DOI:10.32604/biocell.2022.019055

| BIOCELL DOI:10.32604/biocell.2022.019055 |  |
| viewpoint |
From organ-on-a-chip towards body-on-a-chip
Hongik University, Seoul, 04066, Korea
*Address correspondence to: Jong Hwan Sung, jhsung22@hongik.ac.kr
Received: 31 August 2021; Accepted: 13 December 2021
Abstract: Organ-on-a-chip technology aims to reproduce the key physiological features of human organs and tissues, even complex actions of multi-organ interaction. While organ-on-a-chips at single-organ level has made notable achievement during the last decade, multi-organ-on-a-chips, which manifests unique advantages, has started gathering attention only recently. In this viewpoint, we discuss the current status of organ-on-a-chip technology, with a specific emphasis on multi-organ-on-a-chip. Key technological advances contributing to the maturation of the field, and challenges that need to be addressed before wider adoption in relevant fields are discussed. We will share our perspectives on how the multi-organ-on-a-chip can improve the drug development process.
Keywords: Multi-organ-on-a-chip; Disease models; Drug development; In vitro model system
The difficulties associated with drug development (Scannell et al., 2012) mainly comes from the fact that the model systems used in the process show limited physiological relevance to humans. Cell-based in vitro systems can partially reflects cellular physiology, often decaying rapidly after separation from the body (Freires et al., 2017), and animal to human extrapolation is often inaccurate (Seok et al., 2013; Pound and Ritskes-Hoitinga, 2018).
As suggested by recent review articles, organ-on-a-chip technology is considered as a promising solution to this problem (Ingber, 2020; Jalili-Firoozinezhad et al., 2021). Organ-on-a-chips, also known as tissue chips or microphysiological systems (MPS), offer advantages over conventional cell-based in vitro models. The precise control over three-dimensional geometries, flow of fluids, and transport of molecules in microscales allows recreation of the tissue microenvironment. It allows manipulation of experimental conditions, enabling hypothesis-driven research. For example, the effect of fluid flow on cell physiology can be studied by applying flow with controlled flow rates (Chi et al., 2015; Lee et al., 2019). The most prominent advantage is that it can connect multiple organs and allow communication between them. So-called multi-organ-on-a-chip, also often termed body-on-a-chip or human-on-a-chip, enables realization of complex and dynamic interactions between multiple organs, analogous to what happens in the human body (Lee and Sung, 2018a; Sung et al., 2019b; Sung, 2021).
The conception of organ-on-a-chip technology came with the development of soft lithography, which uses elastic silicone materials, polydimethylsiloxane (PDMS), to create microscale devices (McDonald et al., 2000) in the field of microfluidics, which uses precise handling of microscale liquids. The application of microfluidics in biological context was demonstrated in early 2000s (Allen and Bhatia, 2003; Sia and Whitesides, 2003).
The concept of multi-organ-on-a-chip was conceived in its preliminary form as macroscale bioreactors fluidically connected using pumps to transfer cell culture media between different organ modules (Shuler et al., 1996). Termed by the authors as a ‘cell culture analog’, this system was used to probe the metabolism-dependent toxicity of naphthalene toxicology (Sweeney et al., 1995). With the advance of microfluidic technology, the cell culture analog became microscale, fabricated on a silicon wafer and connected via microfluidic channels (Sin et al., 2004; Viravaidya et al., 2004). These proof-of-concept studies proved that organ-organ interaction can be reproduced in vitro (Sung and Shuler, 2009; Tatosian and Shuler, 2009; Sung et al., 2010).
Advancing from these studies, a seminal paper was published in 2010, where recapitulation of organ-level functionality of the lung by creating alveolar-capillary interface and applying cyclic stretching was demonstrated, to study pathological mechanism underlying pulmonary diseases (Huh et al., 2010). Collaborative multi-year research funding effort from multiple institutes such as US Food and Drug Administration (FDA), National Institutes of Health (NIH), and Defense Advanced Research Projects Agency (DARPA), was initiated in US, and similar funding effort was initiated in Europe as well (Low et al., 2021). Recent research works are aimed towards building multi-organ systems for mimicking complex pathophysiological processes in the body (Miller and Shuler, 2016).
In addition to the funding effort tin the public sector, recent effort in commercialization in the industrial sector is also noteworthy (Ewart et al., 2017; Ribas et al., 2018; Low et al., 2021). Many US and Eurpose-based companies, including Mimetas, TissUse, CNBio, Emulate, and Hesperos are collaborating with major pharmaceutical companies seeking ways to incorporate organ-on-a-chip technologies in drug development process.
Key Technologies of Organ-on-a-Chip
Early works on organ-on-a-chip focused on development of microfluidic devices or fabrication methods for recreating the tissue microenvironment. For example, a novel method for fabricating silicon substrate or soft hydrogel into 3D villi structure of intestinal epithelium was developed to culture gut epithelial cell line in 3D shape (Sung et al., 2011; Esch et al., 2012). Cyclic stretching of a porous membrane structure mimicking the alveolar-capillary interface using pneumatic vacuum control enabled realization of breathing motion (Huh et al., 2010) and peristaltic movement of the intestine (Kim et al., 2012). Control of convection and diffusion process at microscale enabled spontaneous formation of perfusable, vascular structure within extracellular matrix (ECM) (Kim et al., 2013).
One of the most important technological advances required for organ-on-a-chip systems are improving the physiological relevance of the system, by incorporating stem cells, primary cells, or even organoids (Kasendra et al., 2018). Some obstacles need to be overcome, such as the high cost of cells and media, and insufficient supply with inconsistent quality of cell phenotypes. The use of organoids can be limited, since organoids are self-organized multicellular 3D tissue models and it is difficult to create barrier structures commonly seen in the epithelial tissue of the intestine, skin, and alveolar.
3D bioprinting is another area that has made significant contribution to organ-on-a-chip field. Early organ-on-a-chip works relied heavily on photolithography-based fabrication methods, which is useful for fabricating inorganic materials, but inadequate for mimicking the extracellular matrix (ECM) environment of in vivo tissues. The ability to fabricate natural and synthetic hydrogels in microscale structures has offered great opportunities for organ-on-a-chip fields (Zhang et al., 2016).
Real-time, noninvasive detection in organ-on-a-chip devices is important, and various optical and electrochemical sensing methodologies have been integrated with organ-on-a-chip systems (Choi et al., 2016). Thorough validation with conventional detection methods and standardization efforts may be needed. Acquisition of a large set of data will also allow a deeper insight in the interpretation of the experimental results, particularly when combined with artificial intelligence technology (de Chiara et al., 2021).
Organ-on-a-chips offer the possibilities that conventional in vitro methods cannot. Multi-organ-on-a-chips enable simulation of dynamic, complex interaction of multiple organs, which is impossible to achieve with conventional static systems, as illustrated in Fig. 1 (Lee and Sung, 2018a; Sung, 2021). Early proof-of-concept studies demonstrated that pharmacokinetic-pharmacodynamic (PK-PD) profiles of drugs can be simulated (Sung et al., 2010). A suitable mathematical framework may be necessary to interpret the chip data and extrapolation to humans (Sung et al., 2018; Sung et al., 2019a). More recent studies have demonstrated the practical ability of multi-organ-on-a-chips to predict the PK profiles of drugs (Maass et al., 2017; Tsamandouras et al., 2017; Edington et al., 2018; Herland et al., 2020).
The multi-organ-on-a-chip can be a powerful in vitro platform for modeling complex diseases. Many diseases, including obesity, diabetes, metabolic syndromes, and immune-related diseases show complex mechanisms involving different organs, and in many cases conventional static in vitro systems are inadequate for modeling such diseases. Multi-organ-on-a-chips can be useful for screening therapeutics for such diseases, as well as probing the unknown mechanisms of disease progression. For example, non-alcoholic fatty liver disease (NALFD) manifests its symptoms with excessive fat accumulation in the liver, but treatment of NAFLD may be possible by improving the barrier integrity of the gut epithelium (Lee and Sung, 2018b; Jeon et al., 2021). Lastly, incorporation of patient-originated cells into the multi-organ-on-a-chip will lead to personalized humans-on-a-chip.
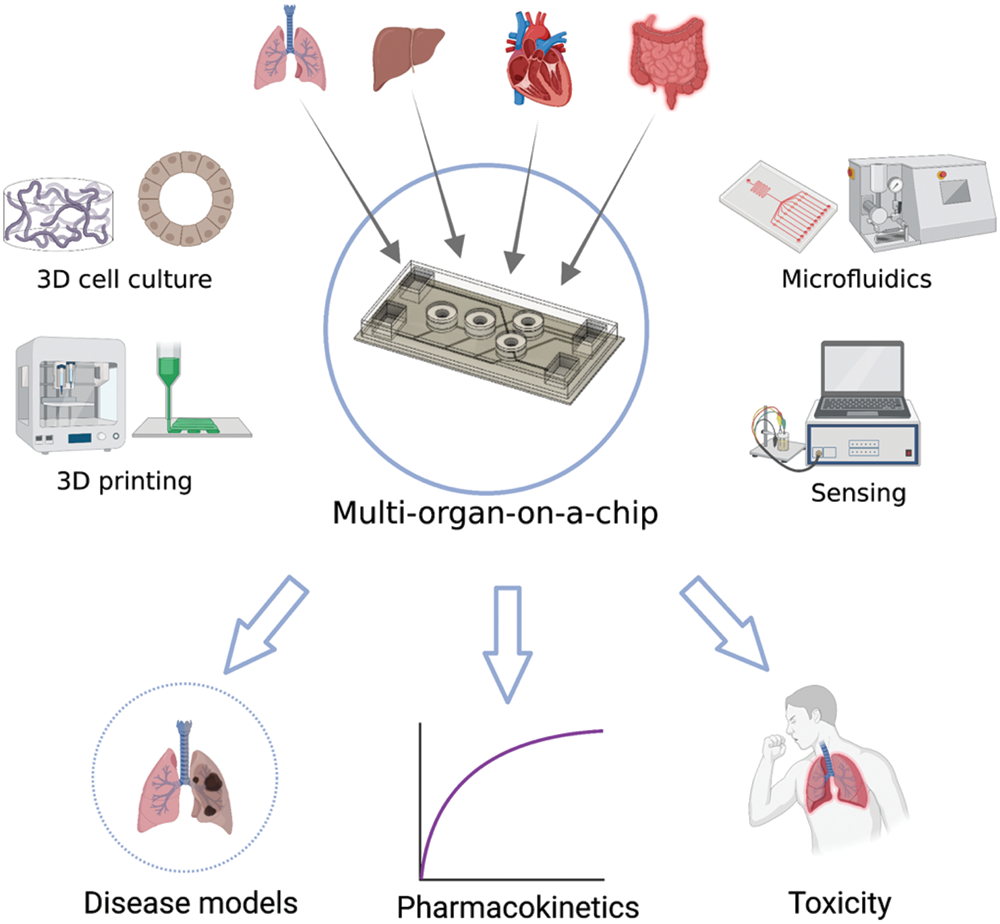
Figure 1: Schematics of the concept of multi-organ-on-a-chip with constituting technical components and potential applications. The image was created with https://BioRender.com.
Several challenges remain before wider adoption of this technology will be seen. Validation of physiological relevance has always been a major target that many researchers have been addressing. Recent progress seems promising, as evidences are accumulating that shows pathophysiological processes can be simulated (Benam et al., 2020; Tang et al., 2020), with increasing availability of stem cells, organoids, and primary cells. Improvement of chip fabrication material is necessary. While polydimethylsiloxane (PDMS) is ideal material with many advantages in lab scale, industrial applications will require material with easier mass production and reduced adsorption. Connecting multiple organ modules may require complex fluidic system for recirculation, where novel perfusion methods such as pneumatic pumping may be useful (Rebelo et al., 2016), or gravity-induced perfusion (Lee et al., 2017). This may offer additional advantages by eliminating the need for tubes, and consequently reducing the possibility of bubbles and dead volumes in the system.
As the multi-organ-on-a-chip systems grow in its complexity with increasing number of organs, the importance of mathematical framework for designing and interpreting the system will also increase. One aspect that needs special attention is how to ‘scale’ different organs to reflect how the human body works. As allometric scaling law suggests, organisms with different masses have different organ masses, as well as other physiological parameters (West et al., 1997). Since the multi-organ-on-a-chip is an extremely miniaturized version of the human body in terms of its size, direct application of allometric scaling law may not be sufficient and rigorous mathematical approach for chip design may be needed (Abaci and Shuler, 2015). At least for the early phase of multi-organ-on-a-chip technology, it will be important to accurately define which physiological functionalities need to be mimicked. For example, the intestine chip may be needed to model the absorption of drugs, or inflammation in response to external stimulation, or in vitro milieu for commensal bacteria. Depending on which aspect is being focused on, the optimal ratio of the intestine relative to organs, for example the liver may be different (Lee et al., 2017; Maass et al., 2017). Despite these challenges remaining, the future multi-organ-on-a-chip technology looks very promising, and recent progress in commercialization has been impressive with major pharmaceutical companies actively participating in the movement.
Authors’ Contribution: The author confirms sole responsibility for the following: study conception and design, data collection, analysis and interpretation of results, and manuscript preparation.
Funding Statement: This work was supported by National Research Foundation of Korea, under Grant (Basic Research Lab, 2019R1A4A1025958), and by the Ministry of Trade, Industry & Energy (MOTIE), Republic of Korea, under the Technology Innovation Program, Grant 20008414 (Development of intestine-liver-kidney multiorgan tissue chip mimicking absorption distribution metabolism excretion of drug), Hongik University Research Fund.
Conflicts of Interest: The author declares that they have no conflicts of interest to report regarding the present study.
Abaci HE, Shuler ML (2015). Human-on-a-chip design strategies and principles for physiologically based pharmacokinetics/pharmacodynamics modeling. Integrative Biology 7: 383–391. DOI 10.1039/C4IB00292J. [Google Scholar] [CrossRef]
Allen JW, Bhatia SN (2003). Formation of steady-state oxygen gradients in vitro: Application to liver zonation. Biotechnology and Bioengineering 82: 253–262. DOI 10.1002/(ISSN)1097-0290. [Google Scholar] [CrossRef]
Benam KH, Novak R, Ferrante TC, Choe Y, Ingber DE (2020). Biomimetic smoking robot for in vitro inhalation exposure compatible with microfluidic organ chips. Nature Protocols 15: 183–206. DOI 10.1038/s41596-019-0230-y. [Google Scholar] [CrossRef]
Chi M, Yi B, Oh S, Park DJ, Sung JH, Park S (2015). A microfluidic cell culture device (muFCCD) to culture epithelial cells with physiological and morphological properties that mimic those of the human intestine. Biomedical Microdevices 17: 9966. [Google Scholar]
Choi JR, Song H, Sung JH, Kim D, Kim K (2016). Microfluidic assay-based optical measurement techniques for cell analysis: A review of recent progress. Biosensors and Bioelectronics 77: 227–236. DOI 10.1016/j.bios.2015.07.068. [Google Scholar] [CrossRef]
de Chiara F, Ferret-Minana A, Ramon-Azcon J (2021). The synergy between organ-on-a-chip and artificial intelligence for the study of NAFLD: From basic science to clinical research. Biomedicines 9: 248. DOI 10.3390/biomedicines9030248. [Google Scholar] [CrossRef]
Edington CD, Chen WLK, Geishecker E, Kassis T, Soenksen LR et al. (2018). Interconnected microphysiological systems for quantitative biology and pharmacology studies. Scientific Reports 8: 4530. DOI 10.1038/s41598-018-22749-0. [Google Scholar] [CrossRef]
Esch MB, Sung JH, Yang J, Yu C, Yu J, March JC, Shuler ML (2012). On chip porous polymer membranes for integration of gastrointestinal tract epithelium with microfluidic ‘body-on-a-chip’ devices. Biomedical Microdevices 14: 895–906. DOI 10.1007/s10544-012-9669-0. [Google Scholar] [CrossRef]
Ewart L, Fabre K, Chakilam A, Dragan Y, Duignan DB et al. (2017). Navigating tissue chips from development to dissemination: A pharmaceutical industry perspective. Experimental Biology and Medicine 242: 1579–1585. DOI 10.1177/1535370217715441. [Google Scholar] [CrossRef]
Freires IA, Sardi JC, de Castro RD, Rosalen PL (2017). Alternative animal and non-animal models for drug discovery and development: Bonus or burden? Pharmaceutical Research 34: 681–686. DOI 10.1007/s11095-016-2069-z. [Google Scholar] [CrossRef]
Herland A, Maoz BM, Das D, Somayaji MR, Prantil-Baun R et al. (2020). Quantitative prediction of human pharmacokinetic responses to drugs via fluidically coupled vascularized organ chips. Nature Biomedical Engineering 4: 421–436. DOI 10.1038/s41551-019-0498-9. [Google Scholar] [CrossRef]
Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, Ingber DE (2010). Reconstituting organ-level lung functions on a chip. Science 328: 1662–1668. DOI 10.1126/science.1188302. [Google Scholar] [CrossRef]
Ingber DE (2020). Is it time for reviewer 3 to request human organ chip experiments instead of animal validation studies? Advanced Science 7: 2002030. DOI 10.1002/advs.202002030. [Google Scholar] [CrossRef]
Jalili-Firoozinezhad S, Miranda CC, Cabral JMS (2021). Modeling the human body on microfluidic chips. Trends in Biotechnology 39: 838–852. DOI 10.1016/j.tibtech.2021.01.004. [Google Scholar] [CrossRef]
Jeon JW, Lee SH, Kim D, Sung JH (2021). In vitro hepatic steatosis model based on gut-liver-on-a-chip. Biotechnology Progress 37: e3121. [Google Scholar]
Kasendra M, Tovaglieri A, Sontheimer-Phelps A, Jalili-Firoozinezhad S, Bein A et al. (2018). Development of a primary human small intestine-on-a-chip using biopsy-derived organoids. Scientific Reports 8: 2871. DOI 10.1038/s41598-018-21201-7. [Google Scholar] [CrossRef]
Kim HJ, Huh D, Hamilton G, Ingber DE (2012). Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab on a Chip 12: 2165–2174. DOI 10.1039/c2lc40074j. [Google Scholar] [CrossRef]
Kim S, Lee H, Chung M, Jeon NL (2013). Engineering of functional, perfusable 3D microvascular networks on a chip. Lab on a Chip 13: 1489–1500. DOI 10.1039/c3lc41320a. [Google Scholar] [CrossRef]
Lee DW, Choi N, Sung JH (2019). A microfluidic chip with gravity-induced unidirectional flow for perfusion cell culture. Biotechnology Progress 35: e2701. DOI 10.1002/btpr.2701. [Google Scholar] [CrossRef]
Lee DW, Ha SK, Choi I, Sung JH (2017). 3D gut-liver chip with a PK model for prediction of first-pass metabolism. Biomedical Microdevices 19: 100. DOI 10.1007/s10544-017-0242-8. [Google Scholar] [CrossRef]
Lee SH, Sung JH (2018a). Organ-on-a-chip technology for reproducing multiorgan physiology. Advanced Healthcare Materials 7: 1700419. DOI 10.1002/adhm.201700419. [Google Scholar] [CrossRef]
Lee SY, Sung JH (2018b). Gut-liver on a chip toward an in vitro model of hepatic steatosis. Biotechnology and Bioengineering 115: 2817–2827. DOI 10.1002/bit.26793. [Google Scholar] [CrossRef]
Low LA, Mummery C, Berridge BR, Austin CP, Tagle DA (2021). Organs-on-chips: Into the next decade. Nature Reviews Drug Discovery 20: 345–361. DOI 10.1038/s41573-020-0079-3. [Google Scholar] [CrossRef]
Maass C, Stokes CL, Griffith LG, Cirit M (2017). Multi-functional scaling methodology for translational pharmacokinetic and pharmacodynamic applications using integrated microphysiological systems (MPS). Interative Biology 9: 290–302. DOI 10.1039/C6IB00243A. [Google Scholar] [CrossRef]
McDonald JC, Duffy DC, Anderson JR, Chiu DT, Wu H, Schueller OJ, Whitesides GM (2000). Fabrication of microfluidic systems in poly(dimethylsiloxane). Electrophoresis 21: 27–40. DOI 10.1002/(ISSN)1522-2683. [Google Scholar] [CrossRef]
Miller PG, Shuler ML (2016). Design and demonstration of a pumpless 14 compartment microphysiological system. Biotechnology and Bioengineering 113: 2213–2227. DOI 10.1002/bit.25989. [Google Scholar] [CrossRef]
Pound P, Ritskes-Hoitinga M (2018). Is it possible to overcome issues of external validity in preclinical animal research? Why most animal models are bound to fail. Journal of Translational Medicine 16: 304. DOI 10.1186/s12967-018-1678-1. [Google Scholar] [CrossRef]
Rebelo SP, Dehne EM, Brito C, Horland R, Alves PM, Marx U (2016). Validation of bioreactor and human-on-a-chip devices for chemical safety assessment. Advances in Experimental Medicine and Biology 856: 299–316. DOI 10.1007/978-3-319-33826-2. [Google Scholar] [CrossRef]
Ribas J, Pawlikowska J, Rouwkema J (2018). Microphysiological systems: Analysis of the current status, challenges and commercial future. Microphysiological Systems 2: 1. DOI 10.21037/mps.2018.10.01. [Google Scholar] [CrossRef]
Scannell JW, Blanckley A, Boldon H, Warrington B (2012). Diagnosing the decline in pharmaceutical R&D efficiency. Nature Reviews Drug Discovery 11: 191–200. DOI 10.1038/nrd3681. [Google Scholar] [CrossRef]
Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV et al. (2013). Genomic responses in mouse models poorly mimic human inflammatory diseases. Proceedings of the National Academy of Sciences of the United States of America 110: 3507–3512. DOI 10.1073/pnas.1222878110. [Google Scholar] [CrossRef]
Shuler ML, Ghanem A, Quick D, Wong MC, Miller P (1996). A self-regulating cell culture analog device to mimic animal and human toxicological responses. Biotechnology and Bioengineering 52: 45–60. DOI 10.1002/(ISSN)1097-0290. [Google Scholar] [CrossRef]
Sia SK, Whitesides GM (2003). Microfluidic devices fabricated in poly(dimethylsiloxane) for biological studies. Electrophoresis 24: 3563–3576. DOI 10.1002/(ISSN)1522-2683. [Google Scholar] [CrossRef]
Sin A, Chin KC, Jamil MF, Kostov Y, Rao G, Shuler ML (2004). The design and fabrication of three-chamber microscale cell culture analog devices with integrated dissolved oxygen sensors. Biotechnology Progress 20: 338–345. DOI 10.1021/(ISSN)1520-6033. [Google Scholar] [CrossRef]
Sung JH (2021). Multi-organ-on-a-chip for pharmacokinetics and toxicokinetic study of drugs. Expert Opinion on Drug Metabolism & Toxicology 17: 969–986. DOI 10.1080/17425255.2021.1908996. [Google Scholar] [CrossRef]
Sung JH, Kam C, Shuler ML (2010). A microfluidic device for a pharmacokinetic-pharmacodynamic (PK-PD) model on a chip. Lab on a Chip 10: 446–455. DOI 10.1039/b917763a. [Google Scholar] [CrossRef]
Sung JH, Shuler ML (2009). A micro cell culture analog (microCCA) with 3-D hydrogel culture of multiple cell lines to assess metabolism-dependent cytotoxicity of anti-cancer drugs. Lab on a Chip 9: 1385–1394. DOI 10.1039/b901377f. [Google Scholar] [CrossRef]
Sung JH, Wang Y, Shuler ML (2019a). Strategies for using mathematical modeling approaches to design and interpret multi-organ microphysiological systems (MPS). APL Bioengineering 3: 021501. DOI 10.1063/1.5097675. [Google Scholar] [CrossRef]
Sung JH, Wang YI, Kim JH, Lee JM, Shuler ML (2018). Application of chemical reaction engineering principles to ‘body-on-a-chip’ systems. AIChE Journal 64: 4351–4360. DOI 10.1002/aic.16448. [Google Scholar] [CrossRef]
Sung JH, Wang YI, Narasimhan Sriram N, Jackson M, Long C et al. (2019b). Recent advances in body-on-a-chip systems. Analytical Chemistry 91: 330–351. DOI 10.1021/acs.analchem.8b05293. [Google Scholar] [CrossRef]
Sung JH, Yu J, Luo D, Shuler ML, March JC (2011). Microscale 3-D hydrogel scaffold for biomimetic gastrointestinal (GI) tract model. Lab on a Chip 11: 389–392. DOI 10.1039/C0LC00273A. [Google Scholar] [CrossRef]
Sweeney LM, Shuler ML, Babish JG, Ghanem A (1995). A cell culture analogue of rodent physiology: Application to naphthalene toxicology. Toxicology in Vitro 9: 307–316. DOI 10.1016/0887-2333(95)00007-U. [Google Scholar] [CrossRef]
Tang H, Abouleila Y, Si L, Ortega-Prieto AM, Mummery CL, Ingber DE, Mashaghi A (2020). Human organs-on-chips for virology. Trends in Microbiology 28: 934–946. DOI 10.1016/j.tim.2020.06.005. [Google Scholar] [CrossRef]
Tatosian DA, Shuler ML (2009). A novel system for evaluation of drug mixtures for potential efficacy in treating multidrug resistant cancers. Biotechnology and Bioengineering 103: 187–198. DOI 10.1002/bit.22219. [Google Scholar] [CrossRef]
Tsamandouras N, Chen WLK, Edington CD, Stokes CL, Griffith LG, Cirit M (2017). Integrated gut and liver microphysiological systems for quantitative in vitro pharmacokinetic studies. AAPS Journal 19: 1499–1512. DOI 10.1208/s12248-017-0122-4. [Google Scholar] [CrossRef]
Viravaidya K, Sin A, Shuler ML (2004). Development of a microscale cell culture analog to probe naphthalene toxicity. Biotechnology Progress 20: 316–323. DOI 10.1021/(ISSN)1520-6033. [Google Scholar] [CrossRef]
West GB, Brown JH, Enquist BJ (1997). A general model for the origin of allometric scaling laws in biology. Science 276: 122–126. DOI 10.1126/science.276.5309.122. [Google Scholar] [CrossRef]
Zhang YS, Arneri A, Bersini S, Shin SR, Zhu K et al. (2016). Bioprinting 3D microfibrous scaffolds for engineering endothelialized myocardium and heart-on-a-chip. Biomaterials 110: 45–59. DOI 10.1016/j.biomaterials.2016.09.003. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |