DOI:10.32604/biocell.2022.016714

| BIOCELL DOI:10.32604/biocell.2022.016714 |  |
| Article |
A549/DDP derived exosomes can affect cisplatin chemosensitivity via transporting CXCR4 to A549 cells
1Department of Radiation Oncology, Changzhou Cancer Hospital Affiliated to Soochow University, Changzhou, 213032, China
2Department of Cancer Radiotherapy, The First Affiliated Hospital of USTC, Division of Life Sciences and Medicine, University of Science and Technology of China (Anhui Provincial Cancer Hospital), Hefei, 230031, China
3Department of Neurology, The Affiliated Changzhou No. 2 People’s Hospital of Nanjing Medical University, Changzhou, 213001, China
*Address correspondence to: Jianfang Liu, jianfangliu1982@163.com; Yayun Cui, yayuncui@163.com
#These authors contributed equally to this work
Received: 19 March 2021; Accepted: 31 May 2021
Abstract: The resistance of cancer cells to the anti-cancer drugs is the most important reason that affecting the efficacy of the non-small cell lung cancer (NSCLC) chemotherapy; thus, to explore the underlying mechanism of drug resistance of NSCLC medications is urgently needed for improving the therapeutic efficacy of current anti-NSCLC chemotherapies. The aim of the present study is to explore the roles of exosomes in the chemosensitivity of A549 cells and the related mechanism. A549 cells and cisplatin resistant cell line A549/DDP derived exosomes were isolated, and the expressions of CXCR4 were compared. Then, after cisplatin treatment, A549 cells were treated with exosomes, and the proliferation, apoptosis, migration, and invasion of the cells were examined. Finally, the tumorigenic effect of A549/DDP derived exosomes were also evaluated by cisplatin treated xenograft tumor mice models in vivo. We found that A549/DDP derived exosomes increased the proliferation, migration, and invasion, and inhibited the apoptosis and cisplatin sensitivity of A549 cells. CXCR4 was also significantly increased in cells treated with A549/DDP derived exosomes. Furthermore, A549/DDP derived exosomes may also decrease the chemosensitivity of NSCLC cells to cisplatin in vivo. Our data suggested that A549/DDP derived exosomes can affect the chemosensitivity of A549 cells to cisplatin, possibly by transporting CXCR4 to A549 cells. Our data may provide novel evidence for the investigation of drug resistance of NSCLC.
Keywords: Exosome; Drug resistance; CXCR4; NSCLC cells
Lung cancer is the most common type of cancer, and it is also the leading causes of cancer-related motility worldwide (Toda et al., 2020; Yang et al., 2019; Zhao et al., 2020). 85% of lung cancer patients suffered from the non-small cell lung cancer (NSCLC) (Incharoen et al., 2019; Toruner et al., 2020). Results of previous studies suggested that NSCLC may develop from a series of abnormal genetic changes in the epithelial cells of lung tissue, which can further lead to the uncontrolled growth of the cells and metathesis of the tumor (Cancer Genome Atlas Research Network, 2012; Zhu et al., 2015). In current clinical application, chemotherapy is the most commonly used method for the treatment of non-mutant NSCLC. However, the therapeutic efficacy of current anti-NSCLC chemotherapies remains unsatisfactory, and the long-term prognosis of patients with NSCLC remains poor (Xu et al., 2019a; Xu et al., 2019b). It is believed that the resistance of cancer cells to the anti-cancer drugs is the most important reason that limits the efficacy of the anti-NSCLC chemotherapy (Jin et al., 2019; Xu et al., 2019a). Thus, to explore the underlying mechanism of drug resistance of NSCLC treatments is urgently needed for improving the therapeutic efficacy of current anti-NSCLC chemotherapies.
Exosomes are a group of nanovesicles with the size of 50–150 nm, which are produced and secreted by all types of cells. Exosomes can be detected in all human body fluids, such as blood, urine etc. Exosomes have been identified as efficient communication system between tumor cells, which play important role in promoting tumor occurrence and development (Graner et al., 2018). The contents of exosomes include nucleic acids, proteins, lipids, and other soluble factors (Whiteside, 2016). Exosome-related RNA (miRNA, lncRNA, circRNA), protein, DNA and even metabolites can change the progress of tumor cells (Zhang and Yu, 2019). In addition, exosomes also have a strong influence on drug resistance through multiple mechanisms (Olejarz et al., 2020). Therefore, tumor exosomes may be a promising research area for inhibiting tumor development, and potential biomarkers in cancer therapy.
It has been proved that circulating tumor exosomes may transfer the nucleic acids or the proteins into different type of cells and participate in the process of carcinogenesis in different cancers (Li et al., 2020a; Schuldner et al., 2019; Xiao et al., 2019). For example, exosomes derived from cancer initiating cells can promote the metastasis of pancreatic cancer through the CD44v6/Tspan8 signaling pathway (Sun et al., 2019). Analysis of circulating tumor DNA/RNA in glioma patients shows that circulating exosomal-derived RNA can better predict tumor progression and resistance to treatment compared with cell-free (cfDNA) (Pasqualetti et al., 2019). Knockdown of syntaxin 6 significantly reduced the exosome secretion in enzalutamide resistant cells, which regulates enzalutamide resistance in prostate cancer (Peak et al., 2020). Leukemic cells selectively transport miR-320 to exosomes through HNRNPA1, which are endocytosed by surrounding bone marrow mesenchymal stromal cells, thereby inhibiting osteogenesis and promoting the growth of tumor cells (Gao et al., 2019b). These studies suggesting that exosomes contribute to the development and drug resistance of cancer. Further exploration of its molecular mechanism may provide a potential target for tumor treatment.
C-X-C chemokine receptor type 4 (CXCR4) is a specific receptor of stromal-derived-factor-1, and it is also a G protein-coupled receptor composed of 352 amino acids (Jubb et al., 2019; Niimi et al., 2020). It has been proved that CXCR4 plays an important role in tumorigenesis, development, and metastasis (Ding et al., 2020; Oum et al., 2020). CXCR4 is overexpressed in more than 23 different types of human cancers, including kidney cancer, lung cancer, liver cancer, breast cancer, etc. (Chatterjee et al., 2014). The high expression of CXCR4 promotes the aggressiveness of tumor cells, enhances chemoresistance and inhibits cell apoptosis. Clinically, high CXCR4 expression may be related to the poor prognosis of lung cancer patients (Zhu et al., 2020). It has been reported that the CXCR4-inhibiting nanocomplex reduces tumor fibrosis and promotes cytotoxic T lymphocytes penetration, regulating the lung immune response, thereby further enhancing the anti-PD-L1 immune response (Li et al., 2020b). However, the roles of exosome-related CXCR4 in chemotherapy resistance of lung cancer remains unclear.
In the present study, we will investigate whether exosomes affect the chemosensitivity of NSCLC cells by transporting CXCR4. Our results may provide theoretical basis for investigation of drug resistance of NSCLC.
NSCLC cell line A549 and NSCLC cisplatin resistance cell line A549/DDP were purchased from Cell bank of Chinese Academy of Sciences (Shanghai, China). Both A549 and A549/DDP cells were cultured in RPMI-1640 medium (Invitrogen, USA) supplemented with 10% fetal bovine serum, and 1% penicillin and streptomycin (Invitrogen, USA) at 37°C in a 5% CO2 atmosphere. A549/DDP cells were supplemented with 2 µg/mL cisplatin to maintain the drug-resistant phenotype. Cells were passaged every other day, and cells in the logarithmic growth phase were used for further experiments.
Exosome isolation and characterization
To obtain A549 and A549/DDP-released exosome, cells were collected and centrifuged. ExoPrep reagent was added to the supernatant and incubated on ice for 1 h. The mixture was then centrifuged, and the exosome pellet was re-suspended in 100 μL phosphate-buffered saline (PBS). The exosome samples were assessed using Western blot and transmission electron microscope.
Transmission electron microscopy (TEM)
TEM was conducted to identify the purified exosome. Ten microliter of exosome suspension was absorbed onto carbon-coated cooper grids (200 mesh) for 1 min in a dry environment. The grids were washed with distilled water for 2 min each and then negatively stained with 2% uranyl acetate solution for 1 min. After air dry, the grids were visualized using a transmission electron microscope operated at 80 kV.
SiRNA-CXCR4 and siRNA-NC were purchased from GenePharma (Shanghai, China). A549/DDP cells were seeded onto six-well plates for 24 h, then, transfected with siRNA-CXCR4 and siRNA-NC using Lipofectamine 2000 (Invitrogen; Carlsbad, CA, USA) following the instructions of manufacturers. Cells were collected for further experiments after 48 h of transfection.
Cell viability was measured using Cell Counting Kit-8 (CCK-8) assay (Promega, USA) in 96-well plates. 10% CCK-8 solution was added to each well and the plate was incubated at 37°C for 2 h. The absorbance at 450 nm was measured.
The cell apoptosis was estimated using flow cytometry. cells were washed twice with cold phosphate-buffered saline (PBS) and re-suspended in PBS. Then, cells were double-stained with Annexin V-FITC and propidium iodide (PI) following the instructions of manufacturer. Apoptosis was analyzed with a BD Biosciences FACSCalibur Flow Cytometer (BD Biasciences, USA) and the percentage of apoptotic cells was determined.
Cells were seeded into 6-well plates. Scratch wounds were made through the cell monolayer with a 200 μL pipette tip in each well, and the wells were washed with phosphate buffer solution (PBS) to remove the detached cells. Images were taken under a light microscope and wound distance was measured in three locations for each well. Images are shown as representatives of three independent experiments.
Cell invasion was evaluated by using 24-well transwell filters (8 μm pores, Millipore, USA). Briefly, the upper chambers were coated with 100 μL of diluted Matrigel (BD, USA) and exposed to UV light for 2 h. Cells were suspended at 3 × 105 cells/mL in serum-free RPMI-1640 medium and a total of 100 μL were added to the upper chamber. The lower chamber of each well contained 1 mL of RPMI-1640 medium containing 10% FBS. After 24 h incubation, the cells that penetrated into the lower chamber were fixed, stained with crystal violet, and counted in five randomly selected microscopic fields of each well. Images are shown as representatives of three independent experiments.
Quantitative Real-Time PCR (RT-qPCR)
Trizol reagent was added to cells or exosomes to extract total RNA. OD260/280 was measured to ensure the quality of the extracted RNA, and 2% denaturing agarose gel electrophoresis was applied to evaluate the RNA integrity. Reverse transcription of total RNA to cDNA was performed using SuperScript First strand cDNA synthesis kit (Life Technologies, USA) according to the manufacturer’s instructions. Gene expression was then measured using RT-qPCR with SYBR-Green Master Mix kit (Life Technologies, USA). GAPDH was used as the housekeeping gene. The 2−ΔΔCt method was used to calculate the relative expression of mRNA to GAPDH. All the primers used for the amplification of the genes are listed in Table 1.

Cells were washed with ice-cold PBS and total proteins were extracted with RIPA lysis buffer. Tissue samples were homogenized in the buffer and subsequently centrifuged to obtain the total protein supernatants. The concentration of total protein was determined using BCA Protein Assay Kit (Pierce, USA). Equal amounts of protein extracts were separated on a 10% sodium dodecyl sulfate polyacrylamide gels (SDS–PAGE) and transferred to polyvinylidene difluoride (PVDF) membranes. The membranes were blocked in Tris buffered saline Tween 20 (TBST) solution containing 5% skimmed milk at room temperature for 2 h, and then incubated with primary antibodies at 4°C overnight. The protein bands were detected with horseradish peroxidase (HRP)-conjugated secondary antibodies and visualized using enhanced chemiluminescence detection system (Millipore, USA). Images are shown as representatives of three independent experiments.
6-week-old C57BL/6 nude mice were purchased from Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences. All animal experiments were completed at the Experimental Animal Center of the University of Science and Technology of China. The mice were housed under a 12-h light-dark cycle, 25–27°C, and a humidity of about 40%. The mice were allowed to eat and drink freely. This study was approved by the Animal Ethics Committee of The First Affiliated Hospital of USTC (Anhui Provincial Cancer Hospital). The animal study has been performed in accordance with the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. A549 cells were treated with or without A549/DDP-released exosomes. Then, 3 × 106 cells were subcutaneously injected into 6-week-old C57BL/6 nude mice and mice were treated with DDP. The formation of the tumors was examined weekly. At week 5 post-implantation, mice were anesthetized by an intraperitoneal injection of pentobarbital sodium (100 mg/kg) followed by cervical dislocation and the volume as well as the weight of the tumors were measured (n = 6 mice per group).
All experiments were carried out in triplicate. All data were analyzed using SPSS 19.0 software. The values were presented as mean ± SD. Statistical significance was analyzed using paired Student’s t-test. P-value below 0.05 is generally considered statistically significant.
Isolation and characterization of exosomes
Exosomes derived from A549 and A549/DDP cells were isolated by sequential centrifugation and ExoPrep reagent. The isolated exosomes were characterized by TEM and Western blot analysis. In TEM analysis, we observed intact circular particles, ranging in diameter from about 30 to 150 nm (Fig. 1A). Western blotting results showed that the vesicles were highly positive for ALIX and CD63, which are identified as specific markers for exosomes (Fig. 1B).
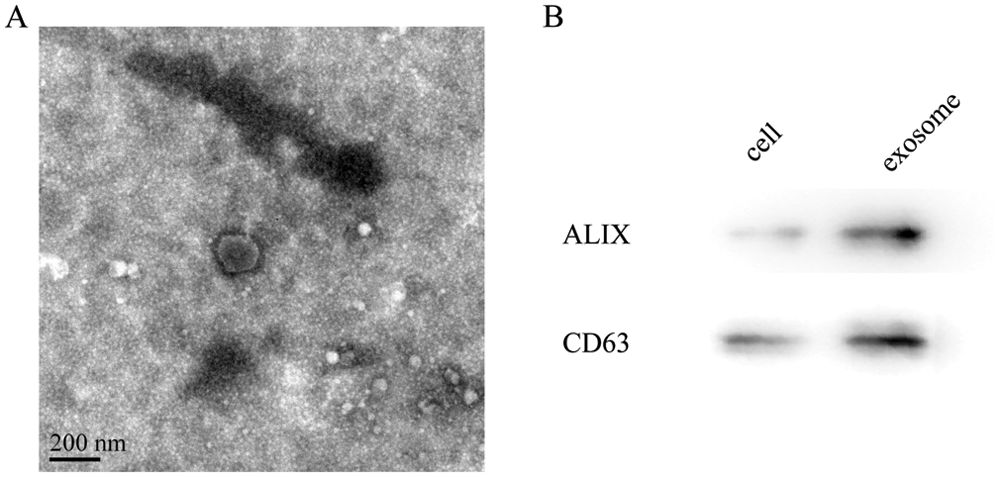
Figure 1: Characterization of exosomes from A549/DDP cells. (A) Representative TEM images of exosomes derived from A549 cells showing exosome morphology and size range. Scale bar: 200 nm. (B) The expression of exosome marker protein by Western blot.
A549/DDP exosomes inhibited the anti-proliferative and pro-apoptotic effects of DDP in A549 cells
Cell viability was analyzed using CCK8 assay. As shown in Fig. 2A, cisplatin decreased the viability of A549 cells, and exosomes + cisplatin treated A549 cells has shown a time-dependent increase in cell proliferation compared with cisplatin treated cells (Fig. 2A, P < 0.01). Next, flow cytometry was used to detect the apoptotic extent in tumor cells. The results showed that cisplatin increased the apoptosis in A549 cells and exosome decreased the apoptosis of cisplatin treated A549 cells (Fig. 2B, P < 0.01). Then, we investigated the effect of A549/DDP exosomes on the expression of apoptosis related proteins, Bcl-2, Bax, and Cleaved-caspase 3, using RT-qPCR and Western blotting assays. Our data suggested that cisplatin decreased the expression of Bcl-2 and increased the expression of Bax and Cleaved-caspase-3; while exosomes+cisplatin showed opposite trends both in mRNA and protein levels (Figs. 2C and 2D, P < 0.01). These were consistent with the results in Fig. 2B.

Figure 2: A549/DDP exosomes inhibited the anti-proliferative and pro-apoptotic effects of DDP in A549 cells. (A) Cell proliferation was measured by CCK8 assay. (B) Cell apoptosis was measured by flow cytometry assay. (C) The mRNA expressions of Bcl-2, Bax and Cleaved-caspase-3 were examined using RT-qPCR. (D) The protein expressions of Bcl-2, Bax and Cleaved-caspase-3 were examined by Western blotting, GAPDH was used as the loading control. All data shown are mean ± SD from three separate experiments. **P < 0.01.
A549/DDP exosomes inhibited the anti-migration and anti-invasion effects of DDP in A549 cells
The effects of exosomes derived from A549/DDP on cell migration and invasion of cisplatin treated A549 cells were examined. As shown in Fig. 3A, cisplatin decreased the migration ability of A549 cells, and when A549 cells were treated with cisplatin + exosomes, the migration was increased comparing with the cisplatin group. A similar result was observed in the transwell invasion assay, in exosomes + cisplatin treated cells presented a higher invasive potential comparing with the cisplatin treated group (Fig. 3B, P < 0.01). Similarly, metastasis-associated proteins (ZEB1 and MMP-9) were investigated by RT-qPCR and Western blotting assays. As shown in Figs. 3C and 3D, cisplatin decreased the mRNA and protein expression of ZEB1 and MMP-9, while exosomes + cisplatin increased the expression of ZEB1 and MMP-9, compared with cells treated with cisplatin only.

Figure 3: A549/DDP exosomes inhibited the anti-migration and anti-invasion ability of DDP in A549 cells. (A) Scratch wound assay was used to assess the A549 migration. (B) Invasion ability of A549 cells was measured using the transwell assay. (C) The mRNA expression of ZEB1 and MMP-9 were examined using RT-qPCR. (D) The protein expression of ZEB1 and MMP-9 were examined using Western blotting, GAPDH was used as the loading control. All data shown are mean ± SD from three separate experiments. *P < 0.05, **P < 0.01.
Exosomal transfer of CXCR4 is functionally relevant
Then, the expression level of CXCR4 in exosomes derived from A549 and A549/DDP cells were detected by RT-qPCR and Western blot. As shown in Figs. 4A and 4B, the mRNA and protein expression of CXCR4 was significantly increased in exosomes derived from A549/DDP cells compared with the A549 cells. Moreover, Figs. 4C and 4D showed that CXCR4 mRNA and protein expressions increased significantly in A549 cells treated with A549/DDP derived exosomes. These results suggested that A549/DDP exosomes may transport CXCR4 to A549 cells.

Figure 4: A549/DDP exosomes transport CXCR4 to A549 cells. (A) Expression of CXCR4 mRNA level in exosomes. (B) Expression of CXCR4 protein level in exosomes. (C) Expression of CXCR4 mRNA level in A549 cells. (D) Expression of CXCR4 protein level in A549 cells, GAPDH was used as the loading control. Data are mean ± SD from three separate experiments. **P < 0.01, ***P < 0.001.
Knockdown of CXCR4 attenuate effects of exosomes derived from A549/DDP in cisplatin treated A549 cells
To investigate whether CXCR4 related to the effects of exosomes derived from A549/DDP in cisplatin treated A549 cells, the expression of CXCR4 was reduced in A549/DDP cells. Our results showed that the mRNA and protein expression of CXCR4 were notably decreased in cells transfected with siRNA-CXCR4 compared with those transfected with siRNA-NC (Figs. 5A and 5B, P < 0.001 or P < 0.0001). Then, the cell proliferation in A549 cells was examined using CCK-8 assay. DDP + exosomes demonstrated a markedly increased of cell proliferation when compared with the DDP group. However, knockdown the expression of CXCR4 reversed this pro-proliferation effect (Fig. 5C). Consistently, knockdown the expression of CXCR4 also increased the rate of apoptosis reduced by DDP+ exosomes (Fig. 5D, P < 0.0001).

Figure 5: Knockdown of CXCR4 attenuate effects of exosomes derived from A549/DDP in cisplatin treated A549 cells. (A) Expression of CXCR4 mRNA was detected by RT-qPCR. (B) Expression of CXCR4 protein was detected by Western blot. (C) Cell proliferation was measured by CCK8 assay. (D) Cell apoptosis was measured by flow cytometry assay. All data shown are mean ± SD from three separate experiments. ***P < 0.001, ****P < 0.0001.
A549/DDP exosomes inhibited the anti-tumor effects of DDP in xenograft tumor mice models
Finally, the in vivo effects of exosomes derived from A549/DDP were also investigated by nude mice xenograft tumor models. As shown in Figs. 6A and 6B, the average volume of the tumors was significantly increased in exosomes+cisplatin group compared with the cisplatin group at different time points. Moreover, after the mice were sacrificed, the average weight of the tumors that measured at the end of the experiment was also significantly larger in exosomes+cisplatin group compared with the cisplatin group (Fig. 6C, P < 0.01).

Figure 6: Effects of exosomes derived from A549/DDP on the tumor growth of A549 cells in cisplatin treated xenograft tumor mice models in vivo. (A) The average volume of the tumors during the 5-week period of tumor formation. (B) Images of the dissected tumors from the nude mice at the end of the study. (C) The average weight of the tumors at the end of the study. n = 6 mice per group. * or ** means significance between DDP-treated group and DDP+exosomes-treated group. *P < 0.05, **P < 0.01.
NSCLC is one of the leading causes of death worldwide (Toruner et al., 2020; Xu et al., 2019a). Although patients with NSCLC have benefited from chemotherapy with the improved overall survival, current chemotherapeutic agents have serious side effects in patients. Therefore, the discovery and development of the specific molecular targets necessitate modulation in anti-cancer therapy.
Tumor cells are known to release exosomes into the circulation (Gao et al., 2019a; Lin et al., 2019). In the present study, we isolated and characterized A549 and A549/DDP-derived exosomes using Western blot and TEM analysis. Consistent with the general observation, exosomes isolated have an intact circular membrane, ranging from 30–150 nm in a diameter. Exosomal membranes were also enriched with exosome specific markers. Our data confirmed that the microvesicles released from NSCLC cells were exosomes in terms of their size, morphology, and protein components.
Exosomes and their cargo can be transferred from one cell to another, mediating cell-to-cell communication and subsequently regulating cell biological functions (Gao et al., 2019b; Schuldner et al., 2019). It has been previously reported that exosomes can activate NSCLC cells by interaction with them (Navarro et al., 2019). Exosomes have also been associated with the chemosensitivity of different type of cancer cells. Thus, we addressed the questions whether cisplatin resistant NSCLC cells-derived exosomes could have effects on chemosensitivity of NSCLC cells to cisplatin. In the present study, the effects of A549/DDP derived exosomes on the proliferation, apoptosis, migration, invasion, and cisplatin sensitivity of A549 cells were examined. We observed that A549/DDP derived exosomes significantly inhibited the inhibitory effect of cisplatin on tumor cell proliferation, migration, and invasion, and also inhibited the promotion of apoptosis. In addition, we also found that exosomes have the same effect on the expression levels of apoptosis and metastasis-related proteins. These results suggested that A549/DDP derived exosomes can inhibit the effects of cisplatin and increase the cisplatin resistance of A549 cells.
CXCR4 was known to participate in the cisplatin resistance of NSCLC cells (Asare-Werehene et al., 2020; Federici et al., 2014; Wang et al., 2018). However, it is unclear whether exosomes affect the chemotherapy resistance of lung cancer by transporting CXCR4. Interestingly, we observed that the expression of CXCR4 was significantly increased in exosomes derived from A549/DDP cells compared with A549 cells, and when A549 cells were treated with A549/DDP derived exosomes, the levels of CXCR4 was significantly increased. These results suggested that CXCR4 was involved in the cisplatin resistance of A549 cells and can be transferred by exosomes. Next, to investigate whether CXCR4 participates in the cisplatin resistance promotion effect of A549/DDP exosomes to A549 cells. We knocked down the expression of CXCR4 in A549 cells. Our results showed that knocking down the expression of CXCR4 can reverse the inhibitory effects of exosomes on the anti-proliferative, anti-migration, anti-invasion and pro-apoptotic of cisplatin in A549 cells. These results suggest that A549/DDP exosomes may affect the drug sensitivity of lung cancer cells by transporting CXCR4.
Furthermore, we also confirmed the tumorigenic behaviors of A549/DDP derived exosomes in cisplatin treated xenograft tumor mice models in vivo. Our data collectively suggested that A549/DDP derived exosomes may affect the chemosensitivity of A549 cells by increasing the proliferation, migration and invasion ability and inhibiting the apoptosis of A549 cells.
In conclusion, we observed that A549/DDP derived exosomes can affect the chemosensitivity of A549 cells to cisplatin, possibly by transporting CXCR4 to A549 cells. Our data provide new evidence for the investigation of drug resistance of NSCLC.
Availability of Data and Materials: The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Author Contribution: The authors confirm contribution to the paper as follows: study conception and design: Yayun Cui, Mingming Fang; data collection: Mingming Fang, Ning Ge, Jianfang Liu; analysis and interpretation of results: Ning Ge, Jianfang Liu; draft manuscript preparation: Mingming Fang, Yayun Cui. All authors reviewed the results and approved the final version of the manuscript.
Ethics Approval: This study was approved by the Animal Ethics Committee of The First Affiliated Hospital of USTC (Anhui Provincial Cancer Hospital). The animal study has been performed in accordance with the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health.
Funding Statement: This study was supported by The Fundamental Research Funds for the Central Universities [No. WK9110000071]; Youth Fund of Anhui Cancer Hospital [Nos. 2018YJQN019, 2020YJQN007].
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Asare-Werehene M, Nakka K, Reunov A, Chiu CT, Lee WT et al. (2020). The exosome-mediated autocrine and paracrine actions of plasma gelsolin in ovarian cancer chemoresistance. Oncogene 39: 1600–1616. [Google Scholar]
Cancer Genome Atlas Research Network (2012). Comprehensive genomic characterization of squamous cell lung cancers. Nature 489: 519. [Google Scholar]
Chatterjee S, Azad BB, Nimmagadda S (2014). The intricate role of CXCR4 in cancer. Advances in Cancer Research 124: 31–82. [Google Scholar]
Ding X, Huang R, Zhong Y, Cui N, Wang Y et al. (2020). CTHRC1 promotes gastric cancer metastasis via HIF-1α/CXCR4 signaling pathway. Biomedicine & Pharmacotherapy 123: 109742. [Google Scholar]
Federici C, Petrucci F, Caimi S, Cesolini A, Logozzi M et al. (2014). Exosome release and low pH belong to a framework of resistance of human melanoma cells to cisplatin. PLoS One 9: e88193. [Google Scholar]
Gao ML, Yin BC, Ye BC (2019a). Construction of a DNA-AuNP-based satellite network for exosome analysis. Analyst 144: 5996–6003. [Google Scholar]
Gao X, Wan Z, Wei M, Dong Y, Zhao Y et al. (2019b). Chronic myelogenous leukemia cells remodel the bone marrow niche via exosome-mediated transfer of miR-320. Theranostics 9: 5642. [Google Scholar]
Graner MW, Schnell S, Olin MR (2018). Tumor-derived exosomes, microRNAs, and cancer immune suppression. Seminars in Immunopathology 40: 505–515. [Google Scholar]
Incharoen P, Charonpongsuntorn C, Saowapa S, Sirachainan E, Dejthevaporn T et al. (2019). Role of BIM deletion polymorphism and BIM expression as predictive biomarkers to maximize the benefit of EGFR-TKI treatment in EGFR-positive NSCLC. Asian Pacific Journal of Cancer Prevention 20: 3581. [Google Scholar]
Jin M, Shi C, Yang C, Liu J, Huang G (2019). Upregulated circRNA ARHGAP10 predicts an unfavorable prognosis in NSCLC through regulation of the miR-150-5p/GLUT-1 axis. Molecular Therapy-Nucleic Acids 18: 219–231. [Google Scholar]
Jubb B, Lewis M, Mcfadyen L, Simpson P, Mori J et al. (2019). Incidence of CXCR4 tropism and CCR5-tropic resistance in treatment-experienced participants receiving maraviroc in the 48-week MOTIVATE 1 and 2 trials. Antiviral Chemistry and Chemotherapy 27: 2040206619895706. [Google Scholar]
Li T, Sun X, Chen L (2020a). Exosome circ_0044516 promotes prostate cancer cell proliferation and metastasis as a potential biomarker. Journal of Cellular Biochemistry 121: 2118–2126. [Google Scholar]
Li Z, Wang Y, Shen Y, Qian C, Oupicky D, Sun M (2020b). Targeting pulmonary tumor microenvironment with CXCR4-inhibiting nanocomplex to enhance anti-PD-L1 immunotherapy. Science Advances 6: eaaz9240. [Google Scholar]
Lin Q, Qu M, Zhou B, Patra HK, Sun Z et al. (2019). Exosome-like nanoplatform modified with targeting ligand improves anti-cancer and anti-inflammation effects of imperialine. Journal of Controlled Release 311: 104–116. [Google Scholar]
Navarro A, Molins L, Marrades RM, Moises J, Viñolas N et al. (2019). Exosome analysis in tumor-draining pulmonary vein identifies NSCLC patients with higher risk of relapse after curative surgery. Cancers 11: 249. [Google Scholar]
Niimi K, Kohara M, Sedoh E, Fukumoto M, Shibata S et al. (2020). FOXO1 regulates developmental lymphangiogenesis by upregulating CXCR4 in the mouse-tail dermis. Development 147: dev181545. [Google Scholar]
Olejarz W, Dominiak A, Żołnierzak A, Kubiak-Tomaszewska G, Lorenc T (2020). Tumor-derived exosomes in immunosuppression and immunotherapy. Journal of Immunology Research 2020: 6272498. [Google Scholar]
Oum YH, Shetty D, Yoon Y, Liang Z, Voll RJ et al. (2020). A benzenesulfonamide derivative as a novel PET radioligand for CXCR4. Bioorganic & Medicinal Chemistry 28: 115240. [Google Scholar]
Pasqualetti F, Restante G, Gonnelli A, Rofi E, Molinari A et al. (2019). Dabrafenib treatment in a patient with BRAF V600E ganglioglioma: Circulating exosome-derived cancer RNA supports treatment choice and clinical monitoring. Neuro-Oncology 21: 1610. [Google Scholar]
Peak TC, Panigrahi GK, Praharaj PP, Su Y, Shi L et al. (2020). Syntaxin 6-mediated exosome secretion regulates enzalutamide resistance in prostate cancer. Molecular Carcinogenesis 59: 62–72. [Google Scholar]
Schuldner M, Dörsam B, Shatnyeva O, Reiners KS, Kubarenko A et al. (2019). Exosome-dependent immune surveillance at the metastatic niche requires BAG6 and CBP/p300-dependent acetylation of p53. Theranostics 9: 6047. [Google Scholar]
Sun H, Rana S, Wang Z, Zhao K, Schnölzer M et al. (2019). The pancreatic cancer-initiating cell marker CD44v6 affects transcription, translation, and signaling: Consequences for exosome composition and delivery. Journal of Oncology 2019: 3516973. [Google Scholar]
Toda K, Ueyama M, Tanaka S, Tsukayama I, Mega T et al. (2020). Ellagitannins from Punica granatum leaves suppress microsomal prostaglandin E synthase-1 expression and induce lung cancer cells to undergo apoptosis. Bioscience, Biotechnology, and Biochemistry 84: 757–763. [Google Scholar]
Toruner GA, Tang Z, Tang G, Medeiros LJ, Hu S (2020). Low ALK FISH positive metastatic non-small cell lung cancer (NSCLC) patients have shorter progression-free survival after treatment with ALK inhibitors. Cancer Genetics 241: 57–60. [Google Scholar]
Wang J, Jia H, Zhang B, Yin L, Mao F et al. (2018). HucMSC exosome-transported 14-3-3ζ prevents the injury of cisplatin to HK-2 cells by inducing autophagy in vitro. Cytotherapy 20: 29–44. [Google Scholar]
Whiteside TL (2016). Tumor-derived exosomes and their role in cancer progression. Advances in Clinical Chemistry 74: 103–141. [Google Scholar]
Xiao F, Xiao S, Xue M (2019). miR-139 controls viability of ovarian cancer cells through apoptosis induction and exosome shedding inhibition by targeting ATP7A. OncoTargets and Therapy 12: 10727. [Google Scholar]
Xu Q, Lin D, Li X, Xiao R, Liu Z et al. (2019a). Association between single nucleotide polymorphisms of NOTCH signaling pathway-related genes and the prognosis of NSCLC. Cancer Management and Research 11: 6895. [Google Scholar]
Xu R, Cao X, Zhang B, Wang J, Wang L, Sun W (2019b). BLACAT1 is negatively associated with prognosis in patients with NSCLC and inhibits cell progression, metastasis and epithelial-mesenchymal transition through down-regulating Wnt/beta-catenin signaling pathway. European Review for Medical and Pharmacological Sciences 23: 6217–6225. [Google Scholar]
Yang X, Xiang M, Geng L, Wen Y, Du X (2019). Anlotinib combined with S-1 in the third-line treatment of stage IV non-small cell lung cancer: Study protocol for phase II clinical trial. Asian Pacific Journal of Cancer Prevention 20: 3849. [Google Scholar]
Zhang L, Yu D (2019). Exosomes in cancer development, metastasis, and immunity. Biochimica et Biophysica Acta (BBA)–Reviews on Cancer 1871: 455–468. [Google Scholar]
Zhao J, Zhang Q, Zou G, Gao G, Yue Q (2020). Arenobufagin, isolated from toad venom, inhibited epithelial-to-mesenchymal transition and suppressed migration and invasion of lung cancer cells via targeting IKKβ/NFκB signal cascade. Journal of Ethnopharmacology 250: 112492. [Google Scholar]
Zhu J, Chen M, Chen N, Ma A, Zhu C et al. (2015). Glycyrrhetinic acid induces G1-phase cell cycle arrest in human non-small cell lung cancer cells through endoplasmic reticulum stress pathway. International Journal of Oncology 46: 981–988. [Google Scholar]
Zhu Q, Luo R, Gu J, Hou Y, Chen Z et al. (2020). High CXCR4 expression predicts a poor prognosis in resected lung adenosquamous carcinoma. Journal of Cancer 11: 810. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |