DOI:10.32604/biocell.2022.018762

| BIOCELL DOI:10.32604/biocell.2022.018762 |  |
| Article |
The molecular characteristics of soybean ARR-B transcription factors
1Heilongjiang Bayi Agricultural University, Daqing, 163319, China
2National Coarse Cereals Engineering Technology Research Center, Daqing, 163319, China
*Address correspondence to: Gaobo Yu, yugaobo81@163.com; Yuxian Zhang, zyx_lxy@126.com
Received: 16 August 2021; Accepted: 15 October 2021
Abstract: The Type-B authentic response regulator (ARR-Bs) gene family is one of the important plant-specific transcription factor families involved in variety of physiological processes. However, study of ARR-Bs gene family in soybean is limited. Genome-wide analysis and expression profiling of the ARR-Bs gene family were performed in the soybean genome. 31 ARR-Bs genes (namely GmARR-B1-31) were identified, containing conserved catalytic domains with protein lengths and molecular weights ranging from 246 to 699 amino acids (aa) and 28.30 to 76.86 kDa, respectively. Phylogenetic analysis grouped ARR-Bs genes into three clusters—Cluster I, Cluster II, and Cluster III—which included 15, 12, and 4 genes, respectively, and were asymmetrically distributed on 17 chromosomes. Tissue specific expression analysis of GmARR-Bs family revealed a high transcription level in flowers, roots and seeds. The subcellular localization of GmARR5, GmARR14 were observed in the nucleus, and the promoter region of them included low-temperature responsive element (LTR), the C-repeat/dehydration responsive element (DRE), MBS, ABRE, MYB, MEJA and TCA-elements, which possibly participate in abiotic stress and hormones responses. qRT-PCR analysis showed that the expression of GmARR-B genes were affected by different abiotic stresses, especially cold stress and salt stress, and GmARR-B5 and GmARR-B14 were significantly induced by cold stress. This suggested that ARR-Bs genes were involved in multiple abiotic stress response pathways and acted as a positive factor under cold stress.
Keywords: Soybean; Type-B authentic response regulator (B-ARRs); Abiotic stress
Soybean (Glycine max (L.) Merrill) is one of the significant cash crops worldwide. It can not only be an excellent source of protein, oil, and specialized metabolites such as isoflavonoids and saponins, but also improve soil by microbial nitrogen fixation (Abdelghany et al., 2020; Hina et al., 2018). Therefore, it is necessary to promote soybean yield. However, soybean often suffers from a variety of adverse environments, including salt, drought, cold, heat and pathogens, which limits the yield of soybean (Shu et al., 2015). Transcription factors are molecular switches that control gene expression, so they play an important role in plant response to abiotic stress (Kidokoro et al., 2014).
In the past decades, a large number of identified and characterized transcription factors (VOZ (Li et al., 2020), bZIP (Zhang et al., 2020b), WRKY (Yu et al., 2016), GRF (Chen et al., 2019), etc.) in soybean, which are widely associated with improving abiotic stress resistance, can effectively promote the growth and development of soybean and increase yield (Hwang and Back, 2019; Tolosa and Zhang, 2020). In previous studies, It has been demonstrated that the type-B authentic response regulators (type-B ARRs) gene family is one of the large plant-specific transcription factors family, which play an essential roles in response to abiotic stresses (D’Agostino, 2000). There are 11 genes and a potential pseudogene in the Arabidopsis genome encoding type-B ARR proteins (ARR 1, 2, 10–14, 18–21, 23) (Takatoshi et al., 2004; Horák et al., 2008; Pils and Heyl, 2009). Each of type-B ARRs contains the two common and characteristic domains, namely, “receiver domain” and “GARP motif”, respectively. The receiver domain is implicated in phosphorylation, the C-terminal extension carrying a highly conserved phospho-accepting REC domain that was responsible for the regulation of transcription of cytokinin-related genes (Sakai et al., 2001). The C-terminal region is highly variable, but they all have a small conserved motif composed of 60 amino acids, which is closely related to the Myb repeat of real mammals (Safi et al., 2017), whereas the GARP motif serves as a specific DNA binding domain (Hosoda et al., 2002). The 11 type-B ARRs of Arabidopsis share a common structural but fall into three subfamilies based on phylogenetic analysis, Subfamily Ӏ (7 members, AtARR 1, 2, 10–12, 14, 18), Subfamily II (3 members, AtARR 13, 21, 23) and Subfamily III (2 members, AtARR 19, 20) response regulators (Tajima et al., 2004). The Subfamily 1 has been most extensively characterized.
The expression pattern in Arabidopsis and genetic analysis indicate that at least five members, ARR 1 (Sakai et al., 2010), ARR2 (Lohrmann et al., 2001), ARR 10 (Yokoyama et al., 2007), ARR 11, and ARR 12, of Subfamily Ӏ mediate cytokinin signaling, acting as transcriptional regulators (Mason et al., 2005). Cytokinin signal transduction is mediated by a multistep phosphorelay two-component signal transduction system (TCS), including cytokinin receptors, phosphotransferases, and type-B response regulators (Kakimoto, 2003; Werner and Schmülling, 2009). According to this model of Argyros (Argyros et al., 2008) and Ishida (Ishida et al., 2008), the transcription of many genes that response to cytokinin is positive mediated by the type-B ARRs, for the last step of the TCS process, and type-B ARRs plays a role at the top of the transcription cascade. In addition to participating in cytokinin response, previous studies have shown that type-B ARR gene family is also involved in cell response to ethylene and environmental stress through TCS (Schaller et al., 2011; Hutchison, 2002; Hwang et al., 2002). In Arabidopsis, ARR 1, ARR 10 and ARR 12 have been shown to play a role in cold and drought signaling, and their mutants showed dwarf characteristics (Nguyen et al., 2016; Kang et al., 2013); ARR 2 functions in ethylene signal transduction, and induces plant immunity to a bacterial pathogen via TGA 1/NPR dependent salicylic acid signaling pathway (Carella et al., 2015; Shi et al., 2012). It was also found that type-A ARRs played a role in abiotic stresses resistance in rice, ABA, dehydration, salt and cold could induce the expression of OsRR 6 gene (Jain et al., 2006). What’s more interesting is that type-B ARRs has been revealed to promote the expression of type-A ARR directly through phosphorylation, or rapidly and briefly induce type-A ARRs through cytokinin treatment (Birkenbihl et al., 2005; Buechel et al., 2010). And type-A ARRs was regulated by type-B ARRs to improve crop cold tolerance (Jeon et al., 2010; Jeon and Kim, 2013; Kim and Jeon, 2013). In agreement with this, drought-responsive expression of ARR 22, type-C ARR, occurs in cytokinin receptor-dependent and receptor-independent pathways. overexpression of ARR 22 can also increase cold stress resistance (Kang et al., 2013; Wallmeroth et al., 2019). To sum up, type-B ARRs positively participated in plant growth and respond to environmental stress directly or indirectly.
Because of their apparent multifunctionality, type-B ARR genes are promising targets for developing crop varieties that are better adapted to abiotic stresses. Type-B ARRs have been identified and applied to enhance abiotic stress resistance in many plants, including Arabidopsis thaliana, Populus, rice, pear, and peach (Schaller et al., 2008; Yu et al., 2020; Huang et al., 2019; Ni et al., 2017; Zeng et al., 2017). Here, we also performed a genome-wide search of type-B ARR gene family members in soybean. Identification and characterization of type-B ARRs in soybean could provide a new insight for further study on stress resistance function and regulation mechanism of soybean type-B ARRs from the perspective of functional differentiation and evolution.
Identification and nomenclature of the type-B authentic response regulator genes in soybean
To identify type-B ARR family members of soybean from Soybean Database, Arabidopsis ARR-Bs gene sequence was acquired from TAIR database (http://www.arabidopsis.org/), which was used as a comparative sequence to search type-B ARR family members of soybean from NCBI (http://www.ncbi.nlm.nih.gov/), phytozome (https://phytozome.jgi.doe.gov/pz/portal.html#) and plantTFDB (http://planttfdb.cbi.pku.edu.cn/). The type-B ARR domain of the ARR proteins (PF 00072 and PF 00049) (http://pfam.xfam.org) as queries to Hmmer software searched for GmARR genes at E-value ≤ 1 × 10−10. The sequence with E < 1 × 10−10 and amino acid number >200 aa was selected. Using the unique gene name and the characteristic functional domains of ARRs, the potential GmARR-B genes were confirmed on the SMART (http://smart.embl-heidelberg.de) website (Ivica et al., 2015). A total of 31 non-redundant candidate GmARR-B genes were retained for further analysis. For the nomenclature of each putative gene, the prefix “Gm” (for Glycine max (L.) Merrill) was added to ARR followed by type “B” and Arabic numbers. 31 type-B GmARR genes were named from GmARR-B 1 to GmARR-B 31 according to their physical locations (from top to bottom) on chromosomes 1–20.
Phylogenetic tree of GmARR-B genes, conserved motif and exon/intron structure analysis
The full-length amino acid sequences of Arabidopsis (Arabidopsis thaliana) type-B ARRs were acquired using NCBI BLASTp tools from the NCBI database (https://www.ncbi.nlm.nih.gov/). To identify orthologs of the type-B ARR genes in soybean genome, the amino acid sequence of Arabidopsis type-B ARRs were used to search the publicly available databases at NCBI (http://www.ncbi.nlm.nih.gov/) and Phytozome (http://phytozome.jgi.doe.gov/pz/portal.html). The protein sequences were imported into Mega v7.0 software, multiple sequence alignment was implemented by clusalw method, and phylogenetic analysis was carried out by neighbor connection (NJ) algorithm. For this, a Poisson model was applied, with bootstrapping (1000 replicates) and pairwise deletion for gaps/missing data (Luo et al., 2017), divided GmARR-B genes into different subgroups. The conserved functional motifs within GmARR-B gene domains were identified using the MEME v4.9 program (http://meme.sdsc.edu) (Bailey et al., 2009). The length of the motif was fixed to 6–100 amino acids. To detect motifs ZOOPs model was used, which considers that the motif occurrence can be zero or 1 in a sequence. Maximum 10 motifs were searched (Bailey et al., 2009). The exon/intron structures of individual GmARR-B family genes were obtained through the Gene Structure Display Server (http://gsds.cbi.pku.edu.cn) (Guo et al., 2007) by aligning the coding or cDNA sequences with their corresponding genomic DNA sequences from Phytozome v9.1 (http://www.phytozome.net/) (Goodstein et al., 2012).
Analysis of chromosome distribution, Cis-regulatory elements and gene duplication and synteny
The chromosome distribution of soybean ARR-B genes was obtained from Phytozome, GmARR-B genes were mapped onto the 17 soybean chromosomal linkage groups according to their physical position (bp). MapInspect was used to map chromosome location. The 1,500 bp upstream of the transcription start site of all GmARR-B genes insoybean was obtained from Phytozome v9.1 (http://www.phytozome.net/), and the cis-regulatory elements were identified using the PlantCARE program (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al., 2002).
We attempted to identify the duplication event and analyzed the synteny relationships among the ARR-Bs of A. thaliana, and soybean. And duplicated genes were obtained from Plant genome duplication database (PGDD) (http://chibba.agtec.uga.edu/duplication/) by downloading the dataset of duplicated blocks in soybean genome (Lee et al., 2013). Duplicated GmARR-B gene pairs were searched in the dataset. Similarly, syntenic blocks between soybean and A. thaliana were also searched. A total number of 19 chromosomes (A. thaliana-5, soybean-14) with a total number of 29 ARR-Bs (A. thaliana-22, soybean-7) were used to map the synteny relationships. TBtools (Chen et al., 2020) was used to map gene replication.
Physical and chemical properties and subcellular localization of GmARR-Bs protein
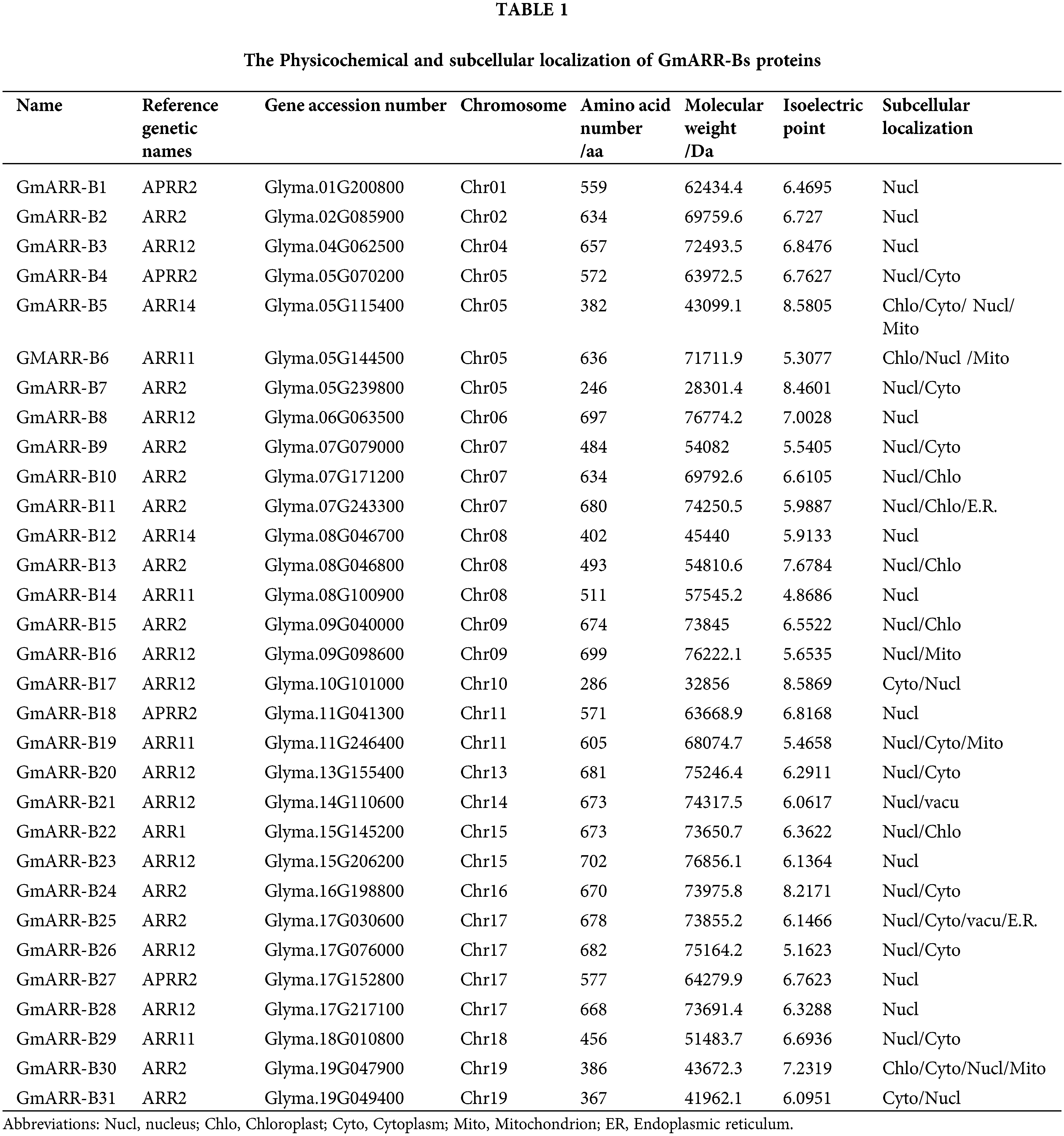
Physico-chemical features of the GmARR-B proteins, including length, molecular weight, theoretical isoelectric point, instability index were predicted using the protein identification and analysis tools in ExPASy Server10 (https://web.expasy.org/protparam/) (Panu et al., 2012). All selected proteins were subjected to sub-cellular localization prediction using pSORT web server (http://www.genscript.com/psort/wolf_psort.html) to know the relative abundance of these proteins in different sub-cellular compartments.
Secondary structure analysis of proteins
Prediction of secondary structure of GmARR-B proteins by NPS (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl page=/NPSA/npsa_sopma.html) online website (Akbudak and Filiz, 2020).
RNA seq data analysis for soybean expression in diverse tissues
To dissect the expression patterns of GmARR-B genes in the tissue and organ, we used all the genes as obtained for the soybean genomes. The RNA seq data for the expression of GmARR-Bs in different tissues at various developmental stages were obtained from Phytonome (https://phytozome.jgi.doe.gov/pz/portal.html#) databases (Debbie et al., 2007). The RNA sequencing reads have been listed in Suppl. Table S1. The heat maps were constructed using heat maps of the R Packages.
Soybean (Glycine max) seeds with uniform colors and sizes were selected and disinfected with 95% ethyl alcohol and washed with sterile water five times before they were sown. The soybean seeds were sowed with the soil at 25°C (16 h light/8 h dark cycle) in a greenhouse. The treatments for abiotic stress were: (1) cold stress: the seedlings of V 1 stage (The first trefoil stage of vegetative growth period) were transferred to the incubator at 4°C for 8 days. (2) heat stress: the seedlings of V 1 stage were transferred to the incubator at 30°C for 8 days. (3) salt stress: 20 mL of 300 mmol/L NaCl solutions was irrigated into V 1 seedlings for 4 days. (4) alkaline stress: 20 mL of 75 mmol/L NaHCO3 solutions was irrigated into V 1 seedlings for 4 days. (5) ABA stress: 100 μmol/L ABA solution was sprayed on both sides of the leaves of seedlings at V1 stage to drip with the leaves. All experiment involved three biological repeats and each repetition has six soybean plants. The samples were collected from each treatment at 0, 6, 12, 24, 48, and 96 h after stresses and frozen in liquid nitrogen, and stored at −80°C.
RNA extraction and quantitative PCR analysis
We investigated the expression of the 10 ARR genes under ABA, cold, heat, salt, and alkali stress conditions, which are closest to Arabidopsis in the evolutionary tree, in order to confirm if different abiotic stress treatments influenced the expression of type-B GmARR genes (Fig. 1). Using a sterile mortar and pestle, frozen soybean leaves were crushed to a fine powder under liquid nitrogen. TriZol reagent was used to extract total RNA from 30 mg fresh weight of frozen tissue. The cDNA was synthesized using the ReverTra Ace qPCR RT Kit (TAKARA) as directed by the manufacturer. The GmActin1 gene was used as a control, and relative expression levels of genes were calculated by the 2−ΔΔCT method (Livak and Schmittgen, 2001). Each sample was assayed in three repeats, the significance of the differences between the samples was analyzed using paired T-test, with P-values of *P < 0.05 and **P < 0.01 being considered significant.
qRT-PCR primers were designed using Primer 5.0 (Suppl. Table S2).
Plasmid construction and subcellular localization
Two genes with high expression under abiotic stress were selected for subcellular localization test to further clarify whether the protein encoded by the GmARR-B family is embedded within the nucleus or not. Therefore, two genes were subsequently chosen to make a GFP-Gm.05G115400/GFP-Gm.08G100900 (ARR-B 5&14), fusion vector (pYBA1132-Gm_05G115400/Gm.08G100900 (ARR-B 5&14). The free GFP was used as the control. For subcellular localization study, GmARR-Bs were obtained by gene cloning, and cloned into the vector GFP-pYBA 1132 with the target fragment by homologous recombination, transformed into Escherichia coli DH5α. Then the plasmid was transferred into Agrobacterium GV3101 by freeze-thaw method. Agrobacterium tumefaciens were cultured overnight for 24 h, gently centrifuged and diluted in osmotic buffer (10 mM MgSO4·7H2O with 100 μM acetosyringone; both from Sigma (Sigma GmbH, Erlangen, Germany) until optical density (OD) 0.1 and used for infiltration by syringe on the Nicotiana benthamiana leaf surface. Transient expression was visualized through fluorescence microscope 48 h later. An excitation wavelength of 434 nm was used and the emissions were collected between 460 and 490 nm to visualize GFP
Identification and phylogenetic analysis of type-B GmARR genes in soybean
31 soybean type-B ARRs were identified in the soybean genome by queries to conduct sequence homology searches of type-B AtARRs, which is nearly three times as many as Arabidopsis (12). 31 type-B GmARR genes were named from GmARR-B 1 to GmARR-B 31 on the basis of their physical locations (from top to bottom) on chromosomes 1–20. We compared the amino acid sequences of all 31 putative type B-GmARR with each other to determine whether these genes contain key amino acid residues required for ARR function, as shown in Suppl. Fig. S1.
To understand the evolution history of type-B ARR genes and assist in the classification of the type-B ARR gene family in soybean, a phylogenetic tree was constructed from aligned soybean and Arabidopsis ARR protein sequences. A total of 43 type-B ARR genes from soybean (31), Arabidopsis (12) were used for phylogenetic analysis. The maximum likelihood phylogeny indicated that all of the 31 newly uncovered type-B GmARR genes and 12 type-B AtARR genes were divided into three subgroups by distinctive protein structures (Type Ӏ, Type B II, Type III; Fig. 1). The Subgroup I contained 15 GmARR-Bs and 5 AtARR-Bs, which was the largest group in this phylogenetic tree; Subgroup II included 12 GmARR-Bs and 5 AtARR-Bs; Subgroup III consisted of 4 GmARR-Bs and 2 AtARR-Bs. Among them, the distribution of GmARR-Bs in soybean was mostly concentrated in the Ӏ subgroup, and less in the II and III subgroups (Fig. 1). In addition, we also predicted the secondary protein structures of the type-B GmARR genes family in Suppl. Fig. S2.
Analysis of type-B GmARR proteins is shown in Table 1. All of these 31 type-B GmARR genes contained the domains PF00072 and PF00049 based on Pfam and SMART tests. The GmARR-B genes coded 246–699 amino acids, with calculated molecular weights from 28.30 to 76.86 kDa, average molecular weight of 63.13 kDa. At the same time, we also found that the isoelectric points of these nucleic acids ranged from 4.87 to 8.59, and most of these proteins were negatively charged, while only 4 proteins were positively charged (Table 1). Further bioinformatics investigations demonstrated that GmARR-Bs were possibly localized in a few organelles such as the nucleus, cytoplasm, chloroplasts, and mitochondria (Table 1). And most of the type-B GmARRs were located in the nucleus.
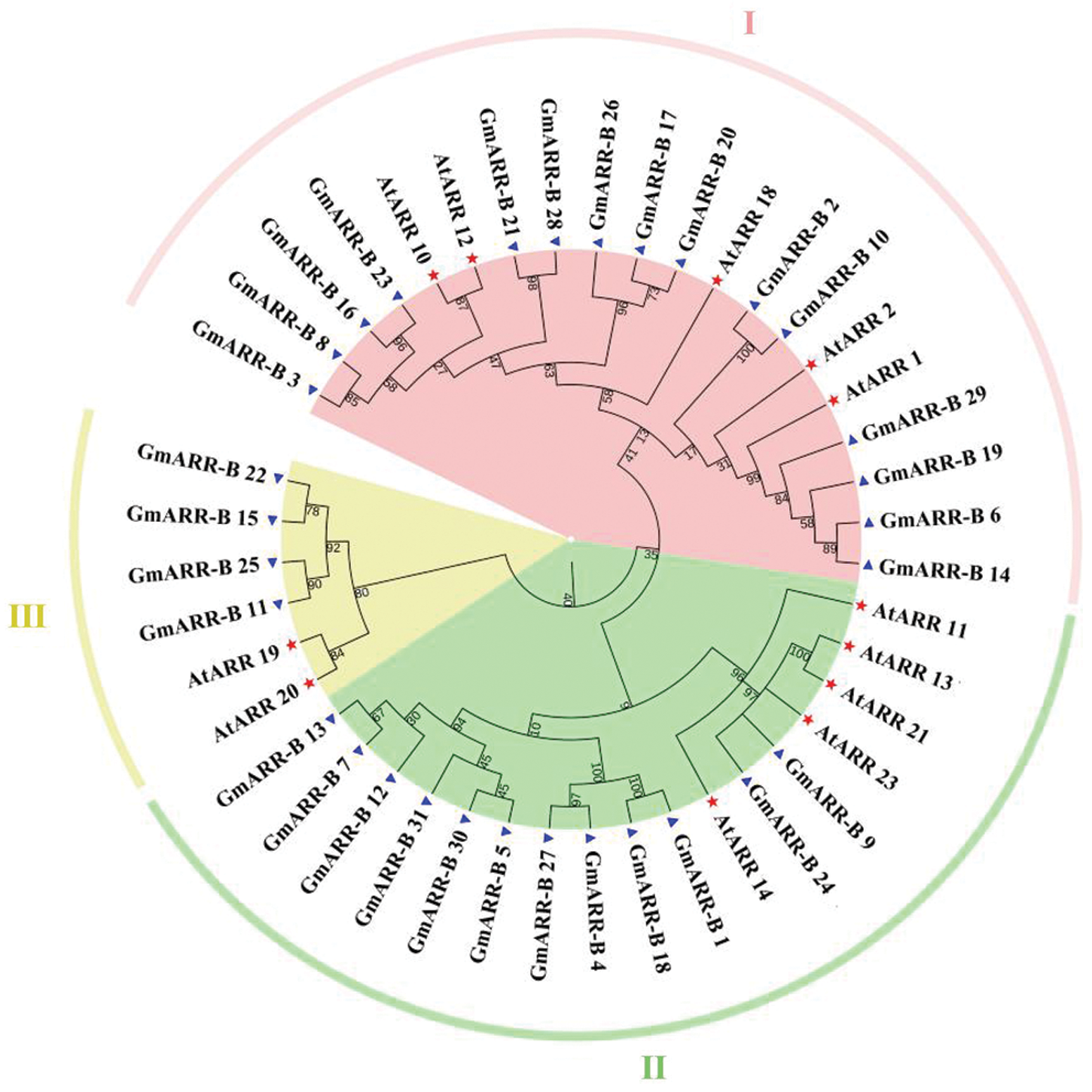
Figure 1: Phylogenetic relationships of the type B authentic response regulators (ARRs). Phylogenetic relationships of the ARR-Bs from soybean (Blue triangle) and Arabidopsis (Red Pentagram). MEGA 7.0 was used to create the phylogenetic tree. The 43 ARR-Bs from two plant species may be classified into three groups, with distinct colored arcs (Note: At stands for Arabidopsis thaliana, while Gm stands for Glycine max.).
Analysis of the structure and conserved domain of type-B GmARR genes
In order to reveal the diversity of the gene sequence of the ARR-B gene family in soybean, exon-intron distribution and 10 different conserved motifs of each gene were analyzed (Fig. 2). The motifs with a size range of 15–20 amino acids were selected for identification, and other details of amino acid sequence conservation and the length of 10 conservative motifs were shown in Suppl. Table S3. The specific motifs of subfamily lead to the difference of gene function of type-B GmARR subfamily to some extent. The motif analysis showed that each group of genes classified by phylogenetic analysis has the similar conserved motif composition in its own group clearly, however, motif conservation differed slightly within each subgroup and was more dramatically across subgroups. We identified ten conserved motifs in all putative 31 type-B GmARRs and named them as Motifs 1 to 10. Motifs 1–7 were present in all of the type-B GmARRs. We also observed motifs that were specific to a subgroup, including Motif 9 and Motif 10 that were only discovered in Group II genes. Similarly, motif 8 was found to exist in Groups Ӏ and III genes. In GmARR-Bs, the similarity of conservative motifs was high, and the homology was relatively close, which suggested that the function was relatively conserved.
The evolution of the genome and the functional divergence of multigene family members are both linked to gene structure. Exonic and introic regions differed among the 31 type-B GmARRs found (Fig. 2). In type-B GmARRs, the number of introns varied from three to ten. Genes were grouped into three categories based on sequence homology, with common gene structural characteristics within each cluster. It was consistent with the results of evolutionary tree analysis. One cluster, which was comprised of Type Ӏ genes, all shared 5 homologous exons and 4 homologous introns, except GmARR-B 17. Be the same with Type Ӏ, the Type III gene cluster (GmARR-B 11, GmARR-B 15, GmARR-B 25 and GmARR-B 22) all shared 5 homologous exons and 4 homologous introns. However, Type II genes did not show such clear exon-intron arrangements across the subgroup, among them, GmARR-B 1, GmARR-B 4, GmARR-B 18 and GmARR-B 27 had the highest number of exons (11), while GmARR-B 7 had the least number of exons. The different gene structures existing in the different phylogenetic groups suggested that genes evolved into diverse exon-intron structures to carry out different functions in the soybean genome, indicating that in the variable cutting process, these genes may be from the same transcript to possess similar characteristics.
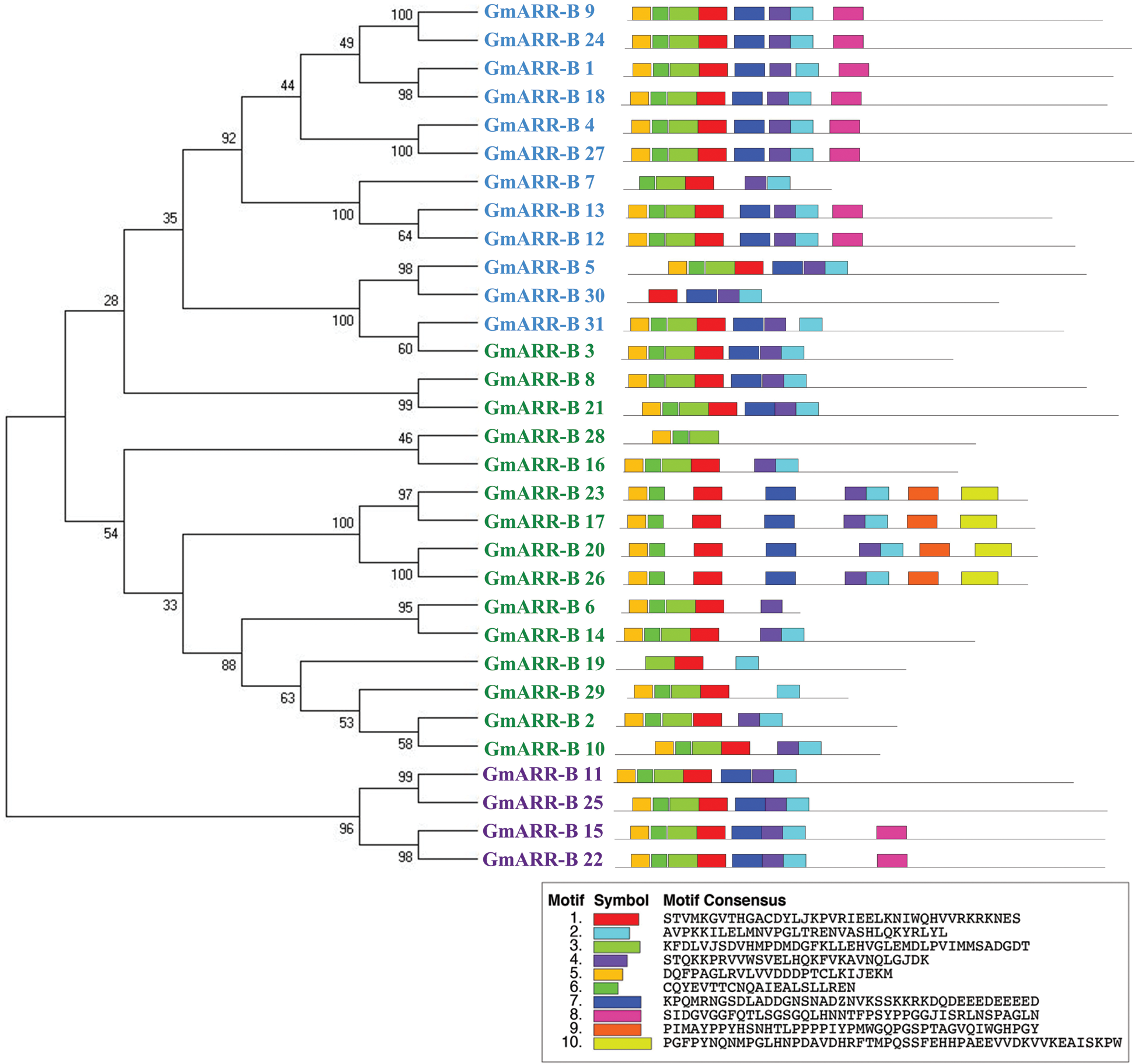
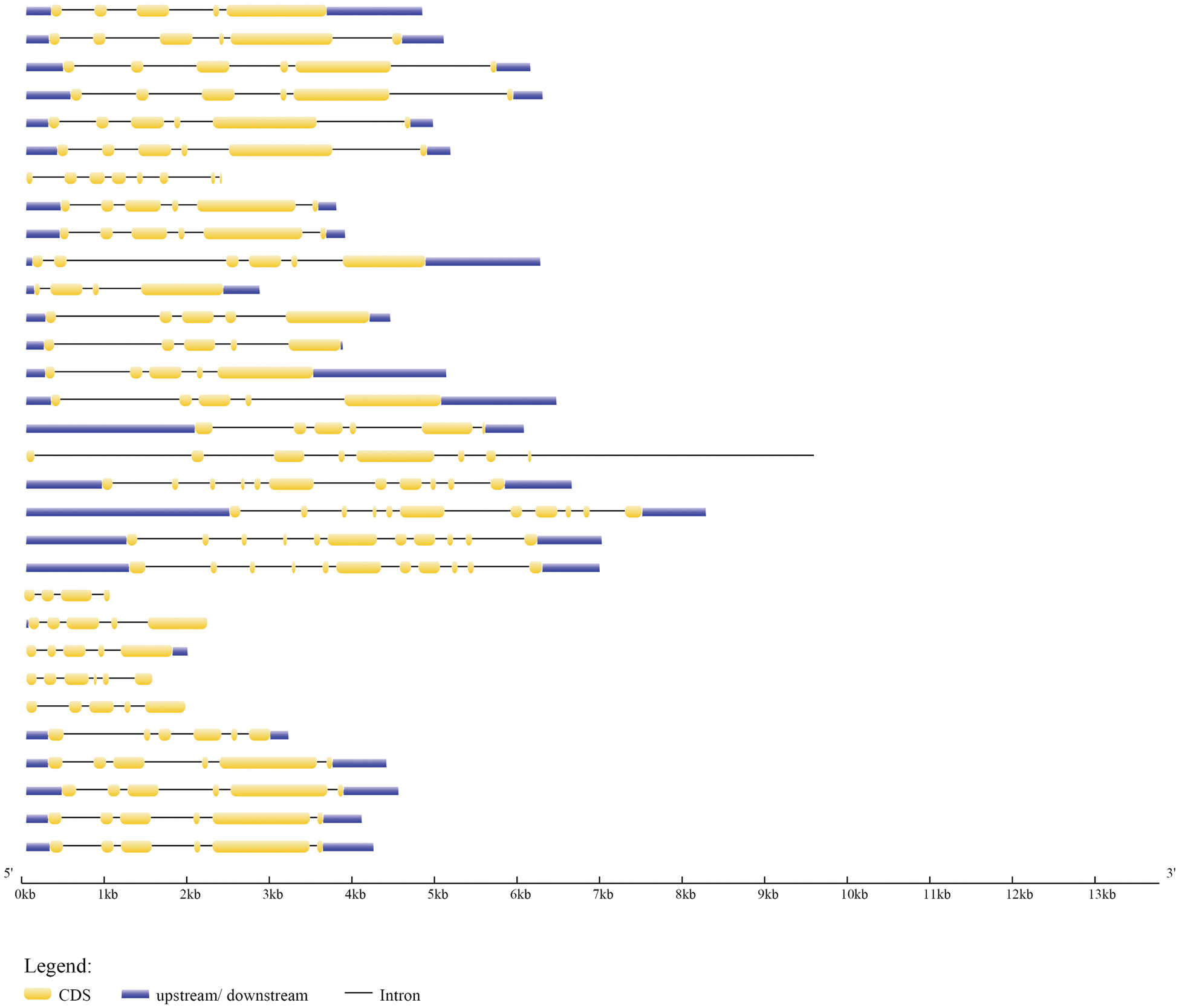
Figure 2: G mARR-Bs phylogenetic connections, conserved domains, and gene structures on the left side of the picture, the three subgroups are represented by different colors. Green is Subgroup 1, blue is Subgroup 2, and purple is Subgroup 3. The schematic representation of motif analysis in the center: The upper panel indicates predicted motifs in type-B GmARRs, represented in different colour using MEME suite v5.1.0. Whereas the lower panel shows the signature of each motif with conserved amino acid residues. Structural elucidation of 31 identified type-B GmARR genes on the right. The structural features of type-B GmARRs are represented in different colors, where Orange indicates an exonic region, blue indicates upstream/downstream region, black indicates intronic region.
Chromosomal location, collinearity, and cis-element analysis of type-B GmARRs
The chromosomal locations and gene structures of all GmARR-Bs were downloaded from soybean genome database. The cross sections of chromosome location and gene structure of all GmARR-B are shown in Suppl. Fig. S3. The 31 GmARR-B genes were widely distributed on 17 chromosomes except chromosome 3, 12, 20. (Fig. 3 and Suppl. Fig. S3). Chromosome 5 and chromosome 17 had the maximum of GmARR-B genes, with 4 members. Three GmARR-B genes were discovered on chromosomes 7 and 8, while two genes were discovered on chromosomes 9, 11, 15 and 19, and the other chromosomes (1, 2, 4, 6, 10, 13, 14, 16 and 18) had only one type-B GmARR gene (Fig. 3).
The generation and maintenance of gene families are mainly rely on gene tandem duplication and segmental genome duplication. We have reason to believe that two homologous genes that are physically closely linked may be produced by tandem replication. As shown in Fig. 3, synteny was used to analyze GmARR-B gene duplication. There were 10 GmARR-Bs including GmARR-B 1, 2, 4-8, 11, 14 and GmARR-B 17 showed collinearity with interior of soybean. There were 7 GmARR-Bs including GmARR-B 5, 9, 11, 14 and GmARR-B 15-17 that showed collinearity with the ARRs of dicots, Arabidopsis.
Each GmARR-B gene was extracted 1.5 kb genomic sequences upstream of its transcriptional start codon for cis-regulatory elements in order to further expound the biological functions and regulation network of GmARR-Bs, and 13 cis-acting regulatory elements related to stress, hormone, and development were found in the upstream sequences. Many elements linked to stress response and plant hormones were found in the promoter region of this GmARR-B family gene. The number of cis-acting regulatory elements on each ARR-B genes varied widely, from 3 to 13. As shown in Fig. 4, cold resistance related LTR, was found in the promoter region of GmARR-B 6, 8, 11, 19–22, 25, 29 and 30; the drought-induced element MYB and MBS were found in the promoter region of GmARR-B 2, 5, 6, 11, 28, 30 and GmARR-B 1, 16, 17, 23. Except for abiotic stress, hormone-responsive elements were predicted, almost all GmARR-B genes contained hormone responsive element ARE, except GmARR-B 8, 9, 16. ABA-responsive elements ABREs were found in the promoter region of GmARR-B 4, 6-10, 16-19, 27 and 31. MeJA-responsive elements were revealed in the promoter region of GmARR-B 2, 3, 5, 9, 10-12, 15, 17 and GmARR-B 19. SA-acting elements were explored in GmARR-B 1, 4, 6, 13, 14, 18, 20 and GmARR-B 24. And there were seven type-B GmARR genes (GmARR-B 1, 4, 9, 15, 16, 20, 23, 26), which were identified as auxin homeostatic elements and five type B GmARR genes (GmARR-B 2, 7, 17, 23, 25) were identified as gibberellin homeostatic elements. In addition, as light-responsive elements, MRE was found in the promoter region of GmARR-B genes.
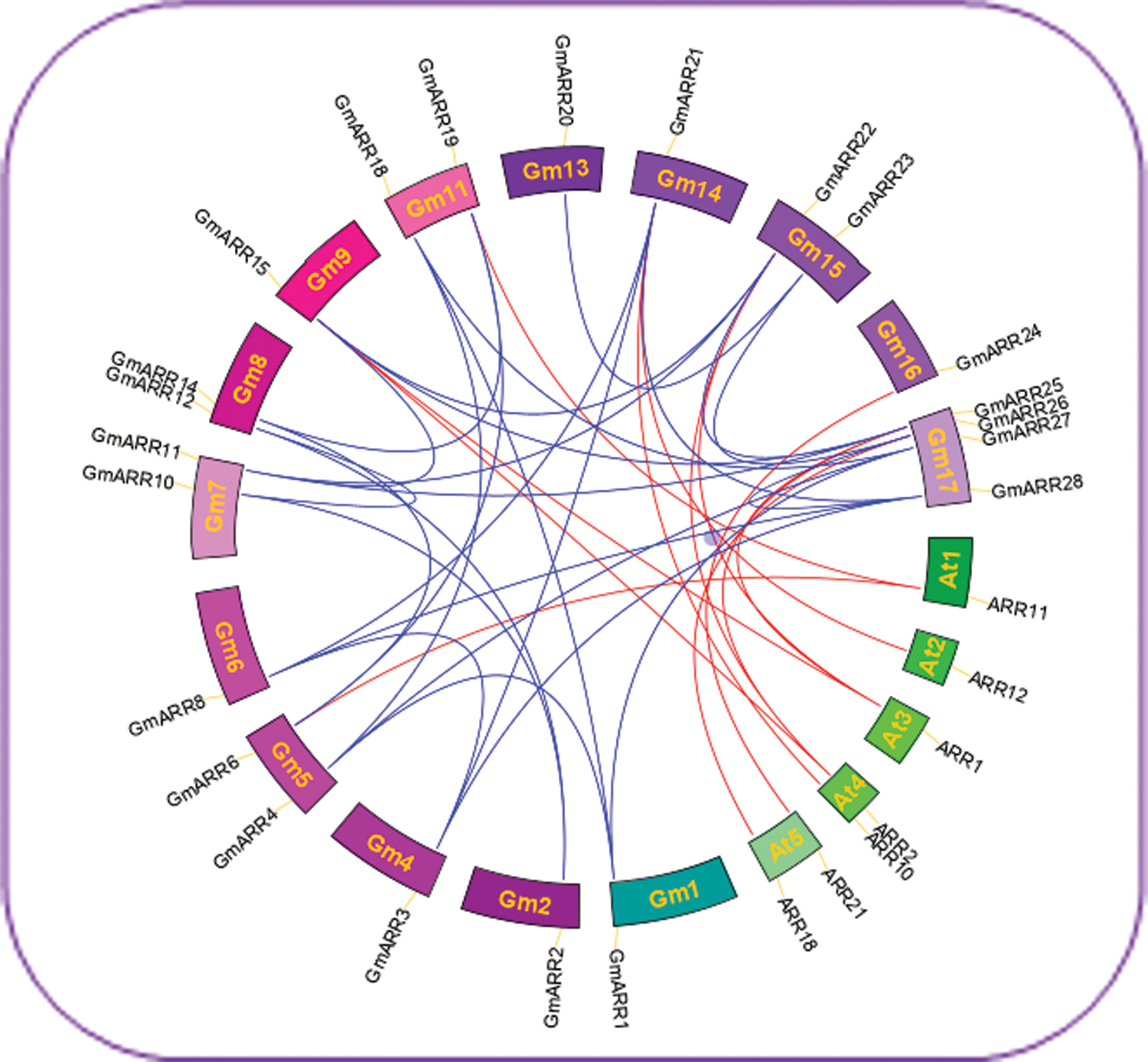
Figure 3: The Circos plot representation for synteny relationship of ARR-B genes in soybean and Arabidopsis using CIRCOS v0.52. Each chromosome is represented by a color box, with the chromosome number shown in each chromosomal box. Megabase is the scale. A short blue line indicates the approximate position of each soybean ARR-B gene. The linkage group with segmentally duplicated ARR-B gene pairs is indicated by red lines in the circle, and segmental duplication areas were found using the Plant Genome Duplication Database.
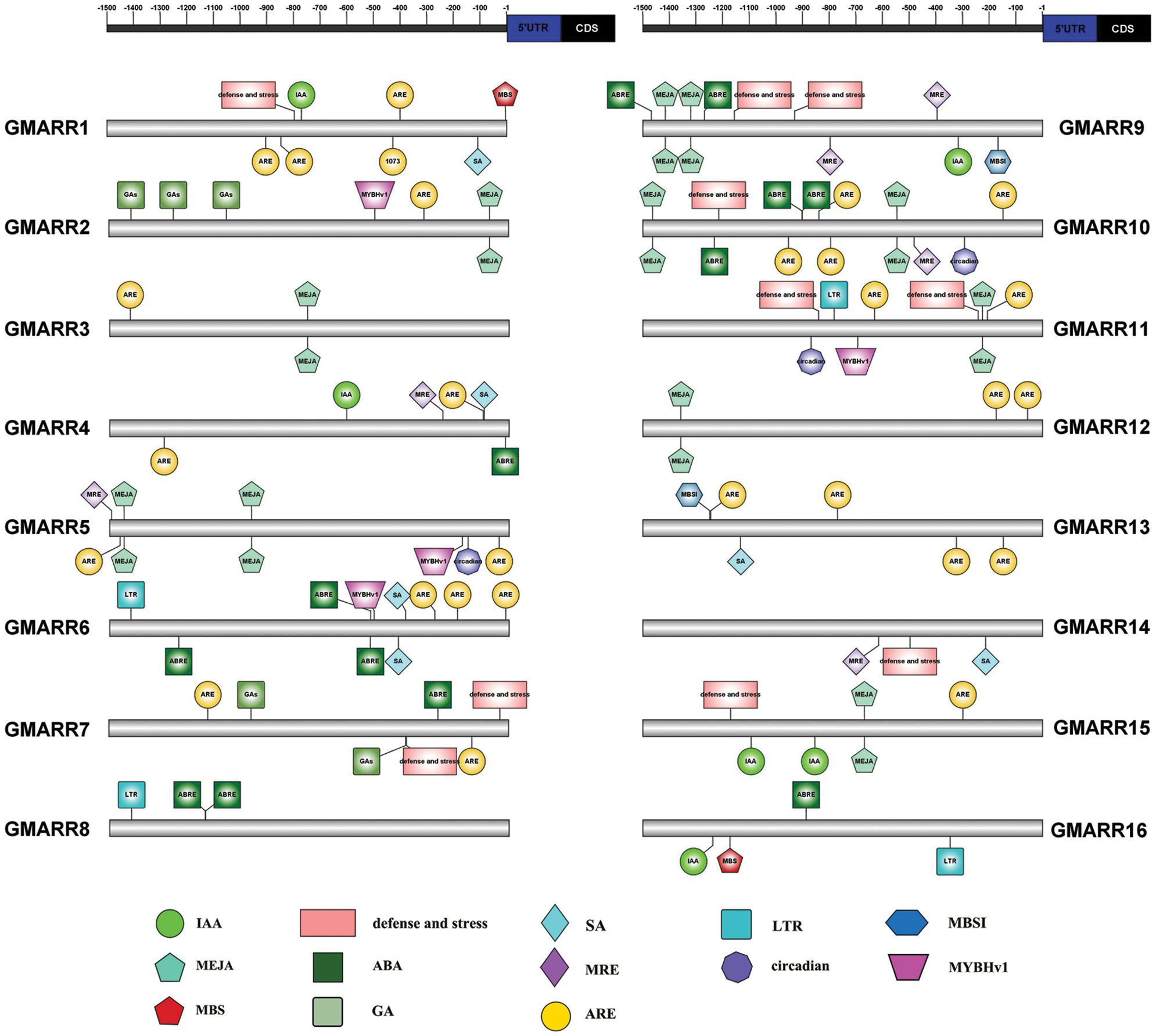
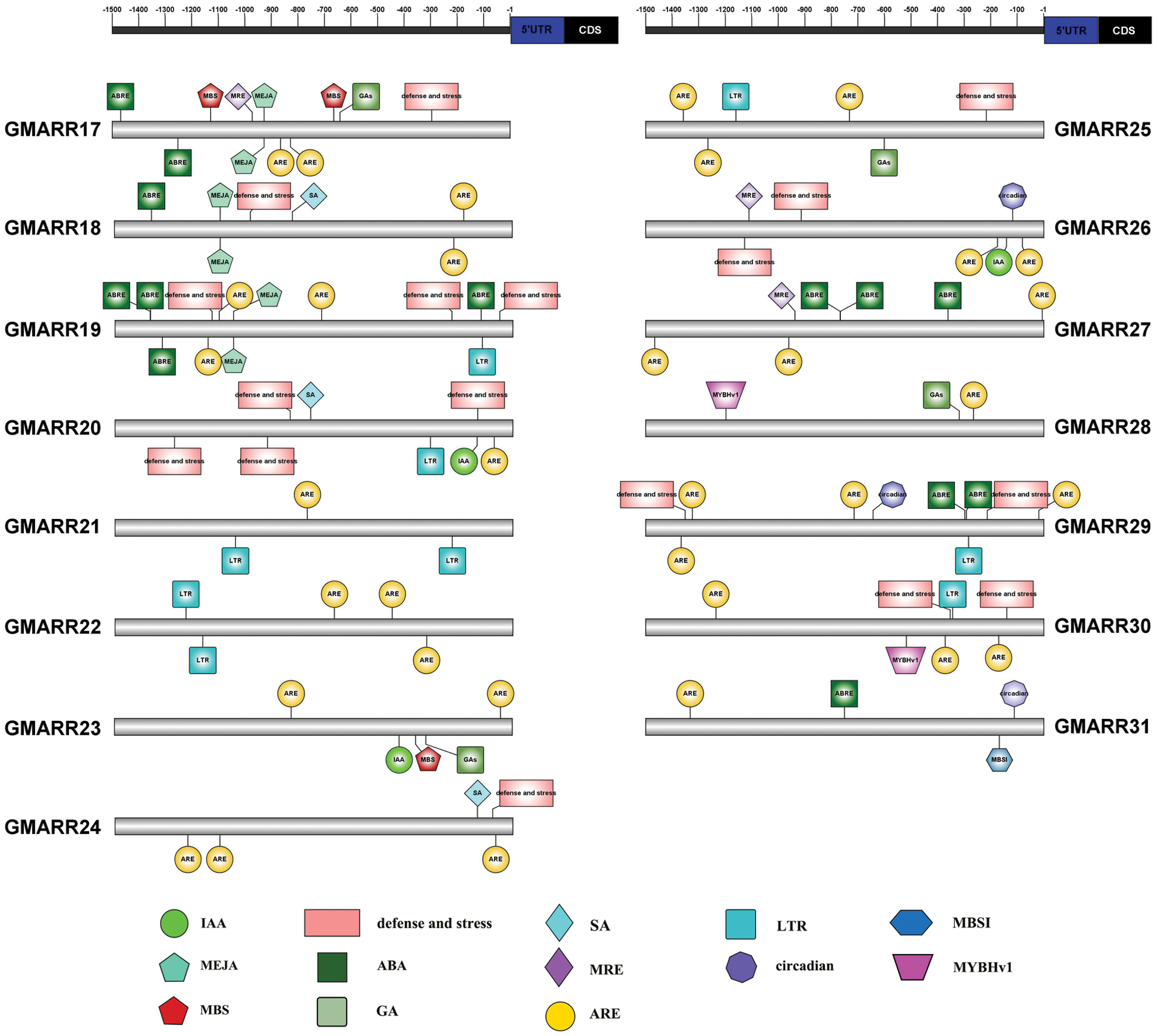
Figure 4: Anticipated cis-elements within the promoter districts (1,500 bp upstream from the transcription start location) of the soybean GmARR-B genes. Each functional element is represented by a different surname, and the corresponding ruler position in the 1.5 kb area before CDS is marked with a line. The action elements corresponding to different figures are shown in the lower part of the figure.
Tissue and organ specific expression of GmARR-B genes in soybean
To dissect the expression patterns of GmARR-B genes in the tissue and organ, the transcription level of GmARR-Bs in eleven diverse tissues were analyzed (Suppl. Table S1). The heat map (Fig. 5) revealed that the expression of 31 GmARR-Bs was high in the apical meristem, seed, root, leaf, nodules, and flower, with the expression patterns largely grouped into three Groups (A–C) based on the hierarchical clustering analysis. Group A contained 11 soybean GmARR-B members (GmARR-B 3, 6, 8, 14, 15, 19, 21–23, 28, 29), and 9 genes of them were members of the Type II subgroup and 2 genes (GmARR-B 15, 22) of them were members of the type III subgroup. All these members showed highly expressed in root and nodules. Interestingly, Group B included 13 soybean GmARR-B members, which were all Type-Ӏ members, and expressed in seed, pod skin and flower. Among them, the expression of GmARR-B 20 was high in pod, while the transcription level of GmARR-B 1 and GmARR-B 12 were high in seed. Group C was consisted of 7 GmARR-B members (GmARR-B 4, 11, 17, 25, 27, 30, 31), most of which showed consistent expression pattern in the majority of tissues. GmARR-B 11 and GmARR-B 17 had significant levels of transcription in the root. The expression of four GmARR-B genes (GmARR-B 4, 27, 30, and 31) was also significantly increased in the shoot apical meristem. Tissue expression patterns particular to subfamilies may be linked to gene functions. The above results show that ARR-B transcription factors may play diverse roles in the growth and developmental regulation of soybean.
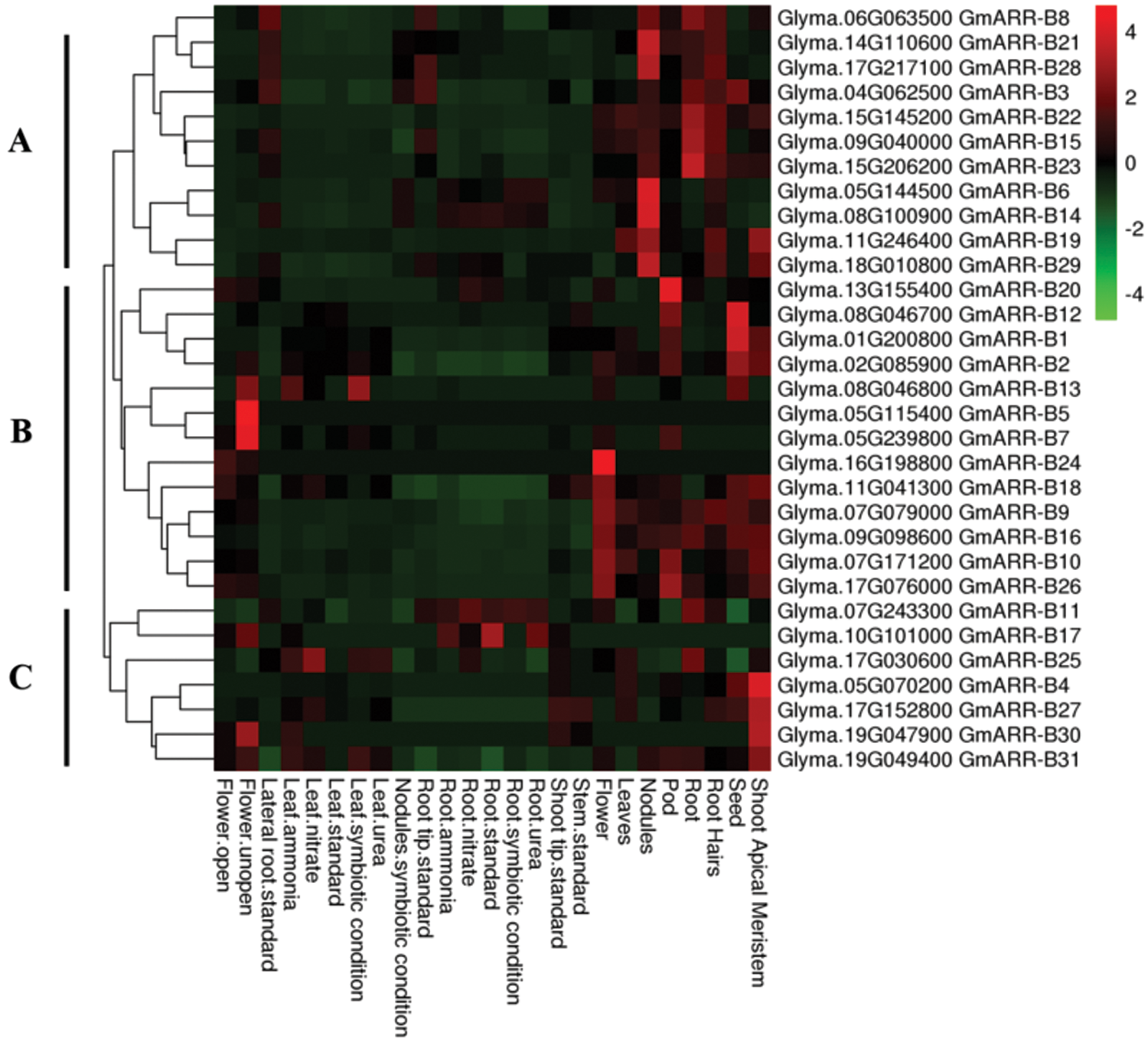
Figure 5: GmARR-B expression profiles in various tissues. The degree of expression is shown by the color bar to the right of the graph, which ranges from light blue to red, suggesting that the amount of expression is growing.
GmARR-Bs were involved in abiotic stress responses
Under ABA, cold, heat, salt and alkali stress conditions, the leaf spots of NBT, DAB and Evans blue staining indicated that, with the extension of stress time, the accumulation of reactive oxygen species and H2O2 in leaves and the degree of cell damage gradually increased (Fig. 6). In addition, compared with the control, the reactive oxygen species content and antioxidant enzyme activity of soybean under ABA, cold, heat, salt and alkali stress increased to some extent with the extension of stress time, and the most significant increase of all indexes was observed after 24 h treatment, which further reflected the stress degree of soybean (Fig. 7).
Quantitative Real-Time PCR was conducted to examine the expression level of 10 GmARR-B qualities beneath abiotic stresses, including ABA, cold, heat, salt, alkali. All the 10 GmARR-B genes responded to the five stresses, but the reaction speeds and intensities were different. It could be seen from Fig. 8, under ABA stress (40 μmol/L), compared with the control, the expression of 10 genes were inhibited after treatment 24 h. However, partially restored to the control level at 48 h, and the transcription level of GmARR-B 5 was significantly induced. For the cold treatment, the expression of GmARR-B 9 and GmARR-B 13 was significantly up-regulated at 12 h, almost all individuals were unequivocally actuated at 24 h and 48 h, except GmARR-B 7, GmARR-B 17 and GmARR-B 30 at 24 h. Under heat conditions, the expression of 10 GmARR-B genes were inhibited at 6 h exposed to heat stress, except for GmARR-B 14, while the transcription of tested GmARR-B genes were induced, and reached the highest level at 24 h, except for GmARR-B 17. Salt stress unequivocally induced the expression of ARR genes, and most GmARR-B members individuals appeared an awfully critical reaction to salt stress, compared with control. All individuals were strongly induced between 6 h and 24 h after treatment, but for the expression of GmARR-B 30. The expression patterns of the GmARR-B qualities amid alkali base treatment displayed self-evident contrasts, showing the utilitarian differences of these genes. For GmARR-B 7, 9, 12, 30 and GmARR-B 31, the gene expression was enhanced and reached the peak at 6 h, but the transcription level of GmARR-B 13 was induced between 24 h and 48 h. And the expression of GmARR-B 17 got the highest transcription level at 24 h. Additionally, the transcription of several GmARR-B genes decreased, such as GmARR-B 5, GmARR-B 14 and GmARR-B 29.
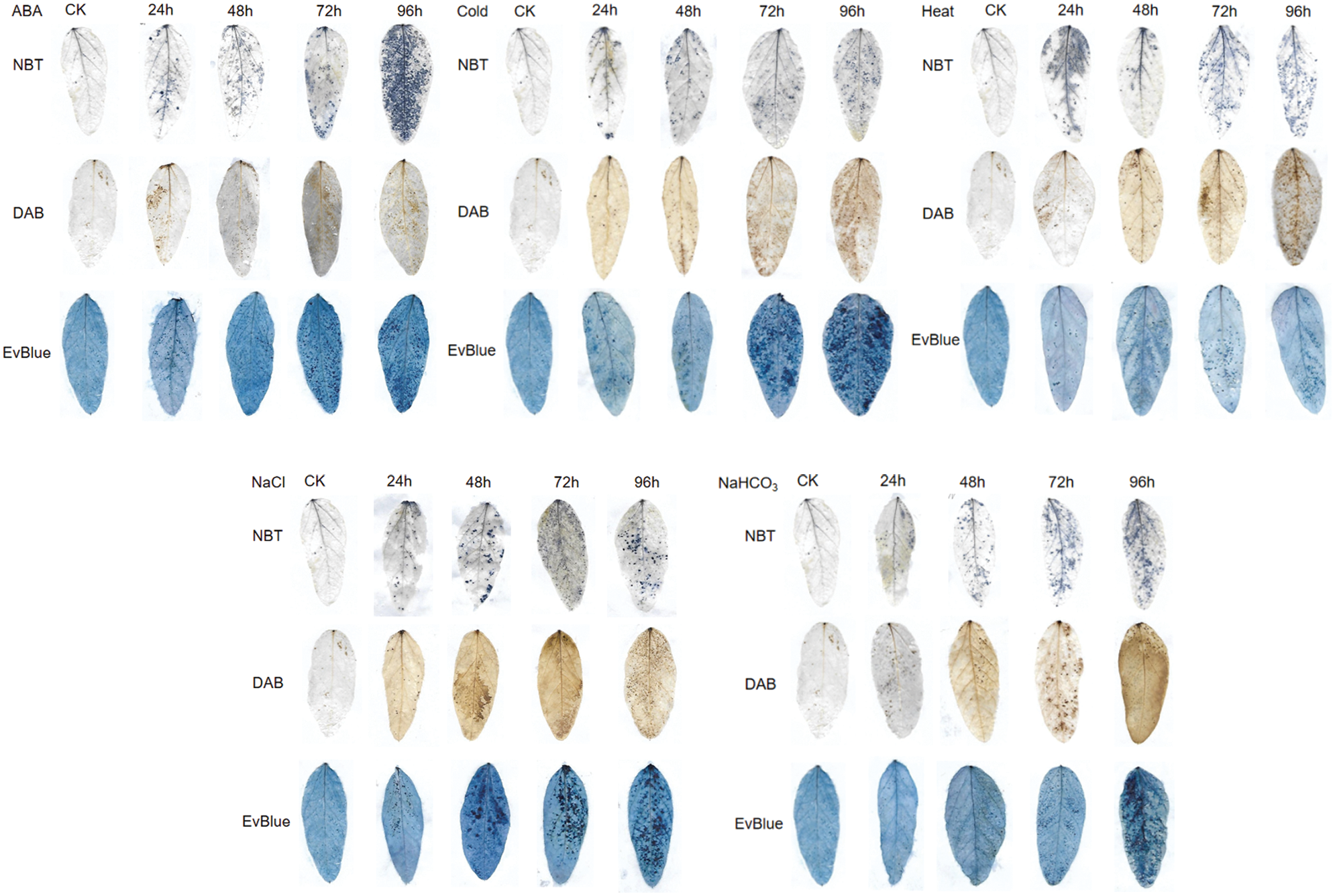
Figure 6: Reactive oxygen species staining of soybean leaves under different abiotic stress.
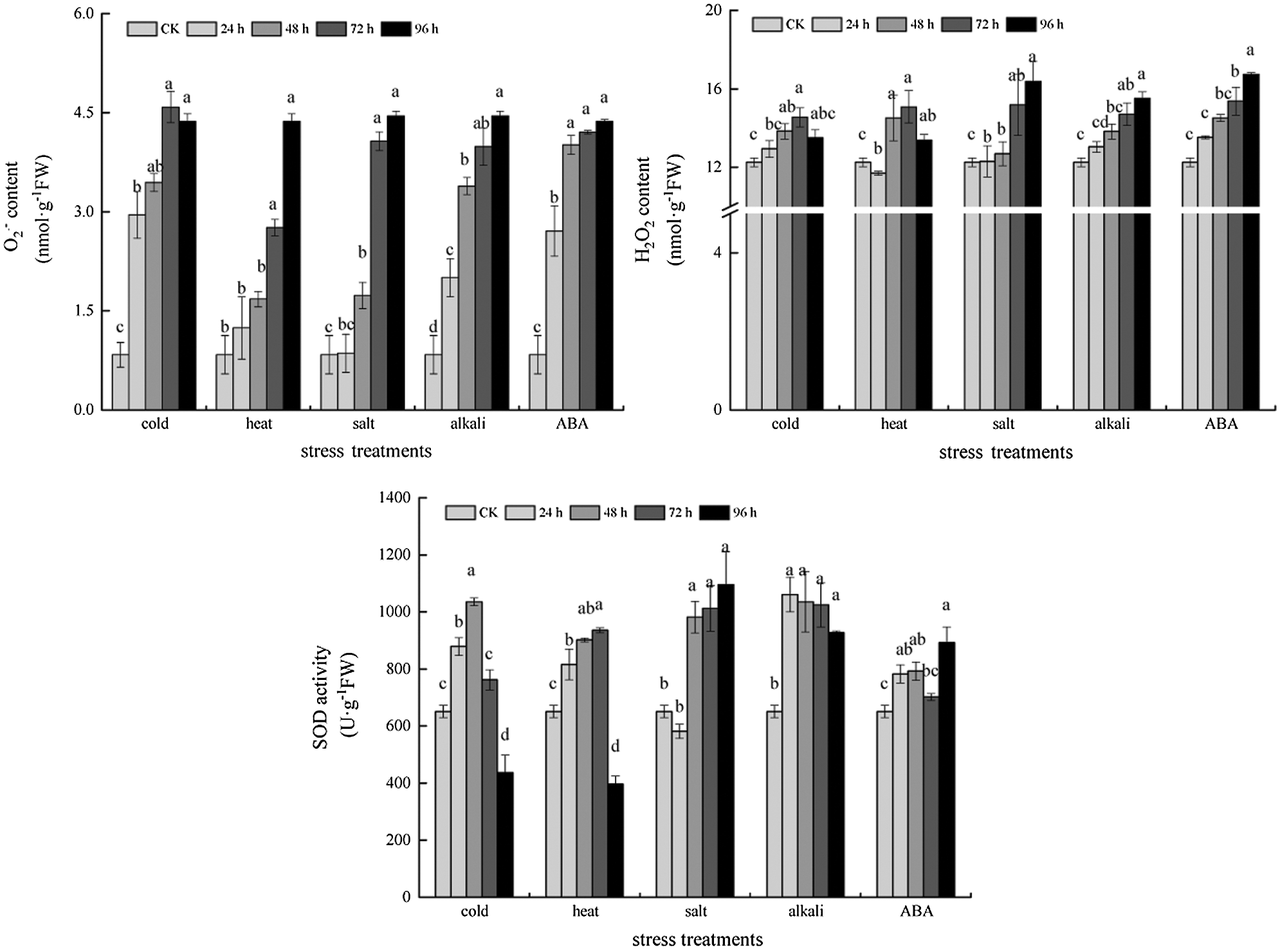
Figure 7: Superoxide anion, hydrogen peroxide content and antioxidant enzyme activity under different stress treatments.
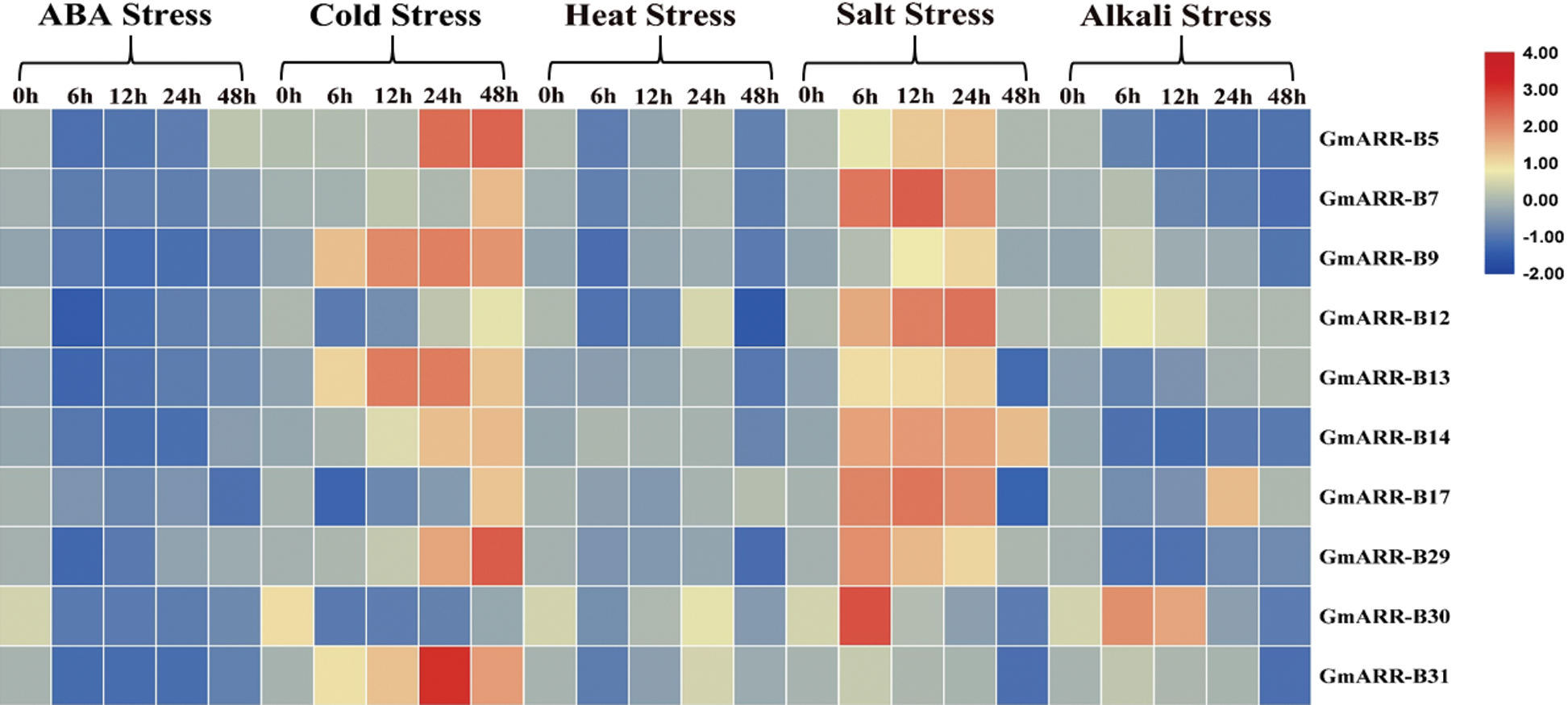
Figure 8: Heat map of the expression patterns of 10 GmARR-Bs under ABA, cold, heat, salt and alkali treatments by quantitative real-time PCR (qRT-PCR) analysis of ARR-B genes in soybean seedlings in response at different treatmenttime points (0, 6, 12, 24 and 48 h). The data comes from three separate biological investigations, and the relative transcription levels of GmARR-B genes were normalized to the expression of GmActin 1.
Subcellular localization of GmARR-B 5 and GmARR-B 14
Prediction analysis indicated that GmARR-Bs exhibited various patterns of subcellular localization (Table 1), although mostly concentrated in the nucleus. GmARR-B 5 and GmARR-B 14 with high expression under abiotic stress were selected to determine if the protein encoded were embedded within the nucleus or not. The free GFP was used as the control. We transiently expressed the GFP fusion protein with these two genes in tobacco leaves. Green fluorescence signals of both fusion proteins with target gene were localized in the nucleus (Fig. 9), consistent with the predicted localization using bioinformatics programs.
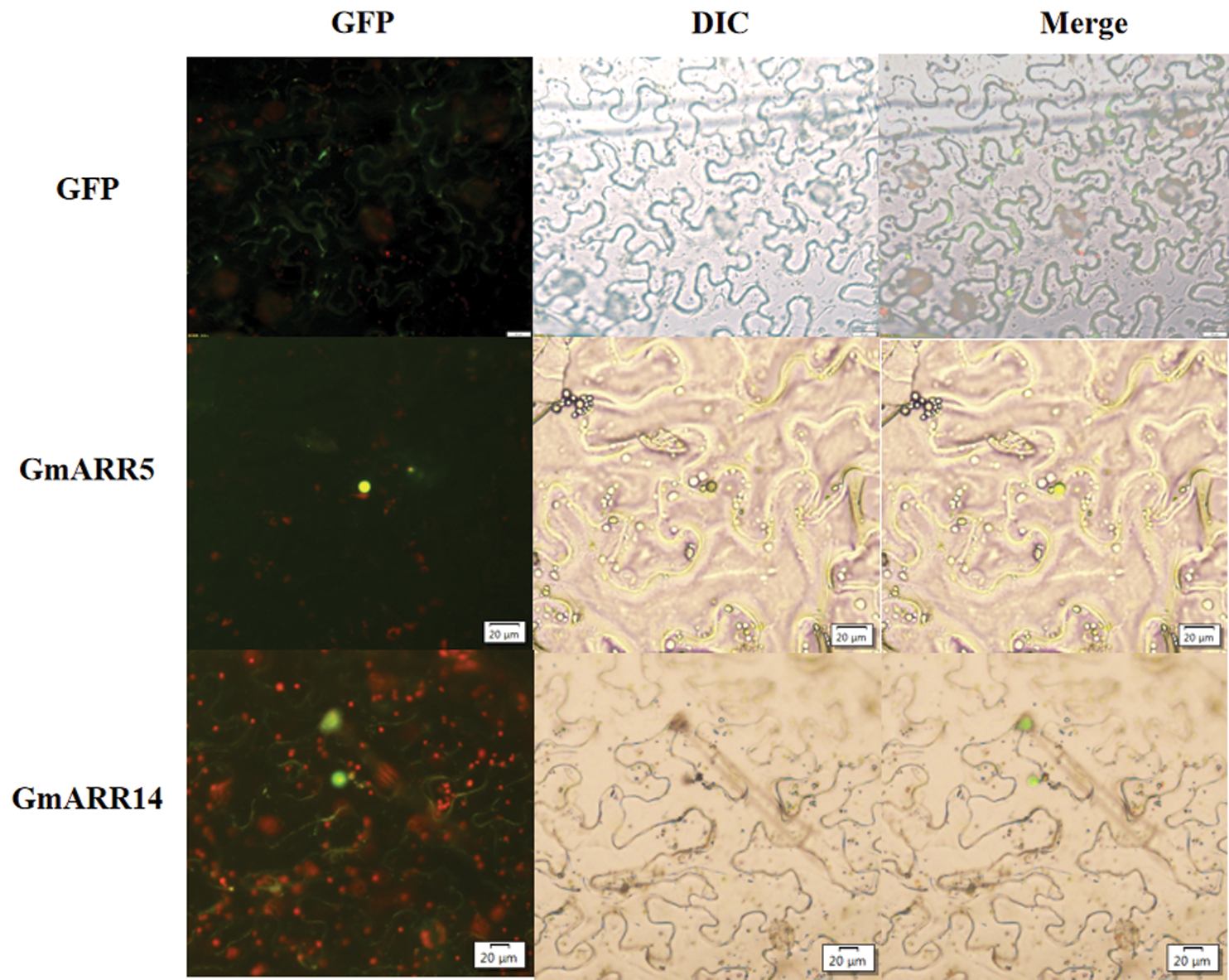
Figure 9: Subcellular localization of GmARR-Bs. GmARR-Bs-GFP fusion constructs were utilized to setting-out the subcellular localization of GmARR-B 5 and GmARR-B 14. Images of GFP were captured with fluorescence microscope and are shown in green. Scale bars, 20 μm).
Soybean is a major leguminous trim; it is of extraordinary centrality of breeding high-yield varieties of this important crop in our changing environment. In order to adapt to the changing environment, a full understanding of the key regulators of stress resistance is essential. As important components of cytokinin signaling pathways, the type-B ARR gene family has been appeared to play imperative parts in plant reaction to different environment stresses (Nakamichi et al., 2009). This paper has systematically analyzed the type-B ARR gene family in soybean, assigned the GmARR-B genes family. The results revealed that theGmARR-B genes family consisted of 31 gene members in soybean, which was largely more than those identified in other species, such as Arabidopsis (12) (Ramírez-Carvajal et al., 2008), rice (13) (Schaller et al., 2007), pear (23) (Ni et al., 2017), and peach (11) (Zeng et al., 2017). In previous studies on TCS system of soybean, 15 type-B response regulatory factors were also found, which were inconsistent with the results of this experiment. This difference may be due to the following reasons: In the process of identification of GmARR-B family members, the former scholars focused on TCS system, and performed similarity search among known.
TCS members’ protein sequence data sets. In this study, we used Arabidopsis ARR-B protein sequence to query soybean proteome data set (Keiichi et al., 2010). Through phylogenetic analysis, we also found that the GmARR-B genes family could be classified into three clades within the neighbor-joining phylogenetic tree (Fig. 1). The results were consistent with B-ARR gene family in Arabidopsis thaliana.
The type-B ARR gene family is an old gene family with developmental inertia. As the ARR genes identified with Arabidopsis (AtARR) and rice (OsRR), in addition to a highly conserved phospho-accepting REC domain, type-B ARR gene family in soybean contained the B motifs (a phosphorylated Asp residue at the center and a short stretch of conserved amino acids) (Hosoda et al., 2002), which was consistent with previous studies in A. thaliana. Genetic structure analysis showed that the exon distribution and location differed across genes in the soybean type-B ARR genes family, but a much higher conservation of motifs (Fig. 2). We also discovered that the expression of GmARR-Bs was conserved in the clade, conserved domain and gene structures between homologous genes. Gene duplication could be a common wonder in eukaryotes, and contributes to biological diversity during evolution (Magadum et al., 2013; Taylor and Raes, 2004). To explore type-B GmARR gene duplication occasions, the location of ARR-B genes within the soybean genome was determined (Fig. 7), and homologous genes of soybean and A. thaliana was compared (Fig. 3). Co-expression analysis, which was a pointer of functional correlation between genes, recommended that a majority of the GmARR-B genes remained functionally correlated, in spite to their sequence divergence. Although it is not uncommon to know that there is functional redundancy in ARR genes of A. thaliana (Nguyen et al., 2016). There were 10 GmARR-Bs (GmARR-B 1, 2, 4–8, 11, 14, 17) showed collinearity with interior of soybean and 7 GmARR-Bs (GmARR-B 5, 9, 11, 14, 15–17) showed collinearity with the ARRs of Arabidopsis (dicot). To sum up, type-B ARR genes might have arisen from a common ancestor, these patterns are almost similar with other species, suggesting that these genes occurred in a common ancestor before Chlamydomonas and exhibited partial or complete divergence from one another between the ARR-B gene family (Pickett and Meeks-Wagner, 1995; Zhang et al., 2020a; Yu et al., 2012).
The subcellular distribution of protein is closely related to its function, which is one of the important characteristics of a protein. The accuracy of distinguishing different proteins by the major subcellular localization of proteins is over 90%. Previous studies suggest that the nucleus is integral in the coordination of various cellular activities, some of which geared at improving the performance under abiotic stress conditions (Hosoda et al., 2002). In this study, we identified 31 genes encoding putative nuclear type-B ARRs in the soybean genome (Table 1), similar with Arabidopsis, nuclear ARRs constitute the largest fraction of the ARR-B gene family in soybean (Tajima et al., 2004). Meanwhile, we transiently expressed GmARR-B 5 and GmARR-B 14 genes with GFP fusion protein in tobacco leave to further to verify this prediction. Green fluorescence signals of both genes fusion proteins were localized in the nucleus (Fig. 9), consistent with the predicted localization using bioinformatics programs, which suggested to function in cellular regulation in relation to stress tolerance. Furthermore, we also identified various types of conserved cis-regulatory elements in the promoter regions of GmARR-Bs to further clarify the roles in response to abiotic stresses.
Cis-acting element analysis of soybean type-B ARR genes showed that these genes were closely related to growth, hormone signal transduction and abiotic stress resistance. The expression pattern of GmARR-B genes in different tissues (Fig. 5) showed that all these members largely grouped into three Groups (A–C) based on the hierarchical clustering analysis. The distinct GmARR-B genes groups exhibited high variation in their expression profiles, dependent on tissue/organ types and developmental stages. It show that ARR-B transcription factors may play diverse roles in the growth and developmental regulation of soybean. Nguyen et al. (2016) reported that the promoter regions of ARR-B are rich in cis-element involved in CTK and drought resistance response, and an Arabidopsis triple loss-of-function mutant of ARR 1, ARR 10, and ARR 12 exhibited reduced stature, enhanced seed size, altered chlorophyll and anthocyanin content, and an aborted primary root with protoxylem but no metaxylem. It further indicated that GmARR-B family was related to the growth and development of plants. In addition, in Arabidopsis, the result of transient assay demonstrated that the ARR 1, ARR 2, and ARR 10 of type-B ARRs were not only involved in cytokinin signaling, but also regulated the mRNA expression of type-A ARRs (Argyros et al., 2008), and in Chinese cabbage, the promoter regions of BrRR 3 and BrRR 17 are rich in cis-element involved in ABA response, BrRR 3 and BrRR 17 strongly and instantly suppressed by ABA treatment (Liu et al., 2014), possibly related to various hormones-related cis-elements in its promoter region (Zhang et al., 2016; Zhang and Huang, 2010) (i.e., ABA, MeJA, IAA, and SA) (Fig. 4). The presence of these cis-regulatory elements including ABRE, MeJA, SA implicates GmARR-B genes in regulating various hormonal signaling pathways related to stress response. This result is consistent with those of the type-B ARR genes obtained in other plant species. What’s more interesting is that the presence of various abiotic stress-specific cis-regulatory elements, including LTR, MYB and MBS, in the promoter region of identified GmARR-Bs could suggest that these genes were involved in stresses tolerance of soybean (Fig. 4). ARR 1, ARR 10, and ARR 12 negatively regulated plant responses to drought, and ARR 8 and ARR 9, as negative regulators, were also involved in plant responses to water stress stimulated by sorbitol (Wohlbach et al., 2008). Low temperature inhibits root growth by reducing auxin accumulation via ARR 1/12 (Zhu et al., 2015; Shi et al., 2012), while overexpression of ARR 5, ARR 7, and ARR 15 enhanced the freezing tolerance of plants (Kieber and Schaller, 2014). In order to further confirm the function of type-B GmARRs, we analyzed 10 ARR-B genes distributed in different parts of cells under different abiotic stresses with qRT analysis, containing ABA, cold, heat, salt and alkali stress. As shown in Fig. 8, all 10 GmARR-Bs responded to the five stress conditions, but the response speeds and intensities were different. The expression of GmARR-Bs reached the peak at 48 h after ABA and cold stress treatment. While the expression of GmARR-Bs was significantly up-regulated at 24 h after cold, heat and salt stress. Genes expression levels coped with different treatments, especially GmARR 5 and GmARR 14, were significantly induced. Our results showed that type-B ARRs might have a positive effect on abiotic stresses tolerance. These results were consistent with the result reported previously about the type-B ARRs involved in environmental stress response of crops such as Arabidopsis and Chinese cabbage.
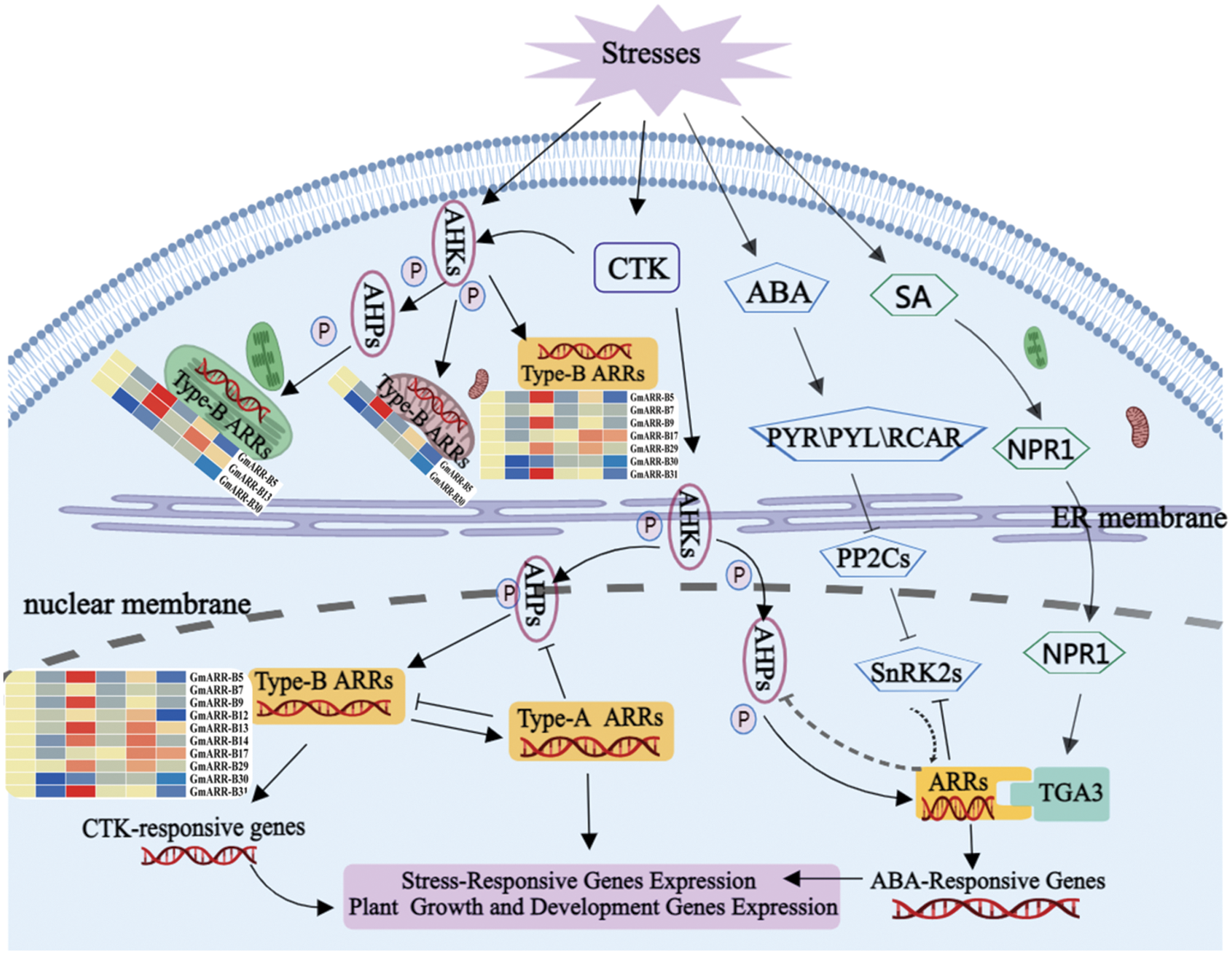
Figure 10: Proposed model for response regulators responding to stress and hormone signals through different regulatory pathways. (Abiotic stressors change phytohormone synthesis and distribution, which then mediates stress responses via hormone signaling components and type-B ARRs. The activation and repression effects are indicated by arrows and bar ends, respectively. MedPeer (http://www.medpeer.cn/) was used to produce this image).
Finally, based on prior studies and the evidence we gathered in this study, we propose a model of ARR response to stresses, depicting a responsive transcriptional network (Fig. 10). Plants have developed their own self-regulation mechanism through physiological and metabolic reactions to deal with different environmental stresses. Extensive studies on the type-B ARR gene family members, in vivo or in vitro, have recommended to be involved in many biological processes, counting cytokinin signaling, plant growth, and stress responses through a chaperone or by isomerization of proline residues during protein folding. Previous studies have shown that cytokinin signaling pathway plays a significant role in plant growth and development (Kieber and Schaller, 2018). Additional recent studies have revealed the role of cytokinins and the cytokinin TCS in abiotic stress responses, including cold and drought stress. As shown in Fig. 10, on the one hand, when plants are exposed to abiotic stresses, AHKs, as stress receptor in cytoplasm, will be stimulated and regulate GmARR-B 5, 13, 30 predicted in cytoplasm and mitochondria, and GmARR-B 5, 7, 9, 17, 29, 30, 31 predicted in chloroplast through phosphorylation. The expression of GmARR-B 5, 9, 17, 29, 31 filled in cytoplasm, was significantly up-regulated under abiotic stress, and the expression of 5 in chloroplast and mitochondria was also significantly increased, especially under low temperature stress. These genes were significantly up-regulated under cold and salt stress to improve stress resistance. On the other hand, type-B ARRs expression will be affected in some degrees by CTK, ABA and SA signaling. Abiotic stress stimulates cytokinin synthesis by binding to the AHK 2/AHK 3/AHK 4 cytokinin receptors within the ER lumen, activating the transmitter domain, which autophosphorylates on a His residue (indicated by p). The phosphate is subsequently transferred to an AHP protein, which travels between the cytoplasm and the nucleus, the mitochondrion, and the chloroplast. The AHPs transfer phosphate to type-B ARRs in the nucleus, and abiotic stress, particularly cold and salt stress, dramatically up-regulated the expression of type-B ARR genes such as GmARR-B 5, 9, 13, 14, and 31, which controlled the expression of numerous target genes, including the type-A ARRs. Plant growth and development related genes, cytokinin response genes and abiotic stress related genes. Then, the type-A ARRs, which are phosphorylated by the AHPs, then feedback to block cytokinin signaling (indicated by ⊥) (Kieber and Schaller, 2014; Xie et al., 2019; Nishiyama et al., 2011). Meanwhile, SnRK2s interacted with ARR in the nucleus, revealing the ABA and CK signaling pathways’ cross-talk. By interacting with type-B ARRs gene in the nucleus, TGA in the SA pathway controls soybean growth and abiotic stress response (Hass et al., 2014). This work adds to the growing body of data suggesting the GmARR-B gene family is involved in plant abiotic stress response.
In this study, 31 type-B ARR genes were identified with soybean Phylogenetic analysis. Gene and protein structure analysis showed that 31 B-type GmARRs could be divided into three subfamilies and mainly distributed in the nuclear region. Exon/intron and motif analysis showed that GmARR-B genes structures and conserved motifs were similar within group but diverse between groups. It was further confirmed that the expansion of type-B GmARR gene family might mainly come from chromosome replication. Expression profiling revealed variable regulation models of GmARR-B genes in different parts of tissues, and the possible transcriptional regulatory networks of GmARR-Bs were further explored. The qRT-PCR analysis revealed that all 10 GmARR-Bs responded to five stresses, although the response speeds and intensities were different, especially for GmARR 5 and GmARR 14, which were significantly induced by cold stress, due to the presence of LTR, ABRE and other stress response elements in the promoter region of the gene family. These findings not only contribute to a better understanding of the Glycine max ARR-B gene family’s complicated regulation, but also give useful information for future soybean functional genomics research.
Acknowledgement: We would like to thank the reviewers and editors for careful reading and helpful comments on this manuscript.
Availability of Data and Materials: All data generated or analysed during this study are included in this published article.
Author Contribution: Conceptualization, GY, YZ; methodology, HL, RC; software, JL; formal analysis, HL; investigation, RC, HL and XJ; resources, YZ; data curation, QC, FC, CR and ZC; writing-original draft preparation, HL; writing-review and editing, YZ; funding acquisition, YZ. All authors have read and agreed to the published version of the manuscript.
Funding Statement: This work was financially supported by the National Modern Agricultural Industrial Technology System (CARS-04-01A).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Abdelghany AM, Zhang S, Azam M, Abd S, Yue SB et al. (2020). Natural variation in fatty acid composition of diverse world soybean germplasms grown in China. Agronomy 10: 24. [Google Scholar]
Akbudak MA, Filiz E (2020). Genome-wide investigation of proline transporter (ProT) gene family in tomato: Bioinformatics and expression analyses in response to drought stress. Plant Physiology and Biochemistry 157: 13–22. [Google Scholar]
Argyros RD, Mathews DE, Chiang YH, Palmer CM, Thibault DM et al. (2008). Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell 20: 2102–2116. [Google Scholar]
Bailey TL, Boden M, Buske FA, Frith M, Grant CE et al. (2009). MEMESUITE: Tools for motif discovery and searching. Nucleic Acids Research 37: W202–208. [Google Scholar]
Birkenbihl RP, Jach G, Saedler H, Huijser P (2005). Functional dissection of the plant-specific SBP-domain: Overlap of the DNA-binding and nuclear localization domains. Journal of Molecular Biology 352: 585–596. [Google Scholar]
Buechel S, Leibfried A, To JP, Zhao Z, Andersen SU et al. (2010). Role of A-type ARABIDOPSIS RESPONSE REGULATORS in meristem maintenance and regeneration. European Journal of Cell Biology 89: 279–284. [Google Scholar]
Carella P, Wilson DC, Cameron RK (2015). Some things get better with age: Differences in salicylic acid accumulation and defense signaling in young and mature Arabidopsis. Frontiers in Plant Science 5: 775. [Google Scholar]
Chen C, Chen H, Zhang Y, Thomas HR, Frank MH et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Molecular Plant 13: 1194–1202. [Google Scholar]
Chen F, Yang Y, Luo X, Zhou W, Dai Y et al. (2019). Genome-wide identification of GRF transcription factors in soybean and expression analysis of GmGRF family under shade stress. BMC Plant Biology 19: 269. [Google Scholar]
D’Agostino IB (2000). Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiology 124: 1706–1717. [Google Scholar]
Debbie W, Ben V, Hardeep N, Ammar R, Wilson GV et al. (2007). An electronic fluorescent pictograph browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. DOI 10.1371/journal.pone.0000718. [Google Scholar] [CrossRef]
Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD et al. (2012). Phytozome: A comparative platform for green plant genomics. Nucleic Acids Research 40: D1178–1186. [Google Scholar]
Guo AY, Zhu QH, Chen X, Luo JC (2007). GSDS: A gene structure display server. Hereditas 29: 1023–1026. [Google Scholar]
Hass C, Lohrmann J, Albrecht V, Sweere U, Hummel F et al. (2014). The response regulator 2 mediates ethylene signalling and hormone signal integration in Arabidopsis. EMBO Journal 23: 3290–3302. DOI 10.1038/sj.emboj.7600337. [Google Scholar] [CrossRef]
Hina A, Singh MB, Bhalla PL (2018). Genomic and molecular analysis of conserved and unique features of soybean PIF4. Scientific Reports 8: 12569. DOI 10.1038/s41598-018-30043-2. [Google Scholar] [CrossRef]
Horák J, Grefen C, Berendzen KW, Hahn A, Stierhof YD et al. (2008). The Arabidopsis thaliana response regulator ARR22 is a putative AHP phospho-histidine phosphatase expressed in the chalaza of developing seeds. BMC Plant Biology 8: 77. [Google Scholar]
Hosoda K, Imamura A, Katoh E, Hatta T, Tachiki M et al. (2002). Molecular structure of the GARP family of plant Myb-related DNA binding motifs of the Arabidopsis response regulators. Plant Cell 14: 2015–2029. [Google Scholar]
Huang X, Song X, Chen R, Zhang B, Li C et al. (2019). Genome-wide analysis of the DREB subfamily in Saccharum spontaneum reveals their functional divergence during cold and drought stresses. Frontiers in Genetics 10: 1326. [Google Scholar]
Hutchison C (2002). Cytokinin signaling in Arabidopsis. Plant Cell 14: S47–59. [Google Scholar]
Hwang I, Chen HC, Sheen J (2002). Two-component signal transduction pathways in Arabidopsis. Plant Physiology 129: 500–515. [Google Scholar]
Hwang OJ, Back K (2019). Melatonin deficiency confers tolerance to multiple abiotic stresses in rice via decreased brassinosteroid levels. International Journal of Molecular Sciences 20: 5173. [Google Scholar]
Ishida K, Yamashino T, Yokoyama A, Mizuno T (2008). Three type-B response regulators, ARR1, ARR10 and ARR12, play essential but redundant roles in cytokinin signal transduction throughout the life cycle of Arabidopsis thaliana. Plant and Cell Physiology 49: 47–57. [Google Scholar]
Ivica L, Tobias D, Peer B (2015). SMART: Recent updates, new developments and status in 2015. Nucleic Acids Research 43: 257–260. DOI 10.1093/nar/gku949. [Google Scholar] [CrossRef]
Jain M, Tyagi AK, Khurana JP (2006). Molecular characterization and differential expression of cytokinin-responsive type-A response regulators in rice (Oryza sativa). BMC Plant Biology 6: 1. [Google Scholar]
Jeon J, Kim J (2013). Arabidopsis response Regulator1 and Arabidopsis histidine phosphotransfer Protein2 (AHP2AHP3, and AHP5 function in cold signaling. Plant Physiology 161: 408–424. [Google Scholar]
Jeon J, Kim NY, Kim S, Kang NY, Novák O et al. (2010). A subset of cytokinin two-component signaling system plays a role in cold temperature stress response in Arabidopsis. Journal of Biological Chemistry 285: 23371–23386. [Google Scholar]
Kakimoto T (2003). Perception and signal transduction of cytokinins. Annual Review of Plant Biology 54: 605–627. [Google Scholar]
Kang NY, Chuloh C, Jungmook K (2013). Inducible expression of Arabidopsis Response Regulator 22 (ARR22a type-C ARR, in transgenic Arabidopsis enhances drought and freezing tolerance. PLoS One 8: e79248. [Google Scholar]
Keiichi M, Takuhiro Y, Tetsuya S, Yamaguchi-Shinozaki K, Shinozaki K et al. (2010). Genome-wide analysis of two-component systems and prediction of stress-responsive two-component system members in soybean. DNA Research 17: 303–324. DOI 10.1093/dnares/dsq021. [Google Scholar] [CrossRef]
Kidokoro S, Watanabe K, Ohori T, Moriwaki T, Maruyama K et al. (2014). Soybean DREB1/CBF-type transcription factors function in heat and drop as well as cold stress responsive gene expression. Plant Journal 81: 505–518. DOI 10.1111/tpj.12746. [Google Scholar] [CrossRef]
Kieber JJ, Schaller GE (2014). Cytokinins. Arabidopsis Book 12: e0168. DOI 10.1199/tab.0168. [Google Scholar] [CrossRef]
Kieber JJ, Schaller GE (2018). Cytokinin signaling in plant development. Development 145: 845. DOI 10.1242/dev.149344. [Google Scholar] [CrossRef]
Kim J, Jeon J (2013). Cold signaling via the two-component signaling system. Molecular Plant 6: 15–17. DOI 10.1093/mp/sss151. [Google Scholar] [CrossRef]
Lee TH, Tang H, Wang X, Paterson AH (2013). PGDD: A database of gene and genome duplication in plants. Nucleic Acids Research 41: D1152–D1158. DOI 10.1093/nar/gks1104. [Google Scholar] [CrossRef]
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research 30: 325–327. DOI 10.1093/nar/30.1.325. [Google Scholar] [CrossRef]
Li B, Zheng JC, Wang TT, Min DH, Wei WL et al. (2020). Expression analyses of soybean VOZ transcription factors and the role of GmVOZ1G in drought and salt stress tolerance. International Journal of Molecular Sciences 21: 2177. [Google Scholar]
Liu Z, Zhang M, Kong L, Lv Y, Zou M et al. (2014). Genome-wide identification, phylogeny, duplication, and expression analyses of two-component system genes in Chinese cabbage (Brassica rapa ssp. pekinensis). DNA Research 4: 379–396. [Google Scholar]
Livak KJ, Schmittgen TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25: 402–408. [Google Scholar]
Lohrmann J, Sweere U, Zabaleta E, Bäurle I, Keitel C et al. (2001). The response regulator ARR2: A pollen-specific transcription factor involved in the expression of nuclear genes for components of mitochondrial complex I in Arabidopsis. Molecular Genetics & Genomics 265: 2–13. [Google Scholar]
Luo M, Cai M, Zhang J, Li Y, Zhang R et al. (2017). Functional divergence and origin of the DAG-like gene family in plants. Scientific Reports 7: 5688. [Google Scholar]
Magadum S, Banerjee U, Murugan P, Gangapur D, Ravikesavan R (2013). Gene duplication as a major force in evolution. Journal of Genetics 92: 155–161. [Google Scholar]
Mason MG, Mathews DE, Argyros DA, Maxwell BB, Kieber JJ et al. (2005). Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell 17: 3007–3018. [Google Scholar]
Nakamichi N, Kusano M, Fukushima A, Kita M, Ito S et al. (2009). Transcript profiling of an Arabidopsis pseudo response regulator arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant and Cell Physiology 50: 447–462. DOI 10.1093/pcp/pcp004. [Google Scholar] [CrossRef]
Nguyen KH, Ha CV, Nishiyama R, Watanabe Y, Leyva-González MA et al. (2016). Arabidopsis type-B cytokinin response regulators ARR1, ARR10, and ARR12 negatively regulate plant responses to drought. Proceedings of the National Academy of Sciences of the United States of America 113: 3090–3095. [Google Scholar]
Ni J, Bai S, Gao L, Qian M, Zhong L et al. (2017). Identification, classification, and transcription profiles of the B-type response regulator family in pear. PLoS One 12: e0171523. [Google Scholar]
Nishiyama R, Watanabe Y, Fujita Y, Le DT, Kojima M et al. (2011). Analysis of cytokinin mutants and regulation of cytokinin metabolic genes reveals important regulatory roles of cytokinins in drought, salt and abscisic acid responses, and abscisic acid biosynthesis. Plant Cell 23: 2169–2183. [Google Scholar]
Panu A, Manohar J, Konstantin A, Baratin D, Csardi G et al. (2012). ExPASy: SIB bioinformatics resource portal. Nucleic Acids Research 40: W597–W603. DOI 10.1093/nar/gks400. [Google Scholar] [CrossRef]
Pickett FB, Meeks-Wagner DR (1995). Seeing double: Appreciating genetic redundancy. Plant Cell 7: 1347–1356. [Google Scholar]
Pils B, Heyl A (2009). Unraveling the evolution of cytokinin signaling. Plant Physiology 151: 782–791. [Google Scholar]
Ramírez-Carvajal G, Morse A, Davis J (2008). Transcript profiles of the cytokinin response regulator gene family in Populus imply diverse roles in plant development. New Phytologist 177: 77–89. [Google Scholar]
Safi A, Medici A, Szponarski W, Ruffel S, Lacombe B et al. (2017). The world according to GARP transcription factors. Current Opinion in Plant Biology 39: 159–167. [Google Scholar]
Sakai H, Aoyama T, Oka A (2010). Arabidopsis ARR1 and ARR2 response regulators operate as transcriptional activators. Plant Journal 24: 703–711. DOI 10.1046/j.1365-313x.2000.00909.x. [Google Scholar] [CrossRef]
Sakai H, Honma T, Aoyama T, Sato S, Kato T et al. (2001). ARR1, a transcription factor for genes immediately responsive to cytokinins. Science 294: 1519–1521. [Google Scholar]
Schaller GE, Doi K, Hwang I, Kieber JJ, Khurana JP et al. (2007). Nomenclature for two-component signaling elements of rice. Plant Physiology 143: 555–557. [Google Scholar]
Schaller GE, Shiu SH, Armitage JP (2011). Two-component systems and their co-option for eukaryotic signal transduction. Current Biology 21: R320–R330. [Google Scholar]
Schaller G, Kieber J, Shiu S (2008). Two-component signaling elements and histidyl-aspartyl phosphorelays. Arabidopsis Book 6: e0112. [Google Scholar]
Shi Y, Tian S, Hou L, Huang X, Zhang X et al. (2012). Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type-A ARR genes in Arabidopsis. Plant Cell 24: 2578–2595. [Google Scholar]
Shu Y, Tao Y, Wang S, Huang L, Yu X et al. (2015). GmSBH1, a homeobox transcription factor gene, relates to growth and development and involves in response to high temperature and humidity stress in soybean. Plant Cell Reports 34: 1927–1937. [Google Scholar]
Tajima Y, Imamura A, Kiba T, Amano Y, Yamashino T et al. (2004). Comparative studies on the type-B response regulators revealing their distinctive properties in the His-to-Asp phosphorelay signal transduction of Arabidopsis thaliana. Plant & Cell Physiology 45: 28–39. [Google Scholar]
Takatoshi K, Koh A, Hitoshi S, Mizuno T (2004). Arabidopsis Response Regulator, ARR22, ectopic expression of which results in phenotypes similar to the wol cytokinin-receptor mutant. Plant & Cell Physiology 45: 1063–1077. DOI 10.1093/pcp/pch128. [Google Scholar] [CrossRef]
Taylor JS, Raes J (2004). Duplication and divergence: The evolution of new genes and old ideas. Annual Review of Genetics 38: 615–643. [Google Scholar]
Tolosa LN, Zhang Z (2020). The role of major transcription factors in solanaceous food crops under different stress conditions: Current and future perspectives. Plants 9: 56. [Google Scholar]
Wallmeroth N, Jeschke D, Slane D, Nägele J, Veerabagu M et al. (2019). ARR22 overexpression can suppress plant two-component regulatory systems. PLoS One 14: e0212056. [Google Scholar]
Werner T, Schmülling T (2009). Cytokinin action in plant development. Current Opinion in Plant Biology 12: 527–538. [Google Scholar]
Wohlbach DJ, Quirino BF, Sussman MR (2008). Analysis of the Arabidopsis histidine kinase ATHK 1 reveals a connection between vegetative osmotic stress sensing and seed maturation. Plant Cell 20: 1101–1117. [Google Scholar]
Xie Z, Nolan TM, Jiang H, Yin Y (2019). AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis. Frontiers in Plant Science 10: 228. [Google Scholar]
Yokoyama A, Yamashino T, Amano Y, Tajima Y, Imamura A et al. (2007). Type-B ARR transcription factors, ARR10 and ARR12, are implicated in cytokinin-mediated regulation of protoxylem differentiation in roots of Arabidopsis thaliana. Plant & Cell Physiology 48: 84–96. [Google Scholar]
Yu H, Kong X, Huang H, Wu W, Park J et al. (2020). STCH4/REIL2 confers cold stress tolerance in Arabidopsis by promoting rRNA processing and CBF protein translation. Cell Reports 30: 229–242.e5. [Google Scholar]
Yu Y, Wang N, Hu R, Xiang F (2016). Genome-wide identification of soybean WRKY transcription factors in response to salt stress. Springerplus 5: 920. [Google Scholar]
Yu Y, Zhang H, Li W, Mu C, Zhang F et al. (2012). Genome-wide analysis and environmental response profiling of the FK506-binding protein gene family in maize (Zea mays L.). Gene 498: 212–222. [Google Scholar]
Zeng J, Zhu X, Haider M, Wang X, Zhang C et al. (2017). Genome-wide identification and analysis of the type-B authentic response regulator gene family in peach (Prunus persica). Cytogenetic and Genome Research 151: 41–49. [Google Scholar]
Zhang MP, Liu YH, Xu W, Smith CW, Murray SC et al. (2020a). Analysis of the genes controlling three quantitative traits in three diverse plant species reveals the molecular basis of quantitative traits. Scientific Reports 10: 10074. [Google Scholar]
Zhang M, Liu Y, Cai H, Guo M, Chai M et al. (2020b). The bZIP transcription factor GmbZIP15 negatively regulates salt- and drought-stress responses in soybean. International Journal of Molecular Sciences 21: 7778. [Google Scholar]
Zhang Y, Lan H, Shao Q, Wang R, Chen H et al. (2016). An A20/AN1-type zinc finger protein modulates gibberellins and abscisic acid contents and increases sensitivity to abiotic stress in rice (Oryza sativa). Journal of Experimental Botany 67: 315–326. [Google Scholar]
Zhang Z, Huang R (2010). Enhanced tolerance to freezing in tobacco and tomato overexpressing transcription factor TERF2/LeERF2 is modulated by ethylene biosynthesis. Plant Molecular Biology 73: 241–249. [Google Scholar]
Zhu J, Zhang KX, Wang WS, Gong W, Liu WC et al. (2015). Low temperature inhibits root growth by reducing auxin accumulation via ARR1/12. Plant and Cell Physiology 56: 727–736. [Google Scholar]
Supplementary Materials
Supplementary Figure 1 Sequence alignment of GmARR-Bs.
Supplementary Figure 2 Prediction of secondary structure of GmARR-Bs gene family proteins.
Supplementary Figure 3 Chromosome location of GmARR-Bs.
Supplementary Table 1 The RNA sequencing reads of GmARR-B genes in the tissues and organs.
Supplementary Table 2 The qRT-PCR primers.
Supplementary Table 3 Details and length of 10 conserved motifs.
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |