 Open Access
Open Access
ARTICLE
Methyltransferase 3A-mediated promoter methylation represses retinoic acid receptor responder 3 expression in basal-like breast cancer
Department of Pancreatic Surgery, West China Hospital, Sichuan University, Chengdu, 610041, China
* Corresponding Author: XUBAO LIU. Email:
BIOCELL 2023, 47(2), 319-328. https://doi.org/10.32604/biocell.2023.025250
Received 01 July 2022; Accepted 31 August 2022; Issue published 18 November 2022
Abstract
Retinoic acid receptor responder 3 (RARRES3) has been characterized as a tumor suppressor in multiple types of cancer. This study aimed to examine the expression profile of RARRES3 across the PAM50 subtypes of breast cancer. The DNA methylation status of RARRES3 was checked in the basal-like subtype, and the underlying mechanisms of its dysregulation were explored. RNA-sequencing (seq) and methylation data from The Cancer Genome Atlas were used for in-silico analysis. Basal-like representative SUM149 and MDA-MB-468 cell lines were used for in vitro and in vivo studies. Compared to tumor-adjacent normal tissues, only the basal-like tumor tissues had significantly downregulated RARRES3 expression. The methylation level of four CpG sites in the promoter region showed a strong negative correlation with RARRES3 expression. The gene coding for DNA methyltransferase 3A (DNMT3A) had consistent positive correlations with the methylation of the CpG sites. Chromatin Immunoprecipitation-quantitative polymerase chain-reaction and bisulfite sequencing PCR showed that DNMT3A could bind to the promoter region of RARRES3 and promote methylation of the CpG sites within the region. DNMT3A knockdown significantly restored RARRES3 expression at the mRNA and protein level in the two cell lines. CCK-8, colony formation, and flow cytometric analysis showed that RARRES3 overexpression attenuated the growth-promoting effects of DNMT3A overexpression and also weakened the DNMT3A overexpression-induced activation of ERK1/2 and PI3K/AKT signaling. In summary, this study revealed that DNMT3A enhances promoter methylation of the RARRES3 gene and suppresses its transcription in basal-like breast cancer. The DNMT3A-RARRES3 signaling pathway might be a potential target for the treatment of this tumor subtype.Keywords
Retinoic acid receptor responder 3 (RARRES3), which is also known as phospholipase A and acyltransferase 4, retinoid-inducible gene 1 (RIG1) or tazarotene-induced gene 3, has been characterized as a tumor suppressor in multiple types of cancers, including liver cancer (Wei et al., 2017), gastric cancer (Tang et al., 2021), bladder cancer (Lu et al., 2022), and breast cancer (Dydensborg et al., 2009). RARRES3 retards breast cancer cell proliferation by suppressing Wnt/beta-catenin signaling and inhibiting EGFR-mediated signal transductions, including phosphorylation of extracellular signal-regulated kinase 1/2 (ERK1/2) and PI3K/AKT (Hsu et al., 2015). It also suppresses breast cancer lung metastasis by inhibiting the adhesion of the tumor cells to the lung parenchyma and promoting tumor cell differentiation (Dydensborg et al., 2009; Morales et al., 2014).
However, RARRES3 expression is usually downregulated in cancers due to complex epigenetic mechanisms. In human liver cancer, histone methyltransferase G9a can epigenetically silence RARRES3 by promoting H3K9 di-methylation (Wei et al., 2017). In human bladder cancer, H3K36me2 demethylase KDM2A is usually upregulated (Lu et al., 2022). Its upregulation decreased H3K36me2 enrichment in the RARRES3 promoter (Lu et al., 2022). RARRES3 mRNA stability and translation might be hampered by several microRNAs, such as miR-545 (Liu et al., 2016), miR-BART6-3p (Lu et al., 2017), and miR-1286 (Wei et al., 2019).
Breast cancer is a group of heterogeneous tumors with great variations in genetic and phenotypic features (Turashvili and Brogi, 2017). Traditional Prediction Analysis of Microarray 50 (PAM50) defines five intrinsic tumor subtypes, including luminal A, luminal B, human epidermal growth factor receptor 2 [HER2]-enriched (HER2+), basal-like, and normal-like (Parker et al., 2009). The subtypes vary significantly in terms of their molecular dysregulation and biological properties (Caan et al., 2014; Petrovic et al., 2021; Russnes et al., 2017). Patients with different subtypes of tumors have different prognoses (Caan et al., 2014).
Considering the potent suppressive effect of RARRES3 on breast cancer, we examined the expression profile of RARRES3 across the different subtypes of breast cancer using RNA-sequencing (seq) data from The Cancer Genome Atlas (TCGA). Then, we checked its DNA methylation status in the basal-like subtype and explored the underlying mechanisms of its dysregulation.
Data mining from the cancer genome atlas
The RNA-seq and DNA methylation data (methylation 450k, reported as β-value) in TCGA-Breast Cancer (BRCA) was extracted from the Pan-cancer dataset using the portal provided by the UCSC Xena Browser (https://xenabrowser.net/) (Goldman et al., 2020). Gene expression was compared among tumor-adjacent normal tissues and the PAM50 tumor subtypes. We used the log2 (TPM+0.001) data in the database to compare gene expression and calculate correlations.
The basal-like representative SUM149 cell line was purchased from Cobioer (Nanjing, Jiangsu, China). MDA-MB-468 cells were purchased from Procell (Wuhan, Hubei, China). The two cell lines were cultured using the mediums and conditions introduced previously (Giordano et al., 2012). Cells collected in their logarithmic phase of growth were used for experiments. Lentiviral DNMT3A shRNAs were constructed using the pLKO.1-puro plasmids. The following shRNA sequences were used: shDNMT3A#1, 5′-CCACCAGAAGAAGAGAAGAAT-3′; shDNMT3A#2, 5′-CCCAAGGTCAAGGAGATTATT-3′; shDNMT3A#3, 5′-CCGGCTCTTCTTTGAGTTCTA-3′. Lentiviral DNMT3A (NM_022552) and RARRES3 (NM_004585) overexpressing particles were generated using pLV-Puro. Lentivirus used for infection was generated by co-transfecting the recombinant pLKO.1 shRNA plasmids or pLV-Puro overexpressing plasmids with psPAX2 packaging plasmid and pMD2.G envelope plasmid to 293T cells. Supernatants were collected 48 h after transfection, passed through a 0.45-μm filter, and then concentrated. Cells were infected at a multiplicity of infection (MOI) of 10.
Traditional western blotting was performed following the methods introduced previously (Wu et al., 2021). In brief, cell samples (MDA-MB-468 and SUM149 cells) were collected and lysed. The protein concentration was measured using a bicinchoninic acid (BCA) protein assay kit. Nearly 30 μg of samples were loaded to each lane, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and transferred onto a polyvinylidene fluoride membrane (Millipore, Billerica, MA, USA). The membranes were blocked and incubated with primary antibodies at 4°C overnight. The following antibodies and dilutions were used: anti-DNMT3A (1:1000, #32578, Cell Signaling Technology, Danvers, MA, USA); anti-RARRES3 (1:1000, PA5-115825, Thermo Fisher Scientific, Waltham, MA, USA); anti-cyclin D1 (1:2000, 26939-1-AP, Proteintech, Wuhan, Hubei, China), anti-p21 (1:2000, 10355-1-AP, Proteintech), anti-p-ERK1/2 (1:2000, #4370, Cell Signaling Technology); anti-ERK1/2 (1:2000, #4695, Cell Signaling Technology); anti-p-AKT (1:5000, 66444-1-Ig, Proteintech), anti-Akt (1:10000, 60203-2-Ig, Proteintech) and anti-β-actin (1:2000, 20536-1-AP, Proteintech). The membranes were thoroughly washed and incubated with HRP-conjugated secondary antibodies for 2 h at room temperature. Then, the protein bands were developed using enhanced chemiluminescence (ECL) (BeyoECL Star, Beyotime, Shanghai, China).
Quantitative real-time-polymerase chain reaction (qRT-PCR) assays
Gene expression was assessed by qRT-PCR assays, performed as previously described (Wu et al., 2021). In brief, total RNA was isolated from cell samples (MDA-MB-468 and SUM149 cells) and subjected to reverse transcription. The cDNA was then used as the template for PCR. Real-time amplification was performed using LightCycler 480 SYBR Green I Master on a LightCycler 480 Real-Time PCR System (Roche Diagnostics, Basel, Switzerland). Gene expression was normalized with GAPDH. Relative gene expression was calculated using the 2−ΔΔCT method. The following primers were used for amplification: DNMT3A, forward: 5′-CCTCTTCGTTGGAGGAATGTGC-3′; reverse: 5′-GTTTCCGCACATGAGCACCTCA-3′; RARRES3, forward: 5′-GCAGGAACTGTGAGCACTTTGTC-3′; reverse: 5′-GCAACAACCAGGATTCCAAGCG-3′; GAPDH, forward: 5′-GTCTCCTCTGACTTCAACAGCG-3′; reverse: 5′-ACCACCCTGTTGCTGTAGCCAA-3′.
Bisulfite sequencing PCR (BSP)
BSP assay was conducted following the method described previously (Masalmeh et al., 2021). In brief, genomic DNA was extracted from cell samples and treated with sodium bisulfite using the EZ DNA Methylation-Gold kit (Zymo Research, Irvine, CA, USA). Then, the converted DNA was subjected to PCR assay using bisulfite-specific primers designed by the MethPrimer software (forward: 5′-GGAGGTAGATTATAAGGTTAAGAGAT-3′; reverse: 5′- AAAACTAATCATTTCTTACACAAACCATA-3′), that cover the cg05817709 sites (Suppl. Fig. S1). Then, PCR products were purified using a QIAquick PCR purification kit (QIAGEN, Germany) and cloned into the pGEM-T Easy Vector (Promega, Madison, WI, USA). Six insert-positive bacterial clones were selected for sequencing.
Chromatin immunoprecipitation (ChIP)-qPCR
ChIP was conducted using a commercial ChIP Assay Kit (Beyotime), following standard protocols (Gade and Kalvakolanu, 2012). In brief, MDA-MB-468 and SUM149 cells were lysed After 48 h of lentiviral infection for DNMT3A knockdown. Immunoprecipitation was performed using anti-DNMT3A (#32578, Cell Signaling Technology) or normal rabbit IgG (negative control). Then, the immunoprecipitated chromatin samples were purified and used as the template for subsequent qPCR assays. The following primers were used: primer set 1 (not covering any CpG sites), F: 5′-GACCAGTGCAATGGAGACA-3′, R: 5′-GGTAACAGAAAGGCAGGAAGA-3′; primer set 2 (covering four potential DNMT3A binding CpG sites), F: 5′-GAGGCAGGAGAATCACTTGAA-3′, R: 5′-TGAGACAGAGTTTCGCTCTTG-3′.
SUM149 or MDA-MB-468 cells with DNMT3A or RARRES3 overexpression alone or in combination were seeded into the 24-well plates at a density of 5 × 103 cells/well. Cells were cultured in normal conditions for 14 days. After fixing with 4% paraformaldehyde, the cell colonies were stained with 0.1% crystal violet for 20 min at room temperature. The bottom side of the plates was scanned, and the number of colonies was counted under a light microscope.
Flow cytometric analysis of cell cycle distribution
After 48 h of lentiviral infection, SUM149 or MDA-MB-468 cells were harvested, fixed, and stained with Pharmingen PI/RNase Staining buffer (cat. no. 550825; BD Biosciences, San Jose, CA, USA) for 20 min at room temperature according to the manufacturer’s instructions. Then, cells were detected using a BD FACSCalibur (BD Biosciences). Data analysis was performed using NovoExpress software (v.1.5.4, Agilent, Santa Clara, CA, USA).
Animal studies were conducted in Jinruijie Biotechnology Service Center, Chengdu, China and were approved by the ethics committee of the institution (Approval no. 20220126SCU). All animal-related procedures were conducted following the Guide for the Care and Use of Laboratory Animals. Female athymic nude mice (BALB/c-nu/nu), approximately 5–6 weeks old (18–20 g), were purchased from Vital River Laboratory Animal Technology (Beijing, China) and raised in a specific-pathogen-free environment. After treating as indicated, 5 × 106 cells in 0.2 mL PBS and Matrigel matrix (BD Biosciences) mix (v/v, 1:1) were injected subcutaneously into the fourth mammary fat pad. Tumor volume was measured using a caliper twice weekly. Mice were euthanized by CO2 asphyxiation on day 35 after tumor cell inoculation. The xenograft tumors were excised, fixed in 4% paraformaldehyde, embedded in paraffin, and sectioned for IHC staining of Ki-67.
Results were collected and integrated using GraphPad 8.01. Quantitative data were represented as mean ± SD. An unpaired T-test was utilized to compare two groups. The correlation was assessed by Pearson’s r. One-way ANOVA with post-hoc Dunnett’s multiple comparisons test was performed to assess the differences in multiple groups. P < 0.05 was considered significantly different.
RARRES3 expression is negatively correlated with the methylation status of CpG sites within its promoter region
We used RNA-seq data from the TCGA database and compared RARRES3 expression across the tumor-adjacent normal tissues, and the five PAM50 subtypes of primary tumors (Fig. 1A). Compared to normal tissues adjacent to the tumor (n = 113), only the basal-like (basal) tumor tissues had significantly downregulated RARRES3 expression (Fig. 1A). Then, we used available methylation 450k data in the basal cases (n = 84), and checked the correlation between the methylation status (β-value) of five CpG sites in the RARRES3 gene promoter and gene body (Fig. 1B). Pearson’s correlation showed that the methylation level of four CpG sites in the promoter region, including cg18112681, cg05817709, cg25599242, and cg04999352 showed strong negative correlations (r ≤ 0.6) with RARRES3 expression (Figs. 1B–1F). A weak positive correlation (r = 0.246) was observed between the methylation level of cg13899097 (within the gene body) and RARRES3 expression (Fig. 1G).
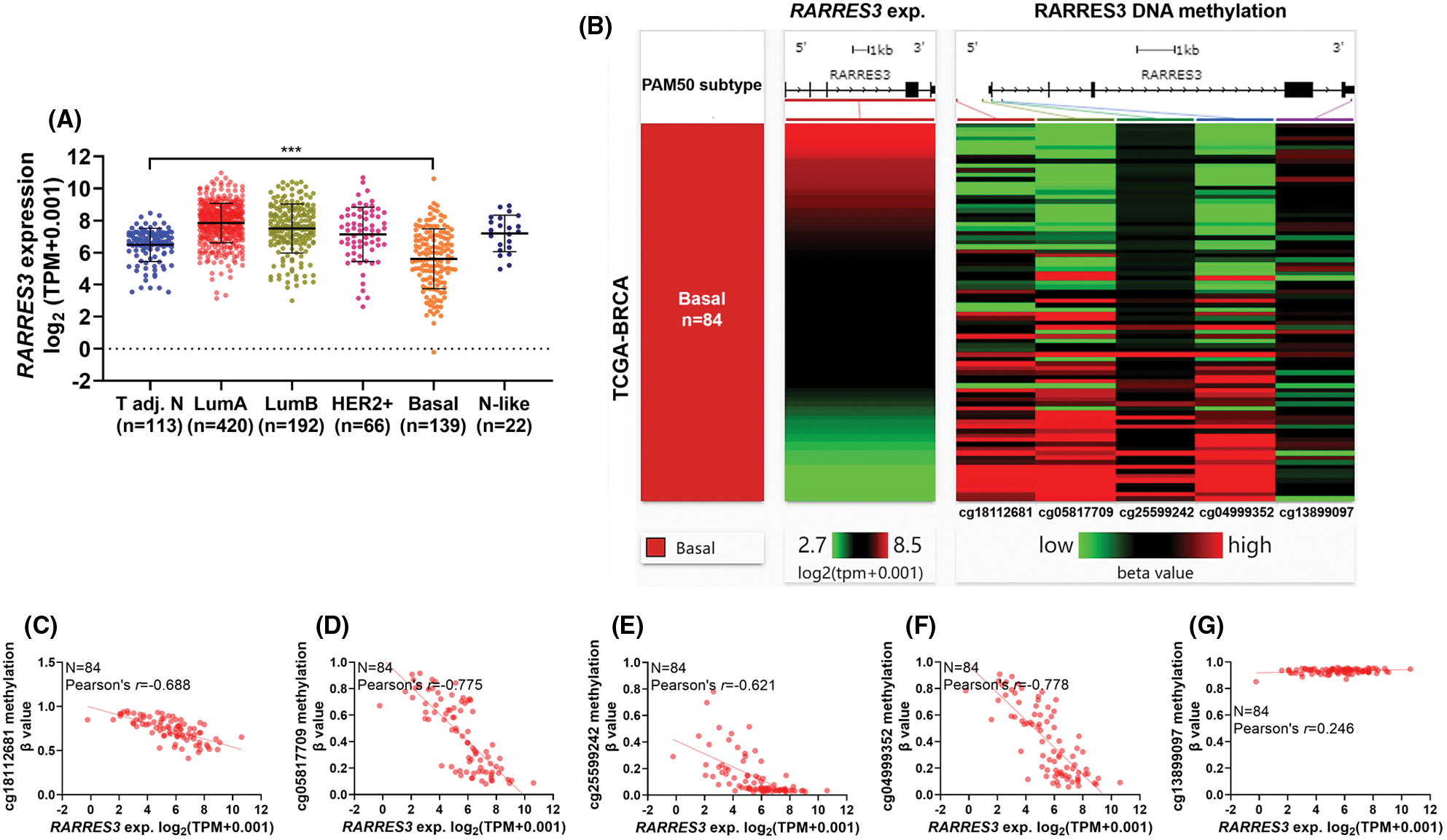
Figure 1: RARRES3 expression is negatively correlated with the methylation status of CpG sites within its promoter region. (A) Plot chart showing RARRES3 expression in tumor-adjacent normal tissues (n = 113) and the PAM50 tumor subgroups in The Cancer Genome Atlas-Breast Cancer (TCGA-BRCA) extracted from Pan-cancer database. (B) A heatmap was generated to show the correlation between the expression of RARRES3 and the methylation levels (β value) of five CpG sites in the RARRES3 gene promoter and gene body in primary basal-like tumors. Only the primary tumors with RNA-seq and DNA methylation 450k data simultaneously (n = 84) were included for analysis. (C–G) Plot charts were generated to show the correlation between RARRES3 expression and the methylation level of cg18112681 (C), cg05817709 (D), cg25599242 (E), cg04999352 (F) or cg13899097 (G) in primary basal-like tumors. Pearson’s correlation was conducted. ***P < 0.001.
DNA methyltransferase 3A-mediated promoter methylation represses RARRES3 expression in basal-like breast cancer
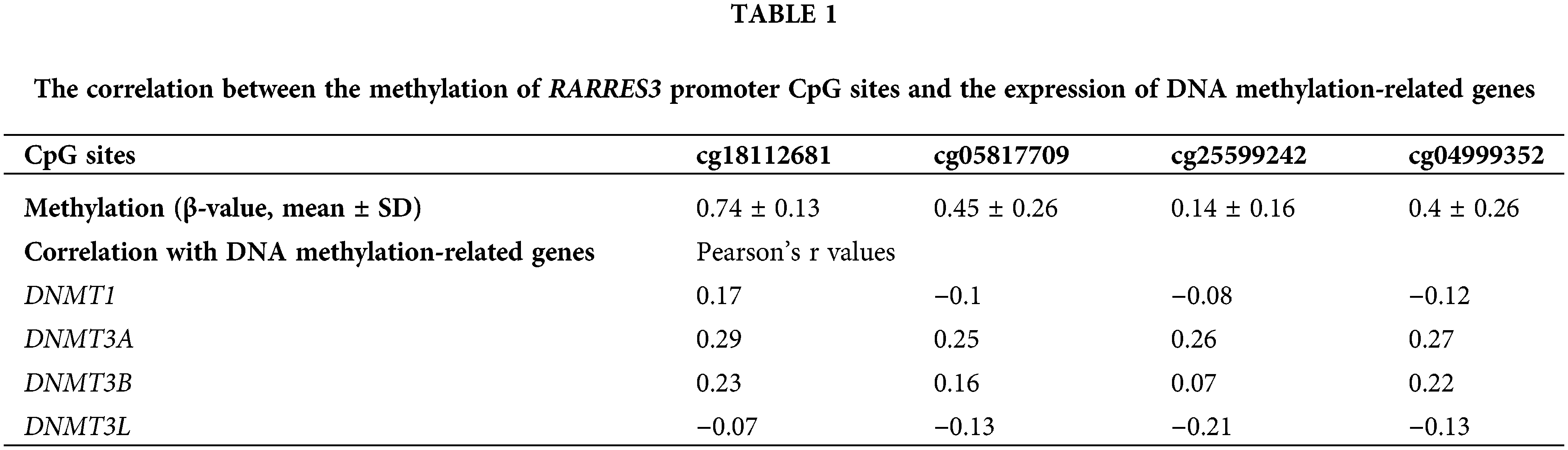
To explore the critical enzymes regulating RARRES3 promoter methylation, we checked the correlation between the methylation of the CpG sites and the expression of DNA methylation-related genes, including DNMT1, DNMT3A, DNMT3B, and DNMT3L (Table 1) and found that only DNMT3A had a consistent positive correlation with the methylation of the CpG sites.
Therefore, we hypothesized that DNMT3A might be a critical enzyme suppressing RARRES3 expression in basal-like breast cancer. To validate this hypothesis, we used two basal-like tumor cell lines (SUM149 and MDA-MB-468) as the in vitro models. These two cell lines were infected with lentiviral DNMT3A shRNA (Figs. 2A, 2C, and 2D). DNMT3A knockdown significantly restored RARRES3 expression at the mRNA and protein levels (Figs. 2B, 2C, and 2E). We checked the promoter sequence of the RARRES3 promoter and confirmed the location of the four CpG sites (upstream = 1297, downstream = 500, Supple. Fig. S1, Fig. 2F). We then performed ChIP-qPCR to detect the specific binding of DNMT3A to the RARRES3 promoter region. Two primer sets were designed: primer set 1 does not cover any CpG site and primer set 2 covers four CpG sites within the promoter (Fig. 2F). Results showed a significant amplification of amplicon 2 (covering the four CpG sites), but not amplicon 1 in the samples precipitated by anti-DNMT3A (Figs. 2G and 2H). Moreover, DNMT3A knockdown significantly hampered the amplification using primer set 2 (Figs. 2G and 2H). The BSP assay confirmed that DNMT3A knockdown reduced the methylation of the DNMT3A promoter in both SUM149 and MDA-MB-468 cells (Figs. 2I and 2J).
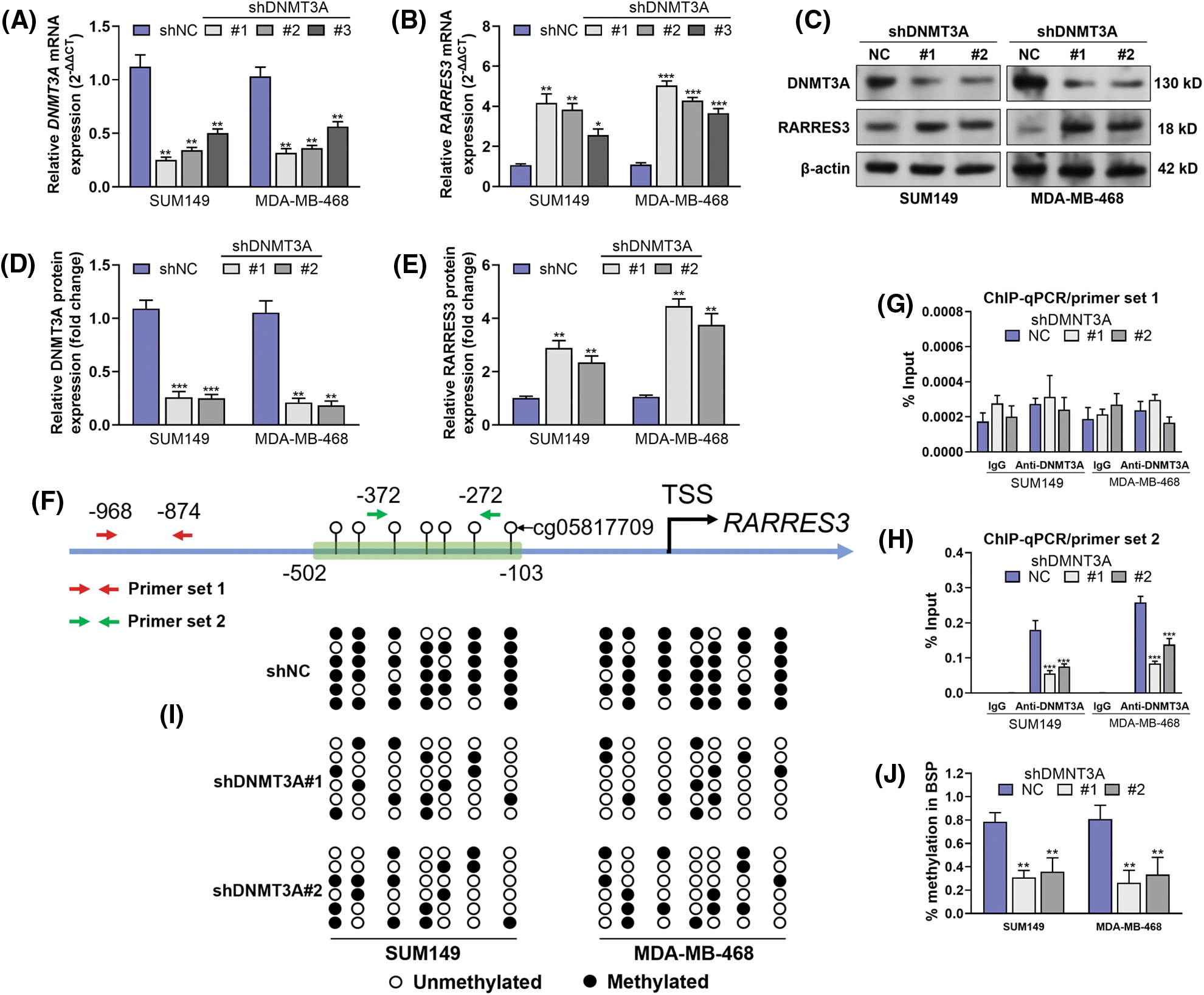
Figure 2: DNA methyltransferase 3A (DNMT3A)-mediated promoter methylation represses RARRES3 expression in basal-like breast cancer. (A–E) Quantitative real-time polymerase chain reaction (qRT-PCR) (A and B) and western blot (C–E) analyses were conducted to check the expression of DNMT3A and RARRES3 at the mRNA and protein levels in SUM149 and MDA-MB-468 cells 48 h after infection of lentiviral DNMT3A shRNA (#1: shDNMT3A#1, #2: shDNMT3A#2; #3: shDNMT3A#3) or the negative control. (F) The promoter segments of RARRES3. The CpG sites covered by BSP primers were labeled within the green frame. Primer set 1 and set 2 were used for chromatin immunoprecipitation (ChIP)-qPCR assays. (G and H) The ChIP-qPCR assay was performed to show the enrichment of RARRES3 promoter segments covering (using primer set 2) (G) or not covering (using primer set 1) (H) the predicted DNMT3A binding sites in SUM149 and MDA-MB-468 cells with or without DNMT3A knockdown. ChIP was conducted using anti-DNMT3A. (I and J) Bisulfite sequencing PCR (BSP) assay was performed to detect the changes in CpG sites (corresponding to the sites within the green box in panel F) methylation status upon DNMT3A knockdown in SUM149 and MDA-MB-468 cells. Data are represented as mean ± SD of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
DNA methyltransferase 3A facilitates basal-like tumor cell proliferation by inhibiting RARRES3
On confirming that RARRES3 could be epigenetically silenced by DNMT3A, we hypothesized that DNMT3A might exacerbate malignant behaviors of basal-like tumor cells via repressing RARRES3 expression. DNMT3A overexpression (Figs. 3A, 3C and 3D) elevated cyclin D1 expression but reduced p21 expression (Figs. 3C, 3F and 3G). In comparison, RARRES3 overexpression (Figs. 3B, 3C, and 3E) weakened the effects of DNMT3A overexpression on cyclin D1 and p21 expression (Figs. 3C, 3F and 3G). DNMT3A overexpression significantly promoted the proliferation (Figs. 3H and 3I), colony formation (Figs. 3J–3L), and cell-cycle progression (Figs. 3M–3O) of SUM149 and MDA-MB-468 cells. Notably, RARRES3 overexpression attenuated the growth-promoting effects mediated by DNMT3A overexpression (Figs. 3H–3O).
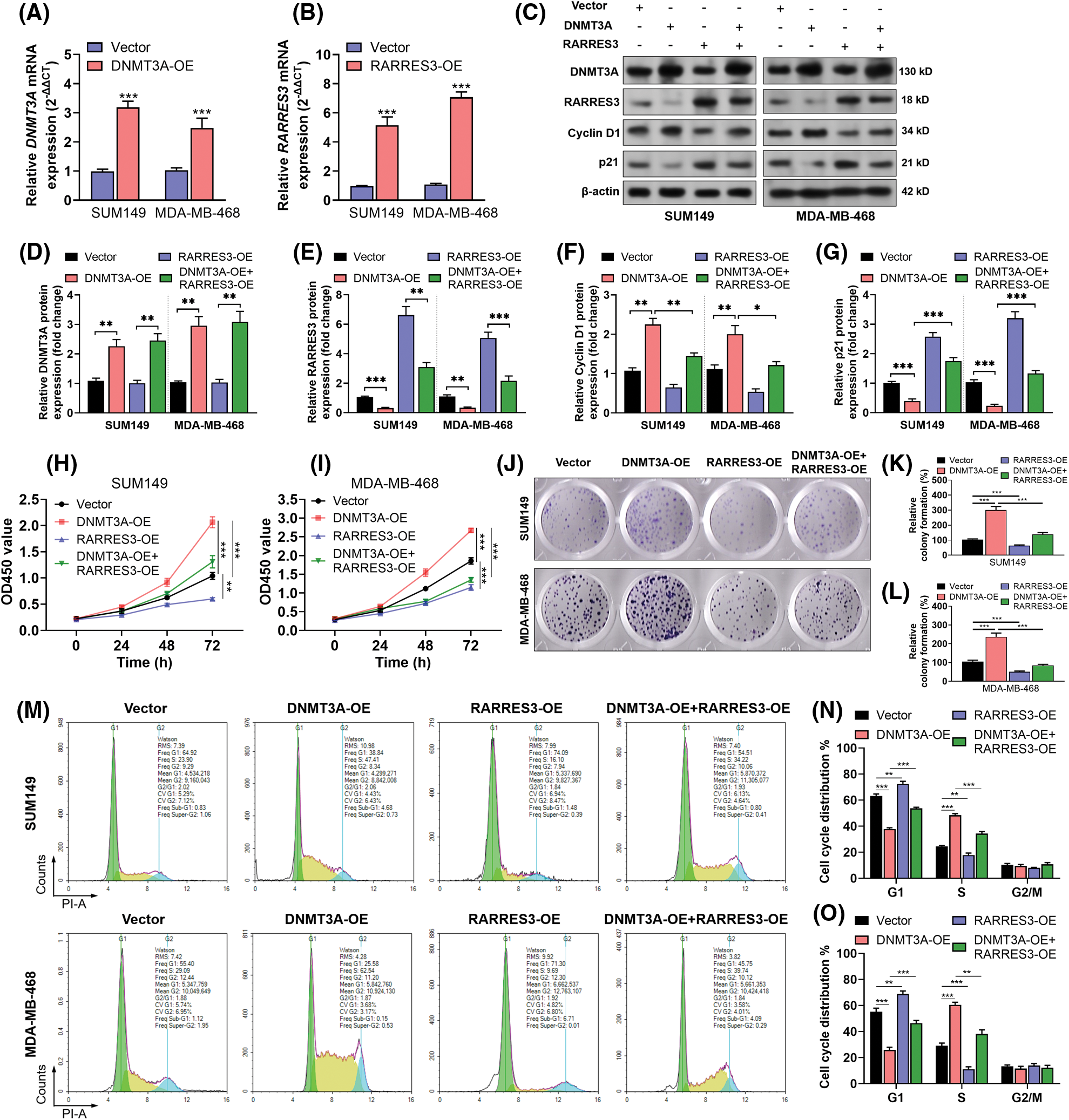
Figure 3: The DNA methyltransferase 3A (DNMT3A)-retinoic acid receptor responder 3 (RARRES3) axis modulates basal-like tumor cell proliferation in vitro. (A and B) Quantitative real-time polymerase chain reaction (qRT-PCR) (A and B) was conducted to check the expression of DNMT3A (A) and RARRES3 (B) at the mRNA and protein levels in SUM149 and MDA-MB-468 cells 48 h after lentiviral infection for DNMT3A or RARRES3 overexpression. (C–G) Western blot analysis was performed to check the expression of DNMT3A, RARRES3, cyclin D1, and p21 in SUM149 and MDA-MB-468 cells 48 h after lentiviral DNMT3A or RARRES3 overexpression alone or in combination. Quantitation of protein expression was performed in (D–G) based on three repeats. (H–O) CCK8 analysis of the vitality (H and I), colony formation assay (J–L), and flow cytometric analysis of cell cycle distribution (M–O) of SUM149 and MDA-MB-468 cells with DNMT3A or RARRES3 overexpression alone or in combination. Data are represented as mean ± SD from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
To verify the growth-regulating effect of the DNMT3A-RARRES3 axis in vivo, we generated MDA-MB-468 cell-based xenograft tumor models in nude mice. According to the tumor growth curve, DNMT3A overexpression significantly enhanced tumor growth (Figs. 4A and 4B). RARRES3 overexpression significantly weakened the growth-promoting effects of DNMT3A overexpression (Figs. 4A and 4B). IHC staining results of the proliferation marker Ki-67 are consistent with the tumor growth curves (Fig. 4C). Furthermore, western blotting results showed that DNMT3A overexpression could elevate the expression of phosphorylated ERK1/2 (p-ERK1/2) and phosphorylated AKT (p-AKT) in the tumor tissues. These trends could be partially reversed by restoring RARRES3 expression (Fig. 4D).
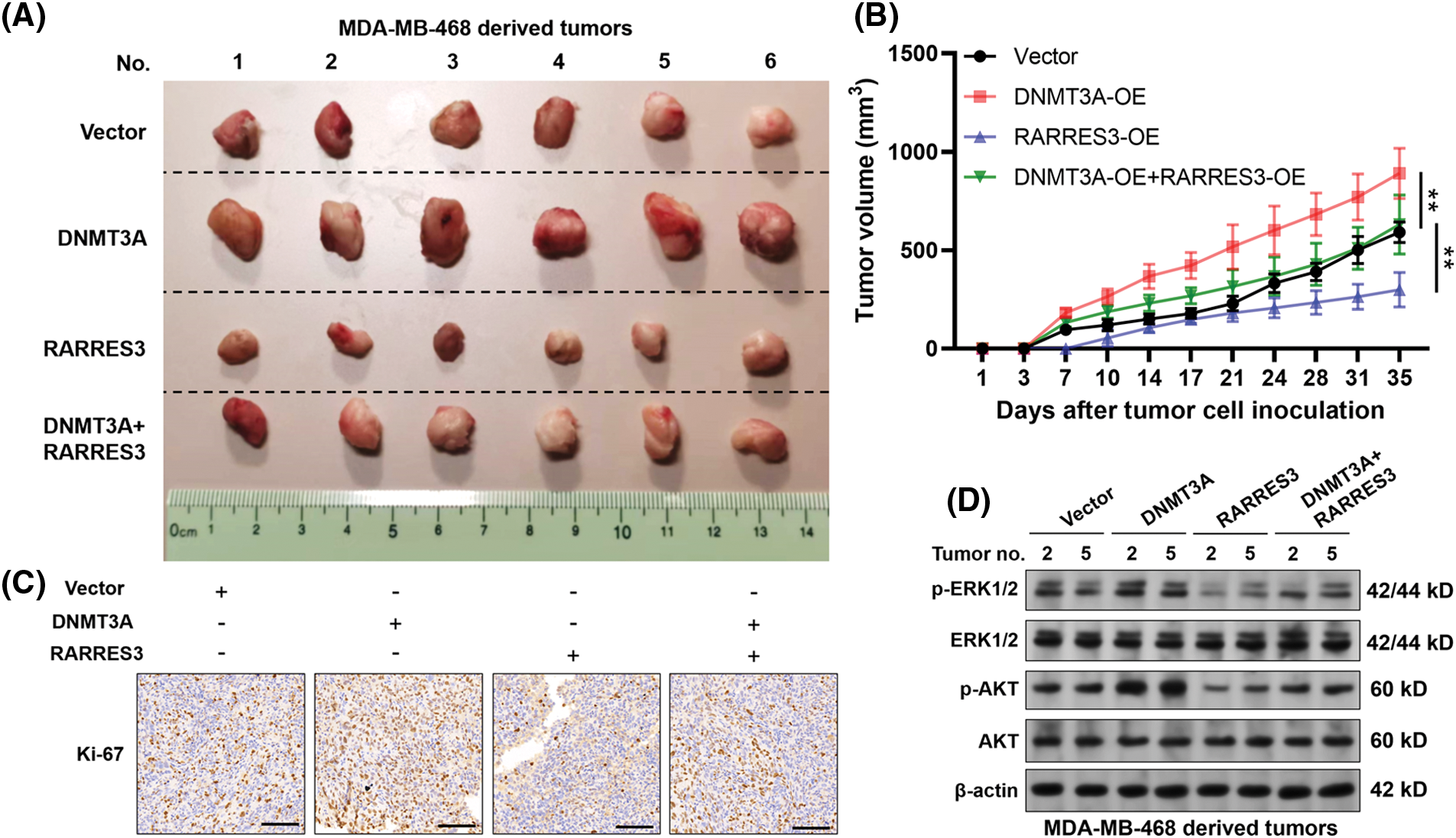
Figure 4: The DNA methyltransferase 3A(DNMT3A)-retinoic acid receptor responder 3 (RARRES3) axis modulates basal-like tumor growth in vivo. (A) Xenograft tumors derived from MDA-MB-468 cells with DNMT3A or RARRES3 overexpression alone or in combination, 7 weeks post-inoculation. (B) The growth curves of the tumors were measured twice a week and are expressed as changes in tumor volume (mm3). (C) Tumors were stained with antibodies to detect the proliferative marker Ki-67. Scale bar: 100 μM. (D) Western blotting assays were conducted to check the expression of p-ERK1/2, total ERK1/2, p-AKT, and total AKT in representative tumors (2 and 5 represent the corresponding tumor tissues in panel A). Data are represented as mean ± SD. **P < 0.01. OE: overexpression.
RARRES3 has been found to act as a downstream target of the tumor protein p53 (TP53) and can inhibit tumor cell growth, induce apoptosis, and promote terminal differentiation of keratinocytes via multiple signaling pathways (Hsu and Chang, 2015). Therefore, restoration of its expression might be a potential strategy for cancer treatment. One previous study confirmed that RARRES3 expression varied significantly between ER-positive and ER-negative tumors. Loss of function of RARRES3 in the ER-negative cases stimulates tumor invasion and promotes metastasis to the lung (Morales et al., 2014). These findings suggest that RARRES3 might have subtype-specific regulations.
In this study, we explored the expression profile of RARRES3 in the PAM50 subtypes of breast cancer and confirmed its downregulation in the basal-like subtype. Most of the basal-like breast tumors are triple-negative (lack of expression of estrogen receptor (ER), progesterone receptor (PR), and HER2) (Rody et al., 2011). Histologically, basal-like tumors are usually high grade, with high mitotic indices, pushing borders of invasion, and atypical medullary features (Milioli et al., 2017; Wang et al., 2022). These features make them highly aggressive, with limited therapeutic responses. The loss of RARRES3 expression in the ER-negative tumor cases might be associated with a reduction in the erythroid transcription factor, GATA differentiation genes such as GATA3 (Dydensborg et al., 2009), and an increase of the pluripotency gene EZH2 (Morales et al., 2014). In the non-basal-like breast cancers, GATA3 gene expression is preserved (Yu et al., 2019), while that of EZH2 is not elevated to the level as high as in basal-like cases (Hirukawa et al., 2018). These mechanisms help to explain the specific loss of RARRES3 in basal-like tumors.
Promoter methylation is an important mechanism of gene downregulation (Tian et al., 2022). Via in-silico analysis using data from TCGA-BRCA, we found that RARRES3 might be epigenetically silenced by promoter hypermethylation in basal-like breast tumors. DNMT3A might be a critical mediator of this alteration. DNMT3A and DNMT3B catalyze de novo methylation by transferring a methyl group from S-adenyl methionine to the C-5 position of cytosine residue (Lyko, 2018). Using SUM149 and MDA-MB-468 cells as the in vitro cell models, we confirmed that DNMT3A could bind to the promoter region of RARRES3 and promote methylation of the CpG sites within the region. DNMT3A knockdown significantly restored RARRES3 expression at the mRNA and protein level in the two cell lines.
DNMT3A overexpression has been widely reported in breast cancer (Man et al., 2022). Together with the basic helix-loop-helix transcription factor (MYC), DNMT3A represses the transcription of miR-200b in triple-negative breast cancer cells (Pang et al., 2018). Functionally, miR-200b is a potential tumor suppressor that synergistically repressed multiple tumor-promoting genes, including zinc-finger E-box-binding homeobox factor 1 (ZEB1), Sex determining region Y-box 2 (SOX2), and Cluster of differentiation 133 (CD133) (Pang et al., 2018). MiR-770-5p can restore the expression of E-Cadherin in MDA-MB-231 cells by targeting DNMT3A, thus inhibiting epithelial-to-mesenchymal transition phenotypes, along with motility and invasion (Noyan et al., 2021). In addition, DNMT3A expression might be a biomarker of decitabine sensitivity in triple-negative breast cancer (Yu et al., 2018). Its expression is associated with significantly shorter disease-free survival or overall survival in certain subgroups of patients with breast cancer (Yu et al., 2015).
In vitro and in vivo tumor growth assays revealed that DNMT3A overexpression-associated tumor cell growth could be attenuated by RARRES3 overexpression. The ERK1/2 and PI3K/AKT signaling pathways are predominant in governing cell growth, differentiation, and survival of triple-negative breast cancer (Khan et al., 2019; Umemura et al., 2007; Zhang et al., 2019). Since both DNMT3A and RARRES3 are implicated in ERK1/2 and PI3K/AKT signaling (Hsu et al., 2015; Zhou et al., 2022), we checked their regulatory effects on these signaling pathways in representative tumor samples. DNMT3A overexpression-induced activation of ERK1/2 and PI3K/AKT signaling is weakened by RARRES3 overexpression. These findings indicate that DNMT3A facilitates the proliferation of basal-like breast cancer cells by inhibiting DNMT3A expression and support the notion that DNMT3A and RARRES3 act on some common molecular axes. In addition to gene promoter methylation, we also observed a positive correlation between the methylation of one gene body CpG site (cg13899097) with RARRES3 transcription. Compared to gene promoter methylation, gene body methylation might have different regulations. In some cases, it may promote gene expression (Arechederra et al., 2018). One previous study showed that DNMT3A could facilitate gene transcription via non-promoter DNA methylation (Wu et al., 2010). Therefore, we infer that DNMT3A might also modulate RARRES3 expression via other epigenetic mechanisms, which is worthy of future exploration.
In summary, this study revealed that DNMT3A could enhance promoter methylation of the RARRES3 gene and suppress its transcription in basal-like breast cancer. The DNMT3A-RARRES3 signaling pathway might be a potential target for the treatment of this tumor subtype.
Availability of Data and Materials: All data generated or analyzed during this study are included in this published article (and its supplementary information files).
Author Contribution: Study design: YLT and XBL; experimentation: YLT; data collection: YLT and XBL; analysis of results: YLT and XBL; manuscript preparation: YLT and XBL. All authors reviewed the final version of the manuscript.
Ethics Approval: Animal studies were approved by the Ethics Committee of Jinruijie Biotechnology Service Center, Chengdu, China (Approval No. 20220126SCU).
Funding Statement: This study was supported by the Science & Technology Department of Sichuan Province, China (2022YFS0314).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
Arechederra M, Daian F, Yim A, Bazai SK, Richelme S, Dono R, Saurin AJ, Habermann BH, Maina F (2018). Hypermethylation of gene body CpG islands predicts high dosage of functional oncogenes in liver cancer. Nature Communications 9: 3164. DOI 10.1038/s41467-018-05550-5. [Google Scholar] [CrossRef]
Caan BJ, Sweeney C, Habel LA, Kwan ML, Kroenke CH et al. (2014). Intrinsic subtypes from the PAM50 gene expression assay in a population-based breast cancer survivor cohort: Prognostication of short- and long-term outcomes. Cancer Epidemiology, Biomarkers and Prevention 23: 725–734. DOI 10.1158/1055-9965.EPI-13-1017. [Google Scholar] [CrossRef]
Dydensborg AB, Rose AA, Wilson BJ, Grote D, Paquet M, Giguere V, Siegel PM, Bouchard M (2009). GATA3 inhibits breast cancer growth and pulmonary breast cancer metastasis. Oncogene 28: 2634–2642. DOI 10.1038/onc.2009.126. [Google Scholar] [CrossRef]
Gade P, Kalvakolanu DV (2012). Chromatin immunoprecipitation assay as a tool for analyzing transcription factor activity. Methods in Molecular Biology 809: 85–104. DOI 10.1007/978-1-61779-376-9. [Google Scholar] [CrossRef]
Giordano CR, Mueller KL, Terlecky LJ, Krentz KA, Bollig-Fischer A, Terlecky SR, Boerner JL (2012). A targeted enzyme approach to sensitization of tyrosine kinase inhibitor-resistant breast cancer cells. Experimental Cell Research 318: 2014–2021. DOI 10.1016/j.yexcr.2012.06.001. [Google Scholar] [CrossRef]
Goldman MJ, Craft B, Hastie M, Repecka K, McDade F et al. (2020). Visualizing and interpreting cancer genomics data via the Xena platform. Nature Biotechnology 38: 675–678. DOI 10.1038/s41587-020-0546-8. [Google Scholar] [CrossRef]
Hirukawa A, Smith HW, Zuo D, Dufour CR, Savage P et al. (2018). Targeting EZH2 reactivates a breast cancer subtype-specific anti-metastatic transcriptional program. Nature Communications 9: 2547. DOI 10.1038/s41467-018-04864-8. [Google Scholar] [CrossRef]
Hsu TH, Chang TC (2015). RARRES3 regulates signal transduction through post-translational protein modifications. Molecular & Cellular Oncology 2: e999512. DOI 10.1080/23723556.2014.999512. [Google Scholar] [CrossRef]
Hsu TH, Jiang SY, Chang WL, Eckert RL, Scharadin TM, Chang TC (2015). Involvement of RARRES3 in the regulation of Wnt proteins acylation and signaling activities in human breast cancer cells. Cell Death and Differentiation 22: 1561. DOI 10.1038/cdd.2015.90. [Google Scholar] [CrossRef]
Khan MA, Jain VK, Rizwanullah M, Ahmad J, Jain K (2019). PI3K/AKT/mTOR pathway inhibitors in triple-negative breast cancer: A review on drug discovery and future challenges. Drug Discovery Today 24: 2181–2191. DOI 10.1016/j.drudis.2019.09.001. [Google Scholar] [CrossRef]
Liu Z, Dou C, Yao B, Xu M, Ding L et al. (2016). Ftx non coding RNA-derived miR-545 promotes cell proliferation by targeting RIG-I in hepatocellular carcinoma. Oncotarget 7: 25350–25365. DOI 10.18632/oncotarget.8129. [Google Scholar] [CrossRef]
Lu B, Wei J, Zhou H, Chen J, Li Y, Ye L, Zhao W, Wu S (2022). Histone H3K36me2 demethylase KDM2A promotes bladder cancer progression through epigenetically silencing RARRES3. Cell Death and Disease 13: 547. DOI 10.1038/s41419-022-04983-7. [Google Scholar] [CrossRef]
Lu Y, Qin Z, Wang J, Zheng X, Lu J et al. (2017). Epstein-barr virus miR-BART6-3p inhibits the RIG-I pathway. Journal of Innate Immunity 9: 574–586. DOI 10.1159/000479749. [Google Scholar] [CrossRef]
Lyko F (2018). The DNA methyltransferase family: A versatile toolkit for epigenetic regulation. Nature Reviews Genetics 19: 81–92. DOI 10.1038/nrg.2017.80. [Google Scholar] [CrossRef]
Man X, Li Q, Wang B, Zhang H, Zhang S, Li Z (2022). DNMT3A and DNMT3B in breast tumorigenesis and potential therapy. Frontiers in Cell and Developmental Biology 10: 916725. DOI 10.3389/fcell.2022.916725. [Google Scholar] [CrossRef]
Masalmeh RHA, Taglini F, Rubio-Ramon C, Musialik KI, Higham J et al. (2021). de novo DNA methyltransferase activity in colorectal cancer is directed towards H3K36me3 marked CpG Islands. Nature Communications 12: 694. DOI 10.1038/s41467-020-20716-w. [Google Scholar] [CrossRef]
Milioli HH, Tishchenko I, Riveros C, Berretta R, Moscato P (2017). Basal-like breast cancer: Molecular profiles, clinical features and survival outcomes. BMC Medical Genomics 10: 19. DOI 10.1186/s12920-017-0250-9. [Google Scholar] [CrossRef]
Morales M, Arenas EJ, Urosevic J, Guiu M, Fernandez E et al. (2014). RARRES3 suppresses breast cancer lung metastasis by regulating adhesion and differentiation. EMBO Molecular Medicine 6: 865–881. DOI 10.15252/emmm.201303675. [Google Scholar] [CrossRef]
Noyan S, Andac Ozketen A, Gurdal H, Gur Dedeoglu B (2021). miR-770-5p regulates EMT and invasion in TNBC cells by targeting DNMT3A. Cellular Signalling 83: 109996. DOI 10.1016/j.cellsig.2021.109996. [Google Scholar] [CrossRef]
Pang Y, Liu J, Li X, Xiao G, Wang H et al. (2018). MYC and DNMT3A-mediated DNA methylation represses microRNA-200b in triple negative breast cancer. Journal of Cellular and Molecular Medicine 22: 6262–6274. DOI 10.1111/jcmm.13916. [Google Scholar] [CrossRef]
Parker JS, Mullins M, Cheang MC, Leung S, Voduc D et al. (2009). Supervised risk predictor of breast cancer based on intrinsic subtypes. Journal of Clinical Oncology 27: 1160–1167. DOI 10.1200/JCO.2008.18.1370. [Google Scholar] [CrossRef]
Petrovic I, Milivojevic M, Arsenijevic A, Lazic A, Grujicic N-K, Schwirtlich M, Popovic J, Stevanovic M (2021). Retinoic acid affects basic cellular processes and SOX2 and SOX18 expression in breast carcinoma cells. BIOCELL 45: 1355–1367. DOI 10.32604/biocell.2021.015817. [Google Scholar] [CrossRef]
Rody A, Karn T, Liedtke C, Pusztai L, Ruckhaeberle E et al. (2011). A clinically relevant gene signature in triple negative and basal-like breast cancer. Breast Cancer Research 13: R97. DOI 10.1186/bcr3035. [Google Scholar] [CrossRef]
Russnes HG, Lingjaerde OC, Borresen-Dale AL, Caldas C (2017). Breast cancer molecular stratification: From intrinsic subtypes to integrative clusters. American Journal of Pathology 187: 2152–2162. DOI 10.1016/j.ajpath.2017.04.022. [Google Scholar] [CrossRef]
Tang Y, Wang T, Yu Y, Yan Y, Wu C (2021). Upregulation of HOXC9 generates interferon-gamma resistance in gastric cancer by inhibiting the DAPK1/RIG1/STAT1 axis. Cancer Science 112: 3455–3468. DOI 10.1111/cas.15043. [Google Scholar] [CrossRef]
Tian F, Hu H, Wang D, Ding H, Chi Q, Liang H, Zeng W (2022). Immune-related DNA methylation signature associated with APLN expression predicts prognostic of hepatocellular carcinoma. BIOCELL 46: 2291–2301. DOI 10.32604/biocell.2022.020198. [Google Scholar] [CrossRef]
Turashvili G, Brogi E (2017). Tumor heterogeneity in breast cancer. Frontiers in Medicine 4: 227. DOI 10.3389/fmed.2017.00227. [Google Scholar] [CrossRef]
Umemura S, Yoshida S, Ohta Y, Naito K, Osamura RY, Tokuda Y (2007). Increased phosphorylation of Akt in triple-negative breast cancers. Cancer Science 98: 1889–1892. DOI 10.1111/j.1349-7006.2007.00622.x. [Google Scholar] [CrossRef]
Wang L, Luo Z, Sun M, Yuan Q, Zou Y, Fu D (2022). Identification of a three-gene signature in the triple-negative breast cancer. BIOCELL 46: 595–606. DOI 10.32604/biocell.2022.017337. [Google Scholar] [CrossRef]
Wei L, Chiu DK, Tsang FH, Law CT, Cheng CL, Au SL, Lee JM, Wong CC, Ng IO, Wong CM (2017). Histone methyltransferase G9a promotes liver cancer development by epigenetic silencing of tumor suppressor gene RARRES3. Journal of Hepatology 67: 758–769. DOI 10.1016/j.jhep.2017.05.015. [Google Scholar] [CrossRef]
Wei X, Gu X, Ma M, Lou C (2019). Long noncoding RNA HCP5 suppresses skin cutaneous melanoma development by regulating RARRES3 gene expression via sponging miR-12. OncoTargets and Therapy 12: 6323–6335. DOI 10.2147/OTT. [Google Scholar] [CrossRef]
Wu C, Tuo Y, Hu G, Luo J (2021). miR-183-5p aggravates breast cancer development via mediation of RGS2. Computational and Mathematical Methods in Medicine 2021: 9664195. DOI 10.1155/2021/9664195. [Google Scholar] [CrossRef]
Wu H, Coskun V, Tao J, Xie W, Ge W, Yoshikawa K, Li E, Zhang Y, Sun YE (2010). Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science 329: 444–448. DOI 10.1126/science.1190485. [Google Scholar] [CrossRef]
Yu J, Qin B, Moyer AM, Nowsheen S, Liu T et al. (2018). DNA methyltransferase expression in triple-negative breast cancer predicts sensitivity to decitabine. Journal of Clinical Investigation 128: 2376–2388. DOI 10.1172/JCI97924. [Google Scholar] [CrossRef]
Yu S, Jiang X, Li J, Li C, Guo M, Ye F, Zhang M, Jiao Y, Guo B (2019). Comprehensive analysis of the GATA transcription factor gene family in breast carcinoma using gene microarrays, online databases and integrated bioinformatics. Scientific Reports 9: 4467. DOI 10.1038/s41598-019-40811-3. [Google Scholar] [CrossRef]
Yu Z, Xiao Q, Zhao L, Ren J, Bai X et al. (2015). DNA methyltransferase 1/3a overexpression in sporadic breast cancer is associated with reduced expression of estrogen receptor-alpha/breast cancer susceptibility gene 1 and poor prognosis. Molecular Carcinogenesis 54: 707–719. DOI 10.1002/mc.22133. [Google Scholar] [CrossRef]
Zhang W, Xia D, Li Z, Zhou T, Chen T, Wu Z, Zhou W, Li Z, Li L, Xu J (2019). Aurora-A/ERK1/2/mTOR axis promotes tumor progression in triple-negative breast cancer and dual-targeting Aurora-A/mTOR shows synthetic lethality. Cell Death and Disease 10: 606. DOI 10.1038/s41419-019-1855-z. [Google Scholar] [CrossRef]
Zhou Y, Yang Z, Zhang H, Li H, Zhang M, Wang H, Zhang M, Qiu P, Zhang R, Liu J (2022). DNMT3A facilitates colorectal cancer progression via regulating DAB2IP mediated MEK/ERK activation. Biochimica et Biophysica Acta (BBA)-Molecular Basis of Disease 1868: 166353. DOI 10.1016/j.bbadis.2022.166353. [Google Scholar] [CrossRef]
Supplementary Materials

Figure S1: The promoter segments of RARRES3
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools