 Open Access
Open Access
Identification of STAT5B as a biomarker for an early diagnosis of endometrial carcinoma
1 Chunliu Maternity Hospital District of Dalian Women and Children Medical Center, Dalian, 116033, China
2 Department of Obstetrics and Gynecology, The General Hospital of Northern Theater Command, Shenyang, 110016, China
3 Department of Gynecology, Shaanxi Provincial People’s Hospital, Xi’an, 710068, China
4 Department of Obstetrics and Gynecology, The Second Hospital of Dalian Medical University, Dalian, 116023, China
* Corresponding Author: XUELIAN CHEN. Email:
BIOCELL 2023, 47(10), 2283-2300. https://doi.org/10.32604/biocell.2023.030086
Received 23 March 2023; Accepted 19 June 2023; Issue published 08 November 2023
Abstract
Background: The late detection of endometrial carcinoma (EC) at an advanced stage often results in a poor patient prognosis. It is hence important to identify reliable biomarkers to facilitate early detection of EC. Signal transducer and activator of transcription (STAT) family members play an important role in several tumors, however, their impact on EC development and progression remains unclear. Methods: Machine learning methods were used to investigate the importance of STAT5B in EC. Results: Hence, we explored the UALCAN data mining platform and found that while STAT1 and STAT2 were upregulated, STAT5A, STAT5B, and STAT6 were downregulated in EC. This high expression of STAT5B and STAT6 predicted favorable clinical outcomes, whereas the increased expression of STAT1 and STAT2 predicted poor clinical outcomes. Subsequent pathway enrichment analysis revealed that the STAT family was mainly involved in apoptosis pathway activation, cell cycle disruption, and epithelial–mesenchymal transition. Drug sensitivity analysis demonstrated that STAT5A/5B expression was negatively correlated with drug resistance in EC. Further, the expression of STAT5B mRNA and protein was correlated with several clinicopathological characteristics. Tumor Immune Estimation Resource (TIMER) analysis revealed that STAT5B expression was positively correlated with the abundance of infiltrating CD8+ T cells and neutrophils while its copy number variation was associated with the overall immune cell infiltration. The data on the correlations between STAT5B expression and related genes in uterine corpus endometrial carcinoma (UCEC) in cBio Cancer Portal showed the closest correlation of STAT5B expression with that of KIAA0753 (also known as moonraker and OFIP), followed by COL27A1 in EC. Pathway enrichment analysis further showed that STAT5B-related genes were involved in the mitogen-activated protein kinase (MAPK) and Ras signaling pathways. Conclusion: Collectively, our findings provided new insights into the role of the STAT family in EC. It also highlighted new targets for future research on diagnostic and prognostic markers and STAT5B as a novel marker for drug sensitivity screening.Keywords
The incidence of EC has been reported to increase in recent years globally, especially in developed countries, and it has become a serious threat to the health of women (Crosbie et al., 2022). Currently, surgery supplemented with postoperative radiotherapy and chemotherapy is the main strategy for treating EC. Owing to challenges in early detection, many patients with EC are diagnosed at an advanced stage, which is also associated with a poor prognosis. Thus, there is an urgent need to explore biological indicators with high sensitivity and specificity for guiding early clinical diagnosis and treatment of EC.
Signal transducer and activator of transcription (STAT) is a family of transcription factors including seven main members, namely, STAT1, STAT2, STAT3, STAT4, STAT5A, STAT5B, and STAT6. These are widely expressed in different cells and tissues and perform diverse functions. Accumulating evidence points to the role of the STAT family in tumor proliferation, invasion, metastasis, immune escape, and drug resistance (Galimberti et al., 2022; Golus et al., 2022; Liu et al., 2021b; Mirzaei et al., 2021; Wong et al., 2022; Xin et al., 2021; Yang et al., 2022b; Zhu et al., 2023). However, the expression of STAT family members in EC and their clinical significance remain unclear, especially, since the role of STAT5B in EC has not yet been reported. Studies have demonstrated a relatively complex role of STAT5B in tumors. On one hand, STAT5B could influence tumor development directly by regulating the expressions of target genes related to the cell cycle (Hou et al., 2023), proliferation (Subramaniam et al., 2020), and apoptosis (Zhang et al., 2015). STAT5B could also promote tumor progression by inducing mitochondrial dysfunction, increasing intracellular reactive oxygen species (ROS) levels, and inducing DNA damage (Du et al., 2012). On the other hand, STAT5B also showed an anti-tumor function by participating in immune-related biological processes in specific microenvironments (Dong et al., 2019; Fujimura et al., 2017; Rao et al., 2020). This study analyzed the prognostic significance of STAT5B and associated signaling in EC.
This analysis provided evidence for the role of STATs in EC while alsooffering novel diagnostic and prognostic biomarkers and potential therapeutic targets. The current study was conducted in accordance with the STrengthening the REporting of Genetic Association Studies (STREGA) reporting checklist.
In this study, we systematically evaluated the relationships of the expression of STATs with the diagnosis and prognosis of UCEC employing integrated bioinformatics approaches based on public RNA-sequencing (RNA-seq) data from The Cancer Genome Atlas (TCGA), UALCAN, Sangerbox, HPA, and tools like Kaplan–Meier plotter, GSCALite, GeneMANIA, cBioPortal, Tumor Immune Estimation Resource (TIMER), Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis, and the Database for Annotation, Visualization and Integrated Discovery (DAVID). We also investigated genetic methylation and mutations in STATs, along with their biological functions and impacts on drug sensitivity and patient prognosis. Particularly, this study focused on STAT5B among all STATs, which showed the most significant differential expression in EC as compared to normal tissues. The potential pathological influence of STAT5B and its associations with clinical indicators, immune cell infiltration, biological processes of related genes, signaling pathways, and the ceRNA network were examined.
All the procedures involving human genes in this study were performed in accordance with the Declaration of Helsinki (as revised in 2013). UALCAN is a data mining platform that uses TCGA expression profile data from 31 cancer types to analyze gene expression and the correlation of the gene with multiple clinicopathological parameters (Chandrashekar et al., 2017). We utilized the Clinical Proteomic Tumor Analysis Consortium (CPTAC) database of UALCAN to analyze the protein levels of the STAT family in EC. The TCGA database of UALCAN was employed to analyze DNA methylation levels of the STAT family in EC.
Sangerbox is a platform with vast biological information and a variety of analytical and visualization tools, and convenient data download functions. We used this Sangerbox platform (http://vip.sangerbox.com/) to analyze the expression of STAT5B and its family in different tumor tissues and normal tissues.
The HPA database (http://www.proteinatlas.org/) is a protein database that uses various tissue-based technologies to derive the information between human tissue protein and cell distribution (Shi et al., 2023). We employed the HPA database to search for and extract the immunohistochemical profile of the STAT family factor in the EC tissue and the normal endometrial tissue from its database.
The Kaplan-Meier plotter (http://www.kmplot.com/) is an online bioinformatics tool, and we used it to explore the impact of STAT expression on the survival of UCEC patients (Hou et al., 2017).
GSCALite (http://bioinfo.life.hust.edu.cn/web/GSCALite/) is a web server used for the dynamic analysis and visualization of oncogenic genes related to drug sensitivity and cancer development and progression. GSCALite enables analyses of genomic variation and related pathway activity through its Pathway Activity module (Liu et al., 2018). Moreover, we analyzed the relationship between the expression of STAT family members and the sensitivity to 265 small molecules in the Genomics of Drug Sensitivity in Cancer database using drug sensitivity modules. The Single Nucleotide Variation module was also used to explore the mutation status of the STAT family members in EC. Furthermore, the miRNAs interacting with STATs were predicted using this platform.
GeneMANIA (http://www.genemania.org/) was used to study protein interactions among the STAT family members.
The Cancer Genome Study cBioPortal (https://www.cbioportal.org/) is a widely used data analysis method for analyzing and visualizing TCGA data (Reimer et al., 2021). We used this platform to analyze the data of 546 UCEC patients in the TCGA database and to assess the links between the expression of STAT5B and related genes in UCEC.
The correlation between STAT5B expression and immune cell infiltration in UCEC was explored using TIMER (http://cistrome.org/TIMER/) (Zhang et al., 2021). We employed the Wilcoxon rank-sum test to investigate the relationship between the copy number variation of STAT5B and the immune cell infiltration.
Functional enrichment analysis
We used the DAVID tool (https://david.ncifcrf.gov/) for functional enrichment analysis of STAT5B and its co-expressed genes. We further explored their related GO terms, namely biological process (BP), cellular component (CC), and molecular function (MF), and enriched KEGG pathways. GOTERM_BP_DIRECT, GOTERM_CC_DIRECT, GOTERM_MF_DIRECT, and KEGG_PATHWAY were selected as GO terms for the GO enrichment analysis.
Competitive endogenous RNA analysis
We used Starbase (http://starbase.sysu.edu.cn/) to study long noncoding RNA (lncRNA), microRNA (miRNA), competitive endogenous RNA (ceRNA), RNA-binding proteins, the mRNA interconnections from CLIP-Seq and to predict the functions of lncRNA and protein-coding genes (Kariuki et al., 2023). The mRNA, miRNA, and lncRNA sequencing datasets of UCEC patients were downloaded from the TCGA database. The binding correlations between miRNA and mRNA,miRNA and lncRNA were predicted by the StarBase database. We used the “Limma”, “reshape2”, “ggpubr” and “ggExtra” packages in R for correlation analysis while cytoscape was used for visualizing the results.
Statistical significance was set at a p-value of <0.05 unless otherwise stated. All the statistical analyses were conducted using SPSS (version 26.0; Chicago, IL, USA).
Supplementary figures were placed in results part.
STAT family mRNA expression level among different tumors and their normal tissues
The results of the Sangerbox analysis showed that compared to the normal tissues, STAT1 was highly expressed in most tumors, namely UCEC, BRCA, GBM, LGG, CESC, LUAD, ESCA, stomach and esophageal carcinoma (STES), COAD, PRAD, STAD, HNSC, LUSC, LIHC, SKCM, THCA, OV, PAAD, TGCT, UCS, and LAML. However, its expression was low in High-Risk Wilms Tumor (WT), ACC, and KICH. Further, STAT5A and STAT5B showed low expression in most tumors inclusive of UCEC, BRCA, CESC, LUAD, ESCA, COAD, COADREAD, PRAD, LUSC, WT, BLCA, THCA, OV, UCS, ACC, and KICH. However, they were overexpressed in GBM, LGG, STAD, PRAD, TGCT, UCS, ALL, LAML. Furthermore, STAT6 also showed low expression in most tumors such as LGG, UCEC, BRCA, CESC, LUAD, ESCA, STES, COAD, PRAD, LUSC, LIHC, WT, SKCM, BLCA, THCA, UCS, and ACC. It was overexpressed in KRIP, KIRC, PRAD, and LAML (p < 0.05). In addition, the expression of STAT2, STAT3, and STAT4 showed no significant differences between EC and normal endometrial tissues (Suppl. Fig. S1).
Differential expression of STATs in uterine corpus endometrial carcinoma
UALCAN analysis of the UCEC cohort data from the TCGA database showed that STAT1 (Fig. 1A, p = 1.62E-12) and STAT2 (Fig. 1B, p = 8.85E-04) were significantly overexpressed in UCEC tissues compared to the expression in healthy endometrial tissues. However, the expression of STAT5A (Fig. 1E, p = 3.11E-09), STAT5B (Fig. 1F, p = 2.67E-12), and STAT6 (Fig. 1G, p = 2.19E-10) was significantly downregulated. Further, STAT3 (Fig. 1C, p = 0.22) and STAT4 (Fig. 1D, p = 0.47) expression did not differ significantly in UCEC tissues from normal tissues. At the protein level, STAT1 (Fig. 2A, p = 4.14E-11), STAT2 (Fig. 2B, p = 5.68E-06), STAT3 (Fig. 2C, p = 2.89E-02), and STAT4 (Fig. 2D, p = 2.24E-06) were overexpressed in UCEC tissues compared to normal tissues. Further, the protein levels of STAT5A (Fig. 2E, p = 2.01E-47), STAT5B (Fig. 2F, p = 3.81E-26), and STAT6 (Fig. 2G, p = 2.38E-26) were significantly downregulated in UCEC tissues when compared to those in normal tissues. The immunohistochemical profiles of EC analyzed in the HPA database were basically consistent with the above results (Suppl. Fig. S2). Overall, our results showed a significant difference in the expression of the STAT family factor in EC. We subsequently analyzed whether the STAT family members were prognostic prediction factors for EC.
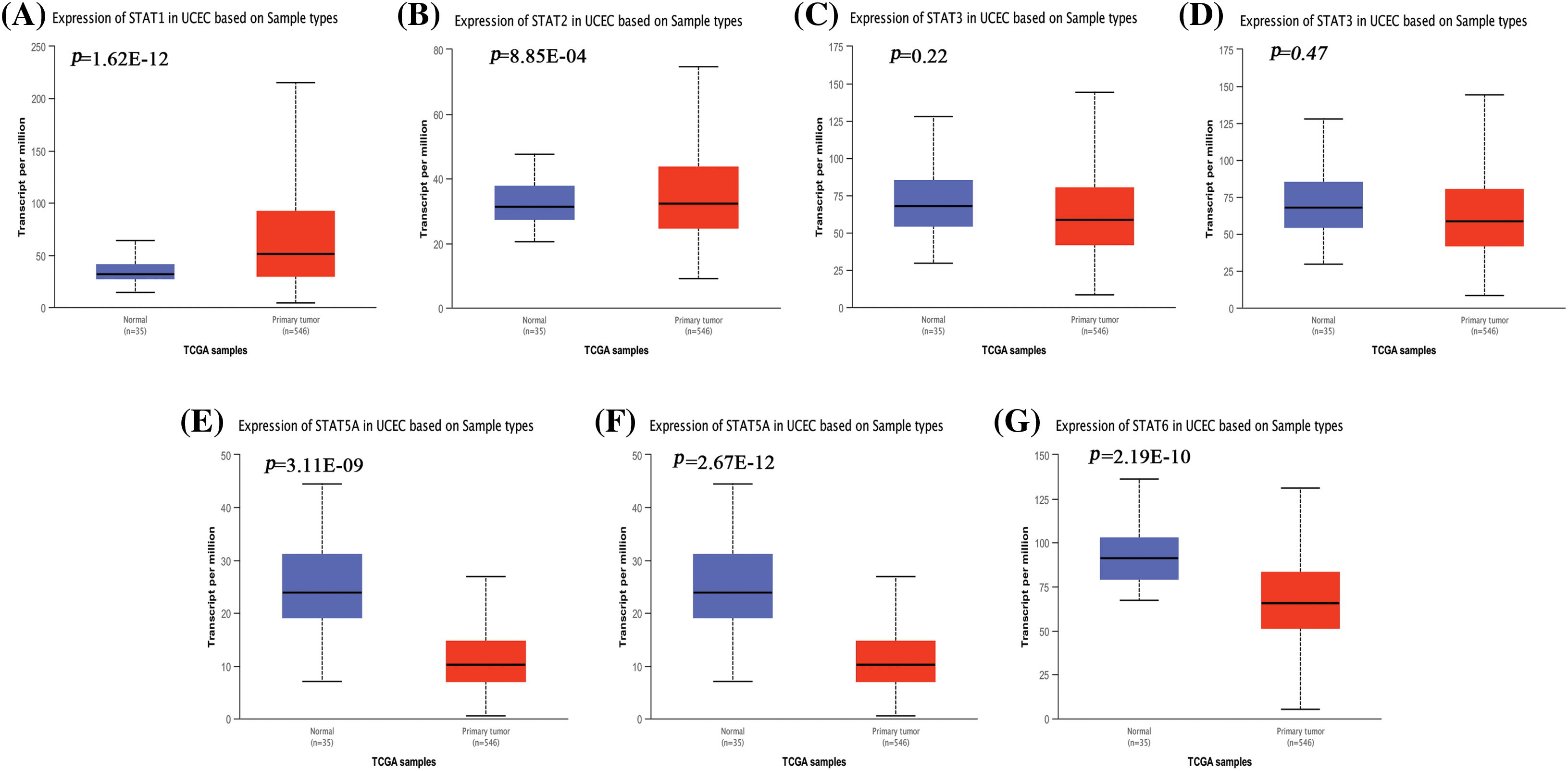
Figure 1: Differential gene expression analysis of STAT1 (A), STAT2 (B), STAT3 (C), STAT4 (D), STAT5A (E), STAT5B (F), and STAT6 (G) in UCEC and normal endometrial tissues from the TCGA database presented as box plots. STAT-signal transducer and activator of transcription; UCEC-uterine corpus endometrial carcinoma; TCGA-the cancer genomic atlas.
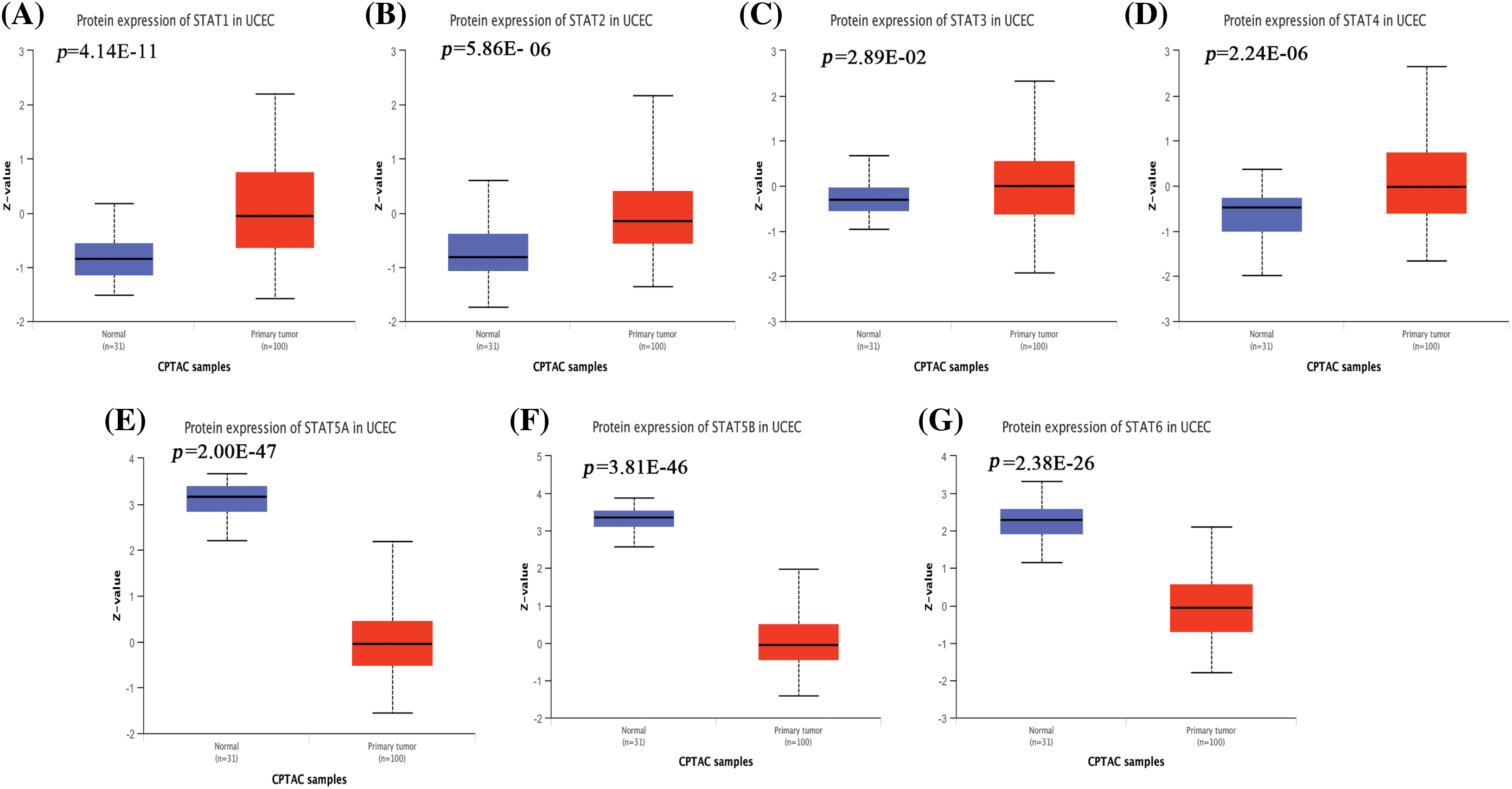
Figure 2: Protein levels of the STAT family in EC. Box plots show the expression levels of STAT1 (A), STAT2 (B), STAT3 (C), STAT4 (D), STAT5A (E), STAT5B (F), and STAT6 (G) in UCEC and normal tissues from the CPTAC database. EC-endometrial carcinoma; STAT-signal transducer and activator of transcription; UCEC-uterine corpus endometrial carcinoma; CPTAC-clinical proteomic tumor analysis consortium.
Prognostic value of the STAT family in uterine corpus endometrial carcinoma
The Kaplan–Meier plot analysis of the expression of STATs with the overall survival of UCEC patients showed that patients with overexpression of STAT1, STAT2, and STAT4 had a worse prognosis. However, the overexpression of STAT5B and STAT6 was associated with a better prognosis. Further, the expression of STAT3 and STAT5A had no effect on the prognosis of patients with UCEC (Fig. 3).
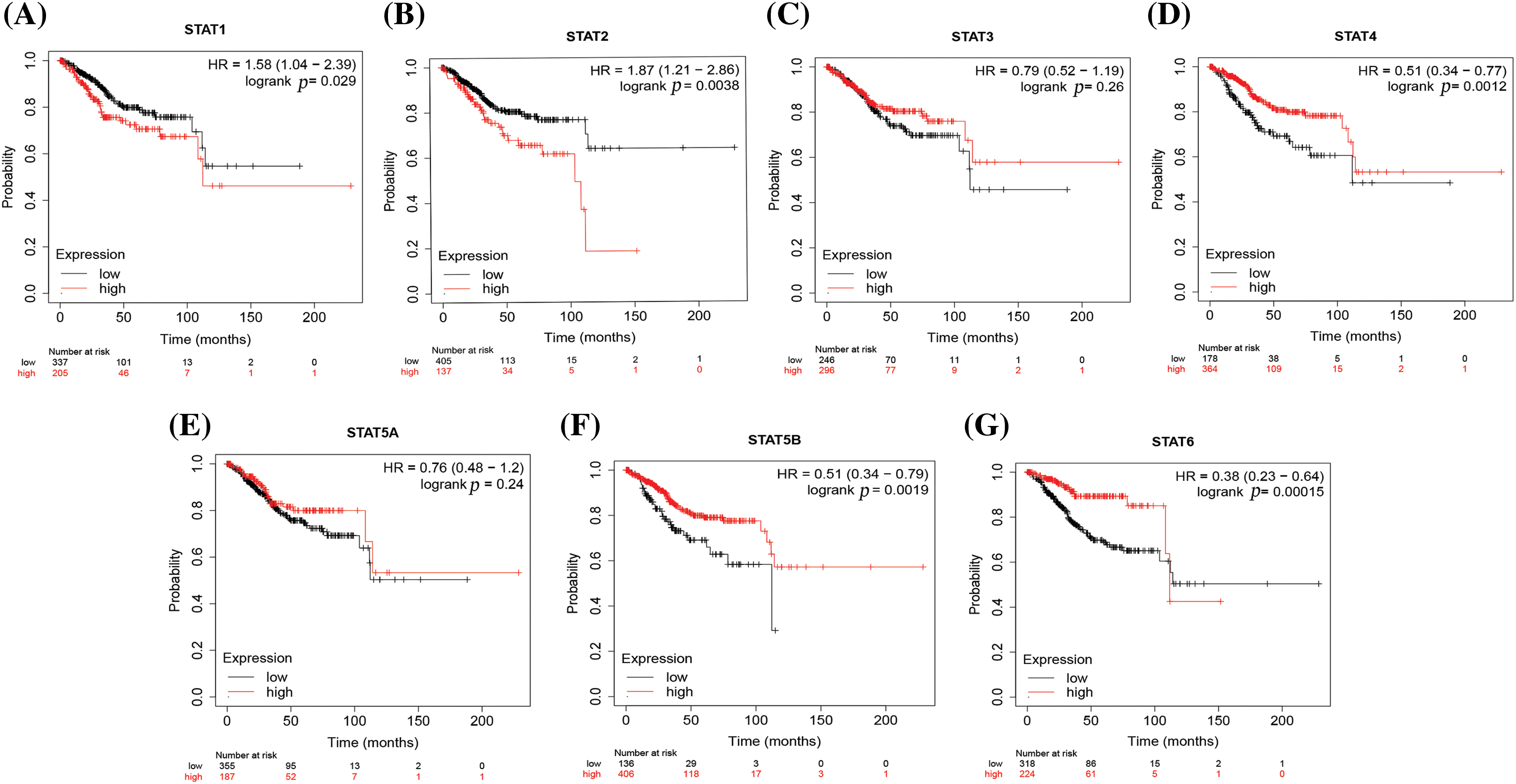
Figure 3: The prognostic value of STATs (in terms of the overall survival) in UCEC. STAT1 (A), STAT2 (B), STAT3 (C), STAT4 (D), STAT5A (E), STAT5B (F), and STAT6 (G); Key: STAT-signal transducer and activator of transcription; UCEC-uterine corpus endometrial carcinoma.
Effects of STAT mutations in uterine corpus endometrial carcinoma
Given that the expression of the STAT family of factors was significantly different in endometrial cancer and was related to the prognosis, we further analyzed the details of promoter methylation, gene mutation type, biological function, drug sensitivity, and related genes.
Methylation profile of STAT family genes
We used the Ualcan online tool to further study the extent of promoter methylation of STAT family genes analyzed in this study. The samples were divided into the normal group (n = 46) and the tumor group (n = 438). The β value of the vertical axis represented the level of methylation from 0 (non-methylation) to 1 (complete methylation). The levels of promoter methylation with STAT2 (Suppl. Fig. S3B, p = 0.03), STAT4 (Suppl. Fig. S3D, p = 1E-12), and STAT6 (Suppl. Fig. S3G, p = 0.02) were significantly downregulated in UCEC when compared to those in the normal group. However, the levels of promoter methylation for STAT5A (Suppl. Fig. S3E, p = 1.62E-12), and STAT5B (Suppl. Fig. S3F, p = 3.07E-10) were upregulated in the UCEC group. The differences between the groups were statistically significant (Suppl. Fig. S3).
Mutations in the STAT family members in uterine corpus endometrial carcinoma
The gene mutation analysis showed that the most common mutation type of the STAT family genes in UCEC patients was missense mutation, followed by nonsense mutation, in-frame deletion, multi-hit mutation, frameshift insertion, splice-site mutation, and frameshift deletion (Fig. 4). For the STAT family members, the mutation rate of STAT1, STAT2, STAT6, STAT5A, and STAT5B was 44%, 38%, 33%, 30%, and 20%, respectively.
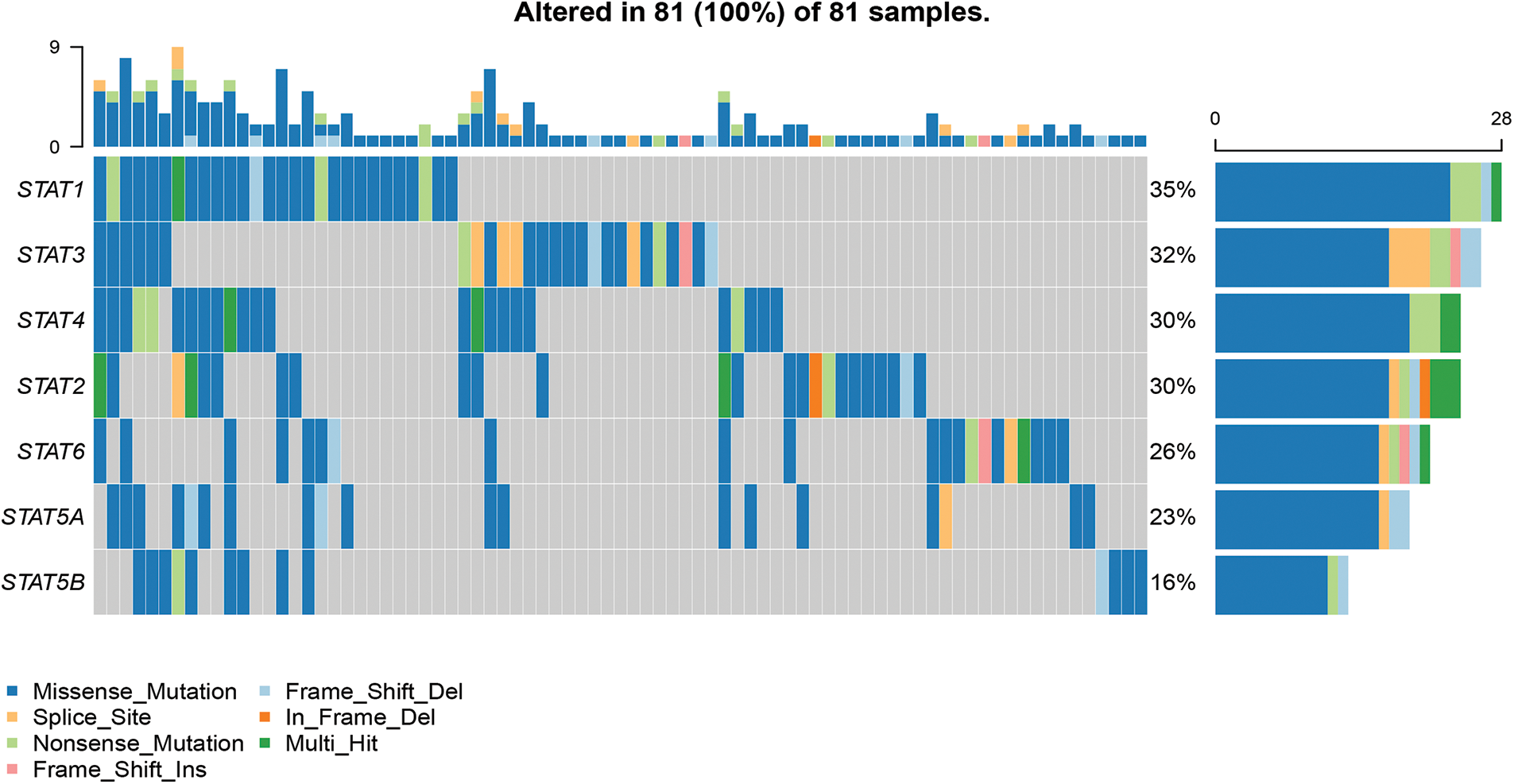
Figure 4: Mutations in the STAT family members in UCEC. The waterfall chart indicates the distribution and classification of mutation types in STATs in a total of 81 UCEC samples; Key: STAT-signal transducer and activator of transcription; UCEC-uterine corpus endometrial carcinoma.
Associations of cancer-related pathways and drug resistance of STATs in uterine corpus endometrial carcinoma
The main cancer-related activities of STATs were apoptotic pathway activation, cell cycle disruption, and induction of epithelial–mesenchymal transition (EMT) (Fig. 5A). Drug sensitivity analysis showed that the expression of STAT5A/5B was negatively related to drug resistance (Fig. 5B).
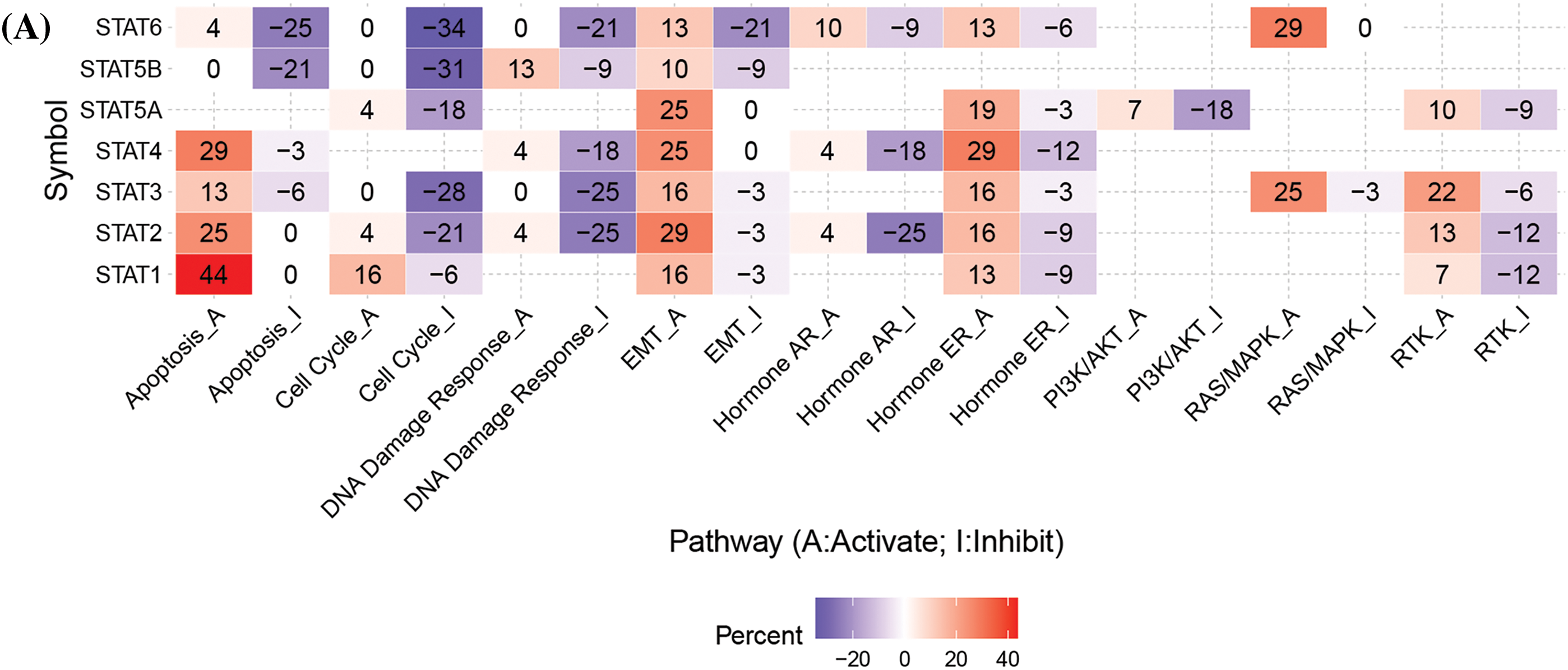
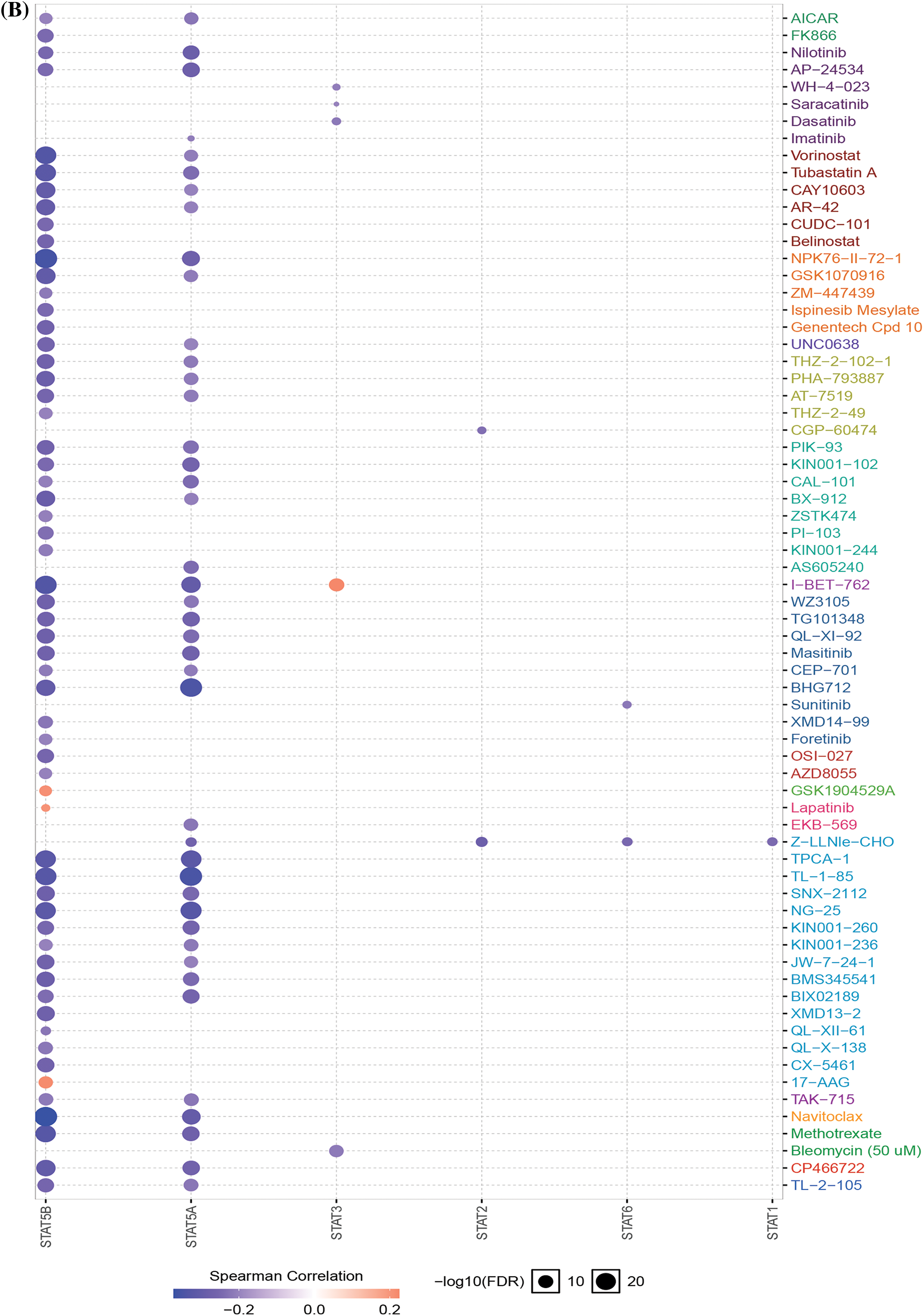
Figure 5: Cancer-related pathways and drug resistance analysis of STATs in UCEC. (A) Association between STATs and cancer-related activities. (B) The role of STATs in drug sensitivity; Key: STAT-signal transducer and activator of transcription; UCEC-uterine corpus endometrial carcinoma.
To explore the role of the STAT family in UCEC, we generated a network to identify the links between adjacent genes and STAT family members. As shown in Fig. 6A, a network diagram of proteins predicted to be able to interact with STATs was developed. GO analysis showed that the main functional processes associated with STAT-related genes included the stem cell factor (SCF)-SCF receptor (KIT) signaling pathways, transmembrane receptor protein tyrosine kinase signaling pathways, PID-KIT pathway, natural killer (NK) cell immune positive adjustment, neurotrophic factor signal transduction pathways, NK cell-mediated cytotoxicity, PID-PI3KCI pathways, antigen receptor-mediated signaling pathways, and lymphocyte differentiation-related processes (Fig. 6B).
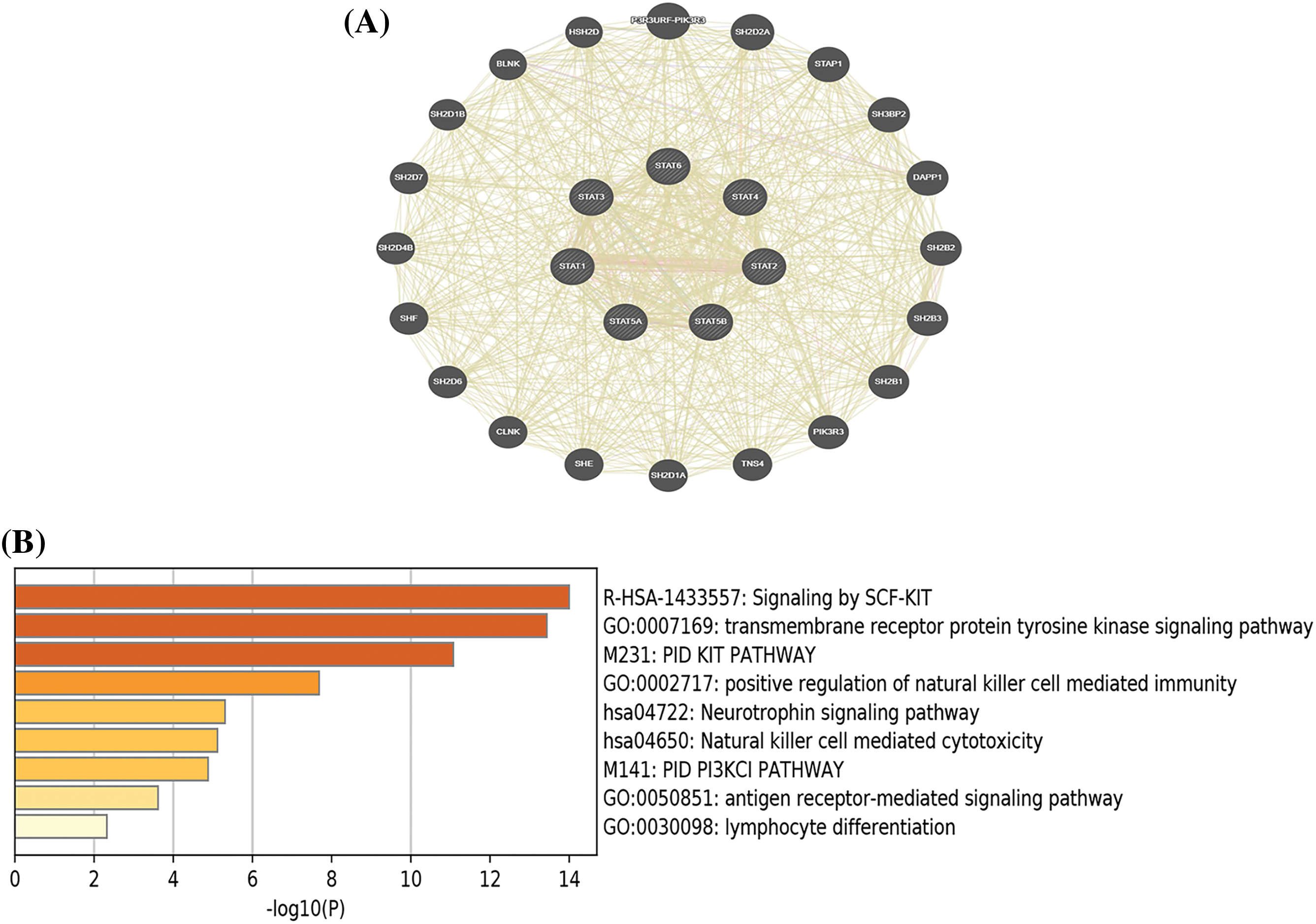
Figure 6: Network of STAT family-related genes and enrichment analysis. (A)Protein–Protein interaction network of the STAT family. (B) The top 10 enriched GO terms of STAT-related genes; Key: STAT-signal transducer and activator of transcription; GO-Gene ontology.
Associations between STAT5B expression and clinicopathological features of uterine corpus endometrial carcinoma patients
Given that an abnormal STAT5B expression showed the most significant association with UCEC prognosis, we further analyzed the correlations between STAT5B expression and clinicopathological indicators of UCEC patients. These features include body weight, age, tumor grade, cancer stage, menopausal status, tissue type, and TP53 mutation status. The results demonstrated that the mRNA levels (Fig. 7, from TCGA data) and protein levels (Fig. 8, from CPTAC data) of STAT5B were significantly correlated with all of these assessed clinicopathological indicators.
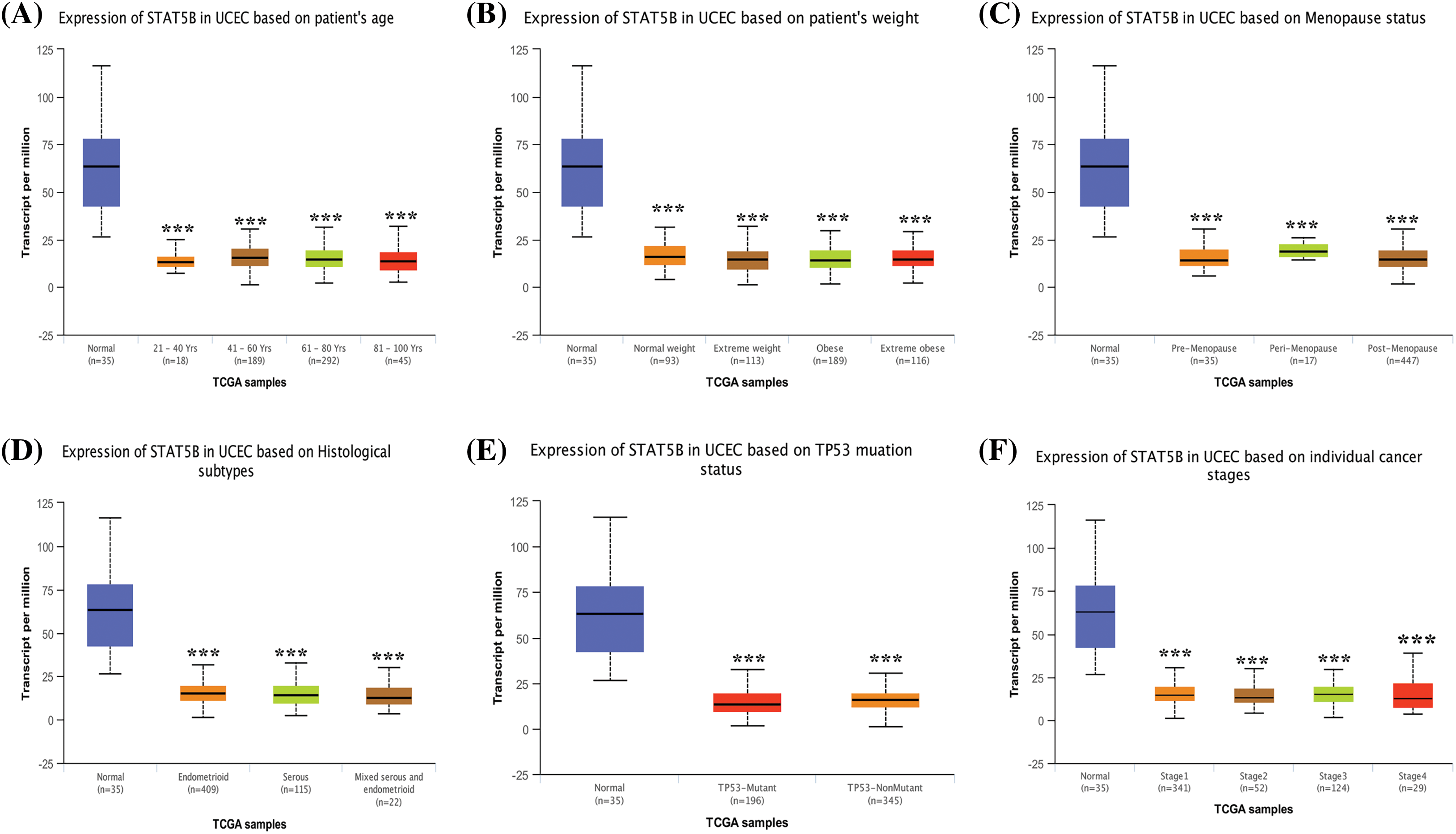
Figure 7: STAT5B mRNA expression level analysis in UCEC patients stratified by weight (A), age (B), menopausal status (C), histological subtype (D), TP53 mutation status (E), and cancer stage (F). ***p < 0.001; Key: STAT-signal transducer and activator of transcription; UCEC-uterine corpus endometrial carcinoma.
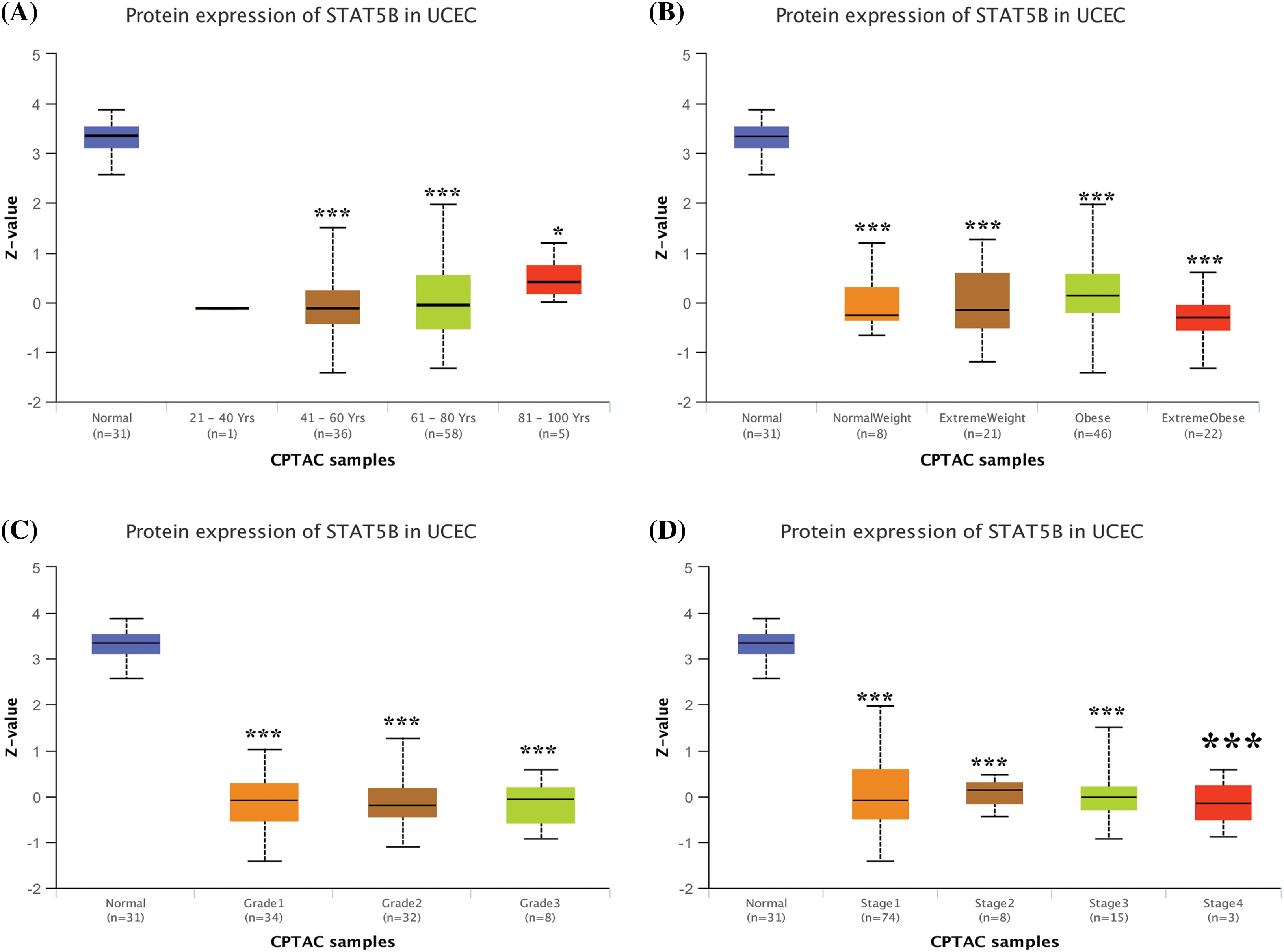
Figure 8: STAT5B protein expression level analysis in UCEC patients stratified by age (A), weight (B), tumor grade (C), and cancer stage (D). *p < 0.05; ***p < 0.001; Key: STAT-signal transducer and activator of transcription; UCEC-uterine corpus endometrial carcinoma.
Association of STAT5B expression with immune cell infiltration in uterine corpus endometrial carcinoma
As shown in Fig. 9A, STAT5B expression was positively correlated with the abundance of CD8+ T cells and neutrophils in UCEC samples, whereas there were no significant correlations seen for B cells, CD4+ T cells, and macrophages. In addition, changes in the somatic copy number of STAT5B inhibited immune cell infiltration (Fig. 9B).
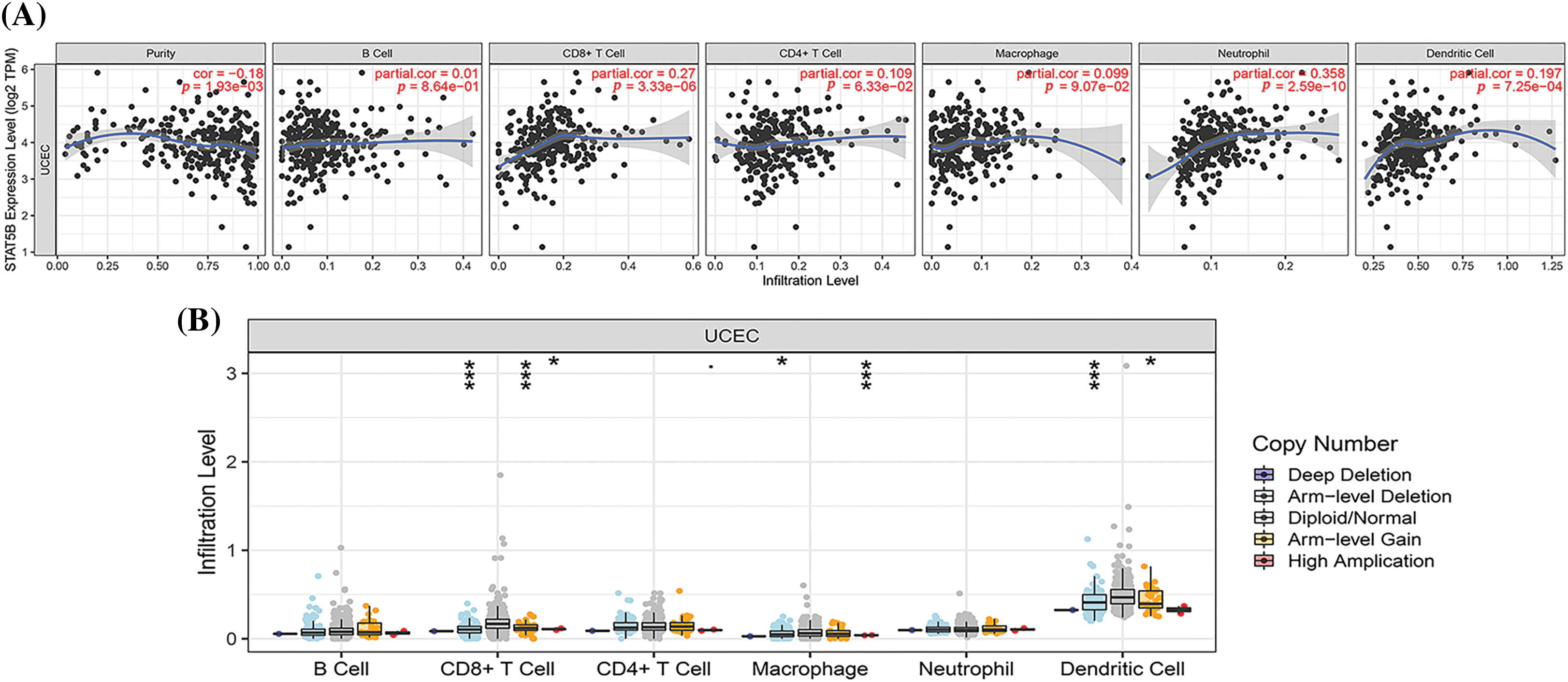
Figure 9: Correlations between STAT5B expression and infiltrating immune cells in UCEC. (A) Correlations between STAT5B expression and immune cell abundance in UCEC. (B) Effect of changes in the somatic copy number of STAT5B and immune cell infiltration in UCEC. *p < 0.05, ***p < 0.001; Key: STAT-signal transducer and activator of transcription; UCEC-uterine corpus endometrial carcinoma.
Enrichment analysis results of STAT5B in uterine corpus endometrial carcinoma
The role of STAT5B in UCEC was further investigated using enrichment analysis (Fig. 10A). We screened STAT5B-related genes, and the top 50 genes showing the highest correlation coefficients were obtained (Figs. 10B and 10C). Specifically, these included KIAA0753 (also known as moonraker and OFIP), PSMB7, COL27A1, COX8A, EZH1, PPP1CA, SHANK3, and ATP6V0B. Among these, KIAA0753 had the strongest positive correlation with STAT5B, whereas PSMB7 showed the strongest negative correlation.
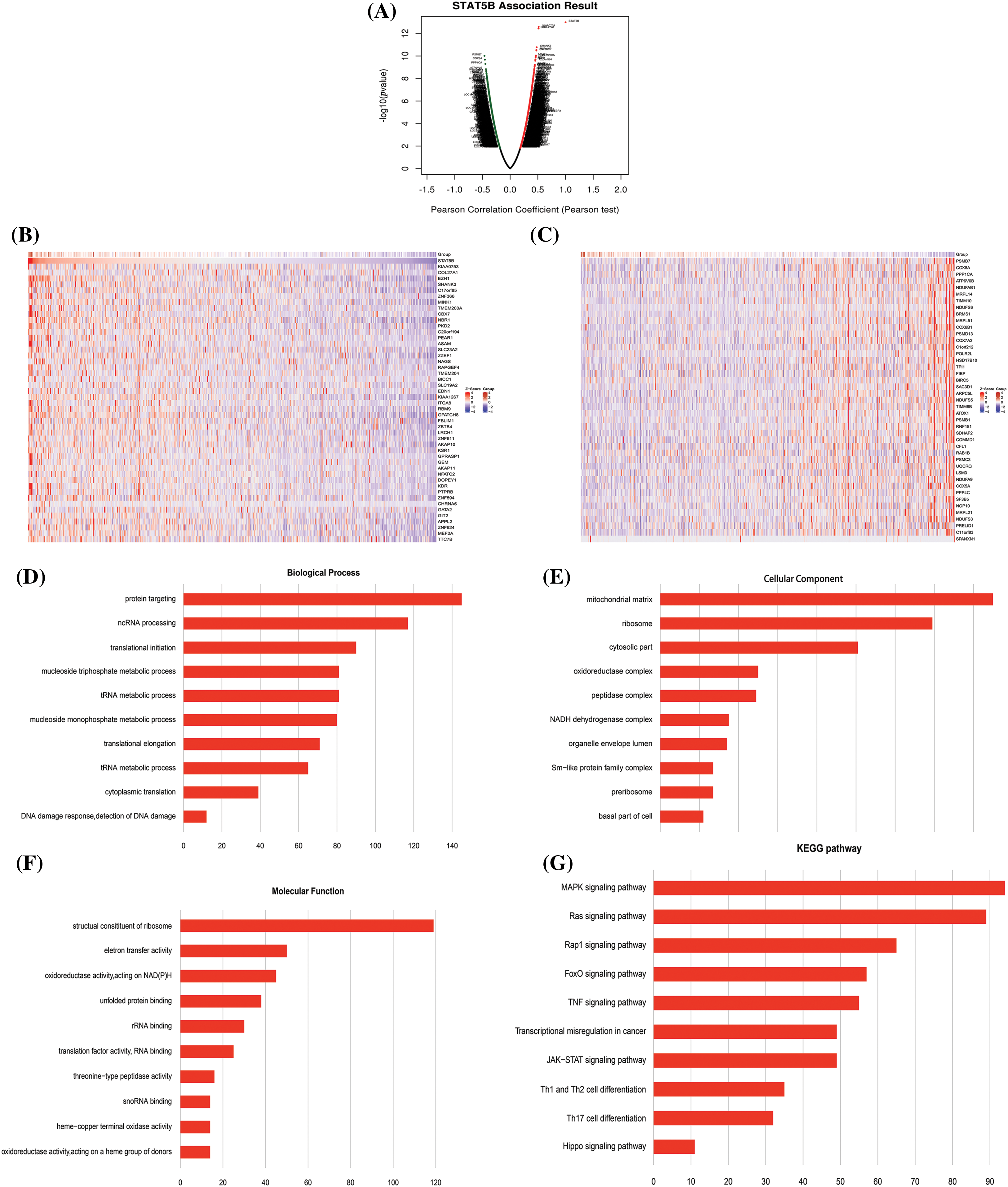
Figure 10: Correlation analysis between differential expression of STAT5B-associated genes and functional enrichment analysis of STAT5B and associated genes in UCEC. (A) Pearson coefficient map of STAT5B-related genes. (B) STAT5B and positively correlated gene expression heat map. (C) STAT5B and negatively correlated gene expression heat map. (D–F) Bar graphs of the top 10 enriched GO terms. (G) Enriched KEGG pathways of STAT5B and associated genes; Key: STAT-signal transducer and activator of transcription; UCEC-uterine corpus endometrial carcinoma; GO-Gene ontology; KEGG-Kyoto Encyclopedia of Genes and Genomes.
We screened the biological processes of targeted protein delivery, non-coding RNA synthesis and processing, and protein translation initiation (Fig. 10D). Further GO analysis demonstrated that STAT5B-related genes were mainly involved in the cellular components of the chondral matrix, mitochondria, and cytoplasm (Fig. 10E). The molecular functions of oxidoreductase activity, acting on NAD (P), and several others were also screened (Fig. 10F). KEGG pathway enrichment analysis showed that STAT5B-related genes were mainly involved in the mitogen-activated protein kinase (MAPK) signaling pathway, Ras signaling pathway, Ras-associated protein-1 (Rap1) signaling pathway, Forkhead box O (FOXO) signaling pathway, tumor necrosis factor (TNF) signaling pathway, cancer transcriptional misregulation, Janus kinase (JAK)-STAT transcriptional misregulation, T-helper cell (Th1) and Th2 cell differentiation, Th17 cell differentiation, and Hippo transcriptional misregulation (Fig. 10G).
The competitive endogenous RNA regulation network of STAT5B
We found four main miRNAs regulating STAT5B using relevant approaches, namely, miR877-5P, miR24-3P, miR200A-3P, and miR141-3P. Further analysis based on 22 lncRNAs regulating these miRNAs developed a ceRNA regulatory network of STAT5B (Suppl. Fig. S4).
EC is one of the three major malignant tumors in the field of gynecology. The incidence of EC is increasing as the living standards and the rate of obesity increase, seriously affecting women’s health globally. It is vital to find prognosis indicators for the early discovery and early treatment of EC. Studies have increasingly shown that the JAK/STAT family is closely related to EC, and is also becoming an important part of finding therapeutic targets in recent years. The JAK/STAT signaling pathway is involved in regulating basic cellular mechanisms such as proliferation, invasion, survival, inflammation, and immunity. Abnormal JAK/STAT signaling leads to cancer progression and metastasis. Accordingly, many proteins of the STAT family have been shown to be abnormally expressed in a variety of malignancies, including breast cancer, osteosarcoma, lung cancer, and bladder cancer. Further, they are considered to be associated with cancer progression and could significantly affect the prognosis of patients (Alsheikh et al., 2021; Assefnia et al., 2014; Galimberti et al., 2022; Golus et al., 2022; Kharma et al., 2014; Liu et al., 2021; Liu et al., 2021b; Mirzaei et al., 2021; Wong et al., 2022; Xin et al., 2021; Yang et al., 2022a; Zhu et al., 2023). Nothwithstanding these results, the role of STAT family members in the development, metastasis, and prognosis of EC has not yet been systematically analyzed.
At present, there is no study focusing on the STAT family factor from the overall perspective. For the first time, we used databases such as UALCAN, Sangerbox, HPA, TIMER, GSCalite, cBio Cancer Portal, Genemania, StarBase, and other databases to analyze the expression level of the STAT family of factors in pan-cancer tissues. We also assessed promoter methylation, mutant frequency, immune checkpoint, prognosis, and possiblly related genes. Our pan-cancer analysis confirmed that while STAT1 was overexpressed in most tumor tissues such as EC tissue, STAT5A, STAT5B, and STAT6 were downregulated in EC. We hypothesize that the STAT family may be a prognostic indicator for EC.
This study showed that STAT1 expression was significantly upregulated in EC at mRNA and protein levels using bioinformatics approaches based on public transcriptome data, Further, the highly expressed STAT1 predicted a poor prognosis of patients with EC, which was in line with the conclusions of previous studies. For instance, Kharma et al. (2014) suggested that STAT1 promoted the proliferation, adhesion, migration, and invasion of high-serous papillary carcinoma cells and that STAT1 was an oncogene. Further, Yang et al. (2020) also proposed that STAT1 could directly bind to the LINC01123 promoter region to activate its transcription and stimulate the progression of EC.
We found no significant difference in the mRNA levels of STAT3 and STAT4 between EC and normal samples, whereas significant differences in the expression of these STATs were detected at the protein level. Several studies have reported a relationship between STAT3 and EC, suggesting that the upregulation of STAT3 phosphorylation could induce the proliferation of EC cells. However, STAT3 downregulation promoted the apoptosis of tumor cells, which was a key to the activation of various growth factors or cytokine signaling pathways. For example, Chen et al. (2007)) found that STAT3 was activated in human EC, and Fan et al. (2021) reported its activation in cervical cancer. In this study, STAT3 expression was significantly related to EC only at the protein level but was not related to the patient prognosis. One explanation for such a discrepancy may be related to the fact that these previous studies focused specifically on the function of STAT3, whereas the current research analyzed its expression in EC and its relationship with prognosis. However, we found no reports on the associations of STAT2, STAT4, or STAT5 with EC to date.
A recent report focused on the relationship between STAT6 and EC. For instance, Jin et al. (2021) studied the effect of the emerging organic pollutant tetrachlorobisphenol A on EC. They indicated that the increased phosphorylation of STAT6 was one of the contributors responsible for the occurrence and development of EC. The inconsistency between this finding and our present study might also be related to the focus on STAT6 phosphorylation and its functions rather than on our overall analysis of STAT6 expression in EC. Therefore, the detailed role of STAT6 in EC requires further investigation.
Many studies have validated DNA methylation as one of the most important forms of epigenetic modifications in tumor formation (Bu et al., 2023; Tang et al., 2023; Wang et al., 2023). Promoter methylation is a kind of DNA methylation system that regulates the transcription of mRNA from a target gene. This study showed that promoter methylation levels of STAT2, STAT4, and STAT6 were reduced in the EC group, while those of STAT5A and STAT5B were elevated in EC group. There were relatively few reports on the STAT family factor methylation status in tumors. For instance, Yoon et al. (2019) studied lung cancer in which methylation of the STAT4 regulating area was found to have significant differences between normal and lung cancer cells, subsequently affecting the complement factor H. The specific mechanism of this finding was not clear. In another study, Zhang et al. (2007) proved that the DNA methylation of the STAT5A gene selectively inhibited the expression of STAT5A and that the STAT5A protein could function as a key tumor inhibitory factor. Another report verified the role of SGI-110 as a new type of small molecular methylation inhibitor in ovarian cancer. They found that SGI-110 could specifically lower methylation, which increased the sensitivity of patients to chemotherapy (Fang et al., 2014). Based on our analyses, it was hypothesized that the increased DNA methylation in EC caused silence of STAT5A/5B, downregulated expression, and reduced cancer suppression, leading to tumor occurrence. The role of the STAT family factor methylation in EC needs further in-depth research.
We further found that the three most common mutation types in the STAT family in EC were missense mutations, nonsense mutations, and frameshift deletions. Previous studies have also shown that mutations in STAT family genes were associated with the development of disease. For example, Liu et al. (2021b) reported that osteosarcoma patients exhibiting a high somatic JAK-STAT mutation frequency had a relatively favorable prognosis. Another study found that STAT1 mutation was related with chronic mucocutaneous candidiasis and pancytopenia (Dabas et al., 2021).
Mutations play an important role in cancer development, and they are also important specific molecular targets of chemotherapy. The JAK-STAT pathway is a particularly effective target for the development of safe and novel anti-cancer drugs. Our drug sensitivity analysis demonstrated that the expression of STAT5A/5B was negatively correlated with drug resistance, suggesting STAT5A/5B as a potential screening indicator for drug sensitivity. Previous studies on the drug sensitivity of STATs were mainly focused on other members of the STAT family. For example, Lee et al. (2019) reviewed the role of STAT3 and oxidative phosphorylation in cancer, and suggested that these factors could be utilized as therapeutic targets by restoring the drug sensitivity of drug-resistant overexpressed oncogenes. More recently, another study found that rilpivirine improved liver fibrosis through the selective STAT1-dependent induction of hepatic stellate cell apoptosis (Martí-Rodrigo et al., 2020). Further, Moon et al. (2015) showed that SMAD3/4 regulated p-STAT3 signal transduction via IL-6 and P21, and highlighted the important role of STAT3 signal transduction in the SMAD3/4-mediated drug sensitivity of chemoresistant colorectal cancer cells. Another study further supported the role of the JAK-STAT pathway in the pathogenesis of NK cell-related malignancies (Dufva et al., 2018). Although these findings have suggested the involvement of the members of the STAT family in drug sensitivity, the role of STAT5 in drug resistance has not been reported. The finding of the present study showed that STAT5A/5B expression was negatively correlated with drug resistance in EC, indicating the need to confirm the potential of STATs as therapeutic targets.
Previous analysis showed that the STAT family was mainly related to the activation of apoptotic pathways, inhibition of the cell cycle, and induction of EMT among cancer-related functions. These findings were similar to another study that found that STAT activity affected changes in the apoptotic sensitivity of melanoma cells (Krasilnikov et al., 2003). In addition, Assefnia et al. (2014) showed that STAT5 specifically bound to the proximal promoter of TRP63 in breast tissue, and influenced cell cycle arrest, apoptosis, survival, and EMT. Hence, dysregulation of STAT expression can contribute to EC occurrence and progression by regulating the activation of apoptotic pathways, disrupting the cell cycle, and inducing EMT. These effects would be based on the associations with SCF-KIT signaling pathways, transmembrane receptor protein tyrosine kinase signaling pathways, PID-KIT pathway, NK cell immune positive adjustment, neurotrophic factor signal transduction pathways, NK cell-mediated cytotoxicity, PID-PI3KCI pathways, antigen receptor-mediated signaling pathways, and lymphoid cell differentiation. We also found several proteins that interacted with the STAT family, offering useful targets for subsequent analysis of the detailed mechanism of action of these interactions in EC.
Given that STAT5B showed the most significant difference in both expression and survival analysis among the members of the STAT family analyzed, we further focused on examining its role in EC. A clear association between STAT5B and the clinicopathological features of patients with UCEC was detected. Further, STAT5B expression was significantly correlated with all the clinicopathological indicators of UCEC patients, which was significantly lower than that in the controls. The significance of STAT5B has also been highlighted in other malignancies, including eosinophilic myeloma, T-cell large granular lymphocytic leukemia, chronic lymphoproliferative disorder of NK cells, and CD8 lymphoma (Cross et al., 2019; Pham et al., 2018; Putri et al., 2019; Shahmarvand et al., 2018). Studies have shown that the mechanism of action of STAT5B in tumors was relatively complicated. For instance, in cancer promotion, STAT5B was expressed in leukemia, prostate cancer, pancreatic tube adenocarcinoma (PDAC), which promoted tumor proliferation, invasion, and metastasis by inducing CD9 to impact ABL-N and Gascibada resistance (Kollmann et al., 2021; Subramaniam et al., 2020). However, a low expression of STAT5B was seen in liver cell carcinoma and Merkel cell carcinoma (MCC) and exerted a cancer-suppressive effect by regulating Treg and tumor infiltration leukocytes (Dong et al., 2019; Fujimura et al., 2017; Rao et al., 2020). However, the role of STAT5B in EC has not yet been reported.
In addition, immune cell infiltration analysis showed that STAT5B expression was positively related to the abundance of CD8+ T cells and neutrophils and that changes in the somatic copy number of STAT5B inhibited immune cell infiltration. A previous study showed that a variety of immune-related pathways were dysregulated in patients with impaired JAK/STAT signal transduction. Moreover, STAT5B inhibited IgE-induced cytokine production and limited IgE production in vivo (Kiwanuka et al., 2021). Further, the activation of STAT5b induced dendritic cell tolerance in another study (Zerif et al., 2020). Despite these findings, further studies are needed to determine the immune infiltration associated mechanism of STAT5B in EC.
We screened genes related to STAT5B expression in UCEC and showed that KIAA0753 was the most strongly correlated gene, followed by COL27A1. KIAA0753 is a centrosome and pericentrosome satellite protein that plays a key role in cartilage calcification and cartilage-to-bone transition (Stephen et al., 2017). COL27A1 encodes the type XXVII collagen α1 chain to provide connective tissue structural support and is reported to be associated with dental and genital abnormalities, Steel syndrome, and esophageal carcinoma in situ (Satoh et al., 2021). Our GO functional enrichment analysis further demonstrated that STAT5B-related genes were involved in the formation of the cartilage matrix, targeted delivery of proteins, synthesis and processing of non-coding RNA, and protein functions. From KEGG pathway analysis, it could be observed that STAT5B-related genes were involved in the MAPK and Ras signaling pathways. These findings have provided a starting point for further investigation of the potential inhibitory mechanism of STAT5B in EC.
In this study, the lncRNA-miRNA-mRNA regulating network of STAT5B included four main miRNAs regulating STAT5B, namely miR877-5P, miR24-3P, miR200A-3P, and miR141-3P. Previous research showed that miR877-5P could inhibit cell progress in lung cancer through the Foxm1 target (Liu et al., 2022). In addition, miR-877-5P has been proven to have anti-tumor effects on breast cancer (Liu et al., 2022). Reports showed that miR-877-5P played an important role in female gingival cell tumors (Yang et al., 2021), live cancer cell carcinoma (Yu et al., 2021), and bladder cancer (Zhou et al., 2021). However, there was no research article found on miR877-5P in EC. Further, miR24-3P was confirmed only to be involved in the development of non-small cell lung cancer (NSCLC) and pancreatic cancer, and was predicted to be used as a cancer suppression agent in pancreatic cancer (Borchardt et al., 2021; Olbromski et al., 2018). While miR200A-3P might be related to lung cancer EMT, it might also be the inhibitory factor of glioblastoma multiforme (Berthois et al., 2014; Sarkar et al., 2020). MiR141-3P was considered to be related to brain metastases with bladder cancer, prostate cancer, and gastric adenocarcinoma (Bergez-Hernández et al., 2022; Minn et al., 2014; Yang et al., 2022a). Among these miRNAs we reported, miR877-5P has great research potential. We intend to further analyze the expression and prognosis of miR877-5P in EC and study how miR877-5P regulates STAT5B in EC.
The lncRNA-miRNA-mRNA network developed in this study also showed that lncRNAs may play an important role in EC. Among the 22 lncRNAs outlined, the roles of some lncRNAs in tumors, such as SNHG15, DLX6-AS1, and PinK1-AS, have already been analyzed. However, there were only a few studies that studied lncRNAs such as RUSC1-AS1, LBx2-As1, Olmalinc, RPARP-AS1, Ac092718.4, PTOV1-AS1, PTOV1-AS2, and CCDC183-AS1. Among these lncRNAs, only DLX6-AS1 and Pink1-AS had been researched in EC so far. The role of other lncRNAs in EC requires further studies.
To summarize, this study suggested that the STAT family was differentially expressed in EC, which was associated with EC prognosis. This was especially seen for STAT5b, which has been previously validated as a tumor suppressor gene in EC. This analysis of the expression and clinical significance of the STAT family utilizing bioinformatics approaches provided new targets for future research to identify novel screening biomarkers, prognostic markers, and therapeutic targets for EC. However, as our study had some limitations, further in vitro or in vivo experiments are needed to verify our results that were generated based on public datasets. We would further verify the expression of STAT5B in EC by collecting clinical data for analysis and explore whether STAT5B is related to pathological factors such as stage, grading, and vascular metastasis. In-depth research on the clinical significance of mirRNA, lncRNA, and circular RNA (circRNA) related to STAT5B is also warranted for discovering specific blockers or small molecule inhibitors of STAT5B for targeted EC therapy.
Acknowledgement: None.
Funding Statement: The authors received no specific funding for this study.
Author Contributions: The authors confirm their contribution to the paper as follows: Conception, design and administrative support: Xuelian Chen; Provision of study materials or patients: All authors; Collection and assembly of data: All authors; Data analysis and interpretation: Xuelian Chen, Yunzheng Zhang, and Junjian He; Manuscript writing: All authors. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The datasets generated during analysis and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
Alsheikh HAM, Metge BJ, Pruitt HC, Kammerud SC, Chen D, Wei S, Shevde LA, Samant RS (2021). Disruption of STAT5A and NMI signaling axis leads to ISG20-driven metastatic mammary tumors. Oncogenesis 10: 45. [Google Scholar] [PubMed]
Assefnia S, Kang K, Groeneveld S, Yamaji D, Dabydeen S, Alamri A, Liu X, Hennighausen L, Furth PA (2014). Trp63 is regulated by STAT5 in mammary tissue and subject to differentiation in cancer. Endocrine-related Cancer 21: 443–457. [Google Scholar] [PubMed]
Bergez-Hernández F, Arámbula-Meraz E, Alvarez-Arrazola M, Irigoyen-Arredondo M, Luque-Ortega F, Martínez-Camberos A, Cedano-Prieto D, Contreras-Gutiérrez J, Martínez-Valenzuela C, García-Magallanes N (2022). Expression analysis of miRNAs and their potential role as biomarkers for prostate cancer detection. American Journal of Mens Health 16: 15579883221120989. [Google Scholar]
Berthois Y, Delfino C, Metellus P, Fina F, Nanni-Metellus I, Al Aswy H, Pirisi V, Ouafik L, Boudouresque F (2014). Differential expression of miR200a-3p and miR21 in grade II-III and grade IV gliomas: Evidence that miR200a-3p is regulated by O6-methylguanine methyltransferase and promotes temozolomide responsiveness. Cancer Biology & Therapy 15: 938–950. [Google Scholar]
Borchardt H, Ewe A, Morawski M, Weirauch U, Aigner A (2021). miR24-3p activity after delivery into pancreatic carcinoma cell lines exerts profound tumor-inhibitory effects through distinct pathways of apoptosis and autophagy induction. Cancer Letters 503: 174–184. [Google Scholar] [PubMed]
Bu Q, Luo X, He L, Ma J, He S et al. (2023). Septin9 DNA methylation as a promising biomarker for cervical cancer. Journal of Obstetrics and Gynaecology 43: 2151356. [Google Scholar] [PubMed]
Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, Varambally S (2017). UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 19: 649–658. [Google Scholar] [PubMed]
Chen CL, Hsieh FC, Lieblein JC, Brown J, Chan C, Wallace JA, Cheng G, Hall BM, Lin J (2007). Stat3 activation in human endometrial and cervical cancers. British Journal Cancer 96: 591–599. [Google Scholar]
Crosbie EJ, Kitson SJ, McAlpine JN, Mukhopadhyay A, Powell ME, Singh N (2022). Endometrial cancer. Lancet 399: 1412–1428. [Google Scholar] [PubMed]
Cross NCP, Hoade Y, Tapper WJ, Carreno-Tarragona G, Fanelli T et al. (2019). Recurrent activating STAT5B N642H mutation in myeloid neoplasms with eosinophilia. Leukemia 33: 415–425. [Google Scholar] [PubMed]
Dabas A, Arora P, Kumar S, Kapoor S, Yadav S (2021). STAT 1 mutation associated with chronic mucocutaneous candidiasis and pancytopenia. Pediatric Allergy and Immunology 32: 798–800. [Google Scholar] [PubMed]
Dong Z, Chen Y, Yang C, Zhang M, Chen A, Yang J, Huang Y (2019). STAT gene family mRNA expression and prognostic value in hepatocellular carcinoma. OncoTargets and Therapy 12: 7175–7191. [Google Scholar] [PubMed]
Du W, Wang YC, Hong J, Su WY, Lin YW, Lu R, Xiong H, Fang JY (2012). STAT5 isoforms regulate colorectal cancer cell apoptosis via reduction of mitochondrial membrane potential and generation of reactive oxygen species. Journal of Cellular Physiology 227: 2421–2429. [Google Scholar] [PubMed]
Dufva O, Kankainen M, Kelkka T, Sekiguchi N, Awad SA et al. (2018). Aggressive natural killer-cell leukemia mutational landscape and drug profiling highlight JAK-STAT signaling as therapeutic target. Nature Communications 9: 1567. [Google Scholar] [PubMed]
Fan JT, Zhou ZY, Luo YL, Luo Q, Chen SB, Zhao JC, Chen QR (2021). Exosomal lncRNA NEAT1 from cancer-associated fibroblasts facilitates endometrial cancer progression via miR-26a/b-5p-mediated STAT3/YKL-40 signaling pathway. Neoplasia 23: 692–703. [Google Scholar] [PubMed]
Fang F, Munck J, Tang J, Taverna P, Wang Y et al. (2014). The novel, small-molecule DNA methylation inhibitor SGI-110 as an ovarian cancer chemosensitizer. Clinical Cancer Research 20: 6504–6516. [Google Scholar] [PubMed]
Fujimura T, Furudate S, Kambayahsi Y, Kakizaki A, Yamamoto Y, Okuhira H, Fujimoto N, Aiba S (2017). Phospho-STAT5B expression is a prognostic marker for merkel cell carcinoma. Anticancer Research 37: 2335–2341. [Google Scholar] [PubMed]
Galimberti S, Baldini C, Baratè C, Fornili M, Balducci S et al. (2022). Myeloid neoplasms and autoimmune diseases: Markers of association. Clinical and Experimental Rheumatology 40: 49–55. [Google Scholar] [PubMed]
Golus M, Bugajski P, Chorbińska J, Krajewski W, Lemiński A, Saczko J, Kulbacka J, Szydełko T, Małkiewicz B (2022). STAT3 and its pathways’ dysregulation-underestimated role in urological tumors. Cells 11: 3024. [Google Scholar] [PubMed]
Hou Y, Huang S, Liu J, Wang L, Yuan Y, Liu H, Weng X, Chen Z, Hu J, Liu X (2023). DOT1L promotes cell proliferation and invasion by epigenetically regulating STAT5B in renal cell carcinoma. American Journal of Cancer Research 13: 276–292. [Google Scholar] [PubMed]
Hou GX, Liu P, Yang J, Wen S (2017). Mining expression and prognosis of topoisomerase isoforms in non-small-cell lung cancer by using Oncomine and Kaplan-Meier plotter. PLoS One 12: e0174515. [Google Scholar] [PubMed]
Jin X, Su H, Xu L, Wang Y, Su R, Zhang Z, Guan G, Li Z (2021). Different co-culture models reveal the pivotal role of TBBPA-promoted M2 macrophage polarization in the deterioration of endometrial cancer. Journal of Hazardous Materials 413: 125337. [Google Scholar] [PubMed]
Kariuki D, Asam K, Aouizerat BE, Lewis KA, Florez JC, Flowers E (2023). Review of databases for experimentally validated human microRNA-mRNA interactions. Database 2023: baad014. [Google Scholar] [PubMed]
Kharma B, Baba T, Matsumura N, Kang HS, Hamanishi J et al. (2014). STAT1 drives tumor progression in serous papillary endometrial cancer. Cancer Research 74: 6519–6530. [Google Scholar] [PubMed]
Kiwanuka KN, Motunrayo Kolawole E, McLeod J.J. A, Baker B, Paez PA et al. (2021). Stat5B is required for IgE-mediated mast cell function in vitro and in vivo. Cellular Immunology 364: 104344. [Google Scholar] [PubMed]
Kollmann S, Grausenburger R, Klampfl T, Prchal-Murphy M, Bastl K et al. (2021). A STAT5B-CD9 axis determines self-renewal in hematopoietic and leukemic stem cells. Blood 138: 2347–2359. [Google Scholar] [PubMed]
Krasilnikov M, Ivanov VN, Dong J, Ronai Z (2003). ERK and PI3K negatively regulate STAT-transcriptional activities in human melanoma cells: Implications towards sensitization to apoptosis. Oncogene 22: 4092–4101. [Google Scholar] [PubMed]
Lee M, Hirpara JL, Eu JQ, Sethi G, Wang L, Goh BC, Wong AL (2019). Targeting STAT3 and oxidative phosphorylation in oncogene-addicted tumors. Redox Biology 25: 101073. [Google Scholar] [PubMed]
Liu CJ, Hu FF, Xia MX, Han L, Zhang Q, Guo AY (2018). GSCALite: A web server for gene set cancer analysis. Bioinformatics 34: 3771–3772. [Google Scholar] [PubMed]
Liu Y, Liao S, Bennett S, Tang H, Song D, Wood D, Zhan X, Xu J (2021b). STAT3 and its targeting inhibitors in osteosarcoma. Cell Proliferation 54: e12974. [Google Scholar] [PubMed]
Liu Z, Wang X, Cao L, Yin X, Zhang Q, Wang L (2022). MicroRNA-877-5p inhibits cell progression by targeting FOXM1 in lung cancer. Canadian Respiratory Journal 2022: 4256172. [Google Scholar] [PubMed]
Liu W, Wang R, Zhang Y, Wang H, Huang Z, Jin T, Yang Y, Sun Y, Cao S, Niu X (2021). Whole-exome sequencing in osteosarcoma with distinct prognosis reveals disparate genetic heterogeneity. Cancer Genetics 256–257: 149–157. [Google Scholar] [PubMed]
Liu H, Xiang L, Mei Y (2022). miR-877-5p inhibits epithelial mesenchymal transformation of breast cancer cells by targeting FGB. Disease Markers 2022: 4882375. [Google Scholar] [PubMed]
Martí-Rodrigo A, Alegre F, Moragrega Á. B, García-García F, Martí-Rodrigo P, Fernández-Iglesias A, Gracia-Sancho J, Apostolova N, Esplugues JV, Blas-García A (2020). Rilpivirine attenuates liver fibrosis through selective STAT1-mediated apoptosis in hepatic stellate cells. Gut 69: 920–932. [Google Scholar]
Minn YK, Lee DH, Hyung WJ, Kim JE, Choi J, Yang SH, Song H, Lim BJ, Kim SH (2014). MicroRNA-200 family members and ZEB2 are associated with brain metastasis in gastric adenocarcinoma. International Journal of Oncology 45: 2403–2410. [Google Scholar] [PubMed]
Mirzaei S, Gholami MH, Mahabady MK, Nabavi N, Zabolian A et al. (2021). Pre-clinical investigation of STAT3 pathway in bladder cancer: Paving the way for clinical translation. Biomedicine and Pharmacotherapy 133: 111077. [Google Scholar] [PubMed]
Moon SU, Kang MH, Sung JH, Kim JW, Lee JO, Kim YJ, Lee KW, Bang SM, Lee JS, Kim JH (2015). Effect of Smad3/4 on chemotherapeutic drug sensitivity in colorectal cancer cells. Oncology Reports 33: 185–192. [Google Scholar] [PubMed]
Olbromski M, Rzechonek A, Grzegrzolka J, Glatzel-Plucinska N, Chachaj A, Werynska B, Podhorska-Okolow M, Dziegiel P (2018). Influence of miR-7a and miR-24-3p on the SOX18 transcript in lung adenocarcinoma. Oncology Reports 39: 201–208. [Google Scholar] [PubMed]
Pham HTT, Maurer B, Prchal-Murphy M, Grausenburger R, Grundschober E et al. (2018). STAT5BN642H is a driver mutation for T cell neoplasia. Journal of Clinical Investigation 128: 387–401. [Google Scholar] [PubMed]
Putri A, Rinaldi I, Louisa M, Koesnoe S (2019). The role of STAT5 in tyrosine kinase inhibitor (IMATINIB) resistance in CML patients. Acta Medica Indonesiana 51: 348–352. [Google Scholar] [PubMed]
Rao J, Wu X, Zhou X, Deng R, Ma Y (2020). TMEM205 is an independent prognostic factor and is associated with immune cell infiltrates in hepatocellular carcinoma. Frontiers in Genetics 11: 575776. [Google Scholar] [PubMed]
Reimer N, Unberath P, Busch H, Ingenerf J (2021). FhirSpark-implementing a mediation layer to bring FHIR to the cBioPortal for cancer genomics. Studies in Health Technology and Iinformatics 281: 303–307. [Google Scholar] [PubMed]
Sarkar A, Rahaman A, Biswas I, Mukherjee G, Chatterjee S, Bhattacharjee S, Mandal DP (2020). TGFβ mediated LINC00273 upregulation sponges mir200a-3p and promotes invasion and metastasis by activating ZEB1. Journal of Cellular Physiology 235: 7159–7172. [Google Scholar] [PubMed]
Satoh C, Kondoh T, Shimizu H, Kinoshita A, Mishima H, Nishimura G, Miyazaki M, Okano K, Kumai Y, Yoshiura KI (2021). Brothers with novel compound heterozygous mutations in COL27A1 causing dental and genital abnormalities. European Journal of Medical Genetics 64: 104125. [Google Scholar] [PubMed]
Shahmarvand N, Nagy A, Shahryari J, Ohgami RS (2018). Mutations in the signal transducer and activator of transcription family of genes in cancer. Cancer Science 109: 926–933. [Google Scholar] [PubMed]
Shi T, Hu Z, Tian L, Yang Y (2023). Pan-cancer landscape of CENPO and its underlying mechanism in LUAD. Respiratory Research 24: 113. [Google Scholar] [PubMed]
Stephen J, Vilboux T, Mian L, Kuptanon C, Sinclair CM et al. (2017). Mutations in KIAA0753 cause Joubert syndrome associated with growth hormone deficiency. Human Genetics 136: 399–408. [Google Scholar] [PubMed]
Subramaniam D, Angulo P, Ponnurangam S, Dandawate P, Ramamoorthy P, Srinivasan P, Iwakuma T, Weir SJ, Chastain K, Anant S (2020). Suppressing STAT5 signaling affects osteosarcoma growth and stemness. Cell Death and Disease 11: 149. [Google Scholar] [PubMed]
Tang H, Guan Y, Yuan Z, Guo T, Tan X, Fan Y, Zhang E, Wang X (2023). Histone demethylase KDM4B contributes to advanced clear cell renal carcinoma and association with copy number variations and cell cycle progression. Epigenetics 18: 2192319. [Google Scholar] [PubMed]
Wang Y, Mao Y, Wang C, Jiang X, Tang Q, Wang L, Zhu J, Zhao M (2023). RNA methylation-related genes of m6A, m5C, and m1A predict prognosis and immunotherapy response in cervical cancer. Annals of Medicine 55: 2190618. [Google Scholar] [PubMed]
Wong GL, Manore SG, Doheny DL, Lo HW (2022). STAT family of transcription factors in breast cancer: Pathogenesis and therapeutic opportunities and challenges. Seminars in Cancer Biology 86: 84–106. [Google Scholar] [PubMed]
Xin S, Fang W, Li J, Li D, Wang C, Huang Q, Huang M, Zhuang W, Wang X, Chen L (2021). Impact of STAT1 polymorphisms on crizotinib-induced hepatotoxicity in ALK-positive non-small cell lung cancer patients. Journal of Cancer Research and Clinical Oncology 147: 725–737. [Google Scholar] [PubMed]
Yang F, Liu XQ, He JZ, Xian SP, Yang PF, Mai ZY, Li M, Liu Y, Zhang XD (2022b). Occludin facilitates tumour angiogenesis in bladder cancer by regulating IL8/STAT3 through STAT4. Journal of Cellular and Molecular Medicine 26: 2363–2376. [Google Scholar] [PubMed]
Yang J, Tian S, Wang B, Wang J, Cao L et al. (2021). CircPIK3C2A facilitates the progression of glioblastoma via targeting miR-877-5p/FOXM1 axis. Frontiers in Oncology 11: 801776. [Google Scholar] [PubMed]
Yang C, Wu S, Mou Z, Zhou Q, Dai X et al. (2022a). Exosome-derived circTRPS1 promotes malignant phenotype and CD8+ T cell exhaustion in bladder cancer microenvironments. Molecular Therapy 30: 1054–1070. [Google Scholar] [PubMed]
Yang Y, Wu J, Zhou H, Liu W, Wang J, Zhang Q (2020). STAT1-induced upregulation of lncRNA LINC01123 predicts poor prognosis and promotes the progression of endometrial cancer through miR-516b/KIF4A. Cell Cycle 19: 1502–1516. [Google Scholar] [PubMed]
Yoon YH, Hwang HJ, Sung HJ, Heo SH, Kim DS, Hong SH, Lee KH, Cho JY (2019). Upregulation of complement factor H by SOCS-1/3–STAT4 in lung cancer. Cancers 11: 471. [Google Scholar] [PubMed]
Yu Y, Bian L, Liu R, Wang Y, Xiao X (2021). Circular RNA hsa_circ_0061395 accelerates hepatocellular carcinoma progression via regulation of the miR-877-5p/PIK3R3 axis. Cancer Cell International 21: 10. [Google Scholar] [PubMed]
Zerif E, Khan FU, Raki AA, Lullier V, Gris D, Dupuis G, Amrani A (2020). Elucidating the role of Ezh2 in tolerogenic function of NOD bone marrow-derived dendritic cells expressing constitutively active Stat5b. International Journal of Molecular Sciences 21: 6453. [Google Scholar] [PubMed]
Zhang H, Ding C, Li Y, Xing C, Wang S, Yu Z, Chen L, Li P, Dai M (2021). Data mining-based study of collagen type III alpha 1 (COL3A1) prognostic value and immune exploration in pan-cancer. Bioengineered 12: 3634–3646. [Google Scholar] [PubMed]
Zhang Y, Liu K, Zhang Y, Qi J, Lu B, Shi C, Yin Y, Cai W, Li W (2015). ABL-N may induce apoptosis of human prostate cancer cells through suppression of KLF5, ICAM-1 and Stat5b, and upregulation of Bax/Bcl-2 ratio: An in vitro and in vivo study. Oncology Reports 34: 2953–2960. [Google Scholar] [PubMed]
Zhang Q, Wang HY, Liu X, Wasik MA (2007). STAT5A is epigenetically silenced by the tyrosine kinase NPM1-ALK and acts as a tumor suppressor by reciprocally inhibiting NPM1-ALK expression. Nature Medicine 13: 1341–1348. [Google Scholar] [PubMed]
Zhou X, Wang F, Wu H, Chen X, Zhang Y et al. (2021). Thymoquinone suppresses the proliferation, migration and invasiveness through regulating ROS, autophagic flux and miR-877-5p in human bladder carcinoma cells. International Journal of Biological Sciences 17: 3456–3475. [Google Scholar] [PubMed]
Zhu M, Li S, Cao X, Rashid K, Liu T (2023). The STAT family: Key transcription factors mediating crosstalk between cancer stem cells and tumor immune microenvironment. Seminars in Cancer Biology 88: 18–31. [Google Scholar] [PubMed]
Supplementary Materials
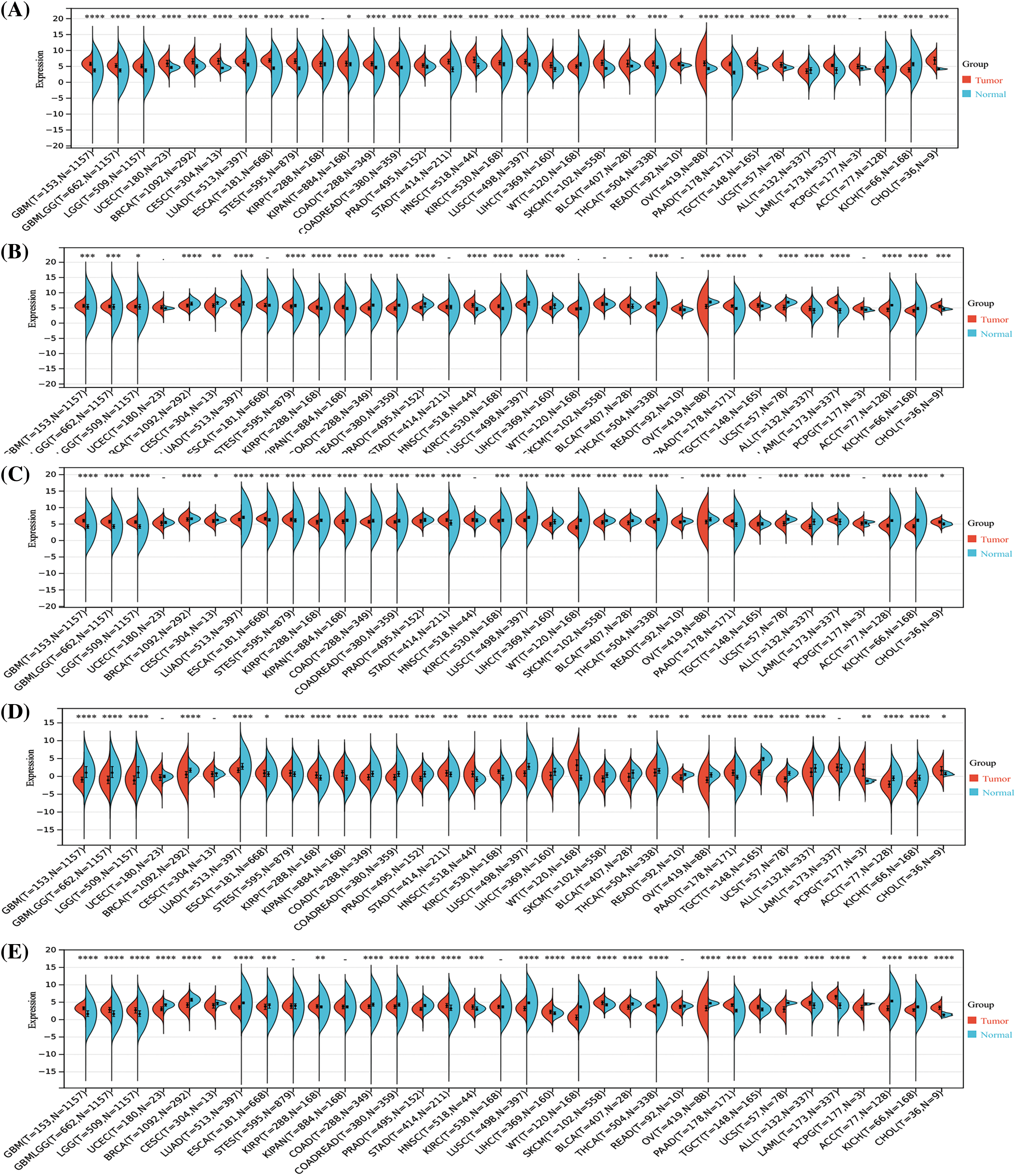
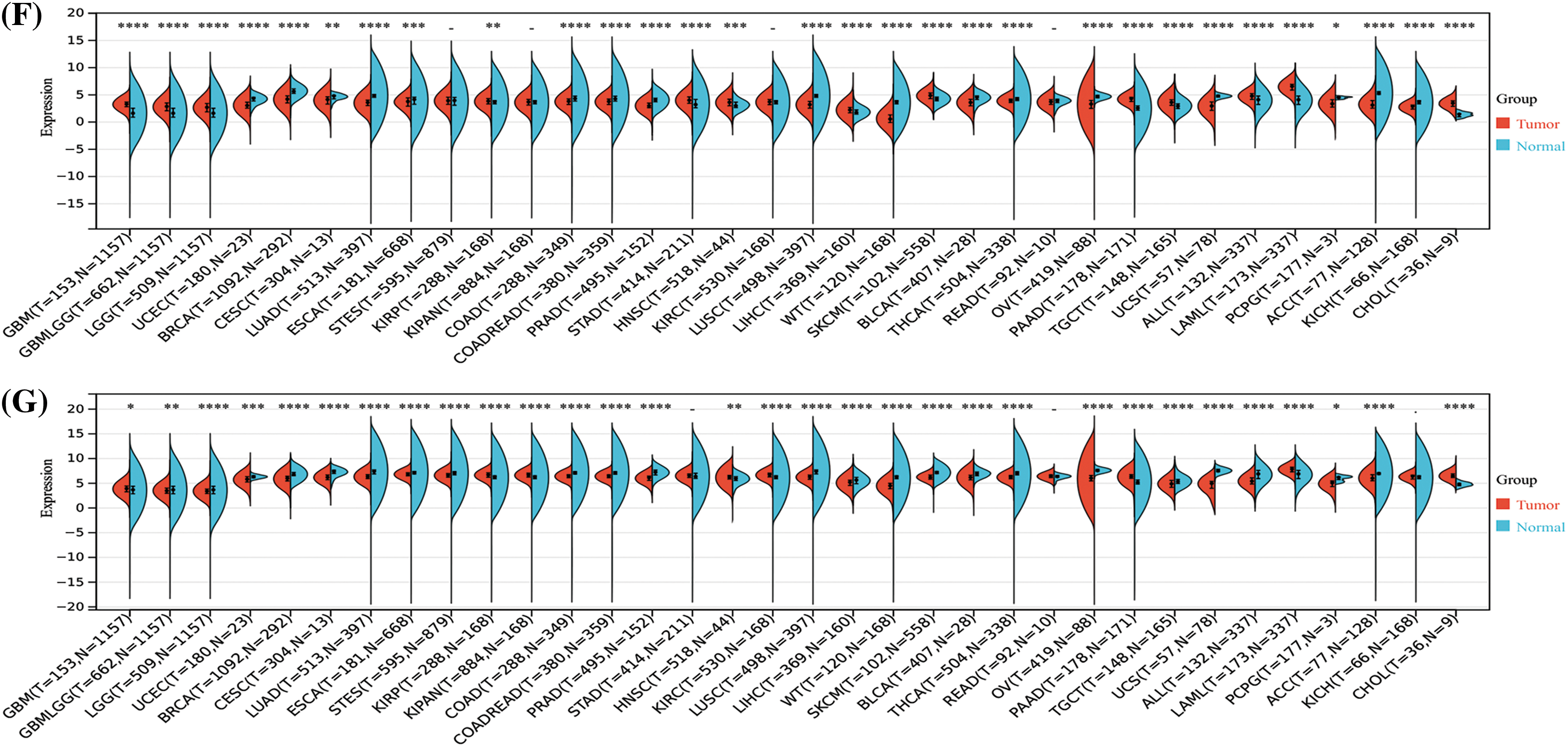
Figure S1: Analysis of the mRNA expression of the signal transducer and activator of transcription (STAT) family among different tumors and normal tissues by the Sangerbox analysis. STAT1 (A), STAT2 (B), STAT3 (C), STAT4 (D), STAT5A (E), STAT5B (F), STAT6 (G) *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
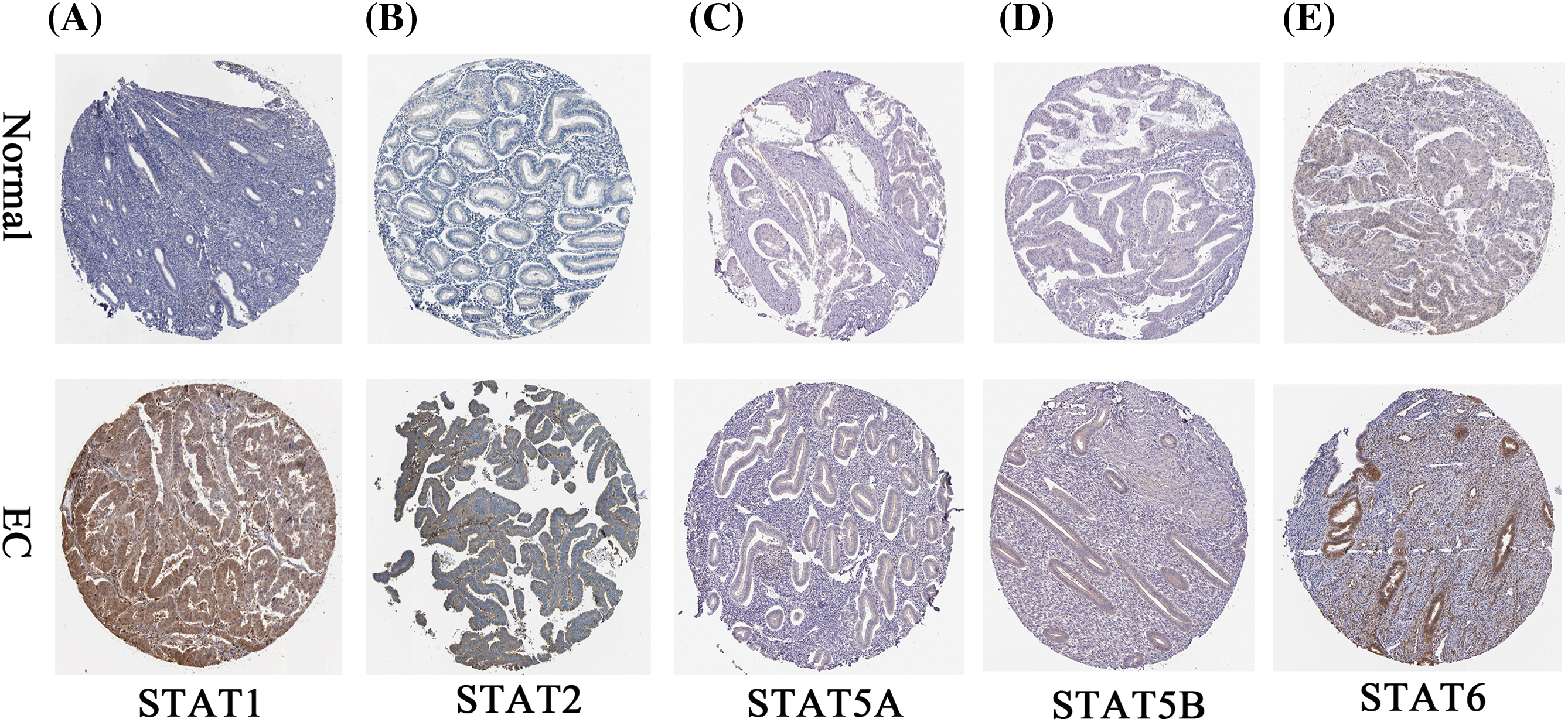
Figure S2: The immunohistochemical profile of the STAT family in EC tissues and normal endometrial tissues. EC-endometrial carcinoma; STAT-signal transducer and activator of transcription.
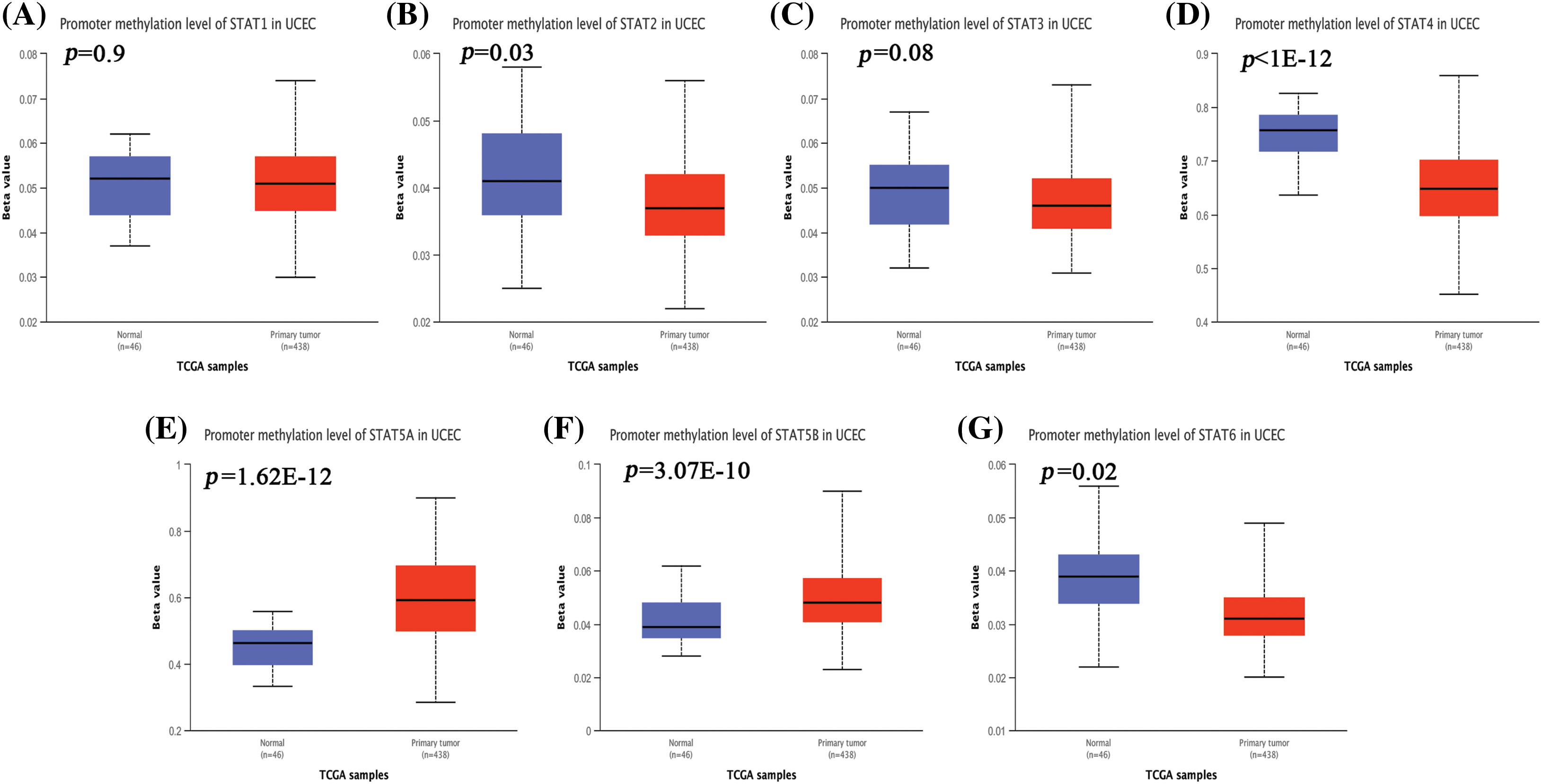
Figure S3: The promoter methylation level of STATs in UCEC. STAT1 (A), STAT2 (B), STAT3 (C), STAT4 (D), STAT5A (E), STAT5B (F), and STAT6 (G); Key: STAT-signal transducer and activator of transcription; UCEC-uterine corpus endometrial carcinoma.
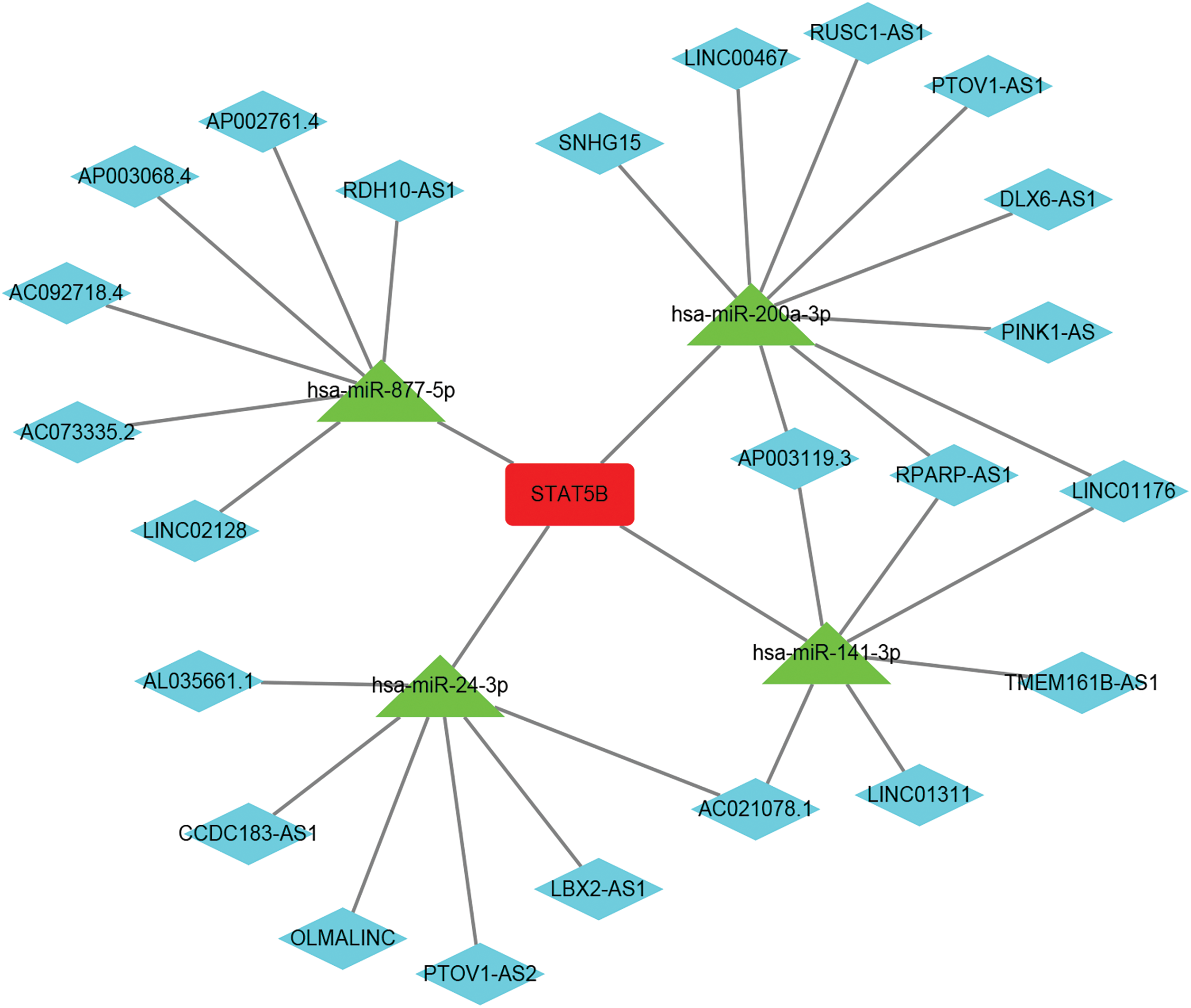
Figure S4: The lncRNA-miRNA-mRNA regulation network of STAT5B (red represents STAT5B, green represents miRNA, and blue represents lncRNA); Key: lncRNA: long non coding RNA; miRNA-microRNA; STAT-signal transducer and activator of transcription; UCEC- uterine corpus endometrial carcinoma.
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools