 Open Access
Open Access
VIEWPOINT
Junctional adhesion molecule-A (JAM-A) in gynecological cancers: Current state of knowledge
Department of Cytobiology and Proteomics, Medical University of Lodz, Lodz, 92-215, Poland
* Corresponding Author: KAMILA CZUBAK-PROWIZOR. Email:
BIOCELL 2023, 47(4), 731-737. https://doi.org/10.32604/biocell.2023.025677
Received 25 July 2022; Accepted 26 December 2022; Issue published 08 March 2023
Abstract
Junctional adhesion molecule-A (JAM-A), also known as the F11 receptor (F11R), is one of the tight junction components. JAM-A is a transmembrane glycoprotein that regulates many cellular processes, i.e., angiogenesis, leukocyte transendothelial migration, intercellular permeability, epithelial-to-mesenchymal transition, and platelet activation. Of note, it is involved in the pathogenesis of various cancer types, including gynecological cancers. Only a few studies are available about this cancer type. Observed aberrant JAM-A expression in gynecological cancers correlates with poor patient prognosis. To the best of our knowledge, conflicting JAM-A roles in various cancer types suggest that its involvement is complex and tumor-type specific. The underlying molecular mechanisms and pathways responsible for JAM-A functions were not fully elucidated and need to be identified. Finding appropriate novel molecular cancer biomarkers may reduce observed very high mortality rates and could contribute to personalized treatment development. The main aim of the present viewpoint article is to report the current knowledge about JAM-A participation in gynecological malignancies.Keywords
Multiprotein complexes found in vertebrate intercellular junctions (i.e., tight junctions, gap junctions, desmosomes, and adherens junctions) facilitate the transmission of information between neighboring cells and cell-cell adhesion (Zihni et al., 2016; Rusu and Georgiou, 2020). Tight junctions (TJs) are structures located directly below the apical surface in the lateral epithelial and endothelial cell membrane (Rusu and Georgiou, 2020). Mainly, TJs are involved in the regulation of cellular processes such as maintaining the apical-basal polarization of cells (intramembrane diffusion barrier) and sealing the space between adjacent cells (paracellular diffusion barrier) (Zihni et al., 2016). TJs also play the role of signaling platform which regulates gene expression (Gonzalez-Mariscal et al., 2014), proliferation (Diaz-Coranguez et al., 2019), and differentiation of cells (Zihni et al., 2014).
Junctional adhesion molecule-A (JAM-A), also known as the F11 receptor (F11R), is one of the TJ components (Kornecki et al., 1990; Martìn-Padura et al., 1998) and is also expressed on circulating platelets and leukocytes (Wang and Chen, 2022). Shortly, JAM-A is a transmembrane glycoprotein that contains an extracellular region at the N-terminus with two immunoglobulin (Ig)-like domains, a transmembrane segment, and a short cytosolic tail at the C-terminus (Martìn-Padura et al., 1998; Prota et al., 2003; Steinbacher et al., 2018).
Moreover, it regulates many cellular processes, including angiogenesis (Naik et al., 2008), cell migration (Azari et al., 2010; Wang and Liu, 2022) and adhesion (Mandell et al., 2005), leukocyte transendothelial migration (Corada et al., 2005; Khandoga et al., 2005), intercellular permeability (Laukoetter et al., 2007), epithelial-to-mesenchymal transition (EMT) (Communal et al., 2020) and platelet activation (Babinska et al., 2002). In the literature, JAM-A is described as a protein involved in the pathogenesis of neurological disorders (Padden et al., 2007), cardiovascular diseases (Babinska et al., 2019; Koenen and Weber, 2022; Rath et al., 2022; Wang and Chen, 2022), rheumatoid arthritis (Fang et al., 2016), inflammatory bowel disease (Vetrano and Danese, 2009), and many types of neoplastic diseases including breast (McSherry et al., 2009; Murakami et al., 2011; Vellanki et al., 2019; Bednarek et al., 2020; Vences-Catalan et al., 2021; Smith et al., 2022), lung (Magara et al., 2017; Zhao et al., 2017), nasopharyngeal (Jiang et al., 2019; Dai et al., 2021), head and neck (Kurose et al., 2016; Kakiuchi et al., 2021), testicular (Tarulli et al., 2013), thyroid (Orlandella et al., 2019), colorectal (Caykara et al., 2019; Lampis et al., 2021), gastric (Huang et al., 2014), endometrial (Koshiba et al., 2009), cervical (Akimoto et al., 2016; Murakami et al., 2021), and ovarian cancer (Boljevic et al., 2019; Communal et al., 2020). The involvement of JAM-A in various malignancies progression and its correlation with poor patient prognosis are reviewed in detail in our previous article (Czubak-Prowizor et al., 2022).
For a long time, the loss of TJ-related proteins was considered necessary in the early stages of cancer metastasis (Lee et al., 2005; Martin and Jiang, 2009; Martin et al., 2010; Suren et al., 2014; Shimada et al., 2017). The overexpression of proteins located in TJs may also activate intracellular signaling pathways responsible for metastasis and tumorigenesis (Leech et al., 2015). Depending on the cancer type, both low and high JAM-A levels correlate with poor clinical prognosis in cancer patients (Czubak-Prowizor et al., 2022). A positive correlation (i.e., JAM-A overexpression was associated with adverse clinical outcomes) was observed in breast cancer (McSherry et al., 2009; Murakami et al., 2011; Leech et al., 2018; Cruz et al., 2021), lung cancer (Magara et al., 2017; Zhao et al., 2017), glioblastoma (Rosager et al., 2017), ovarian cancer (Boljevic et al., 2019; Communal et al., 2020), multiple myeloma (Solimando et al., 2018; Solimando et al., 2020), lymphoma (Xu et al., 2017), oral squamous cell carcinoma (Upadhaya et al., 2019), and cervical adenocarcinoma (Akimoto et al., 2016; Murakami et al., 2021). While a negative correlation (i.e., underexpression of JAM-A was associated with adverse clinical outcomes) was described in colorectal cancer (Lampis et al., 2021), endometrial carcinoma (Koshiba et al., 2009), pancreatic cancer (Fong et al., 2012), and gastric cancer (Huang et al., 2014). The tissue-specific role of JAM-A in the progression and invasiveness of neoplastic diseases is complex and not fully understood; therefore, this protein is still the subject of discussion and research interest.
The following viewpoint article focuses on the current knowledge about JAM-A participation in gynecological malignancies progression and metastasis.
Most gynecological cancer-related deaths in women are caused by epithelial ovarian cancer (EOC) development. The most common and aggressive histological type of EOC is high-grade serous carcinoma of uterine adnexa (HGSC). The main factors contributing to the high mortality rate in EOC patients are the lack of specific symptoms at the initial stages of the disease, which temporizes the diagnosis, the lack of effective screening tests, and the development of chemoresistance. Finding new biomarkers of ovarian cancer is so crucial. Inconsistent data about JAM-A expression in EOC is presented in the literature (Boljevic et al., 2019; Communal et al., 2020). Among other gynecological malignancies, disturbances in JAM-A levels have also been studied in the cervical (Akimoto et al., 2016; Murakami et al., 2021) and endometrial carcinoma (Koshiba et al., 2009).
In 2019, Boljevic et al. (2019) first published results showing the clinical significance of JAM-A gene expression in the pathogenesis of EOC. JAM-A gene expression levels were determined in 44 epithelial ovarian cancer formalin-fixed paraffin-embedded (FFPE) tissue blocks using reverse transcription and quantitative real-time polymerase chain reaction (RT-qPCR) method. In the tested EOC samples, 75% were represented by serous histological type, and approximately 61.5% were advanced International Federation of Gynecologists and Obstetricians (FIGO) stage (III + IV). Authors reported a worse overall survival rate in EOC patients characterized by JAM-A overexpression compared to patients with low JAM-A gene expression. Furthermore, high JAM-A gene expression was related to the advanced FIGO staging system suggesting JAM-A participation in EOC progression. Based on their studies, they proposed the JAM-A gene as a potential prognostic and diagnostic biomarker in EOC (Boljevic et al., 2019).
Unfortunately, these studies did not identify the molecular mechanisms by which JAM-A contributes to the EOC progression, which would confirm and strengthen the obtained results. Additionally, this is a rather retrospective study because researchers used only one laboratory method and then statistically analyzed using receiver operating characteristic (ROC) curve analysis, Fisher’s exact test, Kaplan-Meier method, and univariate Cox regression analysis (Boljevic et al., 2019).
High-grade serous carcinoma of uterine adnexa
High-grade serous carcinoma of uterine adnexa (HGSC) is one of the EOC histotypes. In 2020, Communal et al. (2020) indicated that shorter overall survival and shorter progression-free survival (i.e., poor clinical outcome parameters) were associated with low JAM-A protein expression (revealed using the Kaplan–Meier method and univariate Cox regression analysis). This data was inconsistent with results presented by Boljevic et al. (2019) one year earlier. In this study, JAM-A protein levels were determined using immunofluorescent staining with digital image analysis in HGSC FFPE tissue microarrays (1,526 clinical samples). Based on clinicopathologic characteristics, most of the tested samples were from patients in the III FIGO stage, high-grade serous subtype, and without chemotherapy before the surgery in which the material for the study was collected. Normal fallopian tube tissues (15 samples) from women without gynecological cancer were also included in the study as a control (Communal et al., 2020).
JAM-A expression analysis revealed prominent variability in its expression in HGSC clinical samples. Importantly, statistically significant differences in JAM-A expression between normal vs. cancer tissues were not observed. However, decreased JAM-A expression characterized samples from patients in the III + IV FIGO stage in comparison to the I + II FIGO stage. HGSC tissues indicated cell apico-basal polarity loss (Communal et al., 2020).
Moreover, analysis of proteomic data from the Clinical Proteomic Tumor Analysis Consortium revealed increased EMT in tumor samples characterized by low JAM-A protein levels. It suggested that the EMT-dependent mechanism could be responsible for the observed results (Communal et al., 2020).
The authors also identified the JAM-A protein as a candidate for the novel molecular biomarker of HGSC, based on its high expression on the cell surface (analysis of 26 human HGSC-derived cell lines using the flow cytometric method). Subsequently, protein expression levels were determined in 101 primary HGSC tissues and 15 normal fallopian tube tissues as control using immunofluorescence staining. It was exhibited that in the tested tissue cohort, JAM-A could be a prognostic predictor of poor outcomes (Communal et al., 2020).
Tumor surgical specimens from patients with cervical adenocarcinoma and adenocarcinoma in situ characterized the higher expression of some TJs proteins, exactly JAM-A, claudin-1, claudin-4, and claudin-7 in comparison to non-neoplastic tissues (Akimoto et al., 2016). In this study, 55 cancer patients were included. Selected proteins in tested specimens were stained immunohistochemically. In cervical adenocarcinoma cells, JAM-A and claudin-1 were delocalized from the apical-most part of the intercellular membrane and spread to the whole membrane.
Based on these findings, the authors proposed JAM-A and claudin-1 as potential diagnostic markers of cervical adenocarcinoma, which distinguish cancer cells from the adjacent cervical columnar epithelium (normal cells) with high specificity and sensitivity (ROC curve analysis) (Akimoto et al., 2016).
In a recent study, the same research group from Sapporo (Japan) showed that aberrant JAM-A expression significantly influenced uterine cervical adenocarcinoma progression (i.e., higher malignant potential) through JAM-A interaction with CD155 (also called the poliovirus receptor) (Murakami et al., 2021). Also suggested that cell surface JAM-A and CD155 could be potential therapeutic targets. The authors tested 67 surgical specimens by immunohistochemical staining and in human HCA1 cervical adenocarcinoma cell line after stable JAM-A knockout (KO) determining cell proliferation, colony formation, and collective migration. JAM-A overexpression in tissues from cancer patients correlated with shorter relapse-free survival, and overall survival (poor clinical outcome and prognosis). In the HCA1 cell line, after JAM-A KO was observed, there was a decrease in cell migration ability, cell proliferation, and colony formation. Of note, tested specific antibodies against JAM-A suppressed cell proliferation and indicated that JAM-A loss contributes to intensified sensitivity of the drug against this cancer type (Murakami et al., 2021).
Human endometrial malignancy was the last tested gynecological cancer type in the context of the role of JAM-A in its pathogenesis (Koshiba et al., 2009). Poor clinical outcomes, described as short patient progression-free survival and overall survival (Kaplan–Meier curves), were correlated with low JAM-A levels. Additionally, histologic grade, stage, and myometrial invasion are negatively associated with JAM-A expression. JAM-A could be a prognostic marker of this cancer type because its level is decreased in advanced and high-grade histotypes (Koshiba et al., 2009).
In 3D-cultured human endometrial cancer cells studies, particularly in the KLE cell line, a poorly differentiated adenocarcinoma cell line, JAM-A expression was lower than in the Ishikawa cell line, which is a well-differentiated cancer cell line. Furthermore, the JAM-A mRNA level in the 3D-culture of Ishikawa cells was significantly higher than in these cells cultured in a monolayer (Koshiba et al., 2009). The obtained results show a significant difference between the applied research methods, simultaneously indicating the validity of using 3D culture.
In the described study, JAM-A expression in human tissues was determined immunohistochemically, but in adenocarcinoma cell lines, also by RT-qPCR. The study included 24 normal endometrial tissue samples and endometrial carcinoma tissue specimens from 50 women (Koshiba et al., 2009).
Over the past decades, aberrant JAM-A expression and its function in the pathogenesis of neoplastic diseases, including gynecological cancers, have been the subjects of research interest. To the best of our knowledge, contradictory JAM-A roles in various cancer-type progression suggest that its influence is complex and tumor-type specific. Nowadays, only a few studies are available about the JAM-A role in gynecological cancers (Fig. 1) (Koshiba et al., 2009; Akimoto et al., 2016; Boljevic et al., 2019; Communal et al., 2020; Murakami et al., 2021). To sum up, possible mechanisms by which aberrant JAM-A levels could influence gynecological cancer development and progression are the cell apico-basal polarity loss, JAM-A delocalization from the most apical part of the intracellular membrane to the whole membrane, and increased EMT which takes part in the tumor development and progression. Disturbances of the described processes or their intensification could significantly affect the gynecological cancer progression.
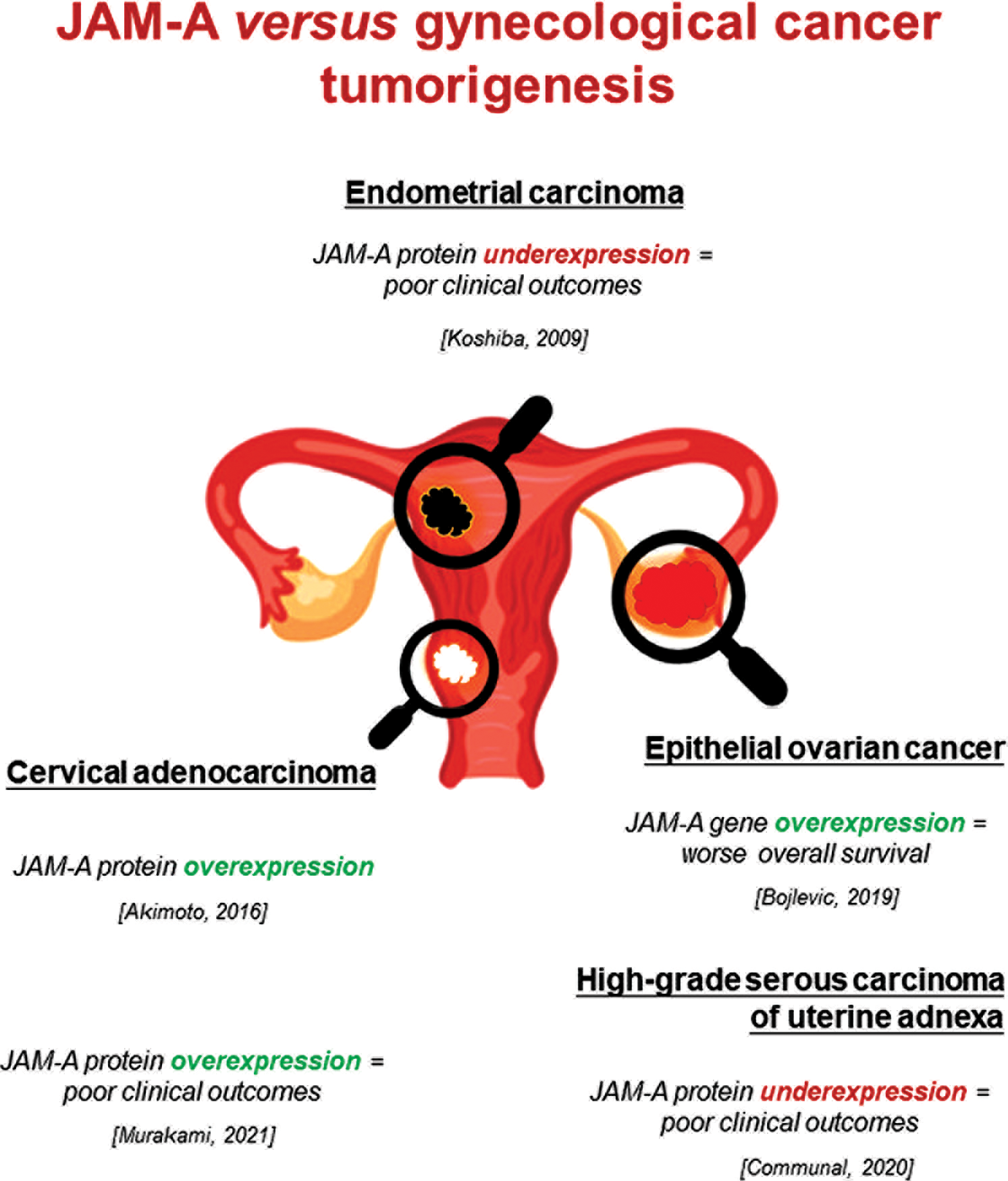
Figure 1: Aberrant expression of JAM-A gene/protein in gynecological cancers and its association with poor clinical parameters. This figure is designed using resources from Freepik.com.
Conflicting data about JAM-A expression level in EOC has been described in the literature (Boljevic et al., 2019; Communal et al., 2020). These studies had some limitations. Boljevic et al. (2019) determined the JAM-A gene expression levels using the RT-qPCR method in only 44 EOC tissue blocks. The tested EOC cohort was not homogeneous (only 26 samples could be HGSC). To strengthen the obtained results, the authors should also check the JAM-A protein level, add a control group to the research, choose a more homogeneous study cohort, and try to identify the possible molecular mechanisms by which JAM-A contributes to the EOC progression. In our opinion, this research is rather a retrospective study because researchers used only one laboratory method on a small EOC cohort and then performed a statistical analysis. One year later, Communal et al. (2020) published their studies determining JAM-A protein levels using immunofluorescent staining of 1,526 clinical samples from patients with HGSC (the most lethal EOC histotype). The authors also included normal fallopian tube tissues as a control cohort. Importantly, statistically significant differences in JAM-A protein expression between normal vs. cancer tissues were not observed—probably because of a significant difference between the size of the study groups (control: 15 samples, cancer: 1,526 samples). Moreover, the authors suggest that the EMT-dependent mechanism could be responsible for the obtained results. These discrepancies may be related to the large differences in the study group, as well as their heterogeneity and the use of only one experimental method.
In conclusion, the role of JAM-A in the tumor progression and metastasis of gynecological cancers is poorly understood and requires further research. Also, the underlying molecular mechanisms and pathways responsible for multiple JAM-A functions were not fully elucidated and need to be identified. Finding appropriate novel molecular cancer biomarkers may reduce observed very high mortality rates and could contribute to personalized treatment development.
Funding Statement: The authors received no specific funding for this study.
Author Contributions: The authors confirm their contribution to the paper as follows: conception and design: KCP, MS; data collection: KCP; draft manuscript preparation: KCP; manuscript revision: MS, KCP. All authors contributed to the writing and approved the final version of the manuscript.
Availability of Data and Materials: Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
Akimoto T, Takasawa A, Murata M, Kojima Y, Takasawa K et al. (2016). Analysis of the expression and localization of tight junction transmembrane proteins, claudin-1, -4, -7, occludin and JAM-A, in human cervical adenocarcinoma. Histology and Histopathology 31: 921–931. https://doi.org/10.14670/HH-11-729 [Google Scholar] [PubMed] [CrossRef]
Azari BM, Marmur JD, Salifu MO, Cavusoglu E, Ehrlich YH, Kornecki E, Babinska A (2010). Silencing of the F11R gene reveals a role for F11R/JAM-A in the migration of inflamed vascular smooth muscle cells and in atherosclerosis. Atherosclerosis 212: 197–205. https://doi.org/10.1016/j.atherosclerosis.2010.05.014 [Google Scholar] [PubMed] [CrossRef]
Babinska A, Clement CC, Przygodzki T, Talar M, Li Y et al. (2019). A peptide antagonist of F11R/JAM-A reduces plaque formation and prolongs survival in an animal model of atherosclerosis. Atherosclerosis 284: 92–101. https://doi.org/10.1016/j.atherosclerosis.2019.02.014 [Google Scholar] [PubMed] [CrossRef]
Babinska A, Kedees MH, Athar H, Sobocki T, Sobocka MB, Ahmed T, Ehrlich YH, Hussain MM, Kornecki E (2002). Two regions of the human platelet F11-Receptor (F11R) are critical for platelet aggregation, potentiation and adhesion. Thrombosis and Haemostasis 87: 712–721. https://doi.org/10.1055/s-0037-1613070 [Google Scholar] [CrossRef]
Bednarek R, Selmi A, Wojkowska D, Karolczak K, Popielarski M, Stasiak M, Salifu MO, Babinska A, Swiatkowska M (2020). Functional inhibition of F11 receptor (F11R/junctional adhesion molecule-A/JAM-A) activity by a F11R-derived peptide in breast cancer and its microenvironment. Breast Cancer Research Treatment 179: 325–335. https://doi.org/10.1007/s10549-019-05471-x [Google Scholar] [PubMed] [CrossRef]
Boljevic I, Malisic E, Milovic-Kovacevic M, Jovanic I, Bukumiric Z, Jankovic R (2019). High expression of junctional adhesion molecule-A is associated with poor survival in patients with epithelial ovarian cancer. The International Journal of Biological Markers 34: 262–268. https://doi.org/10.1177/1724600819850178 [Google Scholar] [PubMed] [CrossRef]
Caykara B, Alsaadoni H, Pence HH, Pence S, Yilmaz Aydogan H, Tastekin D (2019). Investigation of JAM-A (rs790056) and LFA-1 (rs8058823) gene variants in Turkish colorectal cancer patients. Turkish Journal of Gastroenterology 30: 872–876. https://doi.org/10.5152/tjg.2019.19141 [Google Scholar] [PubMed] [CrossRef]
Communal L, Medrano M, Sircoulomb F, Paterson J, Kobel M et al. (2020). Low junctional adhesion molecule-A expression is associated with an epithelial to mesenchymal transition and poorer outcomes in high-grade serous carcinoma of uterine adnexa. Modern Pathology 33: 2361–2377. https://doi.org/10.1038/s41379-020-0586-0 [Google Scholar] [PubMed] [CrossRef]
Corada M, Chimenti S, Cera MR, Vinci M, Salio M et al. (2005). Junctional adhesion molecule-A-deficient polymorphonuclear cells show reduced diapedesis in peritonitis and heart ischemia-reperfusion injury. Proceedings of the National Academy of Sciences of the United States of America 102: 10634–10639. https://doi.org/10.1073/pnas.0500147102 [Google Scholar] [PubMed] [CrossRef]
Cruz RGB, Madden SF, Richards CE, Vellanki SH, Jahns H et al. (2021). Human epidermal growth factor receptor-3 expression is regulated at transcriptional level in breast cancer settings by junctional adhesion molecule-a via a pathway involving beta-catenin and FOXA1. Cancers 13: 871. https://doi.org/10.3390/cancers13040871 [Google Scholar] [PubMed] [CrossRef]
Czubak-Prowizor K, Babinska A, Swiatkowska M (2022). The F11 Receptor (F11R)/Junctional Adhesion Molecule-A (JAM-A) (F11R/JAM-A) in cancer progression. Molecular and Cellular Biochemistry 477: 79–98. https://doi.org/10.1007/s11010-021-04259-2 [Google Scholar] [PubMed] [CrossRef]
Dai BQ, Jiang X, Feng LC (2021). LncRNA TP73-AS1 regulates miR-495 expression to promote migration and invasion of nasopharyngeal carcinoma cells through junctional adhesion molecule A. The Kaohsiung Journal of Medical Sciences 37: 361–370. https://doi.org/10.1002/kjm2.12338 [Google Scholar] [PubMed] [CrossRef]
Diaz-Coranguez M, Liu X, Antonetti DA (2019). Tight junctions in cell proliferation. International Journal of Molecular Sciences 20: 5972. https://doi.org/10.3390/ijms20235972 [Google Scholar] [PubMed] [CrossRef]
Fang TJ, Lin CH, Lin YZ, Li RN, Ou TT, Wu CC, Tsai WC, Yen JH (2016). F11R mRNA expression and promoter polymorphisms in patients with rheumatoid arthritis. International Journal of Rheumatic Diseases 19: 127–133. https://doi.org/10.1111/1756-185X.12663 [Google Scholar] [PubMed] [CrossRef]
Fong D, Spizzo G, Mitterer M, Seeber A, Steurer M, Gastl G, Brosch I, Moser P (2012). Low expression of junctional adhesion molecule A is associated with metastasis and poor survival in pancreatic cancer. Annals of Surgical Oncology 19: 4330–4336. https://doi.org/10.1245/s10434-012-2381-8 [Google Scholar] [PubMed] [CrossRef]
Gonzalez-Mariscal L, Dominguez-Calderon A, Raya-Sandino A, Ortega-Olvera JM, Vargas-Sierra O, Martinez-Revollar G (2014). Tight junctions and the regulation of gene expression. Seminars in Cell & Developmental Biology 36: 213–223. https://doi.org/10.1016/j.semcdb.2014.08.009 [Google Scholar] [PubMed] [CrossRef]
Huang JY, Xu YY, Sun Z, Wang ZN, Zhu Z, Song YX, Luo Y, Zhang X, Xu HM (2014). Low junctional adhesion molecule A expression correlates with poor prognosis in gastric cancer. Journal of Surgical Research 192: 494–502. https://doi.org/10.1016/j.jss.2014.06.025 [Google Scholar] [PubMed] [CrossRef]
Jiang X, Dai B, Feng L (2019). miR-543 promoted the cell proliferation and invasion of nasopharyngeal carcinoma by targeting the JAM-A. Human Cell 32: 477–486. https://doi.org/10.1007/s13577-019-00274-0 [Google Scholar] [PubMed] [CrossRef]
Kakiuchi A, Kakuki T, Ohwada K, Kurose M, Kondoh A et al. (2021). HDAC inhibitors suppress the proliferation, migration and invasiveness of human head and neck squamous cell carcinoma cells via p63mediated tight junction molecules and p21mediated growth arrest. Oncology Reports 45: 46. https://doi.org/10.3892/or.2021.7997 [Google Scholar] [PubMed] [CrossRef]
Khandoga A, Kessler JS, Meissner H, Hanschen M, Corada M, Motoike T, Enders G, Dejana E, Krombach F (2005). Junctional adhesion molecule-A deficiency increases hepatic ischemia-reperfusion injury despite reduction of neutrophil transendothelial migration. Blood 106: 725–733. https://doi.org/10.1182/blood-2004-11-4416 [Google Scholar] [PubMed] [CrossRef]
Koenen RR, Weber C (2022). Jam-A unleashed incites thromboinflammatory coronary artery disease. JACC: Basic to Translational Science 7: 462–464. https://doi.org/10.1016/j.jacbts.2022.03.014 [Google Scholar] [PubMed] [CrossRef]
Kornecki E, Walkowiak B, Naik UP, Ehrlich YH (1990). Activation of human platelets by a stimulatory monoclonal antibody. The Journal of Biological Chemistry 265: 10042–10048. https://doi.org/10.1016/S0021-9258(19)38776-9 [Google Scholar] [CrossRef]
Koshiba H, Hosokawa K, Kubo A, Tokumitsu N, Watanabe A, Honjo H (2009). Junctional adhesion molecule A [corrected] expression in human endometrial carcinoma. International Journal of Gynecological Cancer 19: 208–213. https://doi.org/10.1111/IGC.0b013e31819bc6e9 [Google Scholar] [PubMed] [CrossRef]
Kurose M, Kakuki T, Takano K, Kondo A, Obata K et al. (2016). Junctional adhesion molecule-A in head and neck squamous cell carcinoma. Advances in Oto-Rhino-Laryngology 77: 92–97. https://doi.org/10.1159/issn.0065-3071 [Google Scholar] [CrossRef]
Lampis A, Hahne JC, Gasparini P, Cascione L, Hedayat S et al. (2021). MIR21-induced loss of junctional adhesion molecule A promotes activation of oncogenic pathways, progression and metastasis in colorectal cancer. Cell Death & Differentiation 28: 2970–2982. https://doi.org/10.1038/s41418-021-00820-0 [Google Scholar] [PubMed] [CrossRef]
Laukoetter MG, Nava P, Lee WY, Severson EA, Capaldo CT et al. (2007). JAM-A regulates permeability and inflammation in the intestine in vivo. Journal of Experimental Medicine 204: 3067–3076. https://doi.org/10.1084/jem.20071416 [Google Scholar] [PubMed] [CrossRef]
Lee SK, Moon J, Park SW, Song SY, Chung JB, Kang JK (2005). Loss of the tight junction protein claudin 4 correlates with histological growth-pattern and differentiation in advanced gastric adenocarcinoma. Oncology Reports 13: 193–199. https://doi.org/10.3892/or.13.2.193 [Google Scholar] [CrossRef]
Leech AO, Cruz RG, Hill AD, Hopkins AM (2015). Paradigms lost-an emerging role for over-expression of tight junction adhesion proteins in cancer pathogenesis. Annals of Translational Medicine 3: 184. https://doi.org/10.3978/j.issn.2305-5839.2015.08.01 [Google Scholar] [PubMed] [CrossRef]
Leech AO, Vellanki SH, Rutherford EJ, Keogh A, Jahns H et al. (2018). Cleavage of the extracellular domain of junctional adhesion molecule-A is associated with resistance to anti-HER2 therapies in breast cancer settings. Breast Cancer Research 20: 140. https://doi.org/10.1186/s13058-018-1064-1 [Google Scholar] [PubMed] [CrossRef]
Magara K, Takasawa A, Osanai M, Ota M, Tagami Y et al. (2017). Elevated expression of JAM-A promotes neoplastic properties of lung adenocarcinoma. Cancer Science 108: 2306–2314. https://doi.org/10.1111/cas.13385 [Google Scholar] [PubMed] [CrossRef]
Mandell KJ, Babbin BA, Nusrat A, Parkos CA (2005). Junctional adhesion molecule 1 regulates epithelial cell morphology through effects on 1 integrins and rap1 activity. Journal of Biological Chemistry 280: 11665–11674. https://doi.org/10.1074/jbc.M412650200 [Google Scholar] [PubMed] [CrossRef]
Martin TA, Jiang WG (2009). Loss of tight junction barrier function and its role in cancer metastasis. Biochimica et Biophysica Acta 1788: 872–891. https://doi.org/10.1016/j.bbamem.2008.11.005 [Google Scholar] [PubMed] [CrossRef]
Martin TA, Mansel RE, Jiang WG (2010). Loss of occludin leads to the progression of human breast cancer. International Journal of Molecular Medicine 26: 723–734. https://doi.org/10.3892/ijmm_00000519 [Google Scholar] [PubMed] [CrossRef]
Martìn-Padura I, Lostaglio S, Schneemann M, Williams L, Romano M et al. (1998). Junctional adhesion molecule, a novel member of the immunoglobulin superfamily that distributes at intercellular junctions and modulates monocyte transmigration. The Journal of Cell Biology 142: 117–127. https://doi.org/10.1083/jcb.142.1.117 [Google Scholar] [PubMed] [CrossRef]
McSherry EA, McGee SF, Jirstrom K, Doyle EM, Brennan DJ, Landberg G, Dervan PA, Hopkins AM, Gallagher WM (2009). JAM-A expression positively correlates with poor prognosis in breast cancer patients. International Journal of Cancer 125: 1343–1351. https://doi.org/10.1002/ijc.24498 [Google Scholar] [PubMed] [CrossRef]
Murakami M, Giampietro C, Giannotta M, Corada M, Torselli I et al. (2011). Abrogation of junctional adhesion molecule-A expression induces cell apoptosis and reduces breast cancer progression. PLoS One 6: e21242. https://doi.org/10.1371/journal.pone.0021242 [Google Scholar] [PubMed] [CrossRef]
Murakami T, Takasawa A, Takasawa K, Akimoto T, Aoyama T et al. (2021). Aberrant expression of junctional adhesion molecule-A contributes to the malignancy of cervical adenocarcinoma by interaction with poliovirus receptor/CD155. Cancer Science 112: 906–917. https://doi.org/10.1111/cas.14734 [Google Scholar] [PubMed] [CrossRef]
Naik TU, Naik MU, Naik UP (2008). Junctional adhesion molecules in angiogenesis. Frontiers in Bioscience 13: 258–262. https://doi.org/10.2741/2676 [Google Scholar] [PubMed] [CrossRef]
Orlandella FM, Mariniello RM, Iervolino PLC, Auletta L, De Stefano AE et al. (2019). Junctional adhesion molecule-A is down-regulated in anaplastic thyroid carcinomas and reduces cancer cell aggressiveness by modulating p53 and GSK3 alpha/beta pathways. Molecular Carcinogenesis 58: 1181–1193. https://doi.org/10.1002/mc.23001 [Google Scholar] [PubMed] [CrossRef]
Padden M, Leech S, Craig B, Kirk J, Brankin B, McQuaid S (2007). Differences in expression of junctional adhesion molecule-A and β-catenin in multiple sclerosis brain tissue: Increasing evidence for the role of tight junction pathology. Acta Neuropathologica 113: 177–186. https://doi.org/10.1007/s00401-006-0145-x [Google Scholar] [PubMed] [CrossRef]
Prota AE, Campbell JA, Schelling P, Forrest JC, Watson MJ, Peters TR, Aurrand-Lions M, Imhof BA, Dermody TS, Stehle T (2003). Crystal structure of human junctional adhesion molecule 1: Implications for reovirus binding. Proceedings of the National Academy of Sciences of the United States of America 100: 5366–5371. https://doi.org/10.1073/pnas.0937718100 [Google Scholar] [PubMed] [CrossRef]
Rath D, Rapp V, Schwartz J, Winter S, Emschermann F et al. (2022). Homophilic interaction between transmembrane-JAM-A and soluble JAM-A regulates thrombo-inflammation: Implications for coronary artery disease. JACC: Basic to Translational Science 7: 445–461. https://doi.org/10.1016/j.jacbts.2022.03.003 [Google Scholar] [PubMed] [CrossRef]
Rosager AM, Sorensen MD, Dahlrot RH, Boldt HB, Hansen S, Lathia JD, Kristensen BW (2017). Expression and prognostic value of JAM-A in gliomas. Journal of Neuro-Oncology 135: 107–117. https://doi.org/10.1007/s11060-017-2555-0 [Google Scholar] [PubMed] [CrossRef]
Rusu AD, Georgiou M (2020). The multifarious regulation of the apical junctional complex. Open Biology 10: 190278. https://doi.org/10.1098/rsob.190278 [Google Scholar] [PubMed] [CrossRef]
Shimada H, Abe S, Kohno T, Satohisa S, Konno T et al. (2017). Loss of tricellular tight junction protein LSR promotes cell invasion and migration via upregulation of TEAD1/AREG in human endometrial cancer. Scientific Reports 7: 37049. https://doi.org/10.1038/srep37049 [Google Scholar] [PubMed] [CrossRef]
Smith YE, Wang G, Flynn CL, Madden SF, MacEneaney O et al. (2022). Functional antagonism of junctional adhesion molecule-A (JAM-Aoverexpressed in breast ductal carcinoma in situ (DCISreduces HER2-positive tumor progression. Cancers 14: 1303. https://doi.org/10.3390/cancers14051303 [Google Scholar] [PubMed] [CrossRef]
Solimando AG, Brandl A, Mattenheimer K, Graf C, Ritz M et al. (2018). JAM-A as a prognostic factor and new therapeutic target in multiple myeloma. Leukemia 32: 736–743. https://doi.org/10.1038/leu.2017.287 [Google Scholar] [PubMed] [CrossRef]
Solimando AG, Da Via MC, Leone P, Borrelli P, Croci GA et al. (2020). Halting the vicious cycle within the multiple myeloma ecosystem: Blocking JAM-A on bone marrow endothelial cells restores the angiogenic homeostasis and suppresses tumor progression. Haematologica 105: 1–19. https://doi.org/10.3324/haematol.2019.239913 [Google Scholar] [PubMed] [CrossRef]
Steinbacher T, Kummer D, Ebnet K (2018). Junctional adhesion molecule-A: Functional diversity through molecular promiscuity. Cellular and Molecular Life Sciences 75: 1393–1409. https://doi.org/10.1007/s00018-017-2729-0 [Google Scholar] [PubMed] [CrossRef]
Suren D, Yildirim M, Kaya V, Alikanoglu AS, Bulbuller N, Yildiz M, Sezer C (2014). Loss of tight junction proteins (Claudin 1, 4, and 7) correlates with aggressive behavior in colorectal carcinoma. Medical Science Monitor 20: 1255–1262. https://doi.org/10.12659/MSM.890598 [Google Scholar] [PubMed] [CrossRef]
Tarulli GA, Stanton PG, Loveland KL, Rajpert-De Meyts E, McLachlan RI, Meachem SJ (2013). A survey of Sertoli cell differentiation in men after gonadotropin suppression and in testicular cancer. Spermatogenesis 3: e24014. https://doi.org/10.4161/spmg.24014 [Google Scholar] [PubMed] [CrossRef]
Upadhaya P, Barhoi D, Giri A, Bhattacharjee A, Giri S (2019). Joint detection of claudin-1 and junctional adhesion molecule-A as a therapeutic target in oral epithelial dysplasia and oral squamous cell carcinoma. Journal of Cellular Biochemistry 120: 18117–18127. https://doi.org/10.1002/jcb.29115 [Google Scholar] [PubMed] [CrossRef]
Vellanki SH, Cruz RGB, Richards CE, Smith YE, Hudson L, Jahns H, Hopkins AM (2019). Antibiotic Tetrocarcin-A down-regulates JAM-A, IAPs and induces apoptosis in triple-negative breast cancer models. Anticancer Research 39: 1197–1204. https://doi.org/10.21873/anticanres.13230 [Google Scholar] [PubMed] [CrossRef]
Vences-Catalan F, Rajapaksa R, Kuo CC, Miller CL, Lee A, Ramani VC, Jeffrey SS, Levy R, Levy S (2021). Targeting the tetraspanin CD81 reduces cancer invasion and metastasis. Proceedings of the National Academy of Sciences of the United States of America 118: 24, e2018961118. https://doi.org/10.1073/pnas.2018961118 [Google Scholar] [PubMed] [CrossRef]
Vetrano S, Danese S (2009). The role of JAM-A in inflammatory bowel disease: Unrevealing the ties that bind. Annals of the New York Academy of Sciences 1165: 308–313. https://doi.org/10.1111/j.1749-6632.2009.04045.x [Google Scholar] [PubMed] [CrossRef]
Wang J, Chen X (2022). Junctional adhesion molecules: Potential proteins in atherosclerosis. Frontiers in Cardiovascular Medicine 9: 888818. https://doi.org/10.3389/fcvm.2022.888818 [Google Scholar] [PubMed] [CrossRef]
Wang J, Liu H (2022). The roles of junctional adhesion molecules (JAMs) in cell migration. Frontiers in Cell and Developmental Biology 10: 843671. https://doi.org/10.3389/fcell.2022.843671 [Google Scholar] [PubMed] [CrossRef]
Xu PP, Sun YF, Fang Y, Song Q, Yan ZX et al. (2017). JAM-A overexpression is related to disease progression in diffuse large B-cell lymphoma and downregulated by lenalidomide. Scientific Reports 7: 7433. https://doi.org/10.1038/s41598-017-07964-5 [Google Scholar] [PubMed] [CrossRef]
Zhao C, Wang A, Lu F, Chen H, Fu P, Zhao X, Chen H (2017). Overexpression of junctional adhesion molecule-A and EphB2 predicts poor survival in lung adenocarcinoma patients. Tumour Biology 39: 1010428317691000. https://doi.org/10.1177/1010428317691000 [Google Scholar] [PubMed] [CrossRef]
Zihni C, Balda MS, Matter K (2014). Signalling at tight junctions during epithelial differentiation and microbial pathogenesis. Journal of Cell Science 127: 3401–3413. https://doi.org/10.1242/jcs.145029 [Google Scholar] [PubMed] [CrossRef]
Zihni C, Mills C, Matter K, Balda MS (2016). Tight junctions: From simple barriers to multifunctional molecular gates. Nature Reviews Molecular Cell Biology 17: 564–580. https://doi.org/10.1038/nrm.2016.80 [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools