DOI:10.32604/biocell.2022.017217

| BIOCELL DOI:10.32604/biocell.2022.017217 |  |
| Article |
Pseudogene HMGN2P46 as a microRNA sponge to regulate HMGN2 expression via competing for miR-590-3p in severe acute pancreatitis
1Department of Pathology, Fourth Medical Center of the Chinese PLA General Hospital, Beijing, 100048, China
2Translational Medicine Research Center, Fourth Medical Center and Medical Innovation Research Division of the Chinese PLA General Hospital, Beijing, 100048, China
3Faculty of Hepato-Pancreato-Biliary Surgery, Chinese PLA General Hospital, Beijing, 100853, China
*Address correspondence to: Weimin Li, liweimin@301hospital.com.cn; Jiangyang Lu, yubofan@buaa.edu.cn
Received: 22 April 2021; Accepted: 07 July 2021
Abstract: HMGN2 have functions in inflammatory response. However, the role of HMGN2 in severe acute pancreatitis (SAP) remains unclear. Here, our study was to discuss the role and regulatory mechanism ofHMGN2 in SAP. In this study, the SAP cell model of AR42J was used to study the function and mechanism of HMGN2 in SAP. The protein expression in cells and serums were examined by western blot and ELISA assay. qPCR was used to test the transcriptional RNA level. Cell viability were examined by MTT assay. Luciferase assay was used to evaluate the interaction between gene and gene. Our results showed that HMGN2 was significantly upregulated in SAP patients. The database predicted and luciferase assay data indicated the HMGN2 was directly binding with miR-590-3p. ELISA, MTT and western blot experiments showed that the HMGN2 were promoted the cell proliferation, reduced the inflammation, and repressed the cell autophagy. Mechanism studies showed that the pseudogene HMGN2P46 level was positively correlated with HMGN2 and upregulated HMGN2 expression by competing for miR-590-3p in SAP. Taken together, all over these results showed upregulation of HMGN2 alleviates SAP, this process was regulated by HMGN2P46 competitively binding with miR-590-3p, which may provide a new insight for the treatment and intervention in SAP. Pseudogene HMGN2P46 was a miRNA sponge to regulate HMGN2 level by competing for miR-590-3p to alleviate the process of SAP. It provided a novel strategy for the diagnosis and treatment of severe acute pancreatitis.
Keywords: HMGN2; Pseudogene; Severe acute pancreatitis; Inflammation; miR-590-3p
Severe acute pancreatitis (SAP) is a subtype of acute pancreatitis with multiple organ failure and systemic inflammatory response syndrome (Pezzilli et al., 2015). At present, the incidence of acute pancreatitis continues to increase worldwide, and it is one of the most common gastrointestinal causes of hospitalization in China. Treatment for SAP includes intravenous fluid resuscitation, feeding, prophylactic antibiotics, probiotics, and endoscopic retrograde cholangiopancreatography (ERCP). In addition, interventions such as abstinence from alcohol and smoking cessation are important to reduce readmissions or prevent pancreatitis (Greenberg et al., 2016; Zerem, 2014). Despite improvements in the treatment of SAP, disease-related mortality rates are still as high as 30%–40% (Garg and Singh, 2019). Therefore, the pathogenesis of SAP has been the focus of much research. Current studies suggest that oxidative stress, excessive release of pro-inflammatory cytokines, calcium overload, and dysregulation of autophagy are the main mechanisms that cause disease pathology (Gukovskaya et al., 2017; Mandalia et al., 2018).
HMGN2 is a gene which belongs to the high mobility group chromosome of the histone (HMG) members of the family and is the most abundant non-histone family in the nucleus of vertebrates and invertebrates. Previous studies have suggested that HMGN2 is related to DNA replication and transcriptional activity. Current studies have shown that HMGN2 plays a certain role in mouse embryo transfer (Li et al., 2017a), M2 cell polarization (Wang et al., 2019), and stem cell fate (Garza-Manero et al., 2019). In addition, HMGN2 has a good antibacterial (Li et al., 2017b), anti-inflammatory (Deng et al., 2012) and anticancer activities (Chen et al., 2018; Su et al., 2014). However, the role of HMGN2 in SAP is still unclear.
In recent years, non-coding RNA has received great attention in describing the molecular pathogenesis of SAP. Among them, microRNAs (miRNAs) are evolutionarily conserved pleiotropic small RNAs that inhibit gene expression after transcription. MiRNAs lead to translation inhibition of a large number of genes by sequence-specific binding to the 3’-untranslated regions (UTRs) of homologous mRNAs (Pu et al., 2019). It is reported that miR-590-3p is an interesting gene that play an exactly opposite effect in different cancers (Ge and Gong, 2017; Salem et al., 2018) and also have function in inflammation injury (Liu et al., 2020b). But there have been no related reports in SAP.
Similar to miRNAs, pseudogenes belong to another class of non-coding RNAs, defined as genomic elements that resemble real genes. Pseudogenes are common and abundant in the genome. They originate from the decay of repeated genes in the process of evolution, and are similar to functional genes, but have defects such as termination codon, frameshift and deletion in the coding sequence. Pseudogenes were previously considered biologically insignificant because they retain mutations and stop codons. However, nucleotide sequences within pseudogenes may be conserved due to evolutionary selection pressures, suggesting that they may indeed have an important role (Zheng et al., 2018). Pseudogenes were expressed tissue-specific and abnormally activated in some diseases and malignancies (Grandér and Johnsson, 2015; Tutar et al., 2018), suggesting that may be involved in some diseases and malignancies, although the mechanism remains unclear. It has been shown to pseudogenes act as affinity endogenous RNAs that regulate other RNA transcripts through affinity shared miRNAs (An et al., 2017). Interestingly, the frequency of pseudogenes such as HMG, OCT4 and Nanog was higher than expected (Lum et al., 2000; Pain et al., 2005), suggesting that it may be involved in the coordination and maintenance of related functions of their parent genes. Liu and his teams reported that pseudogene HMGA1P6 acted as a competitive endogenous RNA to promote ovarian cancer cell malignancy by upregulating HMGA1 and HMGA2 expression (Tian et al., 2020). de Martino et al. (2020) showed HMGA1P7 promoted B-cell lymphoma in mice by regulating HMGA1 and other cancer-related genes through miRNA. HMGN2P46 belongs to pseudogenes of HMGN2, but the functions also remain unknown.
In this study, we reported that HMGN2 and its pseudogene HMGN2P46 were abnormally expressed in SAP patient serum. At the same times, the expression levels of the two genes are directly related. In addition, we found that as a competitive endogenous RNA, HMGN2P46 can protect HMGN2 transcription from inhibition by miR-590-3p to play a protective role in the SAP process. This study provides the first evidence of a remarkable interaction among HMGN2, miR-590-3p, and HMGN2P46 and offered a new sight on the treatment of SAP.
10 pairs SAP patient serum and normal serum samples were collected in our hospital from 2017 to 2020. All of SAP samples were provided by an average age of 41.3 years. Normal serum samples were taken from 12 normal volunteers which have a 33.1-year average age. All the serum samples were centrifuged at 4°C, stored in a −80°C ultra-low temperature refrigerator. Each patient signed informed consent and ethics committee documents of Ethics Committee of our hospital to approve this study.
Cell culture, transfection, and SAP model
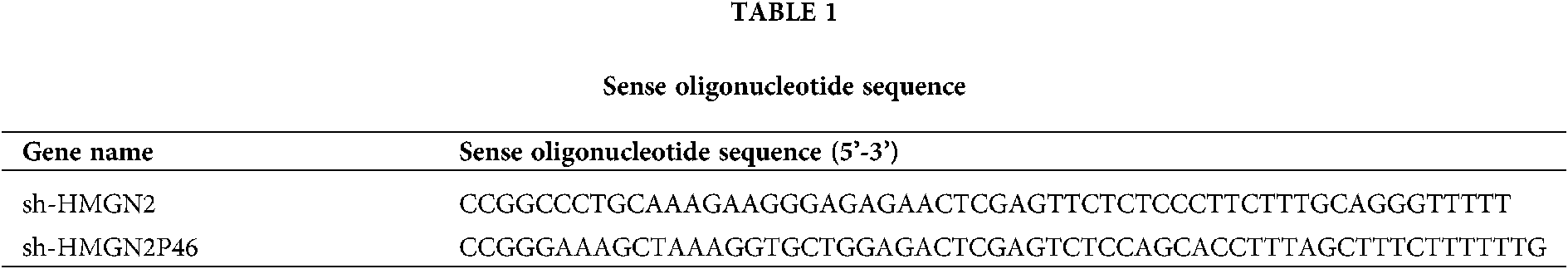
AR42Jcell was maintained with 15% FBS (Hyclone, USA), 100 μg/mL penicillin/streptomycin (Thermo, USA) at 37°C in a CO2 incubator (5% CO2 and 95% air, 95% humidity). The cell was cultured by passage after fusion at 70%–80%. WT/mut HMNG2, WT/mut HMNG2P46, sh-HMGN2, sh-HMGN2P46 and miR-590-3p mimic were designed, synthesized, and encapsulated by company (RiboBio, Guangzhou China). AR42Jcell was transfected with lentivirus plasmid and collected at 72 h after transfection. The AR42J SAP cell model in vitro was stimulated by lipopolysaccharide (LPS) in different concentrations (0, 1, 10, 100 mg/L) and different times (0, 6, 12, 18, and 24 h). The TNF-α level, cell survival and apoptosis were detected by Western blot and MTT assay. No LPS condition cell was used as control. sh-HMGN2 and sh-HMGN2P46 sense Oligonucleotide sequence showed in Table 1.
Enzyme linked immunosorbent assay (ELISA)
The ELISA assay was used to detect the HMGN2 level in SAP patient serums. The quantification of HMGN2 was assayed by Human HMGN2 ELISA Kit (No. JL38663, Jianglaibio, Shanghai China) according to the manufacturer’s instructions. OD values were measured by microplate at the wavelength of 450 nm.
The proliferation of AR42J cells by LPS treatment or plasmid transfection were detected by MTT assay. Firstly, the treated experimental cells were seeded and incubated in 96-well plates at density of 5–8 × 103 cells/well and detected at 12, 24, 48, and 72 h, respectively. Then MTT solution (Thermo, USA) was added and incubated with the cells (dark), 150 μL DMSO (Sigma) was added after 4 h. Finally, the plates were shaken, and the absorbance was detected at 490 nm by a microplate reader (Gene, Hong Kong) after blue formazan crystals dissolved. The cell survival rate (%) = OD (experimental group)/OD (blank control group) × 100%. The experiment was repeated three times.
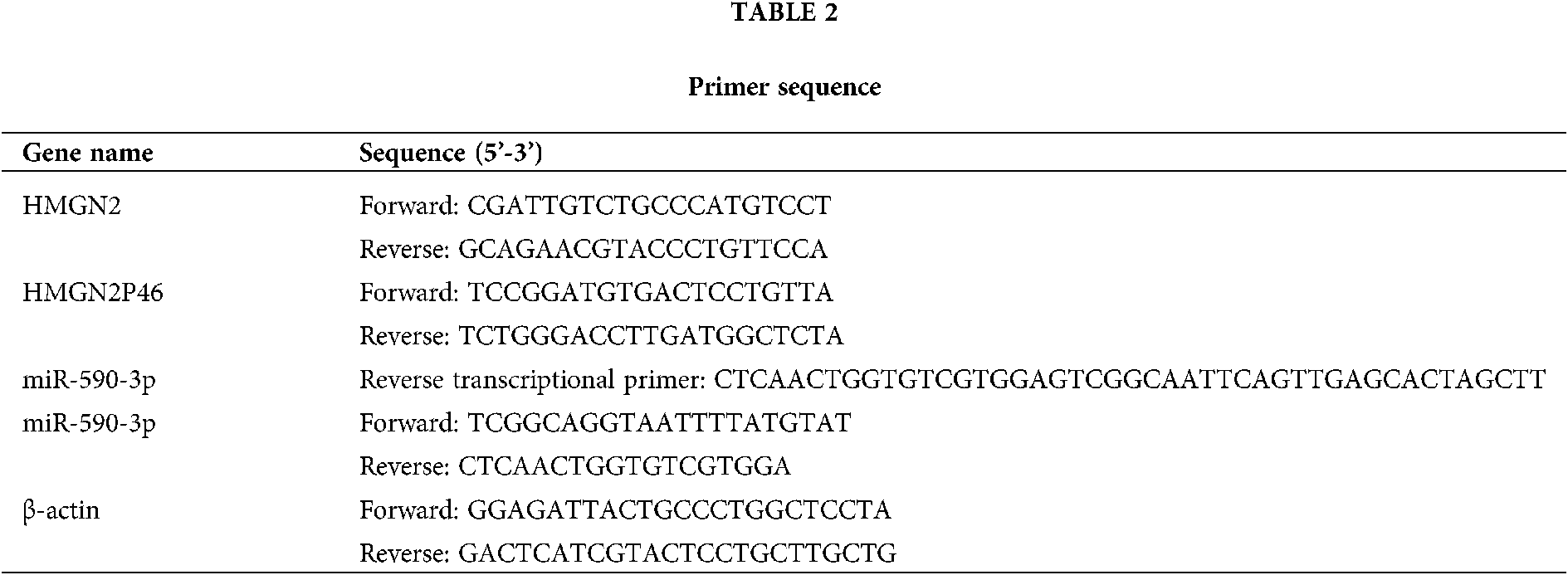
The total RNA was extracted from patient serums and treatment cells by using TRIzol reagent (Thermo, USA) according to the manufacturer’s information. Then the cDNA was synthesized by SuperScript VILO™ cDNA kit (Thermo, USA) following the manufacturer’s manual. The reaction system and conditions of qPCR was referred to SYBR™ Green PCR Supermix kit instructions (Thermo, USA). The primers of HMGN2, HMGN2P46, miR-590-3p, and β-actin were shown in Table 2, β-actin as an internal control.
The total protein was extracted from SAP patient serums and LPS-treated cells. The concentration of protein was quantified by Pierce™ BCA assay kit (Thermo, USA). In western blot analysis process, the total protein (30 μg) was separated by 10% SDS-PAGE Gel. Then the separated proteins were transferred to the PVDF membrane (Millipore, USA). The blots were incubated with HMGN2 (#9437, 1:500), TNF-α (#3707, 1:800), IL-10 (#12163, 1:1000), IL-6 (#12153, 1:1000), PARP1 (#9532, 1:800), cleaved caspase3 (#9664, 1:1000), Bcl2 (#15071, 1:1000), ULK1 (#6439, 1:1000), Beclin1 (#3738, 1:600), SQSTM1 (#88588, 1:900), and β-actin (#4970, 1:1000) (CST, USA) overnight at 4°C. The enhanced chemiluminescence detection kit (CST, USA) was used to visualize the target band after the second antibody incubation (goat anti-rabbit IgG 1:2000, CST, USA).
The potential target miRNAs of HMGN2 were searched in Targetscan, StarBase and miRDB database website. The sequence of HMGN2 and HMGN2P46 with miR-590-3p binding sites were amplified by PCR and cloned to construct HMGN2, HMGN2P46 wild-type (WT) reporter vector, respectively. The mutant miR-590-3p binding sites was inserted into HMGN2, HMGN2P46 mutant-type (mut) reporter vector by site-directed mutagenesis (Thermo, USA), respectively. All the plasmid was constructed by company (RiboBio, Guangzhou, China). In the luciferase reporter assay process, the HEK293 or AR42J cells were cultured in 24-well plates (3–5 × 104 cells/well) and transfected alone or co-transfected with the constructed reporter vector and miR-590-3p mimics. After transfected 48 h, the relative luciferase activity was detected by the Dual-Luciferase Reporter Assay System (Promega, USA).
All the data were analyzed by GraphPad Prism 6 (GraphPad Software, San Diego, CA USA). Two-way ANOVA, One-way ANOVA and Student’s t-test were performed, and the results showed as mean ± SD. P < 0.05 was considered to have statistical significance. All experiments repeated at least three times.
HMGN2 is upregulated during SAP and targeted by miR-590-3p
At the beginning, we detected and found the HMGN2 level was significantly increased in SAP patients’ serum compared with normal serum (Figs. 1a and 1b) (P < 0.05). Besides, the bioinformatics analysis predicted that miR-590-3p may be the target gene of HMGN2 and the luciferase data showed HMGN2 was directly binding with miR-590-3p (Figs. 1c and 1d) (P < 0.05) and negatively regulated by miR-590-3p (Figs. 1e and 1f) (P < 0.05). These data suggested HMGN2 may play an important role in SAP and regulated by miR-590-3p.

Figure 1: HMGN2 increased in SAP patients and regulated by miR-590-3p directly. (a–b) The HMGN2 protein levels in serum of SAP patients and normal serum were detected by ELISA and western blot assay. (c) The potential target miRNAs of HMGN2 in Venn diagram were predicted in Targetscan, StarBase and miRDB database. (d) Relative luciferase activity mediated by reporter constructs harboring the WT or mut 3’-UTR of HMGN2 upon transfection with 100 nMmiR-NC or miR-590-3p in HEK293 cell. (e–f) The HMGN2 level in HEK293 cell were detected by qRT-PCR and western blot assay. β-actin as an internal gene control. *P < 0.05, **P < 0.01. Data are present the mean ± SD of three independent experiments.
HMGN2P46 level is positively correlated with HMGN2 in SAP
Then we speculated that the expression level of miR-590-3p in SAP might be negatively correlated with HMGN2 because of HMGN2 directly targeting miR-590-3p. However, there was no significant correlation between miR-590-3p and HMGN2 protein level (r = 0.1367, P = 0.2368) (Fig. 2a), suggesting that there was other regulatory mechanism may be involved in HMGN2 expression. Studies have shown that pseudogenes play a role in many physiological and pathological processes and participate in the function of genes by regulating the mRNA expression of coding genes with similar sequences (Lum et al., 2000; Pain et al., 2005). In our study, whether there is a pseudogene of HMGN2 to regulate its expression. The corresponding pseudogenes of HMGN2 were searched by NCBI database and selected transcriptional sequences genes. It was found that HMGN2P46 was significant positively related to the expression of HMGN2 (r = 0.7217, P = 0.0001) (Fig. 2b).
HMGN2P46 is a processed pseudogene located at 15q21. It is highly homologous to the coding sequence of HMGN2. The HMGN2P46 lacked the HMGN2 3’-UTR sequence which binding with miR-590-3p, but actually the sequence variations of HMGN2P46 contribute to a new match with miR-590-3p (Fig. 2c) and the luciferase assay data was confirmed this combination (Fig. 2d). In addition, we used the LPS to induce the SAP model in AR42J cell line at different times and concentrations and the results showed the SAP cell model was success (Suppl. Fig. S1). Then we detected the HMGN2, HMGN2P46, and miR-590-3p levels in LPS-induced SAP cell model and the qPCR results showed that HMGN2, HMGN2P46 were upregulated, while miR-590-3p decreased after 10 mg/L LPS stimulated 24 h in AR42J cell (Fig. 2e) (P < 0.05). Besides, further qRT-PCR analysis confirmed miR-590-3p has only effect on the HMGN2P46 mRNA level, the HMGN2P46 level was remarkably repressed by miR-590-3p (Fig. 2f) (P < 0.05). All the data indicated that HMGN2P46 level is positively correlated with HMGN2 and regulated by miR0-590-3p in SAP.

Figure 2: The HMGN2 levels have a significant correlation with HMGN2P46 in SAP. (a) Correlation between HMGN2 protein level and miR-590-3p level. (b) Correlation between HMGN2 protein level and HMGN2P46 level. (c) Bioinformatic analysis of HMGN2 and its pseudogene HMGN2P46. (d) Relative luciferase activity was detected after transfection with 100 nM miR-590-3p upon reporter constructs harboring the WT/mut-HMGN2P46. (e) The expression of HMGN2, HMGN2P46, and miR-590-3p levels in LPS-induced SAP cell model. (f) The HMGN2P46 levels in LPS-induced SAP cell model after transfection with 100 nM miR-590-3p. *P < 0.05, **P < 0.01. Data are present the mean ± SD of three independent experiments.
HMGN2P46 upregulates HMGN2 expression by competing for miR-590-3p in SAP
In order to evaluate the role of HMGN2P46 in regulating HMGN2 expression level, we used the luciferase reporter assay which including wild-type HMGN2 3’-UTR and the results showed the miR-590-3p overexpression significantly downregulated the luciferase signal activity by bind with WT-HMGN2 3’-UTR in both HEK293 and AR42J cells, while WT-HMGN2P46 overexpression significantly increased the luciferase activity because of competing interaction with miR-590-3p to keep HMGN2 level but not mut-HMGN2P46 which unbinding with miR-590-3p (Figs. 3a and 3b) (P < 0.05). Besides, the HMGN2P46 downregulation could decreased the luciferase activity, while anti-miR-590-3p reversed the effect of HMGN2P46 downregulation by binding with miR-590-3p (Figs. 3c and 3d) (P < 0.05). In addition, the western blot results showed that HMGN2P46 inhibition also downregulated the HMGN2 protein level, and anti-miR-590-3p overexpression rescued the effect of HMGN2P46 inhibition via interacting with miR-590-3p to keep more HMGN2 translation (Figs. 3e and 3f) (P < 0.05). All the results suggested that HMGN2P46 upregulated HMGN2 expression by competing interaction with miR-590-3p.

Figure 3: HMGN2P46 competitive binding with miR-590-3p to upregulate HMGN2 expression in SAP. (a–b) The luciferase activity results in both HEK293 and AR42J cells after transfected with control, miR-590-3p, WT-HMGN2P46 or mut-HMGN2P46, and WT-/mut-HMGN2P46 + miR-590-3p, respectively. (c) The mRNA levels of HMGN2 and HMGN2P46 in AR42J cell after HMGN2 and/or HMGN2P46 downregulation were detected by qPCR. (d) The luciferase activity in AR42J cells after transfected with sh-control, sh-HMGN2P46, and sh-HMGN2P46 + anti-miR-590-3p. (e–f) The western blot and statistical results of HMGN2 protein levels in SAP cell model after transfected with sh-control, sh-HMGN2P46, and sh-HMGN2P46 + anti-miR-590-3p. β-actin as an internal gene control. *P < 0.05, **P < 0.01. Data are present the mean ± SD of three independent experiments.
HMGN2 promotes the cell survival in SAP
To explore the effect of HMGN2 and HMGN2P46 on AR42J cell proliferation upon a SAP cell model, the cells were transfected with sh-control, sh-HMGN2, sh-HMGN2P46, sh-HMGN2 + sh-HMGN2P46, and sh-HMGN2P46 + anti-miR-590-3p knockdown plasmid and detected by MTT assay, respectively. The results showed both of HMGN2 and HMGN2P46 repression aggravated LPS-induced AR42J cell injury, while miR-590-3p inhibition could rescued this effect (Fig. 4a) (P < 0.05). Besides, we assessed the effect on cell apoptosis protein levels by western blot and both of HMGN2 and HMGN2P46 downregulation increased SAP AR42J cell apoptosis, while miR-590-3p downregulation revered the further aggravation which caused by HMGN2P46 inhibition (Fig. 4b) (P < 0.05). Furthermore, only WT-HMGN2P46 could affected the HMGN2 and cell apoptosis key protein levels in SAP cell model but not mut-HMGN2P46 (Figs. 4c and 4d) (P < 0.05), indicating the HMGN2P46 as a decoy to interact with miR-590-3p for regulate HMGN2 level and cell proliferation. All data showed that HMGN2 was regulated by the combination HMGN2P46 and miR-590-3p to benefit AR42J cells survival from SAP injury.

Figure 4: The HMGN2 regulated the cell proliferation in SAP. (a) The cell proliferation upon a SAP cell model were detected by MTT assay after transfected with sh-control, sh-HMGN2, sh-HMGN2P46, sh-HMGN2 + sh-HMGN2P46, and sh-HMGN2P46 + anti-miR-590-3p knockdown plasmid. (b) The protein levels of PARP1, cleaved caspase3, and Bcl-2 were assessed by western blot after transfected with a series of knockdown plasmid. (c–d) The HMGN2, PARP1, cleaved caspase3, and Bcl-2 protein levels in SAP cell were detected by western blot after transfected with WT- or mut-HMGN2P46 plasmid. β-actin as an internal control. “*”: compared with sh-control, “Δ”: compared with sh-HMGN2, “ϕ”: compared with sh-HMGN2P46, all mean P < 0.05. Data are present the mean ± SD of three independent experiments.
HMGN2 reduces cell inflammation response in SAP
Excessive release of pro-inflammatory cytokines is one of the pathological mechanisms in SAP, so we detected the effect of HMGN2 on the inflammatory response after HMGN2 inhibition. The results showed that downregulation of HMGN2 or HMGN2P46 significantly repressed anti-inflammatory factor IL-10 level and increased pro-inflammatory cytokines TNF-α and IL-6 levels compared with the control, while the miR-590-3p inhibition significantly decreased the TNF-α and IL-6 levels and upregulated the IL-10 (Figs. 5a and 5b) (P < 0.05). In addition, only WT-HMGN2P46 could decreased the key inflammation response factors TNF-α and IL-6 levels and upregulated the IL-10 level in SAP cell model (Figs. 5c and 5d) (P < 0.05). All the results indicated HMGN2P46 may reduce the release of inflammation factors as a decoy to interact with miR-590-3p for regulate HMGN2 level in SAP.

Figure 5: The HMGN2 affected inflammatory response in SAP cell model. (a–b) The inflammation response of AR42J cell with LPS-induced for 24 h after HMGN2, HMGN2P46, and miR-590-3p downregulation were detected by western blot. (c–d) The inflammatory related factors IL-10, TNF-α, and IL-6 protein levels in SAP cell model after transfected with WT- or mut-HMGN2P46 plasmid were evaluated by western blot. β-actin as an internal control. “*”: compared with sh-control, “Δ”: compared with sh-HMGN2, “ϕ”: compared with sh-HMGN2P46, all mean P < 0.05. Data are present the mean ± SD of three independent experiments.
HMGN2 regulates the autophagy in SAP
Autophagy is one of the pathological mechanisms of SAP, so we also examined the changes of autophagy after HMGN2 inhibition. The results indicated that both HMGN2 and HMGN2P46 inhibition significantly decreased autophagy key factors such as ULK1, SQSTM1, and Beclin1 protein levels compared with the control, especially ULK1, while the downregulation of miR-590-3p significantly reversed this effect (Figs. 6a and 6b) (P < 0.05). Besides, only WT-HMGN2P46 reduced the autophagy factors ULK1, SQSTM1, and Beclin1 protein levels in SAP cell model (Figs. 6c and 6d) (P < 0.05). All the data suggested that HMGN2P46 involved the autophagy processas a decoy to compete with miR-590-3p to increase HMGN2 level in SAP.

Figure 6: The HMGN2 affected the autophagy process in SAP cell model. (a–b) The cell autophagyin SAP cell model after HMGN2, HMGN2P46, and miR-590-3p downregulation were evaluated by western blot. (c–d) The protein levels of autophagy-related factors ULK1, SQSTM1 and Beclin1 in SAP cell model after transfected with WT- or mut-HMGN2P46 plasmid were tested by western blot. β-actin as an internal control. “*”: compared with sh-control, “Δ”: compared with sh-HMGN2, “ϕ”: compared with sh-HMGN2P46, all mean P < 0.05. Data are present the mean ± SD of three independent experiments.
Studies have shown that pseudogenes were conserved in biological evolution, expressed tissue-specific and abnormally activated in partial diseases and cancers (Grandér and Johnsson, 2015; Tutar et al., 2018; Zheng et al., 2018), which means the pseudogenes may play an important role in the progress of some diseases and cancers. Tan et al. (2019) stated PIWI-36712 interacted with pseudogene SEPW1P to inhibit breast cancer progression and chemoresistance. Liu et al. (2020a) reported that AGPG pseudogene regulated cancer glycolytic process by targeting PFKFB3. Yu et al. (2019) showed that pseudogene POU5F1B was increased in cervical cancer and promoted cell proliferation and migration. In other diseases, Wang et al. reported that pseudogene PTENP1 regulated PTEN level to promote differentiation process of osteoclast and reduce the osteoporosis by targeting miR-214 (Wang et al., 2020). Our study focused on the function and mechanism of HMGN2 and its pseudogene HMGN2P46 in SAP.
In our research, HMGN2 upregulated significantly inpatient serum and AR42J cell model of SAP. Besides, HMGN2 directly binding with miR-590-3p, and the expression does not have much correlation with miR-590-3p level but positively correlated with its pseudogene HMGN2P46 in SAP patient serum. Furthermore, the downregulation of HMGN2 and HMGN2P46 significantly inhibited cell growth, promoted inflammatory response and autophagy process in SAP cell model. In molecular mechanisms, the functions of HMGN2P46 were achieved by regulating HMGN2 protein level, which competing interaction with miR-590-3p to increase HMGN2 protein level. Our molecular mechanisms, HMGN2P46 as a decoy competing with miR-590-3p to upregulate HMGN2 expression in SAP, are consistent with previous studies.
HMG family members, which including HMGA, HMGB, and HMGN, are play an important role in inflammation and infection of body. HMGA1 and HMGA2 as two driver of inflammation, aggravated LPS-induced cell inflammation and apoptosis by different pathway (Cai et al., 2020; Li et al., 2020). Besides, HMGB1, as a controlling target for inflammation, is a therapeutic target for severe pulmonary inflammation (Andersson et al., 2020), sepsis and trauma-induced inflammation (Deng et al., 2019). There are a few studies on the anti-inflammatory effects of HMGN. In our results, HMGN2 relieved inflammation in SAP by regulating pseudogene HMGN2P46/miR-590-3p axis, which provide a new sight of HMGN family on pathological mechanism and therapeutic targets in SAP.
Besides, autophagy process is also regulated by HMG family. HMGA1 inhibited autophagy to promote myocardial dysfunction in diabetic cardiomyopathy (Wu et al., 2020). HMGA2 have function in Cr(VI)-induced autophagy (Yang et al., 2017). HGMB1 is a critical regulator of autophagy (Tang et al., 2010), but there are also few reports on autophagy function ofHMGN at present. In our study showed that HMGN2 was regulated by pseudogene HMGN2P46/miR-590-3p axis to promote cell autophagy in SAP. It adds autophagy regulation to HMGN2 and provide a new idea of HMG family on pathological mechanism in SAP.
MiRNAs are evolutionarily conserved small RNAs that inhibit gene expression after transcription. MiRNAs typically lead to translation inhibition or mRNA degradation by interacting with the 3’-UTR sequences of homologous mRNAs (Pu et al., 2019). MiR-590-5p was always showed in many cancers (Ge and Gong, 2017; Salem et al., 2018). Moreover, Ma et al. (2019) reported that miR-590-3p inhibited TNFRAF-6 to attenuate acute kidney injury in septic mice. MiR-590-3p also inhibited the LPS-induced sepsis apoptosis and inflammation (Liu et al., 2020b). In our study, miR-590-3p as a sponge to regulate inflammatory response of SAP and we did not discuss the function of miR-590-3p alone in SAP inflammation. The function of miR-590-3p in SAP need more experiment to confirm.
Taken together, HMGN2P46 as a decoy competing with miR-590-3p to upregulate HMGN2 expression in SAP, thus regulated cell viability, inflammation, and autophagy to attenuate the SAP injury. Our study provides a new insight for understanding HMGN2 roles in SAP and a new therapeutic target for SAP.
Availability of Data and Materials: All data generated or analysed during this study are included in this published articleand its supplementary information files.
Author Contribution: The authors confirm contribution to the paper as follows: study conception and design: Hongqiang Zhao, Jiangyang Lu; data collection: Hongqiang Zhao, Qi Liu; analysis and interpretation of results: Hongqiang Zhao, Qi Liu, Haorun Liu; draft manuscript preparation: Hongqiang Zhao, Weimin Li. All authors reviewed the results and approved the final version of the manuscript.
Ethics Approval: This study and all experiments involved are under approval of the Chinese PLA General Hospital.
Funding Statement: This work was supported by National Natural Science Foundation of China [81901957]; China Postdoctoral Science Foundation [2018M633723].
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
An Y, Furber KL, Ji S (2017). Pseudogenes regulate parental gene expression via ceRNA network. Journal of Cellular and Molecular Medicine 21: 185–192. [Google Scholar]
Andersson U, Ottestad W, Tracey KJ (2020). Extracellular HMGB1: A therapeutic target in severe pulmonary inflammation including COVID-19? Molecular Medicine 26: 42. [Google Scholar]
Cai ZL, Shen B, Yuan Y, Liu C, Xie QW et al. (2020). The effect of HMGA1 in LPS-induced myocardial inflammation. International Journal of Biological Sciences 16: 1798. [Google Scholar]
Chen J, Fan Y, Cui B, Li X, Yu Y et al. (2018). HMGN2: An antitumor effector molecule of γδT cells. Journal of Immunotherapy 41: 118–124. [Google Scholar]
de Martino M, de Biase D, Forzati F, Credendino SC, Palma G et al. (2020). HMGA1-pseudogene7 transgenic mice develop B cell lymphomas. Scientific Reports 10: 7057. [Google Scholar]
Deng LX, Wu GX, Cao Y, Fan B, Gao X et al. (2012). The chromosomal protein HMGN2 mediates the LPS-induced expression of β-defensins in mice. Inflammation 35: 456–473. [Google Scholar]
Deng M, Scott MJ, Fan J, Billiar TR (2019). Location is the key to function: HMGB1 in sepsis and trauma-induced inflammation. Journal of Leukocyte Biology 106: 161–169. [Google Scholar]
Garg PK, Singh VP (2019). Organ failure due to systemic injury in acute pancreatitis. Gastroenterology 156: 2008–2023. [Google Scholar]
Garza-Manero S, Sindi AaA, Mohan G, Rehbini O, Jeantet VH et al. (2019). Maintenance of active chromatin states by HMGN2 is required for stem cell identity in a pluripotent stem cell model. Epigenetics & Chromatin 12: 73. [Google Scholar]
Ge X, Gong L (2017). MiR-590-3p suppresses hepatocellular carcinoma growth by targeting TEAD1. Tumor Biology 39: 1010428317695947. [Google Scholar]
Grandér D, Johnsson P (2015). Pseudogene-expressed RNAs: emerging roles in gene regulation and disease. In: Morris K (ed.Long Non-Coding RNAs in Human Disease. Current Topics in Microbiology and Immunology. Cham: Springer. [Google Scholar]
Greenberg JA, Hsu J, Bawazeer M, Marshall J, Friedrich JO et al. (2016). Clinical practice guideline: Management of acute pancreatitis. Canadian Journal of Surgery 59: 128. [Google Scholar]
Gukovskaya AS, Gukovsky I, Algül H, Habtezion A (2017). Autophagy, inflammation, and immune dysfunction in the pathogenesis of pancreatitis. Gastroenterology 153: 1212–1226. [Google Scholar]
Li DD, Yue L, Yang ZQ, Zheng LW, Guo B (2017a). Evidence for Hmgn2 involvement in mouse embryo implantation and decidualization. Cellular Physiology and Biochemistry 44: 1681–1695. [Google Scholar]
Li H, Shen XF, Zhou XE, Shi YE, Deng LX et al. (2017b). Antibacterial mechanism of high-mobility group nucleosomal-binding domain 2 on the Gram-negative bacteria Escherichia coli. Journal of Zhejiang University–Science B 18: 410–420. [Google Scholar]
Li X, Sun M, Long Y (2020). Cyanidin-3-O-glucoside attenuates lipopolysaccharide-induced inflammation in human corneal epithelial cells by inducing Let-7b-5p-mediated HMGA2/PI3K/Akt pathway. Inflammation 43: 1088–1096. [Google Scholar]
Liu J, Liu ZX, Wu QN, Lu YX, Wong CW et al. (2020a). Long noncoding RNA AGPG regulates PFKFB3-mediated tumor glycolytic reprogramming. Nature Communications 11: 1507. [Google Scholar]
Liu L, Liu F, Sun Z, Peng Z, You T, Yu Z (2020b). LncRNA NEAT1 promotes apoptosis and inflammation in LPS-induced sepsis models by targeting miR-590-3p. Experimental and Therapeutic Medicine 20: 3290–3300. [Google Scholar]
Lum H, Lee KL, Yu G (2000). The chicken genome contains no HMG1 retropseudogenes but a functional HMG1 gene with long introns. Biochimica et BiophysicaActa (BBA)–Gene Structure and Expression 1493: 64–72. [Google Scholar]
Ma J, Li YT, Zhang SX, Fu SZ, Ye XZ (2019). MiR-590-3p attenuates acute kidney injury by inhibiting tumor necrosis factor receptor-associated factor 6 in septic mice. Inflammation 42: 637–649. [Google Scholar]
Mandalia A, Wamsteker EJ, Dimagno MJ (2018). Recent advances in understanding and managing acute pancreatitis. F1000Research 7: F1000 Faculty Rev-1959. [Google Scholar]
Pain D, Chirn GW, Strassel C, Kemp DM (2005). Multiple retropseudogenes from pluripotent cell-specific gene expression indicates a potential signature for novel gene identification. Journal of Biological Chemistry 280: 6265–6268. [Google Scholar]
Pezzilli R, Zerbi A, Campra D, Capurso G, Golfieri R et al. (2015). Consensus guidelines on severe acute pancreatitis. Digestive and Liver Disease 47: 532–543. [Google Scholar]
Pu M, Chen J, Tao Z, Miao L, Qi X et al. (2019). Regulatory network of miRNA on its target: Coordination between transcriptional and post-transcriptional regulation of gene expression. Cellular and Molecular Life Sciences 76: 441–451. [Google Scholar]
Salem M, O’brien JA, Bernaudo S, Shawer H, Ye G et al. (2018). miR-590-3p promotes ovarian cancer growth and metastasis via a novel FOXA2-versican pathway. Cancer Research 78: 4175–4190. [Google Scholar]
Su L, Hu A, Luo Y, Zhou W, Zhang P, Feng Y (2014). HMGN2, a new anti-tumor effector molecule of CD8+ T cells. Molecular Cancer 13: 178. [Google Scholar]
Tan L, Mai D, Zhang B, Jiang X, Zhang J et al. (2019). PIWI-interacting RNA-36712 restrains breast cancer progression and chemoresistance by interaction with SEPW1 pseudogene SEPW1P RNA. Molecular Cancer 18: 9. [Google Scholar]
Tang D, Kang R, Livesey KM, Cheh CW, Farkas A et al. (2010). Endogenous HMGB1 regulates autophagy. Journal of Cell Biology 190: 881–892. [Google Scholar]
Tian X, Song J, Zhang X, Yan M, Wang S et al. (2020). MYC-regulated pseudogene HMGA1P6 promotes ovarian cancer malignancy via augmenting the oncogenic HMGA1/2. Cell Death & Disease 11: 167. [Google Scholar]
Tutar L, Özgür A, Tutar Y (2018). Involvement of miRNAs and pseudogenes in cancer. In: Wu W (ed.MicroRNA and Cancer. Methods in Molecular Biology. New York, NY: Humana Press. [Google Scholar]
Wang CG, Wang L, Yang T, Su SL, Hu YH, Zhong D (2020). Pseudogene PTENP1 sponges miR-214 to regulate the expression of PTEN to modulate osteoclast differentiation and attenuate osteoporosis. Cytotherapy 22: 412–423. [Google Scholar]
Wang X, Chen S, Ren H, Chen J, Li J et al. (2019). HMGN2 regulates non-tuberculous mycobacteria survival via modulation of M1 macrophage polarization. Journal of Cellular and Molecular Medicine 23: 7985–7998. [Google Scholar]
Wu QQ, Liu C, Cai Z, Xie Q, Hu T et al. (2020). High-mobility group AT-hook 1 promotes cardiac dysfunction in diabetic cardiomyopathy via autophagy inhibition. Cell Death & Disease 11: 160. [Google Scholar]
Yang F, Zhao L, Mei D, Jiang L, Geng C et al. (2017). HMGA2 plays an important role in Cr (VI)-induced autophagy. International Journal of Cancer 141: 986–997. [Google Scholar]
Yu J, Zhang J, Zhou L, Li H, Deng ZQ, Meng B (2019). The octamer-binding transcription factor 4 (OCT4) pseudogene, POU domain class 5 transcription factor 1B (POU5F1Bis upregulated in cervical cancer and down-regulation inhibits cell proliferation and migration and induces apoptosis in cervical cancer cell lines. Medical Science Monitor: International Medical Journal of Experimental and Clinical Research 25: 1204. [Google Scholar]
Zerem E (2014). Treatment of severe acute pancreatitis and its complications. World Journal of Gastroenterology 20: 13879. [Google Scholar]
Zheng LL, Zhou KR, Liu S, Zhang DY, Wang ZL et al. (2018). dreamBase: DNA modification, RNA regulation and protein binding of expressed pseudogenes in human health and disease. Nucleic Acids Research 46: D85–D91. [Google Scholar]

SUPPLEMENTARY FIGURE S1: The cell model construction of severe acute pancreatitis in vitro. (a) The TNF-α protein levels in AR42J cell after stimulated by LPS in different concentrations (0, 1, 10, 100 mg/L) for 24 h were detected by Western blot. (b) The TNF-α protein levels in AR42J cell after stimulated by 10 mg/L LPS at different time (0, 6, 12, 18, and 24 h) were assessed by Western blot. (c) The AR42J cell proliferation after stimulated by 10 mg/L LPS treatment at different time was tested by MTT assay. (d) The apoptosis protein levels of PARP1, cleaved capase3, and Bcl-2 in AR42J cell after 10 mg/L LPS treatment were detected by Western blot. β-actin as a negative control. *P < 0.05, **P < 0.01. Data are present the mean ± SD of three independent experiments.
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |