 Open Access
Open Access
ARTICLE
Long non-coding RNA-ATB induces trastuzumab resistance and aggravates the progression of gastric cancer by repressing miR- 200c via ZNF217 elevation
The Second Department of Oncology, The Third Affiliated Hospital of Qiqihar Medical University, Qiqihar, China
* Corresponding Author: Jiazhuang Li,
(This article belongs to the Special Issue: Non-Coding RNAs in the Regulation of Human Cancers)
BIOCELL 2023, 47(10), 2313-2320. https://doi.org/10.32604/biocell.2023.029860
Received 11 March 2023; Accepted 27 May 2023; Issue published 08 November 2023
Abstract
Background: Trastuzumab resistance accounts for chemotherapy failure in gastric cancer patients in clinical practice. The significance of long non-coding RNAs (lncRNAs) in the maintenance of drug resistance in gastric cancer has been already underlined. Method: This study aimed to identify the specific role of lncRNA-ATB in gastric cancer progression and trastuzumab resistance. The downstream miRs of lncRNA-ATB and target genes of miRs were predicted by bioinformatics analysis and verified using dual luciferase reporter assay. Loss- and gain-function assays were performed to explore the roles of lncRNA-ATB, miR-200c, and zinc-finger protein 217 (ZNF217) in the cell functions and trastuzumab resistance of a trastuzumab-resistant gastric cancer cell line (NCI-N87-TR). Result: LncRNA-ATB was upregulated, while miR-200c was downregulated. Depletion of lncRNA-ATB or miR-200c elevation led to a decrease in malignant properties of NCI-N87-TR cells. LncRNA-ATB could negatively target miR- 200c, which in turn inversely targeted and reduced the expression of ZNF217. Silencing of ZNF217 could inhibit cell viability and migration. Conclusion: lncRNA-ATB promoted the progression and trastuzumab resistance of gastric cancer by repressing miR-200c via ZNF217 upregulation.Keywords
As the most commonly seen gastrointestinal tumor subtype, survival improvement in patients who received traditional surgery and chemoradiation for the treatment of gastric cancer remains difficult (Ye et al., 2020). Meanwhile, lymphatic metastasis and distant metastasis serve as the leading causes of death in gastric cancer patients during treatment (Kinami et al., 2021; Lin et al., 2022a). Trastuzumab, a therapeutic monoclonal antibody directed against human epidermal growth factor receptor 2 (HER2), approved for treating patients with HER2 amplification, and has been utilized to treat gastric cancer (Bang et al., 2010). However, drug resistance usually happens during trastuzumab treatment (Gomez-Martín et al., 2014). Commonly, trastuzumab resistance is due to hyperactivated PI3K or PTEN loss mutations in the research of gastric cancer (Roviello et al., 2022). Emerging evidence indicates that epithelial-mesenchymal transition (EMT), a key regulator of distant metastasis, is associated with trastuzumab resistance (Zhou et al., 2018). Therefore, the identification of a new target for gastric cancer progression and trastuzumab resistance is urgently needed.
Long non-coding RNAs (lncRNAs) are long RNA transcripts without protein-coding capability, and possess potential in the modulation of diverse biological processes, such as cell cycle transition, apoptosis, metabolism, and EMT (Ghafouri-Fard and Taheri, 2020; Gugnoni and Ciarrocchi, 2019). LncRNAs may serve as a novel clinical diagnosis marker and target for cancer treatment (Ghafouri-Fard and Taheri, 2020). Prior data have shown lncRNA-ATB acts in tumor metastasis by modulation of the pro-metastatic effect of TGF-β and EMT (Li et al., 2016). More importantly, lncRNAs regulate biological processes via microRNAs (miRs) (Luo et al., 2020). MiR-200c-3p overexpression is capable of limiting the proliferation and invasion ability of gastric cancer cells (Kurata et al., 2018; Zhou et al., 2018). Meanwhile, zinc-finger protein 217 (ZNF217), a direct downstream target of miR-200c, mediates trastuzumab resistance caused by the lncRNA-ATB-miR-200c axis (Shi et al., 2015). These aforementioned reports allowed us to speculate that the potential regulation of lncRNA-ATB in gastric cancer progression and trastuzumab resistance occurs through the miR-200c/ZNF217 axis. Hence, the main objective of the study was to determine if this hypothesis was valid and to further explore the molecular mechanism by which lncRNA-ATB mediates the progression of gastric cancer and drug resistance to trastuzumab through the miR-200c/ZNF217 axis regulation.
The downstream target miRs of lncRNA-ATB and potential target genes of miRs were predicted by lncRInter. Next, the binding sites between miRs and their target genes were predicted using TargetScan and TarBase, respectively. The expression of these genes in gastric cancer samples was identified by GEPIA, and the co-expression of related genes in gastric cancer was predicted by Chipbase v2.0.
The human gastric cancer cell line NCI-N87, obtained from the Typical Culture Preservation Commission Cell Bank of the Chinese Academy of Sciences in Shanghai, China, was cultured in Roswell Park Memorial Institute-1640 medium (Gibco, CA, USA) supplemented with 10% fetal bovine serum (PAN-Biotech, Bavaria, Germany). The cells were incubated at 37°C in a humid atmosphere with 5% CO2. Trastuzumab-resistant NCI-N87 cells (NCI-N87-TR cells) were induced following a previously described method (Guo et al., 2021). For the establishment of resistant cells, trastuzumab (30 μg/mL) was used for 6 months duration.
The NCI-N87 cells were transfected with miR-200c inhibitor, lncRNA-ATB small interfering RNA (siRNA), and ZNF217 siRNA, along with their respective negative controls (NC), obtained from HanBio Technology in Shanghai, China. The transfection was carried out using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) in less than 50% confluent cells. The transfected cells were then cultured in a 37°C incubator with 5% CO2. For transient transfection, Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) was used following the manufacturer’s instructions. Briefly, the cells were seeded in cell culture plates and allowed to reach 50% confluence. Then, a mixture of lncRNA-ATB siRNA, miR-200c inhibitor, si-ZNF217, or negative control siRNA group, along with Lipo2000 transfection reagent (Invitrogen, Carlsbad, CA, USA), was transfected into the cells.
Cell proliferation determination
NCI-N87 cells (8 × 103 cells/ well) were seeded in a 96-well plate and cultured at 37°C with 5% CO2. After culturing for 24, 48, and 72 h, the cultures were examined. Cells were incubated with 10 μL Cell Counting Kit-8 (Abbkine, Wuhan, China) for 60 min. Next, the absorbance rate was tested on a microplate reader at OD 450 nm.
NCI-N87 cells were seeded in a 6-well plate. A scratch was created at the center of each well with the help of the 10 µL sterile pipette tip. The scratch was imaged at 0 and 48 h under a microscope and cell migration was examined.
RNA isolation and quantitation
Total RNA was isolated from NCI-N87 cells using the Total RNA Kit (Omega Bio-tek, Guangzhou, China). The cDNA of lncRNA-ATB, miR-200c, and ZNF217 was synthesized using the ReverTra Ace qPCR RT Kit (Toyobo, Osaka, Japan). Quantitative real-time-polymerase chain reaction (RT-qPCR) was performed using the TransStart® Top Green qPCR SuperMix (Transgen, Beijing, China). For miRNA quantification, Bulge-loopTM miRNA qRT-PCR Primer Sets (including one RT primer and a pair of qPCR primers for each set) specific to miR-200c-3p and U6 were used; these were designed by RiboBio (Guangzhou, China). The results were analyzed using the 2−∆∆CT method.
Protein was extracted using protease inhibitor-containing RIPA buffer (Boster, Wuhan, China) and the concentration was determined utilizing the BCA Kit (Beyotime, Jiangsu, China). Following separation by electrophoresis, the proteins were electro-transferred onto polyvinylidene fluoride membranes (Roche, Mannheim, Germany), which were probed with antibodies (primary and secondary antibodies, all from Proteintech, Wuhan, China). ImageJ software (NIH) was adopted for estimating the intensity of the protein bands. Antibodies used were as follows: anti-ZNF217 antibody (67730-1-lg, 1: 1000), anti-E-cadherin antibody (60335-1-lg, 1: 2000), anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH) antibody (60004-1-lg, 1: 50000), and horse-radish peroxidase (HRP)-conjugated Affinipure goat anti-mouse lgG (H+L) (1: 2000, SA00001-1).
Data were analyzed using GraphPad Prism 9.4 (GraphPad, San Diego, CA, USA). The analysis results were described as the mean ± standard deviation. Statistical comparisons were processed utilizing the Student’s t-test and one-way or two-way ANOVA. Statistical significance was described as p < 0.05.
LncRNA-ATB is elevated in NCI-N87-TR cells
As shown in Figs. 1A, 1B, NCI-N87-TR cells showed markedly higher viability and migration than NCI-N87 cells. Additionally, reduced expression of E-cadherin was observed in NCI-N87-TR cells compared with NCI-N87 cells (Fig. 1C) and elevated lncRNA-ATB was detected (Fig. 1D). The above results proved that a trastuzumab-resistant cell model NCI-N87-TR with elevated lncRNA-ATB expression was successfully constructed.
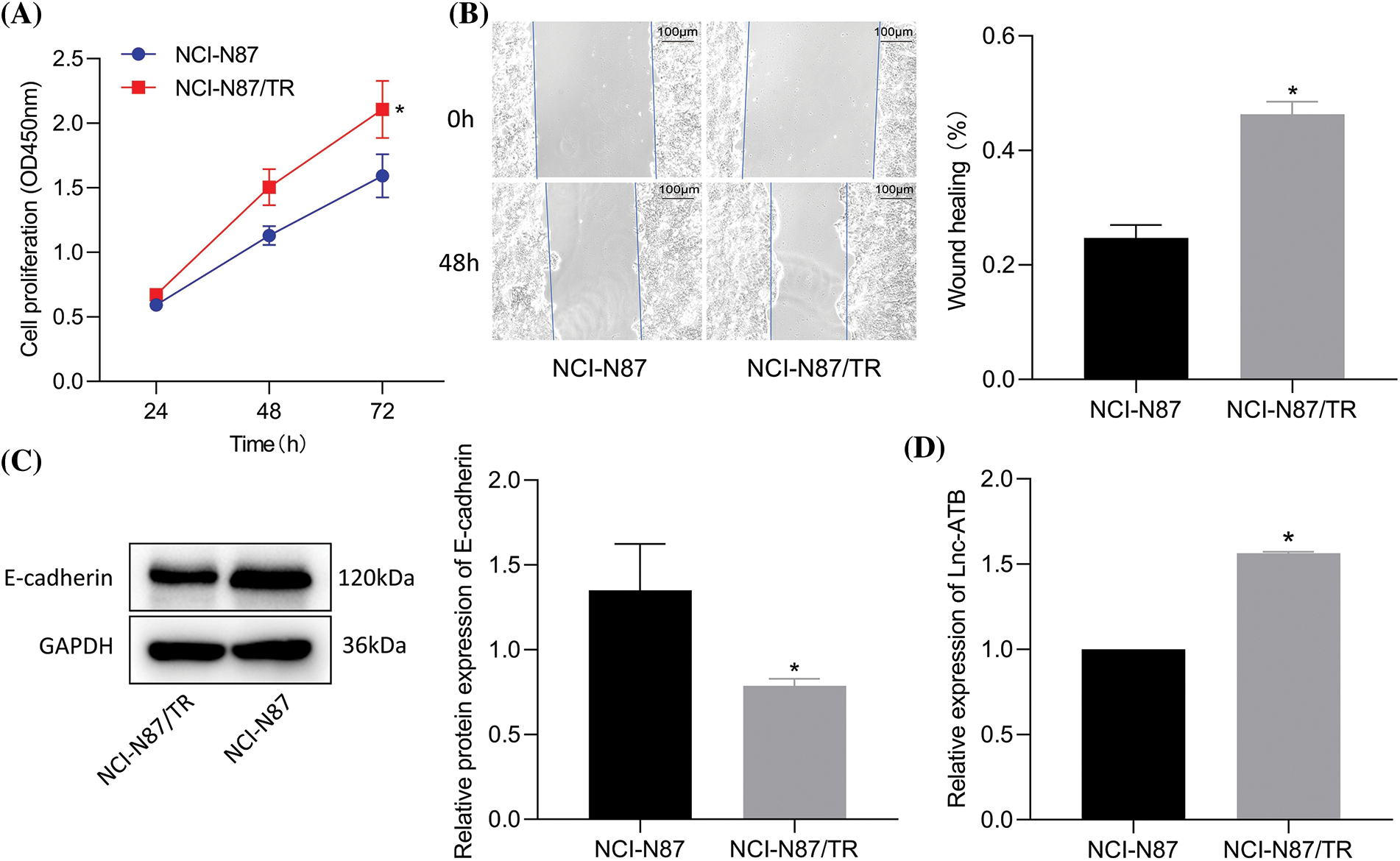
Figure 1: LncRNA-ATB expression is elevated in NCI-N87-TR cells. (A) Viability of NCI-N87 and NCI-N87-TR cells was determined by Cell Counting Kit-8 assay. (B) Migration of NCI-N87 and NCI-N87-TR cells detected by scratch test. (C) The levels of epithelial-mesenchymal transition maker E-cadherin in NCI-N87 and NCI-N87-TR cells was detected by immunoblotting. (D) The expression of lncRNA-ATB in NCI-N87 and NCI-N87-TR cells was determined by quantitative real-time-polymerase chain reaction. *p < 0.05 vs. NCI-N87 cells.
Silencing of lncRNA-ATB suppresses trastuzumab resistance, migration, and viability in NCI-N87/TR cells
LncRNA-ATB was knocked-down (Fig. 2A) and the efficiency was confirmed by RT-qPCR. A superior knockdown efficiency for si-lnc-ATB-2 was noted and was thus selected for subsequent assays. Si-lncRNA-ATB markedly inhibited NCI-N87-TR cell viability and migration compared to si-NC treatment (Figs. 2B, 2C). Furthermore, immunoblotting results revealed that E-cadherin upregulated after lncRNA-ATB-mediated inhibition (Fig. 2D). These data indicate that silencing of lncRNA-ATB could suppress trastuzumab resistance, migration, and viability.
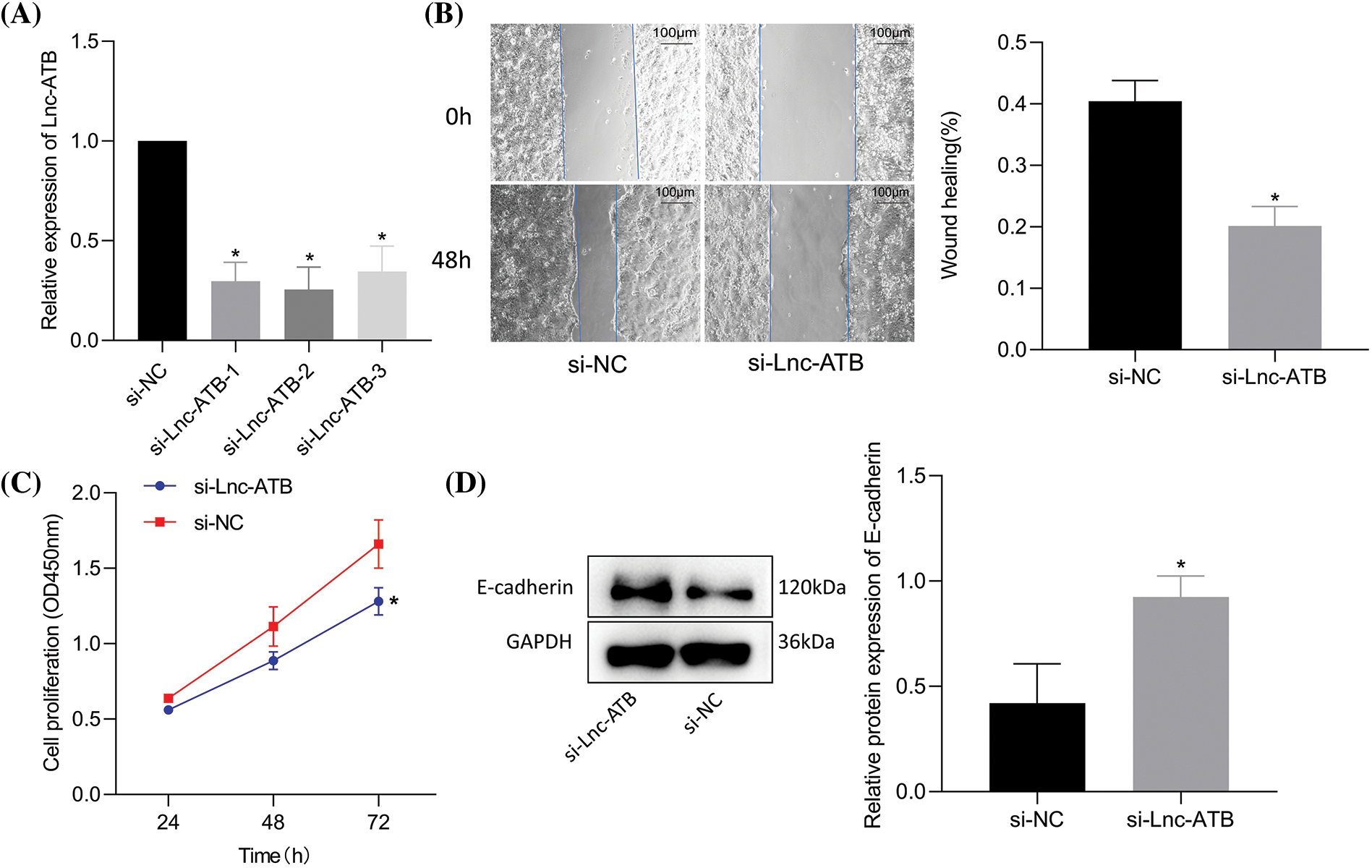
Figure 2: Silencing of lncRNA-ATB suppresses trastuzumab resistance and migration of NCI-N87-TR cells. (A) The knockdown efficiency of lncRNA-ATB in NCI-N87-TR cells was determined by quantitative real-time-polymerase chain reaction. (B) NCI-N87-TR cell viability was determined by Cell Counting Kit-8 assay. (C) Scratch test to detect the migration of NCI-N87-TR cells treated with si-lncRNA-ATB. (D) Determination of E-cadherin expression in NCI-N87-TR cells treated with si-lncRNA-ATB by immunoblotting. *p < 0.05 vs. si-NC.
MiR-200c is negatively targeted by lncRNA-ATB
MiR-200c was identified as a potential downstream target of lncRNA-ATB based on predictions from the RNAInter website (Fig. 3A). RT-qPCR analysis demonstrated a decrease in miR-200c expression in NCI-N87-TR cell lines (Fig. 3B). The predicted binding sites between miR-200c and lncRNA-ATB were confirmed (Shi et al., 2015). Luciferase assays performed in HEK293T cells transfected with miR-200c mimic showed a decrease in luciferase activity of lncRNA-ATB-WT, while no change was observed with lncRNA-ATB-MUT (Shi et al., 2015). Additionally, silencing lncRNA-ATB significantly increased the level of miR-200c (Fig. 3C), indicating the binding interaction between lncRNA-ATB and miR-200c.
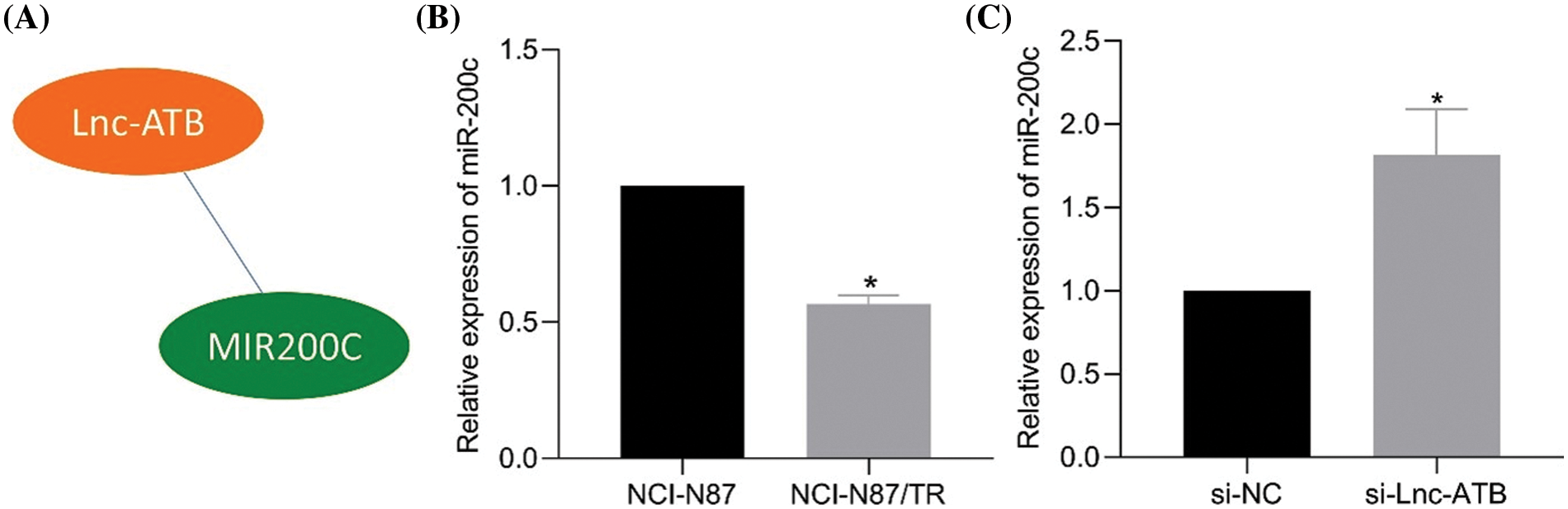
Figure 3: LncRNA-ATB binds to miR-200c and downregulates the expression of miR-200c in NCI-N87/TR cells. (A) The RNAInter website predicts miR-200c as a potential target of lncRNA-ATB. (B) The expression of miR-200c in NCI-N87 and NCI-N87-TR cells was determined by quantitative real-time-polymerase chain reaction (RT-qPCR). (C) The expression of miR-200c in NCI-N87-TR cells after silencing lncRNA-ATB was determined by RT-qPCR. *p < 0.05 compared with NCI-N87, si-NC.
MiR-200c binds to and negatively regulates zinc-finger protein 217 expression
According to the prediction by lncRInter and TargetScan websites, lncRNA-ATB, miR-200c, and ZNF217 form competitive triads (Fig. 4A). ZNF217 expression was elevated in clinical gastric cancer samples from The Cancer Genome Atlas database (Fig. 4B). As expected, elevated ZNF217 expression was detected in NCI-N87-TR cell lines (Fig. 4C). The predicted binding site of miR-200c on ZNF217 was also revealed (Bai et al., 2014). Luciferase assay further revealed a decrease in luciferase activity of ZNF217-WT in HEK293T cells transfected with miR-200c mimic, and the same did not change for ZNF217-MUT (Bai et al., 2014). Furthermore, RT-qPCR and immunoblotting results revealed an increase in the expression of ZNF217 after silencing of miR-200c (Figs. 4D, 4E). Taken together, lncRNA-ATB positively regulated ZNF217 by targeting miR-200c in NCI-N87-TR cells.
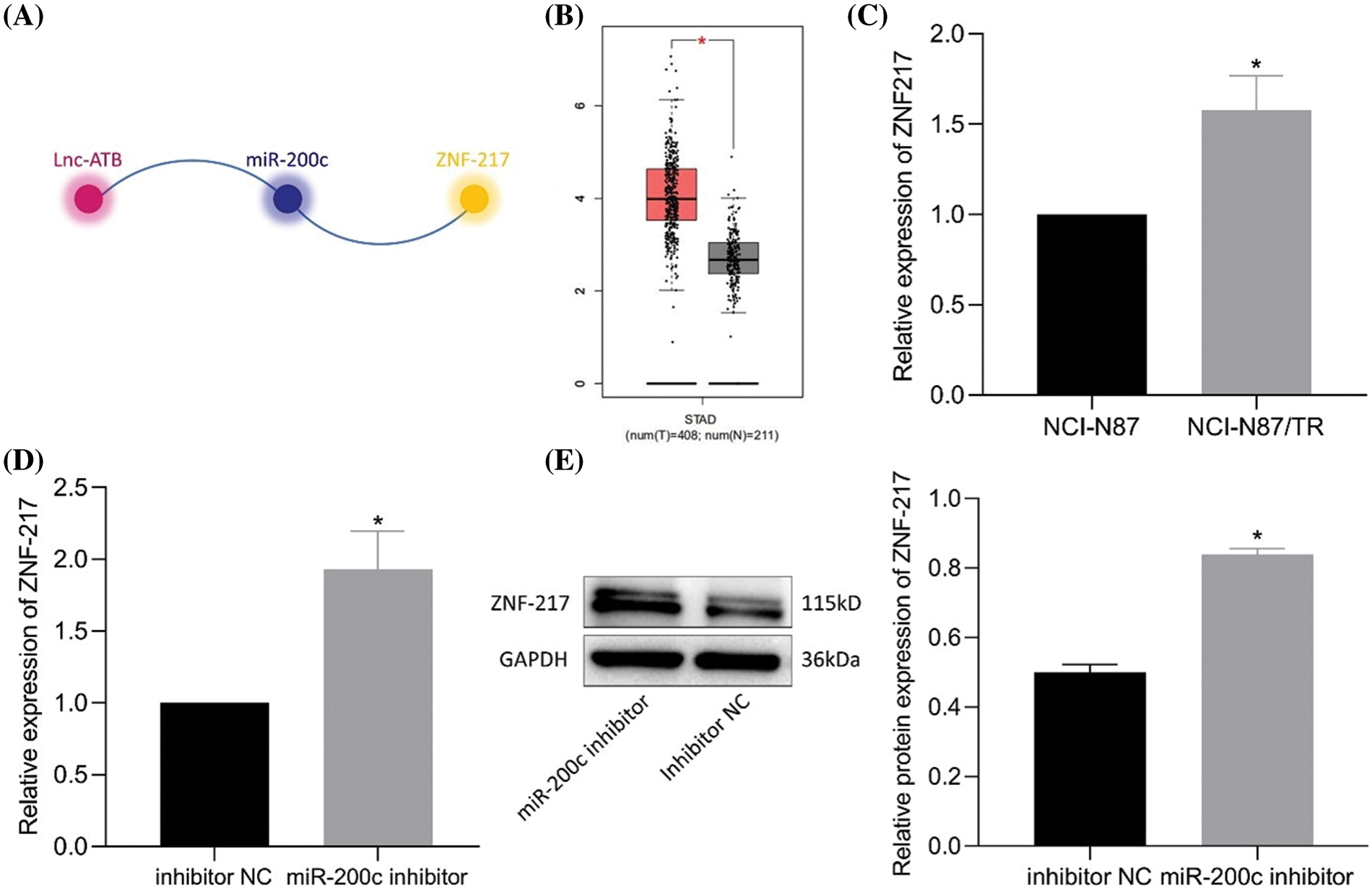
Figure 4: MiR-200c binds to zinc-finger protein 217 (ZNF217) and downregulates the expression of ZNF217. (A) The relationship between lncRNA-ATB, miR-200c, and ZNF217 was predicted by RNAInter and TargetScan. (B) A box plot of the expression of ZNF217 in gastric cancer samples in The Cancer Genome Atlas and analyzed by Gene Expression Profiling Interactive Analysis. (C) The expression of ZNF217 in NCI-N87 and NCI-N87-TR cells was determined by quantitative real-time-polymerase chain reaction (RT-qPCR). (D) Expression of ZNF217 in NCI-N87-TR cells after silencing of miR-200c determined by RT-qPCR. (E) Expression of ZNF217 in NCI-N87-TR cells after silencing of miR-200c determined by immunoblotting. *p < 0.05 vs. NCI-N87 cells or inhibitor NC.
Silencing zinc-finger protein 217 reduces the trastuzumab resistance, migration, and viability of NCI-N87-TR cells
Silencing ZNF217 was validated by RT-qPCR (si-ZNF217-2 showed a better silencing efficiency) (Fig. 5A). Compared to si-NC, si-ZNF217 caused a decrease in cell viability and migration (Figs. 5B, 5C) of NCI-N87-TR cells. Immunoblotting results revealed that si-ZNF217 brought about an elevation in E-cadherin expression (Fig. 5D). Overall, inhibition of ZNF217 contributed to the suppression of trastuzumab resistance, migration, and viability of NCI-N87-TR cells.
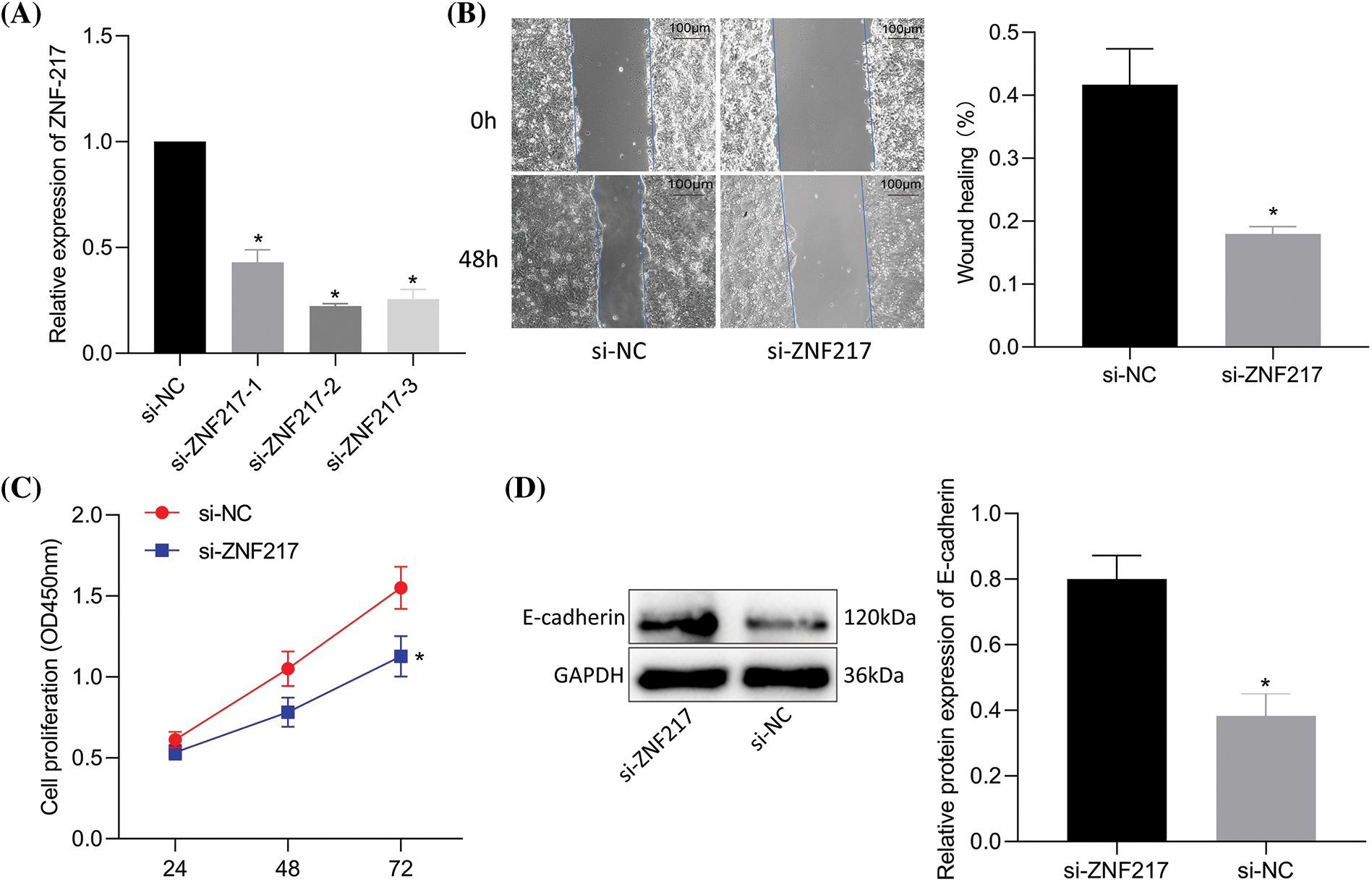
Figure 5: Silencing of zinc-finger protein 217 (ZNF217) suppresses trastuzumab resistance, migration, and viability of NCI-N87/TR cells. (A) The efficiency of ZNF217 knockdown in NCI-N87-TR cells was determined by quantitative real-time-polymerase chain reaction. (B) Scratch test to determine NCI-N87-TR cell migration upon treatment with si-ZNF217. (C) NCI-N87/TR cell viability upon treatment with si-ZNF217 determined by Cell Counting Kit-8 assay. (D) The protein levels of E-cadherin in NCI-N87-TR cells treated with si-ZNF217 were determined by immunoblotting. *p < 0.05 vs. si-NC.
LncRNA-ATB promotes the progression of various tumors, indicating its function as a novel biomarker for clinical diagnosis gastric cancer (Jang et al., 2017; Chen et al., 2018). In this study, we offered evidence that overexpression of lncRNA-ATB could promote the progression of gastric cancer and lead to trastuzumab resistance by regulating the miR-200c-ZNF217 axis (Fig. 6).
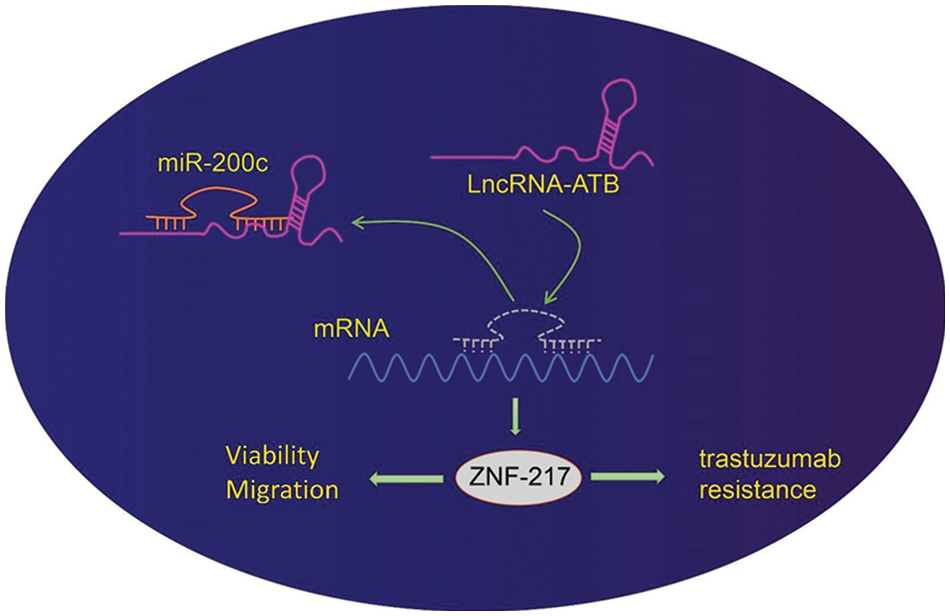
Figure 6: Schematic diagram of the mechanism by which lncRNA-ATB affects gastric cancer progression and trastuzumab resistance. LncRNA-ATB promotes trastuzumab resistance, viability, and migration of gastric cancer cells through the upregulation of zinc-finger protein 217 (ZNF217) by binding to miR-200c.
Preliminary results of this study revealed dramatically upregulated expression of lncRNA-ATB in trastuzumab resistant gastric cancer cells, silencing of which could repress the trastuzumab resistance, migration, and viability of NCI-N87-TR cells. Consistently, lncRNA-ATB is significantly upregulated in gastric cancer tissues, while its poor expression in vitro could inhibit proliferation and cell cycle arrest of gastric cancer cells (Lei et al., 2017). In addition, overexpression of lncRNA-ATB could induce trastuzumab resistance and invasion metastasis cascade in breast cancer by inducing EMT (Shi et al., 2015). Meanwhile, lncRNA-ATB has been shown to promote gastric cancer progression (Shen et al., 2018), and it correlates with poor outcomes in clinical gastric cancer patients (Saito et al., 2015).
Our subsequent experimentation provided evidence supporting the notion that miR-200c is a downstream target of lncRNA-ATB and is negatively regulated by lncRNA-ATB. This observation is consistent with a previous research implicating miR-200c as a target of lncRNA-ATB in the context of silica-induced pulmonary fibrosis (Liu et al., 2017). A recent study identified the specific binding site of miR-200c within lncRNA-ATB, while the inhibition of miR-200c rescued the effects of lncRNA-ATB knockdown on cell viability and apoptosis induction in cholangiocarcinoma cell lines (HuCCT1 and RBE) (Lin et al., 2019). Our study demonstrated that inhibition of miR-200c could counteract the inhibitory effects of lncRNA-ATB silencing on migratory, proliferative, and trastuzumab-resistant properties. MiR-200c can suppress tumor progression in various types of cancer (Mutlu et al., 2016); elevated levels of miR-200c inhibit metastasis and drug resistance by regulating EMT in breast cancer cells (Bai et al., 2014; Liu et al., 2018). In fact, overexpression of miR-200c-3p has been shown to overcome cisplatin resistance in human gastric cancer cells by downregulating the expression of ERCC3 and ERCC4 genes (Li et al., 2019). Compared to NCI-N87 cells, NCI-N87-TR cells, characterized by high expression of EMT markers, exhibited increased migration and invasion, were sensitized to trastuzumab, and exhibited a decrease in the malignant properties upon miR-200c upregulation, which was attributed to the suppression of ZEB1 and ZEB2 (Zhou et al., 2018). Overall, these findings highlight the crucial role of the lncRNA-ATB/miR-200c axis in modulating gastric cancer progression and trastuzumab resistance.
We found that ZNF217 was inversely targeted by miR-200c, and ZNF217 knockdown inhibited migratory, proliferative, and trastuzumab resistance. ZNF217 is a direct downstream target of miR-200c and is required for miR-200c-mediated inhibition of EMT transition in breast cancer cells (Bai et al., 2014). ZNF217 elevation in vitro could promote the malignant properties of colorectal cancer and ovarian clear cell carcinoma cells and facilitate in primary and distant metastasis (Li et al., 2014). Silencing of ZNF217 increases trastuzumab sensitivity and suppresses the invasive capacity of breast cancer cells (Bai et al., 2014). Suppressed tumorigenesis and malignancy in colon cancer have been found to be offset by the restoration of ZNF217 (Zhang et al., 2015). More importantly, the expression of ZNF217 is enhanced in gastric cancer tissues and cells, which can facilitate the progression of gastric cancer (Qiang and Li, 2021). The current study also shows that lncRNA-ATB can increase the expression of ZNF217 by binding to miR-200c. In addition, restoration of ZNF217 expression neutralized lncRNA-ATB-depletion-induced suppressive effects on gastric cancer progression and trastuzumab resistance. Similarly, studies have underlined that lncRNAs can function as miRNA sponges, and thus limit their regulatory effects on the target mRNAs (Entezari et al., 2022; Fang and Xia, 2022; Lin et al., 2022b; Shetty et al., 2022). Much in line with the current results, previously published studies have indicated that lncRNA-ATB contributes to the promotion of trastuzumab resistance and invasion in breast cancer by competitively binding to miR-200c and upregulating ZNF217 (Shi et al., 2015). These findings highlight the role of the lncRNA-ATB/miR-200c/ZNF217 axis in the progression and trastuzumab resistance of gastric cancer.
To conclude, this study reveals that lncRNA-ATB serves as a key modulator of the miR-200c-ZNF217 axis, wherein lncRNA-ATB can bind to miR-200c and thus increase the expression of ZNF217, the miR-200c target. This mechanism promotes the progression and trastuzumab resistance of gastric cancer. The findings unravel the mechanism behind trastuzumab resistance in gastric cancer. Taken together, our data indicate that lncRNA-ATB can be a new effective cancer therapy target against drug resistance of gastric cancer. However, further investigations are required to focus on the key regulator mediating the upregulation of lncRNA-ATB during trastuzumab resistance in gastric cancer to provide a promising way for its treatment.
Acknowledgement: None.
Funding Statement: This study was supported by grants from the Basic Scientific Research Fund for Heilongjiang Provincial Universities in 2018 (2018-KYYWF-0105).
Author Contributions: Jiazhuang Li and Wei Zhang designed the study and performed the experiments, Shoubao Gao and Li Sun collected the data, Qingyang Tai and Ying Liu analyzed the data, and Jiazhuang Li prepared the manuscript. All authors read and approved the final manuscript.
Availability of Data and Materials: The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflict of interest to report regarding the present study.
References
Bai WD, Ye XM, Zhang MY, Zhu HY, Xi WJ et al. (2014). MiR-200c suppresses TGF-β signaling and counteracts trastuzumab resistance and metastasis by targeting ZNF217 and ZEB1 in breast cancer. International Journal of Cancer 135: 1356–1368. https://doi.org/10.1002/ijc.28782 [Google Scholar] [PubMed] [CrossRef]
Bang YJ, Van Cutsem E, Feyereislova A, Chung HC, Shen L et al. (2010). Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGAA phase 3, open-label, randomised controlled trial. Lancet 376: 687–697. https://doi.org/10.1016/S0140-6736(10)61121-X [Google Scholar] [PubMed] [CrossRef]
Chen Y, Wei G, Xia H, Tang Q, Bi F (2018). Long noncoding RNA-ATB promotes cell proliferation, migration and invasion in gastric cancer. Molecular Medicine Reports 17(11940–1946. https://doi.org/10.3892/mmr.2017.8077 [Google Scholar] [PubMed] [CrossRef]
Entezari M, Taheriazam A, Orouei S, Fallah S, Sanaei A et al. (2022). LncRNA-miRNA axis in tumor progression and therapy response: An emphasis on molecular interactions and therapeutic interventions. Biomedicine & Pharmacotherapy 154: 113609. https://doi.org/10.1016/j.biopha.2022.113609 [Google Scholar] [PubMed] [CrossRef]
Fang W, Xia Y (2022). LncRNA HLA-F-AS1 attenuates the ovarian cancer development by targeting miR-21-3p/PEG3 axis. Anticancer Drugs 33: 671–681. https://doi.org/10.1097/CAD.0000000000001288 [Google Scholar] [PubMed] [CrossRef]
Ghafouri-Fard S, Taheri M (2020). Long non-coding RNA signature in gastric cancer. Experimental and Molecular Pathology 113: 104365. https://doi.org/10.1016/j.yexmp.2019.104365 [Google Scholar] [PubMed] [CrossRef]
Gomez-Martín C, Lopez-Rios F, Aparicio J, Barriuso J, García-Carbonero R et al. (2014). A critical review of HER2-positive gastric cancer evaluation and treatment: From trastuzumab, and beyond. Cancer Letters 351: 30–40. https://doi.org/10.1016/j.canlet.2014.05.019 [Google Scholar] [PubMed] [CrossRef]
Gugnoni M, Ciarrocchi A (2019). Long noncoding RNA and epithelial mesenchymal transition in cancer. International Journal of Molecular Sciences 20: 1924. https://doi.org/10.3390/ijms20081924 [Google Scholar] [PubMed] [CrossRef]
Guo R, Zhang D, Zhang C, Yang Y, Liu H, Yang Y, Lin J (2021). Immunotoxin IHP25-BT with low immunogenicity and off-target toxicity inhibits the growth and metastasis of trastuzumab-resistant tumor cells. International Journal of Pharmaceutics 608: 121081. https://doi.org/10.1016/j.ijpharm.2021.121081 [Google Scholar] [PubMed] [CrossRef]
Jang SY, Kim G, Park SY, Lee YR, Kwon SH et al. (2017). Clinical significance of lncRNA-ATB expression in human hepatocellular carcinoma. Oncotarget 8: 78588–78597. https://doi.org/10.18632/oncotarget.21094 [Google Scholar] [PubMed] [CrossRef]
Kinami S, Saito H, Takamura H (2021). Significance of lymph node metastasis in the treatment of gastric cancer and current challenges in determining the extent of metastasis. Frontiers in Oncology 11: 806162. https://doi.org/10.3389/fonc.2021.806162 [Google Scholar] [PubMed] [CrossRef]
Kurata A, Yamada M, Ohno SI, Inoue S, Hashimoto H, Fujita K, Takanashi M, Kuroda M (2018). Expression level of microRNA-200c is associated with cell morphology in vitro and histological differentiation through regulation of ZEB1/2 and E-cadherin in gastric carcinoma. Oncology Reports 39: 91–100. https://doi.org/10.3892/or.2017.6093 [Google Scholar] [PubMed] [CrossRef]
Lei K, Liang X, Gao Y, Xu B, Xu Y, Li Y, Tao Y, Shi W, Liu J (2017). Lnc-ATB contributes to gastric cancer growth through a MiR-141-3p/TGFβ2 feedback loop. Biochemical and Biophysical Research Communications 484: 514–521. https://doi.org/10.1016/j.bbrc.2017.01.094 [Google Scholar] [PubMed] [CrossRef]
Li M, Gao M, Xie X, Zhang Y, Ning J, Liu P, Gu K (2019). MicroRNA-200c reverses drug resistance of human gastric cancer cells by targeting regulation of the NER-ERCC3/4 pathway. Oncology Letters 18: 145–152. https://doi.org/10.3892/ol.2019.10304 [Google Scholar] [PubMed] [CrossRef]
Li J, Meng H, Bai Y, Wang K (2016). Regulation of lncRNA and its role in cancer metastasis. Oncology Reports 23: 205–217. https://doi.org/10.3727/096504016x14549667334007 [Google Scholar] [PubMed] [CrossRef]
Li J, Song L, Qiu Y, Yin A, Zhong M (2014). ZNF217 is associated with poor prognosis and enhances proliferation and metastasis in ovarian cancer. International Journal of Clinical and Experimental Pathology 7: 3038–3047. [Google Scholar] [PubMed]
Lin Z, Wang R, Zhou Y, Wang Q, Yang CY, Hao BC, Ke CF (2022a). Prediction of distant metastasis and survival prediction of gastric cancer patients with metastasis to the liver, lung, bone, and brain: Research based on the SEER database. Annals of Translational Medicine 10: 16. https://doi.org/10.21037/atm-21-6295 [Google Scholar] [PubMed] [CrossRef]
Lin H, Yang L, Tian F, Nie S, Zhou H, Liu J, Chen W (2019). Up-regulated LncRNA-ATB regulates the growth and metastasis of cholangiocarcinoma via miR-200c signals. OncoTargets and Therapy 12: 7561–7571. https://doi.org/10.2147/ott.S217676 [Google Scholar] [PubMed] [CrossRef]
Lin X, Zhuang S, Chen X, Du J, Zhong L et al. (2022b). lncRNA ITGB8-AS1 functions as a ceRNA to promote colorectal cancer growth and migration through integrin-mediated focal adhesion signaling. Molecular Therapy 30: 688–702. https://doi.org/10.1016/j.ymthe.2021.08.011 [Google Scholar] [PubMed] [CrossRef]
Liu B, Du R, Zhou L, Xu J, Chen S et al. (2018). miR-200c/141 regulates breast cancer stem cell heterogeneity via targeting HIPK1/β-catenin axis. Theranostics 8: 5801–5813. https://doi.org/10.7150/thno.29380 [Google Scholar] [PubMed] [CrossRef]
Liu Y, Li Y, Xu Q, Yao W, Wu Q, Yuan J, Yan W, Xu T, Ji X, Chunhui N (2017). Long non-coding RNA-ATB promotes EMT during silica-induced pulmonary fibrosis by competitively binding miR-200c. Biochimica et Biophysica Acta (BBA)-Molecular Basis of Disease 1864: 420–431. https://doi.org/10.1016/j.bbadis.2017.11.003 [Google Scholar] [PubMed] [CrossRef]
Luo X, Wang J, Wei X, Wang S, Wang A (2020). Knockdown of lncRNA MFI2-AS1 inhibits lipopolysaccharide-induced osteoarthritis progression by miR-130a-3p/TCF4. Life Sciences 240: 117019. https://doi.org/10.1016/j.lfs.2019.117019 [Google Scholar] [PubMed] [CrossRef]
Mutlu M, Raza U, Saatci Ö, Eyüpoğlu E, Yurdusev E, Şahin Ö (2016). miR-200c: A versatile watchdog in cancer progression, EMT, and drug resistance. Journal of Molecular Medicine 94: 629–644. https://doi.org/10.1007/s00109-016-1420-5 [Google Scholar] [PubMed] [CrossRef]
Qiang F, Li J (2021). CircCSNK1G1 contributes to the tumorigenesis of gastric cancer by sponging miR-758 and regulating ZNF217 expression. Cancer Management and Research 13: 5027–5038. https://doi.org/10.2147/CMAR.S305767 [Google Scholar] [PubMed] [CrossRef]
Roviello G, Catalano M, Iannone LF, Marano L, Brugia M, Rossi G, Aprile G, Antonuzzo L (2022). Current status and future perspectives in HER2 positive advanced gastric cancer. Clinical and Translational Oncology 24: 981–996. https://doi.org/10.1007/s12094-021-02760-0 [Google Scholar] [PubMed] [CrossRef]
Saito T, Kurashige J, Nambara S, Komatsu H, Hirata H et al. (2015). A long non-coding RNA activated by transforming growth factor-β is an independent prognostic marker of gastric cancer. Annals of Surgical Oncology 22: S915–S922. https://doi.org/10.1245/s10434-015-4554-8 [Google Scholar] [PubMed] [CrossRef]
Shen X, Piao L, Zhang S, Cui Y, Cui Y, et al. (2018). Long non-coding RNA activated by TGF-β expression in cancer prognosis: A meta-analysis. International Journal of Surgery 58: 37–45. https://doi.org/10.1016/j.ijsu.2018.08.004 [Google Scholar] [PubMed] [CrossRef]
Shetty A, Venkatesh T, Kabbekodu SP, Tsutsumi R, Suresh PS (2022). LncRNA-miRNA-mRNA regulatory axes in endometrial cancer: A comprehensive overview. Archives of Gynecology and Obstetrics 306: 1431–1447. https://doi.org/10.1007/s00404-022-06423-5 [Google Scholar] [PubMed] [CrossRef]
Shi SJ, Wang LJ, Yu B, Li YH, Jin Y, Bai XZ (2015). LncRNA-ATB promotes trastuzumab resistance and invasion-metastasis cascade in breast cancer. Oncotarget 6: 11652–11663. https://doi.org/10.18632/oncotarget.3457 [Google Scholar] [PubMed] [CrossRef]
Ye DM, Xu G, Ma W, Li Y, Luo W, Xiao Y, Liu Y, Zhang Z (2020). Significant function and research progress of biomarkers in gastric cancer. Oncology Letters 19: 17–29. https://doi.org/10.3892/ol.2019.11078 [Google Scholar] [PubMed] [CrossRef]
Zhang ZC, Zheng LQ, Pan LJ, Guo JX, Yang GS (2015). ZNF217 is overexpressed and enhances cell migration and invasion in colorectal carcinoma. Asian Pacific Journal of Cancer Prevention 16: 2459–2463. https://doi.org/10.7314/APJCP.2015.16.6.2459 [Google Scholar] [PubMed] [CrossRef]
Zhou X, Men X, Zhao R, Han J, Fan Z et al. (2018). miR-200c inhibits TGF-β-induced-EMT to restore trastuzumab sensitivity by targeting ZEB1 and ZEB2 in gastric cancer. Cancer Gene Therapy 25: 68–76. https://doi.org/10.1038/s41417-017-0005-y [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools