DOI:10.32604/biocell.2022.020229

| BIOCELL DOI:10.32604/biocell.2022.020229 |  |
| Article |
Effects of two vectors on the expression of the NbNAC1 transcription factor and preparation of its polyclonal antibody
College of Horticulture and Plant Protection, Joint International Research Laboratory of Agriculture & Agri-Product Safety, the Ministry of Education of China, Yangzhou University, Yangzhou, 225009, China
*Address correspondence to: Feng Zhu, zhufeng@yzu.edu.cn
Received: 12 November 2021; Accepted: 31 January 2022
Abstract: The NAC (NAM, ATAF, and CUC) superfamily is one of the largest plant-specific families containing transcription factors. An increasing number of studies suggest that NAC1 is involved in plants response to different biotic and abiotic stimulis. Nicotiana benthamiana is a widely used system for evaluating plant-pathogen interactions. In order to study the biochemical function of NbNAC1, NbNAC1 protein and antibody are essential. Therefore, we focused on developing a prokaryotic expression system for producing the Nicotiana benthamiana NbNAC1 protein of in Escherichia coli and the preparation of its polyclonal antibody. Firstly, we constructed two different molecular weight prokaryotic expression vectors: pGE vector with GST tag (pGEX4T-1–NbNAC1) and pET expression vector with His tag (pET28a-NbNAC1). The NbNAC1 protein can be successfully expressed in both vectors. The His-tagged NbNAC1 proteins are insoluble, while the GST-tagged NbNAC1 proteins are partially soluble. We then successfully purified and enriched both proteins. The His-tagged NbNAC1 was chosen to immunize rabbits owing to an unknown protein accompanying the GST-tagged NbNAC1. The anti-NbNAC1 polyclonal antibody had good specificity and could be used in subsequent protein-related studies.
Keywords: NbNAC1 transcription factor; Nicotiana benthamiana; Prokaryotic expression; Purification; Antibodies
Transcription factors (TF) play important roles in plant responses to biotic and abiotic stresses. The NAC (no apical meristem, NAM; Arabidopsis transcription activation factor, ATAF; cup-shaped cotyledon, CUC) superfamily is one of the largest plant-specific TF families. Abundant NAC members have been identified in different plants, such as Arabidopsis, rice, tobacco and grape (Nuruzzaman et al., 2013). Many studies suggest that NAC1 plays an important role in plant stress tolerance. The SlNAC1 gene is noticeably upregulated during Pseudomonas infection (Huang et al., 2013). Silencing of NbNAC1 in Nicotiana benthamiana results in enhanced susceptibility to Pseudomonas bacteria (Huang et al., 2013). The Arabidopsis ortholog of SlNAC1 is induced by Botrytis cinerea infection (Wu et al., 2009). ATAF1 has been demonstrated to be a negative regulator of defense responses to both necrotrophic fungal and bacterial pathogens (Wang et al., 2009). Overexpression of grapevine NAC1 (VvNAC1) promotes tolerance to both necrotrophic and biotrophic pathogens in Arabidopsis (Henanff et al., 2013). Furthermore, grapevine leaves treated with SA or MeJA increases VvNAC1 expression (Henanff et al., 2013). Currently, the molecular mechanism and roles of NAC1 in plant immunity remain unclear.
N. benthamiana is a useful material for studies about plant-microbe interactions and other areas (Bombarely et al., 2012; Goodin et al., 2008). N. benthamiana is susceptible to many well-characterized bacterial, fungal, oomycete, and virus pathogens, as well as certain aphids (Bos et al., 2010; Jonge et al., 2012; Kamoun et al., 1998; Li et al., 2017; Ramsey et al., 2017; Wei et al., 2007; Yoshino et al., 2012; Zhu et al., 2015). The NbNAC1 gene was first cloned by RT-PCR from N. benthamiana in our previous study (Zhu et al., 2019). The NbNAC1 protein and antibody are essential for studying the biochemical function and molecular mechanism of NbNAC1. However, purifying the NbNAC1 protein directly from plants is a time-consuming and laborious process owing to its low expression level and the long growth cycle of plants. Thus, a recombinant DNA technology is utilized for producing the recombinant proteins in microbial systems. Among microorganisms, available host systems include bacteria, filamentous fungi, yeast, and unicellular algae (Zhu et al., 2012). Escherichia coli is a commonly used organism for producing of recombinant proteins because its use as a host organism is well known (Hao et al., 2012). Therefore, in this study we focused on developing a prokaryotic expression system for producing the NbNAC1 protein in E. coli and the preparation of its polyclonal antibody.
Bacterial strains, vectors, and enzymes
E. coli DH5α was used as the host for subcloning and plasmid amplification. E. coli Rosetta (DE3) was used for the expression of the recombinant protein. Two prokaryotic expression vectors with different tags, pET-28a (+) and pGEX-4T-1, were used as the expression plasmid. The pMD-19T vector, BamHI, EcoRI, XhoI and T4 DNA ligase were purchased from Takara (Dalian, China). All other chemical reagents were of analytical grade and manufactured in China.
We cloned the protein coding region of NbNAC1 using two pair primers NbNAC1-F (5'-CGGGATCCATGATCAAAGGCGTACAC-3') and NbNAC1-R (5'- CGCTCGAGGTAAGGTTTCTGCATGTATA-3'), where the underlined bases represent BamHI and XhoI endonuclease sites, respectively, plus NbNAC1-F’ (5'-CCGGAATTCATGATCAAAGGCGTA-3') and NbNAC1-R’ (5'-CCGCTCGAGGTAAGGTTTCTGCATGTATA-3'), where underlined bases represent EcoRI and XhoI endonuclease sites, respectively. The RT-PCR products were purified and recovered using a TIANgel Midi Purification Kit (Tiangen, Beijing, China).
Construction of prokaryotic expression vectors
The resulting PCR products were digested with BamHI and XhoI, and ligated into the pET-28a plasmid at the corresponding restriction sites. The resulting PCR products were digested with EcoRI and XhoI, and ligated into the pGEX-4T-1 plasmid at the corresponding restriction sites. The ligation mixture was transferred into E. coli DH5a competent cells for propagation of the recombinant plasmids. The recombinant plasmids pET-28a-NbNAC1 and pGEX-4T-1-NbNAC1 were confirmed by plasmid PCR, restriction endonucleases digestion, and sequencing.
Small-scale expression of recombinant protein in E. coli
Cultivation of the recombinant strain was carried out in 50 mL Luria-Bertani (LB) media and cultured at 37°C. We used various concentrations of IPTG (0.5 mM, 1 mM, 1.5 mM, and 2 mM) for inducing the recombinant protein. There were no significant differences with varying IPTG concentration. Cell growth at 1 mM IPTG was not affected; so, this concentration was used extensively in the prokaryotic expression experiments. When the optical density of the culture solution at 600 nm reached approximately 0.6, a final concentration of 1.0 mM isopropyl-β-d-thiogalactoside (IPTG) was added. The culture solution was then separated to culture at 18°C, 28°C, or 37°C. After 4 h, the induction was stopped, the cultures were centrifuged at 7992 g for 1 min, and the proteins were extracted from the collected bacteria cells. Finally, these protein samples were assayed with sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE).
Large-scale expression of recombinant protein in E. coli
The recombinant strain was cultured in 1 L of LB medium at 37°C until the optical density of the culture solution at 600 nm reached 0.6. IPTG was added to prepare a 1 mM final concentration, which was cultured for 4 h. The induced culture was next centrifuged at 5550 g for 15 min at 4°C. A 50 mL of protein suspension buffer (200 mM Tris-HCl, 500 mM NaCl, 10% glycerol) was added to resuspend the 35 grams of precipitate. The resuspended culture was sonicated for 10 min (the sonication time was 2 s and the intermittent time was 3 s) with an ultrasonic cell breaker. The sonicated culture was centrifuged at 22733 g for 1 h. The supernatant and precipitate were collected for future use. Finally, the supernatant and precipitate samples were assayed with SDS-PAGE.
Purification of His-tagged protein from inclusion bodies by Ni-NTA resin affinity chromatography
The His-tagged NbNAC1 proteins from inclusion bodies were purified using 5 mL of Ni-NTA resin affinity chromatography (GenScript, Nanjing, China). After disruption and centrifugation, the precipitate was resuspended with a dialysate that was a solution prepared with 8 M urea and protein suspension buffer (200 mM Tris-HCl, 500 mM NaCl, 10% glycerol). The insoluble protein pellet was dialysed. Before purification, insoluble protein pellet needed to be completely dissolved and renatured by dialysis. Further processing was performed according to the manufacturer’s instructions. Each wash fraction was analyzed by SDS–PAGE using 12.5% polyacrylamide separation gel. The collected fractions were then analyzed by SDS–PAGE.
Dialysis of proteins from inclusion bodies
Pre-treatment of the dialysis bag followed the manufacturer’s instructions (Solarbio, Beijing, China) with the prepared 1L for each concentration of dialysis solutions containing 6 M, 4 M, 2 M, 1 M, 0.5 M, 0.2 M, 0.1 M and 0 M urea, respectively. The 10 ml of eluate containing the target protein was placed into the dialysis bag, which was placed into the dialysis solutions from high to low concentrations at 4°C with the dialysis time for each dialysis solutions being over 2 h. Finally, the dialyzed protein was quick-frozen in liquid nitrogen and stored at –80°C.
Purification of GST-tagged soluble protein by Glutathione Resin Affinity chromatography
The GST-tagged soluble NbNAC1 proteins were purified using Glutathione Resin Affinity chromatography (GenScript, Nanjing, China) and processed according to the manufacturer’s instructions. First of all, add 5 mL of GST purification medium to the chromatography column to allow the preservation solution in the purification medium to flow out, and then 5 column volumes of protein suspension buffer were added to process the chromatography column. After that, PMSF was added with a final concentration of 1 mM to the supernatant sample obtained from a large amount of protein induction. Then pass the sample through the column and collect the effluent, moreover repeat the passage through the column 2–3 times. Besides, add 5 column volumes of protein suspension buffer (200 mM Tris-HCl, 500 mM NaCl, 10% glycerol) to the column in order to wash the column to remove the contaminants bound in the GST purification medium. Then use 10 mL of the configured glutathione eluent (10 mM reduced glutathione, 5 mM dithiothreitol) to elute the target protein. Each wash fraction was analyzed by SDS–PAGE using 12.5% polyacrylamide separation gel. The collected fractions were then analyzed by SDS–PAGE.
Concentration of NbNAC1 protein
The purified proteins were concentrated using Amicon® Ultra-15 Centrifugal Filter Devices (Millipore) following the manufacturer’s protocol. The molecular weight cut off level of Amicon® Ultra-15 Centrifugal Filter is 3 kDa. The particle diameter of this ultrafiltration centrifuge tube is between 1.5 nm and 3 nm. The concentration of purified NbNAC1 protein was determined according to the BCA method.
Preparation of polyclonal antibodies
New Zealand big-ear rabbits were first immunized subcutaneously with 1 mg/mL purified His-tagged NbNAC1 proteins using the same volume as Freund’s complete adjuvant. The remaining procedures followed Zhu et al. (2012).
Antibodies titer determination by western blotting
Purified NbNAC1 proteins were analyzed in 12.5 % (v/v) SDS-PAGE. A western blot analysis was performed following Zhu et al. (2012).
The protein coding region of NbNAC1 gene (GenBank accession No. MF033266) had been cloned by RT-PCR from N. benthamiana in our previous study (Zhu et al., 2019). To study the effects of two prokaryotic expression vectors on the expression of the NbNAC1 transcription factor, two pair primers (NbNAC1-F, NbNAC1-R, and NbNAC1-F’, NbNAC1-R’) were respectively used to amplified the NbNAC1 gene with different endonuclease sites. As shown in Fig. 1A, the NbNAC1 gene with BamHI and XhoI endonuclease sites was successfully amplified by the plasmid PCR. In addition, we successfully obtained the NbNAC1 gene with EcoRI and XhoI endonuclease sites (Fig. 1B).
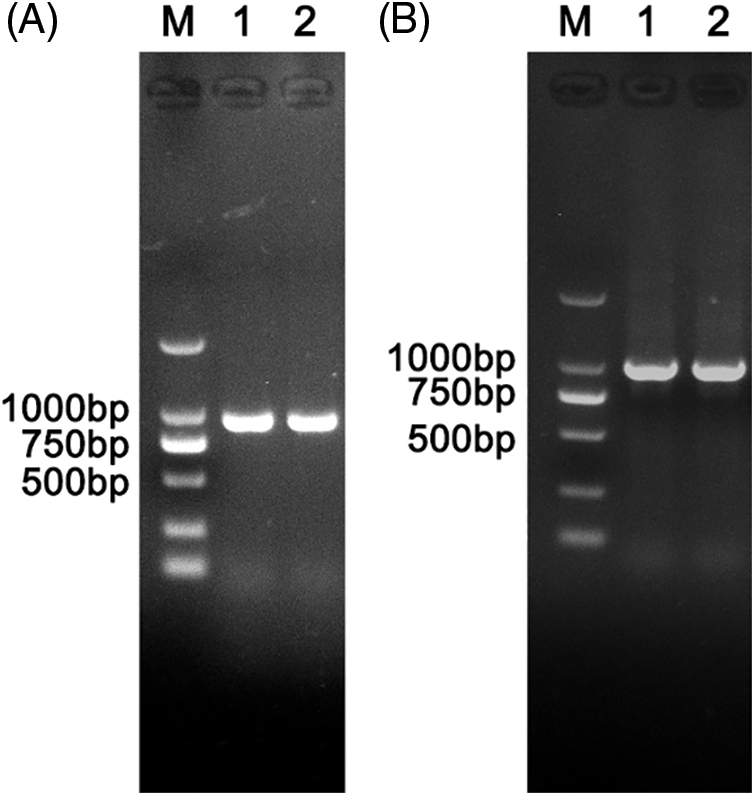
Figure 1: Amplification of the NbNAC1 gene with different endonuclease sites. (A) Amplification of the NbNAC1 gene with BamHI and XhoI endonuclease sites. Lane M: 2 kb DNA Marker. Lanes 1 and 2: amplified products of the NbNAC1 gene with BamHI and XhoI endonuclease sites. (B) Amplification of the NbNAC1 gene with EcoRI and XhoI endonuclease sites. Lane M: 2 kb DNA Marker. Lanes 1 and 2: amplified products of the NbNAC1 gene with EcoRI and XhoI endonuclease sites.
Construction of the prokaryotic expression vectors
The PCR product NbNAC1 with BamHI and XhoI endonuclease sites was digested with BamHI and XhoI, and ligated into the pET-28a plasmid containing the His-tags at the corresponding restriction sites (Fig. 2A). Another PCR product with EcoRI and XhoI endonuclease sites was digested with EcoRI and XhoI, and ligated into the pGEX-4T-1 plasmid with GST-tags at the corresponding restriction sites (Fig. 2B). The ligation mixture was transformed into E. coli DH5a competent cells for propagation of the recombinant plasmid. The recombinant plasmids pET28a-NbNAC1 were confirmed by plasmid PCR (Fig. 3A). Furthermore, the recombinant plasmids pET28a-NbNAC1 were confirmed by restriction endonucleases digestion (Fig. 3B). As shown in Fig. 3B, the restriction endonucleases digestion assay had two bands appearing in the agarose gel. Similar, the recombinant plasmids pGEX4T-1–NbNAC1 were confirmed by plasmid PCR (Fig. 3C) and restriction endonucleases digestion (Fig. 3D). Both pET28a-NbNAC1 and pGEX4T-1–NbNAC1 were transformed into E. coli Rosetta (DE3). The recombinant plasmids pET28a-NbNAC1 and pGEX4T-1-NbNAC1 were further confirmed by sequencing in E. coli Rosetta.
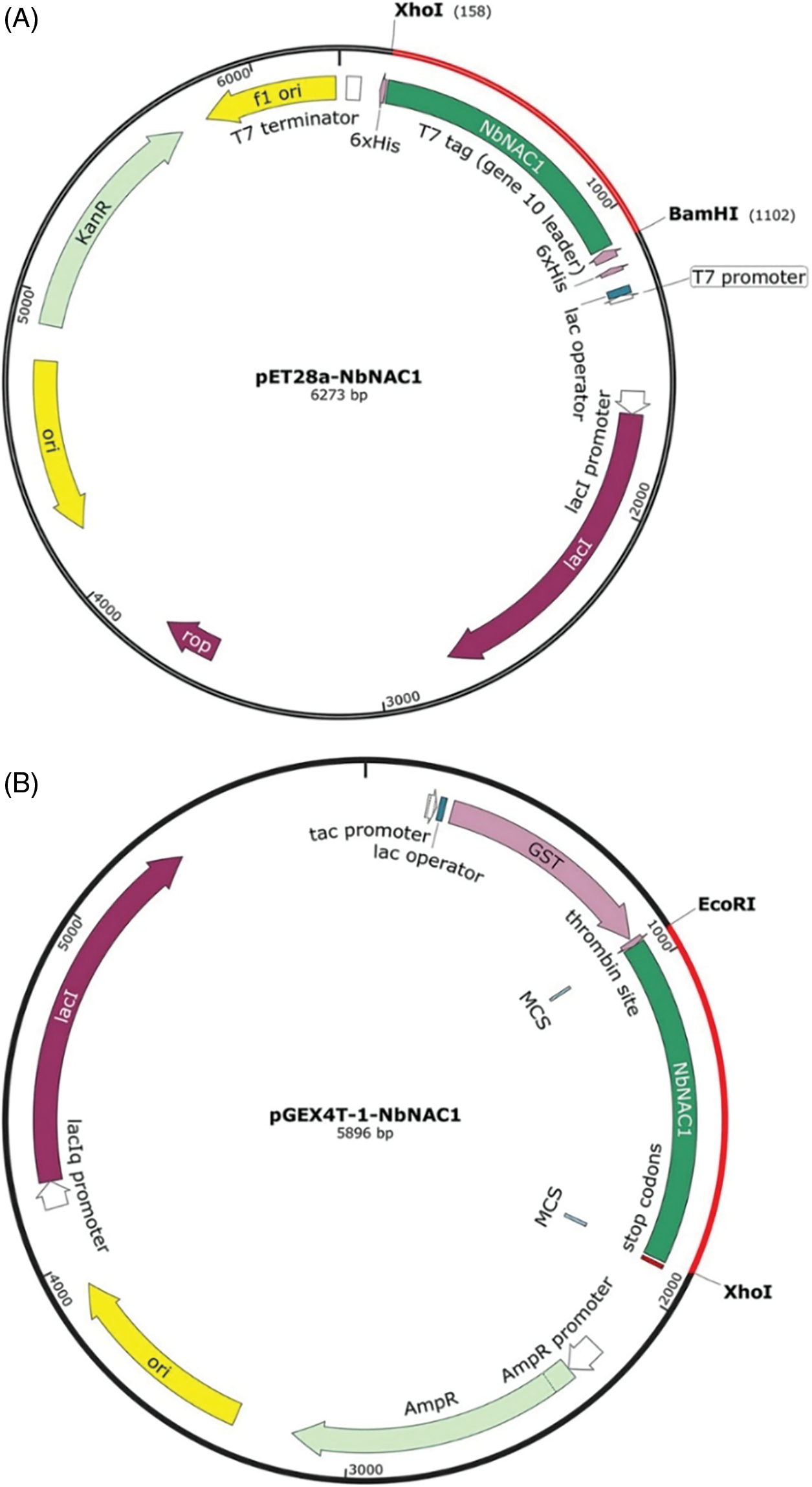
Figure 2: Graphical representation of two different molecular weight prokaryotic expression vectors. (A) Construction of the pET expression vector with the His-tag (pET28a-NbNAC1). The PCR producted NbNAC1 with BamHI and XhoI endonuclease sites was digested with BamHI and XhoI, and ligated into the pET-28a plasmid with the His-tag at the corresponding restriction sites. The resulting expression vector was pET28a–NbNAC1 encoded NbNAC1. (B) Construction of the pGE vector with a GST-tag (pGEX4T-1–NbNAC1). The PCR product NbNAC1 with EcoRI and XhoI endonuclease sites was digested with EcoRI and XhoI, and ligated into the pGEX-4T-1 plasmid with the GST-tag at the corresponding restriction sites. The resulting expression was vector pGEX4T-1–NbNAC1 encoded NbNAC1.
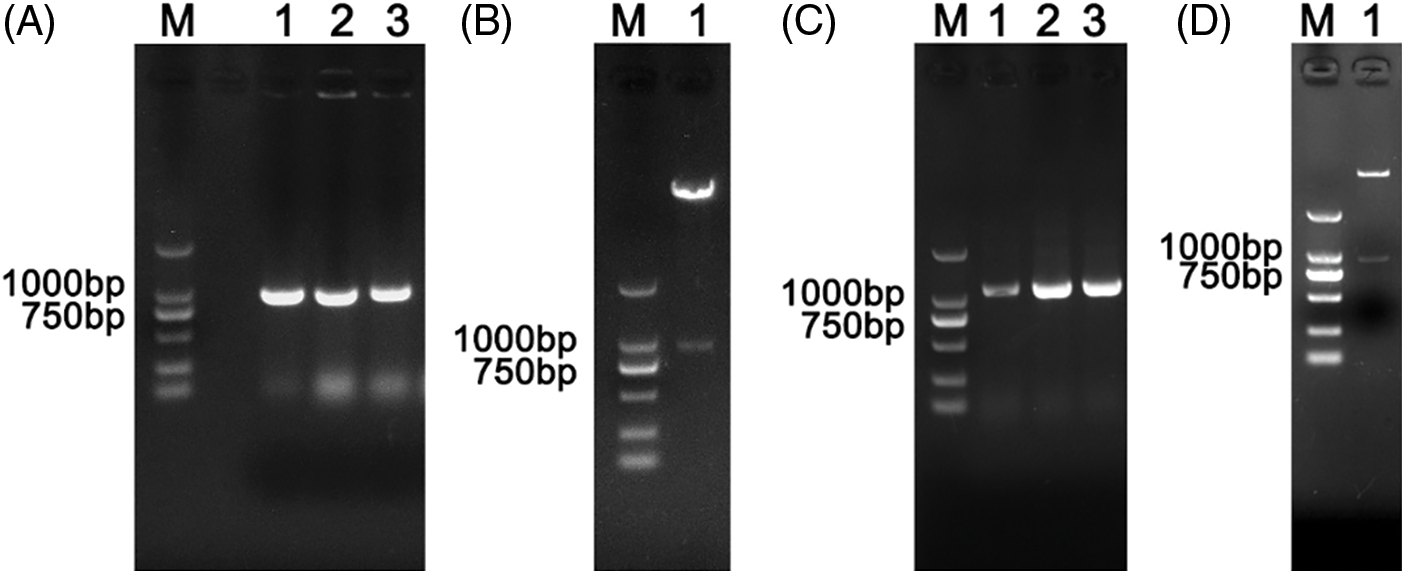
Figure 3: Verification of recombinant plasmids pET-28a-NbNAC1 and pGEX-4T-1-NbNAC1 by plasmid PCR and restriction endonucleases digestion. (A) The recombinant plasmids pET28a-NbNAC1 were confirmed by plasmid PCR. Lane M: 2 kb DNA Marker. Lanes 1, 2, and 3: detection of amplified products of NbNAC1 by plasmid PCR. (B) The recombinant plasmids pET28a-NbNAC1 were confirmed by restriction endonucleases digestion. Lane M: 2 kb DNA Marker. Lane 1: recombinant plasmids pET28a-NbNAC1 digested with BamHI and XhoI. (C) The recombinant plasmids pGEX-4T-1-NbNAC1 were confirmed by plasmid PCR. Lane M: 2 kb DNA Marker. Lanes 1, 2, and 3: detection of amplified products of NbNAC1 by plasmid PCR. (D) The recombinant plasmids pGEX-4T-1-NbNAC1 were confirmed by restriction endonucleases digestion. Lane M: 2 kb DNA Marker. Lane 1: recombinant plasmids pGEX-4T-1-NbNAC1 digested with EcoRI and XhoI.
Small-scale expression of recombinant NbNAC1 protein
The expression of the His-tagged NbNAC1 protein at different temperatures was initially investigated using a small-scale expression system. The molecular weight of the His-tagged NbNAC1 protein is approximately 41 kDa band according to the ORF of the NbNAC1 protein sequence. As shown in Fig. 4A, the SDS-PAGE analysis revealed the target proteins expressed an approximately 41 kDa band for NbNAC1 (IPTG lanes 2, 4, and 6 were induced). However, without the IPTG induction the target proteins were not observed without IPTG induction (lanes 1, 3, and 5). Furthermore, the expression of the recombinant NbNAC1 protein was maximized at 37°C (lane 2), which was much higher than the induced expressions at 28°C (lane 4) and 18°C (lane 6) (Fig. 4A).
We next investigated the expression of the GST-tagged NbNAC1 protein at different temperatures using a small-scale expression system. The molecular weight of the GST-tagged NbNAC1 protein is approximately 62 kDa according to the ORF of NbNAC1 protein sequence. As shown in Fig. 4B, SDS-PAGE analysis revealed that the GST-tagged NbNAC1 proteins expressed an approximately 62 kDa band for NbNAC1 (IPTG lanes 2, 4, and 6 were induced). However, without IPTG induction the GST-tagged NbNAC1 proteins were not observed (lanes 1, 3, and 5). Furthermore, the amount of recombinant NbNAC1 protein at 37°C (lane 6) was higher than at 28°C (lane 4) or 18°C (lane 2) (Fig. 4B).
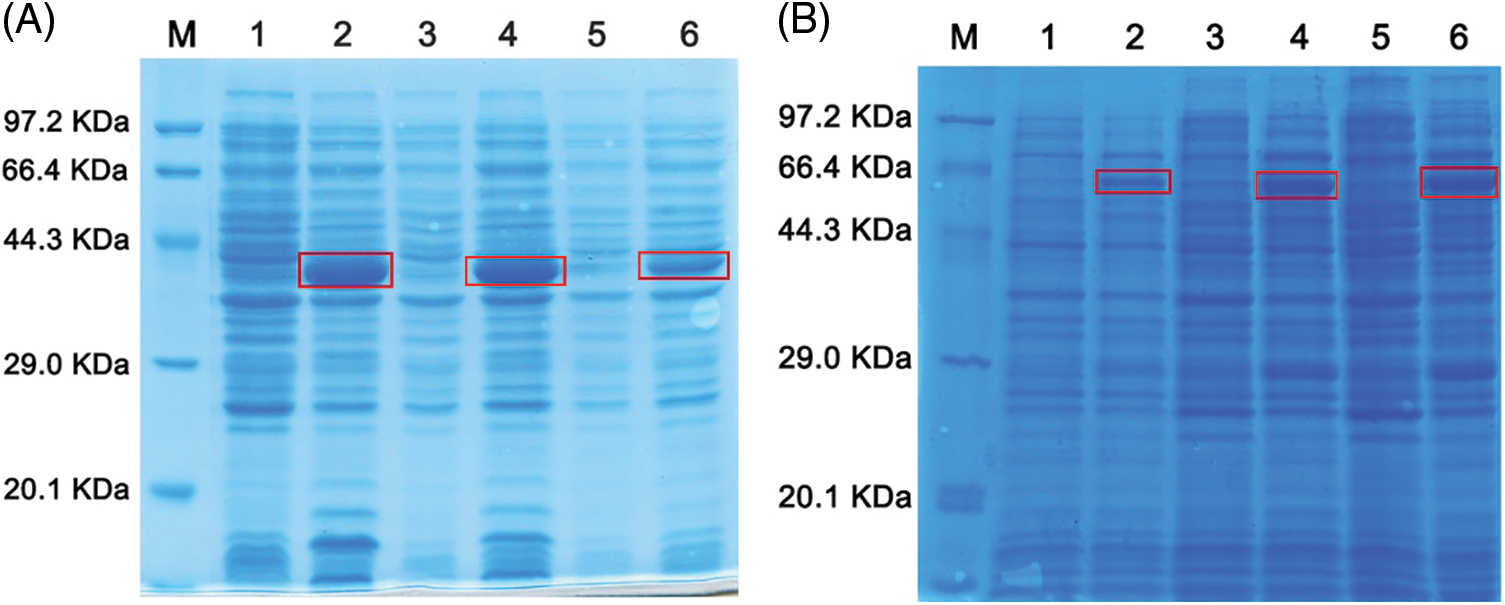
Figure 4: Analysis of the small-scale expression of recombinant NbNAC1 protein by SDS-PAGE. (A) The expression of His-tagged NbNAC1 proteins at different temperatures was investigated using the small-scale expression of recombinant proteins. Lane M: protein molecular weight marker. Lane 1: uninduced at 37°C. Lane 2: IPTG induced at 37°C. Lane 3: uninduced at 28°C. Lane 4: IPTG induced at 28°C. Lane 5: uninduced at 18°C. Lane 6: IPTG induced at 18°C. (B) The expression of GST-tagged NbNAC1 proteins at different temperatures was investigated using the small-scale expression of recombinant proteins. Lane M: protein molecular weight marker. Lane 1: uninduced at 18°C. Lane 2: IPTG induced at 18°C. Lane 3: uninduced at 28°C. Lane 4: IPTG induced at 28°C, Lane 5: uninduced at 37°C. Lane 6: IPTG induced at 37°C.
Detection the solubility of recombinant NbNAC1 protein
To determine whether the recombinant NbNAC1 protein is in inclusion bodies, the solubility of the His-tagged NbNAC1 proteins and GST-tagged NbNAC1 proteins was investigated. As shown in Fig. 5A, the His-tagged NbNAC1 proteins were almost all in the pellet (lanes 2, 4, and 6), but not in the supernatant (lanes 1, 3, and 5). Thus, the His-tagged NbNAC1 proteins are insoluble and belong to inclusion bodies proteins. Likewise, as shown in Fig. 5C, most of the GST-tagged NbNAC1 proteins were still in the pellet (lane 1), but the supernatant also had some of the target proteins (lane 2). Therefore, the GST-tagged NbNAC1 proteins were partially soluble.
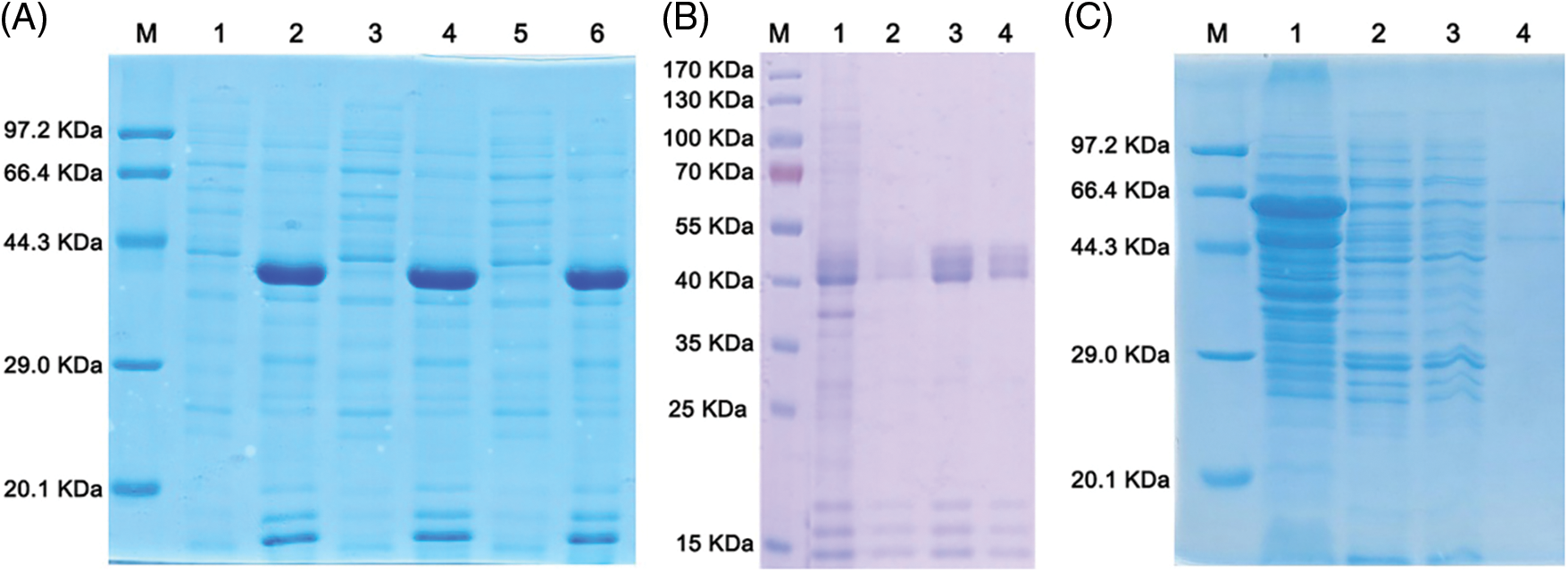
Figure 5: Analysis of the solubility and purification of recombinant NbNAC1 proteins by SDS-PAGE. (A) Analysis of the solubility of the His-tagged NbNAC1 proteins by SDS-PAGE. Lane M: protein molecular weight marker. Lanes 1, 3, and 5 are the same samples: supernatants. Lanes 2, 4, and 6 are the same samples: precipitates. (B) Large-scale expression and purification of His-tagged NbNAC1 proteins. The His-tagged NbNAC1 proteins expressed in E. coli using large-scale expression of recombinant proteins. The His-tagged NbNAC1 proteins were purified by Ni-NTA resin affinity chromatography. Lane M: protein molecular weight marker. Lanes 1–4: different collected samples. Lane 1: flow through fraction. Lane 2: wash solution. Lane 3: elution solution 1 (30 mM imidazole). Lane 4: elution solution 2 (300 mM imidazole). (C) Analysis of the solubility, large-scale expression and purification of pGEX-4T-1-NbNAC1 proteins. The GST-tagged NbNAC1 proteins expressed in E. coli using large-scale expression of recombinant proteins. The GST-tagged NbNAC1 proteins were purified by glutathione resin affinity chromatography. Lanes 1–4: collected different samples. Lane 1: precipitate. Lane 2: supernatant. Lane 3: flow through fraction. Lane 4: elution solution.
Large-scale expression and purification of recombinant NbNAC1 protein
To obtain a large amount of target proteins, the His- and GST-tagged NbNAC1 proteins were induced to express in E. coli using a large-scale expression scheme. The His- and GST-tagged NbNAC1 proteins were purified by Ni-NTA Resin affinity chromatography and glutathione resin affinity chromatography, respectively. As shown in Fig. 5B, high levels of the His-tagged NbNAC1 proteins were observed in the eluent (lanes 3 and 4). Lower level of the GST-tagged NbNAC1 proteins were also found in the eluent (lane 4) (Fig. 5C).
Concentration of recombinant NbNAC1 protein
The purified NbNAC1 protein was next enriched using Amicon® Ultra-15 Centrifugal Filter Devices (Millipore) and following the manufacturer’s protocol. As shown in Fig. 6A, the SDS-PAGE analysis revealed that an approximately 41 kDa single band (lanes 2 and 3) for the His-tagged NbNAC1 proteins. We successfully obtained high concentrations of the His-tagged NbNAC1 proteins (5 mg, 0.5 mg/mL). Similarly, the SDS-PAGE analysis revealed that an approximately 62 kDa band (lanes 1, 2, 3, and 4) for the GST-tagged NbNAC1 proteins (2.5 mg, 1.25 mg/mL) (Fig. 6B). However, there was an approximately 46 kDa contaminant of unknown nature under the target protein (Fig. 6B).
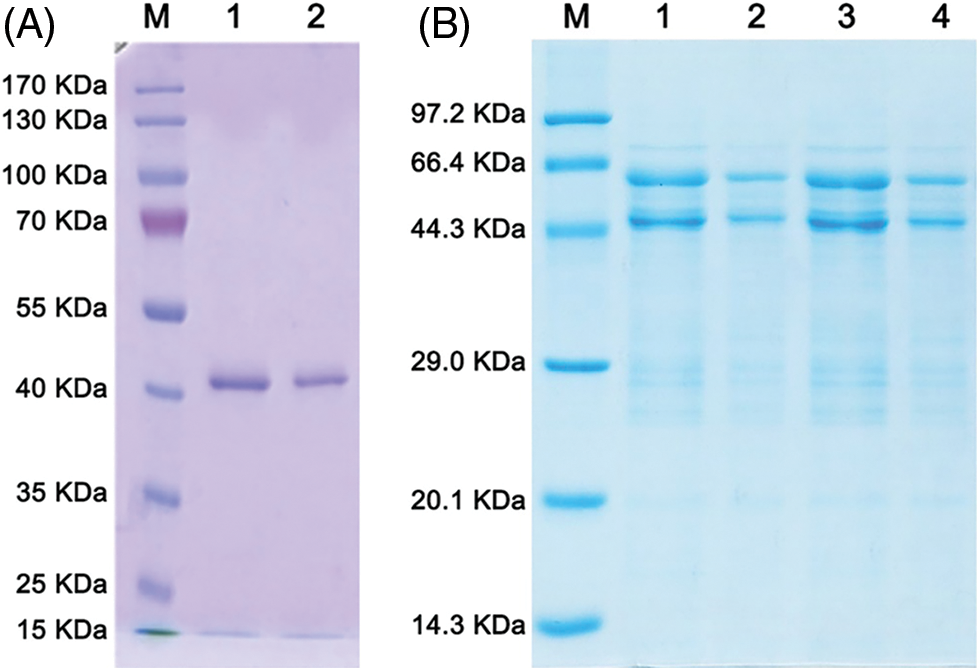
Figure 6: Concentration of recombinant NbNAC1 protein. (A) The His-tagged NbNAC1 proteins were renatured and concentrated with Amicon® Ultra-15 Centrifugal Filter Devices (Millipore). Lane M: protein molecular weight marker. Lanes 1 and 2 are the same samples: concentrated proteins (0.5 mg/mL). (B) The GST-tagged NbNAC1 proteins were concentrated with Amicon® Ultra-15 Centrifugal Filter Devices (Millipore). Lane M: protein molecular weight marker. Lanes 1 and 3 are the same samples: concentrated proteins (0.5 mg/mL). Lanes 2 and 4 are the same samples: concentrated proteins (0.1 mg/mL).
Titer and specificity analysis of NbNAC1 antibodies
Since the GST-tagged NbNAC1 proteins were purified with contaminant of unknown nature, we selected the His-tagged NbNAC1 proteins to immunize rabbits and obtained serum antibodies. The antibodies in different dilutions were tested with the different concentration of the purified His-tagged NbNAC1 proteins. As expected, an approximately 41 kDa band was detected with different dilutions of anti-NbNAC1 antibodies and different concentration of the purified His-tagged NbNAC1 proteins (Fig. 7A). The strongest approximately 41 kDa bands were shown in the Lanes 1, 4, and 7 (loaded the highest concentration of His-tagged NbNAC1 protein) (Fig. 7A). Furthermore, NbNAC1 antibody was used to detect the E. coli crude extract protein after induction of NbNAC1 expression and N. benthamiana crude extract protein. The results suggest that NbNAC1 antibody can also detect the NbNAC1 protein from the E. coli crude extract protein after induction of NbNAC1 expression and N. benthamiana crude extract protein (Fig. 7B). Therefore, our results suggested the anti-NbNAC1 antibodies had a high degree of detectable sensitivity.
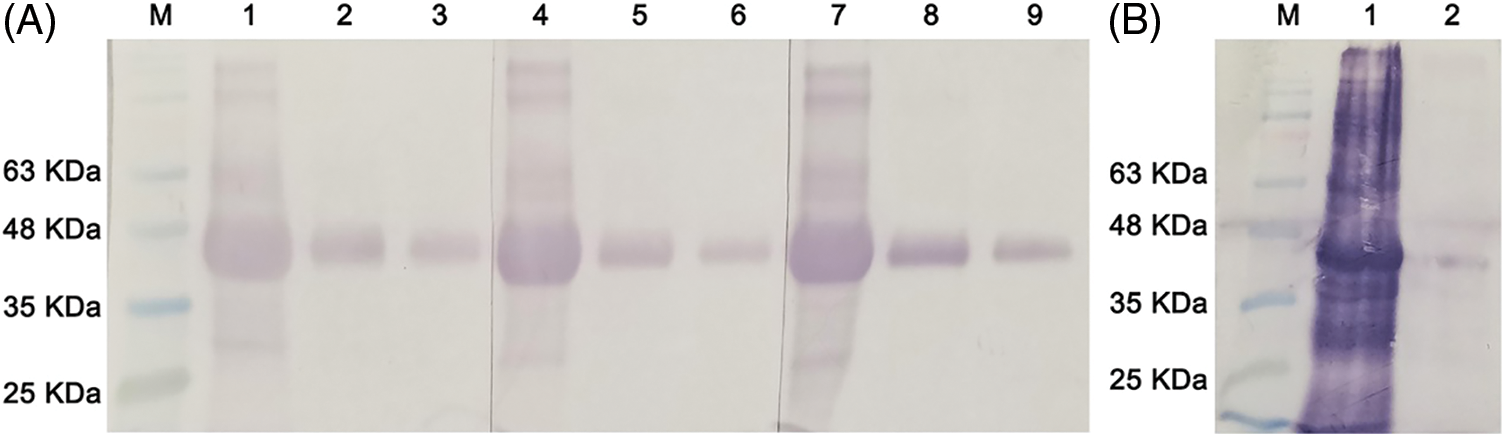
Figure 7: NbNAC1 antibody titer determination by western blotting. (A) Western blot assay was performed with a dilution series of recombinant NbNAC1 protein and 3 different antibody concentrations. The SDS-PAGE resolving gel and stacking gel were prepared, and 10 µL protein was loaded on the gel. After electrophoresis, the proteins were electrophoretically transferred to nitrocellulose membranes. The membranes were incubated with different dilutions of homemade anti-NbNAC1 polyclonal antibodies. After incubation for 2 h at 37°C, the membranes were thoroughly washed and incubated with alkaline phosphatase-conjugated anti-rabbit IgG antibodies for 1 h at 37°C. The membranes were stained in dark conditions with 10 ml of 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium (BCIP/NBT) about ten minutes, followed by the staining solution being poured off and the reaction being terminated with double distilled water containing EDTA. Lane M: protein molecular weight marker. Lanes 1, 4, 7: recombinant NbNAC1 protein is not diluted (0.5 mg/mL); Lanes 2, 5, 8: recombinant NbNAC1 protein is diluted 10 times (0.05 mg/mL); Lanes 3, 6, 9: recombinant NbNAC1 protein is diluted 100 times (0.005 mg/mL). Lanes 1–3, 4–6, 7–9: polyclonal antibody of NbNAC1 diluted into different multiples: Lanes 1–3: 1000×; Lanes 4–6: 500×; Lanes 7–9: 100×, respectively. (B) NbNAC1 antibody was used to detect the E. coli crude extract protein after induction of NbNAC1 expression and N. benthamiana crude extract protein by western blot. Lane M: protein molecular weight marker. Lane 1: E. coli crude extract protein after induction of NbNAC1 expression. Lane 2: N. benthamiana crude extract protein.
NAC transcription factors not only play important roles in plant growth and development (Zhu et al., 2019; Kjaersgaard et al., 2011; Yang et al., 2011; Zhong et al., 2010), but also participate in biotic and abiotic stress tolerance (Nakashima et al., 2012; Tran et al., 2010; Xia et al., 2010a; Tang et al., 2017). For example, NAC transcription factors could enhance plant resistance to droughts (He et al., 2017), low temperatures (Xia et al., 2010b) and high salinity conditions (Xu et al., 2015). However, the molecular mechanism and roles of NAC remain unclear. To further explore how NbNAC1 interacts with other factors to regulate plant resistance, we first needed to express a large number of target proteins using prokaryotic expression technology and then prepare polyclonal serum antibodies.
Prokaryotic expression techniques are often used to study the structure and function of proteins. The usual prokaryotic expression host organism is E. coli and its modified strain, which have simple structures that can easily express high quantities of proteins (Rosano and Ceccarelli, 2014; Chen, 2012). The heterologous expression of plant proteins in E. coli is affected by many factors, such as the expression vector, host bacteria, and induction temperature. In E. coli, temperature is the most important factor for the prokaryotic overexpression of a protein. In prokaryotic expression, proteins that are expressed too fast cannot be folded properly, which can easily form inclusion bodies that and have no biological activities that can affect subsequent studies (Li et al., 2001; Shimada et al., 2005; Wilkinson and Harrison, 1991). Therefore, the key factors for determining the successful purification of proteins are the expression vectors, host strains, expression inducement conditions, and renaturation of inclusion (Dong et al., 2008).
We compared the effects of two prokaryotic expression vectors on the expression of NbNAC1. We constructed two different molecular weight prokaryotic expression vectors: a pGE vector with GST-tag (pGEX4T-1–NbNAC1) and a pET expression vector with His-tag (pET28a-NbNAC1) (Fig. 2). The former has a high molecular weight GST-tag, while the latter has a small His-tag. The reason of choosing a pGE vector with a GST-tag as the prokaryotic expression vector is that the GST tag protein is a soluble protein that can be used to increase the solubility of various exogenous proteins and can be expressed in large quantities within E. coli to increase the expression of target proteins. The pGE vector with the GST-tag (pGEX4T-1–NbNAC1) can be successfully induced to the NbNAC1 protein (Fig. 4B). The expression of GST-tagged NbNAC1 proteins was maximized at 37°C (Fig. 4B). Most of the GST-tagged NbNAC1 proteins were in the pellet, but the supernatant also contained some of the target proteins (Fig. 5C). These results suggested that the GST-tagged NbNAC1 proteins were partial soluble. Therefore, the GST-tagged NbNAC1 proteins can be purified by Glutathione Resin affinity chromatography and concentrated with Amicon® Ultra-15 Centrifugal Filter Devices. SDS-PAGE analysis revealed that the GST-tagged NbNAC1 proteins were successfully purified and enriched (Fig. 6B). However, an unknown protein was also obtained (Fig. 6B). An ideal immunogen is a prerequisite for the preparation of high-quality antibodies. Therefore, the GST-tagged NbNAC1 proteins were not chosen for immunizing rabbits.
The NbNAC1 protein can be successfully expressed using a pET expression vector with a His-tag (pET28a-NbNAC1) (Fig. 4A). The expression level of His-tagged NbNAC1 proteins were maximized at 37°C (Fig. 4A). The His-tagged NbNAC1 proteins were almost entirely contained within the pellet, but not in the supernatant (Fig. 5A). Thus, the His-tagged NbNAC1 proteins are insoluble. Most fusion proteins (His-tagged NbNAC1) are in the form of inclusion bodies. Lower induction temperatures are beneficial for improving the formation of soluble proteins and reducing the expression of insoluble inclusions in the prokaryotic expression system (Wang et al., 2016). However, the His-tagged NbNAC1 proteins are still in the form of inclusion bodies when induced at lower temperatures (Fig. 5A). The His-tagged NbNAC1 proteins cannot achieve this effect at low temperatures, which may be related to the structure of the protein. The proteins in inclusion bodies can be transformed into soluble proteins after denaturation, renaturation, and other treatments. The components of such proteins are uniform, which facilitates the purification and functional analysis of the expression products (Carrió, 2002; Clark, 2001; Li et al., 2007; Tsumoto et al., 2003). Therefore, the His-tagged NbNAC1 proteins were successfully purified, dialyzed, and concentrated in the present study. However, the reason why there were clear contaminants in the low range in Fig. 5B for the purified His-tagged protein but not in Fig. 6 was that it may be caused by the following reasons. For example, the protein was degraded during the experiment or the protein was broken during using ultrasonic breaking causing it difficult to separate the tagged part from the target protein or improper concentration of imidazole made it difficult to remove some impurities. We successfully obtained high concentrations of His-tagged NbNAC1 proteins (0.5 mg/mL). The His-tag has a small molecular weight, a weak immune effect, and does not affect protein activity. Moreover, it is easy to purify and identify in later stage. Therefore, when the antibodies were prepared for rabbit immunity, we used the His-tagged NbNAC1 proteins for immunizing the rabbits and obtaining serum antibodies. Because His-tagged NbNAC1 proteins were injected for antibody production. Therefore, polyclonal antibody probably cross-reacts with His-tagged proteins.
Western blot is a common method for verifying the interactions between specific antibodies and corresponding proteins. It has the advantages of having a sensitive reaction and requiring few materials (Zhu et al., 2012). Thus, antibodies valence was determined by western blotting in this study.
Through this experiment, we found that the anti-NbNAC1 polyclonal antibody had a lower background and clear bands after hybridization with purified NbNAC1 proteins and NbNAC1 protein from the E. coli crude extract protein after induction of NbNAC1 expression and N. benthamiana crude extract protein (Fig. 7). The anti-NbNAC1 polyclonal antibody may be used in subsequent protein-related studies. Our study provides the basis for follow-up studies on the molecular mechanism of NbNAC1 in plant immunity. We are willing to share our antibody upon request.
Availability of Data and Materials: Data supporting this article are details in this manuscript.
Author Contribution: The authors confirm contribution to the paper as follows: study conception and design: Feng Zhu; data collection: Qinqin Zhang, Yangkai Zhou; analysis and interpretation of results: Qiping Zhang, Mengyao Cao, Zhaolin Ji; draft manuscript preparation: Feng Zhu. All authors reviewed the results and approved the final version of the manuscript.
Ethics Approval: The experimental rabbits used in this study were approved by Department of Science and Technology of Zhejiang Province, with License No. SYXK 2017-0009 in August 11, 2017.
Funding Statement: This work was supported by the National Natural Science Foundation of China (31500209); Natural Science Foundation of Jiangsu Province of China, (BK20201431); Agricultural science and technology independent innovation Foundation of Jiangsu Province of China (CX (20) 3128); Open Project Program of Joint International Research Laboratory of Agriculture and Agri-Product Safety, the Ministry of Education of China, Yangzhou University (JILAR-KF202006); Qing Lan Project of Yangzhou University and Yangzhou University of “High-end Talent Support Program”.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Bombarely A, Rosli HG, Vrebalov J, Moffett P, Martin GB (2012). A draft genome sequence of Nicotiana benthamiana to enhance molecular plant-microbe biology research. Molecular Plant Microbe Interactions 25: 1523–1530. [Google Scholar]
Bos JI, Prince D, Pitino M, Maffei ME, Win J et al. (2010). A functional genomics approach identifies candidate effectors from the aphid species Myzus persicae (green peach aphid). PLoS Genetics 6: e1001216. [Google Scholar]
Carrió MM (2002). Biotechnology, construction and deconstruction of bacterial inclusion bodies. Journal of Biotechnology 96: 3–12. [Google Scholar]
Chen R (2012). Bacterial expression systems for recombinant protein production: E. coli and beyond. Biotechnology Advances 30: 1102–1107. [Google Scholar]
Clark ED (2001). Protein refolding for industrial processes. Current Opinion in Biotechnology 12: 202–207. [Google Scholar]
Dong XB, Tang B, Li J, Xu Q, Fang ST et al. (2008). Expression and purification of intact and functional soybean (Glycine max) seed ferritin complex in Escherichia coli. Journal of Microbiology and Biotechnology 18: 299–307. [Google Scholar]
Goodin MM, Zaitlin D, Naidu RA, Lommel SA (2008). Nicotiana benthamiana: Its history and future as a model for plant-pathogen interactions. Molecular Plant Microbe Interactions 21: 1015–1026. [Google Scholar]
Hao YJ, Wei W, Song QX, Chen HW, Zhang YQ et al. (2012). Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant Journal 68: 302–313. [Google Scholar]
He L, Shi XX, Wang YN, Guo Y, Yang KJ et al. (2017). Arabidopsis ANAC069 binds to C [A/G] CG [T/G] sequences to negatively regulate salt and osmotic stress tolerance. Plant Molecular Biology 93: 1–19. [Google Scholar]
Henanff GL, Profizi C, Courteaux B, Rabenoelina F, Gérard C et al. (2013). Grapevine NAC1 transcription factor as a convergent node in developmental processes, abiotic stresses, and necrotrophic/biotrophic pathogen tolerance. Journal of Experimental Botany 64: 4877–4893. [Google Scholar]
Huang W, Miao M, Kud J, Niu X, Ouyang B et al. (2013). A stress-related transcription factor, is finetuned on both the transcriptional and the post-translational level. New Phytologist 197: 1214–1224. [Google Scholar]
Jonge RD, Esse HP, Maruthachalam K, Bolton MD, Santhanam P et al. (2012). Tomato immune receptor Ve1 recognizes effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proceedings of the National Academy Sciences USA 109: 5110–5115. [Google Scholar]
Kamoun S, van WP, Vleeshouwers VG, de Groot KE, Govers F (1998). Resistance of Nicotiana benthamiana to Phytophthora infestans is mediated by the recognition of the elicitor protein INF1. Plant Cell 10: 1413–1426. [Google Scholar]
Kjaersgaard T, Jensen MK, Christiansen MW, Gregersen P, Kragelund BB et al. (2011). Senescence-associated barley NAC (NAM, ATAF1, 2, CUC) transcription factor interacts with radical-induced cell death 1 through a disordered regulatory domain. Journal of Biological Chemistry 286: 35418–35429. [Google Scholar]
Li FF, Zhao N, Li ZH, Xu XB, Wang YQ et al. (2017). PA calmodulin-like protein suppresses RNA silencing and promotes geminivirus infection by degrading SGS3 via the autophagy pathway in Nicotiana benthamiana. PLoS Pathogens 13: e1006213. [Google Scholar]
Li J, Lü CR, Dou L, Dou ZY (2007). Construction of prokaryotic expression vector of mouse Nanog gene and its expression. Scientia Agricultura Sinica 40: 373–378. [Google Scholar]
Li ZY, Liu CP, Zhu LQ, Jing GZ, Zhou JM (2001). The chaperone activity of trigger factor is distinct from its isomerase activity during co-expression with adenylate kinase in Escherichia coli. FEBS Letters 506: 108–112. [Google Scholar]
Nakashima K, Takasaki H, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2012). NAC transcription factors in plant abiotic stress responses. Biochimica et Biophysica Acta 1819: 97–103. [Google Scholar]
Nuruzzaman M, Sharoni AM, Kikuchi S (2013). Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Frontiers in Microbiology 4: 248–263. [Google Scholar]
Ramsey JS, Wilson AC, Vos MD, Sun Q, Tamborindeguy C et al. (2017). Genomic resources for Myzus persicae: EST sequencing, SNP identification, and microarray design. BMC Genomics 8: 423. [Google Scholar]
Rosano GL, Ceccarelli EA (2014). Recombinant protein expression in Escherichia coli: Advances and challenges. Frontiers in Microbiology 5: 172–188. DOI 10.3389/fmicb.2014.00172. [Google Scholar] [CrossRef]
Shimada K, Nagano M, Kawai M, Koga H (2005). Influences of amino acid features of glutathione S-transferase fusion proteins on their solubility. Proteomics 5: 3859–3863. DOI 10.1002/(ISSN)1615-9861. [Google Scholar] [CrossRef]
Tang GY, Shao FX, Xu PL, Shan L, Liu ZJ (2017). Overexpression of a peanut NAC gene AhNAC4, confers enhanced drought tolerance in tobacco. Russian Journal of Plant Physiology 64: 525–535. DOI 10.1134/S1021443717040161. [Google Scholar] [CrossRef]
Tran LS, Nishiyama R, Yamaguchi-Shinozaki K, Shinozaki K (2010). Potential utilization of NAC transcription factors to enhance abiotic stress tolerance in plants by biotechnological approach. GM Crops 1: 32–39. DOI 10.4161/gmcr. [Google Scholar] [CrossRef]
Tsumoto K, Ejima D, Kumagai I, Arakawa T (2003). Practical considerations in refolding proteins from inclusion bodies. Protein Expression and Purification 28: 1–8. DOI 10.1016/S1046-5928(02)00641-1. [Google Scholar] [CrossRef]
Wang QQ, Li YH, Hu LL, Qin YG, Zhao ZY et al. (2016). Prokaryotic expression of cucumber CsERECTA gene and preparation of its polyclonal antibody. Acta Botanica Boreali-Occidentalia Sinica 36: 1031–1038. [Google Scholar]
Wang XE, Basnayake BM, Zhang HJ, Li GJ, Li W et al. (2009). The Arabidopsis ATAF1, a NAC transcription factor, is a negative regulator of defense responses against necrotrophic fungal and bacterial pathogens. Molecular Plant Microbe Interactions 22: 1227–1238. [Google Scholar]
Wei CF, Kvitko BH, Shimizu R, Crabill E, Alfano JR et al. (2007). A Pseudomonas syringae pv. tomato DC3000 mutant lacking the type III effector HopQ1-1 is able to cause disease in the model plant Nicotiana benthamiana. Plant Journal 51: 32–46. [Google Scholar]
Wilkinson DL, Harrison RG (1991). Predicting the solubility of recombinant proteins in Escherichia coli. Nature Biotechnology 9: 443–448. [Google Scholar]
Wu YR, Deng ZY, Lai JB, Zhang YY, Yang CP et al. (2009). Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Research 19: 1279–1290. [Google Scholar]
Xia N, Zhang G, Liu XY, Deng L, Cai GL et al. (2010a). Characterization of a novel wheat NAC transcription factor gene involved in defense response against stripe rust pathogen infection and abiotic stresses. Molecular Biology Reports 37: 3703–3712. [Google Scholar]
Xia N, Zhang G, Sun YF, Zhu L, Xu LS et al. (2010b). TaNAC8, a novel NAC transcription factor gene in wheat, responds to stripe rust pathogen infection and abiotic stresses. Physiological and Molecular Plant Pathology 74: 394–402. [Google Scholar]
Xu ZY, Gongbu ZX, Wang CY, Xue F, Zhang H et al. (2015). Wheat NAC transcription factor TaNAC29 is involved in response to salt stress. Plant Physiology and Biochemistry 96: 356–363. [Google Scholar]
Yang SD, Seo PJ, Yoon HK, Park CM (2011). The Arabidopsis NAC transcription factor VNI2 integrates abscisic acid signals into leaf senescence via the COR/RD genes. Plant Cell 23: 2155–2168. [Google Scholar]
Yoshino K, Irieda H, Sugimoto F, Yoshioka H, Okuno T et al. (2012). Cell death of Nicotiana benthamiana is induced by secreted protein NIS1 of Colletotrichum orbiculare and is suppressed by a homologue of CgDN3. Molecular Plant Microbe Interactions 25: 625–636. [Google Scholar]
Zhong RQ, Lee C, Ye ZH (2010). Global analysis of direct targets of secondary wall NAC master switches in Arabidopsis. Molecular Plant 3: 1087–1103. [Google Scholar]
Zhu F, Che YP, Xu F, Zhou YK, Qian K et al. (2019). Simultaneous silencing of two target genes using virus-induced gene silencing technology in Nicotiana benthamiana. Zeitschrift für Naturforschung C 74: 151–159. [Google Scholar]
Zhu F, Deng XG, Xu F, Jian W, Peng XJ et al. (2015). Mitochondrial alternative oxidase is involved in both compatible and incompatible host-virus combinations in Nicotiana benthamiana. Plant Science 239: 26–35. [Google Scholar]
Zhu F, Xu MY, Wang SD, Jia SD, Zhang P et al. (2012). Prokaryotic expression of pathogenesis related protein 1 gene from Nicotiana benthamiana: Antifungal activity and preparation of its polyclonal antibody. Biotechnology Letters 34: 919–924. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |