 Open Access
Open Access
ARTICLE
The microRNA pathway activation in insect cell model upon Autographa californica multiple nucleopolyhedrovirus infection
1 Key Laboratory of Chemical Biology and Molecular Engineering of Ministry of Education, Institute of Biotechnology, Shanxi University, Taiyuan, 030006, China
2 Department of Medical Laboratory Science, Fenyang College, Shanxi Medical University, Fenyang, 032200, China
* Corresponding Author: YUEJUN FU. Email:
BIOCELL 2023, 47(3), 627-645. https://doi.org/10.32604/biocell.2023.026182
Received 21 August 2022; Accepted 24 October 2022; Issue published 03 January 2023
Abstract
Background: The immune system of insects exerts fundamentally different antiviral mechanisms than mammals. MicroRNAs (miRNAs) play vital roles in developing insect antiviral immunity. MiRNAs expression profiles of insects changed significantly during baculovirus infection. Methods: Differential expression profiles of miRNAs in Spodoptera frugiperda were monitored by next-generation sequencing (NGS) and RT-qPCR during Autographa californica multiple nucleopolyhedrovirus (AcMNPV) infection. The transcription levels of genes were detected by RT-qPCR. The 50% tissue culture infective dose (TCID50) endpoint dilution assay was used to determine the proliferation of progeny virus. Results: NGS revealed that 49 miRNAs were differentially expressed in Sf9 cells, and 10 of them were significantly up- or down-regulated. Though RT-qPCR analysis, we observed the similar trends for the expression patterns of significantly differentially expressed miRNAs from NGS. Moreover, the transcription levels of core genes, Exportin5, Dicer1, and Argonaute1, in miRNA biogenesis pathways were significantly increased after AcMNPV infection. For five selected miRNAs, miR-34-5p could regulate the proliferation of baculovirus progeny virus and energy metabolism. Conclusion: The miRNAs biogenesis pathway in Sf9 cells plays an important role and may be stimulated to resist AcMNPV infection. This work provides evidence for the molecular mechanism of baculovirus-insect interaction and offers novel ideas and directions for green pest control technology.Keywords
MicroRNAs (miRNAs) are evolutionarily conserved, short (19~25 nucleotides) noncoding RNAs that regulate the post-transcriptional silencing of target genes (Lee et al., 1993; Reinhart et al., 2000). Based on the canonical pathway, a miRNA gene is transcribed by RNA polymerase II in the nucleus to produce the primary miRNA (pri-miRNA) transcript, which contains one or more stem-loop structures; pri-miRNA is cleaved by Drosha (a ribonuclease III enzyme) into hairpin precursor miRNA (pre-miRNA), and then it is translocated into the cytoplasm by exportin-5. In the cytoplasm, pre-miRNA is cleaved by dicer, producing a short duplex RNA molecule (Lee et al., 2004; Thakur et al., 2022; Bartel, 2004). Two dicer proteins have been identified in insects: dicer-1 is mainly involved in miRNA biogenesis, and dicer-2 produces siRNAs (Lee et al., 2004). The miRNA duplex triggers the formation of the RNA-induced silencing complex (RISC), and Argonaute protein (Ago) is the critical component of RISC. There are two Ago proteins: Ago1 for miRNA and Ago2 for siRNAs; however, recent research indicates that miRNAs can also be loaded into Ago2 complexes, especially in insects (Yang and Lai, 2011). When the miRNA duplex becomes related to the RISC, one of the strands is degraded, and the other forms the mature miRNA (Yang and Lai, 2011). MiRNAs have been reported in plants and animals, even their virus. MiRNA expression profiles of various insects and diverse functions, such as development and host-virus interactions, were found (Monsanto-Hearne and Johnson, 2018; Shrivastava et al., 2022).
Baculoviruses are large double-stranded DNA viruses that infect insects. Autographa californica multiple nucleopolyhedroviruses (AcMNPV) is the archetypal species of the alphabaculovirus. Spodoptera frugiperda is one of the most important pests for many crops, such as corn, soybeans, and sorghum, especially for maize and cotton in America (Johnson, 1987). It has recently invaded the African and Asian continents and first appeared in Yunnan Province, China, in January 2019 (Li et al., 2020). The interaction between baculoviruses and their insect hosts is complex and varied. After infecting the hosts, both baculoviruses and the insect hosts change in a series of ways to respond to this state. Baculoviruses affect apoptosis, cell cycle, and development of the host (Zhou et al., 2004) and acquire beneficial genes from the host genome for its replication (Thiem, 2009). The hosts also respond to the virus infection simultaneously by regulating the gene expression of baculoviruses, activating some proteins that resist viruses and immune pathways, such as miRNAs pathways (Thiem, 2009; Kemp et al., 2013; Liu et al., 2015; Zhao et al., 2022b).
Based on previous studies, we tried to reveal the role of the miRNA biogenesis pathway in the response of hosts to baculoviruses and to find miRNAs that regulate baculovirus replication. Dubey et al. (2017) reported that miR-2b expression was up-regulated during chikungunya virus (CHIKV) infection in Aedes aegypti. The ubiquitin-related modifier (URM) transcript is negatively regulated by miR-2b to control CHIKV replication (Dubey et al., 2017). The expression level of bantam-3p was increased after AcMNPV infection in Sf9 cells. Virus DNA replication was decreased in the presence of bantam-3p mimic and increased in the presence of bantam-3p inhibitor (Shi et al., 2016). In this study, miRNA expression profiles of Sf9 cells were detected by next-generation sequencing (NGS) during AcMNPV infection. We revealed the miRNA expression in insect cells during baculovirus infection and the critical roles of the miRNA pathways in virus-host interaction. Further, we found the vital function of miR-34-5p, which was down-regulated after AcMNPV infection, in regulating viral replication.
S. frugiperda IPLB-Sf21-AE colonial isolate 9 (Sf9) cells were cultured in a serum-free insect cell culture medium (SFX-Insect, HyClone, US) at 27°C. Recombinant baculovirus AcMNPV-enhanced green fluorescent protein (EGFP), which was constructed by inserting the egfp gene under the locus of the p10 promoter of AcMNPV, was used to infect Sf9 cells. Sf9 cells were infected with five multiplicity of infection (MOI) AcMNPV-EGFP, and were harvested at 72 h post-infection (h p.i.).
Next-generation sequence analysis of Sf9-encoded miRNAs
Sf9 cells were infected with AcMNPV-EGFP and were used for the sequencing of small RNAs (sRNAs). Total RNA was extracted from Sf9 cells, and the small RNA libraries were further sequenced using Illumina Hiseq 2500 as per Illumina protocols by Sangon Biotechnology (Shanghai, China). The reads having lengths between 17–34 bp were considered.
Mature miRNA sequences and pre-miRNA sequences of all available insect species were obtained from the miRBase v22.1 database. The 17–35 bp reads were mapped against known mature sequences of S. frugiperda, and those reads satisfying the set criteria were separated and named based on their subject annotation. The unannotated sRNAs to known sequences of S. frugiperda were mapped to other insects’ miRNAs in miRBase. The highly homologous miRNAs to other insects were considered to be known miRNAs.
The remaining sequences were subjected to novel miRNAs, which were predicted by miRDeep2 v2.0.0.8 (Friedländer et al., 2012). The potential of novel miRNA precursors and sequences to form a hairpin structure was also considered by miRDeep2, and the miRDeep2 score was used to measure the authenticity of novel miRNAs. The name of the novel miRNAs was abbreviated with ‘-’ ligature species name, ‘novel’, number, ‘mature’ or ‘star’, for example, ‘sfr-novel-1-mature’.
MiRNAs expression from NGS was calculated after normalizing the reads. The samples were normalized by calculating reads per million of total sRNA reads (RPM) and were used for comparing the relative abundance of specific miRNAs. The miRNAs in AcMNPV-infected cells were further profiled against mock cells to identify the differentially expressed miRNAs upon AcMNPV-EGFP infection by edgeR (Zhou et al., 2014). The expression value of RMP after normalization was >1 as the filtering condition, and the miRNAs with low expression levels were filtered out.
MiRNAs target genes prediction was performed by the Miranda algorithm (Fahlgren et al., 2007). The threshold parameter set in the prediction process was S ≥ 150, ΔG ≤ −30 kcal/mol, and demanded strict 5′ seed pairing. S refers to single-residue-pair match scores in the matching area, ΔG is Gibbs free energy of miRNA: mRNA binding formation. 3′ untranslated region (3′UTRs) of S. frugiperda were supplied from transcriptome sequencing data (National Center for Biotechnology Information Sequence Read Archive (SRA) accession numbers: SRP075752 (Sf9 cells), SRP075795 (Sf9 cells infected by AcMNPV-EGFP), which have been published by our laboratory (Wei et al., 2017). The targets of selected down- or up-regulated miRNAs were further subjected to analysis by clusterProfiler for Gene Ontology enrichment and KEGG pathway, keeping the threshold P-value < 0.05.
Differential expression of miRNAs analysis by reverse transcription qPCR (RT-qPCR)
The significantly differentially expressed miRNAs from NGS were verified by RT-qPCR. Total RNA was isolated using RNAiso Plus (Takara, Dalian, China). cDNA of mature miRNAs was synthesized by the tailing reaction method using miRNA First Strand cDNA Synthesis kits (Sangon Biotechnology Corp, Shanghai, China). The U6 RNA was included as the reference. The stem-loop RT-qPCR method is widely used to detect and quantify mature miRNAs. For cDNA synthesis of miRNA by stem-loop RT-qPCR method, total RNA was reverse transcribed in combination with the specific stem-loop reverse transcription primer of each miRNA. The 5S rRNA was reverse transcribed by random primer and included as the reference for the respective sample. The sequence of all primers is listed in Suppl. Table 1.
The transcription level of protein genes in the miRNAs biogenesis pathway was validated by reverse transcription qPCR
Total RNA was isolated using RNAiso Plus (Takara Biotechnology, Dalian, China). The cDNA was synthesized using an EasyScript First Strand cDNA Synthesis SuperMix (Transgen, Beijing, China) with oligo dT primer, and the β-actin was used as the endogenous reference gene. The qPCR was conducted with PerfectStartTM Green qPCR SuperMix (Transgene, Beijing, China). The 2−ΔΔCq method was used to calculate the relative level of miRNA or mRNA. The sequence of all primers is listed in Suppl. Table 1.
miR-11-3p, miR-2a-3p, miR-34-5p, miR-92a-3p, and miR-278-3p mimics and inhibitors were chemically synthesized and modified by Sangon Biotechnology (Shanghai, China). The negative control of mimic (mimic NC) and inhibitor (inhibitor NC) were provided by Sangon Biotechnology. The mimics and inhibitors were used to transfect into Sf9 cells by TransIT-Insect Transfection Reagent (Mirus, USA), following the manufacturer’s instructions. Sf9 cells were infected with 5 MOI AcMNPV-EGFP at 8 h after transfection with 5 nM miRNA mimic, 40 nM miRNA inhibitor, 5 nM mimic NC, and 40 nM inhibitor NC, respectively.
Proliferation analysis of progeny virus
Sf9 cells were infected with AcMNPV-EGFP after transfecting with miRNA mimics or inhibitors. The supernatants containing the budded virions (BVs) were harvested and serially diluted 10−1–10−8 times. The proliferation of progeny virus was determined using the 50% tissue culture infective dose (TCID50) endpoint dilution assay (Emery, 1992; Feng et al., 2012). The process is described below. The Sf9 cells were infected with the diluted viruses. The experiment with each concentration gradient was repeated eight times at least. After 5–7 days, the number of green fluorescence in each experimental group was counted, then the TCID50 of each group was calculated.
Determination of glucose consumption, lactic acid content, and ATP content
Supernatants in multiple groups were obtained to determine the contents of glucose and lactic acid using the Glucose content determination Kit and Lactic acid determination Kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China) according to the protocol of the manufacturers. Sf9 cells pellets were obtained to determine the content of ATP using the ATP content determination Kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China), and the protein concentrations were determined with BCA Protein Assay Kit (Solarbio Life Sciences, Beijing, China). The glucose mass in the culture medium was calculated according to the following formula:
Glucose mass (mg) = glucose content (mmol/L) × medium volume (L) × glucose molar mass (180 mg/mmol)
The glucose mass in the fresh culture medium was subtracted from the glucose mass in the medium of the treatment group to obtain glucose consumption.
Statistical analyses were performed by SPSS software. Data were shown as mean ± SD from at least three independent experiments. The averages of the treatment and the control groups were compared using a bidirectional unpaired Student’s t-test. P-value < 0.05 was set to be statistically significant (*P < 0.05, **P < 0.01).
sRNAs of Sf9 cells by next-generation sequencing
We sequenced small RNA libraries generated from uninfected Sf9 cells and AcMNPV-EGFP-infected Sf9 cells by Illumina Hiseq 2500. In total, 26176613 reads were generated from all libraries. After discarding low-quality and junk sequences, 3610334 and 4121022 unique reads without redundant sequences were obtained in size range of 17–35 bp in mock and AcMNPV-EGFP-infected Sf9 libraries, respectively (Table 1).
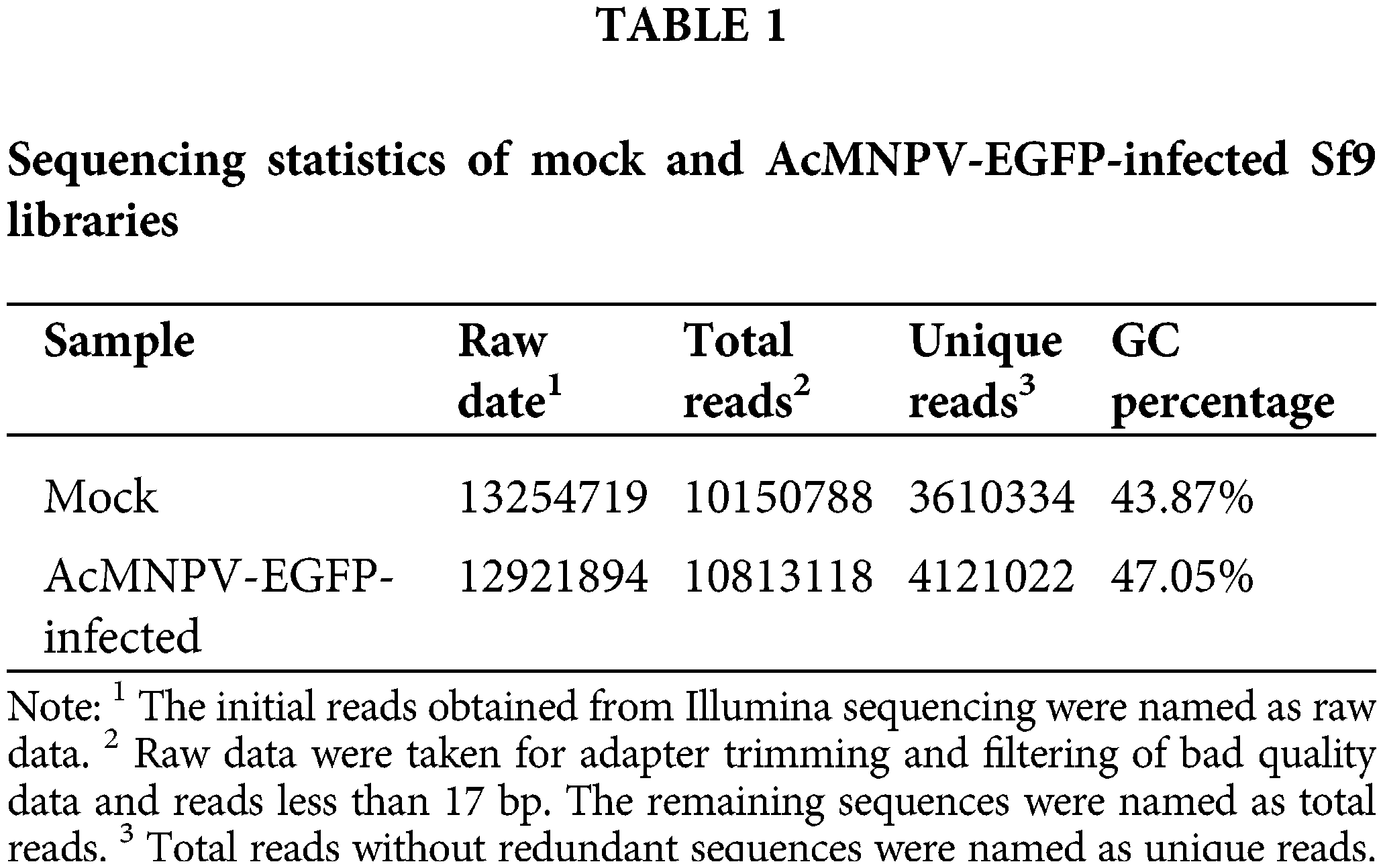
The length distributions of the total small RNA in size ranged from 17 to 35 nt and showed two prominent peaks. One peak was located between 19–22 nt, which is the typical length of mature miRNAs and siRNAs. However, we finally failed to find the miRNA or siRNA encoded by AcMNPV. The other peak was located between 27–30 nt, which may be considered the size of P-element induced wimpy testis (PIWI)-interacting RNAs (piRNAs) (Zharikova and Mironov, 2016). The number of miRNAs and piRNAs represented about 50% and 40% of the total sRNA reads, respectively, which were preponderant (Fig. 1a).
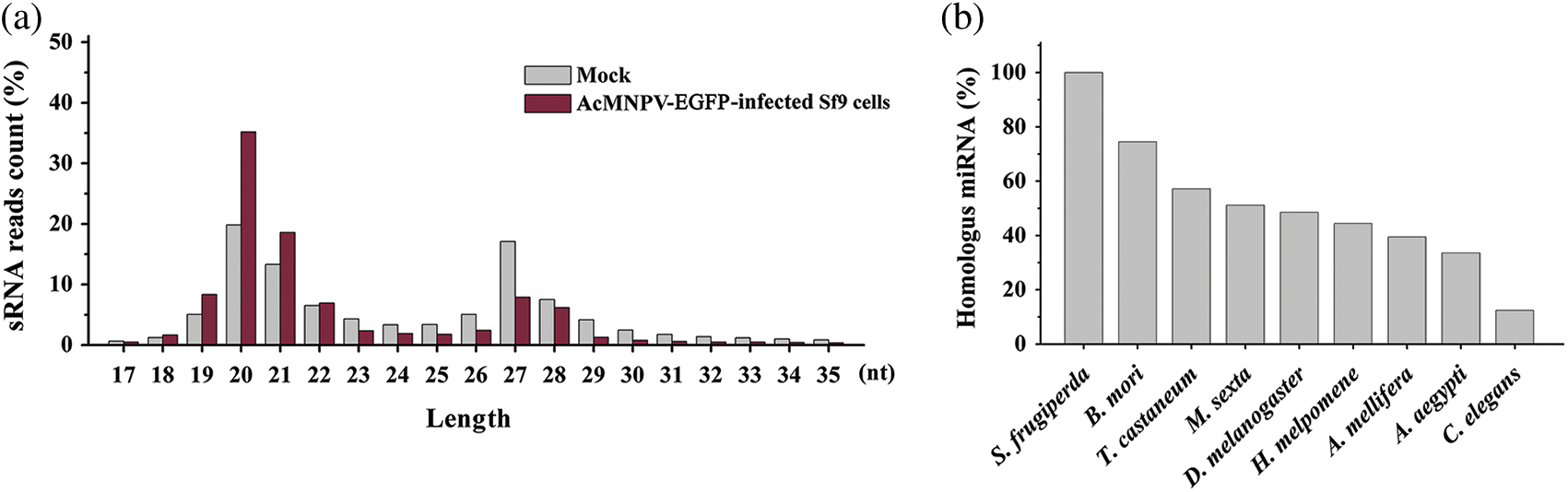
Figure 1: Length distribution of small RNA reads and conservation of homologous miRNAs in all libraries. (a) Length distribution of small RNA reads in mock and AcMNPV-EGFP-infected Sf9 libraries; (b) Percentage of miRNAs in Sf9 cell with homologous miRNAs in B. mori, T. castaneum, M. sexta, D. melanogaster, H. melpomene, A. mellifera, A. aegypti and C. elegans.
Identification of known miRNAs and novel miRNAs
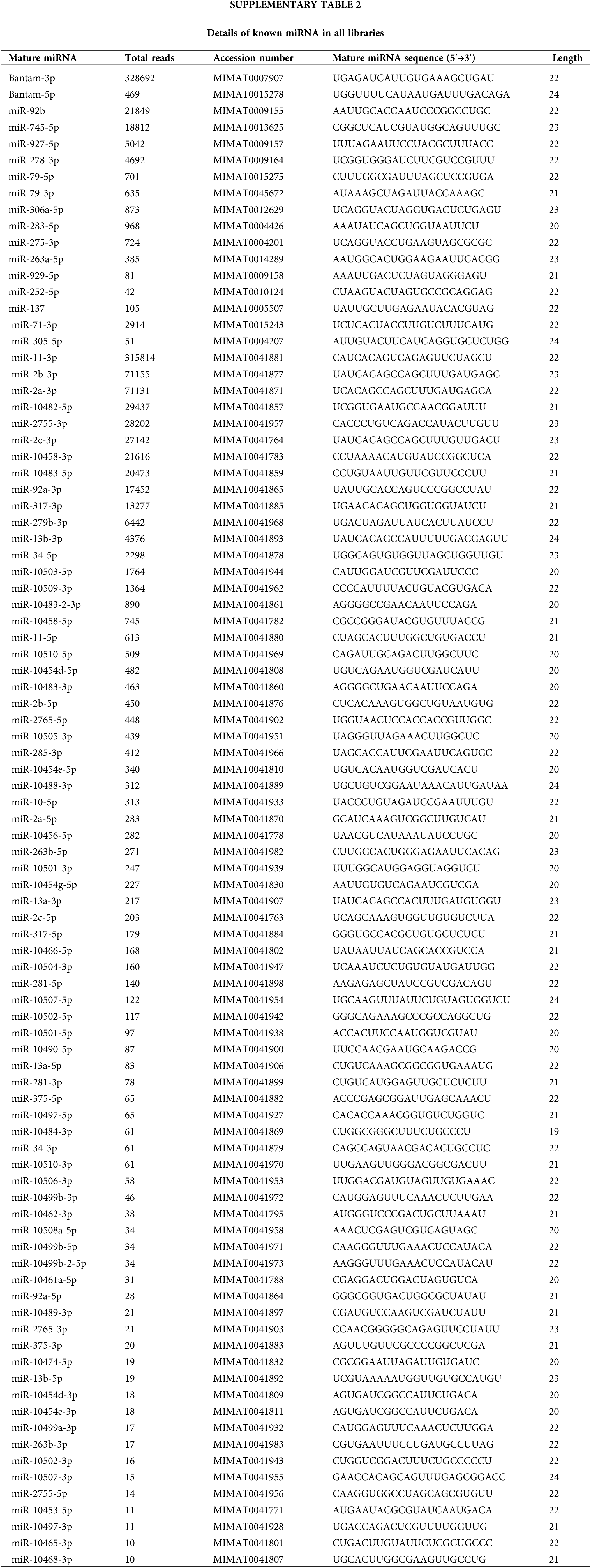
We detected 91 known miRNAs in all libraries. The details are shown in Suppl. Table S2. The top ten known miRNAs with the highest abundance were bantam-3p, miR-11-3p, miR-2b-3p, miR-2a-3p, miR-10482-5p, miR-2755-3p, miR-2c-3p, miR-92b, miR-10458-3p, miR-10483-5p, and some of them were in concurrence with recent reports (Mehrabadi et al., 2013; Kakumani et al., 2015).
The miRDeep2 v2.0.0.8. analysis revealed that 104 novel mature miRNAs were identified as novel miRNAs (Suppl. Table S3). A higher miRDeep score means a higher degree of authenticity for the novel precursor miRNAs, by extension, a higher degree of authenticity for novel miRNAs (Friedländer et al., 2008). Therefore, in Suppl. Table S3, the authenticity of novel miRNAs with higher miRDeep scores is greater than that with lower miRDeep scores, and the authenticity of each novel miRNA needs further experimental verification, such as the northern blot.
Not only the novel miRNAs but also some known miRNAs have homology with other insect species. More than 70% of homologous miRNAs were found in Bombyx mori, almost 60% in Tribolium castaneum, 50% in Manduca sexta and Drosophila melanogaster, 44% in Heliconius melpomene, 39% in Apis mellifera, 33% in Aedes aegypti, only 12% in Caenorhabditis elegans (Fig. 1b). miRNAs in B. mori and T. castaneum were more similar to these in S. frugiperda than other insects.
Differential expression of Sf9 miRNAs upon AcMNPV-EGFP infection
The result of NGS revealed 49 miRNAs differentially expressed in Sf9 cells (P < 0.05) (Fig. 2a). Based on P-value < 0.05 and the absolute value of log2 fold change >1 (difference times >2), we identified five significantly up-regulated miRNAs, which were miR-10461a-5p, miR-275-3p, miR-10453-5p, miR-10484-3p, miR-2765-5p; and five down-regulated miRNAs, including miR-263b-5p, miR-10499b-5p, bantam-5p, miR-745-5p, miR-79-5p. The differential expression level of miRNAs suggested that they might play important roles in host-viruses interaction.
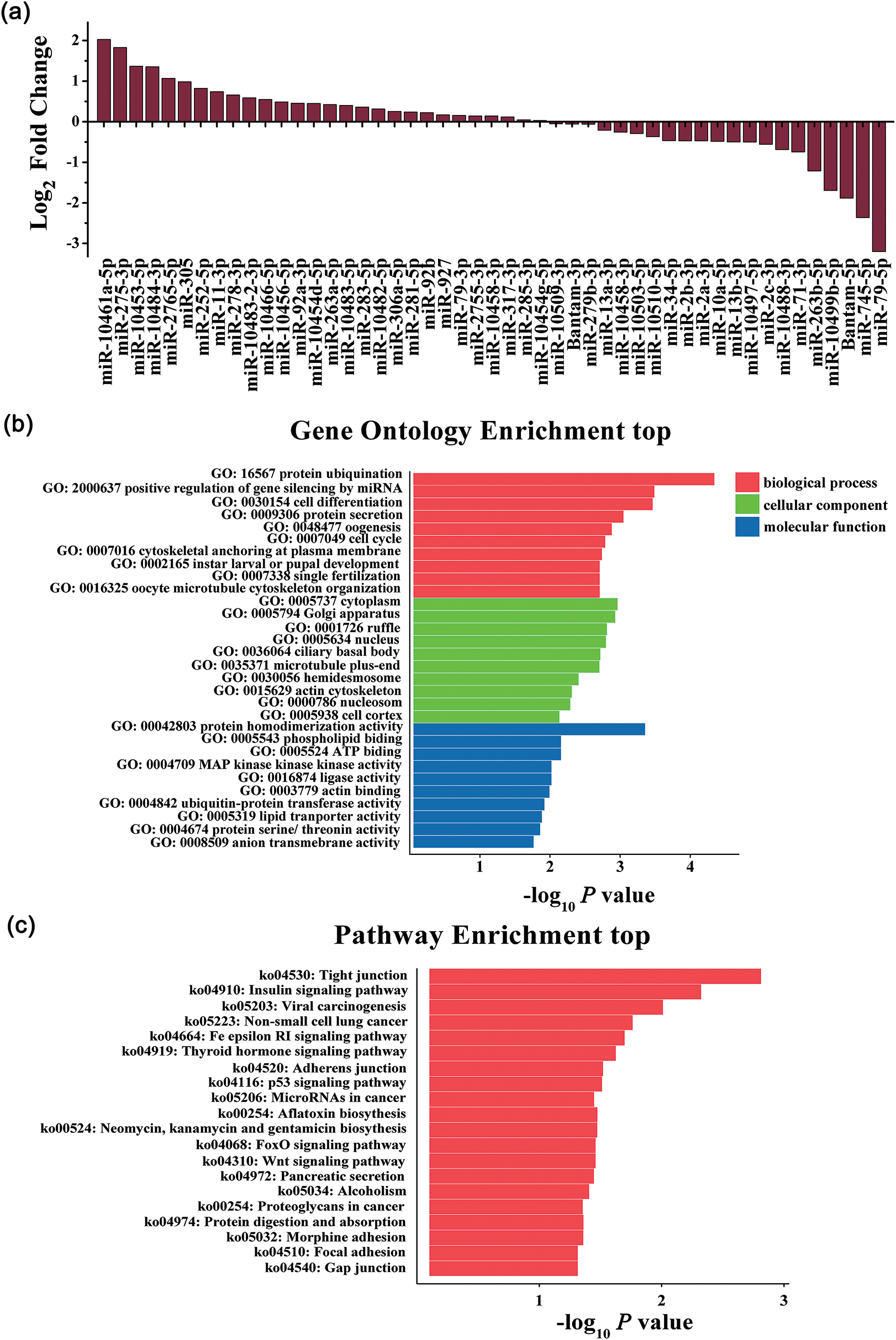
Figure 2: Differential expression of miRNAs detected by NGS and GO enrichment and KEGG pathway analysis of target genes. (a) Log2 Fold change of miRNAs identified in mock and AcMNPV-EGFP infected Sf9 cells by NGS; (b) GO enrichment analysis. Bar charts represented the top 10 of each Gene Ontology term targeted by miRNAs; (c) KEGG pathway analysis. Bar charts represented the top 20 KEGG pathways targeted by miRNAs.
Target gene prediction, GO enrichment, and KEGG pathway analyses
MiRNAs regulate target gene expression through the level of complementary binding between miRNAs sequence and their target mRNA. MiRNAs mainly bind to the 3′UTR of the target mRNA. According to the principle, the differentially expressed miRNAs mentioned above were analyzed for their putative targets by the Miranda algorithm. The predicted targets were involved in a broad range of biological processes. The significant GO terms and KEGG pathways target genes are shown in Figs. 2b and 2c. The most significantly enriched GO terms included nucleus (GO: 0005634) and cytoplasm (GO: 0005737), protein ubiquitination (GO: 0016567), etc. It is worth noting that positive regulation of gene silencing by miRNA (GO: 2000637) in biological process terms was included in GO enrichment. The most significantly enriched KEGG pathways included pathways in the insulin signaling pathway (ko04910), tight junction (ko04530), p53 signaling pathway (ko04115), etc. These results showed that miRNAs played vital roles in host-virus interaction by altering the expression of target genes, and these potential target genes were involved in a variety of biological processes.
Validation of differential expression miRNAs by RT-qPCR
Nine significantly differentially expressed miRNAs (excluding bantam-5p, a miRNA star of bantam) and some other miRNAs (miR-252-5p, miR-92a-3p, miR-92b, miR-11-3p, bantam-3p, miR-2a-3p, miR-34-5p, miR-278-3p) from NGS (Fig. 2a) were verified by RT-qPCR analysis. We observed a similar trend for the expression patterns of these miRNAs measured by RT-qPCR when compared with that in the miRNA NGS data (Fig. 3a). However, miR-278-3p was down-regulated, which was different from that of NGS. miR-34-5p, whose expression was down-regulated in Sf9 cells during AcMNPV-EGFP infection, became the focus of the follow-up studies.
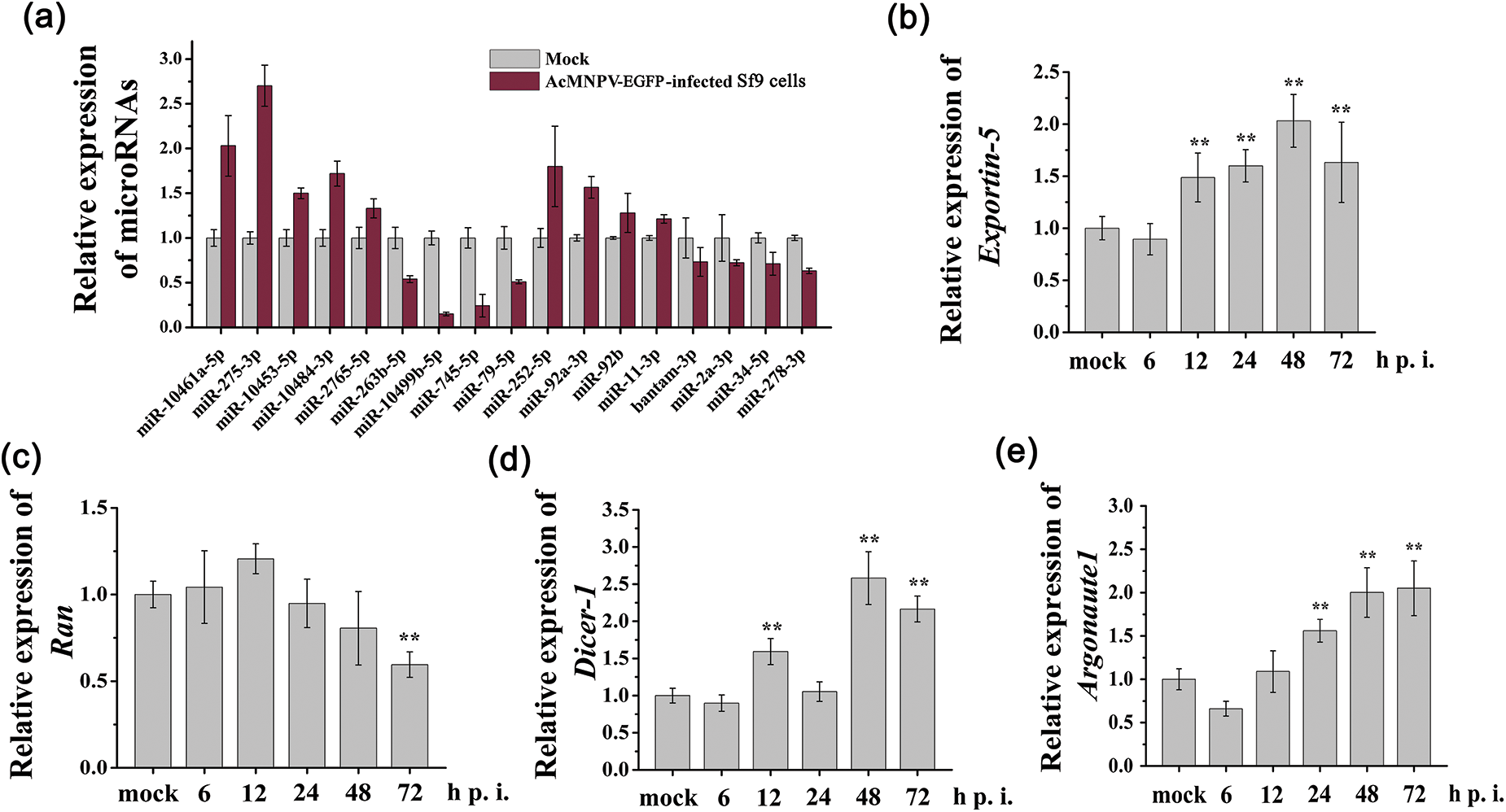
Figure 3: RT-qPCR analysis of relative expression of miRNAs and core genes in miRNAs biogenesis pathway. (a) miRNAs from the sequencing data were verified; (b~e) Relative transcription levels of core genes in miRNAs biogenesis pathway were verified. The asterisks indicated that the t-test showed a signifcant difference relative to the control (**P < 0.01).
Core genes in the miRNAs biogenesis pathway were regulated by AcMNPV-EGFP
As shown in Figs. 3b~3e, the relative transcription level of Exportin5 increased from 12 to 72 h p.i.; the transcription level of Ran remained almost unchanged until 72 h p.i.; the transcription level of Dicer1 was increased at 12, 48, and 72 h p.i.; the transcription level of Argonaute1 significantly increased from 24 to 72 h p.i. These results demonstrated miRNA biogenesis pathway in Sf9 cells was activated during AcMNPV-EGFP infection.
miRNA-34-5p suppressed the replication of progeny virus
According to the above results, the miRNA biogenesis pathway in Sf9 cells was activated to resist AcMNPV infection. MiRNAs play essential roles in insect-baculovirus interaction. However, which miRNAs play key roles in insect-baculovirus interactions? We selected five miRNAs, which are highly expressed in Sf9 cells and are sensitive to AcMNPV infection, and investigated their regulatory effects on the replication of AcMNPV. They were miR-11-3p, miR-2a-3p, miR-34-5p, miR-92a-3p, and miR-278-3p, respectively. Different synthetic miRNAs mimics and inhibitors were used to regulate the expression level of their respective miRNAs in Sf9 cells. The RT-qPCR analysis results showed that miRNAs expression level was significantly up and down-regulated in Sf9 cells by their mimics and inhibitors transfection until 72 h p.i. (Suppl. Fig. S1, Fig. 4b). As shown in Suppl. Figs. S1b, S1c and S1d, in miR-2a-3p, miR-92a-3p, and miR-278-3p mimics and inhibitors groups, the infectious BVs levels exhibited similar to AcMNPV-EGFP groups. It means that miR-2a-3p, miR-92a-3p, and miR-278-3p did not affect the proliferation of BVs. The proliferation levels in the AcMNPV-EGFP, miR-11-3p mimic, and miR-11-3p inhibitor groups showed no significant difference at 24 and 72 h p.i. However, at 48 h p.i., the proliferation level of progeny virus in the miR-11-3p mimic group was higher than that in the other two groups (Suppl. Fig. S1a). It implied that miR-11-3p exerted a particular effect on the BV proliferation.
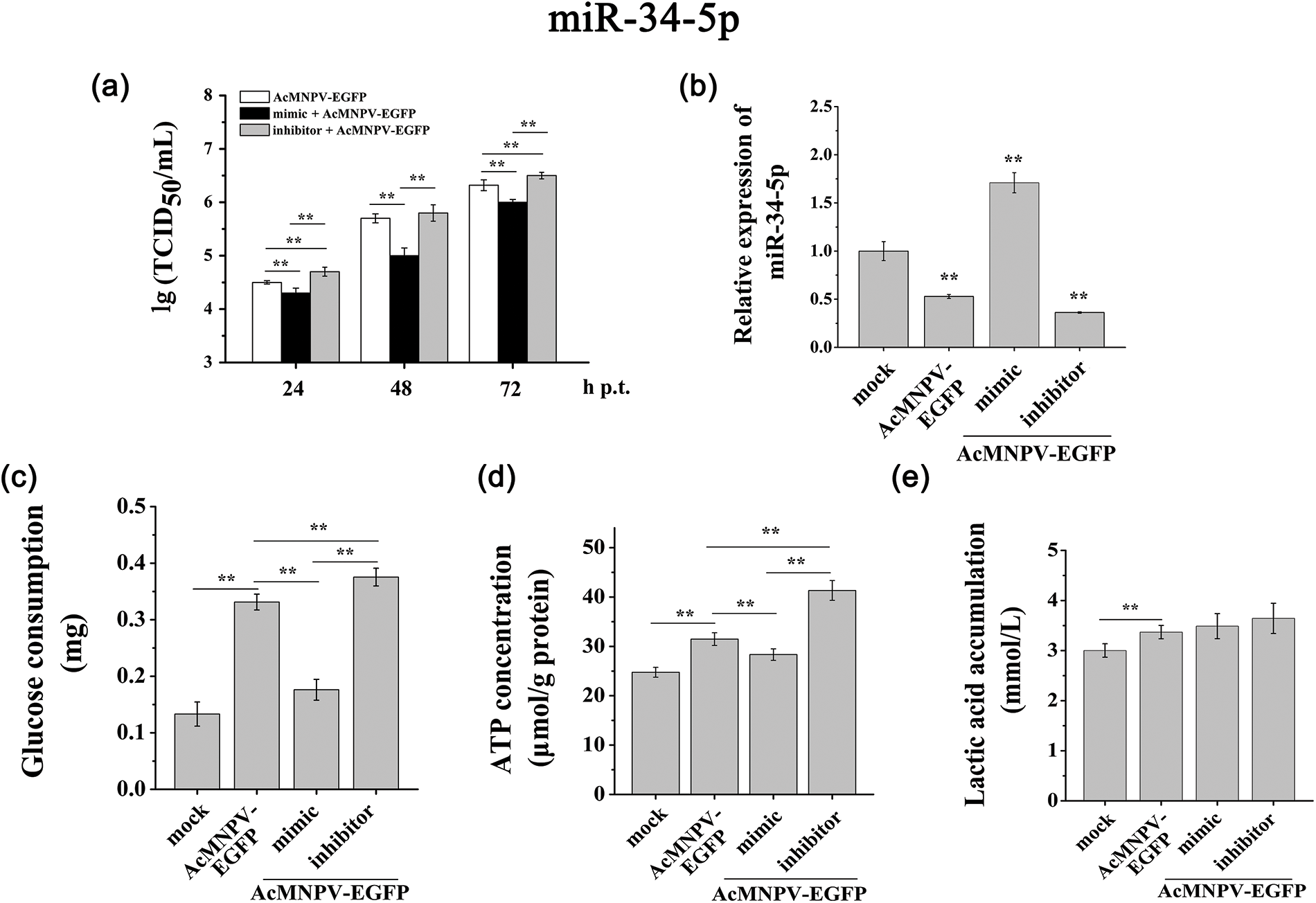
Figure 4: miR-34-5p regulated the proliferation of AcMNPV-EGFP and the energy metabolism of host cells. (a) 50% tissue culture infective dose (TCID50) end-point dilution assay; (b) RT-qPCR analysis of the expression of miR-34-5p after transfection with miR-34-5p mimic and inhibitor; (c~e) Effects of miRNAs on energy metabolism in host Sf9 cells. The effect of miR-34-5p mimic and inhibitor on the glucose consumption (c), ATP content (d) and lactic acid accumulation (e) in host Sf9 cells. The asterisks indicated that the t-test showed a signifcant difference relative to the control (**P < 0.01).
In these five miRNAs, we focused on miR-34-5p. In the miR-34-5p mimic treatment group, the BVs titer showed a distinct down regulation compared to AcMNPV-EGFP group at 24, 48, and 72 h p.i. In contrast, the proliferation levels of progeny virus in the miR-34-5p inhibitor group were increased compared to AcMNPV-EGFP group (Fig. 4a). These phenomena demonstrated that miR-34-5p inhibited the proliferation of progeny virus. When Sf9 cells were infected by AcMNPV, a series of changes were stimulated in the host, including the differential expression of some miRNAs. miR-34-5p inhibits the infectivity of progeny virus, while the virus inhibits the expression of miR-34-5p for its proliferation.
miR-34-5p suppressed the glucose metabolism in host cells
Baculovirus growth relies heavily on host resources. The utilization and distribution of energy in the host can influence viral replication (Feng et al., 2021). Glucose is the most critical energy source of insect cells in culture, and baculovirus uses the host’s glucose to provide energy for its replication through the tricarboxylic acid (TCA) cycle (Kamen et al., 1996; Bernal et al., 2009; Lin et al., 2020). The glucose consumption in the medium was increased significantly during AcMNPV-EGFP infection. At the same time, the ATP content of Sf9 cells showed similar trend to the glucose consumption during AcMNPV-EGFP infection, which was increased significantly (Suppl. Fig. S2). Overexpression of miR-11-3p increased the glucose consumption compared with the AcMNPV-EGFP group, and ATP content and lactic acid accumulation were also increased, although they were still higher than in uninfected group. Down regulation of miR-11-3p showed the opposite effects (Suppl. Fig. S2a). Overexpression of miR-2a-3p inhibited glucose consumption compared with the AcMNPV-EGFP group, and lactic acid accumulation was also decreased. Down regulation of miR-2a-3p showed the opposite effect (Suppl. Fig. S2b). For miR-92a-3p and miR-278-3p, there was no significant difference among mimic, inhibitor, and AcMNPV-EGFP groups in terms of the glucose consumption, ATP generation and lactic acid accumulation (Suppl. Figs. S2c and S2d). These phenomena suggested that miR-11-3p and miR-2a-3p affected the energy metabolism of host cells during AcMNPV infection.
Significantly, overexpression of miR-34-5p inhibited the glucose consumption and ATP content compared with AcMNPV-EGFP group, although the glucose consumption was still higher than in mock group. And down regulation of miR-34-5p showed the opposite effect (Figs. 4c and 4d). Compared with the AcMNPV-EGFP and mock groups, down regulation of miR-34-5p increased lactic acid accumulation, although the accumulation was still higher than that in mock group (Fig. 4e). These phenomena suggested that overexpression of miR-34-5p inhibited glucose utilization by virus, while down regulation of miR-34-5p contributed to lactic acid accumulation.
MiRNAs have been discovered in many insects, including B. mori, D. melanogaster, A. aegypti, etc. (He et al., 2008; Monsanto-Hearne and Johnson, 2018). Moreover, the expression levels of host miRNAs could be altered during virus infection, and the differential expression of the miRNAs represented one aspect of host antiviral responses (Kong et al., 2018). In this study, the length distributions of the total sRNA in size 17–35 nt showed two major peaks. One peak, located between 19–22 nt, was considered to be mature miRNA or siRNA because of the matched size. Theoretically, these miRNA or siRNA contain both host Sf9 cell encoded miRNA and AcMNPV encoded miRNA. According to the method (Singh et al., 2010), we used the BLASTn program to map the unique reads with the AcMNPV genome, 8626 unique reads from infected Sf9 cells mapped onto the AcMNPV genome. At present, five miRNAs encoded by AcMNPV have been found, which are AcMNPV-miR-1, AcMNPV-miR-2, AcMNPV-miR-3, AcMNPV-miR-4, and AcMNPV-miR-5, respectively. Among them, the functions of AcMNPV-miR-1, 3, and 4 have been clarified, respectively (Zhu et al., 2013; Zhu et al., 2016; Jiao et al., 2019; Wang et al., 2021; Zhao et al., 2022a). However, we did not find the miRNA or siRNA encoded by AcMNPV, including these five known AcMNPV-miRs. At the same time, due to the low copy number of miRNA encoded by AcMNPV, we are looking for novel methods to find them. The other peak between 25–30 nt corresponds to the size of piRNAs. PiRNAs have several functions, such as regulation of transposon expression and transposition, regulation of host genes’ expression, the influence on epigenetic mechanisms, histone modifications, and DNA methylation (Zharikova and Mironov, 2016; Kolliopoulou et al., 2019). Moreover, piRNAs have other structural features, such as a bias for U at position 1 and a ‘ping-pong’ signature (Vodovar et al., 2012). Therefore, whether sRNAs in the other peak of 25–30 nt are true piRNAs needs further study and proof. Here, we just focused on miRNAs encoded by Sf9 cells.
We detected 91 known miRNAs and 104 novel miRNAs in sRNA libraries. Upon AcMNPV-EGFP infection, miRNAs showed differential expression. The result revealed 49 miRNAs to be differentially expressed in Sf9 cells. These differentially expressed miRNAs are not the same as the report from Mehrabadi et al. (2013). They identifted 116 known miRNAs in Sf9 cells, and many miRNAs were differentially expressed. However, reads from deep sequencing were mapped to B. mori genome sequences as a primary source of reference because the S. frugiperda genome was not available until 2013 (Mehrabadi et al., 2013). In this study, the S. frugiperda genome (ZJU_Sfru_1.1), S. frugiperda transcriptome (SRP075752) (Wei et al., 2017), and S. frugiperda miRNA in miRbase were selected to be primary references. This is very important, even though the genomes of S. frugiperda and B. mori have high homology. AcMNPV can infect S. frugiperda but has a lower capacity to infect B. mori. Therefore, this work can accurately and effectively reflect the roles of miRNAs in the response of S. frugiperda to AcMNPV infection.
Forty-nine differentially expressed miRNAs were obtained by NGS, 10 of which were significantly differentially expressed. We selected 17 of them for qPCR validation. Except for miR-278-3p, the expression trend of other miRNAs was the same as that of NGS. It is known that S. frugiperda and AcMNPV are very close to B. mori and BmNPV at the genomic levels. Wu et al. (2016a) showed that 38 silkworm miRNAs were differentially expressed after BmNPV infection. The differentially expressed miRNAs of silkworm and S. frugiperda were different. The expression level of miR-278-3p was down-regulated following AcMNPV-EGFP infection by RT-qPCR. Moreover, Wu et al. (2016b) reported that the expression level of B. mori miR-278-3p was down-regulated after BmCPV infection, and miR-278-3p affected the replication of BmCPV by regulating the expression of the IBP2 gene.
The expression of key genes in the miRNA pathway was measured by RT-qPCR. The role of exportin-5 is to transport pre-miRNA from the nucleus to the cytoplasm, and pre-miRNA is cleaved by Dicer1. Ago1 is the key component of RISC. Upon AcMNPV-EGFP infection, the transcriptional levels of Exportin-5, Dicer1, and Ago1 were increased, especially at 48–72 h p.i. There was no significant change in the transcriptional level of Ran. The transcriptional levels of these key genes are similar to previous studies (Karamipour et al., 2019; Rahimpour et al., 2019). These results showed the miRNA pathway in Sf9 cells was more active during AcMNPV infection. MiRNA pathway played important roles in the Sf9-AcMNPV interaction.
Host-encoded miRNAs not only regulate the cellular processes but also play important roles in defending against pathogen infection (Hussain and Asgari, 2014; Asgari, 2015; Shrivastava et al., 2022). Among the many differentially expressed miRNAs, which one or more play key roles in insect-virus interaction? Five miRNAs (miR-11-3p, miR-2a-3p, miR-34-5p, miR-92a-3p, and miR-278-3p) were selected, which are conversed, highly expressed in Sf9 cells and are sensitive to AcMNPV-EGFP infection. Among them, miR-11-3p and miR-92a-3p were up-regulated, and miR-2a-3p, miR-34-5p, and miR-278-3p were down-regulated during AcMNPV-EGFP infection. These miRNAs play regulated roles in insect growth and the immune process. miR-11-3p was found to be involved in DNA damage-induced apoptosis. The pro-apoptotic genes reaper (rpr) and head involution defective (hid) are direct targets of miR-11 in Drosophila (Truscott et al., 2011). miR-2a regulates chitin biosynthesis through phoacetylglucosamine mutase (PAGM) in Nilaparvata lugens (Chen et al., 2013). miR-92a suppresses Drosophila olfactory memory consolidation by constraining the expression of kinesin heavy chain 73 (khc73) (Guven-Ozkan et al., 2020). miR-278-3p encoded by S. frugiperda, and miR-278 encoded by B. mori have the same sequence. Down-regulation of miR-278 in BmCPV-infected silkworms could promote the IBP2 mRNA level, which may play a positive role in BmCPV replication (Wu et al., 2016b). We found that overexpression and inhibition of miR-11-3p, miR-2a-3p, miR-92a-3p, and miR-278-3p did not affect the BVs titer of AcMNPV-EGFP. However, miR-34-5p inhibits the infectivity of progeny virus of AcMNPV-EGFP. MiR-34-5p is highly conserved among multiple species, such as arthropods, nematodes, and mammals (Lau et al., 2001; Cheng et al., 2014). In arthropods and Nematoda, miR-34-5p is involved in aging, stress response, neurodevelopment, and insect immunity and development (Lai et al., 2016; Guo et al., 2017; Ye et al., 2019; Wang et al., 2022). In B. mori, miR-34-5p can regulate larval growth and wing formation by targeting BmE74 and BmCPG4 and directly regulating ecdysone signal and stratum corneum protein (Liu et al., 2020). miR-34-5p can mediate the cross-regulation among JH, 20E, and IIS pathways to regulate the wing polymorphism of brown planthopper (Ye et al., 2019). miR-34-5p can regulate the innate immune pathway of Drosophila by inhibiting the expression of two target genes, Dlg1 and Eip75B (Xiong et al., 2016). A recent study found that miR-34-5p targeted the ecdysone receptor (EcR) to regulate the growth and development of lepidopteran pests (Li et al., 2022). In this study, we first discovered that miR-34-5p could regulate the proliferation of baculovirus progeny virus.
Virus infection has significant impacts on host physiology, establishing optimal conditions for successful propagation (Lin et al., 2020). Viruses rely on host cell metabolism to provide the necessary energy and biosynthetic precursors for successful viral replication (Feng et al., 2021). We commonly think that the modulation of host physiology is for virus multiplication, but on the flip side, it can also be an aspect of host resistance. Virus growth relies heavily on host resources, and the distribution and transfer of energy in hosts are important factors that affect viral replication. MiRNA may be a key factor in regulating host metabolism. Of the five miRNAs that we selected, miR-11-3p, miR-2a-3p, and miR-34-5p affect the glucose consumption, ATP production, and lactic acid accumulation in the host cells. Virus proliferation and energy metabolism of the host may be reciprocal causation and form a complex relationship of virus-host interaction.
The study exhibits the differential expression of miRNAs in S. frugiperda cells regulated by AcMNPV. The expression of some core genes in miRNA biogenesis pathways was significantly increased after AcMNPV-EGFP infection. It’s not hard to deduce that the miRNA biogenesis pathway in Sf9 cells is activated to resist AcMNPV infection. MiR-34-5p inhibits the infectivity of progeny virus. MiR-11-3p, miR-2a-3p, and miR-34-5p affected the energy metabolism of host cells (Fig. 5). In general, the study provides evidence for a possible mechanism of insect-virus interaction. That is, the miRNA biogenesis pathway in hosts plays an extremely significant role in antiviral immunity. In future work, we will focus on the targets of miRNAs in S. frugiperda and provide more ideas and evidence for host-virus interaction mechanism research.
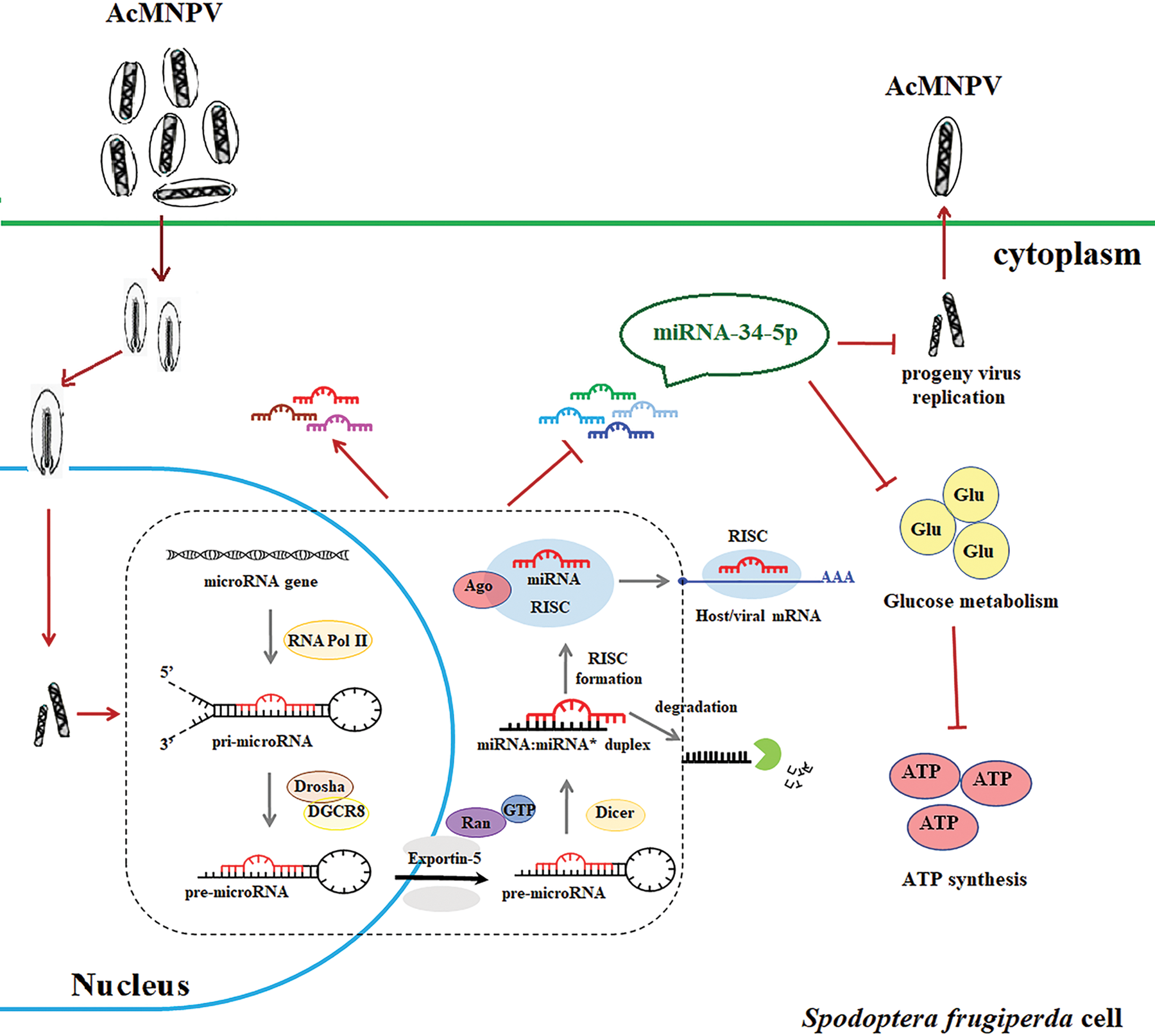
Figure 5: Schematic diagram of the response of the miRNA pathway in S. frugiperda cells upon AcMNPV infection. The diagram shows the key role of the miRNA pathway in response to baculovirus infection. By increasing the expression of some core genes of the miRNAs biogenesis pathway, the miRNA pathway in S. frugiperda cells is activated after AcMNPV infection. It allows the expression of miRNA encoded by S. frugiperda to be up- or down-regulated. Among many differentially expressed miRNAs, miR-34-5p has attracted our attention because of its regulatory effect on viral replication. miR-34-5p inhibits the infectivity of progeny virus and affects the energy metabolism of host cells.
Acknowledgement: This work was supported by the Natural Science Foundation of Shanxi Province and the Central Government Guiding Local Science and Technology Development Fund Project.
Availability of Data and Materials: The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Supplementary Materials: The supplementary material is available online at DOI 10.32604/biocell.2023.026182.
Author Contributions: Conceptualization, methodology, software, validation, formal analysis, investigation, Q.J.; resources, data curation, Q.J. and Y.F.; writing—original draft preparation, Q.J.; writing—review and editing, Y.F.; supervision, Y.F.; project administration, Y.F.; funding acquisition, Y.F. All authors have read and agreed to the published version of the manuscript.
Ethics Approval: Not applicable.
Funding Statement: This research was funded by the Natural Science Foundation of Shanxi Province, Grant No. 201801D121193, and the Central Government Guiding Local Science and Technology Development Fund Project, Grant No. YDZJSX2022A001.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
Asgari S (2015). Regulatory role of cellular and viral microRNAs in insect-virus interactions. Current Opinion in Insect Science 8: 104–110. DOI 10.1016/j.cois.2014.12.008. [Google Scholar] [CrossRef]
Bartel DP (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297. DOI 10.1016/S0092-8674(04)00045-5. [Google Scholar] [CrossRef]
Bernal V, Carinhas N, Yokomizo AY, Carrondo MJ, Alves PM (2009). Cell density effect in the baculovirus-insect cells system: A quantitative analysis of energetic metabolism. Biotechnology and Bioengineering 104: 162–180. DOI 10.1016/j.cell.2009.01.002. [Google Scholar] [CrossRef]
Chen J, Liang Z, Liang Y, Pang R, Zhang W (2013). Conserved microRNAs miR-8-5p and miR-2a-3p modulate chitin biosynthesis in response to 20-hydroxyecdysone signaling in the brown planthopper, Nilaparvata lugens. Biotechnology and Bioengineering 43: 839–848. DOI 10.1016/j.ibmb.2013.06.002. [Google Scholar] [CrossRef]
Cheng CY, Hwang CI, Corney DC, Flesken-Nikitin A, Jiang L, Öner GM, Munroe RJ, Schimenti JC, Hermeking H, Nikitin AY (2014). miR-34 cooperates with p53 in suppression of prostate cancer by joint regulation of stem cell compartment. Cell Reports 6: 1000–1007. DOI 10.1016/j.celrep.2014.02.023. [Google Scholar] [CrossRef]
Dubey SK, Shrinet J, Jain J, Ali S, Sunil S (2017). Aedes aegypti microRNA miR-2b regulates ubiquitin-related modifier to control chikungunya virus replication. Scientific Reports 7: 17666. DOI 10.1038/s41598-017-18043-0. [Google Scholar] [CrossRef]
Emery VC (1992). Baculovirus expression vectors: Choice of expression vector. Methods in Molecular Biology 8: 287–307. DOI 10.1385/0-89603-191-8:287. [Google Scholar] [CrossRef]
Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM et al. (2007). High-throughput sequencing of Arabidopsis microRNAs: Evidence for frequent birth and death of MIRNA genes. PLoS One 2: e219. DOI 10.1371/journal.pone.0000219. [Google Scholar] [CrossRef]
Feng M, Fei S, Xia J, Zhang M, Wu H, Swevers L, Sun J (2021). Global metabolic profiling of baculovirus infection in silkworm hemolymph shows the importance of amino-acid metabolism. Viruses 13. DOI 10.3390/v13050841. [Google Scholar] [CrossRef]
Feng G, Thumbi DK, De Jong J, Hodgson JJ, Arif BM, Doucet D, Krell PJ (2012). Selection and characterization of Autographa californica multiple nucleopolyhedrovirus DNA polymerase mutations. Journal of virology 86: 13576–13588. DOI 10.1128/JVI.01507-12. [Google Scholar] [CrossRef]
Friedländer MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, Rajewsky N (2008). Discovering microRNAs from deep sequencing data using miRDeep. Nature Biotechnology 26: 407–415. DOI 10.1038/nbt1394. [Google Scholar] [CrossRef]
Friedländer MR, Mackowiak SD, Li N, Chen W, Rajewsky N (2012). miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Research 40: 37–52. DOI 10.1093/nar/gkr688. [Google Scholar] [CrossRef]
Guo Y, Li P, Gao L, Zhang J, Yang Z, Bledsoe G, Chang E, Chao L, Chao J (2017). Kallistatin reduces vascular senescence and aging by regulating microRNA-34a-SIRT1 pathway. Aging Cell 16: 837–846. DOI 10.1111/acel.12615. [Google Scholar] [CrossRef]
Guven-Ozkan T, Busto GU, Jung JY, Drago I, Davis RL (2020). miR-92a suppresses mushroom body-dependent memory consolidation in drosophila. eNeuro 7. DOI 10.1523/eneuro.0224-20.2020. [Google Scholar] [CrossRef]
He PA, Nie Z, Chen J, Chen J, Lv Z et al. (2008). Identification and characteristics of microRNAs from Bombyx mori. BMC Genomics 9: 248. DOI 10.1186/1471-2164-9-248. [Google Scholar] [CrossRef]
Hussain M, Asgari S (2014). MicroRNAs as mediators of insect host-pathogen interactions and immunity. Journal of Insect Physiology 70: 151–158. DOI 10.1016/j.jinsphys.2014.08.003. [Google Scholar] [CrossRef]
Jiao Y, Wang J, Deng R, Yu X, Wang X (2019). AcMNPV-miR-3 is a miRNA encoded by Autographa californica nucleopolyhedrovirus and regulates the viral infection by targeting ac101. Virus Research 267: 49–58. DOI 10.1016/j.virusres.2019.05.004. [Google Scholar] [CrossRef]
Johnson JS (1987). Migration and the life history strategy of the fall armyworm, Spodoptera frugiperda in the Western Hemisphere. International Journal of Tropical Insect Science 8: 543–549. DOI 10.1017/S1742758400022591. [Google Scholar] [CrossRef]
Kakumani PK, Chinnappan M, Singh AK, Malhotra P, Mukherjee SK, Bhatnagar RK (2015). Identification and characteristics of microRNAs from army worm, Spodoptera frugiperda cell line Sf21. PLoS One 10: e0116988. DOI 10.1016/j.ibmb.2019.103202. [Google Scholar] [CrossRef]
Kamen AA, Bédard C, Tom R, Perret S, Jardin B (1996). On-line monitoring of respiration in recombinant-baculovirus infected and uninfected insect cell bioreactor cultures. Biotechnology and Bioengineering 50: 36–48. DOI 10.1002/(SICI)1097-0290(19960405)50:1<36::AID-BIT5>3.0.CO;2-2. [Google Scholar]
Karamipour N, Fathipour Y, Talebi AA, Asgari S, Mehrabadi M (2019). The microRNA pathway is involved in Spodoptera frugiperda (Sf9) cells antiviral immune defense against Autographa californica multiple nucleopolyhedrovirus infection. Insect Biochemistry and Molecular Biology 112: 103202. DOI 10.1016/j.ibmb.2019.103202. [Google Scholar] [CrossRef]
Kemp C, Mueller S, Goto A, Barbier V, Paro S et al. (2013). Broad RNA interference-mediated antiviral immunity and virus-specific inducible responses in Drosophila. Journal of Immunology 190: 650–658. DOI 10.4049/jimmunol.1102486. [Google Scholar] [CrossRef]
Kolliopoulou A, Santos D, Taning CNT, Wynant N, Vanden BJ, Smagghe G, Swevers L (2019). PIWI pathway against viruses in insects. Wiley Interdisciplinary Reviews RNA 10: e1555. DOI 10.1002/wrna.1555. [Google Scholar] [CrossRef]
Kong M, Zuo H, Zhu F, Hu Z, Chen L, Yang Y, Lv P, Yao Q, Chen K (2018). The interaction between baculoviruses and their insect hosts. Developmental and Comparative Immunology 83: 114–123. DOI 10.1016/j.dci.2018.01.019. [Google Scholar] [CrossRef]
Lai YW, Chu SY, Wei JY, Cheng CY, Li JC, Chen PL, Chen CH, Yu HH (2016). Drosophila microRNA-34 impairs axon pruning of mushroom body γ neurons by downregulating the expression of ecdysone receptor. Scientific Reports 6: 39141. DOI 10.1038/srep39141. [Google Scholar] [CrossRef]
Lau NC, Lim LP, Weinstein EG, Bartel DP (2001). An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858–862. DOI 10.1126/science.1065062. [Google Scholar] [CrossRef]
Lee RC, Feinbaum RL, Ambros V (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843–854. DOI 10.1016/0092-8674(93)90529-y. [Google Scholar] [CrossRef]
Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW (2004). Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117: 69–81. DOI 10.1016/s0092-8674(04)00261-2. [Google Scholar] [CrossRef]
Li XJ, Wu MF, Ma J, Gao BY, Wu QL et al. (2020). Prediction of migratory outes of the invasive fall armyworm in eastern China using a trajectory analytical approach. Pest Management Science 76: 454–463. DOI 10.1002/ps.5530. [Google Scholar] [CrossRef]
Li L, Zhu B, Sun X, Zheng K, Liang P, Gao X (2022). miR-34-5p, a novel molecular target against lepidopteran pests. Journal of Pest Science. DOI 10.1007/s10340-022-01488-2. [Google Scholar] [CrossRef]
Lin YH, Tai CC, Brož V, Tang CK, Chen P, Wu CP, Li CH, Wu YL (2020). Adenosine receptor modulates permissiveness of baculovirus (budded virus) infection via regulation of energy metabolism in bombyx mori. Frontiers in Immunology 11: 763. DOI 10.3389/fimmu.2020.00763. [Google Scholar] [CrossRef]
Liu W, Liu J, Lu Y, Gong Y, Zhu M et al. (2015). Immune signaling pathways activated in response to different pathogenic micro-organisms in Bombyx mori. Molecular Immunology 65: 391–397. DOI 10.1016/j.molimm.2015.02.018. [Google Scholar] [CrossRef]
Liu Z, Xu J, Ling L, Luo X, Yang D, Yang X, Zhang X, Huang Y (2020). miR-34 regulates larval growth and wing morphogenesis by directly modulating ecdysone signalling and cuticle protein in Bombyx mori. RNA Biology 17: 1342–1351. DOI 10.1080/15476286.2020.1767953. [Google Scholar] [CrossRef]
Mehrabadi M, Hussain M, Asgari S (2013). MicroRNAome of Spodoptera frugiperda cells (Sf9) and its alteration following baculovirus infection. The Journal of General Virology 94: 1385–1397. DOI 10.1099/vir.0.051060-0. [Google Scholar] [CrossRef]
Monsanto-Hearne V, Johnson KN (2018). miRNAs in insects infected by animal and plant viruses. Viruses 10: 354. DOI 10.3390/v10070354. [Google Scholar] [CrossRef]
Rahimpour H, Moharramipour S, Asgari S, Mehrabadi M (2019). The microRNA pathway core genes are differentially expressed during the development of Helicoverpa armigera and contribute in the insect’s development. Insect Biochemistry and Molecular Biology 110: 121–127. DOI 10.1016/j.ibmb.2019.05.005. [Google Scholar] [CrossRef]
Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G (2000). The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403: 901–906. DOI 10.1038/35002607. [Google Scholar] [CrossRef]
Shi X, Ran Z, Li S, Yin J, Zhong J (2016). The effect of microRNA bantam on baculovirus AcMNPV infection in vitro and in vivo. Viruses 8: 136. DOI 10.3390/v8050136. [Google Scholar] [CrossRef]
Shrivastava M, Pandey D, Sharma P, Singh R (2022). MicroRNA regulation and host interaction in response to Aspergillus exposure. BIOCELL 46: 339–356. DOI 10.32604/biocell.2022.016822. [Google Scholar] [CrossRef]
Singh J, Singh CP, Bhavani A, Nagaraju J (2010). Discovering microRNAs from Bombyx mori nucleopolyhedrosis virus. Virology 407: 120–128. DOI 10.1016/j.virol.2010.07.033. [Google Scholar] [CrossRef]
Thakur S, Saini KA, Das J, Saini V, Balhara P, Nanda SJ, Saini VR (2022). miR-153 as biomarker for cancer—functional role as tumor suppressor. BIOCELL 46: 13–26. DOI 10.32604/biocell.2022.016953. [Google Scholar] [CrossRef]
Thiem SM (2009). Baculovirus genes affecting host function. In Vitro Cellular & Developmental Biology Animal 45: 111–126. DOI 10.1007/s11626-008-9170-5. [Google Scholar] [CrossRef]
Truscott M, Islam AB, López-Bigas N, Frolov MV (2011). mir-11 limits the proapoptotic function of its host gene, dE2f1. Genes & Development 25: 1820–1834. DOI 10.1101/gad.16947411. [Google Scholar] [CrossRef]
Vodovar N, Bronkhorst AW, Van Cleef KW, Miesen P, Blanc H, Van Rij RP, Saleh MC (2012). Arbovirus-derived piRNAs exhibit a ping-pong signature in mosquito cells. PLoS One 7: e30861. DOI 10.1371/journal.pone.0030861. [Google Scholar] [CrossRef]
Wang C, Jia Q, Guo X, Li K, Chen W, Shen Q, Xu C, Fu Y (2022). microRNA-34 family: From mechanism to potential applications. The International Journal of Biochemistry & Cell Biology 144: 106168. DOI 10.1016/j.biocel.2022.106168. [Google Scholar]
Wang J, Xing K, Xiong P, Liang H, Zhu M, Zhao J, Yu X, Ning X, Li R, Wang X (2021). Identification of miRNAs encoded by Autographa californica nucleopolyhedrovirus. The Journal of General Virology 102. DOI 10.1099/jgv.0.001510. [Google Scholar] [CrossRef]
Wei L, Cao L, Miao Y, Wu S, Xu S, Wang R, Du J, Liang A, Fu Y (2017). Transcriptome analysis of Spodoptera frugiperda 9 (Sf9) cells infected with baculovirus, AcMNPV or AcMNPV-BmK IT. Biotechnology Letters 39: 1129–1139. DOI 10.1007/s10529-017-2356-8. [Google Scholar] [CrossRef]
Wu P, Jiang X, Guo X, Li L, Chen T (2016a). Genome-wide analysis of differentially expressed microRNA in Bombyx mori infected with nucleopolyhedrosis virus. PLoS One 11: e0165865. DOI 10.1371/journal.pone.0165865. [Google Scholar] [CrossRef]
Wu P, Qin G, Qian H, Chen T, Guo X (2016b). Roles of miR-278-3p in IBP2 regulation and Bombyx mori cytoplasmic polyhedrosis virus replication. Gene 575: 264–269. DOI 10.1016/j.gene.2015.09.009. [Google Scholar] [CrossRef]
Xiong XP, Kurthkoti K, Chang KY, Li JL, Ren X, Ni JQ, Rana TM, Zhou R (2016). miR-34 modulates innate immunity and ecdysone signaling in Drosophila. PLoS Pathogens 12: e1006034. DOI 10.1371/journal.ppat.1006034. [Google Scholar] [CrossRef]
Yang JS, Lai EC (2011). Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Molecular Cell 43: 892–903. DOI 10.1016/j.molcel.2011.07.024. [Google Scholar] [CrossRef]
Ye X, Xu L, Li X, He K, Hua H et al. (2019). miR-34 modulates wing polyphenism in planthopper. PLoS Genetics 15: e1008235. DOI 10.1371/journal.pgen.1008235. [Google Scholar] [CrossRef]
Zhao T, Deng R, Chen M, Wang X (2022b). Identification of differential mRNA and lncRNA expression in AcMNPV-infected Sf9 cells. Biocell 46: 1675–1686. DOI 10.21203/rs.3.rs-61712/v1. [Google Scholar] [CrossRef]
Zhao J, Teng T, Wang J (2022a). An Autographa californica nucleopolyhedrovirus-encoded microRNA, AcMNPV-miR-4, downregulates the expression of host gene alg-2. The Journal of General Virology 103. DOI 10.1099/jgv.0.001769. [Google Scholar] [CrossRef]
Zharikova AA, Mironov AA (2016). piRNAs: Biology and bioinformatics. Molekuliarnaia Biologiia 50: 80–88. DOI 10.7868/S0026898416010225. [Google Scholar] [CrossRef]
Zhou X, Lindsay H, Robinson MD (2014). Robustly detecting differential expression in RNA sequencing data using observation weights. Nucleic Acids Research 42: e91. DOI 10.1093/nar/gku310. [Google Scholar] [CrossRef]
Zhou R, Yu ZH, Li XQ, Jia F, Wu JH, Chen X (2004). Heliocoverpa armigera single nucleocapsid nucleopolyhedrovirus induces Hz-AM1 cell cycle arrest at the G2 phase with accumulation of cyclin B1. Virus Research 105: 113–120. DOI 10.1016/j.virusres.2004.05.001. [Google Scholar] [CrossRef]
Zhu M, Wang J, Deng R, Wang X (2016). Functional regulation of an Autographa californica nucleopolyhedrovirus-encoded MicroRNA, AcMNPV-miR-1, in baculovirus replication. Journal of Virology 90: 6526–6537. DOI 10.1128/JVI.00165-16. [Google Scholar] [CrossRef]
Zhu M, Wang J, Deng R, Xiong P, Liang H, Wang X (2013). A microRNA encoded by Autographa californica nucleopolyhedrovirus regulates expression of viral gene ODV-E25. Journal of Virology 87: 13029–13034. DOI 10.1128/JVI.02112-13. [Google Scholar] [CrossRef]
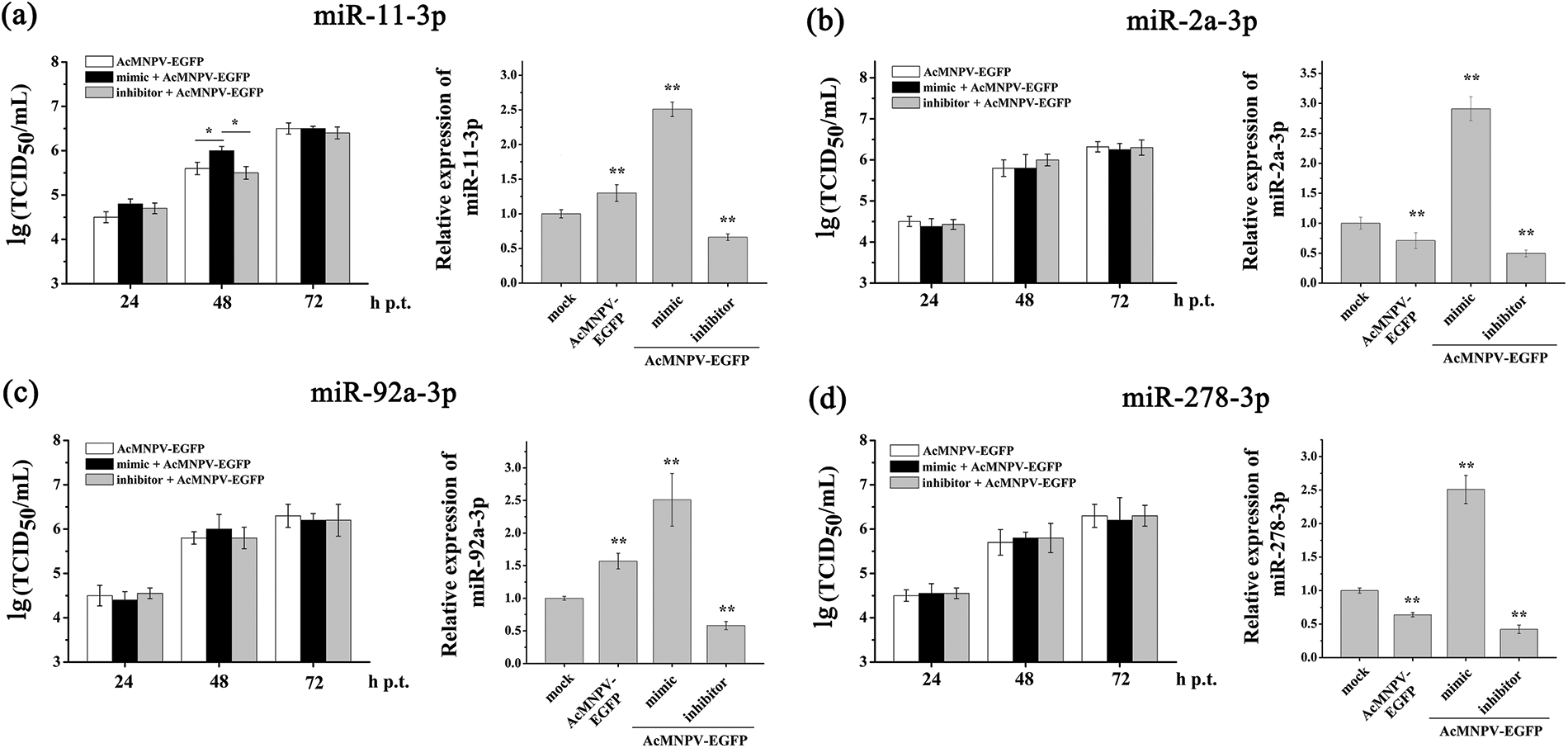
Figure S1: TCID50 analysis of the effects of four pairs of mimic and inhibitor on the BV titer. They were miR-11-3p (a), miR-2a-3p (b), miR-92a-3p (c), and miR-278-3p (d), respectively. Sf9 cells were infected with AcMNPV-EGFP (5 MOI) for 8 h after transfection with the mimic and inhibitor, respectively. The expression of miRNA was analyzed by qPCR after transfection with mimic and inhibitor. The asterisks indicated that the t-test showed a signifcant difference relative to the control (*P < 0.05; **P < 0.01).
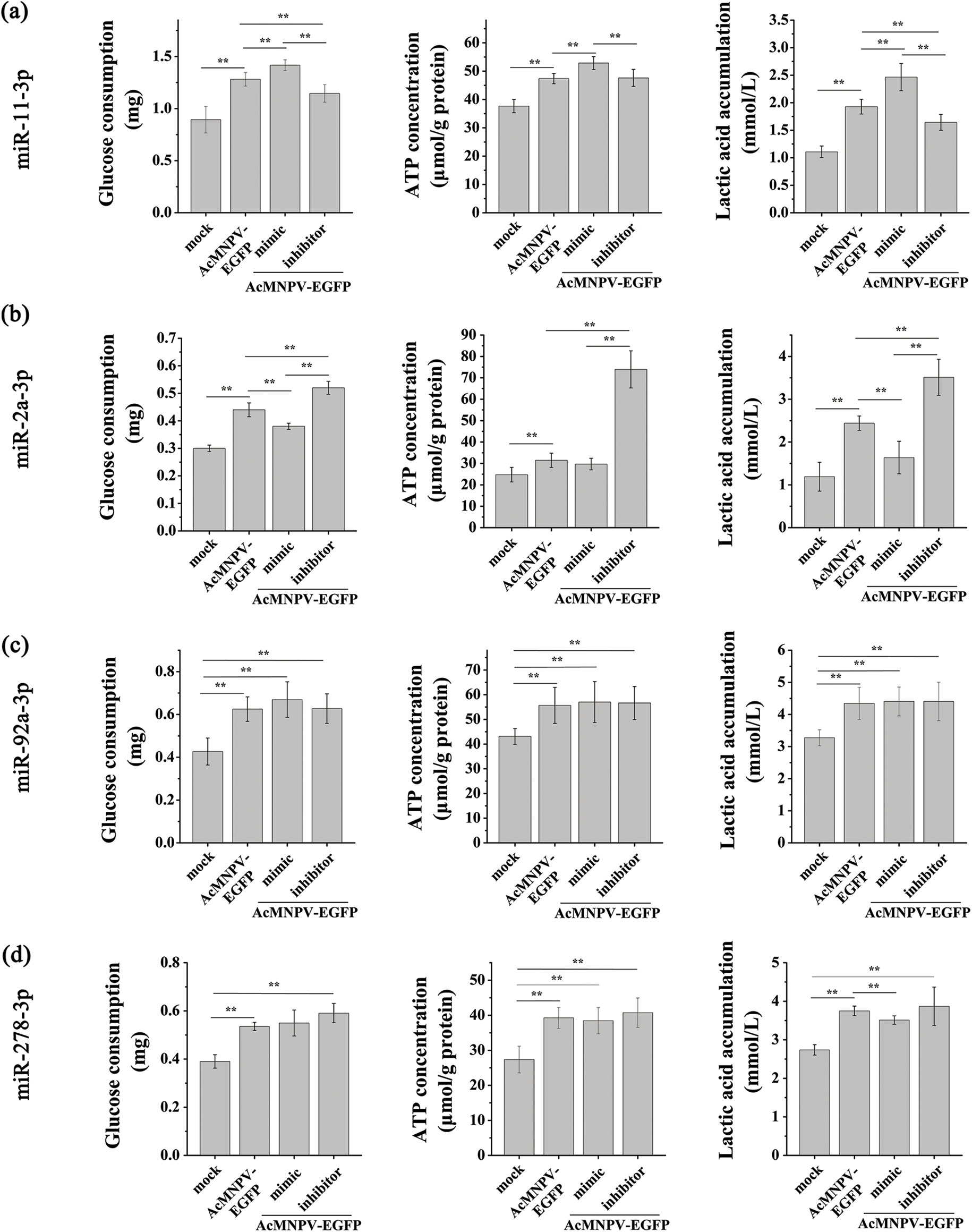
Figure S2: Effects of miRNAs on energy metabolism of host cells. The effect of the four pairs of mimic and inhibitor on the glucose consumption, ATP content, and lactic acid accumulation of host cells. They were miR-11-3p (a), miR-2a-3p (b), miR-92a-3p (c), and miR-278-3p (d), respectively. Sf9 cells were infected with AcMNPV-EGFP (5 MOI) for 8 h after transfection with the mimic and inhibitor, respectively. The asterisks indicated that the t-test showed a signifcant difference relative to the control (**P < 0.01).


Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools