 (Pxmp4) is an essential component of the peroxisome and is responsible for many redox reactions (Reguenga et al., 1999). KEGG pathway analysis showed that Pxmp4 is involved in the metabolism of reactive oxygen species (ROS) through the peroxisome pathway. A recent study has reported that excessive elevation of ROS could lead to oxidative stress and neuron loss in vitro (Angelova and Abramov, 2018). Pxmp4 has an important role in reducing the excess ROS in AD and maintaining redox homeostasis in neuron cells (Singh et al., 2019). The upregulation of peroxisome level in AD mice was shown to improve spatial memory, reduce Aβ deposition, and restore the impaired synaptic function (Inestrosa et al., 2013). In our results, the high expression of Pxmp4 after HPTQ treatment indicates that HPTQ could attenuate oxidative stress and promote the maintenance of redox homeostasis in neuronal cells of AD rats, which is consistent with our previous findings (Cai et al., 2018). The protein regulating synaptic membrane exocytosis protein 4 (Rims4) is critical in regulating neurosynaptic activity (Mittelstaedt et al., 2010). Previous studies have confirmed the important role of Rims4 in neuronal arborization and dendritic spine development as the knockdown of Rims4 protein reduced the complexity of neuronal arborization and caused abnormal axonal and dendritic growth (Alvarez-Baron et al., 2013). Up to now, the relationship between Rims4 and AD has not been studied, but the important role of Rims4 in neuronal axons and dendrites combined with the decrease of Rims4 expression level after HPTQ treatment suggest that Rims4 is a potential therapeutic target of HPTQ in AD treatment. Finally, our results showed S100-A4(S100a4), a protein that belongs to the multifunctional S100 protein family, is another potential therapeutic target (Donato et al., 2013). S100A4 has multiple functions in the nervous system. For example, it promoted neuronal survival in brain injury by regulating neuronal plasticity through the NRG/ErbB4 and ErbB2/Akt pathways (Pankratova et al., 2018). In another study, the interaction of S100a4 with cell surface heparan sulfate proteoglycans could increase the Ca2+ concentration in primary neuronal cells and modulate neuronal plasticity (Kiryushko et al., 2006). In addition, S100A4 could also exert antioxidant effects through the IL-10 receptor/Janus kinase/Signal Transducers and Activation Transcription system (IL-10R/JAK/STAT3) pathway to treat neuronal loss induced by brain injury (Dmytriyeva et al., 2012). Combining the neuroprotective role of S100a4 in neurodegeneration with our proteomics results, we believed that S100a4 could be an important therapeutic target of HPTQ in the treatment of AD.
Previous clinical results and studies have demonstrated that HPTQ can alleviate neuronal damage and improve AD symptoms by modulating multiple cellular mechanisms. However, the molecular targets of HPTQ have not yet been comprehensively studied. Hence, this study analyzed the data obtained from ITRAQ-based proteomic experiments conducted on the HPTQ and the Model groups to identify potential HPTQ therapeutic targets in AD rats. The proteomics analysis suggested that the effect of HPTQ in AD was achieved by improving the deposition of Aβ, regulating autophagy, inhibiting ACHE, regulating neuronal synaptic function and plasticity, reducing ROS damage, and alleviating oxidative stress. Furthermore, our findings suggested that HPTQ could promote protein biosynthesis in neurons and upregulate protein expression in the hippocampus of AD model rats. In conclusion, we analyzed several potential therapeutic targets and therapeutic mechanisms of HPTQ in AD treatment. In light of our findings, we believe that this study could provide a theoretical basis for the use of HPTQ in the treatment of AD.
Funding Statement: This research was supported by the National Natural Science Foundation of China (Grant No. 81873351 and 82205235), the Academic Support Program for High School Discipline (Professional) Top Talent (Grant No. gxbjZD2022024), the Key Project Foundation of Natural Science Research in Universities of Anhui Province in China (grant Nos. KJ2020A0439 and KJ2020A0416), and the Talent Support Program of Anhui University of Traditional Chinese Medicine (2022rcyb028).
Author Contributions: The authors confirm their contribution to the paper as follows: BC, HS, YW, GQ, ZQ, and XL performed the experiments. Proteomic analysis was performed by HS and YW. BC, SY and XL contributed significantly to bioinformatics analysis. XL, QC, and GH performed the experimental validation. XL and GH wrote the manuscript. BC, SY, and HS reviewed and modified the paper.
Availability of Data and Materials: The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Ethics Approval: This experiment was authorized by the Animal Ethics Committee of the Anhui University of Chinese Medicine, on November 7th, 2022. The ethical certification number is AHUCM-rats-2022052.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
Ahmed T, Zahid S, Mahboob A, Farhat SM (2017). Cholinergic system and post-translational modifications: An insight on the role in Alzheimer’s disease. Current 15: 480–494. https://doi.org/10.2174/1570159X14666160325121145 [Google Scholar] [PubMed] [CrossRef]
Alvarez-Baron E, Michel K, Mittelstaedt T, Opitz T, Schmitz F, Beck H, Dietrich D, Becker AJ, Schoch S (2013). RIM3gamma and RIM4gamma are key regulators of neuronal arborization. The Journal of Neuroscience 33: 824–839. https://doi.org/10.1523/JNEUROSCI.2229-12.2013 [Google Scholar] [PubMed] [CrossRef]
Angelova PR, Abramov AY (2018). Role of mitochondrial ROS in the brain: From physiology to neurodegeneration. FEBS Letters 592: 692–702. https://doi.org/10.1002/1873-3468.12964 [Google Scholar] [PubMed] [CrossRef]
Bai B, Wang X, Li Y, Chen PC, Yu K et al. (2020). Deep multilayer brain proteomics identifies molecular networks in Alzheimer’s disease progression. Neuron 105: 975–991.e977. https://doi.org/10.1016/j.neuron.2019.12.015 [Google Scholar] [PubMed] [CrossRef]
Bastrup J, Hansen KH, Poulsen TBG, Kastaniegaard K, Asuni AA, Christensen S, Belling D, Helboe L, Stensballe A, Volbracht C (2021). Anti-Aβ antibody aducanumab regulates the proteome of senile plaques and closely surrounding tissue in a transgenic mouse model of Alzheimer’s disease. Journal of Alzheimer’s Disease 79: 249–265. https://doi.org/10.3233/JAD-200715 [Google Scholar] [PubMed] [CrossRef]
Bian Z, Yamashita T, Shi X, Feng T, Yu H et al. (2021). Accelerated accumulation of fibrinogen peptide chains with Aβ deposition in Alzheimer’s disease (AD) mice and human AD brains. Brain Research 1767: 147569. https://doi.org/10.1016/j.brainres.2021.147569 [Google Scholar] [PubMed] [CrossRef]
Busche MA, Hyman BT (2020). Synergy between amyloid-β and tau in Alzheimer’s disease. Nature Neuroscience 23: 1183–1193. https://doi.org/10.1038/s41593-020-0687-6 [Google Scholar] [PubMed] [CrossRef]
Cai B, Xie D, Wang Y, Li J, Wang T, Ye S, Hua R (2018). Effect of huangpu tongqiao capsule on oxidative stress of hippocampal neurons and mitochondrial apoptotic path-way in rats with Alzheimer’s disease. Chinese Journal of Integrated Traditional and Western Medicine 38: 707–711. https://kns.cnki.net/kcms/detail/11.2787.R.20171205.1116.002.html [Google Scholar]
Cummings JL, Tong G, Ballard C (2019). Treatment combinations for Alzheimer’s disease: Current and future pharmacotherapy options. Journal of Alzheimer’s Disease 67: 779–794. https://doi.org/10.3233/JAD-180766 [Google Scholar] [PubMed] [CrossRef]
Dai L, Wang Q, Lv X, Gao F, Chen Z, Shen Y (2021). Elevated β-secretase 1 expression mediates CD4+ T cell dysfunction via PGE2 signalling in Alzheimer’s disease. Brain, Behavior, and Immunity 98: 337–348. https://doi.org/10.1016/j.bbi.2021.08.234 [Google Scholar] [PubMed] [CrossRef]
Ding Q, Markesbery WR, Cecarini V, Keller JN (2006). Decreased RNA, and increased RNA oxidation, in ribosomes from early Alzheimer’s disease. Neurochemical Research 31: 705–710. https://doi.org/10.1007/s11064-006-9071-5 [Google Scholar] [PubMed] [CrossRef]
Ding Q, Markesbery WR, Chen Q, Li F, Keller JN (2005). Ribosome dysfunction is an early event in Alzheimer’s disease. The Journal of Neuroscience 25: 9171–9175. https://doi.org/10.1523/JNEUROSCI.3040-05.2005 [Google Scholar] [PubMed] [CrossRef]
Dmytriyeva O, Pankratova S, Owczarek S, Sonn K, Soroka V et al. (2012). The metastasis-promoting S100A4 protein confers neuroprotection in brain injury. Nature Communications 3: 1197. https://doi.org/10.1038/ncomms2202 [Google Scholar] [PubMed] [CrossRef]
Donato R, Cannon BR, Sorci G, Riuzzi F, Hsu K, Weber DJ, Geczy CL (2013). Functions of S100 proteins. Current Molecular Medicine 13: 24–57. https://doi.org/10.2174/156652413804486214 [Google Scholar] [CrossRef]
Evans HT, Taylor D, Kneynsberg A, Bodea LG, Gotz J (2021). Altered ribosomal function and protein synthesis caused by tau. Acta Neuropathologica Communications 9: 110. https://doi.org/10.1186/s40478-021-01208-4 [Google Scholar] [PubMed] [CrossRef]
Ferreira-Vieira TH, Guimaraes IM, Silva FR, Ribeiro FM (2016). Alzheimer’s disease: Targeting the cholinergic system. Current Neuropharmacology 14: 101–115. https://doi.org/10.2174/1570159X13666150716165726 [Google Scholar] [PubMed] [CrossRef]
Fjell AM, McEvoy L, Holland D, Dale AM, Walhovd KB, Alzheimer’s Disease Neuroimaging I (2014). What is normal in normal aging? Effects of aging, amyloid and Alzheimer’s disease on the cerebral cortex and the hippocampus. Progress in Neurobiology 117: 20–40. https://doi.org/10.1016/j.pneurobio.2014.02.004 [Google Scholar] [PubMed] [CrossRef]
Huang Y, Mucke L (2012). Alzheimer mechanisms and therapeutic strategies. Cell 148: 1204–1222. https://doi.org/10.1016/j.cell.2012.02.040 [Google Scholar] [PubMed] [CrossRef]
Inestrosa NC, Carvajal FJ, Zolezzi JM, Tapia-Rojas C, Serrano F, Karmelic D, Toledo EM, Toro A, Toro J, Santos MJ (2013). Peroxisome proliferators reduce spatial memory impairment, synaptic failure, and neurodegeneration in brains of a double transgenic mice model of Alzheimer’s disease. Journal of Alzheimer’s Disease 33: 941–959. https://doi.org/10.3233/JAD-2012-120397 [Google Scholar] [PubMed] [CrossRef]
Jiang A, Cai B, Xie D, Ye S, Wang Y, Gao H, Ma J (2019). Effects of Huangpu Tongqiao Capsule on JNK signal pathway proteins and inflammatory factors expression in rats with Alzheimer’s disease. China Journal of Traditional Chinese Medicine and Pharmacy 34: 2441–2444. https://kns.cnki.net/kcms/detail/11.2787.R.20171205.1116.002.html [Google Scholar]
Kabir MT, Uddin MS, Mamun AA, Jeandet P, Aleya L, Mansouri RA, Ashraf GM, Mathew B, Bin-Jumah MN, Abdel-Daim MM (2020). Combination drug therapy for the management of Alzheimer’s disease. International Journal of Molecular Sciences 21: 272. https://doi.org/10.3390/ijms21093272 [Google Scholar] [PubMed] [CrossRef]
Kiryushko D, Novitskaya V, Soroka V, Klingelhofer J, Lukanidin E, Berezin V, Bock E (2006). Molecular mechanisms of Ca2+ signaling in neurons induced by the S100A4 protein. Molecular and Cellular Biology 26: 3625–3638. https://doi.org/10.1128/MCB.26.9.3625-3638.2006 [Google Scholar] [PubMed] [CrossRef]
Knopman DS, Jones DT, Greicius MD (2021). Failure to demonstrate efficacy of aducanumab: An analysis of the EMERGE and ENGAGE trials as reported by Biogen, December 2019. Alzheimer’s & Dementia 17: 696–701. https://doi.org/10.1002/alz.12213 [Google Scholar] [PubMed] [CrossRef]
Lane CA, Hardy J, Schott JM (2018). Alzheimer’s disease. European Journal of Neurology 25: 59–70. https://doi.org/10.1111/ene.13439 [Google Scholar] [PubMed] [CrossRef]
Li N, Hu P, Xu T, Chen H, Chen X, Hu J, Yang X, Shi L, Luo JH, Xu J (2017). iTRAQ-based proteomic analysis of APPSw, ind mice provides insights into the early changes in Alzheimer’s disease. Current Alzheimer Research 14: 1109–1122. https://doi.org/10.2174/1567205014666170719165745 [Google Scholar] [PubMed] [CrossRef]
Liang Z, Zhan Y, Shen Y, Wong CCL, Yates JRIII, Plattner F, Lai KO, Ip NY (2016). The pseudokinase CaMKv is required for the activity-dependent maintenance of dendritic spines. Nature Communications 7: 13282. https://doi.org/10.1038/ncomms13282 [Google Scholar] [PubMed] [CrossRef]
Liu Z, Li T, Li P, Wei N, Zhao Z, Liang H, Ji X, Chen W, Xue M, Wei J (2015). The ambiguous relationship of oxidative stress, tau hyperphosphorylation, and autophagy dysfunction in alzheimer’s disease. Oxidative Medicine and Cellular Longevity 2015: 352723. https://doi.org/10.1155/2015/352723 [Google Scholar] [PubMed] [CrossRef]
Ma K, Chen X, Liu W, Chen S, Yang C, Yang J (2022). CTSB is a negative prognostic biomarker and therapeutic target associated with immune cells infiltration and immunosuppression in gliomas. Scientific Reports 12: 4295. https://doi.org/10.1038/s41598-022-08346-2 [Google Scholar] [PubMed] [CrossRef]
Maina MB, Bailey LJ, Doherty AJ, Serpell LC (2018). The involvement of Abeta42 and tau in nucleolar and protein synthesis machinery dysfunction. Frontiers in Cellular Neuroscience 12: 220. https://doi.org/10.3389/fncel.2018.00220 [Google Scholar] [PubMed] [CrossRef]
Meier S, Bell M, Lyons DN, Rodriguez-Rivera J, Ingram A et al. (2016). Pathological tau promotes neuronal damage by impairing ribosomal function and decreasing protein synthesis. The Journal of Neuroscience 36: 1001–1007. https://doi.org/10.1523/JNEUROSCI.3029-15.2016 [Google Scholar] [PubMed] [CrossRef]
Mittelstaedt T, Alvarez-Baron E, Schoch S (2010). RIM proteins and their role in synapse function. Biological Chemistry 391: 599–606. https://doi.org/10.1515/bc.2010.064 [Google Scholar] [PubMed] [CrossRef]
Monsonego A, Nemirovsky A, Harpaz I (2013). CD4 T cells in immunity and immunotherapy of Alzheimer’s disease. Immunology 139: 438–446. https://doi.org/10.1111/imm.12103 [Google Scholar] [PubMed] [CrossRef]
Nilsson P, Loganathan K, Sekiguchi M, Matsuba Y, Hui K, Tsubuki S, Tanaka M, Iwata N, Saito T, Saido TC (2013). Abeta secretion and plaque formation depend on autophagy. Cell Reports 5: 61–69. https://doi.org/10.1016/j.celrep.2013.08.042 [Google Scholar] [PubMed] [CrossRef]
Pankratova S, Klingelhofer J, Dmytriyeva O, Owczarek S, Renziehausen A, Syed N, Porter AE, Dexter DT, Kiryushko D (2018). The S100A4 protein signals through the ErbB4 receptor to promote neuronal survival. Theranostics 8: 3977–3990. https://doi.org/10.7150/thno.22274 [Google Scholar] [PubMed] [CrossRef]
Poll S, Mittag M, Musacchio F, Justus LC, Giovannetti EA et al. (2020). Memory trace interference impairs recall in a mouse model of Alzheimer’s disease. Nature Neuroscience 23: 952–958. https://doi.org/10.1038/s41593-020-0652-4 [Google Scholar] [PubMed] [CrossRef]
Reguenga C, Oliveira ME, Gouveia AM, Eckerskorn C, Sa-Miranda C, Azevedo JE (1999). Identification of a 24 kDa intrinsic membrane protein from mammalian peroxisomes. Biochimica et Biophysica Acta 1445: 337–341. https://doi.org/10.1016/S0167-4781(99)00061-5 [Google Scholar] [PubMed] [CrossRef]
Rhein V, Song X, Wiesner A, Ittner LM, Baysang G et al. (2009). Amyloid-β and tau synergistically impair the oxidative phosphorylation system in triple transgenic Alzheimer’s disease mice. Proceedings of the National Academy of Sciences of the United States of America 106: 20057–20062. https://doi.org/10.1073/pnas.0905529106 [Google Scholar] [PubMed] [CrossRef]
Schliebs R, Arendt T (2011). The cholinergic system in aging and neuronal degeneration. Behavioural Brain Research 221: 555–563. https://doi.org/10.1016/j.bbr.2010.11.058 [Google Scholar] [PubMed] [CrossRef]
Sharma P, Srivastava P, Seth A, Tripathi PN, Banerjee AG, Shrivastava SK (2019). Comprehensive review of mechanisms of pathogenesis involved in Alzheimer’s disease and potential therapeutic strategies. Progress in Neurobiology 174: 53–89. https://doi.org/10.1016/j.pneurobio.2018.12.006 [Google Scholar] [PubMed] [CrossRef]
Simurda T, Brunclikova M, Asselta R, Caccia S, Zolkova J et al. (2020). Genetic variants in the FGB and FGG genes mapping in the beta and gamma nodules of the fibrinogen molecule in congenital quantitative fibrinogen disorders associated with a thrombotic phenotype. International Journal of Molecular Sciences 21: 4616. https://doi.org/10.3390/ijms21134616 [Google Scholar] [PubMed] [CrossRef]
Singh A, Kukreti R, Saso L, Kukreti S (2019). Oxidative stress: A key modulator in neurodegenerative diseases. Molecules 24: 1583. https://doi.org/10.3390/molecules24081583 [Google Scholar] [PubMed] [CrossRef]
Veitch DP, Weiner MW, Aisen PS, Beckett LA, Cairns NJ et al. (2019). Understanding disease progression and improving Alzheimer’s disease clinical trials: Recent highlights from the Alzheimer’s Disease Neuroimaging Initiative. Alzheimer’s & Dementia 15: 106–152. https://doi.org/10.1016/j.jalz.2018.08.005 [Google Scholar] [PubMed] [CrossRef]
Vorhees CV, Williams MT (2006). Morris water maze: Procedures for assessing spatial and related forms of learning and memory. Nature Protocols 1: 848–858. https://doi.org/10.1038/nprot.2006.116 [Google Scholar] [PubMed] [CrossRef]
Wang D, Zhang J, Jiang W, Cao Z, Zhao F, Cai T, Aschner M, Luo W (2017). The role of NLRP3-CASP1 in inflammasome-mediated neuroinflammation and autophagy dysfunction in manganese-induced, hippocampal-dependent impairment of learning and memory ability. Autophagy 13: 914–927. https://doi.org/10.1080/15548627.2017.1293766 [Google Scholar] [PubMed] [CrossRef]
Wang G, Zhou P, Xie D, Ye S, Gao H, Wang Y, Fang Z, Cai B (2020). Effects of huangpu tongqiao capsules on EGFR-PLCγ signal pathway of hippocampus in rats with Alzheimer’s disease. China Journal of Chinese Materia Medica 45: 2165–2171. https://doi.org/10.19540/j.cnki.cjcmm.20191219.401 [Google Scholar] [PubMed] [CrossRef]
Xu M, Deng J, Xu K, Zhu T, Han L et al. (2019). In-depth serum proteomics reveals biomarkers of psoriasis severity and response to traditional Chinese medicine. Theranostics 9: 2475–2488. https://doi.org/10.7150/thno.31144 [Google Scholar] [PubMed] [CrossRef]
Xu H, Zhou W, Zhan L, Sui H, Zhang L, Zhao C, Lu X (2020). The ZiBuPiYin recipe regulates proteomic alterations in brain mitochondria-associated ER membranes caused by chronic psychological stress exposure: Implications for cognitive decline in Zucker diabetic fatty rats. Sedentary Life and Nutrition 12: 23698–23726. https://doi.org/10.18632/aging.103894 [Google Scholar] [PubMed] [CrossRef]
Yang QS, Wu JH, Li CY, Wei YR, Sheng O et al. (2012). Quantitative proteomic analysis reveals that antioxidation mechanisms contribute to cold tolerance in plantain (Musa paradisiaca L.; ABB Group) seedlings. Molecular & Cellular Proteomics: MCP 11: 1853–1869. https://doi.org/10.1074/mcp.M112.022079 [Google Scholar] [PubMed] [CrossRef]
Ye S, Cai B, Zhou P, Wang G, Gao H, Hua R, You L, Yao Y, Wang Y, Shen G (2020). Huang-pu-tong-qiao formula ameliorates tau phosphorylation by inhibiting the CaM-CaMKIV pathway. Evidence-Based Complementary and Alternative Medicine 2020: 8956071. https://doi.org/10.1155/2020/8956071 [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools