 Open Access
Open Access
ARTICLE
SMC1A served as a potential therapeutic target to regulate malignant phenotypes of cervical cancer
Department of Gynecology, Hebei General Hospital, Shijiazhuang, 050051, China
* Corresponding Author: CONGWEI DAI. Email:
BIOCELL 2023, 47(11), 2471-2484. https://doi.org/10.32604/biocell.2023.029617
Received 28 February 2023; Accepted 26 July 2023; Issue published 27 November 2023
Abstract
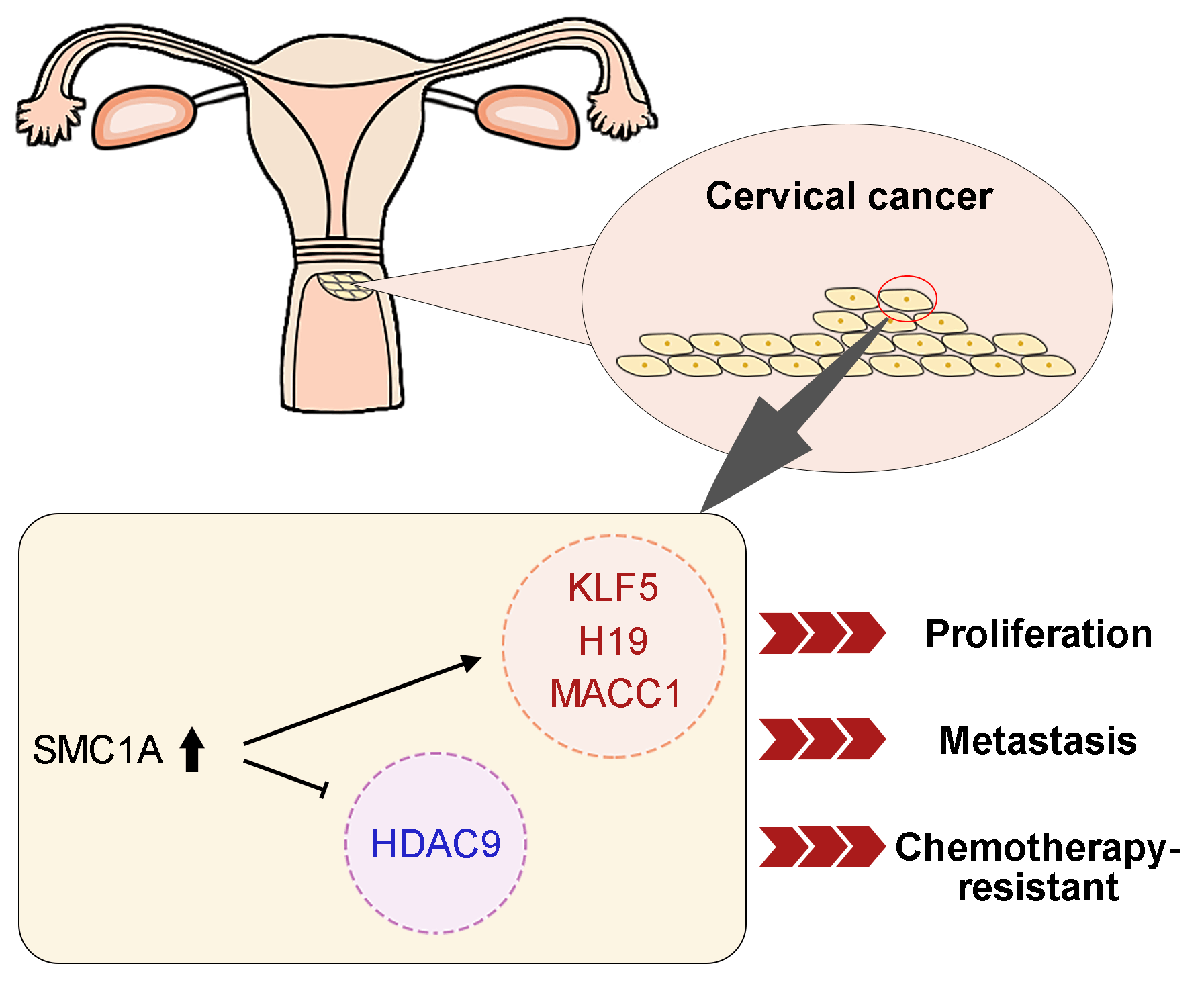
Introduction: Structural maintenance of chromosome 1A (SMC1A) is a crucial compound of the cohesin complex. It has been reported to regulate the epithelial-mesenchymal transition (EMT) process in multiple cancers. Objectives: The present study aims to further clarify the role of SMC1A in cervical cancer. Methods: We analyzed data from four datasets and confirmed that SMC1A showed high expression in cervical cancer samples and was related to poor prognosis of patients with cervical cancer. Cell proliferation of SiHa and C-33A with knockdown of SMC1A was assessed using CCK-8 and colony formation assay. The migration and invasion were estimated by wound healing assay and Transwell assay separately. The effect of SMC1A on the chemosensitivity of cisplatin and paclitaxel in cervical cancer cells was detected by flow cytometry assay. Results: Results of the immunohistochemistry (IHC) assay confirmed that the expression of SMC1A was increased in tumor tissues. The cell viability was remarkably suppressed in SiHa and C-33A by knocking down the expression of SMC1A. The increase of E-cadherin expression and decrease of N-cadherin and Snail expression verified that inhibition of SMC1A suppressed the EMT process of cervical cancer cells. Further, cell migration, and invasion were significantly repressed by the absence of SMC1A. Cisplatin and paclitaxel are effective chemotherapeutic agents used in the treatment of cervical cancer. Silencing of SMC1A remarkably promoted the apoptosis induced by cisplatin and paclitaxel, revealing that the chemotherapy resistance to cisplatin and paclitaxel in cervical cancer could reduce by knocking down SMC1A. Further, metastasis associated with colon cancer 1 (MACC1) was identified as the downstream factor of SMC1A. Its upregulation reversed the proliferation and the EMT process induced by SMC1A silencing. Conclusion: Therefore, our study concluded that SMC1A serves as a therapeutic molecular target to regulate the malignant phenotypes of cervical cancer.Graphic Abstract
Keywords
Cervical cancer is a malignant gynecological tumor with high incidence and has emerged as a severe public health problem (Arbyn et al., 2020). For instance, there is a report documenting that 13.3 out of every 100,000 women suffer from cervical cancer around the world (Singh et al., 2022). Approximately a third of the patients with cervical cancer are distributed in China and India, and cases in China in 2018 were more than those in India (Arbyn et al., 2020). Given these grim figures, it is necessary and urgent to explore a molecular target as an adjuvant therapy to improve the prognosis of cervical cancer.
Effective cancer treatments include surgery, chemotherapy, and radiotherapy (Mandal et al., 2022). Cisplatin, one of the most widely used chemotherapeutic drugs, has shown great efficacy in treating different types of cancer, including cervical cancer (Xie et al., 2021). Paclitaxel is a compound extracted from Taxus brevifolia (Chowdhury et al., 2019). It is a well-known anticancer drug that can be used with cisplatin as a treatment approach for various cancers (Della Corte et al., 2020; Dan et al., 2021). However, due to the frequent use of cisplatin and paclitaxel in cancer treatment, the chemotherapy resistance to paclitaxel has increased in cancer cells (Ashrafizadeh et al., 2021). This has seriously affected the prognosis of many cancers. Consequently, exploring new targeted therapies is necessary to overcome drug resistance.
Structural maintenance of chromosome 1A (SMC1A) is a crucial subunit in the sister chromatid cohesion complex (Gregson et al., 2002). SMC1A is a cohesion member, which has been demonstrated to be indispensable for human cell bipolar mitosis (Díaz-Martínez et al., 2010). In addition, SMC1A modulates the functional connection between the enhancer and promoter of its target gene, further affecting the transcription of the target gene (Dowen et al., 2013). Recently, it has been clarified that the proliferation of lung cancer cells is partially regulated by SMC1A, which arrests the process of the cell cycle (Zhang et al., 2013). There are reports documenting that the alteration of SMC1A expression is crucial in affecting the epithelial-mesenchymal transition (EMT) of breast cancer (Zhang et al., 2022). High expression of SMC1A was reported to facilitate the malignant phenotypes and tumorigenesis of colorectal cancer (Zhou et al., 2017; Sarogni et al., 2019), and was associated with radioresistance of prostate cancer (Yadav et al., 2019; Kumar and Clair, 2021). Further, previous studies suggest that SMC1A showed high expression in cervical cancer (Narayan et al., 2007). However, till date, no report reveals the biological function of SMC1A in cervical cancer. Hence, this study aims to investigate whether SMC1A regulates the malignant phenotypes of cervical cancer.
This study explored the effects of SMC1A on the cell proliferation, migration, invasion, and EMT process of cervical cancer by the depletion of SMC1A in two cervical cancer cell lines. Subsequently, metastasis associated in colon cancer 1 (MACC1) was identified to mediate the regulation of SMC1A in terms of the biological behaviors of cervical cancer cells. Further, the chemotherapy resistance to cisplatin and paclitaxel was also influenced by the downregulation of SMC1A. According to our results, SMC1A might serve as an effective molecule involved in regulating cervical cancer.
The datasets GSE9750, GSE63514, GSE39001, GSE7803 GSE113678, GSE113942, and GSE151666 were downloaded from the Gene Expression Omnibus (GEO) repository (https://www.ncbi.nlm.nih.gov/gds). We used the UALCAN online tool (http://ualcan.path.uab.edu/analysis.html) to analyze the correlation between the expression of SMC1A and different clinical characteristics of cervical cancer along with its expression in multiple cancers (Chandrashekar et al., 2017, 2022). The correlation between SMC1A expression level and disease-free interval (DFI) of patients with cervical cancer was estimated using Gene Set Cancer Analysis (GSCA) (http://bioinfo.life.hust.edu.cn/GSCA/#/). Differentially upregulated genes in the four datasets (GSE9750, GSE63514, GSE39001, and GSE7803) were screened according to the following thresholds: adjusted p < 0.01 and log-fold change (FC) ≥ 1. An online tool (https://www.xiantao.love/) was used to produce the Venn diagram. The potential downstream factors from GSE113678 were screened using p < 0.05 and |logFC| > 1 as filter criteria. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were performed to classify these genes.
We collected healthy cervical tissue (n = 3) and cervical cancer tissue (n = 7) for detecting the expression of SMC1A. These tissues were embedded in paraffin and cut into 5-μm-thick sections, followed by deparaffinizing with xylene, and then rehydrating with graded ethanol. The slides were placed in a heated antigen retrieval solution and heated for 10 min, and then were incubated with 3% H2O2 for 15 min at room temperature. After blocking by 1% bovine serum albumin (BSA) for 1 min, the sections were incubated with anti-SMC1A (Cat. no. 21695-1-AP, 1:50 dilution, Proteintech, Wuhan, China) at 4°C overnight and then treated with HRP labeled goat anti-rabbit IgG (Cat. no. SE134, 1:100 dilution, Solarbio, Beijing, China) for 45 min at room temperature. Subsequently, the slides were dyed by 3,3-diaminobenzidine tetrahydrochloride (Sangon Biotech, Shanghai, China) for 10 min and redyed using hematoxylin (Solarbio, Beijing, China) for 3 min. Finally, they were observed under a BX53 microscope (magnification, ×400, Olympus, Tokyo, Japan).
The human cervical cancer cell lines SiHa (Human Papiloma Virus or HPV+) and C-33A (HPV−) (iCell Bioscience Inc., Shanghai, China) were cultured with minimum essential medium (MEM) (Solarbio, Beijing, China) containing 10% fetal bovine serum (FBS) (Tianhang Biotechnology, Huzhou, China) in an incubator at 37°C. Cells (5 × 106 per well) were seeded in 6-well plates. Two molecules of short hairpin RNAs (shRNAs) targeting SMC1A (shSMC1As) were used to knockdown the expression of SMC1A, and their sequences are as follows: shSMC1A-a: 5′-AGTACAAGATCAACAACAA-3′; shSMC1A-b: 5′-GGAAGAAAGTAGAGACAGA-3′. They were constructed in the vector pRNAH1.1 (GenScript, Nanjing, China) and were transfected into cells using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA), SiHa cells were maintained in a complete medium supplemented with 500 μg/ml G418 (Solarbio, Beijing, China) per well after 24 h transfection to establish the stable cell lines with low expression of SMC1A. The C-33A cells were cultured in a complete medium containing 600 μg/ml G418 after 24 h transfection. Cells were collected 1–2 weeks later for follow-up experiments. Additionally, a commercial MACC1 overexpression plasmid (YouBio, Changsha, China) was transfected into the SMC1A-downregulated cell lines to investigate the role of MACC1 in cervical cancer.
Real-time quantitative PCR assay
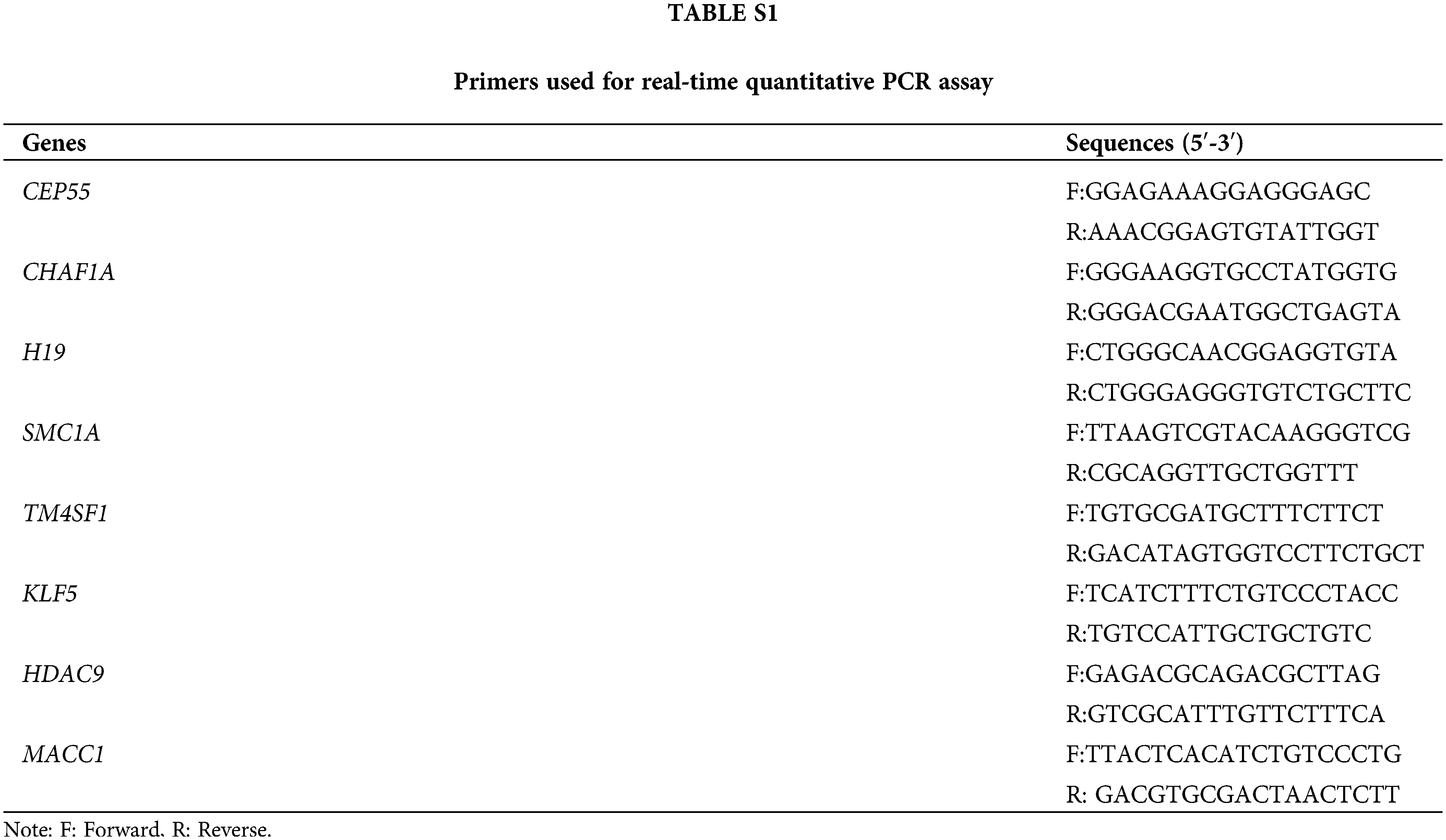
Total RNAs from SiHa and C-33A cells were extracted using TRIpure (BioTeke, Beijing, China). The complementary DNA (cDNA) was synthesized using a BeyoRT II M-MLV reverse transcriptase kit (Beyotime, Shanghai, China) according to the manufacturer’s protocol. The quantitative PCR was performed using 2 × Taq PCR MasterMix (Solarbio, Beijing, China) and SYBR Green (Solarbio, Beijing, China). The primers used in this assay have been listed in Suppl.Table S1.
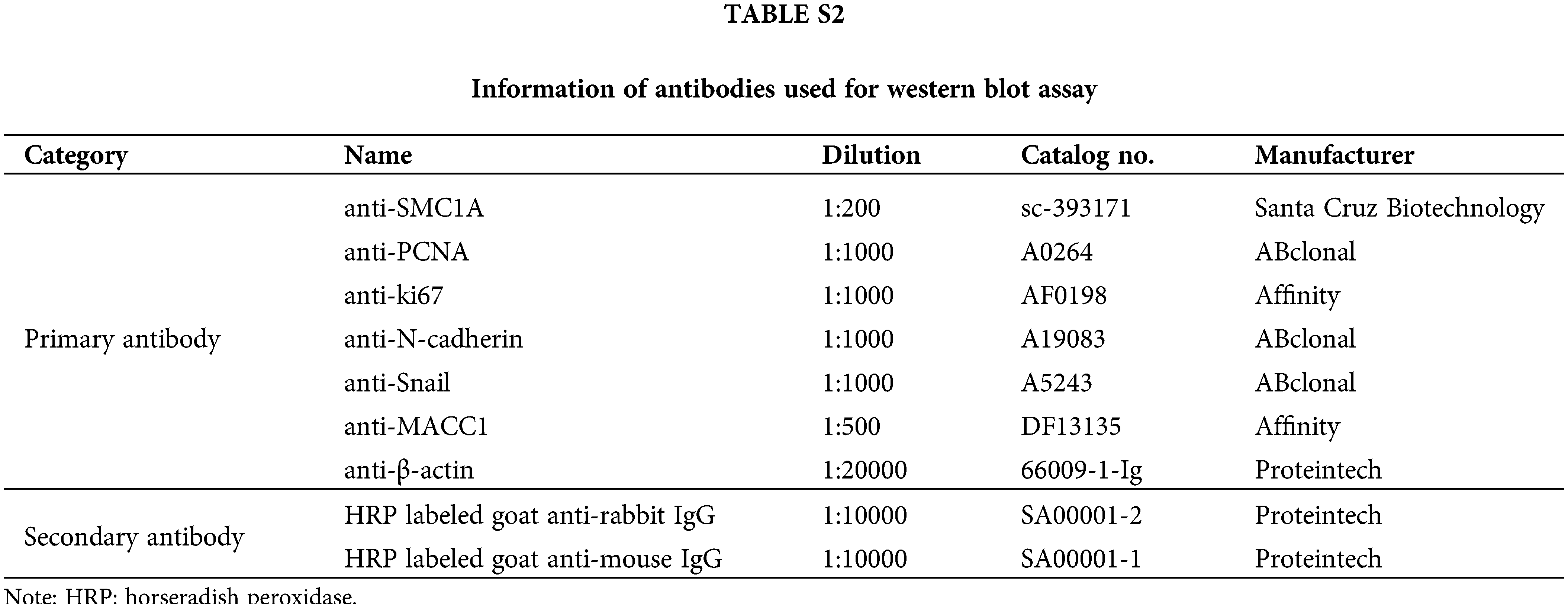
SiHa and C-33A cells were harvested and lysed with RIPA lysate (Beyotime, Shanghai, China) containing 1% phenylmethanesulfonylfluoride (Beyotime, Shanghai, China). Total protein was estimated using an enhanced BCA protein assay kit (Beyotime, Shanghai, China). The equal amounts of protein were separated by sodium dodecyl sulfate-polyacrylamide electrophoresis (SDS-PAGE), and then transferred onto a polyvinylidene difluoride (PVDF) membrane (Thermo Fisher Scientific, Pittsburgh, PA, USA). After blocking in 5% bovine serum albumin (BSA) (Biosharp, Hefei, China) buffer for 1 h, the membrane was probed with primary antibodies overnight at 4°C. Then, the membrane was incubated with secondary antibodies at 37°C for 40 min. The details of these antibodies have been listed in Suppl. Table S2. After reaction with enhanced chemiluminescence (7 Sea Biotech, Shanghai, China) for 5 min, bands on the membrane were observed using a Tanon Image densitometric scanner (Biotanon, Shanghai, China).
Cells (5 × 103 per well) were seeded into 96-well plates in different groups and cultured in an incubator at 37°C for 0, 24, 48, and 72 h, respectively. They were then cultured in 10 μl CCK-8 (KeyGen Biotech, Nanjing, China) reagent for 2 h. To investigate the effects of SMC1A on the chemotherapy sensitivity of cervical cancer cells, we cultured the cells with different concentrations of cisplatin or paclitaxel. Cells were treated with cisplatin (0, 1, 2, 4, 8, 16 μM) or paclitaxel (0, 10, 20, 40, 80 nM) for 48 h. These cells were then incubated with 10 μl CCK-8 reagent for 2 h. The optical density (OD) was evaluated using a microplate reader (BioTek, Winooski, VT, USA) at 450 nm. The formula for calculating the inhibition rate of cervical cancer cells is as follows: inhibition rate = (1 − OD of cells treated with different doses of cisplatin or paclitaxel/OD of untreated cells) × 100%.
When the cell density reached 90%, cells were collected, followed by reseeding 400 cells and their culturing for 14 days. Cell colonies were fixed with paraformaldehyde (Aladdin, Shanghai, China) for 1 min and stained using a Wright—Giemsa Stain Kit (Jiancheng Bioengineering Institute, Nanjing, China) for 5 min. The colony formation rate was calculated using the following formula: colony formation rate = (number of colonies/number of seeded cells) × 100%.
Cells were fixed with 4% paraformaldehyde (Sinopharm, Shanghai, China) for 5 min and incubated with 0.1% tritonX-100 (Beyotime, Shanghai, China) at room temperature for 30 min. After being blocked by 1% BSA (Sangon Biotech, Shanghai, China) for 15 min, cells were incubated with E-cadherin antibody (Cat. no. A20798, 1:100 dilution, ABclonal, Wuhan, China) at 4°C overnight. Subsequently, cells were incubated with fluorescein isothiocyanate (FITC) labeled goat anti-rabbit IgG (Cat. no. ab6717, 1:200 dilution, Abcam, Cambridge, UK) at room temperature for 60 min in the dark, and the nuclei of cells were dyed using 4′,6-diamidino-2-phenylindole (DAPI) (Aladdin, Shanghai, China). The results were observed and captured using a BX53 fluorescence microscope (magnification, ×400, Olympus, Tokyo, Japan).
After the cells were grown to confluence, the medium was replaced with a serum-free medium and treated with mitomycin C (Sigma-Aldrich, St. Louis, MO, USA) for 1 h before detection. Wounds were created using a 200 μl pipette tip and imaged to record the location of cells. Subsequently, cells were continued to be cultured in a serum-free medium for 24 h. Images of cells were captured at 0 h and 24 h using an IX53 inverted microscope (magnification, ×100).
Cells (5 × 104 per well) were seeded into Transwell upper chamber (Labselect, Beijing, China) containing Matrigel (Corning, NY, USA), and 800 μl medium with 10% FBS was supplemented into the lower chamber. The cells were cultured in an incubator with 5% CO2 at 37°C for 24 h. Next, these cells were fixed with 4% formaldehyde (Aladdin, Shanghai, China) for 20 min at 37°C and stained with 0.5% crystal violet (Amresco, Solon, OH, USA) for 5 min. Images of cells that invaded through the Matrigel membrane were captured using an IX53 inverted microscope (magnification, ×200), and the number of invaded cells was calculated.
SMC1A-downregulated cells and cells in shNC and parental groups were treated with 2 μM cisplatin or 80 nM paclitaxel for 48 h. After collecting and cleaning the cells, 5 μl AnnexinV-FITC was added into cell suspension, and then mixed with 5 μl Propidium Iodide (PI). The mixture was incubated at room temperature in the dark for 10 min according to the instructions of the Annexin V-FITC/PI Apoptosis Detection Kit (KeyGen Biotech, Nanjing, China). Cell apoptosis was estimated using the NovoCyteflow cytometer (Agilent, Santa Clara, CA, USA).
In this study, statistical results were presented as mean ± standard deviation (SD). Statistical differences between the two groups were estimated by the Student’s t-test, while differences among three or more groups were examined using analysis of variance (ANOVA). p < 0.05 was regarded as statistically significant.
The association between SMC1A expression and clinical characteristics in cervical cancer
We downloaded four datasets related to cervical cancer, including GSE9750, GSE63514, GSE39001, and GSE7803. The overlap of differentially upregulated genes in cervical cancer was obtained using Venn analysis (Fig. 1A). SMC1A is one of these overlapped genes, and its expression was upregulated in the cervical cancer samples (Fig. 1B). This finding was further demonstrated by the result from the analysis of the cancer genome atlas (TCGA) database (Fig. 1C). Subsequently, compared with patients with lower SMC1A expression, cervical cancer patients with high SMC1A expression had lower survival probability, suggesting that poor prognosis of cervical cancer might be partially related to the upregulation of SMC1A (Fig. 1D). Moreover, analysis results obtained from the UALCAN database showed high SMC1A expression in primary tumors (Fig. 1E). The expression of SMC1A in different characters of cervical cancer has been presented in Figs. 1F–1J. The SMC1A mRNA expression upregulated first and then decreased with the increase of tumor grade or patient’s age (Figs. 1G and 1J). As shown in Fig. 1I, SMC1A expression in patients with N0 and N1 metastasis of cervical cancer was higher than that in normal patients. No clear association was found between SMC1A expression and cancer stages as well as tumor histology of cervical cancer (Figs. 1F and 1H). Additionally, IHC results demonstrated that SMC1A was significantly upregulated in cervical cancer tissues (Fig. 1K). These results validated that SMC1A was highly expressed and possibly played a vital role in cervical cancer.
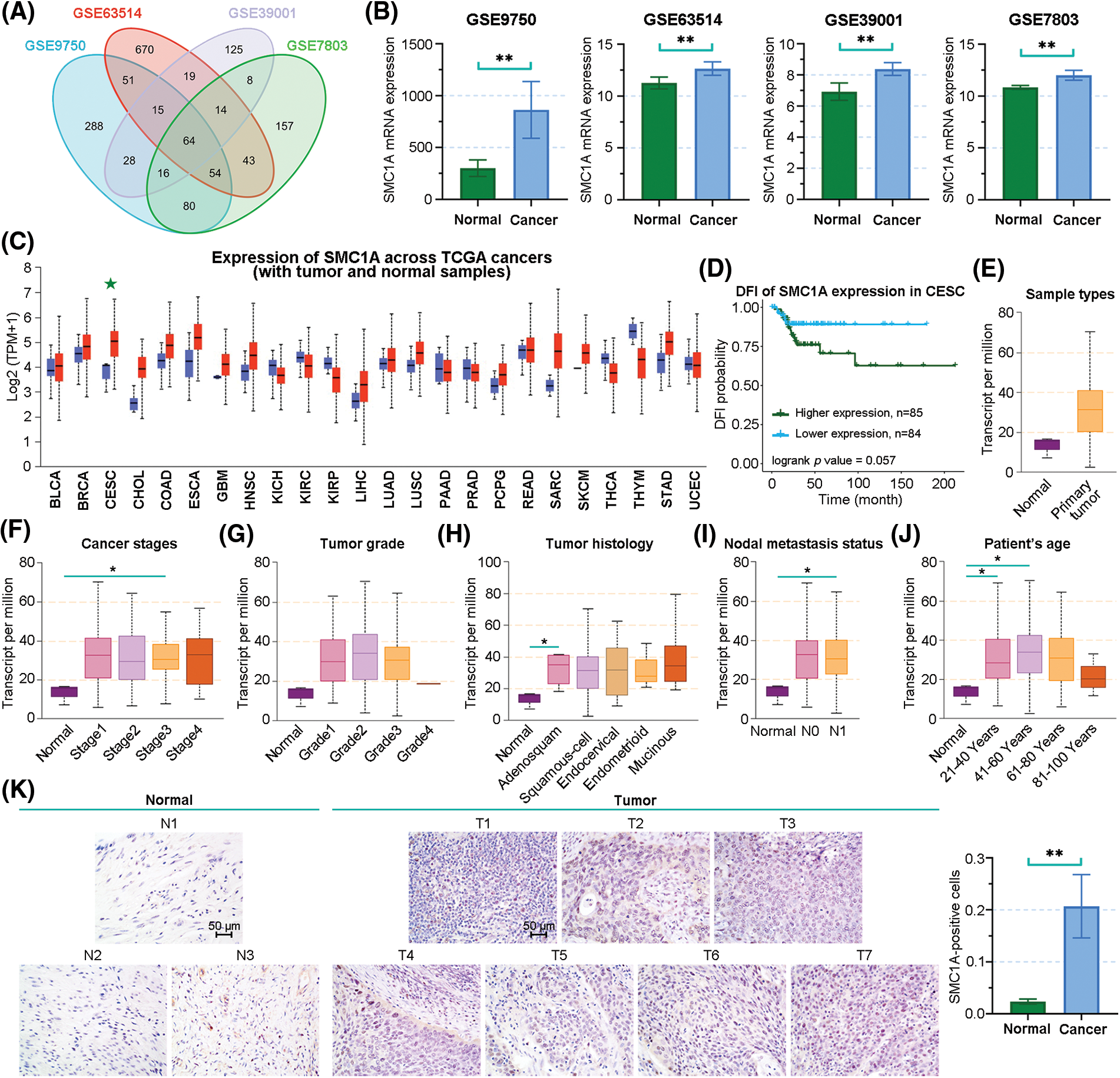
Figure 1: The association between clinical characters and structural maintenance of chromosome 1A (SMC1A) expression in cervical cancer. (A) Venn diagram of the differentially upregulated genes in cervical cancer samples from GSE9750, GSE63514, GSE39001, and GSE7803 datasets. (B) The mRNA expression of SMC1A in the four datasets. (C) SMC1A expression in different cancers from the The Cancer Genome Atlas (TCGA) database detected using UALCAN. (D) The relevance between SMC1A expression and the prognosis of cervical cancer was examined using Gene Set Cancer Analysis (GSCA). The expression of SMC1A in different sample types (E), cancer stages (F), tumor grade (G), tumor histology (H), nodal metastasis status (I), and patient’s age (J) in cervical cancer. (K) The expression of SMC1A was detected in cervical cancer tissue and normal cervical tissue by immunohistochemistry (IHC) assay (N, Normal tissue, n = 3; T, Tumor tissue, n = 7). Scale bars: 50 μm. *p < 0.05, **p < 0.01.
Downregulation of SMC1A inhibits cell proliferation in cervical cancer
Two molecules of shRNAs targeted SMC1A were transfected into SiHa and C-33A cells to knockdown SMC1A. The relative mRNA level of SMC1A was significantly decreased in the two cervical cancer cell lines (Fig. 2A). As shown in Fig. 2B, the protein expression of SMC1A was obviously downregulated by shSMC1A-a and shSMC1A-b in the cancer cells. Additionally, cell proliferation was remarkably suppressed in the SiHa and C-33A cells with depletion of SMC1A at 24, 48, and 72 h (Fig. 2C). Meanwhile, knockdown of SMC1A suppressed the protein expression of ki67 and proliferating cell nuclear antigen (PCNA) (Kayaselçuk et al., 2002) in cervical cancer cells (Fig. 2D). The subsequent colony formation assay verified that silencing of SMC1A significantly repressed the viability of the cancer cells (Fig. 2E). Therefore, SMC1A downregulation effectively suppressed the cell proliferation ability in cervical cancer.
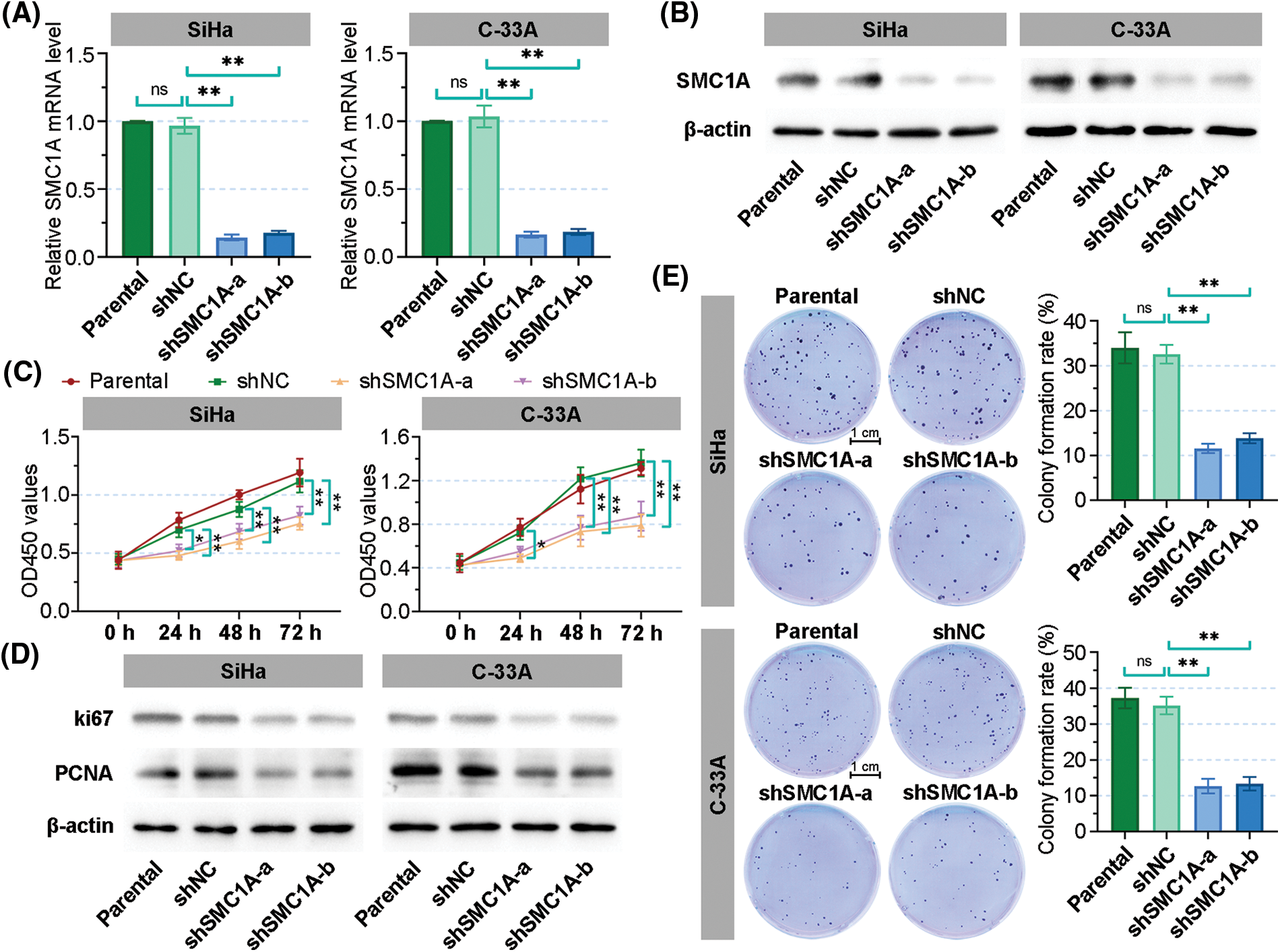
Figure 2: Downregulation of structural maintenance of chromosome 1A (SMC1A) inhibits cell proliferation in cervical cancer. (A) Real-time reverse transcription polymerase chain reaction (RT-qPCR) was used to detect the mRNA expression of SMC1A in SiHa and C-33A cells. (B) The protein expression of SMC1A was assessed by western blotting. (C) Cell viability of SiHa and C-33A cells was examined using the cell counting kit-8 (CCK-8) assay. (D) The protein expression of ki67 and proliferating cell nuclear antigen (PCNA) was evaluated by western blotting. (E) Colony formation assay determined the proliferation of cervical cancer cells transfected by shSMC1As. Scale bars: 1 cm. n = 3. ns, p > 0.05, *p < 0.05, **p < 0.01.
Knockdown of SMC1A suppresses epithelial-mesenchymal transition (EMT), migration, and invasion of cervical cancer cells
Previous reports have identified that EMT acts as an indispensable process to enhance the malignant phenotypes of tumor cells, including migration and invasion (Ribatti et al., 2020). To understand the contribution of SMC1A on EMT in cervical cancer, we then observed cellular characteristics of SiHa and C-33A cells with downregulation of SMC1A. As displayed in Fig. 3A, the parental SiHa cells and negative control cells exhibited an elongated cell morphology. After being transfected by shSMC1A-a and shSMC1A-b, SiHa cells became shorter and rounder. Similarly, the depletion of SMC1A blocked the extension of C-33A cells. This suggested that the EMT process in cervical cancer cells was possibly associated with the alteration of SMC1A expression. Moreover, the changes in the expression of several factors, such as N-cadherin, Snail, and E-cadherin, have been used to estimate the occurrence of EMT (Kim et al., 2006; Hsu et al., 2007). When the expression of SMC1A was knocked down, the protein expression of N-cadherin and Snail declined in the cancer cells (Fig. 3B). In addition, after SMC1A expression was blocked in the cervical cancer cells, a significant increase of E-cadherin expression was observed (Fig. 3C).
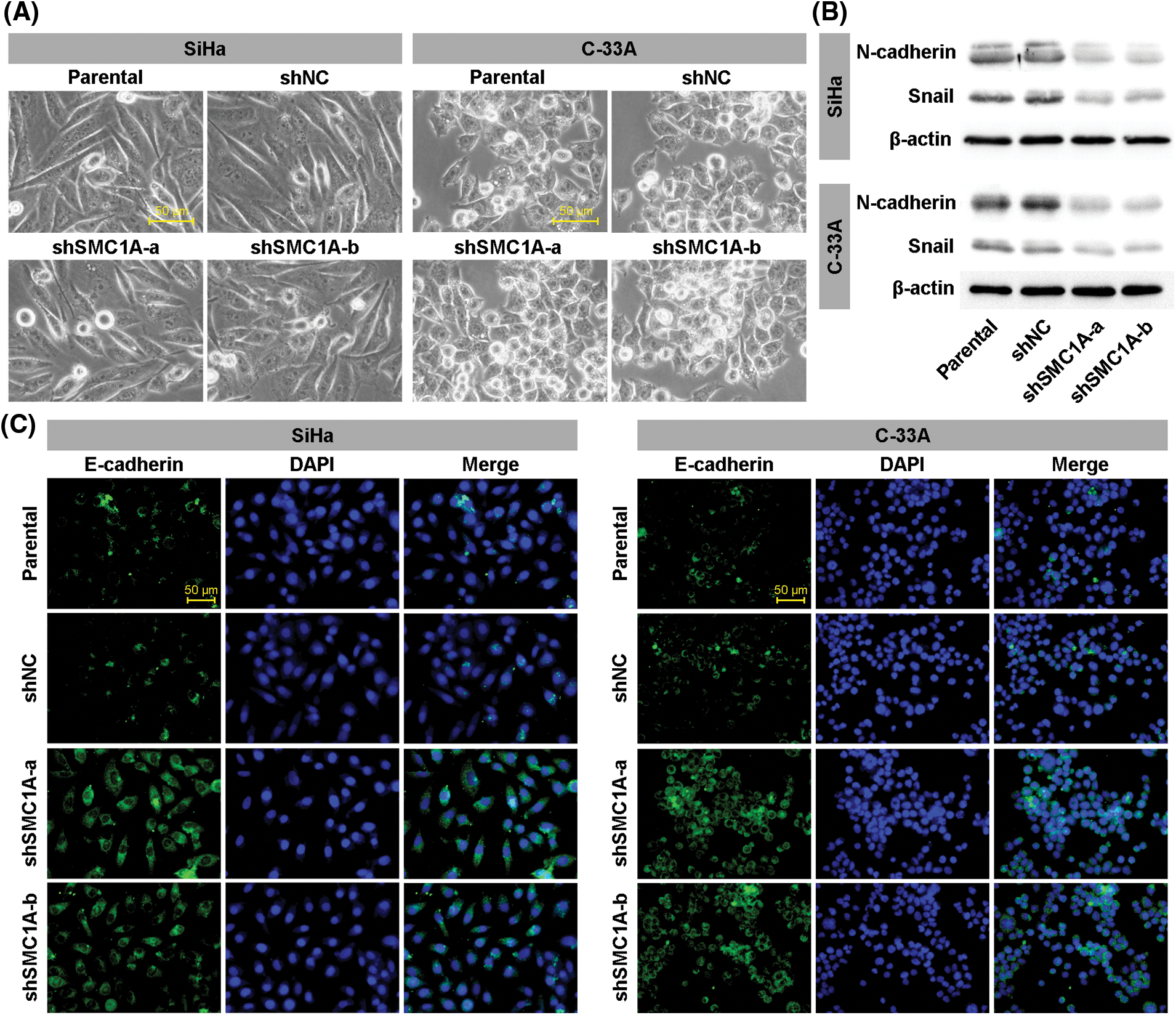
Figure 3: Knockdown of structural maintenance of chromosome 1A (SMC1A) suppresses epithelial-mesenchymal transition (EMT) in cervical cancer. (A) Cell morphology was observed under an inverted microscope. Scale bars: 50 μm. (B) Western blotting was used to examine the protein expression of N-cadherin and Snail. (C) Expression of E-cadherin was estimated using immunofluorescence (IF) staining in SiHa and C-33A cells. Scale bars: 50 μm. n = 3.
Moreover, the wound healing assay revealed that the knockdown of SMC1A obviously inhibited the migration of SiHa and C-33A cells. The migration rate of cervical cancer cells was reduced to less than 40% (Fig. 4A). Meanwhile, the number of invaded cells was significantly decreased in the two cell lines, suggesting that the absence of SMC1A also suppressed the invasion of cervical cancer cells (Fig. 4B). To summarize, these findings supported that the absence of SMC1A suppressed the EMT process of cervical cancer cells, and also inhibited cell migration and invasion in cervical cancer.
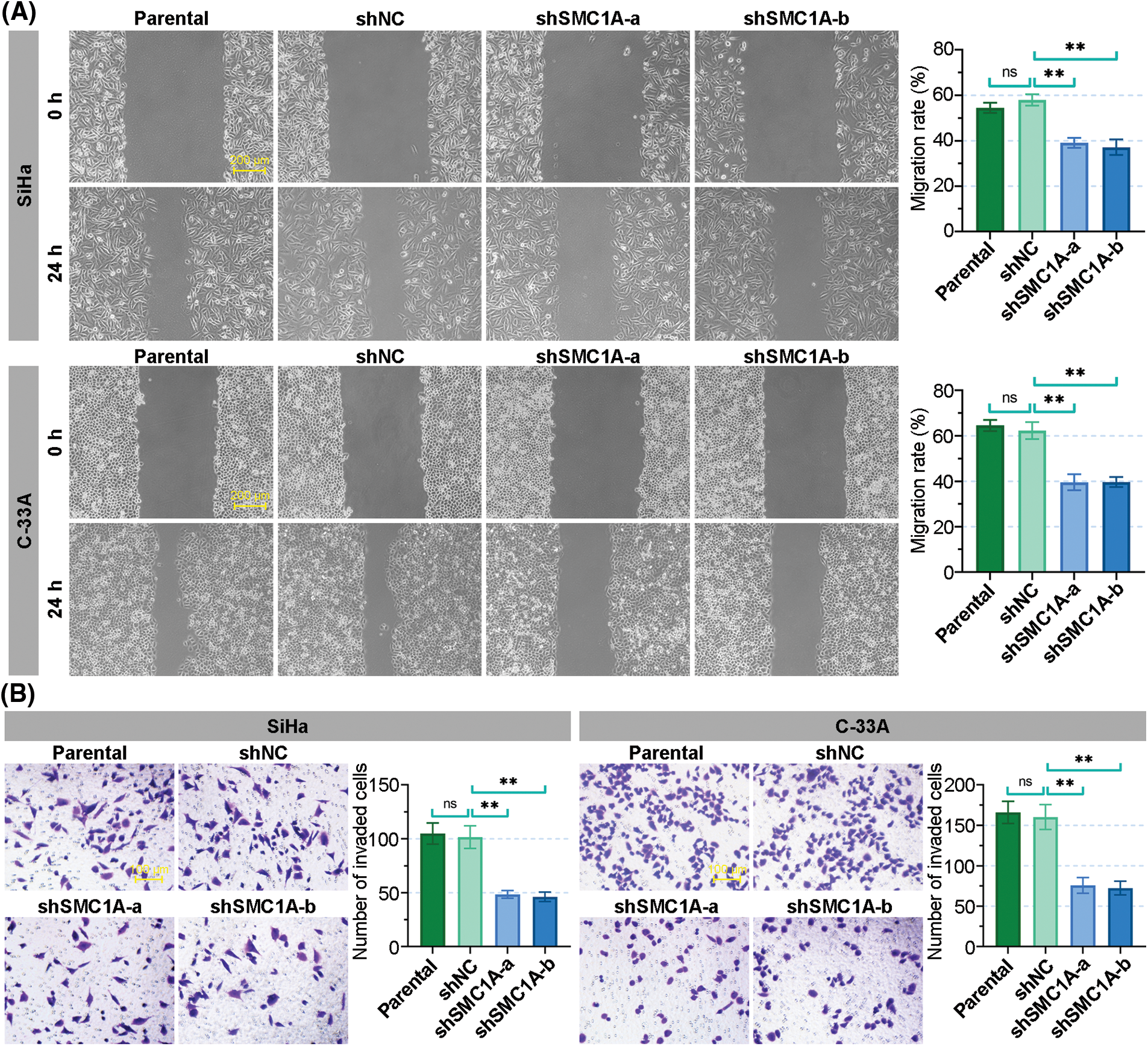
Figure 4: The knockdown of structural maintenance of chromosome 1A (SMC1A) suppresses the migration and invasion of cervical cancer cells. (A) Migration of SiHa and C-33A cells was determined by the wound healing assay. Scale bars: 200 μm. (B) Invasion of cervical cancer cells was examined using transwell assay. Scale bars: 100 μm. n = 3. ns, p > 0.05, *p < 0.05, **p < 0.01.
Silencing of SMC1A improves the chemosensitivity and enhances the apoptosis of cervical cancer cells
There is little literature on the regulation of SMC1A expression on cisplatin and paclitaxel chemotherapy sensitivity in cervical cancer cells. After treatment with different concentrations of cisplatin, the growth of SiHa cells was inhibited and the inhibition rate of the cells with silence of SMC1A was dose-dependently increased. The inhibition rate of C-33A cells was significantly increased by shSMC1A-a and shSMC1A-b compared with the negative control cells at 1, 2, 4, and 8 μM cisplatin treatment (Fig. 5A). When cells were cultured in the medium with different concentration of paclitaxel, their inhibition rate was significantly raised after transfected by the silencing of SMC1A. With the increase of paclitaxel concentration, knockdown of SMC1A showed an obvious inhibitory effect on cervical cancer cells (Fig. 5B). Collectively, reduction of SMC1A might be an effective method to improve the chemosensitivity of cervical cancer cells to cisplatin and paclitaxel.
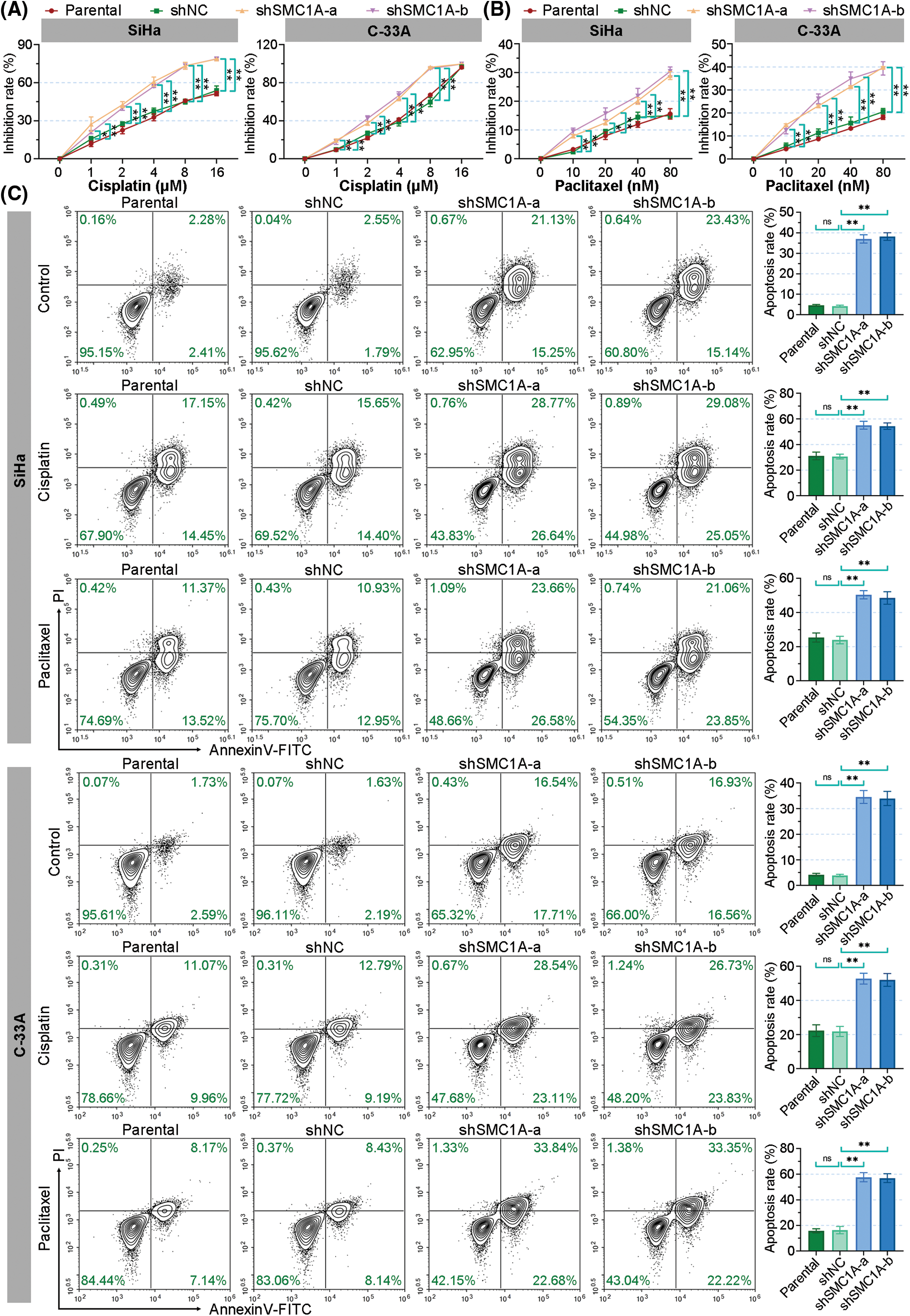
Figure 5: Silencing of structural maintenance of chromosome 1A (SMC1A) improves the chemosensitivity of cervical cancer cells. (A) The resistance of cervical cancer cells to cisplatin was measured using the cell counting kit-8 (CCK-8) assay. (B) The resistance to paclitaxel was assessed using CCK-8 assay. (C) Cell apoptosis induced by cisplatin and paclitaxel was evaluated by flow cytometry. n = 3. ns, p > 0.05, *p < 0.05, **p < 0.01.
Furthermore, the effect of SMC1A on cell apoptosis induced by cisplatin and paclitaxel was estimated by using flow cytometry. As depicted in Fig. 5C, the knockdown of SMC1A significantly increased the apoptosis rate of the SiHa and C-33A cells. When cells were cultured with cisplatin for 48 h, the apoptosis rate of SiHa C-33A cells was increased to over 20%. The absence of SMC1A further facilitated the apoptosis of cervical cancer cells. Meanwhile, paclitaxel also enhanced cell apoptosis in the two cell lines, and the apoptosis rate was significantly upregulated by silencing of SMC1A. Therefore, the downregulation of SMC1A further promoted the apoptosis of cervical cancer cells induced by cisplatin and paclitaxel, indicating that inhibiting SMC1A expression could serve as a molecular therapy approach to assist in the efficacy of cisplatin or paclitaxel in cervical cancer.
The downstream factor MACC1 is decreased by SMC1A downregulation
The dataset GSE113678 was examined for the potential molecules that may be regulated by SMC1A in tumor cells. The top 10 differentially upregulated gene-enriched GO terms with the smallest p-value in the categories of biological process (BP), cellular components (CC), and molecular function (MF) have been presented in Fig. 6A. The KEGG pathway enrichment analysis of the differentially upregulated genes suggested that they were enriched in serval human diseases and cell survival-related signaling pathways (Fig. 6B). Furthermore, the enrichment results in Figs. 6C and 6D revealed that the differentially downregulated genes were involved in the biological behavior of interaction with other molecules. The RNA expression levels of Kruppel-like factor 5 (KLF5), Centrosomal Protein 55 (CEP55), Chromatin Assembly Factor 1 Subunit A (CHAF1A), LINC00008 (H19), Transmembrane 4 L Six Family Member 1 (TM4SF1), Histone Deacetylase 9 (HDAC9) and MACC1 were evaluated using RT-qPCR. As shown in Fig. 6E, the mRNA levels of CEP55, CHAF1A, and TM4SF1 presented no alteration in cancer cells with the knockdown of SMC1A. However, a significant decrease in KLF5, H19, and MACC1 expression was documented in cervical cancer cells due to the knockdown of SMC1A. Interestingly, HDAC9 mRNA expression was markedly increased in cancer cells in the absence of SMC1A. Since MACC1 has been reported to promote malignant phenotypes of cervical cancer, such as migration and invasion (Zhou et al., 2015; Mei et al., 2022), we further explored whether MACC1 mediates the regulation of SMC1A in cervical cancer cells. In the SMC1A-suppressed cells, the protein expression of MACC1 was evidently declined compared with that in shNC-transfected cells (Fig. 6F). Overall, these findings confirmed that MACC1 as a downstream factor was affected by SMC1A.
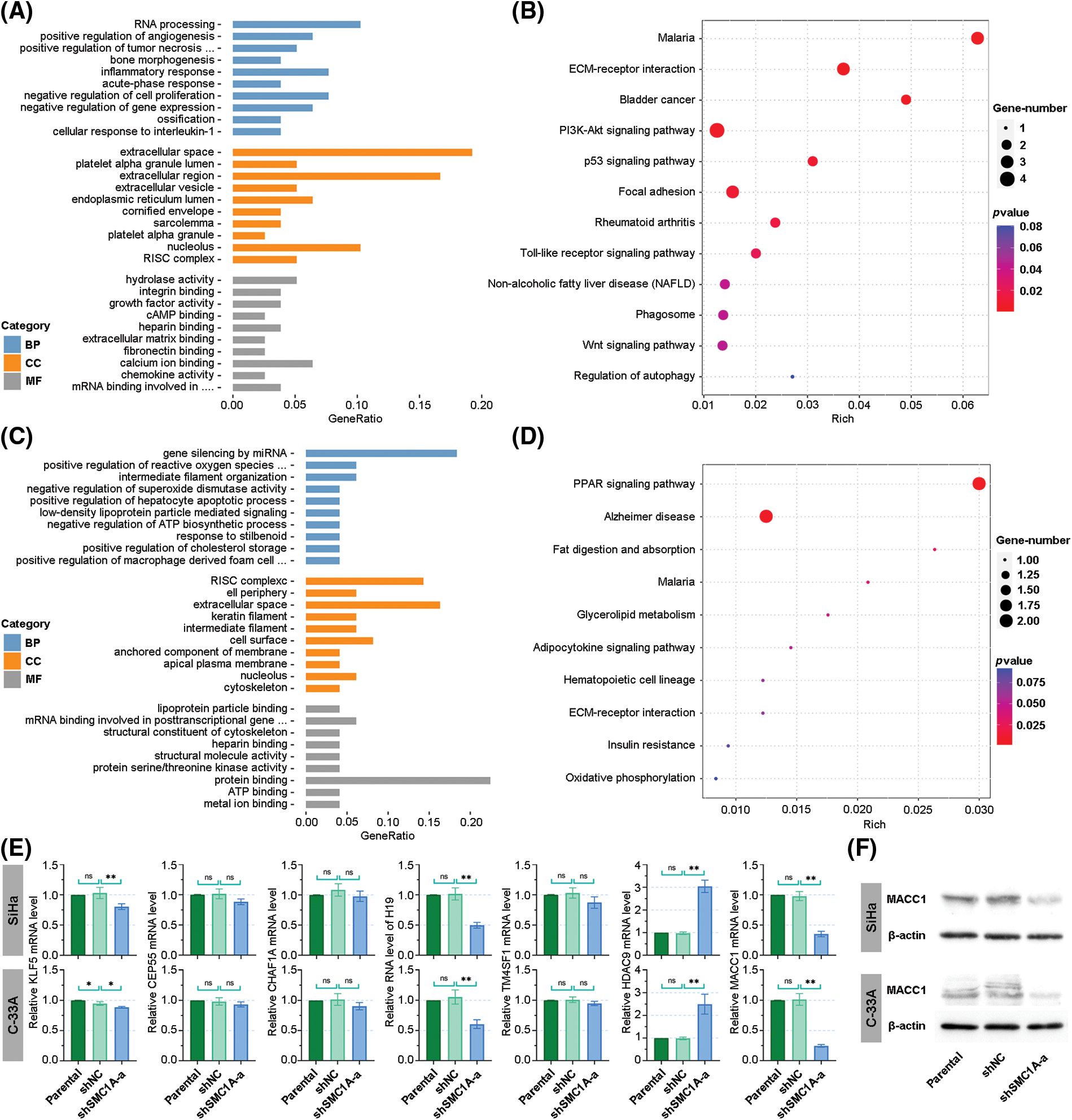
Figure 6: Investigation of downstream factors potentially regulated by structural maintenance of chromosome 1A (SMC1A). (A) Gene ontology (GO) enrichment analysis was performed on genes positively correlated with SMC1A expression. (B) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enrichment analysis of genes positively related to SMC1A expression. (C) GO enrichment analysis of genes negatively associated with SMC1A expression. (D) KEGG pathways enrichment analysis of negatively correlated genes. (E) The RNA levels of potential downstream factors were estimated using real-time reverse transcription polymerase chain reaction (RT-qPCR). (F) The protein expression of metastasis associated in colon cancer 1 (MACC1) was determined by western blotting. BP: Biological process, CC: Cellular components, MF: Molecular function. n = 3. ns, p > 0.05, *p < 0.05, **p < 0.01.
SMC1A affects malignant phenotypes of cervical cancer by regulating MACC1
To further outline whether MACC1 participates in the regulation of SMC1A in malignant phenotype of cervical cancer, MACC1 was overexpressed in SiHa and C-33A cells transfected by shSMC1As. As displayed in Fig. 7A, the protein expression of MACC1 was markedly upregulated in cancer cells. Compared with the proliferation of negative control cells, the silencing of SMC1A significantly suppressed the proliferation of the test cells. As expected, when MACC1 was overexpressed in cancer cells, the downregulation of proliferation induced by shSMC1A was notably reversed at 24, 48, and 72 h after transfection (Fig. 7B). Moreover, the effect of MACC1 on the EMT of cancer cells was examined using immunofluorescence staining. The increase of E-cadherin expression induced by shSMC1As was inhibited in the two cells after transfection of MACC1 overexpression (Fig. 7C). These results demonstrated that MACC1mediated the inhibition of SMC1A decrease on cell proliferation and EMT.
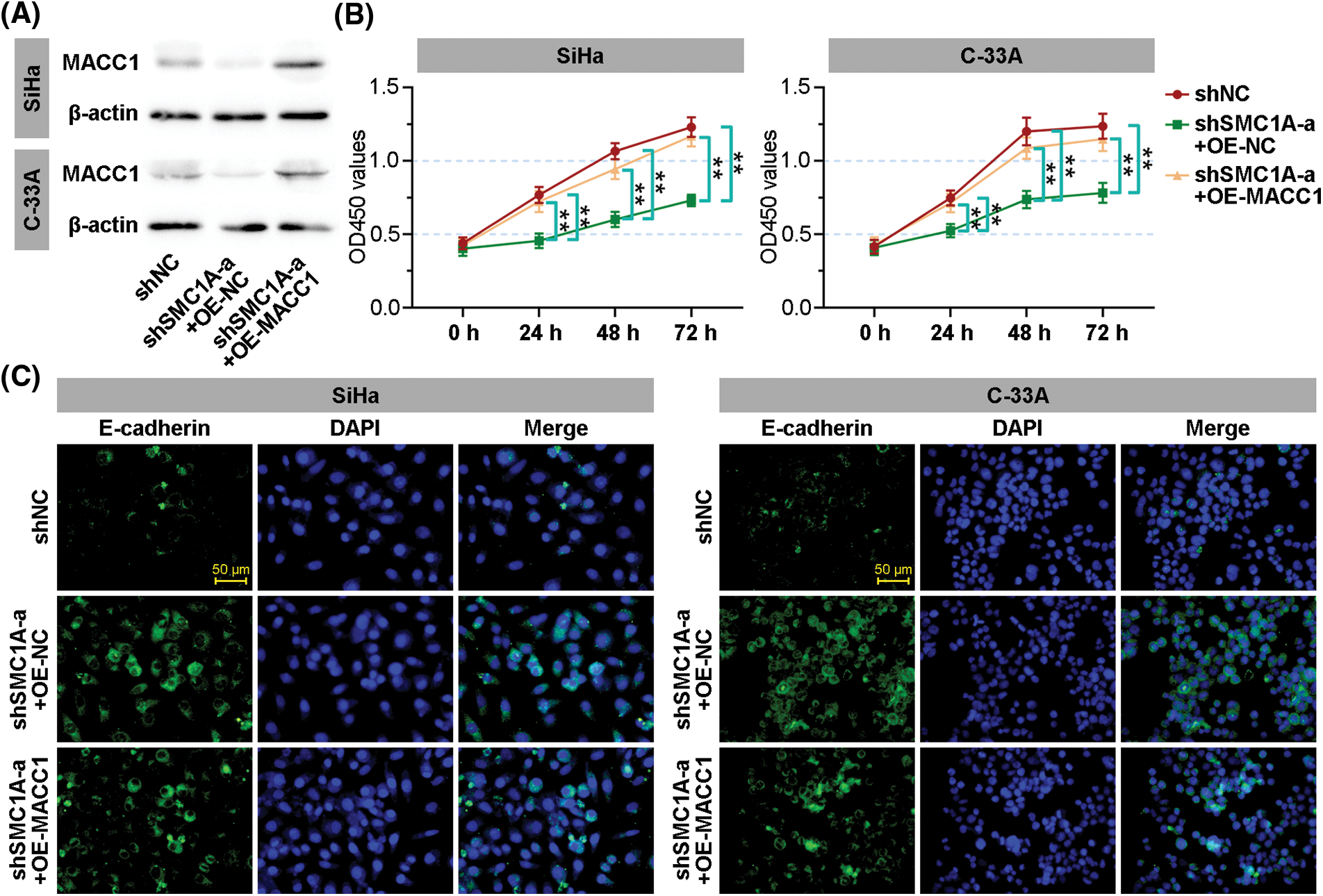
Figure 7: Structural maintenance of chromosome 1A (SMC1A) affects malignant phenotypes of cervical cancer by regulating metastasis associated in colon cancer 1 (MACC1). (A) The protein expression of MACC1 in the SMC1A-downregulated cells with overexpression of MACC1 was detected by western blotting. (B) Cell viability was examined using the cell counting kit-8 (CCK-8) assay. (C) The expression of E-cadherin was assessed by immunofluorescence (IF) staining of the cervical cancer cells. Scale bars: 50 μm. n = 3. **p < 0.01.
Cervical cancer is a gynecological disease that affects many women all over the world (Marth et al., 2017). SMC1A has been reported to regulate the development of several human cancers. In a recent study, SMC1A showed high expression in human colorectal cancer. Further, its overexpression promoted the biological behaviors of colorectal cancer (Sarogni et al., 2019). Additionally, there is literature suggesting that the absence of SMC1A leads to the inhibition of cell proliferation and invasion of lung cancer cells (Zhang et al., 2013). Furthermore, upregulation of SMC1A facilitated cell viability and invasion of cancer cells in colorectal cancer (Zhou et al., 2017). According to these reports, we speculated SMC1A was likely involved in the regulation of cervical cancer. Human papillomavirus (HPV) has been known as the major cause of cervical cancer. In our study, we used two cervical cancer cell lines to investigate the biological function of SMC1A. The SiHa cell line contains the HPV16 genome (Akagi et al., 2014), and C-33A is an HPV-negative cervical cancer cell line. Results obtained from GSE113942 and GSE151666 datasets have been shown in Suppl. Fig. S1, which demonstrated that SMC1A expression levels did not change significantly in HPV-positive and HPV-negative cervical tissues (Xu et al., 2019; Ruiz et al., 2021). Therefore, results in this report demonstrated that the expression of SMC1A was not affected by HPV in cervical cancer. Further, SMC1A was highly expressed in cervical tumor tissues. The proliferation and EMT of cervical cancer were remarkably suppressed by the depletion of SMC1A in cancer cells. SMC1A can be regarded as a potential molecular target to improve the prognosis of cervical cancer.
A previous study has clarified that molecular therapeutic targets for malignant tumors can improve the specificity of therapeutic drugs and minimize the possible side effects of drugs (Zhang et al., 2013). The alteration of Caspase-8 expression partly influenced the chemoresistance of cervical tumors. Its downregulation led to the therapy with CDK9 inhibitor combined with cisplatin overcoming chemotherapy resistance of cervical cancer cells (Mandal et al., 2022). Inhibition of microtubule affinity-regulating kinase (MARK) 2 resulted in enhancing the cytotoxicity of paclitaxel. Additionally, the inhibition of downstream factor histone deacetylase (HDAC) 4 could overcome chemoresistance to paclitaxel in cervical cancer cells (Zeng et al., 2022). Surprisingly, we found that downregulation of SMC1A reduced resistance to the chemotherapeutic drugs cisplatin and paclitaxel. Based on these findings, SMC1A could be used as a novel molecular target to assist therapies for cervical cancer. Increasing the sensitivity of tumor cells to traditional cytotoxic drugs and radiotherapy, so as to reduce the possibility of metastasis and clinical recurrence provides an approach for the improvement of cervical cancer outcomes.
Studies have shown that EMT is a biological process that promotes epithelial cells to acquire mesenchymal characteristics. EMT plays an important role in the invasion and distant metastasis of cancer progression (van Staalduinen et al., 2018; Aiello and Kang, 2019). During this process, the phenotype of epithelial cells becomes shorter and rounded along with weakened cell adhesion (Pastushenko and Blanpain, 2019). Additionally, the reduction of epithelial adhesion proteins such as E-cadherin, and the increase of interstitial molecular markers such as N-cadherin, vimentin, and Snail, are essential hallmarks of the EMT process (Lamouille et al., 2014). A study documented that SMC1A directly targeted the promoter region of Snail to enhance Snail transcription, which facilitated the EMT progression in breast cancer (Zhang et al., 2022). Our study estimated the progression of EMT by observing the phenotype of cervical cancer cells and detecting the alteration of E-cadherin, N-cadherin, and Snail. These results supported that the knockdown of SMC1A promoted the transformation of mesenchymal cells into epithelial cells in cervical cancer via MACC1 regulation.
To explore the underlying mechanism of SMC1A in the regulation of cervical cancer, we screened its downstream factors from the dataset GSE113678. According to the results of GO and KEGG enrichment analyses as well as the transcription levels of potential downstream factors, MACC1 was identified for further exploration of mechanisms. A published genome-wide search demonstrated that MACC1 differently expresses in colorectal cancer samples (Stein et al., 2009). Additionally, downregulation of MACC1 was reported to block migration and invasion of cells and promote apoptosis in cervical cancer. Meanwhile, the AKT/signal transducer and activator of transcription 3 (STAT3) signaling pathway was found to mediate the regulation of MACC1 in cervical cancer (Mei et al., 2022). MACC1 has been identified as a promising molecular target for cervical cancer prognosis (Guo et al., 2014; Wang et al., 2018). In our study, the expression of MACC1 significantly decreased in the SiHa and C-33A cells transfected with knockdown of SMC1A. The proliferation of SMC1A-knockdown cervical cancer cells was significantly enhanced when MACC1 was overexpressed. A remarkable decrease of E-cadherin induced by MACC1 upregulation suggested that MACC1 mediated the regulation of SMC1A in the EMT process of cervical cancer.
Collectively, our study verified that SMC1A was highly expressed in cervical cancer, and it was possibly associated with the poor prognosis of cervical cancer. Knockdown of SMC1A inhibited cell viability and the EMT process of cervical cancer cells by inhibiting the expression of MACC1. The chemotherapy resistance of cells to cisplatin and paclitaxel was also decreased by the silencing of SMC1A. In the future, in vivo experiments are still required to further corroborate the above conclusions.
Acknowledgement: None.
Funding Statement: The authors received no specific funding for this study.
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Weilan Liu, Congwei Dai; data collection: Weilan Liu, Kaiyun Qin; analysis and interpretation of results: Weilan Liu, Yan Jiang, Caifu Zhao; draft manuscript preparation: Weilan Liu. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The data and materials that support the findings of this study are available from the corresponding author upon reasonable request.
Ethics Approval: This study has been approved by the Ethics Committee of the Hebei General Hospital (Approval no. 2023080).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
Aiello NM, Kang Y (2019). Context-dependent EMT programs in cancer metastasis. The Journal of Experimental Medicine 216: 1016–1026. https://doi.org/10.1084/jem.20181827 [Google Scholar] [PubMed] [CrossRef]
Akagi K, Li J, Broutian TR, Padilla-Nash H, Xiao W et al. (2014). Genome-wide analysis of HPV integration in human cancers reveals recurrent, focal genomic instability. Genome Research 24: 185–199. https://doi.org/10.1101/gr.164806.113 [Google Scholar] [PubMed] [CrossRef]
Arbyn M, Weiderpass E, Bruni L, de Sanjosé S, Saraiya M, Ferlay J, Bray F (2020). Estimates of incidence and mortality of cervical cancer in 2018: A worldwide analysis. The Lancet Global Health 8: e191–e203. https://doi.org/10.1016/S2214-109X(19)30482-6 [Google Scholar] [PubMed] [CrossRef]
Ashrafizadeh M, Mirzaei S, Hashemi F, Zarrabi A, Zabolian A et al. (2021). New insight towards development of paclitaxel and docetaxel resistance in cancer cells: EMT as a novel molecular mechanism and therapeutic possibilities. Biomedicine & Pharmacotherapy = Biomedecine 141: 111824. https://doi.org/10.1016/j.biopha.2021.111824 [Google Scholar] [PubMed] [CrossRef]
Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, Varambally S (2017). UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 19: 649–658. https://doi.org/10.1016/j.neo.2017.05.002 [Google Scholar] [PubMed] [CrossRef]
Chandrashekar DS, Karthikeyan SK, Korla PK, Patel H, Shovon AR et al. (2022). UALCAN: An update to the integrated cancer data analysis platform. Neoplasia 25: 18–27. https://doi.org/10.1016/j.neo.2022.01.001 [Google Scholar] [PubMed] [CrossRef]
Chowdhury P, Nagesh PKB, Hatami E, Wagh S, Dan N et al. (2019). Tannic acid-inspired paclitaxel nanoparticles for enhanced anticancer effects in breast cancer cells. Journal of Colloid and Interface Science 535: 133–148. https://doi.org/10.1016/j.jcis.2018.09.072 [Google Scholar] [PubMed] [CrossRef]
Díaz-Martínez LA, Beauchene NA, Furniss K, Esponda P, Giménez-Abián JF, Clarke DJ (2010). Cohesin is needed for bipolar mitosis in human cells. Cell Cycle 9: 1764–1773. https://doi.org/10.4161/cc.9.9.11525 [Google Scholar] [PubMed] [CrossRef]
Dan VM, Raveendran RS, Baby S (2021). Resistance to intervention: Paclitaxel in breast cancer. Mini Reviews in Medicinal Chemistry 21: 1237–1268. https://doi.org/10.2174/1389557520999201214234421 [Google Scholar] [PubMed] [CrossRef]
Della Corte L, Barra F, Foreste V, Giampaolino P, Evangelisti G, Ferrero S, Bifulco G (2020). Advances in paclitaxel combinations for treating cervical cancer. Expert Opinion on Pharmacotherapy 21: 663–677. https://doi.org/10.1080/14656566.2020.1724284 [Google Scholar] [PubMed] [CrossRef]
Dowen JM, Bilodeau S, Orlando DA, Hübner MR, Abraham BJ, Spector DL, Young RA (2013). Multiple structural maintenance of chromosome complexes at transcriptional regulatory elements. Stem Cell Reports 1: 371–378. https://doi.org/10.1016/j.stemcr.2013.09.002 [Google Scholar] [PubMed] [CrossRef]
Gregson HC, van Hooser AA, BallJr AR, Brinkley BR, Yokomori K (2002). Localization of human SMC1 protein at kinetochores. Chromosome Research 10: 267–277. https://doi.org/10.1023/A:1016563523208 [Google Scholar] [PubMed] [CrossRef]
Guo L, Lu W, Zhang X, Luo D, Zhang H (2014). Metastasis-associated colon cancer-1 is a novel prognostic marker for cervical cancer. International Journal of Clinical and Experimental Pathology 7: 4150–4155. [Google Scholar] [PubMed]
Hsu YM, Chen YF, Chou CY, Tang MJ, Chen JH, Wilkins RJ, Ellory JC, Shen MR (2007). KCl cotransporter-3 down-regulates E-cadherin/β-catenin complex to promote epithelial-mesenchymal transition. Cancer Research 67: 11064–11073. https://doi.org/10.1158/0008-5472.CAN-07-2443 [Google Scholar] [PubMed] [CrossRef]
Kayaselçuk F, Zorludemir S, Gümürdühü D, Zeren H, Erman T (2002). PCNA and Ki-67 in central nervous system tumors: Correlation with the histological type and grade. Journal of Neuro-Oncology 57: 115–121. https://doi.org/10.1023/A:1015739130208 [Google Scholar] [PubMed] [CrossRef]
Kim KK, Kugler MC, Wolters PJ, Robillard L, Galvez MG, Brumwell AN, Sheppard D, Chapman HA (2006). Alveolar epithelial cell mesenchymal transition develops in vivo during pulmonary fibrosis and is regulated by the extracellular matrix. Proceedings of the National Academy of Sciences of the United States of America 103: 13180–13185. https://doi.org/10.1073/pnas.0605669103 [Google Scholar] [PubMed] [CrossRef]
Kumar S, Clair DS (2021). Radioresistance in prostate cancer: Focus on the interplay between NF-κB and SOD. Antioxidants 10: 1925. https://doi.org/10.3390/antiox10121925 [Google Scholar] [PubMed] [CrossRef]
Lamouille S, Xu J, Derynck R (2014). Molecular mechanisms of epithelial-mesenchymal transition. Nature Reviews Molecular Cell Biology 15: 178–196. https://doi.org/10.1038/nrm3758 [Google Scholar] [PubMed] [CrossRef]
Mandal R, Raab M, Rödel F, Krämer A, Kostova I et al. (2022). The non-apoptotic function of Caspase-8 in negatively regulating the CDK9-mediated Ser2 phosphorylation of RNA polymerase II in cervical cancer. Cellular and Molecular Life Sciences 79: 597. https://doi.org/10.1007/s00018-022-04598-3 [Google Scholar] [PubMed] [CrossRef]
Marth C, Landoni F, Mahner S, McCormack M, Gonzalez-Martin A, Colombo N (2017). Cervical cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Annals of Oncology 28: iv72–iv83. https://doi.org/10.1093/annonc/mdx220 [Google Scholar] [PubMed] [CrossRef]
Mei J, Zhu C, Pan L, Li M (2022). MACC1 regulates the AKT/STAT3 signaling pathway to induce migration, invasion, cancer stemness, and suppress apoptosis in cervical cancer cells. Bioengineered 13: 61–70. https://doi.org/10.1080/21655979.2021.2006567 [Google Scholar] [PubMed] [CrossRef]
Narayan G, Bourdon V, Chaganti S, Arias-Pulido H, Nandula SV et al. (2007). Gene dosage alterations revealed by cDNA microarray analysis in cervical cancer: Identification of candidate amplified and overexpressed genes. Genes, Chromosomes & Cancer 46: 373–384. https://doi.org/10.1002/gcc.20418 [Google Scholar] [PubMed] [CrossRef]
Pastushenko I, Blanpain C (2019). EMT transition states during tumor progression and metastasis. Trends in Cell Biology 29: 212–226. https://doi.org/10.1016/j.tcb.2018.12.001 [Google Scholar] [PubMed] [CrossRef]
Ribatti D, Tamma R, Annese T (2020). Epithelial-mesenchymal transition in cancer: A historical overview. Translational Oncology 13: 100773. https://doi.org/10.1016/j.tranon.2020.100773 [Google Scholar] [PubMed] [CrossRef]
Ruiz FJ, Inkman M, Rashmi R, Muhammad N, Gabriel N et al. (2021). HPV transcript expression affects cervical cancer response to chemoradiation. JCI Insight 6: e138734. https://doi.org/10.1172/jci.insight.138734 [Google Scholar] [PubMed] [CrossRef]
Sarogni P, Palumbo O, Servadio A, Astigiano S, D’Alessio B et al. (2019). Overexpression of the cohesin-core subunit SMC1A contributes to colorectal cancer development. Journal of Experimental & Clinical Cancer Research 38: 108. https://doi.org/10.1186/s13046-019-1116-0 [Google Scholar] [PubMed] [CrossRef]
Singh D, Vignat J, Lorenzoni V, Eslahi M, Ginsburg O, Lauby-Secretan B, Arbyn M, Basu P, Bray F, Vaccarella S (2022). Global estimates of incidence and mortality of cervical cancer in 2020: A baseline analysis of the WHO global cervical cancer elimination initiative. The Lancet Global Health 11: E197–E206. https://doi.org/10.1016/s2214-109x(22)00501-0 [Google Scholar] [PubMed] [CrossRef]
Stein U, Walther W, Arlt F, Schwabe H, Smith J, Fichtner I, Birchmeier W, Schlag PM (2009). MACC1, a newly identified key regulator of HGF-MET signaling, predicts colon cancer metastasis. Nature Medicine 15: 59–67. https://doi.org/10.1038/nm.1889 [Google Scholar] [PubMed] [CrossRef]
van Staalduinen J, Baker D, Ten Dijke P, van Dam H (2018). Epithelial-mesenchymal-transition-inducing transcription factors: New targets for tackling chemoresistance in cancer? Oncogene 37: 6195–6211. https://doi.org/10.1038/s41388-018-0378-x [Google Scholar] [PubMed] [CrossRef]
Wang S, Zhang Y, Yuan S, Ji X (2018). MicroRNA‐485 targets MACC1 and inhibits cervical cancer cell proliferation and invasion. Molecular Medicine Reports 18: 2407–2416. https://doi.org/10.3892/mmr.2018.9186 [Google Scholar] [PubMed] [CrossRef]
Xie P, Jin Q, Li Y, Zhang J, Kang X, Zhu J, Mao X, Cao P, Liu C (2021). Nanoparticle delivery of a triple-action Pt(IV) prodrug to overcome cisplatin resistance via synergistic effect. Biomaterials Science 10: 153–157. https://doi.org/10.1039/D1BM01556G [Google Scholar] [PubMed] [CrossRef]
Xu J, Liu H, Yang Y, Wang X, Liu P et al. (2019). Genome-wide profiling of cervical RNA-binding proteins identifies human papillomavirus regulation of RNASEH2A expression by viral E7 and E2F1. mBio 10. https://doi.org/10.1128/mBio.02687-18 [Google Scholar] [PubMed] [CrossRef]
Yadav S, Kowolik CM, Lin M, Zuro D, Hui SK, Riggs AD, Horne DA (2019). SMC1A is associated with radioresistance in prostate cancer and acts by regulating epithelial-mesenchymal transition and cancer stem-like properties. Molecular Carcinogenesis 58: 113–125. https://doi.org/10.1002/mc.22913 [Google Scholar] [PubMed] [CrossRef]
Zeng Y, Yin L, Zhou J, Zeng R, Xiao Y et al. (2022). MARK2 regulates chemotherapeutic responses through class IIa HDAC-YAP axis in pancreatic cancer. Oncogene 41: 3859–3875. https://doi.org/10.1038/s41388-022-02399-3 [Google Scholar] [PubMed] [CrossRef]
Zhang X, Dai XY, Qian JY, Xu F, Wang ZW et al. (2022). SMC1A regulated by KIAA1429 in m6A-independent manner promotes EMT progress in breast cancer. Molecular Therapy Nucleic Acids 27: 133–146. https://doi.org/10.1016/j.omtn.2021.08.009 [Google Scholar] [PubMed] [CrossRef]
Zhang YF, Jiang R, Li JD, Zhang XY, Zhao P et al. (2013). SMC1A knockdown induces growth suppression of human lung adenocarcinoma cells through G1/S cell cycle phase arrest and apoptosis pathways in vitro. Oncology Letters 5: 749–755. https://doi.org/10.3892/ol.2013.1116 [Google Scholar] [PubMed] [CrossRef]
Zhou P, Xiao N, Wang J, Wang Z, Zheng S, Shan S, Wang J, Du J, Wang J (2017). SMC1A recruits tumor-associated-fibroblasts (TAFs) and promotes colorectal cancer metastasis. Cancer Letters 385: 39–45. https://doi.org/10.1016/j.canlet.2016.10.041 [Google Scholar] [PubMed] [CrossRef]
Zhou X, Xu CJ, Wang JX, Dai T, Ye YP, Cui YM, Liao WT, Wu XL, Ou JP (2015). Metastasis-associated in colon cancer-1 associates with poor prognosis and promotes cell invasion and angiogenesis in human cervical cancer. International Journal of Gynecological Cancer 25: 1353–1363. https://doi.org/10.1097/IGC.0000000000000524 [Google Scholar] [PubMed] [CrossRef]
Supplementary Materials
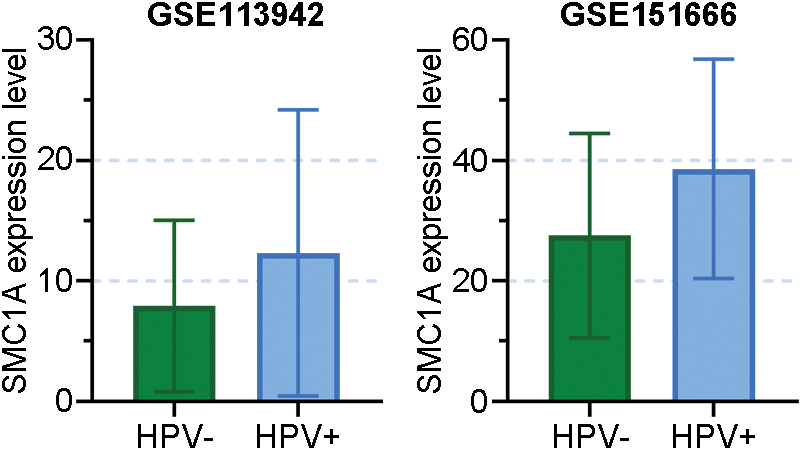
FIGURE S1: Data came from GSE113942 and GSE151666 revealed the SMC1A expression in HPV-positive and HPV-negative cervix tissues.
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools