 Open Access
Open Access
ARTICLE
Single-cell sequencing analysis reveals the molecular mechanism of promotion of SCAP proliferation upon AZD2858 treatment
1 Beijing Laboratory of Oral Health, School of Stomatology, Capital Medical University, Beijing, 100050, China
2 Department of Neurobiology, School of Basic Medical Sciences, Capital Medical University, Beijing, 100069, China
3 Department of VIP Dental Service, School of Stomatology, Capital Medical University, Beijing, 100050, China
4 Laboratory for Oral and General Health Integration and Translation, Beijing Tian Tan Hospital, Capital Medical University, Beijing, 100070, China
* Corresponding Author: JIAN ZHOU. Email:
# These authors contributed equally to this article
(This article belongs to the Special Issue: Cell-Based Regenerative Therapies)
BIOCELL 2023, 47(4), 825-836. https://doi.org/10.32604/biocell.2023.026122
Received 17 August 2022; Accepted 28 November 2022; Issue published 08 March 2023
Abstract
The Wnt/β-catenin signaling pathway is the main target of tooth regeneration regulation. Treatment of cells with AZD2858 stimulates the Wnt/β-catenin signaling pathway, yet the function of this pathway in tooth regeneration remains unclear. Here, we found that AZD2858 promotes the accumulation of β-catenin in the nuclei of stem cells from the apical papilla (SCAPs) and enhances cell proliferation. Single-cell sequencing was performed on SCAPs treated with AZD2858. Eight clusters were identified, namely SCAPs-CNTNAP2, SCAPs-DTL, SCAPs-MYH11, SCAPs-MKI67, SCAPs-CXCL8, SCAPs-TPM2, SCAPs-IFIT2 and SCAPs-NEK10. The pseudo-time trajectory analysis showed that AZD2858 enhanced the evolution of SCAPs from SCAPs-TMP2 clusters to SCAPs-MYH11, SCAPs-CNTNAPs and SCAPs-NEK10 clusters via up-regulation of PRKCA, SMURF2, MAGI2, RBMS3, EXT1, CAMK2D, PLCB4, and PLCB1. These results demonstrate that AZD2858 enhances the proliferation of SCAPs-TPM2 cluster by activating the non-canonical Wnt/β-catenin signaling pathway.Keywords
Tissue engineering with mesenchymal stem cells offers a novel strategy for the regeneration of dental pulp, periodontal tissue, and teeth (Hu et al., 2018). The importance of the Wnt signaling pathway in mesenchymal stem cells for tooth development has been shown in previous studies. Wnt proteins regulate cell growth, development, migration, and differentiation of cells during embryonic development, maintain the multidirectional differentiation ability of stem cells, and determine tissue polarity. In addition to enhancement of the osteogenic differentiation of periodontal fibroblasts, up-regulation of the Wnt signaling pathway also inhibits the differentiation of cementoblasts while promoting their proliferation (Hermans et al., 2021; Duan and Bonewald, 2016; Tamura and Nemoto, 2016; Bao et al., 2021). In the canonical Wnt pathway, Wnt proteins act as ligands to activate Wnt membrane surface receptors, thereby inhibiting the β-catenin degradation complex that includes the GSK3β, and enabling the presence of free β-catenin in the cytoplasm. This results in β-catenin accumulation and promotion of the expression of downstream target genes in the nucleus (Tanaka et al., 2019). The Wnt pathway is also an evolutionarily conserved signaling pathway involved in the regulation of cell differentiation, migration, polarization, and differentiation during embryonic development, and the maintenance of tissue homeostasis. We previously showed that Wnt/β-catenin signaling plays a critical role in mediating the interaction between mesenchymal and epithelial cells during tooth development (Yang et al., 2014). We also showed that Wnt3a, a ligand of the Wnt/β-catenin signaling pathway, recruits endogenous cells that regenerate the neurovascular stroma, and triggers their differentiation into parenchymal odontoblast-like cells that extend the process into the generation of newly formed dentin in the root canal of mandibular incisors in miniature pigs (He et al., 2019). These findings suggest that the Wnt/β-catenin signaling pathway is the main target of tooth regeneration regulation. However, Wnt3a needs to be produced as a recombinant protein for practical applications that are likely to suffer from certain limitations, such as strong immunogenicity and poor specificity. Alternatively, small molecules with rapid, reversible, and safe biological effects have been widely used in stem cell reprogramming and tissue regeneration. The development of small-molecule compounds that specifically target the Wnt/β-catenin signaling pathway to enhance tissue regeneration has thus become a hot topic in recent years.
Several Wnt/β-catenin signaling agonists, such as CHIR99021, tideglusib, and tivantinib, have been reported to promote dentin regeneration. AZD2858 is another Wnt/β-catenin signaling pathway stimulant that inhibits the activity of the axin/APC/GSK-3 complex by suppressing GSK-3, thereby decreasing the phosphorylation of β-catenin and resulting in the accumulation of nuclear β-catenin (Gao et al., 2017). AZD2858 has a favorable biocompatibility profile owing to its low IC50 level. Oral AZD2858 treatment was also shown to promote osteogenic differentiation in rodent bone marrow MSC, bone formation, and callus formation in a fracture (Sisask et al., 2013). This suggests that AZD2858 is also a potent agent to promote dentinogenesis as well as endodontic and periodontal tissue regeneration by activating canonical Wnt signaling. Here, we investigated the effect of AZD2858 treatment on the biological functions of stem cells from the apical papilla (SCAPs). Single-cell sequencing was employed to characterize changes in the signaling cascade and gene expression profiles. Our findings provide valuable insights into the effects of AZD2858 and other Wnt agonists on the role of SCAPs in tooth and periodontal tissue regeneration.
Our study was approved by the ethics committee of Beijing Stomatological Hospital, Capital Medical University (Approval No.: KQYY-201707-002). All procedures involving human participants comply with the ethical standards of the research committee. Informed consent was obtained from all participants.
Sample source and cell culture
Tooth and tissue were obtained with informed patient consent upon approval by the Beijing Stomatological Hospital, Capital Medical University. The crowns of teeth were rubbed gently with 75% ethanol and then stored in sterile 4°C phosphate-buffered saline (PBS) (Invitrogen, Carlsbad, USA) solution after the teeth were separated from the extraction socket. Apical papilla was gently scraped from the apical one-third of immature teeth. Cells were cultured in an incubator set with 5% CO2 and at 37°C. Alpha-modified Eagle’s medium (α-MEM) (Invitrogen, Carlsbad, USA) supplemented with 15% fetal bovine serum (ExCell Bio, Taicang, China), 100 U/mL penicillin, and 100 μg/mL streptomycin (Invitrogen, Carlsbad, USA) was chosen to culture cells, and which was changed every other day. Cells were passaged after reaching 80% confluence. The SCAPs with passage numbers below 15 were used in the following studies. To ensure that the cultured cells were SCAPs, surface antigens of mesenchymal stem cells were identified by flow cytometry (Suppl. Fig. S1).
For 5-ethynyl-20-deoxyuridine (EdU) assays, SCAPs (3 × 104 cells/well) were incubated for 2 h with EdU (Ribobio, Guangzhou, China). After fixation in 4% paraformaldehyde for 30 min and induction with 0.5% Triton X-100 for 20 min at room temperature, cells were treated with 1× Apollo reaction cocktail for 30 min. Subsequently, the DNA was stained with Hoechst 33342 (Thermo Fisher Scientific, Waltham, USA) for 30 min and visualized with a fluorescence microscope.
2 × 105 SCAPs were seeded in each well of a six-well plate and cultured in α-MEM containing 15% fetal bovine serum until confluence. The medium was replaced without serum, then incubated at 37°C for 20 min. Subsequently, 1000 μL pipette tips were used to create three scratches per well, and then washed with PBS. Images of each scratch were taken under an inverted microscope at 0 and 24 h. The pixel areas of the scratches in the images were measured with BioTek Lionheart FX. Finally, the average widths of the scratches were calculated for each group.
The nuclear extracts of SCAPs were prepared with NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Scientific, Waltham, USA, product No. 78833). 2.0 × 105 SCAPs were collected by centrifugation. The pelleted cells were resuspended in 40 μl of Cytoplasmic Extraction Reagent I, incubated on ice for 10 min, mixed with 2.2 μl Cytoplasmic Extraction Reagent II and incubated on ice for 1 min. The lysates were spun at 16,000 g at 4°C for 5 min. The cytoplasmic extract was collected from the supernatant fraction, and the pellets were resuspended in 20 μl of Nuclear Extraction Reagent. The samples were kept on ice and vortexed for 15 s every 10 min, for a total of 40 min. The suspension was then spun at 16,000 g at 4°C for 10 min. The nuclear extract was collected from the supernatant fraction.
Cells were incubated in 60 μl of cell lysis buffer and 1:100 proteinase inhibitor cocktail for 15 min on ice to remove the insoluble fraction. The collected liquid was heated at 70°C in the buffer consisting of 10× sample reduction buffer and 4× lithium dodecyl sulfate sample buffer. Then, the samples were added to SDS-PAGE (10% Bis-Tris gels, Life Technology) for 120 min with 120 V and transferred to nitrocellulose membranes. The membranes were blotted with 5% dehydrated milk and incubated overnight with primary antibodies against GAPDH (ABclonal, Wuhan, China, AC002, 1:10000), β-catenin (ABclonal, Wuhan, China, A19657, 1:1000), HDAC2 (ABclonal, Wuhan, China, A2084, 1:1000), and CDK5 (ABclonal, Wuhan, China, A18080, 1:1000). Membranes were incubated with secondary antibody (IRDye 680LT goat anti-mouse IgG and 800CW goat anti-Rabbit IgG, Odyssey, Jiangsu, China) for 1 h at room temperature. Signals were detected by Infrared Imaging System (Odyssey, Jiangsu, China).
The SCAPs were fixed with 4% paraformaldehyde for 30 min. 0.5% Triton was applied for permeabilization. Cells were blocked with 5% BSA, and incubated with primary antibodies to β-catenin (Abcam, Cambridge, UK, 1:200) for more than 8 h at 4°C. Then the cells were incubated with secondary antibodies (1:500, Thermo Scientific, Waltham, USA) for 1 h in a wet box at room temperature. The cells were next incubated with 4′-6-diamidino-2-phenylindole for 10 min to stain nuclei. Fluorescence images were captured with an Olympus fluorescence microscope (Olympus, Tokyo, Japan).
Single-Cell Sequencing Analysis
10× genomics detection and reverse transcription
Prepared single cell suspensions, 10× barcode gel magnetic beads and oil were added to different chambers of a Chromium Chip B, and GEMs formed through the microfluidic “double cross” system. The GEMs were subjected to reverse transcription and coagulation by PCR. The magnetic beads contained a 30 nt oligo-dT reverse transcription primer, which allowed the poly-A RNA in the cells to be reverse transcribed into cDNA with barcode and UMI information. The magnetic beads were used to purify the cDNA. The purified cDNA was amplified by PCR.
Sequencing library construction
After cDNA amplification was completed, the fragments were digested, and the most suitable fragments were collected with magnetic beads. The read2 sequencing primers were connected through end repair, A and adaptors, and a cDNA library containing P5 and P7 adaptors was constructed by PCR. The library was purified with magnetic beads. A Qubit instrument was used to detect the library concentration, and an Agilent 2100 instrument was used to detect the fragment size.
According to the Illumina User Guide, cluster generation and first-direction sequencing primer hybridization were completed, and the flow cell with the cluster was input into the paired-end program used to perform paired-end sequencing. The sequencing process was controlled by Illumina data collection software, and a real-time data analysis was performed.
Screening of differentially expressed genes
To obtain a list of the differentially expressed genes (DEGs) for each cluster, we only considered expressing more than 30% of the genes in that cluster, against the background of expression of all other cells. For statistical testing, we used the default Wilcoxon test implemented in Seurat. DEGs are defined as genes with a logarithmic change of more than 0.2 and a q value (FDR) of less than 0.05 relative to the background.
Single-cell trajectory analysis and cell-cell interaction analysis
For SCAPs, pseudotime analysis in Monocle was performed on the basis of highly variable genes identified with Seurat. Line plots were constructed in pseudotime order with fitted curves with the “auto” method from the ggplot2 package. Cell-cell interaction analysis was performed with CellChat in R.
Analysis between two groups was performed with unpaired two-tailed Student’s t-tests. For more than two groups, one-way analysis of variance (ANOVA) with Tukey’s multiple comparison post hoc test was performed only if F achieved p < 0.05.
AZD2858 treatment leads to an increase in β-catenin level in SCAPs
Human SCAPs were cultivated in media containing 1, 10, 50, or 100 nM of AZD2858 or LiCl (a GSK-3 non-selective inhibitor). β-catenin levels in whole-cell preparations of SCAPs were found to increase upon treatment with 50 and 100 nM AZD2858 (Figs. 1A and 1B). We also isolated the nuclei from SCAPs and determined β-catenin levels in each isolated fraction to test whether AZD2858 also leads to an increase in β-catenin levels in the nucleus. Western blotting results revealed a significant increase in β-catenin levels in nuclei of SCAPs upon treatment with 10, 50, and 100 nM AZD2858, whereas no statistical difference was detected in the cytoplasm compared to the control group (no AZD2858 treatment) (Figs. 1C and 1D). In addition, immunofluorescence analysis confirmed the accumulation of β-catenin in the nucleus (Figs. 1E and 1F).
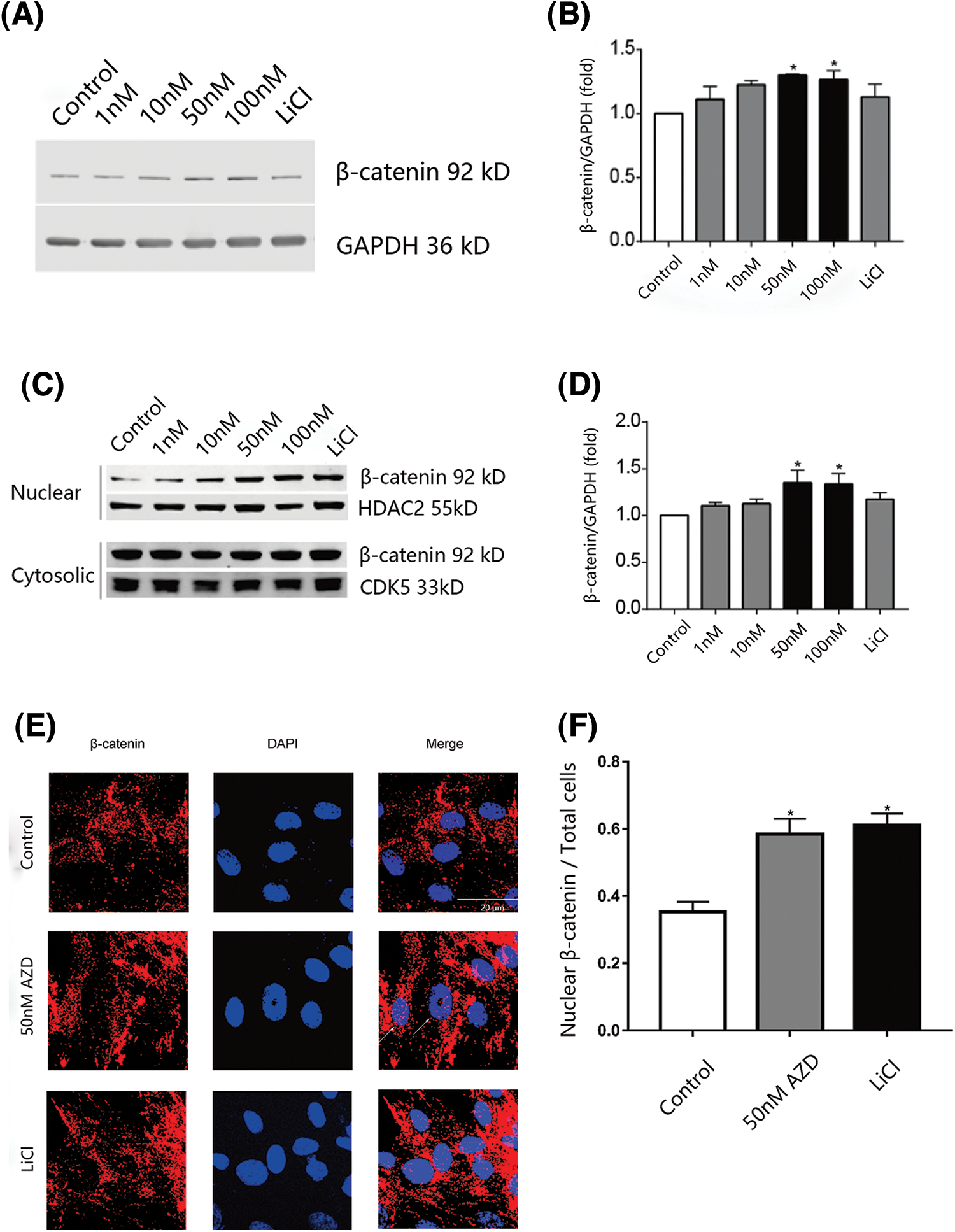
Figure 1: β-catenin nuclear levels are increased in SCAPs treated with AZD2858. Western blot results showed that AZD2858 treatment led to increased expression of β-catenin in SCAPs. GAPDH was used as a reference (A). Quantitative analysis of β-catenin levels based on western blot results (B). AZD2858 treatment was found to increase β-catenin levels in the nuclei of SCAPs. Western blot results indicated that AZD2858 treatment increased the level of nuclear β-catenin in SCAPs. HDAC2 and CDK5 were used as internal nuclear and cytosolic controls, respectively (C). Quantitative analysis of β-catenin levels based on western blot results (D). Immunofluorescence (IF) of β-catenin (red) and 4′-6-diamidino-2-phenylindole (blue) in the control, AZD 50 nM and LiCl groups. The white arrow indicates β-catenin expressed in the nucleus (E). Relative IF expression levels of β-catenin in different groups (F). One-way ANOVA and Student’s t-test were performed to determine statistical significance. All error bars represent SEM (n = 3). *p < 0.05.
AZD2858 treatment enhances the proliferation of SCAPs
We also monitored changes in the proliferation and migration of SCAPs upon AZD2858 treatment. Results of the EdU and CCK8 assays showed that AZD2858 caused an increase in SCAP proliferation (Figs. 2A–2C). The migration ability of SCAPs was not significantly enhanced or weakened by AZD2858 in scratch tests (Fig. 2D), suggesting that AZD2858 is not a chemotactic or chemokinetic agent.
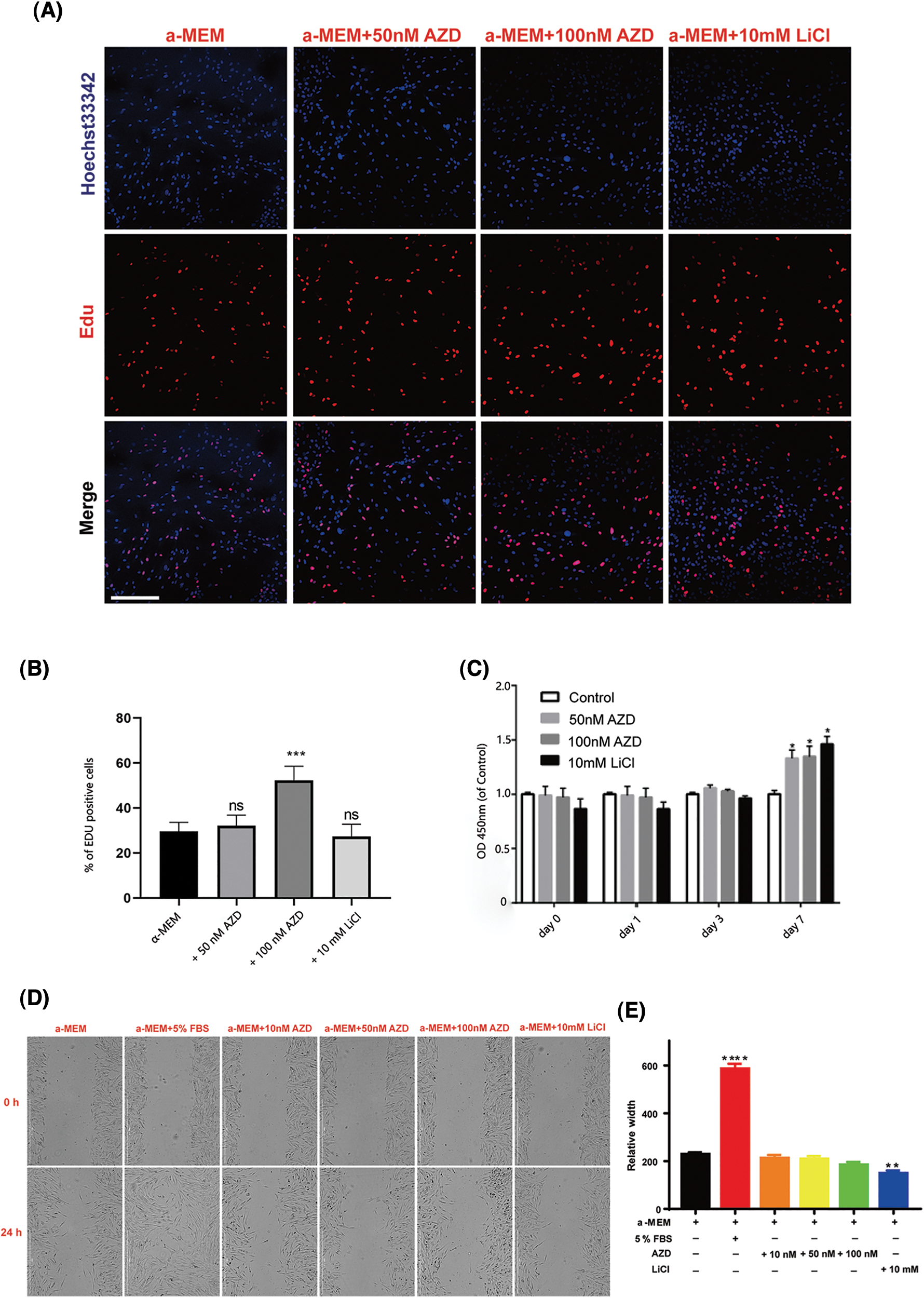
Figure 2: Effects of AZD2858 on the proliferation of SCAPs. EdU assays demonstrated that AZD2858 leads to increased proliferation of SCAPs at 100 nM 24 h after administration (A, B). CCK8 assays showed the effects of different concentrations of AZD2858 and LiCl on SCAP proliferation over 7 days (C). Scratch wound migration assay results showed that AZD2858 had no influence on wound healing (D, E). Scale bar: 100 μm. Values are mean ± SEM of triplicate samples from a representative experiment. One-way ANOVA was performed to compare treatments. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Global transcriptional profiling of SCAPs treated with AZD2858
We performed single-cell sequencing to determine the transcriptional profile of SCAPs treated with AZD2858. SCAPs collected from the teeth of patients were cultured in α-MEM with and without AZD2858, and named SCAP-blank and SCAP-AZD2858 groups, respectively. Cell clusters obtained by dimensionality reduction via principal component analysis were analyzed using the uniform manifold approximation and projection algorithm (UMAP). We successfully removed batch effects from the data using canonical correlation analysis, as evidenced by our observation that the cells from the three samples were almost mixed when projected onto the 2D UMAP space. Genes that showed the highest expression level differences between cells were identified in the eight SCAP clusters obtained. CNTNP2, DTL, MYH11, CXCL8, TPM2, IFIT2, and NEK10 were found to be marker genes in SCAP-AZD2858 and SCAP-blank clusters (Fig. 3A). A total of 24 genes with significant differences in expression were identified, and the expression levels of these genes in each cluster are displayed (Fig. 3B). The distribution of each marker gene in the whole-cell population is also shown in Figs. 3C to 3J.
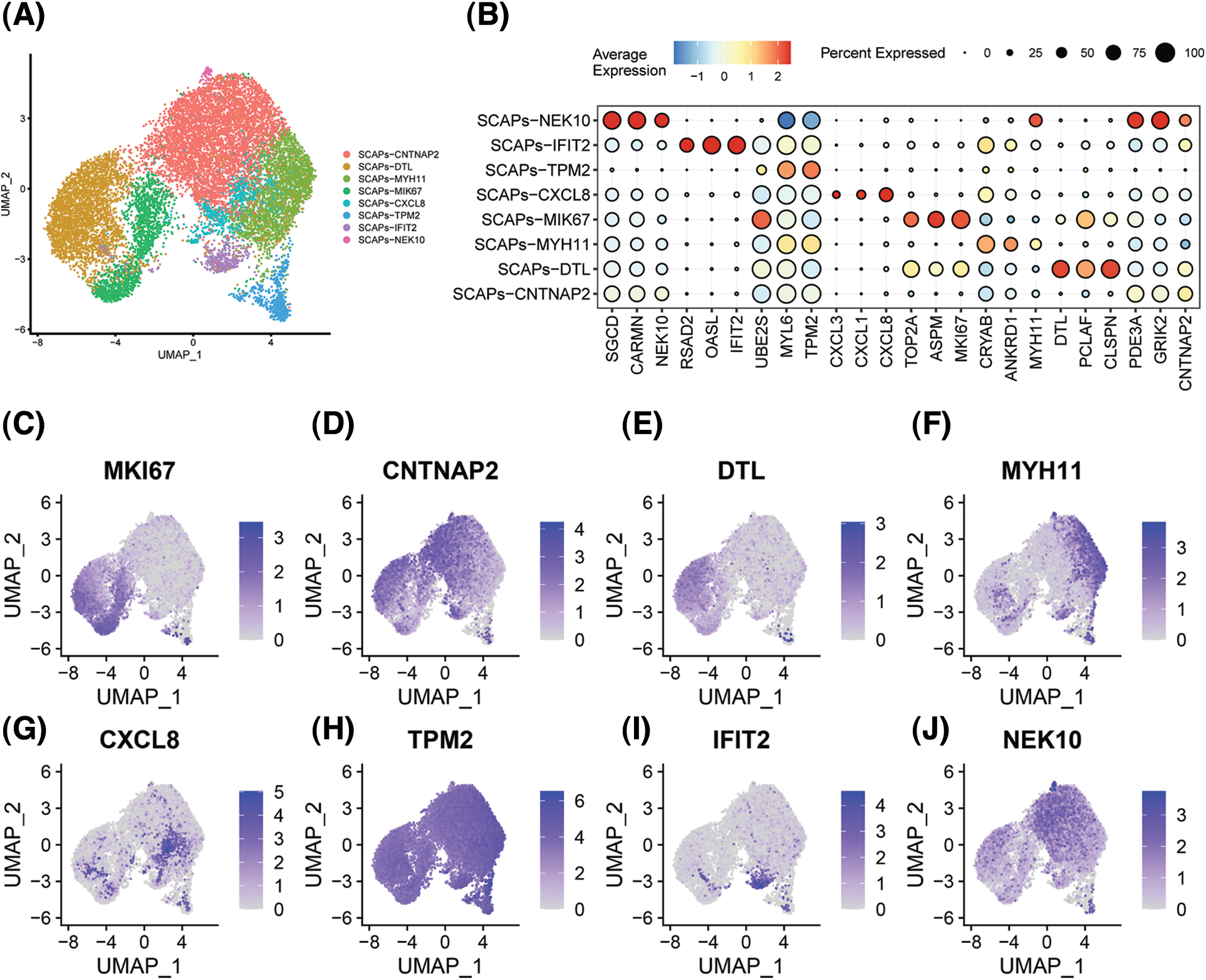
Figure 3: Heterogeneity and representative genes in SCAPs. The overall composition of cells from the SCAPs visualized with UMAP projection after canonical correlation analysis, colored based on clustering results (A). Clustering heat map of the top 3 DEGs for each single cluster. Each column in the figure represents a gene, and each row represents a cluster. Each colored area in the heat map represents the average expression level of the corresponding gene in the cluster. The color key from red to blue indicates high to low expression. The dot size indicates the percentage of cells expressing genes in the right legend (B). The distribution of the top nine expressed genes of whole cells is displayed separately (C to J).
Next, we examined the portion of the main clusters in the SCAP-blank and SCAP-AZD2858 groups (Fig. 4A). The cell-cell interaction analysis results are shown in Fig. 4B. The close interaction between clusters might have been due to the interaction between SCAP-CNTNP2 which acted as a ligand cell and SCAP-CNTNP2 and SCAP-DTL which acted as a receptor cell. Moreover, the SCAPs-TPM2 cluster was observed to include a relatively isolated cell population, which suggests that SCAPs-TPM2 may act as an independent cluster to regulate AZD2858 enhancing the proliferation of SCAPs.
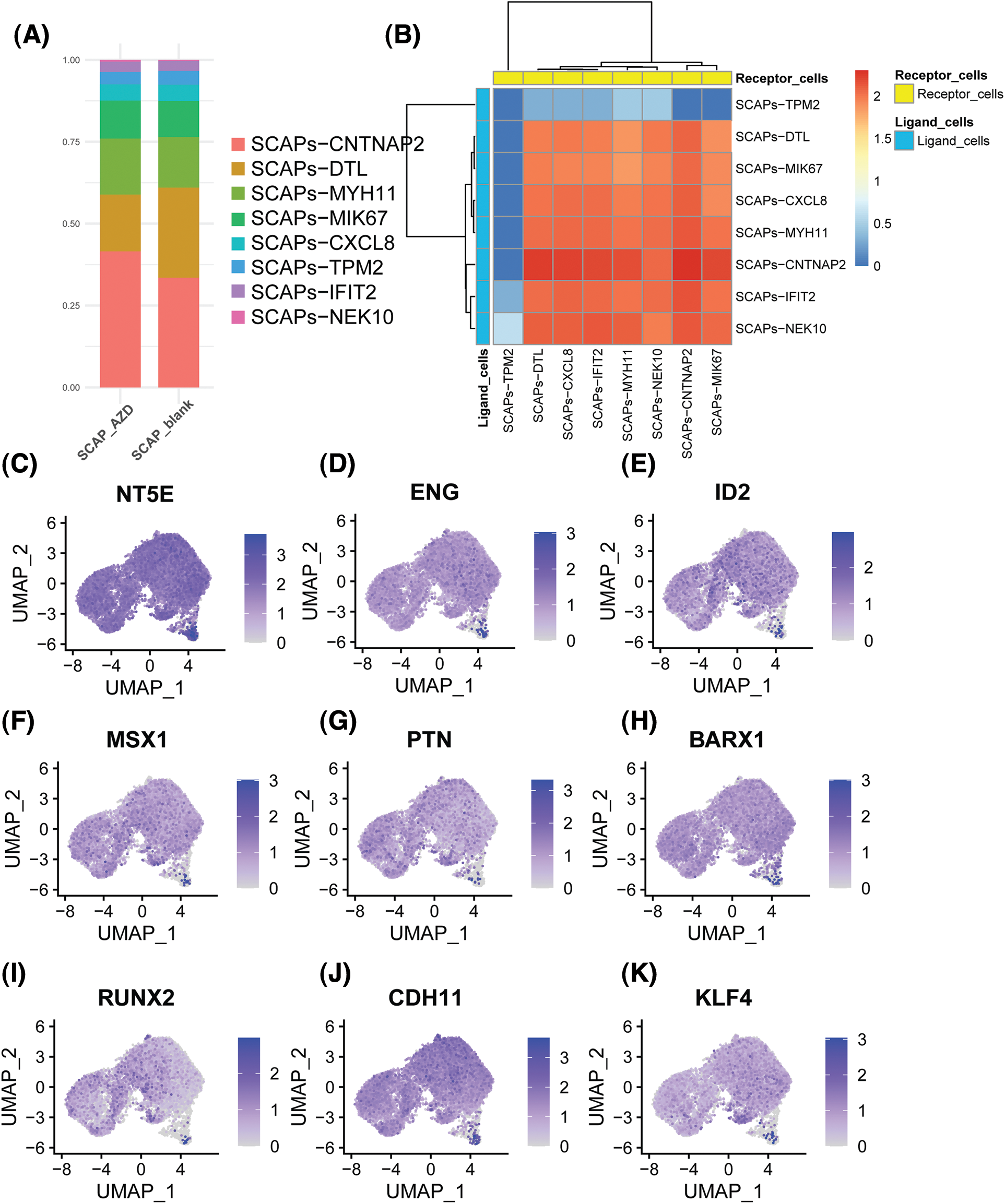
Figure 4: Histogram showing the proportion of each cluster type in the SCAPs-AZD2858 and SCAPs-blank (A). Cell-cell interaction of SCAPs treated with AZD2858. Yellow represents receptor cells, and blue represents ligand cells. The color key from red to blue indicates high to low possibilities (B). The distribution of the expression of stem cell characteristic genes in the whole cell population (C to E). The distribution of the expression of odontogenesis characteristic genes in the whole cell population (F to H). The distribution of the expression of osteogenesis characteristic genes in the whole cell population (I to K).
Next, we determined the expression levels of genes that characterize stemness and odonto/osteogenesis (Figs. 4C to 4K). We found that the stemness genes 5′-nucleotidase ecto (NT5E), endoglin (ENG), and inhibitor of DNA binding 2 (ID2) were expressed at relatively higher levels in the SCAPs-TPM2 cluster. However, msh homeobox 1 (MSX1), pleiotrophin (PTN), BARCX1, RUNX2, CDH11 and KLF4 odonto/osteogenesis genes were found to be only expressed in the SCAP-CNTNP2 cluster.
Single-cell trajectories of SCAPs treated with AZD2858
Monocle was used to determine the temporal order of the SCAPs treated with AZD2858 along a pseudotime trajectory. Unsupervised pseudotime analysis indicated the developmental trajectories of the SCAP-blank and SCAP-AZD groups (Figs. 5A and 5B). Pseudotime ordering yielded the main trajectory with five minor bifurcations. The cell developmental trajectory is shown from right to left along the x-axis. Significant differences in the second bifurcation are observed between the trajectories of SCAP-blank and SCAP-AZD2858 groups. In addition, we observed a possible cell developmental trajectory in the second bifurcation with SCAPs-TMP2, SCAPs-MYH11, SCAPs-CNTNAPs, and SCAPs-NEK10. The trajectories of SCAPs-TMP2, SCAPs-MYH11, SCAPs-CNTNAPs, and SCAPs-NEK10 clusters demonstrated the evolution of different clusters as well (Fig. 5C). Major genes expressed at the second bifurcation of each cluster are shown next to the trajectory map.
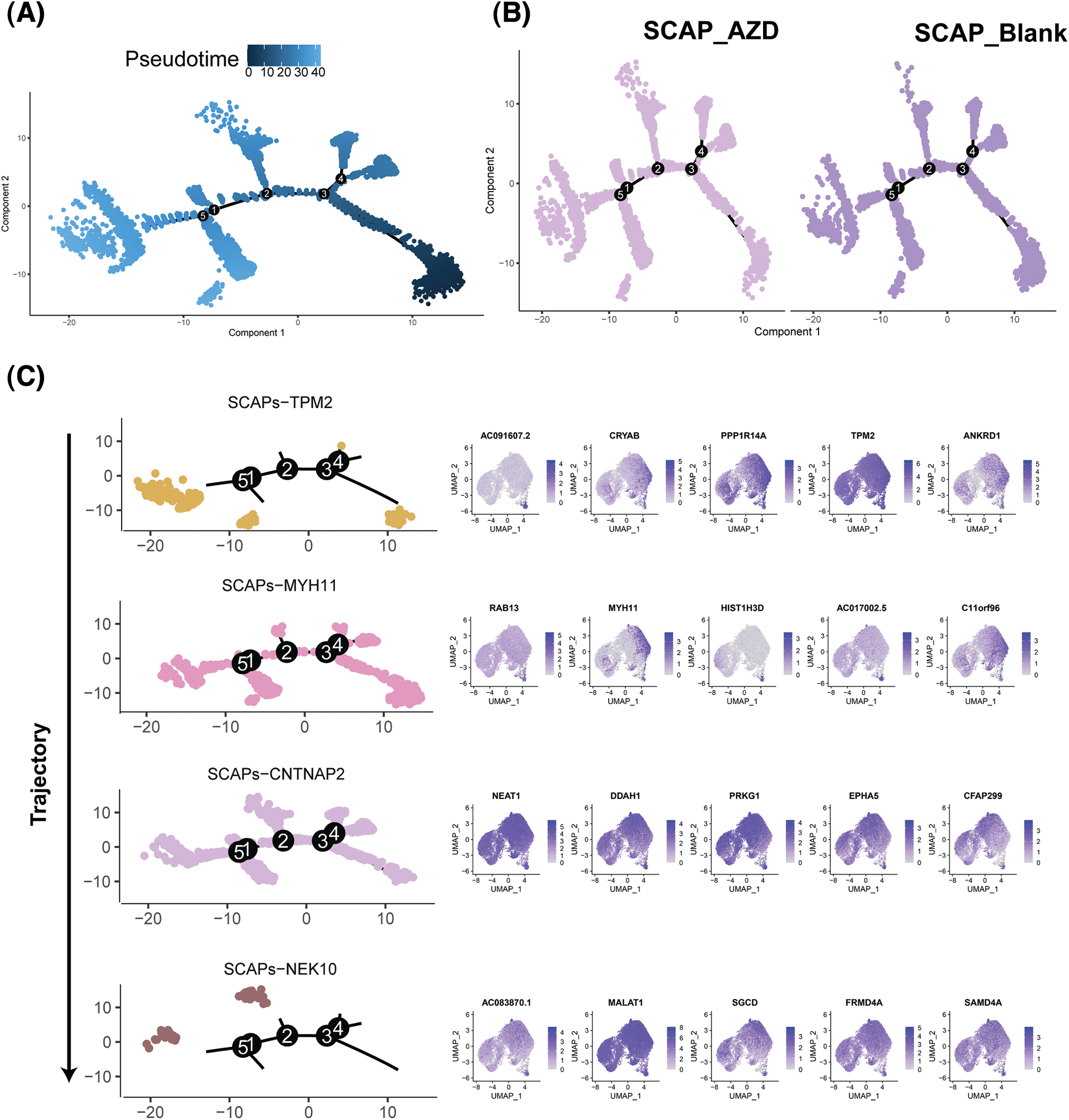
Figure 5: Single-cell trajectories of SCAPs treated with AZD2858. Developmental pseudotime analysis of SCAPs treated with AZD2858. (A) Dark to light blue represents the beginning to the end of the time period. Cells developed along the X-axis from right to left. (B) Developmental trajectories of three groups of cells: SCAP-AZD (left), SCAP-blank (right). (C) Developmental trajectories of the four clusters: SCAPs-TPM2, SCAPs-MYH11, SCAPs-CNTNAP2 and SCAPs-NEK10. The arrow on the left indicates the direction of cell population development. The top five expressed genes in each cluster are shown on the right side of the trajectory diagram.
AZD2858 leads to activation of SCAPs-TMP2 cluster through the canonical Wnt signal pathway
To further investigate the mechanism underlying enhancement of cell proliferation by AZD2858, we determined the up- and down-regulated genes in SCAPs-blank and SCAPs-AZD2858 groups from the cell clusters obtained. Differentially expressed genes between the clusters are displayed in a Venn diagram (Figs. 6A and 6B). Accordingly, PRKCA, SMURF2, MAGI2, RBMS3, EXT1, CAMK2D, PLCB4, and PLCB1 were found to be upregulated in the SCAPs-NEK10 cluster. CTNNB1 was found to be upregulated in the SCAPs-TPM2 cluster. EXT1, PRKCA, PLCB1, and PLCB4 were found to be downregulated in SCAPs-TPM2 and SCAPs-MYH11 clusters. Finally, CAMK2D and RBMS3 were found to be downregulated in the SCAPs-MYH11 cluster. The genes involved in the Wnt signaling pathway were also determined. Accordingly, CTNNB1 (Moroney et al., 2021), EXT1 (Wang et al., 2019), RBMS3 (Wang et al., 2020), MAGI2 (Xu et al., 2021), and MAGI3 (Yang et al., 2021) are involved in the canonical Wnt/β-catenin signaling pathway, whereas PLCB1, PLCB4, PRKCA (Colli et al., 2013), CAMK2D (Zhuang et al., 2017), and SMURF2 (Bernatik et al., 2020) are involved in the non-canonical pathway.
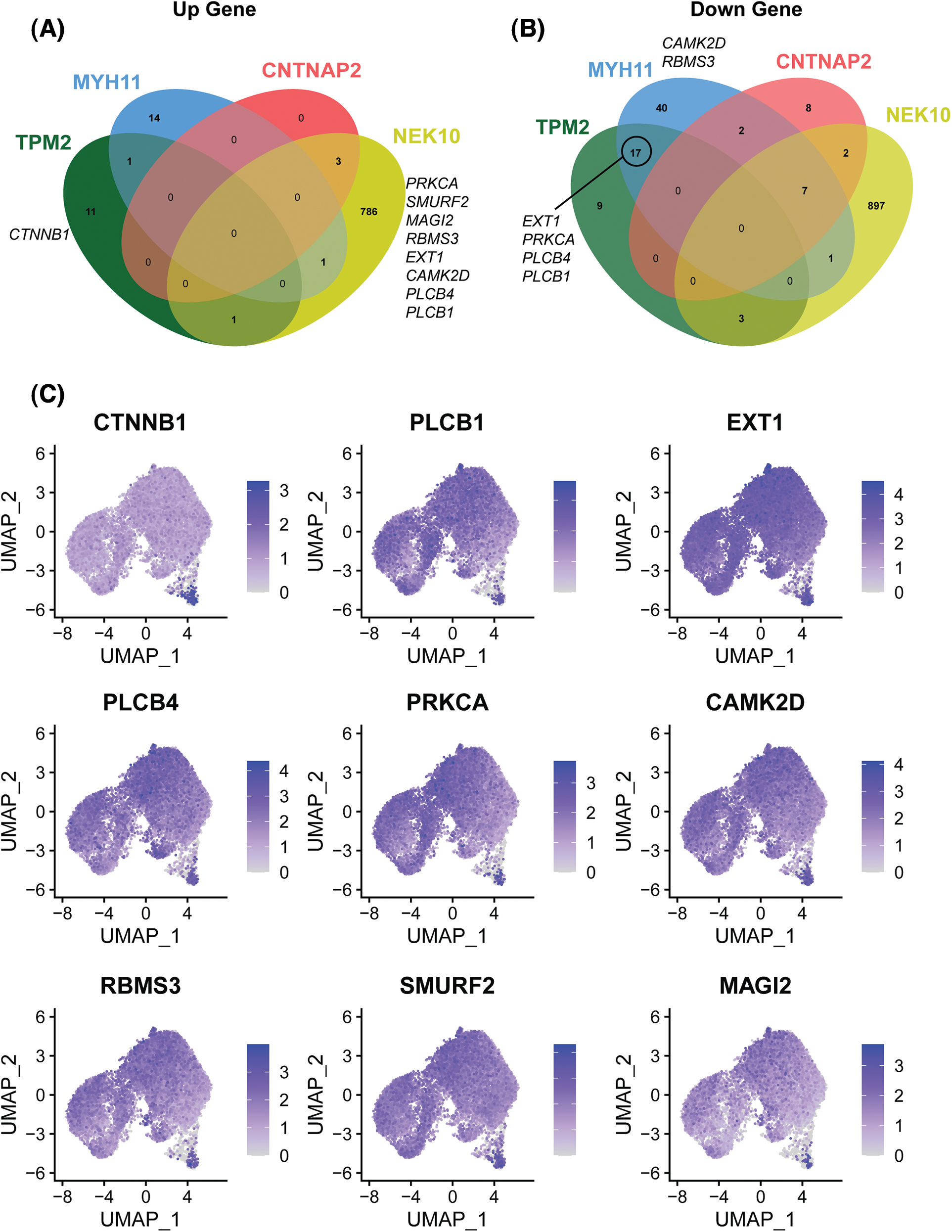
Figure 6: Up- and down-regulated genes of the four clusters associated with the trajectory of cell development between SCAPs-blank and SCAPs-AZD2858 groups (A and B). Distribution of up-and down-regulated expressed genes related to canonical and non-canonical Wnt signaling pathway in whole cells (C).
In conclusion, the developmental trajectory of SCAPs (Fig. 5), and the results of gene expression regulation revealed that the genes expression profile in SCAPs shifted from genes involved in the canonical Wnt/β-catenin signaling pathway to those involved in the non-canonical Wnt/β-catenin signaling pathway during cell development.
The Wnt/β-catenin signaling pathway plays an important role in tooth development (Duan and Bonewald, 2016). This pathway regulates tooth morphology by affecting the proliferation, differentiation, and polarization of odontoblasts and odontogenic mesenchymal cells. Here, we further showed that the Wnt/β-catenin signaling pathway recruits endogenous MSCs, and facilitates tooth regeneration in miniature pigs, suggesting a critical role of Wnt/β-catenin signaling pathway in tissue regeneration.
Small molecules are compounds with molecular weights less than 1000 Da. Compared to biological agents, small molecules can freely pass through the cell membrane, and regulate intracellular signalling pathways. In addition, small molecules exhibit rapid, reversible, and dose-dependent biological effects. In general, the transformation of small molecular structures and optimization of their properties may be easily achieved to increase their drug-likeliness. Time profiles of small molecule release from container materials can be accurately adjusted by modifying their concentration or by combining with other biomaterials. Small molecules also provide advantages including low immunogenicity and toxicity, in addition to easier storage conditions and standardized production procedures. Previous studies have revealed that the Wnt/β-catenin signaling pathway agonists CHIR99021 and Tideglusib induce the formation of thin dentin bridges, which seal pulp perforation and protect the pulp tissue. These molecules were also found to induce dentin regeneration in a rat molar pulp penetration model with a larger tooth size (Zaugg et al., 2020) and pulp penetration hole (Alaohali et al., 2022). In addition, tivantinib, a Wnt/β-catenin signaling pathway agonist used to inhibit the proliferation of liver cancer cells, was found to induce dentin regeneration at the medullary foramen in rats. These studies hint at a novel strategy for tooth regeneration through the activation of the Wnt/β-catenin signaling pathway by small molecule compounds. AZD2858 is a Wnt/β-catenin pathway activator that leads to an increase in β-catenin levels in the nucleus by inhibiting GSK-3β. AZD2858 also promotes osteogenic differentiation of human adipose stem cells (hADSCs) cultured in vitro. Previous in vivo experiments also showed that AZD2858 enhances bone anabolism, and accelerates fracture healing (Sisask et al., 2013). More importantly, AZD2858 was found to show negligible antigenicity at effective concentrations, compared to biological macromolecule alternatives. However, only a few studies have examined the effects of AZD2858 in vivo and in vitro. Cell proliferation is a characteristic of tissue regeneration by stem cells since high proliferation rates provide a cytological foundation for tissue regeneration engineering. Our results suggested that AZD2858 effectively increases the proliferation of dental-derived stem cells, indicating that AZD2858 is a potential regulator of stem cell proliferation.
We also analyzed the expression of genes that characterize stemness and odonto/osteogenesis by conducting single-cell sequencing experiments (Figs. 4C to 4K). The protein encoded by NT5E is a plasma membrane protein that catalyzes the conversion of extracellular nucleotides to membrane-permeable nucleosides, also known as CD73. CD73 is a common surface marker of mesenchymal stem cells (Obara et al., 2016; Zhai et al., 2021). The protein encoded by ENG is involved in the regulation of angiogenesis and is required for the structural integrity of adult vasculature. The role of endoglin in stem cells was first characterized in human hematopoietic stem cells (Pham et al., 2018). Endoglin was subsequently reported to be expressed in mesenchymal stem cells as well. Endoglin has also been identified as a marker for mesenchymal stem cells (Uder et al., 2018; Pham et al., 2018). The protein encoded by ID2 belongs to the inhibitor of the DNA binding family, which includes transcriptional regulators that contain a helix-loop-helix (HLH) domain. ID2 has also been shown to be an early target of osteogenic BMP signaling, and its products may play an important role in regulating BMP-induced bone formation. ID2 is also involved in cell proliferation and differentiation (Peng et al., 2004; Tajima et al., 2007). MSX1 acts as a transcriptional repressor, and plays a vital role in craniofacial development, during odontogenesis particularly (Tucker et al., 1998). In a study of dental pulp mesenchymal stem cells from humans (hDPSCs), MSX1 was shown to be essential for the osteoblast-like differentiation and calcification of hDPSCs in primary teeth (Goto et al., 2016). Therefore, MSX1 is a marker appropriate for the evaluation of the odontogenic and osteogenic abilities of cells. PTN is a heparin-binding growth factor that regulates various intracellular processes such as cell proliferation, survival, growth, differentiation, and migration in several tissues, including neurons and bones. A previous study showed that PTN is synthesized by osteoblasts at an early stage of osteogenic differentiation and can be found in sites of new bone formation, where the PTN is stored in bone matrix (Imai et al., 2021; Tare et al., 2009). In another study, the ALP activity and mineralization ability of DPSCs were found to decrease after PTN depletion, whereas the expression levels of DMP-1 and BSP were also found to decrease. The proliferation of DPSCs was inhibited at 48 and 72 h. Hence, PTN was found to promote the maintenance of DPSCs proliferation and osteo/dentinogenic differentiation potential (Jin et al., 2021). Homeobox protein BarH-like 1 (BARX1) is a transcription factor involved in craniofacial development and odontogenesis (Tucker et al., 1998; Sperber and Dawid, 2008). BARX1 has previously been used as a marker of DPSC presence (Macrin et al., 2019). Cadherin 11 (CDH11) is an integral membrane protein that mediates calcium-dependent cell-cell adhesion. The expression of CDH11 in osteoblastic cell lines, and its upregulation during cell differentiation suggest that CDH11 plays a specific role in bone development and maintenance (di Benedetto et al., 2010). Di Benedetto et al. (2015) found that CDH11 was expressed in undifferentiated DBSCs, and the expression level increased during osteogenesis. KLF transcription factor 4 (KLF4) is a member of the Kruppel-like factor (KLF) family, and plays a key role in proliferation, differentiation, and apoptosis during development. Chen et al. (2009) showed that the expression of KLF4 is closely correlated with growth arrest and the first step of odontoblast and ameloblast differentiation. Huang et al. (2022), Yu et al. (2021), and Tao et al. (2019) found that KLF4 is involved in the regulation of odontoblastic commitment of dental papilla cells and osteoblast/odontoblastic differentiation. Here, we found that these genes are highly expressed in the SCAP-CNTNAP2 cluster. This cluster is likely to play a major role in SCAPs treated with AZD2858, since it accounts for the highest proportion of total cells. Although some of these genes were also highly expressed in individual cells of the SCAP-TMP2 cluster, we reason that this cluster does not play a pivotal role in SCAP treated with AZD2858.
Eight cell clusters were identified by single-cell sequencing of SCAP treated with AZD2858. The SCAPs-TPM2 cluster was particularly interesting, as it appeared as a relatively distinct group of cells. In addition, the SCAPs-TPM2 cluster may act as progenitor cells to influence the differentiation direction of cells in the developmental trajectory of cells.
When SCAPs were treated with AZD2858, the SCAPs-TPM2 cluster was activated and continued to develop along cells in the SCAPS-TPM2, SCAPS-MYH11, SCAPS-CNTNAPS, and SCAPS-NEK10 clusters. Upon activation of SCAPs-TPM2, the canonical Wnt signaling pathway was also promoted. As the cell clusters continued to develop, the non-canonical Wnt signaling pathway was also promoted, resulting in planar cell polarity and promotion of cell proliferation via the Wnt/Ca2+ signaling pathway.
In summary, here, we demonstrate that AZD2858 activates the Wnt/β-catenin pathway and enhances cell proliferation by increasing the accumulation of β-catenin in the nuclei of dental-derived MSCs. The single-cell sequencing results of SCAPs treated with AZD2858 also suggest that AZD2858 activates the SCAPs-TPM2 cluster as a progenitor cell cluster. The non-canonical Wnt signaling pathway was further activated to induce cell development along the trajectories of SCAPS-TMP2, SCAPS-MYH11, SCAPS-CNTNAPS, and SCAPS-NEK10 clusters.
Funding Statement: The authors would like to thank the fund of National Natural Science Foundation of China (82170951) and Beijing Natural Science Foundation (7222079).
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Jian Zhou and Lei Hu; data collection: Yifan Xu and Dongmei Cheng; analysis and interpretation of results: Xin Dong, Liying Lv and Chen Zhang; draft manuscript preparation: Yifan Xu and Dongmei Cheng. All authors reviewed the results and approved the final version of the manuscript.
Ethics Approval: Our study was approved by the ethics committee of Beijing Stomatological Hospital, Capital Medical University (Approval No. KQYY-201707-002). All procedures involving human participants comply with the ethical standards of the research committee. Informed consent was obtained from all participants.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
Alaohali A, Salzlechner C, Zaugg LK, Suzano F, Martinez A, Gentleman E, Sharpe PT (2022). GSK3 inhibitor-induced dentinogenesis using a hydrogel. Journal of Dental Research 101: 46–53. https://doi.org/10.1177/00220345211020652 [Google Scholar] [PubMed] [CrossRef]
Bao J, Yang Y, Xia M, Sun W, Chen L (2021). Wnt signaling: An attractive target for periodontitis treatment. Biomedicine & Pharmacotherapy 133: 110935. https://doi.org/10.1016/j.biopha.2020.110935 [Google Scholar] [PubMed] [CrossRef]
Bernatik O, Paclikova P, Sri Ganji R, Bryja V (2020). Activity of Smurf2 ubiquitin ligase is regulated by the wnt pathway protein dishevelled. Cells 9: 1147. https://doi.org/10.3390/cells9051147 [Google Scholar] [PubMed] [CrossRef]
Castle JC, Loewer M, Boegel S, de Graaf J, Bender C et al. (2014). Immunomic, genomic and transcriptomic characterization of CT26 colorectal carcinoma. BMC Genomics 15: 190. https://doi.org/10.1186/1471-2164-15-190 [Google Scholar] [PubMed] [CrossRef]
Chen Z, Couble ML, Mouterfi N, Magloire H, Chen Z, Bleicher F (2009). Spatial and temporal expression of KLF4 and KLF5 during murine tooth development. Archives of Oral Biology 54: 403–411. https://doi.org/10.1016/j.archoralbio.2009.02.003 [Google Scholar] [PubMed] [CrossRef]
Colli LM, Saggioro F, Serafini LN, Camargo RC, Machado HR, Moreira AC, Antonini SR, de Castro M, Luque RM (2013). Components of the canonical and non-canonical Wnt pathways are not mis-expressed in pituitary tumors. PLoS One 8: e62424. https://doi.org/10.1371/journal.pone.0062424 [Google Scholar] [PubMed] [CrossRef]
di Benedetto A, Brunetti G, Posa F, Ballini A, Grassi FR et al. (2015). Osteogenic differentiation of mesenchymal stem cells from dental bud: Role of integrins and cadherins. Stem Cell Research 15: 618–628. https://doi.org/10.1016/j.scr.2015.09.011 [Google Scholar] [PubMed] [CrossRef]
di Benedetto A, Watkins M, Grimston S, Salazar V, Donsante C, Mbalaviele G, Radice GL, Civitelli R (2010). N-cadherin and cadherin 11 modulate postnatal bone growth and osteoblast differentiation by distinct mechanisms. Journal of Cell Science 123: 2640–2648. https://doi.org/10.1242/jcs.067777 [Google Scholar] [PubMed] [CrossRef]
Duan P, Bonewald LF (2016). The role of the wnt/β-catenin signaling pathway in formation and maintenance of bone and teeth. The International Journal of Biochemistry & Cell Biology 77: 23–29. https://doi.org/10.1016/j.biocel.2016.05.015 [Google Scholar] [PubMed] [CrossRef]
Gao L, Chen B, Li J, Yang F, Cen X, Liao Z, Long X, Castresana JS (2017). Wnt/β-catenin signaling pathway inhibits the proliferation and apoptosis of U87 glioma cells via different mechanisms. PLoS One 12: e0181346. https://doi.org/10.1371/journal.pone.0181346 [Google Scholar] [PubMed] [CrossRef]
Goto N, Fujimoto K, Fujii S, Ida-Yonemochi H, Ohshima H, Kawamoto T, Noshiro M, Shukunami C, Kozai K, Kato Y (2016). Role of MSX1 in osteogenic differentiation of human dental pulp stem cells. Stem Cells International 2016: 8035759. https://doi.org/10.1155/2016/8035759 [Google Scholar] [PubMed] [CrossRef]
He L, Zhou J, Chen M, Lin CS, Kim SG et al. (2019). Parenchymal and stromal tissue regeneration of tooth organ by pivotal signals reinstated in decellularized matrix. Nature Materials 18: 627–637. https://doi.org/10.1038/s41563-019-0368-6 [Google Scholar] [PubMed] [CrossRef]
Hermans F, Hemeryck L, Lambrichts I, Bronckaers A, Vankelecom H (2021). Intertwined signaling pathways governing tooth development: A give-and-take between canonical Wnt and Shh. Frontiers in Cell and Developmental Biology 9: 758203. https://doi.org/10.3389/fcell.2021.758203 [Google Scholar] [PubMed] [CrossRef]
Hu L, Liu Y, Wang S (2018). Stem cell-based tooth and periodontal regeneration. Oral Diseases 24: 696–705. https://doi.org/10.1111/odi.12703 [Google Scholar] [PubMed] [CrossRef]
Huang Z, Yang R, Li R, Zuo Y, Gu F, He M, Bian Z (2022). Mesenchymal Mycn participates in odontoblastic lineage commitment by regulating Krüppel-like Factor 4 (Klf4) in mice. Stem Cell Research & Therapy 13: 78. https://doi.org/10.1186/s13287-022-02749-8 [Google Scholar] [PubMed] [CrossRef]
Imai S, Heino TJ, Hienola A, Kurata K, Büki K, Matsusue Y, Väänänen HK, Rauvala H (2021). Osteocyte-derived HB-GAM (pleiotrophin) is associated with bone formation and mechanical loading. Bone 44: 785–794. https://doi.org/10.1016/j.bone.2009.01.004 [Google Scholar] [PubMed] [CrossRef]
Jin L, Gao F, Zhang L, Wang C, Hu L, Fan Z, Xia D (2021). Pleiotropin enhances the osteo/dentinogenic differentiation potential of dental pulp stem cells. Connective Tissue Research 62: 495–507. https://doi.org/10.1080/03008207.2020.1779238 [Google Scholar] [PubMed] [CrossRef]
Macrin D, Alghadeer A, Zhao YT, Miklas JW, Hussein AM et al. (2019). Metabolism as an early predictor of DPSCs aging. Scientific Reports 9: 2195. https://doi.org/10.1038/s41598-018-37489-4 [Google Scholar] [PubMed] [CrossRef]
Moroney MR, Woodruff E, Qamar L, Bradford AP, Wolsky R, Bitler BG, Corr BR (2021). Inhibiting Wnt/beta-catenin in CTNNB1-mutated endometrial cancer. Molecular Carcinogenesis 60: 511–523. https://doi.org/10.1002/mc.23308 [Google Scholar] [PubMed] [CrossRef]
Obara C, Takizawa K, Tomiyama K, Hazawa M, Saotome-Nakamura A, Gotoh T, Yasuda T, Tajima K (2016). Differentiation and molecular properties of mesenchymal stem cells derived from murine induced pluripotent stem cells derived on gelatin or collagen. Stem Cells International 2016: 9013089. https://doi.org/10.1155/2016/9013089 [Google Scholar] [PubMed] [CrossRef]
Peng Y, Kang Q, Luo Q, Jiang W, Si W et al. (2004). Inhibitor of DNA binding/differentiation helix-loop-helix proteins mediate bone morphogenetic protein-induced osteoblast differentiation of mesenchymal stem cells. The Journal of Biological Chemistry 279: 32941–32949. https://doi.org/10.1074/jbc.M403344200 [Google Scholar] [PubMed] [CrossRef]
Pham H, Tonai R, Wu M, Birtolo C, Chen M (2018). CD73, CD90, CD105 and cadherin-11 RT-PCR screening for mesenchymal stem cells from cryopreserved human cord tissue. International Journal of Stem Cells 11: 26–38. https://doi.org/10.15283/ijsc17015 [Google Scholar] [PubMed] [CrossRef]
Sisask G, Marsell R, Sundgren-Andersson A, Larsson S, Nilsson O, Ljunggren Ö, Jonsson KB (2013). Rats treated with AZD2858, a GSK3 inhibitor, heal fractures rapidly without endochondral bone formation. Bone 54: 126–132. https://doi.org/10.1016/j.bone.2013.01.019 [Google Scholar] [PubMed] [CrossRef]
Sperber SM, Dawid IB (2008). barx1 is necessary for ectomesenchyme proliferation and osteochondroprogenitor condensation in the zebrafish pharyngeal arches. Developmental Biology 321: 101–110. https://doi.org/10.1016/j.ydbio.2008.06.004 [Google Scholar] [PubMed] [CrossRef]
Tajima K, Terai S, Takami T, Kawaguchi K, Okita K, Sakaida I (2007). Importance of inhibitor of DNA binding/differentiation 2 in hepatic stellate cell differentiation and proliferation. Hepatology Research 37: 647–655. https://doi.org/10.1111/j.1872-034X.2007.00089.x [Google Scholar] [PubMed] [CrossRef]
Tamura M, Nemoto E (2016). Role of the Wnt signaling molecules in the tooth. The Japanese Dental Science Review 52: 75–83. https://doi.org/10.1016/j.jdsr.2016.04.001 [Google Scholar] [PubMed] [CrossRef]
Tanaka H, Kawaguchi M, Shoda S, Miyoshi T, Iwasaki R, Hyodo F, Mori T, Hara A, Tomita H, Matsuo M (2019). Nuclear accumulation of β-Catenin in cancer stem cell radioresistance and stemness in human colon cancer. Anticancer Research 39: 6575–6583. https://doi.org/10.21873/anticanres.13873 [Google Scholar] [PubMed] [CrossRef]
Tao H, Lin H, Sun Z, Pei F, Zhang J, Chen S, Liu H, Chen Z (2019). Klf4 promotes dentinogenesis and odontoblastic differentiation via modulation of TGF-β signaling pathway and interaction with histone acetylation. Journal of Bone and Mineral Research 34: 1502–1516. https://doi.org/10.1002/jbmr.3716 [Google Scholar] [PubMed] [CrossRef]
Tare RS, Oreffo ROC, Clarke NMP, Roach HI (2009). Pleiotrophin/Osteoblast-stimulating factor 1: Dissecting its diverse functions in bone formation. Journal of Bone and Mineral Research 17: 2009–2020. https://doi.org/10.1359/jbmr.2002.17.11.2009 [Google Scholar] [PubMed] [CrossRef]
Tucker AS, Matthews KL, Sharpe PT (1998). Transformation of tooth type induced by inhibition of BMP signaling. Science 282: 1136–1138. https://doi.org/10.1126/science.282.5391.1136 [Google Scholar] [PubMed] [CrossRef]
Uder C, Brückner S, Winkler S, Tautenhahn HM, Christ B (2018). Mammalian MSC from selected species: Features and applications. Cytometry Part A 93: 32–49. https://doi.org/10.1002/cyto.a.23239 [Google Scholar] [PubMed] [CrossRef]
Wang X, Cornelis FMF, Lories RJ, Monteagudo S (2019). Exostosin-1 enhances canonical Wnt signaling activity during chondrogenic differentiation. Osteoarthritis Cartilage 27: 1702–1710. https://doi.org/10.1016/j.joca.2019.07.007 [Google Scholar] [PubMed] [CrossRef]
Wang JJ, Liu XY, Du W, Liu JQ, Sun B, Zheng YP (2020). RBMS3 delays disc degeneration by inhibiting Wnt/β-catenin signaling pathway. European Review for Medical and Pharmacological Sciences 24: 499–507. https://doi.org/10.26355/eurrev_202001_20023 [Google Scholar] [PubMed] [CrossRef]
Xu X, Yuan X, Ni J, Guo J, Gao Y, Yin W, Li F, Wei L, Zhang J (2021). MAGI2-AS3 inhibits breast cancer by downregulating DNA methylation of MAGI2. Journal of Cellular Physiology 236: 1116–1130. https://doi.org/10.1002/jcp.29922 [Google Scholar] [PubMed] [CrossRef]
Yang Z, Liu H, Song R, Lu W, Wang H et al. (2021). Reduced MAGI3 level by HPV18E6 contributes to Wnt/β-catenin signaling activation and cervical cancer progression. FEBS Open Bio 11: 3051–3062. https://doi.org/10.1002/2211-5463.13298 [Google Scholar] [PubMed] [CrossRef]
Yang G, Zhou J, Teng Y, Xie J, Lin J, Guo X, Gao Y, He M, Yang X, Wang S (2014). Mesenchymal TGF-β signaling orchestrates dental epithelial stem cell homeostasis through Wnt signaling. Stem Cells 32: 2939–2948. https://doi.org/10.1002/stem.1772 [Google Scholar] [PubMed] [CrossRef]
Yu S, Guo J, Sun Z, Lin C, Tao H et al. (2021). BMP2-dependent gene regulatory network analysis reveals Klf4 as a novel transcription factor of osteoblast differentiation. Cell Death & Disease 12: 197. https://doi.org/10.1038/s41419-021-03480-7 [Google Scholar] [PubMed] [CrossRef]
Zaugg LK, Banu A, Walther AR, Chandrasekaran D, Babb RC, Salzlechner C, Hedegaard MAB, Gentleman E, Sharpe PT (2020). Translation approach for dentine regeneration using GSK-3 antagonists. Journal of Dental Research 99: 544–551. https://doi.org/10.1177/0022034520908593 [Google Scholar] [PubMed] [CrossRef]
Zhai X, Chen K, Yang H, Li B, Zhou T et al. (2021). Extracellular vesicles derived from CD73 modified human umbilical cord mesenchymal stem cells ameliorate inflammation after spinal cord injury. Journal of Nanobiotechnology 19: 274. https://doi.org/10.1186/s12951-021-01022-z [Google Scholar] [PubMed] [CrossRef]
Zhuang X, Zhang H, Li X, Li X, Cong M, Peng F, Yu J, Zhang X, Yang Q, Hu G (2017). Differential effects on lung and bone metastasis of breast cancer by Wnt signalling inhibitor DKK1. Nature Cell Biology 19: 1274–1285. https://doi.org/10.1038/ncb3613 [Google Scholar] [PubMed] [CrossRef]
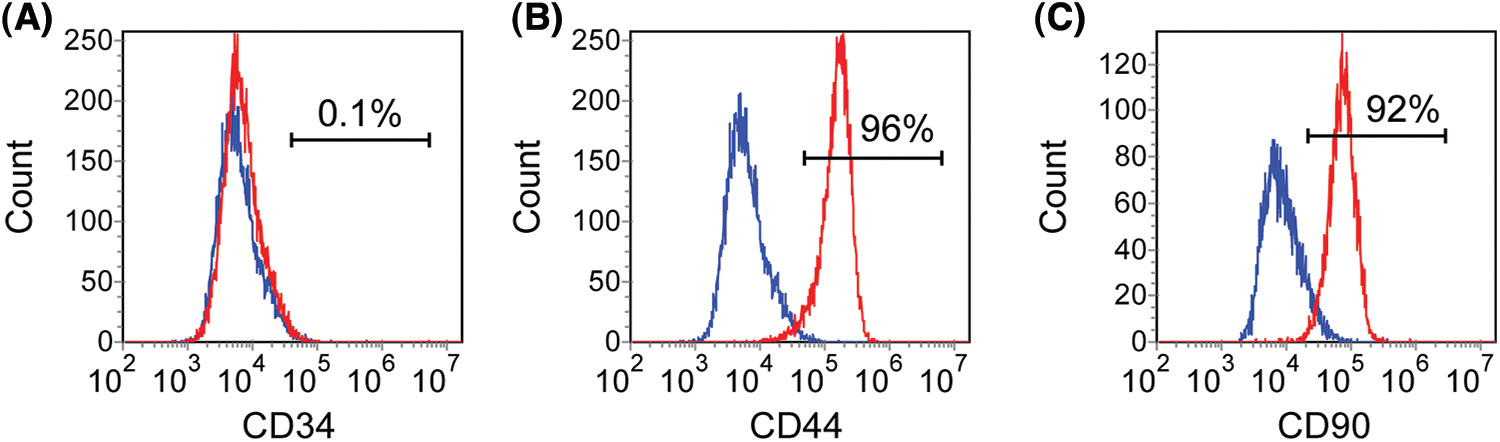
Figure S1: SCAPs cultured in vitro were identified by flow cytometry. CD34, CD44 and CD90 were selected as markers for identification. 1% of SCAP expressed CD34 (A). 92% and 96% of SCAP expressed CD44 and CD90, respectively (B and C).
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools