 Open Access
Open Access
ARTICLE
Research progress and prospects of nucleic acid isothermal amplification technology
1 Suzhou Key Laboratory of Medical Biotechnology, Suzhou Vocational Health College, Suzhou, 215009, China
2 School of Food Science and Engineering, Hainan University, Haikou, 570228, China
3 The Affiliated Infectious Diseases Hospital, Suzhou Medical College of Soochow University, Suzhou, 215100, China
* Corresponding Author: Song-Bai Liu,
# Co-first authors
(This article belongs to the Special Issue: Bioinformatics Study of Diseases)
BIOCELL 2023, 47(11), 2385-2395. https://doi.org/10.32604/biocell.2023.029687
Received 03 March 2023; Accepted 19 April 2023; Issue published 27 November 2023
Abstract
Nucleic acid (DNA and RNA) detection and quantification methods play vital roles in molecular biology. With the development of molecular biology, isothermal amplification of DNA/RNA, as a new molecular biology technology, can be amplified under isothermal condition, it has the advantages of high sensitivity, high specificity, and high efficiency, and has been applied in various fields of biotechnology, including disease diagnosis, pathogen detection, food hygiene and safety detection and so on. This paper introduces the progress of isothermal amplification technology, including rolling circle amplification (RCA), nucleic acid sequence-dependent amplification (NASBA), strand displacement amplification (SDA), loop-mediated isothermal amplification (LAMP), helicase-dependent amplification (HDA), recombinase polymerase amplification (RPA), cross-priming amplification (CPA), and its principle, advantages and disadvantages, and application development are briefly summarized.Keywords
Nucleic acid amplification technology is an important modern tool in biotechnology. With the advancements in molecular biology research, diagnostic methods based on nucleic acid detection have been widely used in medicine, agriculture, environmental monitoring, and other fields (Zhao et al., 2014). At present, the commonly used amplification approach is still polymerase chain reaction (PCR) amplification technology, which requires expensive and large-scale equipment, greatly limiting its application in some studies (Aryan et al., 2010; Bachmann et al., 2009). This limitation however becomes an advantage in the development of isothermal amplification methods, which can be performed at a constant temperature without the need for a thermal cycler device (Zanoli and Spoto, 2013). Isothermal amplification technology is an in vitro nucleic acid amplification technology where the reaction process is continuously maintained at a constant temperature by adding enzymes with different activities and their respective appropriate primers to achieve the rapid amplification of nucleic acids. Compared with other nucleic acid amplification technologies, isothermal amplification has the advantages of being rapid, efficient, and specific and does not require special equipment. Further, isothermal amplification does not require a thermal cycler making it cost-effective and easy to use. Additionally, it is more tolerant to inhibitory components from crude samples in most cases compared to PCR and shows equal or greater sensitivity and reliability (Chow et al., 2008). Isothermal amplification technology is an in vitro nucleic acid amplification technology where the reaction process is continuously maintained at a constant temperature by adding enzymes with different activities and their respective appropriate primers to achieve the rapid amplification of nucleic acids. The RCA technology can rapidly amplifies long single-stranded DNA obtained from tandem template complementary sequences under isothermal conditions; NASBA was designed to amplify single-stranded RNA sequences by simulating retroviral replication; SDA is a chemically modified restriction enzyme recognition sequence is placed on both ends of the target DNA, and the endonuclease opens the strand DNA at its recognition site, which in turn extends the 3′ end of the notch and replaces the next DNA strand; LAMP reaction is usually carried out at 60°C~65°C, the target to be tested is amplified more than 109 times within 1 h, It has good amplification effect; HDA technology using helicase to unwind the double-stranded DNA at a constant temperature. The DNA single-strand binding protein then stabilizes the unsolved single strand to provide a template for primers and DNA polymerase synthesizes complementary double strands, repetition of this cyclic amplification process finally achieves exponential amplification of the target sequence; RPA amplification was performed at 37°C–42°C, which mainly depended on the recombinant enzyme Uvs X, the single-stranded binding protein Gp32 and the strand replacement DNA polymerase Bsu; CPA is performed by stranded displacement of DNA polymerase and does not require an initial denaturation step or addition of nickase, this technique can be divided into single crossing CPA and double crossing CPA. Isothermal amplification techniques are now commonly used in different biotechnological fields due to their rapidity, efficiency, and non-requirement of professional or special equipment. This paper briefly summarizes the reaction principles, advantages and disadvantages, the development and applications of isothermal amplification technologies. It additionally looks at the future development trend of isothermal amplification technology to promote the development of related research fields.
Isothermal Amplification Technology Approaches
Rolling circle amplification technology
The RCA technology, first reported in 1989 (Blanco et al., 1989), is a simple and efficient in vitro nucleic acid amplification technique that rapidly amplifies long single-stranded DNA obtained from tandem template complementary sequences under isothermal conditions (Fire and Xu, 1995; Liu et al., 1996). RCA is an efficient enzymatic isothermal reaction that uses a loop probe as a template to produce long-stranded tandem single-stranded DNA or RNA products in the presence of short-stranded DNA or RNA primers (Xu et al., 2021) (Fig. 1). RCA technology has become an attractive tool for bioassays and analysis given its excellent efficiency, simplicity, and versatility (Mohsen and Kool, 2016; Baner et al., 1998). For example, a DNA fragment construct obtained by target-induced roll loop amplification is complementary to self-cleaving deoxyribozyme (DNAzyme) and releases fluorescent biomolecules. This sensing method can cause multiple signal amplifications with detection limits as low as 0.49 pmol L−1, providing a simple, highly sensitive, and selective low-cost detection platform (Liu et al., 2020). Furthermore, RCA can be combined with other technologies. For example, n recent years, a hybrid method combining RCA and LAMP (RCA-LAMP) was developed to quantify very small amounts of different types of RNAs, such as miRNAs (Hasegawa et al., 2021). An innovative approach entailed an increasingly branched rolling circle amplification (IB-RCA) containing a padlock probe (PP) and a structurally tailored molecular beacon (MB) that was developed for highly sensitive detection of the Kras gene codon 12. This sensing method demonstrates a new concept of IB-RCA amplification albeit in a simple way to efficiently transduce the fluorescence signal to accomplish the highly sensitive and selective detection of oncogenes (Li et al., 2016).
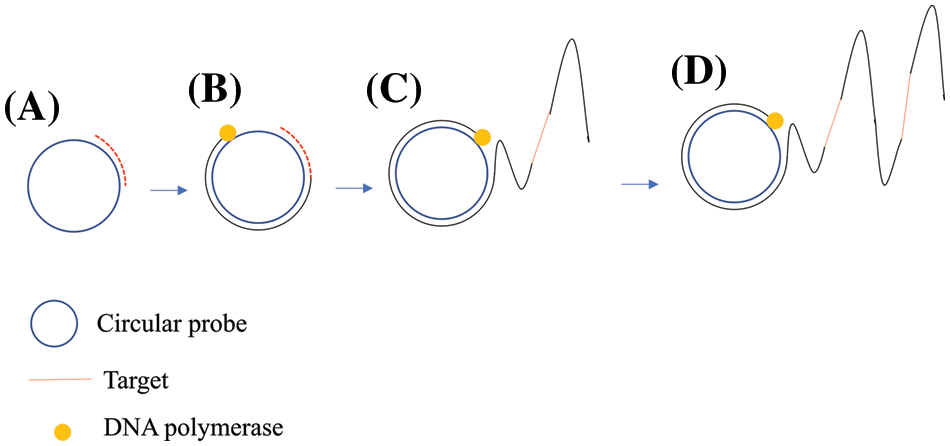
Figure 1: Schematic of the rolling circle amplification (RCA) technique.
Nucleic acid sequence-based amplification
NASBA is an isothermal amplification technique based on a transcription-dependent amplification system and was developed in 1991 (Compton, 1991). NASBA was designed to amplify single-stranded RNA sequences by simulating retroviral replication. The basic principle of NASBA is that the template RNA is recognized by antisense primers and forms complementary DNA strands under the RNA-dependent DNA polymerase activity of reverse transcriptase. The RNA‒DNA complex is then treated by ribonuclease H (RNase H) so that the original RNA strand is degraded. Subsequently, by the DNA-dependent DNA polymerase activity of reverse transcriptase, specific primers containing the T7 promoter recognize the newly synthesized single-stranded DNA strands to form a double-stranded DNA structure containing the T7 promoter, which can be used as a substrate for subsequent cyclic amplification. T7 RNA polymerase enables DNA strands with T7 promoters to serve as templates to synthesize new RNA strands that resemble the original RNA strands (Fig. 2). Usually, NABSA can be carried out at 41°C, and template RNA can be amplified 109 times after 1.5~2 h of amplification with a sensitivity that is comparable to that of RT‒PCR. The end products of NASBA can be detected by gel electrophoresis, real-time fluorescence, colorimetry, and electrochemiluminescence (Cook et al., 2002). For example, real-time isothermal NASBA (RT-NASBA) was applied in the detection of Mycoplasma pneumoniae in one report. The sensitivity of the RT-NASBA and the conventional NASBA assays corresponded to 5 color-changing units (CCU) of M. pneumoniae. These results indicate that the sensitivity of detection of M. pneumoniae by RT-NASBA in respiratory samples might be slightly reduced compared to that detected by conventional NASBA (Loens et al., 2003). In another study, NASBA was employed in the detection of Salmonella enterica cells in liquid whole eggs (Cook et al., 2002). Further, a real-time nucleic acid sequence amplification (RT-NASBA) method was developed for the rapid detection of Japanese encephalitis virus (JEV) that showed stable signals and high specificity within 10 min (Zhou et al., 2021). It was also demonstrated that the sensitivity of NASBA could be substantially increased to 1.5 RNA molecules per microliter by nesting NASBA primers and modifying NASBA internal primers to encode RNA Mango inducer sequences. Since this isothermal nucleic acid detection scheme directly generates a fluorescent reporter, it provides an inexpensive, simple, and specific method for the rapid detection of single molecular weight pathogenic RNAs (Abdolahzadeh et al., 2019).
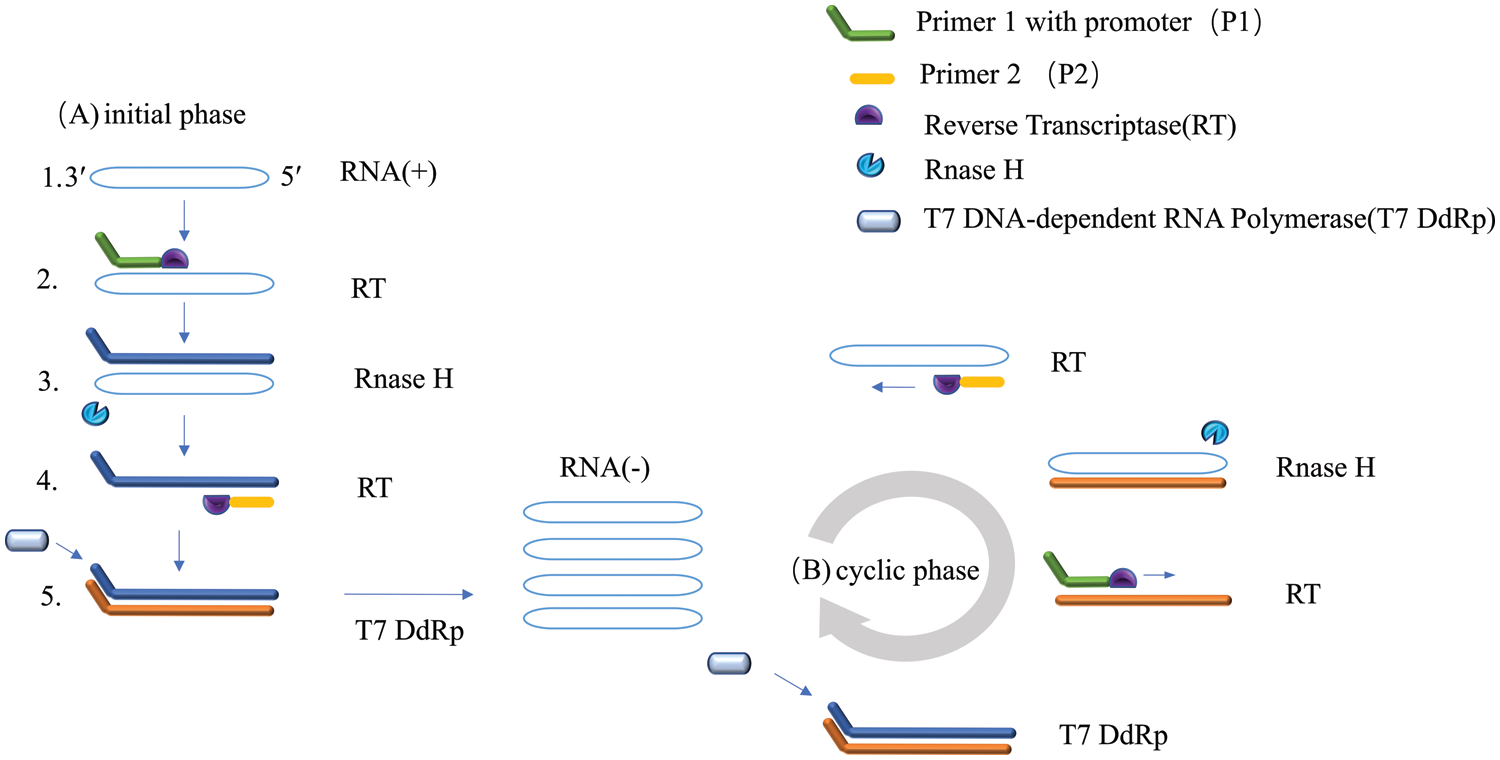
Figure 2: Schematic of the nucleic acid sequence-dependent amplification (NASBA) technology.
Strand displacement amplification technology
SDA is a new in vitro DNA isothermal amplification technique based on an enzymatic reaction first proposed by American scholar (Walker et al., 1992). SDA is an enzymatic in vitro isothermal amplification method for DNA developed in recent years. A chemically modified restriction enzyme recognition sequence is placed on both ends of the target DNA, and the endonuclease opens the strand DNA at its recognition site, which in turn extends the 3′ end of the notch and replaces the next DNA strand (Gill and Ghaemi, 2008; Hellyer and Nadeau, 2014).
The replaced DNA single strand binds to primers and is extended into double strands by DNA polymerase. This process is repeated repeatedly, allowing the target sequence to be amplified efficiently (Fig. 3). An example is that SDA can be used to detect HIV-1 RNA (Mehrpouyan et al., 1997).
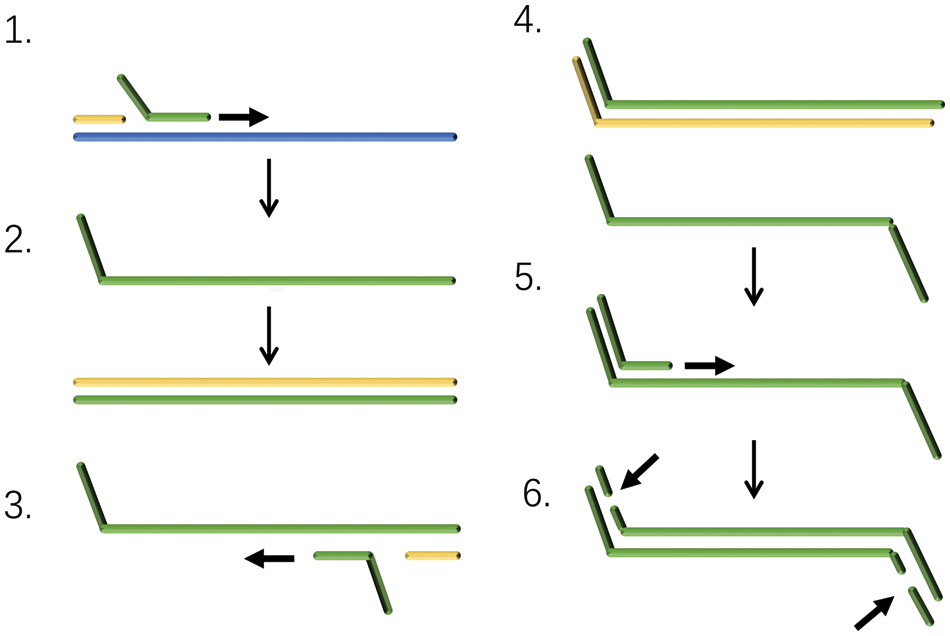
Figure 3: Schematic of strand displacement amplification (SDA) technology.
Loop-mediated isothermal amplification
Notomi et al. (2000) first reported the LAMP technology in 2000. LAMP entails the designing of external primers, the forward external primer (F3) (Wang et al., 2013) and the backward external primer (B3), and a pair of internal primers forward inner primer (FIP) and backward inner primer (BIP) for the target to be tested (Notomi et al., 2000). Due to the strand displacement activity of DNA polymerase, the primer extends along the template and the strand displacement reaction produces target repeat fragments of different lengths. The reaction is usually carried out at 60°C~65°C, the target to be tested is amplified more than 109 times within 1 h, and the sensitivity can reach the detection of 10 copies of the target to be tested. Further, the introduction of an additional pair of ring primers significantly improved the detection sensitivity and reduced the reaction time to less than 30 min (Eboigbodin, 2019).
LAMP is divided into two stages, namely, the initiator synthesis stage and the cyclic amplification stage. The initiator synthesis stage culminates in the formation of stem circular DNA, which serves as a template for the cyclic amplification phase. In the cyclic amplification stage, the displacement reaction occurs under the guidance of the inner primer and Bst DNA synthetase to form stemloop structures of different lengths. The final amplification product is a mixture of stem ring DNA and cauliflower-like DNA, and the product needs to be detected after amplification (Fig. 4). At present, there are three methods for detecting LAMP products. The first is agarose gel electrophoresis detection, the results of which are multiple bands in the shape of ladder bands (Wong et al., 2018). The second method is real-time turbidity detection, the white precipitate generated by amplification has a linear relationship with the amount of DNA. As the whole amplification process is monitored in real-time, it is more efficient and more accurate than endpoint detection methods such as agarose gel electrophoresis. However, this method is suitable for the laboratory and not suitable for on-site detection. The third method is fluorescence colorimetric detection by adding hydroxynaphthol blue to the reaction system. You can directly determine whether the target gene is successfully amplified by observing the color of the LAMP product (Petrusha and Faizuloev, 2020; Goto et al., 2009).
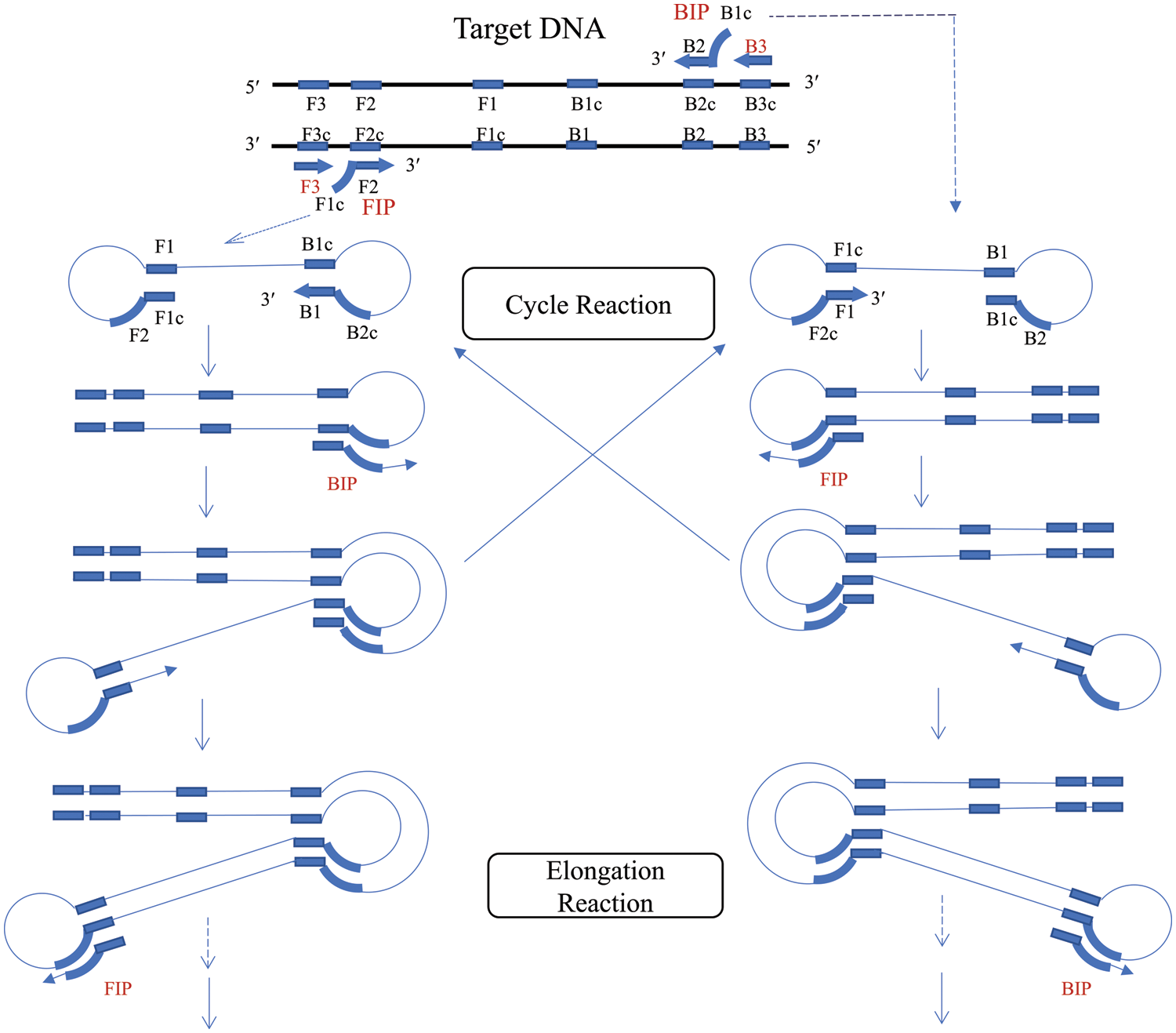
Figure 4: Schematic of the loop-mediated isothermal amplification (LAMP) technique.
In a study in dandelion, based on the nucleotide sequence of nuclear ribosomal DNA (nrDNA), LAMP amplification could be successfully performed by adding purified Taraxacum formosanum (TF) genomic DNA within 45 min under isothermal conditions (Lai et al., 2015). Yuan et al. (2018) developed a method to detect multiple DNA of foodborne pathogens by encapsulating emulsion droplets for LAMP analysis. DNA from different pathogens can be detected by directly observing and analyzing the fluorescence intensity of the resulting droplets, with a detection limit 500 times lower than the bulk phase LAMP. This further demonstrates the ability to quantify bacterial concentrations by detecting bacterial DNA in actual samples and shows great potential in monitoring water resources and assess the pathogenic contamination in samples. Hence, LAMP technology represents a fast, robust, and reliable diagnostic platform for point-of-care testing (Anupama et al., 2019).
Helicase-dependent amplification technology
First proposed in 2004, HDA technology uses helicase to unwind DNA double strands under isothermal conditions, unlike PCR, which requires variable temperatures (Vincent et al., 2004). This technology mimics the natural process of DNA replication in vivo, using helicase to unwind the double-stranded DNA at a constant temperature. The DNA single-strand binding protein (SSB) then stabilizes the unsolved single strand to provide a template for primers and DNA polymerase synthesizes complementary double strands (Yasukawa et al., 2020). Repetition of this cyclic amplification process finally achieves exponential amplification of the target sequence. The difference between HDA technology and traditional PCR is mainly that HDA entails the cyclic production of single-stranded templates by adding helicase and single-stranded binding proteins under isothermal conditions (Fig. 5). This overcomes the shortcomings of traditional PCR that relies on repeated heating and temperature lowering of instruments to obtain single-stranded templates. This HDA technique is simple and efficient and is slightly different from RPA reactions, which support simultaneous amplification reactions in the same tube (Chen et al., 2015).
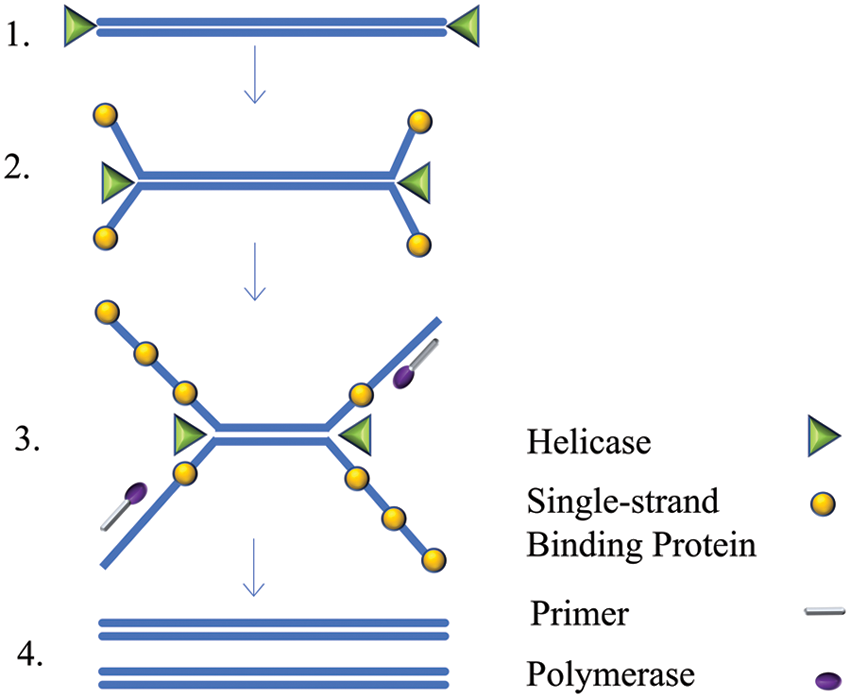
Figure 5: Schematic of the helicase-dependent amplification (HDA) technique.
An example is the reverse transcription-HDA (RT-HDA) technology, which can be used for immediate detection of COVID-19 by combining the power of isothermal amplification and dipstick technology. The RT-HDA coupled dipstick correctly identified all positive and negative specimens, and the results can be visually interpreted using dipstick technology without any specialized equipment. In addition, the RT-HDA coupling test paper can be completed in a short time of approximately 2 h (Shanmugakani and Wu, 2022). Further, the development and optimization of a Si-coated magnetic nanoparticle (Si-MNP) based Si-MNP thermophilic decarboxylase-dependent isothermal amplification (tHDA) method using SYBR Green I for the detection of Staphylococcus aureus-specific DNA fragments has been reported. With a whole detection time of less than 2 h, the approach has good application prospects (Chen et al., 2015).
Recombinase polymerase amplification technology
Piepenburg et al. (2006) developed a new isothermal amplification technology, RPA (Piepenburg et al., 2006). By simulating in vivo amplification of DNA, the fragment of interest was generated within 10 min under isothermal conditions of 37°C~42°C. This technique relies heavily on the recombinant enzyme Uvs X, the single-stranded binding protein Gp32, and the strand-displacing DNA polymerase Bsu. In addition, there is a helper protein Uvs Y in this system. Among these molecules, Uvs X and Uvs Y are encoded by T4 bacteriophages.
The RPA amplification process is similar to the in vivo recombination of simulated DNA (Xu et al., 2014) and is mainly divided into the following four basic steps (Li and Macdonald, 2015): (i) recombinant protease Uvs X forms an enzyme-primer complex with the upstream and downstream primers of oligonucleotides with the assistance of cofactor Uvs Y; (ii) the complex binds to the homologous target sequence and finds and binds to the specific binding site of the target sequence; (iii) the recombinase-oligonucleotide primer complex unwinds the double strand, and the single-stranded binding protein (SSB) binds to the replaced parent chain to prevent further replacement; (iv) under the synergistic effect of strand displacement of the DNA polymerase, the amplification process of specific fragments is initiated. The target sequence can be amplified to the order of 1012 within 30 min, and the amplification product can fully reach the detection level (Singpanomchai et al., 2019) (Fig. 6). Different target molecules have been amplified in a variety of organisms and samples (Lobato and O’Sullivan, 2018).
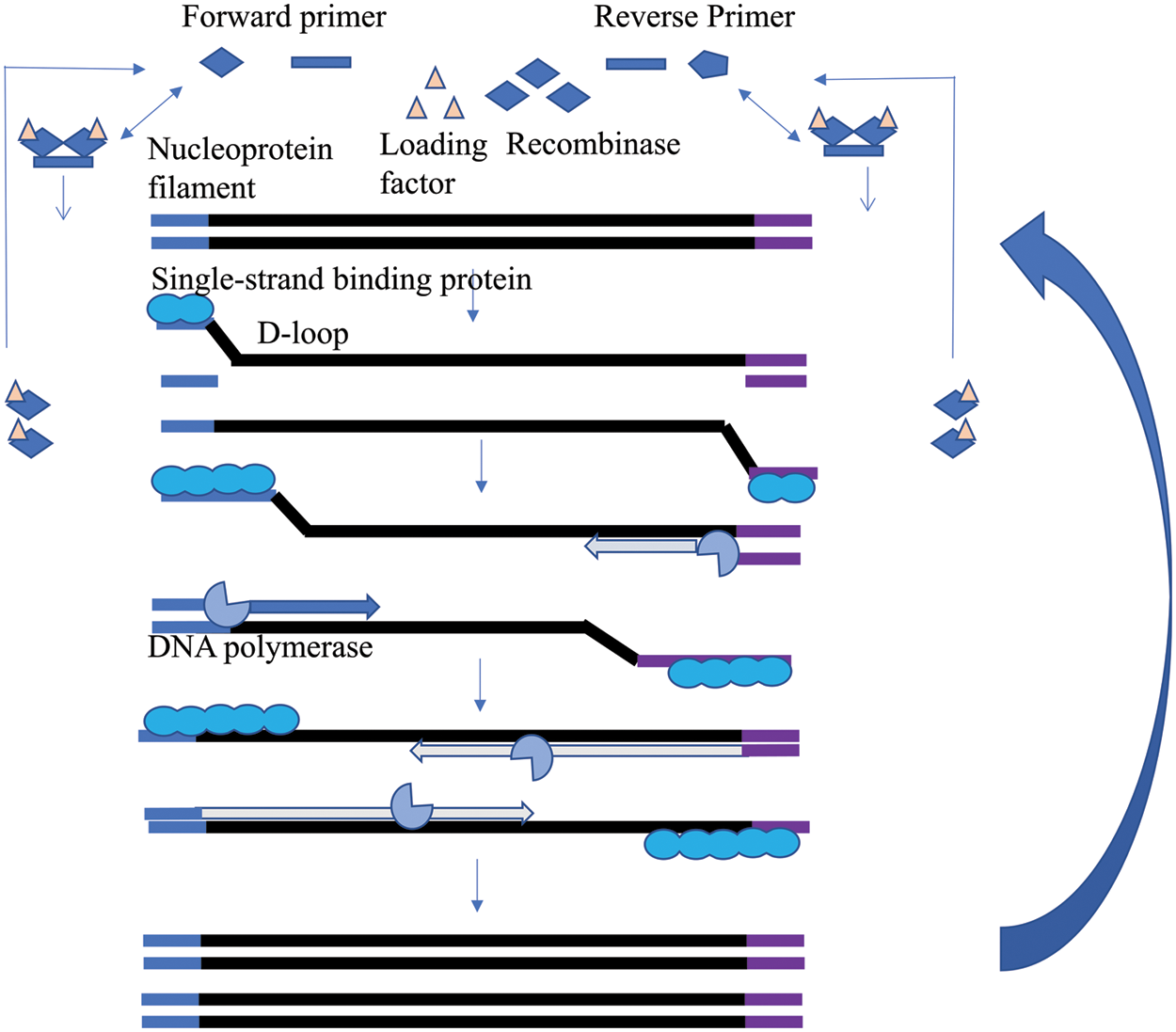
Figure 6: Schematic of the recombinase polymerase amplification (RPA) technique.
RPA technology has the advantages of high sensitivity, specificity, and efficiency and has been widely used in various fields (Zaghloul and El-Shahat, 2014). For instance, Wang et al. (2020) developed real-time RPA and lateral flow strip (LFS)-RPA assays to provide attractive and promising tools for the rapid, convenient, and reliable detection of M. ovipneumoniae in sheep. In another report, Wang et al. (2017) developed and evaluated a rapid real-time reverse transcription recombinase polymerase amplification (RT-RPA) assay for the detection of coxsackievirus A6 (CV-A6). The sensitivity of this assay was 202 copies/reaction and the specificity was 100%. Furthermore, this assay yielded consistent results comparable with a commercial qRT‒PCR diagnostic kit. This approach is therefore potentially useful for the surveillance of CV-A6 infections and its outbreak control. Taking an application in plants, Wang et al. (2020) combined the advantages of RPA and fluorescence detection to establish a rapid, sensitive, specific, and simple detection platform for on-site detection of MON863 maize. Test samples were added directly to the platform after simple pretreatment with the DNA extraction-free method. These results were obtained by real-time monitoring with a portable instrument, which facilitates its use in field sampling. The entire detection process, including sample preparation, RPA, and the identification of amplification results, was accomplished in approximately 10 min. Furthermore, detection was achieved with a simple and inexpensive portable device.
Cross-primer amplification technique
CPA technology developed by Hangzhou Ustar Biologics is the first in vitro nucleic acid amplification technology with independent intellectual property rights in China (Fang et al., 2009; Xu et al., 2012). Depending on the number of cross-primers in the system, this technique can be divided into single crossing CPA and double crossing CPA (Yulong et al., 2010).
Double cross-primer amplification uses two cross-primers and two peel primers. The two cross-primers complementarily bind to the template strand and are extended. The stripping primers then peel off the newly synthesized single strands due to the action of Bst DNA polymerase. Finally, the last two cross-primers synthesize a large number of target fragments due to the action of Bst DNA polymerase using the nascent single strands as templates (Zheng et al., 2020) (Fig. 7).
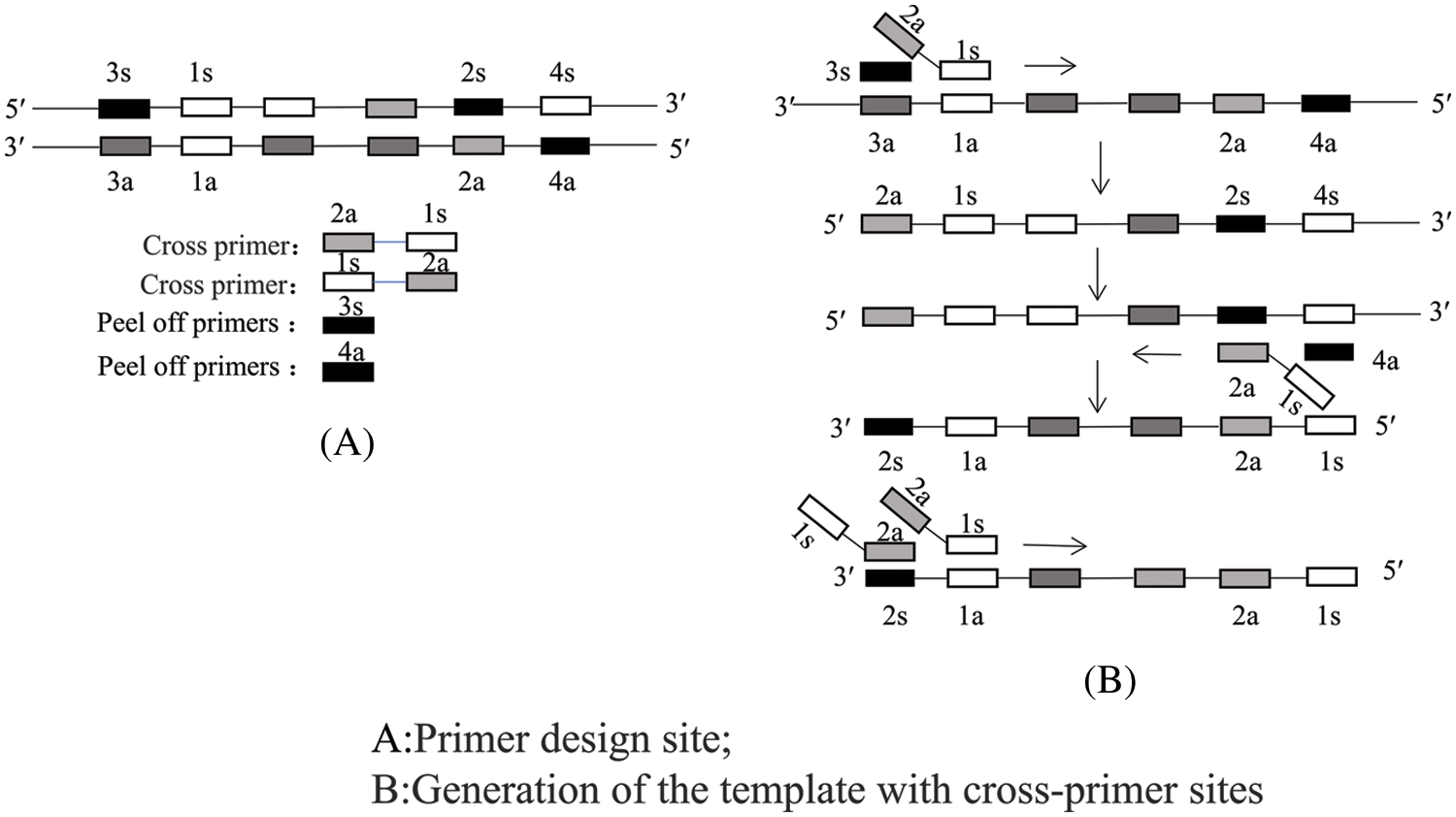
Figure 7: Schematic of the double cross-priming amplification (CPA) technique.
Single cross-primer amplification uses one cross-primer, two peeling primers, and two normal primers. First, the cross-primer binds to the template strand and is extended into a double strand, while the peeling primer separates the new strand from the template due to the action of Bst DNA polymerase. Subsequently, the ordinary primers use the new strand as a template to synthesize two single-stranded DNAs of different lengths (Fig. 8). Finally, these two single strands are used as templates with the cross-primers and ordinary primers being used as primer pairs to form an amplification cycle (Xu et al., 2012).
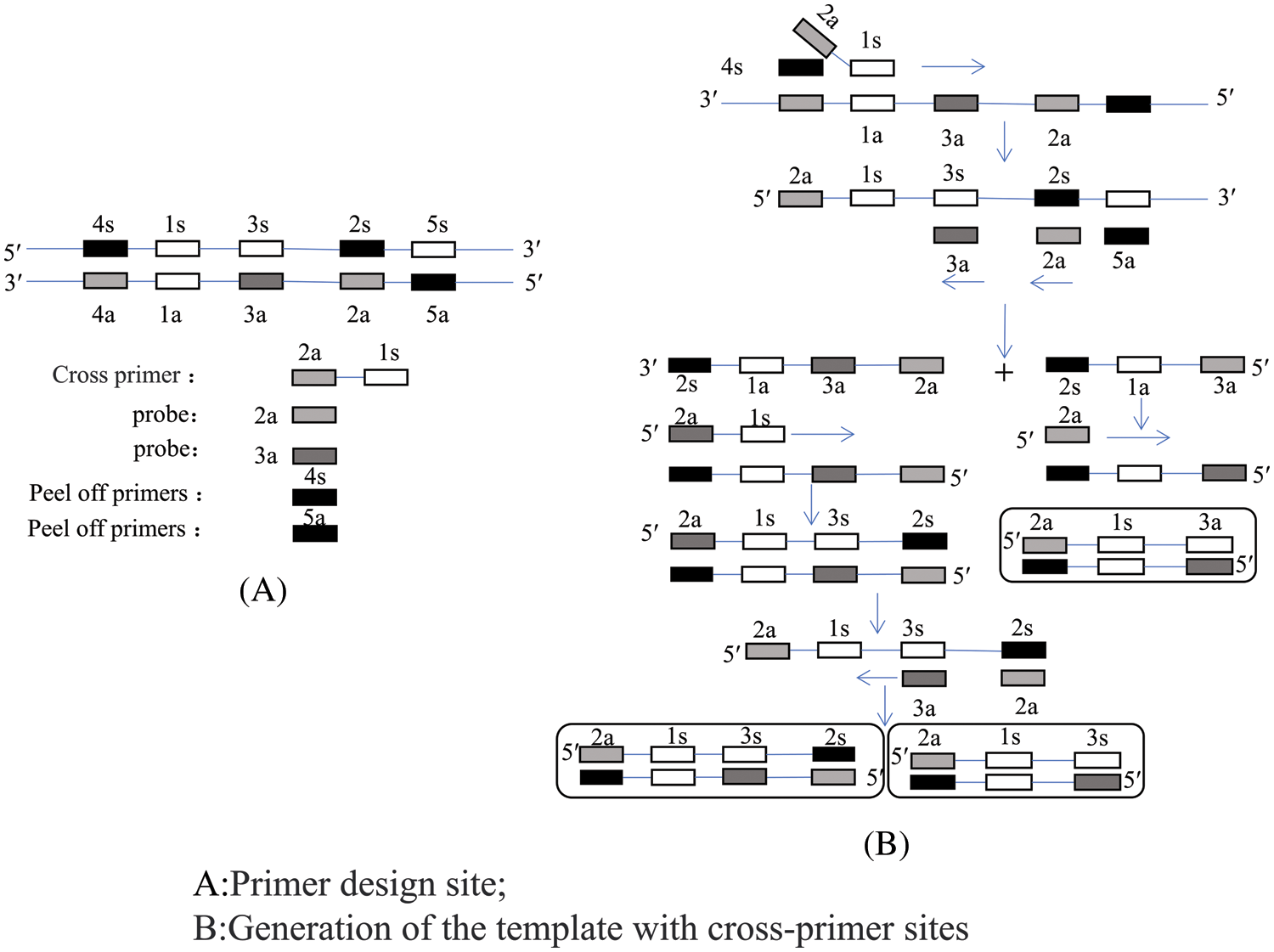
Figure 8: Schematic of a single cross-priming amplification (CPA) technology.
CPA is a class of isothermal nucleic acid amplification reactions that utilize multiple primers and probes, one or more of which is a cross primer (Fang et al., 2009). CPA is performed by stranded displacement of DNA polymerase and does not require an initial denaturation step or addition of nickase. At an assay temperature of 63°C, a high concentration of primers relative to template DNA favors the formation of primer-template hybrids at transient spontaneous denaturation bubbles in the DNA template, rather than reannealing of template strands. Strand displacement is facilitated by the annealing of cross-primers with 5′ ends that are not complementary to the template strand and the binding of displacement primers upstream of the cross-primer (Xu et al., 2012). Compared with LAMP, cross-primed isothermal amplification does not have the limitation of expensive instruments and avoids the generation of harmful substances such as EB generated during ring-mediated isothermal amplification. It also circumvents the high false-positive rate caused by aerosol contamination problems. It is simple to operate, and a large number of single and double-stranded products can be obtained in a water bath at about 63°C for 1 h. Moreover, the results can be obtained quickly by combining CPA with the lateral cross-flow test strip technique (Zhang et al., 2015).
The technique can be used for a variety of tests. For example, cattle, sheep, Arctic fox, and pork samples were identified by CPA combined with colloid gold nucleic acid strips. A simple primer for distinguishing multiple samples was designed using the general single-label CPA primer system, and the species-specific primers of four detection levels were analyzed by the colloidal gold test strip method. This provides a promising tool for detecting the species of different types of meat using a constant temperature amplification technology (Loens et al., 2003). Additionally, a rapid reverse transcription-cross-primer amplification (RT-CPA) combined with lateral flow test strips (LFD) diagnostic method was developed to detect bean pod mottle virus (BPMV) (Yang et al., 2022). The results showed a significantly better performance in detecting field samples of BPMV-infested soybean seeds where the RT-CPA-LFD method was more efficient, simple, and intuitive. In another study, a CPA-based method for the terminator of Agrobacterium tumefaciens nopaline synthase terminator (T-Nos) was established (Zhang et al., 2014). The results showed that the approach could detect rice genomic DNA within 40 min. Further, real-time fluorescence CPA documented comparable sensitivity to conventional real-time PCR and takes less time. As its sensitivity is sufficient to monitor the marker system it provides an attractive method for transgene detection.
Advantages and Disadvantages of Isothermal Amplification Technology
In recent years, the development of isothermal amplification techniques for nucleic acids has replaced traditional PCR amplification techniques (Craw and Balachandran, 2012). The major advantage of these methods is that nucleic acid amplification can be performed at a constant temperature without periodic temperature changes. However, there are some disadvantages, such as annealing of primers at low temperatures without thermal cycling during amplification increases the risk of primer dimer formation. Further, the reaction products are prone to false positive results and there are other issues, which need to be addressed, and further optimized. Some isothermal methods like LAMP have been standardized in molecular biology, while several other methods still need developments for future advances to be applicable. In conclusion, each isothermal amplification method has its advantages and disadvantages. Table 1 below shows the advantages and disadvantages of each method and its application scope. Although the limitations of isothermal methods are less prominent compared to amplification based on non-isothermal PCR, isothermal amplification is expected to have more advantages and minimal limitations to be able to justify itself as an alternative to long-established PCR methods (Asadi and Mollasalehi, 2021).
Polymerase chain reaction (PCR) is one of the most widely used nucleic acid amplification technologies, with high specificity and sensitivity, but this technology relies on expensive thermal cycling apparatus and professional technicians, and is not suitable for on-the-spot detection, Isothermal DNA amplification is emerging as an alternative to PCR-based amplification for point-of-care diagnosis (Kim and Easley, 2011). In recent years, with the rapid development of molecular biology technology techniques, diagnostic methods based on nucleic acid detection have been established and widely used in laboratory testing of human diseases. Deriving benefits from rapid advances in biotechnology, chemistry, and nanotechnology, these isothermal DNA amplification methods have been expanded to detect targets ranging from DNA and RNA to cells, proteins, small molecules, and even ions (Zhao et al., 2015). Isothermal amplification technology is a nucleic acid in vitro amplification technology. The reaction process is always maintained at a constant temperature, and the purpose of rapid amplification of nucleic acids is achieved by adding enzymes with different activities and their specific primers (Qi et al., 2018). Isothermal amplification of nucleic acid is a simple process that can expand the copy number of a specific DNA or RNA fragment at a constant temperature and accumulate nucleic acid sequences quickly and effectively. The ultimate purpose of the establishment of diagnostic testing methods is to apply them to actual testing. Isothermal amplification technology neither requires complex instruments nor does it require thermal denaturation and temperature cycling. Given its advantages of simplicity, speed, high sensitivity, and strong specificity, its development and use are highly expected (Gill and Ghaemi, 2008; Kumar, 2020). It is mainly used in the fields of biological substance detection, biological engineering, medical diagnosis and so on. Isothermal amplification can detect pathogens, toxins and other harmful substances in food, providing a basis for food safety detection; In the aspect of bioengineering, it can be used to detect and screen new genes, DNA synthesis and gene editing: applied in the field of medical diagnosis, such as the detection and diagnosis of viruses and infectious diseases, to provide a basis for clinical diagnosis, etc. In addition, isothermal amplification technology will have greater space for development in the future.
The current nucleic acid isothermal amplification technology has certain advantages over PCR technology in terms of the rapidity in amplification detection (Moore, 2005), instrument miniaturization, reagent cost, visual interpretation results, etc. It is emerging as more promising for wide use at grassroot levels and on-site detection. However, a few issues remain to be addressed. Firstly, the LAMP (Nagamine et al., 2001; Suzuki et al., 2010), HDA (Vincent et al., 2004; Dean et al., 2002), SDA, RCA (Liu et al., 1996) and CPA (Kumar, 2020) methods necessitate more primers and have higher requirements for primer design, which limits their application. Secondly, achieving the detection of multiple or multitarget isothermal amplification under single-tube and closed-tube conditions with high sensitivity and specificity is also a technical problem. This needs to be addressed urgently, otherwise, it will limit the application scope of isothermal amplification technology. Although isothermal amplification technology has advantages that ordinary PCR amplification cannot overcome, it also has some of these mentioned disadvantages. However, with further research, the understanding of the field of molecular biology is getting clearer, and it is believed that more and more perfect new constant temperature amplification technologies will emerge shortly. Compared with traditional PCR approaches, isothermal amplification provides faster and more convenient nucleic acid detection using a simple heating device and efficient amplification at a constant reaction temperature. When isothermal amplification is combined with microfluidics, a more efficient detection platform can be built (Kim and Easley, 2011).
Acknowledgement: None.
Funding Statement: This work was supported by grants from Jiangsu Higher Education Institution Innovative Research Team for Science and Technology (2021), the Key Technology Program of Suzhou People’s Livelihood Technology Projects (Grant Nos. SKY2021029, SZS2020311), the Open Project of Jiangsu Biobank of Clinical Resources (TC2021B009), and the Qing-Lan Project of Jiangsu Province in China (2021, 2022).
Author Contributions: The authors confirm their contribution to the paper as follows: study conception and design: Song-Bai Liu, Ping Xu, Xiangbin Xu; data collection: Shuhui Wu, Ping Xu; analysis and interpretation of results: Shuhui Wu, Ping Xu; draft manuscript preparation: Song-Bai Liu, Xiangbin Xu. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
Abdolahzadeh A, Dolgosheina EV, Unrau PJ (2019). RNA detection with high specificity and sensitivity using nested fluorogenic Mango NASBA. RNA 25: 1806–1813. [Google Scholar] [PubMed]
Anupama KP, Chakraborty A, Karunasagar I, Karunasagar I, Maiti B (2019). Loop-mediated isothermal amplification assay as a point-of-care diagnostic tool for vibrio parahaemolyticus: Recent developments and improvements. Expert Review of Molecular Diagnostics 19: 229–239. [Google Scholar] [PubMed]
Ao W, Aldous S, Woodruff E, Hicke B, Rea L, Kreiswirth B, Jenison R (2012). Rapid detection of rpoB gene mutations conferring rifampin resistance in Mycobacterium tuberculosis. Journal of Clinical Microbiology 50: 2433–2440. [Google Scholar] [PubMed]
Aryan E, Makvandi M, Farajzadeh A, Huygen K, Bifani P, Mousavi SL, Fateh A, Jelodar A, Gouya MM, Romano M (2010). A novel and more sensitive loop-mediated isothermal amplification assay targeting IS6110 for detection of Mycobacterium tuberculosis complex. Microbiological Research 165: 211–220. [Google Scholar] [PubMed]
Asadi R, Mollasalehi H (2021). The mechanism and improvements to the isothermal amplification of nucleic acids, at a glance. Analytical Biochemistry 631: 114260. [Google Scholar] [PubMed]
Bachmann LH, Johnson RE, Cheng H, Markowitz LE, Papp JR, Hook III EW (2009). Nucleic acid amplification tests for diagnosis of Neisseria gonorrhoeae oropharyngeal infections. Journal of Clinical Microbiology 47: 902–907. [Google Scholar] [PubMed]
Baner J, Nilsson M, Mendel-Hartvig M, Landegren U (1998). Signal amplification of padlock probes by rolling circle replication. Nucleic Acids Research 26: 5073–5078. [Google Scholar] [PubMed]
Blanco L, Bernad A, Lázaro JM, Martín G, Garmendia C, Salas M (1989). Highly efficient DNA synthesis by the phage phi 29 DNA polymerase. Symmetrical mode of DNA replication. The Journal of Biological Chemistry 264: 8935–8940. [Google Scholar] [PubMed]
Chen X, Wu X, Gan M, Xu F, He L, Yang D, Xu H, Shah NP, Wei H (2015). Rapid detection of Staphylococcus aureus in dairy and meat foods by combination of capture with silica-coated magnetic nanoparticles and thermophilic helicase-dependent isothermal amplification. Journal of Dairy Science 98: 1563–1570. [Google Scholar] [PubMed]
Chow WHA, McCloskey C, Tong Y, Hu L, You Q, Kelly CP, Kong H, Tang YW, Tang W (2008). Application of isothermal helicase-dependent amplification with a disposable detection device in a simple sensitive stool test for toxigenic Clostridium difficile. The Journal of Molecular Diagnostics 10: 452–458. [Google Scholar] [PubMed]
Compton J (1991). Nucleic acid sequence-based amplification. Nature 350: 91–92. [Google Scholar] [PubMed]
Cook N, Ellison J, Kurdziel AS, Simpkins S, Hays JP (2002). A nucleic acid sequence-based amplification method to detect Salmonella enterica serotype enteritidis strain PT4 in liquid whole egg. Journal of Food Protection 65: 1177–1178. [Google Scholar] [PubMed]
Craw P, Balachandran W (2012). Isothermal nucleic acid amplification technologies for point-of-care diagnostics: A critical review. Lab on a Chip 12: 2469–2486. [Google Scholar] [PubMed]
Dean FB, Hosono S, Fang L, Wu X, Faruqi AF et al. (2002). Comprehensive human genome amplification using multiple displacement amplification. Proceedings of the National Academy of Sciences of the United States of America 99: 5261–5266. [Google Scholar] [PubMed]
Eboigbodin KE (2019). Application of loop-mediated isothermal amplification assay for the detection of chlamydia trachomatis and neisseria gonorrhoeae. Methods in Molecular Biology 2042: 19–25. [Google Scholar] [PubMed]
Fang R, Li X, Hu L, You Q, Li J et al. (2009). Cross-priming amplification for rapid detection of Mycobacterium tuberculosis in sputum specimens. Journal of Clinical Microbiology 47: 845–847. [Google Scholar] [PubMed]
Fire A, Xu SQ (1995). Rolling replication of short DNA circles. Proceedings of the National Academy of Sciences of the United States of America 92: 4641–4645. [Google Scholar] [PubMed]
Gill P, Ghaemi A (2008). Nucleic acid isothermal amplification technologies: A review. Nucleosides, Nucleotides & Nucleic Acids 27: 224–243. [Google Scholar]
Goto M, Honda E, Ogura A, Nomoto A, Hanaki K (2009). Colorimetric detection of loop-mediated isothermal amplification reaction by using hydroxy naphthol blue. BioTechniques 46: 167–172. [Google Scholar] [PubMed]
Hasegawa T, Hapsari D, Iwahashi H (2021). RNase H-dependent amplification improves the accuracy of rolling circle amplification combined with loop-mediated isothermal amplification (RCA-LAMP). PeerJ 9: e11851. [Google Scholar] [PubMed]
Hellyer TJ, Nadeau JG (2014). Strand displacement amplification: A versatile tool for molecular diagnostics. Expert Review of Molecular Diagnostics 4: 251–261. [Google Scholar]
Hønsvall BK, Robertson LJ (2017). From research lab to standard environmental analysis tool: Will NASBA make the leap? Water Research 109: 389–397. [Google Scholar]
Kim J, Easley CJ (2011). Isothermal DNA amplification in bioanalysis: Strategies and applications. Bioanalysis 3: 227–239. [Google Scholar] [PubMed]
Kumar Y (2020). Isothermal amplification-based methods for assessment of microbiological safety and authenticity of meat and meat products. Food Control 121: 107679. [Google Scholar]
Lai GH, Chao J, Lin MK, Chang WT, Peng WH, Sun FC, Lee MS, Lee MS (2015). Rapid and sensitive identification of the herbal tea ingredient Taraxacum formosanum using loop-mediated isothermal amplification. International Journal of Molecular Sciences 16: 1562–1575. [Google Scholar] [PubMed]
Lannutti L, Mira A, Basualdo M, Rodriguez G, Erler S, Silva V, Gisder S, Genersch E, Florin-Christensen M, Schnittger L (2020). Development of a loop-mediated isothermal amplification (LAMP) and a direct LAMP for the specific detection of Nosema ceranae, a parasite of honey bees. Parasitology Research 119: 3947–3956. [Google Scholar] [PubMed]
Li J, Macdonald J (2015). Advances in isothermal amplification: Novel strategies inspired by biological processes. Biosensors & Bioelectronics 64: 196–211. [Google Scholar]
Li H, Xu J, Wang Z, Wu ZS, Jia L (2016). Increasingly branched rolling circle amplification for the cancer gene detection. Biosensors & Bioelectronics 86: 1067–1073. [Google Scholar]
Liu D, Daubendiek SL, Zillman MA, Ryan K, Kool ET (1996). Rolling circle DNA synthesis: Small circular oligonucleotides as efficient templates for DNA polymerases. Journal of the American Chemical Society 118: 1587–1594. [Google Scholar] [PubMed]
Liu C, Han J, Zhou L, Zhang J, Du J (2020). DNAzyme-based target-triggered rolling-circle amplification for high sensitivity detection of microRNAs. Sensors 20: 2017. [Google Scholar] [PubMed]
Lobato IM, O’Sullivan CK (2018). Recombinase polymerase amplification: Basics, applications and recent advances. Trends in Analytical Chemistry 98: 19–35. [Google Scholar] [PubMed]
Loens K, Ieven M, Ursi D, Beck T, Overdijk M, Sillekens P, Goossens H (2003). Detection of Mycoplasma pneumoniae by real-time nucleic acid sequence-based amplification. Journal of Clinical Microbiology 41: 4448–4450. [Google Scholar] [PubMed]
Mehrpouyan M, Bishop JE, Ostrerova N, Van Cleve M, Lohman KL (1997). A rapid and sensitive method for non-isotopic quantitation of HIV-1 RNA using thermophilic SDA and flow cytometry. Molecular and Cellular Probes 11: 337–347. [Google Scholar] [PubMed]
Mohsen MG, Kool ET (2016). The discovery of rolling circle amplification and rolling circle transcription. Accounts of Chemical Research 49: 2540–2550. [Google Scholar] [PubMed]
Moore P (2005). PCR: Replicating success. Nature 435: 235–238. [Google Scholar]
Nagamine K, Watanabe K, Ohtsuka K, Hase T, Notomi T (2001). Loop-mediated isothermal amplification reaction using a nondenatured template. Clinical Chemistry 47: 1742–1743. [Google Scholar] [PubMed]
Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T (2000). Loop-mediated isothermal amplification of DNA. Nucleic Acids Research 28: E63. [Google Scholar] [PubMed]
Petrusha OA, Faizuloev EB (2020). Detection methods for results of a loop-mediated isothermal amplification of DNA. Klinicheskaia Laboratornaia Diagnostika 65: 67–72. [Google Scholar] [PubMed]
Piepenburg O, Williams CH, Stemple DL, Armes NA, Haber J (2006). DNA detection using recombination proteins. PLoS Biology 4: e204. [Google Scholar] [PubMed]
Qi H, Yue S, Bi S, Ding C, Song W (2018). Isothermal exponential amplification techniques: From basic principles to applications in electrochemical biosensors. Biosensors & Bioelectronics 110: 207–217. [Google Scholar]
Seo JH, Park BH, Oh SJ, Choi G, Kim DH, Lee EY, Seo TS (2017). Development of a high-throughput centrifugal loop-mediated isothermal amplification microdevice for multiplex foodborne pathogenic bacteria detection. Sensors & Actuators B Chemical 246: 146–153. [Google Scholar]
Shanmugakani RK, Wu M (2022). An isothermal amplification-coupled dipstick for the rapid detection of COVID-19. Journal of Medical Microbiology 71(4): 001519. [Google Scholar]
Shi C, Liu Q, Ma C, Zhong W (2014). Exponential strand-displacement amplification for detection of microRNAs. Analytical Chemistry 86: 336–339. [Google Scholar] [PubMed]
Singpanomchai N, Akeda Y, Tomono K, Tamaru A, Santanirand P, Ratthawongjirakul P (2019). Naked eye detection of the Mycobacterium tuberculosis complex by recombinase polymerase amplification-SYBR green I assays. Journal of Clinical Laboratory Analysis 33: e22655. [Google Scholar] [PubMed]
Spargo CA, Fraiser MS, Van Cleve M, Wright DJ, Nycz CM, Spears PA, Walker GT (1996). Detection of M. tuberculosis DNA using thermophilic strand displacement amplification. Molecular and Cellular Probes 10: 247–256. [Google Scholar] [PubMed]
Suzuki R, Ihira M, Enomoto Y, Yano H, Maruyama F, Emi N, Asano Y, Yoshikawa T (2010). Heat denaturation increases the sensitivity of the cytomegalovirus loop-mediated isothermal amplification method. Microbiology and Immunology 54: 466–470. [Google Scholar] [PubMed]
Vincent M, Xu Y, Kong H (2004). Helicase-dependent isothermal DNA amplification. EMBO Reports 5: 795–800. [Google Scholar] [PubMed]
Walker GT (1993). Empirical aspects of strand displacement amplification. PCR Methods & Applications 3: 1–6. [Google Scholar]
Walker GT, Little MC, Nadeau JG, Shank DD (1992). Isothermal in vitro amplification of DNA by a restriction enzyme/DNA polymerase system. Proceedings of the National Academy of Sciences of the United States of America 89: 392–396. [Google Scholar] [PubMed]
Wang YW (2016). Signal amplification methods. In: Clinical Virology Manual, pp. 161–172. Fifth edition. Hoboken: Wiley Online Library. [Google Scholar]
Wang K, Wu Y, Yin D, Tang S, Hu G, He Y (2017). Development and evaluation of a rapid recombinase polymerase amplification assay for detection of coxsackievirus A6. Archives of Virology 162: 287–290. [Google Scholar] [PubMed]
Wang X, Xie S, Chen X, Peng C, Xu X, Wei W, Ma T, Cai J, Xu J (2020). A rapid and convenient method for on-site detection of MON863 maize through real-time fluorescence recombinase polymerase amplification. Food Chemistry 324: 126821. [Google Scholar] [PubMed]
Wang Q, Zhou Y, Li S, Zhuo C, Xu S, Huang L, Yang L, Liao K, Xu SY (2013). Real-time fluorescence loop mediated isothermal amplification for the detection of Acinetobacter baumannii. PLoS One 8: e66406. [Google Scholar] [PubMed]
Wong YP, Othman S, Lau YL, Radu S, Chee HY (2018). Loop-mediated isothermal amplification (LAMPA versatile technique for detection of micro-organisms. Journal of Applied Microbiology 124: 626–643. [Google Scholar] [PubMed]
Xu L, Duan J, Chen J, Ding S, Cheng W (2021). Recent advances in rolling circle amplification-based biosensing strategies–A review. Analytica Chimica Acta 1148: 238187. [Google Scholar] [PubMed]
Xu G, Hu L, Zhong H, Wang H, Yusa S, Weiss TC, Romaniuk PJ, Pickerill S, You Q (2012). Cross priming amplification: Mechanism and optimization for isothermal DNA amplification. Scientific Reports 2: 246. [Google Scholar] [PubMed]
Xu C, Li L, Jin W, Wan Y (2014). Recombinase polymerase amplification (RPA) of CaMV-35S promoter and nos terminator for rapid detection of genetically modified crops. International Journal of Molecular Sciences 15: 18197–18205. [Google Scholar] [PubMed]
Yang QQ, Zhao XX, Wang D, Zhang PJ, Hu XN, Wei S, Liu JY, Ye ZH, Yu XP (2022). A reverse transcription-cross-priming amplification method with lateral flow dipstick assay for the rapid detection of Bean pod mottle virus. Scientific Reports 12: 681. [Google Scholar] [PubMed]
Yasukawa K, Yanagihara I, Fujiwara S (2020). Alteration of enzymes and their application to nucleic acid amplification (Review). International Journal of Molecular Medicine 46: 1633–1643. [Google Scholar] [PubMed]
Yuan H, Chao Y, Li SS, Tang MYH, Huang Y, Che Y, Wong AST, Zhang T, Shum HC (2018). Picoinjection-enabled multitarget loop-mediated isothermal amplification for detection of foodborne pathogens. Analytical Chemistry 90: 13173–13177. [Google Scholar] [PubMed]
Zhao Y, Xia Z, Zhang H, Wei L, Huang X (2010). Rapid and sensitive detection of Enterobacter sakazakii by cross-priming amplification combined with immuno-blotting analysis. Molecular and Cellular Probes 24: 396–400. [Google Scholar]
Zaghloul H, El-Shahat M (2014). Recombinase polymerase amplification as a promising tool in hepatitis C virus diagnosis. World Journal of Hepatology 6: 916–922. [Google Scholar] [PubMed]
Zanoli L, Spoto G (2013). Isothermal amplification methods for the detection of nucleic acids in microfluidic devices. Biosensors 3: 18–43. [Google Scholar] [PubMed]
Zhai L, Liu H, Chen Q, Lu Z, Zhang C, Lv F, Bie X (2019). Development of a real-time nucleic acid sequence-based amplification assay for the rapid detection of Salmonella spp. from food. Brazilian Journal of Microbiology 50: 255–261. [Google Scholar] [PubMed]
Zhang X, Du XJ, Guan C, Li P, Zheng WJ, Wang S (2015). Detection of Vibrio cholerae by isothermal cross-priming amplification combined with nucleic acid detection strip analysis. Molecular and Cellular Probes 29: 208–214. [Google Scholar] [PubMed]
Zhang F, Wang L, Fan K, Wu J, Ying Y (2014). The detection of T-Nos, a genetic element present in GMOs, by cross-priming isothermal amplification with real-time fluorescence. Analytical and Bioanalytical Chemistry 406: 3069–3078. [Google Scholar] [PubMed]
Zhao Y, Chen F, Li Q, Wang L, Fan C (2015). Isothermal amplification of nucleic acids. Chemical Reviews 115: 12491–12545. [Google Scholar] [PubMed]
Zhao X, Dong T, Yang Z, Pires N, Høivik N (2012). Compatible immuno-NASBA LOC device for quantitative detection of waterborne pathogens: Design and validation. Lab on a Chip 12: 602–612. [Google Scholar] [PubMed]
Zhao X, Lin CW, Wang J, Oh D H (2014). Advances in rapid detection methods for foodborne pathogens. Journal of Microbiology and Biotechnology 24: 297–312. [Google Scholar] [PubMed]
Zheng F, Li S, Wang S, Feng T, Jiang Z, Pan J (2020). Cross-priming isothermal amplification combined with nucleic acid test strips for detection of meat species. Analytical Biochemistry 597: 113672. [Google Scholar] [PubMed]
Zhou D, Wang S, Yang K, Liu X, Liu W et al. (2021). Rapid and simultaneous detection of Japanese encephalitis virus by real-time nucleic acid sequence-based amplification. Microbial Pathogenesis 150: 104724. [Google Scholar] [PubMed]
Zhu Y, Wang H, Wang L, Zhu J, Jiang W (2016). Cascade Signal amplification based on copper nanoparticle-reported rolling circle amplification for ultrasensitive electrochemical detection of the prostate cancer biomarker. ACS Applied Materials & Interfaces 8: 2573–2581. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF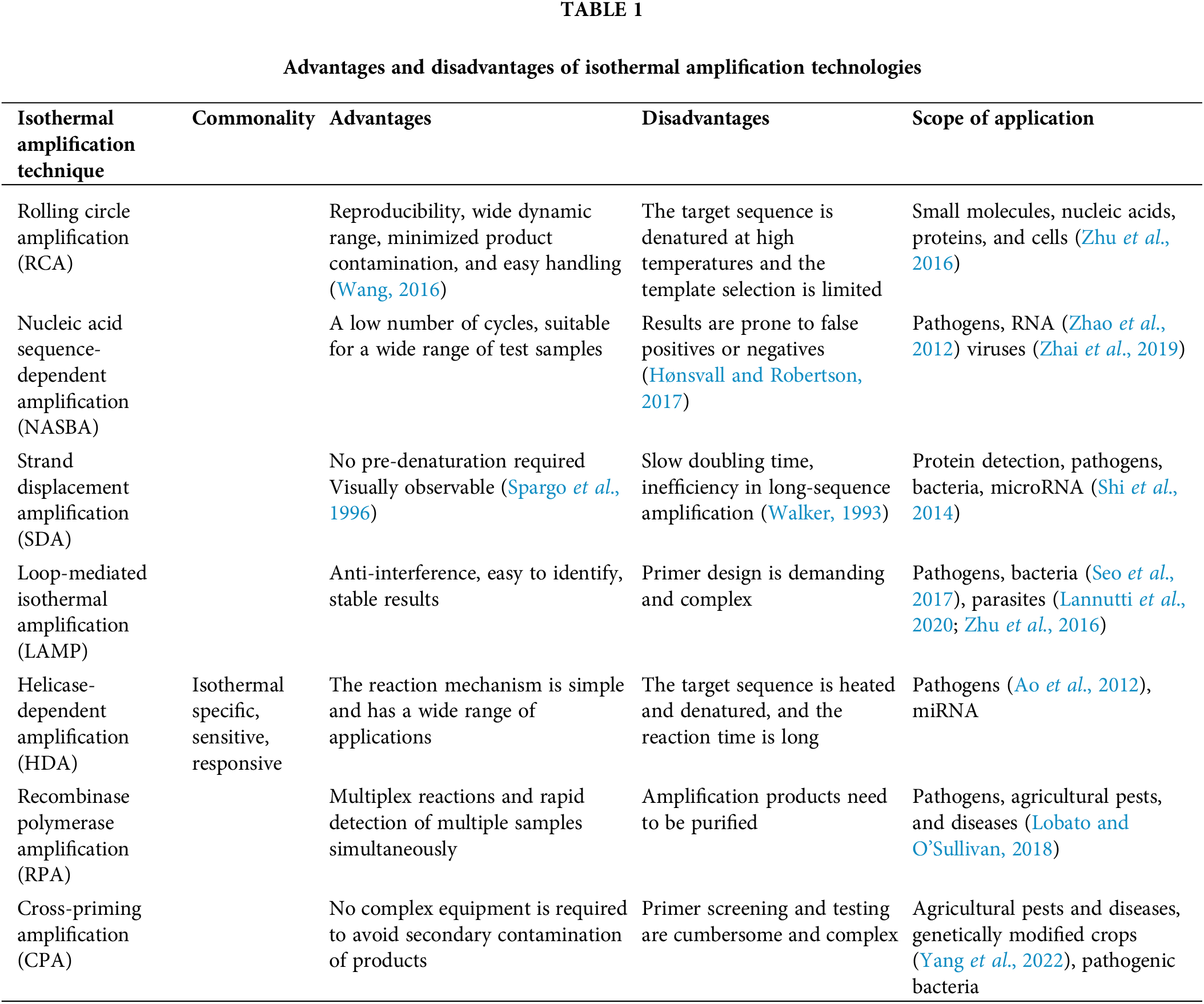
 Downloads
Downloads
 Citation Tools
Citation Tools