 Open Access
Open Access
ARTICLE
Columbianetin acetate inhibits the occurrence and development of pancreatic cancer cells by down-regulating the expression of Meiotic nuclear divisions 1
1 Research Center of Clinical Laboratory Science, Bengbu Medical College, Bengbu, 233030, China
2 Department of Biochemistry and Molecular Biology, School of Laboratory Medicine, Bengbu Medical College, Bengbu, 233030, China
3 Department of Genetics, School of Life Sciences, Bengbu Medical College, Bengbu, 233000, China
* Corresponding Authors: JING ZHANG. Email: ; CHAOQUN LIAN. Email:
(This article belongs to the Special Issue: Cellular and Molecular Toxicology in Reproductive and Developmental Biology)
BIOCELL 2023, 47(2), 297-307. https://doi.org/10.32604/biocell.2023.023553
Received 01 May 2022; Accepted 19 July 2022; Issue published 18 November 2022
Abstract
Columbianetin acetate (CE) is one of the effective components of Angelica pubescens. So far, the specific role and molecular mechanism of CE in pancreatic cancer are not clear. Thus, this study aimed to explore the specific mechanism of CE on pancreatic cancer. The target genes combined with CE were predicted through the PharmMapper database and the 3D molecular structure of CE. Then, the Cancer Genome Atlas (TCGA) and Cistrome data browser (DB) databases were used to screen Meiotic nuclear divisions 1 (MND1)-related genes, transcription factors, and transcription factor data sets, and the intersection of the above data sets. The “limma” package in the R and gene expression profiling interactive analysis (GEPIA) databases were used to analyze the correlation and survival difference between the target genes and MND1 to predict the degree of association between CE and MND1. Western blotting and RT-PCR experiments revealed the regulatory relationship among CE, E2F1, and MND1 at the cellular level. The specific effects of CE on pancreatic cancer cells were explored through CCK8, wound healing, migration, and flow cycle experiments. E2F1, also the predictive transcription factor of MND1, was also the predictive target protein of CE. At the same time, E2F1 and MND1 were closely related in pancreatic tissue. In the cell function experiment, CE and interference with E2F1 expression could reduce the gene and protein expression of MND1, which was closely associated with cell proliferation, migration, and cycle development. Similarly, interfering with the expression of mnd1 can also inhibit the further development of tumor cells. CE may inhibit the development of pancreatic cancer cells by reducing the expression of MND1. This implies that CE may be a potential novel agent for the treatment of pancreatic cancer.Keywords
Pancreatic cancer is a disease with a disproportionate incidence rate and mortality. The five-year survival rate is less than 9%, and the mortality rate is one of the highest among all tumor diseases (Chen et al., 2016; Siegel et al., 2019). Surgical resection can significantly improve the five-year survival rate of patients. However, only a small number of people can benefit from this treatment. The main reason is that there is no effective early diagnosis method; the disease develops rapidly and is easy to relapse and metastasize (Kamisawa et al., 2016; Gillen et al., 2010; Siegel et al., 2014; Singhi et al., 2019). In recent years, many specific drugs for pancreatic cancer have been developed; however, pancreatic cancer cells have many molecular subtypes and unique biological characteristics, which lead to the varying effectiveness of drugs in the treatment of patients (Hidalgo, 2010). Therefore, it is necessary to develop new small-molecule drugs and early diagnostic markers to improve the prognosis of patients with pancreatic cancer.
Columbianetin acetate (CE) is one of the most effective active ingredients in Angelica pubescens and belongs to the coumarin family; it is also a natural anti-inflammatory active ingredient. CE can be absorbed by colon cancer cells by passive diffusion. In animal experiments, intestinal epithelial cells can be well absorbed and fully distributed in peripheral blood by oral administration (Yang et al., 2008; Hou et al., 2020; Ge et al., 2019; Yang et al., 2019). So far, although there are few reports on the specific effects of CE on tumors, the above-discussed studies have shown that CE is easy to metabolize and has anti-inflammatory effects, which may be more helpful in treating tumor diseases.
E2F transcription factor 1 (E2F1), a transcription factor activator, can bind to the dimerization partner on DNA and participate in regulating downstream genes and proteins (Chellappan et al., 1991). In a variety of tumor cells, E2F1 shows overexpression and abnormal amplification, and the accompanying gene changes often promote the occurrence and development of tumor cells (Tarangelo et al., 2015), such as promoting tumor metastasis, drug resistance, and proliferation and immune regulation (Fang et al., 2020; Chun et al., 2020).
Meiotic nuclear divisions 1 (MND1) is a meiotic protein that can promote homologous chromosome pairing. The interaction of MND1 and homologous pairing protein 2 (hop2) contributes to the telomere prolongation and cell proliferation of tumor cells. In related reports, MND1 plays an important role in the prognosis of lung adenocarcinoma and promotes further development of lung adenocarcinoma cells (Kang et al., 2015; Arnoult and Karlseder, 2014; Zhang et al., 2021; Wei et al., 2021). So far, no specific effect of MND1 on pancreatic cancer cells has been reported.
In this study, E2F1, a gene related to CE and MND1, was screened through bioinformatic analysis, which revealed the possible role of CE in regulating the expression of MND1 through E2F1. Furthermore, in cell-based experiments, CE could inhibit the occurrence and development of pancreatic cancer cells by reducing MND1, thus providing a new idea for the prognosis of patients with pancreatic cancer.
The small-molecule drug CE (CAS number 23180-65-6) was purchased from Grint, Wuhan. E2F1 and Vinculin (catalog numbers AF6377 and AF5122, respectively) were purchased from Affinity Biosciences, USA. MND1 (11636-1-AP) was obtained from the Proteintech group. The cell cycle and apoptosis analysis kit were from Shanghai Beibo Biology. Cell counting kit-8 (CCK8, Beyotime, Shanghai, China) was used for cell viability analysis.
Prediction of targeted binding proteins through PharmMapper online tool
PharmMapper database is an online analysis tool that can predict targeted binding proteins through 3D small-molecule structure. The older version includes more than 7000 target pharmacophores and corresponding protein target markers, while the new version of the database has nearly more than 23236 proteins, including 16159 drug pharmacophore models, due to continuous updating, with a strong and accurate ability to predict small-molecule targeted proteins (Wang et al., 2017). For an online prediction of targeted proteins, we downloaded the 3D molecular structure of columbianetin acetate in the pharmMapper database.
The cistrome data browser (DB)
DB is a resource of human-derived chip-seq, DNase-seq, and ATAC-seq chromatin map analysis and maps the genome-wide location of transcription factor binding sites, histone post-translational modifications, and accessible regions of chromatin endonuclease activity. It contains about 47000 human and mouse samples and about 24000 newly collected data sets. The toolkit, helpful for biomedical research (Zheng et al., 2019), was used to search online for MND1-related transcription factors.
Gene expression profiling interactive analysis (GEPIA)
GEPIA is a network tool for the online analysis of gene expression in different tumor groups. It combines TCGA and GTEX databases and uses an interactive and customizable method of gene expression, including differential expression analysis, correlation analysis, and survival analysis that help biologists and clinicians to deeply mine gene functions (Tang et al., 2017). In this study, the correlation between MND1 and E2F1 and the survival analysis of MND1 were analyzed directly online.
The cancer genome atlas (TCGA)
TCGA is a free and public tumor database which aims to study the whole genome spectrum related to cancer. So far, developers have collected whole genome sequencing data of more than 30 tumors and relevant clinical patient data, and this has greatly promoted clinicians and scientific research staff to improve diagnostic methods and treatment standards (Tomczak et al., 2015). From the TCGA database, pancreatic cancer-related RNA-seq and 178 clinical patient data were downloaded, which also included survival time and survival status.
Human pancreatic cancer cell lines 8988 and PANC1 were mainly purchased from the cell bank of the Chinese Academy of Sciences (Shanghai, China). The cell lines were mainly cultured in a DMEM medium (Gibco, USA) containing 10% fetal bovine serum (FBS) at 37°C and a 5% CO2 incubator.
Real-time polymerase chain reaction (PCR) analysis
In a six-well plate, 1.5–2.0 × 106 pancreatic cancer cells were plated and incubated overnight and then treated with CE and MND1 small interfering RNA for 24 h. The cells were lysed with Trizol reagent for 5–10 min, and a total of 2 µg RNA was extracted with the corresponding RNA kit (Invitrogen). The extracted RNA was then reverse transcribed into cDNA using a reverse transcription kit. Then, the synthesized real-time PCR primers of MND1 and GAPDH were mixed with Q-SYBR GREEN Supermix, (Bio-Rad) and the reaction was conducted in real-time fluorescence quantitative PCR instrument for 1–2 h.
The cell lines of the dosing group were cultured for 24–36 h, while those of the interference group were cultured for about 48–60 h. The number of seed cells in the six-well plates remained the same, at about 1.5–2.0 × 106 cells. RIPA and protease inhibitors were used to lyse the cells, shaking once every ten minutes for a total of three times. The extracted protein was separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, then transferred to PVDF membrane and blocked with a rapid blocking solution for 15 min. The membrane was then incubated with the primary antibody in the working solution and kept at 4°C for 12 h. The next day, the membrane was washed three times with Tris-buffered saline with Tween 20 and incubated with the secondary antibody working solution for 2 h at room temperature.
In the beginning, the number of seeded cells remained the same. The culture time of the dosing group and the interference group was 24–36 h and 36–48 h, respectively. Pancreatic cancer cells were added with different concentrations of CE and small interfering RNA and incubated for 36 h. After digestion with trypsin, the cell suspension was extracted, centrifuged, and the cells were resuspended with buffer. The propidium iodide dye was added and incubated at 37°C for 30 min. Finally, the changes cell cycle were detected by flow cytometry.
The number of cells in different groups was about 2000 per hole. Normal cells were cultured in 96-well plates, and different concentrations of CE were added to each well for 24, 48, and 72 h, respectively. At intervals, 5% CCK8 reagent was added. After incubation for 3–4 h, the absorbance value (OD) was measured by an enzyme labeling instrument. After the interference, the cells were cultured in 96 wells for different periods, and the specific CCK8 experiment was conducted as described above.
First, 1.5–2.0 × 106 cells were added to each well in the six-well plate for overnight culture, and then the drug was added at different concentrations and interfering RNA for treatment until 24 h later. Under different treatment groups, three traces were drawn in each well with the same gun head, and the scratch images of 0 h were observed and recorded through an inverted microscope. Then, the scratch images at 24 h were recorded. Finally, the cell migration rates of different groups were analyzed by image J.
At first, 1.5–2.0 × 106 cells were seeded per well in six-well plates and cultured for 24 h, different concentrations of CE and interfering RNA were added, and then cultured for 24–36 h. The cells were digested with trypsin and washed with PBS 3–4 times to completely remove the serum. Then, about 2 × 104 cells were transferred to the chamber and cultured for 24–36 h. The chamber medium of the dosing group contained different concentrations of drug CE, which was then stained, and the pictures of migrating cells were captured under the microscope. The number of migrating cells was calculated by image J.
The “limma” package in R language was used for difference, and correlation analysis, the “beeswarm” package and Cytoscape software were used to draw honeybee map and targeted gene matrix map, and GraphPad prism 4.0 was used to analyze and draw the difference between the treatment group and the control group. The sample statistics were repeated three times for each data, and the results were based on mean ± SD using the student t-test, one-way ANOVA, or two-way ANOVA. P-value < 0.05 was statistically significant.
Prediction of the targeted genes of CE using the PharmaMapper tool
The 3D molecular structure related to CE was downloaded from PubChem (https://pubchem.ncbi.nlm.nih.gov/) database and uploaded to the PharmMapper database for online analysis of target genes (target mRNA) bound to CE. A total of 68 target genes were obtained (Fig. 1A).
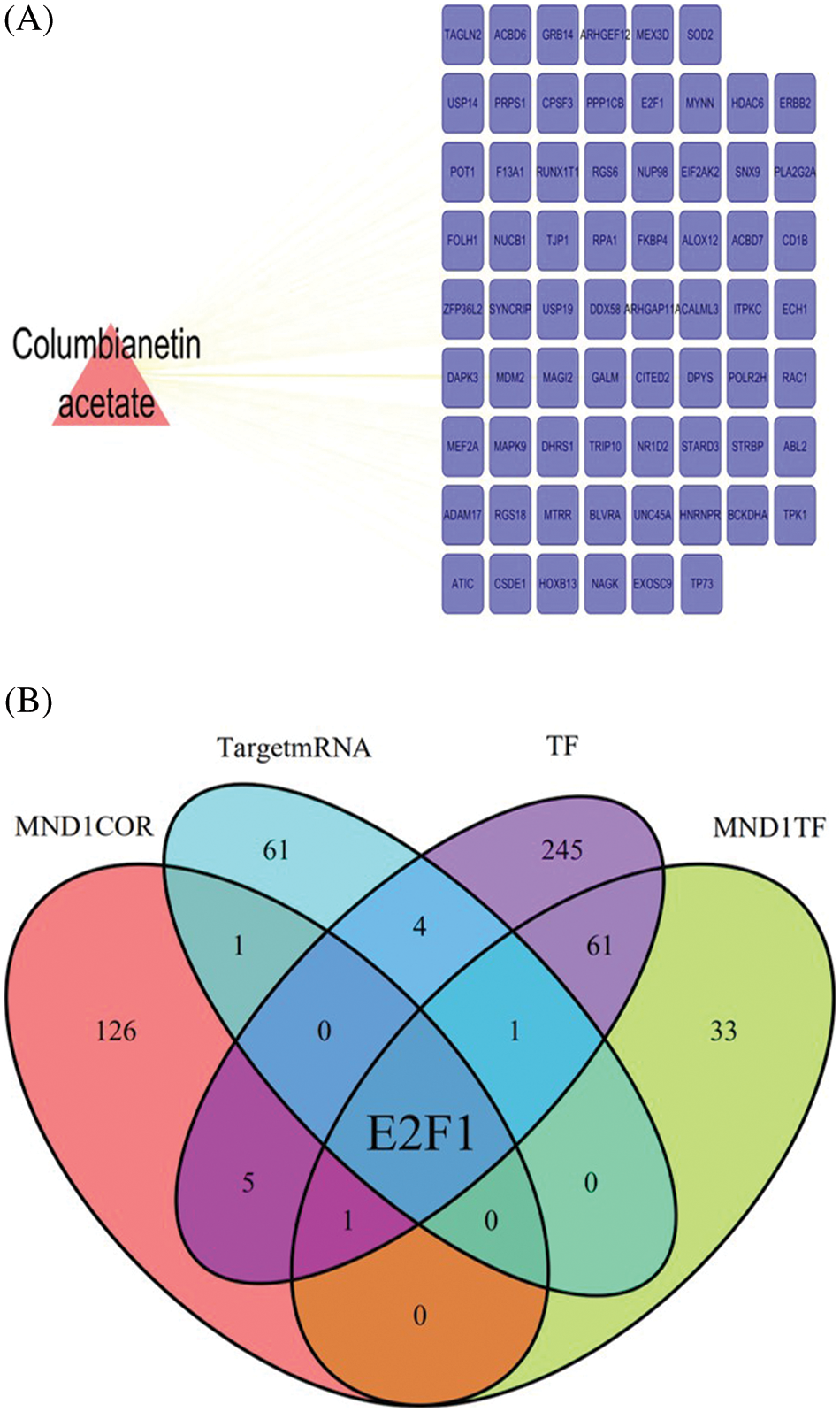
Figure 1: Screening core genes related to Columbianetin acetate and MND1. (A) Based on the PharmMapper database, the target genes of Columbianetin acetate were predicted. (B) Multiple gene sets take intersections, including TargetmRNA, MND1COR, TF, and MND1TF.
Screening core transcription factor E2F1
PAAD-related RNA-seq sequencing data in the TCGA database were downloaded, and 134 MND1-related genes (MND1COR) were screened using the “limma” package in R language, with cor >0.4. Then, the DB database was used to predict the transcription factor (MND1TF) of MND1; the total transcription factor data set (TF) and a total of 97 and 318, were downloaded, respectively. Finally, the intersection of MND1COR, TF, MND1TF, and TargetmRNA was determined to obtain E2F1 (Fig. 1B).
Correlation between meiotic nuclear divisions 1 and E2F transcription factor 1 in pancreatic cancer
Combining the TCGA and GTEX database, the differential expression of MND1 and E2F1 in normal and tumor groups, respectively, was analyzed (Figs. 2A and 2B). The expression of MND1 and E2F1 was generally higher in tumor groups. Online analysis of the correlation degree of MND1 and E2F1 using the GEPIA database revealed that E2F1 and MND1 had a high positive correlation (Fig. 2C). At the same time, the survival differences between high and low expression groups of MND1 were analyzed and revealed that high expression of MND1 was unfavorable to the prognosis of patients (Fig. 2D).
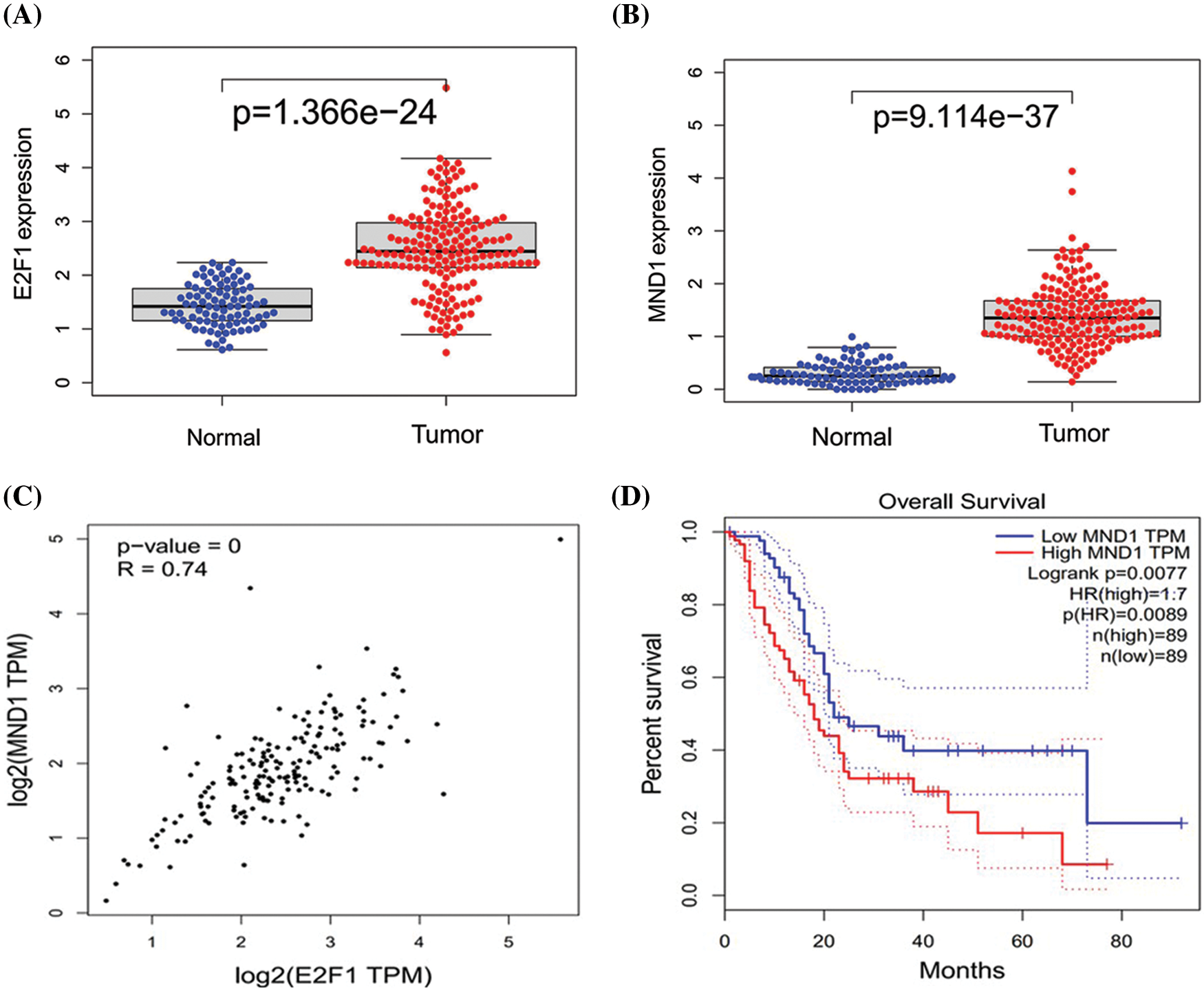
Figure 2: Correlation between MND1 and E2F1. (A, B) Distribution difference of E2F1and MND1 between tumor and normal groups. (C) Correlation linear diagram of E2F1 and MND1. (D) Difference analysis of MND1 between high and low expression groups.
Effect of the addition of columbianetin acetate and interference with E2F transcription factor 1 expression on meiotic nuclear divisions 1
In pancreatic cancer cells, interference with the mRNA and protein expression of E2F1 effectively reduced the expression of MND1 (Figs. 3A and 3B, Figs. 4A and 4B). The addition of different concentrations of small-molecule drug CE gradually decreased the mRNA and protein expression of MND1 in pancreatic cancer cells (Figs. 3C and 3D, Figs. 4C and 4D).
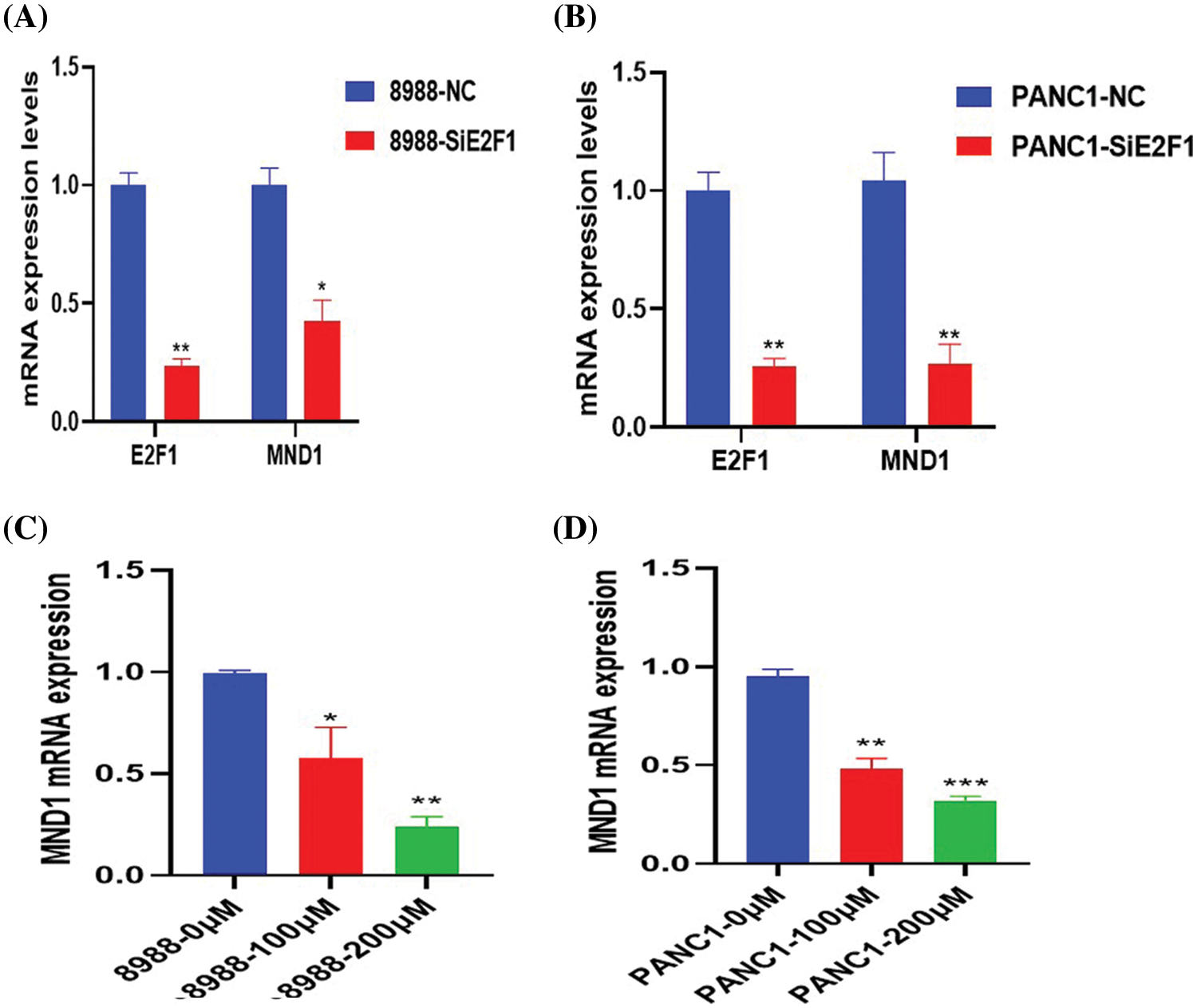
Figure 3: MND1 mRNA expression in pancreatic cancer cells after addition of CE and Small interfering RNA of E2F1 (SiE2F1). (A, B) mRNA expression differences of E2F1 and MND1 in NC and interference groups. (C, D) After treatment with different concentrations of drugs (0, 100 and 200 μM), MND1 expression level in pancreatic cancer cells. Data were represented as mean ± SD (*P < 0.05, **P < 0.01, ***P < 0.001).
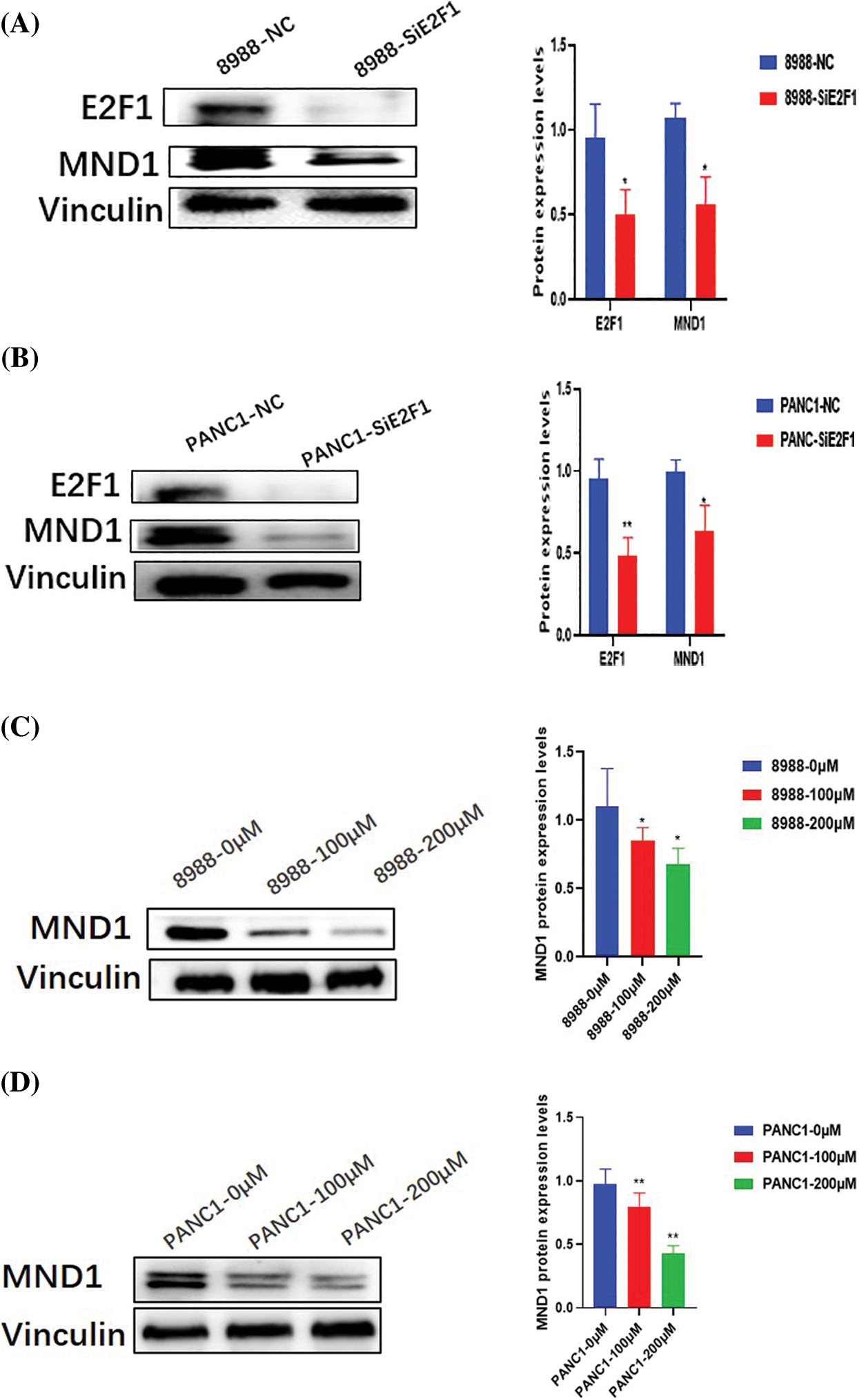
Figure 4: MND1 Protein expression in pancreatic cancer cells after addition of CE and Small interfering RNA of E2F1 (SiE2F1). (A, B) Protein expression differences of E2F1 and MND1 in NC and interference groups. (C, D) After treatment with different concentrations of drugs (0, 100 and 200 μM), MND1 protein expression level in pancreatic cancer cells. Data were represented as mean ± SD (*P < 0.05, **P < 0.01).
Effects of the interfering RNA on the expression of meiotic nuclear divisions 1
RT-PCR and WB experiments reveal that the addition of small interfering RNA for 24–48 h in the interference group led to a decrease in the mRNA and protein levels of MND1 (Figs. 5A–5D).
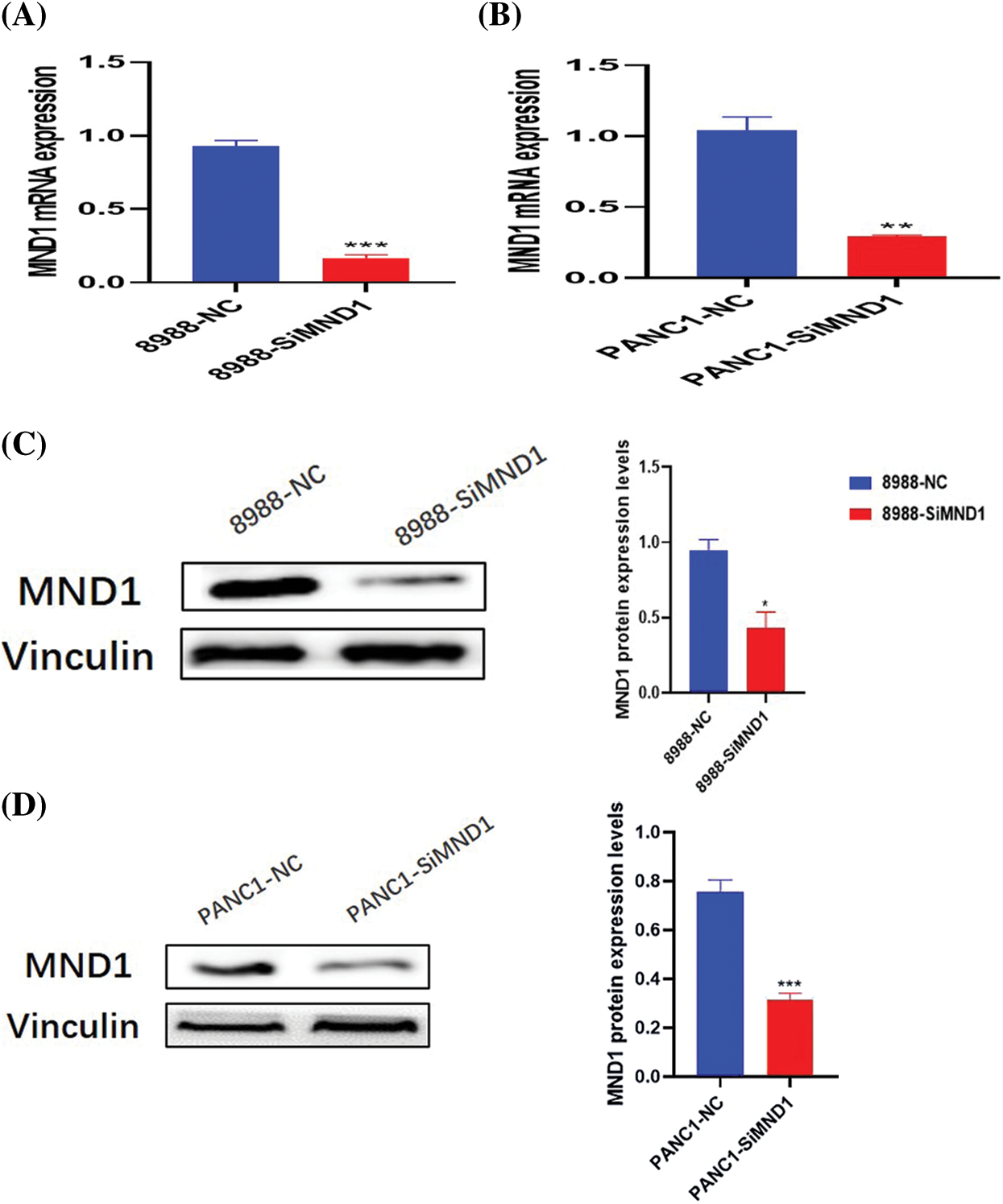
Figure 5: After small interfering RNA treatment, the mRNA and protein expression of mnd1 were different. A–D mRNA (A, B) and protein (C, D) expression levels. Data were represented as mean ± SD (*P < 0.05, **P < 0.01, ***P < 0.001).
Columbianetin acetate and decreased meiotic nuclear divisions 1 inhibit the viability of pancreatic cancer cells
Different concentrations of CE were added to 96-well plates and cultured for 24, 48, and 72 h. CE at 100 and 200 μM could effectively inhibit the viability of pancreatic cancer cells over time (Figs. 6A and 6B). In the interference and control groups, interference with the expression of MND1 could significantly inhibit the viability of pancreatic cancer cells (Figs. 6C and 6D).
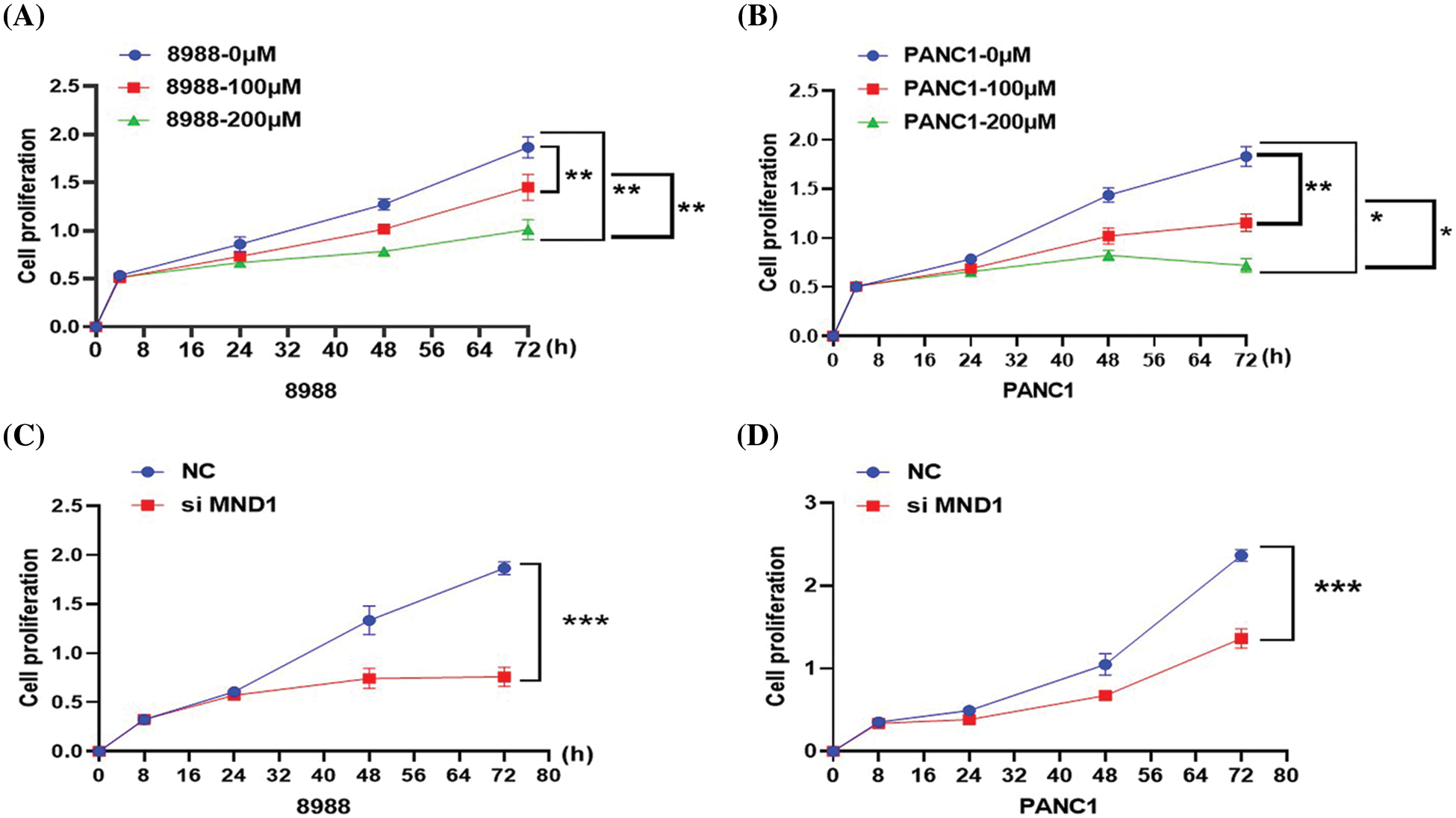
Figure 6: Based on CCK8 assay, effects of CE and MND1 on the proliferation of pancreatic cancer cells. (A, B) Different drug treatment groups. (C, D) MND1 interference group. Data were represented as mean ± SD (*P < 0.05, **P < 0.01, ***P < 0.001).
Columbianetin acetate and meiotic nuclear divisions 1 affect the migration level of pancreatic cancer cells
After treatment with different concentrations of CE and interfering RNA, the migration ability and number of pancreatic cancer cells in the dosing group decreased significantly (Figs. 7A and 7B, Figs. 8A and 8B). Similarly, the migration level of pancreatic cancer cells in the interference group also decreased significantly (Figs. 7C and 7D, Figs. 8C and 8D).
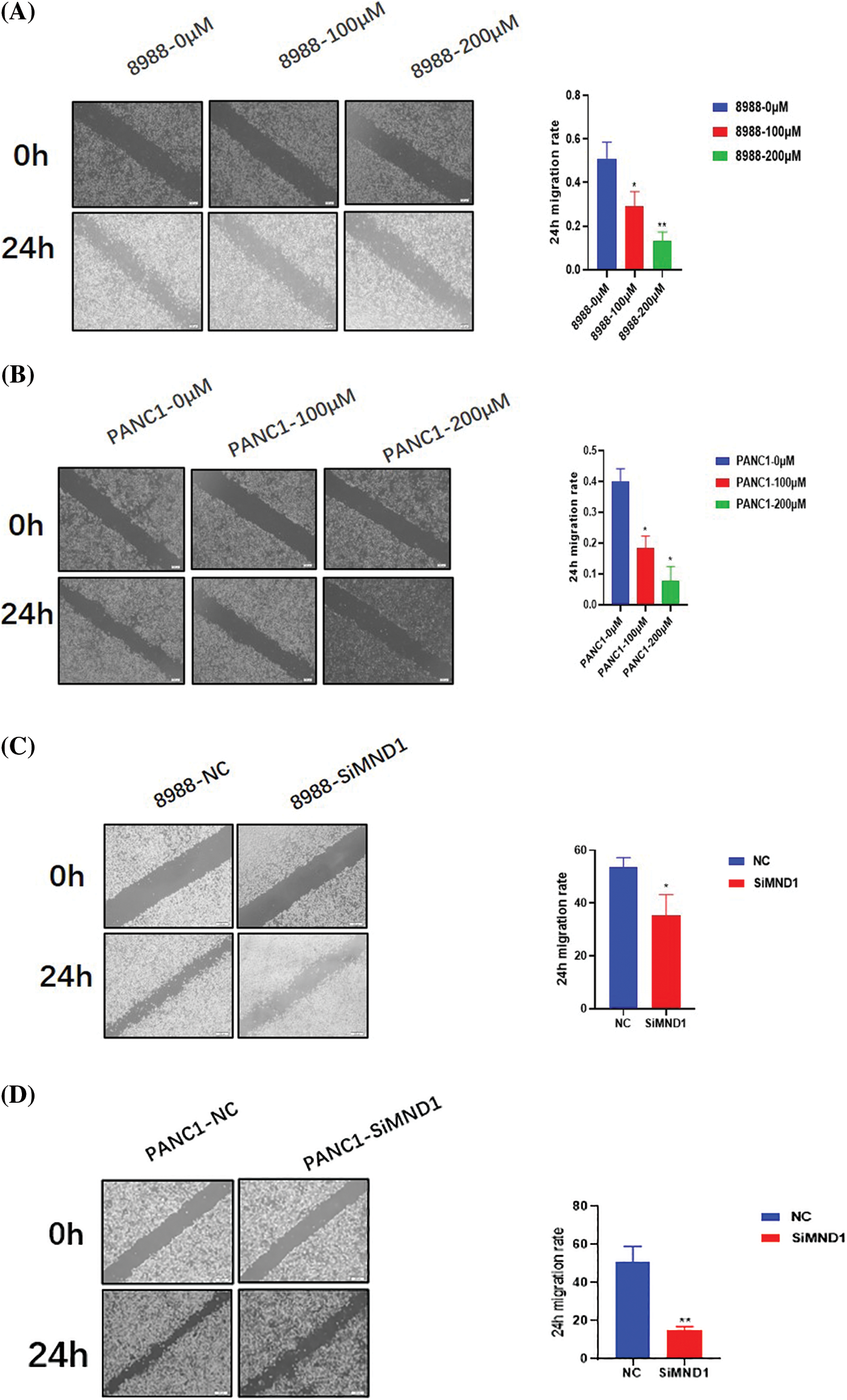
Figure 7: Through the use of wound healing experiment, CE and MND1 regulate the motility of pancreatic cancer cells. (A, B) Different concentrations of CE treatment groups. (C, D) Interference group of MND1. Data were represented as mean ± SD (*P < 0.05, **P < 0.01).
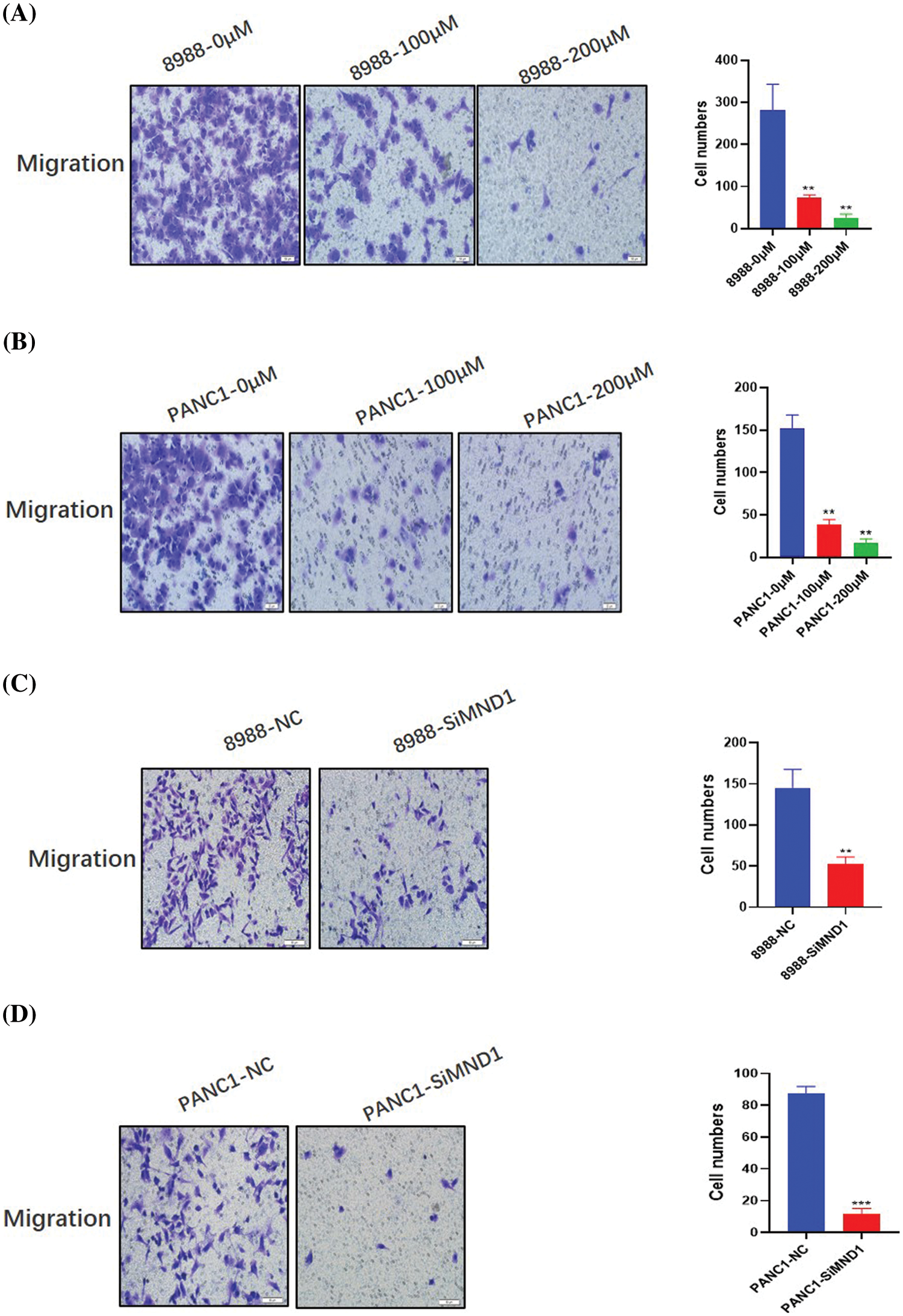
Figure 8: Effects of CE and MND1 on the migration ability of pancreatic cancer cells. (A, B) Different CE concentration groups. (C, D) MND1 down-regulation group. Data were represented as mean ± SD (**P < 0.01, ***P < 0.001).
Columbianetin acetate and meiotic nuclear divisions 1 can effectively affect the cycle development of pancreatic cancer cells
After 36 h of treatment with different concentrations of CE and interfering RNA, the development cycle of different treatment groups was analyzed. The G1 phase in the cells increased with the increase in CE concentration, and the S phase decreased (Figs. 9A and 9B). In the interference group, the G1 phase increased, and the S phase decreased (Figs. 9C and 9D).
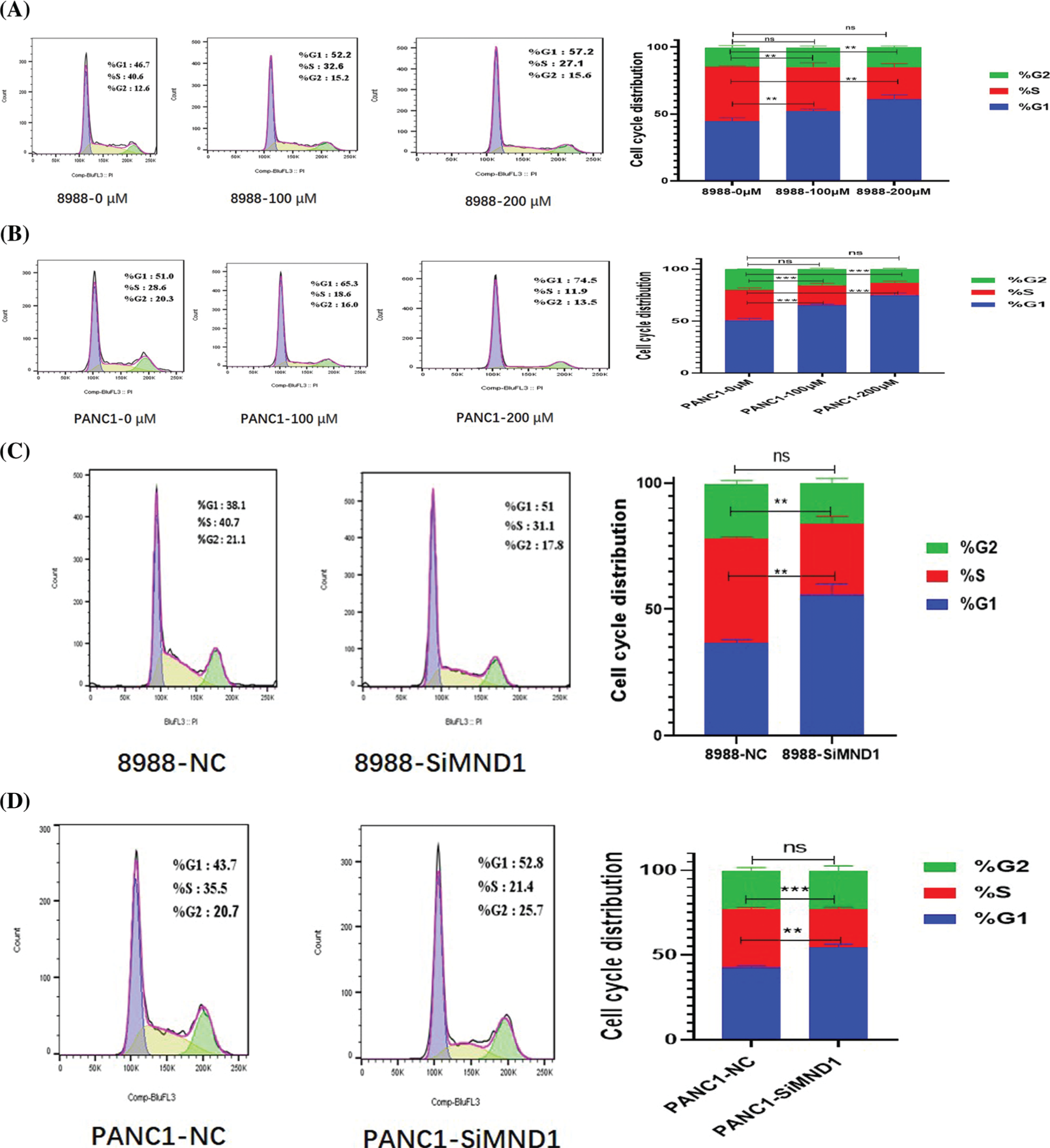
Figure 9: Cell cycle regulation of pancreatic cancer cells by CE and mnd1 based on flow cytometry. (A, B) Different concentration CE groups. (C, D) Interference group.
In recent years, the mechanism of action of small-molecule drugs on tumors has been reported to be increasingly extensive. For example, capsaicin can regulate CDK8/PI3K/Akt/Wnt/β catenin signaling pathway to inhibit breast cancer (Wu et al., 2020), and curcumin inhibits the ERK/NF-κ B pathway to thus hinder the invasion and migration of H2O2-induced pancreatic cancer cells (Cao et al., 2016). Capsaicin and piperine can overcome the resistance of cancer cells to doxorubicin (Li et al., 2018). In terms of tumor immunity, iox1 and doxorubicin are combined for antibody-independent cancer chemotherapy immunotherapy (Liu et al., 2021). CD11b agonists reprogram innate immunity to sensitize pancreatic cancer immunotherapy (Panni et al., 2019), indicating that small-molecule drugs have a broad therapeutic prospect in the treatment of cancer.
As one of the effective components of Angelica pubescens, although the mechanism of action on tumor cells is unclear, CE has a high absorption rate in the small intestine and exerts an anti-inflammatory effect (Yang et al., 2008; Hou et al., 2020; Ge et al., 2019; Yang et al., 2019), which lays a foundation for the treatment of tumor cells. Recently, MND1 was found to be closely related to the prognosis of lung adenocarcinoma and promote the further occurrence and development of lung adenocarcinoma cells (Zhang et al., 2021; Wei et al., 2021). However, the specific impact on pancreatic cancer is not yet clear. Similarly, in pancreatic cancer, the regulatory relationship between CE and MND1 is not clear. Therefore, in this study, we mainly discussed the regulatory relationship between CE and MND1 and the specific effects on pancreatic cancer cells.
The regulatory relationship between CE and MND1 in pancreatic cancer was further explored at both bioinformatics and cellular level. We used the PharmMapper database to predict the target mRNA. At the same time, we downloaded and predicted TF and MND1TFin the DB database. Then, using the RNA-seq sequencing data of PAAD in TCGA, we screened the MND1COR gene set. Taking the intersection of the above four data sets, we found that E2F1 is not only the transcription factor related to MND1 but also the CE binding target gene. E2F1 can regulate the expression of MND1 in lung adenocarcinoma (Zhang et al., 2021). In pancreatic cancer, MND1 and E2F1 are highly expressed in the tumor group, and there is a high positive correlation between MND1 and E2F1, indicating that E2F1 may regulate the expression of MND1 in pancreatic cancer, while there is a binding effect between E2F1 and CE. To further verify if CE may inhibit the expression of MND1 by binding E2F1, we blocked the expression of E2F1 through interfering RNA and found a significant decrease in the expression of MND1. The above findings greatly indicate that CE may bind with E2F1 to regulate the expression of MND1. In the cell experiment, the expression of MND1 decreased CE was added at different concentrations. Later, CCK8, cell healing, transwell migration, and flow cytometry tests revealed that CE could effectively reduce the proliferation, migration, and cycle progression of pancreatic cancer cells. The same experimental results were also observed after interfering with the expression of MND1. These findings suggest that CE may inhibit the occurrence and development of pancreatic cancer by down-regulating MND1 expression.
In conclusion, bioinformatics and cell-level studies revealed that CE could inhibit the proliferation, migration, and cycle progression of pancreatic cancer cells by downregulating the expression of MND1. CE and MND1 can be used as potential therapeutic molecules and early prognostic markers for pancreatic cancer cells.
Availability of Data and Materials: Publicly available datasets were analyzed in this study; these can be found in the GEO database (https://www.NCBI.nlm.nih.gov/geo) and The Cancer Genome Atlas (https://www.portal.gdc.cancer.gov/). The authors confirm that the data supporting the findings of this study are available within the article and its Supplementary materials. A statement that all methods and public data are implemented by relevant guidelines and regulations.
Author’s Contribution: KS, ZZ, and DW provided the idea and design of this article. Clinical data were collected and analyzed by KS, ZZ, and DW. KS and YH prepared the first draft of the article and the drawing charts. CL and JZ reviewed the revised paper. All authors have read and approved the final manuscript.
Ethics Approval: The data used in this study were obtained from publicly available datasets, such as the GEO database (https://www.NCBI.nlm.nih.gov/geo) and The Cancer Genome Atlas (https://www.portal.gdc.cancer.gov/).
Funding Statement: This study was supported by grants from the Key Natural Science Project of Anhui Provincial Education Department (Nos. KJ2018A0221, KJ2020A0578 and KJ2021A0773) and National Innovation Program for College Students (No. 202010367046), Anhui Provincial Undergraduate Innovative Training Program (Nos. S202010367008, S202110367030 and S202110367112).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
Arnoult N, Karlseder J (2014). ALT telomeres borrow from meiosis to get moving. Cell 159: 11–12. DOI 10.1016/j.cell.2014.09.013. [Google Scholar] [CrossRef]
Cao L, Liu J, Zhang L, Xiao X, Li W (2016). Curcumin inhibits H2O2-induced invasion and migration of human pancreatic cancer via suppression of the ERK/NF-κB pathway. Oncology Reports 36: 2245–2251. DOI 10.3892/or.2016.5044. [Google Scholar] [CrossRef]
Chellappan SP, Hiebert S, Mudryj M, Horowitz JM, Nevins JR (1991). The E2F transcription factor is a cellular target for the RB protein. Cell 65: 1053–1061. DOI 10.1016/0092-8674(91)90557-F. [Google Scholar] [CrossRef]
Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J (2016). Cancer statistics in China, 2015. CA: A Cancer Journal for Clinicians 66: 115–132. DOI 10.3322/caac.21338. [Google Scholar] [CrossRef]
Chun JN, Cho M, Park S, So I, Jeon JH (2020). The conflicting role of E2F1 in prostate cancer: A matter of cell context or interpretational flexibility? Biochimica et Biophysica Acta-Reviews on Cancer 1873: 188336. DOI 10.1016/j.bbcan.2019.188336. [Google Scholar] [CrossRef]
Fang Z, Lin M, Li C, Liu H, Gong C (2020). A comprehensive review of the roles of E2F1 in colon cancer. American Journal of Cancer Research 10: 757–768. [Google Scholar]
Ge Y, Chen S, Luo Q, Wang CP, Hao J, He J, Chen X, Yang X, Li J, Chang YX (2019). The tissue distribution of four major coumarins after oral administration of angelicae pubescentis radix extract to rats using ultra-high-performance liquid chromatography. Evidence-Based Complementary and Alternative Medicine 2019: 2365697. DOI 10.1155/2019/2365697. [Google Scholar] [CrossRef]
Gillen S, Schuster T, Meyer Zum Büschenfelde C, Friess H, Kleeff J (2010). Preoperative/neoadjuvant therapy in pancreatic cancer: A systematic review and meta-analysis of response and resection percentages. PLoS Medicine 7: e1000267. DOI 10.1371/journal.pmed.1000267. [Google Scholar] [CrossRef]
Hidalgo M (2010). Pancreatic cancer. The New England Journal of Medicine 362: 1605–1617. DOI 10.1056/NEJMra0901557. [Google Scholar] [CrossRef]
Hou A, Yang L, Zhang J, Wang S, Man W et al. (2020). A strategy for qualitative and quantitative profiling of Angelicae Pubescentis Radix and detection of its analgesic and anti-inflammatory components by spectrum-effect relationship and multivariate statistical analysis. Biomedical Chromatography: 34: e4910. DOI 10.1002/bmc.4910. [Google Scholar] [CrossRef]
Kamisawa T, Wood LD, Itoi T, Takaori K (2016). Pancreatic cancer. Lancet 388: 73–85. DOI 10.1016/S0140-6736(16)00141-0. [Google Scholar] [CrossRef]
Kang HA, Shin HC, Kalantzi AS, Toseland CP, Kim HM, Gruber S, Peraro MD, Oh BH (2015). Crystal structure of Hop2-Mnd1 and mechanistic insights into its role in meiotic recombination. Nucleic Acids Research 43: 3841–3856. DOI 10.1093/nar/gkv172. [Google Scholar] [CrossRef]
Li H, Krstin S, Wang S, Wink M (2018). Capsaicin and piperine can overcome multidrug resistance in cancer cells to doxorubicin. Molecules 23: 557. DOI 10.3390/molecules23030557. [Google Scholar] [CrossRef]
Liu J, Zhao Z, Qiu N, Zhou Q, Wang G, Jiang H, Piao Y, Zhou Z, Tang J, Shen Y (2021). Co-delivery of IOX1 and doxorubicin for antibody-independent cancer chemo-immunotherapy. Nature Communications 12: 2425. DOI 10.1038/s41467-021-22407-6. [Google Scholar] [CrossRef]
Panni RZ, Herndon JM, Zuo C, Hegde S, Hogg GD et al. (2019). Agonism of CD11b reprograms innate immunity to sensitize pancreatic cancer to immunotherapies. Science Translational Medicine 11: 1326. DOI 10.1126/scitranslmed.aau9240. [Google Scholar] [CrossRef]
Siegel R, Ma J, Zou Z, Jemal A (2014). Cancer statistics, 2014. CA: A Cancer Journal for Clinicians 64: 9–29. DOI 10.3322/caac.21208. [Google Scholar] [CrossRef]
Siegel RL, Miller KD, Jemal A (2019). Cancer statistics, 2019. CA: A Cancer Journal for Clinicians 69: 7–34. DOI 10.3322/caac.21551. [Google Scholar] [CrossRef]
Singhi AD, Koay EJ, Chari ST, Maitra A (2019). Early detection of pancreatic cancer: Opportunities and challenges. Gastroenterology 156: 2024–2040. DOI 10.1053/j.gastro.2019.01.259. [Google Scholar] [CrossRef]
Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z (2017). GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Research 45: W98–W102. DOI 10.1093/nar/gkx247. [Google Scholar] [CrossRef]
Tarangelo A, Lo N, Teng R, Kim E, Le L, Watson D, Furth EE, Raman P, Ehmer U, Viatour P (2015). Recruitment of Pontin/Reptin by E2f1 amplifies E2f transcriptional response during cancer progression. Nature Communications 6: 10028. DOI 10.1038/ncomms10028. [Google Scholar] [CrossRef]
Tomczak K, Czerwińska P, Wiznerowicz M (2015). The cancer genome atlas (TCGAAn immeasurable source of knowledge. Contemporary Oncology 19: A68–77. DOI 10.5114/wo.2014.47136. [Google Scholar] [CrossRef]
Wang X, Shen Y, Wang S, Li S, Zhang W, Liu X, Lai L, Pei J, Li H (2017). PharmMapper 2017 update: A web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Research 45: W356–W360. DOI 10.1093/nar/gkx374. [Google Scholar] [CrossRef]
Wei J, Meng G, Wu J, Zhang Q, Zhang J (2021). Genetic network and gene set enrichment analyses identify MND1 as potential diagnostic and therapeutic target gene for lung adenocarcinoma. Scientific Reports 11: 9430. DOI 10.1038/s41598-021-88948-4. [Google Scholar] [CrossRef]
Wu D, Jia H, Zhang Z, Li S (2020). Capsaicin suppresses breast cancer cell viability by regulating the CDK8/PI3K/Akt/Wnt/βcatenin signaling pathway. Molecular Medicine Reports 22: 4868–4876. DOI 10.3892/mmr.2020.11585. [Google Scholar] [CrossRef]
Yang XW, Guo QM, Wang Y (2008). Absorption and transport of 6 coumarins isolated from the roots of Angelica pubescens f. biserrata in human Caco-2 cell monolayer model. Journal of Chinese Integrative Medicine 6: 392–398. DOI 10.3736/jcim20080413. [Google Scholar] [CrossRef]
Yang Y, Zhu R, Li J, Yang X, He J, Wang H, Chang Y (2019). Separation and enrichment of three coumarins from angelicae pubescentis radix by macroporous resin with preparative HPLC and evaluation of their anti-inflammatory activity. Molecules 24: 2664. DOI 10.3390/molecules24142664. [Google Scholar] [CrossRef]
Zhang Q, Shi R, Bai Y, Meng L, Hu J et al. (2021). Meiotic nuclear divisions 1 (MND1) fuels cell cycle progression by activating a KLF6/E2F1 positive feedback loop in lung adenocarcinoma. Cancer Communications 41: 492–510. DOI 10.1002/cac2.12155. [Google Scholar] [CrossRef]
Zheng R, Wan C, Mei S, Qin Q, Wu Q et al. (2019). Cistrome data browser: Expanded datasets and new tools for gene regulatory analysis. Nucleic Acids Research 47: D729–D735. DOI 10.1093/nar/gky1094. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools