 Open Access
Open Access
REVIEW
Cellular and molecular insights into microbiota-mitochondria interplay, therapeutic biomarkers and interventional approaches in COVID-19: A review
1 Institute of Pharmaceutical Research, GLA University, Mathura, India
2 Department of Pharmaceutical Sciences, Maharshi Dayanand University, Rohtak, India
* Corresponding Authors: VIBHAV VARSHNEY. Email: ; AHSAS GOYAL. Email:
BIOCELL 2023, 47(10), 2141-2149. https://doi.org/10.32604/biocell.2023.030853
Received 04 May 2023; Accepted 12 July 2023; Issue published 08 November 2023
Abstract
The persistent global pandemic, COVID-19, stems from the pathogenic influence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), yielding an unprecedented worldwide crisis. With reference to a WHO report, the count of COVID-19 cases had exceeded 754 million by February 03, 2023. Intriguingly, emerging research has spotlighted the intricate interplay of gut microbiota and mitochondrial entities, acting as potent immunomodulatory factors at the cellular and molecular levels. This interconnection operates through a series of dynamic mechanisms. SARS-CoV-2 infection perturbs the delicate equilibrium of gut microbiota, leading to dysbiosis—a signature biomarker. This imbalance is intrinsically linked to exacerbated COVID-19 progression. Mechanistically, this microbial dysbiosis triggers aberrant immune responses, marked by cytokine storms, while also inducing mitochondrial dysfunction. Mitochondrial activities crucial to energy production and immunoregulation are compromised. This dual perturbation of microbiota and mitochondria contributes to disease severity. This review provides a comprehensive overview of these interconnected mechanisms, illuminating how microbiota-mitochondria interplay serves as both a diagnostic biomarker and a promising therapeutic target in the realm of COVID-19.Keywords
SARS-CoV-2 (formerly known as 2019-nCoV) virus is the cause of the highly contagious COVID-19 respiratory disease. In late 2019, the virus was first reported in China and rapidly spread worldwide through close human interactions to become a pandemic (Hu et al., 2021). The worldwide economy, societies, and people are all highly affected by COVID-19. Over 220 million people were infected in the COVID-19 pandemic, and over 4.5 million have died since September 2021 (Mehta et al., 2021). The pandemic has also disrupted healthcare systems, caused widespread economic hardship, and led to social and political unrest in some areas. Gastrointestinal symptoms such as diarrhea, nausea, and vomiting are observed in many COVID-19 patients (Mehta et al., 2021). The human intestinal microbiota contains over 1500 microbial species that are distributed in hundreds of phyla (Mazzarelli et al., 2022). The microbiota is composed of trillions of microbes, including not only bacteria but also fungi, archaea, viruses, or protozoans. It is an extremely diverse ecosystem inhabited by a variety of microbes that colonize specific locations (De and Dutta, 2022). The gut microbiota (GM) has a lot of important functions in humans, which include the development of the innate and adaptive immune system, maintenance of immunological tolerance, protection against bacterial infections by colonizing mucosal surfaces, the creation of various antibacterial agents, and sustaining healthy and normal gut physiology (Belkaid and Hand, 2014). Further, the microbiota plays a crucial role in digestion and metabolism, while controlling epithelial cell differentiation and proliferation, altering insulin resistance as well as increasing secretion, thus affecting brain-body communication and hence the psychological and neurological functions of the host (Nagata et al., 2023). The GM dysregulation affects the health of the host in many ways, such as the gut-brain axis, and the absorption of energy, short-chain fatty acids (SCFAs), choline, bile acids, to name a few. Nevertheless, there is still a long way to go before we fully understand the mechanisms associated with GM in terms of diseases in humans (Gomaa, 2020; Chen et al., 2021).
The mitochondrion is a double-membrane organelle that performs a variety of functions, including steroid hormone and heme production, calcium and iron buffering, reactive oxygen species (ROS) signaling, and energy supply, and is involved in cell death and survival signaling (Srinivasan et al., 2021). The mitochondria, cellular metabolic generators, are crucial in regulating how cells proliferate, die, and maintain their redox state (Handy and Loscalzo, 2012). Cardiolipin and mitochondrial DNA (mtDNA) entry into the matrix of the cell and out of the cell could function as damage-associated molecular patterns (DAMPs), which might elicit an innate inflammatory response (Grazioli and Pugin, 2018).
In mammalian cells, the mitochondrion is a substantial generator of ROS. Therefore, the cytokine storm observed in COVID-19 includes the mitochondrion (Alfarouk et al., 2021). Many research investigations have demonstrated how impaired mitochondria affect the immunological outcome. For instance, according to a recent study, it was reported that during COVID-19, the pro-inflammatory mediators interleukin (IL)-6 and IL-12 and several chemokines such as chemokine (C-X-C motif) ligand (CXCL)-20, chemokine (C-C motif) ligand (CCL)3, CCL8, and CCL4 were produced at elevated levels in human alveolar epithelial cells due to dysfunctional mitochondria (Wang et al., 2019; Burtscher et al., 2020; Chen et al., 2020). All these cytokines were found to promote hyperinflammation. The production of tumor necrosis factor (TNF-α) and IL-1β results from the enormous rise in ROS that also causes such hyper-inflammation (Costela-Ruiz et al., 2020) The human microbiota is primarily located in the respiratory and gastrointestinal tracts (GIT), which is also a prime site to get SARS-CoV-2 infection given the high expression of angiotensin-converting enzyme 2 (ACE-2) and type 2 transmembrane protease serine (TMPRSS2), which mediate the viral entry into a host (Ni et al., 2020; Wiersinga et al., 2020). There is growing evidence that the hyper-inflammatory condition, i.e., a “cytokine storm,” which causes significant systemic disturbances, is responsible for the accelerated progression of COVID-19 disease (Norsa et al., 2020; Chaves Andrade et al., 2020).
Microbiota: A Key Player in the Mitigation of COVID-19
The term “Gut microbiome” is a group of more than 1014 types of microbes found in the GIT, including bacteria, fungi, viruses, and eukaryotes (Zhang et al., 2022). Commensal microbiota modulates the innate immune response and helps prevent respiratory infections by providing an anti-viral response before viral infection; this phenomenon is known as “colonization resistance” (Wang et al., 2022). Gut microbiota performs various functions, one of which is to increase the resistance to infections by influencing type-1 interferon receptors located in respiratory epithelial cells, which respond promptly to viral infection via INF-α and INF-Secretin. Further, in CD4+, CD8+, and T-lymphocytes, (Jandhyala, 2015), the gut microbiota increases the expression of pro-IL-1β, pro-IL-18, and NLRP3 and suppresses that of IFN-YRI and MHC-1 molecules (de Oliveira et al., 2021). The health and nutrition of the gut flora are also essential (Valdes et al., 2018). They can metabolize indigestible substances like dietary fiber and endogenous intestinal mucus, which results in the formation of short-chain fatty acids (SCFA) such as metabolites like butyrate, acetate, and propionate and gas (Liu et al., 2022a). These metabolites maintain the integrity of the intestinal epithelial barrier (Chen et al., 2021).
Interestingly, a study conducted in the Chinese province of Guangdong revealed that COVID-19 suppresses the diversity of this commensal microbiota. To put it another way, opportunistic microorganisms proliferate at a predominant level while in contrast, the abundance of beneficial microbiota decreases (González et al., 2021; Wang et al., 2022). By downregulation of the immunomodulatory activity of the commensal microbiota, which favors dissemination and severity, these opportunistic microbiotas promote hyperinflammation via cytokine storm, often called systemic inflammatory response syndrome (SIRS) or inflammatory factor storm. This leads to microbiota dysbiosis, increased inflammation and GIT symptoms (Chen et al., 2021).
SARS-CoV-2 enters the epidermal cell by binding to ACE-2 and TMPRSS-2 surface receptors present in an epidermal cell, where the virus has undergone self-replication (Yeoh et al., 2021) This lead to the production and release of a large amount of viral proteins or formation of oligomers (processes for replication to utilize small building blocks to generate protein oligomers that assemble in multiple ways, thereby diversifying protein function and regulation) from infected epithelial cells. The innate cells (macrophages and dendritic cells) bind to pathogen associate molecular patterns (PAMP) via pattern-recognition receptors (PRR), which can be found associated with subcellular compartments, such as the cellular and endosomal membranes, the cytosol. Further, RIG-1 like receptor (RLR), Toll-like receptor (TLR), and NOD-like receptor (NLR) in secreted forms extracellularly present in the bloodstream and interstitial fluids recognize distinct microbial components and directly activate immune cells (Medzhitov and Janeway, 1997; Niles et al., 2021; Wicherska-Pawłowska et al., 2021; Zhang et al., 2022). The activation of these receptors triggers the release of proinflammatory factors, including interferon (IFN), interferon-stimulated gene (ISG), and intracellular inflammatory factors such as IL-6, IL-1β, TNF-α, and IFN. (Zuo et al., 2020; Rocchi et al., 2022). As a result, infected cells are targeted by a “suicide attack,” which inadvertently causes collateral damage to healthy cells and tissue. This damage contributes to increased vascular permeability, disrupted circulation, and increased susceptibility to opportunistic pathogen invasion. Additionally, danger-associated molecular patterns (DAMP) activate the RLR and NLR receptors, further amplifying the production of pro-inflammatory factors (Belančić, 2020; Ngo and Gewirtz, 2021). On the luminal surface of murine intestinal epithelial cells, B°AT1 or SLC6A19, a sodium-dependent neutral amino acid transporter, aids in the formation of tight junction (TJ) that reduces pro-inflammatory cytokine storm and modulates mucosal cell autophagy via the mechanistic Target of Rapamycin (mTOR) signaling pathway (Fig. 1). However, when a SARS-CoV-2 infection occurs, the ACE-2 and B°AT1 receptor chaperones co-internalize and the total level of B°AT1 on the luminal surface is downregulated, which facilitates the microbiota dysbiosis and enhances the cytokine storm (Wang et al., 2022).
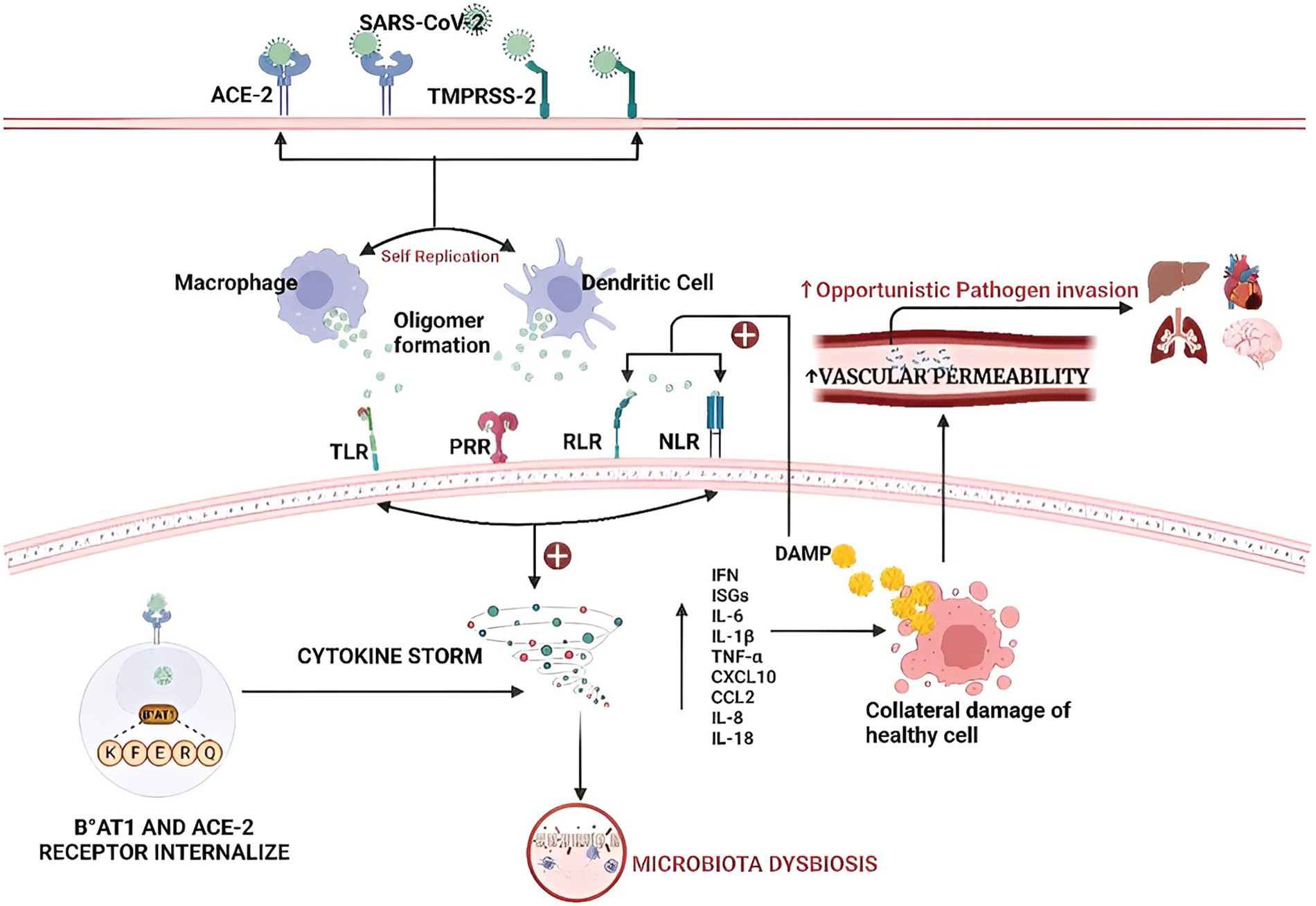
Figure 1: Microbiota dysbiosis in the presence of coronavirus: Invasion of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) via angiotensin-converting enzyme 2 (ACE-2) and type 2 transmembrane protease serine (TMPRSS2) surface receptors present in the epidermal cells. Further, it leads to the formation of oligomers in dendritic cells and macrophages. Further, it activates the RIG-1-like receptor (RLR), Toll-like receptor (TLR), and NOD-like receptor (NLR) resulting in increased production of inflammatory factors that cause the imbalance in microbiota and are responsible for the death of neighboring healthy cells and increased vascular permeability. Abbriviations: SARS-CoV-2: Severe acute respiratory syndrome coronavirus 2; ACE-2: angiotensin-converting enzyme 2; TMPRSS2: Type 2 transmembrane protease serine; RLR: RIG-1-like receptor; TLR: Toll-like receptor; NLR: NOD-like receptor; B°AT1: Sodium-dependent neutral amino acid transporter; IFN: Interferon; ISG: Interferon-stimulated gene; IL: Interleukin; TNF-α: Tumor necrosis factor α; CXCL: Chemokine (C-X-C motif) ligand; CCL: Chemokine (C-C motif) ligand.
Alteration of Mitochondrial Function in SARS-CoV-2 Infection
The mitochondrial antiviral signaling protein MAVS is an important part of the body’s antiviral defense mechanism, which is anchored to the mitochondrial membrane. In contrast, the RLR receptor stimulates MAVS to form oligomers when the SARS-CoV-2 virus enters the cytoplasm. Subsequently, E3 ligase, TNF, tumor necrosis factor receptor–associated factor (TRAF)3, and TRAF6 are among the downstream signaling effectors recruited by MAVS to induce significant amounts of antiviral defense by activating the nuclear factor-κB (NF-κB) pathway (Scott, 2010; Park, 2018; Burtscher et al., 2020). Viruses like the hepatitis C virus, the serine protease NS3/A4, the hepatitis A virus 3ABC protease, and now corona viruses target this pathway by cleaving MAVS and removing it from the mitochondrial outer membrane. This inhibits the pathway and further lowers the cellular capacity of cells to trigger a viral response (Burtscher et al., 2020; Bhowal et al., 2022).
Cells must maintain the precise level of ROS and RNS since they are necessary for signaling (Kaundal et al., 2021). The mitochondria are the major source of cellular ROS, the concentration of which is directly proportional to the activity of the electron transport chain, etc., Alfarouk et al., 2021). A report stated that SARS-CoV-2 triggered COVID-19 hyperinflammation and resulted in mitochondrial redox imbalance (Saleh et al., 2020). The invasion of SARS-CoV-2 infection enhances a series of subsequent events in cells expressing ACE-2 and TMPRSS2. These downstream events activate the NOD-like receptor protein 3 (NLRP-3)-mediated inflammasome signaling pathway, which enhances ROS development, activates the electron transport chain, and increases mt-ROS-a production, resulting in mitochondrial DNA damage (Srinivasan et al., 2021; Chen et al., 2023). Additionally, SARS-CoV-2 infects mitochondria and forms double-membrane vesicles that destabilize the integrity of the mitochondrial membrane. As a result, cardiolipin and mitochondrial DNA are liberated into the cytosol through the mitochondrial membrane, acting as DAMPs. This increases chemokine activity and upregulates the expression of pro-inflammatory genes like TNF-α, IL-6, and TNF-α (Sharma and Sarode, 2022). Subsequently, they switch to a glycolytic metabolism with impaired mitochondria that produce mtROS and further recruit hypoxia-inducible factor-1-α (HIF-1α) (Bhowal et al., 2022). This further activates the TLR-9 and NF-κB pathways, increasing the production of inflammatory cytokines (TNF-α, IL-α), which change the dynamics of mitochondria and, ultimately, induce cell death and the progression of COVID-19 disease (Ganji and Reddy, 2021). Additionally, the inflammasome is also formed by the recognition of PAMPs receptors by bacterial toxins, flagellin proteins, muramyl dipeptide, viral or bacterial RNA and DNA, fungal fragments, fungal mannan, zymosan, and protozoa-derived hemozoin. Various classes of patterns recognition receptors (PRRs), including Toll-like receptors (TLR), NOD-like receptors (NLR), C-type lectin receptors (CLR), and Rig-1 Like receptors (RLR) recognize distinct microbial components and directly activate immune cells (Wicherska-Pawłowska et al., 2021). The TLR and NLR pathways generate downstream signaling and activate the NF-κB pathway and an inflammatory response to elicit pro-inflammatory cytokines and accelerate mitochondrial ROS production (Li and Chang, 2021). TLR activation also decreases the expression of the uncoupling protein-2 (UCP-2). Further, it increases etc. activity, which further increases mitochondrial ROS production, and stimulation of the TLR pathway. This induces TRAF6 protein that translocates to mitochondria and associates subsequently with ECSIT (evolutionarily conserved signaling intermediate in Toll) (Saint-Georges-Chaumet and Edeas, 2016; Pierelli et al., 2017). This protein promotes mitochondrial etc. assembly, accelerates ROS production, and exacerbates inflammation and COVID-19 disease progression (Valdés-Aguayo et al., 2021). The mitochondrial dysbiosis in the presence of coronavirus has been illustrated in Fig. 2.
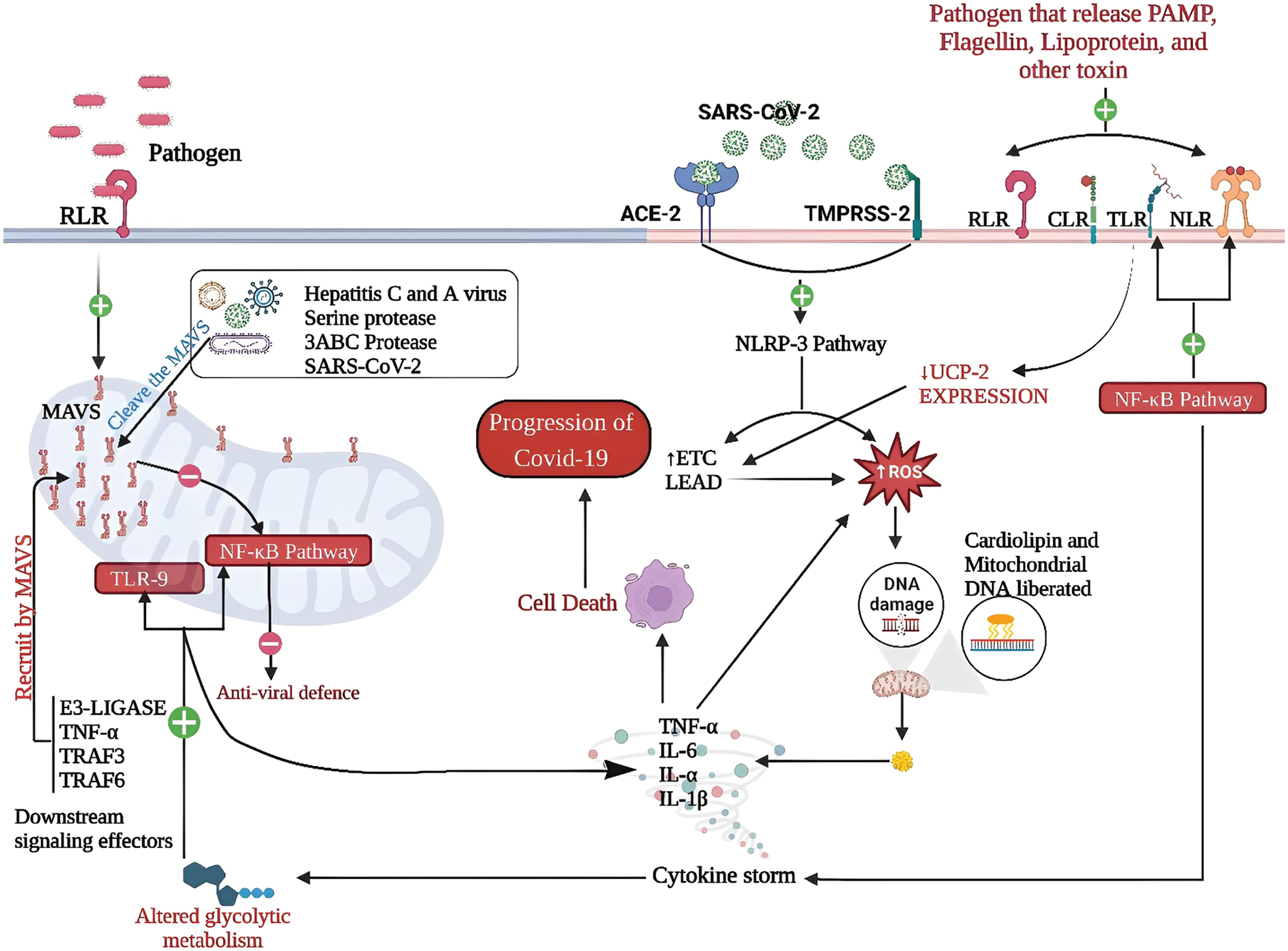
Figure 2: Mitochondrial dysbiosis in the presence of coronavirus: The mitochondrial protein MAVS anchors the membrane, defending against viruses. RLR receptor activation prompts MAVS oligomerization. MAVS recruits E3 ligase, TNF, TRAF3, and TRAF6 to activate NF-κB, bolstering antiviral defenses. However, hepatitis C and hepatitis A viruses cleave MAVS, disrupting the response. SARS-CoV-2 infection via ACE-2 and TMPRSS2 activates NLRP-3, causing mitochondrial redox imbalance and DNA damage. Cardiolipin and mitochondrial DNA release into the cytosol act as DAMPs, increasing inflammation. Altered mitochondrial dynamics, elevated mtROS, and TLR/NF-κB activation induce cell death and worsen COVID-19. The recognition of PAMPs by TLRs and NLRs triggers NF-κB, inflammation, and mitochondrial ROS. Decreased UCP-2 and increased, etc. activity worsen mitochondrial ROS, amplifying inflammation and COVID-19 severity.
Abbriviations: SARS-CoV-2: Severe acute respiratory syndrome coronavirus 2; ACE-2: angiotensin-converting enzyme 2; TMPRSS2: Type 2 transmembrane protease serine; RLR: RIG-1-like receptor; TLR: Toll-like receptor; NLR: NOD-like receptor; IL: Interleukin; TNF-α: Tumor necrosis factor α; TRAF: Tumor necrosis factor receptor–associated factor; NLRP: NOD-like receptor protein; UCP: Uncoupling protein; etc.: Electron transport chain; PAMP: Pathogen associate molecular patterns; ROS: Reactive oxygen speices; MAVS: Mitochondrial antiviral signaling protein; NF-κB: Nuclear factor-κB.
Microbiota-Mitochondria Inter-Talk
The metabolic functions of nutrients are similar in both the microbiome and mitochondria. In addition, they have many structural and functional similarities and several proteins that regulate related or even the same metabolic processes (Fehér et al., 2022). According to recent findings, the microbiota interacts with the host cell, particularly by interacting with the mitochondria and microbial metabolites that also affect mitochondrial function and biogenesis (Zhu et al., 2022). Further, the mitochondria are responsible for influencing the intestinal functional effector cells, such as immune cells, enterochromaffin cells, and epithelial cells. This, in turn, influences the composition and activity of gut microbiota due to their ability to trigger an innate immune response when bacterial infection or cellular damage is detected (Clark and Mach, 2017). Certain gut microbiota compositions have also been associated with polymorphism of mitochondrial genes, including the nds and cytb genes or the D-Loop area in the mitochondrial genome (Franco-Obregón and Gilbert, 2017).
The mitochondria can alter the immune response and lead to a rise in inflammation when infected with bacterial or viral pathogens, resulting in microbiota dysbiosis (Saleh et al., 2020). In addition, beneficial gut microbiota products, including SCFA, are present in the feces, and secondary bile acid may influence mitochondrial function relating to the production of energy, mitochondria biogenesis, redox balance, and inflammatory cascade (Clark and Mach, 2017). SCFAs (such as butyrate, acetate, and succinate) promote mitochondrial activity and neuroimmune regulation in health and disease. Additionally, SCFA is known to activate the enzyme AMP kinase, which is responsible for mitochondrial genesis (Hu et al., 2020; Shandilya et al., 2022).
Microbiota- Mitochondria: Therapeutic Biomarkers to Prevent and Treat COVID-19
The gut microbiome plays a key role in immunomodulation. The microbiota may suggest strategies for controlling the gut microbiome to treat, prevent, or at least reduce the impact of COVID-19. These include therapies like fecal microbiota transplantation (FMT), probiotics, prebiotics, and the direct use of bacterial components (Hussain et al., 2021).
Probiotics show an immunomodulating function in the cytokine storm—IL-6, IL-1β, IL-15, IL-17 IFN-γ, and TNF-α (Singh and Rao, 2021). The Lactobacillus family of probiotic microorganisms alters the immune response to protect an individual against viral respiratory infections. In order to shield the host from viral respiratory infections, certain strains of lactic bacteria may alter the immune response. The clinical investigator efficacy of Lactobacillus brevis CD2 and BLIS K12 in preventing secondary bacterial pneumonia in patients with serious COVID-19 is being evaluated in a Phase II randomized clinical trial (ClinicalTrials.gov Identifier: NCT05175833). Further, the effectiveness of the probiotic Lactobacillus rhamnosus GG is being evaluated in a randomized trial which is conducted at Duke University Hospital in preventing COVID-19 transmission and the onset of symptoms in exposed household contacts (ClinicalTrials.gov Identifier No. NCT04399252) (Sundararaman et al., 2020; Wang et al., 2022). Additionally, some of the randomized controlled trials registered on ClinicalTrials.gov investigating the application of probiotics for the prevention/ treatment of COVID-19 are underway or in the recruiting stage (Brahma et al., 2022). It has also been reported in clinical studies that oral administration of L. plantarum increases cytotoxic CD8 (+) T cells, augments granulocyte-induced phagocytosis, and increases the anti-inflammatory cytokines (IL-4 and IL-10) by reducing the formation of proinflammatory cytokines (IFN-γ and TNF-α) (Nayebi et al., 2022). Interestingly, empirical evidence of the antiviral activity of certain probiotic stains against other coronaviruses has already been established. Thus, the advantage of probiotics for COVID-19 prevention and adjunctive therapy could be considered in the future as mentioned in Table 1 (Santacroce et al., 2021).
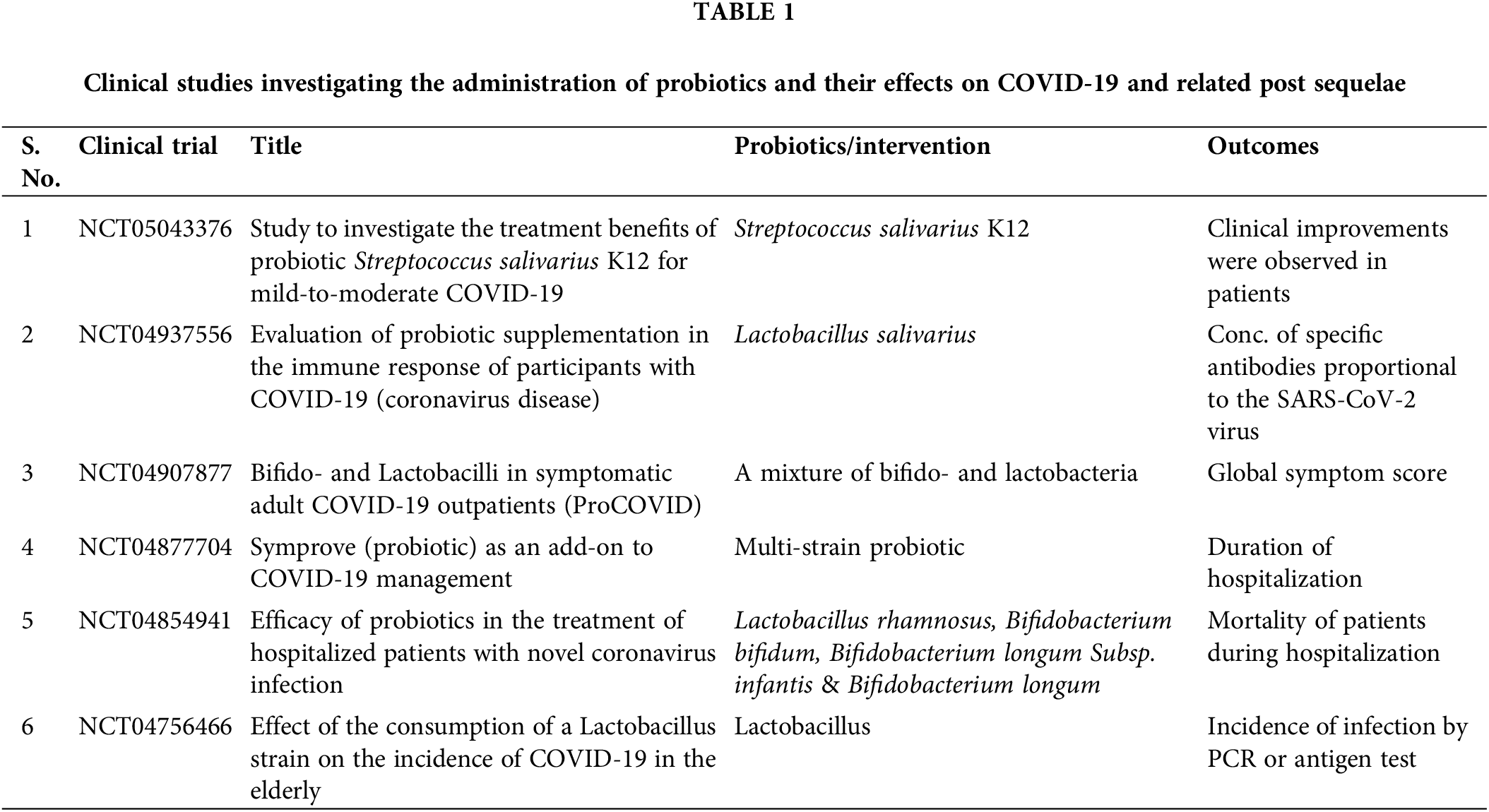
Tocilizumab, an anti-IL-6 monoclonal antibody effective in the treatment of arthritis is being studied in clinical trials for patients affected with SARS-CoV-2 (Castelnovo et al., 2021). Several studies demonstrated that post-FMT decreased the number of IL-6 and other inflammatory cytokines. For example, the microbial ecosystem that can produce IL-10 and support the proliferation of regulatory T cells was restored in a patient with inflammatory bowel disease (IBD) receiving FMT (Lopez and Grinspan, 2016; Kazemian et al., 2021). In general, these data support the use of FMT in seriously ill COVID-19 individuals, either as a treatment or an adjuvant to suppress cytokine storms (Nejadghaderi et al., 2021). Recently, research has been conducted to confirm that FMT is effective as an immunomodulatory risk factor for the progression of COVID-19 disease related to increased cytokine storms and inflammation (ClinicalTrials.gov Identifier No. NCT04824222) (Wang et al., 2022).
Prebiotics are substances found in food that induce beneficial bacteria to grow or function in the gastrointestinal tract (Davani-Davari et al., 2019). Prebiotics may affect COVID-19 by proliferation and survival of probiotic bacteria, enhancing gut diversity of microbial flora and immunity among an adult population (Olaimat et al., 2020). These prebiotics promote the SCFAs-producing microbial gut population, leading to improved inflammatory response and the immune system. This is because the immune responses are known to be controlled by SCFAs (Markowiak-Kopeć and Śliżewska, 2020). It is well documented that SCFAs, particularly butyrate can strengthen the gut barrier function and thus reduce the inflammatory response. Compared to FMT, treatment with prebiotics and probiotics is safer and more effective in terms of their preparation and administration (Cristofori et al., 2021).
Characterization of COVID-19 microbiota in the airway plays an important role in developing strategies for the management of the pathogenesis of COVID-19 (Liu et al., 2022b). Physicians recommend probiotics or prebiotics to restore the gut flora, which act as anti-inflammatory immunomodulators to improve mitochondrial health. Useful therapeutic strategies can be used to specifically target to improve mitochondrial health or reduce the generation of ROS, which was increased during the impaired mitochondrial metabolic pathways. The circulating cell-free mitochondria in human blood are an intriguing novel pathway with promising potential as a biomarker, therapy target, and therapeutic agent (Olaimat et al., 2020; Stier, 2021).
The interplay between the microbiota and mitochondria has been recognized as an important factor in various physiological processes, including the immune system and its responses. In the context of COVID-19, it has been suggested that dysregulation of this interplay may contribute to disease severity. Recent research has shown that changes in the gut microbiota may be linked to mitochondrial dysfunction, which could potentially contribute to COVID-19 pathogenesis. Additionally, mitochondrial dysfunction has been implicated in developing acute respiratory distress syndrome (ARDS), a common complication of severe COVID-19. Therapeutic biomarkers and interventional approaches targeting the microbiota-mitochondria axis may offer potential strategies for treating COVID-19. For example, probiotics and prebiotics have been presented to have a beneficial effect on the gut microbiota and may therefore help to mitigate COVID-19-associated dysbiosis. In addition, interventions aimed at improving mitochondrial function, such as using mitochondrial-targeted antioxidants, may also be beneficial in treating COVID-19. In conclusion, the microbiota-mitochondria interplay plays a crucial role in COVID-19 pathogenesis. However, further research is needed to understand the mechanisms underlying this relationship completely and to develop targeted therapeutic interventions that can improve patient outcomes. To summarize, the potential of microbiota and mitochondrial-targeted interventions offers promising avenues for managing COVID-19.
Acknowledgement: The authors are thankful to BioRender for providing the platform to create professional science figures.
Funding Statement: The authors received no specific funding for this study.
Author Contributions: The manuscript’s conception and design were created by Ahsas Goyal and Vibhav Varshney. The first draft preparation and data curation were carried out by Prashant Singh Kushwah and Neetu Agrawal. Editing and reviewing the final text were done by Govind Singh. All authors approved the final version of the manuscript.
Availability of Data and Materials: Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
Alfarouk KO, Alhoufie STS, Hifny A, Schwartz L, Alqahtani AS et al. (2021). Of mitochondrion and COVID-19. Journal of Enzyme Inhibition and Medicinal Chemistry 36: 1258–1267. https://doi.org/10.1080/14756366.2021.1937144 [Google Scholar] [PubMed] [CrossRef]
Belančić A (2020). Gut microbiome dysbiosis and endotoxemia-Additional pathophysiological explanation for increased COVID-19 severity in obesity. Obesity Medicine 20: 100302. https://doi.org/10.1016/j.obmed.2020.100302 [Google Scholar] [PubMed] [CrossRef]
Belkaid Y, Hand TW (2014). Role of the microbiota in immunity and inflammation. Cell 157: 121–141. https://doi.org/10.1016/j.cell.2014.03.011 [Google Scholar] [PubMed] [CrossRef]
Bhowal C, Ghosh S, Ghatak D, De R (2022). Pathophysiological involvement of host mitochondria in SARS-CoV-2 infection that causes COVID-19: A comprehensive evidential insight. Molecular and Cellular Biochemistry 478: 1325–1343. https://doi.org/10.1007/s11010-022-04593-z [Google Scholar] [PubMed] [CrossRef]
Brahma S, Naik A, Lordan R (2022). Probiotics: A gut response to the COVID-19 pandemic but what does the evidence show? Clinical Nutrition ESPEN 51: 17–27. https://doi.org/10.1016/j.clnesp.2022.08.023 [Google Scholar] [PubMed] [CrossRef]
Burtscher J, Cappellano G, Omori A, Koshiba T, Millet GP (2020). Mitochondria: In the cross fire of SARS-CoV-2 and immunity. iScience 23: 101631. https://doi.org/10.1016/j.isci.2020.101631 [Google Scholar] [PubMed] [CrossRef]
Castelnovo L, Tamburello A, Lurati A, Zaccara E, Marrazza MG et al. (2021). Anti-IL6 treatment of serious COVID-19 disease. Medicine 100: e23582. https://doi.org/10.1097/MD.0000000000023582 [Google Scholar] [PubMed] [CrossRef]
Chaves Andrade M, Souza de Faria R, Avelino Mota Nobre S (2020). COVID-19: Can the symptomatic SARS-CoV-2 infection affect the homeostasis of the gut-brain-microbiota axis? Medical Hypotheses 144: 110206. https://doi.org/10.1016/j.mehy.2020.110206 [Google Scholar] [PubMed] [CrossRef]
Chen X, Chen R, Jin R, Huang Z (2020). The role of CXCL chemokine family in the development and progression of gastric cancer. International Journal of Clinical and Experimental Pathology 13: 484–492. [Google Scholar] [PubMed]
Chen Y, Ye X, Escames G, Lei W, Zhang X et al. (2023). The NLRP3 inflammasome: Contributions to inflammation-related diseases. Cellular & Molecular Biology Letters 28: 51. https://doi.org/10.1186/s11658-023-00462-9 [Google Scholar] [PubMed] [CrossRef]
Chen Y, Zhou J, Wang L (2021). Role and mechanism of gut microbiota in human disease. Frontiers in Cellular and Infection Microbiology 11: 625913. https://doi.org/10.3389/fcimb.2021.625913 [Google Scholar] [PubMed] [CrossRef]
Clark A, Mach N (2017). The crosstalk between the gut microbiota and mitochondria during exercise. Frontiers in Physiology 8: 319. https://doi.org/10.3389/fphys.2017.00319 [Google Scholar] [PubMed] [CrossRef]
Costela-Ruiz VJ, Illescas-Montes R, Puerta-Puerta JM, Ruiz C, Melguizo-Rodríguez L (2020). SARS-CoV-2 infection: The role of cytokines in COVID-19 disease. Cytokine & Growth Factor Reviews 54: 62–75. https://doi.org/10.1016/j.cytogfr.2020.06.001 [Google Scholar] [PubMed] [CrossRef]
Cristofori F, Dargenio VN, Dargenio C, Miniello VL, Barone M, Francavilla R (2021). Anti-inflammatory and immunomodulatory effects of probiotics in gut inflammation: A door to the body. Frontiers in Immunology 12: 23−30. https://doi.org/10.3389/fimmu.2021.578386 [Google Scholar] [PubMed] [CrossRef]
Davani-Davari D, Negahdaripour M, Karimzadeh I, Seifan M, Mohkam M, Masoumi S, Berenjian A, Ghasemi Y (2019). Prebiotics: Definition, types, sources, mechanisms, and clinical applications. Foods 8: 92. https://doi.org/10.3390/foods8030092 [Google Scholar] [PubMed] [CrossRef]
De R, Dutta S (2022). Role of the microbiome in the pathogenesis of COVID-19. Frontiers in Cellular and Infection Microbiology 12: 736397. https://doi.org/10.3389/fcimb.2022.736397 [Google Scholar] [PubMed] [CrossRef]
de Oliveira GLV, Oliveira CNS, Pinzan CF, de Salis LVV, de B. Cardoso CR (2021). Microbiota modulation of the gut-lung axis in COVID-19. Frontiers in Immunology 12: 635471. https://doi.org/10.3389/fimmu.2021.635471 [Google Scholar] [PubMed] [CrossRef]
Fehér J, Élő Á, István L, Nagy ZZ, Radák Z, Scuderi G, Artico M, Kovács I (2022). Microbiota mitochondria disorders as hubs for early age-related macular degeneration. Geroscience 44: 2623–2653. https://doi.org/10.1007/s11357-022-00620-5 [Google Scholar] [PubMed] [CrossRef]
Franco-Obregón A, Gilbert JA (2017). The microbiome-mitochondrion connection: Common ancestries, common mechanisms, common goals. mSystems 2: e00018–17. https://doi.org/10.1128/mSystems.00018-17 [Google Scholar] [PubMed] [CrossRef]
Ganji R, Reddy PH (2021). Impact of COVID-19 on mitochondrial-based immunity in aging and age-related diseases. Frontiers in Aging Neuroscience 12: 13541. https://doi.org/10.3389/fnagi.2020.614650 [Google Scholar] [PubMed] [CrossRef]
Gomaa EZ (2020). Human gut microbiota/microbiome in health and diseases: A review. Antonie Van Leeuwenhoek 113: 2019–2040. https://doi.org/10.1007/s10482-020-01474-7 [Google Scholar] [PubMed] [CrossRef]
González S, Selma-Royo M, Arboleya S, Martínez-Costa C, Solís G et al. (2021). Levels of predominant intestinal microorganisms in 1 month-old full-term babies and weight gain during the first year of life. Nutrients 13: 2412. https://doi.org/10.3390/nu13072412 [Google Scholar] [PubMed] [CrossRef]
Grazioli S, Pugin J (2018). Mitochondrial damage-associated molecular patterns: From inflammatory signaling to human diseases. Frontiers in Immunology 9: 832. https://doi.org/10.3389/fimmu.2018.00832 [Google Scholar] [PubMed] [CrossRef]
Handy DE, Loscalzo J (2012). Redox regulation of mitochondrial function. Antioxidants & Redox Signaling 16: 1323–1367. https://doi.org/10.1089/ars.2011.4123 [Google Scholar] [PubMed] [CrossRef]
Hu B, Guo H, Zhou P, Shi ZL (2021). Characteristics of SARS-CoV-2 and COVID-19. Nature Reviews. Microbiology 19: 141–154. https://doi.org/10.1038/s41579-020-00459-7 [Google Scholar] [PubMed] [CrossRef]
Hu S, Kuwabara R, de Haan BJ, Smink AM, de Vos P (2020). Acetate and butyrate improve β-cell metabolism and mitochondrial respiration under oxidative stress. International Journal of Molecular Sciences 21: 1542. https://doi.org/10.3390/ijms21041542 [Google Scholar] [PubMed] [CrossRef]
Hussain I, Cher GLY, Abid MA, Abid MB (2021). Role of gut microbiome in COVID-19: An insight into pathogenesis and therapeutic potential. Frontiers in Immunology 12: 765965. https://doi.org/10.3389/fimmu.2021.765965 [Google Scholar] [PubMed] [CrossRef]
Jandhyala SM (2015). Role of the normal gut microbiota. World Journal of Gastroenterology 21: 8787. https://doi.org/10.3748/wjg.v21.i29.8787 [Google Scholar] [PubMed] [CrossRef]
Kaundal RK, Kalvala AK, Kumar A (2021). Neurological implications of COVID-19: Role of redox imbalance and mitochondrial dysfunction. Molecular Neurobiology 58: 4575–4587. https://doi.org/10.1007/s12035-021-02412-y [Google Scholar] [PubMed] [CrossRef]
Kazemian N, Kao D, Pakpour S (2021). Fecal microbiota transplantation during and post-COVID-19 pandemic. International Journal of Molecular Sciences 22: 1–14. https://doi.org/10.3390/ijms22063004 [Google Scholar] [PubMed] [CrossRef]
Li P, Chang M (2021). Roles of PRR-mediated signaling pathways in the regulation of oxidative stress and inflammatory diseases. International Journal of Molecular Sciences 22: 7688. https://doi.org/10.3390/ijms22147688 [Google Scholar] [PubMed] [CrossRef]
Liu Q, Mak JWY, Su Q, Yeoh YK, Lui GCY et al. (2022a). Gut microbiota dynamics in a prospective cohort of patients with post-acute COVID-19 syndrome. Gut 71: 544–552. https://doi.org/10.1136/gutjnl-2021-325989 [Google Scholar] [PubMed] [CrossRef]
Liu TFD, Philippou E, Kolokotroni O, Siakallis G, Rahima K, Constantinou C (2022b). Gut and airway microbiota and their role in COVID-19 infection and pathogenesis: A scoping review. Infection 50: 815–847. https://doi.org/10.1007/s15010-021-01715-5 [Google Scholar] [PubMed] [CrossRef]
Lopez J, Grinspan A (2016). Fecal microbiota transplantation for inflammatory bowel disease. Gastroenterologia y Hepatologia 12: 374–379. [Google Scholar]
Markowiak-Kopeć P, Śliżewska K (2020). The effect of probiotics on the production of short-chain fatty acids by human intestinal microbiome. Nutrients 12: 1107. https://doi.org/10.3390/nu12041107 [Google Scholar] [PubMed] [CrossRef]
Mazzarelli A, Giancola ML, Fontana A, Piselli P, Binda E et al. (2022). Gut microbiota composition in COVID-19 hospitalized patients with mild or severe symptoms. Frontiers in Microbiology 13: 1049215. https://doi.org/10.3389/fmicb.2022.1049215 [Google Scholar] [PubMed] [CrossRef]
Medzhitov R, JanewayJr CA (1997). Innate immunity: Impact on the adaptive immune response. Current Opinion in Immunology 9: 4–9. https://doi.org/10.1016/S0952-7915(97)80152-5 [Google Scholar] [PubMed] [CrossRef]
Mehta OP, Bhandari P, Raut A, Kacimi SEO, Huy NT (2021). Coronavirus disease (COVID-19Comprehensive review of clinical presentation. Frontiers in Public Health 8: 582932. https://doi.org/10.3389/fpubh.2020.582932 [Google Scholar] [PubMed] [CrossRef]
Nagata N, Takeuchi T, Masuoka H, Aoki R, Ishikane M et al. (2023). Human gut microbiota and its metabolites impact immune responses in COVID-19 and its complications. Gastroenterology 164: 272–288. https://doi.org/10.1053/j.gastro.2022.09.024 [Google Scholar] [PubMed] [CrossRef]
Nayebi A, Navashenaq JG, Soleimani D, Nachvak SM (2022). Probiotic supplementation: A prospective approach in the treatment of COVID-19. Nutrition and Health 28: 163–175. https://doi.org/10.1177/02601060211049631 [Google Scholar] [PubMed] [CrossRef]
Nejadghaderi SA, Nazemalhosseini-Mojarad E, Asadzadeh Aghdaei H (2021). Fecal microbiota transplantation for COVID-19; a potential emerging treatment strategy. Medical Hypotheses 147: 110476. https://doi.org/10.1016/j.mehy.2020.110476 [Google Scholar] [PubMed] [CrossRef]
Ngo VL, Gewirtz AT (2021). Microbiota as a potentially-modifiable factor influencing COVID-19. Current Opinion in Virology 49: 21–26. https://doi.org/10.1016/j.coviro.2021.04.005 [Google Scholar] [PubMed] [CrossRef]
Ni W, Yang X, Yang D, Bao J, Li R et al. (2020). Role of angiotensin-converting enzyme 2 (ACE2) in COVID-19. Critical Care 24: 422. https://doi.org/10.1186/s13054-020-03120-0 [Google Scholar] [PubMed] [CrossRef]
Niles MA, Gogesch P, Kronhart S, Ortega Iannazzo S, Kochs G, Waibler Z, Anzaghe M (2021). Macrophages and dendritic cells are not the major source of pro-inflammatory cytokines upon SARS-CoV-2 infection. Frontiers in Immunology 12: 647824. https://doi.org/10.3389/fimmu.2021.647824 [Google Scholar] [PubMed] [CrossRef]
Norsa L, Indriolo A, Sansotta N, Cosimo P, Greco S, D’Antiga L (2020). Uneventful course in patients with inflammatory bowel disease during the severe acute respiratory syndrome coronavirus 2 outbreak in Northern Italy. Gastroenterology 159: 371–372. https://doi.org/10.1053/j.gastro.2020.03.062 [Google Scholar] [PubMed] [CrossRef]
Olaimat AN, Aolymat I, Al-Holy M, Ayyash M, Abu Ghoush M, Al-Nabulsi AA, Osaili T, Apostolopoulos V, Liu SQ, Shah NP (2020). The potential application of probiotics and prebiotics for the prevention and treatment of COVID-19. NPJ Science of Food 4: 1–7. https://doi.org/10.1038/s41538-020-00078-9 [Google Scholar] [CrossRef]
Park HH (2018). Structure of TRAF family: Current understanding of receptor recognition. Frontiers in Immunology 9: 1999. https://doi.org/10.3389/fimmu.2018.01999 [Google Scholar] [PubMed] [CrossRef]
Pierelli G, Stanzione R, Forte M, Migliarino S, Perelli M, Volpe M, Rubattu S (2017). Uncoupling protein 2: A key player and a potential therapeutic target in vascular diseases. Oxidative Medicine and Cellular Longevity 2017: 1–11. https://doi.org/10.1155/2017/7348372 [Google Scholar] [PubMed] [CrossRef]
Rocchi G, Giovanetti M, Benedetti F, Borsetti A, Ceccarelli G, Zella D, Altomare A, Ciccozzi M, Guarino MPL (2022). Gut microbiota and COVID-19: Potential implications for disease severity. Pathogens 11: 1050. https://doi.org/10.3390/pathogens11091050 [Google Scholar] [PubMed] [CrossRef]
Saint-Georges-Chaumet Y, Edeas M (2016). Microbiota-mitochondria inter-talk: Consequence for microbiota-host interaction. Pathogens and Disease 74: ftv096. https://doi.org/10.1093/femspd/ftv096 [Google Scholar] [PubMed] [CrossRef]
Saleh J, Peyssonnaux C, Singh KK, Edeas M (2020). Mitochondria and microbiota dysfunction in COVID-19 pathogenesis. Mitochondrion 54: 1–7. https://doi.org/10.1016/j.mito.2020.06.008 [Google Scholar] [PubMed] [CrossRef]
Santacroce L, Inchingolo F, Topi S, Del Prete R, di Cosola M, Charitos IA, Montagnani M (2021). Potential beneficial role of probiotics on the outcome of COVID-19 patients: An evolving perspective. Diabetes and Metabolic Syndrome: Clinical Research and Reviews 15: 295–301. https://doi.org/10.1016/j.dsx.2020.12.040 [Google Scholar] [PubMed] [CrossRef]
Scott I (2010). The role of mitochondria in the mammalian antiviral defense system. Mitochondrion 10: 316–320. https://doi.org/10.1016/j.mito.2010.02.005 [Google Scholar] [PubMed] [CrossRef]
Shandilya S, Kumar S, Kumar Jha N, Kumar Kesari K, Ruokolainen J (2022). Interplay of gut microbiota and oxidative stress: Perspective on neurodegeneration and neuroprotection. Journal of Advanced Research 38: 223–244. https://doi.org/10.1016/j.jare.2021.09.005 [Google Scholar] [PubMed] [CrossRef]
Sharma NK, Sarode SC (2022). Do compromised mitochondria aggravate severity and fatality by SARS-CoV-2? Current Medical Research and Opinion 38: 911–916. https://doi.org/10.1080/03007995.2022.2065140 [Google Scholar] [PubMed] [CrossRef]
Singh K, Rao A (2021). Probiotics: A potential immunomodulator in COVID-19 infection management. Nutrition Research 87: 1–12. https://doi.org/10.1016/j.nutres.2020.12.014 [Google Scholar] [PubMed] [CrossRef]
Srinivasan K, Pandey AK, Livingston A, Venkatesh S (2021). Roles of host mitochondria in the development of COVID-19 pathology: Could mitochondria be a potential therapeutic target? Molecular Biomedicine 2: 38. https://doi.org/10.1186/s43556-021-00060-1 [Google Scholar] [PubMed] [CrossRef]
Stier A (2021). Human blood contains circulating cell-free mitochondria, but are they really functional? American Journal of Physiology. Endocrinology and Metabolism 320: E859–E863. https://doi.org/10.1152/ajpendo.00054.2021 [Google Scholar] [PubMed] [CrossRef]
Sundararaman A, Ray M, Ravindra PV, Halami PM (2020). Role of probiotics to combat viral infections with emphasis on COVID-19. Applied Microbiology and Biotechnology 104: 8089–8104. https://doi.org/10.1007/s00253-020-10832-4 [Google Scholar] [PubMed] [CrossRef]
Valdes AM, Walter J, Segal E, Spector TD (2018). Role of the gut microbiota in nutrition and health. BMJ 361: 36–44. https://doi.org/10.1136/bmj.k2179 [Google Scholar] [PubMed] [CrossRef]
Valdés-Aguayo JJ, Garza-Veloz I, Badillo-Almaráz JI, Bernal-Silva S, Martínez-Vázquez MC et al. (2021). Mitochondria and mitochondrial DNA: Key elements in the pathogenesis and exacerbation of the inflammatory state caused by COVID-19. Medicina 57: 928. https://doi.org/10.3390/medicina57090928 [Google Scholar] [PubMed] [CrossRef]
Wang D, Yang L, Yu W, Wu Q, Lian J et al. (2019). Colorectal cancer cell-derived CCL20 recruits regulatory T cells to promote chemoresistance via FOXO1/CEBPB/NF-κB signaling. Journal for ImmunoTherapy of Cancer 7: 215. https://doi.org/10.1186/s40425-019-0701-2 [Google Scholar] [PubMed] [CrossRef]
Wang B, Zhang L, Wang Y, Dai T, Qin Z, Zhou F, Zhang L (2022). Alterations in microbiota of patients with COVID-19: Potential mechanisms and therapeutic interventions. Signal Transduction and Targeted Therapy 7: 143. https://doi.org/10.1038/s41392-022-00986-0 [Google Scholar] [PubMed] [CrossRef]
Wicherska-Pawłowska K, Wróbel T, Rybka J (2021). Toll-like receptors (TLRsNOD-like receptors (NLRsand RIG-I-like receptors (RLRs) in innate immunity. TLRs, NLRs, and RLRs ligands as immunotherapeutic agents for hematopoietic diseases. International Journal of Molecular Sciences 22: 13397. https://doi.org/10.3390/ijms222413397 [Google Scholar] [PubMed] [CrossRef]
Wiersinga WJ, Rhodes A, Cheng AC, Peacock SJ, Prescott HC (2020). Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19). JAMA 324: 782–793. https://doi.org/10.1001/jama.2020.12839 [Google Scholar] [PubMed] [CrossRef]
Yeoh YK, Zuo T, Lui GCY, Zhang F, Liu Q et al. (2021). Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut 70: 698–706. https://doi.org/10.1136/gutjnl-2020-323020 [Google Scholar] [PubMed] [CrossRef]
Zhang F, Lau RI, Liu Q, Su Q, Chan FKL, Ng SC (2022). Gut microbiota in COVID-19: Key microbial changes, potential mechanisms and clinical applications. Nature Reviews. Gastroenterology & Hepatology 20: 323–337. https://doi.org/10.1038/s41575-022-00698-4 [Google Scholar] [PubMed] [CrossRef]
Zhu Y, Li Y, Zhang Q, Song Y, Wang L, Zhu Z (2022). Interactions between intestinal microbiota and neural mitochondria: A new perspective on communicating pathway from gut to brain. Frontiers in Microbiology 13: 798917. https://doi.org/10.3389/fmicb.2022.798917 [Google Scholar] [PubMed] [CrossRef]
Zuo T, Zhang F, Lui GCY, Yeoh YK, Li AYL et al. (2020). Alterations in gut microbiota of patients with COVID-19 during time of hospitalization. Gastroenterology 159: 944.e8–955.e8. https://doi.org/10.1053/j.gastro.2020.05.048 [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools