 Open Access
Open Access
ARTICLE
Characteristics and expression of the TCP transcription factors family in Allium senescens reveal its potential roles in drought stress responses
1 College of Life Science, Institute of Life Science and Green Development, Hebei University, Baoding, 071000, China
2 Hebei Innovation Center for Bioengineering and Biotechnology, Hebei University, Baoding, 071002, China
3 College of Plant Protection, China Agricultural University, Beijing, 100193, China
* Corresponding Authors: Jianfeng Liu, ; Guixia Liu,
# These authors contributed equally to this work
(This article belongs to the Special Issue: Physiology and Molecular Biology of Plant Stress Tolerance)
BIOCELL 2023, 47(4), 905-917. https://doi.org/10.32604/biocell.2023.026930
Received 08 October 2022; Accepted 28 November 2022; Issue published 08 March 2023
Abstract
Allium senescens, is an important economic and ecological grassland plant with drought-resistant characteristics. A TCP protein transcription factor is important in the regulation of plant development and adverse responses. However, the mechanism by which TCP transcription functions in drought resistance in Allium senescens is still not clear. Here, we obtained a total of 190,305 transcripts with 115,562 single gene clusters based on RNA-Seq sequencing of Allium senescens under drought stress. The total number of bases was 97,195,096 bp, and the average length was 841.06 bp. Furthermore, we found that there were eight genes of the TCP family that showed an upregulated expression trend under drought stress in Allium senescens. We carried out an investigation to determine the evolution and function of the AsTCP family and how they produce an effect in drought resistance. The 14 AsTCP genes were confirmed and divided into class I and class II containing CIN and CYC/TBI subfamilies, respectively. We also found that the expression of AsTCP17 was remarkably upregulated with drought treatment. Besides, the transformation of AsTCP17 in Arabidopsis revealed that the protective enzymes, namely polyphenol oxidase (POD) and superoxide dismutase (SOD), were increased by 0.4 and 0.8 times, respectively. Chlorophyll content was also increased, while the H2O2 and malondialdehyde (MDA) contents were decreased. Staining assays with 3,3′- diaminobenzidine (DAB) also suggested that the AsTCP17 downregulates reactive oxygen species (ROS) accumulation. In addition, overexpression of the AsTCP17 affected the accumulation of drought-related hormones in plants, and the synthesis of ABA. The expression of AtSVP and AtNCED3, related ABA synthesis pathway genes, indicated that the level of expression of AtSVP and AtNCED3 was obviously enhanced, with the overexpression of line 6 showing a 20.6-fold and 7.0-fold increase, respectively. Taken together, our findings systematically analyze the AsTCPs family at the transcriptome expression level in Allium senescens, and we also demonstrated that AsTCP17 protein, as a positive regulator, was involved in drought resistance of Allium senescens. In addition, our research contributes to the comprehensive understanding of the drought stress defense mechanism in herbaceous plants.Keywords
Allium senescens Linn is a perennial plant, with strong stress resistance, simple reproduction and significant environmental and ecological benefits, as well as being edible, and of medicinal and ornamental value (Dandona et al., 2010; Lee et al., 2021). Allium senescens is an important vegetable in the world, and as a popular vegetable, is often used as seasoning because of its unique taste (Bio Sigui Bruno et al., 2020). Its young leaves are also a kind of wild grazing forage and can be directly eaten or further processed into pickles. In addition, Allium senescens as a perennial plant that has strong vitality with a formidable root system under drought stress (Kukushkina and Fomina, 2021). However, at present, the research on Allium senescens mainly focuses on its nutritional value and ecological greening value, but its molecular mechanism of drought resistance is scarcely reported (Beretta et al., 2017). Therefore, taking Allium senescens as our research object, an in-depth analysis of its drought resistance mechanism will provide an important theoretical reference for the drought-resistance molecular breeding of Liliaceae plants.
Drought stress is an important environmental factor that significantly inhibits plant growth and development (Wang et al., 2018). Plants have formed powerful strategies to cope with the effects of drought stress. Recently, studies have revealed that plants adjust to drought stress by regulating the expression of drought-tolerance-related key genes, which is important for the molecular breeding of drought-resistant plants (Aimar et al., 2011; Alvarez et al., 2014). Furthermore, the findings revealed that plants could enhance their drought resistance by activating key transcription factors, leading to changes in hormones, key proteins, defense enzymes, and essential metabolites, all of which could protect plants from drought-induced stress (Jongrungklang et al., 2011; Chen et al., 2017). Therefore, the defensive strategy of plants for drought resistance is a complicated regulatory mechanism. However, whether the transcription factor genes in Allium senescens are involved in its drought resistance mechanism is still unclear.
Transcription factor genes play important roles in plant defenses to biotic and abiotic stress and growth period development. Studies have found that the TEOSINTE BRANCHED1/CYCLOIDEA/PROLIFERATING CELL FACTOR1 transcription factor family (TCPs) contains a basic helixloop-helix (bHLH) and a TCP domain (Cubas et al., 1999). The conserved TCP domain is an important structure that helps TCP transcription factors to be divided into TCP-P or PCF (class I) and TCP-C (class II) subfamilies and plays a key role in protein-protein interactions and DNA binding (Martin-Trillo and Cubas, 2010; Danisman et al., 2012). There are 24 TCPs from Arabidopsis thaliana, 28 TCPs from Oryza sativa, 38 TCPs from Gossypium raimondii and 23 TCPs from Phalaenopsis equestris that have been successfully cloned and identified, respectively (Yao et al., 2010; Ma et al., 2014; Liu et al., 2019). Recently, a few studies have further shown that TCP proteins can regulate plants to adapt to drought stress. Transformation of ZmTCP42 into Arabidopsis could cause sensitivity to ABA hormone at the seed germination period, and further increase drought resistance (Ding et al., 2019). Moreover, the overexpression of OsTCP19 in Arabidopsis increased drought tolerance by mediating ABI4 synthesis (Mukhopadhyay and Tyagi, 2015). Transformed for the Osa-miR319a gene, a TCP gene function target site, showed an improved growth performance and exhibited strong drought- and salt-resistance via enhancing leaf wax, water retention, and decreased Na+ absorption (Cakir et al., 2016). Taken together, the TCP proteins could extensively regulate plant development and drought response. The function of TCP proteins in regulating for drought-resistance in Allium senescens has been scarcely reported.
Above all, research on the TCP family is mainly based on model plants including Oryza sativa, Arabidopsis thaliana and Glycine max. More attention is paid to the aspects of plant organ morphogenesis, cell expansion, and regulation of hormone pathways (Danisman et al., 2012). There is little research on the function of the TCP family in Allium senescens. In particular, the molecular strategy of TCP-related drought resistance in Allium senescens is not clear enough and the understanding of the main genes that relate to drought resistance is not comprehensive. Therefore, it is necessary to comprehensively study the molecular mechanisms of the TCP family and find out the key genes for drought resistance in Allium senescens. The objective of our study was to identify gene expression and the functional characteristics of the AsTCP gene family under drought stress, based on transcriptome analysis, and to further obtain and identify the key genes related to drought-resistance, which will supply a comprehensive understanding of the drought-resistance molecular mechanism and play a key role in obtaining potential genes to improve the drought resistance of Allium senescens.
Plant materials and growth conditions
Some 4-5-year-old Allium senescens plants were obtained from the BaShang grasslands, Zhangjiakou, Hebei Province, China (115.833307′N, 41.649034′E). The region has an average elevation of about 1,700 m, with the highest elevation of about 2,400 m. The plants were further transferred to pots in a greenhouse and subjected to drought stress by 20% PEG-6000.
Arabidopsis (ecotype Columbia, Col-0) seeds are planted directly in vermiculite, and then cultivated in a greenhouse, under conditions of 4500 lux supplementary light, 60% humidity, 16 h of light at 25°C, and 8 h dark. The development/growth period of Arabidopsis was maintained with regular watering using Hoagland nutrient solutions at two to three-day intervals.
Analysis of the transcriptome data of the Allium senescens
During the vegetative growth period of Allium senescens, it germinated and grew for about 20 days, and was subjected to conditions of drought (by 20% PEG-6000) and non-drought (normal condition) for 15 days. For transcriptome analysis, the seedlings with the same growth were screened and the middle and upper parts of leaves were cut to 1.0 g, with three repeats. The RNA library construction, checking, and all sequencing were performed using Majorbio of Shanghai with HiSeq 2500. For our project without reference genomes, clean reads were obtained and spliced with Trinity software to obtain reference sequences for subsequent analysis. The primary sequencings were further analyzed to obtain clean reads. In addition, the contig was acquired to use as a control sequence. The unigene of all transcript gene was regarded as the longest transcript. The gene-related functional annotation was analyzed by using the Shanghai Majorbio Cloud.
Analysis of the functional annotation, classification, and metabolic pathway for the transcriptome data
All genes’ transcripts acquired from the transcriptome sequencing of Allium senescens were analyzed with NR (Non-redundant protein database), SwissProt (SwissProt protein database), Pfam (Protein families database), Cluster of Orthologous Groups (COG), Kyoto Encyclopedia of Genes and Genomes (KEGG), and Gene Ontology (GO). The functional annotation information from all six databases was obtained and counted.
Identification of the TCP family members of Allium senescens
The AsTCPs genes were screened from the whole genome database of Allium senescens to obtain candidate sequences containing TCP structural domains, and all candidate sequences were then submitted to http://pfam.xfam.org/search (Pfam) and NCBI to validate the conserved structural domains. After eliminating the false-positive sequences without TCP special structural domains, the remaining candidate sequences were all TCP genes of Allium senescenss with the identification on ExPASy (http://www.expasy.org/tools/).
Analysis of the structure and homology of the AsTCP family genes
The WOLF PSORT (https://wolfpsort.hgc.jp/) software was also used to assess the subcellular sites of the AsTCP family. We identified the conserved domain of AsTCP family according to the Conserved Domain Search Service software in NCBI. The amino acid sequences of the Allium senescens TCP members were analyzed by MEME software to clarify the sequence characteristics and distribution features of the conserved motifs.
The sequence homology comparison and conserved structural domain analysis of the Allium senescens TCP genes were further performed using the DNAMAN8.0 software. We compared multiple sequences of Arabidopsis and Allium senescens AsTCP proteins after downloading the sequences of all Arabidopsis TCP proteins isolated from the Arabidopsis Information Resource (TAIR). We performed a phylogenetic analysis by the maximum likelihood method and embellished the phylogenetic tree with the iTOL tool to help in the classification of members of the AsTCP protein family.
AsTCPs gene expression pattern under abiotic stress
The levels of expression of the AsTCPs genes were identified by the qRT-PCR method. The Allium senescenss were maintained by hydroponics and divided into five portions in hydroponic vials and incubated at 22°C in a greenhouse after the roots were washed and fixed in hydroponic baskets. And then the five portions of Allium senescenss were treated with water (control), 200 mmol/L NaCl, 20% PEG6000, 100 μmol/L ABA and 200 μmol/L MeJA after 5–6 weeks of growth, respectively. Samples were collected from each of the five portions at 0, 6, 12, and 48 h after treatment, and then RNA was extracted by the trizol method, and reverse transcribed into cDNA. The primers of qRT-PCR for the AsTCPs were made by Primer-BLAST software, and the actin gene from Allium senescens was used as a reference gene (Suppl. Table S1), for which 3 biological repeats were set up and repeated three times. The RNA extraction reagent is Trizol, the reverse transcription kit is 2×Fast Super EvaGreen qPCR Master Mix kit. The temperature and time were set as 96°C for 5°C min, and then by 45 cycles with 95°C for about 15 s, and 57°C for 30 s. The levels of expression of the AsTCPs gene were calculated according to the 2−ΔΔCt method. All primers of AsTCPs and ABA-related genes are listed in Suppl. Table S1.
Development of the transgenic Arabidopsis with AsTCP17
To generate the recombinant AsTCP17 overexpression vector, the obtained open reading frame (ORF) of AsTCP17 was cloned into the expression pCam-AsTCP17 vector driven by the cauliflower mosaic virus 35S promoter (Suppl. Fig. S1A). Then, the recombinant vectors were transferred into wild-type Arabidopsis thaliana (Ecotype Columbia) using Agrobacterium-mediated transformation method to generate the construct, AsTCP17-OE. Transformation of the empty vector plant was used as the wild-type plant. After infection by Agrobacterium containing AsTCP17, T1, T2, and T3 generation plants were successfully obtained based on the Hygromycin medium (Suppl. Fig. S1B), and the T3 generation was used to detect the drought resistance index in our study.
Detection of the defense enzymes in the overexpression of AsTCP17 in Arabidopsis
The 4-5-week-old Arabidopsis with drought stress for 48 h were rapidly stored at −80°C for stabilizing and measuring the enzymes related to the drought-resistance physiological index. All detection of defense enzyme kits were provided by the Nanjing Jiancheng Bioengineering Institute. China, and were used for SOD detection (No. A001-1-1), POD detection (No. A048-3-1), CAT detection (No. A007-1-1), MDA detection (No. A003-1-2), and PRO detection (No. A107-1-1). To further measure ROS in transgenic Arabidopsis, histochemical staining was used according to a previously reported method (Fryer, 2002). The leaves of the transformed Arabidopsis and wild type were respectively stained by Diaminobenzidine (DAB) buffer (pH 3.8, 1 mg/ml) at 37°C in the dark condition. The leaves with the DAB-stained were further set in 75% alcohol to delete chlorophyll, and then photographed for the stained performance.
Measurement of the hormonse in the transgenic Arabidopsis
The related drought-resistance hormones, such as ABA, SA, IAA, and JA were detected by Zoonbio Biotechnology. The leaves extract was vibrated at 4°C for 40 min (12,000 rpm), with further adding dichloromethane (10 mL) in samples, and vibrated at 13,000 rpm for 15 min at 4°C. The supernatant was filtered using Chromosep C18 columns and then fractions were eluted with 10 ml 100% (v/v) methanol. It was then dried by N2 and set in 100 μL 0.1% methane acid by a 0.22 μm filter membrane. The product of samples was analyzed with a ZORBAX SB-C18 column (Agilent Technologies) of the high-performance liquid chromatography-tandem mass spectrometry (HPLC-MS/MS) method. The solvents for the mobile phase were ultrapure water and 0.1% methanoic acid, methanol and 0.1% methanoic acid, and the injected volumes were 2.5 μL. Mass spectrometer (MS) treatment conditions were the voltage at 4000 V, the pressure of nebulizer, air curtain, aux gas of respectively 60, 20, and 75 psi, and the temperature of atomizing was set at 350°C.
Level of expression of the related ABA biosynthesis genes
With analyses of the ABA biosynthetic metabolic pathway, we can infer that AtSVP (short vegetative phase), AtLOS5 (molybdenum cofactor sulfurase), AtNCED3 (nine-cis-epoxycarotenoid dioxygenase 3) and AtAO (aldehyde oxidase) are the main ABA biosynthesis-related genes. Therefore, AtSVP, AtLOS5, AtNCED3 and AtAO sequences were derived based on the Arabidopsis genome database (accession number SRP121096). Four genes were detected after overexpression of the AsTCP17 gene in Arabidopsis by qRT-PCR. The primers for the genes were designed as presented in Table 1. The method is the same as the for AsTCPs gene expression analysis given earlier.
The physiological index and relative expression level of key genes were considered as mean±SD from three independent repeats. Data analysis was carried out using Origin software (version 8.5). Data were considered to be significantly different at **p < 0.01 or *p < 0.05.
Quality identification of the transcriptome data from Allium senescens
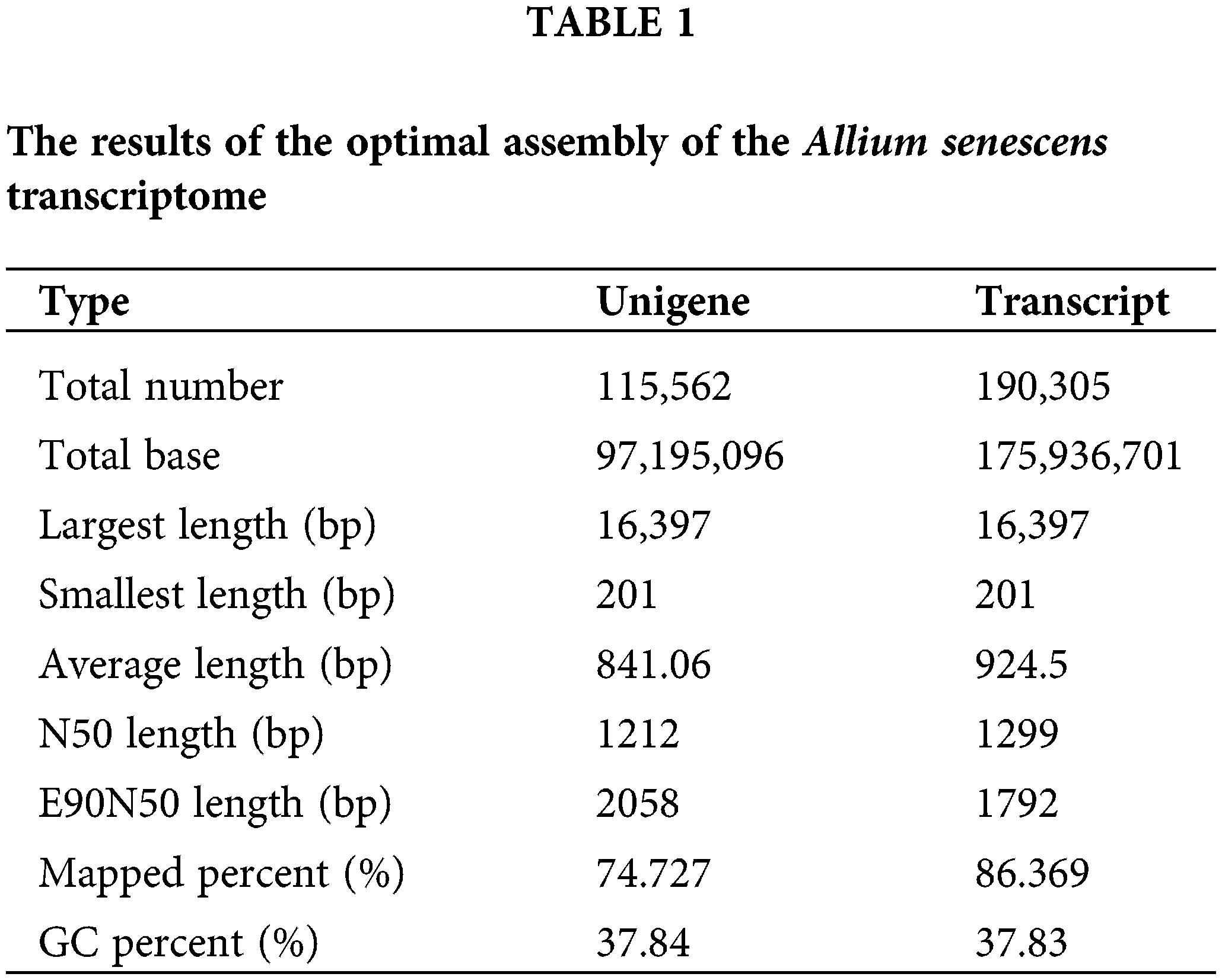
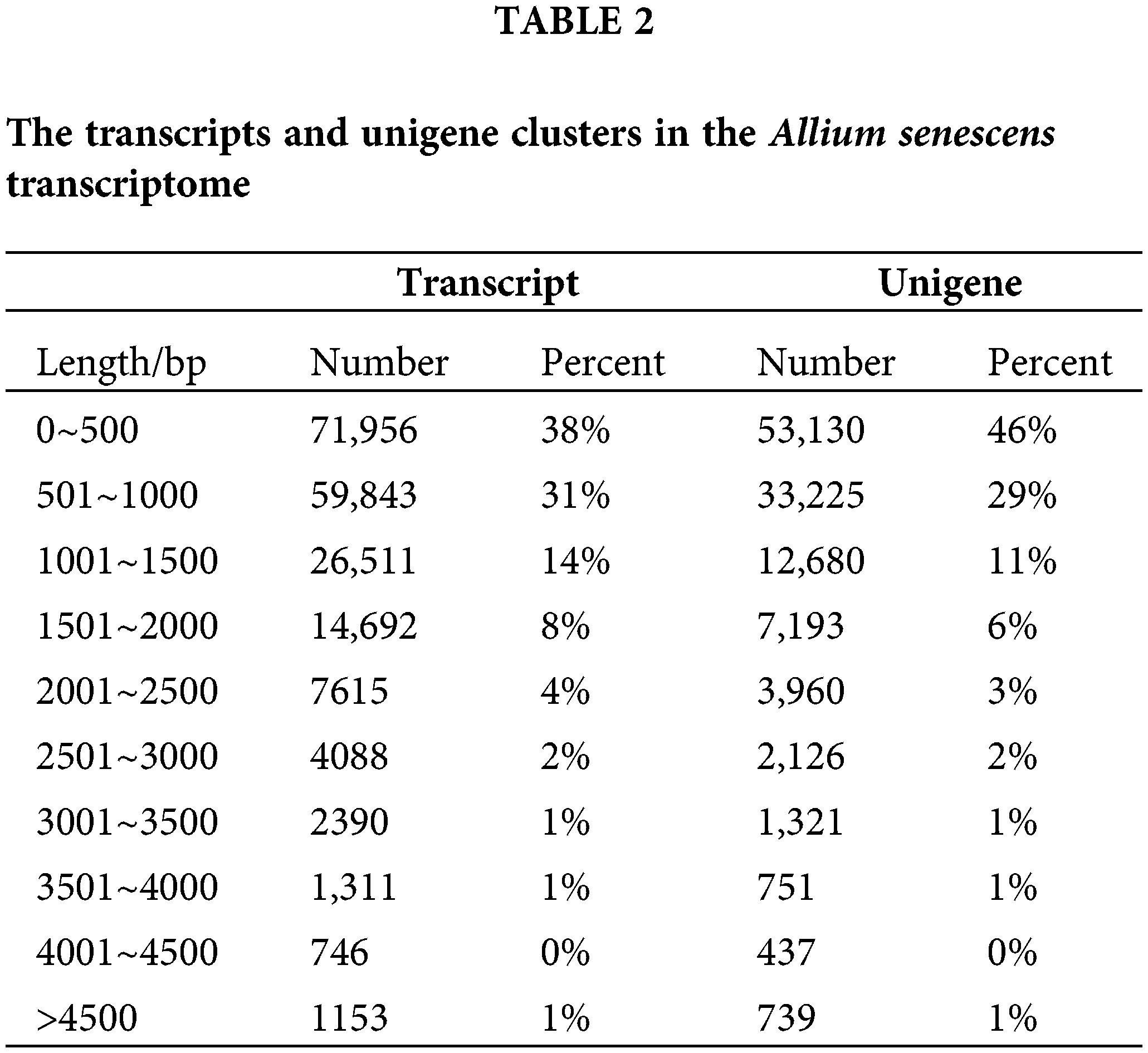
We found that a total of 175,936,701 clean paired-end reads were assembled into 97,195,096 unigenes containing an N50 length of 1299 bp and a GC base share of 37.83% based on transcriptome data (Table 1). Of these, the percentage of transcripts with sequence lengths of 0–500, 501–1000, and 1001–1500 bp were 38%, 31%, and 14%, respectively. The percentage of single gene clusters with sequence lengths of 0–500, 501–1000, and 1001–1500 bp were 46%, 29%, and 11%, respectively (Table 2). The results showed that our transcriptome data is credible.
Major databases analysis of transcriptome data from Allium senescens
Six databases were further used as the references for the functional annotation of the Allium senescens unigenes, including the NR database for 35,269 single gene clusters, the SwissProt database for 26,284 single gene clusters, the Pfam database for 26,617 single gene clusters, the COG database for 31,490 single gene clusters, the GO database to 28,201 single gene clusters, and the KEGG database to 16,201 single gene clusters (Fig. 1A). Of these, 10,608 unigenes were annotated in all six databases, showing that the sequencing data quality is sufficient for further subsequent experimental purpose. In addition, the annotation results of the three major databases, COG, GO and KEGG, were developed for the in-depth understanding of the unigenes results of Allium senescens (Fig. 1B).
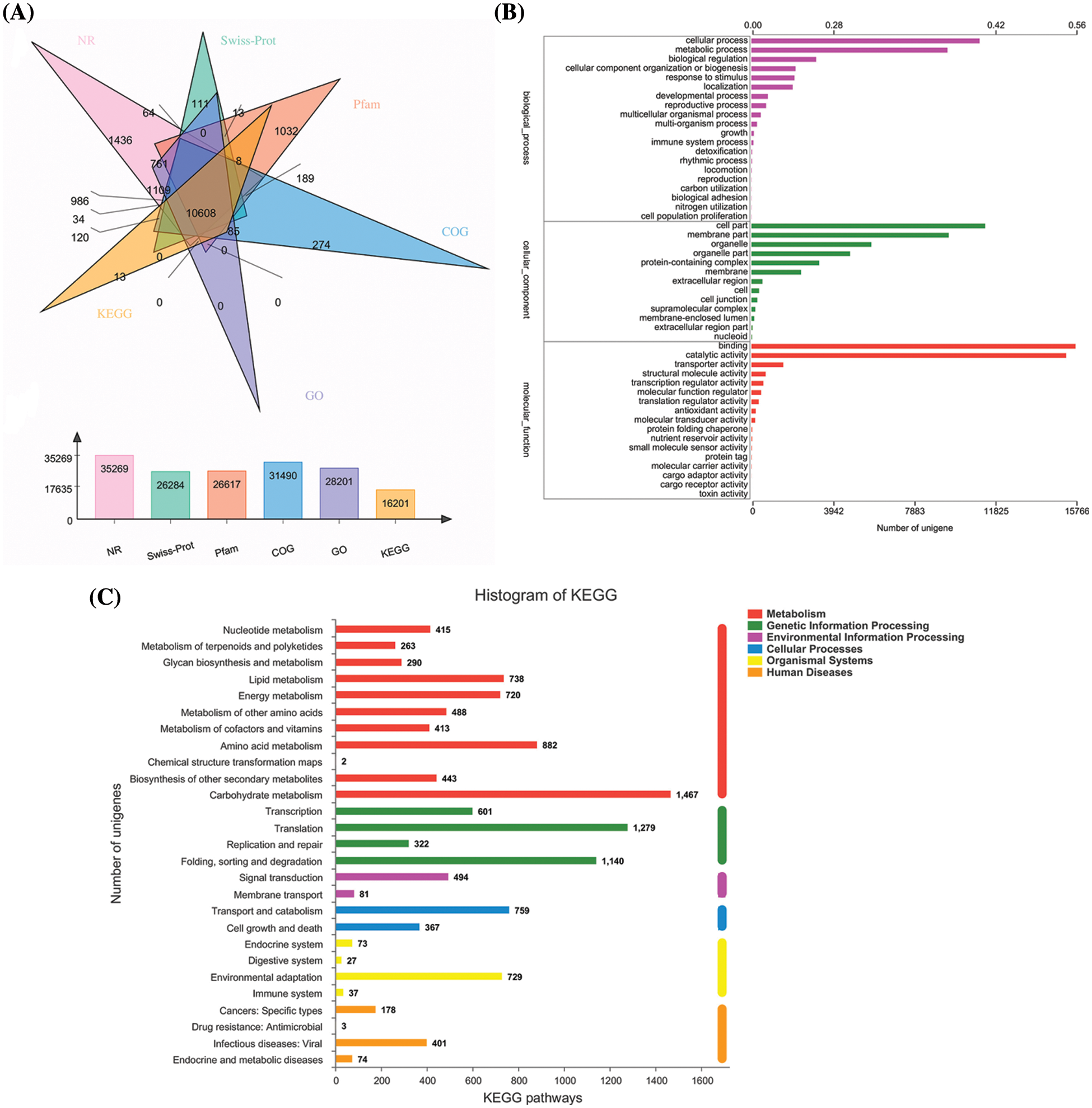
Figure 1: Six major databases used to carry out functional annotation (A) the GO analysis of Unigene (B) and KEGG pathway of Allium senescens (C).
We further obtained unigenes based on the KEGG database as a basis for functional classification. A total of 16,201 unigenes were categorized into six pathways: Cellular Processes, Genetic Function Processing, Metabolism Function, Human Diseases and Organismal Systems, and Environmental Information Processing, respectively, which were further refined into 27 metabolic pathways. The highest number of differentially expressed genes was 1467 for carbohydrate metabolism, followed by 1279 for amino acid metabolism (Fig. 1C).
Identification of the transcription factor in Allium senescens
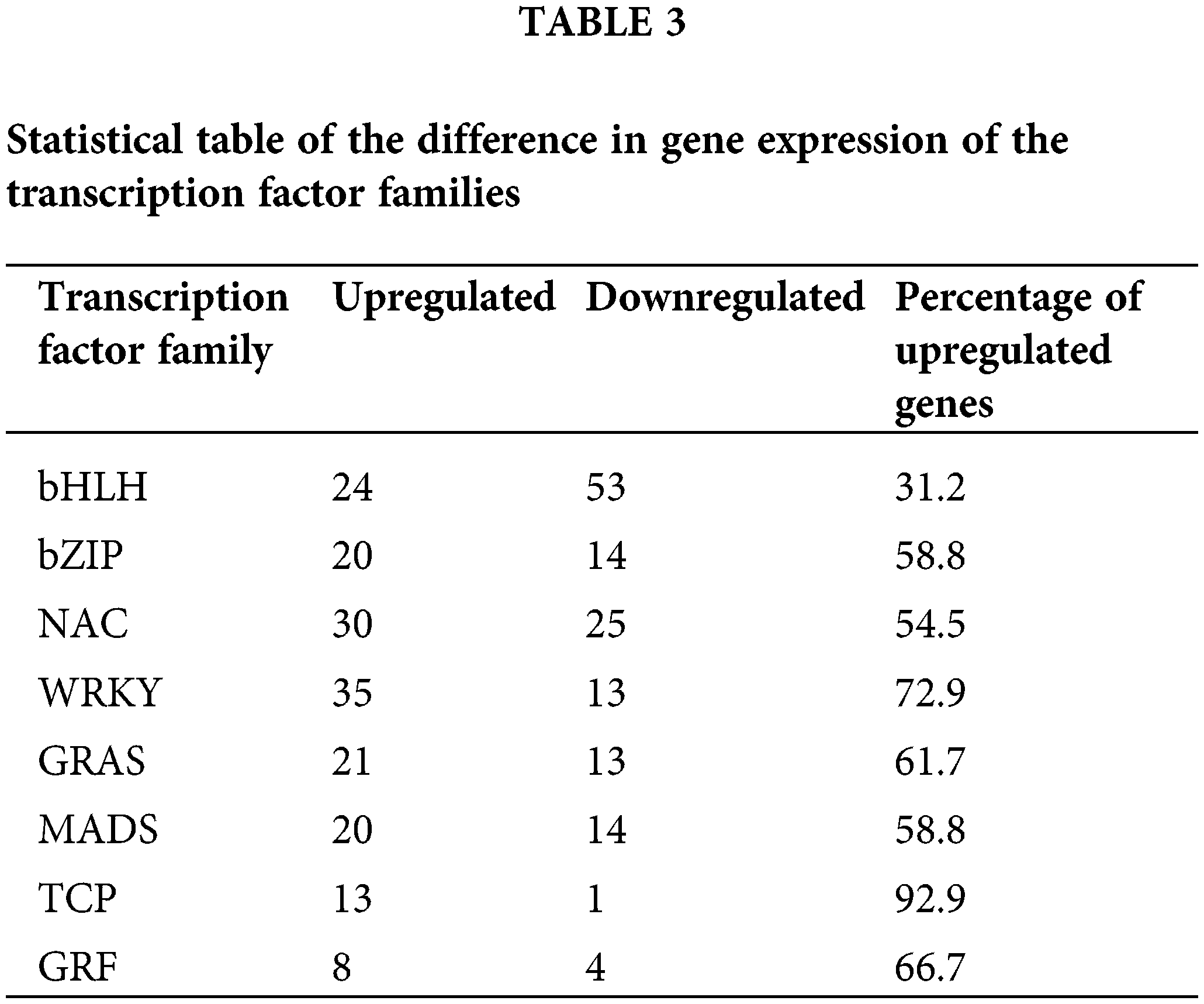
Transcription factors are involved in the complex process of regulating transcription initiation in plants. A total of 8 transcription factor families were selected in the Allium senescens. The families were as follows: the bHLH (77), bZIP (34), NAC (55), WRKY (48), GRAS (34), MADS (34), TCP (14) and GRF (12). All the transcription factor families possibly played a critical role in the drought-stress response. Of all the transcription factor families, the gene expression difference indicated that the TCP family was almost all upregulated with drought induction, with the percentage of upregulated genes being up to 92.9% (Table 3). Therefore, the TCP transcription factor family has become the focus of our research.
Identification of AsTCP family genes in Allium senescens
The gene members of the TCP family of Allium senescens were further identified, and 14 TCP family genes were obtained and named according to the TCP gene family reported in the model plant Arabidopsis thaliana. Based on the results of the functional, structural domain and sequence Blast comparison analysis, seven belonged to the class I PCF subfamily, two to the class II CYC/TBI subfamily, and five to the class II CIN subfamily (Table 4).
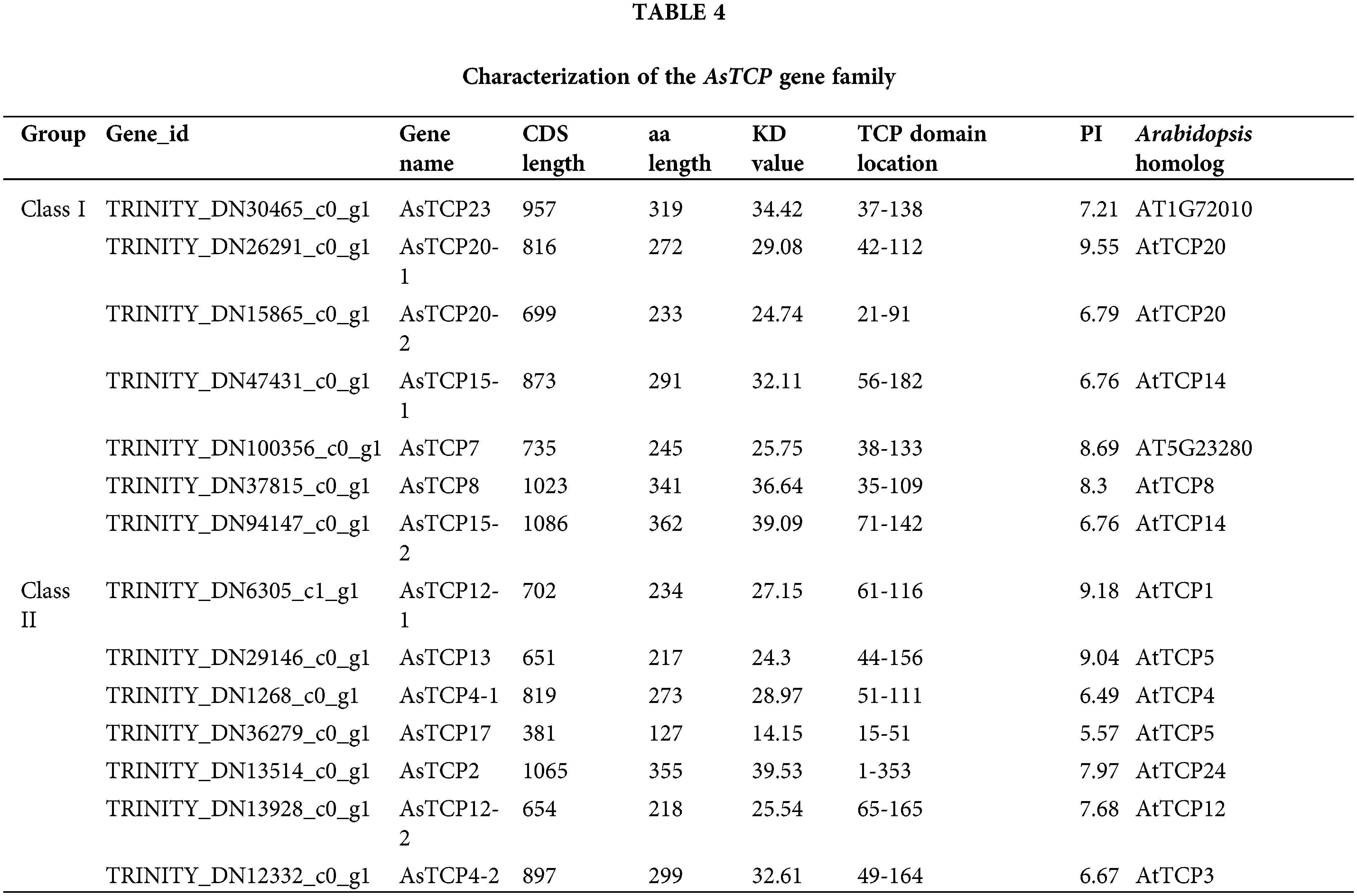
Bioinformatics analysis of AsTCP genes family
A subcellular localization prediction analysis revealed that the AsTCPs protein is predicted to be located in the nucleus (Fig. 2A). Ten conserved motifs of the AsTCPs proteins were identified, and most of them contained motif 5, which is the most important motif of TCP (Fig. 2B). To further analyze the homologous evolution of the AsTCP family genes with other species and construct a phylogenetic tree, we downloaded the TCP transcription factor family protein sequences of Arabidopsis, Zea mays and Allium senescens. The evolutionary tree diagram reveals that the 14 TCP transcription factors of the Allium senescens are distributed in three subfamilies, CYC/TB1, CIN and PCF. There are seven in the PCF subfamily of Class I, two in the CYC/TB1 subfamily of Class II, and five in the CIN subfamily of Class II. The AsTCP showed significant clustering with the AtTCP and ZmTCP family genes, from which it can be inferred that the Allium senescens TCP genes may have similar biological characteristics as in Arabidopsis and Zea mays (Fig. 2C).
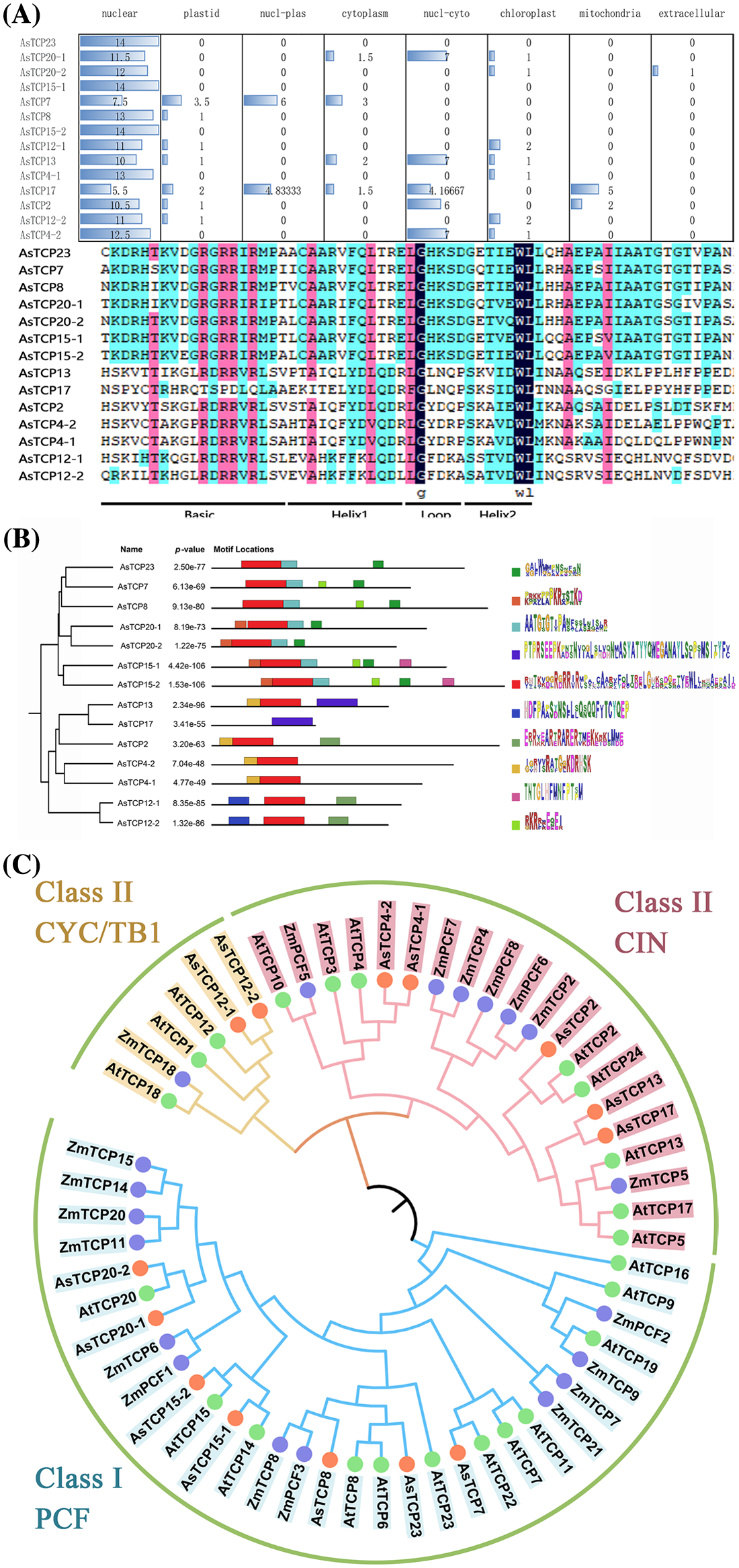
Figure 2: Subcellular localization and homology analysis (A) Motif analysis. (B) and Phylogenetic analysis of TCP family genes (C).
Identification and overexpression of AsTCP17 in Arabidopsis
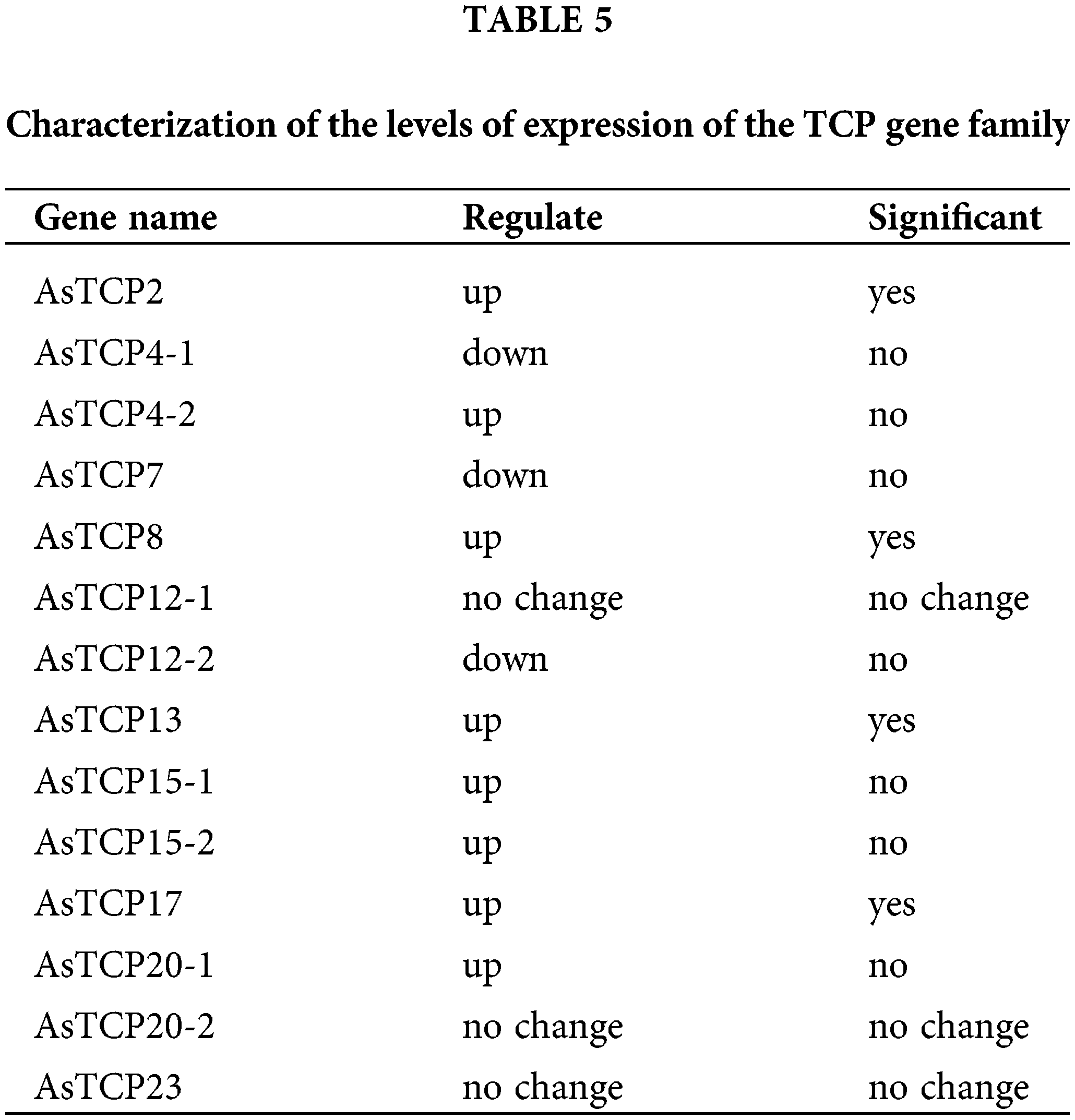
To further identify the function of AsTCPs, we tested the level of expression under drought stress. We found that the level of expression of AsTCP12-1/20-2/23 was not different before or after drought treatment of the plants based on the transcriptome data analysis. In contrast, the other TCP genes showed obvious changes in expression after drought stress, and the transcript level of AsTCP2/8/13/17 genes was significantly increased than that of WT plants (Table 5). The results showed that AsTCP2/8/13/17 genes may be the essential genes that confer drought resistance in Allium senescens.
Based on the results of the RNA-seq analysis, four essential genes, AsTCP2/8/13/17 showed upregulated expression, and were further analyzed by Real-time PCR analysis under PEG, NaCl, ABA and MeJA treatments. The results for the four genes indicated that AsTCPs were upregulated under four treatment conditions, among which the AsTCP17 gene was most significantly and rapidly upregulated after 6 h following treatment. The expression of the AsTCP2/8/13 genes frequently fluctuated without any apparent pattern (Fig. 3), which indicated that the AsTCP17 is likely to be the key gene in Allium senescens plants resistance to drought stress.
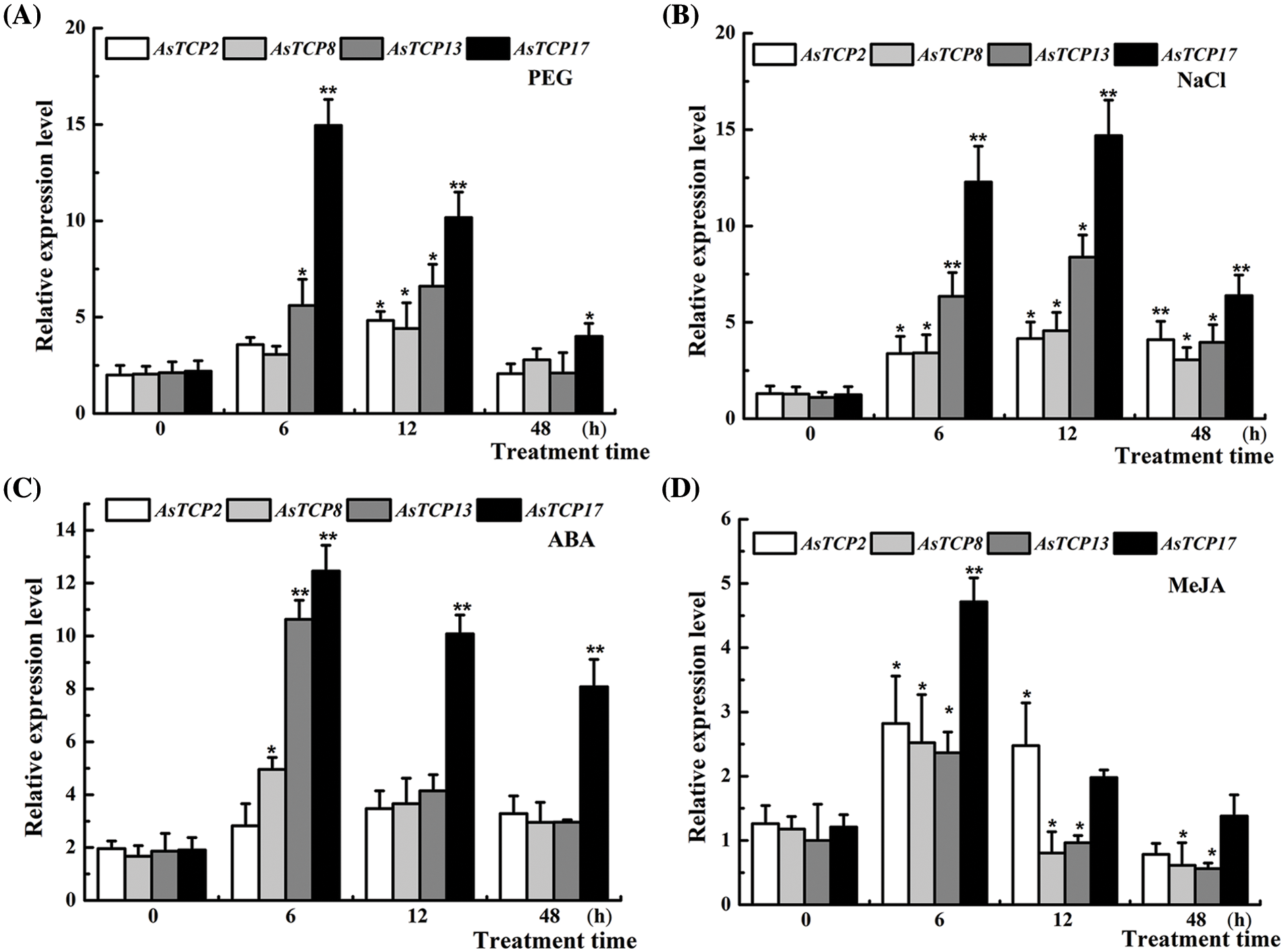
Figure 3: Expression of AsTCP2/8/13/17 under different PEG, NaCl, ABA and MeJA treatments. Data shows the means ± standard deviation (SD) of three replicates. * and ** represent p < 0.05 and p < 0.01, respectively.
Transformation of AsTCP17 enhanced susceptibility of Arabidopsis to drought and defense enzyme
For investigating whether AsTCP17 is involved in the drought tolerance process in Allium senescenss, we constructed an AsTCP17 gene overexpression vector and successfully obtained the T0 generation plants after infesting Arabidopsis with Agrobacterium tumefaciens. We determined the expression of AsTCP17 in the OE1, OE6, and OE8 lines and observed that the transcript level of the AsTCP17 gene was obviously enhanced in the transgenic Arabidopsis (OE1, OE6, and OE8), indicating that the trans-AsTCP17 Arabidopsis plants were successfully obtained (Fig. 4A).
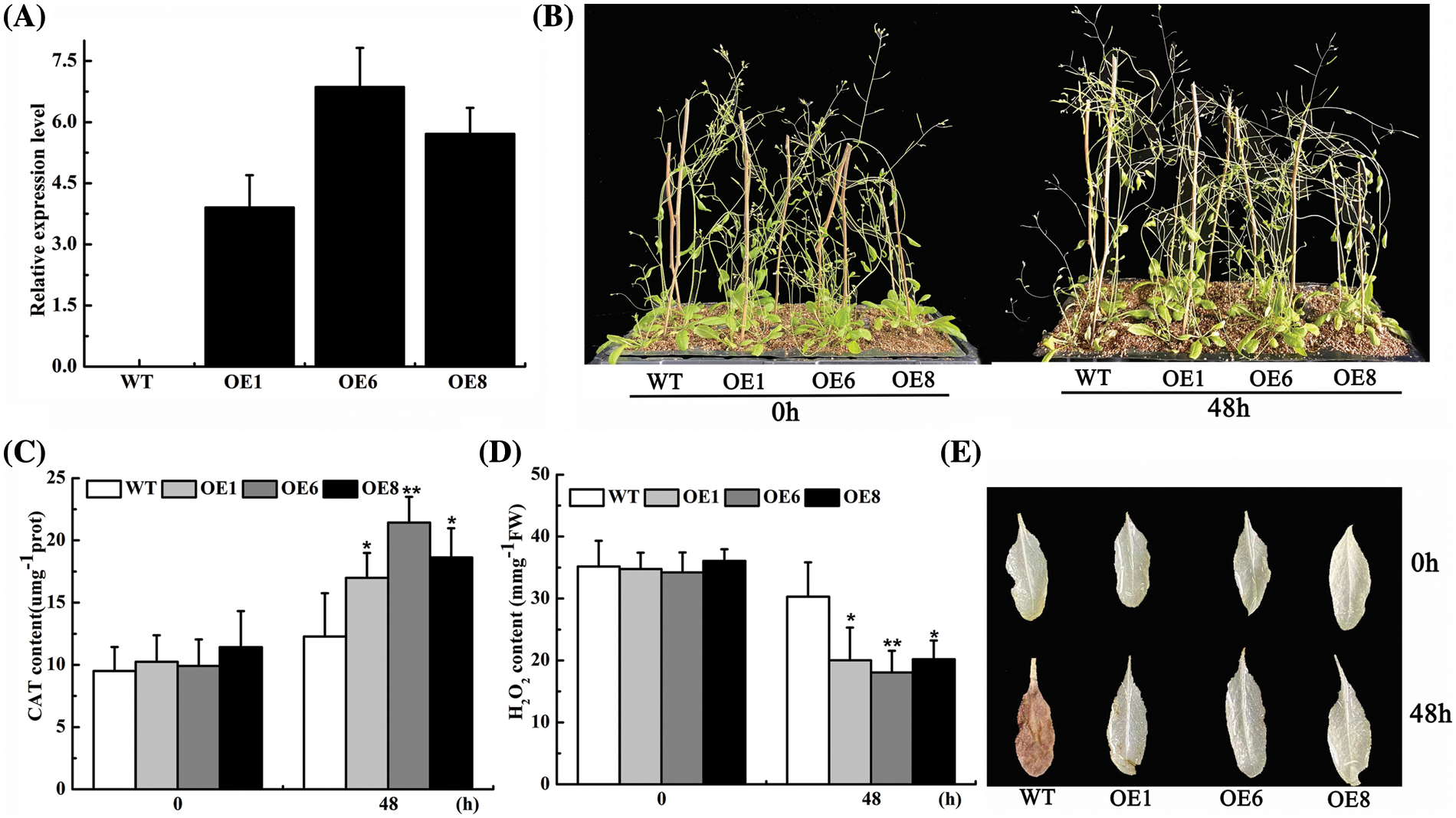
Figure 4: Identification of ROS analysis of the transgenic Arabidopsis. (A) Expression of AsTCP17 in wild-type and transgenic Arabidopsis thaliana of T3 generation. (B) Phenotypic analysis of Arabidopsis thaliana. (C) CAT content. (D) H2O2 content. (E) Visualize ROS content in leaves by DAB staining. Data shows the means ± standard deviation (SD) of three replicates. * and ** represent p < 0.05 and p < 0.01, respectively.
We further identified the drought tolerance of OEs and wild-type plants. The water deficit treatment of OEs and WT plants revealed that rosette leaf wilt was more serious in the wild-type Arabidopsis than in transgenic plants, from which it can be deduced that the transgenic plants showed better adaptation under water deficit conditions (Fig. 4B). The results confirmed that transgenic plants can significantly increase the drought tolerance of Arabidopsis.
In addition, the CAT content of the transgenic lines all increased significantly 0.5-fold, up to the highest by application of PEG for 12 h. (Fig. 4C). H2O2 content further revealed that the H2O2 content of the transformed lines was much lower than those of WT (Fig. 4D). When leaves of the transgenic lines were observed by DAB staining, the reddish-brown color of the leaves deepened as the treatment time increased, while the leaves of the transformed lines were remarkably lighter (Fig. 4E). Our results indicated that the overexpression of AsTCP17 in Arabidopsis increased the CAT content and improved the drought tolerance of Arabidopsis, while it reduced the content of reactive oxygen species.
Identification of the defense enzyme in transgenic Arabidopsis
The plant scavenging system helps to maintain low levels of reactive oxygen species, which can inhibit lipid peroxidation of membranes in adverse conditions and allows plants to withstand drought stress. SOD, POD, and MDA are the necessary antioxidant enzymes in the ROS system. As presented in Fig. 5, the differences in MDA, SOD, and POD activities, and chlorophyll contents between WT and transgenic Arabidopsis were not significant under normal treatment conditions, whereas significant differences in each index were observed after 48 h water deficit treatment. Among them, the peak values of POD and SOD in transgenic lines were increased by 0.8 and 0.4 times (Figs. 5A and B), while MDA was reduced by approximately 0.5-fold in the OEs (Fig. 5C), and chlorophyll content was much higher in the OEs lines than in the wild-type under drought stress (Fig. 5D). Taken together, our findings indicated that the drought-related defense enzymes were more active in the transgenic Arabidopsis which confirmed that AsTCP17 is a positive regulator of drought resistance.
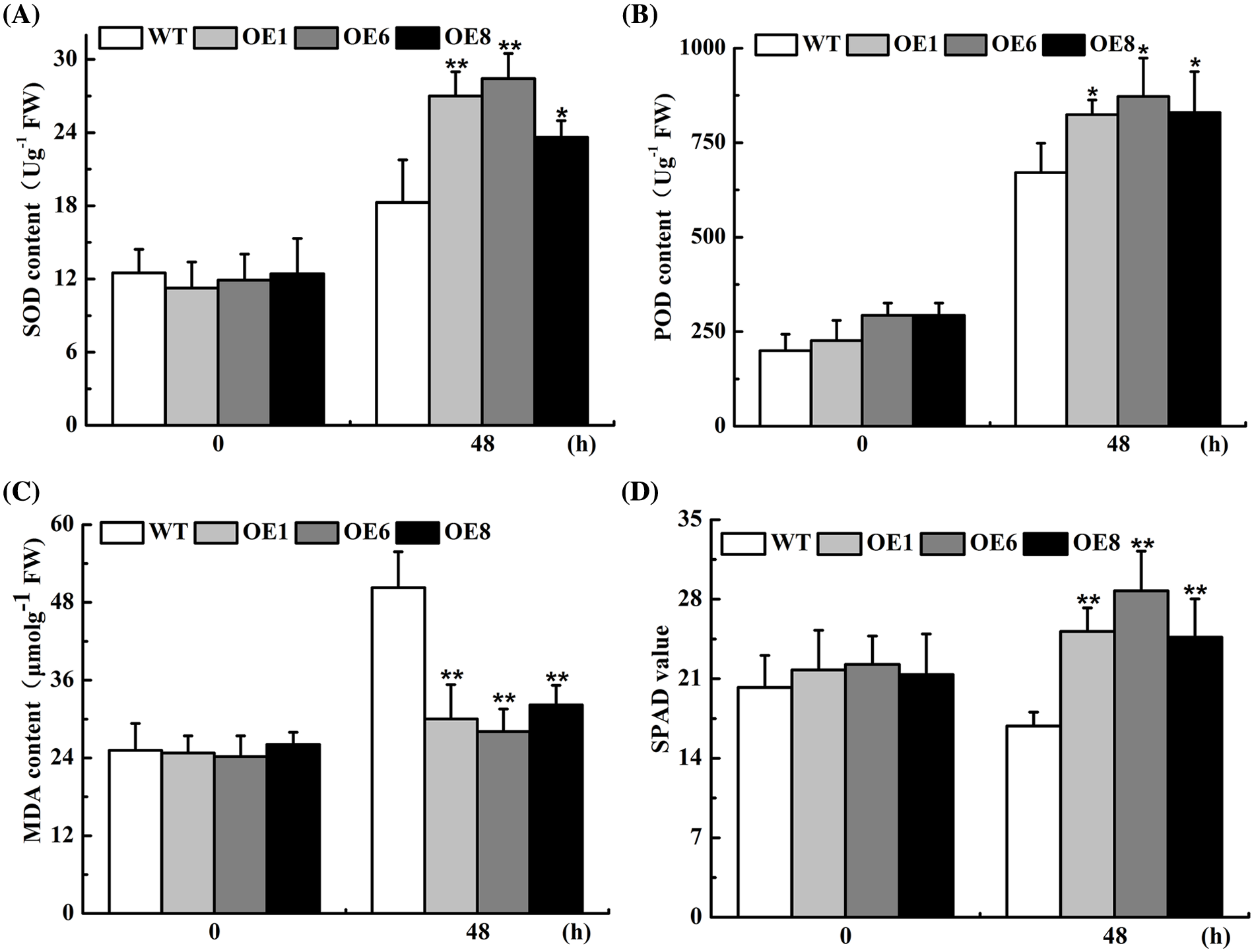
Figure 5: The content of protection enzymes in transgenic lines (OEs) with drought treatment. (A) MDA. (B) POD. (C) SOD. (D) SPAD. Data shows the means ± standard deviation (SD) of three replicates. * and ** represent p < 0.05 and p < 0.01, respectively.
Overexpression of AsTCP17 affected hormone content of the transgenic Arabidopsis
To further analyze the drought-resist mechanism of the transformed Arabidopsis, we compared the drought-related hormones in leaves of the transformed and non-transformed plants under PEG treatment. The results showed that IAA, SA, JA, and ABA in the overexpression lines were a little higher than those of Col-0. However, no significant differences were found under normal conditions (Fig. 6). ABA content in each transgenic line increased by 57.56%, 140.57% and 106.60%, and the SA content increased by 35.56%, 36.29%, and 8.13% compared to Col-0 under drought treatment (Fig. 6). Therefore, overexpression of the AsTCP17 gene affected the synthesis and accumulation of drought-related hormones in plants, especially the synthesis of ABA.
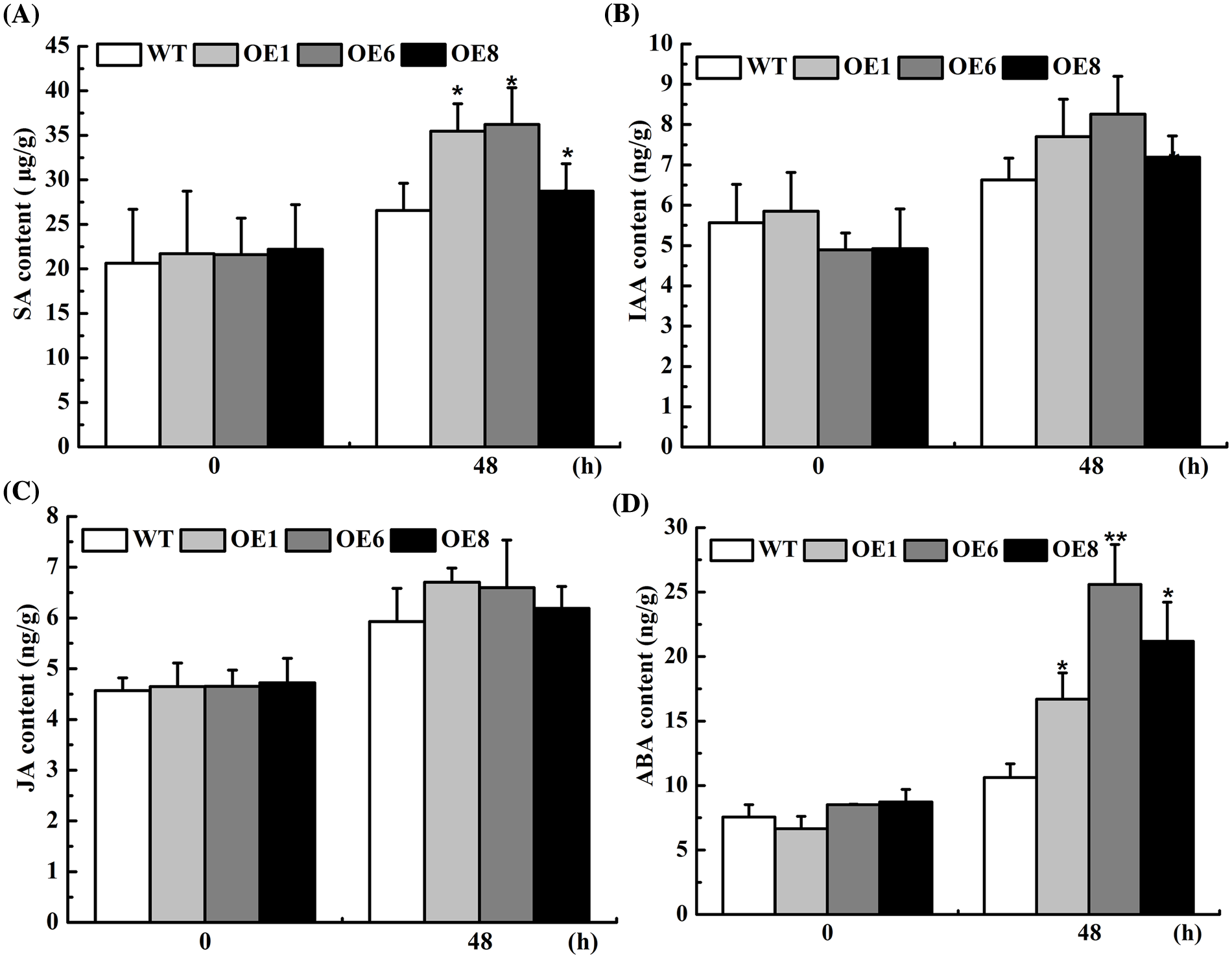
Figure 6: Content of hormones in transformed plants with AsTCP17. (A) SA hormones content. (B) IAA hormones. (C) JA hormones. (D) ABA hormones. Data shows the means ± standard deviation (SD) of three replicates. * and ** represent p < 0.05 and p < 0.01, respectively.
Overexpression of AsTCP17 in Arabidopsis improved genes of the ABA pathway
AtSVP, AtLOS5, AtNCED3 and AtAO are important genes for the ABA synthesis. The expression level of key ABA synthesis genes confirmed that AtSVP and AtNCED3 expression was obviously increased in the overexpression lines, with the OE6 line showing a 20.6-times and 7.0-times increase, respectively, while AtLOS5 and AtAO showed a non-significant increase (Fig. 7). Our findings demonstrated that overexpression of the AsTCP17 gene led to increased levels of expression of AtSVP and AtNCED3, especially AtSVP, which further proved that AsTCP17 is involved in ABA synthesis pathway by regulating the transcription level of AtSVP and AtNCED3.
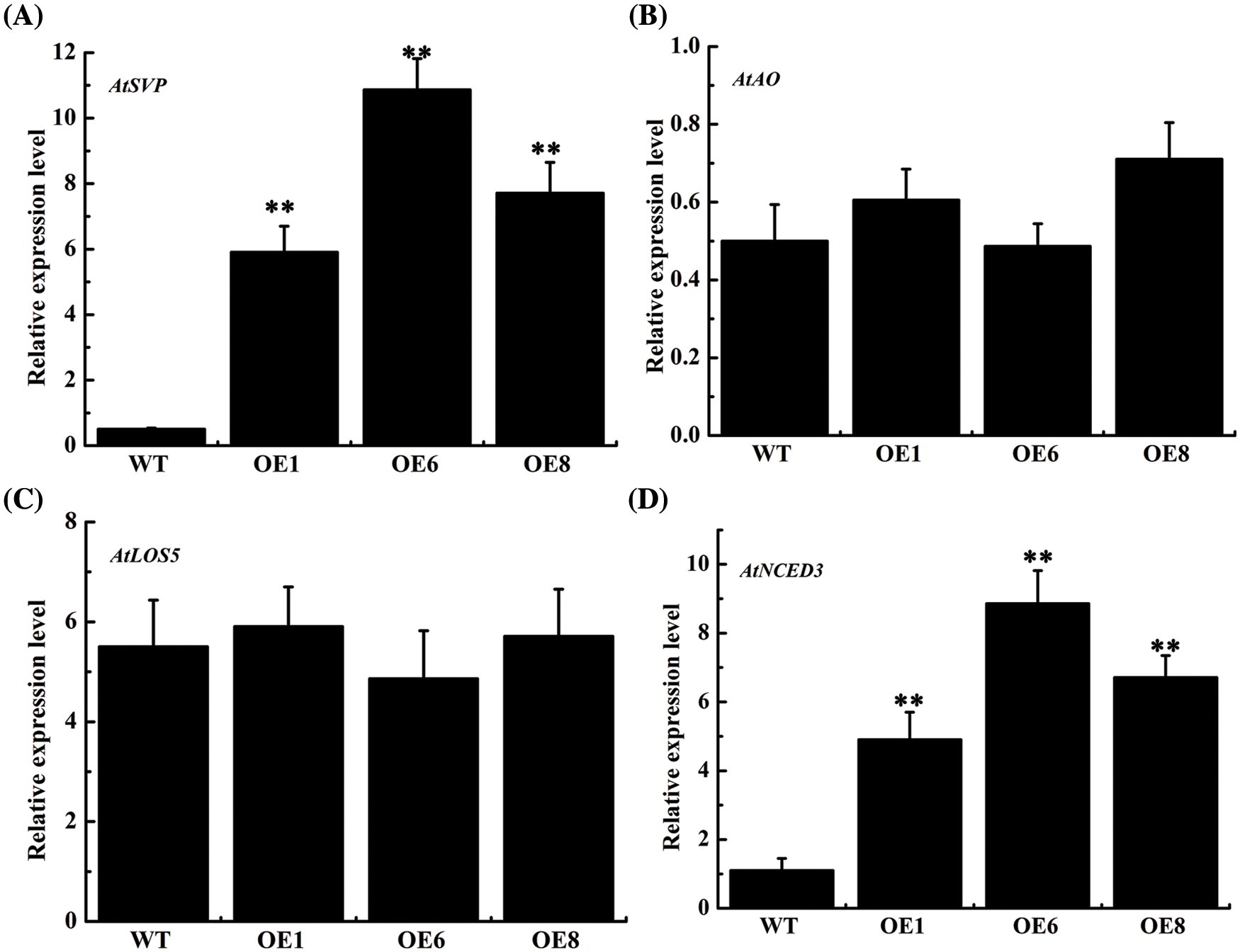
Figure 7: Expression of (A) AtSVP. (B) AtAO. (C) AtLOS5. (D) AtNCED3 associated with the ABA biosynthetic pathways. Data shows the means ± standard deviation (SD) of three replicates. ** represent p < 0.01, respectively.
Allium senescens is an important component part with the functions of the greening of urban and landscaping constructions in urban landscape systems. It also has obvious characteristics of drought resistance and light resistance. Thus, it is feasible for it to be planted at an elevation of 2000 m, whether in grasslands, meadows, or mountain slopes (Arimboor and Arumughan, 2012). In this study, we found that AsTCP17 was upregulated to increase the drought-resistance of Allium senescens, by identifying AsTCP17 expression levels, cell membrane permeability, protection enzymes, and drought-related hormones. Moreover, AsTCP17 can upregulate the AtSVP expression, a key transcription factor gene in the ABA pathway, to enhance the drought-tolerance of the Allium senescens.
The TCP family could activate regulation-related genes involved in a multitude of biotic and abiotic stress responses (Krasensky and Jonak, 2012). The ZmTCP42 of maize acts as a key gene, which plays an important role in the drought-resist of maize (Ding et al., 2019). Overexpression of VuTCP9 in cowpeas (Vigna unguiculata) resulted in greater relative water content (RWC) and less production of ROS, and exhibited strong resistance to salinity and drought tolerance (Mishra et al., 2021). In our study, the AsTCP17 expression was increased under drought stress, such that we postulate that the AsTCP17 protein was involved in the drought resistance of Allium senescens.
Plants could accumulate defense enzyme species and organelles damage when subjected to drought (Sies et al., 2022), while plants that are well-equipped a with defense system could decrease drought stress via multiple biochemical and physiological reactions (Bauer, 2021). In this study, the accumulation of protection-related enzymes was enhanced to a greater degree in the AsTCP17 overexpressed transgenic Arabidopsis than in the wild-type. In addition, previous studies have shown that plants ‘defense is activated by the production of hormone species, mainly represented by polyphenol oxidase, peroxidase, and protease inhibitors (Sies, 2017). Taken together, we conclude that the reason why the protection-related enzymes in transgenic plants changed might be that the expression of AsTCP17 affects a key signaling pathway.
Hormones are believed to play an important role in the complicated signaling process that is initiated in response when plants are subjected to drought stress (Rudu et al., 2009, Antualpa et al., 2017). A study on the root system proteomics of two different cultivated wheat varieties (Triticum aestivum) showed that there was an obvious increase in the number of ABA-responsive proteins for drought resistance of roots, in response to drought stress (Alvarez et al., 2014). In our study, while the ABA content was remarkably increased, there were no significant changes in the JA, IAA, and SA contents of transgenic plants that had been subjected to drought stress. Therefore, the promotion of ABA synthesis might be due to the overexpression of AsTCP17 in transgenic plants. Overall, the ABA biosynthesis pathway is activated, and ABA content is increased, to alleviate drought which induced the AsTCP17 expression.
Abscisic acid (ABA) is a 15-carbon sesquiterpenoid derived from the oxidative cleavage of epoxycarotenoids (Jie et al., 2017). In this study, we further identified the expression pattern of ABA genes, AtSVP, AtAO, AtLOS5 and AtNCED3, of which the level of expression of AtSVP in transgenic Arabidopsis showed the most obvious increase. Therefore, we think that there is a correlation between AsTCP17 expression and the AtSVP expression, which needs to be further studied. The findings indicate that the AsTCP17 gene could be a critical regulator of drought resistance in Allium senescens, and also provide important information about the theoretical mechanism for further studies on molecular mechanisms of drought tolerance in Allium senescens.
Resistance to drought in perennial herbs is usually overlooked and few works have been carried out that specifically investigate the mediation of transcription factors for Allium senescens’ resistance to drought stress. Our findings showed that AsTCP17, as a transcription factor, was upregulated in Allium senescens in response to drought treatment, and is the first evidence of the molecular mechanism that reveals the function of AsTCP17 in responding to drought stress in Allium senescens. Our results could supply the important drought-tolerance genes to the molecular breeding of perennial herbs, and also provide a deeper understanding of about plant-adversity interaction.
Funding Statement: This work was supported by the Hebei Grass Industry Innovation Team of the Modern Agricultural Industry Technology System (HBCT2018050204).
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Xiaohong Fu, Jie Zhao; data collection: Yunmei Zhang; analysis and interpretation of results: Ziyi Wang, Chenxing He, Yibei Jiang; draft manuscript preparation: Jianfeng Liu, Guixia Liu and Dandan Cao. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
Aimar D, Calafat M, Andrade AM, Carassay L, Bouteau F, Abdala G, Molas ML (2011). Drought effects on the early development stages of Panicum virgatum L.: Cultivar differences. Field Crops Research 120: 262–270. https://doi.org/10.1016/j.biombioe.2014.03.004 [Google Scholar] [CrossRef]
Alvarez S, Choudhury SR, Pandey S (2014). Comparative quantitative proteomics analysis of the ABA response of roots of drought-sensitive and drought-tolerant wheat varieties identifies proteomic signatures of drought adaptability. Journal of Proteome Research 13: 1688–1701. https://doi.org/10.1021/pr401165b [Google Scholar] [PubMed] [CrossRef]
Antualpa K, Marcelo SA, Moreira A (2017). Salivary steroids hormones, well-being, and physical performance during an intensification training period followed by a tapering period in youth rhythmic gymnasts. Physiology Behavior 179: 1–8. https://doi.org/10.1016/j.physbeh.2017.05.021 [Google Scholar] [PubMed] [CrossRef]
Arimboor R, Arumughan C (2012). Effect of polymerization on antioxidant and xanthine oxidase inhibitory potential of sea buckthorn (H. rhamnoides) proanthocyanidins. Journal of Food Science 77: 1036–1041. https://doi.org/10.1111/j.1750-3841.2012.02884.x [Google Scholar] [PubMed] [CrossRef]
Bauer G (2021). Inhibition of membrane-associated catalase, extracellular ROS/RNS signaling and aquaporin/H2O2-Mediated intracellular glutathione depletion cooperate during apoptosis induction in the human gastric carcinoma cell line MKN-45. Antioxidants 10: 1585. https://doi.org/10.3390/ANTIOX10101585 [Google Scholar] [PubMed] [CrossRef]
Beretta HV, Bannoud F, Insani M, Berli F, Cavagnaro PF (2017). Relationships among bioactive compounds content and the antiplatelet and antioxidant activities of six sllium vegetable species. Food Technology and Biotechnology 55: 266–275. https://doi.org/10.17113/tb.55.02.17.4722 [Google Scholar] [CrossRef]
Bio Sigui Bruno B, Ayemene Cedrick Ardin K, Kan Kouassi Parfait K, Doudjo S (2020). Effects of onion bulb processing conditions on drying characteristics, physicochemical and functional properties profile of onion (Allium cepa L.) powder. Journal of Food Science 85: 3345–3354. https://doi.org/10.1111/1750-3841.15415 [Google Scholar] [PubMed] [CrossRef]
Cakir BC, Arican E, Zhang B (2016). Small RNA and degradome deep sequencing reveals drought and tissue-specific microRNAs and their important roles in droughtsensitive and drought-tolerant tomato genotypes. Plant Biotechnology Journal 14: 1727–1746. https://doi.org/10.1111/pbi.12533 [Google Scholar] [PubMed] [CrossRef]
Chen XH, Xu Y, Liu H, Li Q, Kang XK (2017). Root/Stem anatomical characteristics of four Malus plants in western Sichuan plateau and their drought adaptation strategy. Acta Botanica Boreali-Occidentalia Sinica 37: 1296–1302. https://doi.org/10.7606/j.issn.1000-4025.2017.07.1296 [Google Scholar] [CrossRef]
Cubas P, Lauter N, Doebley J, Coen E (1999). The TCP domain: A motif found in proteins regulating plant growth and development. The Plant Journal 18: 215–222. https://doi.org/10.1046/J.1365-313X.1999.00444.X [Google Scholar] [PubMed] [CrossRef]
Dandona P, Ghanim H, Chaudhuri A, Dhindsa S, Kim SS (2010). Macronutrient intake induces oxidative and inflammatory stress: Potential relevance to atherosclerosis and insulin resistance. Experimental and Molecular Medicine 42: 245–253. https://doi.org/10.3858/emm.2010.42.4.033 [Google Scholar] [PubMed] [CrossRef]
Danisman S, Wal F, Dhondt S, Waites R, Immink R (2012). Arabidopsis class I and class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiology 159: 1511–1523. https://doi.org/10.1104/pp.112.200303 [Google Scholar] [PubMed] [CrossRef]
Ding SC, Cai ZZ, Du HW, Wang HW (2019). Genome-wide analysis of TCP family genes in Zea mays. identified a role for ZmTCP42 in drought tolerance. International Journal of Molecular Sciences 20: 2762. https://doi.org/10.3390/ijms20112762 [Google Scholar] [PubMed] [CrossRef]
Fryer MJ (2002). Imaging of photo-oxidative stress responses in leaves. Journal of Experimental Botany 53: 1249–1254. https://doi.org/10.1093/jxb/53.372.1249 [Google Scholar] [CrossRef]
Jie F, Hua W, Ma S, Xiang D, Liu R, Xiong L (2017). OsJAZ1 attenuates drought resistance by regulating JA and ABA signaling in rice. Frontiers in Plant Science 8: 2108. https://doi.org/10.3389/fpls.2017.02108 [Google Scholar] [PubMed] [CrossRef]
Jongrungklang N, Toomsan B, Vorasoot N, Jogloy S, Boote KJ, Hoogenboom G, Patanothai A (2011). Rooting traits of peanut genotypes with different yield responses to pre-flowering drought stress. Field Crops Research 120: 262–270. https://doi.org/10.1016/j.fcr.2010.10.008 [Google Scholar] [CrossRef]
Krasensky J, Jonak C (2012). Drought, salt, and temperature stress-induced metabolicre arrangements and regulatory networks. Journal of Experimental Botany 63: 1593–1608. https://doi.org/10.1093/jxb/err460 [Google Scholar] [PubMed] [CrossRef]
Kukushkina T, Fomina T (2021). The content of biologically active substances in the green biomass of perennial onions (Allium L.). Agrarian Bulletin of the Technogenic 207: 85–92. https://doi.org/10.32417/1997-4868-2021-207-04-85-92 [Google Scholar] [CrossRef]
Lee JY, Seo KH, Lee EY, Ji YJ, Lee YJ, Kang MH, Seong HA, Kim HD (2021). Antioxidant and anti-obesity potentials of Korean-Native wild vegetables (Allium species). Horticulturae 7: 541. https://doi.org/10.3390/horticulturae7120541 [Google Scholar] [CrossRef]
Liu MM, Wang MM, Yang J, Wen J, Du H (2019). Evolutionary and comparative expression analyses of TCP transcription factor gene family in land plants. International Journal of Molecular Sciences 20: 3591. https://doi.org/10.3390/ijms20143591 [Google Scholar] [PubMed] [CrossRef]
Ma J, Wang Q, Sun R, Xie F, Jones DC, Zhang B (2014). Genome-wide identification and expression analysis of TCP transcription factors in Gossypium raimondii. Scientific Reports 4: 6645. https://doi.org/10.1038/srep06645 [Google Scholar] [PubMed] [CrossRef]
Martin-Trillo M, Cubas P (2010). TCP genes: A family snapshot ten years later. Trends in Plant Science 15: 31–39. https://doi.org/10.1016/j.tplants.2009.11.003 [Google Scholar] [PubMed] [CrossRef]
Mishra S, Sahu G, Shaw BP (2021). Insight into the cellular and physiological regulatory modulations of class-I TCP9 to enhance drought and salinity stress tolerance in cowpea. Physiologia Plantarun 6: 1–15. https://doi.org/10.1111/PPL.13542 [Google Scholar] [PubMed] [CrossRef]
Mukhopadhyay P, Tyagi AK (2015). OsTCP19 influences developmental and abiotic stress signaling by modulating ABI4-mediated pathways. Science Letter 5: 9998. https://doi.org/10.1038/srep09998 [Google Scholar] [PubMed] [CrossRef]
Rudu I, Weiler EW, KPczyńska E (2009). Do stress-related phytohormones, abscisic acid and jasmonic acid play a role in the regulation of Medicago sativaL. somatic embryogenesis. Plant Growth Regulation 59: 159–169. https://doi.org/10.1007/s10725-009-9399-3 [Google Scholar] [CrossRef]
Sies H (2017). Hydrogen peroxide as a central redox signaling molecule in physiological oxidative stress: Oxidative eustress. Redox Biology 11: 613–619. https://doi.org/10.1016/j.redox.2016.12.035 [Google Scholar] [PubMed] [CrossRef]
Sies H, Belousov VV, Chandel NS, Davies MJ, Jones DP, Mann GE, Murphy MP, Yamamoto M, Winterbourn C (2022). Defining roles of specific reactive oxygen species (ROS) in cell biology and physiology. Nature Reviews Molecular Cell Biology 23: 1–17. https://doi.org/10.1038/S41580-022-00456-Z [Google Scholar] [PubMed] [CrossRef]
Wang M, Jiang B, Peng QW, Liu WR, He XM, Liang ZJ, Lin Y (2018). Transcriptome analyses in different cucumber cultivars provide novel insights into drought stress responses. Journal of Molecular Sciences 19: 2067. https://doi.org/10.3390/ijms19072067 [Google Scholar] [PubMed] [CrossRef]
Yao X, Ma H, Wang J, Zhang D (2010). Genome-wide comparative analysis and expression pattern of TCP gene families in Arabidopsis thaliana and Oryza sativa. Journal of Integrative Plant Biology 49: 885–897 https://doi.org/10.1111/j.1744-7909.2007.00509.x [Google Scholar] [CrossRef]
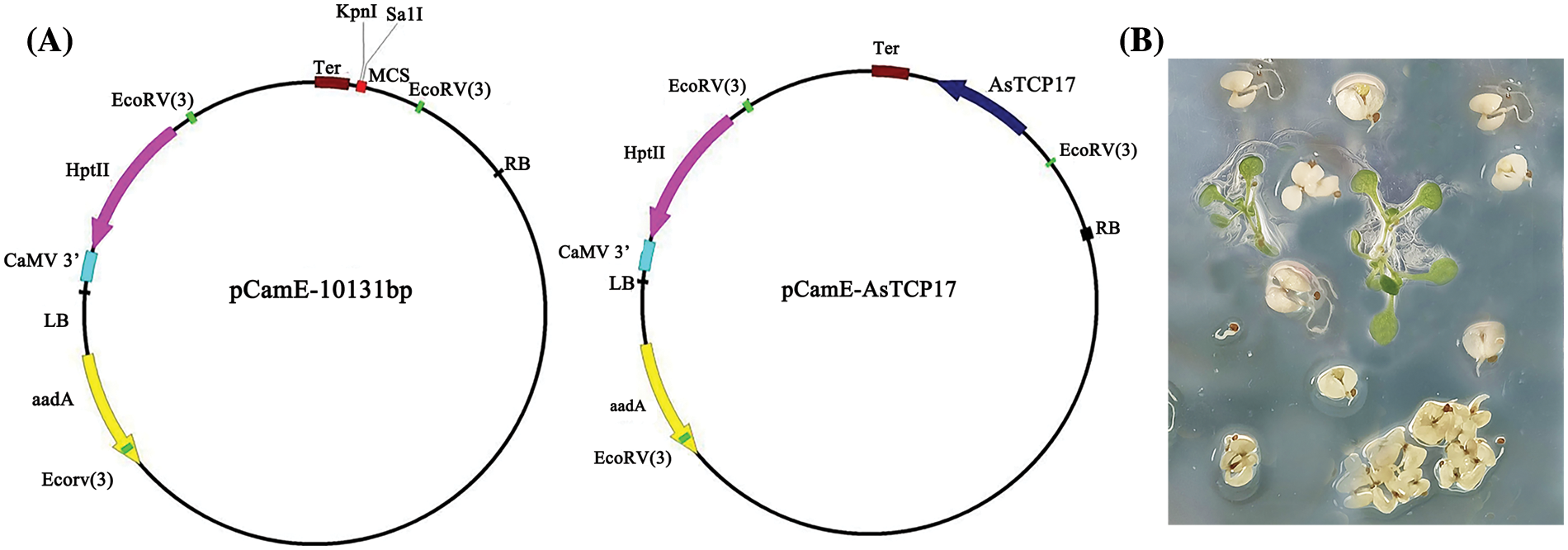
Figure S1: The map of pCam-AsTCP17 vector (A) and transgenic lines selected on the Hygromycin medium (B).

Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools