DOI:10.32604/biocell.2021.016450

| BIOCELL DOI:10.32604/biocell.2021.016450 |  |
| Article |
A global study of transcriptome dynamics in the venom gland of Solenopsis invicta Buren during laboratory rearing
1State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-products, Institute of Plant Protection and Microbiology, Zhejiang Academy of Agricultural Sciences, Hangzhou, 310021, China
2Integrated Plant Protection Center, Lishui Academy of Agricultural and Forestry Sciences, Lishui, 323000, China
3College of Ecology, Lishui University, Lishui, 323000, China
*Address correspondence to: Yaobin Lu, luybcn@163.com; Jun Huang, junhuang1981@aliyun.com
Received: 07 March 2021; Accepted: 09 April 2021
Abstract: Venom plays several important roles in the life of the fire ant, Solenopsis invicta Buren. Laboratory rearing significantly affected the toxin component of S. invicta. However, the molecular mechanism of the change of venomous components when kept in the laboratory is unclear. In this study, RNA sequencing technique (RNA-Seq) was performed to explore differentially expressed genes in the venom gland of S. invicta at 0, 10, and 60 days after laboratory rearing. The RNA-Seq results showed that the expression of a large number of genes changed. The DEGs were involved in multiple pathways, including proteolysis, serine-type endopeptidase, and allergen. Furthermore, RNA-Seq and qRT-PCR data revealed that the expression of some genes related to proteolysis and allergen significantly decreased. Thus, our data generated new data relating to toxin-component and the transcriptome dynamics in the venom gland of S. invicta during laboratory rearing.
Keywords: Solenopsis invicta Buren; RNA-Seq; Venom; Laboratory rearing
Ants (Formicidae) belong to the class Insecta and the order Hymenoptera, which comprises the families Vespidae (wasps), Formicidae (ants), Apidae (bees), among others (Wilson et al., 2010). The family Formicidae consists of more than 13,000 species of ants, most of which have a specialized venom apparatus to sting victims and prey, inoculating a cocktail of pharmacologically and biologically active substances for defense and predation (Carvalho et al., 2019; Touchard et al., 2016).
The venom composition of ants varies significantly between the different ant subfamilies (Hoffman, 2010). Up to now, the composition of venoms from several ant species has been investigated at pharmacological and molecular levels (Hoffman, 2010). The venoms of fire ants are composed of over 90% of piperidinic alkaloids, while the remaining fraction is constituted by amines, formic acid, hydrocarbons, peptides, and proteins (Aili et al., 2014; Pluzhnikov et al., 2014; Hoffman, 2010). Some toxin-like proteins and peptides have been identified using conventional molecular biological and biochemical methods (Inagaki et al., 2004; Inagaki et al., 2008; Aili et al., 2016). Toxin peptides from ants follow commonness with linear and short polycationic peptides and responsible for hemolysis, histamine release, cell lysis, and antimicrobial action (Palma and Brochetto-Braga, 1993; Schumacher et al., 1992). However, the number of identified toxin-like proteins and peptides is so limited in ants.
Solenopsis invicta Buren, a predatory ant species in the subfamily Myrmicinae, is distributed mainly throughout tropical and subtropical regions of the Americas, China, India, Africa, Pacific islands (Pandey et al., 2019; Kruse et al., 2020). The venom of S.invicta has also been the focus of a series of studies (Chen et al., 2019; Eliyahu et al., 2011; Hoffman et al., 1988; Kruse et al., 2020). Some proteins, including phospholipase A/B, the pheromone binding protein, the antigen 5/pathogenesis-related protein, among others, were identified and sequenced up to now (Das et al., 2018; Dos Santos Pinto et al., 2012; Hoffman et al., 1988; Kruse et al., 2020; Monsalve et al., 2020). The proteomic profile of the venom from the fire ant S.invicta was analyzed, which permitted the identification of many toxic proteins such as allergens, neurotoxins, proteins that cause tissue damage/inflammation, proteins influencing the homeostasis of the victims, and proteins that promote venom diffusion (Dos Santos Pinto et al., 2012). A large number of red fire ants die when kept in the laboratory for long periods (Liu et al., 2015). Moreover, our recent work has shown that the level of venomous alkaloid components decreased continuously to a very low level when the fire ants were maintained in the laboratory (Liu et al., 2015). However, it is unclear whether laboratory rearing of the S. invicta affects other venomous components of venom and the molecular mechanism of the change of venomous components when kept in the laboratory. In the current study, RNA sequencing (RNA-Seq) was performed to explore differentially expressed genes in the venom gland of the fire ant S. invicta at 0, 10, and 60 days after laboratory rearing. Along with RNA sequencing and transcriptome annotation, analysis of global patterns of gene expression and functional categorization were performed. Furthermore, characteristics of some putative toxin candidates are predicted and discussed.
Adult fire ant S. invicta was collected from the scientific research base of South China Agricultural University, Zengcheng, Guangdong Province, China, as described in one of our previous reports (Liu et al., 2015). The 0.5 m × 0.5 m × 0.5 m well-developed ant nest derived from a scientific research base was placed in a Fluon storage box (ICI, Wilmington, DE). After two days, the colonies were forced to transfer to smaller Fluon storage boxes using the “Drop method”. Then, a small amount of soil from the nest is then added to the storage box to build an artificial nest. During laboratory rearing at room temperature, clean water, and 10% honey water were provided using the test tube. Three to five individuals of Tenebrio molitor were thrown in the nest per day. After dissecting in ultrapure water at 4°C, the intact venom glands of the fire ant S. invicta at 0, 10, and 60 days after laboratory rearing were frozen with liquid nitrogen and stored at −80°C.
RNA isolation and quantitative real-time PCR (qPCR) analysis
Total RNA was extracted using the RNAprep Pure Plant Plus Kit (DP441, TIANGEN, China) according to the instructions.
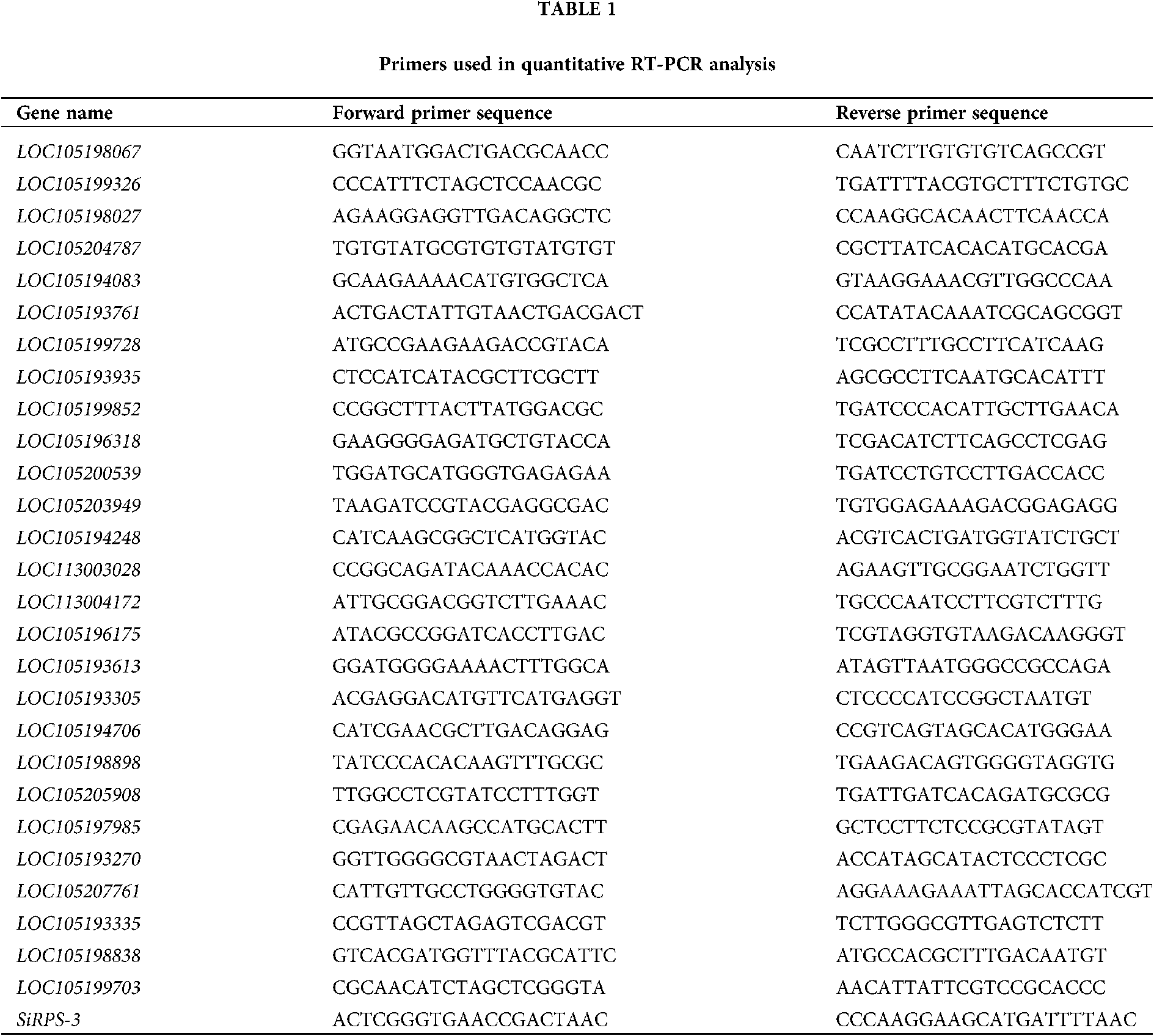
The expression of genes, including 12 DEGs randomly selected, 15 DEGs related to proteolysis and venom allergen, was analyzed by real-time quantitative RT-PCR. cDNAs were synthesized with iScript Reverse Transcription Supermix for RT-qPCR. The quantitative real-time PCR (qPCR) was performed using the fluorescent intercalating dye SYBR Green in a detection system (MJ Research, Opticon 2). Two-step RT-PCR procedure was achieved using a method described previously, as follows: 95°C for 30 s; 40×: 95°C for 10 s, 60°C for 30 s, plate read; melt curve analysis spanning 65–95°C, increment 0.5°C for 0:05 + plate read (Li et al., 2005). The ribosomal protein S3 (SiRPS-3) was employed as standard control (Lucas et al., 2015). All the gene-specific primers were designed using primer3 software (https://primer3.ut.ee/) and listed in Table 1.
Library construction and sequencing
RNA-Seq was performed by Norminkoda Biotechnology Co. Ltd., Wuhan, China. Sequencing libraries were generated using NEBNext1 Ultra™ RNA.
Library Prep Kit for Illumina1 (NEB, USA) following the manufacturer's instruction. Briefly, mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. Fragmentation was carried out using divalent cations under elevated temperature in NEBNext First Strand Synthesis Reaction Buffer. Then, first-strand cDNA was synthesized using a random hexamer primer and M-MuLV Reverse Transcriptase (RNase H-). Second strand cDNA synthesis was subsequently performed using DNA Polymerase I and RNase H. Remaining overhangs were converted into blunt ends via exonuclease/polymerase activities. After adenylation of 3’ ends of DNA fragments, NEBNext Adaptor with hairpin loop structure were ligated to prepare for hybridization. After purification with AMPure XP beads (Beckman Coulter, Beverly, USA) to select cDNA fragments of preferentially 250–300 bp in length, 3 µL USER Enzyme (NEB, USA) was used with size-selected. After incubation of the adaptor-ligated cDNA at 37°C for 15 min followed by 5 min at 95°C, PCR was performed with Phusion High-Fidelity DNA polymerase, Universal PCR primers, and Index (X) Primer. Finally, PCR products were purified using AMPure XP beads, and library quality was assessed on the Agilent 2100 system. Subsequently, preliminary quantification and library attenuation was performed using Qubit 2.0. The fragment size was detected using Agilent 2000.
The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS(Illumia) according to the manufacturer’s instructions. Then, the library preparations were sequenced on an Illumina Hiseq platform and 125 bp/150 bp paired-end reads were generated.
Quality control and reads mapping to the reference genome
Clean reads were obtained by removing reads containing unknown nucleotides larger than 5%, reads containing adaptors, and low-quality reads (the rate of reads of which quality value ≤ 10 is more than 50%) from raw reads. All the subsequent analysis was achieved based on the clean data with high quality. Then, reference genome (https://www.ncbi.nlm.nih.gov/bioproject/?term=49629%5BUID%5D) and gene model annotation files were downloaded from the genome website directly. Paired-end clean reads were aligned to the reference genome using STAR.
Quantification of gene expression level and differential expression analysis
The reads numbers mapped to each gene were counted using HTseq. Then, FPKM (expected number of Fragments Per Kilobase of transcript sequence per Millions of base pairs sequenced) of each gene was calculated based on the length of the gene and reads count mapped to this gene.
Differential expression analysis was performed using the DEGSeq. The resulting P-values were adjusted using Benjamini and Hochberg’s approach for controlling the false discovery rate. Genes with an adjusted P-value <0.05 and absolute log2FC (fold change (condition 2/condition 1) for a gene) >1 by DESeq2 were assigned as differentially expressed.
GO enrichment analysis of differentially expressed genes
Gene Ontology (GO) annotation was performed by the alignment between genes and the GO database. The website tool WEGO (http://wego.genomics.org.cn/cgi-bin/wego/index.pl) was used to perform GO functional classification of all genes and to view the distribution of genes functions at the macro level. The GO enrichment analysis of the DEGs was performed by hypergeometric test in R (Bouzid et al., 2014). R function dhyper was used to calculate P-values, the GO terms with P-value less than 0.05 were considered to be enrichment terms.
Mapping of RNA-Seq data and evaluation of differentially expressed genes
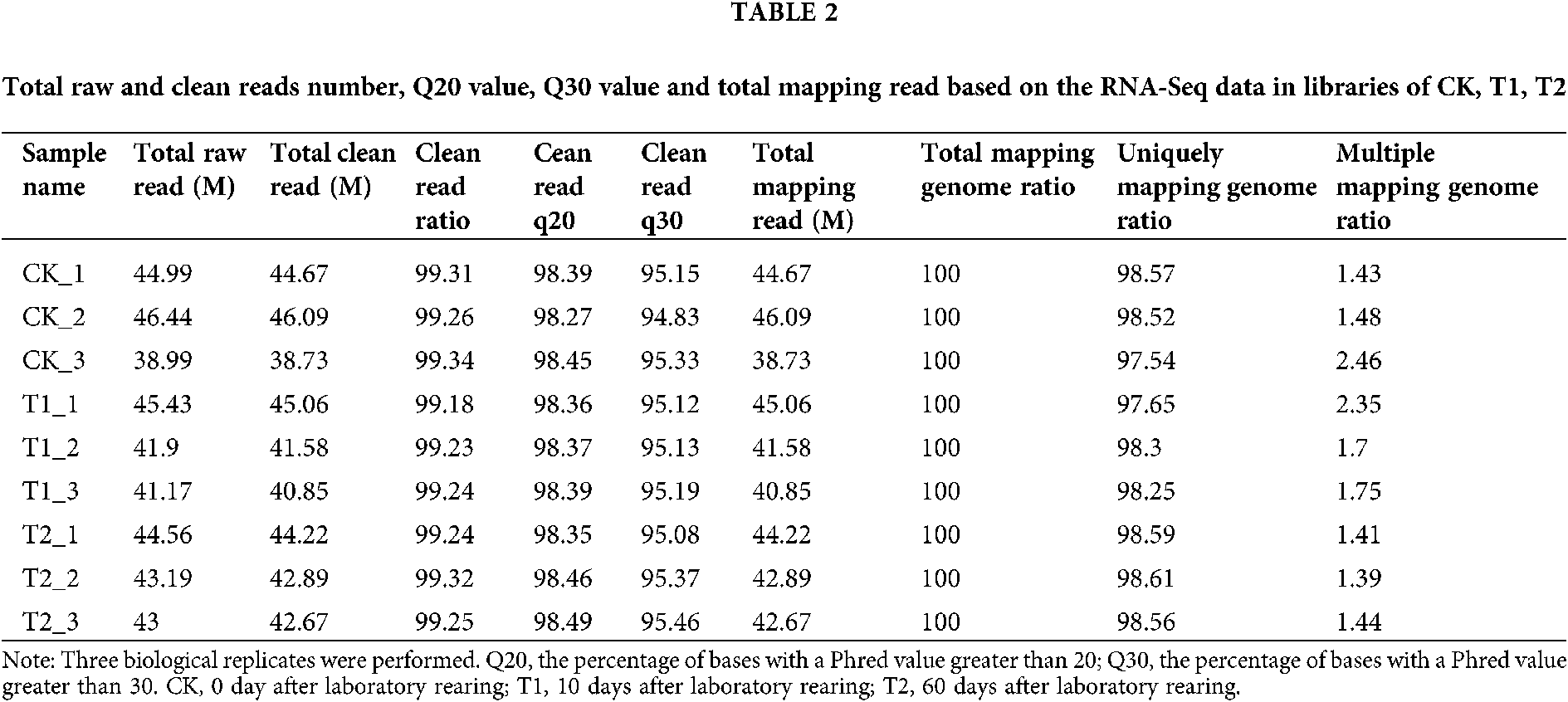
To explore differentially expressed genes in the venom gland of the fire ant S. invicta after laboratory rearing, RNA sequencing (RNA-Seq) analysis was performed. Three cDNA libraries were constructed using mRNA isolated from the venom gland of S. invicta at 0, 10, and 60 days after laboratory rearing (DALR) (namely CK, T1, T2), respectively, and large-scale sequenced. The RNA-Seq data was submitted to Bioproject of NCBI under Accession No. PRJNA682652 (https://www.ncbi.nlm.nih.gov/bioproject/?term=prjna675709). As shown in Table 2, 38.73–46.09 million clean reads were acquired in each individual sample. Among them, 97.54–98.61% clean reads were uniquely mapped in the S. invicta genome using STAR.
To reveal the overall trend of separation of the samples and variability among the sample groups, the principal component analysis (PCA) was performed. As shown in Fig. 1A, the three biological repetitions for each sample were gathered, which manifests the repeatability and reliability of our RNA-Seq analysis. To identify the differentially expressed genes (DEGs) between these three groups, the DEGSeq was employed. The DEGs were defined as the fold change of FPKM expression values were at least 2 in either direction when adjusted P < 0.05. A total of 397 DEGs (153 up-regulated and 244 down-regulated), 1201 DEGs (843 up-regulated and down-regulated), and 1017 DEGs (247 up-regulated and 770 down-regulated) were detected in “0 DALR vs. 60 DALR”, “0 DALR vs. 10 DALR” and “10 DALR vs. 60 DALR”, respectively (Figs. 1B and 1C). Moreover, the Venn diagram shows that 41 commonly DEGs exist in “0 DALR vs. 60 DALR”, “0 DALR vs. 10 DALR”, and “10 DALR vs. 60. DALR” (Fig. 1C). To verify the RNA-Seq data, the transcriptional level of twelve DEGs randomly chosen was analyzed. As shown in Fig. 1D, real-time quantitative PCR analyses showed that the relative expression patterns of the genes were consistent with RNA-Seq data, which demonstrates that the RNA-Seq data are reliable.

Figure 1: Transcripts detected in the venom gland of S. invicta on different days during laboratory rearing. (A) PCA analysis of the transcriptional profiles of CK, T1, and T2. (B) Venn diagram of the differentially expressed genes (DEGs) among “0 DALR vs. 60 DALR”, “0 DALR vs. 10 DALR” and “10 DALR vs. 60 DALR”. (C) Volcano plots of quantitative transcriptome analysis. The log2 (fold change) is plotted on the x-axis, and the negative log10 (FDR) (P-value) is plotted on the y-axis. The red and green circles represent significantly up-regulated genes and down-regulated genes, respectively (the absolute value of log2 (fold change) no less than 1 and FDR less than 0.05). (D) Validation of the expression profile of RNA-Seq (FPKM) by qRT-PCR. Twelve DEGs were randomly selected and validated by qRT-PCR to confirm their expression. Fold-change of FPKM (RNA-Seq) and expression level (qRT-PCR) after laboratory rearing compared to control (i.e., a fold-change of 1 indicates no change compared to control) was calculated. Three biological replicates were performed. Means ± SE are shown. CK, 0 days after laboratory rearing; T1, 10 days after laboratory rearing; T2, 60 days after laboratory rearing.
Gene ontology (GO) enrichment analysis of differentially expressed genes (DEGs)
To obtain ontology of defined terms concerning gene product properties, gene ontology (GO) enrichment analysis of DEGs was performed. As shown in Fig. 2, the most enrichment three GO subcategories of “0 DALR vs. 10 DALR” are the oxidation-reduction process, proteolysis, and transmembrane transport. Similarly, for “0 DALR vs. 60 DALR”, the most enrichment three GO subcategories are proteolysis, serine-type endopeptidase activity, and oxidation-reduction process (Fig. 2). On the other hand, the most enrichment three GO subcategories of “10 DALR vs. 60 DALR” are the oxidation-reduction process, proteolysis, and iron ion binding (Fig. 2). It is apparent that the genes related to the oxidation-reduction process and proteolysis are those with the most significant differential expression in all GO subcategories among the four samples.

Figure 2: Gene Ontology (GO) classification of the differentially expressed genes (DEGs) among “0 DALR vs. 60 DALR”, “0 DALR vs. 10 DALR”, and “10 DALR vs. 60 DALR”. CK, 0 days after laboratory rearing; T1, 10 days after laboratory rearing; T2, 60 days after laboratory rearing.
Analysis of DEGs related to cysteine-type peptidase activity, aspartic-type endopeptidase activity, serine-type peptidase activity and metallopeptidase
Toxin peptide is one group of key components of ant venom responsible for hemolysis, histamine release, cell lysis, and antimicrobial actions (Palma and Brochetto-Braga, 1993; Schumacher et al., 1992). Moreover, the GO enrichment analysis of DEGs revealed that the expression of the genes related to proteolysis, including cysteine-type peptidase activity, aspartic-type endopeptidase activity, serine-type peptidase activity, and metallopeptidase, significantly changed during laboratory rearing. So, we next analyzed the expression patterns of the DEGs in these pathways.
The hierarchical clustering of all DEGs related to cysteine-type peptidase activity, aspartic-type endopeptidase activity, serine-type peptidase activity, and metallopeptidase was performed, and the distance metric for gene expression patterns was determined using Pearson correlation. The hierarchical clustering analysis of these DEGs indicates that the expression of some genes (Class II) significantly increased after laboratory rearing. However, the other genes (Classes I, III, and IV) show different expression patterns (Fig. 3A). In detail, the expression of genes (Class IV) at 10 DALR is significantly lower than that at 0 DALR (Fig. 3A). And the expression of genes (Class I and IV) at 60 DALR is significantly lower than that at 0 DALR (Fig. 3A). It is interesting that the expression of LOC105207761, LOC105193270, LOC105197985, LOC105205908, LOC105198898, LOC105194706, LOC105193305, LOC105193613, LOC105196175, LOC113004172, LOC113003028, LOC105194248, and LOC105206642 gradually decreased during laboratory rearing. So, we deduce that proteins with peptidase activity, encoded by these 12 DEGs, are relevant to the change of toxin-component of S. invicta during laboratory rearing. Moreover, the qRT-PCR also showed that the transcript levels of LOC105207761, LOC105193270, LOC105197985, LOC105205908, LOC105198898, LOC105194706, LOC105193305, LOC105193613, LOC105196175, LOC113004172, LOC113003028, LOC105194248, and LOC105206642 significantly decreased during laboratory rearing (Fig. 3B).

Figure 3: The expression patterns of the DEGs, related to proteolysis. (A) Hierarchical clustering analysis of the differentially expressed genes (DEGs) related to proteolysis, including cysteine-type peptidase activity, aspartic-type endopeptidase activity, serine-type peptidase activity and metallopeptidase. (B) Quantitative RT-PCR analysis of expression of the DEGs related to proteolysis, including cysteine-type peptidase activity, aspartic-type endopeptidase activity, serine-type peptidase activity and metallopeptidase. The expression of the DEGs was analyzed in the venom gland of S. invicta on different days after laboratory rearing. Fold-change of expression level after laboratory rearing compared to control (i.e., a fold-change of 1 indicates no change compared to control) was calculated. Three biological replicates were performed. Means ± SE are shown. Significance of difference was analyzed by Duncan’s test (*P < 0.05; **P < 0.05). CK, 0 days after laboratory rearing; T1, 10 days after laboratory rearing; T2, 60 days after laboratory rearing.
Analysis of DEGs related to venom allergen
Allergen proteins are a key component of ants’ venom. Some genes related to allergen were detected by the RNA-Seq analysis. The laboratory rearing affected the expression of these genes, such as LOC105198838 and LOC105193335 (Fig. 4A). Moreover, the transcript levels of LOC105199703, LOC105198838, and LOC105193335 were examined by qRT-PCR analysis. As shown in Fig. 4B, laboratory rearing has no effect on the expression of LOC105199703. However, the transcript levels of LOC105198838 and LOC105193335 significantly decreased during laboratory rearing.

Figure 4: The expression patterns of the DEGs, related to venom allergen. (A) The expression level of LOC105199703, LOC105198838, and LOC105193335 according to the RNA-Seq analysis. (B) Quantitative RT-PCR analysis of expression of LOC105199703, LOC105198838 and LOC105193335. The expression of LOC105199703, LOC105198838 and LOC105193335 was analyzed in the venom gland of S. invicta at different days after laboratory rearing. Fold-change of expression level after laboratory rearing compared to control (i.e., a fold-change of 1 indicates no change compared to control) was calculated. Three biological replicates were performed. Means ± SE are shown. Significance of difference was analyzed by Duncan’s test (*P < 0.05; **P < 0.05). CK, 0 days after laboratory rearing; T1, 10 days after laboratory rearing; T2, 60 days after laboratory rearing.
Solenopsis invicta Buren has a specialized venom apparatus to sting victims and prey, inoculating a cocktail of biological and pharmacologically active substances (alkaloids, peptides, proteins, among others) for defense and predation (Touchard et al., 2016). A large number of alkaloids and some toxin-like proteins and peptides have been identified in the venom of S. invicta (Inagaki et al., 2004; Inagaki et al., 2008; Aili et al., 2016). Our previous study has revealed that laboratory rearing has a great influence on the venom of S. invicta: the contents of venomous alkaloid components decreased when the fire ants were maintained in the laboratory (Liu et al., 2015). However, the molecular mechanism of the change of venomous components when kept in the laboratory is unknown. Herein, we performed an RNA sequencing technique (RNA-Seq) to detect the transcriptome dynamics in the venom gland of S. invicta when kept in the laboratory. A large number of differentially expressed genes (DEGs) involved in multiple pathways were identified among these samples through the comparative analysis of their transcriptomes. The GO analysis showed that the oxidation-reduction processes were two of the most significantly differential GO subcategories among the three samples, which indicates that proteins encoded by these genes may be relevant to the change of ants’ venomous components when kept in the laboratory. However, a few DEGs related to alkaloids were detected (data not shown), which is a slight inconformity with our previous study that the level of venomous alkaloid components decreased when the fire ants were maintained in the laboratory (Liu et al., 2015). We deduce that laboratory rearing may affect the synthesis of alkaloids at the post-transcriptional level.
To date, many toxin-like peptides and proteins from the venom glands of the ants have been detected. Several hydrolases, oxidoreductases, proteases, Kunitz-like polypeptides, and the less abundant inhibitor cysteine knot (ICK)-like (knottin) neurotoxins and insect defensin have been revealed in the venom of ants using proteomic analysis (Bouzid et al., 2014; Torres et al., 2014). Our RNA-Seq analysis revealed that the expression of the genes related to proteolysis, including cysteine-type peptidase activity, aspartic-type endopeptidase activity, serine-type peptidase activity, and metallopeptidase, significantly changed after laboratory rearing. The qRT-PCR analysis showed that the transcript levels of LOC105207761, LOC105193270, LOC105197985, LOC105205908, LOC105198898, LOC105194706, LOC105193305, LOC105193613, LOC105196175, LOC113004172, LOC113003028, LOC105194248 and LOC105206642 significantly decreased after laboratory rearing. The proteolytic enzyme encoded by these genes may be the vitally venomous components or be responsible for generating the toxin-like peptides in the venom gland of the fire ant S. invicta. Allergens are also conserved venom components in the fire ant S. invicta, from which the allergen nomenclature is derived (Hoffman, 2010). The transcript levels of LOC105198838 and LOC105193335 encoded venom protein Sol g II significantly decreased after laboratory rearing, which indicates that these genes participate in the change of venomous components when kept in the laboratory.
In conclusion, a comprehensive characterization of DEGs related to proteolysis and allergen in the venom gland of S. invicta at 0, 10, and 60 days after laboratory rearing by the RNA-Seq technique has generated new data relating to venomous components and the transcriptome dynamics in the venom gland of S. invicta when kept in the laboratory.
Availability of Data and Materials: All data generated or analyzed during this study are included in this manuscript.
Author Contribution: The authors confirm contribution to the paper as follows: study conception and design: Limin Chen, Jun Huang and Yaobin Lu; data collection: Limin Chen, Xiaowei Li, Tianjun He, Quancong Wu and Tingting Chen; analysis and interpretation of results: Limin Chen, Jun Huang and Yaobin Lu; draft manuscript preparation: Limin Chen and Yaobin Lu. All authors reviewed the results and approved the final version of the manuscript.
Animal Welfare Note: Procedures involving animals and their care were conducted in conformity with institutional guidelines, and was approved by Animal Care and Use Committee of the Zhejiang Academy of Agricultural Sciences.
Ethics Approval: This study was approved by the Institutional Review Board (IRB) Institutional of Plant Protection and Microbiology, Zhejiang Academy of Agricultural Sciences. The approval code is 2020AWE061801. The approval date is June 19, 2020.
Funding Statement: This study was funded by Zhejiang Provincial Basic Public Welfare Research Program under Grant No. LGN21C140005.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Aili SR, Touchard A, Escoubas P, Padula MP, Orivel J et al. (2014). Diversity of peptide toxins from stinging ant venoms. Toxicon 92: 166–178. [Google Scholar]
Aili SR, Touchard A, Koh JMS, Dejean A, Orivel J et al. (2016). Comparisons of protein and peptide complexity in poneroid and formicoid ant venoms. Journal of Proteome Research 15: 3039–3054. [Google Scholar]
Bouzid W, Verdenaud M, Klopp C, Ducancel F, Noirot C, Vetillard A (2014). De Novo sequencing and transcriptome analysis for Tetramorium bicarinatum: A comprehensive venom gland transcriptome analysis from an ant species. BMC Genomics 15: 987. [Google Scholar]
Carvalho DB, Fox EGP, Santos DGD, Sousa JS, Freire DMG et al. (2019). Fire ant venom alkaloids inhibit biofilm formation. Toxins 11: 420. [Google Scholar]
Chen J, Zhao Y, Li XC, Zhao JH (2019). Pyridine alkaloids in the venom of imported fire ants. Journal of Agricultural and Food Chemistry 67: 11388–11395. [Google Scholar]
Das T, Alabi I, Colley M, Yan F, Griffith W et al. (2018). Major venom proteins of the fire ant Solenopsis invicta: Insights into possible pheromone-binding function from mass spectrometric analysis. Insect Molecular Biology 27: 505–511. [Google Scholar]
Dos Santos Pinto JR, Fox EG, Saidemberg DM, Santos LD, da Silva Menegasso AR et al. (2012). Proteomic view of the venom from the fire ant Solenopsis invicta Buren. Journal of Proteome Research 11: 4643–4653. [Google Scholar]
Eliyahu D, Ross KG, Haight KL, Keller L, Liebig J (2011). Venom alkaloid and cuticular hydrocarbon profiles are associated with social organization, queen fertility status, and queen genotype in the fire ant Solenopsis invicta. Journal of Chemical Ecology 37: 1242–1254. [Google Scholar]
Hoffman DR (2010). Ant venoms. Current Opinion in Allergy and Clinical Immunology 10: 342–346. [Google Scholar]
Hoffman DR, Dove DE, Jacobson RS (1988). Allergens in Hymenoptera venom. XX. Isolation of four allergens from imported fire ant (Solenopsis invicta) venom. Journal of Allergy and Clinical Immunology 82: 818–827. [Google Scholar]
Inagaki H, Akagi M, Imai HT, Taylor RW, Kubo T (2004). Molecular cloning and biological characterization of novel antimicrobial peptides, pilosulin 3 and pilosulin 4, from a species of the Australian ant genus Myrmecia. Archives of Biochemistry and Biophysics 428: 170–178. [Google Scholar]
Inagaki H, Akagi M, Imai HT, Taylor RW, Wiese MD et al. (2008). Pilosulin 5, a novel histamine-releasing peptide of the Australian ant, Myrmecia pilosula (Jack Jumper Ant). Archives of Biochemistry and Biophysics 477: 41–416. [Google Scholar]
Kruse B, Anderson J, Simon LV (2020). Fire ant Bites. Treasure Island (FLStatPearls Publishing. [Google Scholar]
Li XB, Fan XP, Wang XL, Cai L, Yang WC (2005). The cotton ACTIN1 gene is functionally expressed in fibers and participates in fiber elongation. Plant Cell 17: 859–875. [Google Scholar]
Liu XF, Chen L, Li JK (2015). Dynamic analysis of venomous alkaloids in workers of the red imported fire ant, Solenopsis invicta (Hymenoptera: Formicidae) maintained in the laboratory. Acta Entomologica Sinica 58: 22–27. [Google Scholar]
Lucas C, Nicolas M, Keller L (2015). Expression of foraging and Gp-9 are associated with social organization in the fire ant Solenopsis invicta. Insect Molecular Biology 24: 93–104. [Google Scholar]
Monsalve RI, Gutiérrez R, Hoof I, Lombardero M (2020). Purification and molecular characterization of phospholipase, antigen 5 and hyaluronidases from the venom of the Asian hornet (Vespa velutina). PLoS One 15: e0225672. [Google Scholar]
Palma MS, Brochetto-Braga MR (1993). Biochemical variability between venoms from different honey-bee (Apis mellifera) races. Comparative Biochemistry and Physiology Part C: Pharmacology, Toxicology and Endocrinology 106: 423–427. [Google Scholar]
Pandey M, Addesso KM, Archer RS, Valles SM, Baysal-Gurel F et al. (2019). Worker size, geographical distribution, and introgressive hybridization of invasive Solenopsis invicta and Solenopsis richteri (Hymenoptera: Formicidae) in Tennessee. Environmental Entomology 48: 727–732. [Google Scholar]
Pluzhnikov KA, Kozlov SA, Vassilevski AA, Vorontsova OV, Feofanov AV, Grishin EV (2014). Linear antimicrobial peptides from Ectatomma quadridens ant venom. Biochimie 107: 211–215. [Google Scholar]
Torres AF, Huang C, Chong CM, Leung SW, Prieto-da-Silva AR, Havt A, Quinet YP, Martins AM, Lee SM, Rádis-Baptista G (2014). Transcriptome analysis in venom gland of the predatory giant ant Dinoponera quadriceps: insights into the polypeptide toxin arsenal of hymenopterans. PLoS One 9: e87556. [Google Scholar]
Touchard A, Aili SR, Fox EG, Escoubas P, Orivel J et al. (2016). The biochemical toxin arsenal from ant venoms. Toxins 8: 30. [Google Scholar]
Schumacher MJ, Schmidt J, Egen NB, Dillon KA (1992). Biochemical variability of venoms from individual European and Africanized honeybees (Apis mellifera). Journal of Allergy and Clinical Immunology 90: 59–65. [Google Scholar]
Wilson R, Gullan PJ, Cranston PS (2010). The Insects: An Outline of Entomology, vol. 14, 4th ed., Wiley: Hoboken, NJ, USA. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |