 Open Access
Open Access
ARTICLE
SIRT2 interacts with DDX24 to promote nasopharyngeal carcinoma growth
Department of Radiotherapy, The Second Affiliated Hospital of Guangxi Medical University, Nanning, China
* Corresponding Author: WENQI LIU. Email:
BIOCELL 2023, 47(11), 2445-2452. https://doi.org/10.32604/biocell.2023.042512
Received 02 June 2023; Accepted 08 September 2023; Issue published 27 November 2023
Abstract
Background: Nasopharyngeal carcinoma (NPC) is one of the most prevalent cancers in Southeast Asia. Sirtuin 2 (SIRT2) is a member of the NAD+-dependent deacetylase family and has been shown to play important roles in numerous biological processes. However, Its function in NPC remains uncertain. The primary aim of this study is to clarify the role of SIRT2 in NPC. Methods: In this research, we examined the effect of SIRT2 silencing on NPC cell proliferation and colony formation using vitro NPC cell lines. Co-immunoprecipitation and mass spectrometry was applied to identify SIRT2-interacting proteins in NPC cells. Results: In comparison to nasopharyngeal epithelial NP69 cells, SIRT2 was up-regulated in multiple NPC cell lines, particularly in CNE2 cells. SIRT2 knockdown abrogated CNE2 cell proliferation and colony formation, whereas SIRT2 overexpression promoted HNE1 cell proliferation and colony formation. The SIRT2-interacting proteins were gathered in gene expression and regulation processes including RNA processing and translation. Among the SIRT2-interacting proteins, there were multiple DEAD-box (DDX) family members. Of note, silencing of DDX24 phenocopied the effect of SIRT2 knockdown on NPC growth. Overexpression of DDX24 restored SIRT2-depleted CNE2 cells to proliferative and colony formation. Conclusions: Our study indicates that SIRT2 can interact with DDX24 to enhance NPC growth. The clinical relevance of SIRT2 and DDX24 in NPC warrants further investigation.Graphic Abstract
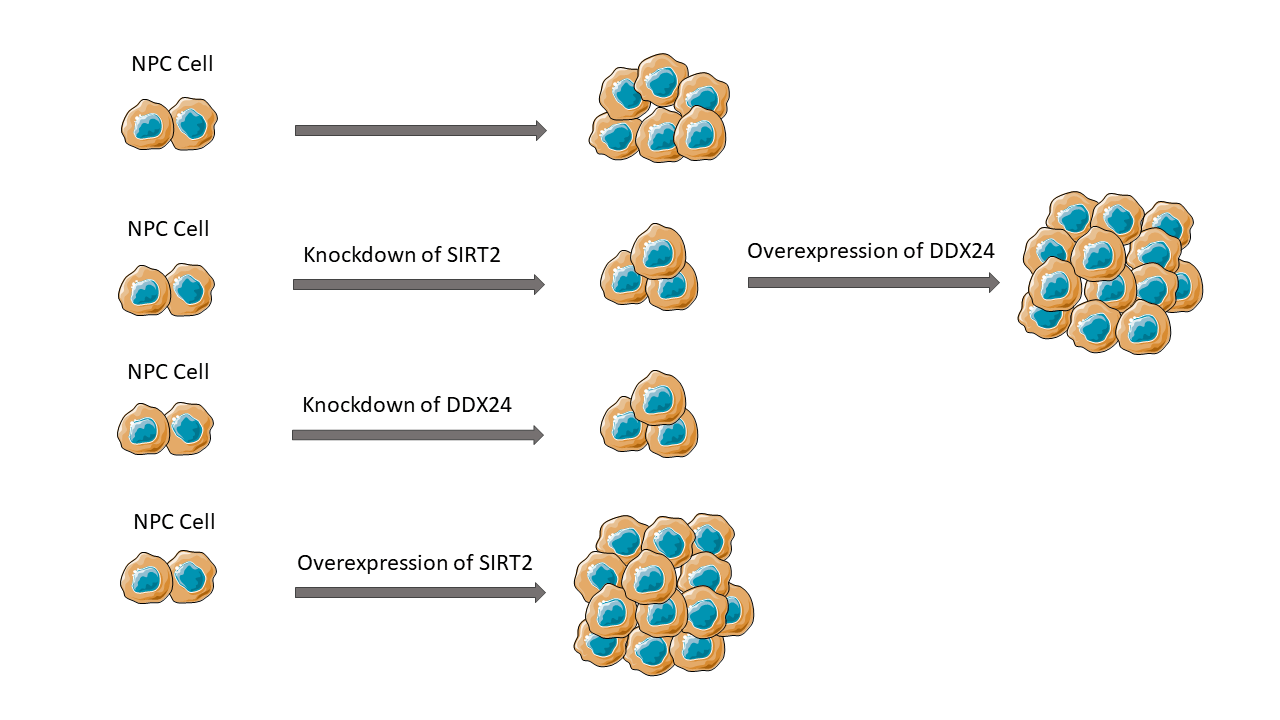
Keywords
Nasopharyngeal carcinoma (NPC) is one of the most common malignancies in Southeast Asia (Wong et al., 2021). Currently, platinum-based chemotherapy and radiation therapy are the main treatment options for NPC (Chen et al., 2019). However, primary or acquired chemoresistance and radiation resistance leads to unsatisfactory therapeutic outcomes (Yuan et al., 2022; Guo et al., 2021). In addition, patients in different regions seem to show different responses to treatment. Induction chemotherapy can improve the prognosis of endemic nasopharyngeal carcinoma, but not in non-endemic (Ou et al., 2016; Cao et al., 2017). This suggests that there are many factors contributing to the occurrence and development of NPC. Understanding the molecular pathways governing NPC malignant phenotypes is important for development of effective therapeutic strategies. Currently, the predictive role and related therapeutic approaches of tumor biomarkers, such as neutrophil to lymphocyte ratio, levels of EBV DNA, and microbiome are in progress (Orlandi et al., 2019).
Sirtuins (SIRTs) are a family of NAD+-dependent deacetylases that participate in a variety of biological processes including tumor development and growth (Jaiswal et al., 2022; Ji et al., 2022). Among the 7 mammalian SIRTs (SIRT1-7), SIRT1 and SIRT6 have been shown to inhibit NPC metastasis (Cheng, 2022; Chen et al., 2021). Unlike the 2 tumor-suppressive SIRTs, SIRT2 plays an oncogenic role in multiple malignancies such as osteosarcoma (Tian et al., 2022), prostate cancer (Lin et al., 2023), and endometrial cancer (Zhao et al., 2022). A recent study has indicated that SIRT2 can promote the resistance to lapatinib, a targeted tyrosine kinase inhibitor, in NPC (Aimjongjun et al., 2019). However, the part of SIRT2 in NPC has not been completely elucidated.
DDX24 is one of the DEAD-box (DDX) protein family of RNA helicases (Rocak and Linder, 2004). There are many aspects of RNA metabolism in which DDX proteins can participate, such as transcription, RNA degradation, pre-mRNA processing, RNA export, translation, and ribosomal biogenesis (Naineni et al., 2023). Thus, DDX proteins serve as important regulators of gene expression. Accumulating evidence has established a link between DDX24 and cancer progression. For instance, Liu et al. (2022) proved that expression of DDX24 is elevated in hepatocellular carcinoma (HCC) and improves proliferation and migration of HCC cells. Shi et al. (2016) showed that DDX24 is overexpressed in cancer cells and can suppress p53 activities to enhance tumor cell growth.
In this work, we explored the influence of SIRT2 in regulating NPC aggressive phenotype and identified an interaction between SIRT2 and DDX24. The mechanism underlying SIRT2-mediated NPC growth was uncovered.
Human nasopharyngeal epithelial cell line (NP69), as well as the NPC cell lines (CNE1, CNE2, and HNE1) were cultured at 37°C in a 5% CO2 incubator using Dulbecco’s modified eagle medium (DMEM) (Solebo, Beijing, China) complemented with 10% fetal bovine serum (Sigma-Aldrich, St. Louis, MO, USA).
Plasmids, small interfering RNAs (siRNAs), and cell transfection
We cloned the full-length coding sequence of SIRT2 into pcDNA3.1 myc-His vector to express Myc-tagged SIRT2. For overexpression studies, human SIRT2 and DDX24 cDNA was inserted into the pcDNA3.1(+) vector. The SIRT2- and DDX24-expressing plasmids (1 μg for each plasmid) were transfected into cancer cells through the Lipofectamine 3000 Transfection Reagent (Invitrogen, Carlsbad, CA, USA). For knockdown studies, CNE2 cells were inoculated in 6-well plates (1.5 × 106 cells/well) and incubated for 24 h. Transfection of indicated siRNAs (30 nM for each) into cells by HiPerFect Transfection Reagent (Qiagen, Hilden, Germany). The transfection efficiency exceeded 85%. The SIRT2 and DDX24 siRNA sequences are 5′-AAUCUCCACAUCCGCAGGCTT-3′ and 5′-GCAGUCAAGCUGTGGCAAATT-3′, respectively. The siRNA sequences for targeting other DDX genes are available on request. After 48 h, SIRT2 and DDX24 mRNA levels were evaluated by quantitative real-time PCR (qRT-PCR) analysis.
Extraction of total RNA from the cells studied using Trizol reagent (Invitrogen) following the manufacturer’s protocol. RNA samples were transcribed to cDNA using the Superscript III Reverse Transcriptase Kit (Invitrogen). The SYBR Premix Ex Taq (TaKaRa, Tokyo, Japan) was used for qRT-PCR. The cycling conditions are indicated as below 95°C pre-denaturation for 5 min and 38 cycles of 95°C for 15 s and 60°C for 40 s. The experiment was repeated three times independently for each sample, using GAPDH as the internal reference. The results were analyzed based on the 2−ΔΔCt formula (Schmittgen and Livak, 2008). The upstream and downstream primers were 5′-CCGGCCTCTATGACAACCTA-3′ and 5′-GGAGTAGCCCCTTGTCCTTC-3′ for SIRT2; 5′-TCCCAATGATTCATGCGGTG-3′ and 5′-CAGGCAATGCATCAGACTCA-3′ for DDX24; and 5′-ACCACAGTCCATGCCATCAC-3′ and 5′-TCCACCACCCTGTTGCTGTA-3′ for GAPDH. The primer sequences for other DDX genes tested in this study are available on request.
Cells were lysed in radio-immunoprecipitation assay (RIPA) buffer supplemented with protease and phosphatase inhibitors (Thermo Fisher Scientific, Waltham, MA, USA). Protein concentration was determined using a protein assay kit (Beyotime, Beijing, China). Lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes. Membranes were blocked with 5% fat-free milk and then incubated at 4°C with the antibodies recognizing SIRT2, DDX24, Myc tag, and GAPDH (Cell Signaling Use Technology, Danvers, MA, USA). After washing, the membranes were incubated with horseradish peroxidase-coupled secondary antibodies (Beyotime, Beijing, China). Protein bands were visualized with chemiluminescence (Solebo, Beijing, China).
The co-immunoprecipitation experiments were carried out with the Immunoprecipitation Kit (Absin, Shanghai, China) following the manufacturer’s protocol. Briefly, CNE2 cells overexpressing Myc-tagged SIRT2 (1 × 107 cells) were lysed in RIPA buffer containing protease and phosphatase inhibitors and precleared with protein A/G agarose beads at 4°C for 1 h. The lysates were incubated with anti-Myc antibody (Cell Signaling Technology, Danvers, MA, USA) or control IgG overnight at 4°C, followed by incubation with protein A/G agarose beads at 4°C for 3 h. Immunoprecipitates were eluted from the agarose beads and subjected to Western blot analysis or mass spectrometry analysis.
Protein samples were resolved by SDS-PAGE and stained with a coomassie stain. Proteins in the gels were digested with trypsin to generate peptides. The peptides were then analyzed on a mass spectrometer. Protein identification was performed by searching against a UniProt protein database. The identified proteins were subjected to Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG; https://www.kegg.jp/) analyses. Significant enrichment of genes in the GO biological processes and KEGG pathways were determined. Fisher’s exact test was used in GO and KEGG enrichment analysis. The GO categories and KEGG pathways with p-values < 0.05 were considered statistically significant.
CNE2 cells transfected with indicated constructs were seeded in 12-well plates at 1 × 104 cells/well. The cells were collected after being cultured for 2–6 days. The number of living cells at each point in time was determined, and the cell proliferation curve was plotted.
Colony formation assay was performed as previously described (Ling et al., 2022). CNE2 cells transfected with indicated constructs were plated in 12-well plates (400 cells/well). Colonies were cultured for 10–14 days, then dyed with crystalline violet and counted.
EdU cell proliferation assay was carried out as previously described (Wang et al., 2022). After transfecting with indicated constructs CNE2 cells were plated on cover slides in 6-well plates (1 × 104 cells/well). EdU working solution (10 μM; Beyotime) was added and incubated for further 2 h. After fixation and permeabilization, cells were incubated with the Click reaction solution in the dark for 30 min. Hoechst 33342 solution was then added to stain nuclei. Counted the number of EdU-positive cells under a fluorescence microscope.
All data are reported as the mean ± standard deviation (SD). Two groups were compared using the Student’s t-test, while multiple groups were compared with one-way analysis of variance. A p-value < 0.05 denotes statistical significance.
Depletion of SIRT2 inhibits the proliferation of NPC cells
Compared to NP69 nasopharyngeal epithelial cells, SIRT2 expression increased but varied among NPC cell lines (p = 0.0004, p < 0.0001 and p < 0.0001, respectively) (Figs. 1A and 1B). Particularly, CNE2 cells exhibited more abundant expression of SIRT2 than the other NPC cell lines tested. To ascertain the role of SIRT2 plays in NPC biology, we knocked down SIRT2 using specific siRNAs. CNE2 cells transfected with SIRT2-targeting siRNAs had significantly lower levels of SIRT2 than control siRNA-transfected cells (p = 0.0033) (Figs. 1C and 1D).
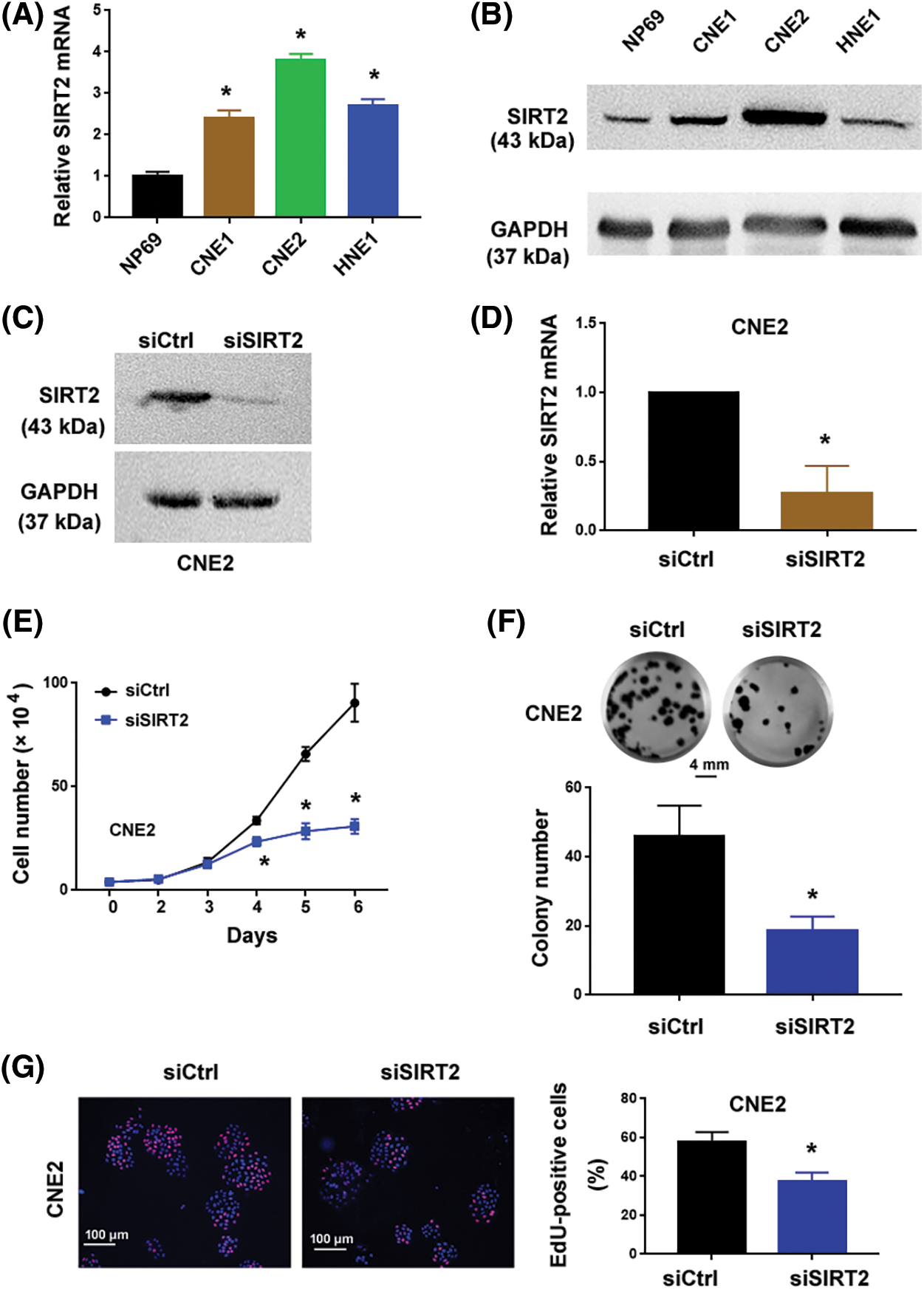
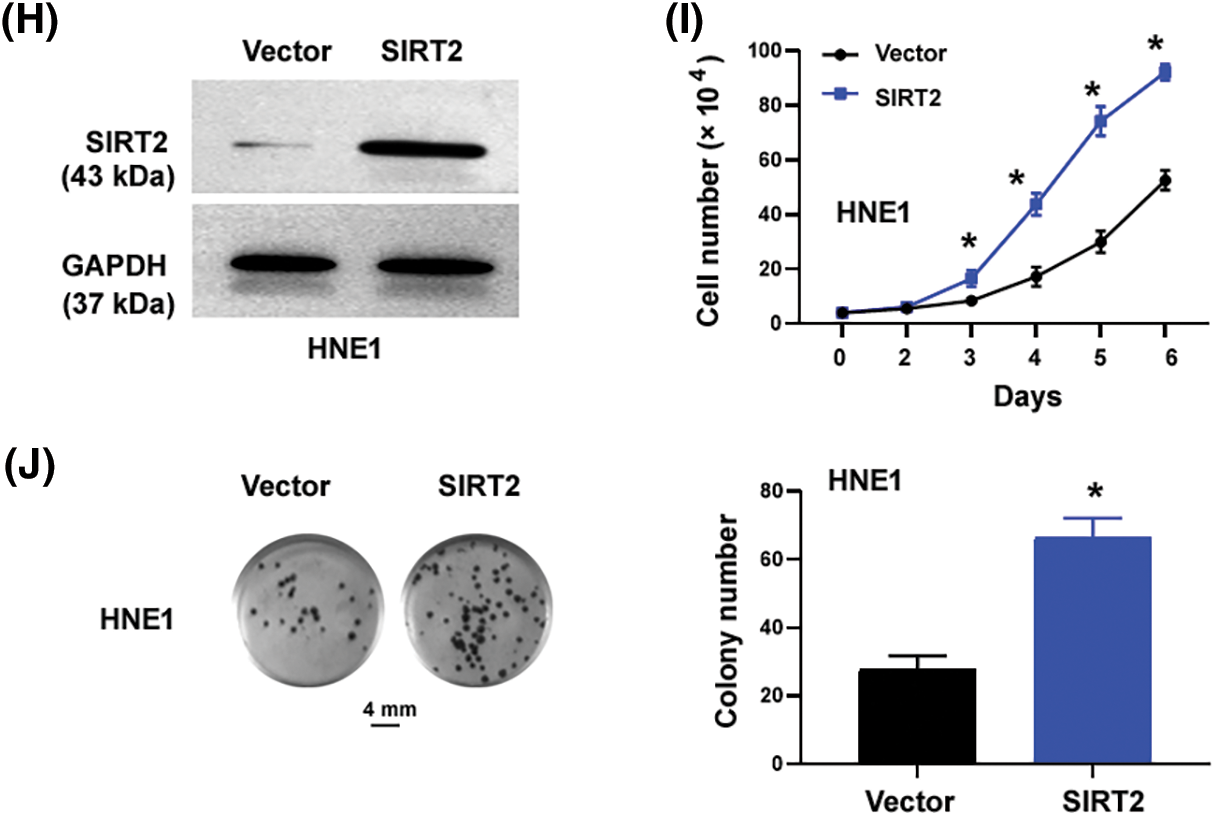
Figure 1: SIRT2 promotes the proliferation of NPC cells. (A) Quantification of SIRT2 mRNA levels in different cell lines. (B) Western blot analysis of SIRT2 protein in different cell lines. (C and D) Measurement of SIRT2 protein (C) and mRNA (D) levels in CNE2 cells transfected with control siRNA (siCtrl) or SIRT2 siRNA (siSIRT2). (E) Cell proliferation assay in CNE2 cells transfected with indicated siRNAs. (F) Colony formation assay in CNE2 cells transfected with indicated siRNAs. Scale bar, 4 mm. (G) EdU incorporation assay in CNE2 cells transfected with indicated siRNAs. Scale bar, 100 μm. (H) Western blot analysis of SIRT2 protein levels in HNE1 cells transfected with indicated plasmids. (I) Cell proliferation assay in HNE1 cells transfected with indicated plasmids. (J) Colony formation assay in HNE1 cells transfected with indicated plasmids. Scale bar, 4 mm. Data represent the mean ± SD of three replicates. *p < 0.05 by the Student’s t-test.
The cell proliferation curve (Fig. 1E) showed that SIRT2-depleted CNE2 cells proliferated more slowly than control cells at day4 (p = 0.0032), day5 (p = 0.0002) and day6 (p = 0.0005). Colony formation assays determined that SIRT2 knockdown cells generated fewer colonies than control cells (p = 0.0083) (Fig. 1F). Furthermore, EdU cell proliferation assays confirmed that the percentage of EdU-positive proliferative cells was obviously reduced in the SIRT2 knockdown group relative to the control group (p = 0.0082) (Fig. 1G). We also performed SIRT2 overexpression studies in HNE1 cells with relatively low levels of endogenous SIRT2. Ectopic expression of SIRT2 enhanced the proliferation and colony formation of HNE1 cells (Figs. 1H–1J). Taken together, SIRT2 is essential for the proliferation of NPC cells.
Protein interaction partners of SIRT2 in NPC cells
To investigate the mechanism by which SIRT2 regulates NPC growth, we performed co-immunoprecipitation and mass spectrometry to identify SIRT2-interacting proteins. For overexpression of SIRT2, the plasmid expressing Myc-tagged SIRT2 was transfected to CNE2 cells (Fig. 2A). Immunoprecipitation followed by SDS-PAGE revealed a specific band at the expected size of Myc-SIRT2 in Myc-SIRT2-overexpressing cells but not vector-transfected cells (Fig. 2B), suggesting an efficient immunoprecipitation of Myc-SIRT2. Co-immunoprecipitated proteins were subjected to in-gel digestion and protein identification by mass spectrometry. GO analysis represented that SIRT2-interacting proteins were enriched in gene expression and regulation processes including gene expression, RNA processing, and translation (Fig. 2C).
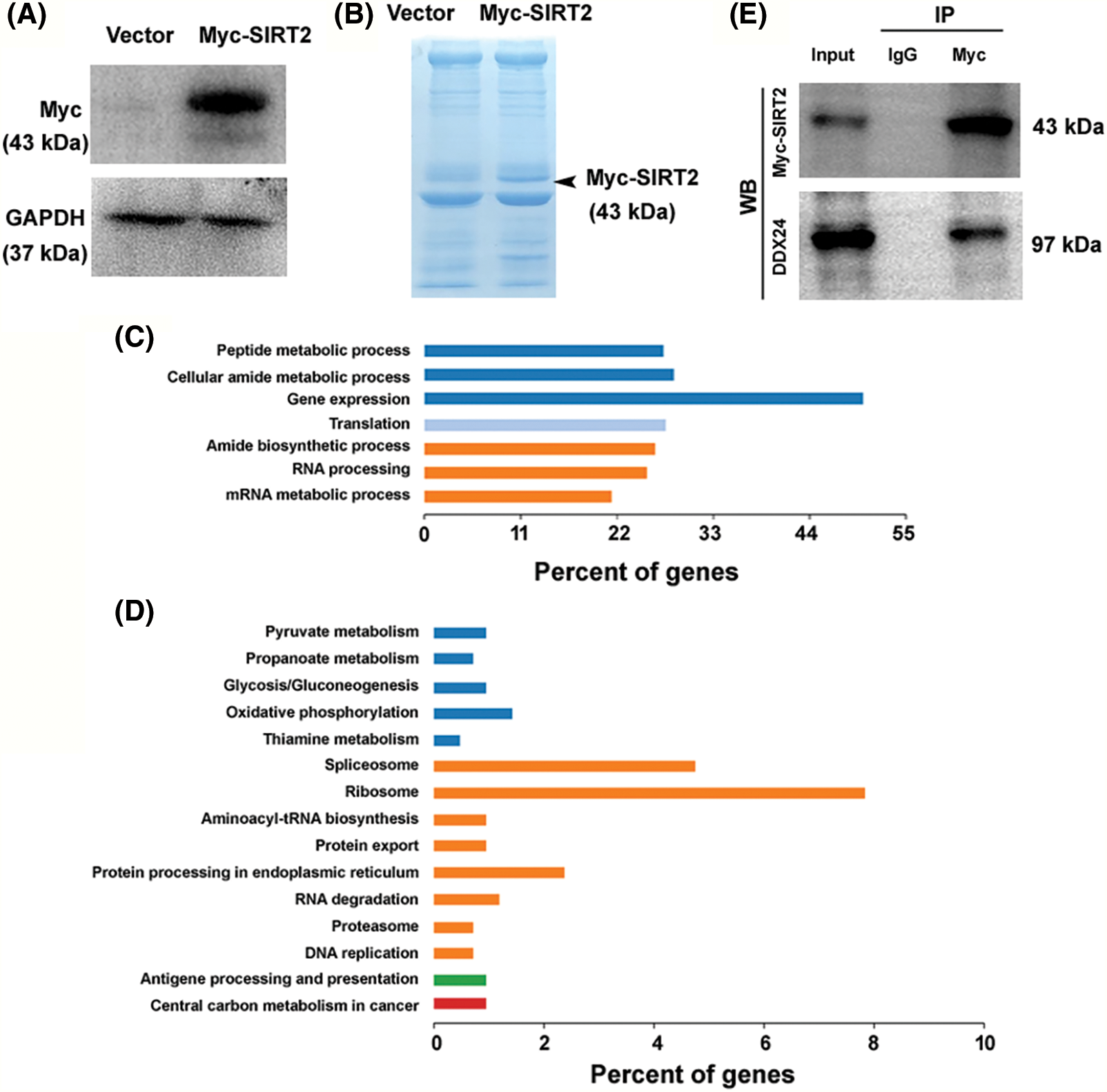
Figure 2: Protein interaction partners of SIRT2 in CNE2 cells. (A) Overexpression of Myc-tagged SIRT2 in CNE2 cells. (B) Myc immunoprecipitates from CNE2 cells transfected with vector or Myc-tagged SIRT2-expressing plasmids were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and stained with coomassie blue stain. (C) Analysis of SIRT2-interacting proteins enriched in GO biological processes. (D) Enrichment of SIRT2-interacting proteins in specific KEGG pathways. (E) Co-immunoprecipitation assay performed in CNE2 cells transfected with Myc-tagged SIRT2-expressing plasmids.
Moreover, KEGG pathway analysis demonstrated an enrichment of SIRT2-interacting proteins in the ribosome and spliceosome pathways (Fig. 2D). These results suggest that SIRT2 may modulate gene expression via regulation of RNA processing and translation. Among the SIRT2-interacting proteins, there were multiple DDX family members including DDX1, DDX5, DDX17, DDX18, DDX24, DDX27, DDX46, DDX54, DDX55, and DDX56. Given the importance of DDX proteins in the regulation of RNA metabolism and gene expression (Rocak and Linder, 2004; Naineni et al., 2023), we speculated that SIRT2-mediated NPC growth might be ascribed to the interaction with DDX proteins.
SIRT2 interacts with DDX24 to promote NPC growth
The 10 candidate DDX genes (i.e., DDX1, DDX5, DDX17, DDX18, DDX24, DDX27, DDX46, DDX54, DDX55, and DDX56) were knocked down to determine their roles in NPC cells. Co-immunoprecipitation assay validated the association between SIRT2 and DDX24 (Fig. 2E). Silencing of DDX24 notably inhibited CNE2 cell proliferation and colony formation (p = 0.0097) (Figs. 3A–3C), which was similar to the effects elicited by SIRT2 knockdown (Fig. 1). Depletion of other DDX genes tested did not influence the growth of CNE2 cells (Suppl. Fig. S1). Most importantly, overexpression of DDX24 (Fig. 3D) restored the proliferation and colony formation of SIRT2-depleted CNE2 cells (Figs. 3E and 3F). These results suggest that the interaction between SIRT2 and DDX24 contributes to NPC cell growth.
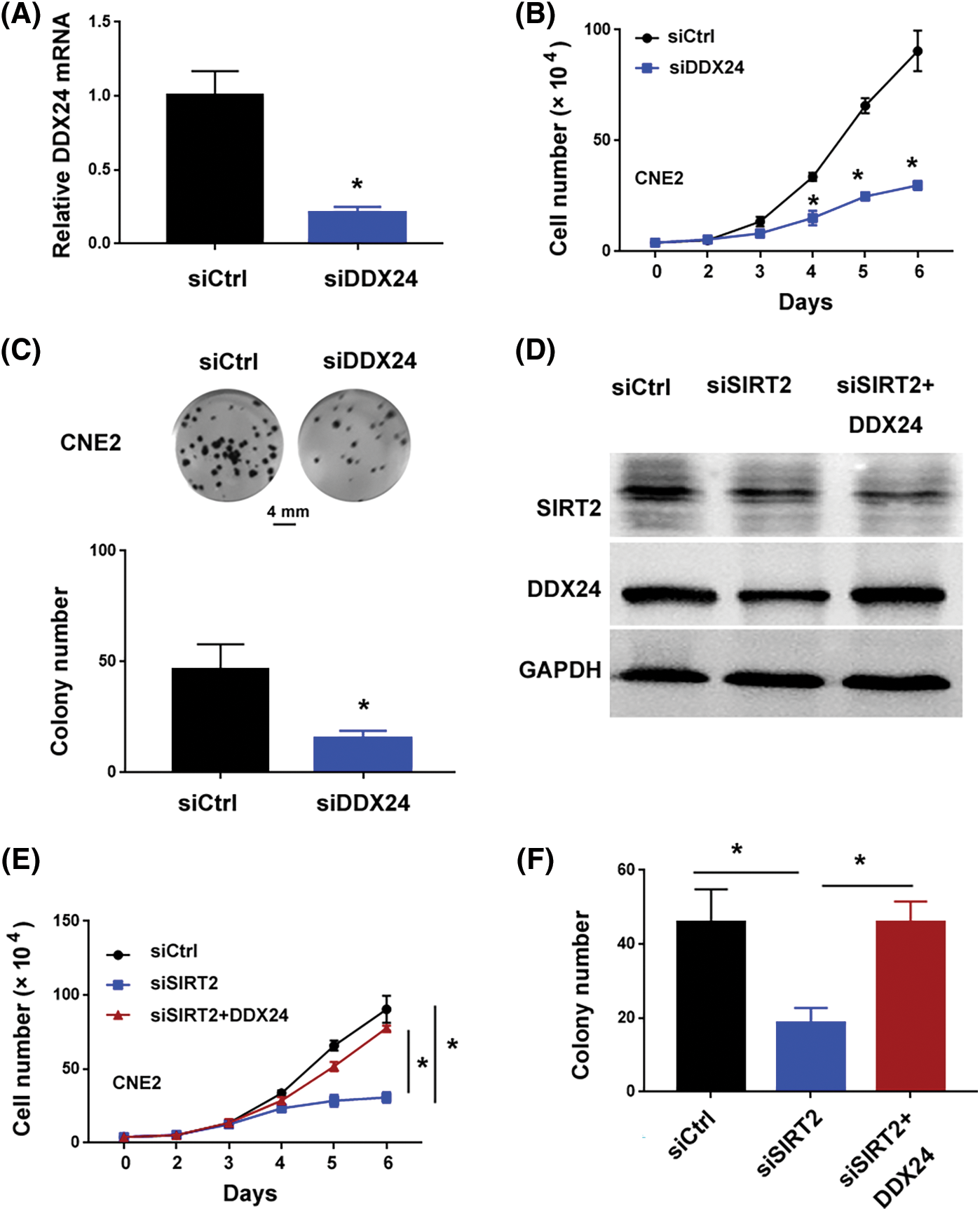
Figure 3: SIRT2 interacts with DDX24 to promote NPC growth. (A) Measurement of DDX24 mRNA in CNE2 cells transfected with control siRNA (siCtrl) or DDX24 siRNA (siDDX24). (B) Cell proliferation assay performed in CNE2 cells transfected with indicated siRNAs. (C) Colony formation assay performed in CNE2 cells transfected with indicated siRNAs. (D) Western blot analysis of SIRT2 and DDX24 protein in CNE2 cells transfected with indicated constructs. (E) Cell proliferation assay performed in CNE2 cells transfected with indicated constructs. (F) Colony formation assay performed in CNE2 cells transfected with indicated constructs. Data represent the mean ± SD of three replicates. *p < 0.05.
Our study shows that SIRT2 is enhanced in NPC cells and contributes to NPC growth. Targeted reduction of SIRT2 decreases the proliferation and colony formation of NPC cells. In contrast, overexpression of SIRT2 promotes NPC cell proliferation. The growth-modulating activity of SIRT2 is causally linked to the interaction between SIRT2 and DDX24. Ectopic expression of DDX24 rescues the proliferation of SIRT2-depleted NPC cells. These findings indicate that SIRT2 can interact with DDX24 to regulate NPC growth. SIRT2 is a member of the classic type III deacetylases and has the capacity to deacetylate histone and non-histone proteins (Wang et al., 2019; Jablonska et al., 2022). Dysregulation of SIRT2 is frequently observed in malignant diseases (Damodaran et al., 2017; Chen et al., 2013; Kim et al., 2011). Chen et al. (2013) stated that SIRT2 is upregulated in HCC and significantly correlated with advanced tumor stage, microscopic vascular invasion, and shorter overall survival. Damodaran et al. (2017) found that SIRT2 expression is reduced in prostate cancer relative to benign tissues and correlates with worse clinicopathologic outcomes. Our research illustrates that SIRT2 expression is elevated in NPC cells compared to nasopharyngeal epithelial cells, suggesting its involvement in NPC progression. However, additional studies are necessary to explore the expression and clinical significance of SIRT2 in NPC.
The effect of SIRT2 in tumor progression depends on cancer type. In some cancer types such as osteosarcoma (Tian et al., 2022) and endometrial cancer (Zhao et al., 2022), SIRT2 can promote tumor progression. However, in other types of cancer for instance colorectal cancer (Wang et al., 2020) and lung cancer (Xu et al., 2015), SIRT2 plays an opposite role. Our data show that knockdown of SIRT2 antagonizes CNE2 cell proliferation and colony formation. Consist with our results, a previous study reported that inhibition of SIRT2 activity increases lapatinib cytotoxicity against NPC cells (Aimjongjun et al., 2019). These findings demonstrate that SIRT2 plays an oncogenic function in NPC.
By performing co-immunoprecipitation and mass spectrometry, we find that SIRT2-interacting proteins are enriched in gene expression and regulation processes including gene expression, RNA processing, and translation. Of note, the SIRT2-interacting proteins include several DDX family members including DDX24. DDX proteins are known to modulate gene expression and translation through alteration of RNA metabolism and transport (Rocak and Linder, 2004; Naineni et al., 2023). These data suggest that SIRT2 might interact with DDX proteins to regulate gene expression, consequently contributing to NPC growth. In agreement with this hypothesis, we show that knockdown of DDX24 phenocopies the effects of SIRT2 silencing on NPC cells. Most importantly, overexpression of DDX24 reverses the suppression of proliferation and colony formation caused by SIRT2 knockdown. It has been documented that DDX24 can promote HCC cell proliferation and migration (Liu et al., 2022). According to another study, DDX24 protein is important for HCC growth and metastasis by decreased TRIM27-induced ubiquitination-related degradation and stabilizing its protein level (Zhang et al., 2022). Our data provide first evidence that DDX24 functions as an oncogene in NPC. Its depletion leads to a remarkable inhibition of NPC cell colony formation and proliferation. In SIRT2-overexpressing CNE2 cells, DDX24 protein levels are raised, but the mRNA abundance remains unchanged. We speculated that the increased protein levels of DDX24 might be the result of SIRT2-mediated stabilization.
Studies are currently underway to address this issue. This study has some limitations. Firstly, the clinical significance of SIRT2 and DDX24 in NPC is not clarified. Secondly, it remains to be determined how SIRT2 increases the protein levels of DDX24 in NPC cells. Finally, the mechanism underlying DDX24-induced NPC growth needs further investigation.
In summary, this work demonstrates that SIRT2 is upregulated in NPC cells and contributes to NPC cell proliferation and colony formation. SIRT2-mediated regulation of NPC growth depends on its interaction with DDX24 protein. The clinical significance of SIRT2 and DDX24 in NPC warrants further exploration.
Acknowledgement: None.
Funding Statement: This work was supported by 2017 Guangxi Appropriate Technology Development and Application Project (S2017013), the Project of Guangxi Health Department (Grant Nos. Z20190059 and Z20181011).
Author Contributions: The authors confirm their contribution to the paper as follows: study conception and design: Haiying Yue, Jian Li, Wenqi Liu; data collection: Xue Ou, Qiulu Zhong, Yiting Xie; Danjing Luo; Yihe Li; Chunxiao Liang; Xuemei Xu; Songnan Du; analysis and interpretation of results: Huijun Zhu, Qinghua Du, Xiangde Li; draft manuscript preparation: Haiying Yue, Chunhui Wang. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
Aimjongjun S, Mahmud Z, Jiramongkol Y, Alasiri G, Yao S, Yagüe E, Janvilisri T, Lam EW (2019). Lapatinib sensitivity in nasopharyngeal carcinoma is modulated by SIRT2-mediated FOXO3 deacetylation. BMC Cancer 19: 1106. [Google Scholar] [PubMed]
Cao SM, Yang Q, Guo L, Mai HQ, Mo HY et al. (2017). Neoadjuvant chemotherapy followed by concurrent chemoradiotherapy versus concurrent chemoradiotherapy alone in locoregionally advanced nasopharyngeal carcinoma: A phase III multicentre randomised controlled trial. European Journal of Cancer 75: 14–23. [Google Scholar] [PubMed]
Chen YP, Chan ATC, Le QT, Blanchard P, Sun Y, Ma J (2019). Nasopharyngeal carcinoma. The Lancet 394: 64–80. [Google Scholar]
Chen J, Chan AW, To KF, Chen W, Zhang Z et al. (2013). SIRT2 overexpression in hepatocellular carcinoma mediates epithelial to mesenchymal transition by protein kinase B/glycogen synthase kinase-3β/β-catenin signaling. Hepatology 57: 2287–2298. [Google Scholar] [PubMed]
Chen F, Ma X, Liu Y, Ma D, Gao X, Qian X (2021). SIRT6 inhibits metastasis by suppressing snail expression in nasopharyngeal carcinoma cells. International Journal of Clinical and Experimental Pathology 14: 63–74. [Google Scholar] [PubMed]
Cheng Y (2022). miR-135b-5p targets SIRT1 to inhibit deacetylation of c-JUN and increase MMP7 expression to promote migration and invasion of nasopharyngeal carcinoma cells. Molecular Biotechnology 64: 693–701. [Google Scholar] [PubMed]
Damodaran S, Damaschke N, Gawdzik J, Yang B, Shi C, Allen GO, Huang W, Denu J, Jarrard D (2017). Dysregulation of Sirtuin 2 (SIRT2) and histone H3K18 acetylation pathways associates with adverse prostate cancer outcomes. BMC Cancer 17: 874. [Google Scholar] [PubMed]
Guo Z, Wang YH, Xu H, Yuan CS, Zhou HH, Huang WH, Wang H, Zhang W (2021). LncRNA linc00312 suppresses radiotherapy resistance by targeting DNA-PKcs and impairing DNA damage repair in nasopharyngeal carcinoma. Cell Death & Disease 12: 69. [Google Scholar]
Jablonska B, Adams KL, Kratimenos P, Li Z, Strickland E, Haydar TF, Kusch K, Nave KA, Gallo V (2022). SIRT2 promotes white matter oligodendrogenesis during development and in models of neonatal hypoxia. Nature Communications 13: 4771. [Google Scholar] [PubMed]
Jaiswal A, Zhu XD, Ju ZY, Saretzki G (2022). Mitochondrial sirtuins in stem cells and cancer. The FEBS Journal 289: 3393–3415. [Google Scholar] [PubMed]
Ji Z, Liu GH, Qu J (2022). Mitochondrial sirtuins, metabolism, and aging. Journal of Genetics and Genomics 49: 287–298. [Google Scholar] [PubMed]
Kim HS, Vassilopoulos A, Wang RH, Lahusen T, Xiao Z et al. (2011). SIRT2 maintains genome integrity and suppresses tumorigenesis through regulating APC/C activity. Cancer Cell 20: 487–499. [Google Scholar] [PubMed]
Lin R, Yang Y, Wu E, Zhou M, Wang S, Zhang Q (2023). Sirt2 promotes cell proliferation and migration through mediating Erk1/2 activation and lactosylceramide accumulation in prostate cancer. The Prostate 83: 71–81. [Google Scholar] [PubMed]
Ling Y, Liang G, Lin Q, Fang X, Luo Q et al. (2022). circCDYL2 promotes trastuzumab resistance via sustaining HER2 downstream signaling in breast cancer. Molecular Cancer 21: 8. [Google Scholar] [PubMed]
Liu T, Gan H, He S, Deng J, Hu X et al. (2022). RNA helicase DDX24 stabilizes LAMB1 to promote hepatocellular carcinoma progression. Cancer Research 82: 3074–3087. [Google Scholar] [PubMed]
Naineni SK, Robert F, Nagar B, Pelletier J (2023). Targeting DEAD-box RNA helicases: The emergence of molecular staples. Wiley Interdisciplinary Reviews-RNA 14: e1738. [Google Scholar] [PubMed]
Orlandi E, Iacovelli NA, Tombolini V, Rancati T, Polimeni A, de Cecco L, Valdagni R, de Felice F (2019). Potential role of microbiome in oncogenesis, outcome prediction and therapeutic targeting for head and neck cancer. Oral Oncology 99: 104453. [Google Scholar] [PubMed]
Ou D, Blanchard P, El Khoury C, de Felice F, Even C et al. (2016). Induction chemotherapy with docetaxel, cisplatin and fluorouracil followed by concurrent chemoradiotherapy or chemoradiotherapy alone in locally advanced non-endemic nasopharyngeal carcinoma. Oral Oncology 62: 114–121. [Google Scholar] [PubMed]
Rocak S, Linder P (2004). Dead-box proteins: The driving forces behind RNA metabolism. Nature Reviews Molecular Cell Biology 5: 232–241. [Google Scholar] [PubMed]
Schmittgen TD, Livak KJ (2008). Analyzing real-time pcr data by the comparative C(T) method. Nature Protocols 3: 1101–1108. [Google Scholar] [PubMed]
Shi D, Dai C, Qin J, Gu W (2016). Negative regulation of the P300-P53 interplay by DDX24. Oncogene 35: 528–536. [Google Scholar] [PubMed]
Tian Y, Liu R, Hou X, Gao Z, Liu X, Zhang W (2022). SIRT2 promotes the viability, invasion and metastasis of osteosarcoma cells by inhibiting the degradation of snail. Cell Death & Disease 13: 935. [Google Scholar]
Wang J, Tan L, Yu X, Cao X, Jia B, Chen R, Li J (2022). lncRNA ZNRD1-AS1 promotes malignant lung cell proliferation, migration, and angiogenesis via the miR-942/TNS1 axis and is positively regulated by the m6A reader YTHDC2. Molecular Cancer 21: 229. [Google Scholar] [PubMed]
Wang Y, Yang J, Hong T, Chen X, Cui L (2019). SIRT2: Controversy and multiple roles in disease and physiology. Ageing Research Reviews 55: 100961. [Google Scholar] [PubMed]
Wang B, Ye Y, Yang X, Liu B, Wang Z et al. (2020). SIRT2-dependent IDH1 deacetylation inhibits colorectal cancer and liver metastases. EMBO Reports 21: e48183. [Google Scholar] [PubMed]
Wong KCW, Hui EP, Lo KW, Lam WKJ, Johnson D et al. (2021). Nasopharyngeal carcinoma: An evolving paradigm. Nature Reviews Clinical Oncology 18: 679–695. [Google Scholar] [PubMed]
Xu W, Jiang K, Shen M, Qian Y, Peng Y (2015). SIRT2 suppresses non-small cell lung cancer growth by targeting JMJD2A. Biological Chemistry 396: 929–936. [Google Scholar] [PubMed]
Yuan L, Li S, Chen Q, Xia T, Luo D et al. (2022). EBV infection-induced GPX4 promotes chemoresistance and tumor progression in nasopharyngeal carcinoma. Cell Death & Differentiation 29: 1513–1527. [Google Scholar]
Zhang H, Liu Y, Wang W, Liu F, Wang W, Su C, Zhu H, Liao Z, Zhang B, Chen X (2022). ALKBH5-mediated m6A modification of lincRNA LINC02551 enhances the stability of DDX24 to promote hepatocellular carcinoma growth and metastasis. Cell Death & Disease 13: 926. [Google Scholar]
Zhao N, Guo Y, Liu P, Chen Y, Wang Y (2022). Sirtuin 2 promotes cell stemness and Mek/Erk signaling pathway while reduces chemosensitivity in endometrial cancer. Archives of Gynecology and Obstetrics 305: 693–701. [Google Scholar] [PubMed]
Supplementary Materials
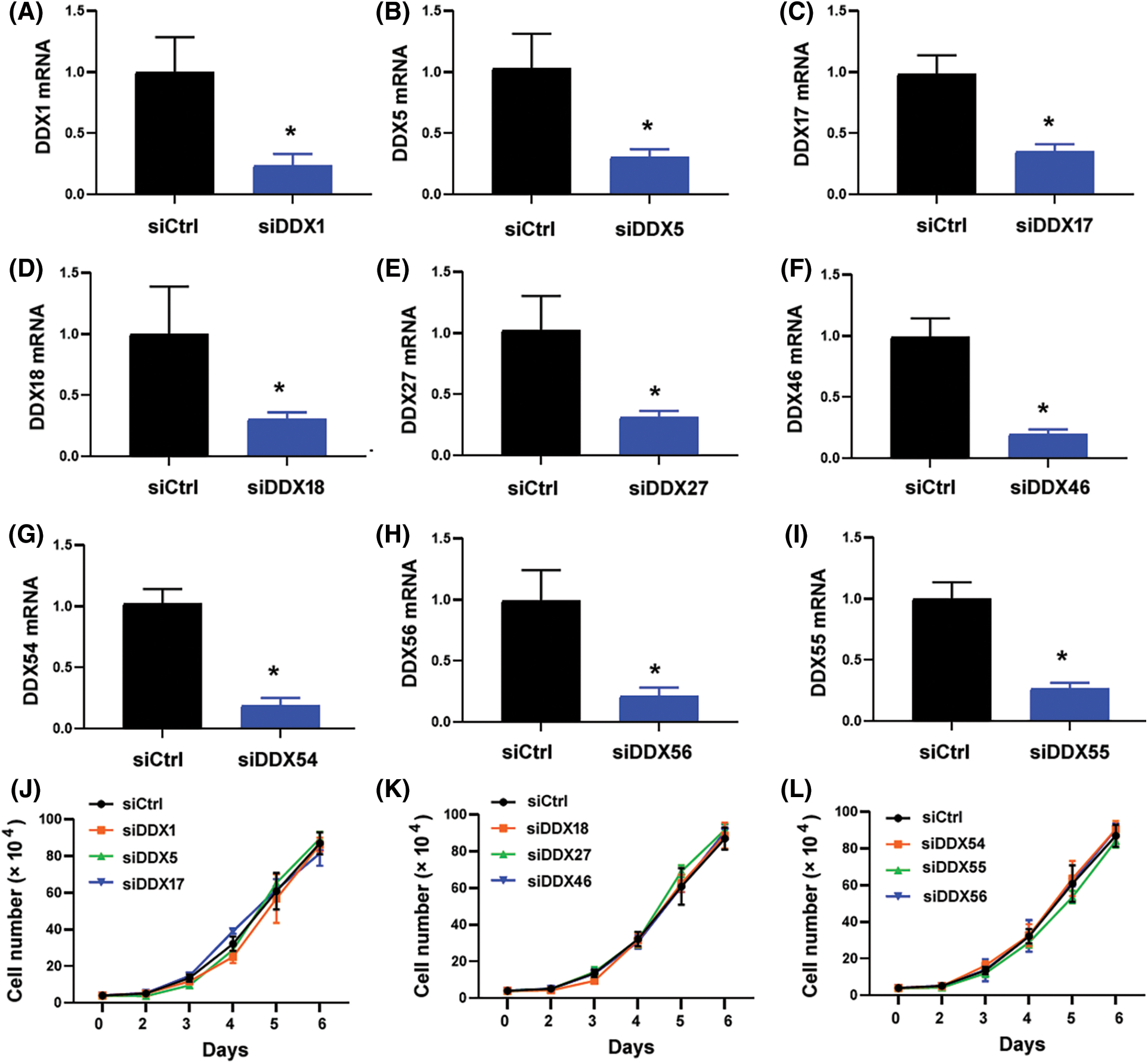
Figure S1: Effect of knockdown of DDX genes on the proliferation of CNE2 cells. (A–I) Quantitative real-time PCR analysis of indicated DDX genes in CNE2 cells transfected with control siRNA (siCtrl) or DDX-targeting siRNAs. Data represent the mean ± SD of three replicates. *p < 0.05. (J–L) Cell proliferation assay performed in CNE2 cells transfected with indicated siRNAs. No significant changes in cell proliferation relative to siCtrl-transfected cells were observed.
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools