DOI:10.32604/biocell.2022.016636

| BIOCELL DOI:10.32604/biocell.2022.016636 |  |
| Article |
Uncoupling tumor necrosis factor-α and interleukin-10 at tumor immune microenvironment of breast cancer through miR-17-5p/MALAT-1/H19 circuit
1Pharmaceutical Biology Department, Faculty of Pharmacy and Biotechnology, German University in Cairo, Cairo, 11835, Egypt
2School of Life and Medical Sciences, University of Hertfordshire Hosted by Global Academic Foundation, New Administrative Capital, 11578, Egypt
3Breast Surgery Department, National Cancer Institute, Cairo University, Cairo, Egypt
4General Surgery Department, Faculty of Medicine, Cairo University, Cairo, Egypt
5Department of Phytochemistry, National Research Centre, Giza, 12622, Egypt
6Biochemistry Department, Faculty of Pharmacy and Biotechnology, German University in Cairo, Cairo, 11835, Egypt
*Address correspondence to: Mohamed Z. Gad, mohamed.gad@guc.edu.eg; Rana Ahmed Youness, r.youness@gaf.edu.eg; r.youness@herts.ac.uk
Received: 14 April 2021; Accepted: 01 June 2021
Abstract: Triple Negative Breast Cancer (TNBC) immunotherapy has recently shown promising approach. However, some TNBC patients presented with resistance. One of the reasons was attributed to the excessive release of cytokines at the tumor microenvironment (TME) such as Tumor necrosis factor alpha (TNF-α) and Interleukin-10 (IL-10). Fine regulation of these cytokines’ levels via non-coding RNAs (ncRNAs) might alleviate the immune quiescent nature of TME at TNBC tumors. However, the extrapolation of ncRNAs as therapeutic tools is highly challenging. Therefore, disentanglement the nature for the isolation of natural compounds that could modulate the ncRNAs and their respective targets is an applicable translational therapeutic approach. Hence, this study aimed to targeting the chief immune suppressive cytokines at the TME (TNF-α and IL-10) via ncRNAs and to examine the effects of Rosemary aerial parts extract on the expression levels of these ncRNAs in TNBC. Results revealed miR-17-5p as a dual regulator of TNF-α and IL-10. Moreover, an intricate interaction has been shown between miR-17-5p and the oncogenic lncRNAs: MALAT1 and H19. Knocking down of MALAT1 and/or H19 caused an induction in miR-17-5p and reduction in TNF-α and IL-10 expression levels. miR-17-5p was found to be down-regulated, while TNF-α, IL-10, MALAT1 and H19 were up-regulated in BC patients. Forced expression of miR-17-5p in MDA-MB-231 cells reduced TNF-α, IL-10, MALAT1 and H19 expression levels, as well as several BC hallmarks. In a translational approach, ursolic acid (UA) isolated from rosemary induced the expression of miR-17-5p, MALAT1 and decreased H19 expression levels. In conclusion, this study suggests miR-17-5p as a tumor suppressor and an immune-activator miRNA in BC through tuning up the immunological targets TNF-α, IL-10 at the TME and the oncological mediators MALAT1 and H19 lncRNAs.
Keywords: Breast cancer, Tumor microenvironment, Cytokines, TNF-α, IL-10, miR-17-5p, MALAT1, H19, lncRNAs, miRNA; Ursolic acid
Triple negative breast cancer (TNBC) is considered the most aggressive breast cancer (BC) subtype (Goldhirsch et al., 2013; Nafea et al., 2020). TNBC patients have higher risk of metastasis and poor overall survival than other BC patients (Dent et al., 2007; Abdel-Latif and Youness, 2020). New immunotherapeutic approaches such as immune-checkpoint inhibitors (ICIs) are promising therapeutic modalities for TNBC patients. However, upon application of ICIs in the clinics, immune-resistance cases have been emerged (Bayraktar et al., 2019; Liu et al., 2019; Seto et al., 2019). Tumor microenvironment (TME) has been demonstrated as one of the main players of immune-resistance experienced by TNBC patients (Fares et al., 2019). TNBC patients’ TME comprises an array of immunomodulatory cytokines such as tumor necrosis factor alpha (TNF-α) and Interleukin 10 (IL-10). Both cytokines are notoriously known for having a potent detrimental impact on the host immune-surveillance (Pileczki et al., 2012; Liubomirski et al., 2019a; Liubomirski et al., 2019b).
TNF-α is known to stimulate the recruitment and activity of certain immune-suppressive cell populations, like T regulatory cells (Tregs), regulatory B lymphocytes (Bregs) and myeloid-derived suppressor cells (MDSCs) in different cancer types (Chen et al., 2007; Schioppa et al., 2011; Zhao et al., 2012). All these cell types are crucial negative modulators of tumor immune surveillance. Moreover, TNF-α can have a pro-malignant effect and when silenced, cell proliferation, invasion and motility were abolished and apoptosis was induced in BC cells (Pileczki et al., 2012; Qiao et al., 2016). Nevertheless, TNF-α activates NF-κB and p38/MAPK pathways which stimulate signal transducer and activator of transcription 3 (STAT3) (Cai et al., 2017), a known transcription factor classified as an oncogene (Bromberg et al., 1999). TNF-α plays an important role in the TME both as a membrane-integrated protein and in its soluble form generated after proteolytic cleavage (Balkwill, 2006).
IL-10 is also a well-established immune suppressor mediator that reduces the antigen presentation capacity of dendritic cells and macrophages, thus inhibiting the production of several cytokines which have important role in tumor immunosurveillance (Bogdan et al., 1991; Higgins et al., 1992). Even though TNF-α and IL-10 have been extensively studied in different types of malignancies, yet their role in TNBC is still unexplored. Considering this, it was vital to investigate their regulation in TNBC hypothesizing that dual targeting of TNF-α and IL-10 in TNBC might provoke synergistic action that improves the host anti-tumor immune response and halts BC progression.
To dually target more than one gene, the use of non-coding RNAs (ncRNAs) could be employed. ncRNAs are multi-functional endogenous molecules that have the ability to simultaneously target more than one transcript (Youness and Gad, 2019). One of the ncRNAs types, microRNAs (miRNAs) could co-operatively regulate a coding gene which is either acting as a tumor suppressor or an oncogene (Youness et al., 2016b). However, a new frontier in the field of ncRNAs is the mutual regulation of miRNAs with other ncRNAs such as long ncRNAs (lncRNAs). lncRNAs are transcripts longer than 200 nucleotides (nt) that are not translated into proteins. LncRNAs can be present in the cytoplasm and/or nucleus of cells and functions as an epigenetic regulator of gene expression by binding to miRNA (acting as miRNA sponge). Furthermore, lncRNAs can act as a signaling, decoy, guiding or scaffold molecule by binding to transcription factors and co-activators to regulate protein activity and signaling (Wang et al., 2020). The crosstalk between miRNAs and lncRNAs has been gaining research attention in the past decade (Youness et al., 2019; Nafea et al., 2020). One of the controversial miRNAs that could act as a tumor suppressor miRNA or as an oncomiR, according to the cellular context and cancer type, is miR-17-5p (Pang et al., 2019; Song et al., 2020). Recently, miR-17-5p has been reported to have a mutual relationship with several lncRNAs such as MIR17HG (Xu et al., 2019a).
The nuclear lncRNA, metastasis-associated lung adenocarcinoma transcript 1 (MALAT1, also known as nuclear enriched abundant transcript 2, NEAT2), is one of the most expressed lncRNAs in normal tissues (Hutchinson et al., 2007). To date, there are more than 889 publications related to MALAT1 in cancer (the PubMed search words “MALAT1 cancer” generated 889 results as of 21 February, 2021) making MALAT1 one of the most studied lncRNAs in many types of cancer. However, the study between MALAT1 and miR-17-5p in BC has never been investigated. Another important lncRNA is H19. H19 has diverse functionally; it mainly acts as a sponger or competitive endogenous RNA (ceRNA) for its targeted miRNAs, such as miR-874, miR-675, miR-200, miR-107, miR194, miR-130a, miR196b, let-7b (Zhang et al., 2013; Fang et al., 2018; Luo et al., 2019). However, there is no information linking H19 with miR-17-5p in BC.
Nonetheless, several studies have shown that phytochemicals exert their medicinal effects through regulating ncRNAs (Babashah et al., 2018, Shaalan et al., 2018; Awad et al., 2019; Kang, 2019; Mekky et al., 2019; Ahmed Youness et al., 2020). One of the phytoconstituents that showed success in inhibiting oncogenesis through targeting miRNAs is ursolic acid (UA) (Xiang et al., 2014; Kim et al., 2018). From the abundant cheap plants in Egypt that have high UA content is Rosmarinus officinalis L. (Rosemary). Several studies have reported that Rosemary inhibits cell proliferation, decreases cell viability, induces apoptosis, and even enhances the effects of some chemotherapeutic drugs in various BC cell lines (Cheung and Tai, 2007; Yesil-Celiktas et al., 2010; Gonzalez-Vallinas et al., 2014; Gonzalez-Vallinas et al., 2015). One of its validated anticancer mechanisms is inhibiting Akt/mTOR pathway (Stemke-Hale et al., 2008). Inhibition of Akt phosphorylation was reported to directly affect TNF-α and IL-10 production (Alotaibi et al., 2018). However, a Rosemary effect on TME cytokines in TNBC has never been investigated and especially its inter-connecting relationship with their regulatory ncRNAs.
In the current work, we investigated the role of miR-17-5p in regulating TNF-α and IL-10 signaling, and its interaction with lncRNAs such as MALAT1 and H19. Moreover, we here report the regulatory role of UA isolated from Rosmarinus officinalis L. (Rosemary) on miR-17-5p and its interactive network of coding and non-coding targets with the aim of finding a potential, easily-available and economic immunomodulatory/anti-cancer therapeutic compound for TNBC patients.
Human tissue samples collection
This study included 40 BC female patients that were subjected to conservative mastectomy or lumpectomy surgery in Kasr El-Aini and National Cancer Institute hospitals. Tumors and their noncancerous adjacent tissues were resected. Tissues were immediately frozen in liquid nitrogen and stored at –80°C. Pathology experts confirmed the pathological identity of all samples. The mean age of recruited patients was 47.5 years (Range 34–68) (Table 1, Supplementary Table S1). All patients agreed and signed an informed consent. The study was approved from the ethical committee of the German University in Cairo, Cairo University and National Cancer Institute hospital and was performed in compliance with the ethical standards of the declaration of Helsinki.

MDA-MB-231 TNBC cell line was obtained from VACSERA, Cairo, Egypt. Cells were cultured in DMEM (Dulbecco’s Modified Eagle Medium) (Serox GmbH, Germany) containing fetal bovine serum (FBS) (10%) (Lonza, Germany) and penicillin/streptomycin (1%) (Lonza, Germany) at 37°C under 5% CO2 (Youness et al., 2019). Cells were grown in monolayer and passaged routinely 1–2 times a week as previously described (Youness et al., 2021).
Aerial parts of Rosmarinus officinalis L. (Rosemary) were collected from Al-Orman Garden, Egypt in May 2019. It was authenticated by Dr. M. Gebali, Taxonomy specialist at National Research Centre, Giza, Egypt. A voucher specimen of the authenticated plant (voucher No. 125) was deposited at herbarium of Pharmaceutical Biology department, Faculty of Pharmacy and Biotechnology, German University in Cairo, Cairo, Egypt.
The aerial parts of rosemary (5 kg) were powdered and exhaustively extracted using 85% ethanol (10L) by cold maceration for 3 days. This process was repeated for three times. The total extract was evaporated under reduced pressure using rotary evaporator (BUCHI, Rotavapor, R-210; Switzerland) at temperature 50°C–60°C to yield brownish semi-solid residue (210 g). This residue was suspended in distilled water, and subjected to liquid-liquid fractionation. Then fractionated using solvents with increasing polarity. The obtained residues were washed with acetone and lyophilized to obtain crude contents yielding 39 g and 45 g of dichloromethane and ethyl acetate fractions, respectively.
UPLC/PDA/ESI/MS analysis of the active ethyl acetate fraction
The analysis of the ethyl acetate fraction was done using mass spectrometric tool (UPLC-PDA-ESI- MS) through Waters ACQUITY Xevo TQD system, which consisted of an ACQUITY UPLC H-Class system and Xevo TQD triple-quadrupole tandem mass spectrometer with an electrospray ionization (ESI) interface (Waters Corp., Milford, MA, USA). Acquity BEH C18 column of 1.7 μm particle size with dimensions 100 mm × 2.1 was used for the separation (Waters, Ireland). The mobile phase consisted of 0.1% formic acid in water to represent the (A) and 0.1% formic acid in acetonitrile to represent (B) using a gradient elution as follows: 0–4 min, 15% B; 4–8 min, 20% B; 8–30 min, 55% B; 30–35 min, 90% B; and 35–40 min, 15% B. The used flow rate was 200 μl/min and injection volume was 10 μl. The eluted compounds were detected at mass ranges from 100 to 1,000 m/z. MS scan was carried out at the following conditions: capillary voltage, 3.5 kV; detection at cone voltages, (20 V–95 V); radio frequency (RF) lens voltage, 2.5 V; source temperature, 150°C; and desolvation gas temperature, 500°C. Nitrogen was used as desolvation and cone gas at a flow rate of 1,000 and 20 L/h, respectively. System operation and data acquisition were controlled using Mass Lynx 4.1 software (Waters).
Isolation of Ursolic Acid (UA) from bioactive ethyl acetate fraction
The EA fraction (45 g) was applied over an open column (64 × 5.5 cm) ID packed with polyamide 350 g as stationary phase. Elution was done using methanol CH3OH:H2O gradient to ensure a complete elution process. The fraction exhibited a major blue spot on thin layer chromatography (TLC) using CH3OH:H2O: nitrous acid 6:3:1 under short wavelength Ultraviolet (UV) (254 nm). It then turned into yellow upon exposure to NH3 vapor. Further column was built up for purification of the major spot, then subjected to HPLC/PDA/ESI/MS analysis to identify its fragmentation pattern. Structure was elucidated and confirmed using Bruker 400 MHz Avance III HD, Switzerland. 1H-NMR (400 MHz) and 13C-NMR (100 MHz) spectra were recorded in deuterated DMSO using TMS as an internal standard and chemical shift values were expressed in δ ppm.
Three different target prediction algorithms TargetScan (http://www.targetscan.org/vert_72/), miRwalk (http://mirwalk.umm.uni-heidelberg.de), and starbase (http://starbase.sysu.edu.cn) were used to search for miRNAs that dually target IL-10 and TNF-α genes.
Oligonucleotides transfection in MDA-MB-231 cells
miR-17-5p mimics, MALAT1 siRNAs and H19 siRNAs were purchased from Qiagen, Germany. Cells were trypsinised, counted and seeded into six-well plates the day before transfection to ensure 80-90% cell confluence on the day of transfection (Youness et al., 2016a). Transfection into cells was performed using HiPerfect® Transfection Reagent (Qiagen, Germany) in accordance with the manufacturer’s advised procedure as previously described (Youness et al., 2016a; Youness et al., 2016b; Youness et al., 2018). All Experiments were run in triplicates and repeated at least three times.
Total RNA extraction from BC tissues and MDA-MB-231 cells
Biazol reagent (Hangzhou bioer technology Co., Ltd., China) was utilized for total RNA extraction from BC patients’ tissues and the TNBC cell lines. The resultant yield of RNA was assessed using a spectrophotometer.
Quantitative real-time polymerase chain reaction analysis (qRT-PCR)
Reverse transcription of TNF-α, IL-10, MALAT1, H19, β-actin and 18s rRNA mRNAs into complementary DNA (cDNA) was performed using a high-capacity cDNA Reverse-Transcription Kit (ABI, California, USA) according to the company's instruction. TaqMan_MicroRNA Reverse-Transcription Kit (ABI) was used to reverse-transcribe extracted miRNAs and specific primers for hsa-miR-17-5p and RNU6B into single-stranded cDNA. Relative expression of TNF-α, IL-10, MALAT1 and H19 was calculated after normalization to β-actin and 18s rRNA as reference genes. miR-17-5p and RNU6B (for normalization) cDNA were quantified using a TaqMan Real-Time qPCR on StepOne™ Systems (ABI, California, USA) (Youness et al., 2019). Relative expression was calculated using the 2–ΔΔCT method. All PCR reactions were run in triplicates and repeated at least three times.
Measurement of TNF-α and IL-10 cytokines
Cytokine levels were measured by ELISA technique. After transfection of MDA-MB-231 cell lines with miR-17-5p for 48 h, supernatants were collected and stored at –20°C until assayed for cytokines by ELISA (Thermo scientific, USA) as per manufacturer’s protocol.
Cellular viability was assessed using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. MDA-MB-231 cells were seeded in 200 µl of culture media (full DMEM) per well in a 96-well plate and incubated 24 h prior to transfection with miR-17-5p mimics (according to the HiPerfect protocol; Qiagen GmbH) or treatment with the natural compounds. At 48 h post-transfection/treatment, 20 µl MTT solution (Sigma Aldrich, Germany) (5 mg/ml MTT in phosphate-buffered saline) was added to each well. Subsequent to incubation for 5 h, the formed formazan (MTT metabolic product) was re-suspended in 200 µl dimethyl sulfoxide (Sigma Aldrich, Germany). Colorimetric measurements and absorbance were performed using Wallac 1420 Victor 2 Multilabel Counter (Perkin Elmer Inc., Waltham, MA, USA) (Youness et al., 2016a). All cell viability experiments were performed in quadrates and repeated five times.
Migration assay (Wound-healing assay)
Cell migration was determined by wound healing assay post-transfection with miR-17-5p oligonucleotides or post-treatment with rosemary or Ursolic acid. Cells were seeded into a 6-well plate. Upon reaching 80%–90% confluence, the cell layer was scratched with a 10 μl pipette tip and washed with culture medium twice and cultured again for up to 48 h. Finally, the scratch wounds were visualized and imaged under a microscope (ZEISS Axio Observer Z1, Jena, Germany) (Youness et al., 2016b). All experiments were performed in triplicates and repeated at least three times. Cells were photographed at 0 h and after 24 h of culture to record the wound width.
We utilized the colony formation assay to determine the TNBC cell proliferative ability post-transfection using miR-17-5p oligonucleotides or post-treatment with Ursolic acid. MDA-MB-231 cells were transfected with miR-17-5p mimics (Qiagen, Germany). The transfected cells were then allowed to grow in 6-well plates and maintained in medium containing 10% FBS, and the medium was replaced after 6–7 days. After 14 days, cells were fixed with 4% formaldehyde and stained with 0.1% crystal violet at room temperature for 30 min. Colonies were counted and compared (Youness et al., 2019). All experiments were performed in triplicates.
Data are expressed as mean ± standard error of the mean (SEM) of at least 3 independent experiments. The statistical analysis was done by using Student’s t-test and P value < 0.05 was considered significant. While, one-way analysis of variance (One-way ANOVA) was performed for the purpose of comparison of more than two groups. Data was analyzed using GraphPad Prism 8.2.1 software.
Selection of miR-17-5p using bioinformatic analysis
Based on bioinformatic analysis, miR-17-5p was selected as the candidate miRNA that has the potential to dually target TNF-α and IL-10 with high binding scores (Figs. 1a and 1b, respectively). Different online algorithms were used. After the search was executed, results were sorted according to their mirSVR score. mirSVR is a regression model that computes a weighted sum of a number of sequence and context features of the predicted miRNA-mRNA duplex. Using miRbase.org database (http://www.mirbase.org), miR-17-5p accession number and mature sequence were retrieved (Table 2). In an attempt to investigate its relationship with other ncRNAs by bioinformatic analysis, miR-17-5p was found to hit the lncRNAs MALAT1 and H19 3’UTR sequence with high scores (Figs. 1c and 1d, respectively). It is also worth mentioning that the binding of miR-17-5p to H19 and MALAT have been experimentally validated in other studies (Jia et al., 2019; Xu et al., 2019b).

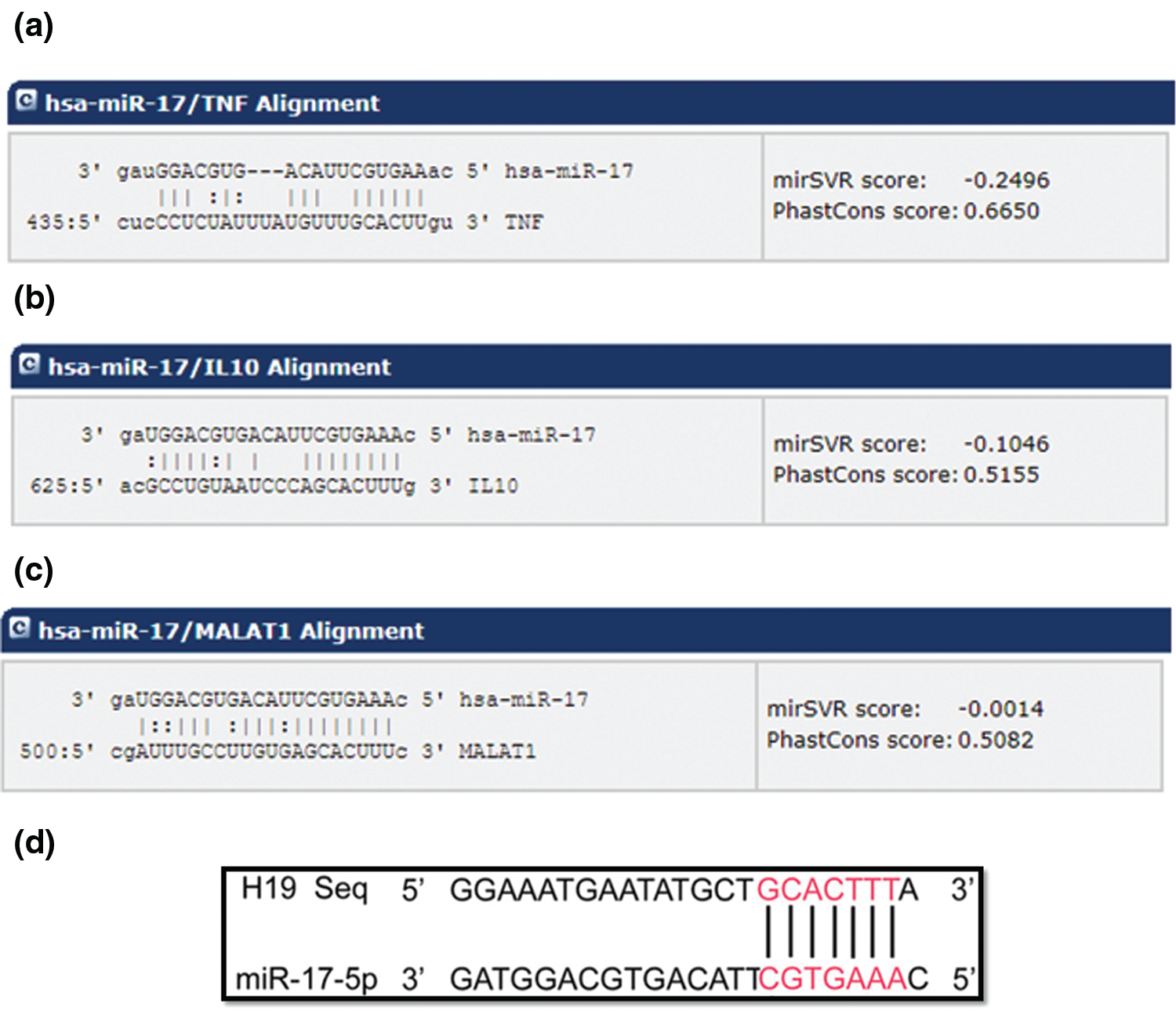
Figure 1: Predicted target region-seed sequence binding for miR-17-5p on the 3’UTR of TNF-α, IL-10, MALAT1 and H19. The panels show the binding regions on the 3’UTR of the target transcripts, TNF-α, IL-10, MALAT1 and H19 aligned with the seed sequence of miR-17-5p, as predicted by different software. The lines indicate complementarity between the binding region of the mRNA and the seed sequence of the miRNA, also shown are the mirSVR scores. These features include three types: (1) duplex features which includes base pairing at the seed region, and 3’end of the mRNA, (2) sequence features that include A/U composition near the target sites and secondary structure accessibility, and (3) global features such as length of the UTR, relative position of the target site in the UTR and conservation score. mirSVR down-regulation scores are calibrated to correlate linearly with the extent of down-regulation. (a) Alignment of the TNF-α transcript with hsa-miR-17. (b) Alignment of IL-10 target transcript with hsa-miR-17. (c) Alignment of the MALAT1 transcript with hsa miR-17 (d) Alignment of H19 transcript with miR-17-5p.
BC patients express high levels of TNF-α and IL-10
To study the expression of TNF-α and IL-10, we first evaluated both cytokines expression in 40 BC tumor samples using qRT-PCR. The expression levels of TNF-α and IL-10 were higher in BC tissues compared with their normal BC samples counterparts (P < 0.0001, Fig. 2a), (P = 0.0001, Fig. 2b), respectively.
BC patients express low levels of miR-17-5p and high levels of MALAT1 and H19 lncRNAs
To screen for the ncRNA transcription levels in BC, miR-17-5p, MALAT1 and H19 expression levels were evaluated in tumor breast tissues of the 40 BC patients. miR-17-5p was significantly down-regulated in BC tissues compared to normal tissues (P = 0.00930, Fig. 2c). On the other hand, MALAT1 and H19 levels were significantly upregulated in BC tissues compared to their normal counterparts (P = 0.0009, P = 0.0079, Figs. 2d and 2e).

Figure 2: Screening of TNF-α, IL-10, miR-17-5p, MALAT-1 and H19 lncRNAs in BC patients. TNF-α, IL-10, miR-17-5p, MALAT-1 and H19 lncRNAs expression level were quantified in 40 BC tissues and in normal tissues using qRT-PCR and were normalized to levels of 18S rRNA as an internal control, while miR-17-5p was normalized to RNU6B. (a) and (b) significant up-regulation of TNF-α and IL-10 in BC tissues; respectively compared to its normal counterparts. (c), (d), and (e) significant down-regulation of miR-17-5p, and upregulation of MALAT1 and H19; respectively in BC tissues compared to its normal counterparts. Student t test was performed. Data are presented as mean ± SEM of three independent experiments; ****= P ≤ 0.0001, *** = P < 0.001, **= P < 0.01 compared with normal tissues.
miR-17-5p is a tumor suppressor in MDA-MB-231 cells
Since literature concerning miR-17-5p role in cancer was controversial, it was essential to examine its impact in BC. Several BC hallmarks were assessed. Ectopic expression of miR-17-5p in MDA-MB-231 led to a significant decrease in cellular viability (P < 0.0001, Fig. 3a), clonogenicity (P = 0.0003, Fig. 3b) and cellular migration capacity of MDA-MB-231 cells compared to mock cells (P = 0.0433, Fig. 3c).

Figure 3: Impact of miR-17-5p transfection on MDA-MB-231 cellular viability, clonogenicity and migration. (a) miR-17-5p mimic decreased MDA-MB-231 cellular viability tested by MTT assay (b) miR-17-5p mimics decreased MDA-MB-231 cell migration capacity measured by wound healing assay. (c) significant reduction of TNBC cells migration capacity by miR-17-5p mimics transfection assessed as % closure of wound. Student t test was performed. Data are presented as mean ± SEM of three independent experiments; *= P < 0.05 compared with control group.
miR-17-5p transfection decreased TNF-α and IL-10 mRNA and protein levels in TNBC cells
Efficient transfection of miR-17-5p oligonucleotides was successfully confirmed in MDA-MB-231 cells (P = 0.0062, Fig. 4a), which resulted in a significant decrease in TNF-α and IL-10 transcripts (P = 0.0080 and P = 0.0054, Figs. 4b and 4c) and protein levels (P < 0.0001 and P = 0.0001, respectively, Figs. 4d and 4e) compared to mock cells.

Figure 4: The impact of miR-17-5p transfection on TNF-α and IL-10 transcript and protein levels in MDA-MB-231 cells. miR-17-5p levels was determined in MDA-MB-231 cells using qRT-PCR 48 h post transfection. miR-17-5p expression was normalized to RNU6B as an internal control. After ectopic expression of miR-17-5p in MDA-MB-231 cells, TNF-α and IL-10 transcript levels were assessed using qRT-PCR and normalized to β-actin as an internal control and their protein levels in the cellular supernatant was quantified using respective ELISA kits and represented as OD 450nm. (a) Levels of miR-17-5p showed 10,000 folds increase in mimicked MDA-MB-231 cells compared to mock cells. (b) and (c) TNF-α and IL-10 transcript levels, respectively were significantly decreased compared to mock post miR-17-5p transfection. (d) and (e) TNF-α and IL-10 protein levels were significantly decreased compared to mock post transfection. Student t test was performed. Data are presented as mean ± SEM of three independent experiments; ****= P ≤ 0.0001, *** = P < 0.001, **= P < 0.01 compared with control group.
miR-17-5p transfection decreased MALAT1 and H19 mRNA levels in TNBC cells
As noticed with TNF-α and IL-10 transcripts, miR-17-5p mimic resulted in a significant decrease in MALAT1 (P = 0.0002, Fig. 5a), and H19 (P = 0.0009, Fig. 5b) expression levels compared to mock cells.

Figure 5: Impact of miR-17-5p on MALAT-1 and H19 transcription levels in MDA-MB-231 cells. MALAT1 and H19 expression levels were assessed post miR-17-5p transfection using qRT-PCR and normalized to β-actin as an internal control. (a–b) MALAT1 and H19 expression levels were decreased post-transfection of miR-17-5p compared to mock cells. Student t test was performed. Data are presented as mean ± SEM of three independent experiments; *** = P < 0.001 compared with control group.
MALAT1 siRNAs transfection decreased TNF-α and IL-10 expression levels
To study the relationship between lncRNA MALAT1 and miR-17-5p target genes; TNF-α and IL-10, cells were transfected with MALAT siRNAs. Efficient knockdown of MALAT1 was first confirmed (P = 0.0223, Fig. 6a), then the influence of MALAT1 silencing on the cytokines levels was investigated. A significant decrease in TNF-α (P = 0.0263, Fig. 6b) and IL-10 (P = 0.0057, Fig. 6c) transcript levels compared to their respective mock cells was observed.

Figure 6: Impact of MALAT1 siRNAs on TNF-α and IL-10 expression (a) efficient transfection of the MALAT1 siRNA was confirmed by a significant decrease in lncRNA MALAT1 expression levels. (b–c) knocking down of MALAT1 caused a significant decrease in TNF-α and IL-10 transcript levels in MDA-MB-231 cells.
H19 siRNAs transfection decreased TNF-α and IL-10 expression levels
Similarly, the effect of lncH19 knocking down was investigated. Efficient knockdown of H19 was first confirmed using qPCR (P = 0.0017, Fig. 7a), which caused a significant decrease in TNF-α (P < 0.0001, Fig. 7b), and IL-10 (P = 0.0032, Fig. 7c) transcript levels.

Figure 7: Impact of H19 siRNAs on TNF-α and IL-10 expression (a) efficient transfection of the H19 siRNA was confirmed by a significant decrease in lncRNA H19 expression levels. (b–c) knocking down of H19 caused a significant decrease in TNF-α and IL-10 transcript levels in MDA-MB-231 cells.
MALAT1 siRNAs transfection decreased H19 and increased miR-17-5p expression levels
To consolidate the mutual relationship between MALAT1 and H19, H19 transcript levels were assessed in response to MALAT1 siRNAs transfection. It was noticeable, that Knocking down of MALAT1 in the MDA-MB-231 cells resulted in a significant decrease in lncRNA H19 transcript levels (P = 0.0072, Fig. 8a). On the other hand knocking down of MALAT-1 resulted in a significant increase in miR-17-5p levels (P = 0.0033, Fig. 8b) confirming their mutual relationship.

Figure 8: Impact of MALAT1 siRNAs on H19 and miR-17-5p expression (a) and (b) efficient silencing of the MALAT1 resulted in a significant decrease in lncRNA H19 expression levels and a significant increase in the miR-17-5p expression levels.
H19 siRNAs transfection decreased MALAT1 and increased miR-17-5p expression levels
Similar results to those of MALAT1 siRNAs transfection was observed with H19 siRNAs; where H19 knocking down resulted in a significant decrease in lncRNA MALAT1 (P = 0.0488, Fig. 9a) and increase in miR-17-5p transcript levels (P < 0.0001, Fig. 9b).

Figure 9: Impact of H19 siRNAs on MALAT1 and miR-17-5p expression (a) and (b) efficient silencing of the H19 resulted in a significant decrease in lncRNA MALAT1 and an increase miR-17-5p expression levels.
Rosemary fractions inhibited cellular viability in TNBC cells
Further approach was taken to investigate the ability of natural compounds to regulate ncRNAs and their respective target cytokines. Rosemary is known for its potential anticancer effect. Fractionation of rosemary aerial parts extract was first performed using dichloromethane and ethyl acetate (EA). Cellular viability of MDA-MB-231 in response to rosemary fractions exposure was evaluated relative to vehicle control cells. Both fractions led to significant decrease in cellular viability, either with diff concentrations (30μM-60μM) of dichloromethane fraction (P ≤ 0.0001, Fig. 10a) or EA fraction (P ≤ 0.0001, Fig. 10b).
Characterization of metabolites of bioactive fraction using HPLC/PDA/ESI/ MS
HPLC-PDA-ESI-MS was used to characterize phenolic metabolites within the bioactive fraction Fig. S1). Metabolites profiling revealed the presence of the phenolic acid, ursolic acid, as the major peak with peak area of 2.5%. The MS, 1H-NMR and 13C-NMR spectral data was compared closely with that of the reported by the literature (Annan et al., 2011).
Ursolic acid (UA) Inhibited cellular viability, colony Formation and migration of TNBC cells
In these experiments, UA was chosen as a major active constituent of the rosemary extract. To functionally analyze the UA effect on TNBC cell lines, MDA-MB-231 cells were treated with different concentrations of UA (20 μM–100 μM). MTT assay was performed and cellular viability was calculated relative to mock cells. A significant decrease in cellular viability was observed upon UA treatment compared to vehicle control (P < 0.0001, Fig. 10c). To investigate the long-term effects on MDA-MB-231 TNBC cell lines, colony forming assay was performed. UA led to a dose-dependent decrease in number of colonies reaching to significance only with the highest concentration (100 μM) (P = 0.0358, Fig. 10d). Cellular migration was calculated as percentage closure of wound. UA treatment resulted in a marked decrease in cellular migration of MDA-MB-231 cells (P = 0.0414, Fig. 10e).

Figure 10: Impact of treatment of rosemary fractions on MDA-MB-231 cellular viability and purified Ursolic acid on MDA-MB-231 cellular viability, clonogenicity and cellular migration. The cytotoxic effect of treatment of (a) Dichloromethane rosemary fraction on MDA-MB-231 was measured using MTT assay where a concentration dependent cytotoxic effect was observed. (b) Ethyl acetate rosemary fraction also resulted in a significant decrease in cellular viability in TNBC cell lines. (c) Ursolic acid showed cytotoxic effects within 24 h of treatment in a dose-dependent manner. (d) Ursolic acid treatment resulted in a decrease in colony forming ability at highest concentration (100 μM) only in MDA-MB-231. (e) a significant reduction of MDA-MB-231 cells migration capacity after Ursolic acid treatment. Student t test was performed. Data are presented as mean ± SEM of three independent experiments: ****= P ≤ 0.0001, **= P < 0.01, compared with control group. One-way analysis of variance (ANOVA) was performed. Data are presented as mean ± SEM of three independent experiments; **** = P < 0.0001.
Ursolic acid increased miR-17-5p, MALAT1, TNF-α and decreased H19 and IL-10 expression levels
MDA-MB-231 cells were treated with 60 µM UA then miR-17-5p, MALAT1 and H19 expression levels were assessed using qRT-PCR. Results showed a significant increase in miR-17-5p (P < 0.0001, Fig. 11a), a significant increase in MALAT1 (P < 0.0001, Fig. 11b), and a significant decrease in H19 (P = 0.0094, Fig. 11c) expression levels. For cytokines, TNF-α expression showed an increase (P = 0.0003, Fig. 11d) while IL-10 expression was decreased (P < 0.0001, Fig. 11e) upon UA treatment.

Figure 11: Impact of Ursolic acid on miR-17-5p, MALAT1, H19, TNF-α and IL-10 expression levels Expression levels in MDA-MB-231 upon Ursolic acid treatment were determined by qRT-PCR 48 h post-treatment, levels were normalized to RNU6B as an internal control. (a) a significant increase in miR-17-5p expression. (b) a significant increase in MALAT1 expression. (c) a significant decrease in H19 expression. (d) a significant increase in TNF-α expression. (e) a significant decrease in IL-10 expression. Student t test was performed. Data are presented as mean ± SEM of three independent experiments: ****= P ≤ 0.0001, **= P < 0.01, compared with control group.
TNBC tumors are the most immunogenic tumors if compared to other BC subtypes (Liu et al., 2018). This fact affected the treatment paradigm of TNBC patients which shifted the clinicians towards immunotherapeutic alternatives such as immune checkpoint inhibitors (ICIs) (Abdel-Latif and Youness, 2020). Despite the success of ICIs, resistant cases started to appear in the clinics. Several studies referred to the role of tumor microenvironment (TME) in this resistance. TME includes an array of immune-modulatory cytokines such as IL-10 and TNF-α that act as a formidable barrier in eliminating the tumor cells by the tumor infiltrating lymphocytes (Ahmad et al., 2018; Cruceriu et al., 2020). Single targeting of either TNF-α or IL-10 using siRNAs or CRISPR Cas technology has not been therapeutically efficient when translated to the clinics (Mumm et al., 2011; Alotaibi et al., 2018; Zhang et al., 2018; Cruceriu et al., 2020).
ncRNAs have recently been casted as prominent regulators of the immune cells. However, their role in regulating the non-cellular components of the TME has seldom been investigated. Therefore, this study focused on unraveling the ncRNAs circuit that dually target IL-10 and TNF-α in TNBC. Bioinformatics analysis showed that miR-17-5p could potentially target TNF-α and IL-10. miR-17-5p is one of the miRNAs that has been studied in different malignancies in a paradoxical manner (Fan et al., 2014; Chen et al., 2016; Zhang et al., 2017; Cai et al., 2018). Nonetheless, miR-17-5p is known to have a potential crosstalk with other classes of ncRNAs such as lncRNAs (Chen et al., 2015; Peng et al., 2015). Consequently, we extended the study to explore the manipulation of TNF-α and IL-10 with miR-17-5p/MALAT1/H19 interacting network in the TNBC. We found that miR-17-5p expression was significantly downregulated in BC tissues, while TNF-α, IL-10, MALAT1 and H19 were significantly upregulated. Transfection of miR-17-5p in MDA-MB-231 cells resulted in a significant down regulation of TNF-α and IL-10. This goes in line with a study performed by Akhtar N et al. (2016) where miR-17-5p was found to act as a negative regulator of TNF-α signaling in rheumatoid arthritis synovial fibroblasts by modulating the protein ubiquitin processes. Yet, our study is considered the first to validate such reciprocal relationship between miR-17-5p and TNF-α in TNBC. The same concept applies to IL-10 as well. Although no literature has explored such interaction but a similar pattern of results was obtained for miR-106a, which share the same seed sequence with miR-17-5p, on IL-10 (Gruszka and Zakrzewska, 2018). Sharma et al. (2009) reported that miR-106a suppress IL-10 through regulating its transcription factor specificity protein 1 (Sp1). miR-17-92 cluster is also known to regulate Sp1. Therefore, one of the mechanisms by which miR-17-5p could suppress IL-10 expression levels is likely through Sp1.
Upon forcing the expression of miR-17-5p in MDA-MB-231 cells, a drastic reduction of MALAT1 and H19 expression levels was observed. On the other hand, knocking down MALAT1 and H19 increased miR-17-5p expression levels. Collectively, this shows that there is a ceRNA network between the ncRNAs of this study where MALAT1 and H19 act as molecular sponge to attract miR-17-5p repressing its consequent inhibitory action on the immune-suppressive TNF-α and IL-10. It is worth mentioning that our results showed that ectopic expression of miR-17-5p or knocking down of either MALAT1 or H19 were able to reduce TNF-α and IL-10 expression levels in TNBC cells, thus highlighting a prominent alleviation of the immune suppressive TME in TNBC cells. Furthermore, we have identified that miR-17-5p can repress the oncogenic lncRNAs MALAT1 and H19 expression.
These findings highlight miR-17-5p as an immune-activator miRNA in TNBC through repressing the immune-suppressive cytokines; TNF-α and IL-10. Furthermore, miR-17-5p is the main player taking the upper hand in miR-17-5p/MALAT1/H19 regulatory axis. We were intrigued to further take these findings forward to a more translational approach. Here we report that rosemary EA fraction significantly reduced cellular viability of MDA-MB-231 cells more efficiently that dichloromethane fraction. This could be supported by another study which showed that rosemary extract inhibited phosphorylation/activation of Akt in TNBC cells (Jaglanian and Tsiani, 2020). UA was purified from the rosemary EA fraction for closer investigation on TNBC hallmarks (El-Askary et al., 2019). UA treatment was able to reduce the cellular viability, clonogenicity as well as migration capacity of the TNBC cells. This was supported by the of ability of UA to reduce Akt expression in MDA-MB-231 cell line (Luo et al., 2017). Treatment of MDA-MB-231cells with UA also modulated the expression of ncRNAs by increasing the miR-17-5p, MALAT1 and reducing H19 expression levels. As with the majority of studies, the current study is subjected to limitations that need to be addressed in future research. First, an array of cytokines such as IL-2, IL-6, IL-8 and IL-12 could be evaluated to identify a cytokine signature for TME of TNBC patients. Secondly, bridging the impact of miR-17-5p, MALAT-1 and H19 on the cellular components of the TME such as CTLs and NK cells with the non-cellular components such as cytokines will contribute to a deeper understanding to the role of miR-17-5p at the TME of TNBC patients. Therefore, our future perspective is to explore the role of miR-17-5p, MALAT-1 and H19 lncRNAs on adative and innate immune ligands and their intrinsic role on tumor associated immune cells such as CTLs, NK cells and macrophages.
In conclusion, this study casts a new light on the role of miR-17-5p in TNBC as a tumor suppressor and an immune-activator miRNA by alleviating the TME immune-suppressive cytokines TNF-α and IL-10. Moreover, it highlights its interplay with the oncogenic lncRNAs MALAT1 and H19, repressing their expression levels. Ursolic acid has antioncogenic and immunomodulatory properties in TNBC partially through modulating the studied ncRNA network. Therefore, this study reports a novel therapeutic nutri-genetic immunotherapeutic approach for TNBC patients.
Availability of Data and Materials: The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors’ Contribution: The authors confirm contribution to the paper as follows: study conception and design: M.Z. Gad, R.A. Youness, H. Handoussa; data collection: R.A. Soliman, H. Handoussa. M. Abdelmohsem; samples and clinical data collection: T. Manie, E. Khallaf; analysis and interpretation of results: R.A. Soliman, H. Handoussa, R.A. Youness, M.Z. Gad; Experimental Design: R.A. Soliman, R.A. Youness; draft manuscript preparation: R.A. Soliman, R.A. Youness, H. Handoussa, M. ElShazly, M.Z. Gad All authors reviewed the results and approved the final version of the manuscript.
Ethics Approval: The study was approved from the ethical committee of the German University in Cairo (Ethical Approval No. 210, Date of Approval: 12-01-2019) and Cairo University (Ethical Approval No. N-42-2019, Date of Approval: 23-08-2019). All experiments were performed in compliance with the ethical standards of the declaration of Helsinki.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Abdel-Latif M, Youness RA (2020). Why natural killer cells in triple negative breast cancer? World Journal of Clinical Oncology 11: 464–476. DOI 10.5306/wjco.v11.i7.464. [Google Scholar] [CrossRef]
Ahmad N, Ammar A, Storr SJ, Green AR, Rakha E, Ellis IO, Martin SG (2018). IL-6 and IL-10 are associated with good prognosis in early stage invasive breast cancer patients. Cancer Immunology, Immunotherapy 67: 537–549. DOI 10.1007/s00262-017-2106-8. [Google Scholar] [CrossRef]
Ahmed Youness R, Amr Assal R, Mohamed Ezzat S, Zakaria Gad M, Abdel Motaal A (2020). A methoxylated quercetin glycoside harnesses HCC tumor progression in a TP53/miR-15/miR-16 dependent manner. Natural Product Research 34: 1475–1480. DOI 10.1080/14786419.2018.1509326. [Google Scholar] [CrossRef]
Akhtar N, Singh AK, Ahmed S (2016). MicroRNA-17 suppresses TNF-alpha signaling by interfering with TRAF2 and cIAP2 association in rheumatoid arthritis synovial fibroblasts. Journal of Immunology 197: 2219–2228. DOI 10.4049/jimmunol.1600360. [Google Scholar] [CrossRef]
Alotaibi MR, Hassan ZK, Al-Rejaie SS, Alshammari MA, Almutairi MM, Alhoshani AR, Alanazi WA, Hafez MM, Al-Shabanah OA (2018). Characterization of apoptosis in a breast cancer cell line after IL-10 Silencing. Asian Pacific Journal of Cancer Prevention 19: 777–783. [Google Scholar]
Annan K, Jackson N, Dickson RA, Sam GH, Komlaga G (2011). Acaricidal effect of an isolate from Hoslundia opposita vahl against Amblyomma variegatum (Acari: Ixodidae). Pharmacognosy Research 3: 185–188. DOI 10.4103/0974-8490.85004. [Google Scholar] [CrossRef]
Awad AR, Youness RA, Ibrahim M, Motaal AA, El-Askary HI, Assal RA, Gad MZ (2019). An acetylated derivative of vitexin halts MDA-MB-231 cellular progression and improves its immunogenic profile through tuning miR-20a-MICA/B axis. Natural Product Research 35: 3126–3130. DOI 10.1080/14786419.2019.1686372. [Google Scholar] [CrossRef]
Babashah S, Bakhshinejad B, Birgani MT, Pakravan K, Cho WC (2018). Regulation of MicroRNAs by phytochemicals: A promising strategy for cancer chemoprevention. Current Cancer Drug Targets 18: 640–651. DOI 10.2174/1568009617666170623124710. [Google Scholar] [CrossRef]
Balkwill F (2006). TNF-alpha in promotion and progression of cancer. Cancer and Metastasis Reviews 25: 409–416. DOI 10.1007/s10555-006-9005-3. [Google Scholar] [CrossRef]
Bayraktar S, Batoo S, Okuno S, Gluck S (2019). Immunotherapy in breast cancer. Journal of Carcinogenesis 18: 2. DOI 10.4103/jcar.JCar_2_19. [Google Scholar] [CrossRef]
Bogdan C, Vodovotz Y, Nathan C (1991). Macrophage deactivation by interleukin 10. Journal of Experimental Medicine 174: 1549–1555. DOI 10.1084/jem.174.6.1549. [Google Scholar] [CrossRef]
Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE,Jr (1999). Stat3 as an oncogene. Cell 98: 295–303. DOI 10.1016/S0092-8674(00)81959-5. [Google Scholar] [CrossRef]
Cai N, Hu L, Xie Y, Gao JH, Zhai W, Wang L, Jin QJ, Qin CY, Qiang R (2018). MiR-17-5p promotes cervical cancer cell proliferation and metastasis by targeting transforming growth factor-beta receptor 2. European Review for Medical and Pharmacological Sciences 22: 1899–1906. [Google Scholar]
Cai X, Cao C, Li J, Chen F, Zhang S, Liu B, Zhang W, Zhang X, Ye L (2017). Inflammatory factor TNF-alpha promotes the growth of breast cancer via the positive feedback loop of TNFR1/NF-kappaB (and/or p38)/p-STAT3/HBXIP/TNFR1. Oncotarget 8: 58338–58352. DOI 10.18632/oncotarget.16873. [Google Scholar] [CrossRef]
Chen C, Lu Z, Yang J, Hao W, Qin Y, Wang H, Xie C, Xie R (2016). MiR-17-5p promotes cancer cell proliferation and tumorigenesis in nasopharyngeal carcinoma by targeting p21. Cancer Medicine 5: 3489–3499. DOI 10.1002/cam4.863. [Google Scholar] [CrossRef]
Chen CL, Tseng YW, Wu JC, Chen GY, Lin KC, Hwang SM, Hu YC (2015). Suppression of hepatocellular carcinoma by baculovirus-mediated expression of long non-coding RNA PTENP1 and MicroRNA regulation. Biomaterials 44: 71–81. DOI 10.1016/j.biomaterials.2014.12.023. [Google Scholar] [CrossRef]
Chen X, Baumel M, Mannel DN, Howard OM, Oppenheim JJ (2007). Interaction of TNF with TNF receptor type 2 promotes expansion and function of mouse CD4+CD25+ T regulatory cells. Journal of Immunology 179: 154–161. DOI 10.4049/jimmunol.179.1.154. [Google Scholar] [CrossRef]
Cheung S, Tai J (2007). Anti-proliferative and antioxidant properties of rosemary Rosmarinus officinalis. Oncology Reports 17: 1525–1531. DOI 10.3892/or.17.6.1525. [Google Scholar] [CrossRef]
Cruceriu D, Baldasici O, Balacescu O, Berindan-Neagoe I (2020). The dual role of tumor necrosis factor-alpha (TNF-alpha) in breast cancer: Molecular insights and therapeutic approaches. Cellular Oncology 43: 1–18. DOI 10.1007/s13402-019-00489-1. [Google Scholar] [CrossRef]
Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P, Narod SA (2007). Triple-negative breast cancer: Clinical features and patterns of recurrence. Clinical Cancer Research 13: 4429–4434. DOI 10.1158/1078-0432.CCR-06-3045. [Google Scholar] [CrossRef]
El-Askary H, Handoussa H, Badria F, El-Khatib AH, Alsayari A, Linscheid M W, Abdel Motaal A (2019). Characterization of hepatoprotective metabolites from Artemisia annua and Cleome droserifolia using HPLC/PDA/ESI/MS-MS. Revista Brasileira de Farmacognosia 29: 213–220. DOI 10.1016/j.bjp.2018.10.001. [Google Scholar] [CrossRef]
Fan M, Sethuraman A, Brown M, Sun W, Pfeffer LM (2014). Systematic analysis of metastasis-associated genes identifies miR-17-5p as a metastatic suppressor of basal-like breast cancer. Breast Cancer Research and Treatment 146: 487–502. DOI 10.1007/s10549-014-3040-5. [Google Scholar] [CrossRef]
Fang Y, Hu J, Wang Z, Zong H, Zhang L, Zhang R, Sun L (2018). LncRNA H19 functions as an Aquaporin 1 competitive endogenous RNA to regulate microRNA-874 expression in LPS sepsis. Biomedicine & Pharmacotherapy 105: 1183–1191. DOI 10.1016/j.biopha.2018.06.007. [Google Scholar] [CrossRef]
Fares CM, van Allen EM, Drake CG, Allison JP, Hu-Lieskovan S (2019). Mechanisms of resistance to immune checkpoint blockade: Why does checkpoint inhibitor immunotherapy not work for all patients? American Society of Clinical Oncology Educational Book 39: 147–164. DOI 10.1200/EDBK_240837. [Google Scholar] [CrossRef]
Goldhirsch A, Winer EP, Coates AS, Gelber RD, Piccart-Gebhart M et al. (2013). Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Annals of Oncology 24: 2206–2223. DOI 10.1093/annonc/mdt303. [Google Scholar] [CrossRef]
Gonzalez-Vallinas M, Molina S, Vicente G, Sanchez-Martinez R, Vargas T, Garcia-Risco MR, Fornari T, Reglero G, Ramirez de Molina A (2014). Modulation of estrogen and epidermal growth factor receptors by rosemary extract in breast cancer cells. Electrophoresis 35: 1719–1727. DOI 10.1002/elps.201400011. [Google Scholar] [CrossRef]
Gonzalez-Vallinas M, Reglero G, Ramirez de Molina A (2015). Rosemary (Rosmarinus officinalis L.) extract as a potential complementary agent in anticancer therapy. Nutrition and Cancer–An International Journal 67: 1221–1229. DOI 10.1080/01635581.2015.1082110. [Google Scholar] [CrossRef]
Gruszka R, Zakrzewska M (2018). The oncogenic relevance of miR-17-92 cluster and its paralogous miR-106b-25 and miR-106a-363 clusters in brain tumors. International Journal of Molecular Sciences 19: 879. DOI 10.3390/ijms19030879. [Google Scholar] [CrossRef]
Higgins TL, Estafanous FG, Loop FD, Beck GJ, Blum JM, Paranandi L (1992). Stratification of morbidity and mortality outcome by preoperative risk factors in coronary artery bypass patients. A clinical severity score. JAMA 267: 2344–2348. DOI 10.1001/jama.1992.03480170070031. [Google Scholar] [CrossRef]
Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A (2007). A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics 8: 39. DOI 10.1186/1471-2164-8-39. [Google Scholar] [CrossRef]
Jaglanian A, Tsiani E (2020). Rosemary extract inhibits proliferation, survival, Akt, and mTOR signaling in triple-negative breast cancer cells. International Journal of Molecular Sciences 21: 810. DOI 10.3390/ijms21030810. [Google Scholar] [CrossRef]
Jia J, Zhang X, Zhan D, Li J, Li Z, Li H, Qian J (2019). LncRNA H19 interacted with miR-130a-3p and miR-17-5p to modify radio-resistance and chemo-sensitivity of cardiac carcinoma cells. Cancer Medicine 8: 1604–1618. DOI 10.1002/cam4.1860. [Google Scholar] [CrossRef]
Kang H (2019). MicroRNA-mediated health-promoting effects of phytochemicals. International Journal of Molecular Sciences 20: 2535. DOI 10.3390/ijms20102535. [Google Scholar] [CrossRef]
Kim K, Shin EA, Jung JH, Park JE, Kim DS, Shim BS, Kim SH (2018). Ursolic acid induces apoptosis in colorectal cancer cells partially via upregulation of microRNA-4500 and inhibition of JAK2/STAT3 phosphorylation. International Journal of Molecular Sciences 20: 114. DOI 10.3390/ijms20010114. [Google Scholar] [CrossRef]
Liu D, Jenkins RW, Sullivan RJ (2019). Mechanisms of resistance to immune checkpoint blockade. American Journal of Clinical Dermatology 20: 41–54. DOI 10.1007/s40257-018-0389-y. [Google Scholar] [CrossRef]
Liu Z, Li M, Jiang Z, Wang X (2018). A comprehensive immunologic portrait of triple-negative breast cancer. Translational Oncology 11: 311–329. DOI 10.1016/j.tranon.2018.01.011. [Google Scholar] [CrossRef]
Liubomirski Y, Lerrer S, Meshel T, Morein D, Rubinstein-Achiasaf L, Sprinzak D, Wiemann S, Korner C, Ehrlich M, Ben-Baruch A (2019a). Notch-mediated tumor-stroma-inflammation networks promote invasive properties and CXCL8 expression in triple-negative breast cancer. Frontiers in Immunology 10: 125. DOI 10.3389/fimmu.2019.00804. [Google Scholar] [CrossRef]
Liubomirski Y, Lerrer S, Meshel T, Rubinstein-Achiasaf L, Morein D, Wiemann S, Korner C, Ben-Baruch A (2019b). Tumor-stroma-inflammation networks promote pro-metastatic chemokines and aggressiveness characteristics in triple-negative breast cancer. Frontiers in Immunology 10: e12532. DOI 10.3389/fimmu.2019.00757. [Google Scholar] [CrossRef]
Luo H, Wang J, Liu D, Zang S, Ma N, Zhao L, Zhang L, Zhang X, Qiao C (2019). The lncRNA H19/miR-675 axis regulates myocardial ischemic and reperfusion injury by targeting PPARalpha. Molecular Immunology 105: 46–54. DOI 10.1016/j.molimm.2018.11.011. [Google Scholar] [CrossRef]
Luo J, Hu YL, Wang H (2017). Ursolic acid inhibits breast cancer growth by inhibiting proliferation, inducing autophagy and apoptosis, and suppressing inflammatory responses via the PI3K/AKT and NF-kappaB signaling pathways in vitro. Experimental and Therapeutic Medicine 14: 3623–3631. DOI 10.3892/etm.2017.4965. [Google Scholar] [CrossRef]
Mekky RY, El-Ekiaby N, El Sobky SA, Elemam NM, Youness RA, El-Sayed M, Hamza MT, Esmat G, Abdelaziz AI (2019). Epigallocatechin gallate (EGCG) and miR-548m reduce HCV entry through repression of CD81 receptor in HCV cell models. Archives of Virology 164: 1587–1595. DOI 10.1007/s00705-019-04232-x. [Google Scholar] [CrossRef]
Mumm JB, Emmerich J, Zhang X, Chan I, Wu L et al. (2011). IL-10 elicits IFNgamma-dependent tumor immune surveillance. Cancer Cell 20: 781–796. DOI 10.1016/j.ccr.2011.11.003. [Google Scholar] [CrossRef]
Nafea H, Youness RA, Abou-Aisha K, Gad MZ (2020). LncRNA HEIH/miR-939-5p interplay modulates triple-negative breast cancer progression through NOS2-induced nitric oxide production. Journal of Cellular Physiology 236(7): 5362–5372. DOI 10.1002/jcp.30234. [Google Scholar] [CrossRef]
Pang F, Liu C, Cui Y, Gong K, Liu G, Bian Y, Gao X, Zhang D (2019). miR-17-5p promotes proliferation and migration of CAL-27 human tongue squamous cell carcinoma cells involved in autophagy inhibition under hypoxia. International Journal of Clinical and Experimental Pathology 12: 2084–2091. [Google Scholar]
Peng H, Ishida M, Li L, Saito A, Kamiya A et al. (2015). Pseudogene INTS6P1 regulates its cognate gene INTS6 through competitive binding of miR-17-5p in hepatocellular carcinoma. Oncotarget 6: 5666–5677. DOI 10.18632/oncotarget.3290. [Google Scholar] [CrossRef]
Pileczki V, Braicu C, Gherman CD, Berindan-Neagoe I (2012). TNF-alpha gene knockout in triple negative breast cancer cell line induces apoptosis. International Journal of Molecular Sciences 14: 411–420. DOI 10.3390/ijms14010411. [Google Scholar] [CrossRef]
Qiao Y, He H, Jonsson P, Sinha I, Zhao C, Dahlman-Wright K (2016). AP-1 is a key regulator of proinflammatory cytokine TNFalpha-mediated triple-negative breast cancer progression. Journal of Biological Chemistry 291: 5068–5079. DOI 10.1074/jbc.M115.702571. [Google Scholar] [CrossRef]
Schioppa T, Moore R, Thompson RG, Rosser EC, Kulbe H, Nedospasov S, Mauri C, Coussens LM, Balkwill FR (2011). B regulatory cells and the tumor-promoting actions of TNF-alpha during squamous carcinogenesis. Proceedings of the National Academy of Sciences 108: 10662–10667. DOI 10.1073/pnas.1100994108. [Google Scholar] [CrossRef]
Seto T, Sam D, Pan M (2019). Mechanisms of primary and secondary resistance to immune checkpoint inhibitors in cancer. Medical Science (Basel) 7: 14. [Google Scholar]
Shaalan YM, Handoussa H, Youness RA, Assal RA, El-Khatib AH, Linscheid MW, Tayebi HMEl, Abdelaziz AI (2018). Destabilizing the interplay between miR-1275 and IGF2BPs by Tamarix articulata and quercetin in hepatocellular carcinoma. Natural Product Research 32: 2217–2220. DOI 10.1080/14786419.2017.1366478. [Google Scholar] [CrossRef]
Song J, Liu Y, Wang T, Li B, Zhang S (2020). MiR-17-5p promotes cellular proliferation and invasiveness by targeting RUNX3 in gastric cancer. Biomedicine & Pharmacotherapy 128: 110246. DOI 10.1016/j.biopha.2020.110246. [Google Scholar] [CrossRef]
Stemke-Hale K, Gonzalez-Angulo AM, Lluch A, Neve RM, Kuo WL et al. (2008). An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Research 68: 6084–6091. DOI 10.1158/0008-5472.CAN-07-6854. [Google Scholar] [CrossRef]
Sharma A, Kumar M, Aich J, Hariharan M, Brahmachari SK, Agrawal A, Ghosh B (2009). Posttranscriptional regulation of interleukin-10 expression by hsa-miR-106a. Proceedings of the National Academy of Sciences of the United States of America 106(145761–5766. DOI 10.1073/pnas.0808743106. [Google Scholar] [CrossRef]
Wang J, Zhao L, Shang K, Liu F, Che J, Li H, Cao B (2020). Long non-coding RNA H19, a novel therapeutic target for pancreatic cancer. Molecular Medicine 26: 30. DOI 10.1186/s10020-020-00156-4. [Google Scholar] [CrossRef]
Xiang F, Pan C, Kong Q, Wu R, Jiang J, Zhan Y, Xu J, Gu X, Kang X (2014). Ursolic acid inhibits the proliferation of gastric cancer cells by targeting miR-133a. Oncology Research 22: 267–273. DOI 10.3727/096504015X14410238486685. [Google Scholar] [CrossRef]
Xu J, Meng Q, Li X, Yang H, Xu J et al. (2019a). Long noncoding RNA MIR17HG promotes colorectal cancer progression via miR-17-5p. Cancer Research 79: 4882–4895. DOI 10.1158/0008-5472.CAN-18-3880. [Google Scholar] [CrossRef]
Xu Y, Zhang Q, Lin F, Zhu L, Huang F, Zhao L, Ou R (2019b). Casiopeina IIgly acts on lncRNA MALAT1 by miR175p to inhibit FZD2 expression via the Wnt signaling pathway during the treatment of cervical carcinoma. Oncology Reports 42(4): 1365–1379. DOI 10.3892/or.2019.7268. [Google Scholar] [CrossRef]
Yesil-Celiktas O, Sevimli C, Bedir E, Vardar-Sukan F (2010). Inhibitory effects of rosemary extracts, carnosic acid and rosmarinic acid on the growth of various human cancer cell lines. Plant Foods for Human Nutrition 65: 158–163. DOI 10.1007/s11130-010-0166-4. [Google Scholar] [CrossRef]
Youness RA, Assal RA, Abdel Motaal A, Gad MZ (2018). A novel role of sONE/NOS3/NO signaling cascade in mediating hydrogen sulphide bilateral effects on triple negative breast cancer progression. Nitric Oxide 80: 12–23. DOI 10.1016/j.niox.2018.07.004. [Google Scholar] [CrossRef]
Youness RA, El-Tayebi HM, Assal RA, Hosny K, Esmat G, Abdelaziz AI (2016a). MicroRNA-486-5p enhances hepatocellular carcinoma tumor suppression through repression of IGF-1R and its downstream mTOR, STAT3 and c-Myc. Oncology Letters 12: 2567–2573. DOI 10.3892/ol.2016.4914. [Google Scholar] [CrossRef]
Youness RA, Gad AZ, Sanber K, Ahn YJ, Lee GJ, Khallaf E, Hafez HM, Motaal AA, Ahmed N, Gad MZ (2021). Targeting hydrogen sulphide signaling in breast cancer. Journal of Advanced Research 27: 177–190. DOI 10.1016/j.jare.2020.07.006. [Google Scholar] [CrossRef]
Youness RA, Gad MZ (2019). Long non-coding RNAs: Functional regulatory players in breast cancer. Noncoding RNA Research 4: 36–44. DOI 10.1016/j.ncrna.2019.01.003. [Google Scholar] [CrossRef]
Youness RA, Hafez HM, Khallaf E, Assal RA, Abdel Motaal A, Gad MZ (2019). The long noncoding RNA sONE represses triple-negative breast cancer aggressiveness through inducing the expression of miR-34a, miR-15a, miR-16, and let-7a. Journal of Cellular Physiology 234: 20286–20297. DOI 10.1002/jcp.28629. [Google Scholar] [CrossRef]
Youness RA, Rahmoon MA, Assal RA, Gomaa AI, Hamza MT, Waked I, El Tayebi HM, Abdelaziz AI (2016b). Contradicting interplay between insulin-like growth factor-1 and miR-486-5p in primary NK cells and hepatoma cell lines with a contemporary inhibitory impact on HCC tumor progression. Growth Factors 34: 128–140. DOI 10.1080/08977194.2016.1200571. [Google Scholar] [CrossRef]
Zhang L, Yang F, Yuan JH, Yuan SX, Zhou WP, Huo XS, Xu D, Bi HS, Wang F, Sun SH (2013). Epigenetic activation of the MiR-200 family contributes to H19-mediated metastasis suppression in hepatocellular carcinoma. Carcinogenesis 34: 577–586. DOI 10.1093/carcin/bgs381. [Google Scholar] [CrossRef]
Zhang Y, Wei C, Guo CC, Bi RX, Xie J, Guan DH, Yang CH, Jiang YH (2017). Prognostic value of microRNAs in hepatocellular carcinoma: A meta-analysis. Oncotarget 8: 107237–107257. DOI 10.18632/oncotarget.20883. [Google Scholar] [CrossRef]
Zhang Z, Lin G, Yan Y, Li X, Hu Y, Wang J, Yin B, Wu Y, Li Z, Yang XP (2018). Transmembrane TNF-alpha promotes chemoresistance in breast cancer cells. Oncogene 37: 3456–3470. DOI 10.1038/s41388-018-0221-4. [Google Scholar] [CrossRef]
Zhao X, Rong L, Zhao X, Li X, Liu X et al. (2012). TNF signaling drives myeloid-derived suppressor cell accumulation. Journal of Clinical Investigation 122: 4094–4104. DOI 10.1172/JCI64115. [Google Scholar] [CrossRef]

Figure S1: HPLC-PDA-ESI-MS chromatogram of rosemary EA fraction represents different phenolic compounds extracted.

 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |