 Open Access
Open Access
ARTICLE
Overexpression of a sugarcane ScCaM gene negatively regulates salinity and drought stress responses in transgenic Arabidopsis thaliana
1 College of Tea and Food Science; Fujian Provincial Key Laboratory of Eco-Industrial Green Technology, Wuyi University, Nanping, 354300, China
2 Key Laboratory of Sugarcane Biology and Genetic Breeding, Ministry of Agriculture; Key Laboratory of Genetics, Breeding and Multiple Utilization of Crops,
Ministry of Education, College of Agriculture, Fujian Agriculture and Forestry University, Fuzhou, 350002, China
3 College of Agriculture, Yulin Normal University, Yulin, 537000, China
* Corresponding Authors: Hui Ling, ; Jun Luo,
(This article belongs to the Special Issue: Physiology and Molecular Biology of Plant Stress Tolerance)
BIOCELL 2023, 47(1), 215-225. https://doi.org/10.32604/biocell.2022.022477
Received 11 March 2022; Accepted 20 May 2022; Issue published 26 September 2022
Abstract
Calmodulin (CaM) proteins play a key role in signal transduction under various stresses. In the present study, the effects of a sugarcane ScCaM gene (NCBI accession number: GQ246454) on drought and salt stress tolerance in transgenic Arabidopsis thaliana and Escherichia coli cells were evaluated. The results demonstrated a significant negative role of ScCaM in the drought and salt stress tolerance of transgenic lines of A. thaliana, as indicated by the phenotypes. In addition, the expression of AtP5CS and AtRD29A, two genes tightly related to stress resistance, was significantly lower in the overexpression lines than in the wild type. The growth of E. coli BL21 cells expressing ScCaM showed weaker tolerance under mannitol and NaCl stress. Taken together, this study revealed that the ScCaM gene plays a negative regulatory role in both mannitol and NaCl stresses, and it possibly exerts protective mechanisms common in both prokaryotes and eukaryotes under stress conditions.Keywords
Calmodulin (CaM) is an important calcium-binding protein ubiquitous in eukaryotes and is the most important receptor for Ca2+ (Nelson and Chazin, 1998). CaM consists of 149 amino acid residues and belongs to the classical EF-hand family with a similar EF-hand domain at both the N- and C-terminals (Gifford et al., 2007). The spherical end of every EF-hand can bind to two Ca2+, and therefore, one CaM can bind to four Ca2+. CaM itself does not have enzymatic activity and cannot play a direct regulatory role but may interact with the downstream CaM-binding protein (CaMBP) after binding to Ca2+ to activate enzymatically active proteins, thereby regulating the development of plant cells and their response to external stimuli (Hoeflich and Ikura, 2002; Poovaiah et al., 2013; Rhoads and Friedberg, 1997).
To date, CaM genes have been identified and analyzed in different plants such as Arabidopsis thaliana (McCormack and Braam, 2003), rice (Oryza sativa) (Chinpongpanich et al., 2012), tomato (Solanum lycopersicum) (Munir et al., 2016), wild tomato (S. pennellii) (Shi and Du, 2020), cabbage (Brassica rapa L. ssp. pekinensis) (Nie et al., 2017), and apple (Malus x domestica) (Boonburapong and Buaboocha, 2007). CaM genes respond to external stimuli, such as high and low temperatures, pathogens, and hormones (Park et al., 2004; Townley and Knight, 2002), and there has been rapid progress in research on the involvement of Ca2+/CaM in plant stress physiology. Currently, Ca2+/CaM research has received extensive attention, and research progress in various crops has been achieved. CaM in Arabidopsis thaliana responds to high-temperature stress (Zhang et al., 2009). Ca2+-CaM is involved in abscisic acid (ABA)-induced antioxidant defense, and the cross-talk between Ca2+-CaM and hydrogen peroxide (H2O2) plays a pivotal role in the ABA signaling pathway (Hu et al., 2007). In rice, CaM was found to regulate glutamate decarboxylase (GAD) activity to respond to anoxia (Reggiani et al., 1995). Zhu et al. (2021) found that the mildew resistance locus O4 interacts with CaM/calmodulin-like (CML) and is involved in root gravity response. In plants, when responding to salt stress, CaM-dependent calcineurin functions within transduction pathways required for salt stress adaptation (Snedden and Fromm, 2001). Specific CaM isoforms are involved in response to pathogens in plants (Heo et al., 1999). In planta expression disclosed that CaM is localized to the cytoplasm and nuclei of the plant cell and suppresses plant defenses against such as hydrogen peroxide (H2O2) accumulation and callose deposition (Fu et al., 2022).
Sugarcane is not only the most important cash crop globally, but it is also an important energy crop. It accounts for 92% of the total sugar production in China, and its production and yield rank 4th after that of grains, oilseeds, and cotton, conferring a special economic status (Su et al., 2020a; Zhang et al., 2018). In China, 85% of sugarcane is planted on saline-alkali soil and early sloping land, and frequent droughts, frost damage, and increasing incidences of pest infestations and diseases in recent years, especially smut, have resulted in low yields of sugarcane in many parts of China. Therefore, the tolerance or resistance of sugarcane to abiotic environmental stresses can be enhanced to improve the yield of sugarcane.
Our group previously cloned the ScCaM gene (NCBI accession number: GQ246454) from sugarcane and confirmed that this gene may receive calcium ion signals on the cell membrane and may be involved in the responses to plant growth and development (Liu et al., 2021). However, no further study of this CaM gene has been carried out yet. In the present study, we successfully transferred this gene into Arabidopsis and obtained five T3 transgenic Arabidopsis lines. Hence, we intended to screen the lines of positive transgenic ScCaM Arabidopsis and evaluate the function of ScCaM overexpressing transgenic Arabidopsis in salinity and drought stress tolerance from the perspectives of seed germination, phenotypic observation, and also at the molecular level. We performed prokaryotic expression analysis and plate stress experiments on the ScCaM protein expressed in transgenic Arabidopsis. Furthermore, the role of ScCaM genes and their potential underlying mechanisms in response to high salinity and drought stresses were examined. This study is expected to provide reference values for functionally validated gene resources for the genetic improvement of sugarcane for stress resistance.
Acquisition of ScCaM-overexpressing transgenic Arabidopsis thaliana
Plant material and growth conditions: the Arabidopsis species used were Colombia ecotype. Seed culture conditions were 22°C, 16 h/d light, 8 h/d dark cycle, 60% relative humidity, and 2200 L× light intensity, and plants were grown in pots at 22°C, 12 h/d light, 12 h/d dark cycle, and 60% relative air humidity.
Genetic transformation of the sugarcane ScCaM gene into A. thaliana and screening of homozygous plants: the eukaryotic expression vector pBWA(V)HS-35S-ScCaM was constructed by introducing the ScCaM coding region into the overexpression vector pBWA(V)HS-35S using the in-fusion cloning method. The recombinant plasmid was validated and then transformed into Agrobacterium tumefaciens GV3101. After Arabidopsis thaliana had bolted, Agrobacterium containing the pBWA(V)HS-35S-ScCaM recombinant plasmid was titrated and used to infect wild-type Col-0 flowers. Next, the flowers were stored for 24 h in the dark, followed by a long period of irradiation (15 h light, 10,000–12,000 L×, 50%–60% relative humidity, and 21°C–24°C temperature) until pod maturity. T3 generation homozygous strains of transgenic ScCaM Arabidopsis were finally obtained using hygromycin B screening and polymerase chain reaction (PCR) amplification assays.
Screening and characterization of T3 generation ScCaM-overexpressing Arabidopsis plants
Screening culture was performed by using Murashige and Skoog (MS) medium (Murashige and Skoog 4.4 g/L, agar 8 g/L, adjusted to pH5.8) containing 100 g/L of hygromycin B, and three seedlings were transplanted to one planting pot when they grew to the 3–4-leaf stage. On reaching the 5–8-leaf stage, RNA from plant leaf was extracted to detect whether the transgenic plants were successfully transformed to overexpress ScCaM. HYG-F/R. For performing RT-PCR, pBWA(V)HS-35S-ScCaM-F/R were used as primers, pBWA(V)HS-35S-ScCaM plasmid was used as a positive control, while water was used as a blank control. cDNA obtained following RNA reverse transcription was used as a template for PCR amplification and electrophoresis. The PCR procedure was as follows: pre-denaturation at 95°C for 5 min, 35 cycles of denaturation at 95°C for 30 s, denaturation at 55°C for 30 s, and extension at 72°C for 30 s, and a final extension at 72°C for 10 min, and holding at 4°C. The primer sequences and RT-PCR system are shown in Suppl. Tables S1 and S2, respectively.
Salinity stress treatment of ScCaM-overexpressing Arabidopsis thaliana
Seeds of T3 generation transgenic overexpressing Arabidopsis thaliana and wild-type Col-0 Arabidopsis thaliana were sterilized using 2% sodium hypochlorite, and the seeds were grown on MS plates containing 0, 100, or 150 mM sodium chloride (NaCl) after sterilization and incubated continuously for 12 d, during which germination potential was observed, and germination rate was determined. A line graph of germination rate was plotted using Origin 8.0 software. Arabidopsis plants were grown for about 4 weeks, and those with uniform growth were selected and subjected to salinity stress. Specifically, 15 mL of 300 mM NaCl was poured on the Arabidopsis plants once every two days for 12 d. Wild-type Col-0 plants were used as controls, their phenotypes were observed, and photographs were acquired. At 12 d of treatment, Arabidopsis leaf RNA was extracted using Trizol and reversed transcribed into cDNA using the Novozymes HiScript® Q RT SuperMix for qPCR (+gDNA wiper) kit for further expression analysis of resistance-related genes.
Drought stress treatment of ScCaM-overexpressing Arabidopsis thaliana
Sterilized ScCaM-overexpressing T3 generation Arabidopsis and wild-type Col-0 Arabidopsis seeds were spotted on MS medium plates containing 0, 200, or 300 mM mannitol and incubated continuously for 12 d. Germination potential was observed, and the germination rate was determined. A line graph of germination rate was plotted using Origin 8.0 software. When the Arabidopsis plants were grown for about 4 weeks, the plants with uniform growth were selected for drought stress treatment. Specifically, the conditions were natural drought (no watering) for 11 d and rehydration for 5 and 11 d, and wild-type Col-0 plants were used as the controls, during which their phenotypes were observed and photographs were acquired. At 11 d of treatment, Arabidopsis leaf RNA was extracted and reverse transcribed into cDNA for further expression analysis of resistance-related genes.
Expression analysis of resistance-related genes in ScCaM-overexpressing Arabidopsis thaliana
Quantitative reverse transcription-PCR (RT-qPCR) was used to analyze the expression of some Arabidopsis stress-related genes under salinity and drought stress, including genes related to reactive oxygen species (ROS) scavenging (AtSOD, AtCAT, and AtAPX) (Yang et al., 2015), ABA response (AtNCED3, AtP5CS, and AtRD29A) (Qiu et al., 2020), salinity stress (AtSOS3), and drought stress (AtCDPK1) (Yang et al., 2015), with Atactin2 as an internal reference gene (Zhu, 2002). Primer sequences are listed in Suppl. Table S1. The qRT-PCR steps were as follows: 50°C, 2 min; 95°C, 2 min; 40 cycles of 95°C, 10 min; 95°C, 15 s; 60°C, 1 min. Relative gene expression was calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001). Significance (p < 0.05) was calculated using one-way ANOVA. Duncan’s new multiple range test was calculated using DPS 9.50 software, and data are expressed as the mean ± standard error (SE). Histograms were constructed using Origin 8.0 software.
Expression analysis of ScCaM in Escherichia coli cells under salinity and drought stress
We constructed the prokaryotic expression vector ScCaM-pGEX-4T-1 and determined the optimal conditions for ScCaM expression in E. coli cells as 16°C, 20 h, and induced by 0.5 mM isopropyl-β-D-thiogalactoside (IPTG) (Su et al., 2020b). In this study, we first tested whether the optimal temperature and IPTG concentration could induce the expression of ScCaM at the protein level. Following the successful induction of ScCaM-pGEX-4T-1, recombinant E. coli cells were subjected to salinity and drought stress experiments to analyze the tolerance of ScCaM protein to salinity and drought in E. coli cells. The procedure was as follows: pGEX-4T-1 and pGEX-4T-1-ScCaM were transformed in prokaryotic expression strain E. coli BL21 (DE3), and single colonies of recombinant bacteria were cultured in ampicillin (70 µg/mL)-containing LB broth. The cells were incubated at 37°C with shaking at 200 rpm until OD600 = 0.6. Subsequently, 0.5 mM IPTG (to analyze the expression of TCGT transcript) was added, and the cells were incubated at 16°C in a 200 rpm shaking incubator for induction. The bacterial suspensions were collected at intervals of 0, 2, 8, and 20 h. The bacteria were then collected via centrifugation at 10,000 g for 10 min at 26°C, mixed with 6× protein loading buffer, boiled at 100°C for 5 min, and centrifuged again at 10,000 g for 5 min at room temperature. Next, 10 µL of supernatant was resolved by 12% sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis, followed by coomassie brilliant blue staining and photograph acquisition. The above bacterial broth collected after 20 h of IPTG induction was diluted to the same concentration using LB broth, i.e., OD600 = 0.7, following which the broth was diluted to 10−3 and 10−4 and used as a stock solution. The diluted broth was spotted onto LB agar plates containing 70 µg/mL ampicillin, incubated overnight at 37°C, and photographed for retention.
Screening and characterization of T3 generation ScCaM-overexpressing Arabidopsis plants
As shown in Suppl. Fig. S1, T3 generation ScCaM-overexpressing Arabidopsis seeds were essentially homozygous by screening cultures on MS medium plates exhibiting hygromycin resistance.
Fig. 1 shows the amplification products of HYG and ScCaM genes in Arabidopsis plants. As shown in Figs. 1A and 1C, a single band of approximately 750 bp in size was amplified when the overexpression plasmid was used as a template and generally matched the length of the vector HYG tag. A single band consistent with the overexpression plasmid was identified in samples 4 to 9, and no band was identified in samples 1 to 3. As shown in Figs. 1B and 1D, a single band was amplified when the overexpression plasmid was used as a template, which was approximately 450 bp in size and generally matched the length of the ScCaM open reading frame. The results indicate that pBWA(V)HS-35S-ScCaM was successfully expressed in the Arabidopsis transgenic strain, and the selected Arabidopsis plants could be used for the next step of analysis for salinity and drought tolerance.

Figure 1: Identification of ScCaM gene positive transgenic Arabidopsis thaliana.
Note: (A and C) PCR detection electropherogram of overexpression vector pBWA(V)HS-35S HYG tag; (B and D) PCR detection electropherogram of ScCaM gene; M: 2000 bp marker; CK: blank control; PC: PCR amplification product of recombinant plasmid pBWA(V)HS-35S-ScCaM; lanes 1~3: PCR amplification products of wild-type Arabidopsis thaliana plants; lanes 4~6: PCR amplification products of transgenic Arabidopsis thaliana T3-1 with ScCaM gene; lanes 7~9: PCR amplification products of transgenic Arabidopsis thaliana T3-5 with ScCaM gene.
Germination status of T3 generation transScCaM Arabidopsis seeds under salinity and drought stresses
Selected transgenic ScCaM T3 generation lines, T3-1 and T3-5, and wild-type Col-0 Arabidopsis seeds were grown on MS blank medium plates and MS medium plates containing 100 mM NaCl, 150 mM NaCl, 200 mM mannitol, and 300 mM mannitol, respectively. The germination rate and growth after 12 days of continuous incubation are shown in Fig. 2, according to which both transgenic lines T3-1 and T3-5 were able to germinate on MS medium plates. The germination rate of Col-0 seeds was 95.24%, and all germinated plants were green, suggesting that they were able to grow normally (Figs. 2A and 2B). The germination rates differed in T3-1, T3-5, and wild-type Col-0 seeds on MS medium plates containing different concentrations of NaCl and mannitol, but the differences were generally small (Figs. 2C–2F). Thus, overexpression of the ScCaM gene did not affect the tolerance of transgenic Arabidopsis to salinity and drought stresses during seed germination.
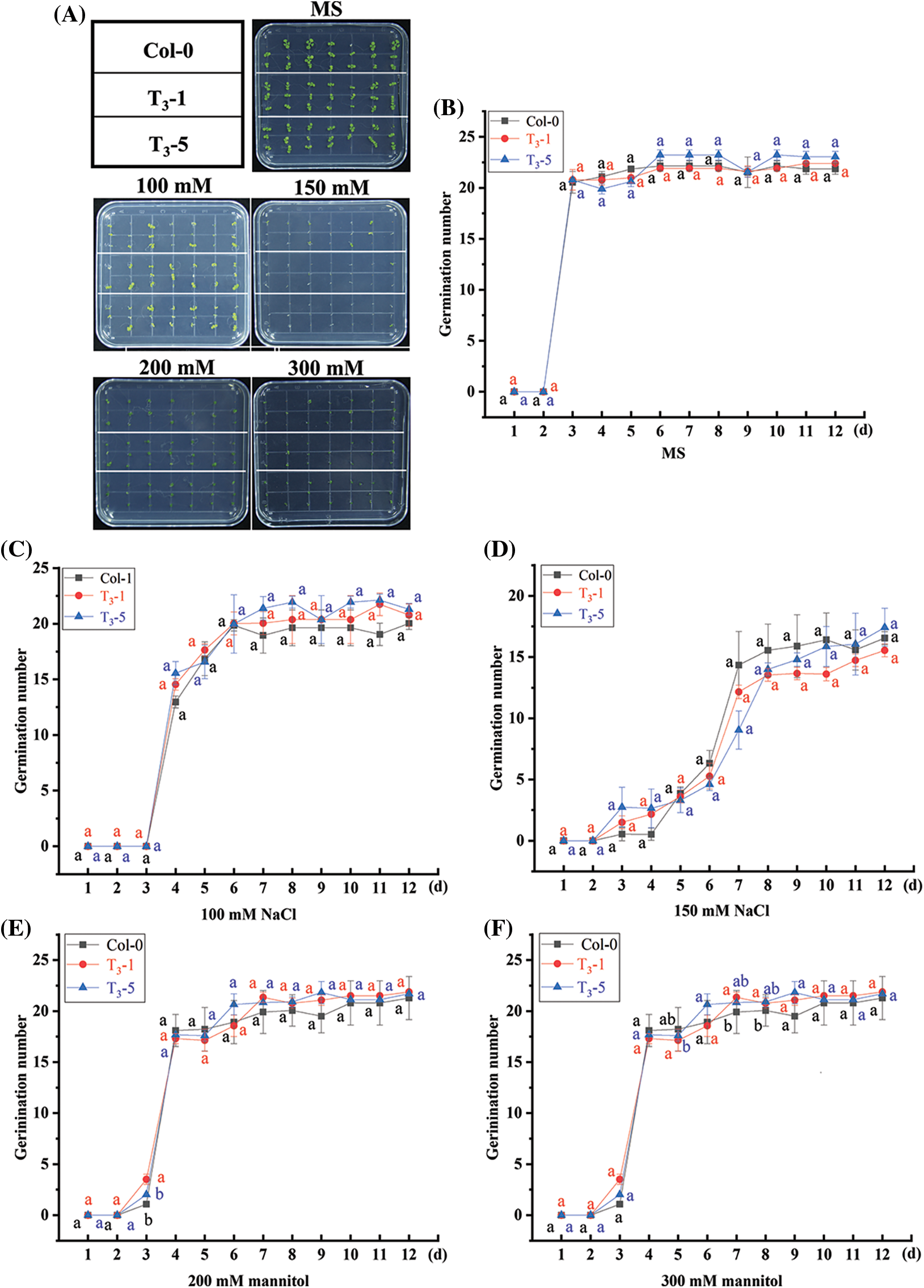
Figure 2: Germination of T3 generation seeds of transgenic Arabidopsis thaliana overexpressing ScCaM gene under drought and salt stress.
Note: (A) The growth status of Arabidopsis thaliana seeds on MS blank medium plates and MS medium plates containing 100 mM NaCl, 150 mM NaCl, 200 mM mannitol, and 300 mM mannitol for 12 days; (B), (C), (D), (E) and (F) are line graphs of germination rate of Arabidopsis seeds cultured on MS blank medium plates and MS medium plates containing 100 mM NaCl, 150 mM NaCl, 200 mM mannitol, and 300 mM mannitol for 12 days, respectively. The experiments were randomized, two replicates were used to avoid plate effects, and the germination number was calculated from the multiple plates. Significance (p < 0.05) was calculated using one-way ANOVA. Duncan's new multiple range test was calculated using DPS 9.50 software, and data are expressed as the mean ± standard error (SE).
Phenotypic analysis of ScCaM transgenic Arabidopsis lines under salinity and drought stresses
To investigate the salinity and drought tolerance of transgenic ScCaM Arabidopsis plants, we treated the plants with 300 mM NaCl solution in water and performed natural drought treatments. As shown in Fig. 3A, there was no significant difference in the growth of Col-0, T3-1, and T3-5 Arabidopsis plants before salinity stress, and after 7 days and 12 days of salinity stress, chlorosis was observed in the leaves of Col-0, T3-1, and T3-5 plants; however, it was more severe in the leaves of T3-1 and T3-5 plants than in the leaves of Col-0 plants. As shown in Fig. 3B, there was no significant difference in the growth of Col-0, T3-1, and T3-5 Arabidopsis plants before drought treatment, and after 11 days of drought treatment, chlorosis was observed in the leaves of Col-0 and ScCaM-transformed T3-1 and T3-5 plants; however, chlorosis was more severe in the leaves of T3-1 and T3-5 plants than in the leaves of Col-0 plants. Thus, the transgenic ScCaM Arabidopsis plants were less tolerant to salinity and drought stress than the Col-0 plants.

Figure 3: Phenotypic analysis of T3 lines ScCaM overexpressing transgenic Arabidopsis thaliana under salt and drought stress.
Note: (A) Phenotypes of 4-weeks-old plants of the transgenic line (T3-1 and T3-5) and the wild type (Col-0) before and after NaCl treatment. (B) Phenotypes of 4-weeks-old plants of the transgenic line (T3-1 and T3-5) and the wild type (Col-0) before and after drought treatment. Each plot contained three Arabidopsis thaliana plants.
Expression analysis of stress resistance-related genes in ScCaM overexpressing transgenic Arabidopsis plants under salinity and drought stresses
We analyzed the expression of relevant stress resistance genes to further clarify the molecular mechanisms underlying drought and salinity tolerance in ScCaM overexpressing transgenic Arabidopsis.
As shown in Fig. 4A, the expression of AtNCED3, AtP5CS, and AtRD29A was significantly downregulated in ScCaM overexpressing transgenic Arabidopsis T3-1 and T3-5 lines after 12 d of salinity stress, which was lower than that of the wild type, respectively. However, the expression of the ROS scavenger gene, AtSOD, and salinity stress response-related gene, AtSOS3, was significantly upregulated in the T3-1 line, which was 4.28-and 2.04-fold higher than that of the control, respectively. Interestingly, the expression of this gene was not significantly different in the T3-5 line compared with the control, and the expression of AtCAT, AtAPX, and AtCDPK1 was not significantly different in Col-0, T3-1, and T3-5 plants.
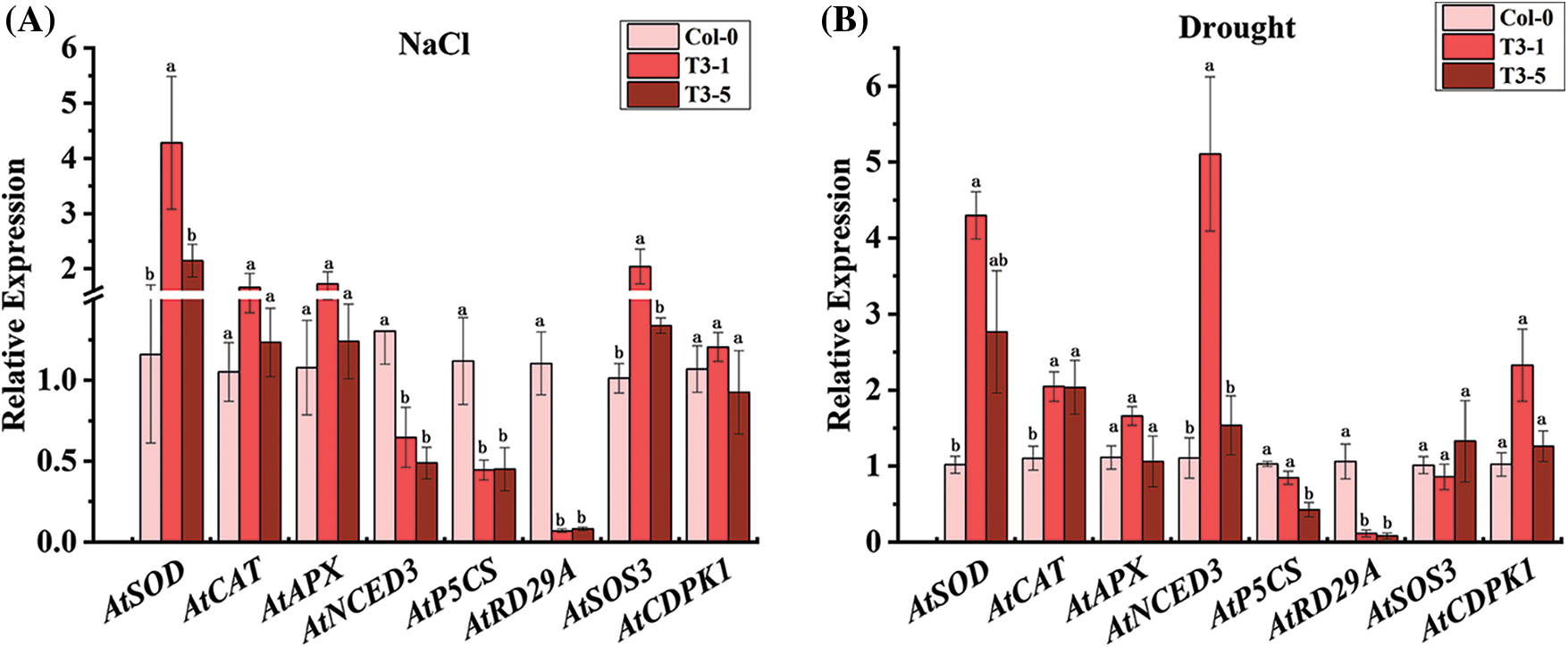
Figure 4: Expression analysis of stress-related genes by qRT-PCR in transgenic Arabidopsis thaliana plants overexpressing ScCaM under NaCl and drought stress.
Note: Atactin2 was used as the reference gene. All data points shown are mean ± SE (n = 3). Different lowercase letters indicate a significant difference, as determined by Duncan’s new multiple range test (p < 0.05).
As shown in Fig. 4B, after 12 d of drought stress, the expression of AtP5CS and AtRD29A was significantly downregulated in both T3-1 and T3-5 plants, which was lower than that of the control, respectively. The ROS scavenging enzyme-coding genes, AtSOD and AtCAT, were significantly upregulated in both the T3-1 and T3-5 lines, which were higher than that of the wild type, respectively. The ABA pathway-related gene, AtNCED3, was significantly upregulated in the T3-1 line, which was 5.11-fold higher than that of the wild type, and the expression of this gene was not significantly different in the T3-5 line compared with the wild type. The expression of AtAPX, AtSOS3, and AtCDPK1 was not significantly different in Col-0, T3-1, and T3 -5 plants.
Overall, the expression of AtNCED3, AtP5CS, and AtRD29A was significantly downregulated, and the expression of AtSOD and AtSOS3 was significantly upregulated in the transgenic lines under salinity stress. However, the expression of AtP5CS and AtRD29A was significantly downregulated, and the expression of AtSOD, AtCAT, and AtNCED3 was significantly upregulated in the transgenic lines under drought stress. Notably, the expression of AtP5CS and AtRD29A was suppressed in both T3-1 and T3-5 lines under salinity and drought stresses, and it is speculated that ScCaM attenuated the tolerance of the transgenic plants to salinity and drought mainly by reducing the expression of AtP5CS and AtRD29A genes.
Expression of ScCaM in Escherichia coli BL21 (DE3) strain
To find the role of the ScCaM gene in salinity and drought resistance in prokaryotes, ScCaM protein expression was induced as previously described (Liu et al., 2021), and stress experiments in LB plates with different concentrations of NaCl and mannitol were performed. As shown in Fig. 5, the accumulation of recombinant protein was observed after 8 h and 20 h induction by 0.5 mM IPTG. Based on EXPASY ProtParam, the estimated molecular weight of ScCaM protein was 16.83 kDa, and the molecular weight of GST protein was 26 kDa; the target protein of GST-ScCaM of about 43 kDa molecular weight was induced and was used for further plate stress experiments.

Figure 5: Prokaryotic expression of the pGEX-4T-1-ScCaM fusion protein.
Note: M: Protein marker; 1~4: control induction for 0, 2, 8, and 20 h; 5~8: recombinant strain pGEX-4T-1-ScCaM induction for 0, 2, 8, and 20 h.
Analysis of ScCaM expression in Escherichia coli cells under salinity and drought stresses
Plate stress experiments for the prokaryotic expression of the recombinant bacterium BL21 with pGEX-4T-1-ScCaM are shown in Fig. 6. According to Fig. 6A, after 12 h of incubation at 37°C, there was no significant difference in the growth of pGEX-4T-1-ScCaM and pGEX-4T-1 cells on LB plates containing 70 μg/mL ampicillin. Besides, pGEX-4T-1-ScCaM and pGEX-4T-1 cells did not differ in growth on 100 mM NaCl and 500 mM NaCl stress plates, i.e., both could grow at 100 mM NaCl with no significant difference in growth, whereas neither could grow on 500 mM NaCl stress plates. However, the growth of both pGEX-4T-1 and pGEX-4T-1-ScCaM cells was inhibited on 300 mM NaCl stress plates, with the growth status of pGEX-4T-1-ScCaM being significantly weaker than that of pGEX-4T-1. It is thus hypothesized that ScCaM attenuated the tolerance of E. coli to NaCl stress.

Figure 6: The spot assays used for monitoring the growth performance of BL21/pGEX-4T-1 and BL21/pGEX-4T-1-ScCaM transformed E. coli cells on LB plates under NaCl and mannitol stress.
Note: The cultures were adjusted to OD600 = 0.7 after IPTG induction; 10 µL from 10−3 (left side of the red line on the plate) and 10−4 (right side of the red line on plate) dilutions of the culture were spotted onto LB medium plate. The growth performance of BL21/pGEX-4T-1 and BL21/pGEX-4T-1-ScCaM cells on LB plates without supplements or stresses were used as controls. (A) The growth performance of BL21/pGEX-4T-1 and BL21/pGEX-4T-1-ScCaM cells on LB plates with 100 mM, 300 mM, and 500 mM NaCl supplements are used as treatments. (B) The growth performance of BL21/pGEX-4T-1 and BL21/pGEX-4T-1-ScCaM cells on LB plates with 500, 750, and 1000 mM mannitol supplements are used as treatments.
Fig. 6B showed that the growth of pGEX-4T-1-ScCaM and pGEX-4T-1 cells on LB plates with 70 μg/mL ampicillin did not differ much after 12 h of incubation at 37°C. Non-recombinant pGEX-4T-1 could grow on 500, 750, and 1000 mM mannitol stress plates. In contrast, pGEX-4T-1-ScCaM cells only grew on 500 mM mannitol stress plates, and there was no colony on 750 mM mannitol or 1000 mM mannitol stress plates; besides, the growth of pGEX-4T-1-ScCaM cells on 500 mM mannitol stress plates was weaker than that of pGEX-4T-1 cells. Thus, it was hypothesized that ScCaM also attenuated the tolerance of E. coli to mannitol stress.
Calcium (Ca2+), as an important intracellular second messenger, was related to various biological programs (Perochon et al., 2011). CaMs proteins, which contain four EF-hand, are vital transducers of Ca2+ signals. It is worth noting that CaMs lack effector domains and they function as sensor relays via Ca2+-dependent interaction with the downstream effector proteins (Perochon et al., 2011). Current studies have shown that CaMs bind and regulate a vast array of target proteins in plants, including transcription factors, metabolic enzymes, kinases, and so on (Astegno et al., 2016; Vandelle et al., 2018). In addition, CaM proteins are involved in many cellular responses through binding to Ca2+ and play an important role, especially in abiotic stresses (Galon et al., 2010; Poovaiah et al., 2013).
Previous studies have shown that drought and salt stresses changed the concentration of Ca2+ (Knight et al., 1997). Calcium signaling has been implicated in the transduction of drought- and salt-stress signals in plants and may play a role in a number of responses to drought stress and water potential changes (Johansson et al., 1996; Takahashi et al., 1997). CaM is considered to regulate several intracellular proteins involved in modulating intracellular Ca2+ levels and provides a negative feedback mechanism for calcium signaling (Kovacs et al., 2010). Shen et al. (2020) found that HvCaM1 negatively regulates salt tolerance via modulation of the expression of HvHKT1s and HvCAMTA4. On the contrary, OsCaM1-1 overexpression in the transgenic rice mitigated salt-induced oxidative damage (Kaewneramit et al., 2019; Yuenyong et al., 2018). In this study, the experiments of eukaryotic overexpression and prokaryotic expression were conducted to explore the function of ScCaM under NaCl and drought stresses. The above results indicated that ScCaM plays a negative regulatory role in response to NaCl and drought stresses. CaMs play both positive and negative roles in plant immunity (Cheval et al., 2013; Poovaiah et al., 2013). Similarly, calcium/calmodulin-dependent protein kinase is negatively and positively regulated by CaM (Miller et al., 2013). In Arabidopsis, CaM2, CaM3, CaM5, and CaM7 were found to negatively regulate Ca2+ influx under heat shock conditions (Niu et al., 2020). Many studies have also suggested a positive regulatory role for CaM. For instance, a previous study found that overexpression of GmCaM4 in soybean enhanced resistance to pathogens and tolerance to salinity stress (Rao et al., 2014). Zhou et al. (2016) demonstrated that the expression of AtCaM1/4 was upregulated under salt stress, and it resulted in enhanced salt tolerance in transgenic A. thaliana. Overexpression of EcCaM in A. thaliana was found to make the plant more tolerant to polyethylene glycol (PEG)-induced drought and salt stress (NaCl) (Jamra et al., 2021). Noman et al. (2019) even thought that GmCaMTA12 may enhance the tolerance of Arabidopsis plants under drought stress through Ca-CaM-CAMTA-mediated stress regulatory mechanisms.
Interestingly, when the germination rate of Arabidopsis seeds overexpressing GmCaMTA12 was determined after treatment with mannitol, it was observed that the germination rate of transgenic Arabidopsis seeds was significantly higher than that of the wild type (Noman et al., 2019). In the present study, the growth potential of ScCaM transgenic Arabidopsis seeds under NaCl and mannitol stress was weaker than that of the wild type, and ScCaM overexpressing transgenic Arabidopsis plants were less tolerant to salinity and drought stresses than the wild type, whereas the expression of AtP5CS and AtRD29A, two genes tightly related to stress resistance, were also significantly lower than that in the wild type. High expression of AtSOD and AtCAT genes mitigates the negative effects of ROS and enhances the tolerance of plants under abiotic stress (Ju et al., 2020; Wang et al., 2021). CaM can regulate ROS-scavenging enzymes (SOD and CAT) (Ming and Zhong-Guan, 1995; Yang and Poovaiah, 2002). However, in this study, a sensitive phenotype was observed. Based on these results, we thus suppose that high expression of SOD/CAT protects cells against ROS, but low expression of the P5CS gene synthesizes less proline as osmotic substances (Jain et al., 2015). The sensitivity of plants is mainly due to the lack of osmotic substances. AtRD29A was found to function in ABA-related response to stress (Liu et al., 2020). We thus speculated that overexpression of ScCaM may inhibit the expression of AtRD29A by inhibiting the ABA signal transduction. Our previous study showed that overexpression of the allogenic ScCaM gene had effects on the growth of transgenic plants (Liu et al., 2021). In this study, we found that the transgenic plants also had a specific phenotype after stress. Hence, a working model was thus proposed in this study (Fig. 7). Based on the model, we can notice that ScCaM may influence the catabolism of osmotic substances and the ABA signal transduction. However, the relationship between growth and stresses in transgenic plants still requires further studies. This may be the reason why ScCaM transgenic Arabidopsis plants were less tolerant to salinity and drought stresses. Interestingly, overexpression of ScCaM in E. coli cells also expressed weaker tolerance under salinity and drought stresses.

Figure 7: A working model of the function of sugarcane calmodulin (ScCaM) in negatively regulating the salinity and drought stress in Arabidopsis (Jain et al., 2015).
Taken together, the present study indicates that the ScCaM gene plays a negative regulatory role under salinity and drought stresses, possibly through influencing the protective mechanisms common in both prokaryotes and eukaryotes under stress conditions.
In summary, ScCaM is a negative regulator in response to salinity and drought stress, and it is speculated that ScCaM mainly reduces the expression of AtP5CS genes, thus weakening the tolerance to salinity and drought stresses in transgenic plants and E. coli cells. However, the specific regulatory mechanism still needs to be investigated. The present study lays the foundation for further analysis of the functional role of ScCaM and its regulatory mechanism in sugarcane. Once this regulatory mechanism is understood, it should facilitate the application of transgenic ScCaM sugarcane in the near future.
Availability of Data and Materials: All relevant data supporting the conclusions of this article are mentioned in this article.
Author Contribution: J.L., Y.R., J.F., and J.L. conceived, designed, and initiated the project. J.L., C.Z., G.W., and X.F. prepared materials. J.L., Y.R., J.F., C.Z., G.W., X.F., and N.H. performed experiments and contributed to data analysis and validation. J.L., J.L., H.L., W.S., and Y.Q. drafted the manuscript. J.L., J.L., H.L., W.S., and Y.Q. revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Ethics Approval: Not applicable.
Funding Statement: This research was funded by the Natural Science Foundation of Fujian Province, China (2018J01470 and 2021J01137), Scientific research projects of introducing talents in Wuyi University (YJ202109), Special fund for scientific and technological innovation of Fujian Agriculture and Forestry University (CXZX2020081A) and China Agriculture Research System of MOF and MARA (CARS-17). The funders had no role in the study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
Astegno A, La Verde V, Marino V, Dell’Orco D, Dominici P (2016). Biochemical and biophysical characterization of a plant calmodulin: Role of the N-and C-lobes in calcium binding, conformational change, and target interaction. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics 1864: 297–307. DOI 10.1016/j.bbapap.2015.12.003. [Google Scholar] [CrossRef]
Boonburapong B, Buaboocha T (2007). Genome-wide identification and analyses of the rice calmodulin and related potential calcium sensor proteins. BMC Plant Biology 7: 4. DOI 10.1186/1471-2229-7-4. [Google Scholar] [CrossRef]
Cheval C, Aldon D, Galaud JP, Ranty B (2013). Calcium/calmodulin-mediated regulation of plant immunity. Biochimica et Biophysica Acta 1833: 1766–1771. DOI 10.1016/j.bbamcr.2013.01.031. [Google Scholar] [CrossRef]
Chinpongpanich A, Limruengroj K, Phean OPS, Limpaseni T, Buaboocha T (2012). Expression analysis of calmodulin and calmodulin-like genes from rice, Oryza sativa L. BMC Research Notes 5: 625. DOI 10.1186/1756-0500-5-625. [Google Scholar] [CrossRef]
Fu J, Shi Y, Wang L, Tian T, Li J, Gong L, Zheng Z, Jing M, Fang J, Ji R (2022). Planthopper-secreted salivary calmodulin acts as an effector for defense responses in rice. Frontiers in Plant Science 13: 841378. DOI 10.3389/fpls.2022.841378. [Google Scholar] [CrossRef]
Galon Y, Finkler A, Fromm H (2010). Calcium-regulated transcription in plants. Molecular Plant 3: 653–669. DOI 10.1093/mp/ssq019. [Google Scholar] [CrossRef]
Gifford JL, Walsh MP, Vogel HJ (2007). Structures and metal-ion-binding properties of the Ca2+-binding helix-loop-helix EF-hand motifs. Biochemical Journal 405: 199–221. DOI 10.1042/bj20070255. [Google Scholar] [CrossRef]
Heo WD, Lee SH, Kim MC, Kim JC, Chung WS et al. (1999). Involvement of specific calmodulin isoforms in salicylic acid-independent activation of plant disease resistance responses. PNAS 96: 766–771. DOI 10.1073/pnas.96.2.766. [Google Scholar] [CrossRef]
Hoeflich KP, Ikura M (2002). Calmodulin in action: Diversity in target recognition and activation mechanisms. Cell 108: 739–742. DOI 10.1016/s0092-8674(02)00682-7. [Google Scholar] [CrossRef]
Hu X, Jiang M, Zhang J, Zhang A, Lin F, Tan M (2007). Calcium-calmodulin is required for abscisic acid-induced antioxidant defense and functions both upstream and downstream of H2O2 production in leaves of maize (Zea mays) plants. New Phytologist 173: 27–38. DOI 10.1111/j.1469-8137.2006.01888.x. [Google Scholar] [CrossRef]
Jain R, Chandra A, Venugopalan V, Solomon S (2015). Physiological changes and expression of SOD and P5CS genes in response to water deficit in sugarcane. Sugar Tech 17: 276–282. DOI 10.1007/s12355-014-0317-2. [Google Scholar] [CrossRef]
Jamra G, Agarwal A, Singh N, Sanyal SK, Kumar A, Pandey GK (2021). Ectopic expression of finger millet calmodulin confers drought and salinity tolerance in Arabidopsis thaliana. Plant Cell Reports 40: 2205–2223. DOI 10.1007/s00299-021-02743-z. [Google Scholar] [CrossRef]
Johansson I, Larsson C, Ek B, Kjellbom P (1996). The major integral proteins of spinach leaf plasma membranes are putative aquaporins and are phosphorylated in response to Ca2+ and apoplastic water potential. Plant Cell 8: 1181–1191. DOI 10.1105/tpc.8.7.1181. [Google Scholar] [CrossRef]
Ju YL, Min Z, Yue XF, Zhang YL, Zhang JX, Zhang ZQ, Fang YL (2020). Overexpression of grapevine VvNAC08 enhances drought tolerance in transgenic Arabidopsis. Plant Physiology and Biochemistry 151: 214–222. DOI 10.1016/j.plaphy.2020.03.028. [Google Scholar] [CrossRef]
Kaewneramit T, Buaboocha T, Sangchai P, Wutipraditkul N (2019). OsCaM1-1 overexpression in the transgenic rice mitigated salt-induced oxidative damage. Biologia Plantarum 63: 335–342. DOI 10.32615/bp.2019.039. [Google Scholar] [CrossRef]
Knight H, Trewavas AJ, Knight MR (1997). Calcium signalling in Arabidopsis thaliana responding to drought and salinity. Plant Journal 12: 1067–1078. DOI 10.1046/j.1365-313x.1997.12051067.x. [Google Scholar] [CrossRef]
Kovacs E, Tóth J, Vértessy BG, Liliom K (2010). Dissociation of calmodulin-target peptide complexes by the lipid mediator sphingosylphosphorylcholine: Implications in calcium signaling. Journal of Biological Chemistry 285: 1799–1808. DOI 10.1074/jbc.M109.053116. [Google Scholar] [CrossRef]
Liu J, Zhang C, Su W, Wu G, Fu X, Que Y, Luo J (2021). Identification, expression, and functional characterization of ScCaM in response to various stresses in sugarcane. Agronomy 11: 2153. DOI 10.3390/agronomy11112153. [Google Scholar] [CrossRef]
Liu Y, Peng L, Gao X, Liu Y, Liu Z, Li X, Yang Y, Wang J (2020). AtPPRT3, a novel E3 ubiquitin ligase, plays a positive role in ABA signaling. Plant Cell Reports 39: 1467–1478. DOI 10.1007/s00299-020-02575-3. [Google Scholar] [CrossRef]
Livak KJ, Schmittgen TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408. DOI 10.1006/meth.2001.1262. [Google Scholar] [CrossRef]
McCormack E, Braam J (2003). Calmodulins and related potential calcium sensors of Arabidopsis. New Phytologist 159: 585–598. DOI 10.1046/j.1469-8137.2003.00845.x. [Google Scholar] [CrossRef]
Miller JB, Pratap A, Miyahara A, Zhou L, Bornemann S, Morris RJ, Oldroyd GE (2013). Calcium/Calmodulin-dependent protein kinase is negatively and positively regulated by calcium, providing a mechanism for decoding calcium responses during symbiosis signaling. Plant Cell 25: 5053–5066. DOI 10.1105/tpc.113.116921. [Google Scholar] [CrossRef]
Ming G, Zhong-Guan L (1995). Calmodulin-binding proteins from Zea mays germs. Phytochemistry 40: 1335–1339. DOI 10.1016/0031-9422(95)00381-G. [Google Scholar] [CrossRef]
Munir S, Khan MR, Song J, Munir S, Zhang Y, Ye Z, Wang T (2016). Genome-wide identification, characterization and expression analysis of calmodulin-like (CML) proteins in tomato (Solanum lycopersicum). Plant Physiology and Biochemistry 102: 167–179. DOI 10.1016/j.plaphy.2016.02.020. [Google Scholar] [CrossRef]
Nelson MR, Chazin WJ (1998). An interaction-based analysis of calcium-induced conformational changes in Ca2+ sensor proteins. Protein Science 7: 270–282. DOI 10.1002/pro.5560070206. [Google Scholar] [CrossRef]
Nie S, Zhang M, Zhang L (2017). Genome-wide identification and expression analysis of calmodulin-like (CML) genes in Chinese cabbage (Brassica rapa L. ssp. pekinensis). BMC Genomics 18: 842. DOI 10.1186/s12864-017-4240-2. [Google Scholar] [CrossRef]
Niu WT, Han XW, Wei SS, Shang ZL, Wang J et al. (2020). Arabidopsis cyclic nucleotide-gated channel 6 is negatively modulated by multiple calmodulin isoforms during heat shock. Journal of Experimental Botany 71: 90–104. DOI 10.1093/jxb/erz445. [Google Scholar] [CrossRef]
Noman M, Jameel A, Qiang WD, Ahmad N, Liu WC, Wang FW, Li HY (2019). Overexpression of GmCAMTA12 enhanced drought tolerance in Arabidopsis and soybean. International Journal of Molecular Sciences 20: 4849. DOI 10.3390/ijms20194849. [Google Scholar] [CrossRef]
Park HC, Kim ML, Kang YH, Jeon JM, Yoo JH et al. (2004). Pathogen- and NaCl-induced expression of the SCaM-4 promoter is mediated in part by a GT-1 box that interacts with a GT-1-like transcription factor. Plant Physiology 135: 2150–2161. DOI 10.1104/pp.104.041442. [Google Scholar] [CrossRef]
Perochon A, Aldon D, Galaud JP, Ranty B (2011). Calmodulin and calmodulin-like proteins in plant calcium signaling. Biochimie 93: 2048–2053. DOI 10.1016/j.biochi.2011.07.012. [Google Scholar] [CrossRef]
Poovaiah BW, Du L, Wang H, Yang T (2013). Recent advances in calcium/calmodulin-mediated signaling with an emphasis on plant-microbe interactions. Plant Physiology 163: 531–542. DOI 10.1104/pp.113.220780. [Google Scholar] [CrossRef]
Qiu JR, Huang Z, Xiang XY, Xu WX, Wang JT et al. (2020). MfbHLH38, a Myrothamnus flabellifolia bHLH transcription factor, confers tolerance to drought and salinity stresses in Arabidopsis. BMC Plant Biology 20: 542. DOI 10.1186/s12870-020-02732-6. [Google Scholar] [CrossRef]
Rao SS, El-Habbak MH, Havens WM, Singh A, Zheng D, Vaughn L, Haudenshield JS, Hartman GL, Korban SS, Ghabrial SA (2014). Overexpression of GmCaM4 in soybean enhances resistance to pathogens and tolerance to salt stress. Molecular Plant Pathology 15: 145–160. DOI 10.1111/mpp.12075. [Google Scholar] [CrossRef]
Reggiani R, Aurisano N, Bertani A (1995). Involvement of calcium and calmodulin in protein and amino acid metabolism in rice roots under anoxia. Plant and Cell Physiology 36: 1525–1529. DOI 10.1093/oxfordjournals.pcp.a078917. [Google Scholar] [CrossRef]
Rhoads AR, Friedberg F (1997). Sequence motifs for calmodulin recognition. The FASEB Journal 11: 331–340. DOI 10.1096/fasebj.11.5.9141499. [Google Scholar] [CrossRef]
Shen Q, Fu L, Su T, Ye L, Huang L, Kuang L, Wu L, Wu D, Chen ZH, Zhang G (2020). Calmodulin HvCaM1 negatively regulates salt tolerance via modulation of HvHKT1s and HvCAMTA4. Plant Physiology 183: 1650–1662. DOI 10.1104/pp.20.00196. [Google Scholar] [CrossRef]
Shi J, Du X (2020). Identification, characterization and expression analysis of calmodulin and calmodulin-like proteins in Solanum pennellii. Scientific Reports 10: 7474. DOI 10.1038/s41598-020-64178-y. [Google Scholar] [CrossRef]
Snedden WA, Fromm H (2001). Calmodulin as a versatile calcium signal transducer in plants. New Phytologist 151: 35–66. DOI 10.1046/j.1469-8137.2001.00154.x. [Google Scholar] [CrossRef]
Su W, Huang L, Ling H, Mao H, Huang N et al. (2020a). Sugarcane calcineurin B-like (CBL) genes play important but versatile roles in regulation of responses to biotic and abiotic stresses. Scientific Reports 10: 167. DOI 10.1038/s41598-019-57058-7. [Google Scholar] [CrossRef]
Su W, Ren Y, Wang D, Su Y, Feng J, Zhang C, Tang H, Xu L, Muhammad K, Que Y (2020b). The alcohol dehydrogenase gene family in sugarcane and its involvement in cold stress regulation. BMC Genomics 21: 521. DOI 10.1186/s12864-020-06929-9. [Google Scholar] [CrossRef]
Takahashi K, Isobe M, Knight MR, Trewavas AJ, Muto S (1997). Hypoosmotic shock induces increases in cytosolic Ca2+ in tobacco suspension-culture cells. Plant Physiology 113: 587–594. DOI 10.1104/pp.113.2.587. [Google Scholar] [CrossRef]
Townley HE, Knight MR (2002). Calmodulin as a potential negative regulator of Arabidopsis COR gene expression. Plant Physiology 128: 1169–1172. DOI 10.1104/pp.010814. [Google Scholar] [CrossRef]
Vandelle E, Vannozzi A, Wong D, Danzi D, Digby AM, Dal Santo S, Astegno A (2018). Identification, characterization, and expression analysis of calmodulin and calmodulin-like genes in grapevine (Vitis vinifera) reveal likely roles in stress responses. Plant Physiology and Biochemistry 129: 221–237. DOI 10.1016/j.plaphy.2018.06.003. [Google Scholar] [CrossRef]
Wang Y, Dai X, Xu G, Dai Z, Chen P, Zhang T, Zhang H (2021). The Ca(2+)-CaM signaling pathway mediates potassium uptake by regulating reactive oxygen species homeostasis in tbacco roots under low-K+ stress. Frontiers in Plant Science 12: 658609. DOI 10.3389/fpls.2021.658609. [Google Scholar] [CrossRef]
Yang C, Zhou Y, Fan J, Fu Y, Shen L et al. (2015). SpBADH of the halophyte Sesuvium portulacastrum strongly confers drought tolerance through ROS scavenging in transgenic Arabidopsis. Plant Physiology and Biochemistry 96: 377–387. DOI 10.1016/j.plaphy.2015.08.010. [Google Scholar] [CrossRef]
Yang T, Poovaiah BW (2002). Hydrogen peroxide homeostasis: Activation of plant catalase by calcium/calmodulin. PNAS 99: 4097–4102. DOI 10.1073/pnas.052564899. [Google Scholar] [CrossRef]
Yuenyong W, Chinpongpanich A, Comai L, Chadchawan S, Buaboocha T (2018). Downstream components of the calmodulin signaling pathway in the rice salt stress response revealed by transcriptome profiling and target identification. BMC Plant Biology 18: 335. DOI 10.1186/s12870-018-1538-4. [Google Scholar] [CrossRef]
Zhang J, Zhang X, Tang H, Zhang Q, Hua X et al. (2018). Allele-defined genome of the autopolyploid sugarcane Saccharum spontaneum L. Nature Genetics 50: 1565–1573. DOI 10.1038/s41588-018-0237-2. [Google Scholar] [CrossRef]
Zhang W, Zhou RG, Gao YJ, Zheng SZ, Xu P, Zhang SQ, Sun DY (2009). Molecular and genetic evidence for the key role of AtCaM3 in heat-shock signal transduction in Arabidopsis. Plant Physiology 149: 1773–1784. DOI 10.1104/pp.108.133744. [Google Scholar] [CrossRef]
Zhou S, Jia L, Chu H, Wu D, Peng X, Liu X, Zhang J, Zhao J, Chen K, Zhao L (2016). Arabidopsis CaM1 and CaM4 promote nitric oxide production and salt resistance by inhibiting s-nitrosoglutathione reductase via direct binding. PLoS Genetics 12: e1006255. DOI 10.1371/journal.pgen.1006255. [Google Scholar] [CrossRef]
Zhu JK (2002). Salt and drought stress signal transduction in plants. Annual Review of Plant Biology 53: 247–273. DOI 10.1146/annurev.arplant.53.091401.143329. [Google Scholar] [CrossRef]
Zhu L, Zhang XQ, Ye D, Chen LQ (2021). The mildew resistance locus O4 interacts with CaM/CML and is involved in root gravity response. International Journal of Molecular Sciences 22: 5962. DOI 10.3390/ijms22115962. [Google Scholar] [CrossRef]

SUPPLEMENTARY FIGURE S1: Selection of T3 generation transgenic overexpressing ScCaM Arabidopsis seeds on plates containing hygromycin.


Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools