DOI:10.32604/biocell.2022.019724

| BIOCELL DOI:10.32604/biocell.2022.019724 |  |
| Article |
A potential impact of A Disintegrin and Metalloproteinase Domain-Like Protein Decysin-1 (ADAMDEC1) on clear cell renal cell carcinoma propagation
Foundation of Research and Science Development, Warsaw, 01-793, Poland
*Address correspondence to: Magdalena Rudzińska-Radecka, magdalena.radecka@fundacjabirn.pl; magdda.rudzinska@gmail.com
Received: 11 October 2021; Accepted: 27 December 2021
Abstract: Clear cell renal cell carcinoma (KIRC) is the most common and aggressive malignancy subtype of renal neoplasm that arises from proximal convoluted tubules. It is characterized by poor clinical outcomes and high mortality of patients due to the lack of specific biomarkers for varying stages of the disease and no effective treatment. Proteases are associated with the development of several malignant tumors in humans by their ability to degrade extracellular matrices, facilitating metastasis. Herein, differentially expressed genes in KIRC cases compared to healthy kidneys were screened out from the Gene Expression Profiling Interactive Analysis (GEPIA) database. This data was applied to determine the most elevated protease in KIRC and as a result, A Disintegrin and Metalloproteinase Domain-Like Protein Decysin-1 (ADAMDEC1) was selected. This expression pattern was exclusive for KIRC and not observed for papillary and chromophobe renal cell carcinomas, in which ADAMDEC1 was at the same level in tumors and non-cancer specimens. Furthermore, the ADAMDEC1 significant increase was detected in the fourteen other human malignancies compared to healthy samples, which suggested its strong involvement in cancer development. Next, GEPIA and Pathology Atlas correlated ADAMDEC1 high expression with more advanced tumor grade and shorter survival of KIRC patients. Xena Functional Genomics Explorer presented that ADAMDEC1 could be hypermethylated in some tumor cases and one somatic mutation in the gene sequence was detected. Finally, a Search Tool for the Retrieval of Interacting Genes/Proteins; STRING base was utilized to predict the interactions of ADAMDEC1 with other molecules and construct the signaling network. In summary, ADAMDEC1 showed the tremendous potential to be the predictive marker for the KIRC and its development. Therefore, this review with data analysis can be a good base for further in vitro and in vivo research that experimentally can confirm the ADAMDEC1 as prognostic biomarkers and therapeutic target of KIRC.
Keywords: A Disintegrin and Metalloproteinase Domain-Like Protein Decysin-1; ADAMDEC1; Clear cell renal cell carcinoma; Patient specimens; Databases
Abbreviations
| RCC: | renal cell carcinoma |
| KIRC: | clear cell renal cell carcinoma |
| ADAMDEC1: | A Disintegrin and Metalloprotease-Like Decysin |
| GEPIA: | Gene Expression Profiling Interactive Analysis database |
| OSCC: | squamous cell carcinoma |
| VHL: | von Hippel-Lindau |
| VEGF: | vascular endothelial growth factor |
| BRCA: | breast invasive carcinoma |
| CESC: | cervical squamous cell carcinoma and endocervical adenocarcinoma |
| DLBC: | lymphoid neoplasm diffuse large B-cell lymphoma |
| ESCA: | esophageal carcinoma |
| GBM: | glioblastoma multiforme |
| HNSC: | head and neck squamous cell carcinoma |
| LUAD: | lung adenocarcinoma lung squamous cell carcinoma |
| LUSC: | ovarian serous cystadenocarcinoma |
| OV: | pancreatic adenocarcinoma |
| PAAD: | skin cutaneous melanoma |
| SKCM: | stomach adenocarcinoma |
| UCEC: | testicular germ cell tumors and uterine corpus endometrial carcinoma |
| KICH: | kidney chromophobe |
| KIRP: | kidney renal papillary cell carcinoma |
| PPI: | protein-protein interaction |
Proteases, enzymes that break the protein peptide bonds, are located in the cytoplasm, mitochondria, lysosome, and extracellular matrix (King et al., 2014). By their action mechanism, they can be classified into six groups: serine, cysteine, threonine, aspartate, glutamic acid proteases and metalloproteases (Eatemadi et al., 2017).
They are responsible for non-specific degradative functions but also can catalyze specific proteolytic processing. Thus proteases are relevant in the control of multiple biological processes in living organisms. Hence, proteases contribute to (i) the fate, localization, and activity of many proteins, (ii) protein-protein interactions, (iii) generation of new bioactive molecules, (iv) cellular information processing (v) molecular signal pathways (López-Otín and Bond, 2008). As a result, proteases influence transcription, cell proliferation and differentiation, tissue morphogenesis/remodeling, neurogenesis, proteostasis, angiogenesis, ovulation, fertilization, wound healing, inflammation, autophagy, apoptosis and others (Cabral-Pacheco et al., 2020; Rudzińska et al., 2020; Rudzińska et al., 2021; Bond, 2019). Consistent with these essential roles in cell behavior and survival, alterations in proteolytic balance underlie multiple pathological conditions such as neurodegenerative disorders, inflammatory and cardiovascular diseases as well as cancer (Martinelli and Rugarli, 2010; Bühling et al., 2006; Klingler and Hardt, 2012; Yang et al., 2009). Many human tumors show the up-regulated protease level at an early stage of development, so proteases are thought to be crucial in tumor angiogenesis, invasion, and infiltration (Koblinski et al., 2000).
A Disintegrin and Metalloprotease-Like Decysin 1 (ADAMDEC1) is a unique, highly conserved secreted metalloprotease with a very rare zinc-binding motif (HEXXHXXGXXD) within the metalloprotease domain (Mueller et al., 1997). It contains pro- and catalytic domains but has a truncated disintegrin domain and lacks the typical transmembrane domain and the cytoplasmic tail (Mueller et al., 1997). The recent studies point to several vital roles of ADAMDEC1 in human diseases, including atherosclerosis (Verdugo et al., 2013), pulmonary sarcoidosis (Crouser et al., 2009), osteoarthritis (Galligan et al., 2007), Crohn’s disease (Smith et al., 2015), gastrointestinal malignancies (Oh et al., 2017) and glioblastoma (Vauléon et al., 2012). However, the natural role of ADAMDEC1 remains unknown, although its characteristic features suggest that it plays a fundamental role in the physiology of mammals.
Renal cell carcinoma (RCC) accounts for about 3% of adult malignancies, with an accelerating trend (increasing 2%–3% per decade) (Padala et al., 2020). Clear cell renal cell carcinoma (KIRC) is the most common subtype characterized by diagnostic in advanced stage and high drug-resistant nature (Makhov et al., 2018). This type of cancer does not show up early warning signs, so understanding the molecular pathogenesis of RCC and refining the diagnostic and treatment options are highly desirable (Motzer et al., 1997). The cancer propagation is attributed to proteolytic degradation of the extracellular matrix initiated by all classes of proteases (Skrzydlewska et al., 2005). It was already presented that various proteases can contribute to RCC malignancies (Drendel et al., 2017; Rudzińska et al., 2020).
Here, all up-regulated proteases were screened in 623 cases of KIRC based on the Gene Expression Profiling Interactive Analysis (GEPIA) database. Analysis elucidated ADAMDEC1 as the strongest accelerated protease in KIRC compared to healthy kidney tissues. This expression pattern was exclusive for KIRC subtype and distinguished it of chromophobe (KICH) and papillary renal (KIRP) cell carcinomas, which presented a similar ADAMDEC1 level in normal and tumor specimens. Furthermore, according to Pathology Atlas and GEPIA, the ADAMDEC1 expression level increased gradually as the tumor stage and correlated with the shorter survival of KIRC patients. Interestingly, ADAMDEC1 was elevated in other 14 types of tumors, including glioblastoma, breast, cervical, lung and others.
Additionally, Xena Functional Genomics Explorer showed that ADAMDEC1 expression could be modified by hypermethylation and one somatic mutation was detected in the gene sequence. Finally, a Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) base was used to predict the interactions of ADAMDEC1 with other proteins and visualize the signaling network.
This work points out the perspective role of ADAMDEC1 in KIRC and its particular implementation of patient outcomes. In addition, ADAMDEC1 showed the potential for experimental analysis to confirm its diagnostic value and molecular target for KIRC.
Characterization of ADAMDEC1 structure and function
ADAMDEC1 mRNA sequence was discovered in dendritic cells of human tonsils and categorized into the ADAM family (Mueller et al., 1997). However, the protein is short and consists 470 aa. Its structure is untypical since ADAMDEC1 is missing the cytoplasmic tail, transmembrane and cysteine-rich domains, thus consisting of only a signaling peptide, prodomain, metalloprotease and disintegrin-like domains (Fig. 1) (Mueller et al., 1997). Additionally, analyzing the canonical structure of ADAM enzymes-histidine (H) repeat motif (HEXXHXXGXXH) within the zinc-binding site of the metalloprotease domain, in ADAMDEC1, the third histidine is replaced by aspartate (D) serving in the motif sequence of HEXXHXXGXXD (Mueller et al., 1997; Lund et al., 2013). This rare structure was connected with the reduced catalytic activity, diminished substrate specificity and enhanced resistance to the regulation by endogenous inhibitors of metalloproteinases (TIMPs) (Lund et al., 2013; Lund et al., 2015).
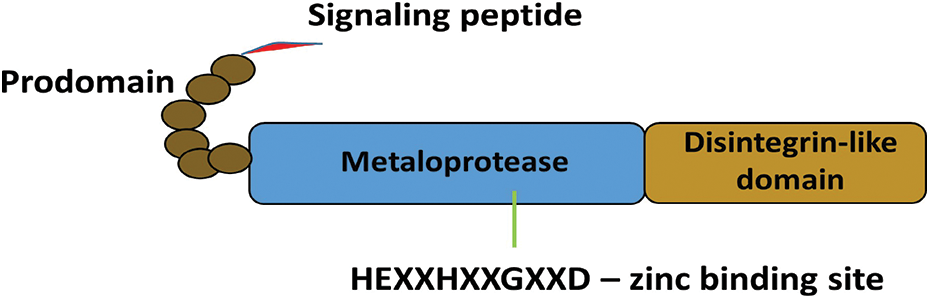
Figure 1: Schematic structure of ADAMDEC1. The unique structure ADAMDEC1 consists of a signaling peptide, prodomain, metalloprotease and disintegrin-like domains.
Physiologically, the highest expression of ADAMDEC1 was found in the gastrointestinal, expressed predominantly in the macrophages in the lamina propria. The lower intensity of ADAMDEC1 was detected in seen, lymph nodes, tonsils, spleen, urinary bladder and placenta (Mueller et al., 1997; Kumagai et al., 2021).
The whole-genome transcriptomic analysis associated ADAMDEC1 expression deviations with Crohn’s disease (de Bruyn et al., 2014), rheumatoid arthritis (Galligan et al., 2007) and atherosclerosis (Papaspyridonos et al., 2006).
ADAMDEC1 downregulation in the inflamed ileal biopsy samples collected from patients with Crohn’s disease compared to healthy controls was detected. This correlation was independent of the degree of mucosal inflammation, thus underlying ADADEC1 role in the pathogenesis of this disease (de Bruyn et al., 2014).
About 24 times enhanced ADAMDEC1 level was detected in fibroblast-like synovial cells isolated from joint tissue taken from patients with rheumatoid arthritis (Galligan et al., 2007). This observation was confirmed by transcriptional datasets generated from the synovial tissue rheumatoid arthritis and osteoarthritis (Li et al., 2019).
Next, the enhanced presence of ADAMDEC1 in the circulating monocytes was found to have the third significant positive association with the occurrence of atherosclerotic plaques in the common carotid artery. Thus, it can suggest the potential function of ADAMDEC1 in atherosclerosis and plaque instability development (Verdugo et al., 2013).
In the context of cancerogenesis, the drastic reduction of ADADEC1 in the colorectal adenoma compared to the normal colorectal tissues was associated with poor prognosis of patients. However, it was not connected with the loss of macrophages in tissues, verified by CD68 expression (Kalmár et al., 2019).
Analysis of patients with craniopharyngioma revealed accelerated ADAMDEC1 expression in collected specimens compared to normal brain tissues. The primary craniopharyngioma-derived cells treated with an anti-estrogen drug, tamoxifen, reduced tumor cell proliferation and also the expression of ADAMDEC1 mRNA/protein levels (Xu et al., 2012).
A potential mechanistic role for ADAMDEC1 in developing oral squamous cell carcinoma (OSCC) has been shown in experiments based on EGF-receptor (EGFR) activation. An ectopic expression of EGFR is detected in approximately 90% of OSCC, and it is a factor that can drive an aggressive phenotype and contribute to decreased response to radiotherapy. The thrombin-stimulated platelets were shown to secrete ADAMDEC1, resulting in the cleavage of the platelet membrane-bound pro-EGF. The soluble EGF then resulted in a migratory and invasive phenotypic shift in the OSCC via EGFR signaling (Chen et al., 2018).
Finally, Jimenez-Pascual et al. (2019), Vauléon et al. (2012) identified ADAMDEC1 as one of the top up-regulated ADAM genes in glioblastoma multiforme (GBM), which correlates with poor patients prognoses. The study showed that ADAMDEC1 rapidly solubilizes the membrane-bound FGF2 and stimulates FGFR1 expressed on the GBM stem cells. Further, the transcription factor Zinc finger E-box-binding homeobox 1 (ZEB1) was activated and consequently induced ADAMDEC1 expression by inhibiting miR-203 (functioning as the tumor-suppressor) and creating a positive feedback loop. Targeting this signaling pathway decreased self-renewal and tumor growth, which highlighted the potential role of ADAMDEC1 in cancer propagation (Jimenez-Pascual et al., 2019).
RCC is originated from the renal epithelium and accounts for >90% of all kidney malignancies. The disease encompasses >10 histological and molecular subtypes, of which KIRC is most common (70%–80%; accounts for most cancer-related deaths), next, papillary (10%–15%), chromophobe (5%–10%) and collecting duct (1%) RCC (Hsieh et al., 2017; Harbin et al., 2015; Muglia and Prando, 2015). These three subtypes are the most frequent histological groups that together represent more than 90% of all RCC (Muglia and Prando, 2015).
Approximately 2%–3% of all RCC are hereditary and several autosomal dominant syndromes (e.g., Hippel–Lindau disease) are described. Well-validated targets, including VHL, VEGFR and mTOR and pathways such as HGF/c-MET and Wnt/β-catenin, are strongly related to RCC pathogenesis (Maher, 2018).
RCC is a disease with complex etiologies, which the combined effect of multiple genes may cause. In this context, VHL (von Hippel-Lindau), p53, p16, p21 and p27 were shown as the primary tumor suppressor genes in RCC, in which VHL and p53 were certified to result in the development of RCC (Liu et al., 2015). Dysfunction of VHL will lead to constitutively aberrant activation of the hypoxic response, like upregulation of vascular endothelial growth factor (VEGF), which is considered to play essential roles in tumor development and angiogenesis (Zhang and Zhang, 2018). p53 is shown to suppress tumor growth and induce cell apoptosis in KIRC (Zhao et al., 2016).
Recently the genetic profiling of human RCC has increasingly been used to identify the potential of epigenetic regulatory mechanisms. It was shown that single-stranded molecules (lncRNA, miRNA) (Trilla-Fuertes et al., 2020; Rudzinska et al., 2021) and DNA methylation (Lasseigne and Brooks, 2018) are deeply involved in renal pathogenesis.
DNA methylation involves the covalent transfer of a methyl group to the C-5 position of the cytosine DNA ring by DNA methyltransferases and causes gene silencing. The genome-wide DNA methylation study in RCC identified increased global methylation in more aggressive cancer types and a potential risk factor associated with malignant transformation. The variations in sequence hypermethylation were dependents on medical aspects, such as aggressive cancer, tumor size, etc.
The potential therapeutic targets, such as small-molecule multikinase inhibitors that target VEGF receptors (sunitinib and sorafenib), the anti-VEGF antibody bevacizumab, and a mammalian target of rapamycin inhibitor temsirolimus, still are not efficient enough in the KIRC currency and chemioresistance is often observed (Larkin et al., 2009; Rudzińska et al., 2021; Penticuff and Kyprianou, 2015). It is connected with the fact that molecular mechanisms underlying this disease have not been fully elucidated. In addition, more than 50% of KIRC are detected incidentally because of a lack of effective diagnostics in the early stages of the disease; it leads to metastasis and increases the mortality rate (Czarnecka et al., 2014). This scenario emphasizes the need to discover new biomarkers for RCC subtypes with the tumor grade-dependent manner. Furthermore, exploring molecular biology of KIRC can deepen the knowledge about its mechanism and identify the novel molecular targets.
ADAMDEC1 Expression Level in Renal Cell Carcinomas and Other Human Tumors
ADAMDEC1 expression level analysis
The online database Gene Expression Profiling Interactive Analysis (GEPIA; http://gepia.cancer-pku.cn/index.html; Tang et al., 2017) is a communal network server that includes a transcriptome sequencing dataset of 9736 tumors together with 8587 adjacent normal tissues from TCGA and Genotype-Tissue Expression (GTEx) data sets. Here, the elevated expression of proteases was analyzed in 623 KIRC cases, compared to 100 non-tumoral kidney samples. Correlation analysis was performed on the selected gene pairwise, and ANOVA test was used to calculate the statistical significance (p < 0.05 was considered as significant).
ADAMDEC1 expression level was analyzed in 16 other human tumors at the same way.
Protease list was downloaded from MEROPS The Peptidase Database (https://www.ebi.ac.uk/merops/; Rawlings et al., 2021).
ADAMDEC1 expression level with tumor grade information was collected from Pathology Atlas (https://www.proteinatlas.org/humanproteome/pathology; Uhlen et al., 2017). Data were analyzed using a non-parametric U-Mann–Whitney test (GraphPad, Prism 6.00 for Windows, Graf Pad software, San Diego, CA, USA). Data are reported as violin plots and p-value < 0.05 was considered statistically significant.
Correlation with survival of patients
By GEPIA the ADAMDEC1 ectopic expression was correlated with survival of KIRC patients. The Kaplan-Meier plots log-rank allowed for identification of predictive value of analyzed gene.
Hypermethylation and somatic mutation analysis
By USC Xena Browser (https://xenabrowser.net/; Goldman et al., 2020), the hypermethylation status of ADAMDEC1 and somatic mutations were analyzed. DNA methylation profile was detected using Infinium Human Methylation 450 and Human Methylation 27 techniques, whereas the somatic mutations were detected using the MuTect2 tool.
The potential interactions of ADAMDEC1 with other molecules was visualized by The Search Tool for the Retrieval of Interacting Genes - STRING database (https://string-db.org/; Szklarczyk et al., 2021). PPI map was constructed, with the active interaction sources, such as experiments, databases, textminig, co-expression, gene fusion, and co-occurrence.
ADAMDEC1 is the strongest overexpressed protease in KIRC and up-regulated transcript in 14 other human tumors compared to non-tumor controls
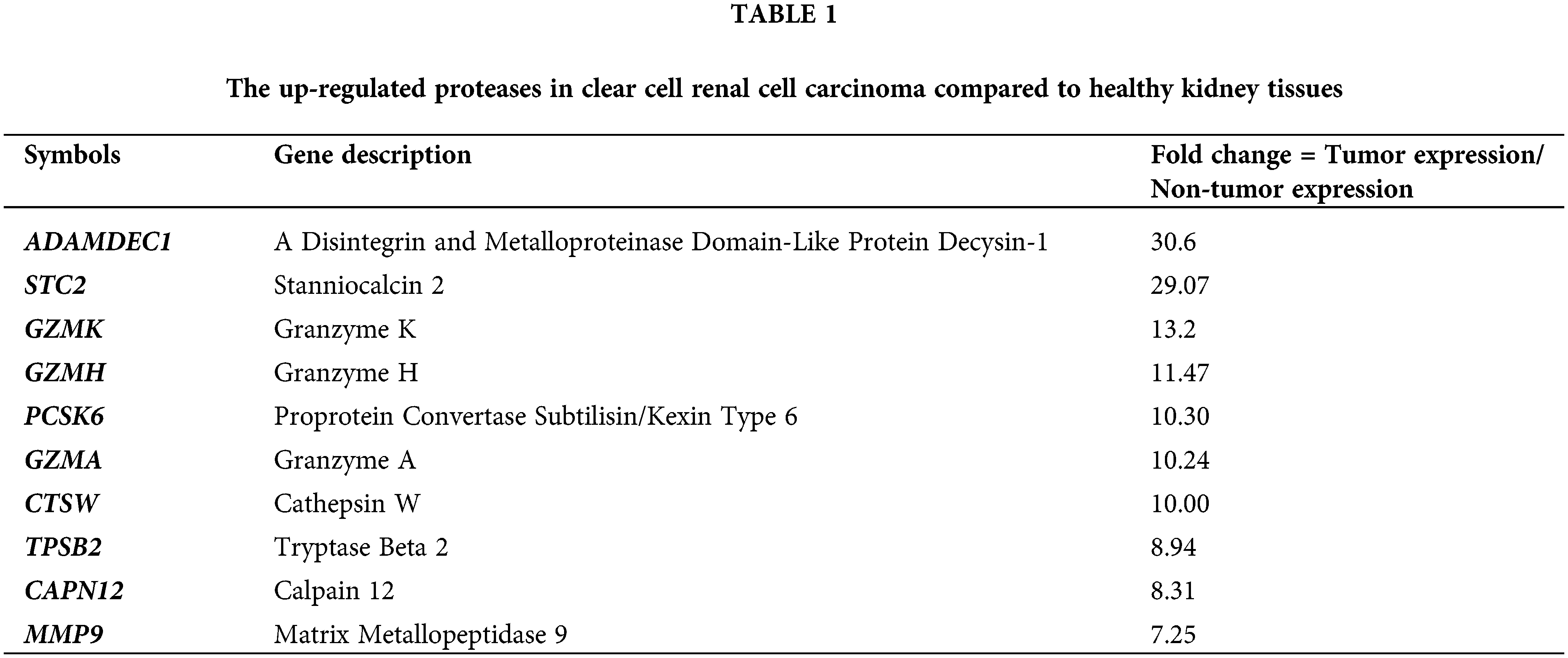
Using the GEPIA database, all significantly up-regulated genes in 623 KIRC cases compared to the healthy cohort (100 samples) were listed. Next, by MEROPS The Peptidase Database, all changed proteases were selected and a shortlist of twentieth strongest changed proteases is included below (Table 1).
The strongest changed ADADEC1 was taken for further analysis and firstly, the significant (p < 0.05) evaluation of ADAMDEC1 expression in KIRC samples compared to normal kidney tissues was confirmed (Fig. 2A). Following, the higher ADAMDEC1 expression was detected in 14 other human tumors (Fig. 2B), including: invasive breast carcinoma (BRCA), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), lymphoid neoplasm diffuse large B-cell lymphoma (DLBC), esophageal carcinoma (ESCA), glioblastoma multiforme (GBM), head and neck squamous cell carcinoma (HNSC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), ovarian serous cystadenocarcinoma (OV), pancreatic adenocarcinoma (PAAD), cutaneous skin melanoma (SKCM), stomach adenocarcinoma (STAD), testicular germ cell tumors (TGCT) and uterine corpus endometrial carcinoma (UCEC). Interestingly, ADAMDEC1 was not elevated in kidney chromophobe (KICH) and kidney renal papillary cell carcinoma (KIRP) samples to serve as distinguishing features of the KIRC subtype.
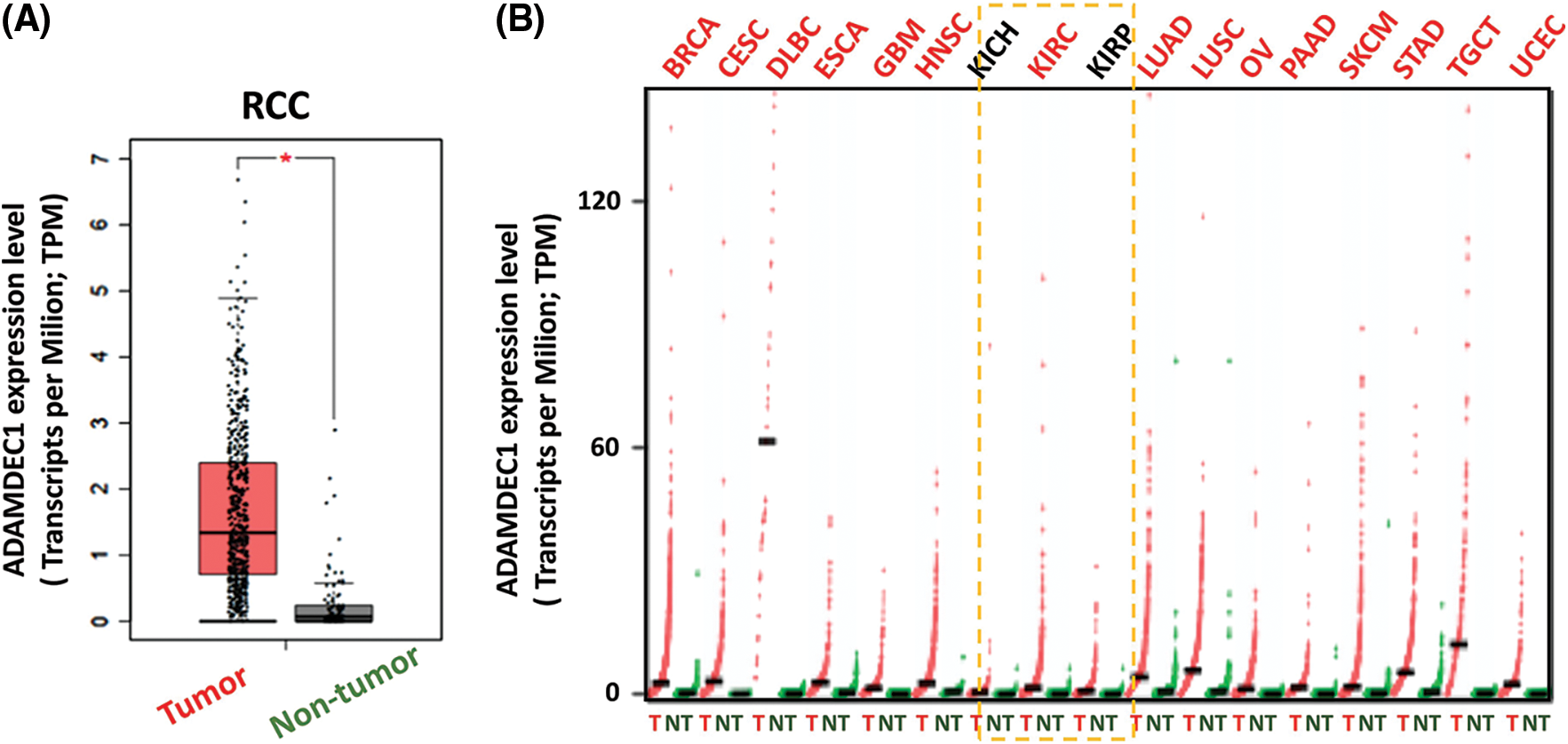
Figure 2: (A) ADAMDEC1 expression level in clear cell renal cell carcinoma samples (KIRC; tumor-red) compared to control kidney tissues (non-tumor-grey); ANOVA test; * p < 0.05. (B) ADAMDEC1 expression level in other human tumors: (BRCA) beast invasive carcinoma, (CESC) cervical squamous cell carcinoma and endocervical adenocarcinoma, (DLBC) lymphoid neoplasm diffuse large B-cell lymphoma, (ESCA) esophageal carcinoma, (GBM) glioblastoma multiforme, (HNSC) head and neck squamous cell carcinoma, (LUAD) lung adenocarcinoma, (LUSC) lung squamous cell carcinoma, (OV) ovarian serous cystadenocarcinoma, (PAAD) pancreatic adenocarcinoma, (SKCM) skin cutaneous melanoma, (STAD) stomach adenocarcinoma, (TGCT) testicular germ cell tumors and (UCEC) uterine corpus endometrial carcinoma–colored red. The lack of significant difference in (KICH) kidney chromophobe and (KIRP) kidney renal papillary cell carcinoma colored black. Analyzed by GEPIA database.
ADAMDEC1 enhanced presence correlates with high-grade KIRC tumor and worse prognosis of patients
Further, ADADEC1 expression was correlated with clinical outcomes of KIRC cases. As a result, the accelerating ADAMDEC1 expression level tendency corresponded to higher tumor stages; from I to IV stages (Fig. 3A). Significant different (*p < 0.05) was noticed between I and III and IV grades. In addition, its enhanced expression notably (p = 0.046) corresponded to a worse survival prognosis of patients (Fig. 3B).
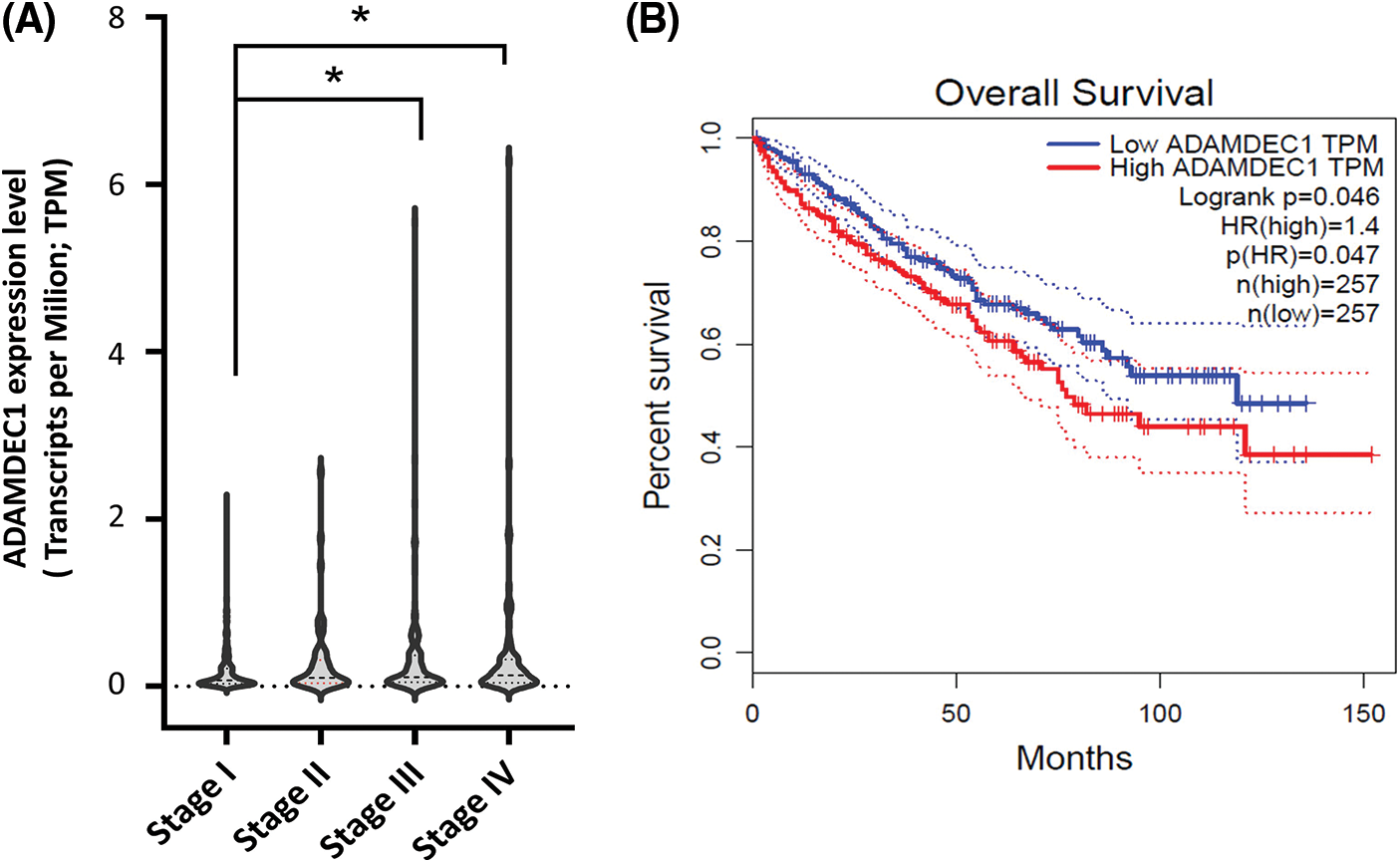
Figure 3: The ADAMDEC1 expression pattern in the clear renal cancer cell (KIRC) and its relation to (A) tumor stages of KIRC patients: I, II, III, and IV staging. According to the American Joint Committee on Cancer Staging (AJCC), Stage I (n = 259) is characterized by the tumor only in the kidney with a max. 7 cm size. Stage II (n = 31): the tumor is found only in the kidney and is larger than 7 cm. Stage III (n = 112): the tumor has grown into major veins within the kidney or perinephric tissue, which is the connective, fatty tissue around the kidneys. However, it has not grown into the adrenal gland on the same side of the body as the tumor. Stage IV (n = 88): the tumor has spread to areas beyond Gerota’s fascia and extends into the adrenal gland on the same body side as the tumor. *p < 0.05 U-Mann-Whitney test. (B) The ADAMDEC1 mRNA expression relation to survival of patients with KIRC; GEPIA database.
ADAMDEC1 expression can be control by hypermethylation and mutations in KIRC cases
To investigate the possible relationship between hypermethylation and ADAMDEC1 expression, an analysis of methylation among KIRC cases was performed using USC Xena browser (Fig. 4C). The hypermethylation was detected in part of both tumors and non-tumor controls.
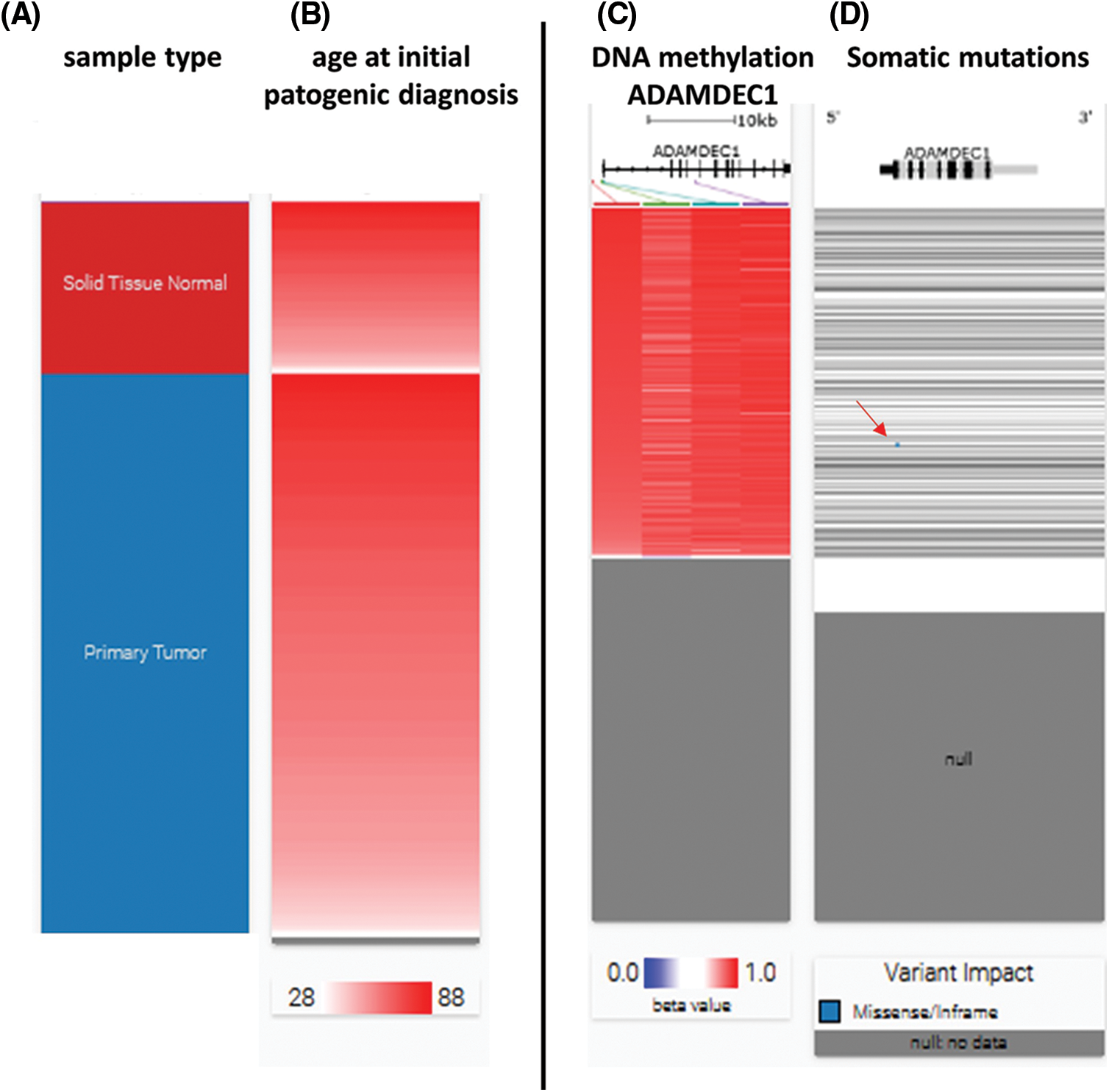
Figure 4: ADAMDEC1 hypermethylation and somatic mutations in clear cell renal cell carcinoma (KIRC) cases. (A) Hierarchical clustering of KIRC tissues–red–solid tissue normal (non-cancer tissue surrounding), blue–primary tumor, (B) age analyzed patients (C) ADAMDEC1 sequence hypermethylation (red); grey–no analyzed samples; hypermethylated sequences localization: (red line: hg38 chr8:24,383,084-24,383,084; green line: hg38 chr8:24,384,181-24,384,182; blue line: hg38 chr8:24,384,596-24,384,597; violet line: hg38 chr8:24,394,941-24,394942). (D) Somatic mutation analyzing. Blue point (a red arrow) indicates missense variant p.W23R/substitution T to C (hg38 ch8:24,384,571) in one KIRC case. Analyzed by USC Xena Browser.
Additionally, the somatic mutations were checked and missense variant p.W23R/substitution T to C (hg38 ch8:24,384,571) in one case was detected (Fig. 4D).
ADAMDEC1 potentially interact with other protein creating a signaling cluster
Proteins and protein-protein interactions form the backbone of the cellular complex machinery. Thus, for the full understanding of biological phenomena, the full network needs to be considered. The Search Tool for the Retrieval of Interacting Genes-STRING database collected scored and integrated all publicly available sources of protein-protein interaction information, and to complemented these with computational predictions. The STRING database uses protein-protein co-occurrence as one type of text mining evidence for protein interactions. Text mining (text data mining) transforms unstructured text into a structured format to identify meaningful patterns and new insights. Text mining systems are designed to extract information from the text in a domain-oriented manner (Franceschini et al., 2013).
Here, STRING was used to seek potential interactions between ADAMDEC1 with other molecules (Fig. 5A). As a result, PPI network was constructed, with the active interaction sources, including experiments, databases, textminig, co-expression, gene fusion, and co-occurrence (Fig. 5B). In the matrix, the nodes correspond to the proteins, and the edges represent the interactions. Using an interaction score threshold of 0.7 (high confidence), the string PPI analysis yielded a highly clustered network (clustering coefficient: 0.73) containing 11 nodes with 19 edges (enrichment p-value < 0.008), false discovery rate = 1.42E-0.5.
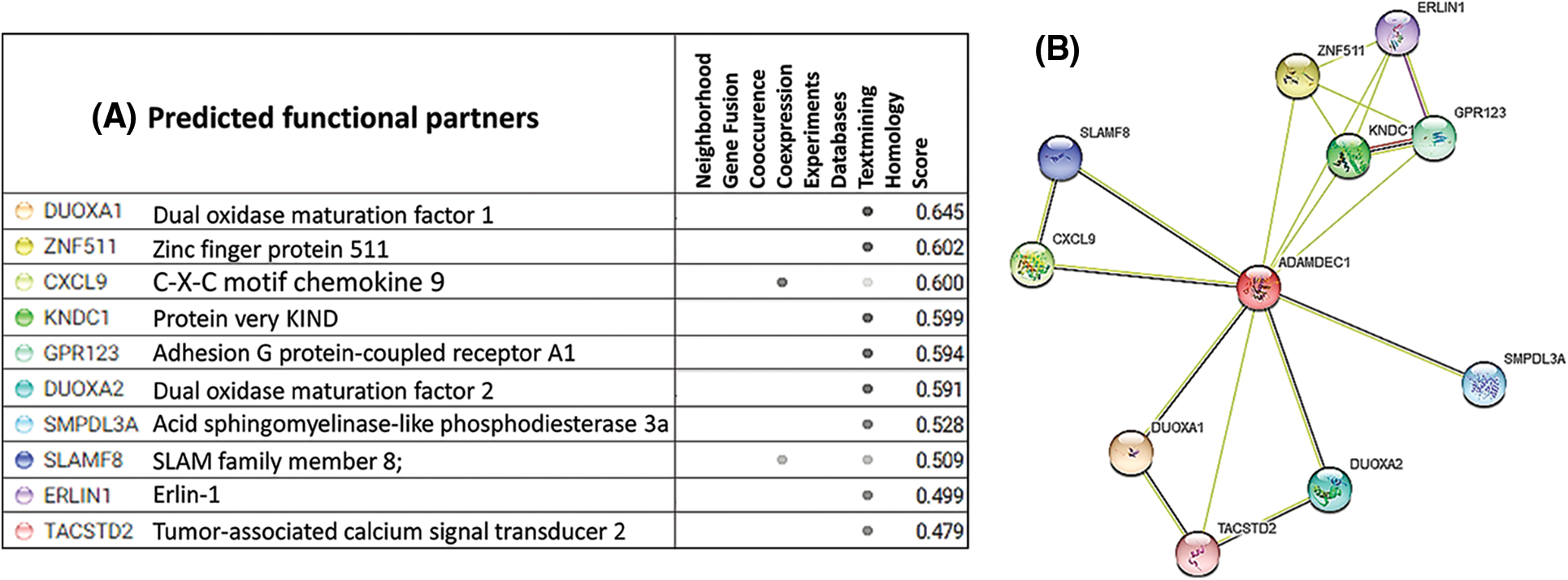
Figure 5: STRING Protein-protein interaction (PPI) analyses. (A) Predicted functional partners of ADAMDEC1 (B) PPI network connectivity of human ADAMDEC1 interactions cluster. The network contains 11 nodes with 19 edges. The confidence score threshold was set at 0.8 (high) for analyses. The gene interaction data used to build the network was based on direct physical interactions that are either experimentally derived or computationally predicted.
As the outcome, the co-expression was found for CXCL9 and SLAMF9. Text mining systems showed the interactions with proteins, including DUOXA1, ZNF511, CXCL9, KNDC1, GPR123, DUOX2A, SMPDL3A, SLAMF8, ERIN1, TACSTD2, with a different score value.
Conclusions and further perspectives
Recently, ADAMDEC1 has been connected with the development of varied human diseases, including cancers. Herein, the presented review with database analysis demonstrates the first evidence for positive relations of ADAMDEC1 expression with KIRC cases and its worse outcomes. It can implicate the potential role of ADAMDEC1 in the pathogenesis of KIRC. The presented findings support the further experimental analysis of ADAMDEC1 as a KIRC biomarker and suggest its essential function in diagnosing and promoting KIRC. If validated, this biomarker can significantly facilitate the introduction of new therapies based on protease inhibition that profoundly affects the efficient treatment of KIRC patients. Additionally, this knowledge can be helpful in research based on specific protease inhibitors against KIRC development.
Author Contribution: The author confirms sole responsibility for the following: study conception and design, data collection, analysis and interpretation of results, and manuscript preparation.
Funding Statement: The author received no specific funding for this study.
Conflicts of Interest: The author declares that they have no conflicts of interest to report regarding the present study.
Bond JS (2019). Proteases: History, discovery, and roles in health and disease. Journal of Biological Chemistry 294: 1643–1651. DOI 10.1074/jbc.TM118.004156. [Google Scholar] [CrossRef]
Bühling F, Groneberg D, Welte T (2006). Proteases and their role in chronic inflammatory lung diseases. Current Drug Targets 7: 751–759. DOI 10.2174/138945006777435362. [Google Scholar] [CrossRef]
Cabral-Pacheco GA, Garza-Veloz I, Castruita-De la Rosa C, Ramirez-Acuña JM, Perez-Romero BA et al. (2020). The roles of matrix metalloproteinases and their inhibitors in human diseases. International Journal of Molecular Sciences 21: 9739. DOI 10.3390/ijms21249739. [Google Scholar] [CrossRef]
Chen R, Jin G, Li W, McIntyre TM (2018). Epidermal Growth Factor (Egf) autocrine activation of human platelets promotes EGF receptor-dependent oral squamous cell carcinoma invasion, migration, and epithelial mesenchymal transition. Journal of Immunology 201: 2154–2164. DOI 10.4049/jimmunol.1800124. [Google Scholar] [CrossRef]
Crouser ED, Culver DA, Knox KS, Julian MW, Shao G et al. (2009). Gene Expression profiling identifies Mmp-12 and ADAMDEC1 as potential pathogenic mediators of pulmonary sarcoidosis. American Journal of Respiratory and Critical Care Medicine 179: 929–938. DOI 10.1164/rccm.200803-490OC. [Google Scholar] [CrossRef]
Czarnecka AM, Kornakiewicz A, Kukwa W, Szczylik C (2014). Frontiers in clinical and molecular diagnostics and staging of metastatic clear cell renal cell carcinoma. Future Oncology 10: 1095–1111. DOI 10.2217/fon.13.258. [Google Scholar] [CrossRef]
de Bruyn M, Machiels K, Vandooren J, Lemmens B, van Lommel L et al. (2014). Infliximab restores the dysfunctional matrix remodeling protein and growth factor gene expression in patients with inflammatory bowel disease. Inflammatory Bowel Diseases 20: 339–352. DOI 10.1097/01.MIB.0000438430.15553.90. [Google Scholar] [CrossRef]
Drendel V, Heckelmann B, Chen CY, Weisser J, Espadas G et al. (2017). Proteome profiling of clear cell renal cell carcinoma in von hippel-lindau patients highlights upregulation of xaa-pro aminopeptidase-1, an anti-proliferative and anti-migratory exoprotease. Oncotarget 8: 100066–100078. DOI 10.18632/oncotarget.21929. [Google Scholar] [CrossRef]
Eatemadi A, Aiyelabegan HT, Negahdari B, Mazlomi MA, Daraee H et al. (2017). Role of protease and protease inhibitors in cancer pathogenesis and treatment. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie 86: 221–231. DOI 10.1016/j.biopha.2016.12.021. [Google Scholar] [CrossRef]
Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ (2013). STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Research 41. DOI 10.1093/nar/gks1094. [Google Scholar] [CrossRef]
Galligan CL, Baig E, Bykerk V, Keystone EC, Fish EN (2007). Distinctive gene expression signatures in rheumatoid arthritis synovial tissue fibroblast cells: Correlates with disease activity. Genes and Immunity 8: 480–491. DOI 10.1038/sj.gene.6364400. [Google Scholar] [CrossRef]
Goldman MJ, Craft B, Hastie M, Repeka K, Mcdade F et al. (2020). Visualizing and interpreting cancer genomics data via the Xena platform. Nature Biotechnology 38: 675–678. DOI 10.1038/s41587-020-0546-8. [Google Scholar] [CrossRef]
Harbin AC, Styskel BA, Patel V, Wang H, Eun DD (2015). Collecting duct renal cell carcinoma found to involve the collecting system during partial nephrectomy: A case report. Journal of Kidney Cancer and VHL 2: 134–139. DOI 10.15586/jkcvhl.2015.37. [Google Scholar] [CrossRef]
Hsieh JJ, Purdue MP, Signoretti S, Swanton C, Albiges L et al. (2017). Renal cell carcinoma. Nature Reviews Disease Primers 3: 131. DOI 10.1038/nrdp.2017.9. [Google Scholar] [CrossRef]
Jimenez-Pascual A, Hale JS, Kordowski A, Pugh J, Silver DJ et al. (2019). ADAMDEC1 maintains a growth factor signaling loop in cancer stem cells. Cancer Discovery 9: 1574–1589. DOI 10.1158/2159-8290.CD-18-1308. [Google Scholar] [CrossRef]
Kalmár A, Nagy ZB, Galamb O, Csabai I, Bodor A et al. (2019). Genome-wide expression profiling in colorectal cancer focusing on lncrnas in the adenoma-carcinoma transition. BMC Cancer 19: 1059. DOI 10.1186/s12885-019-6180-5. [Google Scholar] [CrossRef]
King JV, Liang WG, Scherpelz KP, Schilling AB, Meredith SC et al. (2014). Molecular basis of substrate recognition and degradation by human presequence protease. Structure 22: 996–1007. DOI 10.1016/j.str.2014.05.003. [Google Scholar] [CrossRef]
Klingler D, Hardt M (2012). Targeting proteases in cardiovascular diseases by mass spectrometry-based proteomics. Circulation: Cardiovascular Genetics 5: 265. DOI 10.1161/CIRCGENETICS.110.957811. [Google Scholar] [CrossRef]
Koblinski JE, Ahram M, Sloane BF (2000). Unraveling the role of proteases in cancer. Clinica Chimica Acta 291: 113–135. DOI 10.1016/S0009-8981(99)00224-7. [Google Scholar] [CrossRef]
Kumagai T, Fan S, Smith AM (2021). ADAMDEC1 and its role in inflammatory disease and cancer. Metalloproteinases in Medicine 7: 15–28. DOI 10.2147/MNM.S263813. [Google Scholar] [CrossRef]
Larkin JM, Kipps EL, Powell CJ, Swanton C (2009). Systemic therapy for advanced renal cell carcinoma. Therapeutic Advances in Medical Oncology 1: 15–27. DOI 10.1177/1758834009338430. [Google Scholar] [CrossRef]
Lasseigne DN, Brooks JD (2018). The role of DNA methylation in renal cell carcinoma. Molecular Diagnosis & Therapy 22: 431–442. DOI 10.1007/s40291-018-0337-9. [Google Scholar] [CrossRef]
Li WC, Bai L, Xu Y, Chen H, Ma R et al. (2019). Identification of differentially expressed genes in synovial tissue of rheumatoid arthritis and osteoarthritis in patients. Journal of Cellular Biochemistry 120: 4533–4544. DOI 10.1002/jcb.27741. [Google Scholar] [CrossRef]
Liu X, Wang J, Sun G (2015). Identification of key genes and pathways in renal cell carcinoma through expression profiling data. Kidney & Blood Pressure Research 40: 288–297. DOI 10.1159/000368504. [Google Scholar] [CrossRef]
López-Otín C, Bond JS (2008). Proteases: Multifunctional enzymes in life and disease. Journal of Biological Chemistry 283: 30433–30437. DOI 10.1074/jbc.R800035200. [Google Scholar] [CrossRef]
Lund J, Olsen OH, Sørensen ES, Stennicke HR, Petersen HH et al. (2013). ADAMDEC1 is a metzincin metalloprotease with dampened proteolytic activity. Journal of Biological Chemistry 288: 21367–21375. DOI 10.1074/jbc.M113.474536. [Google Scholar] [CrossRef]
Lund J, Troeberg L, Kjeldal H, Olsen OH, Nagase H et al. (2015). Evidence for restricted reactivity of ADAMDEC1 with protein substrates and endogenous inhibitors. Journal of Biological Chemistry 290: 6620–6629. DOI 10.1074/jbc.M114.601724. [Google Scholar] [CrossRef]
Maher ER (2018). Hereditary renal cell carcinoma syndromes: Diagnosis, surveillance and management. World Journal of Urology 36: 1891–1898. DOI 10.1007/s00345-018-2288-5. [Google Scholar] [CrossRef]
Makhov P, Joshi S, Ghatalia P, Kutikov A, Uzzo RG et al. (2018). Resistance to systemic therapies in clear cell renal cell carcinoma: Mechanisms and management strategies. Molecular Cancer Therapeutics 17: 1355–1364. DOI 10.1158/1535-7163.MCT-17-1299. [Google Scholar] [CrossRef]
Martinelli P, Rugarli EI (2010). Emerging roles of mitochondrial proteases in neurodegeneration. Biochimica et Biophysica Acta 1797: 1–10. DOI 10.1016/j.bbabio.2009.07.013. [Google Scholar] [CrossRef]
Motzer RJ, Russo P, Nanus DM, Berg WJ (1997). Renal cell carcinoma. Current Problems in Cancer 21. DOI 10.1016/s0147-0272(97)80007-4. [Google Scholar] [CrossRef]
Mueller CG, Rissoan MC, Salinas B, Ait-Yahia S, Ravel O et al. (1997). Polymerase chain reaction selects a novel disintegrin proteinase from Cd40-activated germinal center dendritic cells. Journal of Experimental Medicine 186: 655–663. DOI 10.1084/jem.186.5.655. [Google Scholar] [CrossRef]
Muglia VF, Prando A (2015). Renal cell carcinoma: Histological classification and correlation with imaging findings. Radiologia Brasileira 48: 166–174. DOI 10.1590/0100-3984.2013.1927. [Google Scholar] [CrossRef]
Oh BY, Cho J, Hong HK, Bae JS, Park WY et al. (2017). Exome and transcriptome sequencing identifies loss of pdlim2 in metastatic colorectal cancers. Cancer Management and Research 9: 581–589. DOI 10.2147/CMAR.S149002. [Google Scholar] [CrossRef]
Padala SA, Barsouk A, Thandra KC, Saginala K, Mohammed A et al. (2020). Epidemiology of renal cell carcinoma. World Journal of Oncology 11: 79–87. DOI 10.14740/wjon1279. [Google Scholar] [CrossRef]
Papaspyridonos M, Smith A, Burnand KG, Taylor P, Padayachee S et al. (2006). Novel candidate genes in unstable areas of human atherosclerotic plaques. Arteriosclerosis, Thrombosis, and Vascular Biology 26: 1837–1844. DOI 10.1161/01.ATV.0000229695.68416.76. [Google Scholar] [CrossRef]
Penticuff JC, Kyprianou N (2015). Therapeutic challenges in renal cell carcinoma. American Journal of Clinical and Experimental Urology 3: 77–90. [Google Scholar]
Rawlings ND, Barrett AJ, Alex B (2021). MEROPS: The database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Research 42: D343–D350. DOI 10.1093/nar/gkr987. [Google Scholar] [CrossRef]
Rudzinska M, Czarnecka-Chrebelska KH, Kuznetsova EB, Maryanchik SV, Parodi A et al. (2021). Long non-coding Prox1-As1 expression correlates with renal cell carcinoma metastasis and aggressiveness. Non-Coding RNA 7: 25. DOI 10.3390/ncrna7020025. [Google Scholar] [CrossRef]
Rudzińska M, Daglioglu C, Savvateeva LV, Kaci FN, Antoine R et al. (2021). Current status and perspectives of protease inhibitors and their combination with nanosized drug delivery systems for targeted cancer therapy. Drug Design, Development and Therapy 15: 9–20. DOI 10.2147/DDDT.S285852. [Google Scholar] [CrossRef]
Rudzińska M, Parodi A, Maslova VD, Efremov YM, Gorokhovets NV et al. (2020). Cysteine cathepsins inhibition affects their expression and human renal cancer cell phenotype. Cancers 12: 1310. DOI 10.3390/cancers12051310. [Google Scholar] [CrossRef]
Skrzydlewska E, Sulkowska M, Koda M, Sulkowski S (2005). Proteolytic-antiproteolytic balance and its regulation in carcinogenesis. World Journal of Gastroenterology 11: 1251. DOI 10.3748/wjg.v11.i9.1251. [Google Scholar] [CrossRef]
Smith AM, Sewell GW, Levine AP, Chew TS, Dunne J et al. (2015). Disruption of macrophage pro-inflammatory cytokine release in crohn’s disease is associated with reduced optineurin expression in a subset of patients. Immunology 144: 45–55. DOI 10.1111/imm.12338. [Google Scholar] [CrossRef]
Szklarczyk D, Gable AL, Nastou KC, David L, Rebecca K et al. (2021). The STRING database in 2021: customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Research 49: D605–D612. DOI 10.1093/nar/gkaa1074. [Google Scholar] [CrossRef]
Tang Z, Li C, Kang B, Gao G, Li C et al. (2017). GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Research 45: W98–W102. DOI 10.1093/nar/gkx247. [Google Scholar] [CrossRef]
Trilla-Fuertes L, Miranda N, Castellano D, López-Vacas R, Farfán Tello CA et al. (2020). Mirna profiling in renal carcinoma suggest the existence of a group of pro-angionenic tumors in localized clear cell renal carcinoma. PLoS One 15: e0229075. DOI 10.1371/journal.pone.0229075. [Google Scholar] [CrossRef]
Uhlen M, Zhang C, Lee S, Sjöstedt E, Fagerberg L et al. (2017). A pathology atlas of the human cancer transcriptome. Science 357: 1. DOI 10.1126/science.aan2507. [Google Scholar] [CrossRef]
Vauléon E, Tony A, Hamlat A, Etcheverry A, Chiforeanu DC et al. (2012). Immune genes are associated with human glioblastoma pathology and patient survival. BMC Medical Genomics 5: 41. DOI 10.1186/1755-8794-5-41. [Google Scholar] [CrossRef]
Verdugo RA, Zeller T, Rotival M, Wild PS, Münzel T et al. (2013). Graphical modeling of gene expression in monocytes suggests molecular mechanisms explaining increased atherosclerosis in smokers. PLoS One 8: e50888. DOI 10.1371/journal.pone.0050888. [Google Scholar] [CrossRef]
Xu J, Liu L, Zheng X, You C, Li Q (2012). Expression and inhibition of ADAMDEC1 in craniopharyngioma cells. Neurological Research 34: 701–706. DOI 10.1179/1743132812Y.0000000067. [Google Scholar] [CrossRef]
Yang Y, Hong H, Zhang Y, Cai W (2009). Molecular imaging of proteases in cancer. Cancer Growth and Metastasis 2: CGM.S2814. DOI 10.4137/CGM.S2814. [Google Scholar] [CrossRef]
Zhang J, Zhang Q (2018). VHL and hypoxia signaling: Beyond HIF in cancer. Biomedicines 6: 35. DOI 10.3390/biomedicines6010035. [Google Scholar] [CrossRef]
Zhao Z, Chen C, Lin J, Zeng W, Zhao J et al. (2016). Synergy between von hippel-lindau and P53 contributes to chemosensitivity of clear cell renal cell carcinoma. Molecular Medicine Reports 14: 2785–2790. DOI 10.3892/mmr.2016.5561. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |