DOI:10.32604/biocell.2022.016915

| BIOCELL DOI:10.32604/biocell.2022.016915 |  |
| Article |
Single-cell RNA sequencing reveals classical monocytes are the major precursors of rat osteoclasts
1Institute of Biomedical Engineering, West China School of Basic Medical Science and Forensic Medicine, Sichuan University, Chengdu, 610041, China
2Laboratory of Cardiovascular Diseases, Regenerative Medicine Research Center, West China Hospital, Sichuan University, Chengdu, 610041, China
3Laboratory Animal Center, Sichuan University, Chengdu, 610041, China
*Address correspondence to: Liang Li, lilianghx@163.com
#These authors contributed equally to this work
Received: 09 April 2021; Accepted: 06 May 2021
Abstract: To dissect which subset of bone marrow monocyte is the major precursor of osteoclast, 3-month-old rat bone marrow was obtained for single-cell RNA sequencing. A total of 6091 cells were acquired for detailed analysis, with a median number of 1206 genes detected per cell and 17,959 genes detected in total. A total of 19 cell clusters were recognized, with the main lineages identified as B cells, Granulocytes, Monocytes, T cells, Erythrocytes and Macrophages. Monocytes were further divided into classical monocytes and non-classical monocytes. Compared with non-classical monocytes, classical monocytes highly expressed osteoclast differentiation related genes Mitf, Spi1, Fos and Csf1r. Additionally, biological processes of classical monocytes were related to osteoclast differentiation. qPCR revealed differentially expressed genes of classical monocytes played a role in osteoclast differentiation. In conclusion, classical monocytes were identified as the main precursors of osteoclasts in rats, and may contribute to osteoclast differentiation by regulating S100a4, S100a6, S100a10, Fn1, Vcan and Bcl2a1. The results of this study contribute to the understanding of the origin of osteoclasts and may provide potential biomarkers for early diagnosis of bone metabolic diseases, as well as molecular and cellular targets for clinical intervention in bone metabolic diseases.
Keywords: Single-cell RNA sequencing; Monocytes; Subsets; Osteoclast differentiation
Osteoclasts are the main functional cells in bone resorption, which play an important role in bone development, growth, repair and reconstruction (Kurotaki et al., 2020). Understanding the origin of osteoclasts is the key to study the etiology and prevention of bone resorption-related clinical bone metabolic diseases.
Monocytes are cells of the mononuclear phagocyte system which originate from hematopoietic precursor cells (Udagawa et al., 2020). Monocytes can perform multiple functions including supporting tissue homeostasis, mediating host defense, initiating inflammation and differentiating into osteoclasts (Shi and Pamer, 2011).
Monocytes are a heterogeneous population. Through CD14 and CD16, human monocytes can be divided into three subsets: the CD14++CD16– classical monocytes, CD14++CD16+ intermediate monocytes and CD14+CD16++ non-classical monocytes (Ziegler-Heitbrock et al., 2010). Mouse monocytes can also be divided into three subsets: Ly6C+ classical monocytes, Ly6Clow intermediate monocytes, Ly6C– non-classical monocytes (Mildner et al., 2017). However, in the present study, mouse monocytes are usually divided into classical subsets and non-classical subsets, because it is difficult to distinguish the intermediate subsets (Shi and Pamer, 2011). And in rats, two monocyte subsets have also been identified: CD43low classical monocytes, CD43hi non-classical monocytes (Yrlid et al., 2006). Different monocyte subsets are phenotypically and functionally distinct. Classical monocytes have a high phagocytic capacity and are known to be primed for innate immune and migration, while non-classical monocytes intend to secrete inflammatory cytokines and participate in inflammatory response (Chimen et al., 2017; Gren et al., 2015; Wong et al., 2011). Thus, only when we know which subset of monocytes is the main precursor of osteoclasts, can we better intervene the process of bone resorption and effectively prevent clinical bone metabolic diseases.
However, which monocytes subset is the major source of osteoclasts remains elusive. In human, studies have shown that bone marrow CD14+CD16++ non-classical monocytes are more likely to differentiate into osteoclasts than CD14++CD16– classical monocytes (Bolzoni et al., 2017; Chiu et al., 2010). In contrast, several other studies illustrated that CD14++CD16– classical monocytes rather than CD14+CD16++ non-classical monocytes were mainly the osteoclast precursors (Komano et al., 2006; Lari et al., 2009). In mice, Ammari et al. (2018) found that Ly6C+ classical monocytes play a critical role in arthritis bone erosion, and ablation of miR-146a in Ly6C+ classical monocytes increased osteoclast differentiation in vitro. On the other hand, Cambre et al. (2018) revealed mechanical stimulation could induce the recruitment of classical monocytes to differentiate into osteoclasts in mice. However, another study sorted classical monocytes and non-classical monocytes in mice, which found sorted non-classical monocytes displayed an increased capacity to differentiate into osteoclasts (Puchner et al., 2018). For rats, there was little known about the role of the classical monocytes and non-classical monocytes in serving as precursors for osteoclasts. Thus, despite the increasing awareness of heterogeneity of monocytes, the role of classical monocytes and non-classical monocytes in osteoclast formation is still needed to be clarified, especially in rats.
Recent years, single cell RNA sequencing (scRNA-seq) has been widely used to discover rare cell types, understand the differences between subsets, and obtain deeper biological information. At the same time, scRNA-seq can also reveal some special cell functions in cell subsets (Ziegenhain et al., 2017). Single-cell transcriptional profiling had brought about a revolutionary understanding of heterogeneity within monocytes. Except the classical (Mono1) and non-classical subsets (Mono2), scRNA-seq revealed 2 novel human monocyte subsets (Villani et al., 2017). Mono3 was subset associated with cell differentiation and cell proliferation, while Mono4 highly expressed cytotoxic gene including T cell activation genes (Villani et al., 2017). In mice, scRNA-seq also revealed new monocyte subsets which expressed MHCII-associated genes and CD209a (Mildner et al., 2017). Although more studies are needed to confirm and reveal the functional role of new monocyte subsets, these studies indicated that the traditional definition of monocyte subsets may be modified and extended. To the best of our knowledge, scRNA-seq has not been used to identify the subsets of monocytes in rats. Also, there is no study concerning about which monocyte subset is the main precursor of osteoclasts in rats. More importantly, scRNA-seq can identify the functional characteristics of monocyte subsets in vivo by identifying characteristic transcription factors without in vitro culture, so as to explain and clarify the conflicting results of previous studies on osteoclast precursor cells.
Through scRNA-seq, we may modify and extend the monocyte subsets in rats. On the other hand, scRNA-seq could identify which subset of monocytes was the main precursor of osteoclasts more accurately. The aim of this study was to use scRNA-seq to assess the monocyte subsets in rats as well as expound the role of monocyte subsets in osteoclast formation. The results of this study will provide experimental data for the prevention and treatment of bone metabolic diseases.
Experimental animals and sample preparation
The study used healthy 3-month-old Sprague Dawley rats (n = 3) as study subject. All experiments were performed according to the guidelines and protocols approved by the Animal Experiment Ethic Committee of Sichuan University. To acquire bone marrow single cell suspension, femurs was obtained by excising muscle and connective tissue. Then, the bone marrow was flushed out with serum-free medium using a 10 mL syringe. Bone marrow of 3 rats were collected and mixed in a centrifuge tube, centrifuged at 800 rpm for 5 min, then the supernatant was discarded to retain the cell precipitate. Finally, the cell pellet was resuspended in HBSS for subsequent experiments.
Library preparation, sequencing, and pre-processing of scRNA-seq
First, single cell gel bead for cell barcode encoding was prepared. Then, according to the manufacturer’s original protocol, the cDNA library was constructed using the 10 × ChromiumTM Single cell 3’ Library Kit. Then, the single cell library was sequenced on Illumina hiseq using the following reading lengths: 26 bp Read1, 8 bp I7 Index and 98 bp Read2. Cell Ranger (10X Genomics, 2.1.1 version) was used to process Chromium single cell 3’ RNA-seq output. Finally, the Cell Ranger analysis pipeline was used for sample demultiplexing, barcode processing and single cell 3’ gene counting, and the digital gene cell matrix was generated from the data.
Quality control, cell clustering analysis and identification of differentially expressed genes
The Seurat R package was used for quality control and processing of the gene expression matrix. We filter cells based on the number of genes expressed in the cells. Quantified cells were screened out when >500 genes in cell were detected and <25% mitochondrial genes were expressed. The cells were filtered by the Seurat package with mitochondrial genes and ribosomal genes removed, and 6091 high quality cells were retained for further analyses.
After removing the low-quality cells, the expression was homogenized by using the “normalization” function of Seurat software. Then principal component analysis (PCA) using the variable genes as input was performed. The statistically significant 10 PCs were selected as input for t-Distributed Stochastic Neighbor Embedding (tSNE), which visualized the single cells on a two-dimensional space based on expression signatures of the variable genes. By density-based spatial clustering algorithm, cell types were identified on the tSNE map. In addition, the FeaturePlot function and DoHeatMap function was used to visualize the expression of a specific gene in each cluster. Then differentially expressed genes between the specified cell cluster and the remaining cell clusters were identified by meeting P < 0.05 and ≥2-fold differential expression range. A heatmap of the marker genes in each cell cluster was drawn according to the log2FC values.
Through Gene ontology (GO) analysis of differentially expressed genes in each cell cluster, specific biological functions of each cell cluster were investigated. Additionally, potential molecular regulatory pathway in each cell cluster were determined by KEGG pathway analysis. GO terms and KEGG terms with false discovery rate (FDR) less than 0.01 or 0.05 were considered to be significantly enriched.
Flow cytometry analysis and sorting
Bone marrow monocytes were isolated from fresh bone marrow samples using Ficoll density-gradient centrifugation. Then, single-cell suspensions were stained with CD43 PE-conjugated antibody. Because of the low expression of CD43 in classical monocytes, different subsets of monocytes can be circled according to the different fluorescence intensity of CD43.
In vitro osteoclast differentiation induction
Bone marrow monocytes were isolated from fresh bone marrow samples using Ficoll density gradient centrifugation. Then the cells were cultivated 7 days in α-MEM with 1% PenStrep, 12% fetal bovine serum, 30 ng/mL rM-CSF and 50 ng/mL rRANKL to induce osteoclast. Osteoclast differentiation was evaluated by TRAP staining according to the manufacturer’s instructions.
Total RNA was isolated from monocytes, then reverse transcribed into the cDNA by Takara reverse transcription reagent kit. Real-time quantitative PCR (qPCR) was performed to analyze the expression of S100a4, S100a6, S100a10, Fn1, Vcan and Bcl2a1 mRNA according to the manufacturer’s instructions.
Main lineages in rat bone marrow
Following the procedure for tissue separation, single cell isolation and high-throughput single cell RNA sequencing (Fig. 1A), a total of 6091 cells were obtained with a median read of 44876 per cell and a median number of 1206 genes detected per cell. Therefore, 17,959 genes were detected in total with a sequencing saturation rate of 90.2%, indicating that the existing transcripts were almost fully sampled. 19 clusters were recognized using an unsupervised strategy in Seurat (Fig. 1B), with clusters being B cells (clusters 0, 1, 3, 5, 12, 18), granulocytes (clusters 2, 6, 7, 8, 10, 11, 16, 19), monocytes (clusters 4, 9), T cells (clusters 14), erythrocytes (clusters 13, 15) and macrophages (cluster 17) according to their marker genes (Fig. 1C). The heat map of marker gene expression in different cell subsets was shown in Fig. 2. The analysis of marker gene expression revealed some new markers of different lineages.
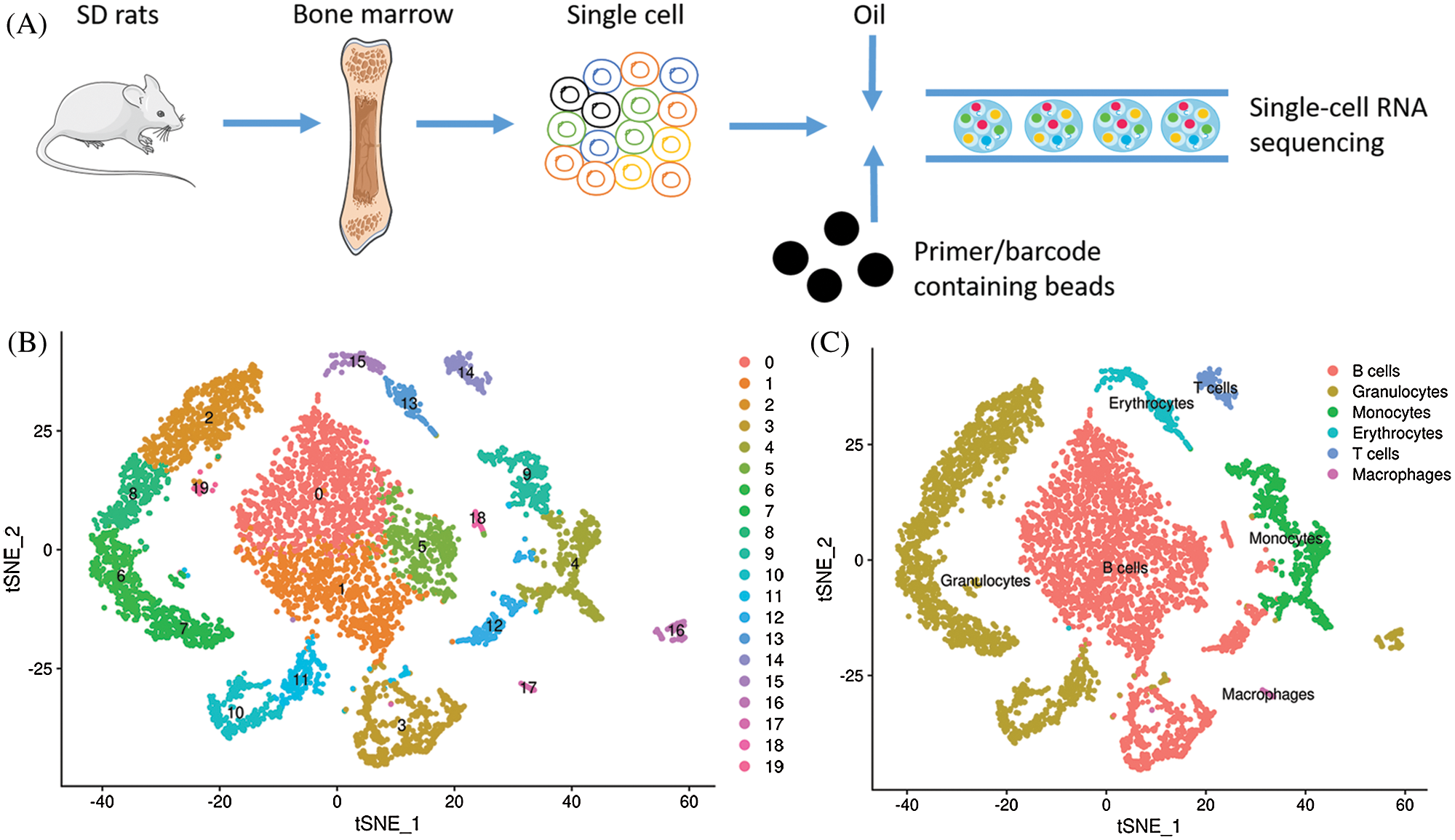
Figure 1: Main cell lineages in rat bone marrow. (A) Workflow procedure for the single-cell RNA sequencing. (B) Clustering of 6091 cells from rat bone marrow. (C) Definition atlas of rat bone marrow (B cells, granulocytes, monocytes, T cells, erythrocytes, macrophages).
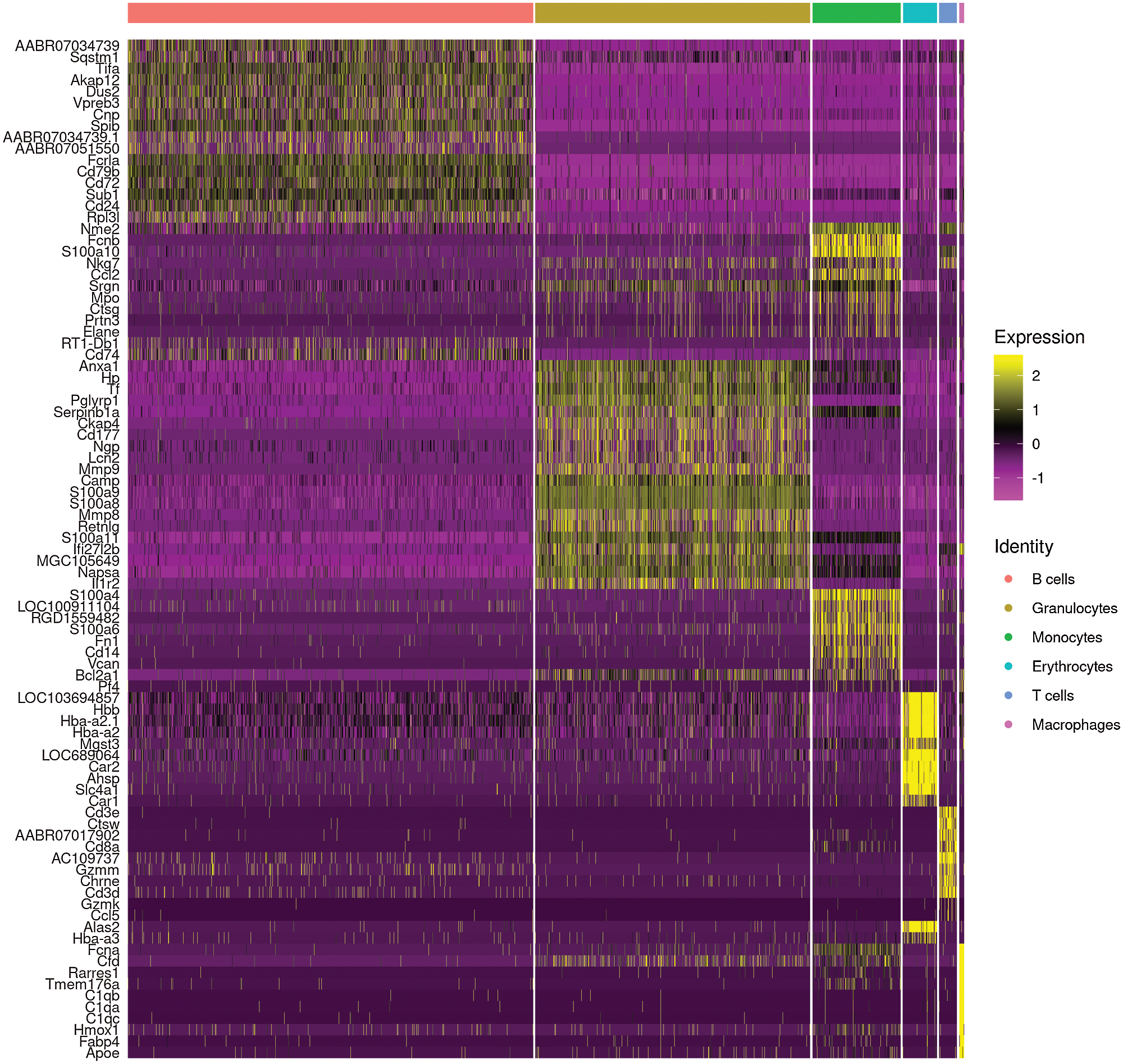
Figure 2: Heatmap of marker genes of main cell lineages.
Identification of subsets of monocytes
The newly developed method for monocyte subsets categorization in rats was based on the differential expression of CD43, and low CD43 expression was found in classical monocytes (Dijkstra et al., 1994). CD14, which was CD antigens on the surface of monocytes, was found to be highly expressed in the classical monocytes (Nasi et al., 2020); CD68, a high affinity for cells of the mononuclear phagocyte system, also highly expressed in the classical monocytes (Athanasou and Quinn, 1990; Fadini et al., 2013). In this study, cluster 4 and cluster 9 were recognized as monocytes. However, these two monocyte subsets were not categorized. Thus, Spn (code CD43), CD14 and CD68 were used to identify the monocyte subsets. As Fig. 3A shows, Dot plot showed Spn, CD14 and CD68 expression in different clusters. Cluster 9 was defined as classical monocytes by the low expression of Spn and high expression of CD14 and CD68, while cluster 4 defined as non-classical monocytes because of its high expression of Spn. As Fig. 3B shows, Violin plot showed selected marker genes expression in different subsets of monocytes. Cluster 9 (classical monocytes) lowly expressed Spn and highly expressed CD14 and CD68 than cluster 4 (non-classical monocytes). Therefore, according to selected marker genes, two subsets were identified within monocytes (Fig. 3C). The newly developed FACS method for monocyte subsets categorization in rats was based on the differential expression of CD43. We further used FACS to verify monocyte subsets in vivo. CD43low (classical monocyte, left) and CD43high (non-classical monocyte, right) were defined in bone marrow derived from rat, which consist with the Single-cell RNA-seq results (Fig. 3D).
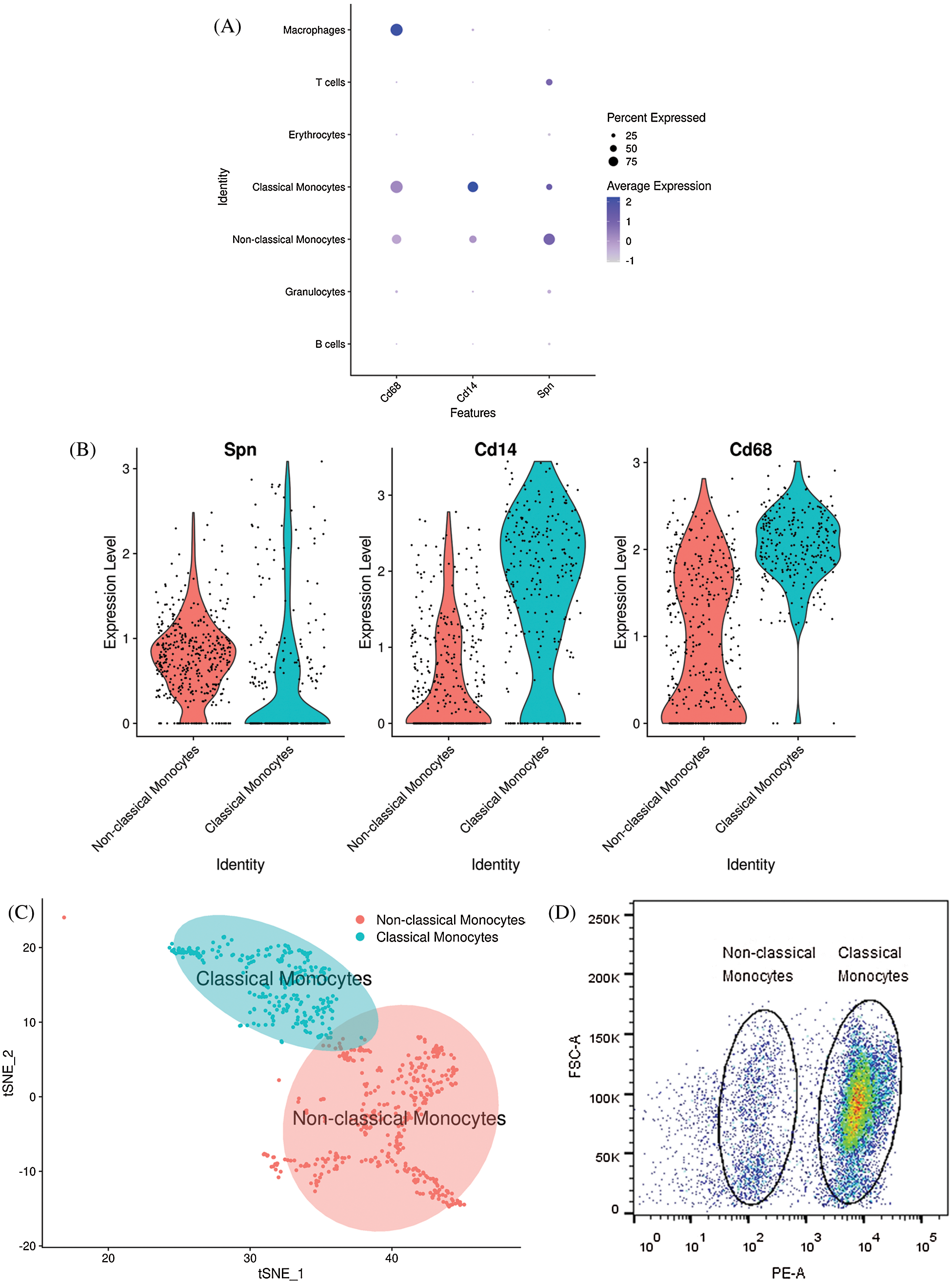
Figure 3: Monocyte marker genes in subsets of monocytes. (A) Dot plot showing monocyte marker genes in all clusters. X-axis, Monocyte marker genes; Y-axis, different clusters; circle size, percentage of Monocyte marker genes-expressing cells. Color, mean values of average gene expression levels in each cluster. (B) Violin plot for monocyte marker genes in different monocyte subsets. (C) Visualization of different subsets of monocytes by t-SNE. (D) Representative FACS images of rat bone marrow-derived monocytes and their subsets analyses. Classical monocytes were defined as CD43low (left circle), and nonclassical monocytes were defined as CD43high (right circle).
Functional exploration of the monocyte subsets in bone marrow derived from rats
To investigate the function and characteristic of the monocyte subsets, KEGG pathway analysis and GO analysis were performed on monocyte subsets and top 20 pathways were listed. The biological processes in classical monocytes in GO analysis were related to osteoclast differentiation (phagocytic vesicle, lysosome, lysosomal membrane), while the top 20 GO pathways in non-classical monocytes were mainly related with the protein metabolism (Fig. 4). The top 20 KEGG pathways in classical monocytes were mainly focused on osteoclast differentiation, rheumatoid arthritis, phagosome and lysosome, which were highly related with the osteoclast differentiation, consistent with the results of the GO analysis (Fig. 5). The pathway analysis suggested classical monocytes had osteoclast differentiation potential. Next, expression of Mitf, Spi1, Fos and Csf1r, which encode transcription factors and cytokine receptor that are associated with and may be necessary for osteoclast differentiation, were evaluated in monocytes subsets. The results revealed that Mitf, Spi1, Fos and Csf1r were highly expressed in classical monocytes (Fig. 6). Thus, these results indicated that classical monocytes were the main osteoclast precursors in rats.
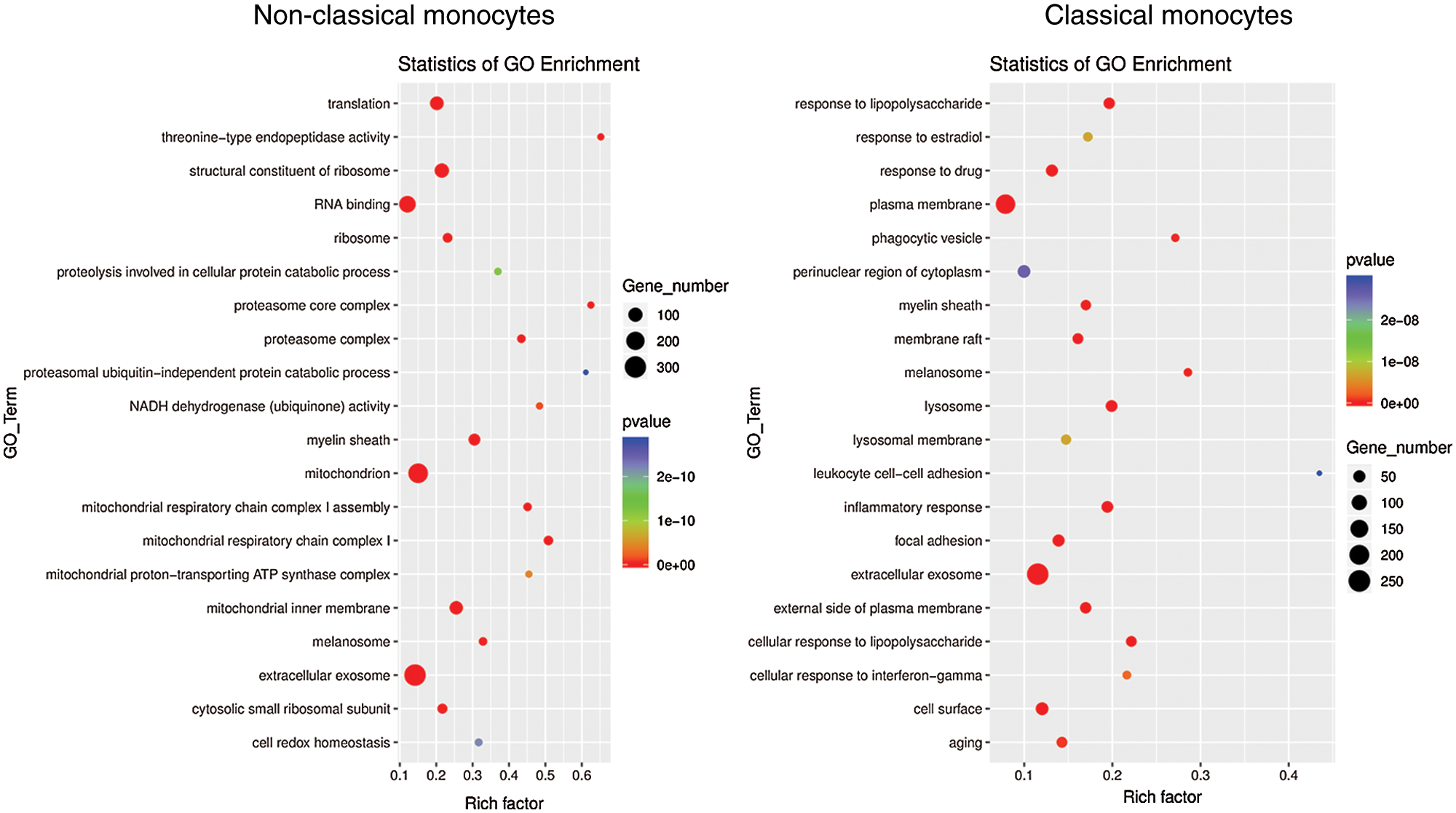
Figure 4: GO analysis of classical monocytes and non-classical monocytes. Dot plot showing selected biological processes in the GO analysis of classical monocytes and non-classical monocytes. X-axis, gene ratio; Y-axis, enriched biological processes terms in monocyte subsets; circle size, gene counts in each term.
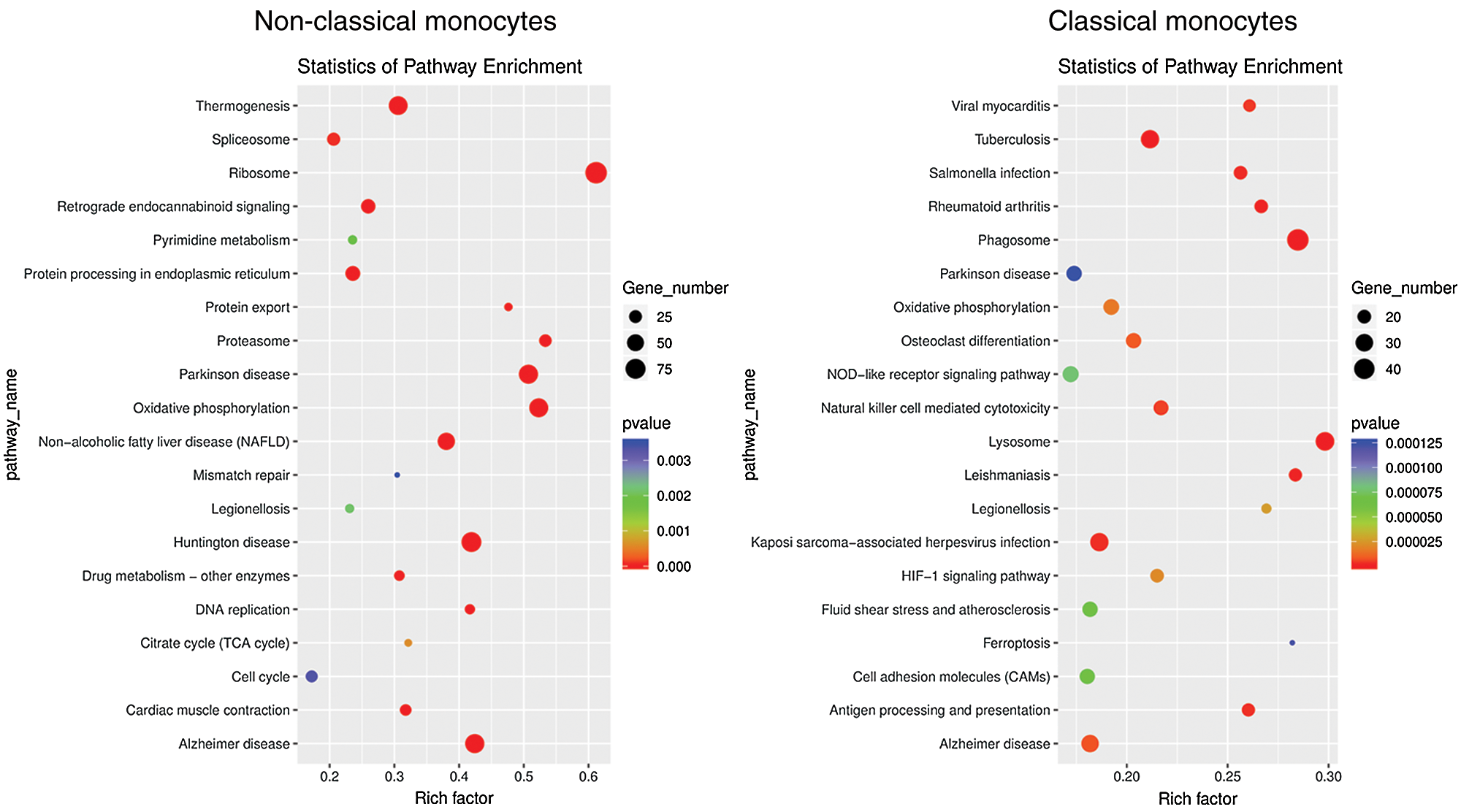
Figure 5: KEGG analysis of classical monocytes and non-classical monocytes. Dot plot showing top 20 biological pathways in the KEGG analysis of classical monocytes and non-classical monocytes. X-axis, gene ratio; Y-axis, enriched biological pathways terms in monocyte subsets; circle size, gene counts in each term.
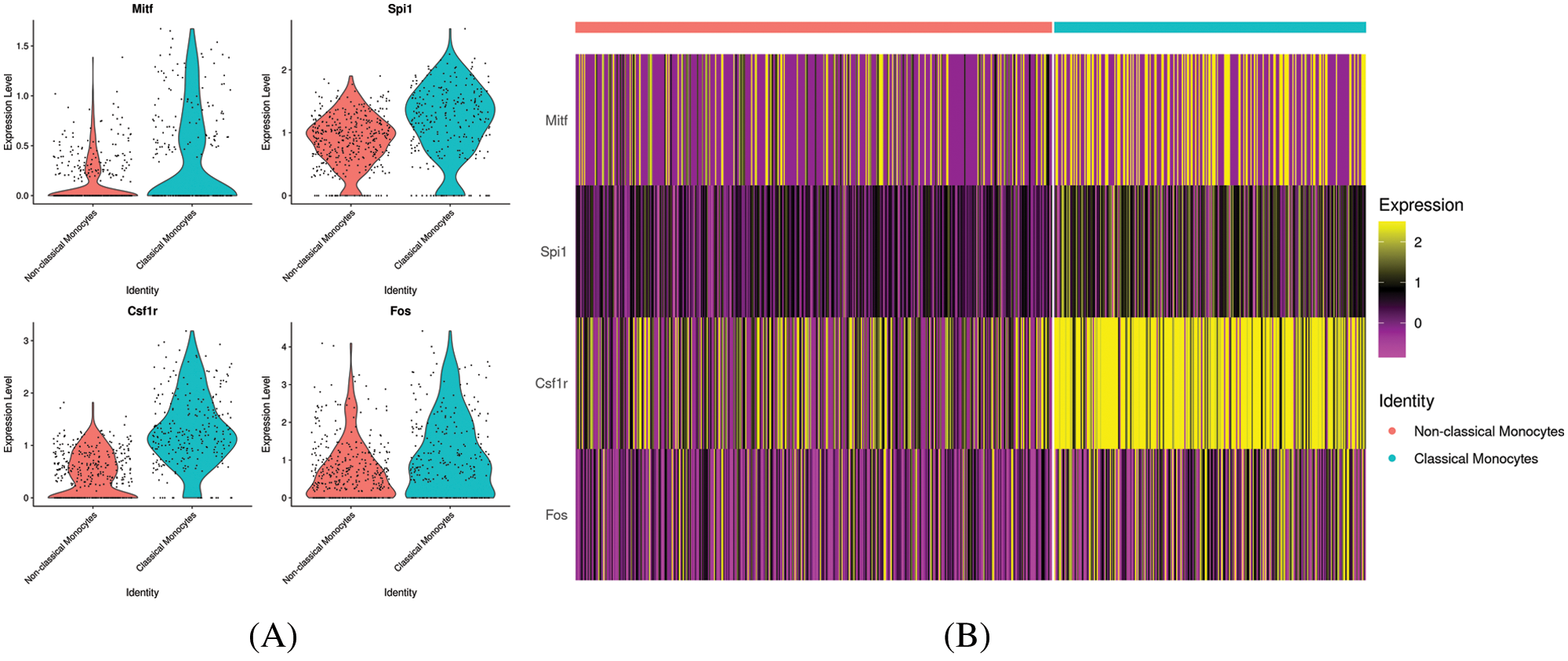
Figure 6: Expression of osteoclast differentiation related genes in classical monocytes and non-classical monocytes. (A) Violin plot for Mitf, Spi1, Fos and Csf1r expression in different monocyte subsets. (B) Heatmap of Mitf, Spi1, Fos and Csf1r in different monocyte subsets.
The differentially expressed genes of the monocyte subsets in bone marrow derived from rats
Differentially expressed genes were identified by comparing cells in a specific cluster with cells in all other clusters. The top 10 differentially expressed genes in non-classical monocytes are shown in Fig. 7A, while the top 10 differentially expressed genes in classical monocytes are shown in Fig. 7B. The non-classical monocytes highly expressed Nme2, Fcnb, S100a10, Nkg7, Ccl2, Srgn, MPO, Ctsg, Prtn3 and Elane, which including some inflammatory cytokines and adhesion molecules. The classical monocytes highly expressed S100a4, LOC100911104, RGD1559482, S100a6, S100a10, Fn1, CD14, Vcan, Bcl2a1 and Fcnb, which mainly composed of S100 molecules and adhesion molecules. Our results generated more accurate features of monocyte subsets in rats, which provide help for the better identification of monocyte subsets.
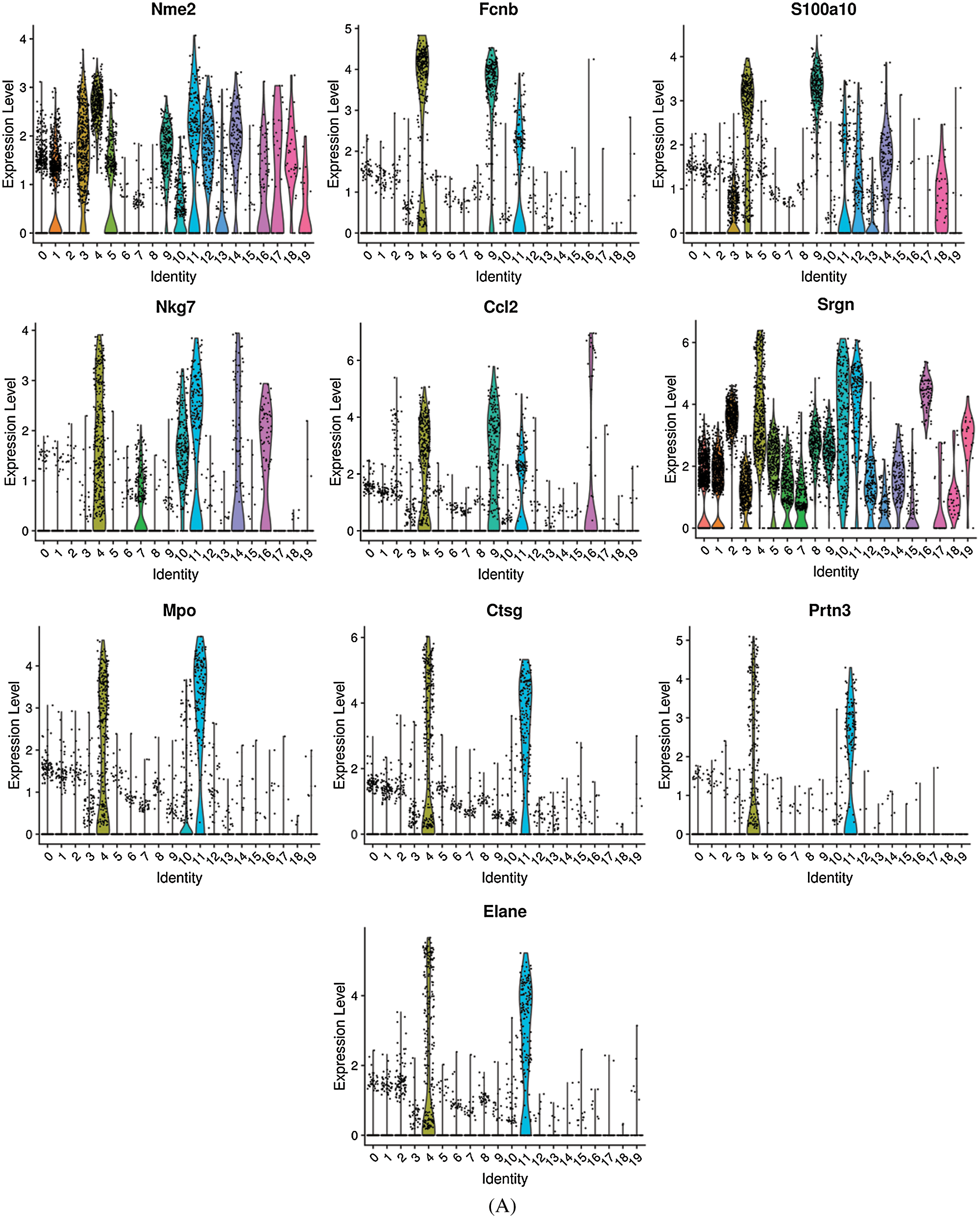
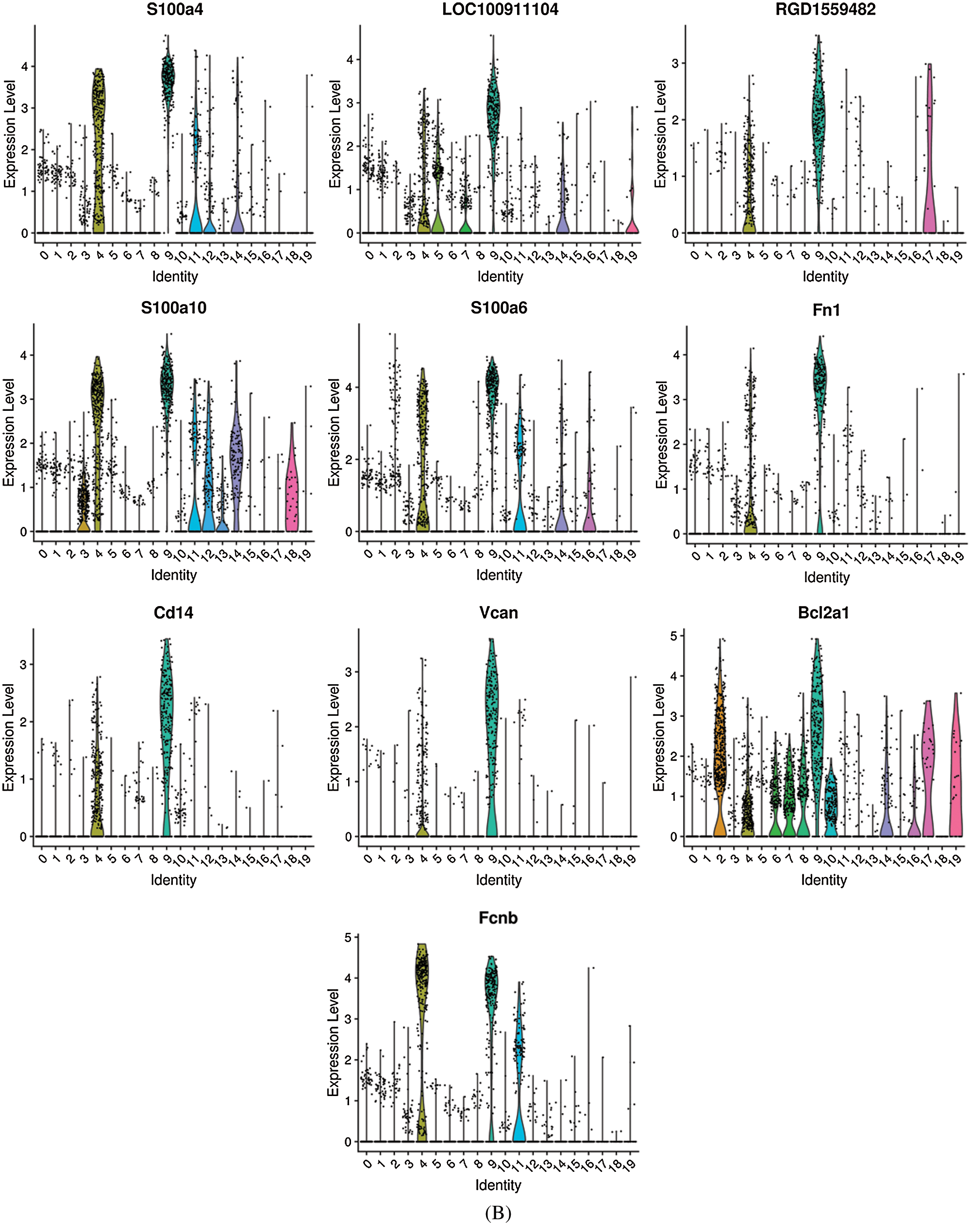
Figure 7: The top 10 differentially expressed genes in non-classical monocytes (A) and classical monocytes (B).
The role of differentially expressed genes of classical monocytes in osteoclast differentiation
Differentially expressed genes of classical monocytes which may be related to osteoclastic differentiation were selected for qPCR analysis. Along with the osteoclast induction, the TRAP positive cells increased, which suggested the osteoclast differentiation of monocytes (Fig. 8A). As Fig. 8B shows, the mRNA expression of S100a4 increased with the days of osteoclast differentiation induction, while S100a6, S100a10, Fn1, Vcan and Bcl2a1 mRNA expression decreased with the days of osteoclast differentiation induction. These selected differentially expressed genes changed their expression during the osteoclast differentiation induction, suggested their potential role involved in the osteoclast differentiation.
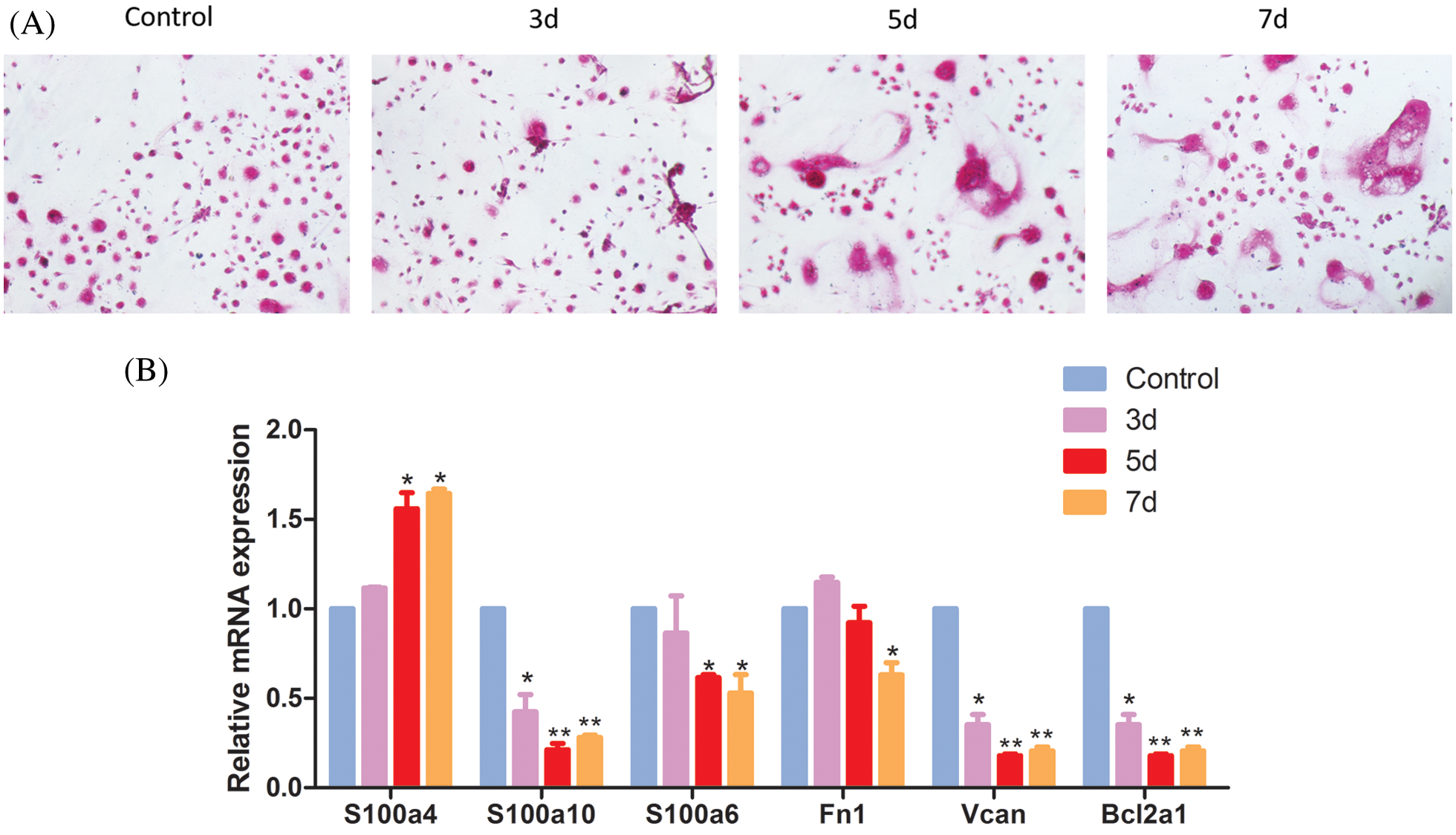
Figure 8: The role of differentially expressed genes of classical monocytes in osteoclast differentiation. (A) TRAP staining of the induced monocytes. Monocytes were cultured with rM-CSF (30 ng/mL) and rRANKL (50 ng/mL) for osteoclast differentiation. The cells were detected for tartrate-resistant acid phosphatase (TRAP) staining on day 7. (B) qPCR for the differentially expressed genes of classical monocytes.
Monocytes are defined by functional and molecular markers (Guilliams et al., 2014). However, it is still unclear whether the existing markers can track more complex internal states of monocytes in rats. To address this question, we detected the subsets of monocytes by analysis of gene expression at single cell resolution and inferring cell types based on cell markers. In our study, two monocyte subsets: classical monocytes (CD43low) and non-classical monocytes (CD43hi) were revealed through the expression of CD43. Although monocyte subsets identified in our study were similar as the previous studies reported, our study had generated more accurate features of monocyte subsets in rats.
Monocytes played an important role in the secretion of inflammatory factors, regulation of innate immunity and osteoclast formation, which may be influenced by the monocyte heterogeneity (Kikuta and Ishii, 2013). In this study, we mainly focused on the role of monocyte subsets in osteoclast formation. To investigate the cell function status, KEGG pathway analysis and GO analysis were performed on monocytes subsets. We found KEGG pathways in classical monocytes focused on osteoclast differentiation, rheumatoid arthritis, phagosome and lysosome, which were highly related with the osteoclast differentiation. The biological processes in classical monocytes in GO analysis were related to osteoclast differentiation (phagocytic vesicle, lysosome, lysosomal membrane), consistent with the results of the KEGG analysis. The pathway analysis suggested classical monocytes had osteoclast differentiation potential. We further derived specific expression signatures for each monocyte subsets, including transcription factors (Mitf, Spi1, Fos) and cytokine receptor (Csf1r), which play key role in the osteoclast differentiation (Kurotaki et al., 2020; Mun et al., 2020; Zhang et al., 2020). We found the Mitf, Spi1, Fos and Csf1r were highly expressed in classical monocytes. Therefore, we speculated that classical monocytes were the main osteoclast precursors in rats. However, detailed mechanistic studies are still needed to reveal the differential functions of these two monocyte subsets.
Considering the key role of classical monocytes in the osteoclast formation, we further explore the role of differentially expressed genes of classical monocytes in osteoclast differentiation. We found mRNA expression of S100a4 in monocytes increased with the days of osteoclast differentiation induction, while S100a6, S100a10, Fn1, Vcan and Bcl2a1 mRNA expression decreased with the days of osteoclast differentiation induction. S100 proteins are multifunctional signaling proteins, which regulate a variety of cellular processes, such as cell growth, cell differentiation, cell migration, and secretion, with remarkable cell and tissue specific expression patterns (Marenholz et al., 2004). As with other ECM components, it is particularly noteworthy that FN1 and Vcan could attract circulating monocytes and play an important role in the adhesion and activation of recruited monocytes by binding to adhesion molecules (Dutta et al., 2017; Wu et al., 2005). Bcl2a1 is an anti-apoptotic activated macrophage protein which highly expressed in osteolysis patients, explaining a possible mechanism about why the particle-laden cells could continuously express macrophage phenotype activation markers (Purdue, 2008). A study exploring gene expression profiles by graph clustering approach and Gene Ontology term enrichment analysis found Bcl2a1 may be associated with osteoclast differentiation (Xiao et al., 2012). Additionally, Bcl2a1 was involved in osteoclast proliferation and apoptosis. Osteoclast proliferation was enhanced by increasing cell cycle and suppressing apoptosis through Bcl2a1 gene regulation (Lee and Lee, 2014). Collectively, S100 proteins, Fn1, Vcan and Bcl2a1 may affect osteoclast formation through their role in cell differentiation, adhesion, proliferation, and apoptosis.
It has been proved that excessive activation of local osteoclasts is involved in focal bone erosion in bone diseases, such as rheumatoid arthritis and osteoporosis (Okamoto et al., 2017). Our study demonstrated that classical monocytes and their differentially expressed genes have a distinct role in regulating osteoclast formation, suggesting that classical monocytes and their differentially expressed genes might be a possible therapeutic target for bone destruction. Therefore, it is intriguing to propose that targeting classical monocytes and their differentially expressed genes simultaneously may have a more apparent inhibitory effect on bone destruction in rheumatoid arthritis or osteoporosis. However, the detailed signal mechanisms of S100a4, S100a6, S100a10, Fn1, Vcan and Bcl2a1 on classical monocytes need to be further studied.
In conclusion, classical monocytes are the main precursor cells of rat osteoclasts, which may participate in osteoclast differentiation by regulating S100A4, S100A6, S100A10, FN1, Vcan and bcl2a1. These findings are helpful to understand the origin of osteoclasts and provide a potential biomarker for early diagnosis of bone diseases.
Availability of Data and Materials: The data that support the findings of this study are available from the corresponding author upon reasonable request.
Author Contribution: Study conception and design: Jirui Wen, Liang Li; data collection: Jirui Wen, Wenchao Wu; analysis and interpretation of results: Min Tang, Mingyue Bao, Xueling He; draft manuscript preparation: Jirui Wen, Xinghong Yao, Liang Li. All authors reviewed the results and approved the final version of the manuscript.
Ethics Approval: This study and all experiments involved are under approval of The Ethical Committee of Sichuan University, with File Code: KS2020038 in March 31, 2020.
Funding Statement: This work was supported by the National Natural Science Foundation of China (Nos. 11572209, 11872260) and National Natural Science Foundation of China (Key Program, No. 11932014).
Conflicts of Interest: The authors declare that they have no competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Ammari M, Presumey J, Ponsolles C, Roussignol G, Roubert C et al. (2018). Delivery of miR-146a to Ly6C(high) monocytes inhibits pathogenic bone erosion in inflammatory arthritis. Theranostics 8: 5972–5985. [Google Scholar]
Athanasou NA, Quinn J (1990). Immunophenotypic differences between osteoclasts and macrophage polykaryons: Immunohistological distinction and implications for osteoclast ontogeny and function. Journal of Clinical Pathologoy 43: 997–1003. [Google Scholar]
Bolzoni M, Ronchetti D, Storti P, Donofrio G, Marchica V et al. (2017). IL21R expressing CD14(+)CD16(+) monocytes expand in multiple myeloma patients leading to increased osteoclasts. Haematologica 102: 773–784. [Google Scholar]
Cambre I, Gaublomme D, Burssens A, Jacques P, Schryvers N et al. (2018). Mechanical strain determines the site-specific localization of inflammation and tissue damage in arthritis. Nature Communications 9: 4613. [Google Scholar]
Chimen M, Yates CM, Mcgettrick HM, Ward LS, Harrison MJ et al. (2017). Monocyte subsets coregulate inflammatory responses by integrated signaling through TNF and IL-6 at the endothelial cell interface. Journal of Immunology 198: 2834–2843. [Google Scholar]
Chiu YG, Shao T, Feng C, Mensah KA, Thullen M et al. (2010). CD16 (FcRγIII) as a potential marker of osteoclast precursors in psoriatic arthritis. Arthritis Research & Therapy 12: R14. [Google Scholar]
Dijkstra CD, Döpp EA, van den Berg TK, Damoiseaux JG (1994). Monoclonal antibodies against rat macrophages. Journal of Immunological Methods 174: 21–23. [Google Scholar]
Dutta B, Park JE, Kumar S, Hao P, Gallart-Palau X et al. (2017). Monocyte adhesion to atherosclerotic matrix proteins is enhanced by Asn-Gly-Arg deamidation. Scientific Reports 7: 5765. [Google Scholar]
Fadini GP, de Kreutzenberg SV, Boscaro E, Albiero M, Cappellari R et al. (2013). An unbalanced monocyte polarisation in peripheral blood and bone marrow of patients with type 2 diabetes has an impact on microangiopathy. Diabetologia 56: 1856–1866. [Google Scholar]
Gren ST, Rasmussen TB, Janciauskiene S, Hakansson K, Gerwien JG, Grip O (2015). A single-cell gene-expression profile reveals inter-cellular heterogeneity within human monocyte subsets. PLoS One 10: e0144351. [Google Scholar]
Guilliams M, Ginhoux F, Jakubzick C, Naik SH, Onai N et al. (2014). Dendritic cells, monocytes and macrophages: A unified nomenclature based on ontogeny. Nature Reviews Immunology 14: 571–578. [Google Scholar]
Kikuta J, Ishii M (2013). Osteoclast migration, differentiation and function: Novel therapeutic targets for rheumatic diseases. Rheumatology 52: 226–234. [Google Scholar]
Komano Y, Nanki T, Hayashida K, Taniguchi K, Miyasaka N (2006). Identification of a human peripheral blood monocyte subset that differentiates into osteoclasts. Arthritis Research & Therapy 8: R152. [Google Scholar]
Kurotaki D, Yoshida H, Tamura T (2020). Epigenetic and transcriptional regulation of osteoclast differentiation. Bone 138: 115471. [Google Scholar]
Lari R, Kitchener PD, Hamilton JA (2009). The proliferative human monocyte subpopulation contains osteoclast precursors. Arthritis Research & Therapy 11: R23. [Google Scholar]
Lee JY, Lee NK (2014). Up-regulation of cyclinD1 and Bcl2A1 by insulin is involved in osteoclast proliferation. Life Sciences 114: 57–61. [Google Scholar]
Marenholz I, Heizmann CW, Fritz G (2004). S100 proteins in mouse and man: from evolution to function and pathology (including an update of the nomenclature). Biochemical and Biophysical Research Communications 322: 1111–1122. [Google Scholar]
Mildner A, Schönheit J, Giladi A, David E, Lara-Astiaso D et al. (2017). Genomic characterization of murine monocytes reveals C/EBPβ transcription factor dependence of Ly6C- cells. Immunity 46: 849–862. [Google Scholar]
Mun SH, Park PSU, Park-Min KH (2020). The M-CSF receptor in osteoclasts and beyond. Experimental & Molecular Medicine 52: 1239–1254. [Google Scholar]
Nasi M, Bianchini E, Lo Tartaro D, De Biasi S, Mattioli M et al. (2020). Effects of whole-body cryotherapy on the innate and adaptive immune response in cyclists and runners. Immunologic Research 68: 422–435. [Google Scholar]
Okamoto K, Nakashima T, Shinohara M, Negishi-Koga T, Komatsu N et al. (2017). Osteoimmunology: The conceptual framework unifying the immune and skeletal systems. Physiological Reviews 97: 1295–1349. [Google Scholar]
Puchner A, Saferding V, Bonelli M, Mikami Y, Hofmann M et al. (2018). Non-classical monocytes as mediators of tissue destruction in arthritis. Annals of the Rheumatic Diseases 77: 1490–1497. [Google Scholar]
Purdue PE (2008). Alternative macrophage activation in periprosthetic osteolysis. Autoimmunity 41: 212–217. [Google Scholar]
Shi C, Pamer EG (2011). Monocyte recruitment during infection and inflammation. Nature Reviews Immunology 11: 762–774. [Google Scholar]
Udagawa N, Koide M, Nakamura M, Nakamichi Y, Yamashita T et al. (2020). Osteoclast differentiation by RANKL and OPG signaling pathways. Journal of Bone and Mineral Metabolism 39: 19–26. [Google Scholar]
Villani AC, Satija R, Reynolds G, Sarkizova S, Shekhar K et al. (2017). Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 356: eaah4573. [Google Scholar]
Wong KL, Tai JJ, Wong WC, Han H, Sem X et al. (2011). Gene expression profiling reveals the defining features of the classical, intermediate, and nonclassical human monocyte subsets. Blood 118: 16–31. [Google Scholar]
Wu YJ, La Pierre DP, Wu J, Yee AJ, Yang BB (2005). The interaction of versican with its binding partners. Cell Research 15: 483–494. [Google Scholar]
Xiao H, Shan L, Zhu H, Xue F (2012). Detection of significant pathways in osteoporosis based on graph clustering. Molecular Medicine Reports 6: 1325–1332. [Google Scholar]
Yrlid U, Jenkins CD, Macpherson GG (2006). Relationships between distinct blood monocyte subsets and migrating intestinal lymph dendritic cells in vivo under steady-state conditions. Journal of Immunology 176: 4155–4162. [Google Scholar]
Zhang Y, Ma C, Liu C, Wu W (2020). NF-κB promotes osteoclast differentiation by overexpressing MITF via down regulating microRNA-1276 expression. Life Sciences 258: 118093. [Google Scholar]
Ziegenhain C, Vieth B, Parekh S, Reinius B, Guillaumet-Adkins A et al. (2017). Comparative analysis of single-cell RNA sequencing methods. Molecular Cell 65: 631–643. [Google Scholar]
Ziegler-Heitbrock L, Ancuta P, Crowe S, Dalod M, Grau V et al. (2010). Nomenclature of monocytes and dendritic cells in blood. Blood 116: 74–80. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |