 Open Access
Open Access
ARTICLE
miR-103-3p regulates the differentiation of bone marrow mesenchymal stem cells in myelodysplastic syndrome
1 School of Medicine, South China University of Technology, Guangzhou, 510006, China
2 Department of Hematology, Guangdong Provincial People’s Hospital, Guangdong Academy of Medical Sciences, Guangzhou, 510080, China
* Corresponding Authors: Xin Du, ; Jianyu Weng,
# These authors contributed equally to this work
§ Present address: Department of Hematology, Guangzhou First People’s Hospital, School of Medicine, South China University of Technology, Guangzhou, 510180, China
(This article belongs to the Special Issue: Decoding Gene (including circRNA, lincRNA miRNA and mRNA) Expression)
BIOCELL 2023, 47(1), 133-141. https://doi.org/10.32604/biocell.2022.022021
Received 17 March 2022; Accepted 24 May 2022; Issue published 26 September 2022
Abstract
The pathogenesis of myelodysplastic syndrome (MDS) may be related to the abnormal expression of microRNAs (miRNAs), which could influence the differentiation capacity of mesenchymal stem cells (MSCs) towards adipogenic and osteogenic lineages. In this study, exosomes from bone marrow plasma were successfully extracted and identified. Assessment of miR-103-3p expression in exosomes isolated from BM in 34 MDS patients and 10 controls revealed its 0.52-fold downregulation in patients with MDS compared with controls (NOR) and was downregulated 0.55-fold in MDS-MSCs compared with NOR-MSCs. Transfection of MDS-MSCs with the miR-103-3p mimic improved osteogenic differentiation and decreased adipogenic differentiation in vitro, while inhibition of miR-103-3p showed the opposite results in NOR-MSCs. Thus, the expression of miR-103-3p decreases in MDS BM plasma and MDS-MSCs, significantly impacting MDS-MSCs differentiation. The miR-103-3p mimics may boost MDS-MSCs osteogenic differentiation while weakening lipid differentiation, thereby providing possible target for the treatment of MDS pathogenesis.Keywords
Bone marrow loss and a high proclivity to leukemic progression are the characteristics of myelodysplastic syndrome (MDS). Recent research has revealed that the mesenchymal niche plays a significant role in the onset and progression of the MDS (Chen et al., 2021; Pronk and Raaijmakers, 2019).
The bone marrow microenvironment (BMME) is a complex network system primarily comprising stromal cells such as mesenchymal stem cells (MSCs), osteoprogenitor cells, vascular endothelial cells, monocytes, macrophages, and cytokines. It maintains normal hematopoietic function by interacting with hematopoietic stem cells (HSCs) to control their proliferation and differentiation (Morrison and Scadden, 2014). Considering that an abnormal BMME is essential for myeloid tumor cell invasion and anti-apoptosis, cloning and amplifying MDS cells, poor hematopoiesis of bone marrow, and disease progression, BMME may be a possible therapeutic target (Matsuda et al., 2004; Mei et al., 2018).
The bone marrow stromal cell (BMSC) population, which makes up a large part of the hematopoietic microenvironment, encompasses a variety of adherent cell types. The three most important types of BMSCs in the BMME are mesenchymal stem cells, endothelial cells, and osteoblasts, and all three play important roles in hematopoietic regulation (Chen et al., 2017; Psaila et al., 2012). Adipocytes, osteoblasts, astrocytes, cardiomyocytes, chondrocytes, hepatocytes, muscles, and neurons can all develop from MSCs (Doan and Chute, 2012). MSCs have a lower ability to differentiate into osteoblasts but a higher ability to differentiate into adipocytes, which could be linked to myeloid malignancies (Woods and Guezguez, 2021). However, more research into this mechanism is needed.
Recent research has suggested that the faulty activities of MDS-MSCs may be related to abnormalities in the expression of microRNA (Meunier et al., 2020). For example, DICER1 expression is downregulated in MDS-MSCs and MSCs from acute myeloid leukemia (AML) patients, and microRNA (miRNA) expression is dysregulated compared with those in controls (NOR-MSCs), and MSCs from both MDS and AML patients are changed, thus limiting stromal assistance to HSCs (Ozdogan et al., 2017; Santamaría et al., 2012). MiR-7977 causes an abnormal decrease in hematopoietic growth factors in MSCs, leading to a reduction in the hematopoietic supportive capacity of CD34+ cells in the bone marrow (Horiguchi et al., 2016). Previous studies have shown that aberrant microRNA expression can affect MDS-MSCs, which can then affect MDS progression.
MicroRNAs (miRNAs) are non-coding RNAs with a length of 18–25 nucleotides and always function as a passive precursor hairpin over target gene expression by degrading or suppressing mRNA translation (Bartel, 2004). Several recent studies have shown the relevance of miRNAs in controlling the hematopoietic microenvironment (Machova et al., 2011). However, the involvement of miRNAs in the shift in the differentiation of MSCs from osteoblasts to adipocytes in MDS is yet to be investigated (Rhyasen and Starczynowski, 2012). We studied the database GSE139471 by GEO-2R (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE139471) and found reduced expression of miR-103-3p in MDS-MSCs.
In the bone marrow supernatant, miRNAs are mostly secreted by MSC-derived exosomes and are involved in cell-to-cell communication, cell signaling, and altering cell or tissue metabolism. The contents of MSC-derived exosomes are not static but depend on MSCs tissue origin, their activities, and their immediate intercellular neighbors. Hence, abnormal miRNA levels in exosomes are of great significance when MSCs are cultured with tumor cells or the in vivo tumor microenvironment (Muntión et al., 2016).
Therefore, in addition to the evidence provided by the database, we detected the expression level of miR-103-3p in exosomes from BM supernatant of MDS patients.
MiR-103 is associated with endometrial cancer, breast cancer, rectal cancer, and other tumors (Chung et al., 2009; Yu et al., 2012; Nonaka et al., 2015) and is also relevant for the proliferation of leukemia cells (Kfir-Erenfeld et al., 2017). MiR-103 plays a key role in facilitating the differentiation of MSCs into adipocytes and promotes preadipocyte ER stress and apoptosis by inhibiting the canonical Wnt/β-catenin pathway (Zhang et al., 2018).
Osteogenic differentiation is regulated by: runt-related transcription factor 2 (RUNX2). RUNX2 is an essential regulator of bone formation and osteogenic differentiation of MSCs (Almalki and Agrawal, 2016). It has a significant impact on the transformation of bone marrow mesenchymal stem cells to osteoblasts (Zhao et al., 2021) and has been identified as a major player in the maintenance of bone homeostasis by promoting osteogenic differentiation of MSCs (Djouad et al., 2017). A number of miRNAs have been recently identified in the regulation of RUNX2 expression/activity, thus affecting the process of osteogenesis. MiRNAs targeting RUNX2 corepressors favor osteogenesis, while those targeting RUNX2 coactivators inhibit osteogenesis (Narayanan et al., 2019).
Adipogenesis relies on the regulation of the expression of several transcription factors, including peroxisome proliferation-activated receptor γ (PPARG). PPARG is one of the master regulators of adipogenic differentiation (Zhuang et al., 2016) and is expressed in both osteoblasts and adipocytes, as well as in MSCs, suggesting its crucial role in regulating adipocyte formation and osteoblast development (Giaginis et al., 2007), and it is considered a validation marker of the adipogenic differentiation of MSCs.
In this study, following our hypothesis that miR-103-3p is related to the function of MDS-MSCs, we detected the expression and level of miR-103-3p and investigated the effect of miR-103-3p on the differentiation of MSCs obtained from the bone marrow of MDS patients.
The MDS patients were recruited from the Department of Hematology, Guangdong Provincial People’s Hospital, Guangdong Academy of Medical Sciences, Guangzhou, Guangdong, China. All the patients diagnosed with MDS were over 18 years of age. Baseline characteristics of the study population included sex, age, peripheral blood cell count, WHO classification, and IPSS-R risk group (Table 1).
Exosome isolation and identification from bone marrow plasma
The Total Exosome Isolation kit (Thermo Scientific, Waltham, MA, USA) was used to extract exosomes from bone marrow plasma. We mixed 1 mL plasma with 0.5 mL PBS and mixed well using a vortex. Then, we added 0.05 mL Proteinase K and 0.2 mL exosome precipitation reagent to the sample and mixed them thoroughly. After incubation of the sample for 10 min at room temperature, we centrifuged the samples for 5 min at room temperature and 10,000 g and discarded the supernatant. The exosomes were trapped in a pellet at the tube’s bottom.
Exosomes are formed when the plasma membrane is doubly invaginated, forming intracellular multivesicular bodies (MVBs) containing intraluminal vesicles. These vesicles are finally released as exosomes with diameters varying from 40 to 160 nm because of MVB fusion to the plasma membrane and exocytosis. Their average diameter is 100 nm. In the realm of exosome research, nanoparticle tracking analysis (NTA) has been recognized as one of the methods for exosome characterization (Kalluri and LeBleu, 2020).
Isolation, culture, and identification of bone marrow mesenchymal stem cells
Fresh bone marrow was used to isolate the mononuclear cells. The cells were then placed in a T75 flask, and the culture medium used was the human MesenCult proliferation kit (STEMCELL Technologies Canada). When the confluence reached 90%, cell passage was accomplished with 0.25% trypsin, usually on the 14th day.
The surface markers of the cultured MSCs were detected by flow cytometry (BD Biosciences, USA). The MSCs were positive for CD73, CD90, and CD105 and negative for CD34, CD45, and CD14 (eBiosciences, USA) (Mushahary et al., 2018), and suitable isotypic controls were used to characterize the MSCs.
MSCs differentiate into adipogenic and osteogenic lineages (Malvicini et al., 2019). A total of 2 × 105 cells were seeded per well in six-well plates containing an osteogenic induction medium, the OriCell® Adult Bone Marrow Mesenchymal Stem Cell Osteogenic Induction and Differentiation Kit (Cyagen), for 21 days; the medium was changed every three days for osteogenic differentiation. On the 21st day, the cells were fixed for 1 h with paraformaldehyde and then stained with Alizarin Red S solution at room temperature for 30 min. An adipogenic induction medium, the OriCell®Human Bone Marrow Mesenchymal Stem Cell Adipogenic Differentiation Basal Medium (Cyagen), was used for adipogenic differentiation. A total of 2 × 105 cells were plated per well in six-well plates containing the full lipid-induced differentiation medium. For each cycle, MSCs were induced to differentiate in medium A for 72 h and then cultured in medium B for 24 h. After 3–5 induction cycles (21 days), the cells were dyed with an oil red solution at room temperature for half an hour.
miR-103-3p mimic, inhibitor, and negative control (mir-NC) were synthesized by Ribo Biotechnology Company, Guangzhou, China. When the MSCs passed to the third generation and reached 60%–70% confluence, Lipofectamine 3000 Transfection Reagent (Thermo Scientific, Waltham, MA, USA) was used to transfect human bone marrow MSCs with miR-103-3p mimic, miR-103-3p inhibitor, and mir-NC all labeled by fluorescent. The transfection working solution was discarded after 48 h, and the MSCs were washed three times with phosphate buffer saline (PBS).
After routine culture for one week, total RNA was extracted from the cells using the miRNeasy Mini Kit (Qiagen, Germany). The extracted RNA was reverse transcribed into cDNA using miRNA 1st strand cDNA synthesis kit AG11717 (Accurate Biology, Hunan, China) following the manufacturer’s instructions. The RNA expression levels of PPARG, RUNX2, and miR-103-3p were determined using quantitative polymerse chain reaction (qPCR). The reference gene for miR-103-3p was U6. The qPCR cycling conditions were as follows: 95°C for 30 s followed by 40 cycles of 95°C for 5 s and 60°C for 30 s.
GraphPad Prism 7.0 (GraphPad Software, San Diego, CA) was used to perform statistical analysis and prepare graphs. The means of the two samples were compared by independent-sample or paired-sample t test, depending on the specific situation.
miR-103-3p expression in exosomes generated from the bone marrow of patients with myelodysplastic syndrome
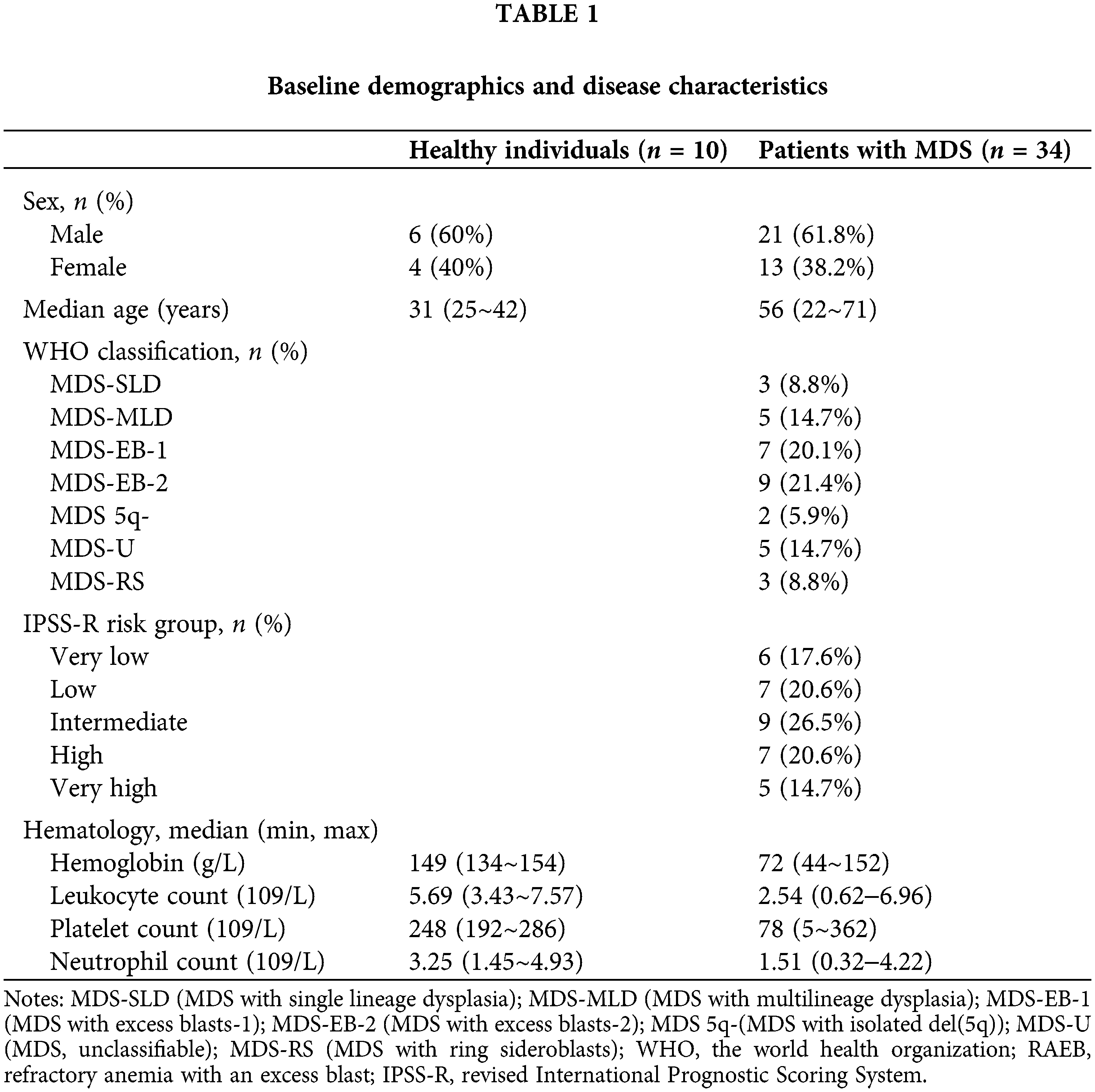
Exosomes were extracted from the BM supernatant of MDS patients, and the exosomal RNA was used for qPCR to detect the expression level of our target microRNA. The expression of miR-103-3p was noticeably reduced (0.52-fold, ΔΔCт = 1.994458422) in exosomes from bone marrow plasma of 34 patients with MDS compared with healthy controls (P ≤ 0.01) (Fig. 1A).
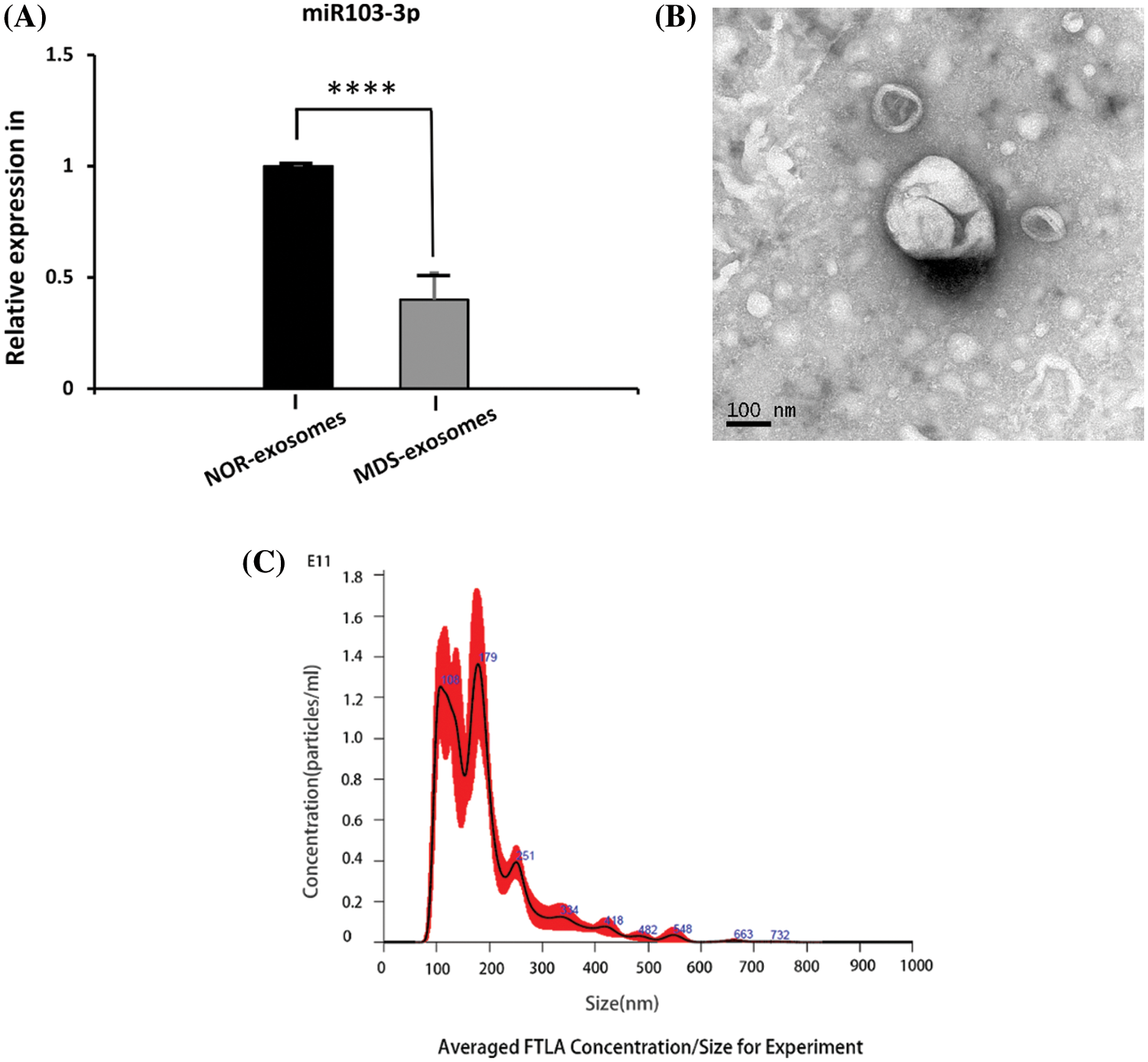
Figure 1: (A) Expression of miR‑103-3p in exosomes derived from the bone marrow plasma samples from 10 healthy controls and 34 patients with MDS ****P ≤ 0.01. (B) Under the electron microscope, exosome structures were observed in all samples. (C) The measured average particle size of the samples was 189.4 ± 2.6 nm, and exosomes were basically in the distribution range of 30–200 nm.
Several methods, including electron microscopy, flow cytometry, and western blot analysis, have been used to characterize exosomes (Mateescu et al., 2017). Herein, electron microscopy revealed exosome structures in all the samples (Fig. 1B). As shown by NTA, exosomes were basically in the distribution range of 30–200 nm (Fig. 1C). The measured average particle size of the exosome was 189.4 ± 2.6 nm.
The expression of miR‑103-3p in mesenchymal stem cells derived from bone marrow
MSCs express CD90, CD73, and CD105, but not CD34, CD14, or CD45, and are plastic-adherent in culture. MDS-MSCs collected from five samples (P3) expressed >90% CD105, CD90, and CD73 surface markers, and did not express CD45, CD34, and CD14 surface markers (Fig. 2A). In early passage (P3), both MDS-MSCs (n = 5) and NOR-MSCs (n = 5) had typical fibroblast-like morphology (Fig. 2B).
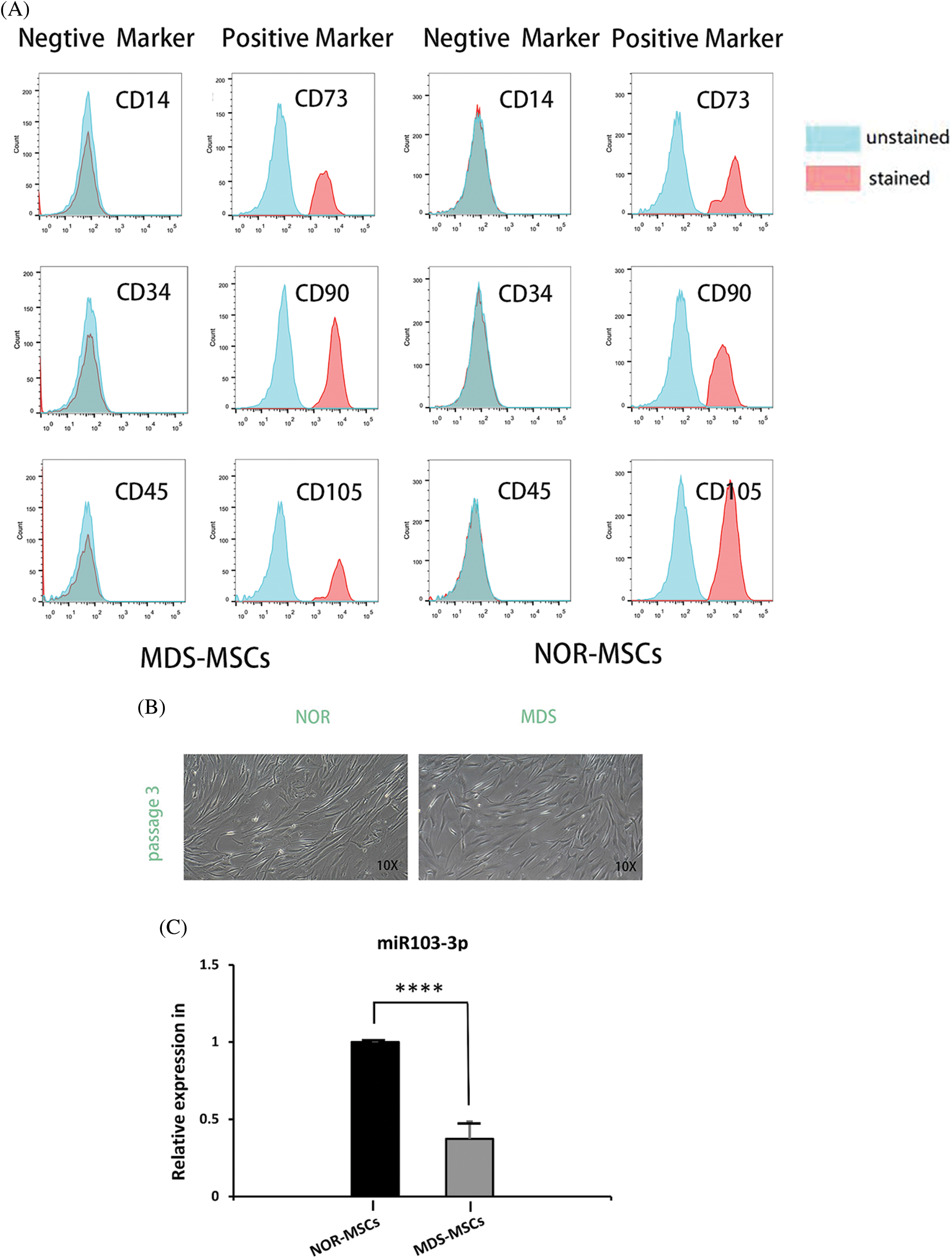
Figure 2: (A) Phenotypic characterization of bone marrow-mesenchymal stem cells (BM-MSCs) from control (NOR-MSCs) and those of myelodysplastic syndrome (MDS) (n = 5, P3). (B) Both MDS-MSCs (n = 5) and NOR-MSCs (n = 5) showed typical fibroblast-like morphology in the early passage (P3). (C) The expression of miR‑103-3p was obviously decreased in the bone marrow of MSCs of patients with MDS, n = 19, ****P ≤ 0.01.
Analysis of the database GSE139471 by GEO-2R (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE139471) revealed that the reduction of miR‑103-3p was the highest and was statistically significant in MDS-MSCs among the 384 microRNAs tested, and quantitative real-time-polymerase chain reaction (qRT-PCR) demonstrated a noticeable decrease in expression of miR‑103-3p (0.55-fold, ΔΔCт = 3.201984678) in BM MSCs of 19 MDS patients, compared with healthy donors (P ≤ 0.01) (Fig. 2C).
Inhibition of adipogenic differentiation due to high expression of miR‑103-3p and enhanced osteogenic differentiation of myelodysplastic syndrome-mesenchymal stem cells
To determine miRNA transfection concentration and overexpression in MDS and downregulated in NOR, we tried transfecting with gradient concentrations (50, 100, 160 nM) miRNA to find the concentration with the highest transfection success rate by measuring fluorescence signal and a transfection concentration of 160 nM of the mimic achieved the highest transfection success rate, that is, more than 90% (Fig. 3A).
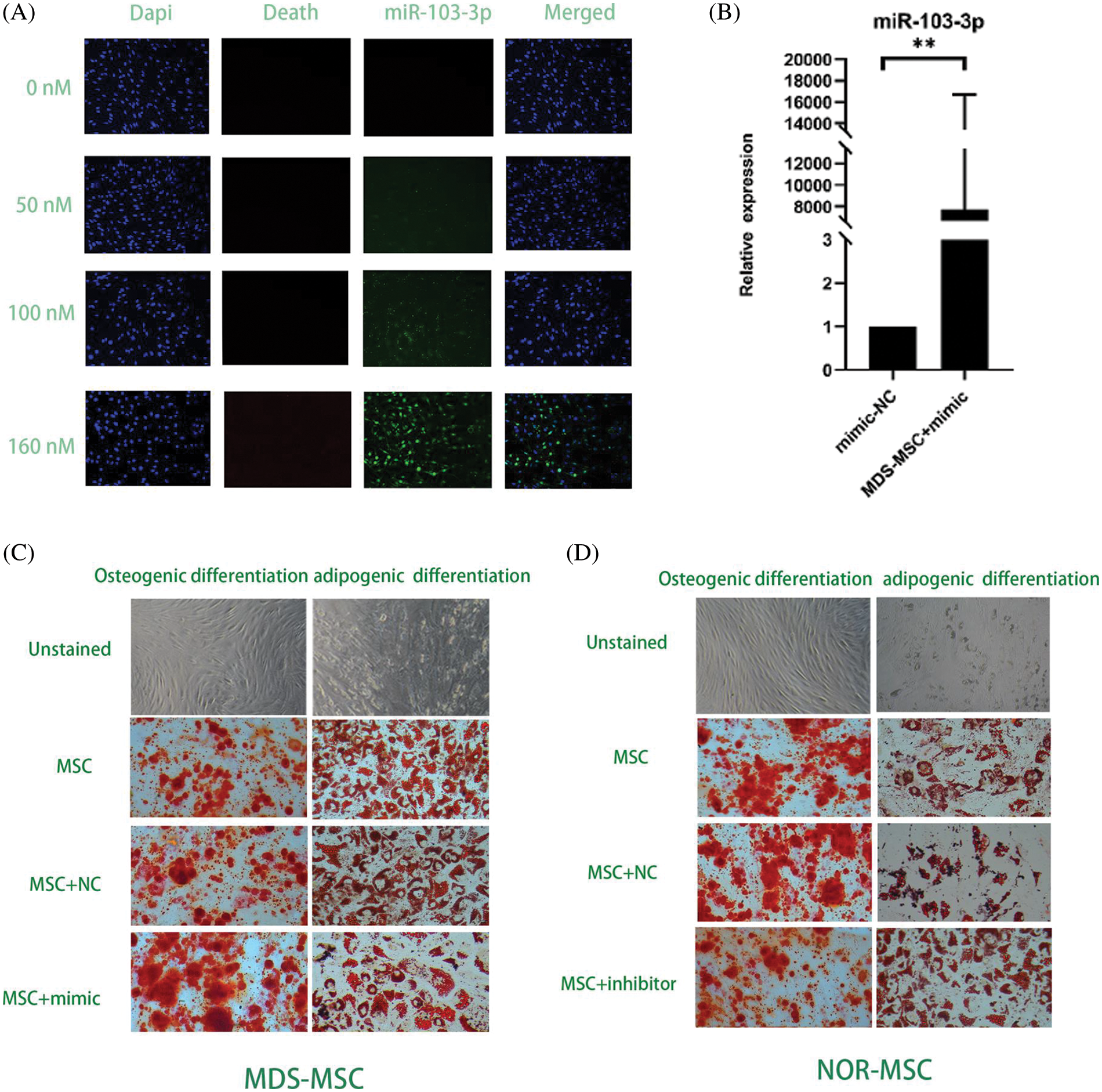
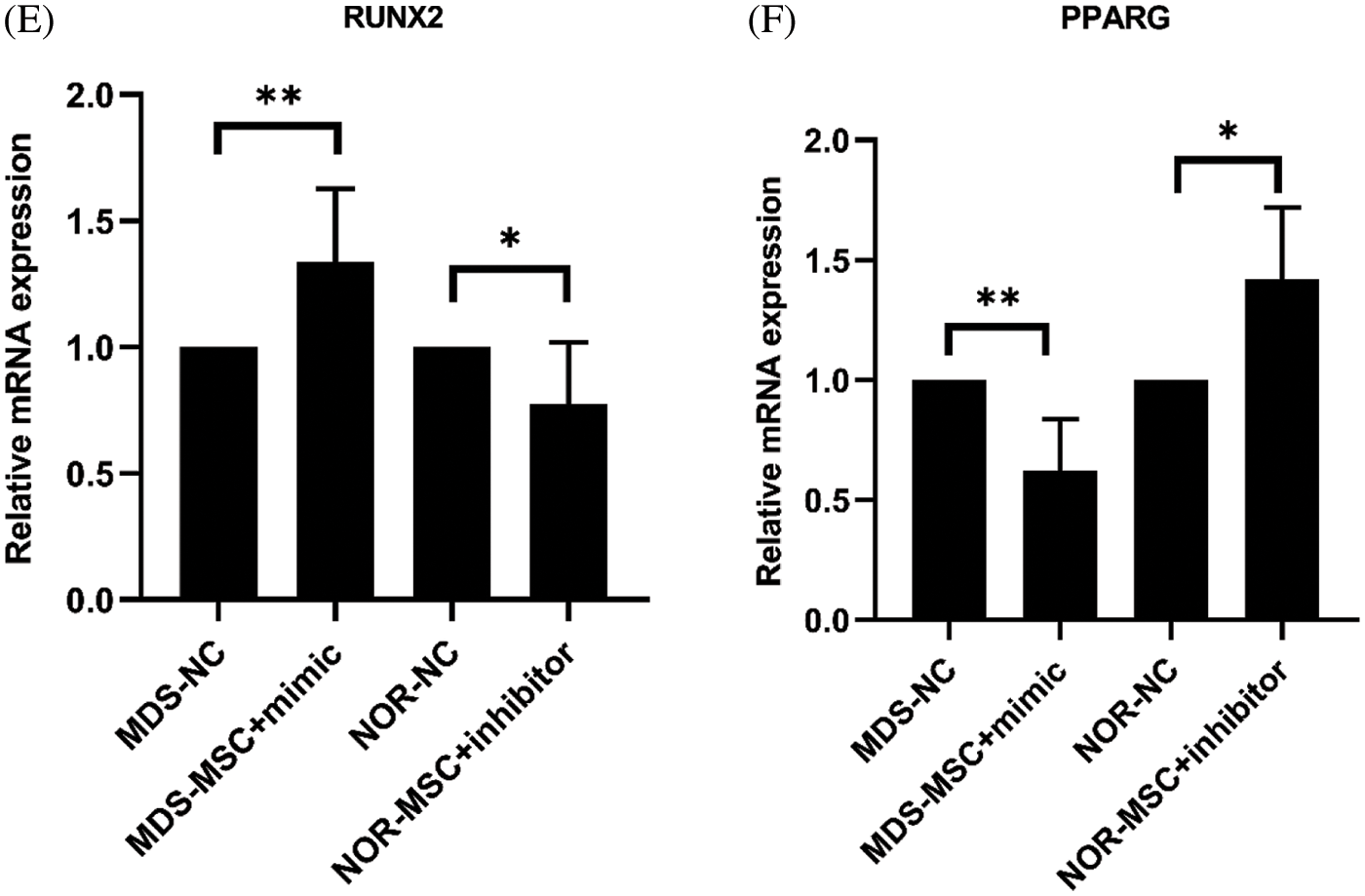
Figure 3: (A) At160 nM concentrations of the miR-103-3p mimic and the inhibitor, the transfection rate of myelodysplastic syndrome-mesenchymal stem cells (MDS-MSCs) was more than 90%. (B) After the transfection, the expression of miR‑103-3p obviously increased in MDS-MSCs; average fold. (C) To compare the degree of differentiation of the cells in each of the groups (as in the figure above), oil red and alizarin red staining were conducted. Compared with the NC group, the alizarin red color in the MDS-MSC+mimic group as greatly enhanced, whereas oil red staining was significantly reduced. (D) Compared with the NC group (control), the NOR-MSC+inhibitor group showed much less calcification. The density of lipid droplets increased considerably. (E) Quantitative real-time polymerase chain (qRT-PCR) analysis of mRNA expression levels of RUNX2 in MDS-MSCs and NOR-MSCs transfected with the miR-103-3p mimic or inhibitor. n = 6, *P ≤ 0.01. (F) Q-PCR analysis of mRNA expression levels of PPARG (coding for peroxisome proliferation-activated receptor γ) in MDS-MSCs and NOR-MSCs transfected with the miR-130a mimic and the miR-130a inhibitor. n = 6, * P ≤ 0.01.
To evaluate whether the aberrant expression of miR-103-3p could affect the function of MSCs in MDS, the miR-103-3p mimic was transfected along with Lipo3000 into 16 MDS-MSCs. After transfection, there was a noticeable improvement in the expression of miR‑103-3p in MDS-MSCs (average fold change 7722.52, P ≤ 0.05) (Fig. 3B). Meanwhile, we transfected the inhibitor of miR-103-3p into five NOR-MSCs.
Additional testing was needed to confirm the impact of miR-103-3p on osteogenic differentiation of MDS-MSCs. Therefore, we overexpressed miR-103-3p in MDS-MSCs and downregulated miR-103-3p in NOR-MSCs. Adipogenic differentiation was significantly inhibited in the MDS-MSCs+mimic group and enhanced in the NOR-MSCs+inhibitor group (Fig. 3D). The opposite was observed for adipogenic differentiation. Meanwhile, the Oil O Red staining revealed decreased lipid content in the MDS-MSCs+mimic group, while alizarin red staining revealed a significant reduction in the formation of mineralized nodules in the NOR-MSC+inhibitor group (Fig. 3C). For lipid formation, staining in MDS-MSCs was stronger than NOR-MSCs, and weaker in bone formation than NOR-MSCs (Figs. 3C and 3D). These data indicated that the high expression of miR-103-3p significantly impeded the advancement of adipogenic differentiation of MDS-MSCs. Overexpression of miR-103-3p inhibited osteogenic differentiation of hMSCs. Following the overexpression of miR-103-3p in MDS-MSCs, there was a significant increase in the expression of RUNX2, whose activity is associated with osteoblast differentiation (Narayanan et al., 2019). However, the expression of PPARG, an indispensable factor of adipogenesis whose pathway promotes the formation of adipocytes (Meyer et al., 2016), decreased slightly (Fig. 3E). Under decreased miR-103-3p expression in NOR-MSCs, the opposite phenomenon was observed (Fig. 3F).
One of the many types of endogenous non-coding, short, small RNAs, known as miRNAs, control the expression of certain genes posttranscriptionally by limiting protein translation or speeding up the degradation of messenger RNA. The main RNA-induced silencing complex is formed by complementary binding with 3′UTR of the target gene mRNA (Bartel, 2004). Over 60% of the human genome contains potential miRNA-binding sites, so the powerful potential of miRNA cannot be ignored (Friedman et al., 2009). We demonstrated that miR-103-3p was poorly expressed in exosomes and MSCs derived from the patients with MDS. However, the knowledge of the pathogenetic role of miR-103-3p in MDS is still lacking. MDS-MSCs may secrete miRNA into microvesicles, a newly discovered method for communication and control between cells (Chen et al., 2010). Therefore, we first detected the expression level of miR-103-3p in exosomes of MDS BM supernatant and observed its reduced expression. Then, the same results were observed when we detected the level of miR-103-3p expression in MDS-MSCs. These results prove that the decrease in miR-103-3p levels in bone marrow was due to the release of MSC-derived exosomes into the BMME. Next, we discussed the role of miR-103-3p in MDS-MSCs in the BMME.
MDS is characterized by ineffective hematopoiesis, which results in considerable morbidity and mortality. MDS-MSCs have been shown to support hematopoiesis through physiological processes, although their role in the pathophysiology of MDS is still unknown. MDS-derived MSCs have much less osteogenic differentiation ability, as evidenced by cytochemical staining and lower expression levels of Osterix and osteocalcin (Geyh et al., 2013). In contrast, MDS-MSCs have a higher proclivity to adipogenic differentiation (Wu et al., 2017). Therefore, through the differentiation experiment to explore the effect of miR-103-3p on the hematopoietic microenvironment of MDS, we could verify the effect of miR-103-3p on the differentiation ability of MDS-MSCs.
Our findings show that downregulation of miR-103-3p markedly decreased calcium deposition and increased lipid content. However, there was a significant increase in calcium deposition and a decrease in lipids in MDS-MSCs transfected with the miR-103-3p mimic. Silencing of miR-103-3p by an inhibitor increased the amount of lipids and decreased calcium deposition. Overexpression of miR-103-3p promoted the expression of the osteoblastic marker RUNX2 and decreased the expression of the adipogenic marker PPARG compared with the cells treated with mimic NC on day 21 after being differentiated. These data suggest that the low expression of miR-103-3p suppressed the osteogenic differentiation of MDS-MSCs and promoted adipogenic differentiation. Histological stainings showed significantly reduced calcium deposition in MDS-MSCs compared with that in NOR-MSCs, while there were significantly more lipid droplets in MDS-MSCs than in NOR-MSCs, which is in agreement with various earlier studies (Geyh et al., 2013; Wu et al., 2017). Osteoblasts support hematopoietic progenitors and comprise a crucial component of the HSC niche (‘endosteal niche’) (Arai et al., 2004; Calvi et al., 2003; Shiozawa et al., 2008; Taichman and Emerson, 1994), and adipogenic differentia which leads to ineffective hematopoiesis (Shiozawa et al., 2008). However, after the after the level of miR-103-3p in MDS was further reduced, the lipid-forming ability of MSCs was weakened, and the osteogenic ability was enhanced. Therefore, we believe that the function of MSCs is improved toward hematopoietic, which is beneficial to the hematopoietic microenvironment of MDS and enhancing the expression of miR-103-3p can enhance the hematopoietic support capacity of MSCs.
In summary, miR-103-3p is poorly expressed in plasma exosomes and MSCs derived from bone marrow of patients with MDS. Promoting the expression of miR-103-3p could stimulate osteogenic differentiation and inhibit adipogenic differentiation of MDS-MSCs in vitro, which can explain the invalid hematopoiesis of MDS. miR-103-3p has not been studied earlier for its role in MDS, and no evidence of hematopoietic association has been reported. Although the specific mechanism is not yet clear, the relationship between miR-103-3p, MDS-MSCs, and exosomes is worth further studies, and abnormal miRNA expression from MDS-MSC-derived exosomes can be considered a new therapeutic approach for the treatment of MDS.
The results showed that the expression level of mir-103-3p in MDS exosomes and mesenchymal stem cells was significantly reduced, and mir-103-3p could significantly affect the differentiation function of MDS-MSCs in vitro, but the effect of mir-103-3p on hematopoietic needs further study.
Acknowledgement: The authors thank the Central Laboratory of Guangdong Provincial People’s Hospital (Guangdong 308 Academy of Medical Sciences).
Availability of Data and Materials: The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Author Contribution: The authors confirm contribution to the paper as follows: study conception and design: Xiaofang Chen, Ningyu Li, Xin Du; data collection: Ningyu Li; analysis and interpretation of results: Ningyu Li; draft manuscript preparation: Ningyu Li, Xiaofang Chen. All authors reviewed the results and approved the final version of the manuscript.
Ethical Approval: All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. The Research Ethics Committee: Guangdong Provincial People’s Hospital, Guangdong Academy of Medical Sciences; Ethical approval code: 2019646H; Date of approval: November 15, 2019.
Informed Consent: Informed consent was obtained from all individual participants included in the study.
Funding Statement: This work was supported by The Nature Science Foundation of China (Nos. 82070176, 82070128, 81900132) and the Medical Science and Technology Research Fund of Guangdong Province (No. A2020585).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
Almalki SG, Agrawal DK (2016). Key transcription factors in the differentiation of mesenchymal stem cells. Differentiation 92: 41–51. DOI 10.1016/j.diff.2016.02.005. [Google Scholar] [CrossRef]
Arai F, Hirao A, Ohmura M, Sato H, Matsuoka S, Takubo K, Ito K, Koh GY, Suda T (2004). Tie2/angiopoietin-1 signaling regulates hematopoietic stem cell quiescence in the bone marrow niche. Cell 118: 149–161. DOI 10.1016/j.cell.2004.07.004. [Google Scholar] [CrossRef]
Bartel DP (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297. DOI 10.1016/S0092-8674(04)00045-5. [Google Scholar] [CrossRef]
Calvi LM, Adams GB, Weibrecht KW, Weber JM, Olson DP et al. (2003). Osteoblastic cells regulate the haematopoietic stem cell niche. Nature 425: 841–846. DOI 10.1038/nature02040. [Google Scholar] [CrossRef]
Chen TS, Lai RC, Lee MM, Choo AB, Lee CN, Lim SK (2010). Mesenchymal stem cell secretes microparticles enriched in pre-microRNAs. Nucleic Acids Research 38: 215–224. DOI 10.1093/nar/gkp857. [Google Scholar] [CrossRef]
Chen X, Li N, Weng J, Du X (2021). Senescent mesenchymal stem cells in myelodysplastic syndrome: Functional alterations, molecular mechanisms, and therapeutic strategies. Frontiers in Cell and Developmental Biology 8: 617466. DOI 10.3389/fcell.2020.617466. [Google Scholar] [CrossRef]
Chen X, Yi H, Zhang B, Liu Y, Wang S, Liao T, Deng Z, Zeng F, Jing Z, He L (2017). Caffeic acid phenethyl ester promotes haematopoietic stem/progenitor cell homing and engraftment. Stem Cell Research & Therapy 8: 255. DOI 10.1186/s13287-017-0708-x. [Google Scholar] [CrossRef]
Chung TK, Cheung TH, Huen NY, Wong KW, Lo KW et al. (2009). Dysregulated microRNAs and their predicted targets associated with endometrioid endometrial adenocarcinoma in Hong Kong women. International Journal of Cancer 124: 1358–1365. DOI 10.1002/ijc.24071. [Google Scholar] [CrossRef]
Djouad F, Ipseiz N, Luz-Crawford P, Scholtysek C, Krönke G, Jorgensen C (2017). PPARβ/δ: A master regulator of mesenchymal stem cell functions. Biochimie 136: 55–58. DOI 10.1016/j.biochi.2016.11.011. [Google Scholar] [CrossRef]
Doan PL, Chute JP (2012). The vascular niche: Home for normal and malignant hematopoietic stem cells. Leukemia 26: 54–62. DOI 10.1038/leu.2011.236. [Google Scholar] [CrossRef]
Friedman RC, Farh KK, Burge CB, Bartel DP (2009). Most mammalian mRNAs are conserved targets of microRNAs. Genome Research 19: 92–105. DOI 10.1101/gr.082701.108. [Google Scholar] [CrossRef]
Geyh S, Oz S, Cadeddu RP, Fröbel J, Brückner B et al. (2013). Insufficient stromal support in MDS results from molecular and functional deficits of mesenchymal stromal cells. Leukemia 27: 1841–1851. DOI 10.1038/leu.2013.193. [Google Scholar] [CrossRef]
Giaginis C, Tsantili-Kakoulidou A, Theocharis S (2007). Peroxisome proliferator-activated receptors (PPARs) in the control of bone metabolism. Fundamental & Clinical Pharmacology 21: 231–244. DOI 10.1111/j.1472-8206.2007.00486.x. [Google Scholar] [CrossRef]
Horiguchi H, Kobune M, Kikuchi S, Yoshida M, Murata M et al. (2016). Extracellular vesicle miR-7977 is involved in hematopoietic dysfunction of mesenchymal stromal cells via poly(rC) binding protein 1 reduction in myeloid neoplasms. Haematologica 101: 437–447. DOI 10.3324/haematol.2015.134932. [Google Scholar] [CrossRef]
Kalluri R, LeBleu VS (2020). The biology, function, and biomedical applications of exosomes. Science 367: 6977. DOI 10.1126/science.aau6977. [Google Scholar] [CrossRef]
Kfir-Erenfeld S, Haggiag N, Biton M, Stepensky P, Assayag-Asherie N, Yefenof E (2017). miR-103 inhibits proliferation and sensitizes hemopoietic tumor cells for glucocorticoid-induced apoptosis. Oncotarget 8: 472–489. DOI 10.18632/oncotarget.13447. [Google Scholar] [CrossRef]
Machová PK, Lopotová T, Klamová H, Burda P, Trněný M, Stopka T, Moravcová J (2011). Expression patterns of microRNAs associated with CML phases and their disease related targets. Molecular Cancer 10: 41. DOI 10.1186/1476-4598-10-41. [Google Scholar] [CrossRef]
Malvicini R, Santa-Cruz D, Pacienza N, Yannarelli G (2019). OCT4 silencing triggers its epigenetic repression and impairs the osteogenic and adipogenic differentiation of mesenchymal stromal cells. International Journal of Molecular Sciences 20: 3268. DOI 10.3390/ijms20133268. [Google Scholar] [CrossRef]
Mateescu B, Kowal EJ, van Balkom BW, Bartel S, Bhattacharyya SN et al. (2017). Obstacles and opportunities in the functional analysis of extracellular vesicle RNA-an ISEV position paper. Journal of Extracellular Vesicles 6: 1286095. DOI 10.1080/20013078.2017.1286095. [Google Scholar] [CrossRef]
Matsuda M, Morita Y, Hanamoto H, Tatsumi Y, Maeda Y, Kanamaru A (2004). CD34+ progenitors from MDS patients are unresponsive to SDF-1, despite high levels of SDF-1 in bone marrow plasma. Leukemia 18: 1038–1040. DOI 10.1038/sj.leu.2403301. [Google Scholar] [CrossRef]
Mei Y, Zhao B, Basiorka A A, Yang J, Cao L, Zhang J, List A, Ji P (2018). Age-related inflammatory bone marrow microenvironment induces ineffective erythropoiesis mimicking del(5q) MDS. Leukemia 32: 1023–1033. DOI 10.1038/leu.2017.326. [Google Scholar] [CrossRef]
Meunier M, Guttin A, Ancelet S, Laurin D, Zannoni J et al. (2020). Extracellular vesicles from myelodysplastic mesenchymal stromal cells induce DNA damage and mutagenesis of hematopoietic stem cells through miRNA transfer. Leukemia 34: 2249–2253. DOI 10.1038/s41375-020-0738-8. [Google Scholar] [CrossRef]
Meyer MB, Benkusky NA, Sen B, Rubin J, Pike JW (2016). Epigenetic plasticity drives adipogenic and osteogenic differentiation of marrow-derived mesenchymal stem cells. Journal of Biological Chemistry 291: 17829–17847. DOI 10.1074/jbc.M116.736538. [Google Scholar] [CrossRef]
Morrison SJ, Scadden DT (2014). The bone marrow niche for haematopoietic stem cells. Nature 505: 327–334. DOI 10.1038/nature12984. [Google Scholar] [CrossRef]
Muntión S, Ramos TL, Diez-Campelo M, Rosón B, Sánchez-Abarca LI et al. (2016). Microvesicles from mesenchymal stromal cells are involved in HPC-microenvironment crosstalk in myelodysplastic patients. PLoS One 11: e0146722. DOI 10.1371/journal.pone.0146722. [Google Scholar] [CrossRef]
Mushahary D, Spittler A, Kasper C, Weber V, Charwat V (2018). Isolation, cultivation, and characterization of human mesenchymal stem cells. Cytometry Part A 93: 19–31. DOI 10.1002/cyto.a.23242. [Google Scholar] [CrossRef]
Narayanan A, Srinaath N, Rohini M, Selvamurugan N (2019). Regulation of RUNX2 by MicroRNAs in osteoblast differentiation. Life Sciences 232: 116676. DOI 10.1016/j.lfs.2019.116676. [Google Scholar] [CrossRef]
Nonaka R, Miyake Y, Hata T, Kagawa Y, Kato T et al. (2015). Circulating miR-103 and miR-720 as novel serum biomarkers for patients with colorectal cancer. International Journal of Oncology 47: 1097–1102. DOI 10.3892/ijo.2015.3064. [Google Scholar] [CrossRef]
Ozdogan H, Gur Dedeoglu B, Oztemur Islakoglu Y, Aydos A, Kose S et al. (2017). DICER1 gene and miRNA dysregulation in mesenchymal stem cells of patients with myelodysplastic syndrome and acute myeloblastic leukemia. Leukemia Research 63: 62–71. DOI 10.1016/j.leukres.2017.10.006. [Google Scholar] [CrossRef]
Pronk E, Raaijmakers M (2019). The mesenchymal niche in MDS. Blood 133: 1031–1038. DOI 10.1182/blood-2018-10-844639. [Google Scholar] [CrossRef]
Psaila B, Lyden D, Roberts I (2012). Megakaryocytes, malignancy and bone marrow vascular niches. Journal of Thrombosis and Haemostasis 10: 177–188. DOI 10.1111/j.1538-7836.2011.04571.x. [Google Scholar] [CrossRef]
Rhyasen GW, Starczynowski DT (2012). Deregulation of microRNAs in myelodysplastic syndrome. Leukemia 26: 13–22. DOI 10.1038/leu.2011.221. [Google Scholar] [CrossRef]
Santamaría C, Muntión S, Rosón B, Blanco B, López-Villar O et al. (2012). Impaired expression of DICER, DROSHA, SBDS and some microRNAs in mesenchymal stromal cells from myelodysplastic syndrome patients. Haematologica 97: 1218–1224. DOI 10.3324/haematol.2011.054437. [Google Scholar] [CrossRef]
Shiozawa Y, Havens AM, Pienta KJ, Taichman RS (2008). The bone marrow niche: habitat to hematopoietic and mesenchymal stem cells, and unwitting host to molecular parasites. Leukemia 22: 941–950. DOI 10.1038/leu.2008.48. [Google Scholar] [CrossRef]
Taichman RS, Emerson SG (1994). Human osteoblasts support hematopoiesis through the production of granulocyte colony-stimulating factor. The Journal of Experimental Medicine 179: 1677–1682. DOI 10.1084/jem.179.5.1677. [Google Scholar] [CrossRef]
Woods K, Guezguez B (2021). Dynamic changes of the bone marrow niche: Mesenchymal stromal cells and their progeny during aging and leukemia. Frontiers in Cell and Developmental Biology 9: 714716. DOI 10.3389/fcell.2021.714716. [Google Scholar] [CrossRef]
Wu Y, Aanei CM, Kesr S, Picot T, Guyotat D, Campos Catafal L (2017). Impaired expression of focal adhesion kinase in mesenchymal stromal cells from low-risk myelodysplastic syndrome patients. Frontiers in Oncology 7: 164. DOI 10.3389/fonc.2017.00164. [Google Scholar] [CrossRef]
Yu D, Zhou H, Xun Q, Xu X, Ling J, Hu Y (2012). microRNA-103 regulates the growth and invasion of endometrial cancer cells through the downregulation of tissue inhibitor of metalloproteinase 3. Oncology Letters 3: 1221–1226. DOI 10.3892/ol.2012.638. [Google Scholar] [CrossRef]
Zhang Z, Wu S, Muhammad S, Ren Q, Sun C (2018). miR-103/107 promote ER stress-mediated apoptosis via targeting the Wnt3a/β-catenin/ATF6 pathway in preadipocytes. Journal of Lipid Research 59: 843–853. DOI 10.1194/jlr.M082602. [Google Scholar] [CrossRef]
Zhao W, Yang H, Chai J, Xing L (2021). RUNX2 as a promising therapeutic target for malignant tumors. Cancer Management and Research 13: 2539–2548. DOI 10.2147/cmar.s302173. [Google Scholar] [CrossRef]
Zhuang H, Zhang X, Zhu C, Tang X, Yu F, Shang GW, Cai X (2016). Molecular mechanisms of PPAR-γ governing MSC osteogenic and adipogenic differentiation. Current Stem Cell Research & Therapy 11: 255–264. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools