 Open Access
Open Access
ARTICLE
dbSCI: A manually curated database of SARS-CoV-2 inhibitors for COVID-19
Institute of Biomedical Informatics, School of Software, Bioinformatics Center, Henan Provincial Engineering Center for Tumor Molecular Medicine, School of Basic Medical Sciences, Academy for Advanced Interdisciplinary Studies, Henan University, Kaifeng, 475004, China
* Corresponding Author: XIANGQIAN GUO. Email:
(This article belongs to the Special Issue: Bioinformatics Study of Diseases)
BIOCELL 2023, 47(2), 367-371. https://doi.org/10.32604/biocell.2023.025310
Received 05 July 2022; Accepted 22 August 2022; Issue published 18 November 2022
Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the pathogen of the ongoing coronavirus disease 2019 (COVID-19) global pandemic. Here, by centralizing published cell-based experiments, clinical trials, and virtual drug screening data from the NCBI PubMed database, we developed a database of SARS-CoV-2 inhibitors for COVID-19, dbSCI, which includes 234 SARS-CoV-2 inhibitors collected from publications based on cell-based experiments, 81 drugs of COVID-19 in clinical trials and 1305 potential SARS-CoV-2 inhibitors from bioinformatics analyses. dbSCI provides four major functions: (1) search the drug target or its inhibitor for SARS-CoV-2, (2) browse target/inhibitor information collected from cell experiments, clinical trials, and virtual drug screenings, (3) download, and (4) submit data. Each entry in dbSCI contains 18 types of information, including inhibitor/drug name, targeting protein, mechanism of inhibition, experimental technique, experimental sample type, and reference information. In summary, dbSCI provides a relatively comprehensive, credible repository for inhibitors/drugs against SARS-CoV-2 and their potential targeting mechanisms and it will be valuable for further studies to control COVID-19.Keywords
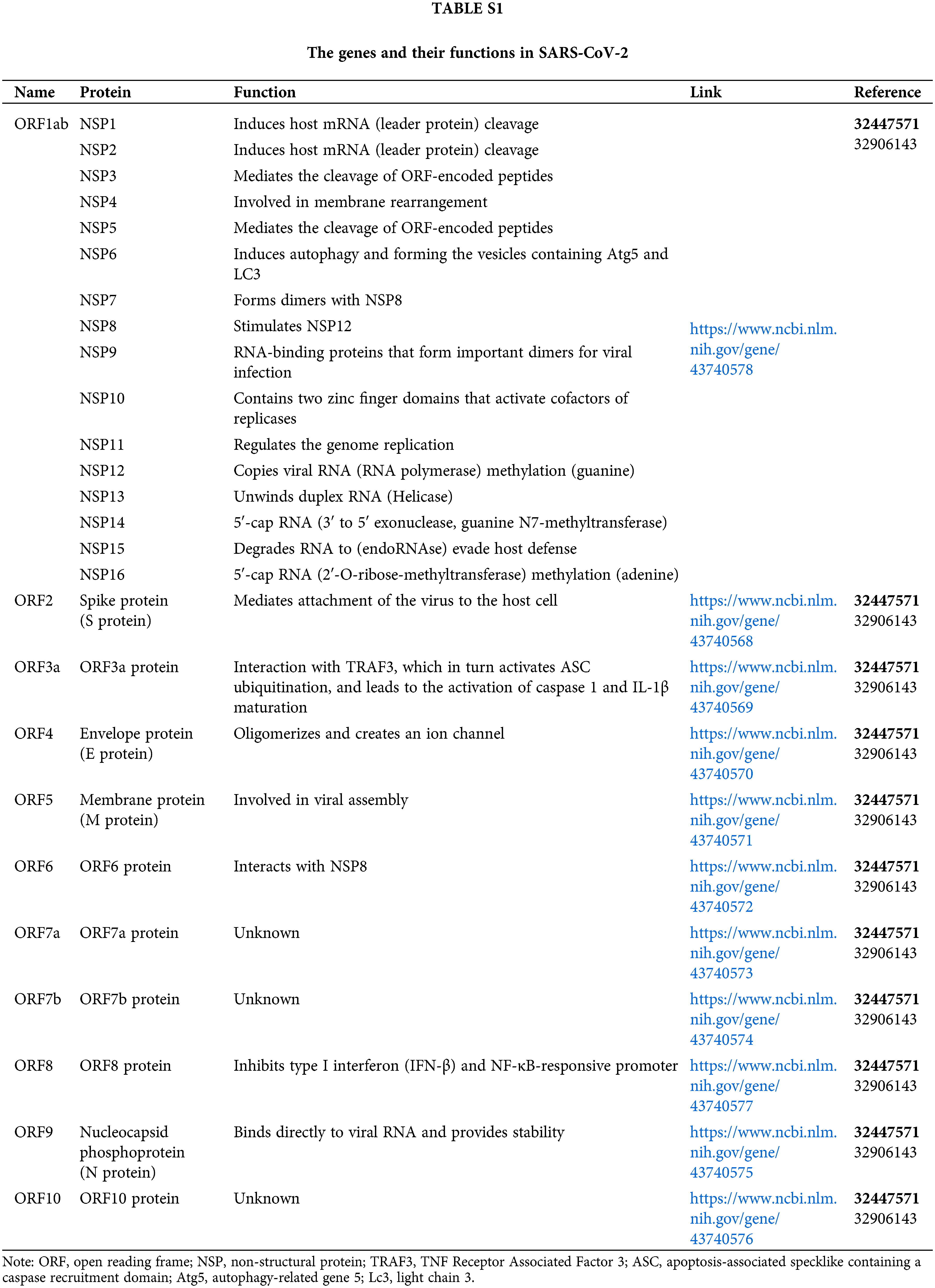
The coronavirus disease 2019 (COVID-19) pandemic is caused by a novel family member of coronaviruses named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). COVID-19 has been threatening global public health for more than two years (Feikin et al., 2022). Until now, there are 572,239,451 confirmed cases and 6,390,401 confirmed deaths worldwide, areas, or territories by 30 July 2022 (WHO, 2022). The morbidity rate of COVID-19 is still high because of the late entry of vaccines and the lack of specific potent drugs (Angrup et al., 2020). SARS-CoV-2 is a single-stranded RNA (ssRNA) virus, and its genome is 29903 nucleotides (nt) long (Bai et al., 2022). There are six functional open reading frames (ORFs) arranged from 5′ to 3′: ORF1a/ORF1b (replicase), ORF2 (spike protein, S), ORF4 (envelope protein, E), ORF5 (membrane protein, M), ORF9 (nucleocapsid protein, N) (Finkel et al., 2021). All genes and their functions in SARS-CoV-2 are mentioned in Suppl. Table S1 (Yoshimoto, 2020). The S1 subunit of S protein promotes angiotensin-converting enzyme-2 (ACE2)-mediated virus attachment while the S2 subunit enhances membrane fusion (Ziegler et al., 2020). Besides, the N protein is formed of a serine-rich linker region sandwiched between the N terminal domain (NTD) and C terminal domain (CTD), which plays a crucial role in viral entry and its post-entry processing (Peng et al., 2020). The E protein contains an NTD, a hydrophobic domain, and CTD and forms viroporins needed for viral assembly (Jackson et al., 2022). The M protein has a hydrophilic C terminal and amphipathic N terminal; its long form facilitates spike incorporation, and the interaction with E promotes virion production. Moreover, several putative ORFs encode accessory proteins (Lu et al., 2021).
Drug development requires strict and extensive in vitro and in vivo experiments as well as multiple clinical trials before clinical application (Ng, 2000). Generally, de novo drug design via structure-based drug repurposing by in silico simulations is a promising approach often used by researchers to identify possible drugs for viral diseases (Marston et al., 2014). Repurposing the approved drugs has been achieving promising results for sudden major health emergencies because the drugs with known modes of action, biotoxicity, and side effects can be more easily translated (Schein, 2020). Through the joint efforts of scientists all over the world to fight SARS-CoV-2, more and more data are now becoming available to allow researchers to explore new potential therapies. The rationale of drug design for COVID-19 is to target viral proteins to inhibit the biological functions of the virus (Lou et al., 2014), directly or indirectly (Martinez, 2020). The potential drug targets for SARS-CoV-2 include spike protein, NSP3 (papain-like proteinase), NSP5 (3C-like proteinase), NSP12 (RNA-dependent RNA polymerase), NSP15 (endoRNAse), etc. (Ou et al., 2020; Quimque et al., 2021). Antiviral compounds for SARS-CoV-2 have also been reported (Yoshimoto, 2020), such as alprostadil, brequinar, and cepharanthine. To accelerate the drug development of COVID-19, we constructed a DataBase for SARS-CoV-2 Inhibitors called dbSCI. In this database, we have collected and curated more than 2000 published potential inhibitors/drugs against COVID-19. This elaborate database specially designed for COVID-19 is expected to facilitate researchers in designing new drugs against SARS-CoV-2 infection.
To construct dbSCI, a total of 22,521 articles were identified upon searching publications up to 16 March 2022 using keywords “((COVID-19) or (SARS-CoV-2)) AND ((drug) OR (inhibitor))” in the PubMed database (https://www.ncbi.nlm.nih.gov/pubmed/). The eligibility criteria for researches were: (1) availability of full-text; (2) study reporting inhibitors or drugs for COVID-19 or SARS-CoV-2; (3) published papers. The exclusion criteria were as follows: (1) not written in English; (2) reviews, comments, case reports, letters, and meta-analyses; (3) studies about diseases other than COVID-19; (4) studies not on inhibitors or drugs. All evaluations were independently conducted by two individuals in parallel. Any conflicts were discussed by all authors to reach a consensus. Finally, the relative information about inhibitors for SARS-CoV-2 from in vitro/in vivo studies, clinical trials, or computational simulation studies from the publications were collected.
Finally, 236 SARS-CoV-2 inhibitors from cell-based experiments, 86 drugs from COVID-19 clinical trials, and 1341 potential SARS-CoV-2 inhibitors from bioinformatics analysis were included. A total of 2777 manually curated entries were obtained, and each entry encompasses comprehensive information, including inhibitor/drug name, targeting protein, the functional mechanism of inhibitor, experimental technique, experimental sample type (cell line and/or tissue), reference information (PubMed ID, year of publication, title of paper). Thereafter, we developed dbSCI using Hypertext Markup Language (HTML5), Java Server Pages (JSP), and JavaScript. The server end is constructed by Java, and the backend database uses SQL Server to store all curated information. dbSCI is currently hosted on a windows server and adopts Apache Tomcat as the application container. dbSCI is freely available at http://bioinfo.henu.edu.cn/COVID/COVIDIH.html.
dbSCI provides a very convenient interface for users to search, browse, download, and submit data (Fig. 1). On the “search” webpage, users can easily search for the inhibitor or its target using the protein target name or inhibitor/drug name; then, comprehensive information on the target/inhibitor will be presented. In addition, users can browse all inhibitors or their targets on three-tab pages, including cell-based experiments, clinical trials, and computational simulations. Users can directly obtain the PubMed reference number (PMIDs), drug name, target(s), clinical trials, and some other information from the website. Meanwhile, the related pharmacological information, including half maximal inhibitory concentration (IC50), half maximal effective concentration EC50, 50% cytotoxic concentration (CC50) (from in vitro studies), and the effectiveness (positive or negative, from clinical trials) as well as the imperative information of International Union of Pure and Applied Chemistry (IUPAC) name, an identifier from the database of chemical (CID) and simplified molecular input line entry system (SMILES) (from computational simulation studies) are also shown on the browser page. Thus, the user can easily obtain this information from different studies and compare them. By clicking “more” in the last column with column name “Detail” on the “search” and “Browse” pages, additional information will be presented. For instance, if the user wants to query a potential drug, such as chloroquine, the user can input the drug name “chloroquine” in the “Search” page. Then, the output page will list all potential entries related to your input “chloroquine,” including the PMID of the related article, the drug target for SARS-CoV-2, and IC50, EC50, and CC50 from in vitro studies. Moreover, any clinical trials or bioinformatics analysis of chloroquine will be shown on the same page as well, including the effectiveness of this drug in clinical trials and bioinformatics results. Furthermore, the user can download the complete data from the download page. Researchers can also de novo submit new data into the database through the submission page provided by dbSCI. All submissions will be quality checked by our research team before being deposited into the dbSCI database.
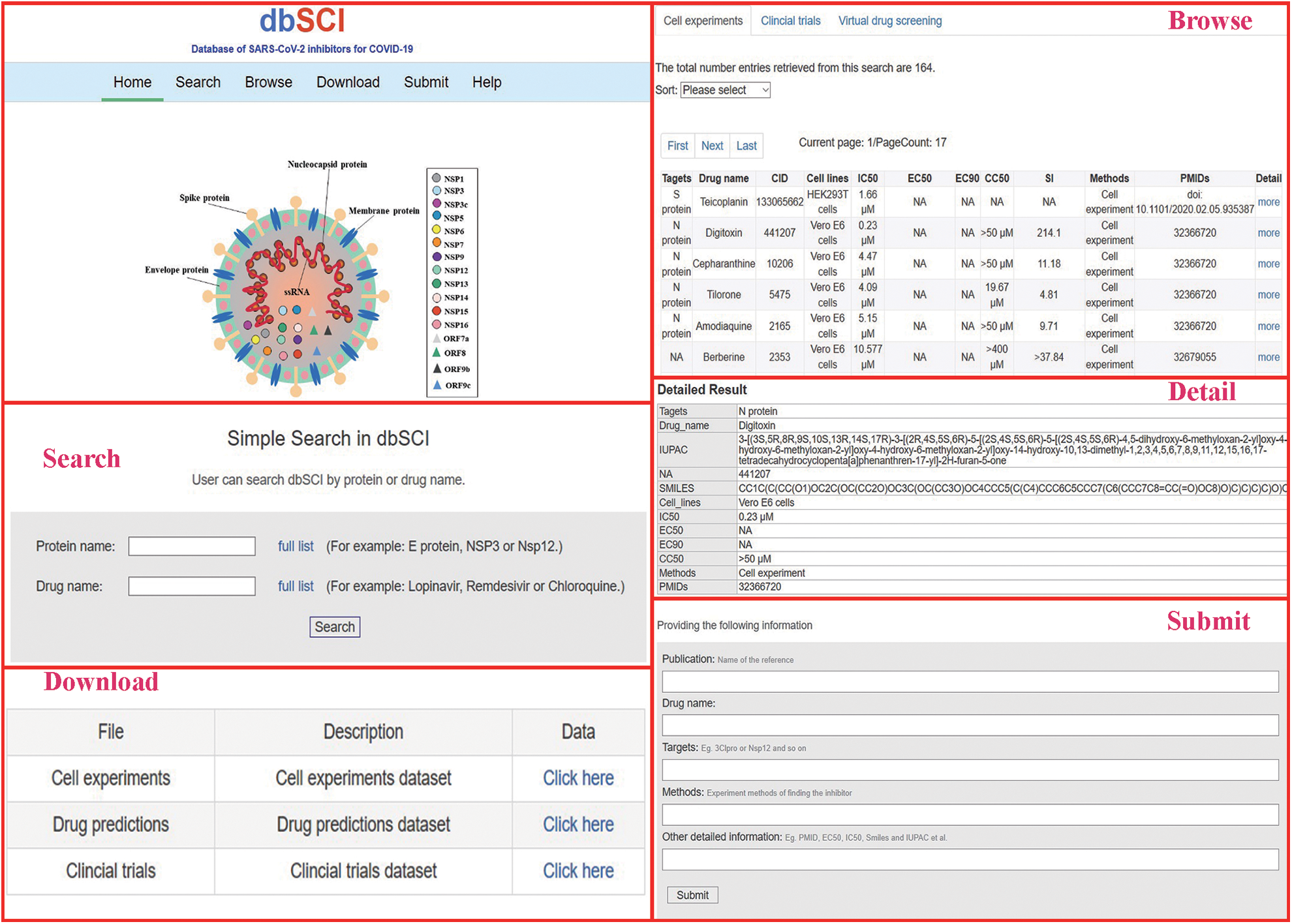
Figure 1: A diagram of dbSCI main interface and subpages.
Currently, there are several databases reporting drug screening for COVID-19, such as COVID19db (Zhang et al., 2022), DockCoV2 (Chen et al., 2021), and CoV-RDB (Tzou et al., 2020). Compared with these existing databases, dbSCI contains all papers reporting inhibitors for SARS-CoV-2, which can be a good supplement for other databases. In the meanwhile, dbSCI has some limitations. First, dbSCI only contains the published papers for COVID-19, while a large number of the preprints of COVID-19 have been excluded. Second, dbSCI offers limited online visualization functions, like the structure visualization of potential drugs and target proteins of SARS-CoV-2. Yu et al. (2020) found that luteolin can bind with the main protease of SARS-CoV-2 by the molecular docking function of AutoDock. Meanwhile, Oso et al. (2021) reported that some natural products (resveratrol, kaempferol, and quercetin) had higher binding affinities for the main protease of SARS-CoV-2 and lower acute oral toxicity compared with chloroquine based on ADMET analysis. In the future, we will update dbSCI to implement these functions by collecting data from PubChem and designing the model for molecular docking and virtual screening (Ferreira and Andricopulo, 2019; Trott and Olson, 2010).
In this study, we manually retrieved and curated the entries of inhibitors or drugs against SARS-CoV-2 for COVID-19 from published literature and developed the dbSCI database. The dbSCI website is updated monthly to offer long-term service. dbSCI provides a relatively comprehensive, credible repository for inhibitors/drugs against SARS-CoV-2 and their potential targeting mechanisms and it will be valuable for basic and clinical researchers to control COVID-19.
Availability of Data and Materials: All data generated or analyzed during this study are included in published articles (and their supplementary information files).
Author Contribution: Study concept and design: Qiang Wang, Longxiang Xie, and Xiangqian Guo. Acquisition and analysis of data: Qiang Wang, Guo Zhao, Baoping Zheng, Xuan Li, Xixi Yu, Zulipinuer Wusiman, Longxiang Xie, and Xiangqian Guo. Design and implementation of the database: Qiang Wang, Guo Zhao, Longxiang Xie, Xuan Li, Xixi Yu, and Xiangqian Guo. Design and implementation of software: Qiang Wang, Guo Zhao, Longxiang Xie, and Xiangqian Guo. Draft of the manuscript: Qiang Wang, Guo Zhao, Longxiang Xie, and Xiangqian Guo.
Ethics Approval: Not applicable.
Funding Statement: This work was supported by the Innovation and Entrepreneurship Training Program for College Students in Henan University (20221022009), the Key Scientific Research Project of Henan Province (22A310012), supporting program for Central Plain Young Top Talents (ZYQR201912176), the program from Academy for Advanced Interdisciplinary Studies of Henan University (Y21008L).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
Angrup A, Kanaujia R, Ray P, Biswal M (2020). Healthcare facilities in low- and middle-income countries affected by COVID-19: Time to upgrade basic infection control and prevention practices. Indian Journal of Microbiology 38: 139–143. DOI 10.4103/ijmm.IJMM_20_125. [Google Scholar] [CrossRef]
Bai C, Zhong Q, Gao GF (2022). Overview of SARS-CoV-2 genome-encoded proteins. Science China Life Science 65: 280–294. DOI 10.1007/s11427-021-1964-4. [Google Scholar] [CrossRef]
Chen TF, Chang YC, Hsiao Y, Lee KH, Hsiao YC, Lin YH, Tu YE, Huang HC, Chen CY, Juan HF (2021). DockCoV2: A drug database against SARS-CoV-2. Nucleic Acids Research 49: D1152–D1159. DOI 10.1093/nar/gkaa861. [Google Scholar] [CrossRef]
Feikin DR, Higdon MM, Abu-Raddad LJ, Andrews N, Araos R et al. (2022). Duration of effectiveness of vaccines against SARS-CoV-2 infection and COVID-19 disease: Results of a systematic review and meta-regression. Lancet 399: 924–944. DOI 10.1016/S0140-6736(22)00152-0. [Google Scholar] [CrossRef]
Ferreira LLG, Andricopulo AD (2019). ADMET modeling approaches in drug discovery. Drug Discovery Today 24: 1157–1165. DOI 10.1016/j.drudis.2019.03.015. [Google Scholar] [CrossRef]
Finkel, Y., Mizrahi, O., Nachshon, A., Weingarten-Gabbay, S., Morgenstern, D et al. (2021). The coding capacity of SARS-CoV-2. Nature, 589: 125–130. [Google Scholar]
Jackson CB, Farzan M, Chen B, Choe H (2022). Mechanisms of SARS-CoV-2 entry into cells. Nature Reviews Molecular Cell Biology 23: 3–20. DOI 10.1038/s41580-021-00418-x. [Google Scholar] [CrossRef]
Lou Z, Sun Y, Rao Z (2014). Current progress in antiviral strategies. Trends in Pharmacological Science 35: 86–102. DOI 10.1016/j.tips.2013.11.006. [Google Scholar] [CrossRef]
Lu S, Ye Q, Singh D, Cao Y, Diedrich JK, Yates JRIII, Villa E, Cleveland DW, Corbett KD (2021). The SARS-CoV-2 nucleocapsid phosphoprotein forms mutually exclusive condensates with RNA and the membrane-associated M protein. Nature Communication 12: 1–15. DOI 10.1038/s41467-020-20768-y. [Google Scholar] [CrossRef]
Marston HD, Folkers GK, Morens DM, Fauci AS (2014). Emerging viral diseases: Confronting threats with new technologies. Science Translational Medicine 6: 253ps10. DOI 10.1126/scitranslmed.3009872. [Google Scholar] [CrossRef]
Martinez MA (2020). Compounds with therapeutic potential against novel respiratory 2019 coronavirus. Antimicrobial Agents and Chemotherapy 64: e00399-20. DOI 10.1128/AAC.00399-20. [Google Scholar] [CrossRef]
Ng TL (2000). Clinical drug evaluation: The regulatory perspectives. Annas of the Academy of Medicine, Singapore 29: 616–620. [Google Scholar]
Oso BJ, Olaoye IF, Omeike SO (2021). Molecular docking and ADMET prediction of natural compounds towards SARS Spike glycoprotein-human angiotensin-converting enzyme 2 and SARS-CoV-2 main protease. Archives of Razi Institute 76: 453–459. DOI 10.22092/ari.2020.351202.1517. [Google Scholar] [CrossRef]
Ou X, Liu Y, Lei X, Li P, Mi D et al. (2020). Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nature Communications 11: 1620. DOI 10.1038/s41467-020-15562-9. [Google Scholar] [CrossRef]
Peng Y, Du N, Lei Y, Dorje S, Qi J, Luo T, Gao GF, Song H (2020). Structures of the SARS-CoV-2 nucleocapsid and their perspectives for drug design. The EMBO Journal 39: e105938. DOI 10.15252/embj.2020105938. [Google Scholar] [CrossRef]
Quimque MTJ, Notarte KIR, Fernandez RAT, Mendoza MAO, Liman RAD et al. (2021). Virtual screening-driven drug discovery of SARS-CoV2 enzyme inhibitors targeting viral attachment, replication, post-translational modification and host immunity evasion infection mechanisms. Journal of Biomolecular Structure & Dynamics 39: 4316–4333. DOI 10.1080/07391102.2020.1776639. [Google Scholar] [CrossRef]
Schein CH (2020). Repurposing approved drugs on the pathway to novel therapies. Medical Research Reviews 40: 586–605. DOI 10.1002/med.21627. [Google Scholar] [CrossRef]
Trott O, Olson AJ (2010). AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry 31: 455–461. DOI 10.1002/jcc.21334. [Google Scholar] [CrossRef]
Tzou PL, Tao K, Nouhin J, Rhee SY, Hu BD, Pai S, Parkin N, Shafer RW (2020). Coronavirus antiviral research database (CoV-RDBAn online database designed to facilitate comparisons between candidate anti-coronavirus compounds. Viruses 10: 1006. DOI 10.3390/v12091006. [Google Scholar] [CrossRef]
WHO. Coronavirus disease (COVID-19) pandemic. https://www.who.int/emergencies/diseases/novel-coronavirus-2019. [Google Scholar]
Yoshimoto FK (2020). The proteins of severe acute respiratory syndrome coronavirus-2 (SARS CoV-2 or n-COV19the cause of COVID-19. Protein Journal 39: 198–216. DOI 10.1007/s10930-020-09901-4. [Google Scholar] [CrossRef]
Yu R, Chen L, Lan R, Shen R, Li P (2020). Computational screening of antagonists against the SARS-CoV-2 (COVID-19) coronavirus by molecular docking. International Journal of Antimicrobial Agents 56: 106012. DOI 10.1016/j.ijantimicag.2020.106012. [Google Scholar] [CrossRef]
Zhang W, Zhang Y, Min Z, Mo J, Ju Z et al. (2022). COVID19db: A comprehensive database platform to discover potential drugs and targets of COVID-19 at whole transcriptomic scale. Nucleic Acids Research 50: D747–D757. DOI 10.1093/nar/gkab850. [Google Scholar] [CrossRef]
Ziegler CGK, Allon SJ, Nyquist SK, Mbano IM, Miao VN et al. (2020). SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell 181: 1016–1035. DOI 10.1016/j.cell.2020.04.035. [Google Scholar] [CrossRef]
Supplementary Materials
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools