 Open Access
Open Access
MiR-19a-3p/PTEN axis regulates the anticancer effect of circHIAT1 in breast cancer in vitro
1 Department of General Surgery, Lanzhou University Second Hospital, Lanzhou, 730000, China
2 Department of General Surgery, Affiliated Hospital of Yunnan University, Kunming, 650051, China
* Corresponding Author: XIAODONG HE. Email:
# These authors contributed equally
(This article belongs to the Special Issue: Non-Coding RNAs in the Regulation of Human Cancers)
BIOCELL 2023, 47(10), 2301-2312. https://doi.org/10.32604/biocell.2023.029935
Received 15 March 2023; Accepted 26 June 2023; Issue published 08 November 2023
Abstract
Objective: Breast cancer is a major cancer threatening the health of women globally. To elucidate the effect of the circHIAT1/miR-19a-3p/phosphatase and tensin homolog (PTEN) axis on regulating the malignant phenotype of breast cancer cells. Methods: The mRNA expression pattern of circHIAT1, miR-19a-3p, and PTEN was checked by real-time quantitative polymerase chain reaction. Then, the knockdown assay was carried out to explore the effect of circHIAT1 and miR-19a-3p on breast cancer. The relative cell experiments, including MTT assay, scratch assay, transwell invasion assay, and flow cytometry analysis, were conducted to verify the influence of circHIAT1 and miR-19a-3p on breast cancer cells. Results: The levels of PTEN and circHIAT1 were reduced, while that of miR-19a-3p was elevated in breast cancer tissues and cells. MiR-19a-3p was proved to be the target gene of circHIAT1 via a dual luciferase experiment, which could also modulate the PTEN mRNA level. Overexpression of circHIAT1 was able to undermine the growth, migratory ability, and invasiveness in breast cancer cells, which could be antagonized by miR-19a-3p mimic. The inhibition of miR-19a-3p in vitro also impaired the malignancy of breast cancer, which depended on the modulation of PTEN expression. Conclusion: CircHIAT1 controls the PTEN expression level in cells of breast cancer by negatively regulating miR-19a-3p. This mechanism controls the growth, invasion, and migration of breast cancer.Keywords
Breast cancer is regarded as one of the leading health threats in women. Cases among Chinese women account for approximately 18% of breast cancer deaths globally (Cao et al., 2021). At present, novel therapeutic approaches have been formulated for treating early breast cancers in developed countries; however, it is a challenge to treat breast cancer patients with advanced metastasis. Meanwhile, the current use of chemotherapeutics can only prolong the survival time of patients in the short term, with widely reported cancer recurrence (Trayes and Cokenakes, 2021). Drug resistance is a common cause of death among breast cancer patients during chemotherapy and the use of targeted drugs (Wein and Loi, 2017; Koual et al., 2020). Therefore, the investigation of molecular mechanisms involved in the growth, survival, and migration of breast cancer cells could lay the foundations for novel therapies.
In recent decades, cancer researchers have focused on the roles of non-coding RNAs in tumor genesis and progression. Circular RNAs (circRNAs) were first discovered in 1979, and in the following 40 years circRNAs have been extensively studied as transcriptional regulators or molecular sponges in pathogenesis, including cardiovascular diseases, metabolic diseases, tumors, and other diseases (Hsu and Coca-Prados, 1979). CircRNAs are covalently enclosed endogenous molecules in eukaryotes produced as transcriptional byproducts (Kristensen et al., 2019). Due to its circular structure, circRNAs are relatively stable and can facilitate the continuous regulation of the biological processes in cells (Kristensen et al., 2018). Previous studies have unveiled versatile functions of circRNAs in tumors, such as encoding small peptides (Wu et al., 2020; Lei et al., 2020) and acting as molecular sponges for microRNA (miRNAs) (Zhang et al., 2021; Misir et al., 2020). CircRNAs participate in the regulation of diverse cellular events during the occurrence and development of breast cancer. Zhang and Mao (2021) discovered that circ 0000442 inhibits the cell growth of breast cancer by modulating the miR-148B-3p/phosphatase and tensin homolog (PTEN)/PI3K/Akt axis through molecular sponge action. Zhu et al. (2021) found that circSLC8A1 sponges miR-671 to control the malignancy of breast cancer cells, and circSLC8A1 overexpression can inhibit tumor growth.
PTEN (phosphatase and tensin homolog) gene encodes lipid phosphatase and is recognized as one of the most frequently mutated genes in cancers (Georgescu et al., 2000). PTEN gene expression is defective in approximately 40% of breast cancers, and PTEN inactivation is associated with poor clinical outcomes due to the failure of conventional therapy (Li et al., 2017; Dillon and Miller, 2014). The inactivation of PTEN in breast cancer facilitates the progression of breast cancer on multiple levels. Li et al. (2019) found that PTEN was suppressed by miR-497-5-P to promote the proliferation of breast cancer cells and tissues. Similarly, downregulated PTEN has a close correlation to metastasis and the increased migration activity of breast cancer cells (Lu et al., 2021; Zhai et al., 2022; Li et al., 2018). Likewise, Treeck et al. (2020) found that functional loss of PTEN in breast cancer cells downregulates estrogen receptor β and induces estrogen resistance (Treeck et al., 2020). Gao et al. (2019) also showed that the loss of PTEN activity enhances AKT phosphorylation to sustain cell proliferation and inhibit apoptosis of breast cancer, leading to resistance to chemotherapy. Similarly, Wang et al. (2016) demonstrated that the PTEN/Akt/P27 signaling pathway mediates adriamycin resistance in breast cancer cells. Therefore, the upstream regulation mechanism of the PTEN gene is worthy of further study. The knowledge regarding the regulatory effect of circRNAs on PTEN expression in breast cancer can suggest important targets for systemic intervention.
This study reports the overexpression of miR-19a-3p in breast cancer tissues and cells and reduced levels of circHIAT1 and PTEN. The function of the cicrHIAT1/miR-19a-3p/PTEN axis in regulating the malignant phenotype of breast cancer cells was validated through overexpression and knockdown experiments.
This study collected breast cancer tissues and para-cancerous specimens from breast cancer patients (N = 50) visiting Yunnan University Affiliated Hospital during 2021–2022. The acquisition of all clinical materials had been approved by the Ethics Committee of Yunnan University Affiliated Hospital (approval number: 2020092). In addition, all study participants provided informed consent. All the sample handling and data processing steps were following the Declaration of Helsinki.
The culture method of MCF-7, HS578T, and MCF-10A cells involved the usage of the T25 cell culture flask in the ultra-clean cell culture hood. Phosphate buffered saline (PBS; 5 mL 1×) was added to the culture flask, followed by 1 mL 0.25% trypsin digestion. Then 3–4 mL of complete medium was added to terminate the digestion. 0.6–0.8 ml cell suspension was placed into a new culture flask with 10 mL medium. The flask was placed horizontally at a constant temperature incubator (37°C, 5% CO2) and the medium was changed every 2–3 days. Cells were maintained in Dulbecco’s modified Eagle medium supplemented with 10% FBS (Yeasen, Beijing, China), and cells were used for experiments with 15 passages. All the cell lines procured from Hongshun Biotechnology (Shanghai, China) were authenticated using STR profiling and tested to be mycoplasma-free by the supplier.
All the expression plasmid, miRNA control, and inhibitors were procured from RiboBio Biotechnology (Guangzhou, China). In a six-well plate, 2.5 × 105 cells were seeded, and transfection was performed when the cells occupied 80% confluency. Serum-free medium (150 μL) was mixed with 9 μL. Lipofectamine® 2000 Reagent (Thermo Fisher Scientific, Shanghai, China) and 2.5 μg plasmid or 150 nM of miRNA molecules. After mixing thoroughly, the mixture was kept at ambient temperature for 5 min. Then, the transfection mixture was added to the cell culture. Experiments were performed 3 days after the transfection.
Real-time quantitative polymerase chain reaction (qRT-PCR)
Cells were mixed with 1 mL Trizol reagent (Thermo Fisher Scientific, Shanghai, China), mixed well, and centrifuged at 12000 g/min at 4°C for 5 min. The supernatant was mixed with an equal volume of isopropyl alcohol for 10 min before being centrifuged at 12000 g/min at 4°C for 10 min. The upper layer of clear solution was discarded, and the precipitation was washed with 75% ethanol (7500 g/min, 4°C, 5 min). The remaining RNA samples were dissolved using DEPC water and the concentration was measured using a spectrophotometer. Then, cDNA was prepared using a reverse transcription kit (Tiangen, Tianjin, China), followed by qRT-PCR on the QuantStudio platform (Applied Biosystems, NY, USA) using the SYBR Green qPCR quantification kit (VAZYME, Shanghai, China).
The sterile, precooled PBS was utilized to rinse the treated cells two times. Cells were subsequently scraped and centrifuged (6000 g/min, 4°C, 5 min). RIPA buffer (Yeasen, Beijing, China) containing phenylmethylsulfonyl fluoride solution (100:1) was added to lyse the cells, and then centrifuged at 13000 g/min at 4°C for 10 min. The collected supernatant samples were loaded to SDS-PAGE gels at 80 V for 160 min. The gel samples were transferred to a PVDF membrane and blocked using 5% BSA solution at ambient temperature for 120 min. The primary antibodies against PTEN, phosphoinositide 3-kinases (PI3K), serine/threonine kinase (AKT), p-PI3K, p-AKT, and actin (1:1000, Abcam) were added and incubated at 4°C overnight. After further labeling with secondary antibody (1:5000, ab214880, Abcam), the membrane was washed with PBST buffer, and the signal was developed using LightShift Chemiluminescent kit (Yeasen, Beijing, China).
Dual-luciferase reporter assay
Passive lysis buffer (100 µL of 1×; Luc-Pair Duo-Luciferase Assay Kit, GeneCopoeia, Nanjing, China) was added to each well of 96-well plates, and the solution was slowly shaken at indoor temperature for 15 min. Then, the cells were centrifuged for 10 min at 12000 rpm/min at 4°C. Luciferase assay reagent II (LAR II; 100 µL) was added to each well and incubated. After 15 min, the firefly luciferase and Renilla luciferase activities were assessed using the Luc-Pair Duo-Luciferase Assay Kit (GeneCopoeia, Nanjing, China). The experiment was repeated for three times.
Cell counting kit-8 (CCK-8) assay
Cells treated in different ways were inoculated in 96 culture plate (1500 cells per well) and maintained in the incubator for a different duration. CCK-8 reagent (10 µL; Yeasen, Beijing, China) was then loaded into each well and 3-h incubation (37°C, 5% CO2). The data were recorded at 450 nm (OD) using a microplate detector.
Flow cytometry assay for cell apoptosis
The cell suspension was prepared using the enzymatic digestion method to ensure cell dispersion. Annexin V-FITC Assay Kit (BD Bioscience, CA, USA) was utilized for 1× binding buffer preparation, and the reagent was put into the binding buffer. The suspended cells were hatched with 500 μL pre-cooled reagent mixture for 20 min at a concentration of 105–106 cells/mL. Apoptotic events were detected by flow cytometry (Attune NxT, Thermo Fisher Scientific, CA, USA).
The trypsinized breast cancer cells were suspended in a complete culture medium and inoculated into a 6-well plate at a rate of 700 cells/well. Cells were maintained in the incubator for 14 days. After the colonies were formed, the cells were immobilized and photographed under a microscope.
After being seeded in the 6 well plates, the cells were cultured until their confluence reached 80%. A 200 μL sterile pipette was utilized to create a scratch in the central area of each hole. Then the cells were incubated at 37°C for 24 h, and an inverted light microscope (Leica AM6000 microscope) was used to take cell images.
Matrigel matrix glue was applied in the Transwell chamber and air-dried at 4°C. Three hundred microliters of medium without serum containing 2.5 × 105 cells were added into the upper well, and 500 µL complete medium was loaded in the lower well. After 24-h culture, invading cells on the membrane were stained with 0.1% crystal violet (Yeasen, Beijing, China) for 30 min. After staining, 33% acetic acid was used to decolorize, and the images were recorded under a microscope.
BALB/C nude female mice (5–6-week-old) were obtained from the Department of Experimental Animal model at Yunnan University and raised under pathogen-free environments. The mixture of 1 × 106 MCF-7 cells and 0.2 mL PBS was administered subcutaneously into the right flank of each mouse. On day 21, following cell injection, mice were sacrificed, and the xenograft tissues were isolated for weight measurement and qRT-PCR analysis. All experimental procedures were conducted in accordance with Animal Ethics Procedures and Guidelines of the Animal Center, Yunnan University Animal Ethics Committee (Ethics Review Number: YNU20220296).
GraphPad Prism 8.0 version was adopted for data handling and statistical comparisons. Student’s t-test was taken to compare the difference between two groups, while the one-way ANOVA approach was utilized in the statistical evaluation of three or more groups. p-value < 0.05 was regarded as the threshold of significance.
Clinical relevance of miR-19a-3p and phosphatase and tensin homolog in breast cancer
To validate the significance of miR-19a-3p in breast cancer, TCGA (https://portal.gdc.cancer.gov/) breast ductal carcinoma miRNA-seq data of the BCGSC project level 3 were searched, and the clinical information of the patients were gathered. The result showed an elevation of miR-19a-3p level in tumor specimens of breast cancer patients, which was in connection with the progression from T1 to T2 stage of breast cancer (Fig. 1A). Further analysis using TCGA breast ductal carcinoma dataset of RNA-seq revealed decreased level of PTEN in breast cancer tissues, while aggregated miRNA-seq data analysis indicated an overall higher level of miR-19a-3p in these specimens (Figs. 1B and 1C).
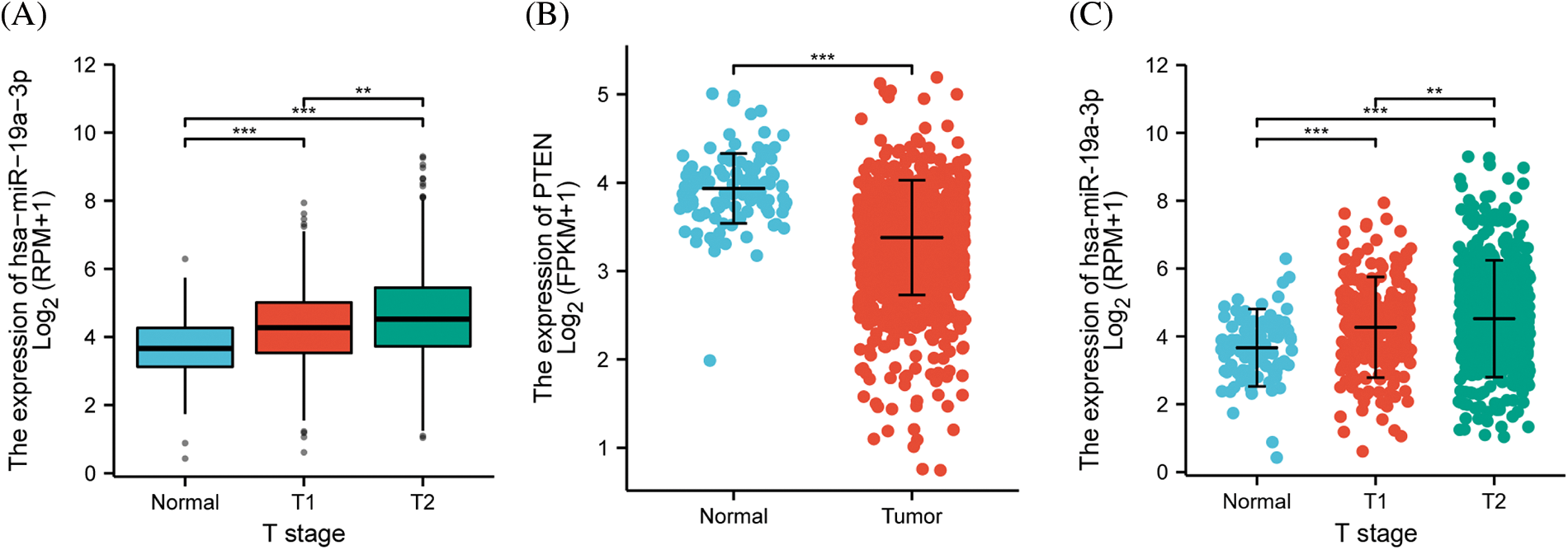
Figure 1: The contents of miR-19a-3p and phosphatase and tensin homolog (PTEN) in tumor samples. (A) Scatter plot of the relationship between miR-19a-3p expression and breast cancer clinical stage using breast ductal carcinoma dataset from TCGA database. The number of samples at the normal stage, T1 stage, and T2 stage was 102, 279, and 623, respectively. (B, C) The scatter plots of PTEN levels in RNA-seq data and miR-19a-3p levels in miRNA-seq data. ***p < 0.0001; **p < 0.01. The number of normal and tumor samples for differential expression analysis of PTEN was 113 and 1109, respectively. The number of normal and tumor samples used for differential expression analysis of miR-19a-3p were 104 and 1103, respectively.
CircHIAT1 competitively binds to miR-19a-3p
To further explore the upstream regulatory mechanism of miR-19a-3p, TargetScan analysis was employed, and the circRNA regulator of miR-19a-3p was predicted. A complementary sequence was found between miR-19a-3p and CircHIAT1 (Fig. 2A). Then, a dual luciferase reporter assay using the vector containing the wild type (WT) or mutated binding sequence (MUT) was performed. MiR-19a-3p mimic suppressed the activity of the circHIAT1-WT reporter but not of the cricHIAT1-MUT reporter (Fig. 2B), exhibiting the interaction of miR-19a-3p and CircHIAT1 via the predicted binding site. The overexpression of circHIAT1 was able to repress the miR-19a-3p level (Fig. 2C) in MCF-7 cells. The in vivo and vitro relative expression of circHIAT1 and miR-19a-3p was further examined. In comparison to healthy tissues (control) and the cell line (MCF-10A), circHIAT1 showed significant downregulation in tumor tissues (BRCA) and MCF-7 and HS578T (Fig. 2D). On the contrary, miR-19a-3p was elevated both in breast cancer tissues and cell lines (Fig. 2E). These data imply that circHIAT1 competitively binds to miR-19a-3p and modulates its expression.
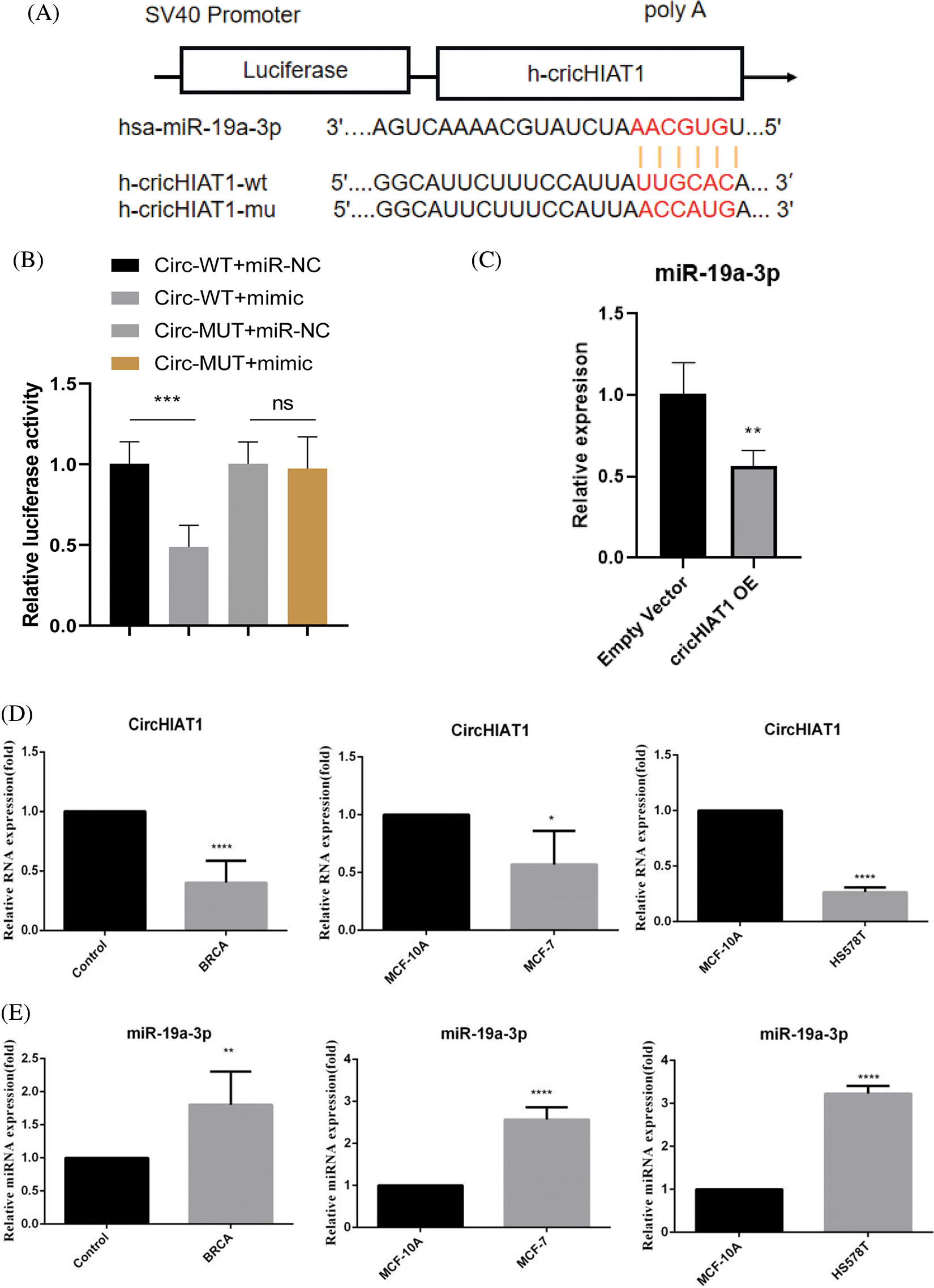
Figure 2: The interaction between circHIAT1 and miR-19a-3p. (A) Binding sites of circHIAT1 and miR-19a-3p predicted by the TargetScan database. (B) Dual luciferase reporter experiment for detecting circHIAT1 and miR-19a-3p interaction. MCF-7 cells were transfected with circHIAT1-WT or MUT report in cells of miR-19a-3p mimic or miR-NC. (C) MCF-7 cells were transfected with circHIAT1 expression vector (circHIAT1-OE) or empty vector, and the miR-19a-3p level was examined by qRT-PCR. (D, E) Analysis of circHIAT1 and miR-19a-3p levels in breast cancer specimens (BRCA) and cancerous cell lines (MCF-7 and HS578T), as well as their normal counterpart (control and MCF-10A). ****p < 0.0001; ***p < 0.001; **p < 0.01; *p < 0.05.
CircHIAT1/miR-19a-3p axis modulates the malignancy of breast cancer cell
We next examined the function of the circHIAT1/miR-19a-3p axis in regulating the malignant phenotype of breast cancer. After being transfected, CCK-8 proliferation was employed, which demonstrated the proliferation-suppressive effect of circHIAT1 overexpression, and the co-transfection of miR-19a-3p mimic reversed this impact of circHIAT1 overexpression (Fig. 3A). Similarly, circHIAT1 overexpression inhibited the MCF-7 cell colony formation, and miR-19a-3p mimic revoked the influence of circHIAT1 overexpression (Fig. 3B). Meanwhile, apoptosis detection by Annexin V and PI staining revealed an increased apoptotic event upon circHIAT1 overexpression. The concomitant administration of miR-19a-3p mimic suppressed the apoptosis induction effect of circHIAT1 (Fig. 3C). To further assess cell migration and invasion phenotype in all groups, scratch assay and Transwell experiment were performed. The migration and invasion capacity of MCF-7 cells were largely restrained by circHIAT1 overexpression, but rescued by the co-transfection of miR-19a-3p mimic (Figs. 3D and 3E). Together, the data indicated that circHIAT1 act as an inhibitor and suppress miR-19a-3p expression in breast cancer.
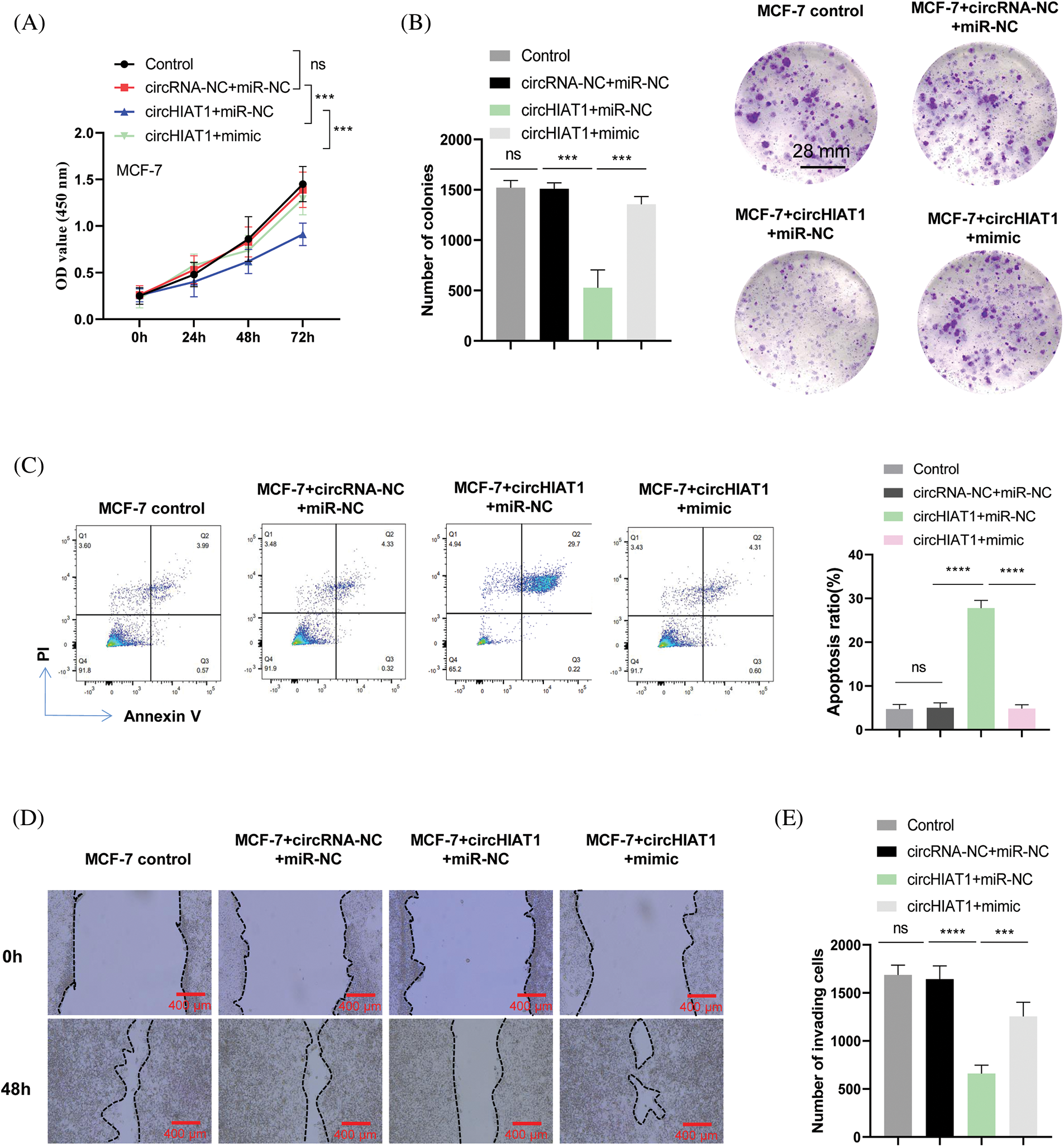
Figure 3: CircHIAT1/miR-19a-3p axis modulates the malignancy of breast cancer cells. MCF-7 cells were transfected with empty vector and miRNA mimic control (circRNA-NC+miR-NC), circHIAT1 expression vector and miRNA mimic control (circHIAT1+miR-NC), or circHIAT1 expression vector and miR-19a-3p mimic (circHIAT1+mimic). (A) Proliferation assay, (B) colony formation assay, (C) annexin V and PI staining of apoptosis, (D) scratch assay, and (E) transwell assay. ****p < 0.0001; ***p < 0.001.
MiR-19a-3p negatively modulated phosphatase and tensin homolog levels in breast cancer
To survey the downstream target of miR-19a-3p, we used TargetScan online resources to predict the mRNA targets, which revealed the complementary binding sites between miR-19a-3p and the 3′ untranslated region (3′UTR) of PTEN mRNA (Fig. 4A). Based on the dual luciferase reporter assay results, miR-19a-3p mimic could restrain the vitality of the WT reporter of PTEN (PTEN-WT), and the mutation of the binding sequence (PTEN-MUT) abrogated the effect of miR-19a-3p mimic (Fig. 4B). Further, PTEN mRNA level showed significant downregulation in BRCA, MCF-7, and HS578T compared to controls (Figs. 4C and 4D). Immunohistochemical staining in the breast cancer tissue and adjacent tissues also exhibited that the protein level of PTEN was higher in the normal tissues (Fig. 4E). MiR-19a-3p mimic significantly reduced the expression level of PTEN in MCF-7, while miR-NC had no effect (Fig. 4F). These data indicate miR-19a-3p suppresses PTEN expression level in the breast cancer cell.
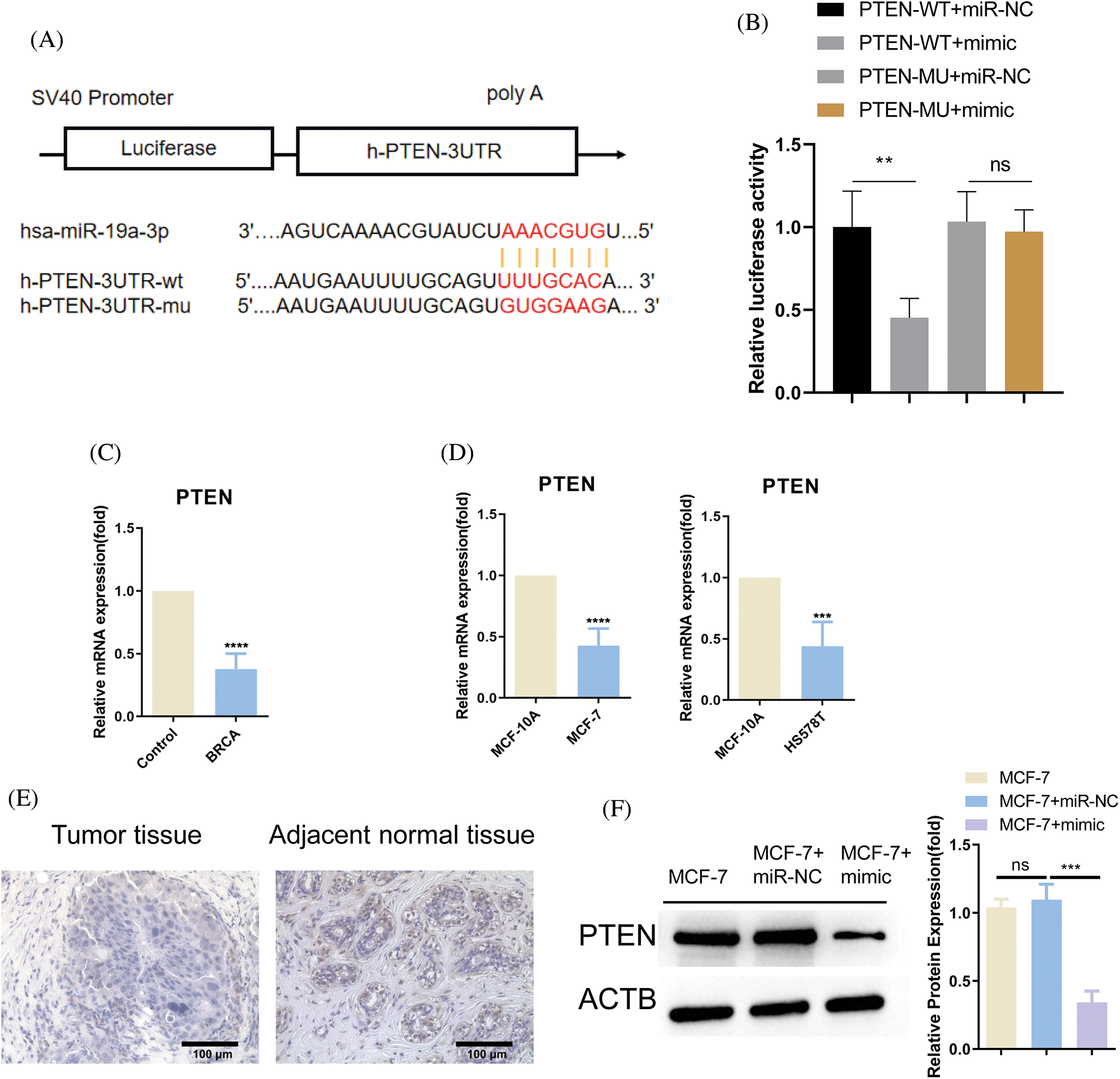
Figure 4: MiR-19a-3p negatively regulates phosphatase and tensin homolog (PTEN). (A) Binding sites of PTEN mRNA 3′-UTR and miR-19a-3p predicted utilizing the TargetScan database. (B) Dual luciferase reporter assay for detecting the interaction of PTEN mRNA and miR-19a-3p. MCF-7 cells were transfected with PTEN-WT or MUT report in the group of miR-19a-3p mimic or miR-NC. (C, D) Analysis of PTEN level in breast cancer specimens (BRCA) and cancerous cell lines (MCF-7 and HS578T), as well as their normal counterpart (control and MCF-10A). (E) Immunohistochemical staining of PTEN in the breast cancer tissue and adjacent tissues. (F) Western blot was performed to detect PTEN expression in MCF-7 cells as well as in MCF-7 cells transfected with miR-19a-3p mimc or miR-NC. ****p < 0.0001; ***p < 0.001; **p < 0.01.
CircHIAT1/miR-19a-3p axis regulates phosphatase and tensin homolog expression and the activity of the PI3K/AKT pathway
Since PTEN is one of the target genes of miR-19a-3p, a well-established inhibitor of PI3K kinase (Izzo et al., 2021; Kechagioglou et al., 2014), we speculated whether circHIAT1/miR-19a-3p axis regulates the presentation of PTEN and the activation status of PI3K/AKT pathway. After the cells were transfected, western blotting was performed to check the PTEN expression and the phosphorylation level of PI3K and AKT. CircHIAT1 overexpression was able to enhance the level of PTEN, while that was repressed by miR-19a-3p mimic co-transfection. In agreement with the PTEN changes, the phosphorylation status of PI3K and AKT was largely inhibited upon circHIAT1 overexpression, and miR-19a-3p mimic co-transfection rescued their phosphorylation levels (Fig. 5).
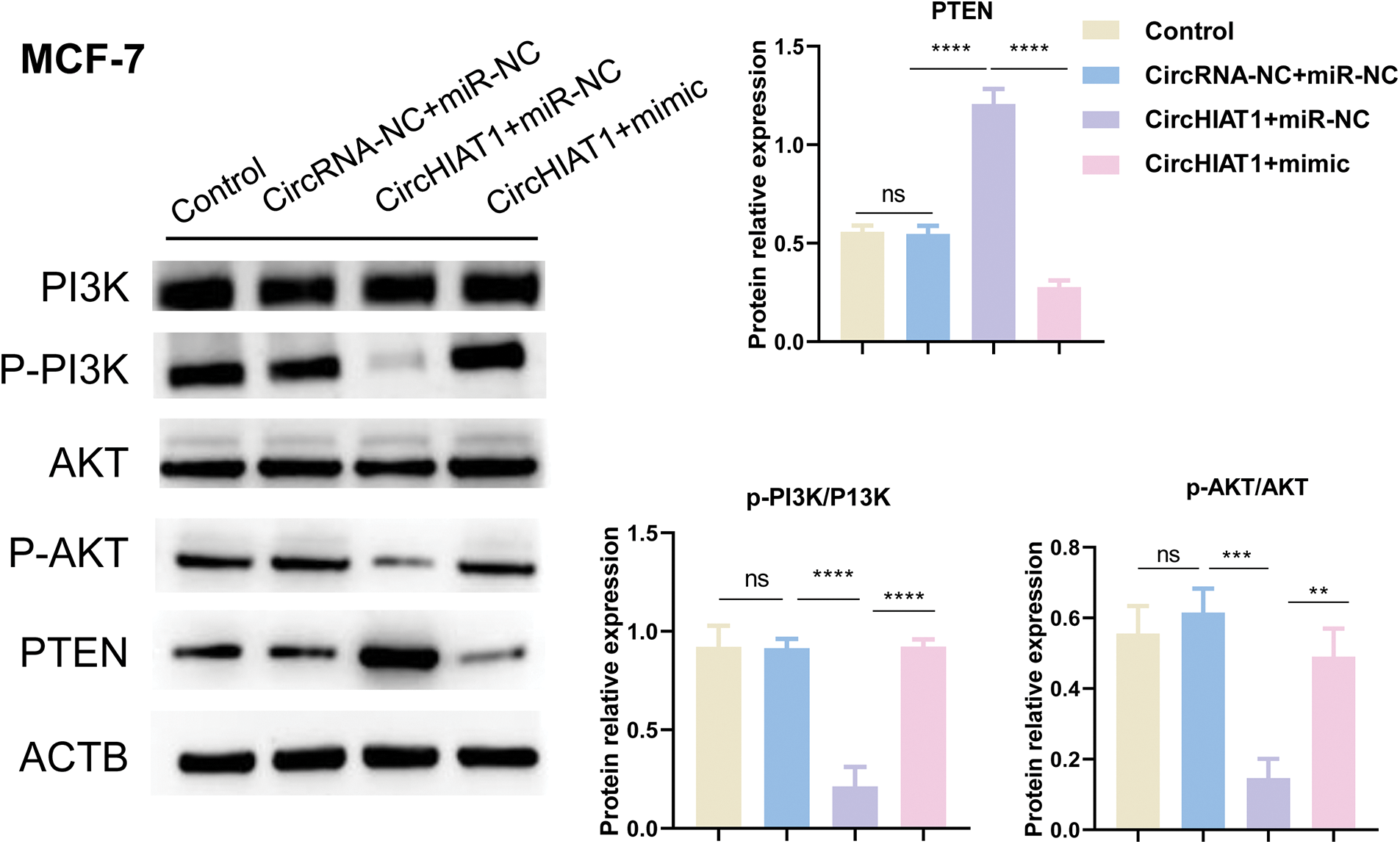
Figure 5: CircHIAT1/miR-19a-3p axis regulates phosphatase and tensin homolog (PTEN) and the activity of the PI3K/AKT pathway. MCF-7 cells were transfected with empty vector and miRNA mimic control (circRNA-NC+miR-NC), circHIAT1 expression vector and miRNA mimic control (circHIAT1+miR-NC), or circHIAT1 expression vector and miR-19a-3p mimic (circHIAT1+mimic). Western blot was performed to detect PTEN expression and the phosphorylation level of PI3K and AKT. ****p < 0.0001; ***p < 0.001; **p < 0.01.
MiR-19a-3p controls the malignancy of breast cancer via targeting phosphatase and tensin homolog
To show the functional roles of miR-19a-3p/PTEN axis and PI3K signaling pathway in the malignancy of breast cancer cells, MCF-7 cells were transfected with inhibitor control and control siRNA (inhibitor-NC and siRNA NC), miR-19a-3p inhibitor+siRNA NC, miR-19a-3p inhibitor+PTEN siRNA, or miR-19a-3p inhibitor+PTEN siRNA+PI3K inhibitor (GDC-0941). As expected, the promte effect of miR-19a-3p on cell growth was revealed (Figs. 6A and 6B). Silencing PTEN could rescue cell proliferation when miR-19a-3p was promoted, and the addition of PI3K inhibitor revoked the effect of PTEN silencing (Figs. 6A and 6B). Transwell invasion assay demonstrated that miR-19a-3p also suppressed cell invasion and silencing PTEN promoted cell invasive ability; the simultaneous treatment of PI3K inhibitor abolished the effect of PTEN silencing (Fig. 6C). Overall, these results highlighted the critical role of miR-19a-3p in regulating the PTEN/PI3K/AKT pathway and the malignant phenotype of breast cancer.
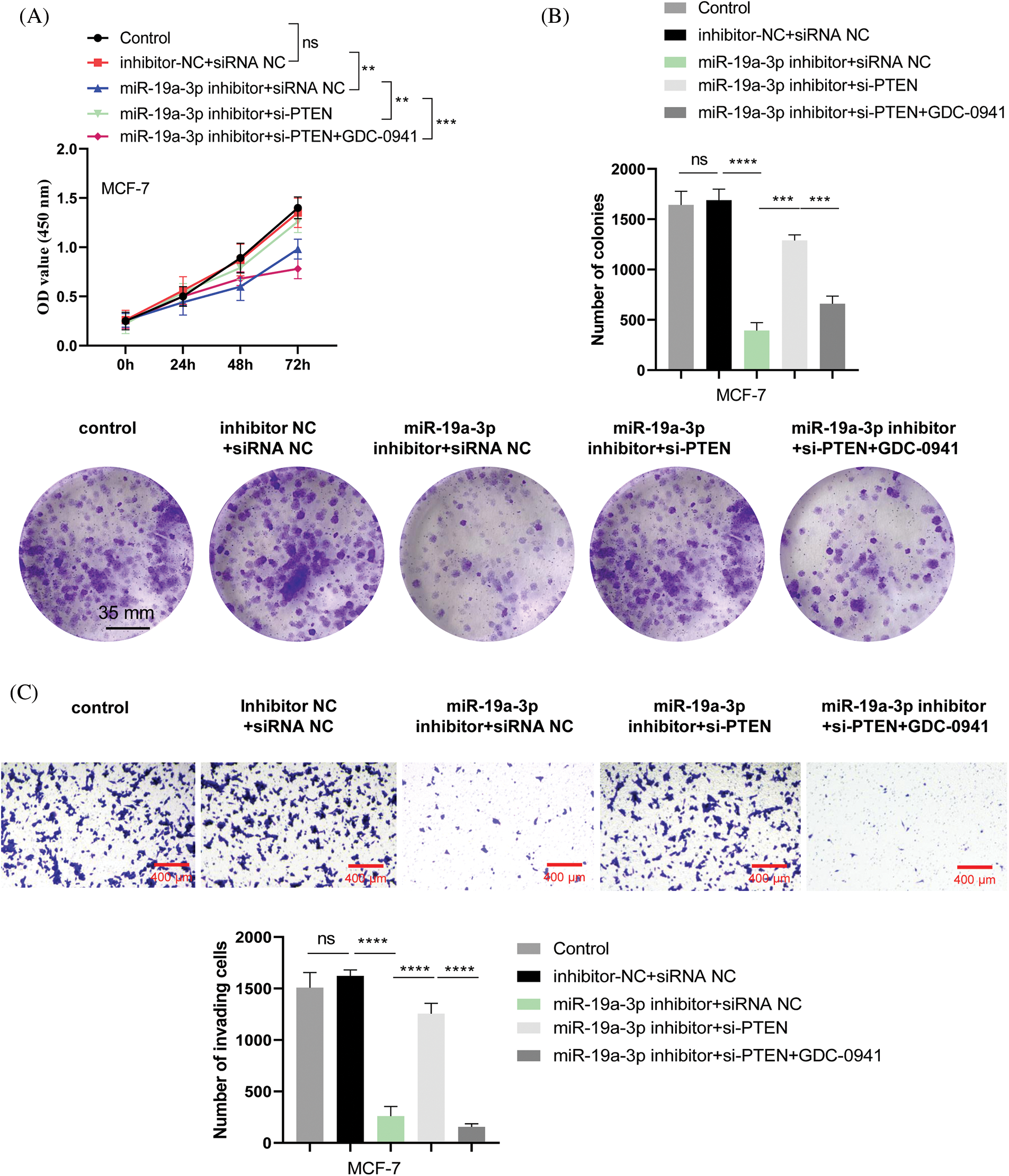
Figure 6: MiR-19a-3p modulates the malignancy of breast cancer via targeting phosphatase and tensin homolog (PTEN) and PI3K signaling pathway. MCF-7 cells were transfected with inhibitor control and control siRNA (inhibitor-NC and siRNA NC), miR-19a-3p inhibitor+siRNA NC, miR-19a-3p inhibitor+PTEN siRNA, or miR-19a-3p inhibitor+PTEN siRNA+PI3K inhibitor (GDC-0941). (A) Cell counting kit-8 proliferation assay, (B) colony formation assay, and (C) transwell invasion assay were performed in the above experimental groups. ****p < 0.0001; ***p < 0.001; **p < 0.01.
CircHIAT1 overexpression suppresses the development of breast cancer
We further evaluated the impact of circHIAT1 overexpression on the tumor development of breast cancer using mice as the xenograft model. Nude mice were injected with control MCF-7 cells, cells expressing empty vector (circRNA-NC) or cells with circHIAT1 expression. MCF-7 cells with circHIAT1 overexpression significantly suppressed the tumor formation of MCF-7 cells (Figs. 7A and 7B). Besides, in the tumor xenograft samples, the overexpression of circHIAT1 was associated with the abnormal expression of miR-19a-3p and PTEN (Figs. 7C–7E). These data further support the tumor-suppressive role of circHIAT1 in breast cancer by targeting the miR-19a-3p/PTEN axis.
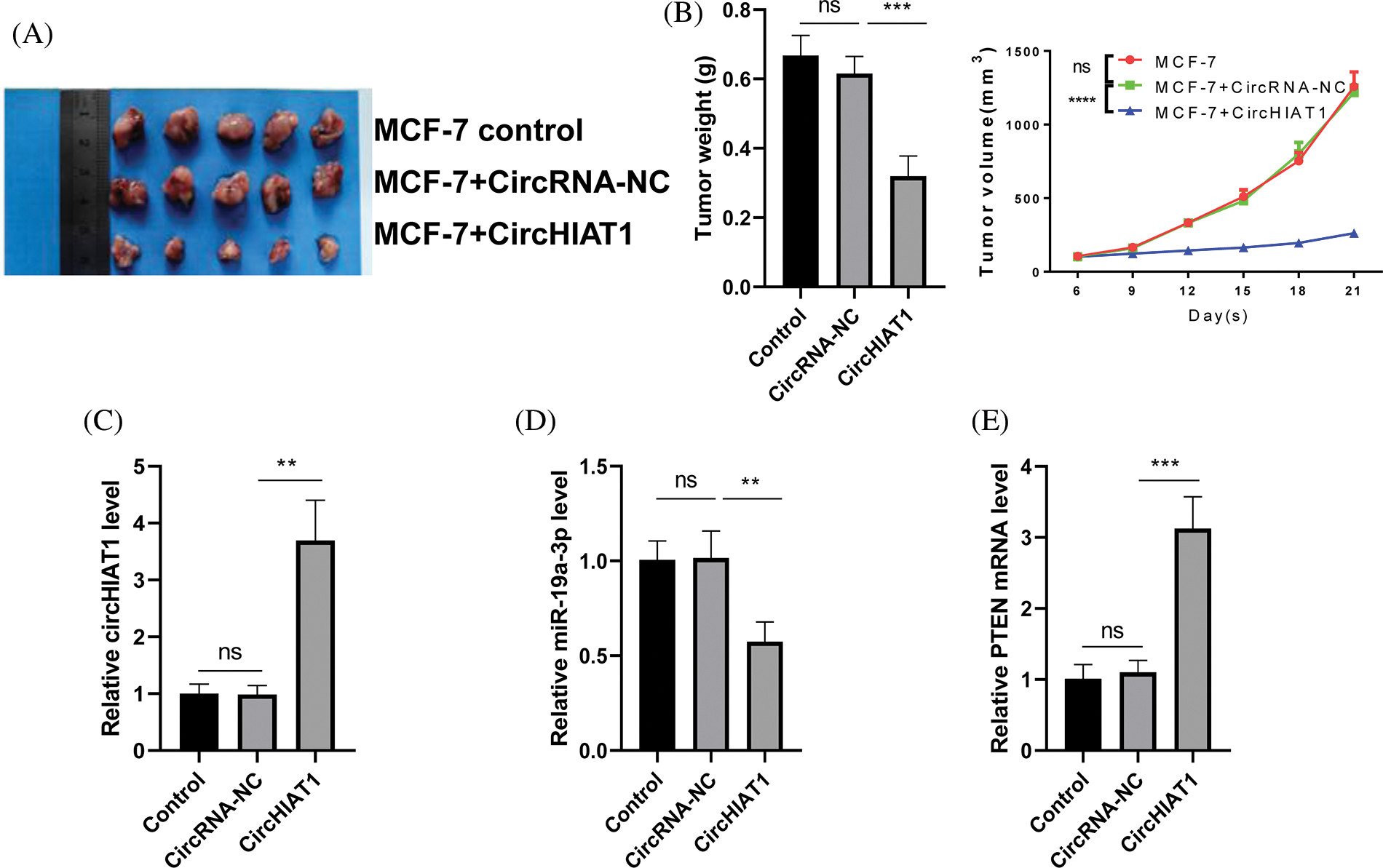
Figure 7: CircHIAT1 overexpression suppresses the development of breast cancer.Nude mice were injected with control MCF-7 cells, cells expressing empty vector (circRNA-NC), or cells with circHIAT1 expression. (A) Xenograft tumor images of each experimental group. (B) Quantization of the weight and volume curves of a xenograft tumor. (C) CircHIAT1 expression. (D) MiR-19a-3p expression, and (E) PTEN mRNA levels were detected in the xenograft samples. ****p < 0.0001; ***p < 0.001; **p < 0.01.
As a tumor suppressor, PTEN is a high-risk factor for lymph node metastasis and the recurrence of breast cancer when its expression decreases (Izzo et al., 2021; Kechagioglou et al., 2014). PTEN can regulate aggressive cellular behaviors, including growth and metastasis. At the same time, as an upstream regulator of the cyclin gene family, PTEN is closely related to tumor cell cycle regulation (Izzo et al., 2021; Kechagioglou et al., 2014; Liu et al., 2016). This study validated the lowered expression of PTEN both in tissues and cells of breast cancer. Further, our database prediction and dual-luciferase reporter experiment revealed that PTEN could be targeted by miR-19a-3p. Moreover, the cell growth and invasion were attenuated by up-regulation of miR-19a-3p, but rescued by PTEN silencing. Therefore, our data identified miR-19a-3p as a tumor-promoting factor and the upstream regulator of PTEN.
MiR-19a-3p was proved to be upregulated in various cancers, such as bladder urothelial carcinoma (Xu et al., 2021), multiple myeloma (Li et al., 2021), and colorectal cancer (Yu et al., 2020). The increased expression level of miR-19a-3p can also be found in the plasma of patients and can be used as a new biomarker in the diagnosis of early non-invasive gastric cancer and colorectal cancer (Yu et al., 2020; Cheng et al., 2020). Whereas whether the high level of miR-19a-3p can also be detected in the blood sample of breast cancer patients requires further investigation. Besides, the expression of miR-19a-3p in bone metastatic prostate cancer tissues and cells is less (Wa et al., 2018). Thus, there may exist different mechanisms in the regulation of miR-19a-3p in tumors of different tissue origins.
PTEN is a canonical inhibitor of PI3K kinase and the PI3K/AKT signaling pathway (Izzo et al., 2021; Kechagioglou et al., 2014). PTEN inactivation combined with the constitutive activation of PI3K/AKT signaling promotes the resistance of breast cancer cells to CDK4/6 inhibitors in vitro and in vivo. Mechanistically, PTEN loss leads to the export of p27 from the nucleus, which in turn activates cyclin dependent kinase 4 (CDK4) and CDK2. In addition, the inactivation of PTEN can also cause resistance to PI3Kα inhibitors (Costa et al., 2020). In triple-negative breast cancer, PTEN inactivation was associated with larger tumor size, multiple lymph node metastases, and greater cell invasiveness (Chai et al., 2022). Our data also displayed that the miR-19a-3p/PTEN axis modulates the invasive behavior of breast cancer, while miR-19a-3p inhibitor suppressed the invasion of breast cancer cells, and PTEN silencing antagonized the effect of miR-19a-3p inhibitor. This finding is consistent with the previous report that PTEN functions as a negative regulator of cell migration and invasion (Dayoub et al., 2022; Zhang and Zheng, 2021). Since PTEN is also found to regulate the epithelial-mesenchymal transition (EMT) in cancer (Fedorova et al., 2022), it remains to be determined whether the miR-19a-3p/PTEN axis modulates the EMT status of breast cancer.
The molecular interplay of circRNA/miRNA/mRNA has been proven to modulate cancer development by an increasing number of studies (Yang et al., 2020; Verduci et al., 2019). CircRNAs are stable RNA molecules engaged in different aspects of cellular activities (Yang et al., 2020; Verduci et al., 2019; Cheng et al., 2020). In this study, we demonstrated that circHIAT1 functions as a molecular sponge to negatively target miR-19a-3p and repress its activity (Fig. 8). The adsorption of miR-19a-3p by circHIAT1 promotes the expression of PTEN. In breast cancer cells, the reduced expression of circHIAT1 causes the release of more miR-19a-3p molecules to suppress PTEN expression, thereby promoting the malignancy of breast cancer cells. These findings are consistent with the reports regarding the tumor-suppressor roles of circHIAT1 in gastric cancer and prostate cancer (Quan et al., 2020; Aghajani Mir et al., 2022). Nevertheless, the mechanisms governing the downregulation of circHIAT1 in different cancers warrant further investigation.
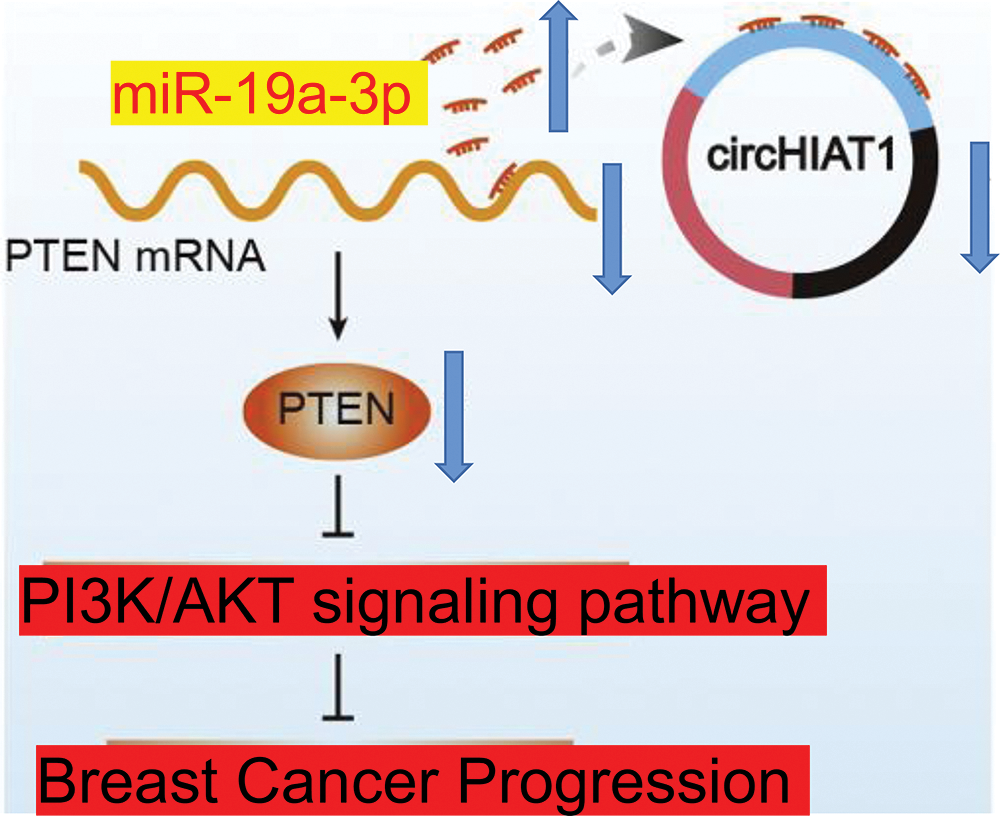
Figure 8: The schematic diagram of the role of circHIAT1/miR-19a-3p/PTEN in the malignancy of breast cancer. circHIAT1 functions as a molecular sponge to adsorb miR-19a-3p and repress its activity. The adsorption of miR-19a-3p by circHIAT1 promotes the expression of phosphatase and tensin homolog (PTEN). In cells, the reduced circHIAT1 expression causes the release of more miR-19a-3p molecules to suppress PTEN, thereby accelerating the malignancy of breast cancer cells.
Our study delineates the molecular interplay among circHIAT1, miR-19a-3p, and PTEN in breast cancer and provides a novel understanding of the downregulation of PTEN by non-coding RNAs. Since circRNA is a class of stable RNA, we speculate that circHIAT1 may be used for the prediction of aggressiveness and PTEN expression in breast cancer. Future works are required to elucidate the mechanisms by which circHIAT1 expression becomes suppressed in breast cancer.
Acknowledgement: Not Applicable.
Funding Statement: The authors received no specific funding for this study.
Author Contributions: Analysis and interpretation of results: Chao Niu and Ruofei Sun; Data analysis; Xiaogang Li and Bo Li; Draft manuscript preparation: Chao Niu and Ruofei Sun. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The data are available from the corresponding author on reasonable request.
Ethics Approval: All experimental procedures were conducted in accordance with Animal Ethics Procedures and Guidelines of the Animal Center, Yunnan University Animal Ethics Committee (Ethics Review Number: YNU20220296).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
Aghajani Mir M, Dinmohammadi H, Moudi E, Motamed N, Daraei A (2022). Clinical values of expression signature of circCDR1AS and circHIAT1 in prostate cancer: Two circRNAs with regulatory function in androgen receptor (AR) and PI3K/AKT signaling pathways. Journal of Clinical Laboratory Analysis 36: e24220. https://doi.org/10.1002/jcla.24220 [Google Scholar] [PubMed] [CrossRef]
Cao W, Chen HD, Yu YW, Li N, Chen WQ (2021). Changing profiles of cancer burden worldwide and in China: A secondary analysis of the global cancer statistics 2020. Chinese Medical Journal 134: 783–791. https://doi.org/10.1097/CM9.0000000000001474 [Google Scholar] [PubMed] [CrossRef]
Chai C, Wu HH, Abuetabh Y, Sergi C, Leng R (2022). Regulation of the tumor suppressor PTEN in triple-negative breast cancer. Cancer Letters 527: 41–48. https://doi.org/10.1016/j.canlet.2021.12.003 [Google Scholar] [PubMed] [CrossRef]
Cheng Y, Su Y, Wang S, Liu Y, Jin L et al. (2020). Identification of circRNA-lncRNA-miRNA-mRNA competitive endogenous RNA network as novel prognostic markers for acute myeloid leukemia. Genes 11: 868. https://doi.org/10.3390/genes11080868 [Google Scholar] [PubMed] [CrossRef]
Cheng J, Yang A, Cheng S, Feng L, Wu X, Lu X, Zu M, Cui J, Yu H, Zou L (2020). Circulating miR-19a-3p and miR-483-5p as novel diagnostic biomarkers for the early diagnosis of gastric cancer. Medical Science Monitor 26: e923444. https://doi.org/10.12659/MSM.923444 [Google Scholar] [PubMed] [CrossRef]
Costa C, Wang Y, Ly A, Hosono Y, Murchie E, Walmsley CS, Huynh T, Healy C, Peterson R, Yanase S (2020). PTEN loss mediates clinical cross-resistance to CDK4/6 and PI3Kα inhibitors in breast cancer. Cancer Discovery 10: 72–85. https://doi.org/10.1158/2159-8290.CD-18-0830 [Google Scholar] [PubMed] [CrossRef]
Dayoub A, Fokin AI, Lomakina ME, James J, Plays M et al. (2022). Inactivation of PTEN and ZFHX3 in mammary epithelial cells alters patterns of collective cell migration. International Journal of Molecular Sciences 24: 313. https://doi.org/10.3390/ijms24010313 [Google Scholar] [PubMed] [CrossRef]
Dillon LM, Miller TW (2014). Therapeutic targeting of cancers with loss of PTEN function. Current Drug Targets 15: 65–79. https://doi.org/10.2174/1389450114666140106100909 [Google Scholar] [PubMed] [CrossRef]
Fedorova O, Parfenyev S, Daks A, Shuvalov O, Barlev NA (2022). The role of PTEN in epithelial-mesenchymal transition. Cancers 14: 3786. https://doi.org/10.3390/cancers14153786 [Google Scholar] [PubMed] [CrossRef]
Gao C, Yuan X, Jiang Z, Gan D, Ding L, Sun Y, Zhou J, Xu L, Liu Y, Wang G (2019). Regulation of AKT phosphorylation by GSK3β and PTEN to control chemoresistance in breast cancer. Breast Cancer Research and Treatment 176: 291–301. https://doi.org/10.1007/s10549-019-05239-3 [Google Scholar] [PubMed] [CrossRef]
Georgescu MM, Kirsch KH, Kaloudis P, Yang H, Pavletich NP, Hanafusa H (2000). Stabilization and productive positioning roles of the C2 domain of PTEN tumor suppressor. Cancer Research 60: 7033–7038. https://doi.org/10.1002/1097-0142(20001215)89:12<2677::AID-CNCR22>3.0.CO;2-L [Google Scholar] [CrossRef]
Hsu MT, Coca-Prados M (1979). Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 280: 339–340. https://doi.org/10.1038/280339a0 [Google Scholar] [PubMed] [CrossRef]
Izzo L, Messineo D, DI Cello P, Nicolanti V, Sterpetti A, Izzo S, Izzo P (2021). Correlation between onco-suppressors PTEN and NM23 and clinical outcome in patients with T1 breast cancer. In Vivo 35: 169–174. https://doi.org/10.21873/invivo.12245 [Google Scholar] [PubMed] [CrossRef]
Kechagioglou P, Papi RM, Provatopoulou X, Kalogera E, Papadimitriou E, Grigoropoulos P, Nonni A, Zografos G, Kyriakidis DA, Gounaris A (2014). Tumor suppressor PTEN in breast cancer: Heterozygosity, mutations and protein expression. Anticancer Research 34: 1387–1400. https://doi.org/10.1007/s13277-013-1375-x [Google Scholar] [CrossRef]
Koual M, Tomkiewicz C, Cano-Sancho G, Antignac JP, Bats AS, Coumoul X (2020). Environmental chemicals, breast cancer progression and drug resistance. Environmental Health 19: 117. https://doi.org/10.1186/s12940-020-00670-2 [Google Scholar] [PubMed] [CrossRef]
Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J (2019). The biogenesis, biology and characterization of circular RNAs. Nature Reviews Genetics 20: 675–691. https://doi.org/10.1038/s41576-019-0158-7 [Google Scholar] [PubMed] [CrossRef]
Kristensen LS, Hansen TB, Venø MT, Kjems J (2018). Circular RNAs in cancer: Opportunities and challenges in the field. Oncogene 37: 555–565. https://doi.org/10.1038/onc.2017.361 [Google Scholar] [PubMed] [CrossRef]
Lei M, Zheng G, Ning Q, Zheng J, Dong D (2020). Translation and functional roles of circular RNAs in human cancer. Molecular Cancer 19: 30. https://doi.org/10.1186/s12-020-1135-7 [Google Scholar] [CrossRef]
Li Y, Chu J, Li J, Feng W, Yang F et al. (2018). Cancer/testis antigen-Plac1 promotes invasion and metastasis of breast cancer through Furin/NICD/PTEN signaling pathway. Molecular Oncology 12: 1233–1248. https://doi.org/10.1002/1878-0261.12311 [Google Scholar] [PubMed] [CrossRef]
Li Y, Gao S, Xue W, Ma Y, Meng Y, Zhang D (2021). MiR-19a-3p functions as an oncogene by regulating FBXO32 expression in multiple myeloma. Balkan Medical Journal 38: 43–49. https://doi.org/10.4274/balkanmedj.galenos.2020.2020.3.121 [Google Scholar] [PubMed] [CrossRef]
Li S, Shen Y, Wang M, Yang J, Lv M, Li P, Chen Z, Yang J (2017). Loss of PTEN expression in breast cancer: Association with clinicopathological characteristics and prognosis. Oncotarget 8: 32043–32054. https://doi.org/10.18632/oncotarget.16761 [Google Scholar] [PubMed] [CrossRef]
Li X, Wang Q, Rui Y, Zhang C, Wang W, Gu J, Tang J, Ding Y (2019). HOXC13-AS promotes breast cancer cell growth through regulating miR-497-5p/PTEN axis. Journal of Cellular Physiology 234: 22343–22351. https://doi.org/10.1002/jcp.28800 [Google Scholar] [PubMed] [CrossRef]
Liu X, Chen Q, Hu XG, Zhang XC, Fu TW et al. (2016). PTP1B promotes aggressiveness of breast cancer cells by regulating PTEN but not EMT. Tumor Biology 37: 13479–13487. https://doi.org/10.1007/s13277-016-5245-1 [Google Scholar] [PubMed] [CrossRef]
Lu Y, Hu X, Yang X (2021). MiR-934 promotes breast cancer metastasis by regulation of PTEN and epithelial-mesenchymal transition. Tissue & Cell 71: 101581. https://doi.org/10.1016/j.tice.2021.101581 [Google Scholar] [PubMed] [CrossRef]
Misir S, Hepokur C, Aliyazicioglu Y, Enguita FJ (2020). Circular RNAs serve as miRNA sponges in breast cancer. Breast Cancer 27: 1048–1057. https://doi.org/10.1007/s12282-020-01140-w [Google Scholar] [PubMed] [CrossRef]
Quan J, Dong D, Lun Y, Sun B, Sun H, Wang Q, Yuan G (2020). Circular RNA circHIAT1 inhibits proliferation and epithelial-mesenchymal transition of gastric cancer cell lines through downregulation of miR-21. Journal of Biochemical and Molecular Toxicology 34: e22458. https://doi.org/10.1002/jbt.22458 [Google Scholar] [PubMed] [CrossRef]
Trayes KP, Cokenakes SEH (2021). Breast cancer treatment. American Family Physician 104: 171–178. [Google Scholar] [PubMed]
Treeck O, Schüler-Toprak S, Skrzypczak M, Weber F, Ortmann O (2020). Knockdown of PTEN decreases expression of estrogen receptor β and tamoxifen sensitivity of human breast cancer cells. Steroids 153: 108521. https://doi.org/10.1016/j.steroids.2019.108521 [Google Scholar] [PubMed] [CrossRef]
Verduci L, Strano S, Yarden Y, Blandino G (2019). The circRNA-microRNA code: Emerging implications for cancer diagnosis and treatment. Molecular Oncology 13: 669–680. https://doi.org/10.1002/1878-0261.12468 [Google Scholar] [PubMed] [CrossRef]
Wa Q, Li L, Lin H, Peng X, Ren D, Huang Y, He P, Huang S (2018). Downregulation of miR‐19a‐3p promotes invasion, migration and bone metastasis via activating TGF‐β signaling in prostate cancer. Oncology Reports 39: 81–90. https://doi.org/10.3892/or.2017.6096 [Google Scholar] [PubMed] [CrossRef]
Wang DD, Yang SJ, Chen X, Shen HY, Luo LJ, Zhang XH, Zhong SL, Zhao JH, Tang JH (2016). MiR-222 induces Adriamycin resistance in breast cancer through PTEN/Akt/p27(kip1) pathway. Tumor Biology 37: 15315–15324. https://doi.org/10.1007/s13277-016-5341-2 [Google Scholar] [PubMed] [CrossRef]
Wein L, Loi S (2017). Mechanisms of resistance of chemotherapy in early-stage triple negative breast cancer (TNBC). Breast 34: S27–S30. https://doi.org/10.1016/j.breast.2017.06.023 [Google Scholar] [PubMed] [CrossRef]
Wu P, Mo Y, Peng M, Tang T, Zhong Y et al. (2020). Emerging role of tumor-related functional peptides encoded by lncRNA and circRNA. Molecular Cancer 19: 22. https://doi.org/10.1186/s12943-020-1147-3 [Google Scholar] [PubMed] [CrossRef]
Xu G, Li J, Yu L (2021). MiR-19a-3p promotes tumor-relevant behaviors in bladder urothelial carcinoma via targeting THBS1. Computational and Mathematical Methods in Medicine 2021: 2710231. https://doi.org/10.1155/2021/2710231 [Google Scholar] [PubMed] [CrossRef]
Yang Y, Wang YJ, Wang F, Yuan LH, Guo ZQ, Wei ZC, Wang ZR, Wang SW (2020). The roles of miRNA, lncRNA and circRNA in the development of osteoporosis. Biological Research 53: 40. https://doi.org/10.1186/s40659-020-00309-z [Google Scholar] [PubMed] [CrossRef]
Yu FB, Sheng J, Yu JM, Liu JH, Qin XX, Mou B (2020). MiR-19a-3p regulates the Forkhead box F2-mediated Wnt/β-catenin signaling pathway and affects the biological functions of colorectal cancer cells. World Journal of Gastroenterology 26: 627–644. https://doi.org/10.3748/wjg.v26.i6.627 [Google Scholar] [PubMed] [CrossRef]
Zhai Z, Mu T, Zhao L, Li Y, Zhu D, Pan Y (2022). MiR-181a-5p facilitates proliferation, invasion, and glycolysis of breast cancer through NDRG2-mediated activation of PTEN/AKT pathway. Bioengineered 13: 83–95. https://doi.org/10.1080/21655979.2021.2006974 [Google Scholar] [PubMed] [CrossRef]
Zhang M, Bai X, Zeng X, Liu J, Liu F, Zhang Z (2021). CircRNA-miRNA-mRNA in breast cancer. Clinica Chimica Acta 523: 120–130. https://doi.org/10.1016/j.cca.2021.09.013 [Google Scholar] [PubMed] [CrossRef]
Zhang XY, Mao L (2021). Circular RNA Circ_0000442 acts as a sponge of MiR-148b-3p to suppress breast cancer via PTEN/PI3K/Akt signaling pathway. Gene 766: 145113. https://doi.org/10.1016/j.gene.2020.145113 [Google Scholar] [PubMed] [CrossRef]
Zhang Y, Zheng A (2021). MiR-142-5p promotes retinoblastoma cell proliferation, migration and invasion by targeting PTEN. Journal of Biochemistry 170: 195–202. https://doi.org/10.1093/jb/mvaa121 [Google Scholar] [PubMed] [CrossRef]
Zhu Q, Zhang X, Zai HY, Jiang W, Zhang KJ, He YQ, Hu Y (2021). CircSLC8A1 sponges miR-671 to regulate breast cancer tumorigenesis via PTEN/PI3k/Akt pathway. Genomics 113: 398–410. https://doi.org/10.1016/j.ygeno.2020.12.006 [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools