 Open Access
Open Access
ARTICLE
Aryl hydrocarbon receptor nuclear translocator 2 as a prognostic biomarker and immunotherapeutic indicator for clear cell renal cell carcinoma
1 Department of Public Health Laboratory Sciences, University of South China, Hengyang, 421001, China
2 Department of Blood Transfusion, Shenzhen Longhua District Central Hospital, Shenzhen, 518000, China
3 Bioinformatics R&D Department, Hangzhou Mugu Technology Co., Ltd., Hangzhou, 310000, China
* Corresponding Author: XILIN XIAO. Email:
# Equal contribution
(This article belongs to the Special Issue: Bioinformatics Study of Diseases)
BIOCELL 2023, 47(11), 2397-2408. https://doi.org/10.32604/biocell.2023.030281
Received 29 March 2023; Accepted 24 May 2023; Issue published 27 November 2023
Abstract
Background: In many cancer types, aryl hydrocarbon receptor nuclear translocator 2 (ARNT2) has been found to be associated with tumor cell proliferation and prognosis. However, the role of ARNT2 in clear cell renal cell carcinoma (ccRCC) has not been completely elucidated. In this study, the potential role of ARNT2 in ccRCC development was characterized. Methods: A pan-cancer dataset (TCGA-TARGET-GTEx) was accessed from UCSC Xena Data Browser. ARNT2 expression in normal and tumor samples was compared. Univariate Cox regression was performed to evaluate the prognostic value of ARNT2. Single sample gene set enrichment analysis (ssGSEA) was used to estimate the enrichment of functional pathways and gene signatures. CIBERSORT and ESTIMATE methods evaluated the immune infiltration. The ARNT2 expression was determined in ccRCC tissue and cell lines using RT-qPCR and Western blot. Results: ARNT2 expression was significantly dysregulated in 23 out of 30 cancer types. Pan-cancer data revealed a strong correlation between ARNT2 expression and immune modulators, immune cell infiltration, and genomic alternations. In ccRCC patients, the low-ARNT2 expression group had higher immune infiltration, CD8 T cells, and programmed cell death ligand 1 expression, as well as higher enrichment score of immunotherapeutic predictors than those in the high-ARNT2 expression group. Low-ARNT2 expression group was more responsive to immunotherapy. Moreover, low ARNT2 expression was observed in ccRCC tissue and cell lines. Conclusions: Dysregulated ARNT2 expression is involved in cancer development and the modulation of the immune microenvironment. ARNT2 can be potentially used as a prognostic indicator and an immunotherapeutic indicator for ccRCC.Keywords
Supplementary Material
Supplementary Material FileAryl hydrocarbon receptor nuclear translocator (ARNT) and ARNT2 are members of the family of transcription factors responsible for embryonic development (Aitola and Pelto-Huikko, 2003). ARNT2 is structurally similar to ARNT, which can combine with aryl hydrocarbon receptor (AhR), hypoxia-inducible factor (HIF)-α, or drosophila single-minded (SIM) as a heterodimer to bind DNA (Hirose et al., 1996; Michaud et al., 2000). ARNT2 is mainly expressed in the neuroepithelium, adrenal medulla (kidney), and the inner layer of the retina (Aitola and Pelto-Huikko, 2003; Freeburg and Abrahamson, 2004; Jain et al., 1998). Mice with ARNT2-knockout die early in the perinatal period and have impaired hypothalamic development (Hosoya et al., 2001; Keith et al., 2001).
Evidence has shown that ARNT2 expression levels are aberrant in multiple cancer types. In non-small cell lung cancer (NSCLC), ARNT2 expression is downregulated compared to that in normal lung tissues (Yang et al., 2015). ARNT2 knockdown in NSCLC cell lines HCC827 and A549 decreased cell apoptosis and promoted tumor cell growth (Yang et al., 2015), which demonstrated its role as a tumor suppressor in NSCLC. ARNT2 was also significantly downregulated in oral squamous cell carcinoma (OSCC) (Kimura et al., 2016). In vitro experiments found that ARNT2 overexpression could inhibit tumor cell proliferation in OSCC and was associated with tumor size (Kimura et al., 2016). Jia et al. (2019) found downregulated ARNT2 in gastric cancer and its correlation to the poor overall survival of gastric cancer patients. In breast cancer, ARNT2 expression is associated with favorable overall survival and disease-free survival (Martinez et al., 2008). Knockdown of ARNT2 in hepatocellular carcinoma cell lines (HCCLM6) significantly enhances tumor cell proliferation, invasion, and migration (Li et al., 2015). However, ARNT2 expression is not downregulated in all cancer types. Bogeas et al. (2018) revealed that ARNT2 knockdown attenuates the expression of transcription factors involved in the tumorigenicity in glioblastoma (GMB), and suppresses the tumorigenic properties in glioblastoma stem-like cells. Previous findings suggested ARNT2 as a potential biomarker for cancer prognosis, but may function in a different mechanism in different cancer types.
Disturbances in the immune response system of the tumor microenvironment (TME) are involved in the development of cancers, including clear cell renal cell carcinoma (ccRCC) (Kobayashi et al., 2022; Wang et al., 2022). Aryl hydrocarbon receptor (AHR) is closely related to innate and adaptive immunity (Trikha and Lee, 2020). Inhibitory pathways associated with AHR and its metabolites may be activated in the TME, thereby promoting immune escape and progression (Murray et al., 2014). AHR is also involved in a variety of immune cell regulatory processes. The presence of endogenous AHR ligands may promote T cell development in the TME (Funatake et al., 2008). AHR is also a key cofactor involved in interleukin-10 production by natural killer (NK) cells (Wagage et al., 2014). In addition, AHR is a transcription factor that determines monocyte differentiation in mice, and that the activation of AHR promotes monocyte-to-dendritic cell differentiation and interferes with monocyte-to-macrophage differentiation (Goudot et al., 2017). The level of immune cell infiltration in the TME is closely related to patient survival and prognosis (Mei et al., 2020). This study, therefore, investigated the prognostic value of ARNT2 and its immunotherapeutic role in ccRCC.
Up to now, none of the studies have explored the role of ARNT2 in ccRCC. The present research examined pan-cancer data to evaluate the relation between ARNT2 expression and prognosis, immune microenvironment, and genomic variations. Functional pathways related to ARNT2 expression, the association between immune infiltration, and ARNT2 expression in ccRCC were explored. A series of oncogenic pathways were found to be significantly associated with ARNT2 expression. Importantly, ARNT2 exhibited the potential to predict the response to clinical therapies in ccRCC.
Retrieval and processing of pan-cancer data
The pan-cancer dataset (TCGA-TARGET-GTEx) with RNA-sequencing (RNA-seq) data (transcript per million, TPM) and survival data was downloaded from UCSC Xena Data Browser (https://xenabrowser.net/datapages/?cohort=TCGA%20TARGET%20GTEx&removeHub=https%3A%2F%2Fxena.treehouse.gi.ucsc.edu%3A443) (Goldman et al., 2020). TCGA-TARGET-GTEx dataset includes 19131 samples and 60499 genes of pan-cancer from three sources (TCGA, TARGET, and GTEx). The expression data of pan-cancer has already been recomputed and integrated by the UCSC platform. We selected the following sample types for analysis: derived cancer—bone marrow, primary tumor, primary blood derived cancer—peripheral blood, primary solid tumor, normal solid tissue, and primary blood normal tissue. The RNA-seq data of ENSG00000172379 (ARNT2) was extracted and then log2 (TPM+0.001) transformation was performed (Suppl. Table S1). Cancer types with no more than three samples were removed.
Assessing the relation between aryl hydrocarbon receptor nuclear translocator 2 expression and immunological features
Four types of immune modulators, including chemokines, immunostimulators, major histocompatibility complex (MHC), and receptors, were obtained from previous research (Ru et al., 2019). Hmisc R package was employed to perform Pearson correlation analysis of the association between ARNT2 expression with immune modulators, immune checkpoints, and immune cells. The CIBERSORT method was applied to estimate the proportion of 22 immune cells (Chen et al., 2018). The ESTIMATE method was utilized to evaluate stromal infiltration and immune infiltration (Yoshihara et al., 2013).
Evaluating the relation between genomic characteristics and aryl hydrocarbon receptor nuclear translocator 2 expression
The pan-cancer single nucleotide variation data processed by Mutect2 were downloaded from Genomic Data Commons (GDC) Data Portal (https://portal.gdc.cancer.gov/). The R package of maftools (version 2.8.05) (Mayakonda et al., 2018) was used to calculate the score of mutant-allele tumor heterogeneity (MATH), and tumor mutation burden (TMB) for each cancer type. Loss of heterozygosity (LOH) and homologous recombination deficiency (HRD) data were obtained from a previous research (Thorsson et al., 2018). ARNT2 expression and TMB, MATH, LOH, and HRD were subjected to Spearman correlation analysis. A p < 0.05 was considered a significant correlation. The R package of surminer was used to determine the optimal cut-off value of ARNT2 expression in classifying ccRCC samples of kidney renal clear cell carcinoma (KIRC) dataset into high- and low-ARNT2 expression groups.
Analysis of the biological pathways related to aryl hydrocarbon receptor nuclear translocator 2 in kidney renal clear cell carcinoma dataset
Gene sets of Hallmark pathways were downloaded from Molecular Signatures Database (MSigDB) (Liberzon et al., 2015). GSVA R package (Hänzelmann et al., 2013) was used to conduct single sample gene set enrichment analysis (ssGSEA) for each ccRCC sample. Then the association between ssGSEA score and ARNT2 expression was examined by Pearson correlation analysis. Gene sets of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were downloaded from the KEGG database (Kanehisa et al., 2017). The FGSEA algorithm (http://bioconductor.org/packages/fgsea/) was conducted to annotate enriched biological pathways. False discovery rate (FDR) < 0.05 meant significantly enriched in GSEA results. Genes of GOBP_RESPONSE_TO_OXIDATIVE_STRESS pathway were obtained from MSigDB.
Analyzing the potential of aryl hydrocarbon receptor nuclear translocator 2 in predicting the response to clinical therapy
A gene list of T-cell-inflamed gene expression profiles (GEP) was obtained from Ayers et al. (2017). Th1/interferon-gamma (IFN-γ) gene signature was obtained from Danilova et al. (2019). Therapeutic signatures for the response prediction to immunotherapy, targeted therapy, and radiotherapy were obtained from the study by Hu et al. (2021). The gene signature of cytolytic activity was determined by referring to Rooney et al. (2015). SsGSEA was implemented to calculate the enrichment score of the above signatures. To examine the difference in the enrichment score of signatures, the Wilcoxon test was conducted between high- and low-ARNT2 expression groups.
The ccRCC specimens and adjacent normal renal tissues were obtained from 30 patients who underwent surgery at Shenzhen Longhua District Central Hospital from January 2021 to November 2022. Written informed consent was also obtained from all patients, and the study was approved by the Ethics Committee of Shenzhen Longhua District Central Hospital.
The human ccRCC cell lines Caki-1, 769P, and ACHN and the human normal renal tubular epithelial cell line HK-2 were obtained from ATCC. Caki1 cells were cultured in McCoy’s 5a (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS) and 1% antibiotics. 769P cells and ACHN cells were cultured in Dulbecco’s modified Eagle medium supplemented with 10% FBS and 1% antibiotics. HK-2 cells were cultured in keratinocyte serum-free medium (ScienCell Research Laboratories, Inc., Carlsbad, CA, USA) supplemented with 1% keratinocyte growth supplement (ScienCell Research Laboratories, Inc.) and 1% antibiotics (100 μL/mL penicillin and 100 mg/mL streptomycin sulfates). All cells were maintained at 37°C in a humidified atmosphere of 5% CO2.
Real-time-quantitative polymerase chain reaction (RT-qPCR)
Total RNA from patient tissues was extracted using Trizol Universal reagent. Reverse transcriptase was used to convert mRNA into the cDNA template for RT-qPCR. RT‑qPCR was performed on an ABI 7500 Sequence Detection System (Life Technologies, USA) using the corresponding PCR reagent according to the manufacturer’s instructions. The primers used were as follows: ARNT2, 5′-GACTCACTTCCTCGCTCAC-3′ (forward) and 5′-GCCATCCTGTTGCCATCAC-3′ (reverse), β-actin sense 5′-GTGGACATCCGCAAAGAC-3′ (forward), and 5′-AAAGGGTGTAACGCAACTA-3′ (reverse). Relative expression was determined using the 2−∆∆CT method.
Approximately 20 μg of total protein was extracted from ccRCC cells, separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to polyvinylidene fluoride membranes and reacted with primary antibodies against ARNT2 (1:500) and β-actin (1:1000). After extensive washing with PBS containing 0.1% Triton X-100, the membrane was incubated with alkaline phosphatase-coupled goat anti-rabbit antibody for 30 min at room temperature. The reagents were visualized using the One StepTM NBT/BCIP reagent (Thermo Fisher Scientific, Rockford, IL, USA) and detected using an Alpha Imager (Alpha Innotech, San Leandro, CA, USA).
R software (version 4.1) was used as a platform to perform statistical analysis. The current bioinformatics analysis was supported by the Sangerbox platform (Shen et al., 2022) (http://vip.sangerbox.com/). Wilcoxon rank sum test was employed to test the two-group significance. p < 0.05 was determined as statistically significant.
The expression pattern of aryl hydrocarbon receptor nuclear translocator 2 in pan-cancer data
The work flow of this study is shown in Suppl. Fig. S1. We first assessed the expression level of ARNT2 in 30 cancer types and corresponding normal samples. Twenty-three cancer types showed a dysregulated expression level of ARNT2 compared with normal samples (p < 0.05, Suppl. Fig. S2A). Significant overexpression of ARNT2 was observed in 11 cancer types such as low-grade glioma (LGG) (tumor: 6.29 ± 0.60, normal: 5.34 ± 1.41, p = 3.8e-79), breast cancer (BRCA) (tumor: 3.00 ± 1.78, normal: 0.68 ± 1.30, p = 4.9e-79), and skin cutaneous melanoma (SKCM) (tumor: 4.43 ± 1.31, normal: −0.19 ± 0.76, p = 1.5e-55). Meanwhile, ARNT2 was downregulated in another 11 cancer types, such as KIRC (tumor: 2.12 ± 1.44, normal: 3.99 ± 1.51, p = 1.3e-49), colon adenocarcinoma (COAD) (tumor: −0.39 ±1.72, normal: 1.17 ± 1.69, p = 1.7e-33), and cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC) (tumor: 0.30 ± 1.94, normal: 1.62 ± 1.25, p = 4.3e-3). We observed that ARNT2 was relatively high-expressed in the skin, peripheral nervous system, eye, fibroblast, adrenal cortex, central nervous system, kidney, lung, and cervix when compared with other tissues (Suppl. Fig. S2B). In ccRCC, compared with normal samples, a significantly downregulated expression of ARNT2 in tumor samples was observed (Fig. 1A). In different clinical stages of ccRCC, we also observed a mild association that ccRCC samples with late stages or grades showed high ARNT2 expression (Figs. 1B and 1C). Similar results were observed in vitro experiments. In Fig. 1D, ARNT2 mRNA expression had an obvious reduction in ccRCC tumor tissue. In ccRCC cell lines, the protein expression of ARNT2 was downregulated (Fig. 1E).
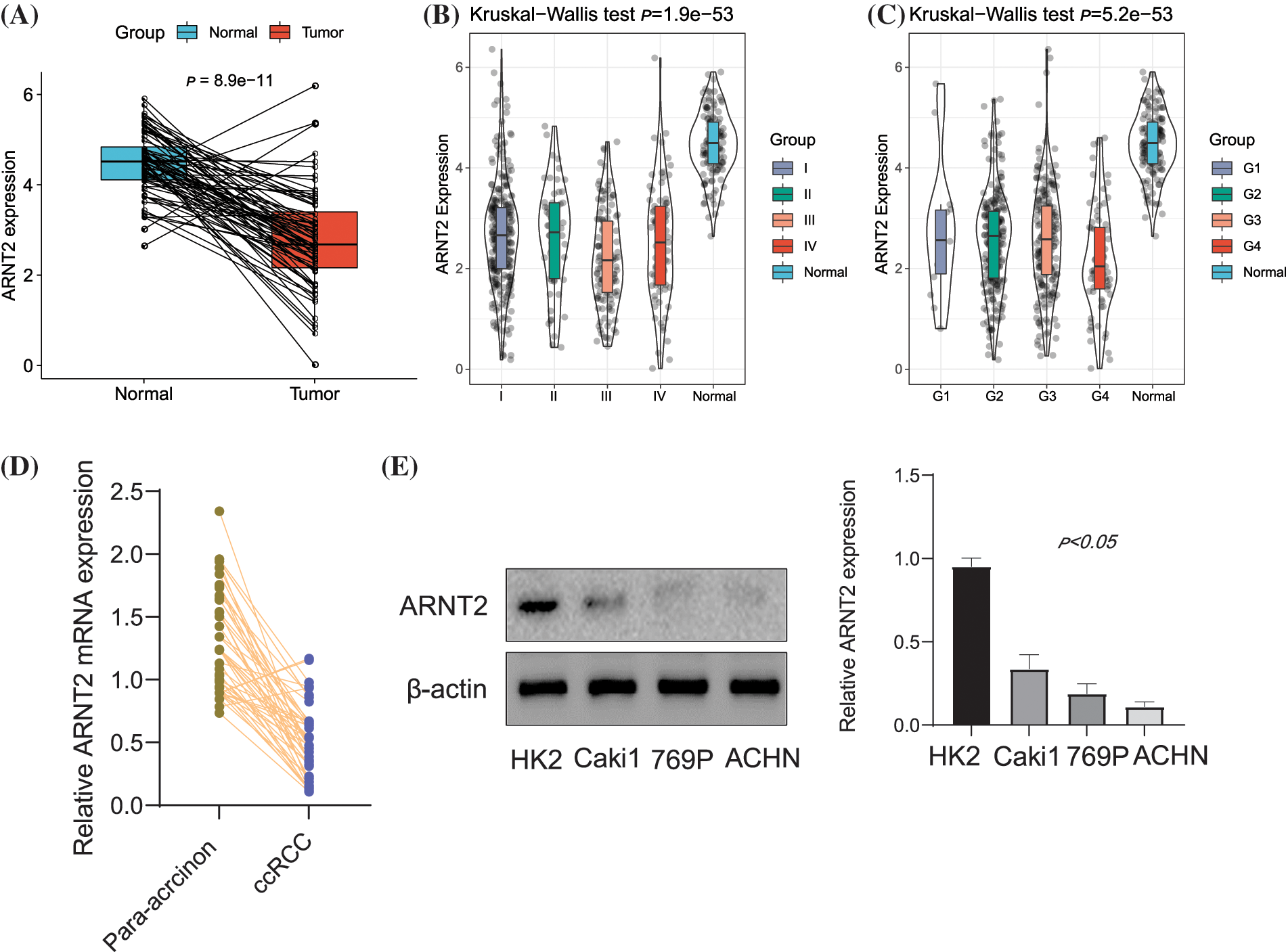
Figure 1: The expression of aryl hydrocarbon receptor nuclear translocator (ARNT2) in kidney renal clear cell carcinoma (KIRC). (A) Comparison of ARNT2 expression in normal and clear cell renal cell carcinoma (ccRCC) samples. (B and C) Comparison of ARNT2 expression in different clinical stages (B) and grades (C). (D) Determination of the mRNA expression in ccRCC tumor tissue by real-time-quantitative polymerase chain reaction. (E) Expression of ARNT2 protein in ccRCC cell lines using Western blot analysis. ns, not significant.
The relation between aryl hydrocarbon receptor nuclear translocator 2 expression and cancer prognosis
Then we assessed the relation between ARNT2 expression and survival time in 30 cancer types. Univariate Cox regression analysis was here to analyze the effect of ARNT2 expression on OS and disease-specific survival (DSS) in different cancer types. As a result, we found that ARNT2 expression was a prognostic factor for overall survival (OS) in pancreatic adenocarcinoma (PAAD) (HR = 0.574), hepatocellular carcinoma (LIHC) (HR = 1.240), head and neck squamous cell carcinoma (HNSC) (HR = 0.872), and KIRC (HR = 0.861) (Suppl. Fig. S3A). As for the effect of ARNT2 expression on DSS, the prognostic value of ARNT2 was shown in PAAD (HR = 0.603), peritoneal carcinomatosis from gastric cancer (PCGC) (HR = 3.160), and KIRC (HR = 0.790) (Suppl. Fig. S3B). From the above results, we speculated that dysregulated ARNT2 expression may be involved in cancerogenesis. Especially, ARNT2 expression was significantly downregulated in KIRC and it was a risk factor for OS and DSS in KIRC.
The association between aryl hydrocarbon receptor nuclear translocator 2 expression and immunity in pan-cancer
Immune response to cancer cells is a crucial factor in determining cancer prognosis. Immune modulators such as cytokines, chemokines, and immune checkpoints play important roles in recruiting immune cells and reconstructing the immune microenvironment. Therefore, from a previous study, we obtained four types of immune modulators, including chemokines, immunostimulators, MHC, and receptors (Ru et al., 2019), and assessed the association between these immune modulators and ARNT2 expression. Pearson correlation coefficients were calculated to be differentially present in different cancer types. A positive correlation was clearly observed in LIHC, COAD, and rectum adenocarcinoma (READ), while a negative correlation was observed in KIRC, BRCA, uterine corpus endometrial carcinoma (UCEC), glioblastoma multiforme (GBM), and LGG (Suppl. Fig. S2C).
Then we evaluated the expression of four important immune checkpoints programmed cell death ligand 1 (PD-L1; CD274), programmed cell death (PD-1; PDCD1), cytotoxic T-lymphocyte–associated antigen 4 (CTLA-4), and lymphocyte activation gene-3 (LAG-3) and their relations to ARNT2 expression. Significant correlations were found in some cancer types, and KIRC was correlated with PD-L1, CTLA-4, and LAG-3 (Figs. 2A–2D). In addition, immune infiltration analysis revealed a significant association of ARNT2 expression with the enrichment of some immune cells in many cancer types (Suppl. Fig. S2D). For example, in most cancer types, ARNT2 expression was negatively related to activated NK cells, CD8 T cells, and activated memory CD4 T cells, but positively correlated with resting mast cells and resting memory CD4 T cells. In KIRC, ARNT2 expression showed a significant relation with most of immune cells, such as a negative correlation with regulatory T cells, CD8 T cells, and follicular helper T cells, and a positive correlation with M2 macrophages, monocytes, and resting mast cells.
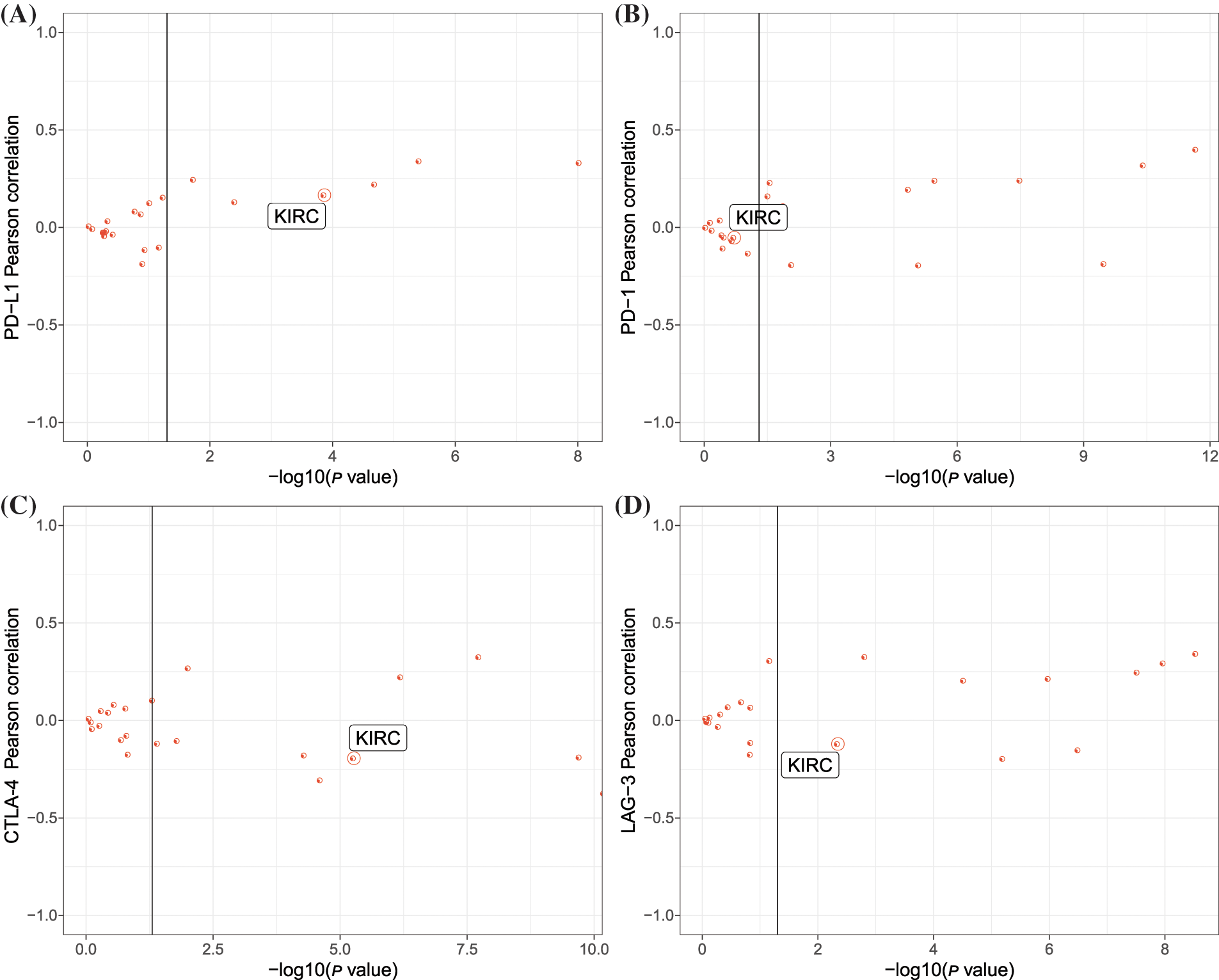
Figure 2: The relation between aryl hydrocarbon receptor nuclear translocator (ARNT2) expression and immunity in kidney renal clear cell carcinoma (KIRC). (A–D) Pearson correlation analysis between ARNT2 expression and four immune checkpoints, including programmed cell death ligand-1 (PD-L1) (A), PD-1 (B), cytotoxic T-lymphocyte–associated antigen 4 (CTLA-4) (C), and lymphocyte activation gene-3 (LAG-3) (D). The y-axis represents −log10 (p value), and the x-axis represents the Pearson correlation coefficients. One red dot indicates one cancer type.
The relation between aryl hydrocarbon receptor nuclear translocator 2 expression and genomic characteristics
Genomic alternations can lead to the aberrant expression of genes that may participate in cancer development. Therefore, we analyzed the relation between ARNT2 expression and different genomic alternations, including TMB, MATH, HRD, and LOH. We found 14 cancer types with a significant correlation between ARNT2 expression and TMB (p < 0.05, Suppl. Fig. S4A). MATH score was associated with ARNT2 expression in six cancer types (Suppl. Fig. S4B). Additionally, ARNT2 expression was observed to be correlated with HRD and LOH in 16 and 14 cancer types, respectively (Suppl. Figs. S4C, S4D). ARNT2 expression in KIRC was negatively correlated with TMB (p < 0.05), HRD (p < 0.001), and LOH (p < 0.01). KIRC samples were then grouped into high- and low-expression ARNT2 groups with the optimal cut-off value determined using the survminer package, and we examined whether there was a difference in the four genomic characteristics in the two expression groups. The low ARNT2 expression group had a higher score of HRD and LOH than in the high ARNT2 expression group (Fig. 3A). Moreover, mutation analysis for KIRC displayed the top 10 mutated genes (Fig. 3B). Notably, PBRM1 was the most frequently mutated in both high- and low-ARNT2 expression groups, but high ARNT2 expression obviously showed more frequent mutations.
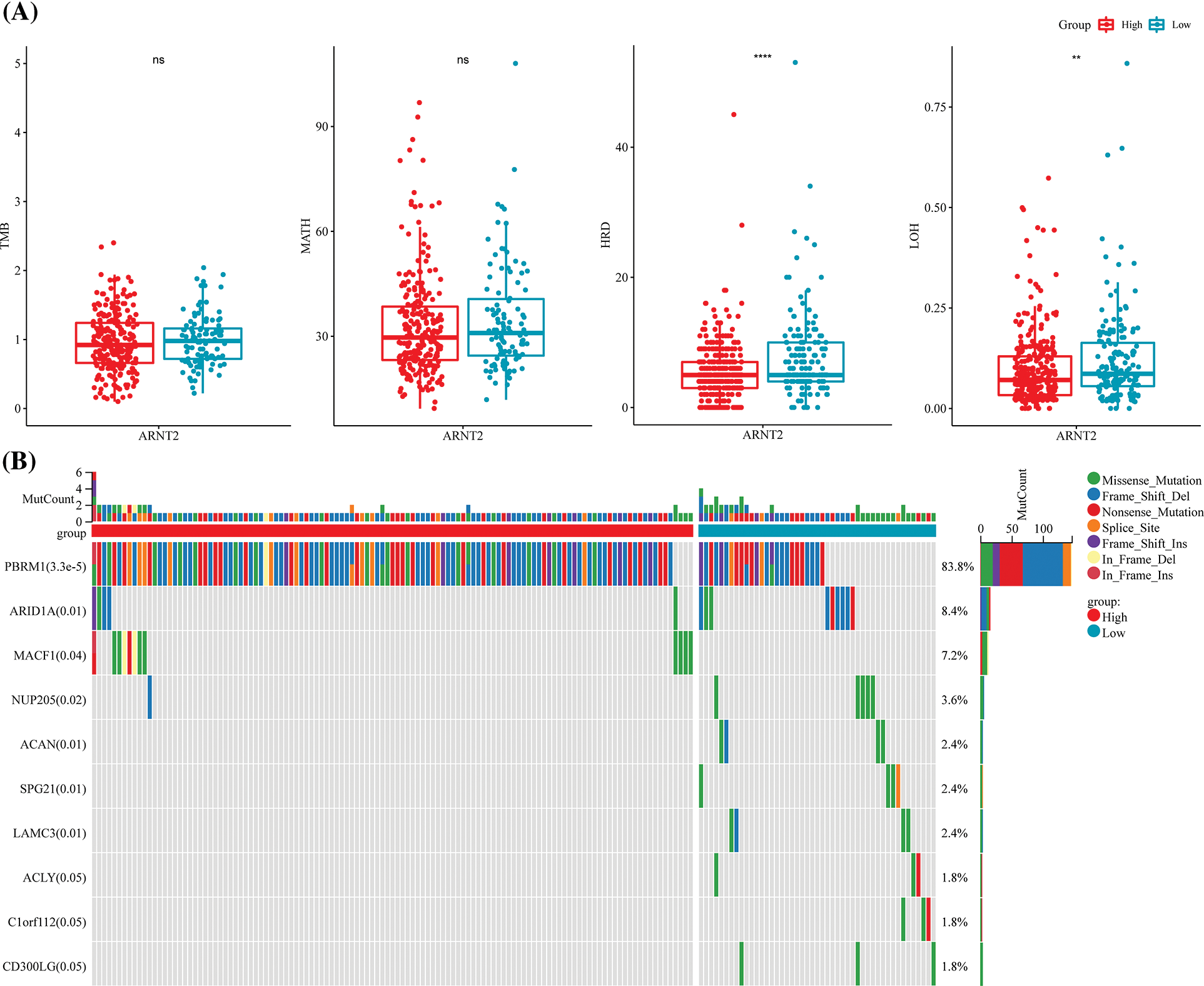
Figure 3: The relation between aryl hydrocarbon receptor nuclear translocator (ARNT2) expression and genomic characteristics. (A) The score of tumor mutation burden (TMB), mutant-allele tumor heterogeneity (MATH), homologous recombination deficiency (HRD), and loss of heterozygosity (LOH) in high- and low-ARNT2 expression groups in kidney renal clear cell carcinoma (KIRC). (B) The top 10 mutated genes in KIRC. The x-axis indicates KIRC samples. ns, not significant.
Functional pathways related to aryl hydrocarbon receptor nuclear translocator 2 expression in kidney renal clear cell carcinoma
To explore pathways related to ARNT2 in KIRC, we calculated the ssGSEA score of hallmark pathways for each KIRC sample. Pearson correlation analysis between ssGSEA score and ARNT2 expression identified the top 10 enriched pathways associated with ARNT2 expression (Fig. 4A). Seven pathways were positively correlated with ARNT2 expressions, such as oxidative phosphorylation, hedgehog signaling, and TGF-β signaling, which have been reported in cancer development. Differentially enriched pathways between high- and low-ARNT2 expression groups were also identified, and the top 15 differential pathways were displayed (Figs. 4B and Suppl. Fig. S5). Cell cycle-related pathways such as P53 signaling pathway, MYC target v2, and DNA repair were significantly enriched in low-ARNT2 expression group. Moreover, GSEA was used to screen enriched KEGG pathways of the high-ARNT2 expression group. We discovered that cellular community and cell motility-related pathways such as regulation of actin cytoskeleton, tight junction, adherens junction, and focal adhesion were significantly accumulated (Fig. 4C).
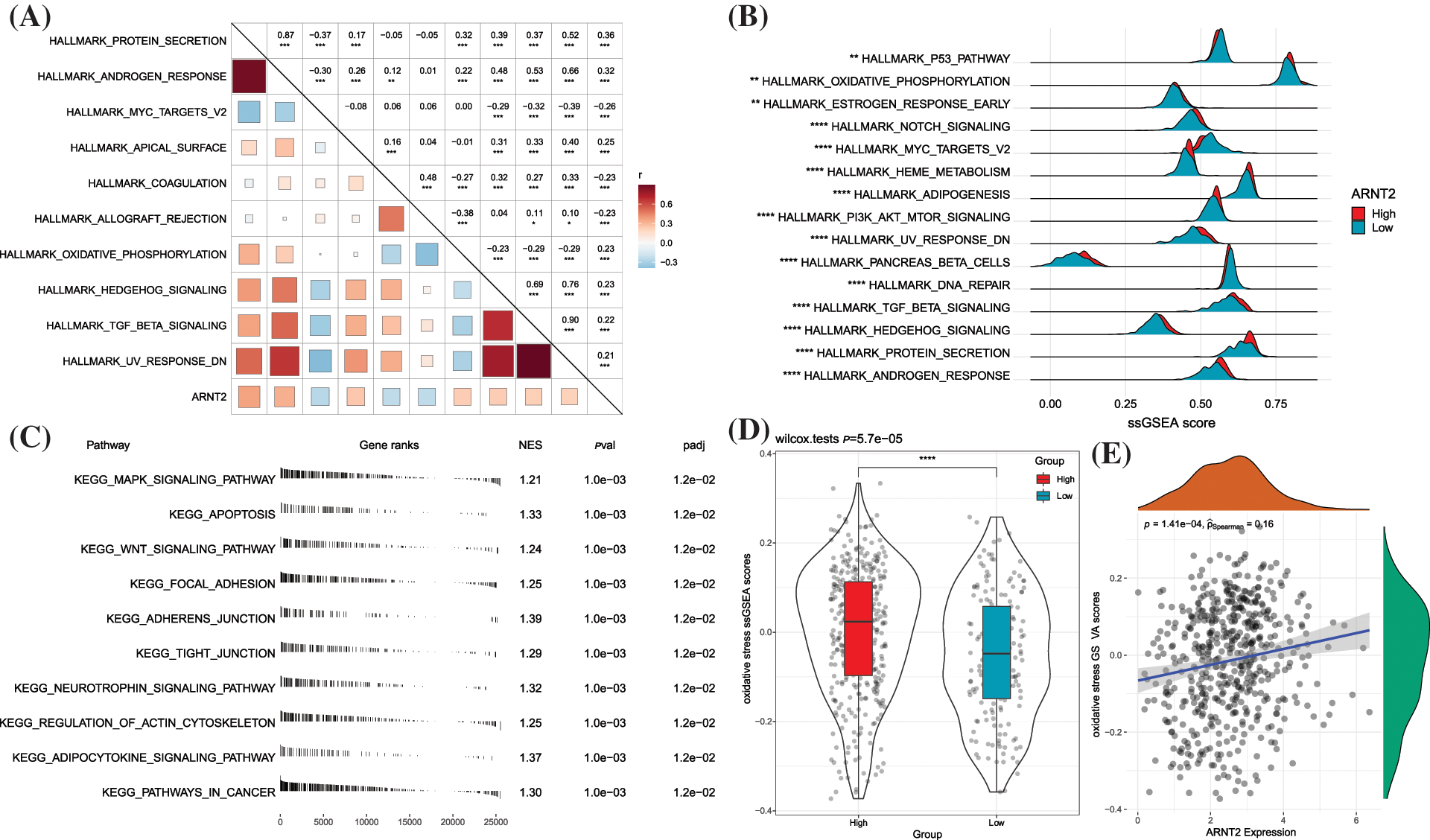
Figure 4: Functional pathways related to aryl hydrocarbon receptor nuclear translocator (ARNT2) expression in kidney renal clear cell carcinoma (KIRC). (A) The top 10 hallmark pathways significantly correlated with ARNT2 expression. (B) The top 10 differential hallmark pathways between high- and low-ARNT2 expression groups. (C) The top 10 enriched pathways in the high-ARNT2 expression group were analyzed by Gene Set Enrichment Analysis. NES, normalized enrichment score. pval, p value. padj, adjusted p value. (D) Gene Set Variation Analysis score of oxidative stress in the two groups. Wilcoxon test was conducted. (E) Pearson correlation analysis between oxidative stress and ARNT2 expression. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Oxidative stress, defined as an excessive increase in reactive oxygen species (ROS), is greatly produced in tumor cells with hyperproliferation. Tumor cells can adapt to the ROS environment by increasing their anti-ROS activity to avoid cell death (Hayes et al., 2020). We then evaluated the relationship between oxidative stress and ARNT2 expression. The high-ARNT2 expression group showed a markedly higher oxidative stress score than the low-ARNT2 expression group (p < 0.0001, Fig. 4D). A significantly positive correlation was also found between oxidative stress and ARNT2 expression (R = 0.16, p = 1.41e-04, Fig. 4E).
The relation between aryl hydrocarbon receptor nuclear translocator 2 expression and immune infiltration in kidney renal clear cell carcinoma
In the pan-cancer analysis, a strong correlation was observed in ARNT2 expression with immune modulators and immune infiltration in many cancer types. This part focused on the specific relationship between immune infiltration and ARNT2 expression in KIRC. Twelve out of 22 immune cells were differentially enriched in high- and low-ARNT2 expression groups, as revealed by CIBERSORT analysis (p < 0.05, Fig. 5A). Specifically, activated NK cells and CD8 T cells showed a greater amount in low-ARNT2 expression group, while a higher distribution of resting memory CD4 T cells, monocytes, and M2 macrophages was observed in the high-ARNT2 expression group. ESTIMATE analysis suggested that immune infiltration was much lower in the high-expression group than the low-expression group (p = 0.0061, Fig. 5B). To determine the immune cells showing an interaction with ARNT2, we delineated an association network among immune cells and ARNT2 based on their correlation coefficients (Fig. 5C). As a result, we found that ARNT2 possibly had a direct interaction with CD8 T cells and monocytes. At the same time, significant correlations were also observed between CD8 T cells and many other immune cells, such as monocytes, resting memory CD4 T cells, and M2 macrophages. The above results suggested that ARNT2 expression had a possible impact on the accumulation of CD8 T cells (Fig. 5D).
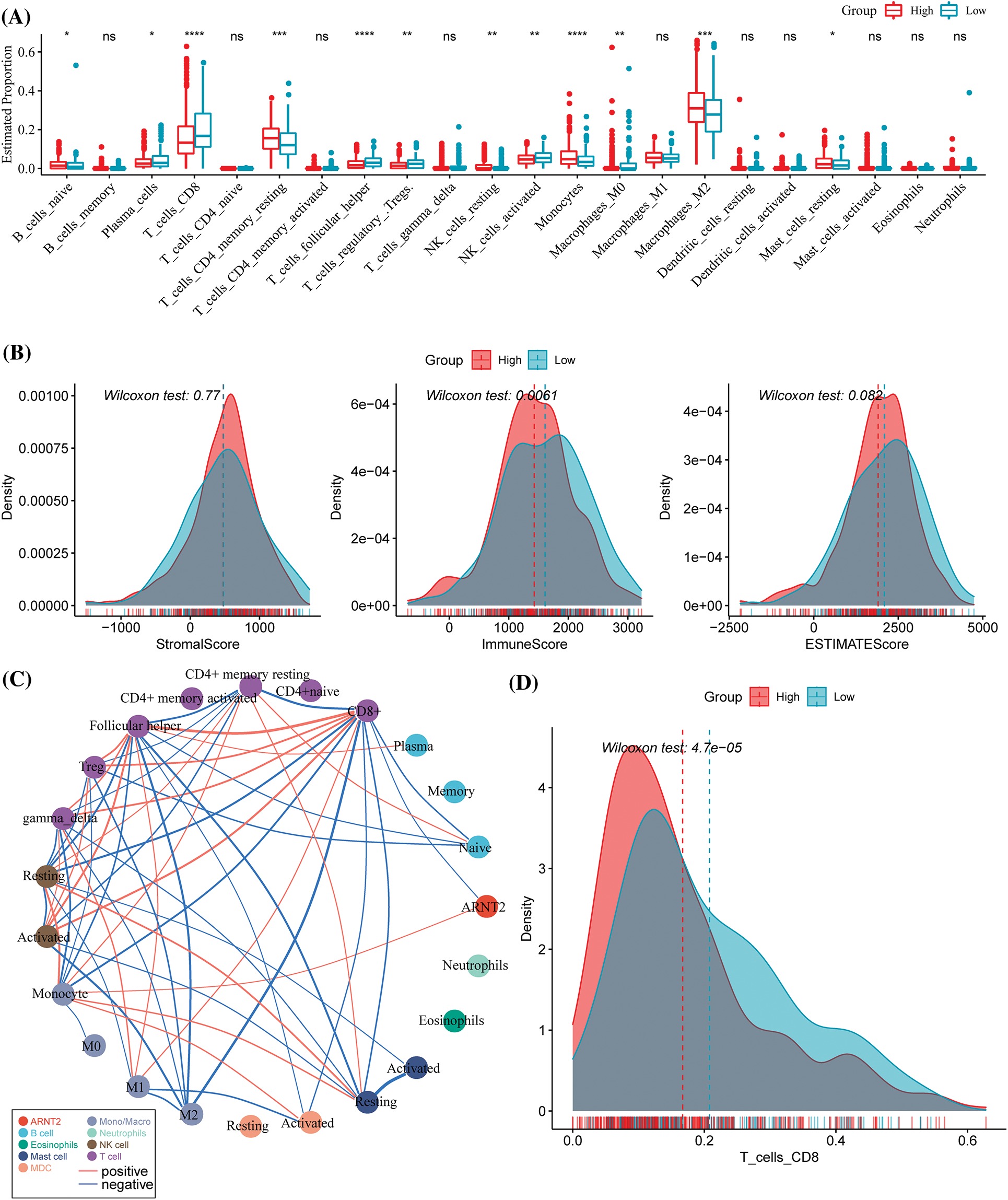
Figure 5: The relation between aryl hydrocarbon receptor nuclear translocator (ARNT2) expression and immune infiltration in kidney renal clear cell carcinoma (KIRC). (A) The estimated proportion of immune cells analyzed by CIBERSORT. (B) ESTIMATE analysis for evaluating stromal and immune infiltration. (C) The network among immune cells and ARNT2. Thicker lines indicate higher absolute correlation coefficients. (D) The distribution of CD8 T cells in the two groups. Wilcoxon test was conducted between two groups. ns, not significant. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
The potential value of aryl hydrocarbon receptor nuclear translocator 2 to predict the response to clinical therapies in kidney renal clear cell carcinoma
Based on the differential immune infiltration between high- and low-ARNT2 expression groups in KIRC, we suspected a difference in response to immunotherapy between the two groups. We calculated the enrichment score of a series of signatures that have been reported to be associated with the response to immunotherapy. The low-ARNT2 expression group had a higher score of cytolytic activity, T-cell-inflamed GEP, and Th1/IFN-γ than the high-expression group (Figs. 6A–6C). Of four important immune checkpoints, LAG3, PDCD1, and CD274 manifested the differential expression between two groups (p < 0.05, Fig. 6D). Among 17 immunotherapy-predicted pathways, 16 pathways were differentially enriched between high- and low-ARNT2 expression groups (p < 0.05, Fig. 6E). Notably, the low-expression group had significantly higher enrichment scores of all 16 pathways than the high-expression group, indicating that low-expression group was more positive to immunotherapy.
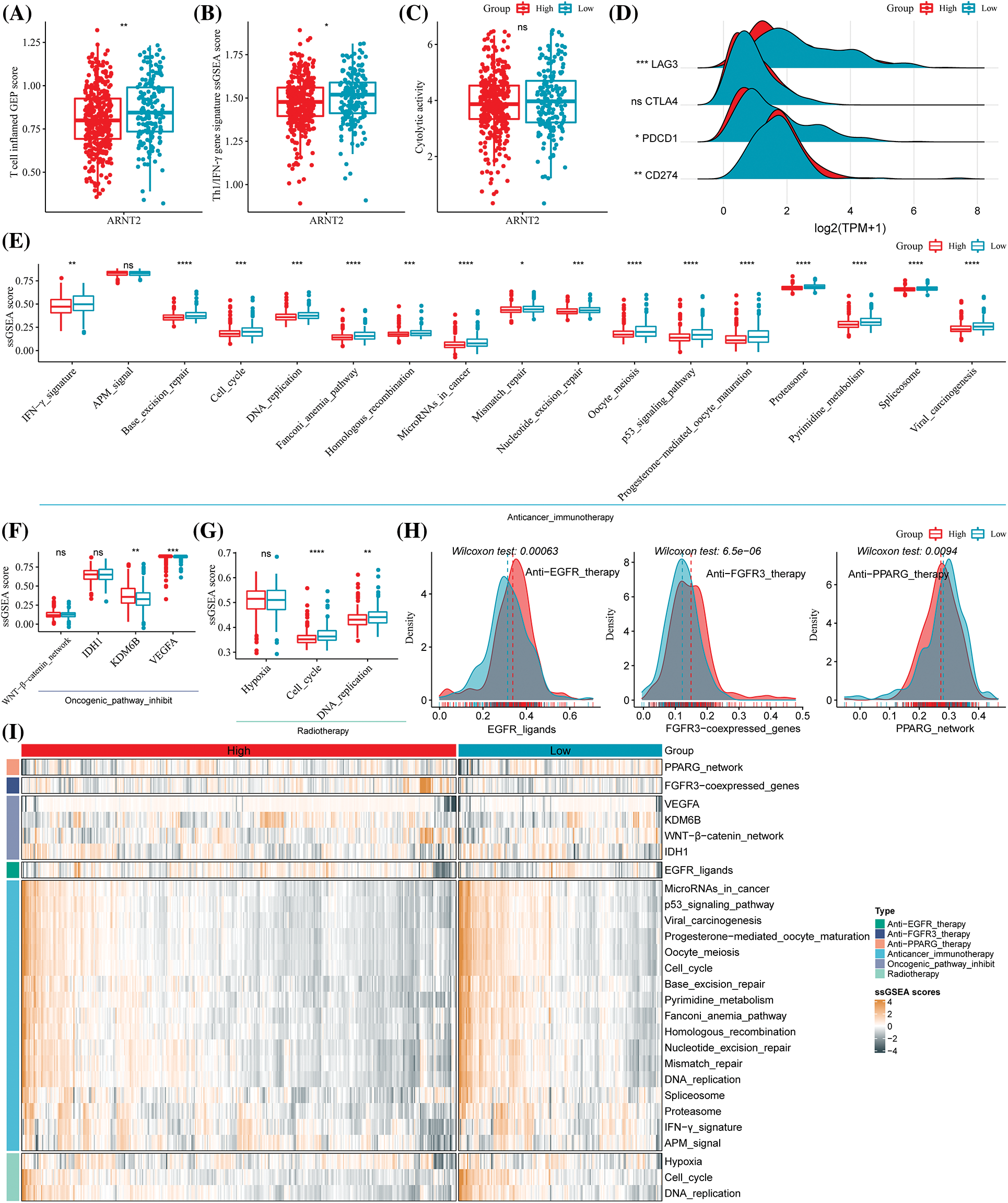
Figure 6: The potential of aryl hydrocarbon receptor nuclear translocator (ARNT2) expression for predicting the response to clinical therapy in kidney renal clear cell carcinoma (KIRC). (A–C) The single sample gene set enrichment analysis (ssGSEA) score of T cell inflamed gene expression profiles (GEP) (A), Th1/IFN-γ (B), and cytolytic activity (C) in high- and low-ARNT2 expression groups. (D) The expression of four immune checkpoints lymphocyte activation gene-3 (LAG3), cytotoxic T-lymphocyte–associated antigen 4 (CTLA4), programmed cell death protein 1 (PDCD1), and CD274 in two groups. (E) The enrichment score of immunotherapy-predicted pathways in two groups. (F) The enrichment score of Wnt-β-catenin, Isocitrate dehydrogenase 1 (IDH1), Lysine demethylase 6B (KDM6B), and vascular endothelial growth factor A (VEGFA) in two groups. (G) The enrichment score of three pathways, including hypoxia, cell cycle, and DNA replication, was related to radiotherapy. (H) The enrichment score of epidermal growth factor receptor (EGFR) ligands, fibroblast growth factor receptor 3 (FGFR3), and peroxisome proliferator-activated receptor gamma (PPARG) in two groups. X-axis indicates the enrichment score, and the y-axis indicates the density of the enrichment score. (I) A heatmap of enrichment score of different indicators predicting the response to different therapies, including anti-EGFR therapy, anti-FGFR3 therapy, anti-PPARG therapy, immunotherapy, oncogenic pathway inhibitors, and radiotherapy. Wilcoxon test was performed between two groups. ns, not significant. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Furthermore, we also evaluated the responses of two groups to targeted therapy and radiotherapy. High- and low-expression groups showed a different enrichment score in lysine-specific demethylase 6B (KDM6B), vascular endothelial growth factor A (VEGFA), cell cycle, DNA replication, EGFR ligands, FGFR3, and PPARG where these genes or pathways are reported to be associated with the efficiency of targeted therapy or radiotherapy (Figs. 6F–6I). Moreover, high- and low-ARNT2 expression also had differential overall survival in KIRC (p = 0.0011, Suppl. Fig. S6). Therefore, we suggested that ARNT2 expression could act as an indicator in the response prediction of immunotherapy, radiotherapy, and targeted therapy.
The role of ARNT2 is implicated in cell proliferation and prognosis of different cancer types, including lung cancer (Yang et al., 2015), oral squamous cell carcinoma (Kimura et al., 2016), gastric cancer (Jia et al., 2019), breast cancer (Martinez et al., 2008), hepatocellular carcinoma (Li et al., 2015), and GBM (Bogeas et al., 2018). In this study, we observed that ARNT2 expression was differential between tumor samples and normal samples in most cancer types. In addition, ARNT2 expression was an independent factor for the prognosis in PAAD, LIHC, HNSC, PCPG, and KIRC, which suggested that ARNT2 possibly played a critical role in cancerogenesis and cancer progression. The results encouraged further analysis to unveil the potential mechanism of ARNT2 in cancer.
TME is comprised of cellular and non-cellular components or extracellular matrix, where the signaling from cancer cells can modulate the components of TME (Wang et al., 2017). Chemokines and their receptors play critical roles in the cellular interaction of TME, which are responsible for immune cell migration, maturation, and differentiation (Bule et al., 2021). The chemokine network is also a potential target for immunotherapy in cancer. Pan-cancer data analysis showed a striking correlation between ARNT2 expression with chemokines and chemokine receptors. For example, a significantly positive correlation was carried out in LIHC, COAD, and READ, and a negative correlation was shown in LGG, GBM, and KIRC. Different cancer types manifested opposite correlations, indicating the different mechanisms of ARNT2 underlying the TME regulation in different cancer types.
In ccRCC samples, ARNT2 expression was negatively related to the enrichment of regulatory T cells, follicular helper T cells, and CD8 T cells, but positively associated with resting memory CD4 T cells, monocytes, and M0 and M2 macrophages. Consistently, the high-ARNT2 expression group also showed a lower enrichment of follicular helper T cells, regulatory T cells, and CD8 T cells, but a higher enrichment of M0, M2 macrophages, monocytes, and resting memory CD4 T cells than in the low-expression group. Overall, the high-ARNT2 expression group had significantly low immune infiltration. Among the above immune cells, the majority of immune cells comprised CD8 T cells and M2 macrophages. Cytolytic CD8 T cells have an anti-cancer immune response and are the backbone of anti-cancer immunotherapy (Raskov et al., 2021). CD8 T cell infiltration has been found to be involved in a favorable cancer prognosis (Ali et al., 2014; Huang et al., 2012). However, with the existence of high PD-1 expression, the overall survival was impaired due to an adaptive immune resistance mechanism (Thompson et al., 2017). In our study, high-ARNT2 expression group had longer overall survival than the low-expression group. Although the low-expression group had high CD8 T cell infiltration, relatively high expression of PD-L1 may result in its worse prognosis. The relation between the immune microenvironment and overall survival was linked to the expression of ARNT2, supporting the premise that ARNT2 is a potentially important modulator in TME.
Functional analysis showed that tumor-related pathways such as TGF-β signaling, Wnt signaling, Hedgehog signaling, and oxidative phosphorylation were more activated in high-ARNT2 expression group than in low-ARNT2 expression group, indicating the possible involvement of ARNT2 in the regulation of these pathways. TGF-β plays a dual role in promoting or suppressing tumor progression. In ccRCC, inhibition of TGF-β can suppress the invasive feature of ccRCC cells (Boström et al., 2013). Wnt signaling also participates in the biological processes in renal cancer, and the inhibition of its signaling transduction can reduce the viability and growth of tumor cells (Xu et al., 2016). Oxidative phosphorylation (OXPHOS), a therapeutic target of cancer cell metabolism, was downregulated or upregulated in cancer (Ashton et al., 2018). Oxidative stress is induced by the accumulation of ROS, and a high concentration of ROS is needed for the proliferation of cancer cells (Jelic et al., 2021). This study observed a positive correlation between ARNT2 expression and oxidative stress, suggesting a potential role of ARNT2 in the process of oxidative stress. However, the specific crosstalk between ARNT2 and oxidative stress needs further experimental clarification.
To assess the value of ARNT2 in the prediction of the therapeutic response in clinical therapy for ccRCC, we collected a series of therapeutic indicators and compared their enrichment in high- and low-ARNT2 expression groups. Evidence presented that higher T-cell-inflamed GEP score and PD-L1 expression were associated with higher response rates to KEYNOTE-028 (a PD-L1 inhibitor) in 20 cancers (Ott et al., 2019). In our results, the low-ARNT2 expression group had significantly higher T-cell-inflamed GEP score and PD-L1 expression than the high-expression group, indicating that the low-expression group could benefit much from taking immune checkpoint blockade therapy. In addition to the differential immune infiltration between high- and low-ARNT2 expression groups in KIRC, the analysis of other 17 immunotherapy-predicted pathways also supports the notion that the low-expression group was more responsive to immunotherapy. Apart from immunotherapy, ARNT2 expression levels were effective in predicting the response to radiotherapy and other targeted therapies such as anti-EGFR therapy, anti-FGFR3 therapy, and anti-PPARG therapy.
This study also had certain limitations. First, the results of this study were mainly based on the analysis of publicly available databases, and more in-depth basic experimental verification should be carried out, such as changes in ARNT2 expression on CD8T cell differentiation and proliferation. Second, the results of targeted therapy are too broad; hence, an in-depth analysis should be carried out to determine specific therapeutic targets that are directly related to ARNT2.
In conclusion, through pan-cancer analysis, our study demonstrated the important role of ARNT2 in cancer progression. We detected a relation between ARNT2 expression and the immune microenvironment, especially in ccRCC. Importantly, we suggest the great potential of ARNT2 in predicting the clinical response to immunotherapy and other targeted therapies for ccRCC.
Acknowledgement: None.
Funding Statement: This study was funded by the Shenzhen Longhua District Medical and Health Institutions Research Fund (Project No. 2022102).
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Renlong Zhou, Shuang Li; data collection: Xilin Xiao; analysis and interpretation of results: Renlong Zhou, Shuang Li, Xilin Xiao; draft manuscript preparation: Shuang Li, Xilin Xiao. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: All data generated or analyzed during this study are included in this published article.
Ethics Approval: Written informed consent was also obtained from all patients, and the study was approved by the Ethics Committee of Shenzhen Longhua District Central Hospital (No. 2022L03556) in March 2022.
Conflicts of Interest: Author Shuang Li is/was employed by Hangzhou Mugu Technology Co. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Materials: The supplementary materials are available online at https://doi.org/10.32604/biocell.2023.030281.
References
Aitola MH, Pelto-Huikko MT (2003). Expression of Arnt and Arnt2 mRNA in developing murine tissues. Journal of Histochemistry & Cytochemistry 51: 41–54. [Google Scholar]
Ali HR, Provenzano E, Dawson SJ, Blows FM, Liu B et al. (2014). Association between CD8+ T-cell infiltration and breast cancer survival in 12,439 patients. Annals of Oncology 25: 1536–1543. [Google Scholar] [PubMed]
Ashton TM, McKenna WG, Kunz-Schughart LA, Higgins GS (2018). Oxidative phosphorylation as an emerging target in cancer therapy. Clinical Cancer Research 24: 2482–2490. [Google Scholar] [PubMed]
Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A et al. (2017). IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. Journal of Clinical Investigation 127: 2930–2940. [Google Scholar] [PubMed]
Bogeas A, Morvan-Dubois G, El-Habr EA, Lejeune FX, Defrance M et al. (2018). Changes in chromatin state reveal ARNT2 at a node of a tumorigenic transcription factor signature driving glioblastoma cell aggressiveness. Acta Neuropathologica 135: 267–283. [Google Scholar] [PubMed]
Boström AK, Lindgren D, Johansson ME, Axelson H (2013). Effects of TGF-β signaling in clear cell renal cell carcinoma cells. Biochemical and Biophysical Research Communications 435: 126–133. [Google Scholar]
Bule P, Aguiar SI, Aires-Da-Silva F, Dias JNR (2021). Chemokine-directed tumor microenvironment modulation in cancer immunotherapy. International Journal of Molecular Sciences 22: 9804. [Google Scholar] [PubMed]
Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA (2018). Profiling tumor infiltrating immune cells with CIBERSORT. Methods in Molecular Biology 1711: 243–259. [Google Scholar] [PubMed]
Danilova L, Ho WJ, Zhu Q, Vithayathil T, de Jesus-Acosta A et al. (2019). Programmed cell death ligand-1 (PD-L1) and CD8 expression profiling identify an immunologic subtype of pancreatic ductal adenocarcinomas with favorable survival. Cancer Immunology Research 7: 886–895. [Google Scholar] [PubMed]
Freeburg PB, Abrahamson DR (2004). Divergent expression patterns for hypoxia-inducible factor-1β and aryl hydrocarbon receptor nuclear transporter-2 in developing kidney. Journal of the American Society of Nephrology 15: 2569–2578. [Google Scholar] [PubMed]
Funatake CJ, Marshall NB, Kerkvliet NI (2008). 2,3,7,8-Tetrachlorodibenzo-p-dioxin alters the differentiation of alloreactive CD8+ T cells toward a regulatory T cell phenotype by a mechanism that is dependent on aryl hydrocarbon receptor in CD4+ T cells. Journal of Immunotoxicology 5: 81–91. [Google Scholar] [PubMed]
Goldman MJ, Craft B, Hastie M, Repečka K, McDade F et al. (2020). Visualizing and interpreting cancer genomics data via the Xena platform. Nature Biotechnology 38: 675–678. [Google Scholar] [PubMed]
Goudot C, Coillard A, Villani AC, Gueguen P, Cros A et al. (2017). Aryl hydrocarbon receptor controls monocyte differentiation into dendritic cells versus macrophages. Immunity 47: 582–596.e6. [Google Scholar] [PubMed]
Hayes JD, Dinkova-Kostova AT, Tew KD (2020). Oxidative stress in cancer. Cancer Cell 38: 167–197. [Google Scholar] [PubMed]
Hirose K, Morita M, Ema M, Mimura J, Hamada H, Fujii H, Saijo Y, Gotoh O, Sogawa K, Fujii-Kuriyama Y (1996). cDNA cloning and tissue-specific expression of a novel basic helix-loop-helix/PAS factor (Arnt2) with close sequence similarity to the aryl hydrocarbon receptor nuclear translocator (Arnt). Molecular and Cellular Biology 16: 1706–1713. [Google Scholar] [PubMed]
Hosoya T, Oda Y, Takahashi S, Morita M, Kawauchi S, Ema M, Yamamoto M, Fujii-Kuriyama Y (2001). Defective development of secretory neurones in the hypothalamus of Arnt2-knockout mice. Genes to Cells 6: 361–374. [Google Scholar] [PubMed]
Hu J, Yu A, Othmane B, Qiu D, Li H et al. (2021). Siglec15 shapes a non-inflamed tumor microenvironment and predicts the molecular subtype in bladder cancer. Theranostics 11: 3089–3108. [Google Scholar] [PubMed]
Huang Y, Wang FM, Wang T, Wang YJ, Zhu ZY, Gao YT, Du Z (2012). Tumor-infiltrating FoxP3+ Tregs and CD8+ T cells affect the prognosis of hepatocellular carcinoma patients. Digestion 86: 329–337. [Google Scholar] [PubMed]
Hänzelmann S, Castelo R, Guinney J (2013). GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 14: 7. [Google Scholar]
Jain S, Maltepe E, Lu MM, Simon C, Bradfield CA (1998). Expression of ARNT, ARNT2, HIF1α, HIF2α and Ah receptor mRNAs in the developing mouse. Mechanisms of Development 73: 117–123. [Google Scholar] [PubMed]
Jelic MD, Mandic AD, Maricic SM, Srdjenovic BU (2021). Oxidative stress and its role in cancer. Journal of Cancer Research and Therapeutics 17: 22–28. [Google Scholar] [PubMed]
Jia Y, Hao S, Jin G, Li H, Ma X, Zheng Y, Xiao D, Wang Y (2019). Overexpression of ARNT2 is associated with decreased cell proliferation and better prognosis in gastric cancer. Molecular and Cellular Biochemistry 450: 97–103. [Google Scholar] [PubMed]
Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K (2017). KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Research 45: D353–D361. [Google Scholar] [PubMed]
Keith B, Adelman DM, Simon MC (2001). Targeted mutation of the murine arylhydrocarbon receptor nuclear translocator 2 (Arnt2) gene reveals partial redundancy with Arnt. Proceedings of the National Academy of Sciences of the United States of America 98: 6692–6697. [Google Scholar] [PubMed]
Kimura Y, Kasamatsu A, Nakashima D, Yamatoji M, Minakawa Y et al. (2016). ARNT2 regulates tumoral growth in oral squamous cell carcinoma. Journal of Cancer 7: 702–710. [Google Scholar] [PubMed]
Kobayashi M, Numakura K, Hatakeyama S, Muto Y, Sekine Y et al. (2022). Severe immune-related adverse events in patients treated with nivolumab for metastatic renal cell carcinoma are associated with PDCD1 polymorphism. Genes 13: 1204. https://doi.org/10.3390/genes13071204 [Google Scholar] [PubMed] [CrossRef]
Li W, Liang Y, Yang B, Sun H, Wu W (2015). Downregulation of ARNT2 promotes tumor growth and predicts poor prognosis in human hepatocellular carcinoma. Journal of Gastroenterology and Hepatology 30: 1085–1093. [Google Scholar] [PubMed]
Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P (2015). The molecular signatures database (MSigDB) hallmark gene set collection. Cell Systems 1: 417–425. [Google Scholar] [PubMed]
Martinez V, Kennedy S, Doolan P, Gammell P, Joyce H et al. (2008). Drug metabolism-related genes as potential biomarkers: Analysis of expression in normal and tumour breast tissue. Breast Cancer Research and Treatment 110: 521–530. [Google Scholar] [PubMed]
Mayakonda A, Lin DC, Assenov Y, Plass C, Koeffler HP (2018). Maftools: Efficient and comprehensive analysis of somatic variants in cancer. Genome Research 28: 1747–1756. [Google Scholar] [PubMed]
Mei J, Xing Y, Lv J, Gu D, Pan J, Zhang Y, Liu J (2020). Construction of an immune-related gene signature for prediction of prognosis in patients with cervical cancer. International Immunopharmacology 88: 106882. [Google Scholar] [PubMed]
Michaud JL, DeRossi C, May NR, Holdener BC, Fan CM (2000). ARNT2 acts as the dimerization partner of SIM1 for the development of the hypothalamus. Mechanisms of Development 90: 253–261. https://doi.org/10.1016/S0925-4773(99)00328-7 [Google Scholar] [PubMed] [CrossRef]
Murray IA, Patterson AD, Perdew GH (2014). Aryl hydrocarbon receptor ligands in cancer: Friend and foe. Nature Reviews Cancer 14: 801–814. [Google Scholar] [PubMed]
Ott PA, Bang YJ, Piha-Paul SA, Razak ARA, Bennouna J et al. (2019). T-cell-inflamed gene-expression profile, programmed death ligand 1 expression, and tumor mutational burden predict efficacy in patients treated with pembrolizumab across 20 cancers: KEYNOTE-028. Journal of Clinical Oncology 37: 318–327. [Google Scholar] [PubMed]
Raskov H, Orhan A, Christensen JP, Gögenur I (2021). Cytotoxic CD8+ T cells in cancer and cancer immunotherapy. British Journal of Cancer 124: 359–367. [Google Scholar] [PubMed]
Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N (2015). Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 160: 48–61. [Google Scholar] [PubMed]
Ru B, Wong CN, Tong Y, Zhong JY, Zhong SSW et al. (2019). TISIDB: An integrated repository portal for tumor-immune system interactions. Bioinformatics 35: 4200–4202. [Google Scholar] [PubMed]
Shen W, Song Z, Xiao Z, Huang M, Shen D et al. (2022). Sangerbox: A comprehensive, interaction-friendly clinical bioinformatics analysis platform. iMeta 1: e36 https://doi.org/10.1002/imt2.36 [Google Scholar] [CrossRef]
Thompson ED, Zahurak M, Murphy A, Cornish T, Cuka N et al. (2017). Patterns of PD-L1 expression and CD8 T cell infiltration in gastric adenocarcinomas and associated immune stroma. Gut 66: 794–801. [Google Scholar] [PubMed]
Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS et al. (2018). The immune landscape of cancer. Immunity 48: 812–830.e14. [Google Scholar] [PubMed]
Trikha P, Lee DA (2020). The role of AhR in transcriptional regulation of immune cell development and function. Biochimica et Biophysica Acta (BBA)—Reviews on Cancer 1873: 188335. [Google Scholar] [PubMed]
Wagage S, John B, Krock BL, Hall AO, Randall LM, Karp CL, Simon MC, Hunter CA (2014). The aryl hydrocarbon receptor promotes IL-10 production by NK cells. Journal of Immunology 192: 1661–1670. [Google Scholar]
Wang B, Song Q, Wei Y, Wu X, Han T, Bu H, Tang S, Qian J, Shao P (2022). Comprehensive investigation into cuproptosis in the characterization of clinical features, molecular characteristics, and immune situations of clear cell renal cell carcinoma. Frontiers in Immunology 13: 948042. [Google Scholar] [PubMed]
Wang M, Zhao J, Zhang L, Wei F, Lian Y et al. (2017). Role of tumor microenvironment in tumorigenesis. Journal of Cancer 8: 761–773. [Google Scholar] [PubMed]
Xu Q, Krause M, Samoylenko A, Vainio S (2016). Wnt Signaling in Renal Cell Carcinoma. Cancers 8: 57. https://doi.org/10.3390/cancers8060057 [Google Scholar] [PubMed] [CrossRef]
Yang B, Yang E, Liao H, Wang Z, Den Z, Ren H (2015). ARNT2 is downregulated and serves as a potential tumor suppressor gene in non-small cell lung cancer. Tumor Biology 36: 2111–2119. [Google Scholar] [PubMed]
Yoshihara K, Shahmoradgoli M, Martínez E, Vegesna R, Kim H et al. (2013). Inferring tumour purity and stromal and immune cell admixture from expression data. Nature Communications 4: 2612. [Google Scholar] [PubMed]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools