 Open Access
Open Access
ARTICLE
Increased MAD2L2 expression predicts poor clinical outcome in Colon Adenocarcinoma
1 School of Basic Medicine, Medical Genetics and Cell Biology, Ningxia Medical University, Yinchuan, 750004, China
2 Key Laboratory of Reproduction and Genetics, Ningxia Medical University, Yinchuan, 750004, China
3 Department of Human Anatomy and Histology Embryology, Ningxia Medical University, Yinchuan, 750004, China
* Corresponding Author: FANG XU. Email:
(This article belongs to the Special Issue: Bioinformatics Study of Diseases)
BIOCELL 2023, 47(3), 607-618. https://doi.org/10.32604/biocell.2023.026445
Received 06 September 2022; Accepted 15 November 2022; Issue published 03 January 2023
Abstract
Background: Colon adenocarcinoma (COAD) is the second leading cause of cancer death worldwide thus, identification of COAD biomarkers is critical. Mitotic Arrest Deficient 2 Like 2 (MAD2L2) is a key factor in mammalian DNA damage repair and is highly expressed in many malignant tumors. This is a comprehensive study of MAD2L2 expression, its diagnostic value, prognostic analysis, potential biological function, and impact on the immune system of patients with COAD. Methods: Gene expression, clinical relevance, prognostic analysis, diagnostic value, GO/KEGG cluster analysis, data obtained from TCGA, and bioinformatics statistical analysis were performed using the R package. Immune responses to MAD2L2 expression in COAD were analyzed using TIMER. The expression of MAD2L2 in HCT116 cells induced by the inflammatory factor TNF-α was detected using Western blot. Results: Our results underscore the clinical diagnostic value and potential biological significance of MAD2L2 in patients with COAD. A high level of MAD2L2 expression has been found in COAD and correlated with tumor status and colon polyps. ROC curve analysis showed that MAD2L2 expression has high diagnostic value in COAD. Analysis of immune infiltration results showed that MAD2L2 expression was positively correlated with neutrophil levels. The western blot results demonstrated that MAD2L2 was dose-dependently present with TNF-α. GO/KEGG revealed that MAD2L2 overexpressed and coexpressed genes were mostly involved in biological functions, including hypoxia response, response to reduced oxygen levels, mitochondrial translation elongation, and other processes. Conclusion: MAD2L2 as a new COAD biomarker contributes to our understanding of how alterations in gene expression and the immunological environment contribute to the development of colon cancer. Following further investigation, MAD2L2 may prove to be a viable target factor for clinical diagnosis and therapy of COAD.Keywords
Colon adenocarcinoma (COAD) is the second leading cause of cancer-related death worldwide, with approximately 1.2 million new cases each year (Malayaperumal et al., 2021). Despite significant advances in multimodal treatment options such as surgery, chemotherapy, radiation, and immunotherapy, the 5-year survival rate for patients with advanced COAD is just 6.6%, and the recurrence rate is substantial (Dulskas et al., 2020). Gene mutations and differences in individual drug responses are barriers to cancer treatment (Zhang et al., 2018; Saito et al., 2021). With the update of the 8th edition of the American Joint Committee on Cancer (AJCC) colorectal cancer (CRC) staging system, molecular tests such as MSI, KRAS, NRAS, and BRAF have been recommended based on high-level “evidence-based medicine” evidence, indicating that colorectal cancer has changed from traditional. Based on the “group” diagnosis and treatment, it has entered the precise “individualized” medical treatment (Weiser, 2018). Key genes involved in carcinogenesis are considered therapeutic targets in precision medicine. As an alternative to traditional colon cancer treatment, targeted therapy is suitable for early diagnosis of COAD (Yu et al., 2020). Anti-vascular endothelial growth factors (anti-VEGF) receptor (Fang et al., 2017), anti-epidermal growth factor receptor (anti-EGFR) (Hong et al., 2020), PD-1 (Zhao et al., 2020), CTLA-4 (Ben et al., 2021), and other genes are now utilized to treat COAD. Although molecular targeted therapy has shown good clinical effects, biomarker screening for the prognosis of COAD is still in the preliminary stage, and does not meet clinical needs. New target molecules are required to effectively treat COAD patients. Consequently, clarifying the development of novel biomarkers as diagnostic and therapeutic targets for COAD is crucial. Mitotic Arrest Deficient 2 Like 2 (MAD2L2) is a structural subunit of mutagenic DNA polymerase and a major component of the shieldin complex. It is involved in a range of biological activities, including DNA damage repair, cell cycle progression, and apoptosis. MAD2L2, as a participant in the DNA damage repair process, is highly expressed in multiple malignant tumors (de Krijger et al., 2021). MAD2L2 is highly expressed in epithelial ovarian cancer, and knockdown of MAD2L2 inhibits the proliferation of ovarian cancer cells, enhances chemosensitivity of ovarian cancer cells, and promotes apoptosis (Karakashev et al., 2020). Knockdown of MAD2L2 leads to reduced colony formation and increased apoptosis in irradiated esophageal squamous cell carcinoma cells and a reduction in tumor weight after radiation in a xenograft nude mouse model (Gu et al., 2019). In breast cancer cells, knockdown of MAD2L2 inhibits cell invasion and migration and promotes expression of transforming growth factor β1 (TGF-β1) (Feng et al., 2016). These results imply that MAD2L2 may have a role in biological processes as well as act as a possible biomarker for the clinical diagnosis and prognostic prediction of COAD (Rimkus et al., 2007). MAD2L2 overexpression was shown to be substantially related with reduced survival in a subset of human colorectal cancers. However, the present investigation did not assess the MAD2L2 gene expression’s unique clinical diagnostic significance in individuals with COAD. We examined COAD gene expression in this research utilizing bioinformatics analysis and data from The Cancer Genome Atlas (TCGA) database. This demonstrates the feasibility of MAD2L2 as a clinical diagnostic marker molecule for COAD. Simultaneously, we used clinical factors and TCGA gene expression data to develop a predictive nomogram that would assist physicians in forecasting the chance of death and directing treatment choices for COAD patients.
Evidence from the TCGA database
Our data were derived from the RNA-seq data obtained from the COAD project of the TCGA database (https://portal.gdc.cancer.gov/). Gene expression, baseline data, clinical logistic correlation, prognostic analysis, clinical diagnostic value, and gene function clustering data were obtained through the R package. Our study excluded samples with insufficient “0” gene expression values and survival information. We retained the RNA-seq and clinical data for further study. A total of 478 COAD patients with comparable clinical symptoms were included in this investigation. Our study conforms with the publishing criteria established by TCGA.
Tumor Immune Estimation Resource (TIMER) (https://cistrome.shinyapps.io/timer/) was utilized to investigate the possibility of a link between MAD2L2 expression and tumor-infiltrating immune cells. We used a genetic module to determine the relationship of MAD2L2 in COAD with tumor-infiltrating immune cells, including CD4+ T cells, dendritic cells, B cells, neutrophils, and macrophages. TIMER developed a graph demonstrating the association between the degree of gene expression and the purity of the tumor. Furthermore, we investigated the connection between MAD2L2 and 24 immune-infiltrating cells in COAD. The markers for 24 immune cells were taken from an article in Immunity (Bindea et al., 2013).
The colon cancer cell line HCT116, used in the experiments, was obtained from the American Type Culture Center. Cells were cultured in DMEM (Vivacell; Shanghai, China) supplemented with 10% FBS (Vivacell; Shanghai, China) and penicillin-streptomycin (10,000 U/ml; Beyotime Institute of Biotechnology). Culture was performed at 37°C and 5% CO2. When HCT116 cells reached 70% growth status, proteins were collected after treatment with TNF-α cytokines at different concentrations (0, 20, 20, 40, 60, 80, 100 ng/ml) for 24 h. TNF-α 50 mg (Peprotech, Suzhou, China).
Western blotting was used to isolate and identify proteins. Total proteins were extracted using RIPA buffer (Beyotime Institute of Biotechnology, Shanghai, China). Protein concentration was then determined using the BCA kit (Nanjing Capgemini Biotechnology Co., Ltd., China). Proteins (10–20 µg/well) were passed through 10% SDS-PAGE at 80V, and then transferred onto 0.2 µm PVDF membranes under wet conditions at 200 mA. The blotted membranes were blocked with 5% skim milk for 1 h at room temperature. The antibody was diluted in primary antibody diluent. Primary antibodies were incubated at room temperature for 3 h. Secondary antibodies were incubated for 1 h at room temperature. Chemiluminescence signal was detected using Pierce™ ECL and analyzed by Image Lab software (version 5.2.1 Bio-Rad Laboratories, Inc., BIO-RAD, CA, USA). The following antibodies were used: primary antibody-TNF-α (cat. 8184s; 1:1,000; cell signaling, USA), MAD2L2 (cat. ab180579; 1:1,000; Abcam, UK), GAPDH (cat. AB-P-R001; 1:1,000; Hangzhou Xianzhi Biotechnology Ltd., China); and secondary antibody-goat anti-rabbit IgG H&L (HRP) (cat. ZB-2306; 1:10,000; Beijing Zhongshan Jinqiao, China).
Transwell assay was used to detect cell invasion and migration. Cell migration was treated by taking cells in logarithmic growth phase and digesting the cells with trypsin. A 24-well plate was taken, 600 ul of complete medium was added to the lower chamber, and the transwell was placed in the plate and incubated for 48 h. Afterwards, 4% paraformaldehyde was added to fix the cells for 20 min, and then the cells were washed 3 times with PBS, stained with crystal violet for 15 min, and photographed and cell counted under a microscope (200X). The cell invasion was treated by first laying down the matrix gel. After that, a 24-well plate was taken, 600 ul of medium containing chemokine was added to the lower chamber, the transwell chambers after gelling were put into the well plate, 100–200 ul of cell suspension was taken and mixed and added to the upper chamber, and the culture was fixed with 4% paraformaldehyde after 48 h, stained with crystalline violet for 15 min, and Matrigel and the non-migrated cells in the upper chamber were gently wiped with cotton swabs, PBS-washed, photographed and cell counted under a microscope (200X). The following materials were mainly used, transwell chambers (Corning Inc., USA), and Matrigel gel (BD Inc., USA).
All statistical analyses were performed using R package (version 3.6.3). To calculate MAD2L2 expression, the Mann–Whitney U test was used. The correlation between clinical features and MAD2L2 expression was analyzed using a binary logistic model. In calculating the 95% CI and HR, the Cox regression module to analyze univariate and multivariate models was used. To compare numerous clinical parameters and survival rates, a univariate survival analysis was performed on each individual patient. Using multivariate Cox analysis, we were able to determine the expression of MAD2L2 as well as the presence of additional pathological and clinical variables. The threshold for MAD2L2 expression was established at a p-value of less than 0.05.
In February 2022, after screening, a total of 478 patients with clinical features related to MAD2L2 expression were obtained from the TCGA website. Table 1 lists the detailed clinical characteristics. Among the 478 participants, among those with low MAD2L2 expression, 121 were female (25.3%), and 118 were male (24.7%); among those with high expression of MAD2L2, 105 were female (22%), and 134 were male (28%). The age of all participants was cut off at 65 years. In terms of COAD pathological stage, among patients with low MAD2L2 expression, 39 patients were in Stage I (8.4%), 89 patients were in Stage II (19.1%), 69 patients were in Stage III (14.8%), and 37 patients were in Stage IV (7.9%); among patients with high MAD2L2 expression, 42 patients were in Stage I (9%), 98 patients were in Stage II (21%), 64 patients were in Stage III (13.7%), and 29 patients were in Stage IV (6.2%). History of colonic polyps, low expression of MAD2L2, 149 patients had no history of colonic polyps (36.5%) and 53 patients had a history of polyps (13%); among the highly expressed MAD2L2, 113 patients had no history of colon polyps (27.7%) and 93 patients had a history of colon polyps (22.8%). The median follow-up time among the 478 patients was 27.6 months.

MAD2L2 is expressed at a high level in COAD
MAD2L2 expression was increased in colon cancer, adrenocortical carcinoma, uroepithelial carcinoma of the bladder, invasive breast cancer, squamous and adenocarcinoma of the cervix, bile duct cancer, diffuse large B-cell lymphoma, esophageal cancer, glioblastoma multiforme, squamous cell carcinoma of the head and neck, low-grade (Fig. 1). RNA-seq research indicated that MAD2L2 expression was much greater in COAD cancer tissues than in paraneoplastic tissues in unpaired vs. paired samples (Figs. 2A and 2B). MAD2L2 mRNA expression was considerably elevated in COAD, as revealed by the findings.

Figure 1: Expression level of MAD2L2 gene in different tumors. (A) The expression status of the MAD2L2 gene in different cancers or specific cancer subtypes was analyzed through UCSC XENA. ns, p ≥ 0.05; *, p < 0.05; **, p < 0.01; ***, p < 0.001.

Figure 2: MAD2L2 expression in adenocarcinoma of the colon tissues. (A) Expression of MAD2L2 in normal and malignant tissues. (B) Expression of MAD2L2 in matched tissues. ***, p < 0.001.
High expression of MAD2L2 is associated with poor clinical prognosis
Table 2 summarizes the correlation between MAD2L2 expression and clinical logistic characteristics in patients with COAD. Expression of MAD2L2 was correlated with the history of colon polyps (p < 0.001) but not with other clinical features. Meanwhile, in the box plot of clinical features, we found that MAD2L2 expression correlated with tumor status and history of colon polyps (Figs. 3A–3H). The results of a univariate study using Cox regression revealed that MAD2L2 gene expression was a categorical dependent variable that was related with poor prognostic clinical characteristics (Table 3). The expression of MAD2L2 was significantly correlated with age greater than 65 years (p = 0.028, [CI] = 1.052–2.463). N stage (N1: p = 0.042, [CI] = 1.019–2.771, N2: p < 0.001, [CI] = 2.593–6.329), M stage (M1: p < 0.001, [CI] = 2.683–6.554), pathological stage (Stage III: p = 0.007, [CI] = 1.436–9.448, Stage IV: p < 0.001, [CI] = 3.608–23.936), CEA level (greater than 5: p < 0.001, [CI] = 1.788–5.471) are relevant. These results suggest that high MAD2L2 expression is highly correlated with poor clinical prognosis in COAD. We also examined the effect of high MAD2L2 expression on the migration and invasion of COAD cells HCT116 and found that the migration and invasion expression of HCT116 cells were significantly increased when MAD2L2 was highly expressed (Figs. 3I and 3J). This result indicates further validation that, high MAD2L2 expression causes poor prognosis of COAD.


Figure 3: MAD2L2 expression of patients with COAD according to different clinical characteristics. (A) Age; (B) Status; (C) T stage; (D) N stage; (E) M stage; (F) Pathologic stage; (G) Primary therapy outcome; (H) History of colon polyps; (I) Migration of HCT116 cells in control group and MAD2L2 high expression group; (J) Invasion of HCT116 cells in control group and MAD2L2 high expression group. ns, p ≥ 0.05; ***, p < 0.001.

The gene expression data for MAD2L2 was used to conduct a ROC curve analysis to determine the clinical diagnostic value of this gene. As shown in Fig. 4A, the AUC area of MAD2L2 was 0.93 when compared to paracancerous tissue vs. COAD. Using subgroup analysis, it was discovered that MAD2L2 gene expression was diagnostic in different stages of COAD, with AUC values of 0.922 in paracancerous tissue vs. stage I, 0.744 in T1/T2, 0.699 in T1/T3, 0.744 in T1/T4, and 0.744 in paracancerous tissue vs. N0 stage, paracancerous tissue vs. M0 stage, and paracancerous tissue vs. M1 stage being the most significant (Figs. 4B–4H). The results showed that MAD2L2 has certain diagnostic value in different clinical stages. We developed a nomogram to predict the 1-, 3-, and 5-year survival probability of patients with COAD by integrating the MAD2L2 gene expression level with characteristics related with clinical prognosis and diagnosis (Fig. 5). The score could be used by doctors to forecast patients’ 1-, 3-, and 5-year survival rates. These findings indicate that MAD2L2 could be utilized as a predictor in the diagnosis of COAD at various clinical stages.

Figure 4: Diagnostic value of MAD2L2 expression in colon cancer. (A) ROC curve for MAD2L2 in normal colon tissue and COAD; (B–H)Subgroup analysis for Normal vs. stage I, T1/T2, T1/T3, T1/T4, Normal vs. N0, Normal vs. M0, Normal vs. M1.

Figure 5: Nomogram used to predict the probability of patients with 1-, 3-, and 5-year overall survival. For risk estimation, determine the status of each clinical factor and the expression value of MAD2L2, and draw a straight line up to the point axis to view the points generated by a single factor. Repeat until the score of all factors is determined. Sum the points and find the summation point on the total points axis. The 1-, 3- and 5-year survival probabilities were then obtained by plotting straight down to the risk axis.
MAD2L2 affects the expression of infiltrating immune cells in COAD
Tumor-infiltrating immune cells play a key role in predicting overall survival in patients with COAD. Therefore, using TIMER, we analyzed the correlation between MAD2L2 expression and immune infiltration levels in COAD. As shown in Fig. 6A, MAD2L2 expression was positively correlated with neutrophil levels (p = 7.4 × 10−4). The results suggest that MAD2L2 plays a role in the immune infiltration of COAD. Additionally, we aimed to evaluate whether the tumor immune microenvironment of patients with COAD who expressed high levels of MAD2L2 was distinct from that of patients with COAD who expressed low levels of MAD2L2. The 480 COAD samples were divided into two groups based on MAD2L2 expression, with 240 samples classified as high expression and 240 as low expression. Using the R package, we performed calculations on immune infiltration to determine levels of 24 immune cells. The immune infiltration algorithm applied to 24 immune cell subtypes helped to assess differences in immune cell expression levels between the high and low MAD2L2 expression groups (Fig. 6B). T cells, aDCs, cytotoxic cells, DCs, iDCs, neutrophils, NK CD56 bright cells, NK CD56dim cells, Tcm cells, Th1 cells, Th17 cells, and Tregs were significantly affected by MAD2L2 expression. Compared with the low expression group, T cells, aDCs, cytotoxic cells, DCs, iDCs, neutrophils, NK CD56 bright cells, NK CD56dim cells, Th1 cells, Th17 cells, and TReg increased in the high expression group (p < 0.05), while Tcm decreased (p < 0.05). We also assessed possible correlations between 24 immune cells (Fig. 6C). The generated heatmap revealed associations between the ratios of several tumor-infiltrating immune cell groups ranging from weak to robust. Also, we validated it at the protein level. Using different concentrations of TNF-α acting on colon cancer HCT116 cells, MAD2L2 showed a dose-dependent effect as the concentration of TNF-α increased (Figs. 7A and 7B). As a result, we hypothesize that MAD2L2 expression is associated with tumor immune infiltration throughout the development of COAD and, to a lesser degree, influences immune cell expression in the tumor immune microenvironment.
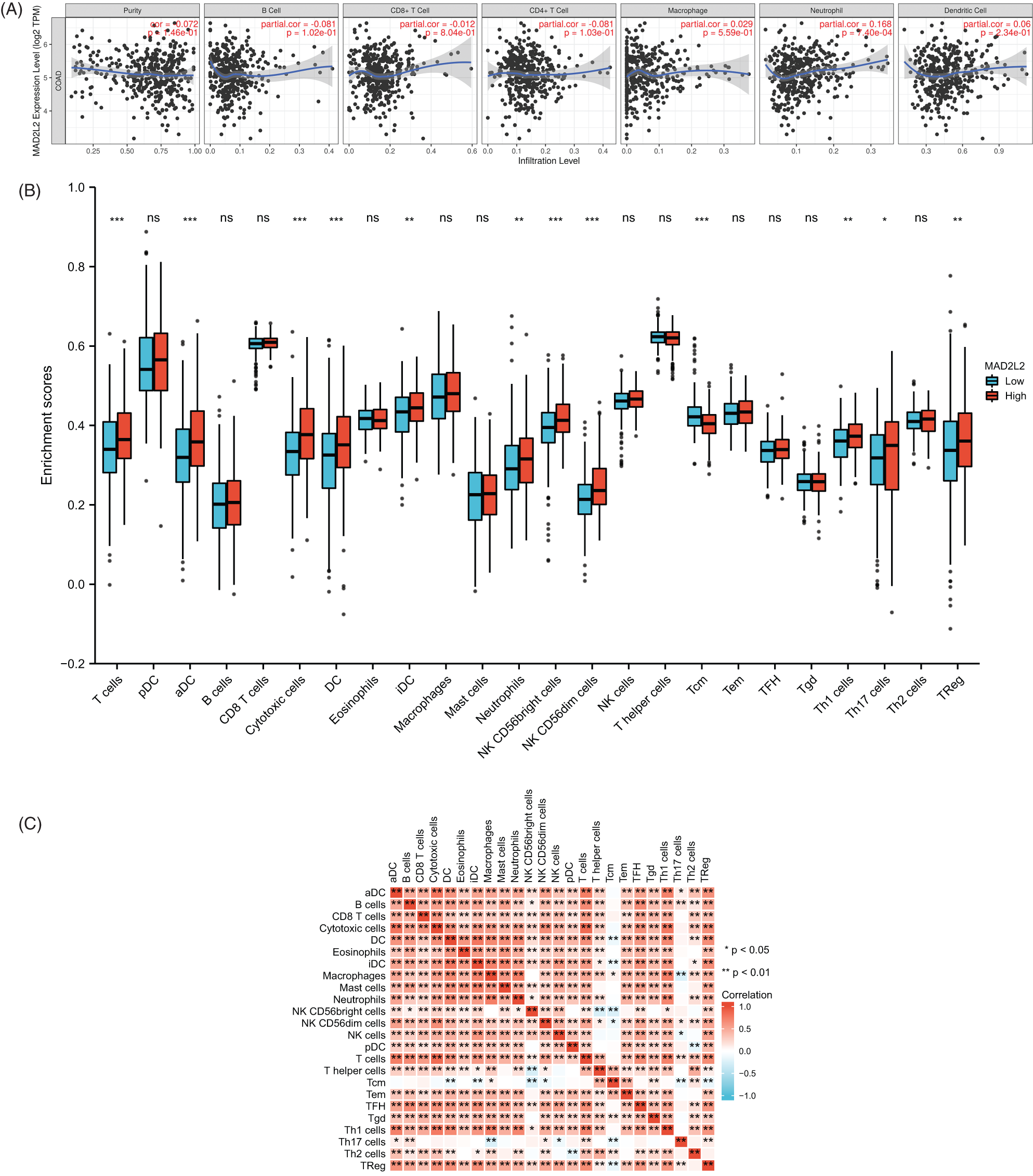
Figure 6: MAD2L2 expression and immunological infiltration are correlated. (A) Correlations between MAD2L2 expression and infiltration levels of the immune system. (B) The proportions of 24 immune cell subtypes in colon cancer samples with high and low MAD2L2 expression. (C) Heatmap depicting the invasion of 24 immune cells into colon cancer samples. ns, p ≥ 0.05; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
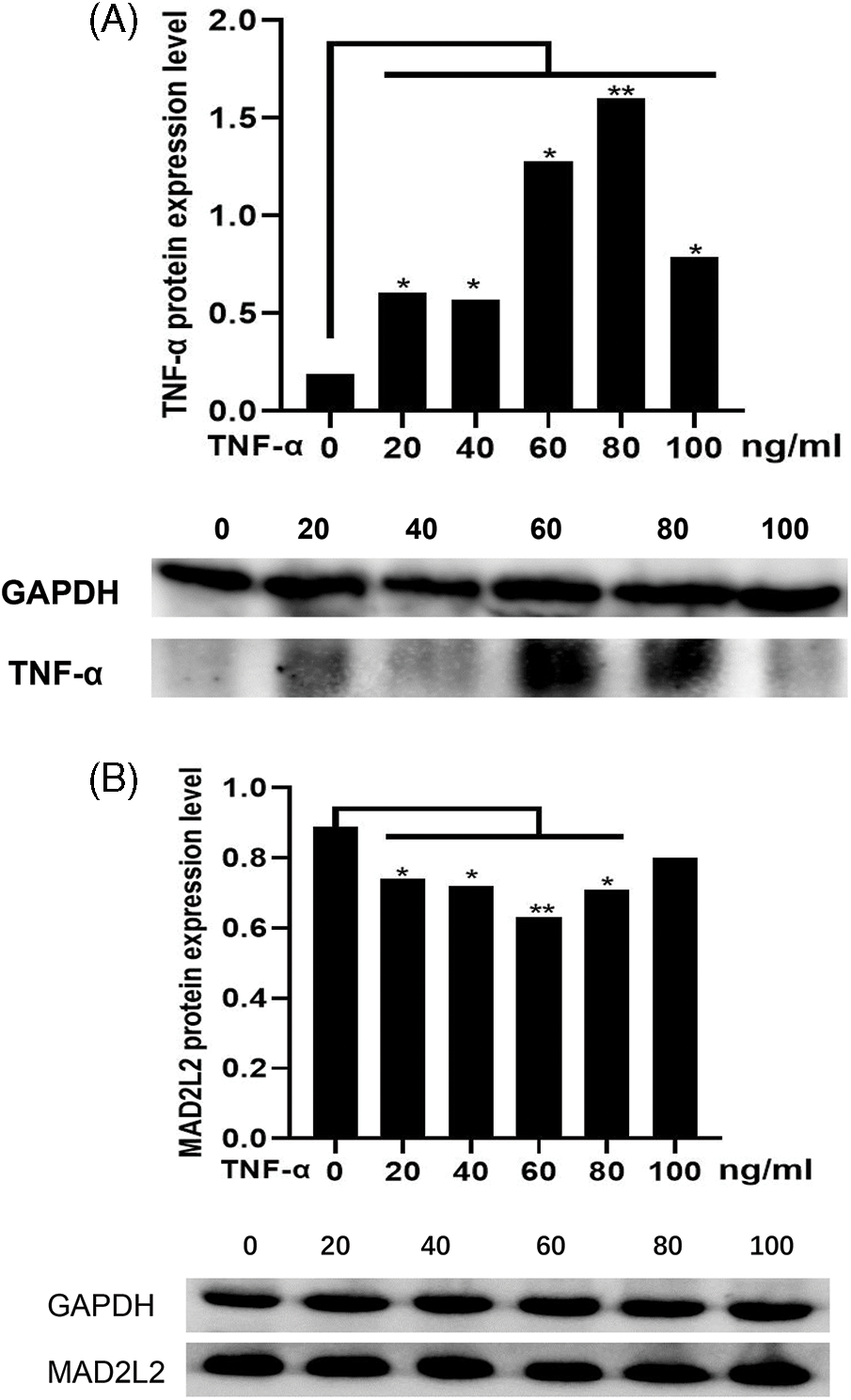
Figure 7: Effect of TNF-α stimulation on MAD2L2 expression. (A) Effect of TNF-α protein expression in COAD under different concentrations of TNF-α. (B) Effect of MAD2L2 protein expression in COAD under different concentrations of TNF-α. *, p < 0.05; **, p < 0.01.
Biological function of MAD2L2 in COAD
To understand the biological significance of MAD2L2 in COAD, we used the R package to analyze the co-expression network of MAD2L2 in COAD data. Based on our findings, we screened and examined the influence of the expression of the top 50 genes positively related with MAD2L2, against the top 50 genes negatively associated with MAD2L2 expression, on overall survival in colon cancer. The results are shown in the heat map in Figs. 8A and 8B. Genetic risk for COAD is predicted to be highest in the top 50 positively associated genes. The top 50 negatively related genes, on the other hand, were the ones that were most likely to be low-risk genes for COAD, according to the analysis. Additionally, this shows that MAD2L2, along with other genes, may be implicated in the overexpression of COAD risk factors and the downregulation of COAD preventative factors, as well as in the occurrence and development of COAD. The Kyoto Encyclopedia of Genes and Genomes (KEGG) Gene Ontology (GO) term annotations showed that MAD2L2 was mainly involved in biological processes when enriched with positively correlated genes, including response to hypoxia, response to decreased oxygen levels, anaphase-promoting complex-dependent catabolic process, response to oxygen levels, mitochondrial translational elongation, cellular components including vesicle coat, Fanconi anemia nuclear complex, ribosome, mitochondrial matrix, mitochondrial inner membrane, pathways including Huntington’s disease and Parkinson’s disease, ubiquitin mediated proteolysis, pathways of neurodegeneration-multiple diseases (Fig. 8C). Histone modification and covalent chromatin modification were inhibited, as were molecular functions such as histone binding, modification-dependent protein binding, histone demethylase activity (H3-K9 specific), methylation-dependent protein binding, and lysine degradation. Biological processes such as histone modification and covalent chromatin modification were also inhibited, as well as pathways such as lysine degradation, which were all inhibited as well (Fig. 8D). From these results, we infer that in COAD, processes such as the hypoxia response, response to reduced oxygen levels, and mitochondrial translation elongation are closely related to MAD2L2 expression.

Figure 8: Genes coexpressed with MAD2L2 in COAD. (A) The top 50 positively correlated genes cotranscript with MAD2L2 in COAD. (B) The top 50 negatively correlated genes cotranscript with MAD2L2 in COAD. (C) BP, CC and KEGG pathway analysis of MAD2L2 positively related genes in COAD. (D) BP, MF and KEGG pathway analysis of MAD2L2 negatively correlated genes in COAD. ***, p < 0.001.
Approximately 70%–80% of colorectal cancers are sporadic tumors (Fearon and Vogelstein, 1990), and their occurrence and development mostly follow the sequence of “adenoma-cancer”. It generally takes 5 to 10 years for the progression from precancerous lesions to cancer, which is the early disease stage. Diagnosis and clinical intervention provide important time windows (Brody, 2015). According to research, colorectal cancer screening, early detection, and treatment may significantly decrease the mortality of individuals with colorectal cancer (Lin et al., 2021). The current work established MAD2L2’s diagnostic and prognostic relevance in COAD, the processes behind its development, and its relationship with immune infiltration and mutation accumulation in hypoxic conditions.
In recent years, many genes important for colorectal cancer progression have been identified (Mo et al., 2015; Kundu et al., 2021; Smeby et al., 2020; Golla et al., 2020; Voutsadakis, 2021; Liu et al., 2019a, 2019b), including KRAS, TP53, APC, PIK3CA, PPARD, and MORC2. With the continuous development of high-throughput technology, microarray analysis has been used to identify unknown colorectal cancer-related oncogenes and biological network analysis (Bogaert and Prenen, 2014). An article published in 2007 indicated that increased MAD2L2 expression was related with a worse outcome in colorectal cancer (Rimkus et al., 2007). Tumors with high expression of MAD2L2 show a marked increase in abnormal mitosis, resulting in chromosomal instability (Varadi et al., 2011). Aberrant expression of MAD2L2 disrupts genome integrity and leads to cancer (Dev et al., 2018). MAD2L2 overexpression promotes the proliferation of human breast cancer cells by promoting abnormal mitosis (Yuan et al., 2006). High MAD2L2 expression is associated with significantly shorter overall survival and progression-free survival in patients with diffuse large B-cell lymphoma (Okina et al., 2015). Consistent with these studies, the results of our RNA-seq data analysis confirmed that expression of MAD2L2 was higher in COAD cancer tissues than in paracancerous tissue in unpaired samples and paired samples, and the mRNA expression of MAD2L2 was significantly upregulated, suggesting that MAD2L2 may be involved in COAD and play an important role in its occurrence and development. In addition, the ROC curve results indicated that MAD2L2 has a high diagnostic value in COAD prognosis.
A study of human colorectal cancer transcripts found that MAD2L2 is not commonly upregulated in colorectal benign adenomas, and its upregulation may represent a relatively late event in the carcinogenesis process (Rimkus et al., 2007). Consistent with this, our investigation discovered a substantial association between MAD2L2 expression and a history of colonic polyps. Meanwhile, in the results of our univariate association analysis, we showed that high MAD2L2 expression was significantly and negatively associated with NCOA3. NCOA3 is a transcriptional co-activator with elevated expression in several tumor types including CRC, which was shown to promote CRC progression by enhancing the Notch signaling pathway (Mo et al., 2015). According to research conducted by Li et al. (2018) overexpression of MAD2L2 may reduce the proliferation, migration, and clonogenicity of CRC cells by triggering the degradation of NCOA3 in the cells. It is suggested that the upregulation of MAD2L2 may not be the cause but a passive consequence of the unstable cancer cell genome adjusting to maintain stability. Cancer-promoting factors such as CHD9, UBN2, and CPEB2, which also support this view, were significantly negatively correlated with the high expression of MAD2L2. When DNA damage occurs, POL3 is normally required in normal cells to complete repair. In contrast to normal cells, cancer cells generally have a penchant for relying on error-repair bypasses, especially the TLS repair bypass, and this pathway is commonly performed by the error-prone and more efficient POLζ, of which MAD2L2 is a subunit (Stodola et al., 2016). We hypothesize that the passive high expression of MAD2L2 in COAD contributes to genomic instability and mutational load tolerance in cancer cells.
MAD2L2 is primarily engaged in biological activities when overexpressed, including the hypoxia response, reaction to decreased oxygen levels, mitochondrial translation elongation, and other processes, as determined by Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) investigations. PRDX2, a typical 2-Cys antioxidant enzyme, plays an important role in scavenging hydrogen peroxide and reactive oxygen species (ROS) levels, thereby protecting cells from oxidative stress (de Franceschi et al., 2011). MAD2L2 can respond to changes in radiation-generated ROS levels by recruiting PRDX2 (Gu et al., 2019). Our findings indicate that MAD2L2 expression is positively linked with PRDX2 expression, and that the glycolytic pathway works as an energy source in cancer cells, resulting in compromised mitochondrial activity and long-term cellular exposure to the potential hazard of ROS. We speculate that MAD2L2 may cooperate with PRDX2 to scavenge overloaded hydrogen peroxide and ROS levels to maintain cancer cell homeostasis. This is more proof supporting MAD2L2’s passive impact.
Equally important, we used the TIMER database to reveal the correlation between MAD2L2 expression and immune infiltration in COAD. We found that MAD2L2 expression was positively correlated with neutrophil levels. When MAD2L2 is highly expressed, T cells, aDCs, cytotoxic cells, DCs, iDCs, neutrophils, NK CD56 bright cells, NK CD56 dim cells, Th1 cells, Th17 cells, and TReg increase. Studies have found that neutrophils are early triggers of various injuries, and their infiltration in intestinal lesions is a marker of the acute phase of ulcerative colitis (Dinallo et al., 2019). Neutrophil elastase, released by neutrophils, is involved in the body’s immune defense and inflammatory response. Elevated levels of neutrophil elastase are highly correlated with COAD invasion and metastasis (Huang et al., 2020). We reasoned that MAD2L2 overexpression might promote neutrophil immune responses and infiltration, thereby triggering tumor immune responses. These results indicate that MAD2L2 expression is required for the control and recruitment of invading immune cells throughout the formation and progression of COAD. However, more cell experiments and the accumulation of clinical data are needed to more accurately understand the interaction between MAD2L2 and neutrophils.
Given that MAD2L2 is significantly expressed in COAD, its increased expression is strongly associated with a poor clinical prognosis and has a high diagnostic value in COAD, suggesting that it may be employed as an independent prognostic factor for COAD patient survival. Additionally, we discovered a strong correlation between MAD2L2 expression and tumor-infiltrating immunity. A further discovery was made using gene set enrichment analysis, which revealed that MAD2L2 has a broad range of potential applications in the occurrence and development of COAD. This is the first study to assess the prognosis of patients with colon cancer from all directions when MAD2L2 expression is abnormal. With a greater knowledge of MAD2L2’s functional range, it may serve as a useful target factor for the clinical diagnosis and prognosis of COAD, perhaps paving the way for biomarker therapy to become the treatment of choice for COAD in the future. Furthermore, even if MAD2L2 can be applied to guide clinical practice in the future, other molecules that interact with MAD2L2 remain to be discovered because it is not a single gene or marker but a set of comprehensive molecular marker signatures.
Availability of Data and Materials: The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Author Contribution: HTS and FX designed the study. HYW performed table production. XL contributed to image drawing. YJH and JL performed download data. FMW and HTS acquired and interpretated the data. HW and HTS assisted with data analysis. All authors read and approved the final manuscript.
Ethics Approval: Not applicable.
Funding Statement: The present study was supported by the Ningxia Hui Autonomous Region Key Research and Development Program (Grant No. 2021BEG03084).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
Ben S, Zhu Q, Chen S, Li S, Du M, Xin J, Chu H, Zhang Z, Wang M (2021). Genetic variations in the CTLA-4 immune checkpoint pathway are associated with colon cancer risk, prognosis, and immune infiltration via regulation of IQCB1 expression. Archives of Toxicology 95: 2053–2063. DOI 10.1007/s00204-021-03040-0. [Google Scholar] [CrossRef]
Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M et al. (2013). Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 39: 782–795. DOI 10.1016/j.immuni.2013.10.003. [Google Scholar] [CrossRef]
Bogaert J, Prenen H (2014). Molecular genetics of colorectal cancer. Annals of Gastroenterology 27: 9–14. [Google Scholar]
Brody H (2015). Colorectal cancer. Nature 521: S1. DOI 10.1038/521S1a. [Google Scholar] [CrossRef]
de Franceschi L, Bertoldi M, de Falco L, Santos Franco S, Ronzoni L, Turrini F, Colancecco A, Camaschella C, Cappellini MD, Iolascon A (2011). Oxidative stress modulates heme synthesis and induces peroxiredoxin-2 as a novel cytoprotective response in β-thalassemic erythropoiesis. Haematologica 96: 1595–1604. DOI 10.3324/haematol.2011.043612. [Google Scholar] [CrossRef]
de Krijger I, Boersma V, Jacobs JJL (2021). REV7: Jack of many trades. Trends in Cell Biology 31: 686–701. DOI 10.1016/j.tcb.2021.04.002. [Google Scholar] [CrossRef]
Dev H, Chiang TW, Lescale C, de Krijger I, Martin AG et al. (2018). Shieldin complex promotes DNA end-joining and counters homologous recombination in BRCA1-null cells. Nature Cell Biology 20: 954–965. DOI 10.1038/s41556-018-0140-1. [Google Scholar] [CrossRef]
Dinallo V, Marafini I, di Fusco D, Laudisi F, Franzè E et al. (2019). Neutrophil extracellular traps sustain inflammatory signals in ulcerative colitis. Journal of Crohn’s and Colitis 13: 772–784. DOI 10.1093/ecco-jcc/jjy215. [Google Scholar] [CrossRef]
Dulskas A, Gaizauskas V, Kildusiene I, Samalavicius NE, Smailyte G (2020). Improvement of survival over time for colorectal cancer patients: A population-based study. Journal of Clinical Medicine 9: 4038. DOI 10.3390/jcm9124038. [Google Scholar] [CrossRef]
Fang X, Hong Y, Dai L, Qian Y, Zhu C, Wu B, Li S (2017). CRH promotes human colon cancer cell proliferation via IL-6/JAK2/STAT3 signaling pathway and VEGF-induced tumor angiogenesis. Molecular Carcinogenesis 56: 2434–2445. DOI 10.1002/mc.22691. [Google Scholar] [CrossRef]
Fearon ER, Vogelstein B (1990). A genetic model for colorectal tumorigenesis. Cell 61: 759–767. DOI 10.1016/0092-8674(90)90186-I. [Google Scholar] [CrossRef]
Feng L, Wei W, Heng Z, Yantao H, Chunbo W (2016). Knockdown of REV7 inhibits breast cancer cell migration and invasion. Oncology Research 24: 315–325. DOI 10.3727/096504016X14666990347590. [Google Scholar] [CrossRef]
Golla JP, Kandyliari A, Tan WY, Chen Y, Orlicky DJ, Thompson DC, Shah YM, Vasiliou V (2020). Interplay between APC and ALDH1B1 in a newly developed mouse model of colorectal cancer. Chemico-Biological Interactions 331: 109274. DOI 10.1016/j.cbi.2020.109274. [Google Scholar] [CrossRef]
Gu C, Luo J, Lu X, Tang Y, Ma Y et al. (2019). REV7 confers radioresistance of esophagus squamous cell carcinoma by recruiting PRDX2. Cancer Science 110: 962–972. DOI 10.1111/cas.13946. [Google Scholar] [CrossRef]
Hong CS, Sun EG, Choi JN, Kim DH, Kim JH et al. (2020). Fibroblast growth factor receptor 4 increases epidermal growth factor receptor (EGFR) signaling by inducing amphiregulin expression and attenuates response to EGFR inhibitors in colon cancer. Cancer Science 111: 3268–3278. DOI 10.1111/cas.14526. [Google Scholar] [CrossRef]
Huang H, Zhang H, Onuma AE, Tsung A (2020). Neutrophil elastase and neutrophil extracellular traps in the tumor microenvironment. Advances in Experimental Medicine and Biology 1263: 13–23. DOI 10.1007/978-3-030-44518-8. [Google Scholar] [CrossRef]
Karakashev S, Fukumoto T, Zhao B, Lin J, Wu S et al. (2020). EZH2 inhibition sensitizes CARM1-high, homologous recombination proficient ovarian cancers to PARP inhibition. Cancer Cell 37: 157–167.e6. DOI 10.1016/j.ccell.2019.12.015. [Google Scholar] [CrossRef]
Kundu S, Ali MA, Handin N, Conway LP, Rendo V, Artursson P, He L, Globisch D, Sjöblom T (2021). Common and mutation specific phenotypes of KRAS and BRAF mutations in colorectal cancer cells revealed by integrative-omics analysis. Journal of Experimental & Clinical Cancer Research 40: 225. DOI 10.1186/s13046-021-02025-2. [Google Scholar] [CrossRef]
Li Y, Li L, Chen M, Yu X, Gu Z et al. (2018). MAD2L2 inhibits colorectal cancer growth by promoting NCOA3 ubiquitination and degradation. Molecular Oncology 12: 391–405. DOI 10.1002/1878-0261.12173. [Google Scholar] [CrossRef]
Lin JS, Perdue LA, Henrikson NB, Bean SI, Blasi PR (2021). Screening for colorectal cancer: Updated evidence report and systematic review for the US preventive services task force. Journal of the American Medical Association 325: 1978–1998. DOI 10.1001/jama.2021.4417. [Google Scholar] [CrossRef]
Liu Y, Deguchi Y, Tian R, Wei D, Wu L et al. (2019a). Pleiotropic effects of PPARD accelerate colorectal tumorigenesis, progression, and invasion. Cancer Research 79: 954–969. DOI 10.1158/0008-5472.CAN-18-1790. [Google Scholar] [CrossRef]
Liu J, Shao Y, He Y, Ning K, Cui X, Liu F, Wang Z, Li F (2019b). MORC2 promotes development of an aggressive colorectal cancer phenotype through inhibition of NDRG1. Cancer Science 110: 135–146. DOI 10.1111/cas.13863. [Google Scholar] [CrossRef]
Malayaperumal S, Sriramulu S, Banerjee A, Makalakshmi MK, Pathak S (2021). Is biotechnological next-generation therapeutics promising enough in clinical development to treat advanced colon cancer? Current Pharmaceutical Biotechnology 22: 1287–1301. DOI 10.2174/1389201021666201126142716. [Google Scholar] [CrossRef]
Mo P, Zhou Q, Guan L, Wang Y, Wang W et al. (2015). Amplified in breast cancer 1 promotes colorectal cancer progression through enhancing notch signaling. Oncogene 34: 3935–3945. DOI 10.1038/onc.2014.324. [Google Scholar] [CrossRef]
Okina S, Yanagisawa N, Yokoyama M, Sakurai Y, Numata Y, Umezawa A, Higashihara M, Murakumo Y (2015). High expression of REV7 is an independent prognostic indicator in patients with diffuse large B-cell lymphoma treated with rituximab. International Journal of Hematology 102: 662–669. DOI 10.1007/s12185-015-1880-3. [Google Scholar] [CrossRef]
Rimkus C, Friederichs J, Rosenberg R, Holzmann B, Siewert JR, Janssen KP (2007). Expression of the mitotic checkpoint gene MAD2L2 has prognostic significance in colon cancer. International Journal of Cancer 120: 207–211. DOI 10.1002/(ISSN)1097-0215. [Google Scholar] [CrossRef]
Saito Y, Koya J, Kataoka K (2021). Multiple mutations within individual oncogenes. Cancer Science 112: 483–489. DOI 10.1111/cas.14699. [Google Scholar] [CrossRef]
Smeby J, Kryeziu K, Berg KCG, Eilertsen IA, Eide PW et al. (2020). Molecular correlates of sensitivity to PARP inhibition beyond homologous recombination deficiency in pre-clinical models of colorectal cancer point to wild-type TP53 activity. eBioMedicine 59: 102923. DOI 10.1016/j.ebiom.2020.102923. [Google Scholar] [CrossRef]
Stodola JL, Stith CM, Burgers PM (2016). Proficient replication of the yeast genome by a viral DNA polymerase. MOJ Bioorganic & Organic Chemistry 291: 11698–11705. DOI 10.1074/jbc.M116.728741. [Google Scholar] [CrossRef]
Varadi V, Bevier M, Grzybowska E, Johansson R, Enquist K et al. (2011). Genetic variation in genes encoding for polymerase ζ subunits associates with breast cancer risk, tumour characteristics and survival. Breast Cancer Research and Treatment 129: 235–245. DOI 10.1007/s10549-011-1460-z. [Google Scholar] [CrossRef]
Voutsadakis IA (2021). The landscape of PIK3CA mutations in colorectal cancer. Clinical Colorectal Cancer 20: 201–215. DOI 10.1016/j.clcc.2021.02.003. [Google Scholar] [CrossRef]
Weiser MR (2018). AJCC 8th edition: Colorectal cancer. Annals of Surgical Oncology 25: 1454–1455. DOI 10.1245/s10434-018-6462-1. [Google Scholar] [CrossRef]
Yu Z, Li X, Duan J, Yang XD (2020). Targeted treatment of colon cancer with aptamer-guided albumin nanoparticles loaded with docetaxel. International Journal of Nanomedicine 15: 6737–6748. DOI 10.2147/ijn.S267177. [Google Scholar] [CrossRef]
Yuan B, Xu Y, Woo JH, Wang Y, Bae YK, Yoon DS, Wersto RP, Tully E, Wilsbach K, Gabrielson E (2006). Increased expression of mitotic checkpoint genes in breast cancer cells with chromosomal instability. Clinical Cancer Research 12: 405–410. DOI 10.1158/1078-0432.CCR-05-0903. [Google Scholar] [CrossRef]
Zhang Y, Li X, Zhou D, Zhi H, Wang P et al. (2018). Inferences of individual drug responses across diverse cancer types using a novel competing endogenous RNA network. Molecular Oncology 12: 1429–1446. DOI 10.1002/1878-0261.12181. [Google Scholar] [CrossRef]
Zhao T, Feng Y, Guo M, Zhang C, Wu Q et al. (2020). Combination of attenuated Salmonella carrying PD-1 siRNA with nifuroxazide for colon cancer therapy. Journal of Cellular Biochemistry 121: 1973–1985. DOI 10.1002/jcb.29432. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools