 Open Access
Open Access
ARTICLE
Correlations among core species corresponding to the clinical staging of periodontitis
1 Central Laboratory, Peking University School and Hospital of Stomatology, Beijing Key Laboratory of Digital Stomatology, National Center of Stomatology, National Clinical Research Center for Oral Diseases, National Engineering Research Center of Oral Biomaterials and Digital Medical Devices, Beijing, China
2 Department of Periodontology, Peking University School and Hospital of Stomatology, Beijing, China
3 Second Clinical Division, Peking University School and Hospital of Stomatology, Beijing, China
4 Department of Geriatric Dentistry, Peking University School and Hospital of Stomatology, Beijing, China
* Corresponding Authors: XIAOPEI CHI. Email: ; YIFEI ZHANG. Email:
# These authors have contributed equally to this work
BIOCELL 2023, 47(2), 431-439. https://doi.org/10.32604/biocell.2023.024948
Received 15 June 2022; Accepted 08 August 2022; Issue published 18 November 2022
Abstract
The correlation between microbiota plays a vital role in the progression of periodontal disease. This study investigated the in situ interaction networks between periodontal pathogens in periodontal and peri-implant disease. We used quantitative real-time polymerase chain reaction and Pearson’s correlation coefficients to quantify the copy numbers and correlations of four oral core species—Fusobacterium nucleatum, Porphyromonas gingivalis, Prevotella intermedia, and Streptococcus gordonii—from 80 subgingival sites (healthy and with periodontitis or gingivitis) in patients with periodontitis, and 68 subgingival sites (healthy and with periodontitis, gingivitis, peri-implantitis, or peri-implant mucositis) in patients with implants. The highest bacterial counts were observed for Porphyromonas gingivalis and Prevotella intermedia at all the sites. Within the same cohorts, the bacterial loads were greater at diseased sites than at healthy sites. Bacterial counts did not differ among clinical sites in the same group (P > 0.05) but differed between periodontitis and peri-implant mucositis sites in the two groups. Porphyromonas gingivalis, F. nucleatum, and Prevotella intermedia had strong correlations at gingivitis and healthy sites and moderate correlations at periodontitis sites in patients with periodontitis. In patients with implants, Prevotella intermedia, F. nucleatum, and S. gordonii had strong correlations only at peri-implantitis sites. Also, based on metagenomic analysis, F. nucleatum and Prevotella intermedia were significantly correlated at the subgingival plaque in peri-implantitis and periodontitis samples. Our results suggest that variations in microbe-microbe interactions in subgingival plaque reflect changes in the progression of periodontal disease, providing a new perspective for understanding the mechanisms of periodontitis and peri-implantitis.Keywords
Periodontal diseases include a range of conditions, from gingivitis to periodontitis, and gingivitis precedes periodontitis. If not treated properly, they eventually lead to tooth loss and even increase the risk of systemic problems (Slots, 2017). Implant-supported reconstruction is an ideal treatment for dental defects caused by severe periodontitis. While if there is no effective maintenance, peri-implant mucositis and peri-implantitis may occur. In peri-implant disease, collagen fibers are non-attached and parallel to the implant surface instead of being perpendicularly arranged from bone to cementum in periodontal disease (Meffert, 1996). Meanwhile, peri-implantitis has a more severe loss of bone than periodontitis because of the different microbiomes and higher microbial diversity in implant sites (Rokaya et al., 2020). All these dental conditions are biofilm-mediated diseases. Oral biofilms are sophisticated structures created by the sequential and ordered interplay of multiple oral bacteria (Kolenbrander et al., 2002). They are recognized as etiologic agents in caries or periodontal/peri-implant disease, which are a result of dysbiosis among plaque biofilm, host, and local microenvironment. Meanwhile, to maintain host-bacteria homeostasis, there is a need for a comprehensive understanding of interactions among microbial species and how they interact to initiate disease.
In one clinical study, Ritz (1967) reported that the dental plaque/biofilm formation due to the deposition of salivary proteins on the gum line, followed by accumulation of Streptococcal species, which covered nearly 90% of the dental biofilm after 2-day formation. As the plaque formation continues, Gram-positive aerobic Streptococcal species mix Gram-positive and Gram-negative anaerobic mcirobiota, including streptococci sp., Actinomyces sp., Veillonella sp., Fusobacteria sp., etc. (Moore et al., 1987). Some research focused only on cataloging the differences in bacterial community members among the healthy and those having gingivitis and periodontitis (Lourenço et al., 2014; Park et al., 2015; Lu et al., 2021). Although the correlation between changes in the structure and function of microbial communities in plaque biofilm and periodontal conditions are strongly implicated, these analyses overlooked that gingivitis and periodontitis are not independent but successive stages of the same disease process (Jeffcoat and Reddy, 1991; Shaw et al., 2016). Later, Nowicki et al. (2018) analyzed the transition of dental conditions from healthy to having gingivitis and identified genes associated with early disease. Nemoto et al. (2021) described a health-to-periodontitis microbiome shift and identified core taxa associated with periodontal disease progression, and Huang et al. (2021) characterized temporal dynamics of the microbiome in gingivitis-to-periodontitis transition. Yu et al. (2019) reported that the composition of the submucosal and subgingival microbiota at periodontitis and peri-implantitis sites was similar in subjects in the same cohort. Nevertheless, these studies did not compare patterns of the bacterial interplay between the periodontal and peri-implant habitats, and the information gap between how the microbial interactions change during disease progression still remains.
To understand whether gingivitis and periodontitis represent a continuum, it would be of interest to evaluate the presence and levels of periodontitis-associated species in gingivitis and health and vice versa. Above all, the conditions involve interaction among microbiota rather than a single bacterium. The species associated with health appear to be species with demonstrated roles during early biofilm colonization in disease sites, such as Streptococcus spp. (Abusleme et al., 2021). Streptococcus gordonii is a commensal bacteria of the oral cavity and pioneer colonizer during the formation of the dental plaque/biofilm (Nairn et al., 2020), which are also critical microbiota related to biofilm formation. Fusobacterium nucleatum, the “bridge” in the oral biofilm development due to its ability to co-aggregate with representatives of all initial and late colonizers (Kolenbrander et al., 2010), was identified as one of the “gingivitis-driver” species in a gingivitis model (Huang et al., 2014), in addition to over-expression of virulence-related genes (Nowicki et al., 2018). Porphyromonas gingivalis, and Prevotella intermedia, as late colonizers, are associated with periodontitis and peri-implantitis (Colombo and Tanner, 2019). So, the interactive relationship between microbiota during biofilm formation plays an important role in disease progression.
In this study, we collected subgingival plaque samples from periodontal healthy, gingivitis, and periodontitis sites in the same cohort and healthy peri-implant, peri-mucositis, and peri-implantitis sites in peri-implantitis patients, representing different stages of periodontal disease progression. Also, we selected S. gordonii, Prevotella intermedia, F. nucleatum, and Porphyromonas gingivalis as representative of different stages of subgingival plaque formation (Kolenbrander et al., 2010) to assess their correlation to disease progression.
Subject enrollment and sampling
Patients with chronic periodontitis were recruited from the Department of Periodontology, Peking University School and Hospital of Stomatology, from December 2013 to May 2015 (Zhang et al., 2017). The inclusion criteria were: good general health and no pregnancy, age 18–65 years (male or female), no antibiotic use in the past three months, and no periodontal therapy in the past year. Subjects who had any systemic condition that could affect the progress of periodontal disease were excluded.
Thirty-four patients with peri-implantitis were recruited from The Second Dental Center, Peking University School, and Hospital of Stomatology, between 2014 and 2017. The inclusion criteria were: good general health and no pregnancy, age 18–65 years, and partial edentulism due to severe periodontitis. Each patient had received at least one dental implant (Straumann, Basel, Switzerland) and one year of systemic periodontal treatment. The subjects had no other oral or systemic disease and had not taken antibiotics for at least three months before sampling.
Two professional dentists examined the periodontal and peri-implant conditions of the participants. Inclusion criteria of gingivitis and peri-implant mucositis were made based on the occurrence of bleeding and the absence of evidence of bone loss. Inclusion criteria of periodontitis and peri-implantitis were made based on the presence of bleeding and/or suppuration on gentle probing, PPD ≥ 5 mm, and attachment loss >5 mm (Berglundh et al., 2018). Diagnostic criteria for the healthy conditions and healthy implants were the absence of inflammation, bleeding, and suppuration on gentle probing, the absence of evidence of bone loss, and the absence of an increase in PPD compared with the previous examination. Eighty subgingival sites (healthy and with periodontitis or gingivitis) in patients with periodontitis and 68 subgingival sites (healthy and with periodontitis, gingivitis, peri-implantitis, or peri-implant mucositis) in patients with implants were collected. The institutional review board of Peking University School and Hospital of Stomatology (Beijing, China) approved the study protocol (No. PKUSSIRB-2012063).
All patients were untreated with periodontal therapy. Before sampling, the patients rinsed their mouths with purified water, and then supragingival plaque and saliva around the sampling position (teeth and implants) were removed with sterile cotton pellets. The subgingival plaque in periodontal pockets was collected with sterile paper points for 10 s. Samples were subsequently placed in 1.5-mL centrifuge tubes with 1 mL TE buffer (20 mM Tris and 2 mM ethylenediaminetetraacetic acid; pH 7.4), transferred to a central laboratory, and frozen at −80°C.
Bacterial genomic DNA was extracted using QIAamp DNA mini kits (Qiagen, Valencia, CA, USA) according to the manufacturer’s protocol. The quality and concentration were tested using the Nanodorp8000 device (Thermo Fisher Scientific, Wilmington, DE, USA). High-quality DNA with an optical density (OD)260/280 ratio of 1.8–2.0 and concentration ≥10 ng/μL was used for further analysis.
Bacterial loads of core species
The bacterial loads of four core species (Porphyromonas gingivalis W83, F. nucleatum ATCC 25586, Prevotella intermedia ATCC 15033, and S. gordonii ATCC 10558) in plaque were determined by quantitative polymerase chain reaction (qPCR). Porphyromonas gingivalis, F. nucleatum, and Prevotella intermedia were cultured in a brain heart infusion medium supplemented with hemin (5 mg/L) and vitamin K (1 mg/mL) at 37°C under anaerobic conditions, and S. gordonii was cultured in a brain heart infusion medium at 37°C in an atmosphere of 5% CO2 and 95% air. Genomic DNA was extracted from the four selected standard strains to quantify the bacterial loads in clinical samples. Then, the standard quantified DNA was subjected to serial tenfold dilution from 10 ng to 10 fg for the plotting of standard curves. The clinical DNA samples and standard bacterial DNA were used as templates for PCR in a reaction volume of 20 μL containing 10 μL Power SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA), 0.5 μL of each primer (10 nM), and 2 μL DNA template. The amplification conditions were 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, and at the four species-specific annealing temperatures for 60 s. Amplification was performed on an ABI QuantStudio real-time PCR system (Thermo Fisher Scientific). The four species-specific primers are listed in Table 1 (Zhuang et al., 2016; Maeda et al., 2003; Suzuki et al., 2004; Park and Kook, 2013 ). All samples were run in triplicate.

Correlation analysis of four species based on metagenomic sequencing
We selected three metagenome data (PRJNA552294, PRJNA230363, PRJDB6966) derived from the subgingival samples collected from periodontitis patients, healthy people, and peri-implantitis patients based on whole genome sequencing (WGS). The first two data were obtained after sequencing on the ILLUMINA Hiseq platform, and the last data (PRJDB6966) was obtained after sequencing on the ILLUMINA Miseq platform. All the reads were subject to quality control as follows: containing less than 3% N bases and more than 50% bases with high quality (>3). The high-quality reads mapped to the human genome (hg19) were removed using the Kneaddata pipeline (http://huttenhower.sph.harvard.edu/kneaddata) with default parameters. Then, all the clean reads were aligned to the NCBI bacteria and Human Oral Microbiome Database using Kraken2 (Wood et al., 2019) with default parameters. The correlation analysis was visualized using Spearman software. P < 0.05 was considered statistically significant.
The bacterial copy numbers were log-10 -transformed for analysis. Statistical analyses were performed using SPSS software Version 25 (SPSS Inc., Chicago, IL, USA), and P < 0.05 was taken to indicate significance. Correlations were assessed using Pearson’s correlation coefficient and the Kruskal–Wallis rank-sum test. The strength of correlation between two bacteria was classified using r-values (0–0.19, very weak; 0.2–0.39, weak; 0.40–0.59, moderate; 0.6–0.79, strong; 0.8–1, very strong) (Ruengsomwong et al., 2016).
Bacterial loads of the four core species at different sites
A total of 80 plaque samples were collected from distinct subgingival sites (58 with periodontitis, 16 with gingivitis, and six healthy) in 43 patients with periodontitis, and 68 plaque samples were collected from distinct subgingival/submucosal sites (27 with periodontitis, seven with gingivitis, five with peri-implantitis, 20 with peri-implant mucositis, and nine with healthy implants) in patients with peri-implantitis.
The log-10–transformed counts of the four target species at each clinical sampling site from the two cohorts (cohort 1, patients with periodontitis; cohort 2, patients with peri-implantitis) are illustrated in Fig. 1. Also, total bacterial loads did not differ within or between the samples from the two cohorts (data not shown). With regard to each target species, no significant difference in the bacterial count was detected within or among different clinical sites in the same cohort.

Figure 1: Bacterial loads of single species in the different clinical sites from patients with periodontitis and peri-implantitis. These findings were based on the qPCR results. All the tests were conducted in triplicate and the species were tested by the Kruskal-Wallis rank sum test. A significant difference was set at P < 0.05 with “*” marked. The black bold boxes represent sites from patients with periodontitis, and the gray boxes represent sites from patients with peri-implantitis.
The bacterial load of Prevotella intermedia was similar in all samples. The bacterial load of Porphyromonas gingivalis was significantly lower at healthy tooth sites in cohort 1 than at sites of periodontitis and peri-implant mucositis in cohort 2. The bacterial load of S. gordonii differed significantly between periodontitis sites in cohort 1 and periodontitis and peri-implant mucositis sites in cohort 2. The same difference was also observed between healthy sites in cohort 1 and peri-implant mucositis sites in cohort 2. The bacterial load of F. nucleatum was greater at gingivitis and periodontitis sites in cohort 1 than that at periodontitis sites in cohort 2. In addition, the F. nucleatum bacterial load was greater at healthy sites in subjects with periodontitis than at periodontitis, gingivitis, and peri-implant mucositis sites in cohort 2.
Bacterial loads of the four core species at the same sites in subjects with periodontitis and those with peri-implantitis
In the same cohorts, the bacterial loads were greater at diseased sites than at healthy sites, but the differences were not significant. At periodontitis sites, the log-10–transformed bacterial loads of F. nucleatum and S. gordonii were significantly greater in subjects with periodontitis than in subjects with implants. Prevotella intermedia showed the greatest bacterial load at all sampling sites, and S. gordonii had the smallest bacterial load, except at peri-implantitis sites in cohort 2.
At peri-implantitis and healthy implant sites from cohort 2, loads of Prevotella intermedia and S. gordonii differed significantly. At periodontitis and gingivitis sites in the two cohorts, loads of these bacteria, but not those of F. nucleatum and Porphyromonas gingivalis, differed significantly. The results also revealed a significant difference for S. gordonii relative to the other three bacteria at peri-implant mucositis sites. At healthy sites in cohort 1, differences were significant for Prevotella intermedia and Porphyromonas gingivalis/S. gordonii (Fig. 2).

Figure 2: Bacterial loads of the four core species in the same clinical sites from patients with periodontitis and peri-implantitis. The results were based on the quantitative real-time polymerase chain reaction. All the dates were conducted in triplicate and tested by the Kruskal-Wallis rank sum test. A significant difference was set at P < 0.05 with “*” marked. The colors red, yellow, green, and blue represent Porphyromonas gingivalis, Streptococcus gordonii, Prevotella intermedia, and Fusobacterium nucleatum. (a) Sites from patients with periodontitis and (b) sites from patients with peri-implantitis.
Correlations among the four core species in different sites from peri-implantitis and periodontitis patients
According to the abundance analysis, we observed a positively correlated network among Porphyromonas gingivalis, F. nucleatum, and Prevotella intermedia in the subgingival microbiota at clinically healthy (unaffected) and diseased periodontal sites in cohort 1. A weak positive correlation was found between S. gordonii and F. nucleatum at periodontitis sites. In cohort 2, F. nucleatum and Prevotella intermedia showed a strong correlation at peri-implant mucositis, peri-implantitis, and healthy implant sites; a moderate correlation was detected between F. nucleatum and Porphyromonas gingivalis/Prevotella intermedia at periodontitis sites. A positive correlation network among S. gordonii, F. nucleatum, and Prevotella intermedia was established only in the peri-implantitis sample group. In the peri-implant healthy and peri-implant mucositis groups, no correlation was found between S. gordonii and F. nucleatum/Prevotella intermedia (Fig. 3).

Figure 3: Correlations among the four core species at different sites in periodontitis and peri-implantitis samples. Correlations were assessed using Pearson’s correlation coefficient and the Kruskal–Wallis rank-sum test based on the results of quantitative real-time polymerase chain reaction. The strength of correlation between two bacteria was classified using r-values (0–0.19, very weak; 0.2–0.39, weak; 0.40–0.59, moderate; 0.6–0.79, strong; 0.8–1, very strong). The clinical collected samples were from healthy, gingivitis, and periodontitis sites from periodontitis patients and gingivitis, periodontitis, healthy implant, peri-mucositis, and peri-implantitis sites from peri-implantitis patients. The different types of lines represent different correlations.
Correlation analysis among the four core species based on metagenomic data
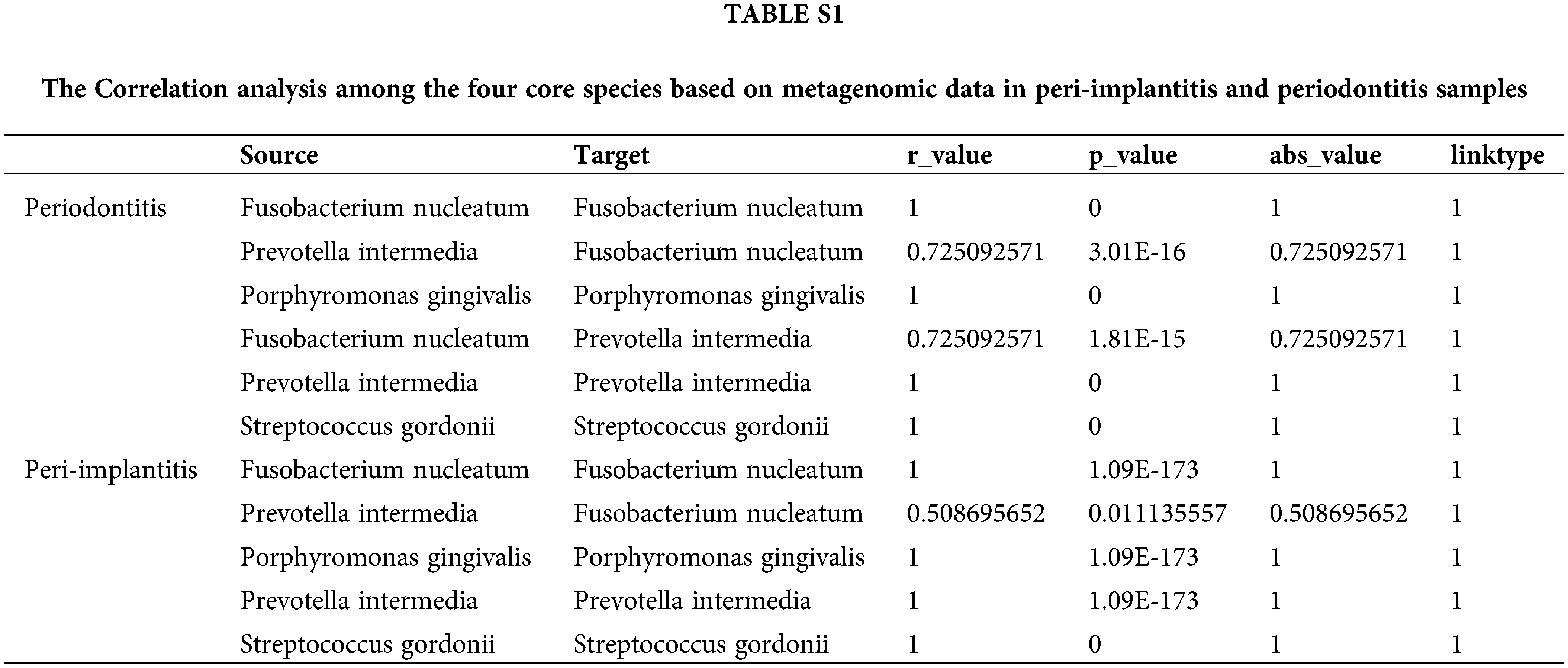
Also, we downloaded data from 92 cases of periodontitis and 24 peri-implantitis to conduct metagenomic analysis; the results showed a significant correlation between F. nucleatum and Prevotella intermedia at the subgingival plaque in peri-implantitis and periodontitis samples (Supple. Table S1).
Many studies have shown that the bacterial community composition and structure in subgingival plaque differ in patients with periodontal disease and healthy individuals (Griffen et al., 2012; Wang et al., 2013; Shi et al., 2015). Certain species have been found to be more frequent and abundant at diseased sites (Gohler et al., 2018; Mullally et al., 2000; Puig-Silla et al., 2017). Prevotella intermedia is one of these bacteria. However, in this study, we found no difference in the frequency or abundance of Prevotella intermedia between affected and unaffected sites or between generalized and localized periodontitis. Our results also revealed that under the same oral conditions (generalized and locally recurrent periodontitis), the affected and unaffected sites tended to have similar bacterial loads. However, patients with local periodontitis had significantly lower bacterial loads than the patients with general chronic periodontitis.
As bacterial interactions can influence biofilm formation, metabolic changes, and physiological function, we hypothesized that variation therein would play an important role in oral health status. In our previous study, we concluded that microbial interactions among key species (F. nucleatum, Porphyromonas gingivalis and Prevotella intermedia) have value in managing the subgingival microbial ecosystem through an in vitro biofilm model (Zhang et al., 2019), and now we further confirmed it through in situ plaque samples. Network analysis has been used to identify interactions among microbiota (i.e., Pearson or Spearman correlation) based on their absolute abundances (Faust and Raes, 2012; Barberan et al., 2012). The positive correlation among Porphyromonas gingivalis, F. nucleatum, and Prevotella intermedia may be due partly to pairwise co-aggregations (Faust and Raes, 2012). Strong (positive or negative) correlations are assumed to have biological, physiological, or ecological significance, possibly resulting from cooperation or competition (Fernandez et al., 2015). The correlation coefficients decreased with the aggravation of periodontal inflammation (healthy–gingivitis–periodontitis). However, only the differences in Porphyromonas gingivalis/F. nucleatum correlation coefficients between healthy and diseased (gingivitis or periodontitis) sample groups were of statistical significance, suggesting that the interplay of Porphyromonas gingivalis and F. nucleatum in the subgingival microbiota is affected during the progression of the disease. Although S. gordonii can also co-aggregate with F. nucleatum and Porphyromonas gingivalis (Park et al., 2005), the correlation between S. gordonii and F. nucleatum was much weaker, and no correlation was detected between S. gordonii and Porphyromonas gingivalis. These observations suggest that the microbial correlations cannot be explained by mere physicochemical colonization but that cell-cell contact contributes to functional interaction (e.g., metabolic communication and/or genetic exchange) (Guo et al., 2014). The variation in microbial correlation may reflect changes in community function. The network structure was consistent at periodontal healthy and diseased sites, i.e., no correlated pair emerged or was lost, and no positive/negative relation changed to the opposite in association with the periodontal condition.
Implant-supported reconstruction is an ideal treatment for dental defects caused by severe periodontitis. However, significantly increased risks of biological complications around dental implants (i.e., peri-implant mucositis and peri-implantitis) have been reported in patients with histories of periodontitis (Sgolastra et al., 2015; Ting et al., 2018). Peri-implant and periodontal diseases have been reported to have different microbial profiles. F. nucleatum and Prevotella intermedia have been reported to be core bacterial species associated with periodontitis and peri-implantitis (Colombo and Tanner, 2019). Therefore, we then analyzed microbial relationships in the subgingival microbiota at healthy and diseased peri-implant sites in a cohort of patients with histories of periodontitis. Our results suggest that they are also related closely to periodontitis and peri-implantitis.
The pairwise interplay of F. nucleatum/Prevotella intermedia and F. nucleatum/Porphyromonas gingivalis at periodontitis sites were consistent in the two cohorts, whereas no correlation between Prevotella intermedia and Porphyromonas gingivalis was found in subjects with implants. Patients in the periodontitis group had generalized active periodontitis, whereas those with peri-implantitis were in the maintenance phase after periodontal therapy with localized recurrence. Thus, the subgingival ecosystem may have differed between cohorts. Implants are thought to accumulate less plaque than teeth, whereas the gingivitis microbiome is more diverse than that seen in peri-implant mucositis (Schincaglia et al., 2017). The relative spatial distribution of Porphyromonas gingivalis and Prevotella intermedia was random (Schillinger et al., 2012), which may explain their dynamic relationship. To determine whether the difference in microbial interplay was affected by variation in bacterial abundance, we compared the absolute abundance of each species between groups and observed no significant change in the abundance of any one of these four species. These results confirm that the changes in interspecies correlations were associated with changes in the microbial ecosystem.
Recent studies have suggested that the microbial relationships shown in correlation interaction networks can be used to determine drivers of disease (Greenblum et al., 2012; Faust and Raes, 2012). To better understand interactions in the human microbiome, most studies have attempted to construct correlation networks with sequencing data. The reads are classified based on similarity to generate profiles (taxonomic or functional) of different microbial samples. This approach is useful for the description of the “social” nature of a microbiome, but the snapshots it provides make little contribution to the description of interactions between community members, and a large amount of metagenomics data renders the recovery of real relationships in the bacterial community difficult (Weiss et al., 2016). Yu et al. (2019) constructed a bacterial occurrence network for periodontal and peri-implant microbiota based on 16S metagenomic sequencing. We found little robust agreement between their results and ours, as the correlations in their network were not sufficiently precise. Although we could not obtain a complete picture of the complex interactions that occur in the periodontal and peri-implant microbiota, our findings add to the above-mentioned results by suggesting the biological relevance of interactions between specific core members of the microbiota. Furthermore, the patterns of the strength of interactions between specific bacterial species are associated with different habitats (periodontal and peri-implant, diseased and healthy); even when the same species are present at similar abundance levels in different habitats, their behavior is not necessarily the same. This difference may be due to differences in the local environment, but it could also reflect differences in the presence or absence of other microorganisms.
There were some limitations in our study. Microbiota, as a whole, interact with each other to influence the progress of the oral disease, including periodontitis and peri-implantitis. We only studied four main pathogenic bacteria; other pathogenic bacteria are also present, for example Treponema denticola and Aggregatibacter actinomycetemcomitans. Other verified samples should be collected, and the clinically isolated strains should be used for in vitro experiments to confirm this conclusion. The influence of bacteria on the progression of periodontal and implant disease in the same patients should be examined to eliminate the differences between individuals.
In summary, the pattern of the correlation network for four core species differed between different periodontal/peri-implant habitats, supporting the hypothesis that the variation in microbiota is relevant to health status. Further studies are required to understand the implications of these interactions and why these differences exist.
Availability of Data: The datasets generated analyzed during the current study are available from the corresponding author on reasonable request.
Author Contribution: Study conceived and design of the experiments: ZYF, CXP; collection of the samples, experimentation and analysis of data: ZQ, ZYF, ZM, LP; manuscript writing and revision: ZQ, ZYF, ZM, CXP. All authors reviewed the results and approved the final draft of the manuscript.
Ethics Approval: The institutional review board of Peking University School and Hospital of Stomatology (Beijing, China) approved the study protocol (No. PKUSSIRB-2012063).
Funding Statement: This study was supported by grants from the Peking University School of Stomatology (PKUSS20170112).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
Abusleme L, Hoare A, Hong BY, Diaz PI (2021). Microbial signatures of health, gingivitis, and periodontitis. Periodontology 2000 86: 57–78. DOI 10.1111/prd.12362. [Google Scholar] [CrossRef]
Barberan A, Bates ST, Casamayor EO, Fierer N (2012). Using network analysis to explore co-occurrence patterns in soil microbial communities. The ISME Journal 6: 343–351. DOI 10.1038/ismej.2011.119. [Google Scholar] [CrossRef]
Berglundh T, Armitage G, Araujo MG, Avila-Ortiz G, Blanco J, Camargo PM (2018). Peri-implant diseases and conditions: Consensus report of workgroup 4 of the 2017 world workshop on the classification of periodontal and peri-implant diseases and conditions. Journal of Clinical Periodontology 45: S286–S291. DOI 10.1111/jcpe.12957. [Google Scholar] [CrossRef]
Colombo APV, Tanner ACR (2019). The role of bacterial biofilms in dental caries and periodontal and peri-implant diseases: A historical perspective. Journal of Dental Research 98: 373–385. DOI 10.1177/0022034519830686. [Google Scholar] [CrossRef]
Faust K, Raes J (2012). Microbial interactions: From networks to models. Nature Reviews Microbiology 10: 538–550. DOI 10.1038/nrmicro2832. [Google Scholar] [CrossRef]
Fernandez M, Riveros JD, Campos M, Mathee K, Narasimhan G (2015). Microbial “social networks”. BMC Genomics 16: S6. DOI 10.1186/1471-2164-16-S11-S6. [Google Scholar] [CrossRef]
Guo L, He X, Shi W (2014). Intercellular communications in multispecies oral microbial communities. Frontiers in Microbiology 5: 328. DOI 10.3389/fmicb.2014.00328. [Google Scholar] [CrossRef]
Griffen AL, Beall CJ, Campbell JH, Firestone ND, Kumar PS, Yang ZK, Podar M, Leys EJ (2012). Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME Journal 6: 1176–1185. DOI 10.1038/ismej.2011.191. [Google Scholar] [CrossRef]
Gohler A, Samietz S, Schmidt CO, Kocher T, Steinmetz I, Holtfreter B (2018). Comparison of oral microbe quantities from tongue samples and subgingival pockets. International Dental Journal 2018: 2048390. DOI 10.1155/2018/2048390. [Google Scholar] [CrossRef]
Greenblum S, Turnbaugh PJ, Borenstein E (2012). Metagenomic systems biology of the human gut microbiome reveals topological shifts associated with obesity and inflammatory bowel disease. PNAS 109: 594–599. DOI 10.1073/pnas.1116053109. [Google Scholar] [CrossRef]
Huang S, He T, Yue F, Xu XJ, Wang LJ et al. (2021). Longitudinal multi-omics and microbiome meta-analysis identify an asymptomatic gingival state that links gingivitis, periodontitis, and aging. mBio 12: e03281-20. DOI 10.1128/mBio.03281-20. [Google Scholar] [CrossRef]
Huang S, Li R, Zeng X, He T, Zhao H et al. (2014). Predictive modeling of gingivitis severity and susceptibility via oral microbiota. ISME Journal 8: 1768–1780. DOI 10.1038/ismej.2014.32. [Google Scholar] [CrossRef]
Jeffcoat MK, Reddy MS (1991). Progression of probing attachment loss in adult periodontitis. Journal of Periodontology 62: 185–189. DOI 10.1902/jop.1991.62.3.185. [Google Scholar] [CrossRef]
Kolenbrander PE, Andersen RN, Blehert DS, Egland PG, Foster JS, Palmer RJJr (2002). Communication among oral bacteria. Microbiology and Molecular Biology Reviews 66: 486–505. DOI 10.1128/MMBR.66.3.486-505.2002. [Google Scholar] [CrossRef]
Kolenbrander PE, Jr Palmer RJ, Periasamy S, Jakubovics NS (2010). Oral multispecies biofilm development and the key role of cell-cell distance. Nature Reviews Microbiology 8: 471–480. DOI 10.1038/nrmicro2381. [Google Scholar] [CrossRef]
Lu C, Chu Y, Liu JR, Liu WY, Ouyang XY (2021). Subgingival microbial profiles of young Chinese adults with stage I/II periodontitis, gingivitis and periodontal health status. Chinese Journal of Dental Research 24: 167–175. DOI 10.3290/j.cjdr.b1965003. [Google Scholar] [CrossRef]
Lourenço TG, Heller D, Silva-Boghossian CM, Cotton SL, Paster BJ, Colombo APV (2014). Microbial signature profiles of periodontally healthy and diseased patients. Journal of Clinical Periodontology 41: 1027–1036. DOI 10.1111/jcpe.12302. [Google Scholar] [CrossRef]
Maeda H, Fujimoto C, Haruki Y, Maeda T, Kokeguchi S, Petelin M, Arai H, Tanimoto I, Nishimura F, Takashiba S (2003). Quantitative real-time PCR using TaqMan and SYBR Green for Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis, Prevotella intermedia, tetQ gene and total bacteria. FEMS Immunology and Medical Microbiology 39: 81–86. DOI 10.1016/S0928-8244(03)00224-4. [Google Scholar] [CrossRef]
Meffert RM (1996). Periodontitis vs. peri-implantitis: The same disease? The same treatment? Critical Reviews in Oral Biology Medicine 7: 278–291. DOI 10.1177/10454411960070030501. [Google Scholar] [CrossRef]
Moore LV, Moore WE, Cato EP, Smibert RM, Burmeister JA, Best AM, Ranney RR (1987). Bacteriology of human gingivitis. Journal of Dental Research 66: 989–995. DOI 10.1177/00220345870660052401. [Google Scholar] [CrossRef]
Mullally BH, Dace B, Shelburne CE, Wolff LF, Coulter WA (2000). Prevalence of periodontal pathogens in localized and generalized forms of early-onset periodontitis. Journal of Periodontal Research 35: 232–241. DOI 10.1034/j.1600-0765.2000.035004232.x. [Google Scholar] [CrossRef]
Nairn BL, Lee GT, Chumber AK, Steck PR, Mire MO, Lima BP, Herzberg MC, Federle MJ (2020). Uncovering roles of Streptococcus gordonii SrtA-Processed proteins in the biofilm lifestyle. Journal of Bacteriology 203: e00544-20. DOI 10.1128/JB.00544-20. [Google Scholar] [CrossRef]
Nowicki EM, Shroff R, Singleton JA, Renaud DE, Wallace D et al. (2018). Microbiota and metatranscriptome changes accompanying the onset of gingivitis. mBio 9: e00575-18. DOI 10.1128/mBio.00575-18. [Google Scholar] [CrossRef]
Nemoto T, Shiba T, Komatsu K, Watanabe T, Shimogishi M et al. (2021). Discrimination of bacterial community structures among healthy, gingivitis, and periodontitis statuses through integrated metatranscriptomic and network analyses. mSystems 6: e0088621. DOI 10.1128/mSystems.00886-21. [Google Scholar] [CrossRef]
Park OJ, Yi H, Jeon JH, Kang SS, Koo KT, Kum KY, Chun J, Yun CH, Han SH (2015). Pyrosequencing analysis of subgingival microbiota in distinct periodontal conditions. Journal of Dental Research 94: 921–927. DOI 10.1177/0022034515583531. [Google Scholar] [CrossRef]
Park SN, Kook JK (2013). Development of Streptococcus gordonii-specific quantitative real-time polymerase chain reaction primers based on the nucleotide sequence of rpoB. Microbiology and Immunology 57: 583–588. DOI 10.1111/1348-0421.12063. [Google Scholar] [CrossRef]
Park Y, Simionato MR, Sekiya K, Murakami Y, James D, Chen W, Hackett M, Yoshimura F, Demuth DR, Lamont RJ (2005). Short fimbriae of Porphyromonas gingivalis and their role in coadhesion with Streptococcus gordonii. Infection and Immunity 73: 3983–3989. DOI 10.1128/IAI.73.7.3983-3989.2005. [Google Scholar] [CrossRef]
Puig-Silla M, Montiel-Company JM, Dasi-Fernandez F, Almerich-Silla JM (2017). Prevalence of periodontal pathogens as predictor of the evolution of periodontal status. Odontology 105: 467–476. DOI 10.1007/s10266-016-0286-x. [Google Scholar] [CrossRef]
Rokaya D, Srimaneepong V, Wisitrasameewon W, Humagain M, Thunyakitpisal P (2020). Peri-implantitis update: Risk indicators, diagnosis, and treatment. European Journal of Dentistry 14: 672–682. DOI 10.1055/s-0040-1715779. [Google Scholar] [CrossRef]
Ritz HL (1967). Microbial population shifts in developing human plaque. Archives of Oral Biology 12: 1561–1568. DOI 10.1016/0003-9969(67)90190-2. [Google Scholar] [CrossRef]
Ruengsomwong S, La-Ongkham O, Jiang JH, Wannissorn B, Nakayama J, Nitisinprasert S (2016). Microbial community of healthy Thai vegetarians and non-vegetarians, their core gut microbiota, and pathogen risk. Journal of Microbiology and Biotechnology 26: 1723–1735. DOI 10.4014/jmb.1603.03057. [Google Scholar] [CrossRef]
Shaw L, Harjunmaa U, Doyle R, Mulewa S, Charlie D et al. (2016). Distinguishing the signals of gingivitis and periodontitis in supragingival plaque: A cross-sectional cohort study in Malawi. Applied and Environmental Microbiology 82: 6057–6067. DOI 10.1128/AEM.01756-16. [Google Scholar] [CrossRef]
Slots J (2017). Periodontitis: Facts, fallacies and the future. Periodontology 2000 75: 7–23. DOI 10.1111/prd.12221. [Google Scholar] [CrossRef]
Suzuki N, Yoshida A, Saito T, Kawada M, Nakano Y (2004). Quantitative microbiological study of subgingival plaque by real-time PCR shows correlation between levels of Tannerella forsythensis and Fusobacterium spp. Journal of Clinical Microbiology 42: 2255–2257. DOI 10.1128/JCM.42.5.2255-2257.2004. [Google Scholar] [CrossRef]
Shi B, Chang M, Martin J, Mitreva M, Lux R et al. (2015). Dynamic changes in the subgingival microbiome and their potential for diagnosis and prognosis of periodontitis. mBio 6: e01926-01914. DOI 10.1128/mBio.01926-14. [Google Scholar] [CrossRef]
Schincaglia GP, Hong BY, Rosania A, Barasz J, Thompson A, Sobue T, Panagakos F, Burleson JA, Dongari-Bagtzoglou A, Diaz PI (2017). Clinical, immune, and microbiome traits of gingivitis and peri-implant mucositis. Journal of Dental Research 96: 47–55. DOI 10.1177/0022034516668847. [Google Scholar] [CrossRef]
Schillinger C, Petrich A, Lux R, Riep B, Kikhney J et al. (2012). Co-localized or randomly distributed? Pair cross correlation of in vivo grown subgingival biofilm bacteria quantified by digital image analysis. PLoS One 7: e37583. DOI 10.1371/journal.pone.0037583. [Google Scholar] [CrossRef]
Sgolastra F, Petrucci A, Severino M, Gatto R, Monaco A (2015). Periodontitis, implant loss and peri-implantitis. A meta-analysis. Clinical Oral Implants Research 26: e8–e16. DOI 10.1111/clr.12319. [Google Scholar] [CrossRef]
Ting M, Craig J, Balkin BE, Suzuki JB (2018). Peri-implantitis: A comprehensive overview of systematic reviews. Journal of Oral Implantology 44: 225–247. DOI 10.1563/aaid-joi-D-16-00122. [Google Scholar] [CrossRef]
Wood DE, Lu J, Langmead B (2019). Improved metagenomic analysis with Kraken 2. Genome Biology 20: 257. DOI 10.1186/s13059-019-1891-0. [Google Scholar] [CrossRef]
Wang J, Qi J, Zhao H, He S, Zhang Y, Wei S, Zhao F (2013). Metagenomic sequencing reveals microbiota and its functional potential associated with periodontal disease. Scientific Reports 3: 1843. DOI 10.1038/srep01843. [Google Scholar] [CrossRef]
Weiss S, Van Treuren W, Lozupone C, Faust K, Friedman J, Deng Y et al. (2016). Correlation detection strategies in microbial data sets vary widely in sensitivity and precision. International Society for Microbial Ecology Journal 10: 1669–1681. DOI 10.1038/ismej.2015.235. [Google Scholar] [CrossRef]
Yu XL, Chan Y, Zhuang L, Lai HC, Lang NP, Keung Leung W, Watt RW (2019). Intra-oral single-site comparisons of periodontal and peri-implant microbiota in health and disease. Clinical Oral Implants Research 30: 760–776. DOI 10.1111/clr.13459. [Google Scholar] [CrossRef]
Zhang Y, Zhen M, Zhan Y, Song Y, Zhang Q, Wang J (2017). Population-genomic insights into variation in Prevotella intermedia and Prevotella nigrescens isolates and its association with periodontal disease. Frontiers in Cellular and Infection Microbiology 7: 409. DOI 10.3389/fcimb.2017.00409. [Google Scholar] [CrossRef]
Zhang Y, Shi W, Song Y, Wang J (2019). Metatranscriptomic analysis of an in vitro biofilm model reveals strain-specific interactions among multiple bacterial species. Journal of Oral microbiology 11: 1599670. DOI 10.1080/20002297.2019.1599670. [Google Scholar] [CrossRef]
Zhuang LF, Watt RM, Mattheos N, Si MS, Lai HC, Lang NP (2016). Periodontal and peri-implant microbiota in patients with healthy and inflamed periodontal and peri-implant tissues. Clinical Oral Implants Research 27: 13–21. DOI 10.1111/clr.12508. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools