DOI:10.32604/biocell.2022.017316

| BIOCELL DOI:10.32604/biocell.2022.017316 |  |
| Review |
Mechanism of tobacco osmotin gene in plant responses to biotic and abiotic stress tolerance: A brief history
1State Key Laboratory of Conservation and Utilization of Subtropical Agro-Bioresources, College of Forestry and Landscape Architecture, South China Agricultural University, Guangzhou, 510642, China
2National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan, 430070, China
*Address correspondence to: Hakim Manghwar, hakim@scau.edu.cn
#These authors contributed equally to this work
Received: 01 May 2021; Accepted: 23 June 2021
Abstract: Plants are recurrently exposed to myriads of biotic and abiotic stresses leading to several biochemical and physiological variations that cause severe impacts on plant growth and survival. To overcome these challenges, plants activate two primary defense mechanisms, such as structural response (cell wall strengthening and waxy epidermal cuticle development) and metabolic changes, including the synthesis of anti-microbial compounds and proteins, especially the pathogenesis-related (PR) proteins. PR proteins are members of a super large family of defense proteins that exhibit antimicrobial activities. Their over-expression in plants provides tolerance to many abiotic and biotic stresses. PR proteins have been classified into 17 families, including PR-5–also called thaumatin-like proteins (TLPs) that involve osmotin and osmotin-like proteins (OLPs). Osmotin was first identified in tobacco (Nicotiana tabacum var. Wisconsin 38), and then its homolog proteins (OLPs) were reported from the whole plant kingdom. Osmotin and OLPs are ubiquitous in all fruits and vegetables. Their expression has been detected in various plant tissues and organs. The phylogenetic tree studies revealed that the osmotin group originated from spermatophytes. Moreover, the atomic structure of OLP has shown similarity to thaumatin and TLPs from monocot and eudicot species, which determines a strong evolutionary pressure in flowering plants for conserving thaumatin fold. This is associated with the role of these proteins against pathogens as defense molecules and to induce stress tolerance to plants against several biotic and abiotic factors. In this review, we have briefly described the development history of osmotin, including its function and mechanism to induce biotic and abiotic stress tolerance to plants.
Keywords: Osmotin; PR-5 proteins; Osmotin-like proteins (OLPs); Biotic stress; Abiotic stress; Plant development
The growth and productivity of crop plants are mainly determined by their susceptibility to environmental stresses. Throughout their life cycle, plants are continually subjected to several biotic and abiotic stresses, which cause biochemical, morphological, and physiological variations that affect normal plant development, productivity, and even survival (Patade et al., 2013; Kumar et al., 2015). The biotic stress factors include bacteria, fungi, and viruses, while drought, salinity, high or low temperature, ultraviolet radiations, and hypoxia are the abiotic factors that provoke stress conditions in plants (Bashir et al., 2020). To overcome these stresses, plants defend themselves by initiating different stress response mechanisms, for example, the structural response that involves cell wall strengthening and waxy epidermal cuticle development.
These responses deliver enough strength and rigidity to the plants that enable them to decrease the effects of damage caused by biotic and abiotic stresses (Bashir et al., 2020). Another stress response mechanism that plants activate is the metabolic modifications, such as the generation of reactive oxygen species (ROS), activation of hypersensitive response, synthesis of anti-microbial compounds, and proteins, particularly the synthesis of PR proteins (Das et al., 2011; Li et al., 2015).
PR proteins are members of a super large protein family in plants–related to defense against abiotic and biotic stresses, and the over-expression of these proteins renders plants tolerant to these stresses (Kaur and Kumar, 2020). Based on the primary structure, amino acid composition, biochemical activities, and serological relationship, PR proteins have been classified into 17 families, i.e., β 1,3 glucanases (PR-2), chitinases (PR-3, 4, 8 and 11), thaumatin-like proteins (PR-5), ribosome-inactivating proteins (PR-10), defensins (PR-12), thionins (PR-13), and lipid transfer proteins (PR-14) (Hakim et al., 2018; Kaur and Kumar, 2020). Among these, pathogenesis-related protein family 5 (PR-5) in particular, is well-known for its role in osmoregulation, plant development, and for its important antimicrobial activities (Mani et al., 2012). The osmotin and osmotin-like proteins (OLPs) among PR-5 have been considered most important in provoking tolerance to several biotic and abiotic stresses in plants (Chowdhury et al., 2017; Bashir et al., 2020). Since the last decade, several studies have been conducted to explore the functions of osmotin and OLPs in a diverse range of plants (Table 1). The over-expression of these genes provides tolerance to drought, salinity, cold, high/low temperatures, plant development, and tolerance to plant diseases caused by fungi, bacteria, or viruses. Most of the research to date has been conducted on the functions and mechanism of osmotin and OLPs. However, in the present study, we have mainly focused on the development history of osmotin and OLPs and have briefly described their function and mechanism for inducing biotic and abiotic stress tolerance in plants.
Development History of Osmotin
Osmotin protein was first discovered in tobacco (Nicotiana tabacum var. Wisconsin 38) cells adapted to a low osmotic potential environment that exhibited salt stress tolerance (Singh et al., 1987). Thus, it was named ‘osmotin’ due to its function in lowering osmotic potential under stress (Viktorova et al., 2012). Later, some homolog proteins were also identified throughout the plant kingdom involving various monocot and dicot plants (Viktorova et al., 2019). Osmotin and its homolog proteins, the OLPs are ubiquitous in all vegetables and fruits (Viktorova et al., 2012). In plants, the expression of these genes has been observed in several organs, such as flowers, leaves, trichomes, grape skin, somatic embryos, and olive seed coat and endosperm. Osmotin has also been found in plant tissues like in the epidermis of corolla, stem, and root (Bashir et al., 2020). However, the studies on phylogenetic tree analysis revealed that the osmotin group emerged from spermatophytes (Faillace et al., 2019). The atomic structure of an OLP, CpOsm from Calotropis procera latex has shown that its three-dimensional structure was very similar to the structure of thaumatin and TLPs from eudicot and monocot species. It indicates a strong evolutionary pressure in flowering plants for conserving thaumatin fold associated with the role of these proteins as defense molecules against pathogens and their function in providing tolerance to plants against different stresses (Ramos et al., 2015).
In fact, osmotin is synthesized in the vacuole possessing a molecular weight of 26.4 kDa in its pre-protein form, but its molecular weight decreases to 24 kDa during maturity. Notably, osmotin is a multifunctional protein with 244 residues. In addition to osmoregulation, it plays a significant role in protecting plants against pathogens. Therefore, it has been classified in the family of PR proteins (PR-5 family) (Abdin et al., 2011; Anu et al., 2015; Su et al., 2017). Osmotin and all other members in the PR-5 family share high amino acid sequence similarity to thaumatin (Cao et al., 2016). Typically, the PR-5 family proteins, also known as TLPs, have a molecular mass ranging from 21 to 26 kDa that usually contain 16-cysteines residues. These residues make 8 disulfide linkages, which are associated with the structural stability of these proteins across a diverse pH range, heat-induced denaturation, and accurate folding (Cao et al., 2016).
As a cationic protein, osmotin has two forms–the one possessing isoelectric point (pI) of 7.8 while the other has pI more than 8.2. The molecular weight of both slightly differs, resulting in water-soluble osmotin I and water-insoluble osmotin II (Noori and Sokhansanj, 2008). In addition, the strong amino acid (Glu, Asp, Asp4) residues exist on the grooves of osmotin proteins, which lead to their acidic nature due to the negative charge (Wan et al., 2017). A recent report on osmotin from Kandelia obovate (KoOsmotin) also indicated a negatively charged molecular surface with a hydrophilic nature of the KoOsmotin protein (Fei et al., 2021). Whereas the OLPs are neutral in nature (Wan et al., 2017). The peptide sequence of osmotin is also similar to the tomato NP protein 24, barley thiamine-like proteins, potato PR protein C, the maize trypsin/a-amylase inhibitor, and tobacco antiviral protein gp 22 (Subramanyam et al., 2011). Osmotin comprises three motifs, which exhibit similar folding to PR-5 proteins like zeamatin and thaumatin (Hakim et al., 2018). However, it does not have glycosylation locations, DNA-binding motifs, and introns (Abdin et al., 2011).
The polypeptide structure of osmotin has shown variation in terms of the presence of C-and N-terminal polypeptide extensions. The C-terminal domain is always supposed to be present in osmotin and other PR-5 proteins (Viktorova et al., 2012), for example, the TlOsm. The presence of a C-terminal elongation helps in the localization of the protein in the vacuole (Viktorova et al., 2019). But some of the proteins in the PR-5 family, such as KoOsmotin (Fei et al., 2021), OLP (Onishi et al., 2006), ZLP (Moeller and Tiffin, 2005; Schnable et al., 2009), TLP (Singh et al., 2013), and zeamatin (Sun et al., 2018) lack C-terminal domain. Several PR-5 proteins that lack C-terminal elongation contain only the N-terminal signal peptide–are mainly considered to be secreted into the extracellular matrix (Onishi et al., 2006) and are localized in the apoplastic space of plants (Tachi et al., 2009). However, the subcellular localization studies performed for plant osmotins have reported their localization on multiple subcellular compartments, including cytoplasm, endoplasmic reticulum, chloroplast, vacuole, and plasma membrane (Le et al., 2018; Fei et al., 2021).
Although extensive research has been conducted on the defense function of osmotin, its complex interplay in gene regulation was not clear until the last decade. Initially, various studies hypothesized that osmotin might be a transcription factor for regulating key genes in response to biotic and abiotic stresses (Barthakur et al., 2001; Husaini and Abdin, 2008). However, later this hypothesis was ruled out by the fact that osmotin did not contain any DNA-binding motifs (Abdin et al., 2011; Fei et al., 2021). Further, the osmotin had also been reported to initiate mitogen-activated protein kinase (MAPK) that was essential for most of the regulatory and signaling processes (Yun et al., 1998). Therefore, researchers have recently demonstrated that osmotin is a key regulator responding to different plant stresses (Le et al., 2018). In addition, osmotin plays a vital role in protecting the structure of native proteins and repairing degraded proteins (Patade et al., 2013).
Role of osmotin in abiotic stress tolerance
Osmotin is a multiple stress-responsive PR-5 protein initially considered as a salt-induced protein in osmotically stressed tobacco cells (Singh et al., 1985; Kumar et al., 2015). Under low water potential conditions, osmotin provides defense to the cell membrane and is involved in osmoregulation and food preservation (Hakim et al., 2018; Bashir et al., 2020). Among all identified PR-5 proteins, osmotin and OLPs are primarily characterized for their function in osmoprotection and anti-fungal activities (Chowdhury et al., 2017). These proteins accumulate under stress conditions that result in the compartmentalization of ions or solutes and initiate anti-pathogen activities (Kumar et al., 2015). To date, the genes that encode osmotins and OLPs have been isolated from various plant species, which are known to exhibit multiple functions to enhance stress tolerance (Le et al., 2018; Li et al., 2020).
After the first discovery from N. tabacum, osmotins and OLPs have now been isolated and cloned in various species of the plant kingdom, as summarized in Table 1. Many studies have reported enhanced tolerance to salinity, cold, drought, as well as some combined stresses by over-expressing osmotin gene in plants (Subramanyam et al., 2012; Le et al., 2018). The ectopic expression of N. tabacum osmotin gene in chilli pepper (Capsicum annum L.) exhibited improved tolerance to salt stress. In addition, transgenic C. annum plants showed increased proline, chlorophyll, relative water contents, and antioxidative enzymes (Subramanyam et al., 2011). In a recent report, Bashir et al. (2021) demonstrated salt tolerance in transgenic olive (Olea europaea L.) plants over-expressing the tobacco osmotin gene. Salt stress tolerance in transgenic olive plants was associated with the induction of the S assimilatory pathway, especially due to an increase in the activity of O-acetyl serine(thiol)lyase (OASTL) (Bashir et al., 2021). Under cold treatment (15°C), the over-expression of N. tabacum osmotin gene improved seed germination rate, and the establishment and growth of seedlings in transgenic tomato (Solanum lycopersicum L.) (Patade et al., 2018). Recently, Fei et al. (2021) has reported the characterization of a novel osmotin gene the KoOsmotin–from Kandelia obovata that induces cold tolerance and helps to enhance the growth of Escherichia coli cells under cold stress. The osmotin from Tripogon loliiformis (TlOsm) provided tolerance to Oryza sativa (rice) plants from cold, salt, and drought stresses. This designates the role of osmotin in response to different abiotic stresses in plants, possibly by the signaling pathway (Le et al., 2018).
Drought is another environmental factor that affects yield in crop plants. Several studies have observed drought tolerance in crop plants by expressing osmotin or OLPs, which enhance physiological responses against drought. Along with drought tolerance, higher grain productivity, transpiration rate, stomatal conductance, and net CO2 assimilation were observed in transgenic soybean (Glycine max L.) expressing OLP gene (SnOLP) (Weber et al., 2014). Tobacco osmotin gene transformation induced drought tolerance in transgenic kiwi (Actinidia deliciosa A. Chev.) and olive plants (Silvestri et al., 2017; Rugini et al., 2018). With higher drought and salt tolerance, over-expressing tobacco osmotin in Mulberry (Morus indica L. cv K2) plants imparted resistance to Fusarium pallidoroseum, Colletotrichum dematium, and Colletotrichum gloeosporioides (Das et al., 2011). The over-expression of SindOLP in sesame enhanced the tolerance toabiotic stress (salinity, drought, and oxidative stress), the charcoal rot pathogen (Macrophomina phaseolina) (Chowdhury et al., 2017). Moreover, osmotins and OLPs also provide a vital role in plant growth and development. For instance, the accumulation of PhOSM in Petunia (Petunia hybrida) flowers, leaves, and roots demonstrates the importance of osmotin in plant development (Byoun and Kim, 1997). Recently, Li et al. (2020) reported that the downregulation of PdOLP1 was involved in wood development, secondary cell wall biosynthesis, higher dry weight biomass, and lignin content in poplar (Populus deltoides Marsh.).
Role of osmotin in biotic stress tolerance
Many osmotins and OLPs have been studied in plants for their role in biotic stress tolerance as presented in Table 1. These proteins possess anti-fungal activities-related to their potential to induce fungal plasma membrane permeabilization (Ramos et al., 2015). The anti-fungal properties of osmotin and OLPs lead to disease resistance in many crop plants, like tomato, potato, rice, and tobacco (Xue et al., 2016; Hakim et al., 2018; Kaur et al., 2020). The CpOsm–an anti-fungal TLP has been reported to inhibit the spore germination of C. gloeosporioides, Neurospora sp., and Fusarium solani (de Freitas et al., 2011). Rather et al. (2015) characterized ObOLP from sweet basil (Ocimum basilicum), which showed anti-fungal activity against Pichia pastoris, Candida albicans, and Saccharomyces cerevisiae. The resistance to potato early blight in susceptible potato plants by over-expressing the OsmWS gene has also been reported (Kaur et al., 2020). Osmotin from tobacco was analyzed for anti-fungal activity, which showed significant effects against Candida tropicalis, C. parapsilosis, Debaryomyces hansenii, Saccharomycodes ludwigii, Hyphopichia burtonii, and Torulaspora globose (Viktorova et al., 2017). Osmotin (OsOSM1) gene over-expression in rice (O. sativa var. Xudao 3) increased resistance to sheath blight, the most destructive rice disease caused by Rhizoctonia solani. The study demonstrated the induction of jasmonic acid-responsive marker genes in response to the upregulation of jasmonic acid in transgenic rice plants. It suggests that the jasmonic acid signaling pathway activation may account for the increased resistance in transgenic lines (Xue et al., 2016). In addition, osmotin has been used to evaluate its role in post-harvest fruit damage by fungal pathogens. The long-term stored kiwifruits of transgenic kiwi plants expressing tobacco osmotin gene exhibited significant resistance to post-harvest damage by Botrytis cinerea and Cadophora luteo-olivacea, which mainly cause gray mould and skin pitting of kiwifruit, respectively (Rugini et al., 2010).
Presently, due to the effects of multiple stress factors in plants, the development of tolerance to individual stresses seems insufficient. Therefore, the introduction of multiple genes is needed to develop biotic and abiotic stress tolerance (Kumar et al., 2016; Sripriya et al., 2017). The combined application of chitinase with osmotin, defensin, PR1, TLP, and glucanase enhanced its anti-fungal activity (Shah et al., 2013; Dolatabadi et al., 2014). Thus, higher resistance to different stresses can be achieved by the combined use of osmotin and other PR proteins. Thus, a combination of osmotin (ap24) and chitinase (chi11) gene expression was reported enhancing the tolerance against sheath blight in transgenic rice (Sripriya et al., 2017). Moreover, the transgenic tomato was developed by combinatorial expression of OLP and Chi11 showing increased tolerance to drought, salt, and fungal (Alternaria solani and Fusarium oxysporun) stresses (Kumar et al., 2016). Thus, the use of multiple genes, such as osmotin or OLPs, along with other stress-tolerant genes can be more useful to confer multiple stress tolerance in plants. With abiotic and biotic stress tolerance, the recent literature highlights the potential of osmotins and OLPs for engineering crops to increase growth and development, productivity, and survival.
Osmotin mode of action in abiotic stress
Osmotin and OLPs exhibit tissue-specific activities and are expressed in response to various environmental and hormonal signals, such as drought, wounding, salinity, ABA, ethylene, and pathogenic (fungi, viruses, bacteria) infection. These signals cause transcriptional and post-transcriptional osmotin mRNA regulation (Noori and Sokhansanj, 2008; Parkhi et al., 2009). Under drought or salinity stresses, osmotin or OLPs maintain cellular osmolarity via compartmentalization of solutes in the vacuole and intercellular space or by structural and metabolic alterations, such as changes in Ca2+ leading to programmed cell death or by the activation of different signaling pathways (Chowdhury et al., 2017), as shown in Fig. 1. The plants respond to salinity stress through gene expression regulation that eventually results in cellular homeostasis renovation, toxin detoxification, and growth recovery (Subramanyam et al., 2011).
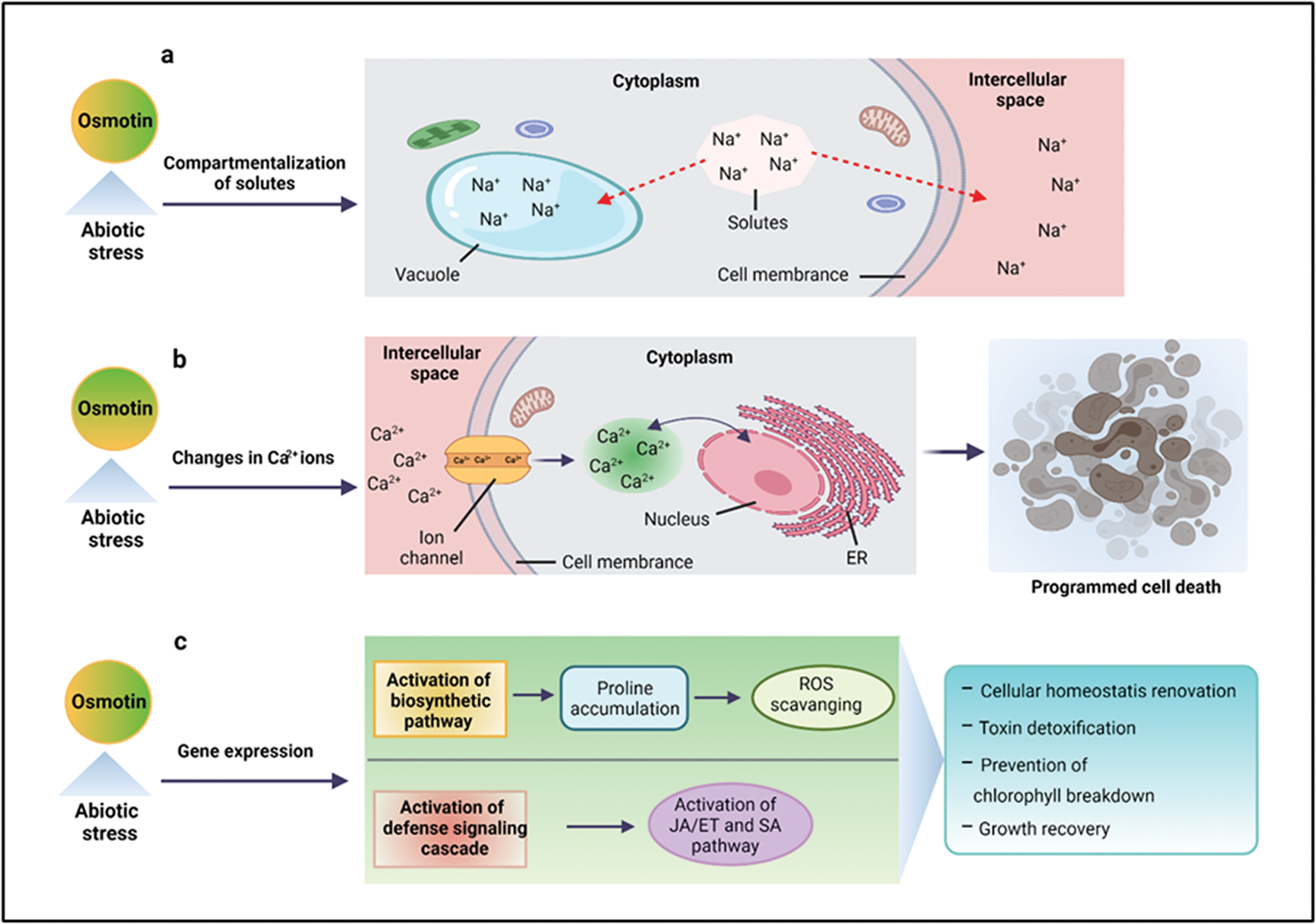
Figure 1: Mechanism of osmotin in abiotic stress tolerance. Osmotin and OLPs confer tolerance to abiotic stresses (salinity, drought, or cold) by compartmentalization of solutes in the vacuole and intercellular space (a), changing Ca2+ regulation that eventually results in programmed cell death (b), and by gene expression regulation, such as the activation of biosynthetic pathway that initiates proline accumulation involved in scavenging of reactive oxygen species (ROS). In addition, it also activates the defense signaling cascade, which is involved in the initiation of JA/ET and SA pathways (c). ER: endoplasmic reticulum, JA: jasmonic acid, ET: ethylene, SA: salicylic acid.
The osmotin gene over-expression triggers downstream gene expression that upregulates the biosynthetic pathway. This pathway is involved in the accumulation of osmolyte proline, which quenches ROS, free radicals, and prevents the breakdown of chlorophyll under higher salt stress conditions (Zhang et al., 2004). The OLP gene is reported to impart increased tolerance to multiple stresses by the integrated activation of multiple components of the defense signaling cascade. For example, the SindOLP gene is involved in the expression of Si-Defensin, Si-Apetala2, and Si-Ethylene-responsive factor, the marker genes for the JA/ET pathway, along with Si-Thaumatin-like protein and Si-Chitinase, the marker genes for the SA pathway (Chowdhury et al., 2017; Pluskota et al., 2019). Moreover, the cell membrane has been considered as the main target of different stresses. Under drought and salt stresses, osmotin provides a defensive role by maintaining cell membrane integrity (Das et al., 2011). For example, OLP-transgenic tomato plants displayed increased thicknesses of the secondary cell wall of vessels and fibers under salt stress–acting as an important mechanism to improve plant resistance to water stress (Kumar et al., 2016).
Osmotin mode of action in biotic stress
Osmotin has been found to have activities against different phytopathogens, such as bacteria and viruses, but it is well-known for its anti-fungal activity against a broad range of fungi. Although the cell wall of fungi has an important role in defining sensitivity to osmotin, the main target position for osmotin and other PR-5 proteins is the fungal plasma membrane (Anžlovar and Dermastia, 2003). To enhance cytotoxicity, osmotin uses an intercellular signal transduction pathway in order to destabilize the protective cell wall barriers (Kumar et al., 2015). After the activation of this pathway, it leads to the stimulation of the MAPK cascade. This causes changes in the cell wall, allowing osmotin to enter the plasma membrane, resulting in the development of a transmembrane pore, which causes membrane leakage and eventually leads to membrane rupture (Singh et al., 2013).
Osmotin is also diffused across the cell wall of fungi by recognizing and binding to the cell wall phosphomannans and enter the cell membrane (Ibeas et al., 2000). The analysis of the interaction between osmotin and the cell surface of pathogens revealed that after treating fungal spores with osmotin the protein is attached to the cell surface components. Where it causes dramatic changes in the permeability of plasma membrane and results in the overflow of cytoplasmic content that shrinks the cell (Ramos et al., 2015), and subsequently it kills the fungus (Roberts and Selitrennikoff, 1990; Sripriya et al., 2017), as presented in Fig. 2. However, the exact mechanism of altering cell membrane permeability by osmotin and other anti-fungal TLPs is unclear. Though, it is considered that due to the negatively charged components in the acidic cleft, these proteins attach to the positively charged membrane proteins like osmoreceptors, ions, or water channels that ultimately lead to a dramatic increase in the water flow across the membrane (Ramos et al., 2015).
In the case of bacteria, osmotin does not exhibit direct anti-bacterial activity, but indirectly it prevents bacterial infection in host tissues. Some bacterial species like Agrobacterium tumefaciens use cytokinins in turning host’s activity to their own favor (Viktorova et al., 2012). Osmotin has a close relationship to cytokinin signal transduction (Kobayashi et al., 2000). It has been assumed that osmotin binding to cytokinin results in the deactivation of cytokinins that prevents bacteria from inducing tumors because the elimination of cytokinin suppresses the proliferation of bacterial infection (Viktorova et al., 2012). Choi et al. (2013) reported that the overexpression of an OLP (CaOSM1) in A. thaliana conferred resistance to a bacterial pathogen Pseudomonas syringae pv. Tomato by accelerating hypersensitive cell death response and accumulating H2O2 that prevented the spread of pathogens to healthy cells. Although, osmotin is mainly characterized for its anti-fungal properties, studies have also reported its anti-viral activity. Osmotin was identified in Samsun NN tobacco leaves after infecting with tobacco mosaic virus (TMV) and induced tolerance to viral infection via a hypersensitive response (Stintzi et al., 1991; LaRosa et al., 1992). Infecting tobacco explants with TMV induced the expression of osmotin in infected leaves (Neale et al., 1990). In addition, OLP was accumulated in habanero pepper (Capsicum chinense) infected with pepper mild mottle virus (PMMoV) (Elvira et al., 2008). Comprehensive research has been conducted on the anti-fungal activity of osmotin and OLPs; however, further research is needed for their anti-bacterial as well as anti-viral properties.
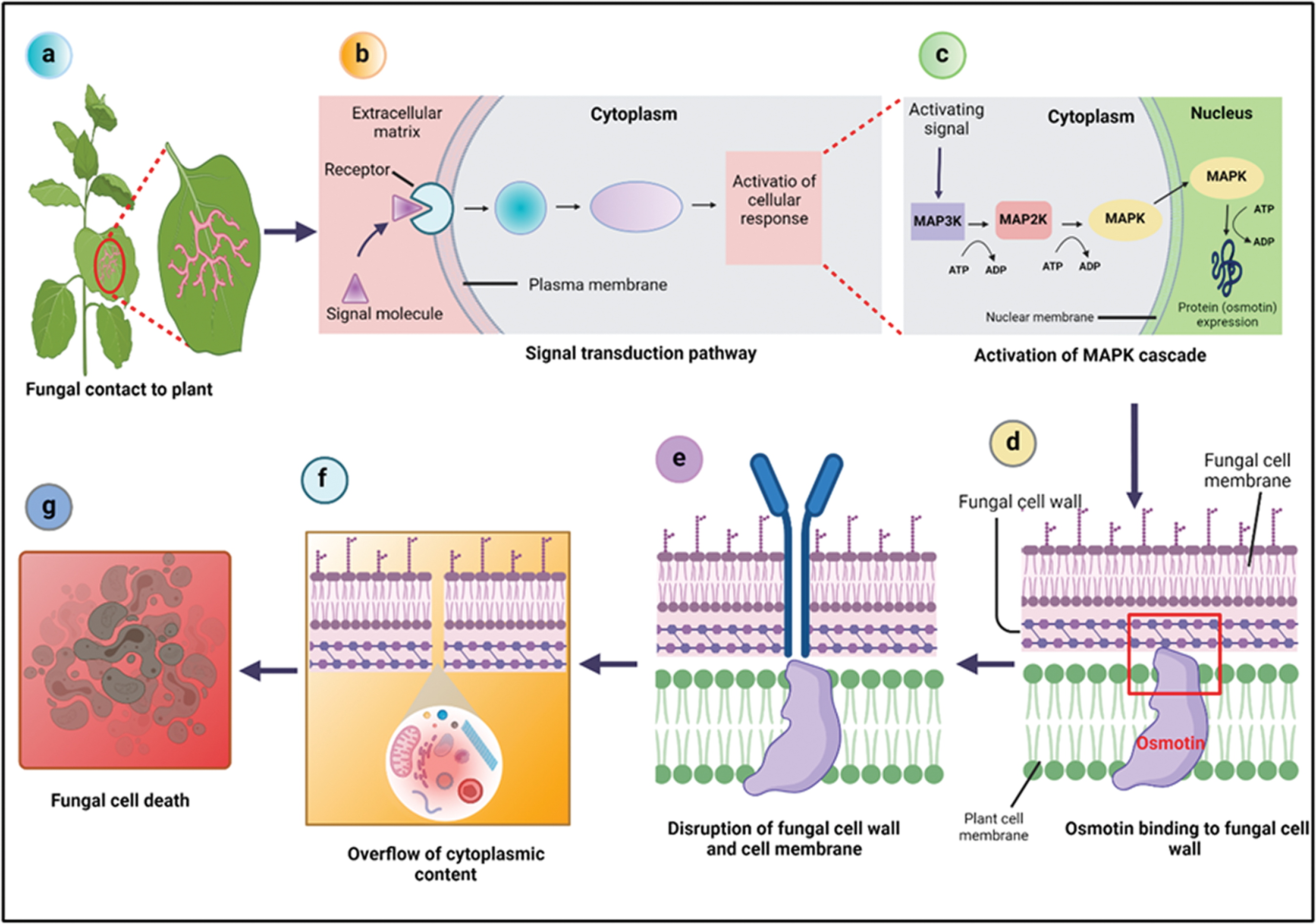
Figure 2: Mechanism of osmotin in biotic (fungal) stress tolerance. After the plant is infected by fungi (a), it initiates signal transduction pathway (b), which leads to the activation of MAPK cascade resulting in protein (osmotin) expression (c). The osmotin binds to the fungal cell wall by recognizing phosphomannans (d) and enters the fungal cell membrane where it causes membrane permeability and eventually leads to membrane disruption (e) that results in the overflow of cytoplasmic contents (f) and eventually kills the fungus (g).
Conclusions and Future Perspective
Osmotin and OLPs belong to the PR-5 family that offer vital functions in imparting tolerance to various stresses. After the first discovery of osmotin from tobacco plants, osmotin and OLPs are identified from the whole plant kingdom. The similarity of OLPs to thaumatin and TLPs determines a strong evolutionary pressure for conserving the thaumatin fold associated with their role as defense molecules against pathogens and confer plant tolerance to abiotic and biotic stresses. Osmotin is a multi-functional key regulator triggering a response to different stresses. It also protects innate protein structures and is involved in normal plant development. Different environmental and hormonal signals, i.e., salinity, low temperature, wounding, drought, ethylene, ABA, and a pathogenic infection regulate the expression of osmotin and OLP genes. Under salt and drought stresses, these proteins maintain cellular osmolarity via solute compartmentalization, metabolic or structural modification, and by activating different signaling pathways. Importantly, inducing tolerance to individual stresses is insufficient; thus, the introduction of multiple genes can be more beneficial for providing strong tolerance to multiple stresses. Therefore, osmotin or OLPs with other key genes can be cloned in crop plants to confer resistance to different stresses and improve important agronomic traits (Viktorova et al., 2017; Fei et al., 2021). Notably, transgenic plants overexpressing these genes can be used in the remediation of salt-contaminated sites (Viktorova et al., 2017). In addition, the development of disease resistance in plants can not only increase crop yield but also overcome environmental pollution by reducing the use of pesticides.
Due to higher anti-fungal activity, osmotin and OLPs have been widely used in the development of transgenic plants to increase the anti-fungal. It indicates the importance of these proteins in the agricultural, and food industry, where they can be used as preservative agents (Viktorova et al., 2012). Moreover, osmotin also has homology to human adiponectin, which is an anti-diabetic and anti-atherosclerotic protein. Both these proteins provide anti-tumor resistance by preventing the activity of p53 gene and caspase enzyme (Das and Chakraborty, 2016; Bashir et al., 2020). Thus, due to the similarity of the osmotin receptor to the adiponectin receptor, it can be applied in the pharmaceutical industry to prepare drugs against insulin resistance, atherosclerosis, obesity, and diabetes.
Authors’ Contribution: The authors confirm contribution to the paper as follows: study conception and manuscript revision: H. Manghwar; draft manuscript preparation: A. Hussain. Both authors reviewed the manuscript and approved the final version of the manuscript.
Funding Statement: The author(s) received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Abdin MZ, Kiran U, Alam A (2011). Analysis of osmotin, a PR protein as metabolic modulator in plants. Bioinformation 5: 336–340. DOI 10.6026/97320630005336. [Google Scholar] [CrossRef]
Annon A, Rathore K, Crosby K (2014). Overexpression of a tobacco osmotin gene in carrot (Daucus carota L.) enhances drought tolerance. In Vitro Cellular & Developmental Biology-Plant 50: 299–306. DOI 10.1007/s11627-013-9590-0. [Google Scholar] [CrossRef]
Anu K, Jessymol KK, Chidambareswaren M, Gayathri GS, Manjula S (2015). Downregulation of osmotin (PR5) gene by virus-induced gene silencing (VIGS) leads to susceptibility of resistant Piper colubrinum Link. to the oomycete pathogen Phytophthora capsici Leonian. Indian Journal of Experimental Biology 53: 329–334. [Google Scholar]
Anžlovar S, Dermastia M (2003). The comparative analysis of osmotins and osmotin-like PR-5 proteins. Plant Biology 5: 116–124. DOI 10.1055/s-2003-40723. [Google Scholar] [CrossRef]
Barthakur S, Babu V, Bansal KC (2001). Over-expression of osmotin induces proline accumulation and confers tolerance to osmotic stress in transgenic tobacco. Journal of Plant Biochemistry Biotechnology 10: 31–37. [Google Scholar]
Bashir MA, Silvestri C, Ahmad T, Hafiz IA, Abbasi NA, Manzoor A, Cristofori V, Rugini E (2020). Osmotin: A cationic protein leads to improve biotic and abiotic stress tolerance in plants. Plants 9: 992. DOI 10.3390/plants9080992. [Google Scholar] [CrossRef]
Bashir MA, Silvestri C, Coppa E, Brunori E, Cristofori V et al. (2021). Response of olive shoots to salinity stress suggests the involvement of sulfur metabolism. Plants 10: 350. [Google Scholar]
Bernardino MC, Couto MLC, Vaslin MF, Barreto-Bergter E (2020). Antiviral activity of glucosylceramides isolated from Fusarium oxysporum against Tobacco mosaic virus infection. PLoS One 15: e0242887. DOI 10.1371/journal.pone.0242887. [Google Scholar] [CrossRef]
Byoun BH, Kim SG (1997). Isolation and molecular characterization of the osmotin gene in Petunia hybrida. Genes & Genomics 19: 340. [Google Scholar]
Cao J, Lv Y, Hou Z, Li X, Ding L (2016). Expansion and evolution of thaumatin-like protein (TLP) gene family in six plants. Plant Growth Regulation 79: 299–307. [Google Scholar]
Choi DS, Hong JK, Hwang BK (2013). Pepper osmotin-like protein 1 (CaOSM1) is an essential component for defense response, cell death, and oxidative burst in plants. Planta 238: 1113–1124. DOI 10.1007/s00425-013-1956-3. [Google Scholar] [CrossRef]
Chowdhury S, Basu A, Kundu S (2017). Overexpression of a new osmotin-like protein gene (SindOLP) confers tolerance against biotic and abiotic stresses in sesame. Frontiers in Plant Science 8: 410. DOI 10.3389/fpls.2017.00410. [Google Scholar] [CrossRef]
Das M, Chauhan H, Chhibbar A, Haq QMR, Khurana P (2011). High-efficiency transformation and selective tolerance against biotic and abiotic stress in mulberry, Morus indica cv. K2, by constitutive and inducible expression of tobacco osmotin. Transgenic Research 20: 231–246. DOI 10.1007/s11248-010-9405-6. [Google Scholar] [CrossRef]
Das S, Chakraborty S (2016). The role of osmotin protein tolerance to biotic and abiotic stress in plants. International Journal of Bioinformatics and Biological Sciences 4: 35. DOI 10.5958/2321-7111.2016.00005.6. [Google Scholar] [CrossRef]
de Freitas CDT, Nogueira FCS, Vasconcelos IM, Oliveira JTA, Domont GB, Ramos MV (2011). Osmotin purified from the latex of Calotropis procera: Biochemical characterization, biological activity and role in plant defense. Plant Physiology and Biochemistry 49: 738–743. DOI 10.1016/j.plaphy.2011.01.027. [Google Scholar] [CrossRef]
Dolatabadi B, Ranjbar GH, Tohidfar M, Dehestani A (2014). Genetic transformation of tomato with three pathogenesis-related protein genes for increased resistance to Fusarium oxysporum f. sp. lycopersici. Journal of Plant Molecular Breeding 2: 1–11. [Google Scholar]
Elvira MI, Galdeano MM, Gilardi P, García-Luque I, Serra MT (2008). Proteomic analysis of pathogenesis-related proteins (PRs) induced by compatible and incompatible interactions of pepper mild mottle virus (PMMoV) in Capsicum Chinese L 3 plants. Journal of Experimental Botany 59: 1253–1265. DOI 10.1093/jxb/ern032. [Google Scholar] [CrossRef]
Faillace GR, Turchetto-Zolet AC, Guzman FL, de Oliveira-Busatto LA, Bodanese-Zanettini MH (2019). Genome-wide analysis and evolution of plant thaumatin-like proteins: A focus on the origin and diversification of osmotins. Molecular Genetics and Genomics 294: 1137–1157. [Google Scholar]
Falcao LL, Silva-Werneck JO, Ramos AD, Martins NF, Bresso E, Rodrigues MA, Bemquerer MP, Marcellino LH (2016). Antimicrobial properties of two novel peptides derived from Theobroma cacao osmotin. Peptides 79: 75–82. DOI 10.1016/j.peptides.2016.03.006. [Google Scholar] [CrossRef]
Fei J, Wang YS, Cheng H, Su YB, Zhong Y, Zheng L (2021). Cloning and characterization of KoOsmotin from mangrove plant Kandelia obovata under cold stress. BMC Plant Biology 21: 1–12. [Google Scholar]
Goel D, Singh AK, Yadav V, Babbar SB, Bansal KC (2010). Overexpression of osmotin gene confers tolerance to salt and drought stresses in transgenic tomato (Solanum lycopersicum L.). Protoplasma 245: 133–141. [Google Scholar]
Hakim UA, Hussain A, Shaban M, Khan AH, Alariqi M et al. (2018). Osmotin: A plant defense tool against biotic and abiotic stresses. Plant Physiology and Biochemistry 123: 149–159. DOI 10.1016/j.plaphy.2017.12.012. [Google Scholar] [CrossRef]
Husaini AM, Abdin MZ (2008). Development of transgenic strawberry (Fragaria ×ananassa Duch.) plants tolerant to salt stress. Plant Science 174: 446–455. DOI 10.1016/j.plantsci.2008.01.007. [Google Scholar] [CrossRef]
Ibeas JI, Lee H, Damsz B, Prasad DT, Pardo JM, Hasegawa PM, Bressan RA, Narasimhan ML (2000). Fungal cell wall phosphomannans facilitate the toxic activity of a plant PR-5 protein. Plant Journal 23: 375–383. DOI 10.1046/j.1365-313x.2000.00792.x. [Google Scholar] [CrossRef]
Kaur A, Kumar A (2020). Cloning and characterization of osmotin (OsmSt) alleles from potato cultivar ‘Kufri Chipsona 1’. bioRxiv. [Google Scholar]
Kaur A, Sudhakara RM, Pati PK, Kumar A (2020). Over-expression of osmotin (OsmWS) gene of Withania somnifera in potato cultivar ‘Kufri Chipsona 1’ imparts resistance to Alternaria solani. Plant Cell, Tissue and Organ Culture 142: 131–142. [Google Scholar]
Kobayashi K, Fukuda M, Igarashi D, Sunaoshi M (2000). Cytokinin-binding proteins from tobacco callus share homology with osmotin-like protein and an endochitinase. Plant and Cell Physiology 41: 148–157. DOI 10.1093/pcp/41.2.148. [Google Scholar] [CrossRef]
Kumar SA, Kumari PH, Jawahar G, Prashanth S, Suravajhala P, Katam R, Sivan P, Rao KS, Kirti PB, Kishor PK (2016). Beyond just being foot soldiers-osmotin like protein (OLP) and chitinase (Chi11) genes act as sentinels to confront salt, drought, and fungal stress tolerance in tomato. Environmental and Experimental Botany 132: 53–65. DOI 10.1016/j.envexpbot.2016.08.007. [Google Scholar] [CrossRef]
Kumar SA, Kumari PH, Kumar GS, Mohanalatha C, Kishor PK (2015). Osmotin: A plant sentinel and a possible agonist of mammalian adiponectin. Frontiers in Plant Science 6: 163. DOI 10.3389/fpls.2015.00163. [Google Scholar] [CrossRef]
LaRosa PC, Chen Z, Nelson DE, Singh NK, Hasegawa PM, Bressan RA (1992). Osmotin gene expression is posttranscriptionally regulated. Plant Physiology 100: 409–415. DOI 10.1104/pp.100.1.409. [Google Scholar] [CrossRef]
Le TT, Williams B, Mundree SG (2018). An osmotin from the resurrection plant Tripogon loliiformis (TlOsm) confers tolerance to multiple abiotic stresses in transgenic rice. Physiologia Plantarum 162: 13–34. DOI 10.1111/ppl.12585. [Google Scholar] [CrossRef]
Li S, Zhang Y, Xin X, Ding C, Lv F et al. (2020). The Osmotin-Like protein gene PdOLP1 is involved in secondary cell wall biosynthesis during wood formation in poplar. International Journal of Molecular Sciences 21: 3993. DOI 10.3390/ijms21113993. [Google Scholar] [CrossRef]
Li XY, Gao L, Zhang WH, Liu JK, Zhang YJ, Wang HY, Liu DQ (2015). Characteristic expression of wheat PR5 gene in response to infection by the leaf rust pathogen, Puccinia triticina. Journal of Plant Interactions 10: 132–141. DOI 10.1080/17429145.2015.1036140. [Google Scholar] [CrossRef]
Mani T, Sivakumar KC, Manjula S (2012). Expression and functional analysis of two osmotin (PR5) isoforms with differential antifungal activity from Piper colubrinum: Prediction of structure-function relationship by bioinformatics approach. Molecular Biotechnology 52: 251–261. DOI 10.1007/s12033-011-9489-0. [Google Scholar] [CrossRef]
Moeller DA, Tiffin P (2005). Genetic diversity and the evolutionary history of plant immunity genes in two species of Zea. Molecular Biology and Evolution 22: 2480–2490. DOI 10.1093/molbev/msi247. [Google Scholar] [CrossRef]
Neale AD, Wahleithner JA, Lund M, Bonnett HT, Kelly A, Meeks-Wagner DR, Peacock WJ, Dennis ES (1990). Chitinase, beta-1, 3-glucanase, osmotin, and extensin are expressed in tobacco explants during flower formation. Plant Cell 2: 673–684. DOI 10.1105/tpc.2.7.673. [Google Scholar] [CrossRef]
Noori SS, Sokhansanj A (2008). Wheat plants containing an osmotin gene show enhanced ability to produce roots at high NaCl concentration. Russian Journal of Plant Physiology 55: 256–258. [Google Scholar]
Onishi M, Tachi H, Kojima T, Shiraiwa M, Takahara H (2006). Molecular cloning and characterization of a novel salt-inducible gene encoding an acidic isoform of PR-5 protein in soybean (Glycine max [L.] Merr.). Plant Physiology and Biochemistry 44: 574–580. DOI 10.1016/j.plaphy.2006.09.009. [Google Scholar] [CrossRef]
Parkhi V, Kumar V, Sunilkumar G, Campbell LAM, Singh NK, Rathore KS (2009). Expression of apoplastically secreted tobacco osmotin in cotton confers drought tolerance. Molecular Breeding 23: 625–639. [Google Scholar]
Patade VY, Khatri D, Kumari M, Grover A, Gupta SM, Ahmed Z (2013). Cold tolerance in Osmotin transgenic tomato (Solanum lycopersicum L.) is associated with modulation in transcript abundance of stress responsive genes. SpringerPlus 2: 1–7. [Google Scholar]
Patade VY, Meena H, Grover A, Gupta SM, Nasim M (2018). Containment evaluation, cold tolerance and toxicity analysis in osmotin transgenic tomato (Solanum lycopersicum L. cv. Pusa Ruby). Biotech 8: 1–9. [Google Scholar]
Pluskota WE, Pupel P, Głowacka K, Okorska SB, Jerzmanowski A, Nonogaki H, Górecki RJ (2019). Jasmonic acid and ethylene are involved in the accumulation of osmotin in germinating tomato seeds. Journal of Plant Physiology 232: 74–81. [Google Scholar]
Ramos MV, de Oliveira RS, Pereira HM, Moreno FB, Lobo MD et al. (2015). Crystal structure of an antifungal osmotin-like protein from Calotropis procera and its effects on Fusarium solani spores, as revealed by atomic force microscopy: Insights into the mechanism of action. Phytochemistry 119: 5–18. DOI 10.1016/j.phytochem.2015.09.012. [Google Scholar] [CrossRef]
Rather IA, Awasthi P, Mahajan V, Bedi YS, Vishwakarma RA, Gandhi SG (2015). Molecular cloning and functional characterization of an antifungal PR-5 protein from Ocimum basilicum. Gene 558: 143–151. DOI 10.1016/j.gene.2014.12.055. [Google Scholar] [CrossRef]
Roberts WK, Selitrennikoff CP (1990). Zeamatin, an antifungal protein from maize with membrane permeabilizing activity. Journal of General Microbiology 136: 1771–1778. DOI 10.1099/00221287-136-9-1771. [Google Scholar] [CrossRef]
Rugini E, Bashir MA, Astolfi S, Cristofori V, Ruggiero B et al. (2018). Transformation of kiwi (Actinidia deliciosa A. Chev.) and olive (Olea europaea L.) with tobacco osmotin gene confers drought-stress tolerance and provides a health-promoting protein source.Proceeding of the 2nd International Conference of Biotechnology and Environment, Alexandria, Egypt. [Google Scholar]
Rugini E, Cristofori V, Martignoni D, Gutierrez-Pesce P, Orlandi S, Brunori E, Biasi R, Muleo R, Magro P (2010). Kiwifruit transgenics for osmotin gene and inoculation tests with Botrytis cinerea and Cadophora luteo-Olivacea. VII International Symposium on Kiwifruit 913: 197–203. [Google Scholar]
Schnable PS, Ware D, Fulton RS, Stein JC, Wei F et al. (2009). The B73 maize genome: Complexity, diversity, and dynamics. Science 326: 1112–1115. DOI 10.1126/science.1178534. [Google Scholar] [CrossRef]
Shah JM, Singh R, Veluthambi K (2013). Transgenic rice lines constitutively coexpressing tlp-D34 and chi11 display enhancement of sheath blight resistance. Biologia Plantarum 57: 351–358. [Google Scholar]
Silvestri C, Celletti S, Cristofori V, Astolfi S, Ruggiero B, Rugini E (2017). Olive (Olea europaea L.) plants transgenic for tobacco osmotin gene are less sensitive to in vitro-induced drought stress. Acta Physiologiae Plantarum 39: 1–9. [Google Scholar]
Singh NK, Bracker CA, Hasegawa PM, Handa AK, Buckel S, Hermodson MA, Pfankoch ED, Regnier FE, Bressan RA (1987). Characterization of osmotin: A thaumatin-like protein associated with osmotic adaptation in plant cells. Plant Physiology 85: 529–536. DOI 10.1104/pp.85.2.529. [Google Scholar] [CrossRef]
Singh NK, Handa AK, Hasegawa PM, Bressan RA (1985). Proteins associated with adaptation of cultured tobacco cells to NaCl. Plant Physiology 79: 126–137. DOI 10.1104/pp.79.1.126. [Google Scholar] [CrossRef]
Singh NK, Kumar KRR, Kumar D, Shukla P, Kirti PB (2013). Characterization of a pathogen induced thaumatin-like protein gene AdTLP from Arachis diogoi, a wild peanut. PLoS One 8: e83963. DOI 10.1371/journal.pone.0083963. [Google Scholar] [CrossRef]
Sripriya R, Parameswari C, Veluthambi K (2017). Enhancement of sheath blight tolerance in transgenic rice by combined expression of tobacco osmotin (ap24) and rice chitinase (chi11) genes. In Vitro Cellular & Developmental Biology-Plant 53: 12–21. [Google Scholar]
Stintzi A, Heitz T, Kauffmann S, Legrand M, Fritig B (1991). Identification of a basic pathogenesis-related, thaumatin-like protein of virus-infected tobacco as osmotin. Physiological and Molecular Plant Pathology 38: 137–146. DOI 10.1016/S0885-5765(05)80131-6. [Google Scholar] [CrossRef]
Su HY, Chou HH, Chow TJ, Lee TM, Chang JS, Huang WL, Chen HJ (2017). Improvement of outdoor culture efficiency of cyanobacteria by over-expression of stress tolerance genes and its implication as bio-refinery feedstock. Bioresource Technology 244: 1294–1303. DOI 10.1016/j.biortech.2017.04.074. [Google Scholar] [CrossRef]
Subramanyam K, Arun M, Mariashibu TS, Theboral J, Rajesh M et al. (2012). Overexpression of tobacco osmotin (Tbosm) in soybean conferred resistance to salinity stress and fungal infections. Planta 236: 1909–1925. DOI 10.1007/s00425-012-1733-8. [Google Scholar] [CrossRef]
Subramanyam K, Sailaja KV, Subramanyam K, Rao DM, Lakshmidevi K (2011). Ectopic expression of an osmotin gene leads to enhanced salt tolerance in transgenic chilli pepper (Capsicum annum L.). Plant Cell, Tissue and Organ Culture 105: 181–192. [Google Scholar]
Sun S, Zhou Y, Chen J, Shi J, Zhao H et al. (2018). Extensive intraspecific gene order and gene structural variations between Mo17 and other maize genomes. Nature Genetics 50: 1289–1295. DOI 10.1038/s41588-018-0182-0. [Google Scholar] [CrossRef]
Tachi H, Fukuda-Yamada K, Kojima T, Shiraiwa M, Takahara H (2009). Molecular characterization of a novel soybean gene encoding a neutral PR-5 protein induced by high-salt stress. Plant Physiology and Biochemistry 47: 73–79. DOI 10.1016/j.plaphy.2008.09.012. [Google Scholar] [CrossRef]
Tong Z, Sun Y, Wang D, Wang L, Li L, Meng X, Feng W, Wurtele ES, Wang X (2016). Identification and functional characterization of HbOsmotin from Hevea brasiliensis. Plant Physiology and Biochemistry 109: 171–180. DOI 10.1016/j.plaphy.2016.09.017. [Google Scholar] [CrossRef]
Viktorova J, Klcova B, Rehorova K, Vlcko T, Stankova L, Jelenova N, Cejnar P, Kundu JK, Ohnoutkova L, Macek T (2019). Recombinant expression of osmotin in barley improves stress resistance and food safety during adverse growing conditions. PLoS One 14: e0212718. DOI 10.1371/journal.pone.0212718. [Google Scholar] [CrossRef]
Viktorova J, Krasny L, Kamlar M, Novakova M, Mackova M, Macek T (2012). Osmotin, a pathogenesis-related protein. Current Protein & Peptide Science 13: 672–681. DOI 10.2174/138920312804142129. [Google Scholar] [CrossRef]
Viktorova J, Rehorova K, Musilova L, Suman J, Lovecka P, Macek T (2017). New findings in potential applications of tobacco osmotin. Protein Expression and Purification 129: 84–93. DOI 10.1016/j.pep.2016.09.008. [Google Scholar] [CrossRef]
Wan Q, Shao HB, Xu ZL, Jia L, Zhang DY, Huang YH (2017). Salinity tolerance mechanism of osmotin and osmotin-like proteins: A promising candidate for enhancing plant salt tolerance. Current Genomics 18: 553–556. DOI 10.2174/1389202918666170705153045. [Google Scholar] [CrossRef]
Weber RL, Wiebke-Strohm B, Bredemeier C, Margis-Pinheiro M, de Brito GG et al. (2014). Expression of an osmotin-like protein from Solanum nigrum confers drought tolerance in transgenic soybean. BMC Plant Biology 14: 1–9. DOI 10.1186/s12870-014-0343-y. [Google Scholar] [CrossRef]
Xue X, Cao ZX, Zhang XT, Wang Y, Zhang YF, Chen ZX, Pan XB, Zuo SM (2016). Overexpression of OsOSM1 enhances resistance to rice sheath blight. Plant Disease 100: 1634–1642. DOI 10.1094/PDIS-11-15-1372-RE. [Google Scholar] [CrossRef]
Yun DJ, Ibeas JI, Lee H, Coca MA, Narasimhan ML, Uesono Y, Hasegawa PM, Pardo JM, Bressan RA (1998). Osmotin, a plant antifungal protein, subverts signal transduction to enhance fungal cell susceptibility. Molecular Cell 1: 807–817. DOI 10.1016/S1097-2765(00)80080-5. [Google Scholar] [CrossRef]
Zhang H, Huang Z, Xie B, Chen Q, Tian X, Zhang X, Zhang H, Lu X, Huang D, Huang R (2004). The ethylene-, jasmonate-, abscisic acid-and NaCl-responsive tomato transcription factor JERF1 modulates expression of GCC box-containing genes and salt tolerance in tobacco. Planta 220: 262–270. DOI 10.1007/s00425-004-1347-x. [Google Scholar] [CrossRef]
Zhao Q, Qiu B, Li S, Zhang Y, Cui X, Liu D (2020). Osmotin-like protein gene from Panax notoginseng is regulated by jasmonic acid and involved in defense responses to Fusarium solani. Phytopathology 110: 1419–1427. DOI 10.1094/PHYTO-11-19-0410-R. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |