 Open Access
Open Access
REVIEW
Recent progress in microbial cell surface display systems and their application on biosensors
College of Chemistry and Pharmaceutical Sciences, Qingdao Agricultural University, Qingdao, 266109, China
* Corresponding Author: LEI HAN. Email:
BIOCELL 2023, 47(6), 1213-1223. https://doi.org/10.32604/biocell.2023.028035
Received 28 November 2022; Accepted 10 February 2023; Issue published 19 May 2023
Abstract
Microbial cell surface display technology is a recombinant technology to express target proteins on the cell membrane, which can be used to redesign the cell surface with functional proteins and peptides. Bacterial and yeast surface display systems are the most common cell surface display systems of prokaryotic and eukaryotic proteins, that are widely applied as the core elements in the field of biosensors due to their advantages, including enhanced stability, high yield, good safety, expression of larger and more complex proteins. To further promote the performance of biosensors, the biomineralized microbial surface display technology was proposed. This review summarized the different microbial surface display systems and the biomineralized surface display systems, where the mechanisms of surface display and biomineralization were introduced. Then we described the recent progress of their applications on biosensors for different types of detection targets. Finally, the outlooks and tendencies were discussed and forecasted with the expectation to provide some general functions and enlightenments to this aspect of research.Keywords
Microbial cell surface display technology is a molecular display technique that employs the gene recombination method to fuse the gene sequences of target proteins and carrier proteins into the microbial host cell (Ding et al., 2019) so that the target proteins can be expressed and localized on the surface of microbial cells, and remain their independent spatial structure and biological activity (Pham and Polakovic, 2020). In the last decade, this ground-breaking technology has been broadly developed (Shibasaki and Ueda, 2014), and research in the field has shown a steady upward trend (Fig. 1a). So far, many proteins have been successfully expressed on cell surfaces of fungi, bacteria, mammals, and plants (Han et al., 2018b; Wang et al., 2021). Therefore, microbial cell display technology has become a helpful tool for displaying proteins on cell surfaces (Ding et al., 2019).
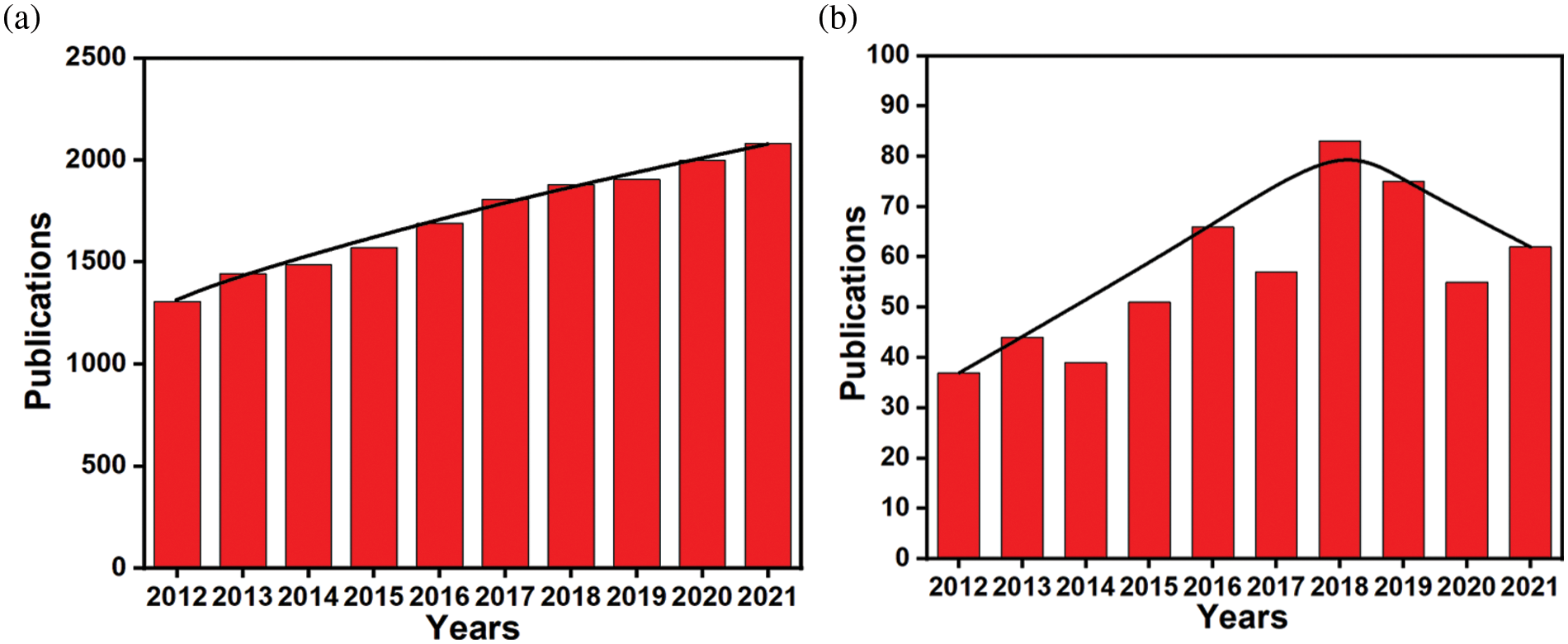
Figure 1: (a) Number of published papers on microbial cell surface display systems by the end of December 2021. Data are from the Web of Science. (b) Number of published papers on biosensors of microbial cell surface display systems by the end of December 2021. Data are from the Web of Science.
Microbial cell surface display technology can realize the functionalization of foreign proteins through a section of carrier protein with secretory transport function and membrane localization (Yang et al., 2019). So, the target foreign protein can be transported and anchored on the cell surface. Hence, the cell surface display system consists of three components: host (microbial cell), carrier (anchoring protein), and passenger (target foreign protein) (Han et al., 2018b). (1) The host cell act as a substrate for fusion proteins and anchoring proteins. (2) The carrier proteins, also called anchoring proteins, are capable of secreting, transporting, and anchoring foreign proteins to the extracellular surface. (3) The passenger protein, also called the target foreign protein, has a specific function. Consequently, it is obligatory to closely coordinate the three mentioned above-mentioned components for the establishment of a proper surface display system. The fusion modes of the target protein sequence and carrier protein sequence mainly include C-terminal fusion and N-terminal fusion (Han et al., 2018b). As the main microbial cell surface display systems, bacterial and yeast surface display systems have been widely used for the display of prokaryotic and eukaryotic proteins, respectively, which have the characteristics of vector diversity, high yield of recombinant proteins, and flexible genetic engineering (Huang et al., 2018).
Up to now, microbial cell surface display technology has been successfully used in many fields, including the construction and high-throughput screening of peptide library, whole-cell adsorbents for heavy metal pollution, development of the live vaccine, recombinant whole-cell catalysis, biofuel cells, biosensors, etc. (Chen et al., 2017; Ueda, 2016). Among them, there have been many advances in cell surface display systems-based biosensors (Fig. 1b). In particular, bacteria and yeast surface display systems have been widely applied in a variety of biosensors (Jahns and Rehm, 2012; Pham and Polakovic, 2020), involving various fields including medical diagnosis, environmental monitoring, food assay, biochemical analysis and so on (Chmielewski et al., 2019; van Bloois et al., 2011). So far, a large number of articles have been published on these aspects. This review mainly introduces bacterial and yeast surface display systems, and their biomineralized systems, the detection methods based on these systems.
Microbial Surface Display System
Bacterial surface display system
The bacterial surface display system takes advantage of the surface display technology to express target proteins on the surface of bacteria for precise functionalization (Chen et al., 2019). This system has been generally used in recombinant bacterial vaccines, biofuel cells, whole-cell catalysts, and bioremediation (Xiang et al., 2019). According to the source of carrier proteins, bacterial surface display systems can be categorized as Gram-negative bacterial surface display systems and Gram-positive bacterial surface display systems (Liu et al., 2020). At present, the carrier proteins are commonly used in the surface display system of Gram-negative bacteria, such as outer membrane proteins (OMPs), lipoproteins, Lpp-OmpT (LOT), surface accessory structure subunits, ice nuclear proteins (INP), and autologous transporters and so on (Hui et al., 2019; Song et al., 2015; Wang et al., 2021; Zhang et al., 2018b). On the other hand, the development of surface display systems for Gram-positive bacteria is not as mature as that for Gram-negative bacteria (Kim et al., 2021).
As for the biosensors, the bacterial surface display system can display the required enzymes on the surface of bacterial cells, making them react on the surface, thus enhancing the sensitivity, stability, and selectivity of biosensors (Han, 2020). For example, the surface display system based on Escherichia coli was applied to construct the acetaldehyde optical biosensor (Liang et al., 2021). The technique mainly fixed the expressed fusion protein (acetaldehyde dehydrogenase, AldDH) onto the outer membrane of E. coli by LOT carrier. AldDH displayed on the surface can catalyze the production of NADH, which can be detected by the spectrometric method. Many relevant works demonstrate that the bacterial surface display system has many advantages (Fig. 2), including (1) bacterial surface is better suited for displaying large-sized proteins, (2) this system is highly productive. (3) This system can proliferate by binary fission because bacterial systems are prokaryotic cells (Park, 2020; van Bloois et al., 2011).
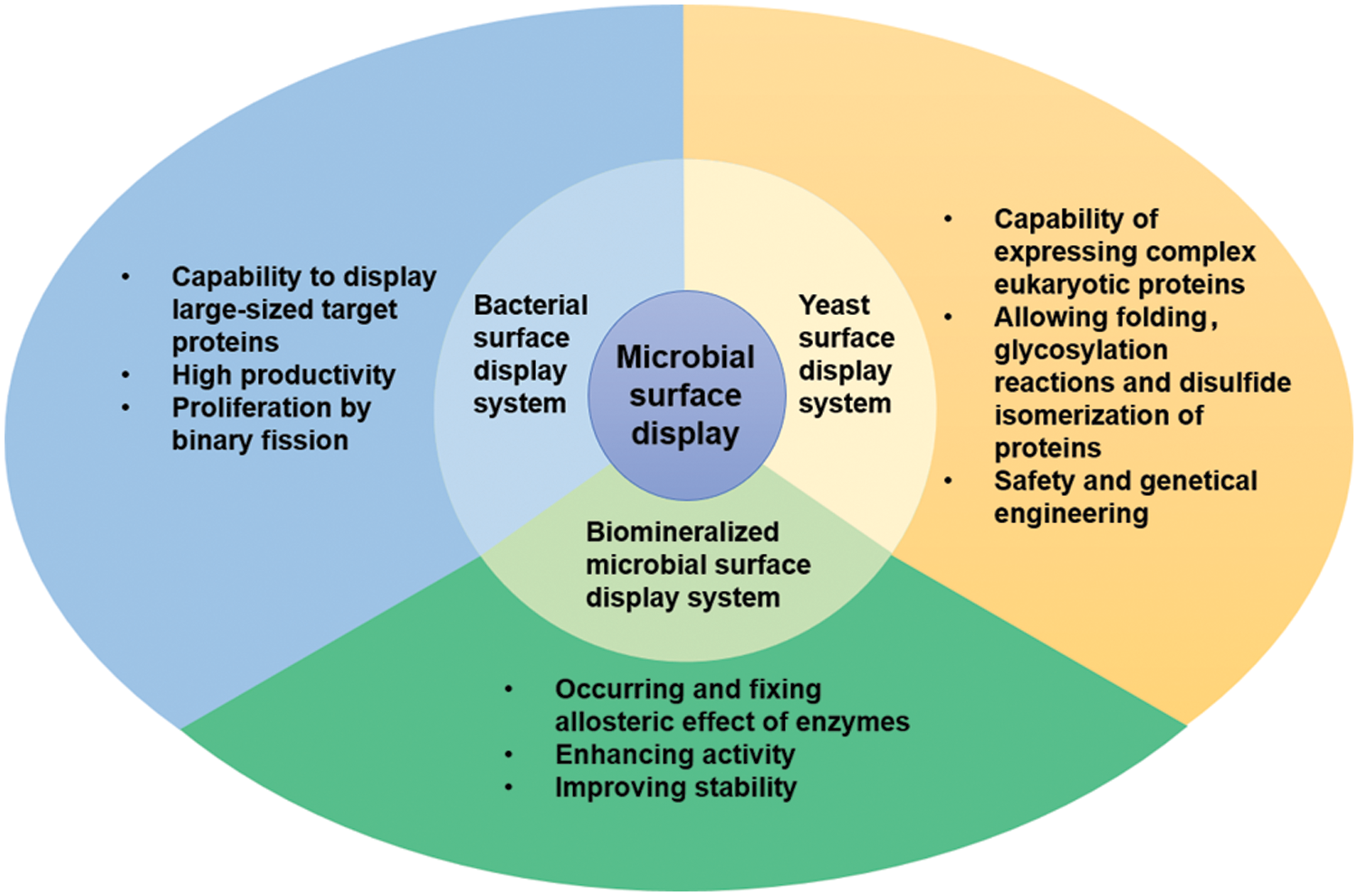
Figure 2: Three kinds of microbial cell surface display systems and their advantages.
The yeast surface display system is an important eukaryotic protein surface display system (Teymennet-Ramirez et al., 2021). The basic principle involves induction and expression of the exogenous target protein gene sequence fused with a carrier gene sequence in yeast cells (Chun et al., 2020), then the fusion protein, guided by signal peptide, is secreted out of the cell. The fused protein can be anchored in the yeast cell wall, thereby immobilizing the foreign protein on the surface of the yeast cell (Fan et al., 2020). The commonly used carriers in yeast cell surface display systems mainly include a-agglutinin, α-agglutinin, Flo1p, etc. (Han et al., 2018b). In recent years, the yeast surface display system has attracted great attention for displaying eukaryotic proteins on the surface of cells (Gal et al., 2016). The yeast surface display system has many advantages, such as superior safety, post-translational modifications, proper folding and glycosylation of proteins, disulfide isomerization of eukaryotic proteins, simplicity of the cell culture and genetic manipulation, and immobilization of protein (Hamilton and Gerngross, 2007; Han et al., 2018b; Park, 2020; Teymennet-Ramirez et al., 2021) (Fig. 2). As the most extensively used yeast on surface display systems, Saccharomyces cerevisiae, a unicellular eukaryote with a cell wall, has been widely applied for construction of the biosensor (Chun et al., 2020; Gal et al., 2016; Liang and Han, 2020a; Zhao et al., 2020).
Biomineralized microbial surface display system
Biomineralization refers to the formation process of inorganic minerals on organisms (Sang et al., 2020). The biggest difference between biomineralization and general mineralization lies in the involvement of biomacromolecules, cells, and other organic substrates (Wei et al., 2019). The characteristic of biological mineralization is the transformation of ions in solution into solid minerals under the control or influence of biological materials at certain physicochemical conditions (Mangano et al., 2019). So, biomineralization is a complex and dynamic process regulated by organic matter, crystal growth mechanism, and external environment. With the increasing attention to biological mineralization, various biological substances (such as the phage, violet membrane, bovine serum albumin, and lysozyme) are used as templates (Liang and Han, 2020b; Zhao et al., 2022) for the synthesis of bio-inorganic hybrid materials (Han and Liu, 2017). Inspired by the bio-templated mineralization described above, researchers have begun to introduce biomineralization into phage surface display systems (Han et al., 2016, 2017). Whereafter, biomineralization was introduced into microbial cell surface display systems, which generated the biomineralized microbial surface display technology.
Biomineralized microbial cell surface display technology is a combination of biomineralization and microbial surface display technology, where the target proteins on the cell surface provide nucleation sites of inorganic crystals to form a bio-inorganic hybrid system. So far, there have been several types of research on the biomineralized microbial surface display system (Bian et al., 2022; Han et al., 2018a; Han and Liu, 2017). The biomineralization can increase the catalytic activity of enzymes on the surface of cells due to allosteric effects (Fig. 2), a phenomenon in which an allosteric effector binds to a site (allosteric site) on an enzyme molecule, causing conformational change, thereby indirectly managing the property of another particular site (active site) on the same enzyme molecule. The above positive performances are reflected in some studies. For example, after Co3(PO4)2·8H2O biomineralization on the cell surface, the displayed enzymes were transformed from the inactive state to the active state by allosteric effects, and the active state was “fixed” (Han and Liu, 2017) (Fig. 3a). The activity of the biomineralized system is increased by about three times, compared with the initial cells (Fig. 3b). In addition, the mineralized cells were also more stable than the original cells (Han and Liu, 2017). In contrast to the biomineralized microbial surface display system, conventional immobilization generally decreases the catalytic activity of enzymes (Sharifi et al., 2018). Therefore, biomineralized microbial surface display systems would become ideal substitutes for conventional immobilized enzymes and whole-cell catalysts.
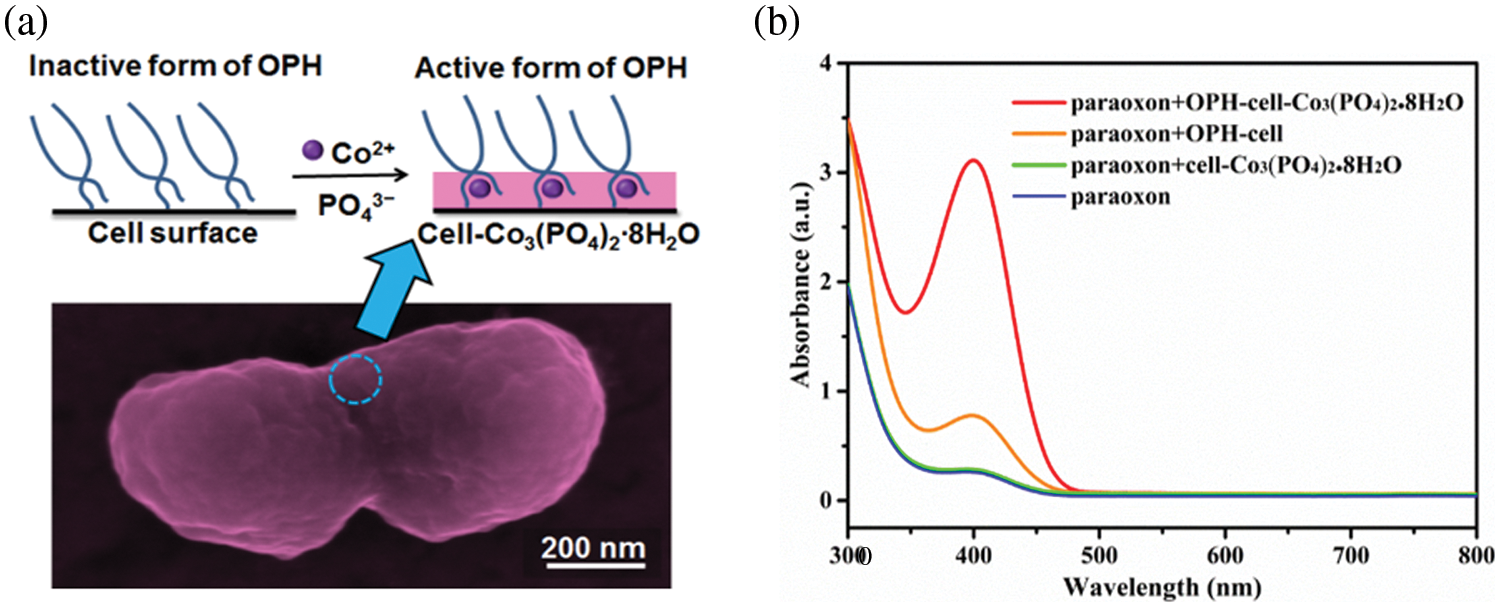
Figure 3: An example of biomineralized microbial surface display systems. (a) Biomineralized organophosphorus hydrolase (OPH)-fused cells exhibit enhanced catalytic activity due to the allosteric effects from “inactive” form to “active” form, where OPH is embedded in inorganic crystal (Co3(PO4)2∙8H2O) (Han and Liu, 2017). (b) The obtained bio-inorganic hybrid whole-cell catalyst shows three times higher activity than the original whole-cell catalyst. Reprinted (adapted) with permission from (Han and Liu, 2017). Copyright (2023) American Chemical Society.
Application of Microbial Cell Surface Display Systems on Biosensors
The biosensor is an assay device that detects various analytes (such as biomacromolecules and small organic molecules) by the biological recognition elements (such as proteins, DNA, and cells) (Park, 2019; Pyun et al., 2005). A biosensor generally consists of three main parts (Fig. 4): receiver, transducer, and other auxiliary equipment (Chen et al., 2023). Among them, the core of the receiver is a molecular recognition element, specifically surface-displayed enzymes. The transducer can capture electrical or optical signals from the enzymatic product by an electrode or optical probe (Hou et al., 2015).
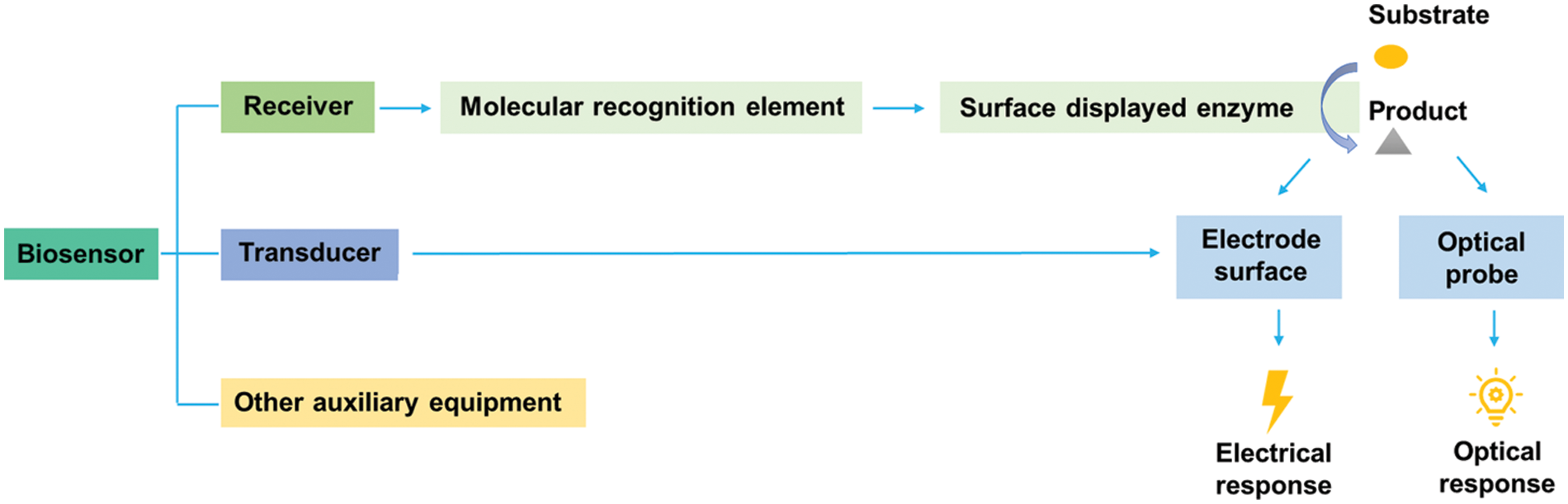
Figure 4: Constituents of biosensors based on microbial surface displayed enzymes.
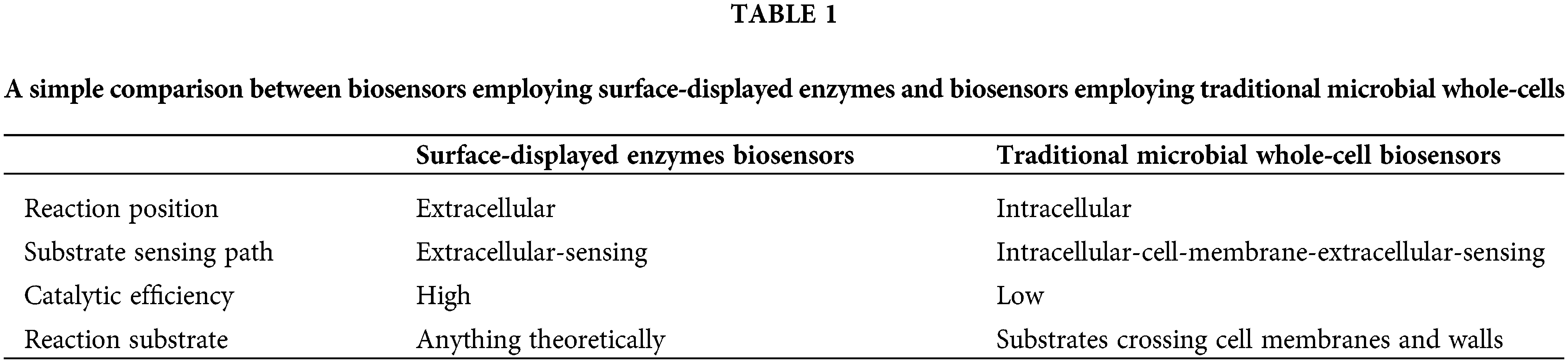
Microbial cell surface display technology is a powerful tool to express and produce proteins on the cell surface and has been widely applied to biosensors. As shown in Table 1, biosensors employing surface-displayed enzymes are superior to those employing traditional microbial whole-cells. For traditional whole-cells, the substrate needs to enter the cell in order to achieve enzyme-based catalytic reactions. By contrast, the enzymes displayed on the cell surface can directly come in contact with the reaction substrate, which greatly improves the catalytic efficiency (Han and Liu, 2017). For the biosensors employing surface-displayed enzymes, the reaction substrate (target) can be anything theoretically. For the traditional microbial whole-cells, the substrate must be able to cross the cell membrane and wall (Ye et al., 2021). Accordingly, by applying microbial cell surface display technology, biosensors can detect various target substances. So far, reported biosensors employing surface-displayed enzymes mainly involve four categories of target substances:
organophosphorus pesticides, phenolic compounds, glucose, and L-glutamate. According to the targets, different types of enzymes are used in surface display systems, such as thelaccase (Li et al., 2021; Park et al., 2019; Wu et al., 2020; Zhang et al., 2018a), organophosphorus hydrolase (Liang et al., 2019), xylanase (Chen et al., 2012), exoglycanase (Chen et al., 2012), and carbonic anhydrase (Fan et al., 2011).
Detection of organophosphorus pesticides
Organophosphorus pesticides (OPP) are phosphorus-based organic compound pesticides that have been widely used in agricultural production (Hassani et al., 2017). In addition, as a strong neurotoxic cholinesterase inhibitor (Tang et al., 2014a), OPP can inhibit hydrolysis of acetylcholine (central nervous system neurotransmitter), causing health problems for animals and humans (Xu et al., 2018). Therefore, it is necessary and urgent to search and employ the detection methods of OPP residue. To overcome the above-mentioned problem, many surface display systems and biomineralized systems are actively studied and applied in biosensors for the detection of OPP (Bian et al., 2022; Han et al., 2018a; Liu et al., 2013; Tang et al., 2014a). In this section, we discuss and provide some recent examples of the OPP biosensor based on display systems.
Direct detection method based on organophosphorus hydrolase
Given that organophosphorus hydrolase (OPH) can hydrolyze a wide range of organophosphorus ester bonds (such as P—S, P—O, and P—F bonds) (Pundir et al., 2019; Tang et al., 2014a), it has been developed as a key element in OPP degradation and biosensing. For example, Tang et al. (2014a) successfully constructed a mutated OPH displayed on the surface of an E. coli strain, using INP as the anchoring motif. Fortunately, the recombinant strain was more robust and stable than the purified OPH, promising for detecting OPP sensitively. Further, they immobilized the fused-OPH on the surface of the ordered mesopore carbons-modified glass carbon electrode to develop a novel microbial biosensor for detecting p-nitrophenyl OPP (Tang et al., 2014b). In addition, the relationship between time and current was observed at different OPP concentrations, then the experimental conditions were optimized. Therefore, a fast and accurate biosensor for detecting OPP was established by applying the microbial cell surface display.
Our team synthesized a suitable composite material, combined with biomineralization and microbial cell surface display, and then constructed an ultra-sensitive electrochemical biosensor to detect OPP residues (Han et al., 2018a). The carbon nanotube@amino acid ionic liquid (CNT@AAIL) composites were introduced into the electrochemical biosensor based on mineralized OPH-fused cells. Considering three reasons for this design: (1) OPH could specifically hydrolyze organophosphorus compounds, (2) to make up for the poor solubility of CNT during electrode modification, AAIL with good fluidity, stability, biocompatibility, and biodegradation was combined for the first time, (3) electrochemical biosensor had the advantage of high sensitivity, fast response speed, and positive real-time detection. Particularly, we used AAIL as a stabilizer and modifier to greatly improve the dispersion and biocompatibility of CNT in the aqueous phase. The obtained CNT@AAIL composite improved the electrical conductivity and electrochemical activity of mineralized OPH-fused cell (M-Cell). Surprisingly, the as-fabricated biosensor was more accurate and had 2–8 orders of magnitude lower detection limit than the OPP analytic method reported. Thus, the anti-interference ability of the OPP biosensor has also been proven to a certain extent.
In previous work, the mineralized OPH-fused cells were prepared by embedding OPH into cobalt phosphate. The synthetic bioinorganic hybrid material was applied to the sensitive paraoxon biosensor. In the sensor, allosteric effects, biomineralization techniques, and cell surface display techniques were combined to enhance OPH activity (Han and Liu, 2017). Therefore, the biomineralized microbial surface display system was promising in the future, especially for the establishment of ultra-sensitive and highly selective biosensors (Han et al., 2018b). After continuous in-depth research, we constructed a portable detecting OPP device (Bian et al., 2022). We took full advantage of structures, enzymes, and mineralized layers of displayed cells as well. To detect pesticides quickly and conveniently, we successfully prepared phosphate-mineralized organophosphorus hydrolase-fused cells (M-OPH) through the combination of biomineralization and microbial surface display techniques. In brief, the portable detection device was constructed by simply precipitating M-OPH on a syringe filter. Small colored impurities of real samples could be filtered out by M-OPH layer-modified filter membrane. In this device, the catalytic activity of OPH was enhanced by the allosteric effect caused by biomineralization; the stability of OPH was also enhanced by the protective effect of inorganic phosphate. With the tandem catalysis of the copper phosphate and OPH, hydrolysates of OPP were further reduced or oxidized to low-toxicity products.
Enzyme inhibition method based on acetylcholinesterase
Acetylcholinesterase (AChE) has been successfully used in OPP detection because organophosphorus is an effective inhibitor of AChE (Qi et al., 2020). For example, Liang’s team developed a fluorescence OPP detection method by combining AChE mutants displayed on the yeast surface and protein-directed electronegative fluorescent gold nanoclusters (Au NCs), which improved the sensitivity of AChE to OPP (Liang and Han, 2020a). Concretely, AChE mutants and wild-type from Drosophila were wonderfully displayed on the surface of S. cerevisiae cells, employing a-agglutinin-mediated cell surface display technology. In addition, the displayed AChE could catalyze the hydrolysis of acetylthiocholine, to produce thiocholine, which could not only bind to Au NCs through Au-S bonds but also absorb Au NCs, resulting in Au NCs aggregation and fluorescence quenching. More importantly, fluorescence detection based on the yeast surface displayed AChE and Au NCs was highly sensitive to minute amounts of OPP, and the detection limit was 2–6 orders of magnitude lower than previously reported methods. Therefore, combined with the enzyme-modified microbial cell surface display system and functional biological nanomaterials, the method had good reliability for the measurement of real samples.
Detection of phenolic compounds based on surface-displayed laccases
Phenolic compounds have a broad range of applications, including production areas, such as energy, food additives, and fine chemicals (Alcazar-Ruiz et al., 2023). Therefore, some phenolic compounds are familiar to us, such as phenol, cresol, thymol, eugenol, aminophenol, nitrophenol, naphthol, carvacrol, chlorophenol, etc. (da Silveira et al., 2015). However, phenolics have different toxicities. Due to their existence in air, water, and food matrices, they pose a significant risk of toxicity to the environment and humans (Govindhan et al., 2015). Therefore, the detection of phenolic compounds is crucial to open up a path for ecological and environmental protection.
Laccases are blue multicopper oxidases, which can oxidize multitudinous aromatic compounds, including the oxidation of phenolic compounds (Akram et al., 2022). They are broadly distributed in bacteria, fungi, plants, and insects (Agarwal et al., 2022). In continuous laccase-based experiments, the immobilized laccase has been inevitably employed (Saoudi and Ghaouar, 2019). Compared to the free laccase, the immobilized laccase has more prominent advantages: (1) increased thermal stability of enzymes, (2) resistance to chemical reagents and extreme conditions, and (3) easy separation from reactants and ability to perform continuous bioreactor operation (Fernandez-Fernandez et al., 2013). Therefore, microbial cell surface display techniques are often used for preparing immobilized laccases. Accordingly, more attention has been paid to biosensors based on laccases displayed in recent years (Ricklefs et al., 2014). For example, Zhang et al. (2018b) developed an electrochemical microbial biosensor. Mechanistically, the sensor could immobilize the bacterial laccase on the E. coli cells and then adsorb modified living cells on the glassy-carbon electrode by combining microbial cell surface display and modified electrodes. In previous work, the live bacterial laccase was directly adsorbed onto the electrode surface and demonstrated the feasibility. However, the live cell activity was not maintained (Zhang et al., 2017). Therefore, Zhang et al. (2017) later designed a laccase-immobilized biosensor by the surface display, and it had been shown to maintain the adsorbed cells’ activity for weeks or even months. Moreover, the electrochemical response of detecting catechol employing the electrochemical microbial biosensor kept a linear relationship within the concentration scope of 5.0 to 300.0 μM under optimum pH. In addition, the proposed biosensor presented certain anti-interference. In another work (Acquaviva et al., 2018), when employed to detect phenolics in real samples, the biosensor showed high accuracy, almost comparable to the results obtained by high-performance liquid chromatography; besides, high reproducibility and stability were also reflected.
Glucose plays an essential role in cell homeostasis and metabolism (Choi and Kim, 2022). It is the energy source and the main energy-supplying substance of living organisms (Zheng et al., 2016). More importantly, glucose levels are closely linked to blood glucose levels (French et al., 2022). Glucose testing is crucial for people as high blood sugar levels can cause a variety of complications, such as diabetes.
About the above problem, Liang’s team gave us appropriate answers using genetically engineered techniques; Liang et al. (2013b) first constructed an E. coli strain displaying glucose dehydrogenase (GDH) from Pseudomonas borealis on its surface with INP as an anchoring motif. In addition, by combining the constructed nanocomposite electrode, a novel glucose electrochemical biosensor was developed. The low detection limit of the prepared biosensor was 4 μM D-glucose. Later, they discovered that by using GDH from Bacillus subtilis for surface display, they could establish GDH mutants with better substrate specificity, stability, and activity (Liang et al., 2013a). In the same year, applying the same combination of the surface display and carbon nanotube-modified electrode, another biosensor for glucose detection was developed (Wang et al., 2013). The difference was that glucose oxidase (GOx) was displayed on the yeast surface with a-agglutinin as the anchoring motif, and the electrode was also changed. The low detection limit was 0.05 mM of D-glucose. This was the first report on yeast surface displaying GOx for glucose detection. More importantly, the GOx-displaying yeast system had high specificity to glucose and good stability over a broad pH range (3.5–11.5), as well as at higher temperatures (56°C). Almost four years later, they made more progress in biosensors for glucose detection. Liu et al. (2017) prepared bi-enzyme-based biosensors by collectively immobilizing GDH-displaying-bacteria and glucoamylase-displaying bacteria. Further, they constructed an electrochemical biosensor sensitive to maltose and glucose and insensitive to other monosaccharides and disaccharides. Interestingly, the sensitivity to detect glucose was 3.75 times higher than maltose at the same concentration. Compared with the biosensor based on free enzymes, the dual-strain modified electrodes showed better performance. The proposed biosensor had a broad dynamic range (0.2–10 mM) and a low detection limit (0.1 mM maltose). Biosensors designed in triplicate had different specific mechanisms but employed the same microbial cell surface display technology, from which the widespread use and great importance of surface display were proved. Later, Zhao et al. (2020) displayed microbial GDH and cholesterol oxidase on the surface of yeast cells and then prepared two biosensors for blood biochemical indicators detection. The yeast cells were immobilized on electrodes to construct electrochemical biosensors for glucose and cholesterol detection. The glucose biosensor efficiently responded at a wide concentration range of 20–600 mg·dL−1.
L-glutamate is a non-essential amino acid that occurs naturally in protein-rich food (Liu et al., 2021). As a functional amino acid, it plays a role in cell metabolism and signaling (Lin et al., 2014). L-glutamate is also a significant excitatory neurotransmitter in the human body. However, excitotoxic processes mediated by glutamate are a major cause of neuropathology (such as stroke) (Hazell, 2007; Sheldon and Robinson, 2007). L-glutamate is also widely served as food flavor enhancers, such as soy sauce, chicken essence, monosodium glutamate (MSG), and some snack flavorings (Kurihara, 2009). Nevertheless, MSG has been associated with negative side effects, especially in animals, including diabetes, obesity, neurotoxic, and hepatotoxic (Kazmi et al., 2017). Therefore, neurodegenerative diseases caused by L-glutamate and the consumption of L-glutamate should be taken seriously. So, an effective and convenient technique for L-glutamate detection is urgently required.
Glutamate oxidase (GluOx) and glutamate dehydrogenase (Gldh) are two enzymes commonly used to detect L-glutamate (Hughes et al., 2016; Mruga et al., 2021). We have learned that Ozel et al. (2014) and Wang et al. (2020) prepared biosensors for detecting L-glutamate based on the fixation of GluOx and nanocomposite materials. However, immobilized enzymes have certain advantages over free enzymes (such as higher activity, greater stability, ability to operate continuously, and more suitable for industrialization) (Fernandez-Fernandez et al., 2013; Ranimol and Sunkar, 2022), but compared with surface display enzymes, immobilization is only a part of microbial cell surface display. Microbial cell surface display techniques can redesign cell surfaces using functional peptides or proteins immobilized on the cell surface to endow cells with some special features (Han et al., 2018b). Therefore, we mainly introduce the employment of surface display technology to detect L-glutamate. In this regard, Liu’s team conducted relevant experiments. First, they reported that Gldh is displayed on the surface of E. coli with the N-terminal region of INP as an anchoring motif (Song et al., 2015). This was the first report on the optical detection of L-glutamate displayed on the bacterial surface. Additionally, the optimal pH value and temperature for the Gldh-displayed-bacterial cell surface were respectively 9.0°C and 70°C. The fused protein maintained almost 100% initial enzyme activity after incubation for 1 month at 4°C. Clearly, the Gldh-fused cell showed high cell activity. Then, based on previous experiments, multiwalled carbon nanotubes, and Gldh-displaying bacteria were modified onto glassy carbon electrodes to prepare L-glutamate biosensors (Liang et al., 2015). The biosensor had a low detection limit (2 μM) and two linear sections of 10 μM–1 mM and 2–10 mM. Thus, the above-discussed L-glutamate biosensor had good selectivity, stability, and anti-interference to accurately detect real samples.
The bacterial surface display system, yeast surface display system, and biomineralized microbial surface display system are reviewed here. The bacterial surface display technology makes full use of the principle of enzyme engineering and opens promising potential. So far, many varieties of anchoring motifs can be used to mediate the bacterial surface display of heterologous proteins. The yeast cell surface display technology has the advantages of display of complex eukaryotic proteins with post-translational modification or larger protein molecules. Both display systems have many merits. For example, cell surface-displayed enzymes can be facilely purified and reused, which is more convenient and economical. The display of enzymes on the surface of the cell can directly react with related substrates, which can dramatically raise catalytic efficiency and expand the types of targets. The cell surface supplies a biologically compatible microenvironment that maintains the stability of enzymes. However, some mechanisms of both systems are still unknown. For example, not much is known about the relationship between the quantity and activity of the enzyme displayed on the cell surface. How are enzymes displayed on the cell surface distributed to the progeny microorganism after the cell division? These issues still need to be explored.
Compared with the above two systems, the seldom-researched biomineralized microbial surface display system is more advanced and efficient and will be the research hotspot with great development potential in the future. By fusing cell surface display with multifunctional inorganic nanomaterials, the biosensor will have higher sensitivity and stability, which will trigger various assay strategies. Theoretically, any enzymes can be displayed after the optimization of the codon. In general, prokaryotic enzymes are displayed on the surface of prokaryotic bacteria (such as E. coli), and eukaryotic enzymes are displayed on the surface of eukaryotic microorganisms (such as S. cerevisiae). So, the enzyme to be displayed depends on the target analyte. The enzyme can be selected according to the target to catalyze the reaction of the target and produce the signal. For example, OPH and AChE were used for surface display to prepare biosensors for organophosphorus detection. Laccases were used to prepare biosensors based on surface display technologies for detecting phenols. Biosensors using GDH, Gox, and glucoamylase displayed on the microbial cell surface were developed to detect glucose. Gldh was used for surface display to prepare biosensors for L-glutamate detection. Therefore, more enzymes are expected to be displayed for more assay targets.
Although the displayed cells are more stable and have better catalytic efficiency than the untreated ones, the present assay methods based on cell surface display systems are still limited to several kinds of assay methods, including electrochemical biosensing, fluorescence, and spectrophotometry. More analytical methods should be combined with cell-surface display techniques. The biomineralized microbial surface display systems will have enormous potential in self-powered sensors, flexible sensors, single-molecule sensors, and in vivo non-invasive microsensors. To some extent, this review provides some information and insights for the research in the field of biosensors based on the microbial surface display system.
Funding Statement: This work was financially supported by the National Natural Science Foundation of China (Grant No. 21705087), Youth Innovation Team Project for Talent Introduction and Cultivation in Universities of Shandong Province (096-1622002), Research Foundation for Distinguished Scholars of Qingdao Agricultural University (663-1117015), the Postgraduate Innovation Program of Qingdao Agricultural University (QNYCX21069), and the National Innovation Training Program for College Students (No. 202210435030).
Author Contributions: The authors confirm their contribution to the paper as follows: study conception and design: H. Chen and L. Han; draft manuscript preparation: H. Chen and L. Han; preparation of figures: Y. Wu and H. Chen; manuscript checking and approval: B. Huang and L. Han. Both authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
Acquaviva A, Jones A, Dennis GR, Shalliker RA, Soliven A (2018). Phenolic profiling of complex tea samples via simultaneous multiplexed detection employing reaction flow HPLC columns and colorimetric post column derivatisation. Microchemical Journal 138: 533–539. https://doi.org/10.1016/j.microc.2018.01.044 [Google Scholar] [CrossRef]
Agarwal N, Solanki VS, Gacem A, Hasan MA, Pare B et al. (2022). Bacterial laccases as biocatalysts for the remediation of environmental toxic pollutants: A green and eco-friendly approach—A review. Water 14: 4068. https://doi.org/10.3390/w14244068 [Google Scholar] [CrossRef]
Akram F, Ashraf S, Haq IU, Shah FI, Aqeel A (2022). Eminent industrial and biotechnological applications of laccases from bacterial source: A current overview. Applied Biochemistry and Biotechnology 194: 2336–2356. https://doi.org/10.1007/s12010-021-03781-9 [Google Scholar] [PubMed] [CrossRef]
Alcazar-Ruiz A, Dorado F, Sanchez-Silva L (2023). Bio-phenolic compounds production through fast pyrolysis: Demineralizing olive pomace pretreatments. Food and Bioproducts Processing 137: 200–213. https://doi.org/10.1016/j.fbp.2022.12.002 [Google Scholar] [CrossRef]
Bian X, Chen D, Han L (2022). Taking full advantage of the structure and multi-activities of mineralized microbial surface-displayed enzymes: Portable three-in-one organophosphate pesticides assay device. Chemical Engineering Journal 429: 132317. https://doi.org/10.1016/j.cej.2021.132317 [Google Scholar] [CrossRef]
Chen YP, Hwang IE, Lin CJ, Wang HJ, Tseng CP (2012). Enhancing the stability of xylanase from Cellulomonas fimi by cell-surface display on Escherichia coli. Journal of Applied Microbiology 112: 455–463. https://doi.org/10.1111/j.1365-2672.2012.05232.x [Google Scholar] [PubMed] [CrossRef]
Chen H, Ullah J, Jia J (2017). Progress in Bacillus subtilis spore surface display technology towards environment, vaccine development, and biocatalysis. Journal of Molecular Microbiology and Biotechnology 27: 159–167. https://doi.org/10.1159/000475177 [Google Scholar] [PubMed] [CrossRef]
Chen T, Wang K, Chi X, Zhou L, Li J et al. (2019). Construction of a bacterial surface display system based on outer membrane protein F. Microbial Cell Factories 18: 70. https://doi.org/10.1186/s12934-019-1120-2 [Google Scholar] [PubMed] [CrossRef]
Chen X, Yao C, Li Z (2023). Microarray-based chemical sensors and biosensors: Fundamentals and food safety applications. TRAC Trends in Analytical Chemistry 158: 116785. https://doi.org/10.1016/j.trac.2022.116785 [Google Scholar] [CrossRef]
Chmielewski M, Kuehle J, Chrobok D, Riet N, Hallek M, Abken H (2019). FimH-based display of functional eukaryotic proteins on bacteria surfaces. Scientific Reports 9: 8410. https://doi.org/10.1038/s41598-019-44883-z [Google Scholar] [PubMed] [CrossRef]
Choi JH, Kim MS (2022). Homeostatic regulation of glucose metabolism by the central nervous system. Endocrinology and Metabolism 37: 9–25. https://doi.org/10.3803/EnM.2021.1364 [Google Scholar] [PubMed] [CrossRef]
Chun J, Bai J, Ryu S (2020). Yeast surface display system for facilitated production and application of phage endolysin. ACS Synthetic Biology 9: 508–516. https://doi.org/10.1021/acssynbio.9b00360 [Google Scholar] [PubMed] [CrossRef]
da Silveira GD, Motta MJ, Müller LS, Lameira O, Athayde ML, Piana M, da Rosa MB, Viana C, de Carvalho LM (2015). Determination of phenolic antioxidants in amazonian medicinal plants by HPLC with pulsed amperometric detection. Journal of Liquid Chromatography & Related Technologies 38: 1259–1266. https://doi.org/10.1080/10826076.2015.1037450 [Google Scholar] [CrossRef]
Ding Z, Guan F, Yu X, Li Q, Wang Q, Tian J, Wu N (2019). Identification of the anchoring protein SpoIIIJ for construction of the microbial cell surface display system in Bacillus spp. International Journal of Biological Macromolecules 133: 614–623. https://doi.org/10.1016/j.ijbiomac.2019.04.033 [Google Scholar] [PubMed] [CrossRef]
Fan S, Liang B, Xiao X, Bai L, Tang X, Lojou E, Cosnier S, Liu A (2020). Controllable display of sequential enzymes on yeast surface with enhanced biocatalytic activity toward efficient enzymatic biofuel cells. Journal of the American Chemical Society 142: 3222–3230. https://doi.org/10.1021/jacs.9b13289 [Google Scholar] [PubMed] [CrossRef]
Fan LH, Liu N, Yu MR, Yang ST, Chen HL (2011). Cell surface display of carbonic anhydrase on Escherichia coli using ice nucleation protein for CO2 sequestration. Biotechnology and Bioengineering 108: 2853–2864. https://doi.org/10.1002/bit.23251 [Google Scholar] [PubMed] [CrossRef]
Fernandez-Fernandez M, Sanroman MA, Moldes D (2013). Recent developments and applications of immobilized laccase. Biotechnology Advances 31: 1808–1825. https://doi.org/10.1016/j.biotechadv.2012.02.013 [Google Scholar] [PubMed] [CrossRef]
French SS, Hudson SB, Webb AC, Knapp CR, Virgin EE, Smith GD, Lewis EL, Iverson JB, DeNardo DF (2022). Glucose tolerance of iguanas is affected by high-sugar diets in the lab and supplemental feeding by ecotourists in the wild. Journal of Experimental Biology 225: jeb243932. https://doi.org/10.1242/jeb.243932 [Google Scholar] [PubMed] [CrossRef]
Gal I, Schlesinger O, Amir L, Alfonta L (2016). Yeast surface display of dehydrogenases in microbial fuel-cells. Bioelectrochemistry 112: 53–60. https://doi.org/10.1016/j.bioelechem.2016.07.006 [Google Scholar] [PubMed] [CrossRef]
Govindhan M, Lafleur T, Adhikari BR, Chen A (2015). Electrochemical sensor based on carbon nanotubes for the simultaneous detection of phenolic pollutants. Electroanalysis 27: 902–909. https://doi.org/10.1002/elan.201400608 [Google Scholar] [CrossRef]
Hamilton SR, Gerngross TU (2007). Glycosylation engineering in yeast: The advent of fully humanized yeast. Current Opinion in Biotechnology 18: 387–392. https://doi.org/10.1016/j.copbio.2007.09.001 [Google Scholar] [PubMed] [CrossRef]
Han MJ (2020). Novel bacterial surface display system based on the Escherichia coli protein MipA. Journal of Microbiology and Biotechnology 30: 1097–1103. https://doi.org/10.4014/jmb.2001.01053 [Google Scholar] [PubMed] [CrossRef]
Han L, Chen D, Li F (2018a). Rational integration of biomineralization, microbial surface display, and carbon nanocomposites: Ultrasensitive and selective biosensor for traces of pesticides. Advanced Materials Interfaces 5: 1801332. https://doi.org/10.1002/admi.201801332 [Google Scholar] [CrossRef]
Han L, Liu A (2017). Novel cell-inorganic hybrid catalytic interfaces with enhanced enzymatic activity and stability for sensitive biosensing of paraoxon. ACS Applied Materials & Interfaces 9: 6894–6901. https://doi.org/10.1021/acsami.6b15992 [Google Scholar] [PubMed] [CrossRef]
Han L, Shao C, Liang B, Liu A (2016). Genetically engineered phage-templated MnO2 nanowires: Synthesis and their application in electrochemical glucose biosensor operated at neutral pH condition. ACS Applied Materials & Interfaces 8: 13768–13776. https://doi.org/10.1021/acsami.6b03266 [Google Scholar] [PubMed] [CrossRef]
Han L, Shi J, Liu A (2017). Novel biotemplated MnO2 1D nanozyme with controllable peroxidase-like activity and unique catalytic mechanism and its application for glucose sensing. Sensors and Actuators B: Chemical 252: 919–926. https://doi.org/10.1016/j.snb.2017.06.096 [Google Scholar] [CrossRef]
Han L, Zhao Y, Cui S, Liang B (2018b). Redesigning of microbial cell surface and its application to whole-cell biocatalysis and biosensors. Applied Biochemistry and Biotechnology 185: 396–418. https://doi.org/10.1007/s12010-017-2662-6 [Google Scholar] [PubMed] [CrossRef]
Hassani S, Momtaz S, Vakhshiteh F, Maghsoudi AS, Ganjali MR, Norouzi P, Abdollahi M (2017). Biosensors and their applications in detection of organophosphorus pesticides in the environment. Archives of Toxicology 91: 109–130. https://doi.org/10.1007/s00204-016-1875-8 [Google Scholar] [PubMed] [CrossRef]
Hazell AS (2007). Excitotoxic mechanisms in stroke: An update of concepts and treatment strategies. Neurochemistry International 50: 941–953. https://doi.org/10.1016/j.neuint.2007.04.026 [Google Scholar] [PubMed] [CrossRef]
Hou C, Fan S, Lang Q, Liu A (2015). Biofuel cell based self-powered sensing platform for L-cysteine detection. Analytical Chemistry 87: 3382–3387. https://doi.org/10.1021/ac504694z [Google Scholar] [PubMed] [CrossRef]
Huang GL, Gosschalk JE, Kim YS, Ogorzalek Loo RR, Clubb RT (2018). Stabilizing displayed proteins on vegetative Bacillus subtilis cells. Applied Microbiology and Biotechnology 102: 6547–6565. https://doi.org/10.1007/s00253-018-9062-x [Google Scholar] [PubMed] [CrossRef]
Hughes G, Pemberton RM, Fielden PR, Hart JP (2016). The design, development and application of electrochemical glutamate biosensors. TRAC Trends in Analytical Chemistry 79: 106–113. https://doi.org/10.1016/j.trac.2015.10.020 [Google Scholar] [CrossRef]
Hui CY, Guo Y, Liu L, Zheng HQ, Wu HM, Zhang LZ, Zhang W (2019). Development of a novel bacterial surface display system using truncated OmpT as an anchoring motif. Biotechnology Letters 41: 763–777. https://doi.org/10.1007/s10529-019-02676-4 [Google Scholar] [PubMed] [CrossRef]
Jahns AC, Rehm BH (2012). Relevant uses of surface proteins—display on self-organized biological structures. Microbial Biotechnology 5: 188–202. https://doi.org/10.1111/j.1751-7915.2011.00293.x [Google Scholar] [PubMed] [CrossRef]
Kazmi Z, Fatima I, Perveen S, Malik SS (2017). Monosodium glutamate: Review on clinical reports. International Journal of Food Properties 17: 1–9. https://doi.org/10.1080/10942912.2017.1295260 [Google Scholar] [CrossRef]
Kim D, Kim W, Kim J (2021). New bacterial surface display system development and application based on Bacillus subtilis YuaB biofilm component as an anchoring motif. Biotechnology and Bioprocess Engineering 26: 39–46. https://doi.org/10.1007/s12257-020-0397-7 [Google Scholar] [PubMed] [CrossRef]
Kurihara K (2009). Glutamate: From discovery as a food flavor to role as a basic taste (umami). American Journal of Clinical Nutrition 90: 719S–722S. https://doi.org/10.3945/ajcn.2009.27462D [Google Scholar] [PubMed] [CrossRef]
Li R, Zhou T, Khan A, Ling Z, Sharma M et al. (2021). Feed-additive of bioengineering strain with surface-displayed laccase degrades sulfadiazine in broiler manure and maintains intestinal flora structure. Journal of Hazardous Materials 406: 124440. https://doi.org/10.1016/j.jhazmat.2020.124440 [Google Scholar] [PubMed] [CrossRef]
Liang B, Han L (2020a). Displaying of acetylcholinesterase mutants on surface of yeast for ultra-trace fluorescence detection of organophosphate pesticides with gold nanoclusters. Biosensors and Bioelectronics 148: 111825. https://doi.org/10.1016/j.bios.2019.111825 [Google Scholar] [PubMed] [CrossRef]
Liang X, Han L (2020b). White peroxidase-mimicking nanozymes: Colorimetric pesticide assay without interferences of O2 and color. Advanced Functional Materials 30: 2001933. https://doi.org/10.1002/adfm.202001933 [Google Scholar] [CrossRef]
Liang B, Lang Q, Tang X, Liu A (2013a). Simultaneously improving stability and specificity of cell surface displayed glucose dehydrogenase mutants to construct whole-cell biocatalyst for glucose biosensor application. Bioresource Technology 147: 492–498. https://doi.org/10.1016/j.biortech.2013.08.088 [Google Scholar] [PubMed] [CrossRef]
Liang B, Li L, Tang X, Lang Q, Wang H et al. (2013b). Microbial surface display of glucose dehydrogenase for amperometric glucose biosensor. Biosensors and Bioelectronics 45: 19–24. https://doi.org/10.1016/j.bios.2013.01.050 [Google Scholar] [PubMed] [CrossRef]
Liang B, Liu Y, Zhao Y, Xia T, Chen R, Yang J (2021). Development of bacterial biosensor for sensitive and selective detection of acetaldehyde. Biosensors and Bioelectronics 193: 113566. https://doi.org/10.1016/j.bios.2021.113566 [Google Scholar] [PubMed] [CrossRef]
Liang B, Wang G, Yan L, Ren H, Feng R, Xiong Z, Liu A (2019). Functional cell surface displaying of acetylcholinesterase for spectrophotometric sensing organophosphate pesticide. Sensors and Actuators B: Chemical 279: 483–489. https://doi.org/10.1016/j.snb.2018.09.119 [Google Scholar] [CrossRef]
Liang B, Zhang S, Lang Q, Song J, Han L, Liu A (2015). Amperometric L-glutamate biosensor based on bacterial cell-surface displayed glutamate dehydrogenase. Analytica Chimica Acta 884: 83–89. https://doi.org/10.1016/j.aca.2015.05.012 [Google Scholar] [PubMed] [CrossRef]
Lin M, Zhang B, Yu C, Li J, Zhang L, Sun H, Gao F, Zhou G (2014). L-glutamate supplementation improves small intestinal architecture and enhances the expressions of jejunal mucosa amino acid receptors and transporters in weaning piglets. PLoS One 9: e111950. https://doi.org/10.1371/journal.pone.0111950 [Google Scholar] [PubMed] [CrossRef]
Liu J, Fan Y, Chen G, Liu Y (2021). Highly sensitive glutamate biosensor based on platinum nanoparticles decorated MXene-Ti3C2Tx for L-glutamate determination in foodstuffs. LWT 148: 111748. https://doi.org/10.1016/j.lwt.2021.111748 [Google Scholar] [CrossRef]
Liu D, Geng H, Zhang Z, Xing Y, Yang D, Liu Z, Wang D (2020). An effective platform for exploring rotavirus receptors by bacterial surface display system. Virologica Sinica 35: 103–109. https://doi.org/10.1007/s12250-019-00174-5 [Google Scholar] [PubMed] [CrossRef]
Liu A, Lang Q, Liang B, Shi J (2017). Sensitive detection of maltose and glucose based on dual enzyme-displayed bacteria electrochemical biosensor. Biosensors and Bioelectronics 87: 25–30. https://doi.org/10.1016/j.bios.2016.07.050 [Google Scholar] [PubMed] [CrossRef]
Liu R, Yang C, Xu Y, Xu P, Jiang H, Qiao C (2013). Development of a whole-cell biocatalyst/biosensor by display of multiple heterologous proteins on the Escherichia coli cell surface for the detoxification and detection of organophosphates. Journal of Agricultural and Food Chemistry 61: 7810–7816. https://doi.org/10.1021/jf402999b [Google Scholar] [PubMed] [CrossRef]
Mangano MG, Hawkes CD, Caron JB (2019). Trace fossils associated with Burgess Shale non-biomineralized carapaces: Bringing taphonomic and ecological controls into focus. Royal Society Open Science 6: 172074. https://doi.org/10.1098/rsos.172074 [Google Scholar] [PubMed] [CrossRef]
Mruga D, Soldatkin O, Paliienko K, Topcheva A, Krisanova N, Kucherenko D, Borisova T, Dzyadevych S, Soldatkin A (2021). Optimization of the design and operating conditions of an amperometric biosensor for glutamate concentration measurements in the blood plasma. Electroanalysis 33: 1299–1307. https://doi.org/10.1002/elan.202060449 [Google Scholar] [CrossRef]
Ozel RE, Ispas C, Ganesana M, Leiter JC, Andreescu S (2014). Glutamate oxidase biosensor based on mixed ceria and titania nanoparticles for the detection of glutamate in hypoxic environments. Biosensors and Bioelectronics 52: 397–402. https://doi.org/10.1016/j.bios.2013.08.054 [Google Scholar] [PubMed] [CrossRef]
Park M (2019). Orientation control of the molecular recognition layer for improved sensitivity: A review. BioChip Journal 13: 82–94. https://doi.org/10.1007/s13206-019-3103-0 [Google Scholar] [CrossRef]
Park M (2020). Surface display technology for biosensor applications: A review. Sensors 20: 2775. https://doi.org/10.3390/s20102775 [Google Scholar] [PubMed] [CrossRef]
Park JH, Kim W, Lee YS, Kim JH (2019). Decolorization of acid green 25 by surface display of CotA laccase on Bacillus subtilis spores. Journal of Microbiology and Biotechnology 29: 1383–1390. https://doi.org/10.4014/jmb.1907.07019 [Google Scholar] [PubMed] [CrossRef]
Pham ML, Polakovic M (2020). Microbial cell surface display of oxidoreductases: Concepts and applications. International Journal of Biological Macromolecules 165: 835–841. https://doi.org/10.1016/j.ijbiomac.2020.09.237 [Google Scholar] [PubMed] [CrossRef]
Pundir CS, Malik A, Preety (2019). Bio-sensing of organophosphorus pesticides: A review. Biosensors and Bioelectronics 140: 111348. https://doi.org/10.1016/j.bios.2019.111348 [Google Scholar] [PubMed] [CrossRef]
Pyun JC, Kim SD, Chung JW (2005). New immobilization method for immunoaffinity biosensors by using thiolated proteins. Analytical Biochemistry 347: 227–233. https://doi.org/10.1016/j.ab.2005.09.008 [Google Scholar] [PubMed] [CrossRef]
Qi L, Wu W, Kang Q, Hu Q, Yu L (2020). Detection of organophosphorus pesticides with liquid crystals supported on the surface deposited with polyoxometalate-based acetylcholinesterase-responsive supramolecular spheres. Food Chemistry 320: 126683. https://doi.org/10.1016/j.foodchem.2020.126683 [Google Scholar] [PubMed] [CrossRef]
Ranimol G, Sunkar S (2022). A comparative study of free laccase and laccase immobilized in copper alginate. Indian Journal of Biochemistry & Biophysics 59: 214–223. https://doi.org/10.56042/ijbb.v59i2.27154 [Google Scholar] [CrossRef]
Ricklefs E, Winkler N, Koschorreck K, Urlacher VB (2014). Expanding the laccase-toolbox: A laccase from Corynebacterium glutamicum with phenol coupling and cuprous oxidase activity. Journal of Biotechnology 191: 46–53. https://doi.org/10.1016/j.jbiotec.2014.05.031 [Google Scholar] [PubMed] [CrossRef]
Sang YH, Pan HH, Tang RK (2020). Condensed-matter chemistry in biomineralization. Progress in Chemistry 32: 1100–1114. https://doi.org/10.7536/pc200431 [Google Scholar] [CrossRef]
Saoudi O, Ghaouar N (2019). Biocatalytic characterization of free and immobilized laccase from Trametes versicolor in its activation zone. International journal of biological macromolecules 128: 681–691. https://doi.org/10.1016/j.ijbiomac.2019.01.199 [Google Scholar] [PubMed] [CrossRef]
Sharifi M, Robatjazi SM, Sadri M, Mosaabadi JM (2018). Covalent immobilization of organophosphorus hydrolase enzyme on chemically modified cellulose microfibers: Statistical optimization and characterization. Reactive & Functional Polymers 124: 162–170. https://doi.org/10.1016/j.reactfunctpolym.2018.01.019 [Google Scholar] [CrossRef]
Sheldon AL, Robinson MB (2007). The role of glutamate transporters in neurodegenerative diseases and potential opportunities for intervention. Neurochemistry International 51: 333–355. https://doi.org/10.1016/j.neuint.2007.03.012 [Google Scholar] [PubMed] [CrossRef]
Shibasaki S, Ueda M (2014). Bioadsorption strategies with yeast molecular display technology. Biocontrol Science 19: 157–164. https://doi.org/10.4265/bio.19.157 [Google Scholar] [PubMed] [CrossRef]
Song J, Liang B, Han D, Tang X, Lang Q, Feng R, Han L, Liu A (2015). Bacterial cell-surface displaying of thermo-tolerant glutamate dehydrogenase and its application in L-glutamate assay. Enzyme and Microbial Technology 70: 72–78. https://doi.org/10.1016/j.enzmictec.2014.12.002 [Google Scholar] [PubMed] [CrossRef]
Tang X, Liang B, Yi T, Manco G, Palchetti I, Liu A (2014a). Cell surface display of organophosphorus hydrolase for sensitive spectrophotometric detection of p-nitrophenol substituted organophosphates. Enzyme and Microbial Technology 55: 107–112. https://doi.org/10.1016/j.enzmictec.2013.10.006 [Google Scholar] [PubMed] [CrossRef]
Tang X, Zhang T, Liang B, Han D, Zeng L, Zheng C, Li T, Wei M, Liu A (2014b). Sensitive electrochemical microbial biosensor for p-nitrophenylorganophosphates based on electrode modified with cell surface-displayed organophosphorus hydrolase and ordered mesopore carbons. Biosensors and Bioelectronics 60: 137–142. https://doi.org/10.1016/j.bios.2014.04.001 [Google Scholar] [PubMed] [CrossRef]
Teymennet-Ramirez KV, Martinez-Morales F, Trejo-Hernandez MR (2021). Yeast surface display system: Strategies for improvement and biotechnological applications. Frontiers in Bioengineering and Biotechnology 9: 794742. https://doi.org/10.3389/fbioe.2021.794742 [Google Scholar] [PubMed] [CrossRef]
Ueda M (2016). Establishment of cell surface engineering and its development. Bioscience Biotechnology and Biochemistry 80: 1243–1253. https://doi.org/10.1080/09168451.2016.1153953 [Google Scholar] [PubMed] [CrossRef]
van Bloois E, Winter RT, Kolmar H, Fraaije MW (2011). Decorating microbes: Surface display of proteins on Escherichia coli. Trends in Biotechnology 29: 79–86. https://doi.org/10.1016/j.tibtech.2010.11.003 [Google Scholar] [PubMed] [CrossRef]
Wang X, Duan J, Cai Y, Liu D, Li X, Dong Y, Hu F (2020). A modified nanocomposite biosensor for quantitative L-glutamate detection in beef. Meat Science 168: 108185. https://doi.org/10.1016/j.meatsci.2020.108185 [Google Scholar] [PubMed] [CrossRef]
Wang H, Lang Q, Li L, Liang B, Tang X, Kong L, Mascini M, Liu A (2013). Yeast surface displaying glucose oxidase as whole-cell biocatalyst: Construction, characterization, and its electrochemical glucose sensing application. Analytical Chemistry 85: 6107–6112. https://doi.org/10.1021/ac400979r [Google Scholar] [PubMed] [CrossRef]
Wang Y, Selvamani V, Yoo IK, Kim TW, Hong SH (2021). A novel strategy for the microbial removal of heavy metals: Cell-surface display of peptides. Biotechnology and Bioprocess Engineering 26: 1–9. https://doi.org/10.1007/s12257-020-0218-z [Google Scholar] [CrossRef]
Wei P, Yuan Z, Jing W, Huang Y, Cai Q et al. (2019). Strengthening the potential of biomineralized microspheres in enhancing osteogenesis via incorporating alendronate. Chemical Engineering Journal 368: 577–588. https://doi.org/10.1016/j.cej.2019.02.202 [Google Scholar] [CrossRef]
Wu Y, Chen Y, Wei N (2020). Biocatalytic properties of cell surface display laccase for degradation of emerging contaminant acetaminophen in water reclamation. Biotechnology and Bioengineering 117: 342–353. https://doi.org/10.1002/bit.27214 [Google Scholar] [PubMed] [CrossRef]
Xiang HY, Wang JF, Yang YF, Lu YC (2019). The research progress of bacterial surface display technology. Progress in Biochemistry and Biophysics 46: 162–168. https://doi.org/10.16476/j.pibb.2018.0181 [Google Scholar] [CrossRef]
Xu YL, Li FY, Ndikuryayo F, Yang WC, Wang HM (2018). Cholinesterases and engineered mutants for the detection of organophosphorus pesticide residues. Sensors 18: 4281. https://doi.org/10.3390/s18124281 [Google Scholar] [PubMed] [CrossRef]
Yang X, Tang H, Song M, Shen Y, Hou J, Bao X (2019). Development of novel surface display platforms for anchoring heterologous proteins in Saccharomyces cerevisiae. Microbial Cell Factories 18: 85. https://doi.org/10.1186/s12934-019-1133-x [Google Scholar] [PubMed] [CrossRef]
Ye M, Ye Y, Du Z, Chen G (2021). Cell-surface engineering of yeasts for whole-cell biocatalysts. Bioprocess and Biosystems Engineering 44: 1003–1019. https://doi.org/10.1007/s00449-020-02484-5 [Google Scholar] [PubMed] [CrossRef]
Zhang Y, Dong W, Lv Z, Liu J, Zhang W, Zhou J, Xin F, Ma J, Jiang M (2018a). Surface display of bacterial laccase CotA on Escherichia coli cells and its application in industrial dye decolorization. Molecular Biotechnology 60: 681–689. https://doi.org/10.1007/s12033-018-0103-6 [Google Scholar] [PubMed] [CrossRef]
Zhang Z, Liu J, Fan J, Wang Z, Li L (2018b). Detection of catechol using an electrochemical biosensor based on engineered Escherichia coli cells that surface-display laccase. Analytica Chimica Acta 1009: 65–72. https://doi.org/10.1016/j.aca.2018.01.008 [Google Scholar] [PubMed] [CrossRef]
Zhang Z, Zhang Z, Hu Y, Liu J, Ni H, Li L (2017). Phenol biosensor based on glassy carbon electrode directly absorbed Escherichia coli cells with surface-displayed bacterial laccase. Procedia Technology 27: 137–138. https://doi.org/10.1016/j.protcy.2017.04.060 [Google Scholar] [CrossRef]
Zhao S, Guo D, Zhu Q, Dou W, Guan W (2020). Display of microbial glucose dehydrogenase and cholesterol oxidase on the yeast cell surface for the detection of blood biochemical parameters. Biosensors 11: 13. https://doi.org/10.3390/bios11010013 [Google Scholar] [PubMed] [CrossRef]
Zhao Y, Liang X, Chen D, Bian X, Liu W, Han L (2022). Denatured proteins show new vitality: Green synthesis of germanium oxide hollow microspheres with versatile functions by denaturing proteins around bubbles. Aggregate 4: e204. https://doi.org/10.1002/agt2.204 [Google Scholar] [CrossRef]
Zheng H, Wang R, Qu J (2016). Effect of different glucose supply conditions on neuronal energy metabolism. Cognitive Neurodynamics 10: 563–571. https://doi.org/10.1007/s11571-016-9401-5 [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools