 Open Access
Open Access
REVIEW
Research progress of TRIMs protein family in tumors
Department of Health Toxicology, School of Public Health, Guilin Medical University, Guilin, 541199, China
* Corresponding Author: XIAONIAN ZHU. Email:
(This article belongs to the Special Issue: Recent Advancement in Cancer Molecular Signaling)
BIOCELL 2023, 47(3), 445-454. https://doi.org/10.32604/biocell.2023.025880
Received 04 August 2022; Accepted 15 November 2022; Issue published 03 January 2023
Abstract
The tripartite motif (TRIMs) protein family has E3 ubiquitin ligase activity among most of its members. They participate in multiple cellular processes and signaling pathways in living organisms, including cell cycle, growth, and metabolism, and mediate chromatin modification, transcriptional regulation, post-translational modification, and cellular autophagy. Previous studies have confirmed that the TRIMs protein family is involved in the development of various cancers and correlated with the prognosis of tumor patients. Here we summarize the biological roles of the TRIMs protein family in cancers.Keywords
A tumor is a complex pathologic process caused by dynamic regulations of multiple factors and various signaling pathways. The mechanisms of tumor occurrence and development have already been widely studied. However, most tumors have inconspicuous or mild symptoms at the early stages that are not easily detected, resulting in missed diagnosis and misdiagnosis. Clinical diagnosis of tumors at the middle and advanced stages always leads to adverse consequences, such as low survival, poor prognosis, and high recurrence. Comprehensive studies on tumor biology have paid close attention to the tripartite motif (TRIMs) protein family, and some of its members are involved in the progression of multiple malignancies. TRIMs play important regulatory roles in the development of liver cancer, lung cancer, colorectal cancer (CRC), breast cancer (BC), and gastric cancer (GC), and are expected to become potential molecular targets for cancer treatment and prognosis (Mandell et al., 2020).
An overview of the tripartite motifs protein family
The TRIMs protein family is referred to as the highly conserved RING-B-box-coiled-coil family (Reymond et al., 2001) that exists in multicellular organisms and is reckoned an E3 ubiquitin ligase-active protein with over 80 family members in the human genome (Mandell et al., 2020). It is characterized by three zinc-binding domains from the N-terminal to the C-terminus, one RING-finger domain (RING-finger domain), one or two zinc-finger namely B-boxes including B1-box and B2-box domains, and highly variable coiled-coil domain (coil-coil region).
The RING domain of TRIMs determines substrate specificity by providing docking sites for E2 conjugates and facilitating ubiquitin ligation to confer TRIMs protein E3 ligase activity, including ubiquitin molecules and ubiquitin-like molecules (UBLs) just like the small ubiquitin-like modifier (SUMO) and neural precursor cell expressed developmentally downregulated protein 8 (Nedd8) molecules (Yang et al., 2020). E3 ligases are in charge of substrate recognition and specificity because they move ubiquitin or UBLs molecules from E2-conjugating enzymes to the substrates (Yang and Xia, 2021). The majority of TRIMs are E3 ligases due to their RING-finger domain; in this way, which can regulate the ubiquitination of different substrates. Interestingly, in some TRIMs that lack the RING domain, their B-box domain can utilize the E3 ubiquitin ligase activity and endow them by offering the E2 ubiquitin-conjugase enzyme a binding site to ubiquitin (Yang et al., 2020), which is similar to the function of RING domain. In humans, there are eight TRIM subgroups with no RING domains, such as TRIM14, TRIM16, TRIM20, TRIM29, TRIM44, TRIM66, TRIM70, TRIM76, TRIM76, and TRIML2, which still have ubiquitin ligase activity because of an occult RING-like fold in their B-box domain (Hatakeyama, 2017).
B-box is further categorized into B-box1 and B-box2 based on the residues of the coordinate zinc ions, and the second binding site of B-box1 is cysteine, while that of B-box2 is histidine, with B-box1 preceding B-box2, but some TRIM proteins contain only one B-box2 (Reymond et al., 2001). Though it is shown to conduce to the human innate immune response to the infection of HIV (Diaz-Griffero et al., 2009), the function of the B-box domain has not yet been completely understood.
The coiled-coil (CC) domain is thought to have enzymatic activity in the interaction of homodimers. It can promote higher-order oligomerization or the formation of heterooligomeric complexes with other TRIMs (Wang and Hur, 2021). The X-ray structures of the CC in all TRIM proteins identified so far show antiparallel arrangements of homodimers, with the catalytic RING domain located on the other side of its rod-shaped central CC domain (Fiorentini et al., 2020). TRIMs share a common overall structure, but the CC has various effects on the overall structure and function of TRIM proteins. In addition, the TRIM family proteins have a complex C-terminal region (C-terminal domain), including the C-terminal subgroup one signature (COS) domain, SPIa and the ryanodine receptor (SPRY) domain, SPRY-associated (PRY) domain, NHL repeats (NHL) domain, bromodomain (BROMO), serine-type IG domain (FIL), fibronectin type III repeat (FNIII), acid-rich region (ACID), Meprin and TRAF homologous domain (MATH), ADP-ribosylation factor family domain (ARF), and transmembrane region (TM) (Hatakeyama, 2017). The SPRY domain is the most common in the human body for mediating protein-protein interactions (Mandell et al., 2020).
Surprisingly, not all TRIMs have all three domains. Some TRIMs can replace one of the three domains but retain the order of the remaining domains. Some TRIMs-like proteins that are not formally grouped into the TRIMs protein family have two of the three domains in the same conserved order (Wang and Hur, 2021).
The TRIMs protein family has been shown to implicate various biological processes, such as development, transcription, and signal transduction (Tocchini and Ciosk, 2015). TRIMs also have been demonstrated to regulate cell proliferation, cell division, and cell metabolism. They can promote or inhibit cell transformation of tumors, mediate chromatin modifications, gene transcription, and post-translational modifications, interact with pathogens, and regulate autophagy (Jaworska et al., 2020).
The tripartite motifs protein family in liver cancer
Hepatocellular carcinoma (HCC) is the sixth most common cancer type and the second leading cause of death worldwide; one of the most common malignant primary liver cancers, accounting for about 85% of cirrhosis-diagnosed patients, and the 5-year survival rate is only 18%. HCC has an insidious onset, high malignancy, and poor prognosis (Hao et al., 2021). Therefore, exploring the occurrence and mechanism of HCC is of great value in improving the prognosis of patients.
Current studies show that in HCC, TRIM3, TRIM16, TRIM26, and TRIM50 of the TRIMs protein family are poorly expressed, while TRIM11, TRIM21, TRIM25, TRIM31, TRIM32, TRIM52, and TRIM66 are over-expressed. This indicates that the TRIMs family proteins play various roles in HCC.
Low expression of TRIM3 was found to be associated with a poor prognosis in patients with HCC (Chao et al., 2014). Further research revealed that TRIM3 over-expression could induce cell cycle arrest at G0/G1 phase and inhibit cell proliferation, migration, invasion, and colony formation (Huang et al., 2017). TRIM16 can inhibit HCC migration, and invasion in vitro and in vivo through down-regulating ZEB2 expression in a proteasome-dependent pathway. Knockdown of TRIM16 can promote HCC migration, invasion, and epithelial-mesenchymal transition (EMT) (Li et al., 2016). EMT is a process of morphological conversion from epithelial to mesenchymal phenotype, providing mobility for cancer cells to generate metastasis (Li et al., 2016). The down-regulation of TRIM26 contributes to a worse prognosis of HCC patients. TRIM26 silencing can promote HCC cell proliferation, colony formation, migration, and invasion (Wang et al., 2015). The proliferation, colony formation and invasion abilities of HCC cells were significantly suppressed after TRIM50 expression was up-regulated. The possible mechanism is that TRIM50 directly targets zinc-finger transcription factor SNAI1 (Snail) for extensive degradation, reverses the Snail-mediated EMT transition, and thus acts to inhibit the development of HCC (Ma et al., 2018). These studies suggest that positive modulation of TRIM3, TRIM16, TRIM26, and TRIM50 may be novel therapeutic strategies for HCC development.
Over-expression of TRIM11 was reported to be closely associated with HCC progression and patient survival (Chen et al., 2017). TRIM11 knockdown reduced the expression of p-PI3K and p-Akt proteins in HCC cells, thus inhibiting the activation of PI3K/Akt signaling. The expression of TRIM11 has a negative relationship with p53 and can play its carcinogenic role in HCC by down-regulating the p53 pathway. TRIM11 can inhibit HCC cell proliferation, migration, invasion, and the EMT process in vitro through the above-mentioned two pathways (Liu et al., 2017; Zhang et al., 2017). TRIM21 was highly expressed in HCC, and TRIM21 genetic ablation protected the liver from oxidative injury and reduced the occurrence of HCC by suppressing the p62-Kelch-like ECH-associated protein 1 (Keap1)-NF-E2-related factor 2 (Nrf2) pathway (Wang et al., 2021). Another study reported that p15INK4b-related sequence/regulation of nuclear pre-mRNA domain-containing protein 1A (RPRD1A) interacted with TRIM21 by increasing the aggregation of p62 and Keap1 to promote nuclear translocation of Nrf2, which further contributes to the progression of HCC (Feng et al., 2021). However, the down-regulation of TRIM21 was associated with a poor prognosis in patients with HCC (Ding et al., 2015). Given the discovery of the different roles of TRIM21 in HCC, more experimental studies are needed to explore the mechanism of TRIM21 in HCC.
TRIM25 may lead to the activation of the Keap1/Nrf2 pathway through direct ubiquitination and degradation of Keap1 by E3 ubiquitin ligase, eliminating the production of reactive oxygen species in response to endoplasmic reticulum stress, thus promoting HCC cell survival and growth (Liu et al., 2020). TRIM31 can promote HCC progression by targeting the tuberous sclerosis complex 1 (TSC1)-TSC2 complex for degradation and further over-activating the mammalian target of rapamycin complex 1 (mTORC1) pathway (Guo et al., 2018a). Another study shows that TRIM31 knockdown directly promotes p53 polyubiquitination and proteasomal degradation at the K48 site, and activates the AMPK pathway, thereby promoting HCC cell resistance to nest loss apoptosis and reverses nest loss apoptosis (Guo et al., 2018b), a type of programmed cell death that occurs when cells lose contact with the extracellular matrix and other cells form. These findings suggest that TRIM31 can be a therapeutic target for metastatic HCC.
Cui et al. (2016) demonstrated that TRIM32 is positively associated with histological grade, tumor size, and hepatitis B surface antigen (HBsAg) in HCC patients. TRIM32 over-expression accelerated the G1-S phase transition of the cell cycle, which in turn promoted the rate of cell proliferation. Additionally, among the drugs for HCC, TRIM32 also induced resistance to oxaliplatin in HCC therapy and predicted a poor prognosis in HCC patients (Cui et al., 2016). Zhang et al. (2018) found that TRIM52 expression was positively correlated with tumor size, tumor lymph node metastasis (TNM) stage, and tumor number. TRIM52 up-regulation promoted cell proliferation, migration, and invasion of HCC cells through the ubiquitination of PPM1A. Furthermore, up-regulation of PPM1A inhibited TRIM52-mediated cell proliferation, migration, and invasion, and enhanced PPM1A activity or expression could be used as a therapeutic strategy to prevent and treat HCC (Zhang et al., 2018). Fan et al. (2019) found that TRIM66 is involved in the regulation of glycogen synthase kinase-3β (GSK-3β) phosphorylation and β-catenin expression. TRIM66 over-expression promoted activation of Wnt/β-catenin signaling, which significantly promoted the proliferation of HCC cells, colony formation, and invasion ability, and thus acted as oncogenic proteins in HCC, indicating the potential of TRIM66 as a target for HCC therapy (Fan et al., 2019).
In conclusion, the TRIMs protein family mainly regulates HCC metastasis to pose important clinical significance in the diagnosis, treatment, and prognosis of HCC.
The tripartite motifs protein family in lung cancer
Lung cancer is one of the most commonly diagnosed cancers in the world and the leading cause of cancer-related deaths, with an estimated 2 million new cases and 1.76 million deaths each year. Lung cancer is classified in terms of pathology into non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC), of which NSCLC is the most common type (Thai et al., 2021). In recent years, progress has been made in the pathogenesis, diagnosis, and treatment of lung cancer, but overall survival still needs to be improved.
The TRIMs protein family shows an important role in lung cancer. Among the TRIMs protein family, TRIM13, TRIM56, and TRIM72 are down-regulated, while TRIM11, TRIM15, TRIM23, TRIM29, TRIM32, TRIM35, TRIM37, TRIM46, TRIM59, and TRIM65 are up-regulated in lung cancer.
TRIM13 over-expression in a xenograft mouse model inhibited tumor growth and induced apoptosis in vivo, and the possible mechanism was to inhibit cell proliferation and induce apoptosis by regulating the nuclear factor kappa B (NF-κB) pathway (Xu et al., 2019). The expression of TRIM56 in lung adenocarcinoma (LUAD) was reduced and associated with a poor prognosis, and the over-expression of TRIM56 inhibited the invasion and migration of LUAD cells (Lu et al., 2021). TRIM72 inhibited tumor progression and stress granule formation by modulating oncogenic protein G3BP2 activity in NSCLC (Li et al., 2021a), which is a potential therapeutic target.
TRIM11 over-expression was observed to significantly reduce lung cancer cell growth and invasiveness, which was correlated with poor outcomes of patients (Wang et al., 2016). While TRIM15 over-expression in NSCLC was associated with a poor prognosis (Han et al., 2019), TRIM15 knockdown reduced tumor cell proliferation and metastasis in vitro and in vivo, whereas ectopic TRIM15 expression promoted cell proliferation and metastasis. The possible mechanism is that TRIM15 promoted NSCLC progression by promoting Keap1 ubiquitination and degradation-mediated Nrf2 stabilization (Liang et al., 2022). TRIM23 over-expression was associated with high expression of NF-κB, poor cell differentiation, poor overall survival (OS), and disease-free survival (DFS), and was associated with a poor prognosis in LUAD patients with platinum-based adjuvant chemotherapy (Zhang et al., 2020a). TRIM29 down-regulation can inhibit cell proliferation, invasion, increase the chemosensitivity of cisplatin in human lung squamous cancer cells, and subsequently exert potent antitumor activity and chemosensitization (Liu et al., 2015). TRIM32 up-regulation was associated with a poor prognosis of lung cancer patients and could promote the proliferation, migration, and invasion of lung cancer cells by activating the Janus kinase 2/signal transducer and activator of transcription 3 (JAK2/STAT3) pathway (Yin et al., 2019). TRIM35 over-expression promoted lung cancer cell proliferation, migration, and invasion and promoted tumor formation in vivo (Zhang et al., 2020b). Knockdown of TRIM37 inhibited NSCLC cell proliferation and tumorigenesis by inhibiting the Wnt/β-catenin signaling pathway (Ding et al., 2018). TRIM46 expression was positively correlated with tumor size, TNM stage, and metastasis, and TRIM46 over-expression increased cell proliferation and cisplatin resistance in LUAD cells by enhancing glycolysis through activating AKT/HK2 signaling (Tantai et al., 2022).
TRIM59 could regulate the initiation of autophagy by negatively regulating the transcription of the beclin1 (BECN1) gene through the NF-κB pathway and trigger the autophagy protein cascade as well as the TNF receptor-related factor 6 (TRAF6)-mediated K63-linked BECN1 ubiquitination, which is related to the inhibitory effect on NF-κB activation observed by TRIM59 (Han et al., 2018). This study indicates the role of TRIM59 in lung cancer by regulating the level of autophagy. Moreover, a recent study showed that tumor-derived exosomal TRIM59 can convert macrophages via regulating ABHD5 proteasomal degradation to activate the NLRP3 inflammasome signaling pathway to promote lung cancer progression by IL-1β secretion (Liang et al., 2020). Knockdown of TRIM65 was confirmed to induce TNRC6A ubiquitination and degradation, regulate miR-138-5p expression to inhibit autophagy, and also attenuate the in vitro and in vivo chemical resistance of NSCLC cells after drug cisplatin treatment, indicating that TRIM65 plays a role and provides new insights into NSCLC autophagy-mediated chemoresistance (Pan et al., 2019).
Taken together, these results suggest that the TRIMs protein family may be a novel and promising target for the prognosis and treatment of lung cancer.
The tripartite motifs protein family in colorectal cancer (CRC)
CRC is the third most common malignancy in the world, and CRC patients always have a high mortality rate and a low survival rate. Despite the significant progress in the development of CRC therapies, new effective molecular biomarkers are still needed (Xie et al., 2020).
Among the TRIMs protein family, TRIM21, TRIM58, and TRIM67 are down-regulated, while TRIM6, TRIM24, TRIM28, and TRIM39 are up-regulated in CRC.
TRIM21 is reported to serve as a therapeutic target for CRC by negatively regulating cell proliferation, adhesion, tissue remodeling, and angiogenesis, as well as a pro-inflammatory response (Zhou et al., 2021b). TRIM58 down-regulation can promote CRC cell invasion and cause a poor prognosis, but TRIM58 over-expression can strongly inhibit CRC cell invasion mainly by decreasing the expression of EMT and matrix metalloproteinase (MMP) genes (Liu et al., 2018). TRIM67 appears as a tumor suppressor in CRC. It is shown to enhance the stability of p53 by disrupting the p53-MDM2 interaction to inhibit the occurrence and progression of CRC (Wang et al., 2019).
In a recent study, TRIM6 over-expression in CRC cells can decrease B-cell translocation gene 2 (BTG2/TIS21) stability by increasing TIS21 ubiquitination through E3-ubiquitin ligase activity and also increase FoxM1 expression and phosphorylation to promote CRC cell proliferation (Zheng et al., 2020). TRIM24 is found to be inversely correlated with the prognosis of CRC patients, and TRIM24 knockdown can inhibit CRC cell proliferation and colony formation. The effect of TRIM24 on CRC cell proliferation is due to its activation of YES-associated protein (YAP) signaling (Xie et al., 2020). Another research indicates that TRIM24 over-expression can facilitate the in vitro and in vivo growth of CRC cells, up-regulate vascular endothelial growth factor expression to consequently stimulate angiogenesis, and promote cell progression via the Wnt/β-catenin signaling (Tian et al., 2022). TRIM28 over-expression is identified to have a relationship with the recurrence and poor prognosis of CRC patients, possibly through its translational repression of the Krüppel associated box (KRAB) domain transcription factor family (Fitzgerald et al., 2018). TRIM39 up-regulation is also associated with a poor prognosis in CRC patients, but its knockdown can inhibit CRC progression (Hu et al., 2021). In addition, it is found that TRIM39 is a positive regulator of autophagosome-lysosome fusion. TRIM39 can inhibit the ubiquitination of Ras-associated protein Rab7 at the K191 residue to promote Rab7 activity, and TRIM39 deficiency inhibits the p53 autophagy flux in a Rab7 activity-dependent manner, thereby inhibiting CRC development.
In summary, the above studies also suggest that TRIMs play an important role in CRC development. They may be explored to be potential biomarkers in CRC treatment.
The tripartite motifs protein family in breast cancer (BC)
BC is one of the most common tumors and is the second leading cause of cancer-related deaths in women worldwide. As observed in the TRIMs protein family, TRIM31, TRIM35, TRIM44, and TRIM72 are down-regulated, while TRIM3, TRIM6, TRIM39, TRIM47, and TRIM59 are up-regulated in BC.
Guo et al. (2021) found that TRIM31 down-regulation was negatively associated with BC progression, and TRIM31 expression was associated with tumor size, Ki67 expression, TNM stage, histological grade, and lymph node infiltration of BC patients. TRIM31 can promote BC cell proliferation, migration, and invasion. TRIM31 may promote BC progression by regulating the ubiquitination of p53 (Guo et al., 2021). Low TRIM35 expression was associated with poor prognosis of BC patients, and TRIM35 inactivated AKT signaling by increasing the ubiquitination of PDK1 to inhibit BC cell proliferation (Wang et al., 2022a). TRIM44 played a role in BC progression by promoting BC cell proliferation and migration through enhancing NF-κB signaling, and it was an independent prognostic factor for distant DFS and OS in patients (Kawabata et al., 2017). Over-expression of TRIM72 inhibited BC cell proliferation and invasion and reduced tumor growth and metastasis in BC xenograft tumor models. Additionally, under a hypoxic tumor microenvironment (TME), decreased TRIM72 expression can induce lactate production and may contribute to the TME to further activate PI3K/Akt/mTOR signaling pathway, which ultimately results in BC progression (Wang et al., 2022b). Thus, TRIM72 could serve as a potential therapeutic target for BC.
It was demonstrated that TRIM3 over-expression was closely associated with low survival in BC patients treated with the estrogen receptor drug tamoxifen (Ye et al., 2021). The up-regulation of TRIM6 expression in BC promoted STIP1 homology and U-Box containing protein 1 (STUB1) degradation through the ubiquitin-dependent proteasome and subsequently activated YAP1 signaling to promote BC growth and migration (Wei et al., 2021). TRIM39 knockdown significantly inhibited BC cell proliferation and arrested the cell cycle at S-phase (Ogura et al., 2022). TRIM47 expression was positively correlated with the shortened DFS in patients with postoperative endocrine therapy, and TRIM47 over-expression activated NF-κB signaling, which contributed to the resistance to endocrine therapy in BC (Azuma et al., 2021). This suggests that TRIM47 can be a potential diagnostic and therapeutic target for endocrine therapy-refractory BC. TRIM59 was shown to promote BC growth, migration, and invasion by inhibiting the p62-selective autophagic degradation of the programmed cell death protein 10 (PDCD10). And then, PDCD10 can mediate RAS homolog family member A (RhoA)-Rho-related coiled-coil kinase (ROCK) 1 signaling to promote BC migration and invasion (Tan et al., 2018). Further research found that depletion of TRIM59 suppressed BC metastasis by promoting RNFT1-induced K63 polyubiquitination and SQSTM1-directed autophagic degradation of PDCD10 (Tan et al., 2019).
These studies suggest that TRIMs act as a significant role in BC and may be a valuable prognostic biomarker for BC patients.
The tripartite motifs protein family in gastric cancer (GC)
GC is the second most common cause of cancer death. Surgery and radiotherapy are the main treatment methods for GC, but they are prone to relapse. Therefore, more diagnostic and therapeutic biomarkers are found to better improve the quality of survival and prognosis of patients with GC (Joshi and Badgwell, 2021).
Among the TRIMs protein family, TRIM3 and TRIM16 are poorly expressed in GC, while TRIM15, TRIM23, TRIM47, and TRIM 54 are over-expressed in GC.
Low TRIM3 expression was shown to have a correlation with shorter OS and was an independent predictor of poor prognosis in GC patients (Farhadi et al., 2022). Low TRIM16 expression in GC was found to increase the expression of three oncogenic proteins, β-catenin, cyclin D, and B-cell lymphoma 2, which was suggested as a possible risk factor contributing to GC progression (Afshar et al., 2021).
The expression of TRIM15 and TRIM 23 were higher in GC tissues than that in normal tissues, and over-expressed TRIM15 and TRIM23 were found to be positively associated with the depth of tumor invasion, lymph node metastasis, distant metastasis, TNM stage and shorter OS in GC patients, which were independent adverse predictors and therapeutic targets (Yao et al., 2018; Zhou et al., 2021a). TRIM47 was significantly correlated with tumor differentiation and the TNM stage of GC patients. The role of TRIM47 in GC development resulted from the regulation of NF-κB, EMT, hypoxia, and apoptosis-related signaling pathways (Xia et al., 2021). TRIM54 showed a negative association with the OS of GC patients and significantly enhanced the proliferation, migration, and invasion of GC cells (Cao et al., 2022).
These researches show a significant relationship between TRIMs family members and the clinicopathological features of GC patients. TRIMs will be used as a prognostic biomarker for GC patients.
The tripartite motifs protein family in other cancers
Some TRIMs family members also take their roles in other cancers. TRIM15 expression is found elevated in pancreatic ductal adenocarcinoma and associated with a poor prognosis of patients. It interacts with apolipoprotein A1 (APOA1) through its PRY/SPRY domain and induces APOA1 degradation through its RING domain to promote pancreatic cancer cell invasion and metastasis (Sun et al., 2021). Down-regulation of TRIM50 in pancreatic cancer is shown to have an association with low survival in patients, and its over-expression suppresses cell proliferation and inhibits the EMT process by degrading Snail1 in pancreatic cancer cells (Li et al., 2021b). TRIM26 expression is up-regulated and plays an oncogenic role in bladder cancer. It promotes cell proliferation, migration, and invasion through the Akt/GSK3/β-catenin pathway (Xie et al., 2021). TRIM22 activates NF-κB signaling to accelerate its degradation by binding to a negative regulator of NF-κB (IKBα) in glioblastoma (Ji et al., 2021).
Great achievements have been made in the diagnosis and treatment of tumors, but more diagnostic value and efficacy evaluation of biomarkers are still needed. It has been confirmed that the TRIMs protein family is closely related to liver cancer, lung cancer, CRC, and GC (Table 1). Through important regulatory signaling pathways such as p53, Wnt/β-catenin, Keap1/Nrf2, and NF-κB signaling pathways, the TRIMs protein family regulates cell proliferation, cell cycle, metastasis, and sensitivity to chemotherapy drugs, thus playing a role in different tumors (Fig. 1). But the mechanism of TRIMs protein family in tumors is not completely clear. Therefore, a more in-depth study and biological mechanisms of the TRIMs protein family in tumors can provide new targets for tumor diagnosis, treatment, and prognosis.
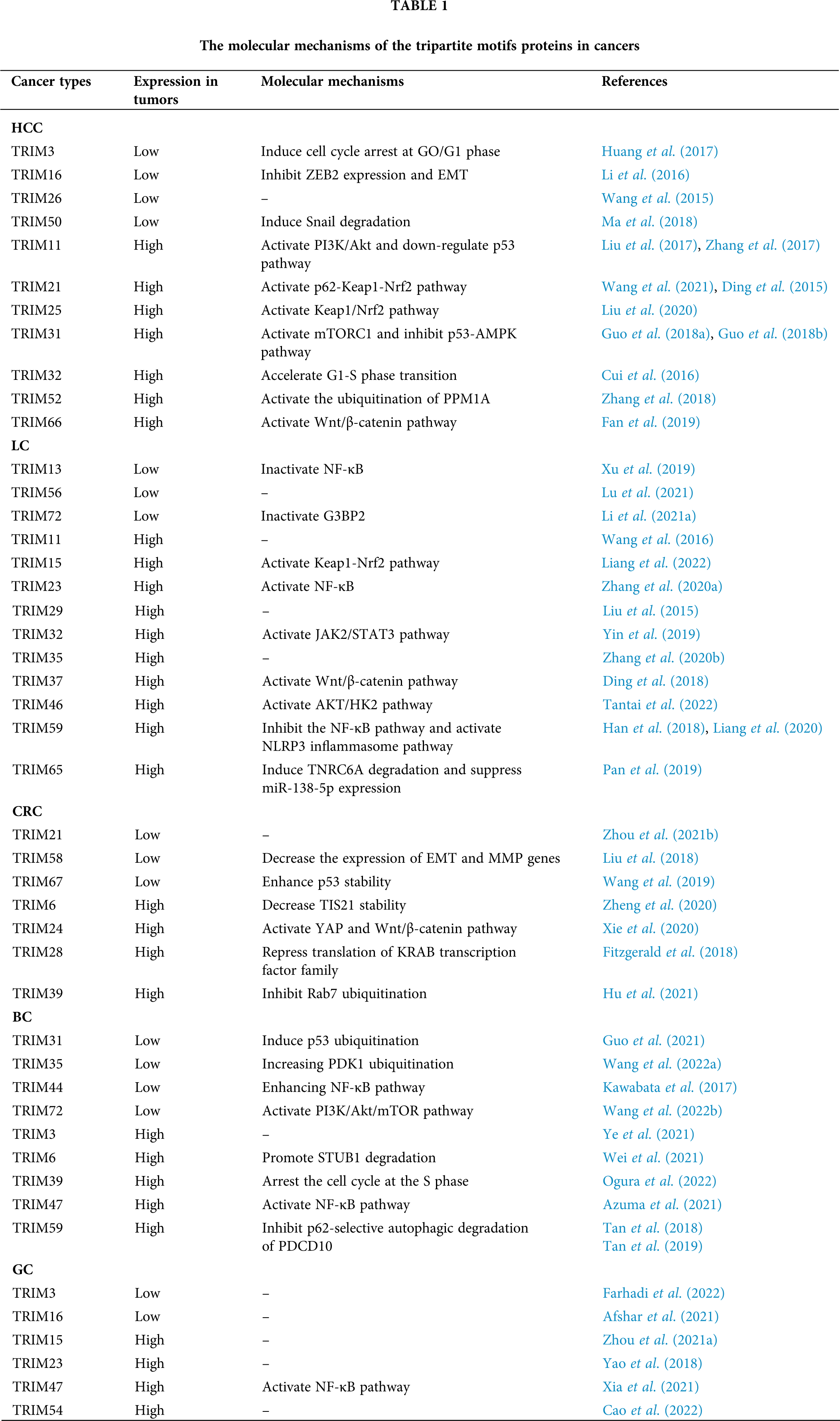
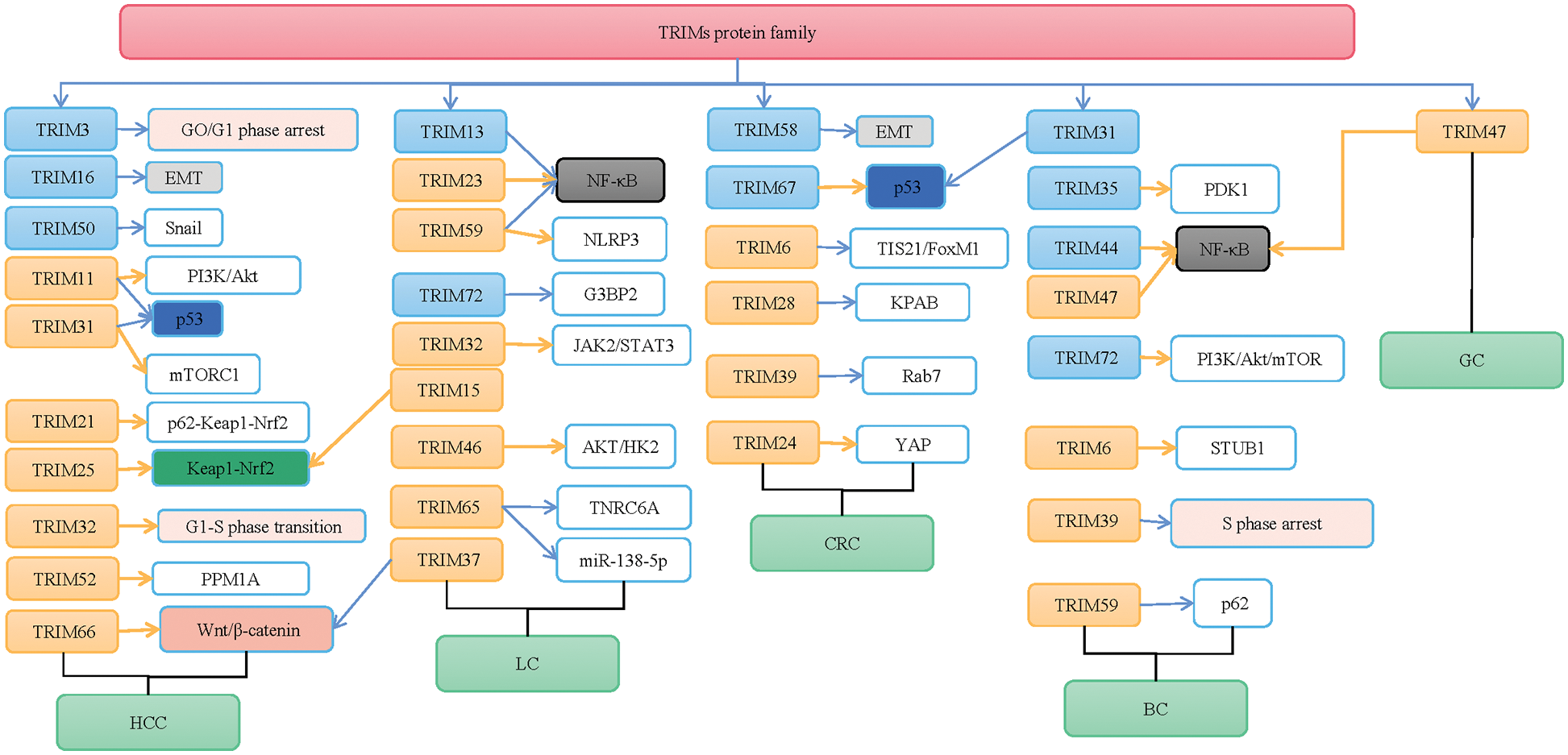
Figure 1: The mechanisms of the tripartite motifs (TRIMs) protein family are involved in human cancers. TRIMs protein family plays an important role in hepatocellular carcinoma (HCC), lung cancer (LC), colorectal cancer (CRC), breast cancer (BC), and gastric cancer (GC) by targeting PI3K/Akt, p53, Kelch-like ECH-associated protein 1 (Keap1)/NF-E2-related factor 2 (Nrf2), mammalian target of rapamycin complex 1 (mTORC1), Wnt/β-catenin, YES-associated protein (YAP), and p62 signaling. Blue arrows indicate the “inhibiting effect” and orange arrows indicate the “promoting effect”.
Author Contribution: The authors confirm their contribution to the paper as follows: study conception and design: YYH, HMW, XNZ; data collection: RYL, SJ, WLX, CY, LX; draft manuscript preparation: YYH, XNZ. All authors compared different studies and approved the final version of the manuscript.
Ethics Approval: Not applicable.
Funding Statement: This work was supported by the National Natural Science Foundation of China (82060607, 81860586) and the Natural Science Foundation of Guangxi Province (2020GXNSFDA297010).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
Afshar J, Mehrzad J, Mehrad-Majd H, Goshayeshi L, Saeidi J (2021). Prognostic significance of tripartite motif containing 16 expression in patients with gastric cancer. Asian Pacific Journal of Cancer Prevention 22: 2445–2451. DOI 10.31557/APJCP.2021.22.8.2445. [Google Scholar] [CrossRef]
Azuma K, Ikeda K, Suzuki T, Aogi K, Horie-Inoue K, Inoue S (2021). TRIM47 activates NF-κB signaling via PKC-ε/PKD3 stabilization and contributes to endocrine therapy resistance in breast cancer. PNAS 118: e2100784118. DOI 10.1073/pnas.2100784118. [Google Scholar] [CrossRef]
Cao H, Li Y, Chen L, Lu Z, You T, Wang X, Ji B (2022). Tripartite motif-containing 54 promotes gastric cancer progression by upregulating K63-linked ubiquitination of filamin C. Asia-Pacific Journal of Clinical Oncology 18: 669–677. DOI 10.1111/ajco.13747. [Google Scholar] [CrossRef]
Chao J, Zhang XF, Pan QZ, Zhao JJ, Jiang SS, Wang Y, Zhang JH, Xia JC (2014). Decreased expression of TRIM3 is associated with poor prognosis in patients with primary hepatocellular carcinoma. Medical Oncology 31: 102. DOI 10.1007/s12032-014-0102-9. [Google Scholar] [CrossRef]
Chen Y, Li L, Qian X, Ge Y, Xu G (2017). High expression of TRIM11 correlates with poor prognosis in patients with hepatocellular carcinoma. Clinics and Research in Hepatology and Gastroenterology 41: 190–196. DOI 10.1016/j.clinre.2016.09.010. [Google Scholar] [CrossRef]
Cui X, Lin Z, Chen Y, Mao X, Ni W et al. (2016). Upregulated TRIM32 correlates with enhanced cell proliferation and poor prognosis in hepatocellular carcinoma. Molecular and Cellular Biochemistry 421: 127–137. DOI 10.1007/s11010-016-2793-z. [Google Scholar] [CrossRef]
Diaz-Griffero F, Qin XR, Hayashi F, Kigawa T, Finzi A, Sarnak Z, Lienlaf M, Yokoyama S, Sodroski J (2009). A B-box 2 surface patch important for TRIM5alpha self-association, capsid binding avidity, and retrovirus restriction. Journal of Virology 83: 10737–10751. DOI 10.1128/JVI.01307-09. [Google Scholar] [CrossRef]
Ding Q, He D, He K, Zhang Q, Tang M, Dai J, Lv H, Wang X, Xiang G, Yu H (2015). Downregulation of TRIM21 contributes to hepatocellular carcinoma carcinogenesis and indicates poor prognosis of cancers. Tumor Biology 36: 8761–8772. DOI 10.1007/s13277-015-3572-2. [Google Scholar] [CrossRef]
Ding Y, Lu Y, Xie X, Sheng B, Wang Z (2018). Silencing TRIM37 inhibits the proliferation and migration of non-small cell lung cancer cells. RSC Advances 8: 36852–36857. DOI 10.1039/C8RA06391E. [Google Scholar] [CrossRef]
Fan W, Du F, Liu X (2019). TRIM66 confers tumorigenicity of hepatocellular carcinoma cells by regulating GSK-3β-dependent Wnt/β-catenin signaling. European Journal of Pharmacology 850: 109–117. DOI 10.1016/j.ejphar.2019.01.054. [Google Scholar] [CrossRef]
Farhadi J, Goshayeshi L, Motavalizadehkakhky A, Mehrzad J, Mehrad-Majd H (2022). Decreased expression of TRIM3 gene predicts a poor prognosis in gastric cancer. Journal of Gastrointestinal Cancer 53: 179–186. DOI 10.1007/s12029-020-00563-0. [Google Scholar] [CrossRef]
Feng X, Jiang T, Yang C, Pang S, Ding Z, Hu H, Wang H, Dong L, Yang N (2021). RPRD1A stabilizes NRF2 and aggravates HCC progression through competing with p62 for TRIM21 binding. Cell Death & Disease 13: 6. DOI 10.1038/s41419-021-04447-4. [Google Scholar] [CrossRef]
Fiorentini F, Esposito D, Rittinger K (2020). Does it take two to tango? RING domain self-association and activity in TRIM E3 ubiquitin ligases. Biochemical Society Transactions 48: 2615–2624. DOI 10.1042/BST20200383. [Google Scholar] [CrossRef]
Fitzgerald S, Espina V, Liotta L, Sheehan KM, O’Grady A, Cummins R, O’Kennedy R, Kay EW, Kijanka GS (2018). Stromal TRIM28-associated signaling pathway modulation within the colorectal cancer microenvironment. Journal of Translational Medicine 16: 89. DOI 10.1186/s12967-018-1465-z. [Google Scholar] [CrossRef]
Guo Y, Li Q, Zhao G, Zhang J, Yuan H et al. (2021). Loss of TRIM31 promotes breast cancer progression through regulating K48- and K63-linked ubiquitination of p53. Cell Death & Disease 12: 945. DOI 10.1038/s41419-021-04208-3. [Google Scholar] [CrossRef]
Guo P, Ma X, Zhao W, Huai W, Li T, Qiu Y, Zhang Y, Han L (2018a). TRIM31 is upregulated in hepatocellular carcinoma and promotes disease progression by inducing ubiquitination of TSC1-TSC2 complex. Oncogene 37: 478–488. DOI 10.1038/onc.2017.349. [Google Scholar] [CrossRef]
Guo P, Qiu Y, Ma X, Li T, Ma X, Zhu L, Lin Y, Han L (2018b). Tripartite motif 31 promotes resistance to anoikis of hepatocarcinoma cells through regulation of p53-AMPK axis. Experimental Cell Research 368: 59–66. DOI 10.1016/j.yexcr.2018.04.013. [Google Scholar] [CrossRef]
Han T, Guo M, Gan M, Yu B, Tian X, Wang JB (2018). TRIM59 regulates autophagy through modulating both the transcription and the ubiquitination of BECN1. Autophagy 14: 2035–2048. DOI 10.1080/15548627.2018.1491493. [Google Scholar] [CrossRef]
Han X, Huang C, Qu X, Liu S, Yang X, Wang Y, Bie F, Liu Q, Du J (2019). Tripartite motif-containing 15 overexpression in non-small cell lung cancer is associated with poor patient prognoses. Journal of Cancer 10: 843–852. DOI 10.7150/jca.27856. [Google Scholar] [CrossRef]
Hao X, Sun G, Zhang Y, Kong X, Rong D, Song J, Tang W, Wang X (2021). Targeting immune cells in the tumor microenvironment of HCC: New opportunities and challenges. Frontiers in Cell and Developmental Biology 9: 775462. DOI 10.3389/fcell.2021.775462. [Google Scholar] [CrossRef]
Hatakeyama S (2017). TRIM family proteins: Roles in autophagy, immunity, and carcinogenesis. Trends in Biochemical Sciences 42: 297–311. DOI 10.1016/j.tibs.2017.01.002. [Google Scholar] [CrossRef]
Hu J, Ding X, Tian S, Chu Y, Liu Z, Li Y, Li X, Wang G, Wang L, Wang Z (2021). TRIM39 deficiency inhibits tumor progression and autophagic flux in colorectal cancer via suppressing the activity of Rab7. Cell Death & Disease 12: 391. DOI 10.1038/s41419-021-03670-3. [Google Scholar] [CrossRef]
Huang XQ, Zhang XF, Xia JH, Chao J, Pan QZ et al. (2017). Tripartite motif-containing 3 (TRIM3) inhibits tumor growth and metastasis of liver cancer. Chinese Journal of Cancer 36: 77. DOI 10.1186/s40880-017-0240-5. [Google Scholar] [CrossRef]
Jaworska AM, Wlodarczyk NA, Mackiewicz A, Czerwinska P (2020). The role of TRIM family proteins in the regulation of cancer stem cell self-renewal. Stem Cells 38: 165–173. DOI 10.1002/stem.3109. [Google Scholar] [CrossRef]
Ji J, Ding K, Luo T, Zhang X, Chen A et al. (2021). TRIM22 activates NF-κB signaling in glioblastoma by accelerating the degradation of IκBα. Cell Death and Differentiation 28: 367–381. DOI 10.1038/s41418-020-00606-w. [Google Scholar] [CrossRef]
Joshi SS, Badgwell BD (2021). Current treatment and recent progress in gastric cancer. CA: A Cancer Journal for Clinicians 71: 264–279. DOI 10.3322/caac.21657. [Google Scholar] [CrossRef]
Kawabata H, Azuma K, Ikeda K, Sugitani I, Kinowaki K, Fujii T, Osaki A, Saeki T, Horie-Inoue K, Inoue S (2017). TRIM44 is a poor prognostic factor for breast cancer patients as a modulator of NF-κB signaling. International Journal of Molecular Sciences 18. DOI 10.3390/ijms18091931. [Google Scholar] [CrossRef]
Li L, Dong L, Qu X, Jin S, Lv X, Tan G (2016). Tripartite motif 16 inhibits hepatocellular carcinoma cell migration and invasion. International Journal of Oncology 48: 1639–1649. DOI 10.3892/ijo.2016.3398. [Google Scholar] [CrossRef]
Li H, Lin PH, Gupta P, Li X, Zhao SL et al. (2021a). MG53 suppresses tumor progression and stress granule formation by modulating G3BP2 activity in non-small cell lung cancer. Molecular Cancer 20: 118. DOI 10.1186/s12943-021-01418-3. [Google Scholar] [CrossRef]
Li R, Zhu L, Peng Y, Zhang X, Dai C, Liu D (2021b). TRIM50 suppresses pancreatic cancer progression and reverses the epithelial-mesenchymal transition via facilitating the ubiquitous degradation of snail1. Frontiers in Oncology 11: 695740. DOI 10.3389/fonc.2021.695740. [Google Scholar] [CrossRef]
Liang M, Chen X, Wang L, Qin L, Wang H, Sun Z, Zhao W, Geng B (2020). Cancer-derived exosomal TRIM59 regulates macrophage NLRP3 inflammasome activation to promote lung cancer progression. Journal of Experimental & Clinical Cancer Research: CR 39: 176. DOI 10.1186/s13046-020-01688-7. [Google Scholar] [CrossRef]
Liang M, Wang L, Sun Z, Chen X, Wang H, Qin L, Zhao W, Geng B (2022). E3 ligase TRIM15 facilitates non-small cell lung cancer progression through mediating Keap1-Nrf2 signaling pathway. Cell Communication and Signaling 20: 62. DOI 10.1186/s12964-022-00875-7. [Google Scholar] [CrossRef]
Liu C, Huang X, Hou S, Hu B, Li H (2015). Silencing of tripartite motif (TRIM) 29 inhibits proliferation and invasion and increases chemosensitivity to cisplatin in human lung squamous cancer NCI-H520 cells. Thoracic Cancer 6: 31–37. DOI 10.1111/1759-7714.12130. [Google Scholar] [CrossRef]
Liu J, Rao J, Lou X, Zhai J, Ni Z, Wang X (2017). Upregulated TRIM11 exerts its oncogenic effects in hepatocellular carcinoma through inhibition of P53. Cellular Physiology and Biochemistry 44: 255–266. DOI 10.1159/000484678. [Google Scholar] [CrossRef]
Liu Y, Tao S, Liao L, Li Y, Li H, Li Z, Lin L, Wan X, Yang X, Chen L (2020). TRIM25 promotes the cell survival and growth of hepatocellular carcinoma through targeting Keap1-Nrf2 pathway. Nature Communications 11: 348. DOI 10.1038/s41467-019-14190-2. [Google Scholar] [CrossRef]
Liu M, Zhang X, Cai J, Li Y, Luo Q, Wu H, Yang Z, Wang L, Chen D (2018). Downregulation of TRIM58 expression is associated with a poor patient outcome and enhances colorectal cancer cell invasion. Oncology Reports 40: 1251–1260. DOI 10.3892/or.2018.6525. [Google Scholar] [CrossRef]
Lu K, Sui Y, Fu L (2021). Identification of TRIM56 as a potential biomarker for lung adenocarcinoma. Cancer Management and Research 13: 2201–2213. DOI 10.2147/CMAR.S288111. [Google Scholar] [CrossRef]
Ma X, Ma X, Qiu Y, Zhu L, Lin Y et al. (2018). TRIM50 suppressed hepatocarcinoma progression through directly targeting SNAIL for ubiquitous degradation. Cell Death & Disease 9: 608. DOI 10.1038/s41419-018-0644-4. [Google Scholar] [CrossRef]
Mandell MA, Saha B, Thompson TA (2020). The tripartite nexus: Autophagy, cancer, and tripartite motif-containing protein family members. Frontiers in Pharmacology 11: 308. DOI 10.3389/fphar.2020.00308. [Google Scholar] [CrossRef]
Ogura T, Azuma K, Takeiwa T, Sato J, Kinowaki K, Ikeda K, Kawabata H, Inoue S (2022). TRIM39 is a poor prognostic factor for patients with estrogen receptor-positive breast cancer and promotes cell cycle progression. Pathology International 72: 96–106. DOI 10.1111/pin.13190. [Google Scholar] [CrossRef]
Pan X, Chen Y, Shen Y, Tantai J (2019). Knockdown of TRIM65 inhibits autophagy and cisplatin resistance in A549/DDP cells by regulating miR-138-5p/ATG7. Cell Death & Disease 10: 429. DOI 10.1038/s41419-019-1660-8. [Google Scholar] [CrossRef]
Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S et al. (2001). The tripartite motif family identifies cell compartments. The EMBO Journal 20: 2140–2151. DOI 10.1093/emboj/20.9.2140. [Google Scholar] [CrossRef]
Sun Y, Ren D, Yang C, Yang W, Zhao J, Zhou Y, Jin X, Wu H (2021). TRIM15 promotes the invasion and metastasis of pancreatic cancer cells by mediating APOA1 ubiquitination and degradation. Biochimica et Biophysica Acta (BBA)-Molecular Basis of Disease 1867: 166213. DOI 10.1016/j.bbadis.2021.166213. [Google Scholar] [CrossRef]
Tan P, He L, Zhou Y (2019). TRIM59 deficiency curtails breast cancer metastasis through SQSTM1-selective autophagic degradation of PDCD10. Autophagy 15: 747–749. DOI 10.1080/15548627.2019.1569951. [Google Scholar] [CrossRef]
Tan P, Ye Y, He L, Xie J, Jing J, Ma G, Pan H, Han L, Han W, Zhou Y (2018). TRIM59 promotes breast cancer motility by suppressing p62-selective autophagic degradation of PDCD10. PLoS Biology 16: e3000051. DOI 10.1371/journal.pbio.3000051. [Google Scholar] [CrossRef]
Tantai J, Pan X, Chen Y, Shen Y, Ji C (2022). TRIM46 activates AKT/HK2 signaling by modifying PHLPP2 ubiquitylation to promote glycolysis and chemoresistance of lung cancer cells. Cell Death & Disease 13: 285. DOI 10.1038/s41419-022-04727-7. [Google Scholar] [CrossRef]
Thai AA, Solomon BJ, Sequist LV, Gainor JF, Heist RS (2021). Lung cancer. Lancet 398: 535–554. DOI 10.1016/S0140-6736(21)00312-3. [Google Scholar] [CrossRef]
Tian H, Zhao H, Qu B, Chu X, Xin X, Zhang Q, Li W, Yang S (2022). TRIM24 promotes colorectal cancer cell progression via the Wnt/β-catenin signaling pathway activation. American Journal of Translational Research 14: 831–848. [Google Scholar]
Tocchini C, Ciosk R (2015). TRIM-NHL proteins in development and disease. Seminars in Cell and Developmental Biology 47–48: 52–59. DOI 10.1016/j.semcdb.2015.10.017. [Google Scholar] [CrossRef]
Wang Y, He D, Yang L, Wen B, Dai J, Zhang Q, Kang J, He W, Ding Q, He D (2015). TRIM26 functions as a novel tumor suppressor of hepatocellular carcinoma and its downregulation contributes to worse prognosis. Biochemical and Biophysical Research Communications 463: 458–465. DOI 10.1016/j.bbrc.2015.05.117. [Google Scholar] [CrossRef]
Wang R, Huang KL, Xing LX (2022a). TRIM35 functions as a novel tumor suppressor in breast cancer by inducing cell apoptosis through ubiquitination of PDK1. Neoplasma 69: 370–382. DOI 10.4149/neo_2021_210823N1205. [Google Scholar] [CrossRef]
Wang HT, Hur S (2021). Substrate recognition by TRIM and TRIM-like proteins in innate immunity. Seminars in Cell and Developmental Biology 111: 76–85. DOI 10.1016/j.semcdb.2020.09.013. [Google Scholar] [CrossRef]
Wang Z, Li H, Wang H, Li X, Zhang Q, Wang H, Li K, Qiu Y (2022b). TRIM72 exerts antitumor effects in breast cancer and modulates lactate production and MCT4 promoter activity by interacting with PPP3CA. Anticancer Drugs 33: 489–501. DOI 10.1097/CAD.0000000000001304. [Google Scholar] [CrossRef]
Wang X, Shi W, Shi H, Lu S, Wang K et al. (2016). TRIM11 overexpression promotes proliferation, migration and invasion of lung cancer cells. Journal of Experimental & Clinical Cancer Research 35: 100. DOI 10.1186/s13046-016-0379-y. [Google Scholar] [CrossRef]
Wang S, Zhang Y, Huang J, Wong CC, Zhai J et al. (2019). TRIM67 activates p53 to suppress colorectal cancer initiation and progression. Cancer Research 79: 4086–4098. DOI 10.1158/0008-5472.CAN-18-3614. [Google Scholar] [CrossRef]
Wang F, Zhang Y, Shen J, Yang B, Dai W et al. (2021). The ubiquitin E3 ligase TRIM21 promotes hepatocarcinogenesis by suppressing the p62-Keap1-Nrf2 antioxidant pathway. Cellular and Molecular Gastroenterology and Hepatology 11: 1369–1385. DOI 10.1016/j.jcmgh.2021.01.007. [Google Scholar] [CrossRef]
Wei C, Wu J, Liu W, Lu J, Li H, Hai B (2021). Tripartite motif-containing protein 6 facilitates growth and migration of breast cancer through degradation of STUB1. European Journal of Histochemistry 65: 3214. DOI 10.4081/ejh.2021.3214. [Google Scholar] [CrossRef]
Xia Y, Wei Z, Huang W, Wei X, He Y (2021). Trim47 overexpression correlates with poor prognosis in gastric cancer. Neoplasma 68: 307–316. DOI 10.4149/neo_2020_200708N706. [Google Scholar] [CrossRef]
Xie X, Li H, Pan J, Han X (2021). Knockdown of TRIM26 inhibits the proliferation, migration and invasion of bladder cancer cells through the Akt/GSK3β/β-catenin pathway. Chemico-Biological Interactions 337: 109366. DOI 10.1016/j.cbi.2021.109366. [Google Scholar] [CrossRef]
Xie W, Zhang Y, Wang B, Hu Y, Zhan B, Wei F, Tang J, Lian J (2020). Tripartite motif containing 24 regulates cell proliferation in colorectal cancer through YAP signaling. Cancer Medicine 9: 6367–6376. DOI 10.1002/cam4.3310. [Google Scholar] [CrossRef]
Xu L, Wu Q, Zhou X, Wu Q, Fang M (2019). TRIM13 inhibited cell proliferation and induced cell apoptosis by regulating NF-κB pathway in non-small-cell lung carcinoma cells. Gene 715: 144015. DOI 10.1016/j.gene.2019.144015. [Google Scholar] [CrossRef]
Yang W, Gu Z, Zhang H, Hu H (2020). To TRIM the immunity: From innate to adaptive immunity. Frontiers in Immunology 11: 02157. DOI 10.3389/fimmu.2020.02157. [Google Scholar] [CrossRef]
Yang L, Xia H (2021). TRIM proteins in inflammation: From expression to emerging regulatory mechanisms. Inflammation 44: 811–820. DOI 10.1007/s10753-020-01394-8. [Google Scholar] [CrossRef]
Yao Y, Liu Z, Guo H, Huang S, Zhong M, Deng J, Xiong J (2018). Elevated TRIM23 expression predicts poor prognosis in Chinese gastric cancer. Pathology-Research and Practice 214: 2062–2068. DOI 10.1016/j.prp.2018.10.010. [Google Scholar] [CrossRef]
Ye R, AiErken N, Kuang X, Zeng H, Shao N, Lin Y, Liu P, Wang S (2021). Tripartite motif-containing 3 (TRIM3) enhances ER signaling and confers tamoxifen resistance in breast cancer. Oncogenesis 10: 60. DOI 10.1038/s41389-021-00350-x. [Google Scholar] [CrossRef]
Yin H, Li Z, Chen J, Hu X (2019). Expression and the potential functions of TRIM32 in lung cancer tumorigenesis. Journal of Cellular Biochemistry 120: 5232–5243. DOI 10.1002/jcb.27798. [Google Scholar] [CrossRef]
Zhang Y, Du H, Li Y, Yuan Y, Chen B, Sun S (2020a). Elevated TRIM23 expression predicts cisplatin resistance in lung adenocarcinoma. Cancer Science 111: 637–646. DOI 10.1111/cas.14226. [Google Scholar] [CrossRef]
Zhang Y, Tao R, Wu SS, Xu CC, Wang JL, Chen J, Yu YS, Tang ZH, Chen XH, Zang GQ (2018). TRIM52 up-regulation in hepatocellular carcinoma cells promotes proliferation, migration and invasion through the ubiquitination of PPM1A. Journal of Experimental & Clinical Cancer Research 37: 116. DOI 10.1186/s13046-018-0780-9. [Google Scholar] [CrossRef]
Zhang J, Xu Z, Yu B, Xu J, Yu B (2020b). Tripartite motif containing 35 contributes to the proliferation, migration, and invasion of lung cancer cells in vitro and in vivo. Bioscience Reports 40: 394. DOI 10.1042/bsr20200065. [Google Scholar] [CrossRef]
Zhang Z, Xu C, Zhang X, Huang L, Zheng C, Chen H, Wang Y, Ju H, Yao Q (2017). TRIM11 upregulation contributes to proliferation, invasion, and EMT of hepatocellular carcinoma cells. Oncology Research 25: 691–699. DOI 10.3727/096504016X14774897404770. [Google Scholar] [CrossRef]
Zheng S, Zhou C, Wang Y, Li H, Sun Y, Shen Z (2020). TRIM6 promotes colorectal cancer cells proliferation and response to thiostrepton by TIS21/FoxM1. Journal of Experimental & Clinical Cancer Research 39: 23. DOI 10.1186/s13046-019-1504-5. [Google Scholar] [CrossRef]
Zhou W, Chen H, Ruan Y, Zeng X, Liu F (2021a). High expression of TRIM15 is associated with tumor invasion and predicts poor prognosis in patients with gastric cancer. Journal of Investigative Surgery 34: 853–861. DOI 10.1080/08941939.2019.1705443. [Google Scholar] [CrossRef]
Zhou G, Wu H, Lin J, Lin R, Feng B, Liu Z (2021b). TRIM21 is decreased in colitis-associated cancer and negatively regulates epithelial carcinogenesis. Inflammatory Bowel Diseases 27: 458–468. DOI 10.1093/ibd/izaa229. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools