 Open Access
Open Access
ARTICLE
Long non-coding RNA DPP10-AS1 represses the proliferation and invasiveness of glioblastoma by regulating miR-24-3p/CHD5 signaling pathway
1 Department of Neurosurgery, The Second Affiliated Hospital of Soochow University, Suzhou, 215004, China
2 Department of Neurosurgery, The First Affiliated Hospital of Bengbu Medical College, Bengbu, 233000, China
* Corresponding Author: JUN DONG. Email:
# These authors contributed equally
BIOCELL 2023, 47(12), 2721-2733. https://doi.org/10.32604/biocell.2023.043869
Received 14 July 2023; Accepted 11 October 2023; Issue published 27 December 2023
Abstract
Objective: This investigation aimed to unveil new prospective diagnosis-related biomarkers together with treatment targets against glioblastoma. Methods: The expression levels of long non-coding RNA (lncRNA) DPP10-AS1 were assessed using real-time quantitative polymerase chain reaction (RT-qPCR) within both the patient tissue specimens and glioblastoma cell lines. The relationship between lncRNA DPP10-AS1 expression in glioblastoma and patient prognosis was investigated. Cell Counting Kit-8 (CCK-8), transwell, and clonogenic experiments were utilized to assess tumor cells’ proliferation, invasiveness, and migratory potentials after lncRNA DPP10-AS1 expression was up or down-regulated. Using an online bioinformatics prediction tool, the intracellular localization of lncRNA DPP10-AS1 and its target miRNA were predicted, and RNA-FISH verified results. A dual-luciferase reporter experiment validated the relationship across miR-24-3p together with lncRNA DPP10-AS1. MiR-24-3p expression within glioblastoma was identified through RT-qPCR, and potential link across miR-24-3p and lncRNA DPP10-AS1 was assessed using Pearson correlation analysis. Moreover, influence from lncRNA DPP10-AS1/miR-24-3p axis upon glioblastoma cell progression was assessed in vivo via a subcutaneous xenograft tumor model. Results: The expression of lncRNA DPP10-AS1 was notably reduced in both surgical specimens of glioblastoma and the equivalent cell lines. Low level of lncRNA DPP10-AS1 in glioblastoma is following poor prognosis. The downregulation of lncRNA DPP10-AS1 in glioblastoma cells resulted in enhanced cellular proliferation, migration, and invasion capabilities, accompanied by downregulated E-cadherin and upregulated vimentin and N-cadherin. Additionally, the observed up-regulation of lncRNA DPP10-AS1 demonstrated a substantial inhibitory function upon proliferation, invasion, and migratory capabilities of LN229 cells. Subcellular localization disclosed that lncRNA DPP10-AS1 had a binding site that interacted with miR-24-3p. Upregulated miR-24-3p was detected in glioblastomas, displaying an inverse correlation with lncRNA DPP10-AS1 expression. MiR-24-3p downstream target has been determined as chromodomain helicase DNA binding protein 5 (CHD5). LncRNA DPP10-AS1 affected the invasion and proliferation of glioblastoma by controlling the miR-24-3p/CHD5 axis. Conclusion: The present study demonstrated that lncRNA DPP10-AS1 can inhibit the invasive, migratory, and proliferative properties of glioblastoma by regulating the miR-24-3p/CHD5 signaling pathway. Consequently, lncRNA DPP10-AS1 has potential as a tumor suppressor and might be utilized for accurate diagnosis and targeted treatments of glioblastomas.Keywords
Abbreviations
| RT-qPCR | Real-time quantitative polymerase chain reaction |
| lncRNA | Long non-coding RNA |
| PBS | Phosphate buffer saline |
| CCK-8 | Cell counting kit-8 |
| CHD5 | Chromodomain helicase DNA binding protein 5 |
| 3′-UTR | 3′-untranslated region |
| FBS | Fetal bovine serum |
| cDNA | Complementary DNA |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase |
| shRNA | Short hairpin RNA |
| BCA | Bicinchoninic Acid |
| TBST | TBS-buffered solution |
| Mut | Mutant |
| Wt | Wild-type |
| SE | Standard error |
| p | p-value |
| OS | Overall survival |
| PFS | Progress-free survival |
Glioblastomas have peak degree for malignancy among gliomas and account for approximately 14.5% of all brain tumors (McKinnon et al., 2021; Grochans et al., 2022). Among the most refractory human cancers, glioblastomas are highly malignant and invasion and have high therapeutic resistance and recurrence. Combined therapies, including surgery, radiochemotherapy, and target and immune therapy, can improve patients’ survival to a certain extent (Rong et al., 2022; Kang et al., 2020), but prognoses remain poor. Hence, it is imperative to do additional research upon pathophysiology of glioblastoma and its associated gene regulatory mechanisms to identify new diagnostic indicators and treatment targets. This will ultimately result in enhanced patient outcomes.
Initially regarded as a transcriptional by-product, long non-coding RNA (lncRNA) contains over 200 nucleotide units and has no biological role because it cannot encode proteins (Yan and Bu, 2021). Nevertheless, lncRNAs play active roles in various biological processes, especially in tumor development, progression, proliferation, metabolism, drug resistance, and metastasis (Tan et al., 2021; Yan and Bu, 2021; Reggiardo et al., 2022). Researchers have studied the impact of lncRNA on the development and progression of glioblastoma. The study conducted by Zhong et al. (2021) demonstrated that lncRNA OXCT1-AS1 suppresses the advancement of glioblastoma through modulating the miR-195-5p/CDC25A axis, serving as a distinctive tumor indicator and a promising new treatment target for glioblastoma. LncRNA PRADX acts as a cancer driver and may serve as a potential therapeutic target for glioblastoma and colon adenocarcinoma (Li et al., 2021).
MiRNA is a family of non-coding RNA with 20–22 nucleotides that engages within functions of lncRNA and can control the expression of genes through bonding onto target mRNA’s 3′-untranslated region (3′-UTR). Various studies have revealed the involvement of several cohorts of lncRNA–miRNA interactions in glioblastoma onset and advancement (Wang et al., 2023; Zhu et al., 2020). LncRNA CRNDE contributes to advancing glioblastomas via the miR-9-5p sponge (Luo et al., 2021). Lnc-TALC is linked to temozolomide resistance through its competitive binding with miR-20b-3p, facilitating c-Met expression (Wu et al., 2019). However, although multiple functions are associated with lncRNA–miRNA complexes, the mechanisms that regulate glioma progression have not been elucidated and have failed to achieve valuable clinical translation.
Therefore, this study aimed to examine the expression of lncRNA DPP10-AS1 in glioblastomas together with eventual impact on the invasive and proliferative properties of glioblastoma. We focused on understanding how lncRNA DPP10-AS1 exerts biological effects and modulates the miR-24-3p/chromodomain helicase DNA binding protein 5 (CHD5) signaling pathway. This research’s findings could offer new perspectives and targets for glioblastoma treatment strategies.
A cohort of 43 individuals diagnosed with glioblastoma, comprising 25 males and 18 females, having mean age of (55.34 ± 6.71) years, were included within study. Surgical samples of glioblastoma and peritumoral tissues were collected. All samples were rinsed with sterile phosphate buffer saline (PBS), cut into small pieces, and soaked in Trizol (Invitrogen, Carlsbad, California, USA). The extraction of total RNA was performed under the directions provided through the manufacturer, and the extracted RNA was then stored at a temperature of −80°C to maintain its integrity for use in experimental procedures. The ethical committee of Soochow University’s Second Affiliated Hospital approved this study (approval number JD-LK-2018-027-02). Both those undergoing treatment and their families provided informed consent.
Human glioblastoma cell lines, including LN229 (ATCC Cat. No. CRL-2611), SNB19 (Cat. No. HTX2394), U251 (China National Collection of Authenticated Cell Cultures, Cat. No. TCHu 58), and U87 (ATCC Cat. No. HTB-14) were maintained in DMEM medium containing 10% fetal bovine serum (FBS) (Gibco, Carlsbad, California, USA). The cells were cultured in a controlled environment through cell incubator (ESPEC BNA-311, Yokohama, Japan) set at 37°C and a carbon dioxide concentration of 5%.
Real-time quantitative polymerase chain reaction (RT-qPCR) assay
Trizol® (Invitrogen, Carlsbad, California, USA) isolated total RNA, consequently undergoing reverse-transcription for complementary DNA (cDNA) through PrimeScript RT reagent (TaKaRa Bio™ Inc., Kyoto, Japan) depending upon kit protocols. RT-qPCR was then executed through SYBR Premix Ex Taq II® reagent (TaKaRa Bio™ Inc., Kyoto, Japan) according to instructions supplied through the manufacturer. The 2−ΔΔCt method was employed for data analysis. Suppl. Table S1 includes the primer sequences utilized in this investigation.
Developing expression vectors and transfecting cells
GenePharma, located in Shanghai, China, developed short hairpin RNA (shRNA) sequences that were particularly customized to target the lncRNA DPP10-AS1. These sequences were then cloned into a plasmid designated pcDNA3.1. through pcDNA3.1 plasmid, the lncRNA DPP10-AS1 overexpression vector was produced. Following confirming the presence of pertinent DNA sequences within a pcDNA3.1 plasmid, stable cell transfection was carried out via Lipofectamine 2000® (Invitrogen™, Carlsbad, California, USA) per provided methods. The positive cells were selected with G418. Infection efficiency was analyzed 72 h after transfection through RT-qPCR.
Cell counting Kit-8 (CCK-8) assessment
Cellular viability evaluation was conducted utilizing CCK-8 testing. The cells were first distributed on a 96-well plate, with individual wells harboring 100 µL culturing media together with a cell population of 1 × 103 cells. Afterward, as directed through the supplier (MedChemExpress, Monmouth Junction, New Jersey, USA), 10% CCK-8 reagent was applied to the cell monolayers in each well at 0, 24, 48, and 72 h. At an optical density of 450 nm, the absorbance was determined through microplate-reader platform (Thermo Fisher Scientific, Carlsbad, California, USA).
An assay for colony formation determined cellular proliferative properties. Cultures were seeded with 800 cells per well for 48 h, then put in a six-well plate and left for two weeks of incubation. After incubation, cellular colonies were treated with a 4% paraformaldehyde solution (ten minutes) to fix them. Following fixation, the colonies were exposed to a 20-min crystal violet stain-step. Subsequently, colonies of a quantity above 50 individual cells were enumerated.
Invasion assay was conducted in Transwell chambers (8 μm; Costar, Corning, New York, USA). To mimic the extracellular matrix, the basement membrane matrix (BD Biosciences™, Franklin Lakes, New Jersey, USA) was placed at the bottom of the top chamber after being diluted within serum-free medium to 1:40 ratio. The upper chamber was then populated with 1 × 104 cells per well. Cells were grown for two days within DMEM enriched through 10% FBS within lower chamber. Post-10-min 4% paraformaldehyde staining, cells at bottom of lower chamber were stained through crystal violet and underwent three PBS washes. Cells were examined and evaluated through inverted microscopy (Olympus™, Tokyo, Japan).
Total cell proteins were extracted using PIPA buffer (Beyotime™, Nanjing, China). The proteomic level was measured through Bicinchoninic Acid (BCA) assessment assembly (Beyotime™, Nanjing, China) depending upon kit protocols. A PVDF membrane (Millipore, Bedford, MA, USA) was then used to transfer 40 μg of the total protein for separation using 10% SDS-PAGE gel electrophoresis. Further, the membrane was blocked for 2 h in 5% BSA and Tween 20-containing TBS-buffered solution (TBST), following the manufacturer’s guidelines. After three TBST washes, the membrane was placed into incubation (120 min) at RT through HRP-conjugated second antibody (ab6721, 1:2000, Abcam, Cambridge, UK). The protein bands were identified using enhanced chemiluminescence with an Amersham Imager 600 (General Electric Company, Boston, USA).
Subcellular localization for lncRNA DPP10-AS1 was predicted using an online bioinformatics assay (Biosearch Technologies™ Novato, California, USA; https://www.biosearchtech.com; Peta-luma, CA). After washing the cells in PBS, they were fixed for 10 min in a 4% formaldehyde solution free of RNases for the RNA-FISH test. After that, PBS with 0.1% Triton X-100 was introduced, whereby cells were allowed to rest on ice for 5 min. After being treated sequentially with acetylation reagent, glycine, and protease K, 400 μL of prehybridization solution was supplemented and incubated (60 min, 42°C). Afterward, cells were hybridized with 400 μL (300 ng/mL) of lncRNA DPP10-AS1-FISH probe (Ribobio, Guangzhou, China) overnight at 4°C. Nuclei were observed using an fluorescent microscope (Olympus, Tokyo, Japan) after being counterstained with DAPI solution (Beyotime, Shanghai, China).
Dual-luciferase reporter assay
Potential miRNA binding sites with lncRNA DPP10-AS1 were predicted through the online prediction software LncBase Predicted (https://diana.e-ce.uth.gr/lncbasev3) database. To produce expected mutant (Mut) or wild-type (Wt) miR-24-3p bonding locations, lncRNA DPP10-AS1 segments were synthesized and cloned into the pGL4.50[luc2/CMV/Hygro] (RiboBio) vector. Through Lipofectamine 2000 transfection kit (Invitrogen, USA), 200 ng of luciferase reporter plasmid carrying lncRNA DPP10-AS1 Wt or Mut 3′UTR was transfected into LN229 cells. Dual-luciferase reporter (Promega™, Madison, Wisconsin, USA) quantified luciferase activity 48 h after transfection, as per kit protocols. Firefly luciferase intensity was standardized using Renilla luciferase activity. There were three biological replicates for every three times the dual-luciferase reporter assay was performed.
Shanghai Experimental Animal Center in China provided the female Balb/c athymic nude mice kept in micro-isolated cages (NASA 1000) with sterile bedding. The Chinese Experimental Animal Association and Soochow University’s ethics committee have authorized the facilities where all animal studies were carried out (SUDA20210708A03). In summary, tumor cells within logarithmic development phase were suspended in PBS and afterward administered through subcutaneous injection into the right axillary region of mice. Each mouse received a dosage of 1 × 106 cells, and the mice (weight = 20 g) used within experiment were across 6 and 8 weeks old. Every 5 days following tumor cell inoculation, the mice were weighed, and the size of the tumor was determined through formula (a2 × b/2), where ‘a’ is the width and ‘b’ is the length. After 30 days following the inoculation of tumor cells, all mice were euthanized while under the influence of general anesthesia. Subsequently, the xenografts were collected, their weights were measured, and photographs were taken. The xenografts’ expression profiles for DPP10-AS1 and miR-24-3p were assessed through the RT-qPCR method.
The data underwent analysis through SPSS 20.0 statistical software. Datasets reflected mean ± standard error (SE). A t-test probed statistical significance across two cohorts. We conducted multiple comparisons through a one-way analysis of variance, while correlation assessment was carried out employing Pearson’s correlation approach. p < 0.05 was deemed to confer statistical significance, with assessments all conducted in triplicate.
Low level of lncRNA DPP10-AS1 in glioblastoma is following poor prognosis
Based upon published glioma lncRNA sequencing data (GSE7696 and TCGA-glioblastoma), an online bioinformatics profile of lncRNA associated with glioblastomas was conducted for probing molecule-based control mechanistics for ncRNA within genesis and advancement for glioblastomas. The screening conditions for vital down-regulated lncRNA were p-value <0.001 and Log2FC < −1. The intersections across two databases resulted in 15 common lncRNA, namely, MIR7-3HG, LINC00320, RFPL1S, HAR1A, EMX2OS, SLC26A4-AS1, DPP10-AS1, DCTN1-AS1, PRKAG2-AS1, OIP5-AS1, MIR124-2HG, LINC00507, LINC00622, and LINC00294 (Fig. 1A). TCGA data evaluations demonstrated expression of only lncRNA DPP10-AS1 correlated with patient prognoses, based upon investigation of lncRNA that may be employed as a therapeutic target for glioblastomas or prognosis evaluation. lncRNA DPP10-AS1 from the transcript of functional gene DPP10, associated with autism, ALS, and neurodegeneration, was selected for further evaluation.
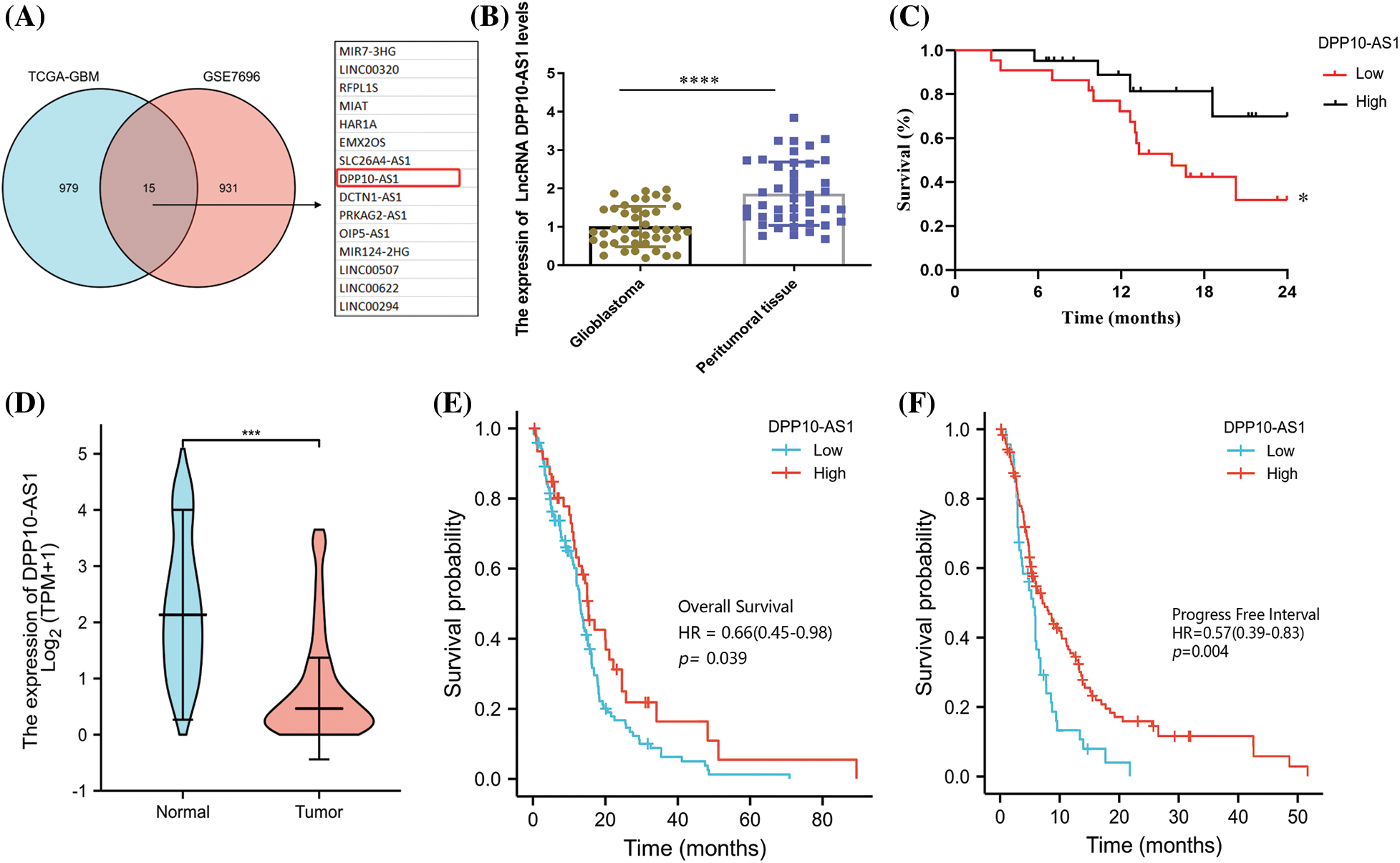
Figure 1: Association of low expression of DPP10-AS1 in glioblastoma tissue with patient survival. (A) GSE7696 and TCGA-glioblastoma screening for significantly different down-regulated lncRNA in glioblastoma. The criteria for evaluation were Log2|FC| >1 and p-value <0.001. (B) RT-qPCR of LncRNA DPP10-AS1 expression in 43 patient glioblastoma tissues. (C) Overall survival (OS) curves of glioblastoma patients with low LncRNA DPP10-AS1 expression and high LncRNA DPP10-AS1 expression. (D) TCGA database analysis of DPP10-AS1 expression. (E, F) Analysis of the TCGA database demonstrating the correlation across patient OS and progress-free survival (PFS) and DPP10-AS1 expression in glioblastoma tissue. *p <0.05; ***p < 0.001; ****p < 0.0001.
To verify the bioinformatics results of lncRNA DPP10-AS1, we collected 43 tumor surgical specimens from patients with glioblastoma and peritumor tissues. To detect the expression of the lncRNA DPP10-AS1 through fluorescence quantitative PCR, total RNA was collected from the specimens. lncRNA DPP10-AS1 level decreased significantly in patient-derived glioblastoma specimens compared with that of peritumor tissues (Fig. 1B). Depending upon median expression level for lncRNA DPP10-AS1, participants were categorized within two cohorts: low- and high-expression cohorts. Patients with low lncRNA DPP10-AS1 expression showed substantially lower overall survival (OS) rates in comparison with individuals having high lncRNA DPP10-AS1 expression, according to the results of a 2-year follow-up (p = 0.0425; Fig. 1C). An examination of the TCGA database was done to determine how lncRNA DPP10-AS1 was expressed in glioblastoma patients. It was observed that lncRNA DPP10-AS1 was not highly expressed in glioblastomas (Fig. 1D) and that patients who had higher levels of lncRNA DPP10-AS1 expression had longer OS times than those who had lower levels (p = 0.039; Fig. 1E). PFS analysis revealed that patients with high lncRNA DPP10-AS1 expression in glioblastoma tissue exhibited substantially longer PFS than patients with low lncRNA DPP10-AS1 expression (p = 0.004; Fig. 1F). Therefore, there is a favorable relationship across lncRNA DPP10-AS1 expression level within glioblastoma tumor tissue together with case prognoses.
The overexpression or knockdown of lncRNA DPP10-AS1 impacted glioblastoma cells’ proliferative, invasive, and migrative properties
Fluorescence quantitative PCR was employed to compare the lncRNA DPP10-AS1 expression levels within glioma cell lines (U251, LN229, SNB19, and U87) into healthy human astrocytes (NHAs). Compared to normal human astrocytes, the data demonstrated a substantial decrease in glioblastoma cells’ expression of the lncRNA DPP10-AS1 (p < 0.05). DPP10-AS1 lncRNA expression levels were significantly high in SNB19 and U251 cells but lower in LN229 and U87 cells (Fig. 2A). Therefore, lncRNA DPP10-AS1 knockdown was performed for SNB19 cells. Specific shRNAs (shRNA-1 and shRNA-2) were used. The transfection of lncRNA DPP10-AS1-targeting shRNAs efficiently suppressed the endogenous expression of lncRNA DPP10-AS1 within SNB19 cells (Fig. 2B). Additionally, the lncRNA DPP10-AS1 expression within LN229 cells was elevated through transfection with the lncRNA DPP10-AS1 overexpression vector (pcDNA-AS1) (p < 0.05, Fig. 2C).
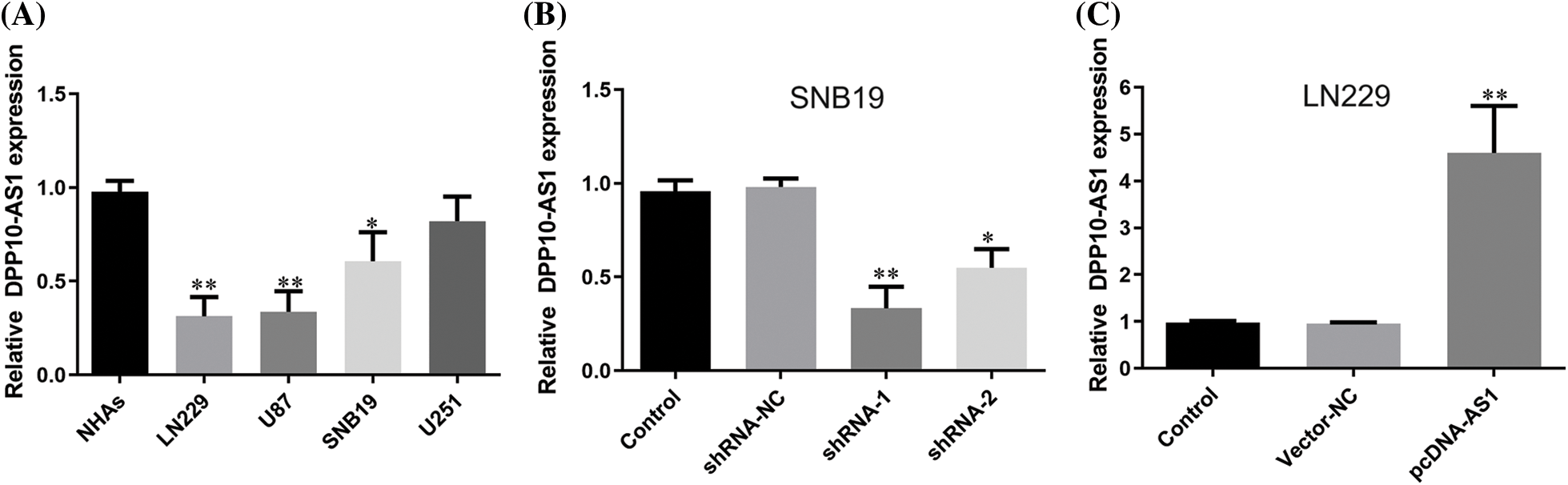
Figure 2: Up- or down-regulation of DPP10-AS1 in glioblastoma cell lines. (A) Fluorescence quantitative PCR analysis for detecting DPP10-AS1 level of glioblastoma cell lines. (B) PCR analysis on DPP10-AS1 knockdown of SNB19 cells. (C) DPP10-AS1 overexpression of LN229 cells. *p < 0.05; **p < 0.01.
CCK-8 data demonstrated SNB19 cell growth to be dramatically suppressed through lncRNA DPP10-AS1 suppression (p < 0.05, Figs. 3A, 3C). In addition, lncRNA DPP10-AS1 overexpression increased LN229 cell proliferation and colony formation (p < 0.05, Figs. 3B, 3D). Additionally, the influence of lncRNA DPP10-AS1 knockdown upon glioblastoma cell invasive and migrative properties was evaluated through Transwell assay, which demonstrated that lncRNA DPP10-AS1 knockdown facilitated SNB19 cell invasive and migrative (Fig. 4A). The upregulation of lncRNA DPP10-AS1 showed a substantial suppression of migration and invasiveness by LN229 cells (p < 0.05, Fig. 4B).
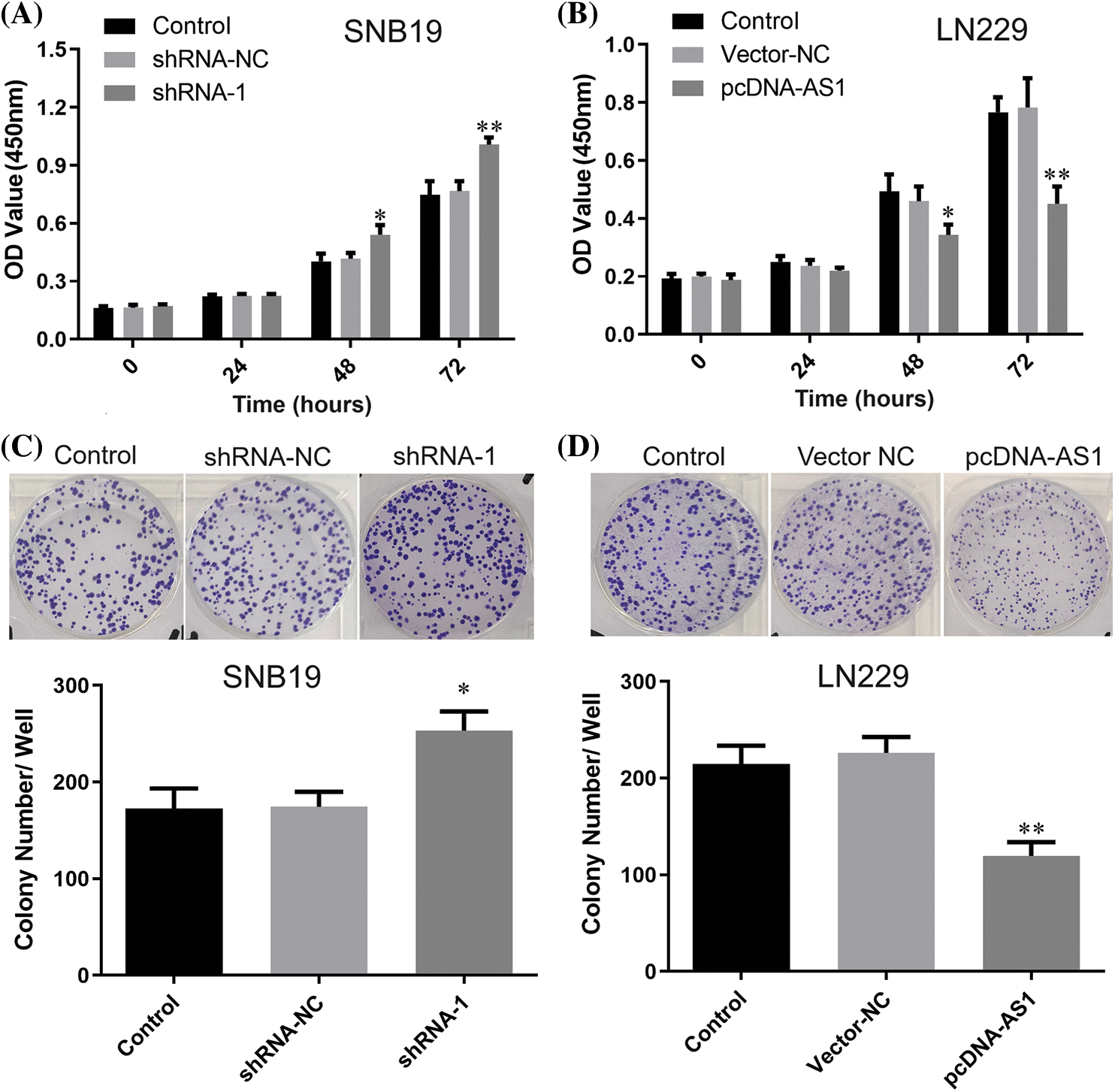
Figure 3: Knockdown or overexpression of lncRNA DPP10-AS1 affected glioblastoma cell proliferation. (A, B) The CCK-8 assay assessed the impact of DPP10-AS1 knockdown or overexpression on the proliferation of SNB19 or LN229 cells. (C, D) Clonogenic assay verified the influence of knockdown or overexpression of DPP10-AS1 upon forming ability of SNB19 or LN229 cells. *p < 0.05; **p < 0.01.
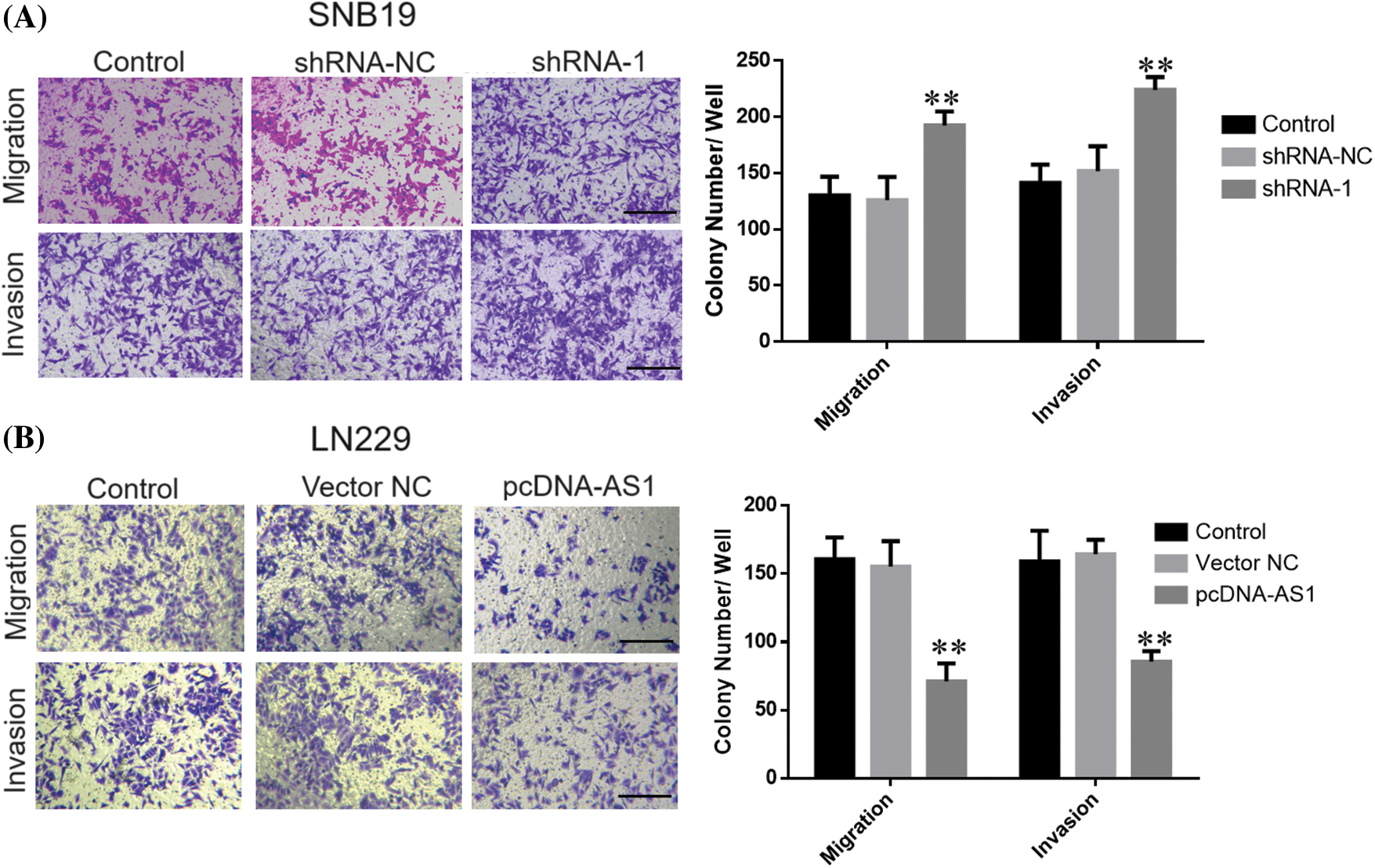
Figure 4: Knockdown or overexpression of LncRNA DPP10-AS1 affected glioblastoma cells’ invasive and migrative abilities. (A) Transwell assay showed DPP10-AS1 knockdown promoted invasiveness and migration of SNB19 cells (bar: 100 μm). (B) DPP10-AS1 overexpression inhibited the invasiveness and proliferation of LN229 cells (bar: 100 μm). **p < 0.01.
In glioblastoma, the lncRNA DPP10-AS1 was found within cytoplasm and functioned as a miR-24-3p sponge
One tool that was utilized to forecast the distribution of lncRNA in five different subcellular components—the cytoplasm, nucleus, ribosomes, and exosomes—was lncLocator (http://www.csbio.sjtu.edu.cn/bioinf/lncLocator/). Findings indicated cytoplasm as main location for lncRNA DPP10-AS1 (Fig. 5A). Similarly, lncRNA DPP10-AS1 overexpression in SNB19 cells demonstrated through RNA-FISH experiment that it was primarily found within cytoplasm (Fig. 5B), and lncRNA DPP10-AS1 had the highest content within cytoplasm (approximately 80%; Fig. 5C). The mechanism of competing endogenous RNA (ceRNA) suggests that lncRNA DPP10-AS1 can function as a competitive endogenous RNA, participating within regulation of downstream target gene expression after directly regulating corresponding intracellular miRNA. This mechanism could be important within development of tumors.
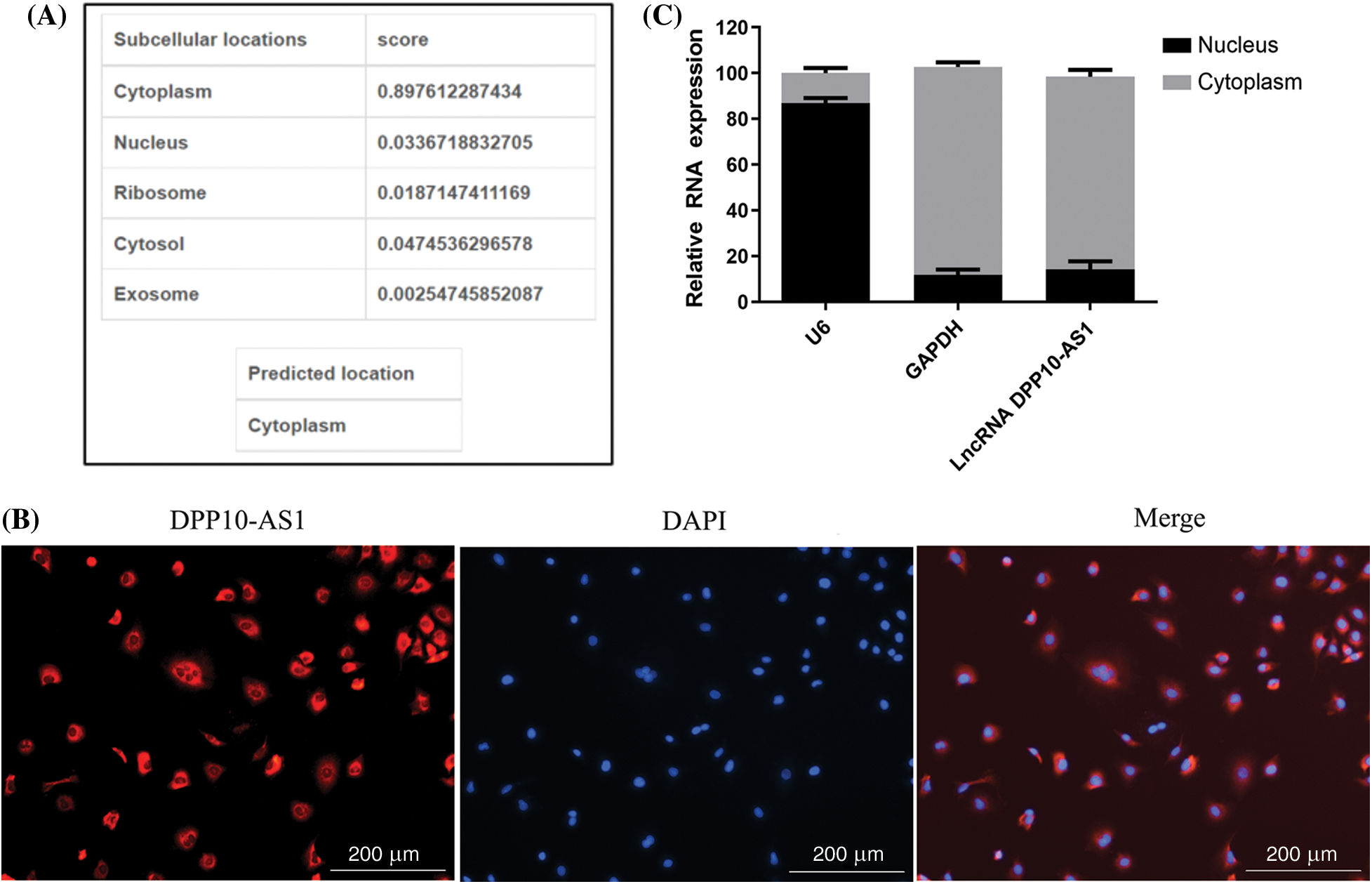
Figure 5: DPP10-AS1 subcellular localization. (A) Bioinformatics prediction of subcellular localization of DPP10-AS1. (B) RNA-FISH analysis on DPP10-AS1 subcellular localization, fluorescence tracing of intracellular DPP10-AS1 distribution (bar: 200 μm). (C) Fluorescence quantitative PCR for subcellular DPP10-AS1 quantification.
We explored an online database for potential miRNA molecules interacting with lncRNA DPP10-AS1. LncBase Predicted v.3 (https://dianalab.e-ce.uth.gr/html/diana/web/index.php?r) and miRcode (http://www.mircode.org/) were utilized. Predicted v.3 predicted 37 miRNAs, while miRcode predicted 22 miRNAs. There are 6 miRNAs for the intersection of predicted miRNA: hsa-miR-128-3p, hsa-miR-338-3p, hsa-miR-211-5p, hsa-miR-204b, hsa-miR-9-5p, and hsa-miR-24-3p (Fig. 6A). We found that knocking down lncRNA DPP10-AS1 can significantly elevated miR-24-3p expression without affecting expression for other miRNA molecules and lncRNA DPP10-AS1 overexpression can significantly decrease miR-24-3p expression (p < 0.01, Figs. 6B, 6C). According to lncRNA DPP10-AS1 and miR-24-3p binding sites predicted online, Wt and Mut luciferase reporter gene vectors were constructed (Fig. 6D). Then, we performed luciferase reporter assay with the two glioblastoma cell lines (SNB19and LN229). When compared to the miRNA-NC cohort, the Wt lncRNA DPP10-AS1 vector’s miR-24-3p mimics dramatically decreased luciferase activity (p < 0.01, Fig. 6E). Nevertheless, no changed luciferase activity was seen within miRNA-NC and miR-24-3p cohorts within Mut-type lncRNA DPP10-AS1 (p > 0.05, Fig. 6F), suggesting that lncRNA DPP10-AS1 can absorb miR-24-3p in SNB19 and LN229 cells.
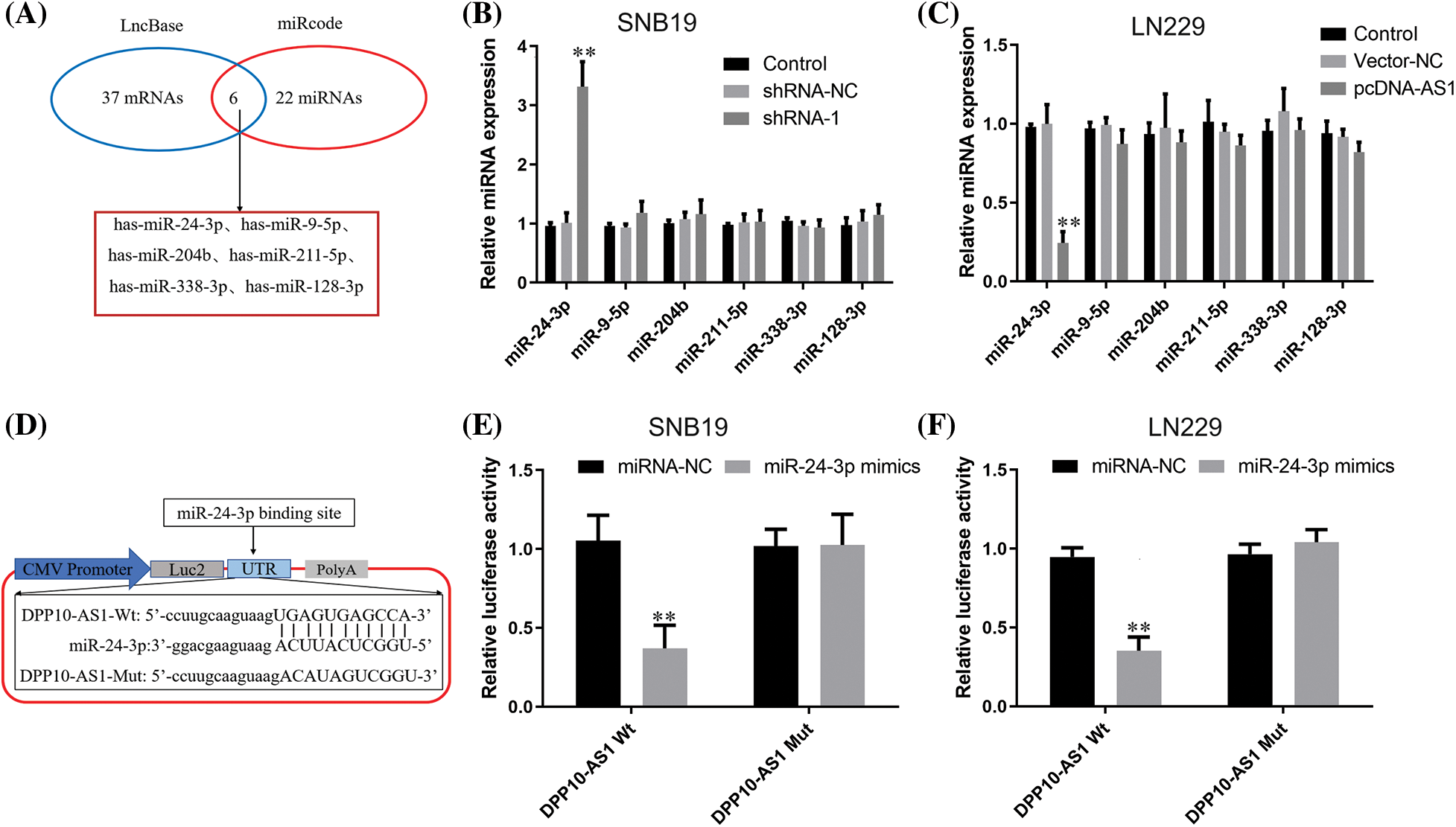
Figure 6: LncRNA DPP10-AS1 can act as a sponge for miR-24-3p in glioblastomas. (A) MiRcode and LncBase Predicted v.3 were utilized to forecast the interacting miRNA of LncRNA DPP10-AS1. The anticipated common miRNA targets of LncRNA DPP10-AS1 are displayed within Venn diagram. (B) Following LncRNA DPP10-AS1 knockdown, the expression of miRNA was measured through RT-qPCR. (C) The expression of miRNA after LncRNA DPP10-AS1 overexpression was measured through RT-qPCR assay. (D) The online predicted lncRNA DPP10-AS1 and miR-24-3p binding sites. (E, F) Detection of DPP10-AS1 and binding activity of miR-24-3p via the luciferase reporter assay. **p < 0.01.
RT-qPCR probed miR-24-3p expression within glioblastoma cultures and clinical tumor samples. MiR-24-3p expression profiles were considerably greater in U87, LN229, SNB19, and U251 cells than in NHAs (p < 0.05, Fig. 7A). When glioblastoma tissues were compared to peritumoral tissues, miR-24-3p was shown to be overexpressed (p < 0.001, Fig. 7B). Pearson’s correlation analysis demonstrated a substantial inverse association across miR-24-3p expression and the lncRNA DPP10-AS1 expression in glioblastoma tissues (p = 0.002, Fig. 7C).
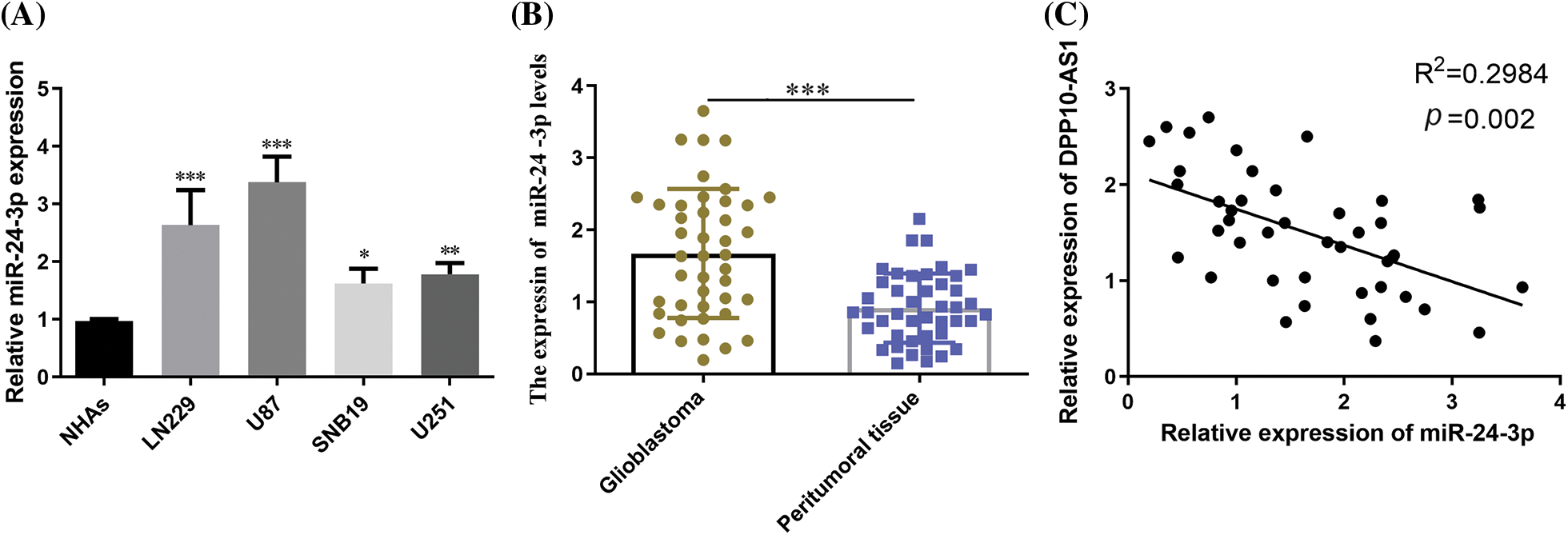
Figure 7: miR-24-3p expression in glioblastoma cell lines and clinical tumor tissues. (A) RT-qPCR examination of glioblastoma cell lines' miR-24-3p expression. (B) The expression of miR-24-3p in glioblastoma tumor tissues. (C) The correlation across miR-24-3p and lncRNA DPP10-AS1 was analyzed using Pearson’s correlation coefficient. *p < 0.05; **p < 0.01; ***p < 0.001.
MiR-24-3p regulated its downstream target gene CHD5
We utilized TargetScan (https://www.targetscan.org/vert_72/) and MiRnada (http://mirtoolsgallery.tech/mirtoolsgallery/node/1055) to forecast potential target genes for probing putative downstream targets for miR-24-3p further. According to the score, FGFR3, CHD5, and RBP4 were the most likely downstream target genes regulated through miR-24-3p. Following that, RT-qPCR examination demonstrated that transfection of miR-24-3p mimics inhibited CHD5 expression (p < 0.05), enhanced RBP4 expression (p < 0.01), exhibiting nil function upon FGFR3 expression in SNB19 cells (with low miR-24-3p expression) (Fig. 8A). In cells of the SNB19 cell line with elevated expression of miR-24-3p, introducing miR-24-3p inhibitor led to notable increase CHD5 expression (p < 0.01). Simultaneously, it did not yield any substantial impact upon expression of FGFR3 and RBP4 (Fig. 8B). The outcomes of the present study suggest that miR-24-3p can modulate the expression of CHD5. Luciferase reporter gene vector was developed based upon predicted binding sequences obtained from an online database. The vector mutant was constructed through site-directed mutagenesis, explicitly targeting the attachment site sequence of CHD5 and miR-24-3p (Fig. 8C). Dataset outcomes for luciferase experiment indicated introduction of the miR-24-3p mimic led to considerable repression in luciferase activity (p < 0.01). Transfecting miR-24-3p mimic, conversely, did not influence luciferase function for CHD5 wild-type (Wt) cells (Fig. 8D). Data obtained through Western blot assessment demonstrated miR-24-3p overexpression led to a substantial reduction within CHD5 expression (p < 0.01, Fig. 8E). While inhibition of miR-24-3p resulted in an upregulation of CHD5 expression.
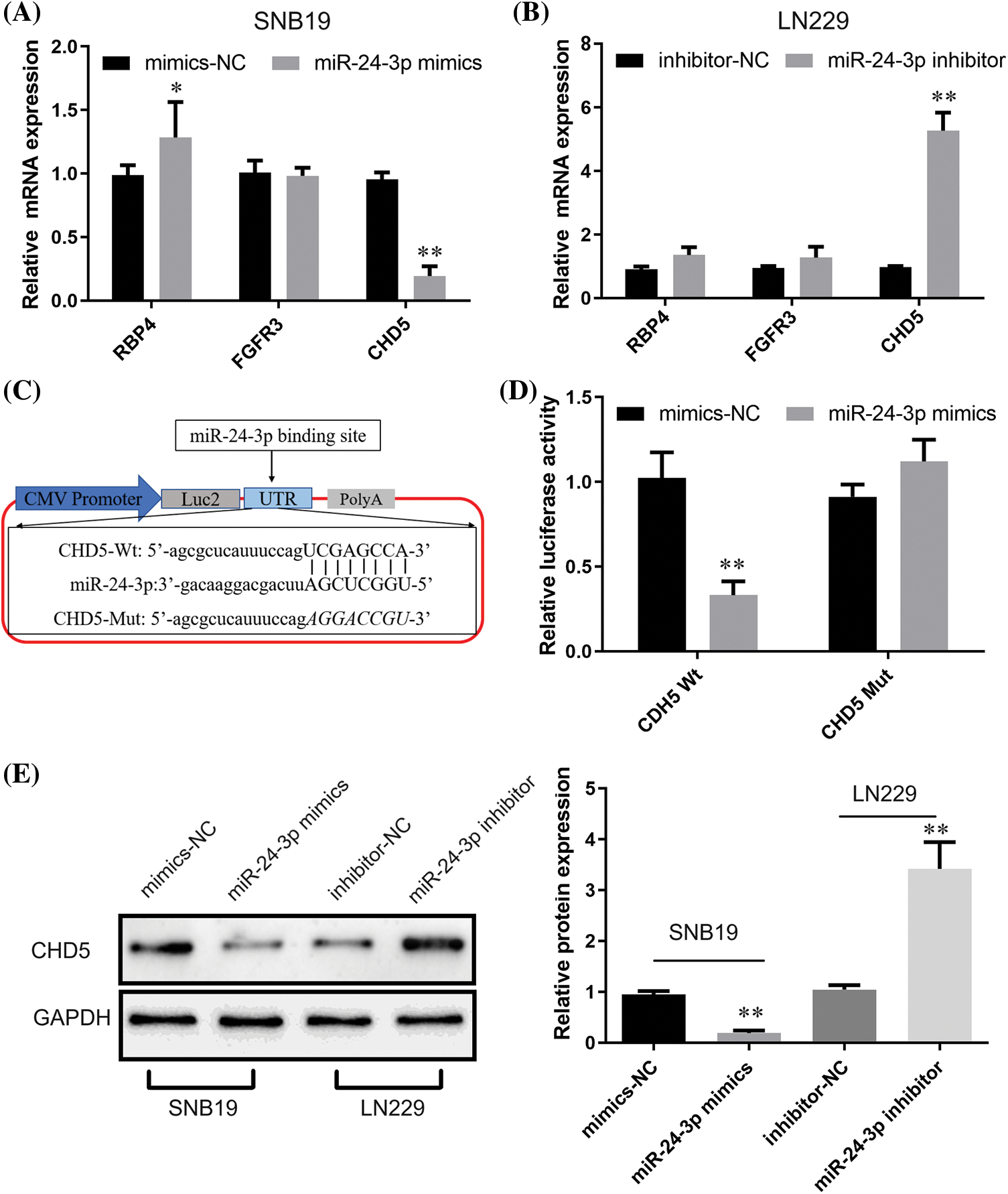
Figure 8: Regulatory effects of miR-24-3p on CHD5. (A, B) The impacts of miR-24-3p up- or down-regulation upon expression of downstream target genes were revealed through RT-qPCR. (C) Bioinformatics prediction of CHD5 and miR-24-3p binding sites. (D) Luciferase reporter gene assay indicated the binding activity of CHD5 and miR-24-3p. (E) The Western blot analysis revealed the impact of downregulating or upregulating miR-24-3p upon expression of CHD5 within experimental setting. *p < 0.05; **p < 0.01.
lncRNA DPP10-AS1 regulated glioblastoma development through the miR-24-3p/CHD5 axis
We performed Western blot analysis to detect whether lncRNA DPP10-AS1 affected CHD5 expression through regulating miR-24-3p in SNB19. For the miR-24-3p inhibitor-NC cohort, lncRNA DPP10-AS1 knockdown dramatically reduced CHD5 expression (p < 0.01). The suppressive potential of shRNA-1 knockdown on CHD5 expression was inhibited through miR-24-3p thwarting transfection within lncRNA DPP10-AS1 knockdown cohort, suggesting that miR-24-3p sponging significantly controlled intracellular expression for lncRNA DPP10-AS1 within glioblastoma cells. CHD5 expression was decreased due to LncRNA DPP10-AS1 knockdown, increasing the intracellular level of miR-24-3p (Fig. 9A).
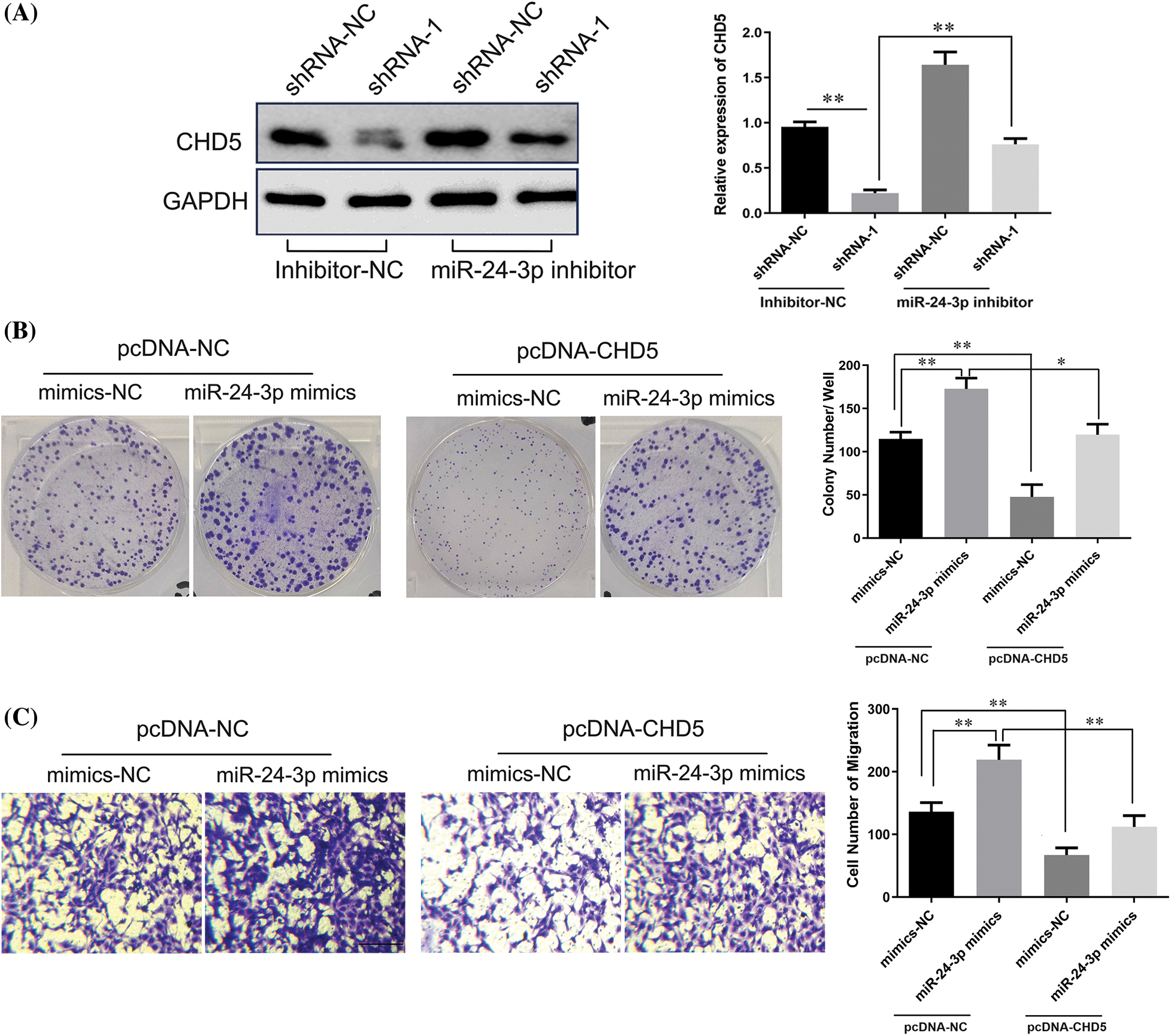
Figure 9: LncRNA DPP10-AS1 regulated CHD5 expression through sponging miR-24-3p. (A) The results obtained from the Western blot analysis demonstrated that DPP10-AS1 had a role in regulating CHD5 expression in SNB19 cells through its interaction with miR-24-3p. (B) Clone formation assay to evaluate the role of miR-24-3p on SNB19 cell proliferation through CHD5. (C) The Transwell experiment was employed to investigate the impact of miR-24-3p on SNB19 cell proliferation and CHD5 regulation (bar: 100 μm). *p < 0.05; **p < 0.01.
Further analysis indicated that miR-24-3p mimic transfection promoted clone formation and the cell migrative abilities of glioblastoma cells. In SNB19 cells, CHD5 overexpression significantly repressed colony formation and cell migrative abilities (p < 0.05). CHD5 overexpression dramatically reduced the impact of overexpression of miR-24-3p on SNB19 cellular migrative and colony formation within miR-24-3p mimic cohort (Figs. 9B, 9C).
In vivo, analysis in a subcutaneous xenografts model further confirmed these findings. Ten days after LN229 cell inoculation, the width and length of each subcutaneous tumor were analyzed every 5 days. Tumor-bearing mice were sacrificed 1 month after tumor cell inoculation, and xenografts were harvested. The weight of the tumor and volume declined significantly within lncRNA DPP10-AS1 overexpression cohort (p < 0.01, Figs. 10A–10C). lncRNA DPP10-AS1 expression level increased significantly within xenografts of lncRNA DPP10-AS1 overexpression cohort (p < 0.01, Fig. 10D), whereas miR-24-3p level decreased significantly (p < 0.01, Fig. 10E).
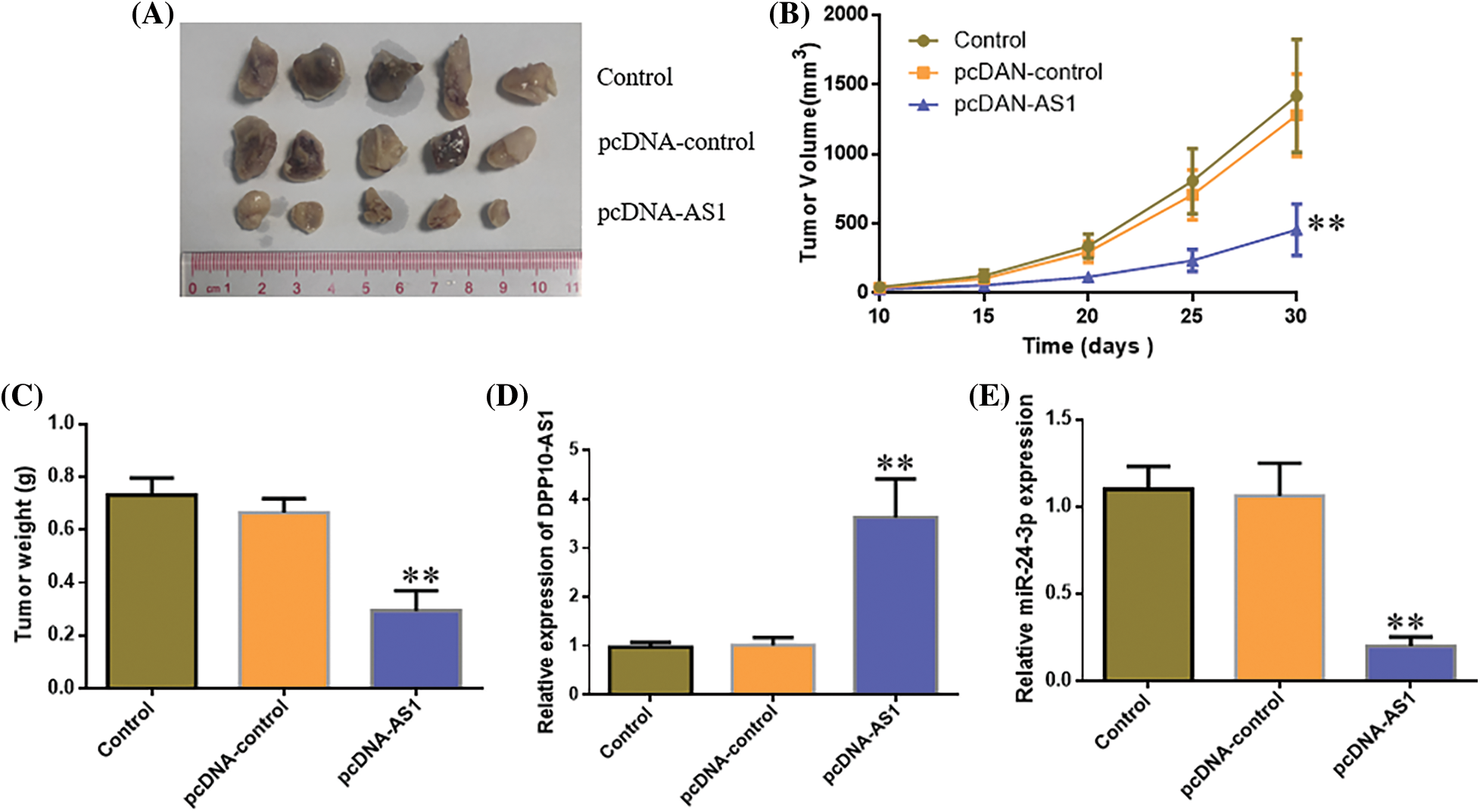
Figure 10: In vivo evaluation of the role of DPP10-AS1 in glioblastoma growth regulation via miR-24-3p. (A) Tumor size in each cohort after tumor cell inoculation for 30 days. (B) Tumor growth curve 10–30 days after inoculation. (C) Tumor weights 30 days after inoculation. (D and E) Real-time quantitative PCR analysis on DPP10-AS1 and miR-24-3p levels in xenografts tumor tissues. **p < 0.01.
Across multiple investigational models of glioblastoma formation, lncRNA DPP10-AS1 was shown to downregulate the transcript level in glioma tissues. lncRNA DPP10-AS1 bound to miR-24-3p competitively and regulated CHD5 expression to suppress the invasive, migrative, and proliferative capabilities of glioblastoma in vitro, indicating potential clinical significance of lncRNA DPP10-AS1 and valuable role in serving as a candidate for glioblastoma diagnosis and treatment.
Aberrantly expressed functional lncRNA was found in cell lines and clinical tumor tissue samples, which is crucial for tumor and tissue remodeling, development, progression, and therapeutic resistance (Peng et al., 2018). LncRNA PLAC2 can interact with the transcriptional activator STAT1 and the promoter of ribosomal protein RPL36 in gliomas, thereby reducing RP36 expression, inducing cell cycle arrest, and inhibiting cell proliferation (Hu et al., 2018). LINC01116 can regulate VEGFA expression in gliomas through miR-31-5p and participate in tumor angiogenesis (Ye et al., 2020). within present research, glioblastomas expressed significantly less lncRNA DPP10-AS1 than peritumor tissues. The in vitro functional studies indicated that the SNB19 cell invasiveness, migration, and proliferation can all be enhanced through lncRNA DPP10-AS1 suppression.
Moreover, this effect has been associated with higher levels of the expression of N-cadherin and vimentin, and decreased levels of E-cadherin. Furthermore, overexpression of the lncRNA DPP10-AS1 can reduce the expression levels of N-cadherin and vimentin, and prevent the migration, proliferation, and invasion of LN229 cells. A robust correlation has been observed in earlier research across development of prostate cancer and lncRNA DPP10-AS1. Tetramethylpyrazine has been demonstrated to lessen the aggressiveness of prostate cancer through altering the DPP10-AS1/CBP/FOXM1 signaling pathway (Zhou et al., 2020). The anti-tumor impact of lncRNA DPP10-AS1 in colorectal cancer cells is achieved by upregulating ADCY1 through the regulation of miR-127-3p, hence impeding cell invasion and proliferation (Liu et al., 2021). LncRNA DPP10-AS1 predicts unfavorable outcomes in lung cancer patients through promoting malignant progression through the epigenetic activation of its cognate gene DPP10 (Tian et al., 2021). LncRNA DPP10-AS1, as a tumor-regulatory ncRNA, has essential biological regulatory roles within development of prostate, colon, and lung cancer. This work uncovers the clinical significance of lncRNA DPP10-AS1 in treating glioblastoma through thoroughly investigating its role and mechanism in glioblastoma development.
LncRNA may act as a miRNA sponge during mRNA translation or stability regulation, affecting the associated signaling pathways. lncRNA DPP10-AS1 was disclosed to be localized in cytoplasm within current studies, which implied that there may exist a miRNA binding site to lncRNA DPP10-AS1. through regulating the concentration of specific miRNA within cytoplasm, the “silencing effect” on its downstream target gene can be relieved, thereby controlling the target gene expression. The luciferase reporter gene experiment revealed that lncRNA DPP10-AS1 has a complementary binding site that complements miR-24-3p, demonstrating the mutual interaction across two. In comparison with NHAs, miR-24-3p level enhanced abnormally in most glioblastoma cell lines (LN229, U87, SNB19, and U251) and exhibited a negative correlation with lncRNA DPP10-AS1 expression. lncRNA DPP10-AS1 inhibited glioblastoma growth via significantly down-regulating miR-24-3p, thus implying that lncRNA DPP10-AS1 can inhibit glioblastoma growth through regulating miR-24-3p, which offered a molecular basis for hindering glioblastoma progression. Specific target genes involved in tumor growth, migration, and invasion can be controlled through miR-24-3p. MiR-24-3p downregulation can inhibit the growth and propensity for prostate cancer metastasis (Kong et al., 2021).
While focusing on ING1 of miR-24-3p overexpression can accelerate the growth of colon cancer (Gao et al., 2020). Additionally, MiR-24-3p facilitated lung cancer cell proliferation, migration, and invasion (Wang et al., 2021). SOX7 and SOX18, two of miR-24-3p’s downstream target genes, are all linked to the development of cancer, including lung, colon, and prostate cancer (Cheng et al., 2021).
The present research reveals CHD5 as a novel gene targeted through miR-24-3p. As mentioned above, the protein exhibits strong tumor-suppressive properties and substantially influences (Xu et al., 2022). CHD5 expression in NSCLC and colorectal cancer can be an independent prognostic biomarker (Zhu et al., 2019; Zhou et al., 2018). In addition, CHD5 expression can be regulated through miRNA in gliomas, influencing the invasion and proliferation of glioma in vitro (Liu et al., 2018). By modulating the miR-301b-3p/CHD5 axis, the lncRNA SLC16A1-AS1 increased radiosensitivity while suppressing cell proliferation and invasiveness in hepatocellular cancer (Pei et al., 2020). As a result, CHD5 plays an essential physiological regulating role in tumor growth. MiR-24-3p can bind to the 3′UTR of CHD5 and regulate its expression. Functional investigations demonstrated that overexpression of CHD5 can effectively inhibit the effect of miR-24-3p overexpression on the migration and proliferation of glioblastomas. Because of this, lncRNA DPP10-AS1 can affect the ability of glioblastoma to proliferate and invade through controlling the miR24-3p/CHD5 axis.
In conclusion, suppression of the lncRNA DPP10-AS1 was detected in cell lines and clinical samples, and it was strongly linked to the prognoses and outcomes of glioblastoma patients. lncRNA DPP10-AS1 controls the miR24-3p/CHD5 axis, inhibiting glioblastoma invasion and proliferation. Hence, it can be concluded that lncRNA DPP10-AS1 has the characteristics of a newly discovered tumor suppressor, making it a promising candidate for both diagnostic and therapeutic interventions. Owing to advancements in RNA-based therapeutics, the regulatory role of lncRNA DPP10-AS1 is an intriguing therapeutic target. RNA-based drugs targeting lncRNA DPP10-AS1 can restore normal glioblastoma expression, suppressing tumor proliferation and invasion. Additionally, given that lncRNA DPP10-AS1 modulates the miR-24-3p/CHD5 axis, it can be used to develop drugs targeting this axis to enhance treatment outcomes further. This approach may improve therapeutic efficacy and prognosis for patients with glioblastoma.
Acknowledgement: None.
Funding Statement: This study was supported through the Natural Science Foundation of Jiangsu Province (No. BK20201172), the Key Project of the Jiangsu Health Commission (No. ZDB2020016), and the Jiangsu Province Key Research and Development Program: Social Development Project (No. BE2021653).
Author Contributions: The authors confirm their contributions to the paper as follows: study conception and design: JS and LX; data collection: YZ and JF; analysis and interpretation of results: XL and JD; draft manuscript preparation: HL. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Ethics Approval: This study was approved through the Ethics Committee of the Second Affiliated Hospital of Soochow University (JD-LK-2018-027-02) with the informed consent of the patients and their families.
Conflicts of Interest: The authors declare that they have no competing interests, and all authors should confirm its accuracy.
References
Cheng Y, Xiong HY, Li YM, Zuo HR, Liu Y, Liao GL (2021). LncRNA HOXA11-AS promotes cell growth through sponging miR-24-3p to regulate JPT1 in prostate cancer. European Review Medical and Pharmacological Sciences 25: 4668–4677. https://doi.org/10.26355/eurrev_202107_26377 [Google Scholar] [PubMed] [CrossRef]
Gao Z, Zhou L, Hua S, Wu H, Luo L, Li L, Wang S, Liu Y, Zhou Z, Chen X (2020). miR-24-3p promotes colon cancer progression through targeting ING1. Signal Transduction and Targeted Therapy 5: 171. https://doi.org/10.1038/s41392-020-0206-y [Google Scholar] [PubMed] [CrossRef]
Grochans S, Cybulska AM, Simińska D, Korbecki J, Kojder K, Chlubek D, Baranowska-Bosiacka I (2022). Epidemiology of glioblastoma multiforme-literature review. Cancers 14: 2412. https://doi.org/10.3390/cancers14102412 [Google Scholar] [PubMed] [CrossRef]
Hu YW, Kang CM, Zhao JJ, Nie Y, Zheng L, Li HX, Li X, Wang Q, Qiu YR (2018). LncRNA PLAC2 down-regulates RPL36 expression and blocks cell cycle progression in glioma through a mechanism involving STAT1. Journal of Cellular and Molecular Medicine 22: 497–510. https://doi.org/10.1111/jcmm.13338 [Google Scholar] [PubMed] [CrossRef]
Kang X, Zheng Y, Hong W, Chen X, Li H, Huang B, Huang Z, Tang H, Geng W (2020). Recent advances in immune cell therapy for glioblastoma. Frontiers in Immunology 11: 544563. https://doi.org/10.3389/fimmu.2020.544563 [Google Scholar] [PubMed] [CrossRef]
Kong M, Li H, Yuan W, Mao L, Chen J (2021). The role of Circ_PRKCI/miR-24-3p within metastasis of prostate cancer. JBUON 26: 949–955. [Google Scholar] [PubMed]
Li Y, Liu X, Cui X, Tan Y, Wang Q, Wang Y, Xu C, Fang C, Kang C (2021). LncRNA PRADX-mediated recruitment of PRC2/DDX5 complex suppresses UBXN1 expression and activates NF-κB activity, promoting tumorigenesis. Theranostics 11: 4516–4530. https://doi.org/10.7150/thno.54549 [Google Scholar] [PubMed] [CrossRef]
Liu Z, Su D, Qi X, Ma J (2018). MiR‐500a‐5p promotes glioblastoma cell proliferative, migrative and invasive through targeting chromodomain helicase DNA binding protein 5. Molecular Medicine Reports 18: 2689–2696. https://doi.org/10.3892/mmr.2018.9259 [Google Scholar] [PubMed] [CrossRef]
Liu G, Zhao H, Song Q, Li G, Lin S, Xiong S (2021). Long non-coding RNA DPP10-AS1 exerts anti-tumor effects on colon cancer via the upregulation of ADCY1 through regulating microRNA-127-3p. Aging 13: 9748–9765. https://doi.org/10.18632/aging.202729 [Google Scholar] [PubMed] [CrossRef]
Luo X, Tu T, Zhong Y, Xu S, Chen X, Chen L, Yang F (2021). ceRNA network analysis shows that lncRNA CRNDE promotes progression of glioblastoma through sponge mir-9-5p. Frontiers in Genetics 12: 617350. https://doi.org/10.3389/fgene.2021.617350 [Google Scholar] [PubMed] [CrossRef]
McKinnon C, Nandhabalan M, Murray SA, Plaha P (2021). Glioblastoma: Clinical presentation, diagnosis, and management. British Medical Journal 374: n1560. https://doi.org/10.1136/bmj.n1560 [Google Scholar] [PubMed] [CrossRef]
Pei S, Chen Z, Tan H, Fan L, Zhang B, Zhao C (2020). SLC16A1-AS1 enhances radiosensitivity and represses cell proliferative and invasive through regulating the miR-301b-3p/CHD5 axis in hepatocellular carcinoma. Environmental Science and Pollution Research International 27: 42778–42790. https://doi.org/10.1007/s11356-020-09998-1 [Google Scholar] [PubMed] [CrossRef]
Peng Z, Liu C, Wu M (2018). New insights into long non-coding RNAs and their roles in glioma. Molecular Cancer 17: 61. https://doi.org/10.1186/s12943-018-0812-2 [Google Scholar] [PubMed] [CrossRef]
Reggiardo RE, Maroli SV, Kim DH (2022). LncRNA biomarkers of inflammation and cancer. Advances in Experimental Medicine and Biology 1363: 121–145. https://doi.org/10.1007/978-3-030-92034-0 [Google Scholar] [CrossRef]
Rong L, Li N, Zhang Z (2022). Emerging therapies for glioblastoma: Current state and future directions. Journal of Experimental & Clinical Cancer Research 41: 142. https://doi.org/10.1186/s13046-022-02349-7 [Google Scholar] [PubMed] [CrossRef]
Tan YT, Lin JF, Li T, Li JJ, Xu RH, Ju HQ (2021). LncRNA-mediated posttranslational modifications and reprogramming of energy metabolism in cancer. Cancer Communications 41: 109–120. https://doi.org/10.1002/cac2.12108 [Google Scholar] [PubMed] [CrossRef]
Tian H, Pan J, Fang S, Zhou C, Tian H, He J, Shen W, Meng X, Jin X, Gong Z (2021). LncRNA DPP10-AS1 promotes malignant processes through epigenetically activating its cognate gene DPP10 and predicts poor prognosis in lung cancer patients. Cancer Biology & Medicine 18: 675–692. https://doi.org/10.20892/j.issn.2095-3941.2020.0136 [Google Scholar] [PubMed] [CrossRef]
Wang F, Gu T, Chen Y, Chen Y, Xiong D, Zhu Y (2021). Long non-coding RNA SOX21-AS1 modulates lung cancer progress upon microRNA miR-24-3p/PIM2 axis. Bioengineered 12: 6724–6737. https://doi.org/10.1080/21655979.2021.1955578 [Google Scholar] [PubMed] [CrossRef]
Wang R, Li Q, Chu X, Li N, Liang H, He F (2023). Sequencing and bioinformatics analysis of lncRNA/circRNA-miRNA-mRNA in glioblastoma multiforme. Metabolic Brain Disease 38: 2289–2300. [Google Scholar] [PubMed]
Wu P, Cai J, Chen Q, Han B, Meng X et al. (2019). Lnc-TALC promotes O6-methylguanine-DNA methyltransferase expression via regulating the c-Met pathway through competitively binding with miR-20b-3p. Nature Communications 10: 2045. https://doi.org/10.1038/s41467-019-10025-2 [Google Scholar] [PubMed] [CrossRef]
Xu L, Shao F, Luo T, Li Q, Tan D, Tan Y (2022). Pan-cancer analysis identifies CHD5 as a potential biomarker for glioma. International Journal of Molecular Sciences 23: 8489. https://doi.org/10.3390/ijms23158489 [Google Scholar] [PubMed] [CrossRef]
Yan H, Bu P (2021). Non-coding RNA in cancer. Essays in Biochemistry 65: 625–639. https://doi.org/10.1042/EBC20200032 [Google Scholar] [PubMed] [CrossRef]
Ye J, Zhu J, Chen H, Qian J, Zhang L, Wan Z, Chen F, Sun S, Li W, Luo C (2020). A novel lncRNA-LINC01116 regulates tumorigenesis of glioma through targeting VEGFA. International Journal of Cancer 146: 248–261. https://doi.org/10.1002/ijc.32483 [Google Scholar] [PubMed] [CrossRef]
Zhong C, Yu Q, Peng Y, Zhou S, Liu Z, Deng Y, Guo L, Zhao S, Chen G (2021). Novel LncRNA OXCT1-AS1 indicates poor prognosis and contributes to tumorigenesis by regulating miR-195/CDC25A axis in glioblastoma. Journal of Experimental & Clinical Cancer Research 40: 123. https://doi.org/10.1186/s13046-021-01928-4 [Google Scholar] [PubMed] [CrossRef]
Zhou Y, Qiu YE, Qin SL, Luo Y, Cui R, Qin J, Chen JJ, Yu MH, Zhong M, Shi XY (2018). Expression of CHD5 may serve as an independent biomarker of prognosis in colorectal cancer via immunohistochemical staining. International Journal of Clinical and Experimental Pathology 11: 2792–2798. [Google Scholar] [PubMed]
Zhou Y, Zhou Z, Ji Z, Yan W, Li H, Yu X (2020). Tetramethylpyrazine reduces prostate cancer malignancy through inactivation of the DPP10‐AS1/CBP/FOXM1 signaling pathway. International Journal of Oncology 57: 314–324. https://doi.org/10.3892/ijo.2020.5036 [Google Scholar] [PubMed] [CrossRef]
Zhu X, Jiang L, Yang H, Chen T, Wu X, Lv K (2020). Analyzing the lncRNA, miRNA, and mRNA-associated ceRNA networks to reveal potential prognostic biomarkers for glioblastoma multiforme. Cancer Cell International 20: 393. https://doi.org/10.1186/s12935-020-01488-1 [Google Scholar] [PubMed] [CrossRef]
Zhu L, Wang R, Zhang L, Zuo C, Zhang R, Zhao S (2019). rs187960998 polymorphism in miR-211 prevents development of human colon cancer through deregulation of 3’UTR in CHD5. Onco Targets and Therapy 12: 405–412. https://doi.org/10.2147/OTT.S180935 [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools