 Open Access
Open Access
ARTICLE
KIF15, a key regulator of nasopharyngeal carcinoma development mediated by the P53 pathway
1 Department of Otolaryngology & Head and Neck, The First Affiliated Hospital of Guangxi Medical University, Nanning, 530021, China
2 Department of Otolaryngology & Head and Neck, The People’s Hospital of Guangxi Zhuang Autonomous Region, Guangxi Academy of Medical Sciences, Nanning, 530021, China
3 Research Center of Medical Sciences, The People’s Hospital of Guangxi Zhuang Autonomous Region, Nanning, 530021, China
* Corresponding Author: GUANGWU HUANG. Email:
# These authors contributed to the work equally and should be regarded as co-first authors
BIOCELL 2023, 47(3), 533-545. https://doi.org/10.32604/biocell.2023.025280
Received 06 July 2022; Accepted 13 September 2022; Issue published 03 January 2023
Abstract
Background: Kinesin family member 15 (KIF15) is a protein that regulates cell mitosis and plays an important role in the development and progression of several types of human cancers. However, the role of KIF15 in the development of nasopharyngeal cancer (NPC) is still unclear. Methods: The differential expression of KIF15 in NPC and para-carcinoma tissues was evaluated based on data collected from Gene Expression Omnibus (GEO) database and immunohistochemical analysis of clinical specimens collected from a patient cohort. Cell lines 5-8F and CNE-2Z were selected for the construction of KIF15‑knockdown cell models. CCK8 assay, flow cytometry, wound healing, Transwell and clone formation assays were used to detect the proliferation, apoptosis, migration, invasion and colony formation of NPC cells in vitro. A mouse xenograft model and the tail intravenous mouse distant transfer model were constructed for in vivo study. Furthermore, the potential molecular mechanisms underlying the effects of KIF15 were explored through western blot analysis, and several in vitro and in vivo functional assays were performed to explore its role in NPC. Results: The results revealed significantly higher expression of KIF15 in NPC tissues compared to para-carcinoma tissues. High levels of KIF15 expression were also associated with short overall survival (OS) and progression-free survival (PFS). Knockdown of the KIF15 gene led to a cell cycle arrest in the growth 2 (G2) phase, inhibition of cell proliferation, migration, invasion, colony formation, and enhanced cell apoptosis. The in vivo murine xenograft experiments showed that down-regulation of the KIF15 gene could inhibit tumor growth and reduce the risk of liver and lung metastasis in NPC. Moreover, the evaluation of the molecular pathway showed that the mitogen-activated protein kinase/P53 pathways might be involved in the KIF15-induced regulation of NPC. Rescue assays indicated that Pifithrin-α could counteract the pro-proliferative and pro-apoptotic effects mediated by KIF15. Conclusion: This work indicated that KIF15 overexpression accelerated the progression of NPC and promoted the development of distant metastases. Therefore, KIF15 may have an important role as a prognostic indicator and a potential drug target for the treatment of NPC.Keywords
Nasopharyngeal carcinoma (NPC) is a common type of head and neck tumor in China, especially in the southern regions. Most of its pathological subtypes are undifferentiated or poorly differentiated squamous cell carcinoma (Tao and Chan, 2007). NPCs tend to spread quickly to regional lymph nodes and distant sites. As a result, approximately 60% to 70% of NPC cases are diagnosed at an advanced stage. The most effective treatment for these patients involves a combination of radiotherapy and chemotherapy (Lee et al., 2017). However, about 20% to 30% of advanced NPCs recur locally and spread to distant sites after receiving conventional treatment (Chen et al., 2012). Therefore, there is a need to understand the molecular mechanisms involved in NPC to explore and develop more efficient treatments.
The growth and metastasis of cancer cells depend on cell mitosis and proliferation (Logan and Menko, 2019). The cytoskeletal protein system plays an important role in maintaining cancer cell morphology, gene translation, protein transport, and tumor signal transduction necessary for cell mitosis (Ong et al., 2020). The kinesin superfamily proteins (KIFs) are an important group of cytoskeletal proteins that were first found in the axons of squid nerve cells. These proteins use the energy generated from the hydrolysis of adenosine triphosphate (ATP) to promote intracellular transport and cell movement. The KIF proteins are classified into 14 groups according to their structural, functional, and phylogenetic features.
The kinesin family member 15 (KIF15) belongs to the kinesin-12 superfamily, also known as the hklp2, forms part of the tetramer spindle motor (Vanneste et al., 2011). Numerous studies have shown that KIF15 participates in and promotes the occurrence and development of malignant tumors. A study in 2014 demonstrated that KIF15 is overexpressed in breast cancer tissues and increases the risk of recurrence in estrogen receptor (ER)-positive breast cancer patients (Zou et al., 2014). Similarly, Gao et al. (2020) also found that KIF15 contributes to the proliferation and migration of breast cancer cells. However, KIF15 was also implicated in the development of other cancers, including pancreatic cancer (Wang et al., 2017), hepatocellular carcinoma (Kitagawa et al., 2020; Matsushita et al., 2020), lung adenocarcinoma (Qiao et al., 2018), gastric cancer (Ding et al., 2020), and malignant peripheral nerve sheath tumors (Terribas et al., 2020).
Although the expression level of KIF15 has been shown to impact the overall survival (OS) in patients diagnosed with NPC (Mi et al., 2022), the pathway by which KIF15 promotes the progression of NPC is still unknown. Therefore, we applied several in vitro and in vivo functional assays to explore its role in the development of NPC and eventually as a potential therapeutic target in NPC.
Evaluation of kinesin family member 15 expression in a public nasopharyngeal carcinoma dataset
The Gene Expression Omnibus (GEO) database was utilized to compare the expression of KIF15 in normal tissues and NPC tissues. The datasets included in this study were GSE12452 and GSE34573, and the expression profiles were tested using the Affymetrix human genome U133 Plus2.0 array platform. We used the “Limma” package in the R statistics software to screen the differentially expressed genes between the NPC tissue and the normal nasopharyngeal tissue samples. A total of 61 samples, including 14 normal nasopharyngeal tissue samples and 47 NPC samples, were included in the chip information analysis. During the chip information analysis, the quality of the original chip data was first evaluated to identify the data that qualified for further analysis. The data were then filtered and analyzed. Probes with an average signal of less than 0.005 were filtered out. The differential gene expression profile was also screened by |fold change| ≥ 1.3 to minimize the risk of false positive results. Hierarchical cluster analysis was also performed, and a p-value below 0.05 was deemed statistically significant. Significant difference analysis and functional analysis of the differentially expressed genes were conducted on the data that met the filtering criteria.
Collection of clinical samples
A total of 132 NPC and 15 para-carcinoma tissue samples were collected from 132 NPC patients between January 2010 and October 2011. The tissue samples were fixed in formalin and embedded in paraffin tissue microarrays provided by Shanghai Xinchao Biotech Company (cat. no. HNasN132Su01). The tissue microarray protocol was approved by the Human Ethics Committee of the Taizhou Hospital of Zhejiang Province. Written informed consent was obtained from all patients participating in the study. All tissue specimens were evaluated by two experienced pathologists, and the clinical stage of NPC patients was defined according to the American Joint Committee on Cancer (AJCC) (7th edition). All patients enrolled in the study received concurrent chemoradiation and were followed up for at least 5 years, except for those who died before.
The human NPC cell lines (CNE-2Z, HONE-1) were obtained from the Institute of Cancer, Central South University. The NPC cell line C666-1 was obtained from the University of Hong Kong, and the 5-8F cell line was obtained from Sun Yat-Sen University. All cell lines were tested for mycoplasma by fluorescent quantitative polymerase chain reaction (PCR) to avoid mycoplasma infection. The cells were propagated in Dulbecco’s modified Eagle medium (DMEM; Gibco, Rockville, MD), with 1% penicillin/streptavidin (Gibco) and 10% fetal bovine serum (FBS, Gibco) in a humidified atmosphere containing 5% carbon dioxide (CO2) at 37°C. The cell culture media were replaced every 72 h.
RNA preparation, reverse transcription, and quantitative real-time PCR (qRT-PCR)
The total RNA was extracted using the TRIzol reagent (Sigma, cat. no. T9424-100m). The cDNA was then synthesized using a reverse transcription kit (Vazyme, cat. no. R123-01) following the manufacturer’s protocol. The qRT-PCR analysis was performed using the AceQ qPCR SYBR Greenmaster mix (Vazyme) on an ABI VII7 System. The livac (2‑ΔΔCq) method was utilized to quantify the gene expression, and glyceraldehyde-3 phosphate dehydrogenase (GAPDH) was used as a reference control. The following PCR primers were used: KIF15 forward, 5′‑CTC TCA CAG TTG AAT GTC CTT G‑3′ and reverse, 5′‑CTC CTT GTC AGC AGA ATG AAG‑3′; GAPDH forward, 5′‑TGA CTT CAA CAG CGA CAC CCA‑3′ and reverse, 5′‑CAC CCT GTT GCT GTA GCC AAA‑3′. The PCR protocol involved one cycle of 1 min at 95°C, 45 cycles of 10 s at 95°C and 30 s at 60°C, 1 cycle of 15 s at 95°C, 1 cycle of 1 min at 55°C and 15 s at 95°C. The PCR products were separated on a 20 g/l agarose gel, stained with ethidium bromide and viewed under ultraviolet light.
Nasopharyngeal Carcinoma microarray and immunohistochemistry (IHC)
The NPC tissue microarray was used for the IHC staining analysis of the KIF15 protein expression. For the IHC staining, the samples were incubated with the rabbit anti-KIF15 antibody, and the horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG H&L was used as a secondary antibody. The Ki‑67 antibody was used in the IHC analysis of tumor sections removed from the mice models. A light microscope was used to capture the IHC images, and the KIF15 expression was scored by summing up the staining intensity score ranging from 0 to 3, with the percentage of positive cells score ranging from 0 to 4. A final score above 3 was considered a high expression of KIF15, while a final score of 3 or less was considered a low expression of KIF15.
The expression level of KIF15 in the NPC cell lines was determined by lysing the collected cells with a lysis buffer according to the manufacturer’s instructions. The final protein concentration was determined using the Bicinchoninic Acid (BCA) Protein Assay kit. The total cellular proteins were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred to a polyvinylidene difluoride membrane. The membranes were then incubated with 5% bovine serum albumin in a tris‑buffered saline solution containing 0.5% Tween‑20 (TBST) for 60 min and then incubated overnight at 4°C with the following primary antibodies: KIF15 antibody (cat. no. FNab04551; Fine Test), B cell lymphoma-2 (BCL-2) antibody (cat. no. 2876; CST), BCL2 associated X (BAX) antibody (2772; CST), cysteine-aspartic acid protease-9 (Caspase-9) antibody (9502; CST), caspase-3 antibody (9662; CST), E-cadherin (20874; Proteintech), N-cadherin (22018; Proteintech), vimentin (10366; Proteintech), Snail family transcriptional repressor 1 (Snai1, 13099; Proteintech), Snai2/Slug (12129; Proteintech), matrix metallopeptidase 9 (MMP9, cat. no. 27306; Proteintech), phosphatidylinositol-3-kinase (PI3K p110; cat. no. ab127617, Abcam), protein kinase B (AKT; cat. no. 9272, CST), p-AKT (cat. no. 4060, CST), extracellular signal-regulated kinase (ERK; cat. no. 4695, CST), p-ERK (cat. no. 4370, CST), P38 (cat. no. ab170099, Abcam), p-P38 (cat. no. ab1278674, Abcam), P53 (cat. no. 10442, Proteintech), p-P53 (cat. no. 28961, Proteintech), and GAPDH (AP0063; Bioworld). The Immobilon Western Chemiluminescent HRP Substrate was used for the visualization of target protein bands. Each membrane was visualized using the ECL‑Plus™ western blotting system. Densitometric analysis was performed using ImageJ.
RNA interference and transfection
The BR-V-108 lentiviral vector system was used in this study for RNA interference and transfection. Three specific short hairpin RNAs (shRNAs) against KIF15 (shKIF15-1, shKIF15-2, and shKIF15-3) and a negative control shRNA (shCtrl) were packaged and purchased from GenePharma Co., Ltd., Shanghai, China. The sequences of shKIFs were as follows: shKIF15-1 (GCT GAA GTG AAG AGG CTC AAA); shKIF15-2 (AGG CAG CTA GAA TTG GAA TCA); shKIF15-3 (AAG CTC AGA AAG AGC CAT GTT). The 5-8F cells were infected with 40 μL of lentivirus at a concentration of 1 × 108 TU/mL, while the CNE-2Z cells were infected with 20 μL according to the multiplicity of infection (MOI). After the transduction into cells, puromycin was added to the culture medium to enrich the stably infected cells. The transfection and knockdown efficiencies were verified by light microscopy, fluorescence microscopy, qRT-PCR, and western blotting analysis.
Cell counting kit-8 (CCK8) assay
The in vitro cell viability was analyzed using a CCK8 assay. The 5-8F and CNE-2Z cells were placed into 96-well plates with a density of 1500 cells per well and cultured for 5 days. Then 10 μL of the CCK8 solution was added to each well of the plate. After 1 to 4 h of incubation, the solution was mixed gently on an orbital shaker for 1 min to ensure a homogeneous color distribution. The absorbance values were measured at 450 nm using a microplate reader every 24 h for 5 days.
The shKIF15 and shCtrl cells were seeded in 6-well plates. The cells were subsequently digested with trypsin, centrifuged at 1300 rpm for 5 min, and washed with D-Hanks at 4°C. The cells were resuspended using 300 μL of binding buffer and stained using 5 μL of Annexin V‑APC and 5 μL of propidine iodide (eBioscience) per sample for 15 min in the dark at room temperature. The results were visualized using the Guava Soft software.
For the colony formation assay, cells were seeded in 6-well plates at a density of 1,000 cells per well and grown for 2 weeks. The culture medium was replaced every 3 days. After that, the cells were fixed with 4% paraformaldehyde (1 mL/well) and subjected to Giemsa staining (500 μl/well) for 20 min. The cells were then washed several times with distilled water (dd H2O). Finally, photographs were taken, and the colonies were counted.
NPC cells in the exponential growth phase were collected and rinsed in cold PBS and fixed with cold 70% ethanol. The cells were then stained with 1 mg/mL propidium iodide and kept away from light for 10 to 15 min. Flow cytometry was performed on the cell populations throughout the different phases of the cell cycle, including growth-0/growth-1 (G0/G1), synthesis (S), and growth-2/mitotic (G2/M) phases. A cell throughput rate of 200 to 350 cells/s was guaranteed at the time of loading.
The cell suspension was added to a 6-well plate at a concentration of 4 × 105 cells per well. Cells were cultured for a day in a serum-free medium, and then wounds were generated using pipette tips. The medium was replenished with DMEM, and the healed wound distance was separately observed at 0, 24, and 48 h after wound generation. The cell migration rate for each group was calculated according to the measured width of the wound.
Cell migration and invasion assay
Transwell chambers were used for the cell migration assays. The Matrigel invasion chambers with an 8.0-μm PET membrane were used for the cell invasion assays. Around 50,000 NPC cells were seeded in the upper compartment using 100 μL of serum-free medium, and the bottom compartment was filled with 600 μL of medium supplemented with 30% FBS. After an incubation period of 48 h at 37°C, the remaining cells in the upper compartment were removed by cotton swabs, while the cells migrating to the bottom were stained with 0.1% crystal violet (400 μL/well) for 5 min. The migrated cells were photographed and counted under the microscope. Cell migration was determined by observing five random fields under a microscope.
In vivo metastasis evaluation in a tumor‑bearing mouse model
All the experimental animal protocols were approved by the Ethics Committee of the People’s Hospital of Guangxi Zhuang Autonomous Region. The shCtrl and shKIF15 groups consisted of 6 BALB/c male nude mice (4 weeks old). The mice were cultured in the laboratory in a 12-h light/12-h dark environment and were fed a standard diet. A suspension of 200 μl of the 5-8F and CNE-2Z cells at a concentration of 2 × 107 cells/mL, transfected with shKIF15 or shCtrl were injected subcutaneously into the back of the nude mice next to the forelimb. The induced tumors were measured every 4 to 5 days post-injection. In vivo images of the mice were acquired using the vivisection imaging system, and the fluorescence intensity was analyzed quantitatively. For the in vivo metastasis evaluation, 1 × 106 treated NPC cells were injected into the nude mice via the tail vein. Six weeks after the injection, the size of the tumor nodules was observed and recorded. All mice were euthanized by injecting 1% pentobarbital sodium (0.2 mg/g) before reaching the humane endpoint. The subcutaneous tumor, liver, and lung tissues were harvested, weighed, and analyzed using IHC and hematoxylin and eosin (H&E) staining.
During the CCK8 and flow cytometry analysis, the P53 inhibitor pifithrin-alpha (PFT-α) was added to the two cell lines at a concentration of 10 μmol/mL to measure the proliferation and apoptosis levels of the NPC cells. Western blotting experiment was subsequently performed to detect the changes in the p53 phosphorylation state.
All data were obtained from no less than three independent experiments. The continuous data were expressed as means ± standard deviations, and the age groups were divided according to the median age of all patients. All statistical analyses were carried out using GraphPad Prism software version 6. The Student’s t-test and chi-square (χ2) test were used to analyze the significant difference before and after the KIF15 knockdown. The Mann-Whitney U test and the Spearman rank correlation analysis were used to compare the clinical data. Survival analysis was performed by plotting a Kaplan-Meier curve, and the log-rank test was used to identify factors influencing survival. For all statistical tests, a p-value below 0.05 was considered statistically significant.
Correlation between the expression of kinesin family member 15 in nasopharyngeal carcinoma tissues and patient survival
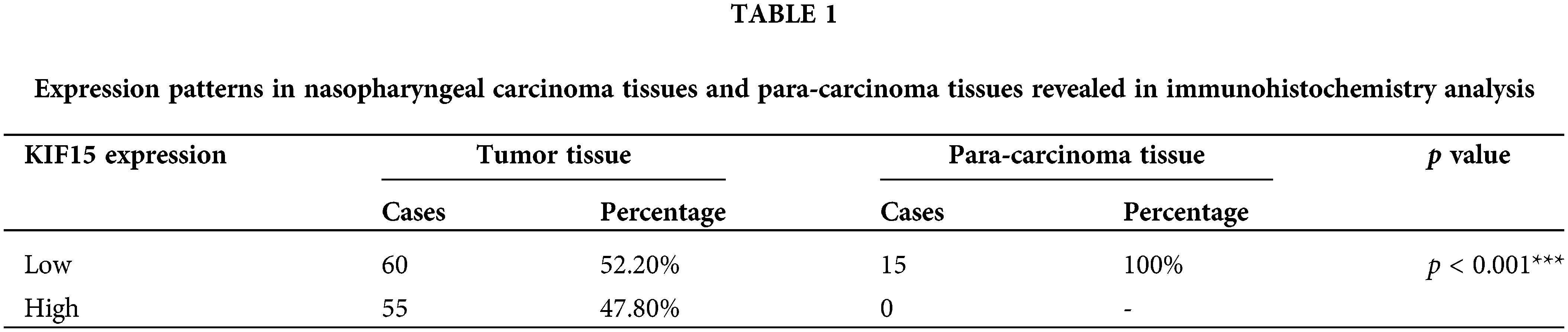
The heat maps and volcano plot displaying the cluster analysis findings for the differentially expressed genes in NPC and normal mucosal tissue samples are shown in Figs. 1A and 1B. The results showed that the KIF15 mRNA expression level in the NPC tissues was significantly higher when compared with that in normal tissue samples (Fig. 1C, p < 0.001). IHC analysis of the KIF15 expression in 132 NPC tissue was performed as described. The K1F15 expression could not be scored in 17 cases due to tissue peeling or poor staining quality. This analysis showed that the protein expression of KIF15 in NPC tissues was significantly higher when compared with that in para-carcinoma tissues (Table 1 and Fig. 1D).
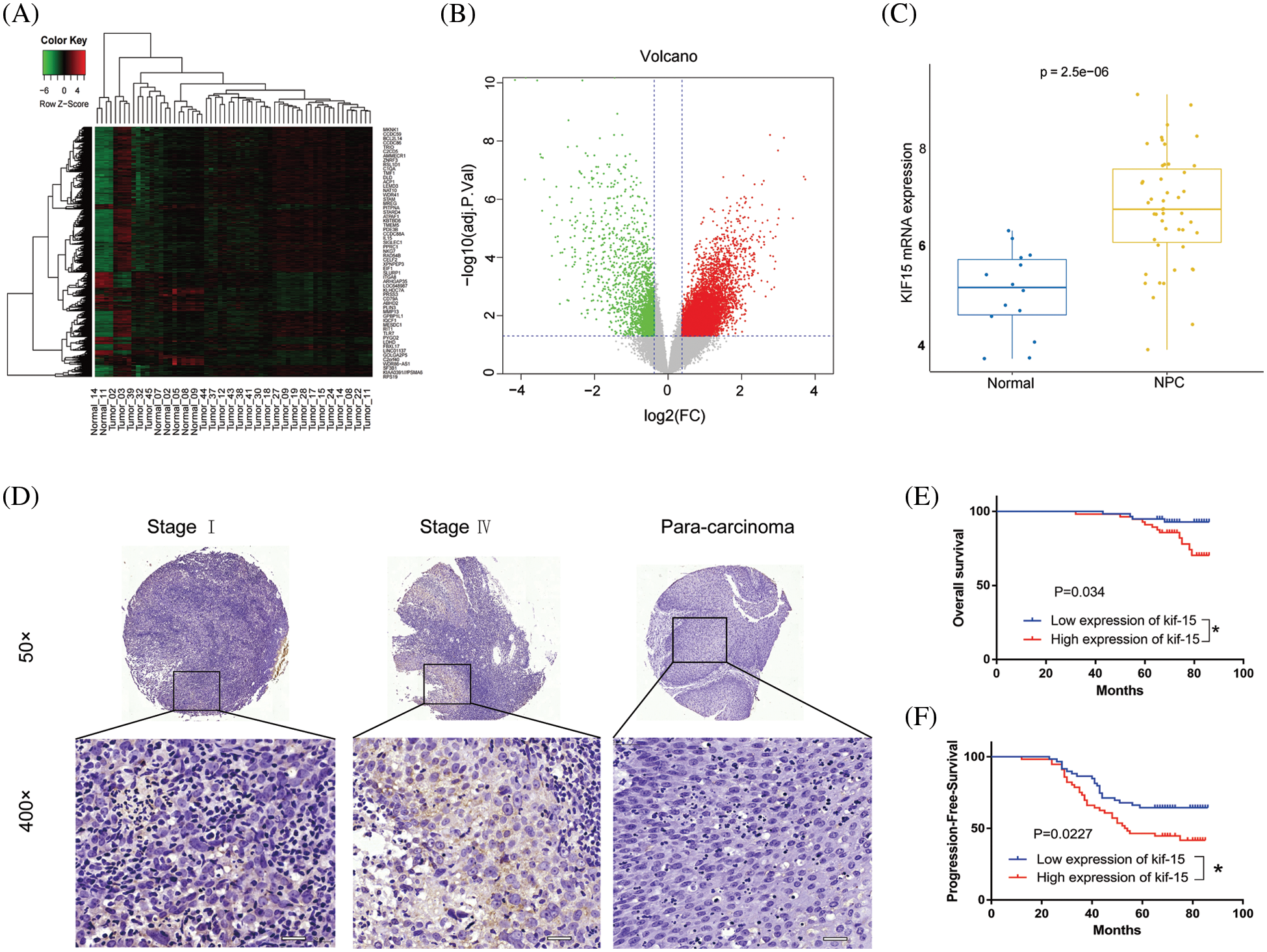
Figure 1: Kinesin family member 15 (KIF15) is highly expressed in nasopharyngeal carcinoma (NPC) tissues and significantly correlated with poor patient survival. (A) Heat map and (B) Volcano map used to show genes that have significantly differentially expressed genes in NPC and normal tissues. (C) The Box plot shows the expression levels of KIF15 in NPC tissues and normal tissue (log2 FC = 1.6370, p < 0.001). (D) Expression levels of KIF15 in NPC tissues and para-carcinoma tissues were detected by immunohistochemical staining, Scale bars: 50 μm. (E) Kaplan–Meier survival curves of overall survival (OS, p < 0.05). (F) Kaplan–Meier survival curves of progression-free survival (PFS, p < 0.05). *p < 0.05.
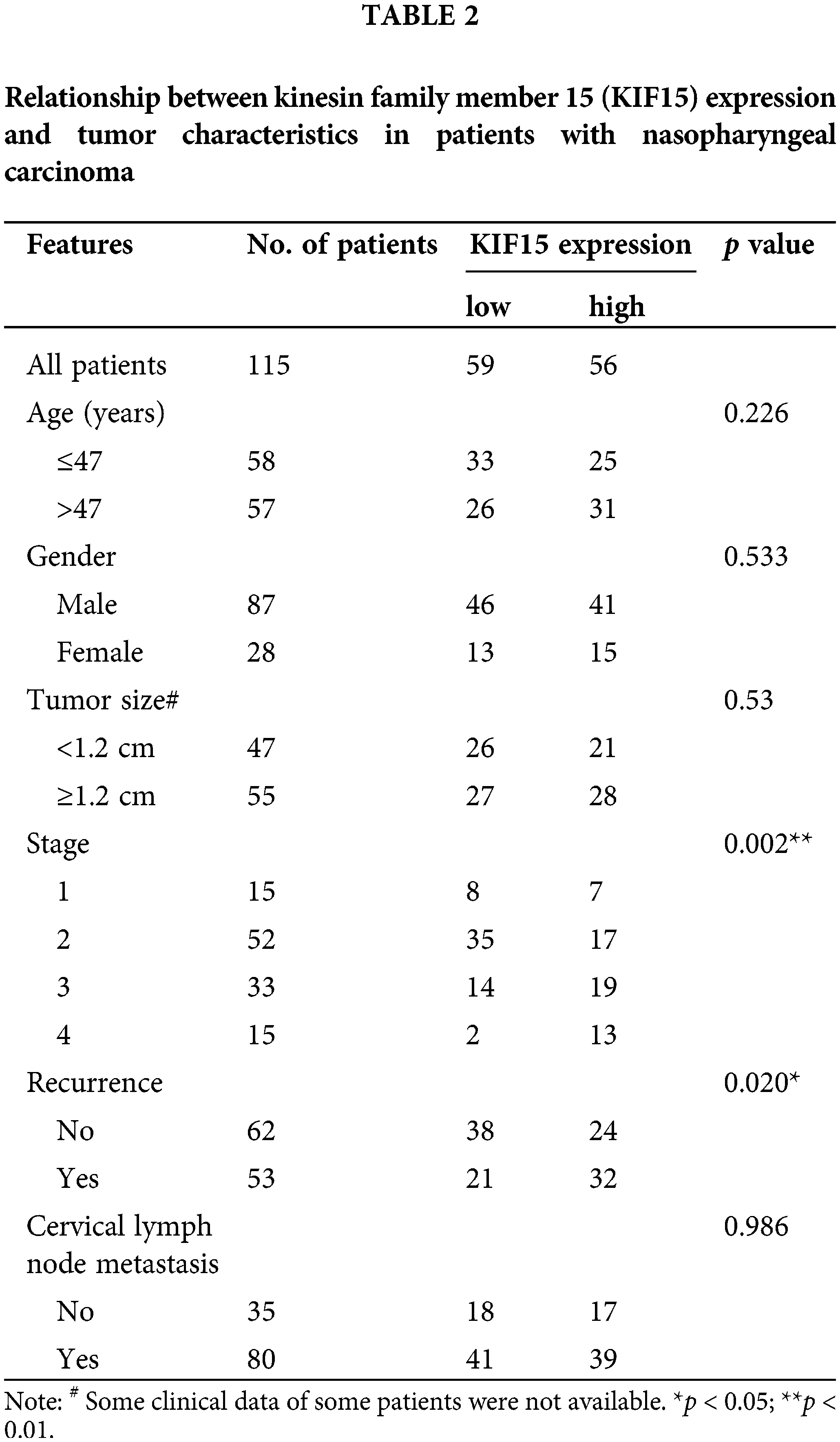
In addition, the protein expression of KIF15 was related to the clinical stage and recurrence but not to age, gender, tumor size, and lymph node metastasis (Table 2). The Spearman rank correlation analysis showed a significant positive correlation between the expression of KIF15 and clinical staging (p = 0.004, two-tailed). Moreover, according to the Kaplan-Meier survival analysis, the expression of KIF15 in NPC was significantly related to OS (Fig. 1E) and progression-free survival (Fig. 1F). These results suggest that high KIF15 expression levels could be used as a poor prognostic indicator for NPC patients.
Construction of kinesin family member 15 knockdown cell models
The qRT-PCR revealed a significantly lower lever of KIF15 in the C666-1 line when compared with the other three cell lines (CNE-2Z, 5-8F, HONE-1). Moreover, the shKIF15-1 RNAi sequence was identified as the most efficient sequence for the KIF15 knockdown (Fig. 2A). The 5-8F and CNE-2Z cell lines were selected for the construction of the KIF15-knockdown cell models. The cells transfected with the corresponding empty vector were used as the negative control (shCtrl). Over 80% transduction efficiency was observed in both the 5-8F and CNE-2Z cells after detecting the fluorescence of the green fluorescent protein (GFP) on the lentivirus vector (Fig. 2B). The qPCR and western blotting analysis confirmed the knockdown of the KIF15 gene in the shKIF15 group (Fig. 2C). These findings indicate that the shKIF15 and shCtrl cell models were successfully constructed for the subsequent studies.
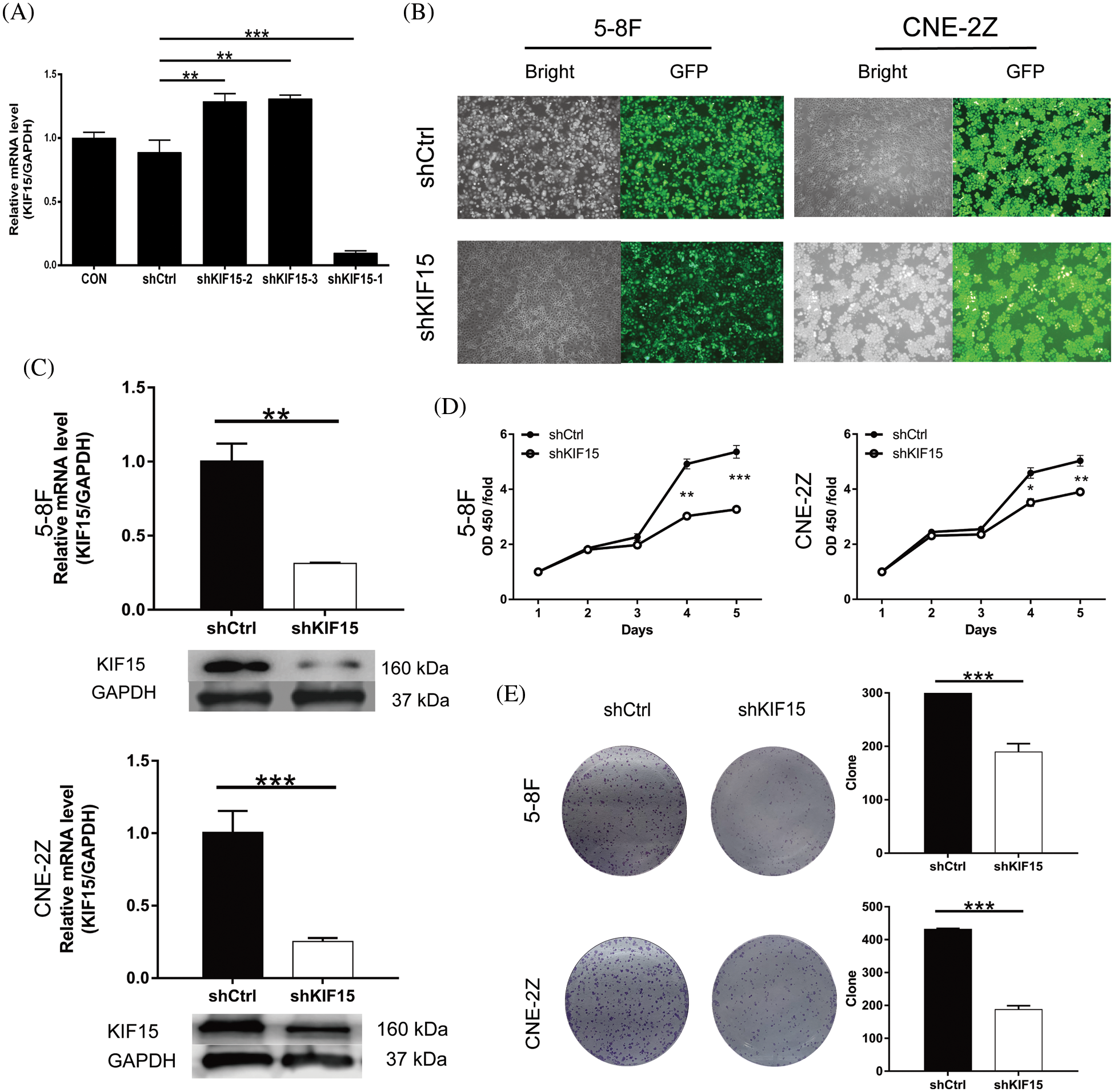
Figure 2: Kinesin family member 15 (KIF15) knockdown inhibits the proliferation of nasopharyngeal carcinoma (NPC) and clone formation in vitro. (A) Reverse transcription and quantitative real-time PCR (qRT-PCR) to evaluate the knockdown efficiency of KIF15 by shRNAs (shKIF15-1, shKIF15-2, and shKIF15-3). (B) Microscopy images illustrating the fluorescence levels used to confirm the shCtrl infection in the 5-8F and CNE2Z cells. (C) The knockdown efficiency of KIF15 in NPC cells was detected by qRT-PCR (upper, p < 0.01 in 5-8F, p < 0.001 in CNE2Z) and western blotting analysis. (D) CCK8 assays were conducted to evaluate the proliferation rate of the 5-8F and CNE-2Z cells with or without KIF15 knockdown (p < 0.001 in 5-8F cells, p < 0.01 in CNE2Z). (E) Colony formation assay illustrating a significant reduction in the cell colony number in the shKIF15 group compared with the shCtrl group, in the 5-8F and the CNE-2Z cells (p < 0.001). *p < 0.05, **p < 0.01, ***p < 0.001.
The biological behavior of NPC cells following the KIF15 knockdown
Compared with the shCtrl group, the cell proliferation of the shKIF15 group was significantly inhibited in the 5-8F and CNE-2Z cell lines (Fig. 2D). The results of the clone formation assay showed that after the KIF15 knockdown, the number of clones in the shKIF15 group was significantly reduced compared with that of the shCtrl group for both the 5-8F and CNE-2Z (p < 0.001, Fig. 2E) cell lines. These findings indicate that the expression of KIF15 in NPC cells may promote tumor cell proliferation and clone formation.
Impact of downregulating the expression of kinesin family member 15 on apoptosis and cell cycle in vitro
In terms of cell apoptosis, flow cytometry indicated that the 5-8F and CNE2Z cells with the KIF15 gene being silenced exhibited a significantly increased apoptotic rate compared with the control group (Figs. 3A and 3B, p < 0.01). After the knockdown of the KIF15 gene, the western blot analysis indicated significantly enhanced expression of BCL-2 and the cleaved caspase-3 proteins in both cell lines (Fig. 3C), but there was no change in the cleaved Caspase-9 protein level. The cell cycle analysis revealed a higher percentage of cells in the G2 phase (Fig. 3D, p < 0.05). These findings suggest that the KIF15 tends to function in late mitosis.
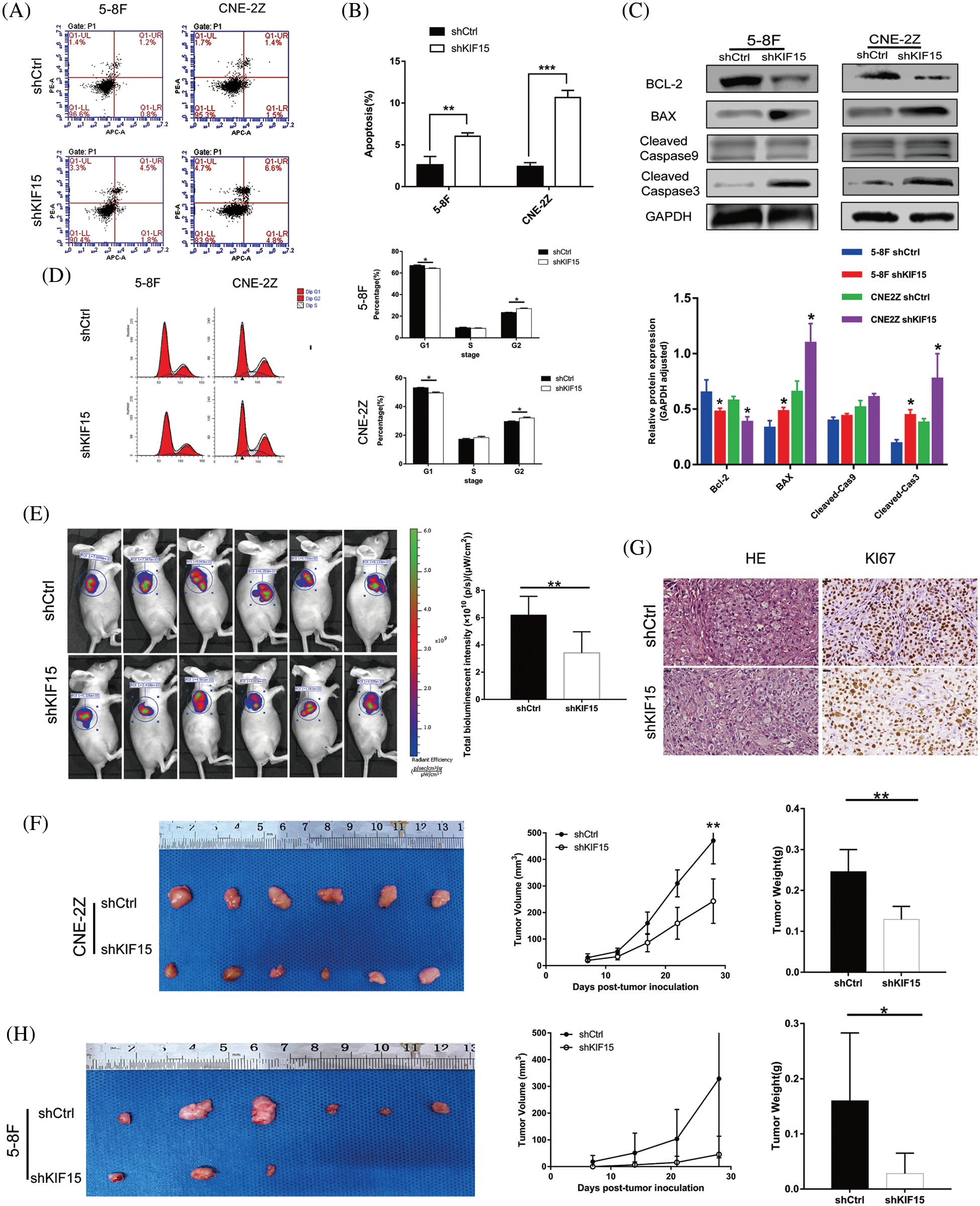
Figure 3: Knockdown of kinesin family member 15 (KIF15) induces in vitro cell apoptosis, affects the cell cycle in vitro, and represses nasopharyngeal carcinoma (NPC) tumor progression in vivo. (A and B) Flow cytometry indicated that 5-8F and CNE-2Z with KIF15 silencing exhibited significantly increased apoptotic rate compared with the control group (p < 0.01); (C) After the knockdown of the KIF15 gene, the western blotting analysis indicated that the B cell lymphoma-2 (BCL-2) protein was down-regulated, while the BCL2 associated X (BAX) and caspase-3 were significantly upregulated in both cell lines. (D) Cell cycle analysis evaluating the percentage of cells in the G1, S, and G2 phases indicates a cell cycle arrest in the G2 phase. (E) A significant reduction in the fluorescence intensity was noted in the shKIF15 group after injecting the CNE-2Z cells (p < 0.01). (F) Exogenous tumor volume and weight were assessed, and the results showed that the tumor growth rate was slowed down by KIF15 knockdown after injecting CNE-2Z cells (p < 0.01). (G) The sections of xenografts were processed for hematoxylin and eosin, and Ki-67 staining in the CNE-2Z group. Ki-67 staining was reduced in the shKIF15 group compared with the shCtrl group, Scale bars: 20 μm. (H) Images of the tumors extracted from the mice. All mice in the shCtrl group formed tumors under the skin after injecting 5-8F cells, while only three mice in the shKIF15 group formed tumors. The mean tumor weight was significantly higher in the shCtrl group when compared with the shKIF15 group (p < 0.05). *p < 0.05, **p < 0.01, ***p < 0.001.
Tumor progression in vivo following the suppression of the KIF15 gene
The mice formed tumors under the skin 4 weeks after injecting them with the 5-8F and CNE-2Z cells treated with shKIF15 or shCtrl. The evaluation of the fluorescence intensity (Fig. 3E), tumor volume, and tumor weight (Fig. 3F) confirmed that the suppression of KIF15 expression could inhibit the development of NPC in mice. In the shKIF15 xenograft models of 5-8F cells, only 3 in 6 nude mice formed tumors under the skin, while all mice in the shCtrl group formed tumors (Fig. 3H). The IHC analysis of the xenograft mice sections showed a higher expression of the Ki-67 biomarker for tumor growth in the shCtrl group compared to the shKIF15 group (Fig. 3G, p < 0.01). These findings confirmed that the knockdown of the KIF15 gene could inhibit NPC development in vivo.
Impact of the KIF15 gene knockdown in regulating the migration and invasion abilities of nasopharyngeal carcinoma cells in vitro and in vivo
In the wound healing assay, compared with the shCtrl group, the cell migration rate of the shKIF15 group of 5-8F cells was reduced by 29% after 48 h from the start (Fig. 4A, p < 0.001), while it was reduced by 8% in CNE2Z cells (Fig. 4A, p < 0.05). The transwell assay showed a higher reduction in the migration and invasion rate of the shKIF15 group when compared with the shCtrl group (Fig. 4B, p < 0.05).
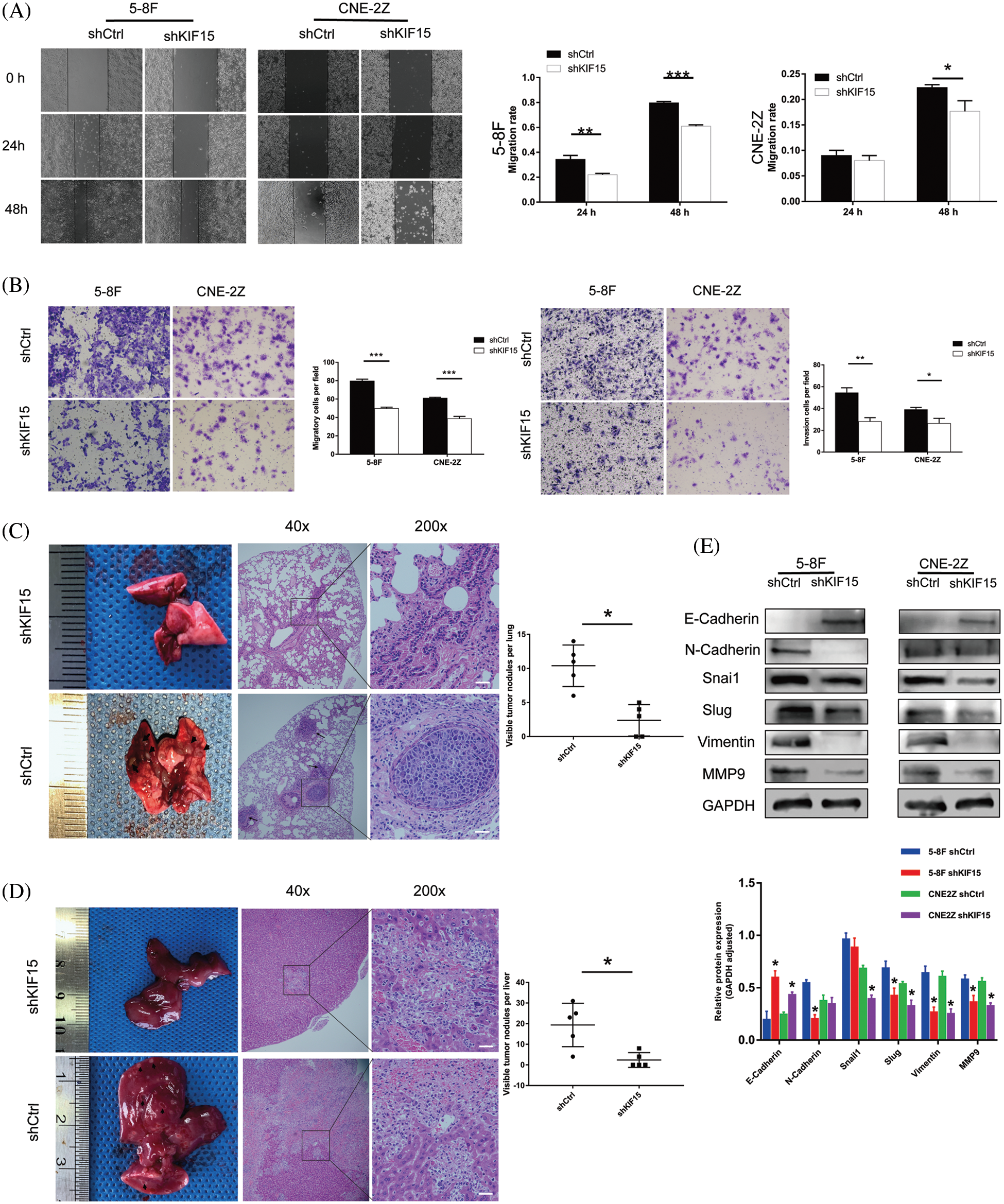
Figure 4: Kinesin family member 15 (KIF15) silencing repressed tumor migration, invasion, and distant metastasis ability in vitro and in vivo. (A) The wound-healing assay was conducted on the short hairpin (sh)KIF15 and shCtrl transfected cells to evaluate their migratory ability. In the 5-8F cells, the migration rate of cells in the shKIF15 group decreased by 24% after 48 h (p < 0.001), while in the CNE-2Z cells, the migration rate decreased by 13% (p < 0.05). (B) A transwell assay was conducted on the transfected cells to evaluate their migration and invasion ability. The migration rate of cells in the shKIF15 group decreased in the 5-8F and CNE-2Z cells (p < 0.001). The same result was obtained in the invasion experiment (p < 0.05). (C) In the tail vein injection metastasis model, all nude mice in the shCtrl group developed metastatic lesions (black arrows) in the lungs within six weeks after the injection of 5-8F cells. However, only some of the mice in the shKIF15 group developed lung metastases. Scale bars: 50 μm (p < 0.05). (D) Images illustrating the development of metastatic liver lesions in some mice. Compared with the shKIF15 group, the shCtrl group had a significantly larger number (p < 0.05) of lesions, ulcers, and cavitations (shown by the black arrow). These lesions had an invasive and necrotic appearance under the microscope. Scale bars: 50 μm. (E) Western blotting was performed to detect the expression of EMT-related proteins in the 5-8F and CNE-2Z cells. *p < 0.05, **p < 0.01, ***p < 0.001.
The in vivo metastasis evaluation of the nude mice in the shCtrl group showed visible nodules on the lung surface of all mice (shown by the black arrow in Fig. 4C, p < 0.05). H&E staining of the lung tissue confirmed that the surface nodules of the lung were metastatic NPCs. Under the microscope, the metastatic nodules in the lungs of the control group increased significantly, and a tumor thrombus in a blood vessel was also observed (lower part of Fig. 4C). At the same time, in the KIF15 knockdown group of the nude mice, the number of nodules on the liver surface was significantly reduced compared to the control group (shown by the black arrow in Fig. 4D, p < 0.05). H&E staining of the liver tissue confirmed that, in the shCtrl group, there were larger areas of necrosis in the liver tissue (lower part of Fig. 4D). These results indicate that under in vivo physiological conditions, KIF15 can promote the invasion and metastasis of NPC cells. At the same time, the suppression of KIF15 expression can inhibit the metastasis of NPC cells in vivo.
The western blotting analysis of the epithelial-mesenchymal transition (EMT) proteins showed that in the 5-8F cell line, N-cadherin was down-regulated, and E-cadherin was upregulated after knocking down the KIF15 gene (Fig. 4E). However, no similar changes were found in the CNE-2Z cell line. Concurrently, the transcription factors Snai2/Slug and vimentin related to metastasis were all down-regulated after knocking down the KIF15 gene. The metal matrix protein 9 (MMP9) was also down-regulated in both cell lines. These findings indicated that the expression of KIF15 in NPC may promote the epithelial matrix transition involved in the development of distant metastasis.
Mechanistic study of the KIF15 gene within the mitogen-activated protein kinase/P53 pathway
The western blotting tests used to identify the differentially expressed proteins in the shCtrl and shKIF15 cells revealed significant upregulation of the p-ERK and p-38 expression in the KIF15 knockdown group (p < 0.05, Fig. 5A). These results suggested that KIF15 could inhibit apoptosis of the NPC cells by regulating the p-ERK/p38 expression within the MAPK pathway. Although a reduction in the AKT protein expression level only occurred in the 5-8F cells after KIF15 knockdown; its phosphorylation level was not significantly altered. At the same time, there was no corresponding change in the PI3K and ERK protein expression (p > 0.05). These findings suggest the KIF15 knockdown upregulated the p-ERK, p38, and p-P53 proteins, which in turn activated the MAPK/P53 signal pathway.
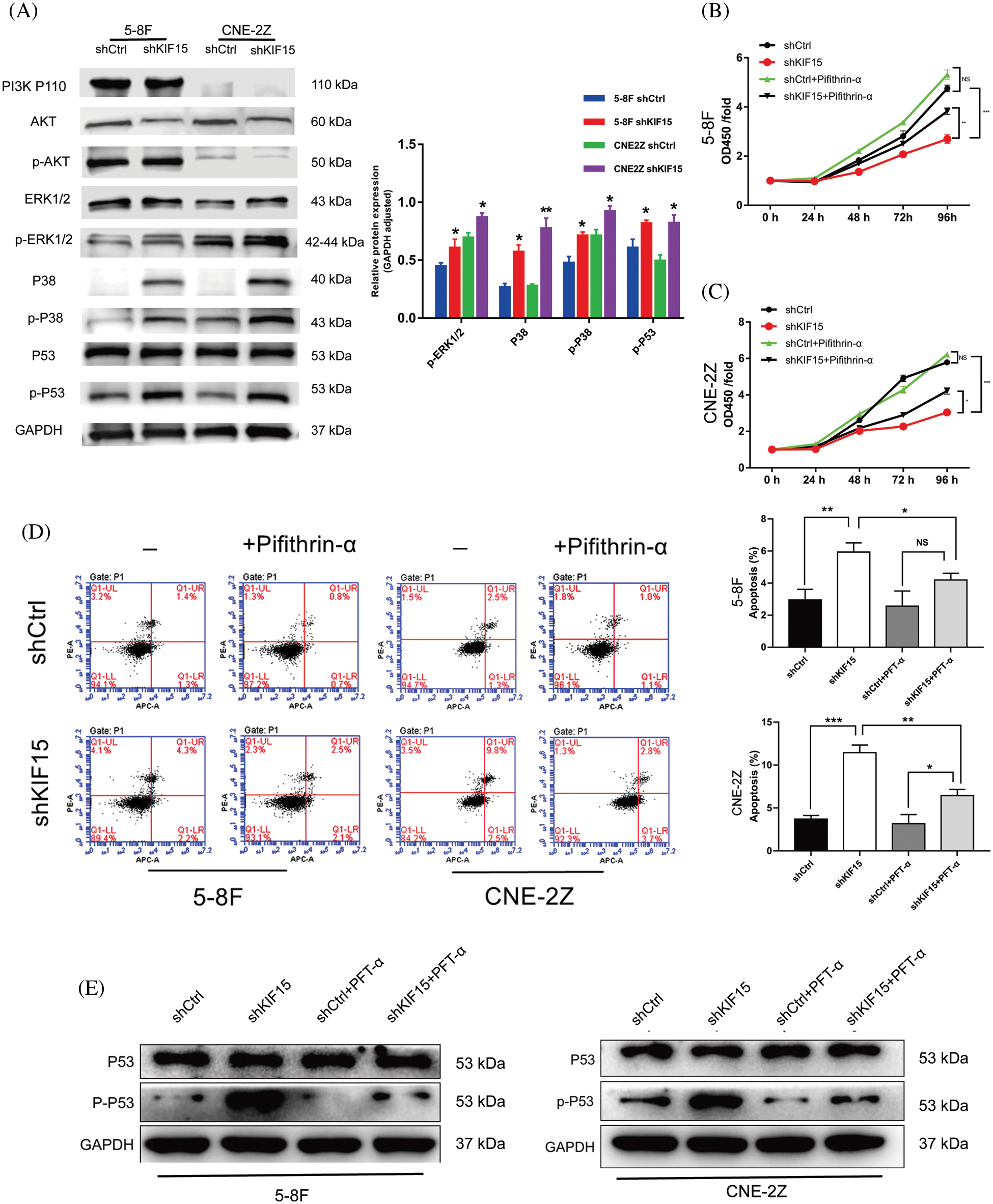
Figure 5: Kinesin family member 15 (KIF15) may deregulate the mitogen-activated protein kinase (MAPK)/P53 signal pathway, thereby promoting the proliferation of nasopharyngeal carcinoma (NPC) cells while also preventing apoptosis. (A) The key proteins involved in the MAPK pathway were evaluated after KIF15 silencing in both cell lines, and the protein expression levels of p-ERK, P38, p-P38, and p-P53 were significantly upregulated (p < 0.05). (B) and (C) illustrate the results of the rescue tests whereby the proliferating cells in both cell lines increased after adding the P53 inhibitor PFT-α (p < 0.05). (D) Flow cytometry analysis illustrating the apoptosis levels of NPC cells after adding PFT-α. (F) After adding PFT-α, the western blotting experiment showed that the phosphorylation level of p53 was significantly inhibited. *p < 0.05, **p < 0.01, ***p < 0.001.
Impact of exogenous P53 inhibitors on cell proliferation and apoptosis
After adding the P53 inhibitor PFT-α, the cell proliferation increased in both the 5-8F and CNE-2Z cells (Figs. 5B and 5C, p < 0.05). The results of the flow cytometry analysis showed that after adding PFT-α, the apoptosis levels of the NPC cells in the shKIF15 group were downregulated compared to that observed before (Fig. 5D). After adding PFT-α, the western blotting experiment showed that the phosphorylation level of p53 was significantly inhibited (Fig. 5E), potentially preventing the P53-mediated apoptosis induced by the release of cytotoxic substances.
The KIF proteins are composed of two heavy chains and two light chains. Their heads have a highly conserved motion domain, which can hydrolyze ATP and provide chemical energy for their mechanical movement along microtubules. In contrast, the tails are combined with a specific cargo through a light chain. The cargo may include vesicles, organelles, or macromolecules (Boleti et al., 1996). More importantly, KIFs can also participate in chromosome congression, alignment, and cytokinesis (Wittmann et al., 2000; Vanneste et al., 2011). They are strictly regulated through temporal synthesis and only present when needed (Gruneberg et al., 2004; Vanneste et al., 2009). However, the dysregulation of KIF proteins may lead to uncontrolled cell growth. As a result, several studies have shown that these proteins are involved in the development of several diseases (Sleiman et al., 2017), including human cancers (Lucanus and Yip, 2018; Li et al., 2020). Therefore, there is a need to understand the functions of kinesin proteins as this may contribute to developing targeted cancer therapies.
In the present study, nasopharyngeal cancer cells were found to have high KIF15 mRNA expression levels. The protein expression of KIF15 in NPC tissues was significantly higher than that in the para-carcinoma tissues. The KIF15 expression was also significantly correlated with cancer grade, recurrence rate, and poor prognosis. A study by Mi et al. (2022) also showed that the expression level of KIF15 correlated with OS in NPC patients. Similar results have also been reported in hepatocellular carcinoma (Kitagawa et al., 2020), lung cancer (Qiao et al., 2018), breast cancer (Zeng et al., 2020), and other tumors (Ding et al., 2020; Wang et al., 2020). These studies confirm that high expression of KIF15 can worsen the prognosis of cancer patients. Based on the clinical findings described above, it was hypothesized that overexpression of KIF15 in NPC may also promote cancer progression. Therefore, 5-8F and CNE-2Z cell lines were selected to construct KIF15 knockdown cell models for the loss-of-function study. The results revealed that the silencing of the KIF15 inhibited cell proliferation, induced cell apoptosis, and caused cell cycle arrest at the G2 phase. As expected, both the in vitro colony formation assay and in vivo xenograft assays showed that the tumor formation ability of the shKIF15 cells was suppressed, which further led to slower xenograft growth.
Invasion and metastasis are important markers of tumor malignancy (Ridley et al., 2003; Elgundi et al., 2019) and are also important causes of treatment failure in NPC (Mao et al., 2016). Increasingly, members of the kinesin family are closely associated with cell invasion and metastasis. The KIF18A expression was associated with worse clinical staging and poorer prognosis in prostate cancer patients, and interference with the KIF18A expression significantly inhibited the proliferation and metastasis of PC-3 cell lines (Zhang et al., 2019a). Li et al. (2020) found that the downregulation of the KIF15 gene in vitro and in hepatocellular carcinoma organoids resulted in a significant reduction in the expression of stemness-related genes. Furthermore, the KIF15 downregulation in human HCC xenograft models delayed tumor initiation, growth, and metastasis. In another study, Wang et al. (2017) found that KIF15 promoted the ability of pancreatic cancer cells to metastasize to the liver and lung in nude mice. In addition, KIF15 also promoted the expression of MMP2 and MMP14, which contributed to the remodeling of the extracellular matrix and the promotion of metastasis of pancreatic cancer cells. In the present study, the knockdown of the KIF15 gene could significantly inhibit the motility, mobility, and invasion of NPC cells, which are necessary for tumor progression and metastasis. This finding explains why patients with high expression levels of KIF15 have a higher risk of recurrence from another perspective.
The in vivo experiments performed in our study also showed that KIF15 could promote distant metastasis of NPC cells to the liver and lungs. Available evidence suggests that KIF15 is involved in the EMT of NPC. MMP9 promotes the hydrolysis of the intercellular adhesion protein E-cadherin, while the transcription factor Snai2 represses E-cadherin protein expression. As a result, E-cadherin is gradually replaced by N-cadherin, which provides greater linkage flexibility. In addition, vimentin contributes to EMT by altering cell shape. These changes eventually lead to cell separation and enhanced cell motility.
Apoptosis is a process that occurs in multicellular when a cell intentionally “decides” to die (Elmore, 2007). Once the apoptosis program is started, various apoptosis-related proteins in the cell gather together and then enter the continuous reaction process. Previous studies have confirmed that KIFs also play an important role in apoptosis (Castillo et al., 2007; Li et al., 2020; Terribas et al., 2020). Therefore, in this study, we decided to analyze the molecular mechanism leading to changes in the apoptosis rate in NPC cells after the knockdown of the KIF15 gene. This research provided biological evidence that the silencing of the KIF15 gene induced cell apoptosis in NPC. In this process, the protein Bcl-2 was significantly down-regulated, while the expression of BAX and caspase-3 were enhanced. Subsequent results of the western blotting analysis confirmed that the MAPK signaling pathways, particularly the ERK/P38 MAPK pathway, may be involved in the development and promotion of NPCs with high KIF15 expression. The rescue experiments with p53, a downstream gene of p38, confirmed that the addition of a p53 inhibitor counteracted the pro-apoptotic effect of KIF15. Extensive research in apoptosis suggests that c-Jun N-terminal kinases and p38 MAPKs play key roles in integrating signals via transcription-dependent and transcription-independent mechanisms (Wagner and Nebreda, 2009). This eventually leads to the activation of caspases. Prior studies have also found that several Bcl-2 family proteins, in both pro- and anti-apoptotic groups, are under the control of p38 MAPK cascades (Green and Llambi, 2015). Based on the above findings, it was reasonable to speculate that KIF15 could promote the progression of NPC via the activation of the MAPK signaling pathways that inhibit apoptosis. However, the role of KIF15 in regulating apoptosis in NPC is still unclear, and further studies will be required.
Induction and adjuvant chemotherapy are essential parts of the treatment in NPC patients (Lee et al., 2017; Zhang et al., 2019b). Docetaxel and paclitaxel are anti-microtubule taxanes that are often used in the first- or second-line treatment of NPCs. Unfortunately, resistance to chemotherapy remains a critical medical challenge, and many KIFs are associated with resistance to docetaxel and paclitaxel (Ganguly et al., 2011; Singel et al., 2013; Khongkow et al., 2016; Lee et al., 2020). Therefore, drugs targeting the KIFs may improve the therapeutic ratio by inhibiting cell proliferation and infiltration while reducing drug resistance. One promising target is KIF11 (Eg5), a kinesin that plays a key role in spindle formation by generating forces that separate the two poles. However, in a phase I trial, Eg5 inhibitors were found to be less effective than expected as cancer cells developed resistance to them by escalating the expression of KIF15 as an alternative to Eg5 (Gomez et al., 2012; Groen, 2013; Dumas et al., 2016). Therefore, the drugs that target KIF15 could be used in combination with Eg5 inhibitors (Hancock, 2014; Sebastian, 2017) for treating NPC in the future. However, further research is required for the development and clinical implementation of drugs targeting the KIF proteins.
This work demonstrated that KIF15 overexpression accelerated the progression of NPCs and promoted the development of distant metastases. Therefore the expression of KIF15 could be used as a new prognostic indicator and a potential therapeutic target for the treatment of NPC.
Acknowledgement: We are very grateful to Professor Sai-Wah Tsao from the University of Hong Kong for generously presenting the NPC cell line C666-1 and Professor Musheng Zeng from Sun Yat-sen University for kindly providing 5-8F.
Availability of Data and Materials: The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Author Contribution: The authors confirm their contribution to the paper as follows: study conception and design: Guangwu Huang; data collection: Yongli Wang, Shenhong Qu; analysis and interpretation of results: Yong Yang, Ying Qin, Fei Liu; draft manuscript preparation: Yongli Wang, Shenhong Qu. All authors reviewed the results and approved the final version of the manuscript.
Ethics Approval: The tissue microarray protocol was approved by the Human Ethics Committee of the Taizhou Hospital of Zhejiang province. Date of adoption: January 26, 2010; Certificate No. 2010.2.8. All patients signed an informed consent approved by the Institutional Review Board.
The animal experimental protocols were approved by the Ethics Committee of the People’s Hospital of Guangxi Zhuang Autonomous Region; Date of adoption: August 21, 2018; Certificate No. Research-2018-47.
Funding Statement: This work was supported by Guangxi Key Research and Development Plan (GuiKe-AB18050011); the Grant of Guangxi Science and Technology Base and Talent Project (GuiKe-AD20297069).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
Boleti H, Karsenti E, Vernos I (1996). Xklp2, a novel Xenopus centrosomal kinesin-like protein required for centrosome separation during mitosis. Cell 84: 49–59. DOI 10.1016/S0092-8674(00)80992-7. [Google Scholar] [CrossRef]
Castillo A, Morse HC, Godfrey VL, Naeem R, Justice MJ (2007). Overexpression of Eg5 causes genomic instability and tumor formation in mice. Cancer Research 67: 10138–10147. DOI 10.1158/0008-5472.CAN-07-0326. [Google Scholar] [CrossRef]
Chen L, Hu CS, Chen XZ, Hu GQ, Cheng ZB et al. (2012). Concurrent chemoradiotherapy plus adjuvant chemotherapy versus concurrent chemoradiotherapy alone in patients with locoregionally advanced nasopharyngeal carcinoma: A phase 3 multicentre randomised controlled trial. The Lancet. Oncology 13: 163–171. DOI 10.1016/S1470-2045(11)70320-5. [Google Scholar] [CrossRef]
Ding L, Li B, Yu X, Li Z, Li X et al. (2020). KIF15 facilitates gastric cancer via enhancing proliferation, inhibiting apoptosis, and predict poor prognosis. Cancer Cell International 20: 125. DOI 10.1186/s12935-020-01199-7. [Google Scholar] [CrossRef]
Dumas ME, Sturgill EG, Ohi R (2016). Resistance is not futile: Surviving Eg5 inhibition. Cell Cycle 15: 2845–2847. DOI 10.1080/15384101.2016.1204864. [Google Scholar] [CrossRef]
Elgundi Z, Papanicolaou M, Major G, Cox TR, Melrose J et al. (2020). ZNF367-induced transcriptional activation of KIF15 accelerates the progression of breast cancer. International Journal of Biological Sciences 16: 2084–2093. DOI 10.7150/ijbs.44204. [Google Scholar] [CrossRef]
Elmore S (2007). Apoptosis: A review of programmed cell death. Toxicologic Pathology 35: 495–516. DOI 10.1080/01926230701320337. [Google Scholar] [CrossRef]
Ganguly A, Yang H, Cabral F (2011). Overexpression of mitotic centromere-associated Kinesin stimulates microtubule detachment and confers resistance to paclitaxel. Molecular Cancer Therapeutics 10: 929–937. DOI 10.1158/1535-7163.MCT-10-1109. [Google Scholar] [CrossRef]
Gao X, Zhu L, Lu X, Wang Y, Li R et al. (2020). KIF15 contributes to cell proliferation and migration in breast cancer. Human Cell 33: 1218–1228. DOI 10.1007/s13577-020-00392-0. [Google Scholar] [CrossRef]
Gomez HL, Philco M, Pimentel P, Kiyan M, Monsalvo ML et al. (2012). Phase I dose-escalation and pharmacokinetic study of ispinesib, a kinesin spindle protein inhibitor, administered on days 1 and 15 of a 28-day schedule in patients with no prior treatment for advanced breast cancer. Anti-Cancer Drugs 23: 335–341. DOI 10.1097/CAD.0b013e32834e74d6. [Google Scholar] [CrossRef]
Green DR, Llambi F (2015). Cell death signaling. Cold Spring Harbor perspectives in Biology 7: a006080. DOI 10.1101/cshperspect.a006080. [Google Scholar] [CrossRef]
Groen A (2013). Microtubule motors: A new hope for kinesin-5 inhibitors? Current Biology 23: R617–618. DOI 10.1016/j.cub.2013.05.049. [Google Scholar] [CrossRef]
Gruneberg U, Neef R, Honda R, Nigg EA, Barr FA (2004). Relocation of Aurora B from centromeres to the central spindle at the metaphase to anaphase transition requires MKlp2. The Journal of Cell Biology 166: 167–172. DOI 10.1083/jcb.200403084. [Google Scholar] [CrossRef]
Hancock WO (2014). Mitotic kinesins: A reason to delve into kinesin-12. Current Biology 24: R968–970. DOI 10.1016/j.cub.2014.09.011. [Google Scholar] [CrossRef]
Khongkow P, Gomes AR, Gong C, Man EP, Tsang JW et al. (2016). Paclitaxel targets FOXM1 to regulate KIF20A in mitotic catastrophe and breast cancer paclitaxel resistance. Oncogene 35: 990–1002. DOI 10.1038/onc.2015.152. [Google Scholar] [CrossRef]
Kitagawa A, Masuda T, Takahashi J, Tobo T, Noda M et al. (2020). KIF15 expression in tumor-associated monocytes is a prognostic biomarker in hepatocellular carcinoma. Cancer Genomics Proteomics 17: 141–149. DOI 10.21873/cgp.20174. [Google Scholar] [CrossRef]
Lee AWM, Tung SY, Ng WT, Lee V, Ngan RKC et al. (2017). A multicenter, phase 3, randomized trial of concurrent chemoradiotherapy plus adjuvant chemotherapy versus radiotherapy alone in patients with regionally advanced nasopharyngeal carcinoma: 10-year outcomes for efficacy and toxicity. Cancer 123: 4147–4157. DOI 10.1002/cncr.30850. [Google Scholar] [CrossRef]
Lee J, Cho YJ, Lee JW, Ahn HJ (2020). KSP siRNA/paclitaxel-loaded PEGylated cationic liposomes for overcoming resistance to KSP inhibitors: Synergistic antitumor effects in drug-resistant ovarian cancer. Journal of Controlled Release 321: 184–197. DOI 10.1016/j.jconrel.2020.02.013. [Google Scholar] [CrossRef]
Li Q, Qiu J, Yang H, Sun G, Hu Y et al. (2020). Kinesin family member 15 promotes cancer stem cell phenotype and malignancy via reactive oxygen species imbalance in hepatocellular carcinoma. Cancer Letters 482: 112–125. DOI 10.1016/j.canlet.2019.11.008. [Google Scholar] [CrossRef]
Logan CM, Menko AS (2019). Microtubules: Evolving roles and critical cellular interactions. Experimental Biology and Medicine 244: 1240–1254. DOI 10.1177/1535370219867296. [Google Scholar] [CrossRef]
Lucanus AJ, Yip GW (2018). Kinesin superfamily: roles in breast cancer, patient prognosis and therapeutics. Oncogene 37: 833–838. DOI 10.1038/onc.2017.406. [Google Scholar] [CrossRef]
Mao YP, Tang LL, Chen L, Sun Y, Qi ZY et al. (2016). Prognostic factors and failure patterns in non-metastatic nasopharyngeal carcinoma after intensity-modulated radiotherapy. Chinese Journal of Cancer 35: 103. DOI 10.1186/s40880-016-0167-2. [Google Scholar] [CrossRef]
Mi J, Ma S, Chen W, Kang M, Xu M et al. (2022). Integrative pan-cancer analysis of KIF15 reveals its diagnosis and prognosis value in nasopharyngeal carcinoma. Frontiers in Oncology 12: 772816. DOI 10.3389/fonc.2022.772816. [Google Scholar] [CrossRef]
Matsushita J, Suzuki T, Okamura K, Ichihara G, Nohara K (2020). Identification by TCGA database search of five genes that are aberrantly expressed and involved in hepatocellular carcinoma potentially via DNA methylation changes. Environmental Health and Preventive Medicine 25: 31. DOI 10.1186/s12199-020-00871-8. [Google Scholar] [CrossRef]
Ong MS, Deng S, Halim CE, Cai W, Tan TZ et al. (2020). Cytoskeletal proteins in cancer and intracellular stress: A therapeutic perspective. Cancers 12: 238. DOI 10.3390/cancers12010238. [Google Scholar] [CrossRef]
Qiao Y, Chen J, Ma C, Liu Y, Li P et al. (2018). Increased KIF15 expression predicts a poor prognosis in patients with lung adenocarcinoma. Cellular Physiology and Biochemistry 51: 1–10. DOI 10.1159/000495155. [Google Scholar] [CrossRef]
Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH et al. (2003). Cell migration: Integrating signals from front to back. Science 302: 1704–1709. DOI 10.1126/science.1092053. [Google Scholar] [CrossRef]
Sebastian J (2017). Dihydropyrazole and dihydropyrrole structures based design of Kif15 inhibitors as novel therapeutic agents for cancer. Computational Biology and Chemistry 68: 164–174. DOI 10.1016/j.compbiolchem.2017.03.006. [Google Scholar] [CrossRef]
Singel SM, Cornelius C, Batten K, Fasciani G, Wright WE et al. (2013). A targeted RNAi screen of the breast cancer genome identifies KIF14 and TLN1 as genes that modulate docetaxel chemosensitivity in triple-negative breast cancer. Clinical Cancer Research 19: 2061–2070. DOI 10.1158/1078-0432.CCR-13-0082. [Google Scholar] [CrossRef]
Sleiman PMA, March M, Nguyen K, Tian L, Pellegrino R et al. (2017). Loss-of-function mutations in KIF15 underlying a braddock-carey genocopy. Human Mutation 38: 507–510. DOI 10.1002/humu.23188. [Google Scholar] [CrossRef]
Tao Q, Chan AT (2007). Nasopharyngeal carcinoma: Molecular pathogenesis and therapeutic developments. Expert Reviews in Molecular Medicine 9: 1–24. DOI 10.1017/S1462399407000312. [Google Scholar] [CrossRef]
Terribas E, Fernandez M, Mazuelas H, Fernandez-Rodriguez J, Biayna J et al. (2020). KIF11 and KIF15 mitotic kinesins are potential therapeutic vulnerabilities for malignant peripheral nerve sheath tumors. Neuro-Oncology Advances 2: i62–i74. DOI 10.1093/noajnl/vdz061. [Google Scholar] [CrossRef]
Vanneste D, Ferreira V, Vernos I (2011). Chromokinesins: Localization-dependent functions and regulation during cell division. Biochemical Society Transactions 39: 1154–1160. DOI 10.1042/BST0391154. [Google Scholar] [CrossRef]
Vanneste D, Takagi M, Imamoto N, Vernos I (2009). The role of Hklp2 in the stabilization and maintenance of spindle bipolarity. Current Biology 19: 1712–1717. DOI 10.1016/j.cub.2009.09.019. [Google Scholar] [CrossRef]
Wagner EF, Nebreda AR (2009). Signal integration by JNK and p38 MAPK pathways in cancer development. Nature Reviews Cancer 9: 537–549. DOI 10.1038/nrc2694. [Google Scholar] [CrossRef]
Wang J, Guo X, Xie C, Jiang J (2017). KIF15 promotes pancreatic cancer proliferation via the MEK-ERK signalling pathway. British Journal of Cancer 117: 245–255. DOI 10.1038/bjc.2017.165. [Google Scholar] [CrossRef]
Wang Q, Han B, Huang W, Qi C, Liu F (2020). Identification of KIF15 as a potential therapeutic target and prognostic factor for glioma. Oncology Reports 43: 1035–1044. DOI 10.3892/or.2020.7510. [Google Scholar] [CrossRef]
Wittmann T, Wilm M, Karsenti E, Vernos I (2000). TPX2, a novel xenopus MAP involved in spindle pole organization. The Journal of Cell Biology 149: 1405–1418. DOI 10.1083/jcb.149.7.1405. [Google Scholar] [CrossRef]
Zeng H, Li T, Zhai D, Bi J, Kuang X et al. (2020). ZNF367-induced transcriptional activation of KIF15 accelerates the progression of breast cancer. International Journal of Biological Sciences 16: 2084–2093. DOI 10.7150/ijbs.44204. [Google Scholar] [CrossRef]
Zhang H, Shen T, Zhang Z, Li Y, Pan Z (2019a). Expression of KIF18A is associated with increased tumor stage and cell proliferation in prostate cancer. Medical Science Monitor 25: 6418–6428. DOI 10.12659/MSM.917352. [Google Scholar] [CrossRef]
Zhang Y, Chen L, Hu GQ, Zhang N, Zhu XD et al. (2019b). Gemcitabine and Cisplatin induction chemotherapy in nasopharyngeal carcinoma. The New England Journal of Medicine 381: 1124–1135. DOI 10.1056/NEJMoa1905287. [Google Scholar] [CrossRef]
Zou JX, Duan Z, Wang J, Sokolov A, Xu J et al. (2014). Kinesin family deregulation coordinated by bromodomain protein ANCCA and histone methyltransferase MLL for breast cancer cell growth, survival, and tamoxifen resistance. Molecular Cancer Research 12: 539–549. DOI 10.1158/1541-7786.MCR-13-0459. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools