 Open Access
Open Access
ARTICLE
Expression and function of long non-coding RNA DLX6-AS1 in endometrial cancer
1 Department of Gynecologic Oncology, The International Peace Maternity and Child Health Hospital, School of Medicine, Shanghai Jiao Tong University, Shanghai, 200030, China
2 Shanghai Key Laboratory of Embryo Original Diseases, Shanghai, 200030, China
3 Shanghai Municipal Key Clinical Specialty, Shanghai, 200030, China
* Corresponding Author: YAN LIANG. Email:
# These authors contributed to the work equally
BIOCELL 2023, 47(4), 869-877. https://doi.org/10.32604/biocell.2023.026037
Received 11 August 2022; Accepted 15 November 2022; Issue published 08 March 2023
Abstract
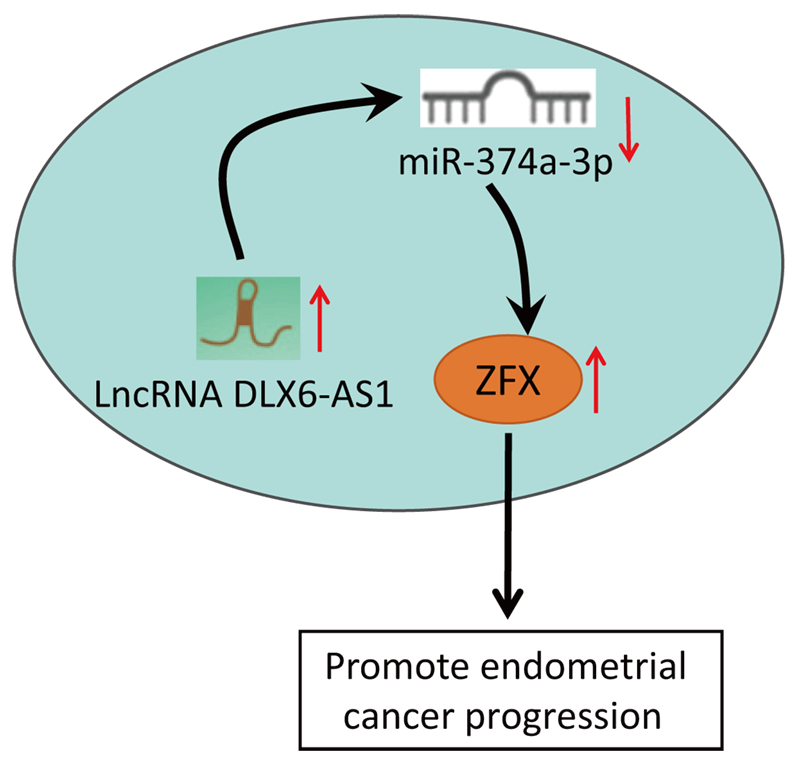
Background: LncRNA DLX6-AS1 has been uncovered to exert effects on various cancers. Nevertheless, the impacts of DLX6-AS1 on endometrial cancer (EC) development remained obscure. The study explored the influence of DLX6-AS1 on EC progression via the microRNA (miR)-374a-3p/zinc-finger protein (ZFX) axis.Methods: EC cell lines were collected and DLX6-AS1, miR-374a-3p, and ZFX levels in EC cell lines were detected. The EC cells were transfected with DLX6-AS1, miR-374a-3p, and ZFX constructs to examine the biological functions of EC cells. The xenograft model was established for detecting tumor growth. Rescue experiments were conducted to verify the interaction of DLX6-AS1, miR-374a-3p, and ZFX in EC cells.Results: DLX6-AS1 and ZFX levels were elevated, while miR-374a-3p exhibited a reduced level in EC cells. Silencing DLX6-AS1 and elevated miR-374a-3p expressions repressed the biological activities of EC cells. Reduced DLX6-AS1 repressed tumor development. MiR-374a-3p silencing reversed the impacts of DLX6-AS1 silencing, while ZFX overexpression abrogated the impacts of miR-374a-3p elevation on EC cell growth. Mechanically, DLX6-AS1 was found to bind to miR-374a-3p, and miR-374a-3p targeted ZFX.Conclusion: DLX6-AS1 depletion restricts the malignant phenotype of EC cells. The study might provide novel therapeutic biomarkers for EC treatment.Graphic Abstract
Keywords
Endometrial cancer (EC) is a common gynecological cancer, and its rates continue to rise worldwide (Passarello et al., 2019). The post-menopausal women are frequently afflicted by EC, accompanied by genital bleeding (Aoki et al., 2020). Risk factors of EC mainly comprise excessive exposure to unopposed estrogen. Age, obesity, hypertension, and diabetes also contribute to triggering the occurrence of EC (Cai et al., 2021). Surgery is the optimal option for treating EC. Other adjuvant EC therapeutic strategies have been applied, including pelvic radiation therapy, vaginal brachytherapy, chemotherapy, and chemotherapy-combined radiation therapy (van den Heerik et al., 2021). The incidence of EC has risen worldwide, and the challenges of EC treatment prevent the standard of care from being delivered (Liu et al., 2020). Therefore, it is necessary to further pave the new pathway for EC treatment.
Long noncoding RNAs (lncRNAs) have been found to exert fundamental impacts on human tumorigenesis and development. DLX6-AS1 is a novel lncRNA that affects the progression of multiple cancers (Xue et al., 2020). The DLX6-AS1 high levels have been validated in EC tissues and cells, and the functional augmentation of DLX6-AS1 can facilitate EC progression (Zhao and Xu, 2020). Nevertheless, studies for directly probing the impacts of DLX6-AS1 in EC are rare. As an oncogene, DLX6-AS1 has a binding relationship with microRNA (miR) and can induce the expression alternation of downstream genes (Zheng et al., 2021). Most miRs have previously been reported to be deregulated in EC (Donkers et al., 2021). For instance, a reduced level of miR-374 and miR-374a in EC cell lines has been validated (Jayaraman et al., 2017; Talkowski et al., 2021). As for miR-374a-3p, it has been revealed that increased expression of miR-374a-3p contributes to restraining the biological activities of non-small-cell lung cancer cells (NSCLCs), (Yang et al., 2020), while the miR-374a-3p function in EC has remained largely unknown. We predicted in our research the binding sites between miR-374a-3p and zinc-finger protein (ZFX). As reported, ZFX is highly expressed in EC tissues, and ZFX overexpression accelerates the progression of EC (Yang et al., 2021). From the above-mentioned studies, it could be deduced that DLX6-AS1, miR-374a-3p, and ZFX play a pivotal function in cancer progression, but the impacts of DLX6-AS1 in EC via the modulation of the miR-374a-3p/ZFX axis remained rarely studied. Thus, this study was conducted to pave the novel pathway for EC treatment modalities.
Ethics statement for animal experiments
The operations of the animal protocol were ratified by the Institutional Animal Care and Use Committee of The International Peace Maternity and Child Health Hospital, School of Medicine, Shanghai Jiao Tong University (approval number: 20200815). Animal experiments were in full compliance with local, national, ethical, and regulatory principles and local licensing arrangements.
Human endometrial endothelial cells (hEEC) and EC cell lines (HEC-1-B, AN3-CA, KLE, HEC1-A, and Ishikawa) were provided by American Type Culture Collection (Manassas, USA). DMEM (Sigma-Aldrich, St Louis, MO, USA) added with 10% fetal bovine serum (FBS, Sigma-Aldrich, St Louis, MO, USA) was adopted for cell culture (Wang et al., 2020).
In Ishikawa cells, short hairpin RNA (sh-DLX6-AS1) and sh-NC were offered by Shanghai Integrated Biotechnology Solutions, Ltd. (Shanghai, China). MiR-374a-3p mimic, miR-374a-3p inhibitor, and their corresponding NCs were offered by RiboBio (Guangzhou, China); (Dai et al., 2020). The ZFX overexpression plasmid pcDNA3.1-ZFX and its corresponding negative control (NC) were procured from GenePharma (Shanghai, China) (Song et al., 2020). Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) was employed for construct transfection. The transfection efficiency was assessed by reverse transcription-quantitative polymerase chain reaction (RT-qPCR).
The groups were: sh-NC group, sh-DLX6-AS1 group, sh-DLX6-AS1 + inhibitor NC group, sh-DLX6-AS1 + miR-374a-3p inhibitor, mimic NC group, miR-374a-3p mimic group, miR-374a-3p mimic + pcDNA3.1 group, and miR-374a-3p mimic + pcDNA3.1-ZFX group.
MTT (3-[4,5-dimethylthiazol-2-yl]-2,5 diphenyl tetrazolium bromide) assay for cell proliferation
Ishikawa cells (3 × 103 cells/well) were incubated in 96-well culture plates for 0, 24, 48, and 72 h and incubated with MTT solution for 4 h. Then, dimethyl sulfoxide (DMSO) was appended, and cells were resuspended until the crystals were completely dissolved. Lastly, the optical density value was measured at 490 nm.
Cell migration and invasion capabilities were tested through the transwell assay in the presence or absence of Matrigel (Corning Incorporated, Corning, NY, USA). Cells were fixed by serum-free medium and incubation in the upward chamber, and DMEM with 10% FBS was filled into the bottom chamber. After hot water immersion for 24-h, cells were fixed to the bottom of the upward chamber, followed by ethanol treatment and 0.1% crystalline violet staining. A microscope was adopted for counting (Dai et al., 2020).
RNA immunoprecipitation (RIP) analysis
Cells were lysed using RIP buffer supplemented with RNAase inhibitors. The cell lysates were subjected to incubation with Protein A/G magnetic agarose beads (Thermo Fisher Scientific, Waltham, MA, USA) pre-coated with anti-Ago2 (Bio-Rad, Hercules, CA, USA) and anti-IgG (Abcam, Cambridge, UK) as the NC. Beads were harvested, and bound RNA was extracted to measure the levels of miR-374a-3p and ZFX mRNA (Li et al., 2020).
Determination of target relation
The DLX6-AS1 binding site to miR-374a-3p was queried by the RNA22 website. The TargetScan was implemented to detect the binding site of ZFX to miR-374a-3p. The miR-374a-3p binding sequence of DLX6-AS1 or ZFX was amplified and inserted into the pGL3 luciferase reporter vector. The recombinant vectors were called DLX6-AS1 wild type (DLX6-AS1-WT) or ZFX-WT. After that, the mutant binding site matching miR-374a-3p of DLX6-AS1 or ZFX was cloned into the pGL3 luciferase reporter vector to generate DLX6-AS1-mutant (MUT) or ZFX-MUT. Ishikawa cells were transfected with the above plasmids (mimic NC and miR-374a-3p mimic), and after 48-h, luciferase activity was determined in different groups by a dual luciferase reporter gene assay system (Li et al., 2019b).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
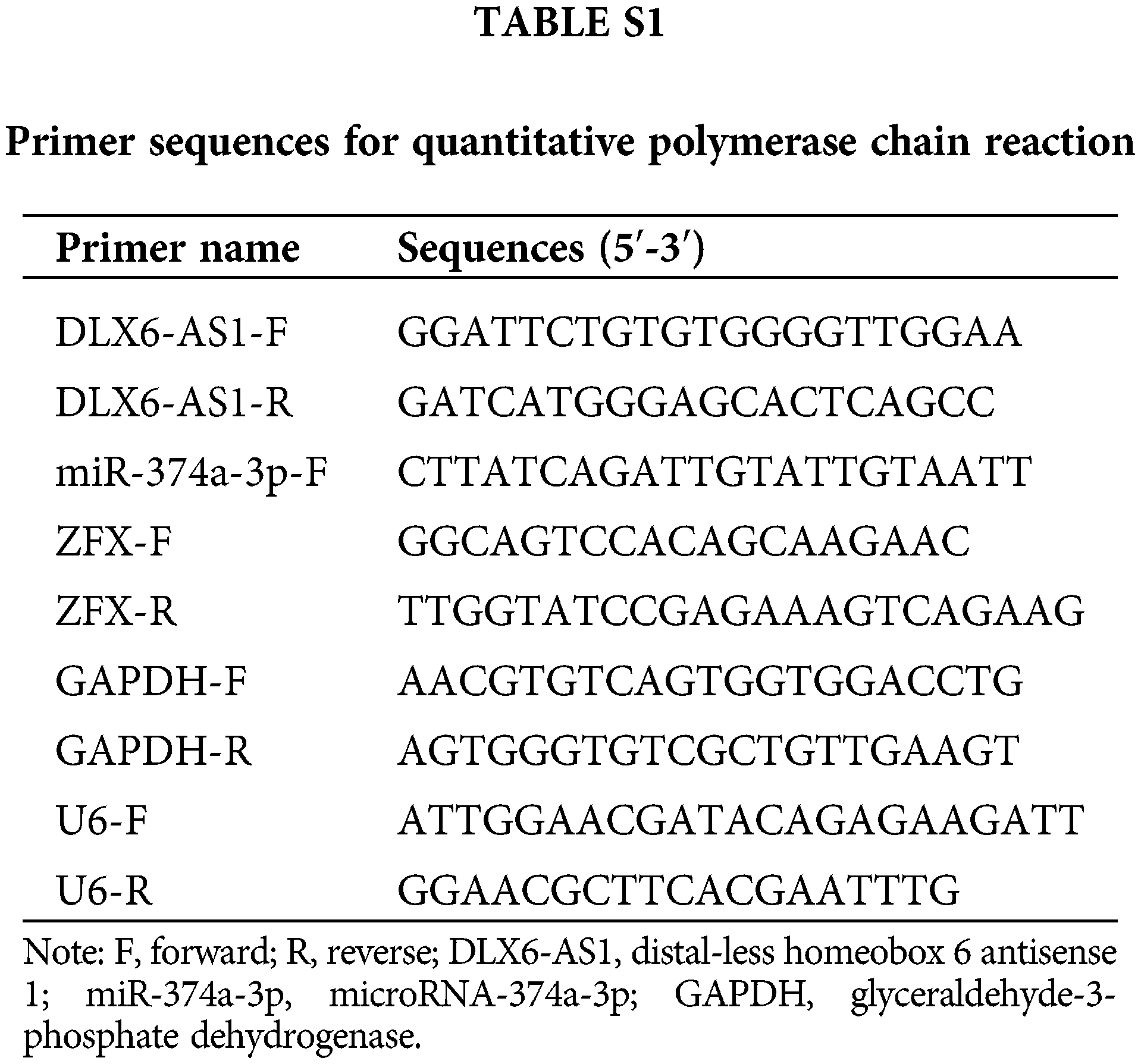
RNA was isolated with Trizol reagent (Life Technologies, Carlsbad, CA, USA). Complementary DNA (cDNA) was synthesized with PrimeScript RT master mix kit (Takara, Dalian, China) together with All-in-OneTM miRNA First strand cDNA Synthesis Kit (GeneCopoeia, Rockville, MD, USA). RT-qPCR reactions were conducted using a SYBR Premix Ex Taq II Kit (Takara) along with a CFX96 real-time PCR detection system (Bio-Rad Laboratories, Hercules, CA, USA). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and U6 served as internal references in this study. All the used primers are listed in Suppl. Table 1. The expression assessment was determined by the 2−ΔΔCt method (He et al., 2020).
Proteins were subjected to denaturation in sample buffer spiked with protease inhibitor mixture, SDS-polyacrylamide gel electrophoresis, followed by transfer to nitrocellulose membranes. The membranes were treated with 1-h blocking with 5% milk and overnight immunoblotting with the primary antibodies (ZFX antibody: 1:1000; GAPDH antibody: 1:2000 (both Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA) at 4°C. The membranes were subjected to 1-h of incubation with a corresponding secondary antibody (1:3000; Solarbio, Beijing, China). Western blots were visualized and quantified (Guo et al., 2015; Bai et al., 2019).
Female BALB/c nude mice were housed at 24 ± 1°C, 50% humidity, 12-h light-dark cycle, with water and food supply. Ishikawa cells infected with lentivirus containing DLX6-AS1 shRNA, ZGX shRNA, or corresponding negative control (NC) lentivirus. After one-week of acclimation, mice were randomly classified into four groups (n = 6/group): sh-DLX6-AS1 group, sh-NC group, sh-ZFX group, and sh-CTR group. Then, 1 × 107 cells stably expressing sh-DLX6-AS1, sh-NC, sh-ZFX, and sh-CTR were injected subcutaneously into the dorsal belly. Tumor volume was calculated every 7 d for 35 d using the formula [volume = (π × length × width2)/6]. All animals were euthanized, and tumor xenografts were obtained and weighed (Wang et al., 2020). Tumor tissues were fixed with 4% paraformaldehyde for subsequent immunohistochemical staining.
All groups of tumor tissues were dehydrated, paraffin-embedded, and sectioned (4 μm). The tissue sections were pre-treated in a microwave oven for 20 min and then incubated with 3% H2O2 for 30 min to block the endogenous peroxidase activity in the sections. Next, the sections were incubated at room temperature with primary antibody against Ki-67 (1 μg/mL, ab15580, Abcam, Cambridge, UK) and Cleaved-caspase-3 (1:200, Cell Signaling Technology, Danvers, MA, USA) for 15 min and subsequently treated with the corresponding secondary antibody (1:2000, ab205718, Abcam, Cambridge, UK) for 30 min at room temperature. After that, the sections were developed with DAB (3,3’-diaminoben-zidine) solution (P0203, Beyotime, Shanghai, China) for 10 min, then counterstained with hematoxylin staining solution (C0107, Beyotime), followed by DPX blocking, and photographing under a microscope. The positive expression rates of Ki-67 and cleaved-caspase-3 in various tumor tissues were also calculated (Luo et al., 2021).
The SPSS 21.0 (SPSS, Inc., NY, USA) was implemented for data analysis. The t-test or one-way ANOVA was used for comparison between groups, as well as Tukey’s test. p < 0.05 was considered statistically significant.
DLX6-AS1 is elevated and miR-374a-3p reduced in EC cells, and DLX6-AS1 targets miR-374a-3p
The determination of DLX6-AS1 and miR-374a-3p levels in EC cell lines disclosed that DLX6-AS1 levels were high in EC cells (Fig. 1A), and miR-374a-3p levels were reduced in EC cell lines (Fig. 1B) compared to those in hEEC cells. These data evidenced the oncogenic character of DLX6-AS1, whereas the tumor suppressive role of miR-374a-3p in EC. Then, Ishikawa cells were screened for follow-up experiments.
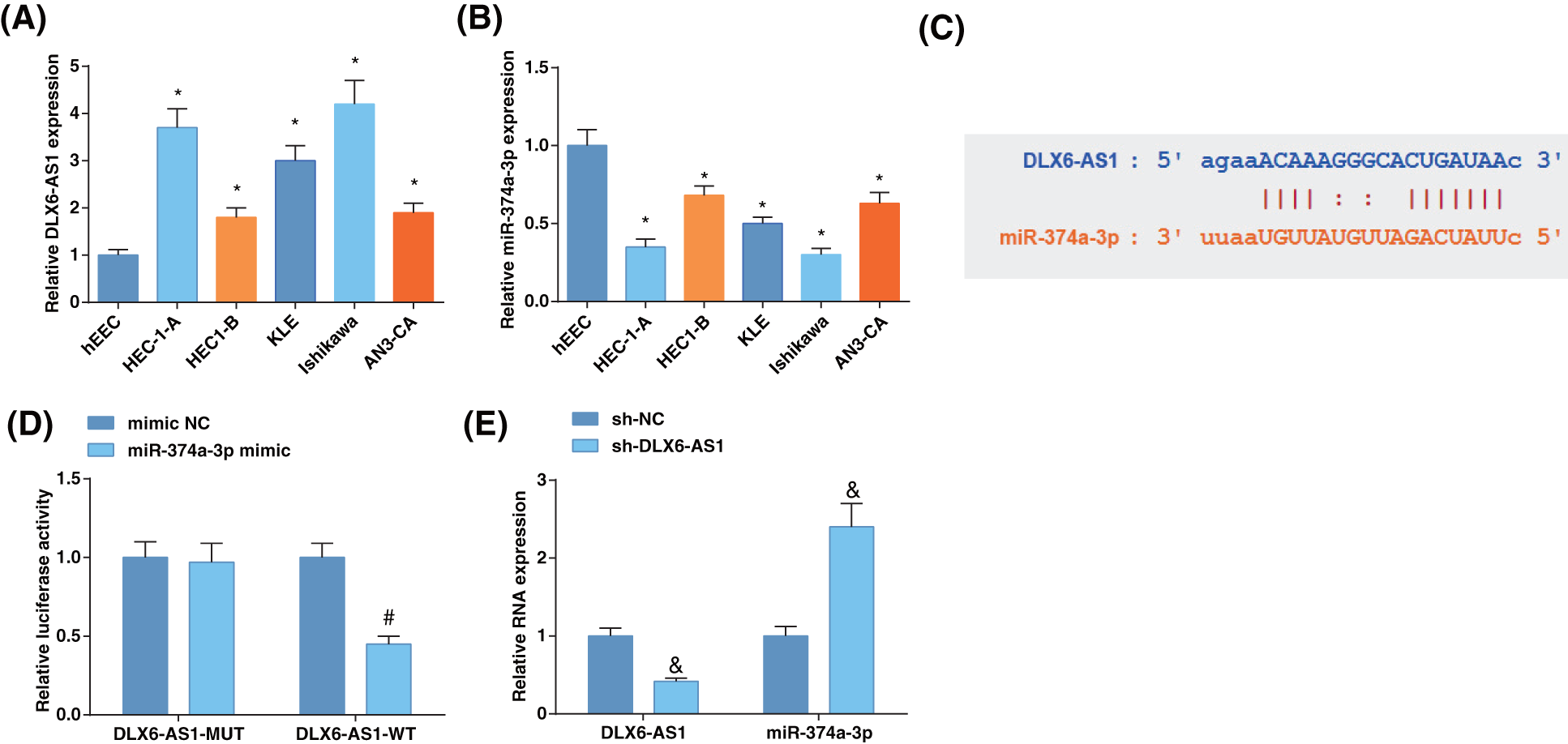
Figure 1: The level of DLX6-AS1 is elevated and miR-374a-3p is reduced in EC cells, and DLX6-AS1 targets miR-374a-3p. (A) DLX6-AS1 levels in EC cell lines and hEEC cells were assessed by reverse transcription-quantitative polymerase chain reaction (RT-qPCR); (B) miR-374a-3p levels in EC cells were assessed by RT-qPCR; (C) Validation of the binding sites of DLX6-AS1 and miR-374a-3p; (D) Confirmation of the targeting relation between DLX6-AS1 and miR-374a-3p; (E) Estimation of miR-374a-3p levels by RT-qPCR. N = 3; *p < 0.05 vs. hEEC cells; #p < 0.05 vs. the mimic NC group; & p < 0.05 vs. the sh-NC group.
The starBase revealed the binding sites of DLX6-AS1 and miR-374a-3p (Fig. 1C), whose relation was confirmed by the luciferase activity assay. MiR-374a-3p-mimic co-transfected with DLX6-AS1-WT showed reduced luciferase activity in Ishikawa cells (Fig. 1D). To unravel the correlation between DLX6-AS1 and miR-374a-3p, we measured DLX6-AS1 and miR-374a-3p levels in cells after sh-DLX6-AS1 transfection. It turned out that miR-374a-3p levels were enhanced after transfection of cells with sh-DLX6-AS1 (Fig. 1E). These data indicated the interaction between DLX6-AS1 and miR-374a-3p.
Inhibition of DLX6-AS1 retards EC cell growth by regulating miR-374a-3p
To determine any correlation between DLX6-AS1 and miR-374a-3p levels with EC cell progression, Ishikawa cells were further treated, and the DLX6-AS1 and miR-374a-3p levels reflected reduced DLX6-AS1 expression after transfection with sh-DLX6-AS1 and sh-DLX6-AS1 + anti-miR-374a-3p (Fig. 2A), indicating the successful transfection. MTT assay revealed that the decrease in DLX6-AS1 after DLX6-AS1 silencing prevented cell proliferation (Fig. 2B), and Transwell assay revealed restricted cell migratory and invasive capacities (Figs. 2C, 2D); However, miR-374a-3p depletion reversed the suppressive impacts of DLX6-AS1 silencing in EC cells growth. Collectively, these findings suggest that DLX6-AS1 depletion inhibited EC cell development by altering miR-374a-3p levels.
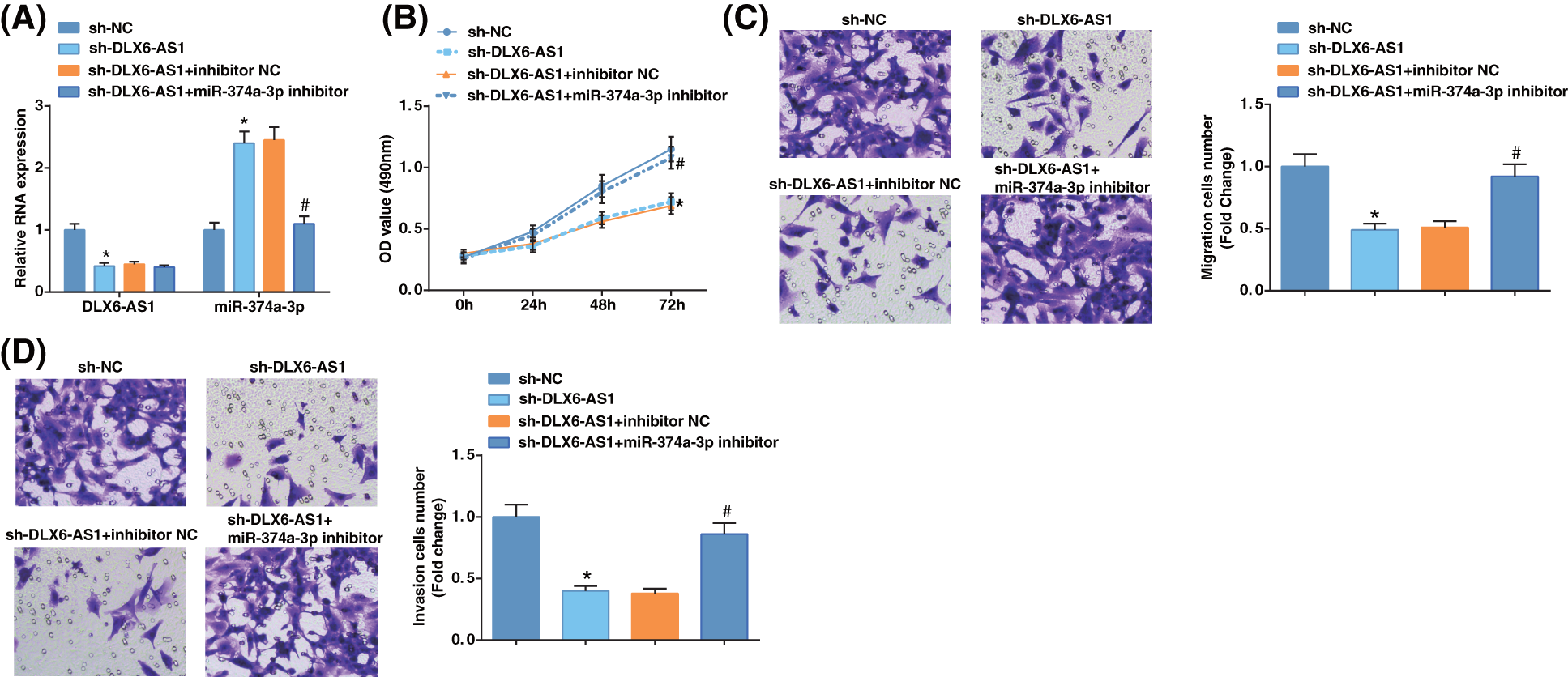
Figure 2: Inhibition of DLX6-AS1 impedes EC cell growth by regulating miR-374a-3p. (A) DLX6-AS1 and miR-374a-3p levels were examined by RT-qPCR in EC cells; (B) The proliferation of EC cells were tested by MTT assays; (C, D) The migration and invasion capabilities of EC cells were tested by Transwell assays. N = 3; *p < 0.05 vs. the sh-NC group; #p < 0.05 vs. the sh-DLX6-AS1 + inhibitor NC group.
MiR-374a-3p targets ZFX in EC cells
ZFX level is elevated in EC (Song et al., 2020). Hence, we examined the ZFX levels in EC cell lines, which suggested augmented ZFX expression in EC cells (Fig. 3A). TargetScan forecasted binding sites of miR-374a-3p and ZFX 3'UTR (Fig. 3B). Luciferase activity assay disclosed that miR-374a-3p could restrict the luciferase activity of ZFX-WT (Fig. 3C). RIP experiments indicated that ZFX and miR-374a-3p levels were high after treatment with Ago2, implying that ZFX targeted the RNA-caused depleted complex (Fig. 3D). The above discoveries demonstrated that miR-374a-3p negatively modulated ZFX in EC.
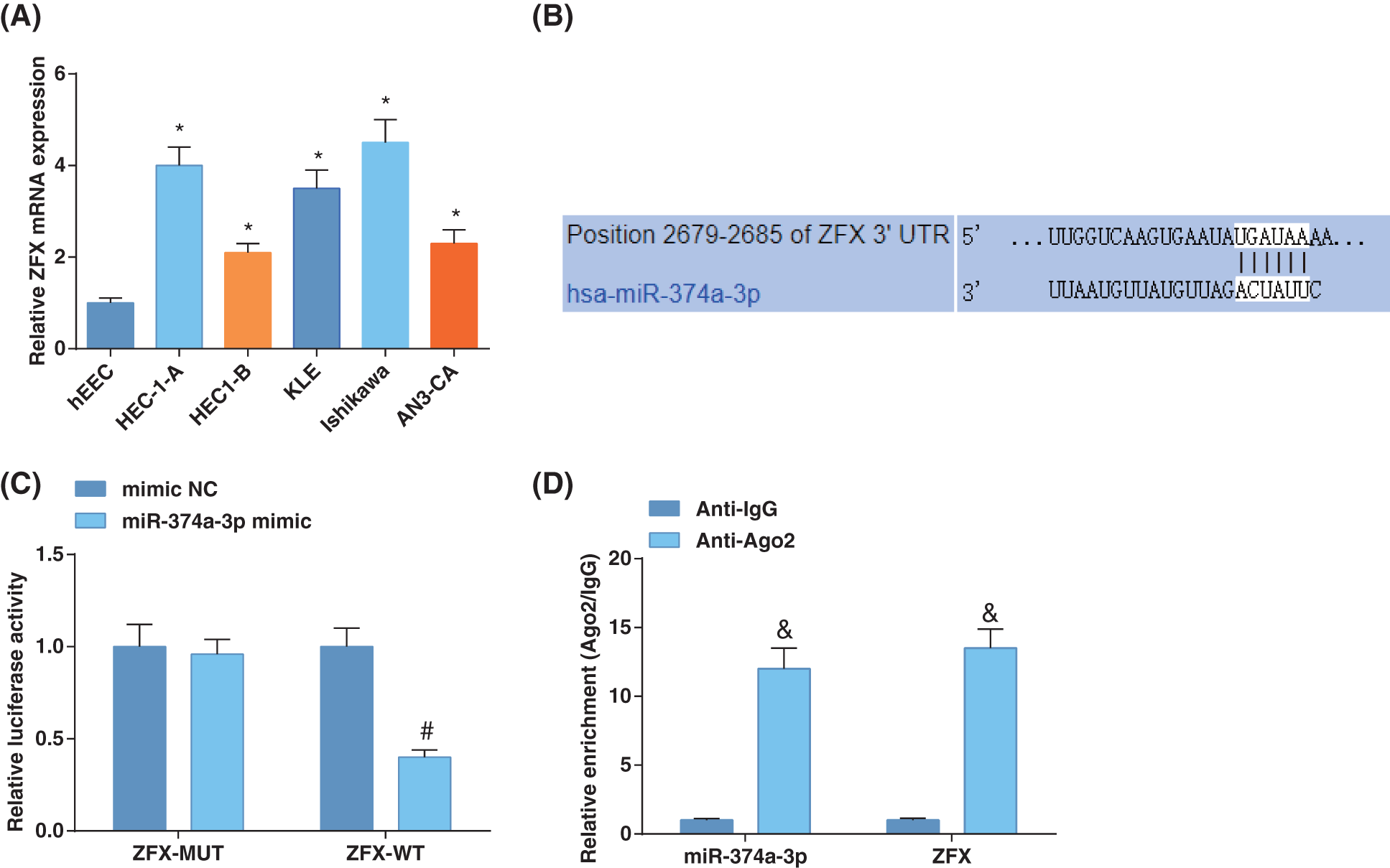
Figure 3: MiR-374a-3p targets ZFX in EC. (A) Zinc-finger-protein (ZFX) levels in EC cells and hEEC cells were estimated by reverse transcription-quantitative polymerase chain reaction; (B) Prediction of the binding site of miR-374a-3p and ZFX; (C) Confirmation of the binding relation between miR-374a-3p and ZFX; (D) The enrichment of ZFX and miR-374a-3p was assessed by RNA immunoprecipitation assay in EC cells. N = 3; *p < 0.05 vs. hEEC cells; #p < 0.05 vs. the mimic NC group; & p < 0.05 vs. the Anti-IgG group.
Zinc-finger-protein overexpression compromises the influence of miR-374a-3p elevation on EC cells
To probe whether miR-374a-3p regulated EC cell progression by targeting ZFX, we first examined miR-374a-3p and ZFX levels in EC cells upon further treatment with miR-374a-3p mimic and ZFX augmentation. The results revealed that miR-374a-3p levels were enhanced, while ZFX expression was reduced after miR-374a-3p mimic treatment (Fig. 4A). After miR-374a-3p augmentation, the growth of EC cells was repressed according to the findings from MTT and Transwell analyses, while ZFX augmentation offset the impacts of elevated miR-374a-3p on decelerating EC cell proliferative, migratory, and invasive capabilities (Figs. 4B–4D). The outcomes implied that miR-374a-3p controlled EC cell growth by targeting ZFX.
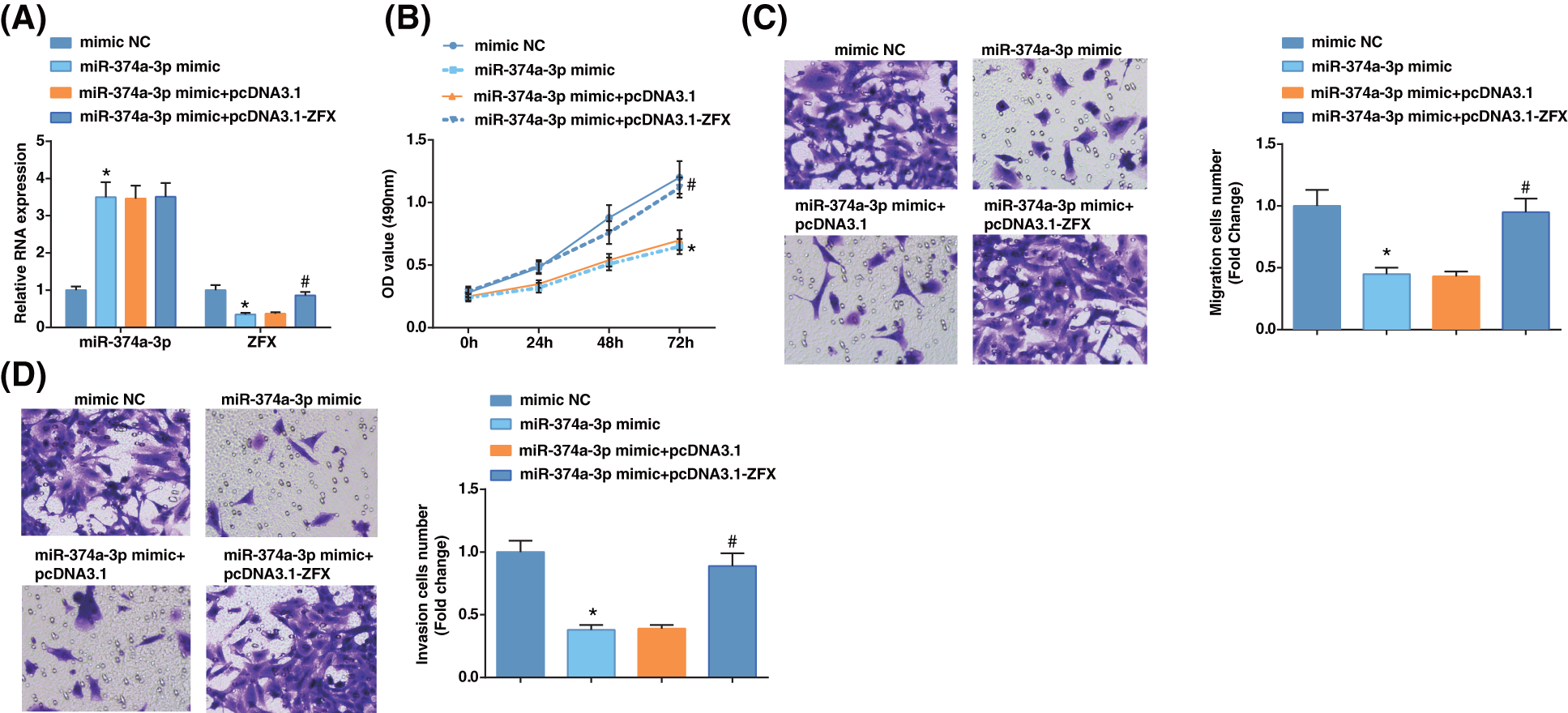
Figure 4: Zinc-finger-protein (ZFX) overexpression reverses the influence of miR-374a-3p elevation on EC cells. (A) miR-374a-3p and ZFX levels in EC cells were tested by reverse transcription-quantitative polymerase chain reaction; (B–D) The biological activities of EC cells were tested by MTT and Transwell assays. N = 3; *p < 0.05 vs. the mimic NC group; #p < 0.05 vs. the miR-374a-3p mimic + pcDNA3.1 group.
DLX6-AS1/miR-374a-3p axis regulates the enrichment of ZFX in EC cells
We then examined the ZFX levels by western blotting after further treatment. Inhibition of DLX6-AS1 decreased ZFX levels, while miR-374a-3p silencing reversed the impacts of silencing DLX6-AS1 on reducing ZFX levels; miR-374a-3p elevation reduced ZFX levels, and ZFX overexpression could reverse the impacts of miR-374a-3p elevation on reducing ZFX levels (Figs. 5A, 5B). These results thus evidenced that the DLX6-AS1/miR-374a-3p axis in EC cells regulated the abundance of ZFX.
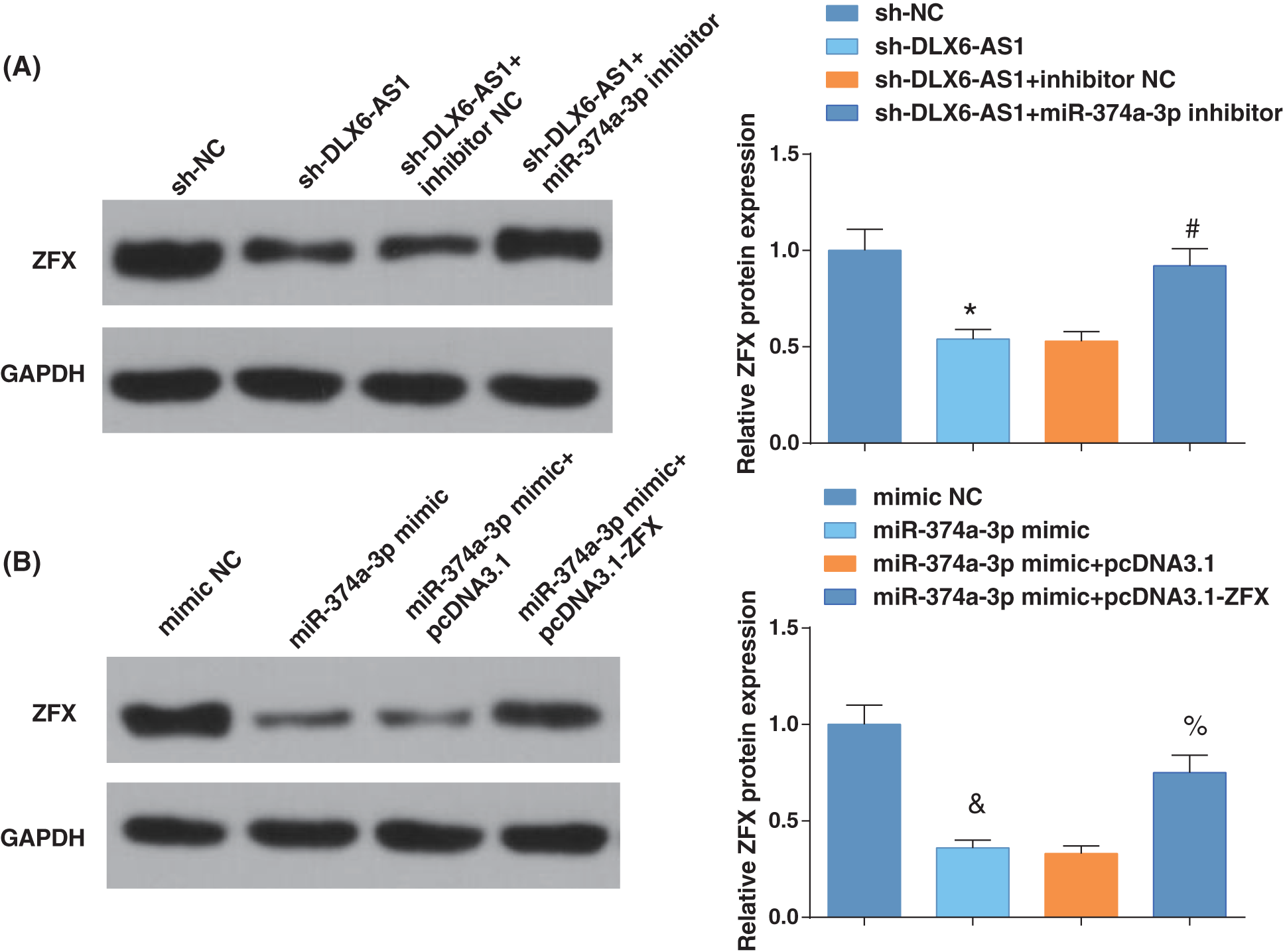
Figure 5: DLX6-AS1/miR-374a-3p axis regulates the enrichment of zinc-finger-protein (ZFX) in EC cells. (A) ZFX expression were tested by western blot analysis; (B) ZFX levels were measured by western blot analysis. N = 3; *p < 0.05 vs. the sh-NC group; #p < 0.05 vs. the sh-DLX6-AS1 + inhibitor NC group; p < 0.05 vs. the mimic NC group; % p < 0.05 vs. the miR-374a-3p mimic + pcDNA3.1 group.
Silencing DLX6-AS1 represses tumor growth in vivo
A xenograft tumor model was constructed to address the impacts of DLX6-AS1 and ZFX in vivo. Sh-DLX6-AS1-or sh-ZFX-injected mice had diminished tumor volume and weight (Figs. 6A, 6B). In the tumor tissues of sh-DLX6-AS1-or sh-ZFX-injected mice positive expression rate of Ki-67 decreased, but the positive expression rate of cleaved caspase3 increased (Fig. 6C). In summary, the low expression of DLX6-AS1 and silencing of ZFX suppressed tumor growth and tumor tissue cell proliferation, and promoted tumor cell apoptosis in vivo.
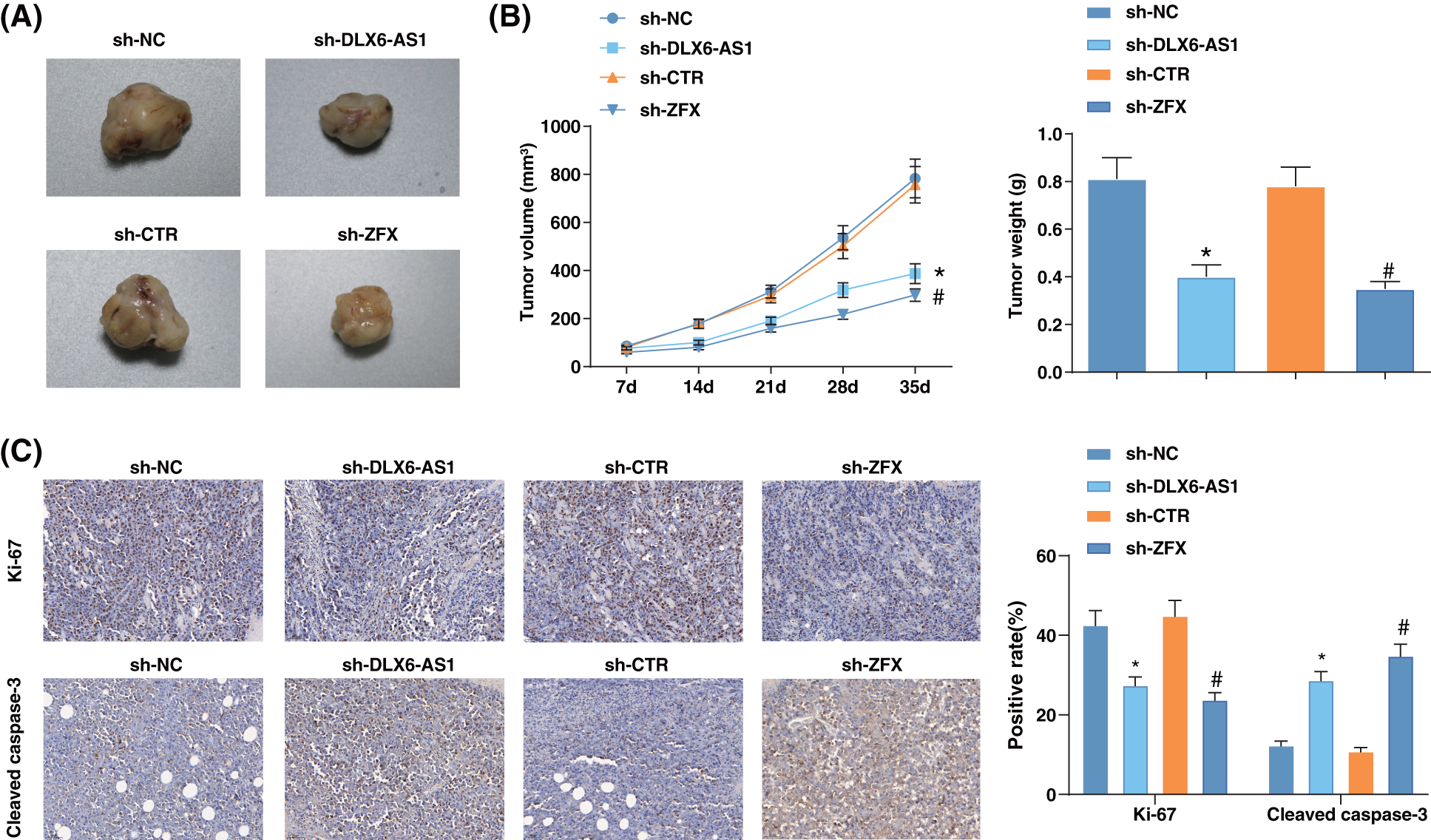
Figure 6: Silencing DLX6-AS1 represses tumor growth in vivo. (A) Representative pictures of tumors in mice; (B) Tumor growth in mice; (C) The positive expression rates of Ki-67 and cleaved-caspase-3 in various tumor tissues were calculated by immunohistochemical staining (n = 6 mice); *p < 0.05 vs. the sh-NC group; #p < 0.05 vs. the sh-CTR group.
Although patients with EC have a good prognosis at an early stage, the high-grade patients suffer greatly. Besides, although several new drugs have been tested, the results remained gloomy, and no effective targeted agents have been approved (van Nyen et al., 2018). To further probe promising therapeutic agents for EC treatment, this study focused on the functions of DLX6-AS1 in the biological activities of EC cells. Collectively, it was manifested that the silenced DLX6-AS repressed the biological behaviors of EC cells via the miR-374a-3p/ZFX axis.
As previously described, DLX6-AS1 affects the modulation of multiple cancers. Hui et al. (2020) have concluded that DLX6-AS1 levels are augmented in ECs, and the depleted DLX6-AS1 weakens the proliferative and invasive activities of EC cells (Zhao and Xu, 2020). The same trend of DLX6-AS1 expression has also been discovered in gastric cancer, and the depleted DLX6-AS1 hinders the activities of gastric cancer cells (Wu et al., 2020). It has been disclosed that DLX6-AS1 is elevated in prostate cancer, while DLX6-AS1 decrement blocks prostate cancer progression by suppressing cell malignant phenotype (Zhu et al., 2021b). Partly inconsistent with previous discoveries, our study demonstrated a high level of DLX6-AS1 in EC cells, and DLX6-AS1 reduction could hinder the malignant phenotypes of EC cells. For the impacts of DLX6-AS1 in tumor growth, Liu et al. (2020) stated a high level of DLX6-AS1 in osteosarcoma, and DLX6-AS1 deficiency also retards in vivo tumor growth (Zhang et al., 2019). Likewise, we observed that the mice injected with si-DLX6-AS1 could block the tumor growth of EC. In terms of the modulating mechanism of DLX6-AS1 and its downstream gene, Li et al. (2019b) illustrated that the repressive effect of DLX6-AS1 deficiency on repressing the activity of liver cancer cells is abolished after miR-424-5p reduction (Li et al., 2019a). Consistently, our study unveiled that miR-374a-3p treatment could reverse the impacts of si-DLX6-AS1 on repressing EC cell growth.
Thereafter, the binding relation between DLX6-AS1 and miR-374a-3p was predicted by the bioinformatics websites. MiR-374a-3p has been validated to be depleted in different cancers and play an antitumor role. For example, the miR-374a-3p level is depleted in NSCLC, and increased miR-374a-3p attenuates the development of NSCLC (Yang et al., 2020). In line with previous discoveries, in this study, miR-374a-3p exerted a low expression in EC cells, and its overexpression contributed to the repression of malignant functions of EC cells. As for the effects of the decrease in miR-374a-3p expression, it has been confirmed that the inhibitor of miR-374a-3p aggravates lipopolysaccharides-stimulated damage in osteoarthritis (Shi and Ren, 2020). Zhu et al. (2021b) have validated that reduction in miR-374a-3p level reverses the inhibitory impacts of sh-KCNMB2-AS1 in hindering bladder cancer progression (Zhu et al., 2021a). The study also confirmed the modulating mechanism of the DLX6-AS1/miR-374a-3p axis. Specifically, miR-374a-3p silencing inverted the impacts of si-DLX6-AS1 in repressing EC cell growth.
We then hypothesized that miR-374a-3p had binding sites with ZFX. Yang et al. (2020) found augmented expression of ZFX in EC tissues (Yang et al., 2021). The same expression tendency of ZFX has also been reported by Pourkeramati et al. (2019) who unraveled increased ZFX levels in breast cancer tissues (Pourkeramati et al., 2019). Functionally, elevated ZFX levels resulted in active pancreatic cancer cells and accelerated tumor growth in vivo (Song et al., 2018). Mechanically, ZFX elevation partly abrogates miR-1296 overexpression-induced functions of blocking the biological function of triple-negative breast cancer cells (Zhou et al., 2021). Consistent with the conclusion, our work disclosed high expression of ZFX in EC, and augmentation of ZFX reversed the repressing effects of increased miR-374a-3p level on EC cell growth repress.
To conclude, our study demonstrates that DLX6-AS1 and ZFX levels are increased in EC cells, while the level of miR-374a-3p was low in EC cells. Functionally, DLX6-AS1 silencing caused repression of biological activities of EC cells through the modulation of the miR-374a-3p/ZFX axis. This work underlines the modulating mechanism of the DLX6-AS1/miR-374a-3p/ZFX axis in EC progression and aids in developing novel biomarkers, new interventions, and treatment for EC, thus exploring underlying therapeutic strategies of EC. Nevertheless, more studies are warranted to further unveil the function of the downstream regulatory factors of DLX6-AS1 in EC. Meanwhile, it would be significant to discuss antitumor drug tolerance effects in terms of the DLX6-AS1/miR-374a-3p/ZFX axis. From the therapeutic angle, it is significant to deliver the lentivirus/drug into the (wild-type) tumor to examine the effects of DLX6-AS1 knockdown on tumor growth.
Funding Statement: This work was supported by Shanghai Municipal Health Commission (Grant/Award No. 20194Y0050).
Author Contributions: Wei Shi contributed to study design; Yan Liang contributed to manuscript editing; Jianxia Lin and Xianjing Xie contributed to experimental studies; Rong Jin contributed to data analysis. All authors read and approved the final manuscript.
Availability of Data and Materials: The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics Approval: The operations of the animal protocol were ratified by the Institutional Animal Care and Use Committee of The International Peace Maternity and Child Health Hospital, School of Medicine, Shanghai Jiao Tong University (approval number: 20200815). Animal experiments were in full compliance with local, national, ethical, and regulatory principles and local licensing arrangements.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
Aoki Y, Kanao H, Wang X, Yunokawa M, Omatsu K, Fusegi A, Takeshima N (2020). Adjuvant treatment of endometrial cancer today. Japanese Journal of Clinical Oncology 50: 753–765. https://doi.org/10.1093/jjco/hyaa071 [Google Scholar] [PubMed] [CrossRef]
Bai Y, Yang Y, Yan Y, Zhong J, Blee AM et al. (2019). RUNX2 overexpression and PTEN haploinsufficiency cooperate to promote CXCR7 expression and cellular trafficking, AKT hyperactivation and prostate tumorigenesis. Theranostics 9: 3459–3475. https://doi.org/10.7150/thno.33292 [Google Scholar] [PubMed] [CrossRef]
Cai Y, Wang B, Xu W, Liu K, Gao Y, Guo C, Chen J, Kamal MA, Yuan C (2021). Endometrial cancer: Genetic, metabolic characteristics, therapeutic strategies and nanomedicine. Current Medicinal Chemistry 28: 8755–8781. https://doi.org/10.2174/0929867328666210705144456 [Google Scholar] [PubMed] [CrossRef]
Dai Q, Deng J, Zhou J, Wang Z, Yuan XF, Pan S, Zhang HB (2020). Long non-coding RNA TUG1 promotes cell progression in hepatocellular carcinoma via regulating miR-216b-5p/DLX2 axis. Cancer Cell International 20: 8. https://doi.org/10.1186/s12935-019-1093-6 [Google Scholar] [PubMed] [CrossRef]
Donkers H, Hirschfeld M, Weiss D, Erbes T, Jaeger M, Pijnenborg JMA, Bekkers R, Galaal K, Consortium E (2021). Usefulness of microRNA detection in the diagnostics of endometrial cancer. Acta Obstetricia et Gynecologica Scandinavica 100: 1148–1154. https://doi.org/10.1111/aogs.14141 [Google Scholar] [PubMed] [CrossRef]
Guo Y, Fan Y, Zhang J, Lomberk GA, Zhou Z et al. (2015). Perhexiline activates KLF14 and reduces atherosclerosis by modulating ApoA-I production. Journal of Clinical Investigation 125: 3819–3830. https://doi.org/10.1172/JCI79048 [Google Scholar] [PubMed] [CrossRef]
He S, Huang Y, Dong S, Qiao C, Yang G, Zhang S, Wang C, Xu Y, Zheng F, Yan M (2020). MiR-199a-3p/5p participated in TGF-beta and EGF induced EMT by targeting DUSP5/MAP3K11 in pterygium. Journal of Translational Medicine 18: 332. https://doi.org/10.1186/s12967-020-02499-2 [Google Scholar] [PubMed] [CrossRef]
Jayaraman M, Radhakrishnan R, Mathews CA, Yan M, Husain S, Moxley KM, Song YS, Dhanasekaran DN (2017). Identification of novel diagnostic and prognostic miRNA signatures in endometrial cancer. Genes & Cancer 8: 566–576. https://doi.org/10.18632/genesandcancer.144 [Google Scholar] [PubMed] [CrossRef]
Li J, He M, Xu W, Huang S (2019a). LINC01354 interacting with hnRNP-D contributes to the proliferation and metastasis in colorectal cancer through activating Wnt/beta-catenin signaling pathway. Journal of Experimental & Clinical Cancer Research 38: 161. https://doi.org/10.1186/s13046-019-1150-y [Google Scholar] [PubMed] [CrossRef]
Li D, Tang X, Li M, Zheng Y (2019b). Long noncoding RNA DLX6-AS1 promotes liver cancer by increasing the expression of WEE1 via targeting miR-424-5p. Journal of Cellular Biochemistry 120: 12290–12299. https://doi.org/10.1002/jcb.28493 [Google Scholar] [PubMed] [CrossRef]
Li Z, Yao H, Wang S, Li G, Gu X (2020). CircTADA2A suppresses the progression of colorectal cancer via miR-374a-3p/KLF14 axis. Journal of Experimental & Clinical Cancer Research 39: 160. https://doi.org/10.1186/s13046-020-01642-7 [Google Scholar] [PubMed] [CrossRef]
Liu MC, Gardner AB, Wolford JE, Tewari KS (2020). Endometrial cancer in the morbidly obese: A review. Current Opinion in Obstetrics & Gynecology 32: 42–50. https://doi.org/10.1097/GCO.0000000000000606 [Google Scholar] [PubMed] [CrossRef]
Luo N, Liu S, Li X, Hu Y, Zhang K (2021). Circular RNA circHIPK3 promotes breast cancer progression via sponging MiR-326. Cell Cycle 20: 1320–1333. https://doi.org/10.1080/15384101.2021.1939476 [Google Scholar] [PubMed] [CrossRef]
Passarello K, Kurian S, Villanueva V (2019). Endometrial cancer: An overview of pathophysiology, management, and care. Seminars in Oncology Nursing 35: 157–165. https://doi.org/10.1016/j.soncn.2019.02.002 [Google Scholar] [PubMed] [CrossRef]
Pourkeramati F, Asadi MH, Shakeri S, Farsinejad A (2019). Differential expression profile of ZFX variants discriminates breast cancer subtypes. Iranian Biomedical Journal 23: 47–56. https://doi.org/10.29252/ibj.23.1.47 [Google Scholar] [CrossRef]
Shi FL, Ren LX (2020). Up-regulated miR-374a-3p relieves lipopolysaccharides induced injury in CHON-001 cells via regulating wingless-type MMTV integration site family member 5B. Molecular and Cellular Probes 51: 101541. https://doi.org/10.1016/j.mcp.2020.101541 [Google Scholar] [PubMed] [CrossRef]
Song N, Zhang Y, Kong F, Yang H, Ma X (2020). HOXA-AS2 promotes type I endometrial carcinoma via miRNA-302c-3p-mediated regulation of ZFX. Cancer Cell International 20: 359. https://doi.org/10.1186/s12935-020-01443-0 [Google Scholar] [PubMed] [CrossRef]
Song X, Zhu M, Zhang F, Zhang F, Zhang Y et al. (2018). ZFX promotes proliferation and metastasis of pancreatic cancer cells via the MAPK pathway. Cellular Physiology and Biochemistry 48: 274–284. https://doi.org/10.1159/000491727 [Google Scholar] [PubMed] [CrossRef]
Talkowski K, Kielbasinski K, Peszek W, Grabarek BO, Boron D, Oplawski M (2021). Salinomycin modulates the expression of mRNAs and miRNAs related to stemness in endometrial cancer. Current Pharmaceutical Biotechnology 22: 317–326. https://doi.org/10.2174/1573403X16666200621160742 [Google Scholar] [PubMed] [CrossRef]
van den Heerik A, Horeweg N, de Boer SM, Bosse T, Creutzberg CL (2021). Adjuvant therapy for endometrial cancer in the era of molecular classification: Radiotherapy, chemoradiation and novel targets for therapy. International Journal of Gynecological Cancer 31: 594–604. https://doi.org/10.1136/ijgc-2020-001822 [Google Scholar] [PubMed] [CrossRef]
van Nyen T, Moiola CP, Colas E, Annibali D, Amant F (2018). Modeling endometrial cancer: Past, present, and future. International Journal of Molecular Sciences 19: 2348. https://doi.org/10.3390/ijms19082348 [Google Scholar] [PubMed] [CrossRef]
Wang Y, Yin L, Sun X (2020). CircRNA hsa_circ_0002577 accelerates endometrial cancer progression through activating IGF1R/PI3K/Akt pathway. Journal of Experimental & Clinical Cancer Research 39: 169. https://doi.org/10.1186/s13046-020-01679-8 [Google Scholar] [PubMed] [CrossRef]
Wu Q, Ma J, Meng W, Hui P (2020). DLX6-AS1 promotes cell proliferation, migration and EMT of gastric cancer through FUS-regulated MAP4K1. Cancer Biology & Therapy 21: 17–25. https://doi.org/10.1080/15384047.2019.1647050 [Google Scholar] [PubMed] [CrossRef]
Xue C, Lv L, Jiang J, Li L (2020). Promising long noncoding RNA DLX6-AS1 in malignant tumors. American Journal of Translational Research 12: 7682–7692. [Google Scholar] [PubMed]
Yang D, Ma X, Xu J, Jia K, Liu X, Zhang P (2021). Zfx-induced upregulation of UBE2J1 facilitates endometrial cancer progression via PI3K/AKT pathway. Cancer Biology & Therapy 22: 238–247. https://doi.org/10.1080/15384047.2021.1883186 [Google Scholar] [PubMed] [CrossRef]
Yang H, Wang Z, Wang Z (2020). Long noncoding RNA KCNMB2-AS1 increases ROCK1 expression by sponging microRNA-374a-3p to facilitate the progression of non-small-cell lung cancer. Cancer Management and Research 12: 12679–12695. https://doi.org/10.2147/CMAR.S270646 [Google Scholar] [PubMed] [CrossRef]
Zhang N, Meng X, Mei L, Zhao C, Chen W (2019). LncRNA DLX6-AS1 promotes tumor proliferation and metastasis in osteosarcoma through modulating miR-641/HOXA9 signaling pathway.Journal of Cellular Biochemistry 120: 11478–11489. https://doi.org/10.1002/jcb.28426 [Google Scholar] [PubMed] [CrossRef]
Zhao H, Xu Q (2020). Long non-coding RNA DLX6-AS1 mediates proliferation, invasion and apoptosis of endometrial cancer cells by recruiting p300/E2F1 in DLX6 promoter region. Journal of Cellular and Molecular Medicine 24: 12572–12584. https://doi.org/10.1111/jcmm.15810 [Google Scholar] [PubMed] [CrossRef]
Zheng Q, Gu X, Yang Q, Chu Q, Dai Y, Chen Z (2021). DLX6-AS1 is a potential biomarker and therapeutic target in cancer initiation and progression. Clinica Chimica Acta 517: 1–8. https://doi.org/10.1016/j.cca.2021.02.006 [Google Scholar] [PubMed] [CrossRef]
Zhou Y, Ma G, Peng S, Tuo M, Li Y, Qin X, Yu Q, Kuang S, Cheng H, Li J (2021). Circ_0000520 contributes to triple-negative breast cancer progression through mediating the miR-1296/ZFX axis. Thoracic Cancer 12: 2427–2438. https://doi.org/10.1111/1759-7714.14085 [Google Scholar] [PubMed] [CrossRef]
Zhu J, Huang Y, Zhang Y, Huang R, Huang C (2021a). KCNMB2-AS1 promotes bladder cancer progression through sponging miR-374a-3p to upregulate S100A10. Frontiers in Genetics 12: 655569. https://doi.org/10.3389/fgene.2021.655569 [Google Scholar] [PubMed] [CrossRef]
Zhu X, Ma X, Zhao S, Cao Z (2021b). DLX6-AS1 accelerates cell proliferation through regulating miR-497-5p/SNCG pathway in prostate cancer. Environmental Toxicology 36: 308–319. https://doi.org/10.1002/tox.23036 [Google Scholar] [PubMed] [CrossRef]
Supplementary Materials
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools