 Open Access
Open Access
ARTICLE
Development of a prognostic signature for esophageal cancer based on a novel 7-DNA damage repair genes signature
Department of Thoracic Surgery, Minhang Hospital, Fudan University, Shanghai, 201199, China
* Corresponding Author: DAOWEN JIANG. Email:
BIOCELL 2022, 46(12), 2601-2613. https://doi.org/10.32604/biocell.2022.021300
Received 06 January 2022; Accepted 04 March 2022; Issue published 10 August 2022
Abstract
Esophageal cancer (EC) was an aggressive malignant neoplasm characterized by high morbidity and poor prognosis. Identifying the changes in DNA damage repair genes helps to better understand the mechanisms of carcinoma progression. In this study, by comparing EC samples and normal samples, we found a total of 132 DDR expression with a significant difference. Moreover, we revealed higher expression of POLN, PALB2, ATM, PER1, TOP3B and lower expression of HMGB1, UBE2B were correlated to longer OS in EC. In addition, a prognostic risk score based on 7 DDR gene expression (POLN, HMGB1, TOP3B, PER1, UBE2B, ATM, PALB2) was constructed for the prognosis of EC. Meanwhile, EC cancer samples were divided into 3 subtypes based on 132 DDR genes expressions. Clinical profile analysis showed cluster C1 and C2 showed a similar frequency of T2, which was remarked higher than that in cluster 3. Moreover, we found the immune cell inflation levels were significantly changed in different subtypes of EC. The infiltration levels of T cell CD8+, B cell and NK cells were greatly higher in cluster 2 than that in cluster 1 and cluster 3. The results showed T cell CD4+ infiltration levels were dramatically higher in cluster 1 than that in cluster 2 and cluster 3. Finally, we perform bioinformatics analysis of DEGs among 3 subtypes of EC and found DDR genes may be related to multiple signaling, such as Base excision repair, Cell cycle, Hedgehog signaling pathway, and Glycolysis/Gluconeogenesis. These results showed DDR genes may serve as new target for the prognosis of EC and prediction of the potential response of immune therapy in EC.Keywords
Esophageal cancer (EC) ranks the eleventh amid most common carcinomas, with an estimated 473,000 cases annually worldwide. And EC is the sixth widely occurred inducer of carcinoma-associated death, approximately 436,000 death tolls (Global Burden of Disease Cancer et al., 2019). EC consists of two major histological subtypes, including adenocarcinoma (EAC) and squamous cell carcinoma (ESCC). ESCC is the main histological type, accounting for 80% of the global EC cases (Cancer Genome Atlas Research et al., 2017; Gu et al., 2020; Freedman et al., 2007). Although the management and treatment of patients with EC have improved, the overall 5-year overall survival (OS) rate (about 10%) and 5-year OS after esophagectomy (within 15% and 40%) are still poor (Huang and Yu, 2018).
Alcohol, tobacco and acetaldehyde related to alcoholic beverages are all carcinogens of EC, leading to the occurrence and progression of ESCC (Ohashi et al., 2015). Aldehyde Dehydrogenase 2 (ALDH2) polymorphism is also the contributor to ESCC development, especially in Asian population (Lewis and Smith, 2005). Obesity and Barrett’s esophagus constitutes the main risk factors for EAC (Turati et al., 2013). The occurrence of EC is the result of a complex process involving changes in multiple genes.
One hallmark of carcinoma is the instability of genomic which is caused by various insults leading to DNA damage. Double strand breaks (DSBs) is the most dangerous and threatening genotoxic injury, resulting in chromosome rearrangement and cell death (da Silva, 2021). Defective components in DNA damage and repair toolset are the fundamental reason for the occurrence and development of different sorts of carcinomas (da Silva, 2021). DNA damage repair pathway work together in concert to eliminate DNA damage and sustain the stability and integrity of the genome (da Silva, 2021). DNA damage repair (DDR) genes exhibit pivotal roles during this process in human cells. As biochemical and mechanistic standards demonstrated, DDR genes mainly consist of 7 functional pathways (Wang et al., 2020a). Dysregulated DDR pathways functions importantly regarding carcinoma risk, carcinoma development and response to treatment (Curtin, 2012). DDR pathways with up-regulation were related to the resistance to DNA-damaging radiotherapy and chemotherapy. In particular, activated DSBs repair genes are one of the factors of tumor resistance to radiotherapy and chemotherapy (Curtin, 2012). DDR has also been shown to take part in the metabolism of carcinomas. The activation of DDR promotes nucleotide synthesis and glucose metabolism, whereas lessening glutamine anaplerosis (Turgeon et al., 2018). In melanoma, the genes with up-regulation in DDR pathways has a relation to tumor metastasis (Broustas and Lieberman, 2014). The mutations of DDR genes have an association with immune-related genes expression in ovarian carcinoma and muscle invasive bladder carcinoma (Vidotto et al., 2019). Herein, identifying the changes in DDR pathway helps to better understand the mechanisms of carcinoma progression. Here, our present article aims to comprehensively investigate the expression alteration of DDR pathway genes in EC. We plan to identify new prognostic model and novel molecular subtypes based on expression of DDR genes in EC. We think our results will prove DDR genes may serve as a newly generated target for the treatment of EC.
The RNA sequence data of Esophageal were obtained from TCGA website (https://portal.gdc.cancer.gov), which included 13 normal samples and 182 EC samples. Meanwhile, the clinical information of these EC patients were also extracted.
Analysis of differential DDR genes
A total of 229 DDR genes were retrieved from the TISIDB (Ru et al., 2019) database (http://cis.hku.hk/TISIDB/index.php).
Construction of the prognostic Model Based on the DDR genes
The prognostic model was established via the “glmnet” R package. The survival analysis was performed with “survminer” R package, and the ROC curve analysis was performed via the “survival” and “timeROC” R packages.
Tumor immune infiltration analysis
To further analyze the differences in immune cell inflation of 3 subtypes of esophageal cancer, the TIMER (Li et al., 2017a) algorithm (https://cistrome.shinyapps.io/timer/) was used to investigate the differences in the proportion of 6 immune cell subsets (Gu et al., 2021).
The KEGG analysis was performed using DAVID system (https://david.ncifcrf.gov/).
R software v4.1 (https://www.r-project.org/) was used for all analyses. P-value < 0.05 was regarded as statistically significant.
Aberrant expression of DNA damage repair genes (DDR) in esophageal cancer
The present study aimed to identify the interaction between 229 DDR genes and esophageal cancers. Totally, the expression of 132 DDR genes were observed to have a remarkably difference between EC and normal samples (Suppl. Table 1). 119 genes were overexpressed in esophageal cancer, whereas another 13 genes were suppressed in EC samples (Fig. 1). Among these DEGS, HAP1, RDM1, RAD54L, UBE2T, EXO1 were the top 5 up-regulated DDR genes, and NEIL1, REV3L, POLI, XPA, PER1 were the top 5 down-regulated DDR genes (Fig. 1).
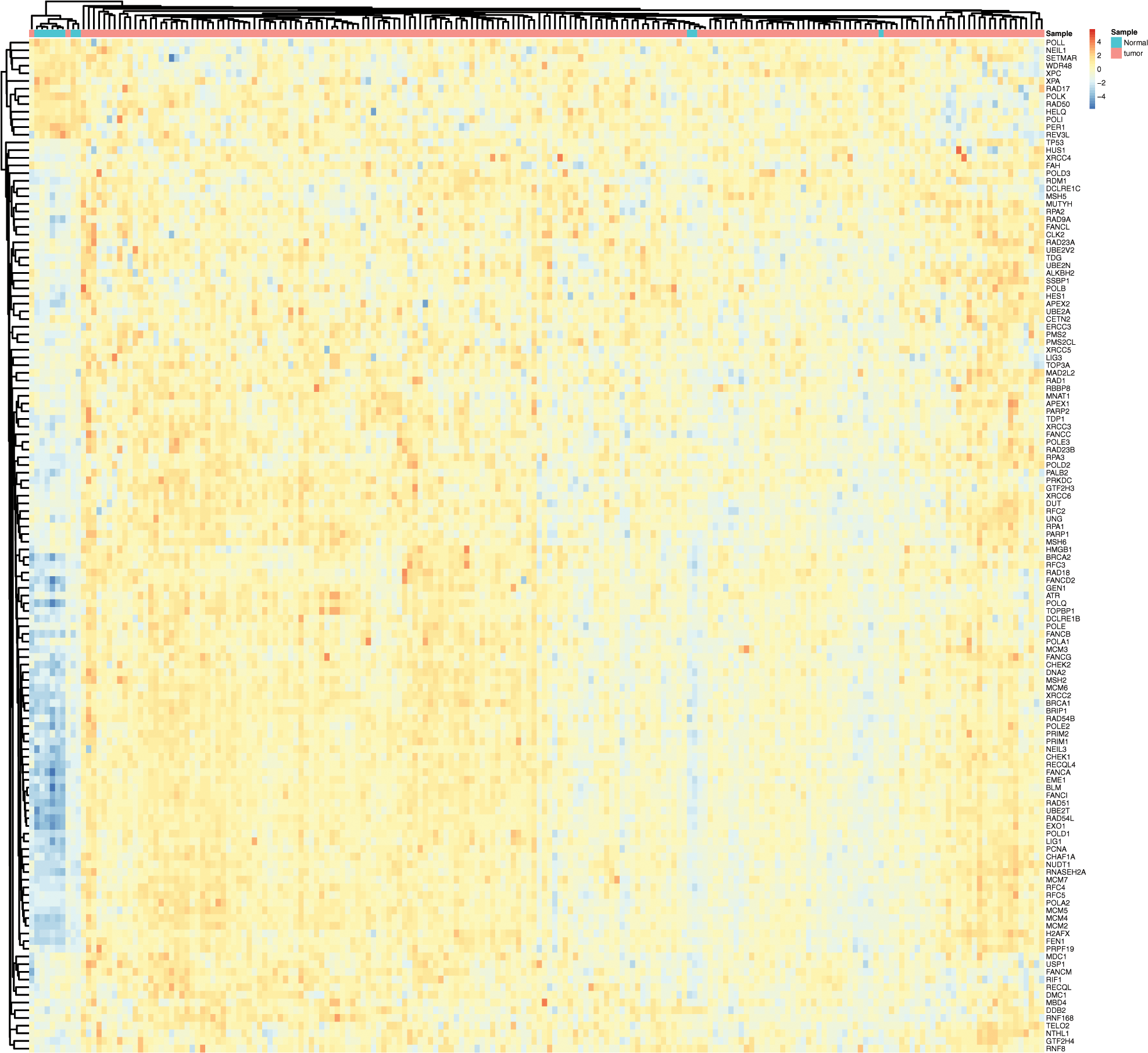
Figure 1: DNA damage repair genes (DDR) was differently expressed in esophageal cancer. The heatmap analysis of DDR genes in EC.
The dysregulation of DNA damage repair genes (DDR) were correlated to shorter OS in esophageal cancer
We next performed Kaplan-Meier analysis to evaluate the association between DDR expression and OS time in EC. As present in Fig. 2, the results showed that higher expression of POLN, PALB2, ATM, PER1, TOP3B, and lower expression of HMGB1, UBE2B were related to longer OS time in EC (Figs. 2A–2G).
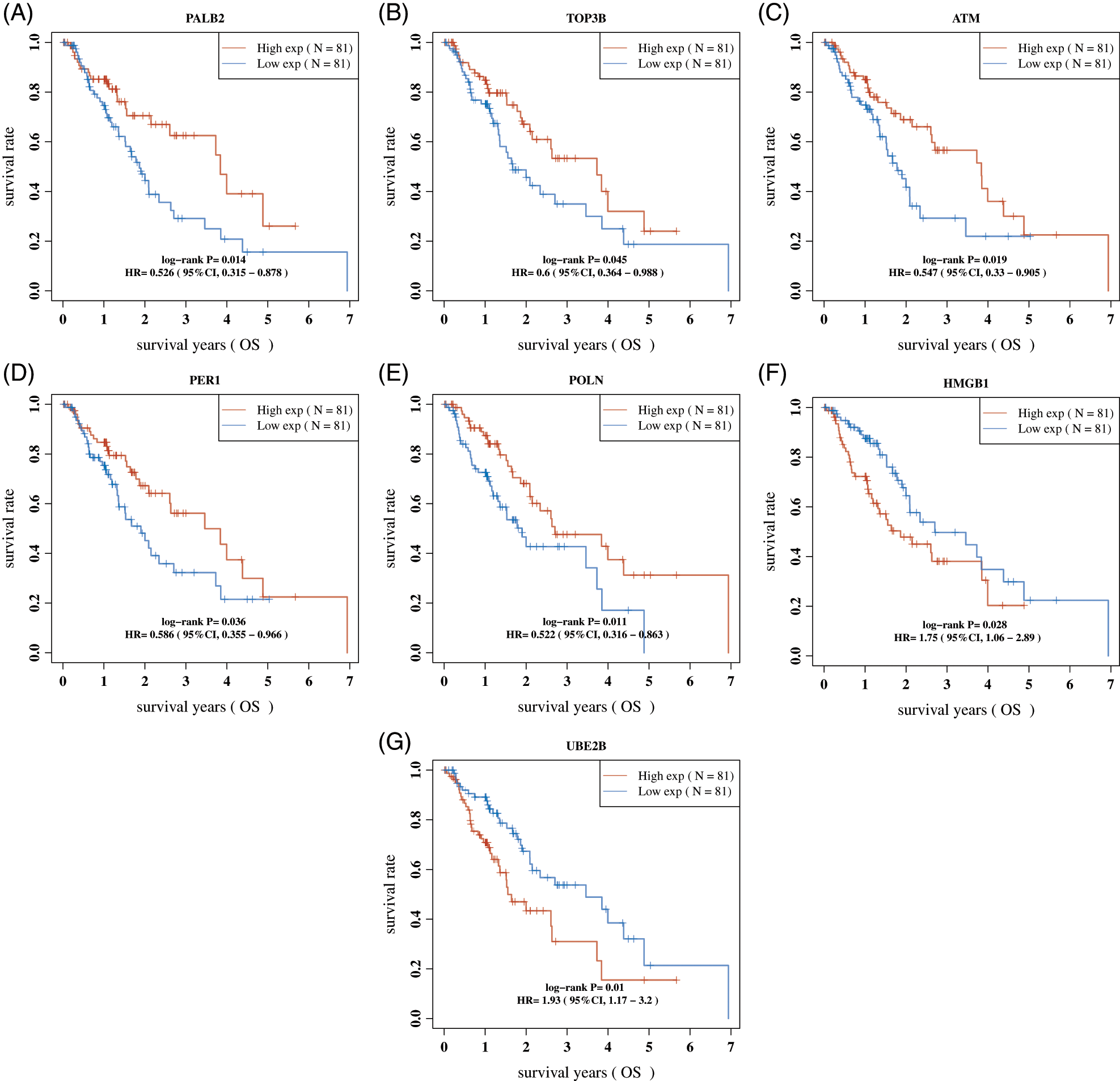
Figure 2: The dysregulation of DDR genes were correlated to shorter OS in Esophageal cancer. (A–G) The dysregulation of were PALB2 (A), TOP3B (B), ATM (C), PER1 (D), POLN (E), HMGB1 (F), and UBE2B (G) correlated to shorter OS in Esophageal cancer.
Construction of a prognostic DDR genes signature
LASSO regression analysis was performed based on DDR expression, and 7 genes were retained according to the optimal lambda value, including POLN, HMGB1, TOP3B, PER1, UBE2B, ATM, PALB2 (Figs. 3A and 3B). Then, a prognostic model based on 7 DDR genes was developed as follows: Riskscore = (−0.4278) * POLN + (0.3241) * HMGB1 + (−0.0328) * TOP3B + (−0.03) * PER1 + (0.8504) * UBE2B + (−0.0913) * ATM + (−0.548) * PALB2. The EC samples were classified into high- and low-risk groups based on the median value as cutoff (Fig. 3C). Kaplan–Meier analysis showed EC patients with higher risk scores had a shorter OS time (Fig. 3D). Next, the AUC for OS prediction was calculated. The AUC of the ROC curve reached 0.779 at 1 year, 0.725 at 3 years, and 0.705 at 5 years (Fig. 3E).
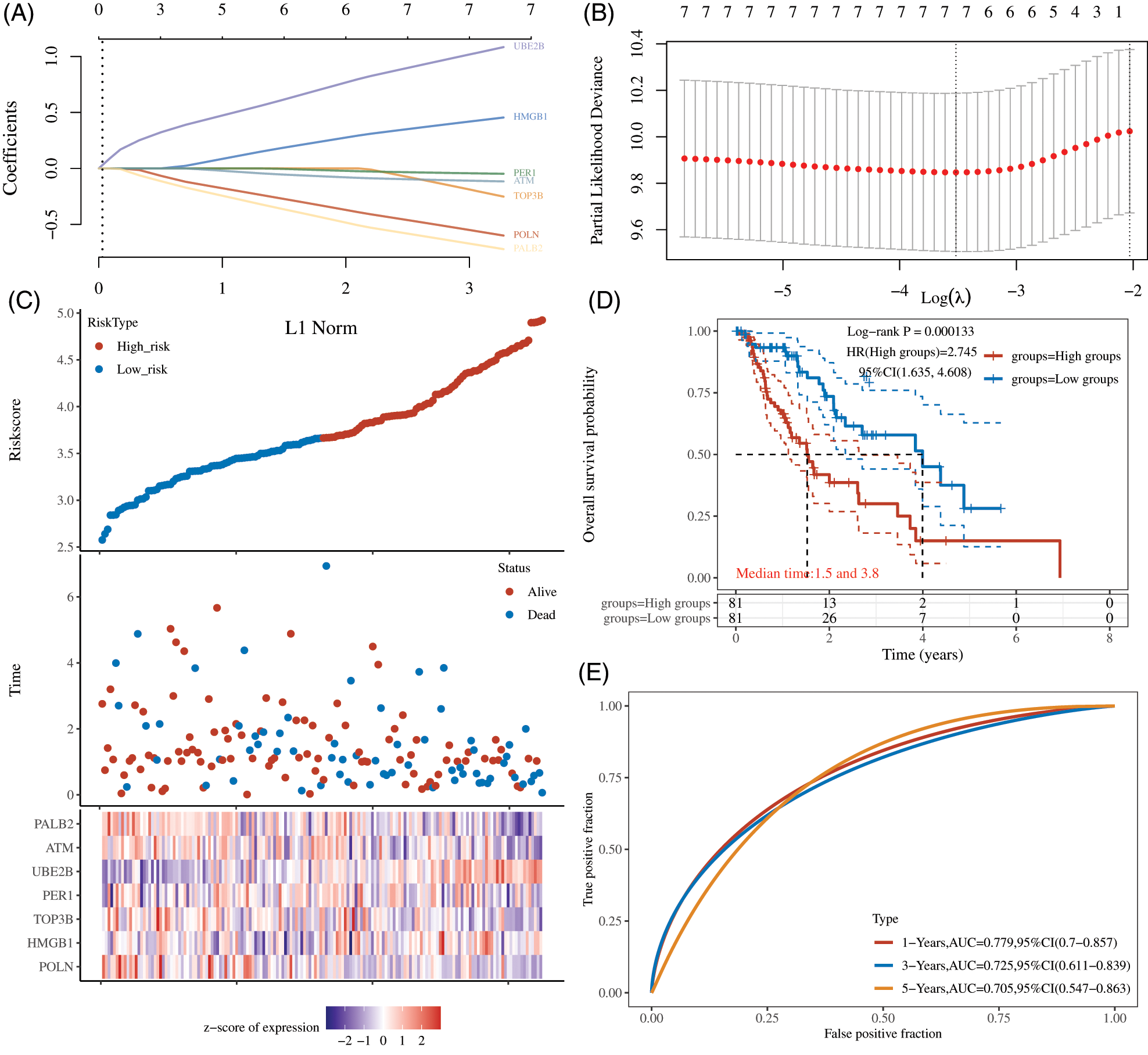
Figure 3: Construction of a Prognostic DDR genes Signature. (A) LASSO coefficients profiles were performed in EC. (B) LASSO regression were calculated using minimum lambda value. (C) The EC samples were classified into high- and low-risk groups. (D) Kaplan–Meier analysis showed EC patients with higher risk scores had a shorter OS time. (E) The AUC for OS prediction was calculated.
Consensus clustering analysis for esophageal cancer based on the expression of DNA damage repair genes
We next performed consensus clustering for Esophageal cancer based on the expression of DNA damage repair genes. k = 3 was identified based on the similarity displayed by the expression levels of 132 DDR genes expressions (Figs. 4A and 4B). EC patients were divided into cluster 1, cluster 2 and cluster 3 (Fig. 4C). There were 81 EC patients in cluster 1, 27 EC patients in cluster 2, 54 EC patients in cluster 3. As present in Fig. 4D, we observed almost 90% of DDR genes expressions were down-regulated in cluster 2, indicating the genome instability may be higher in cluster 2 compared to other groups.
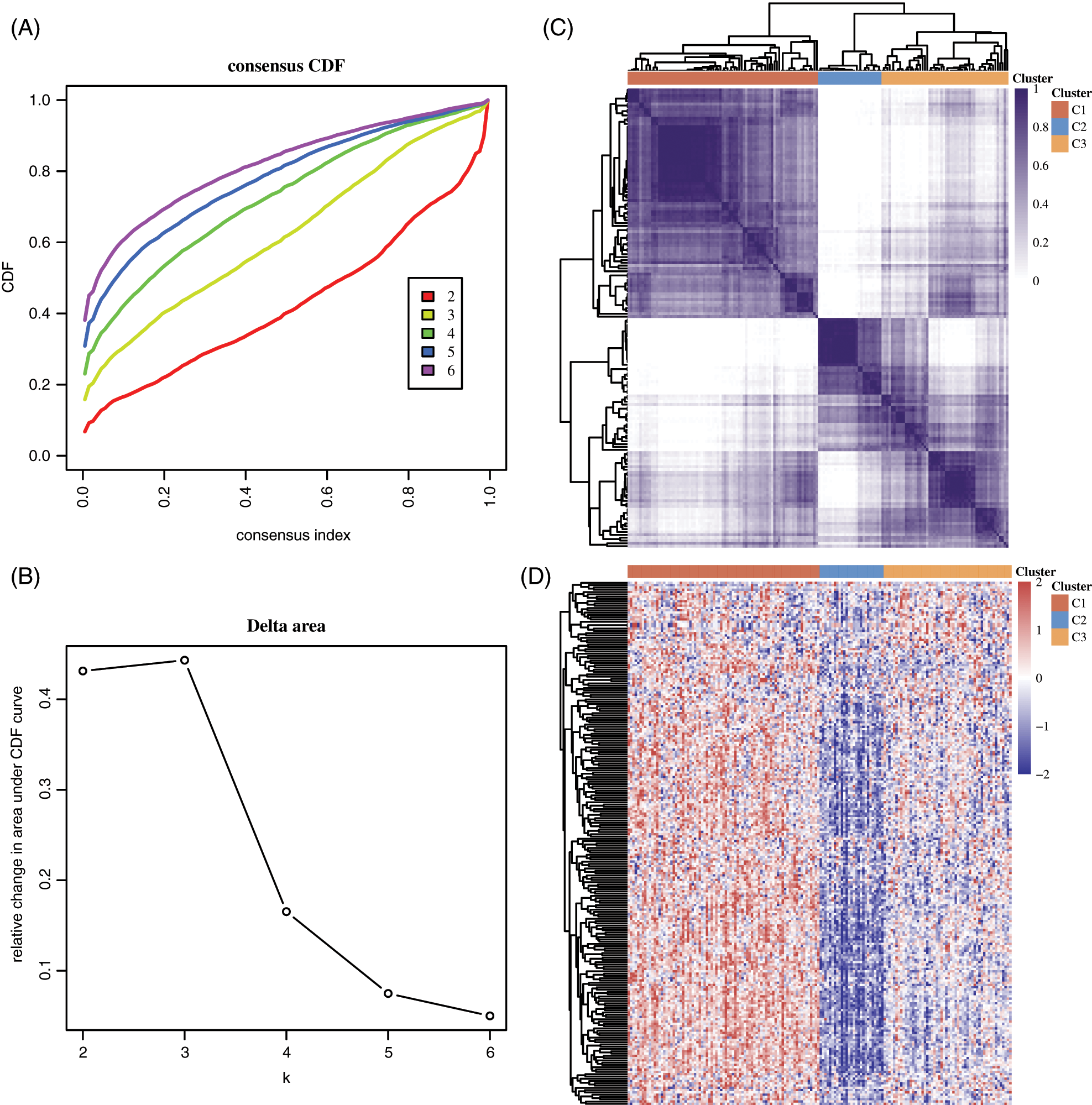
Figure 4: Consensus clustering analysis for esophageal cancer based on the expression of DNA damage repair genes. (A–B) k = 3 was identified based on the similarity displayed by the expression levels of 132 DDR genes expressions. (C) EC patients were divided into cluster 1, cluster 2 and cluster 3. (D) The expression of DDR genes in different clusters of EC was shown.
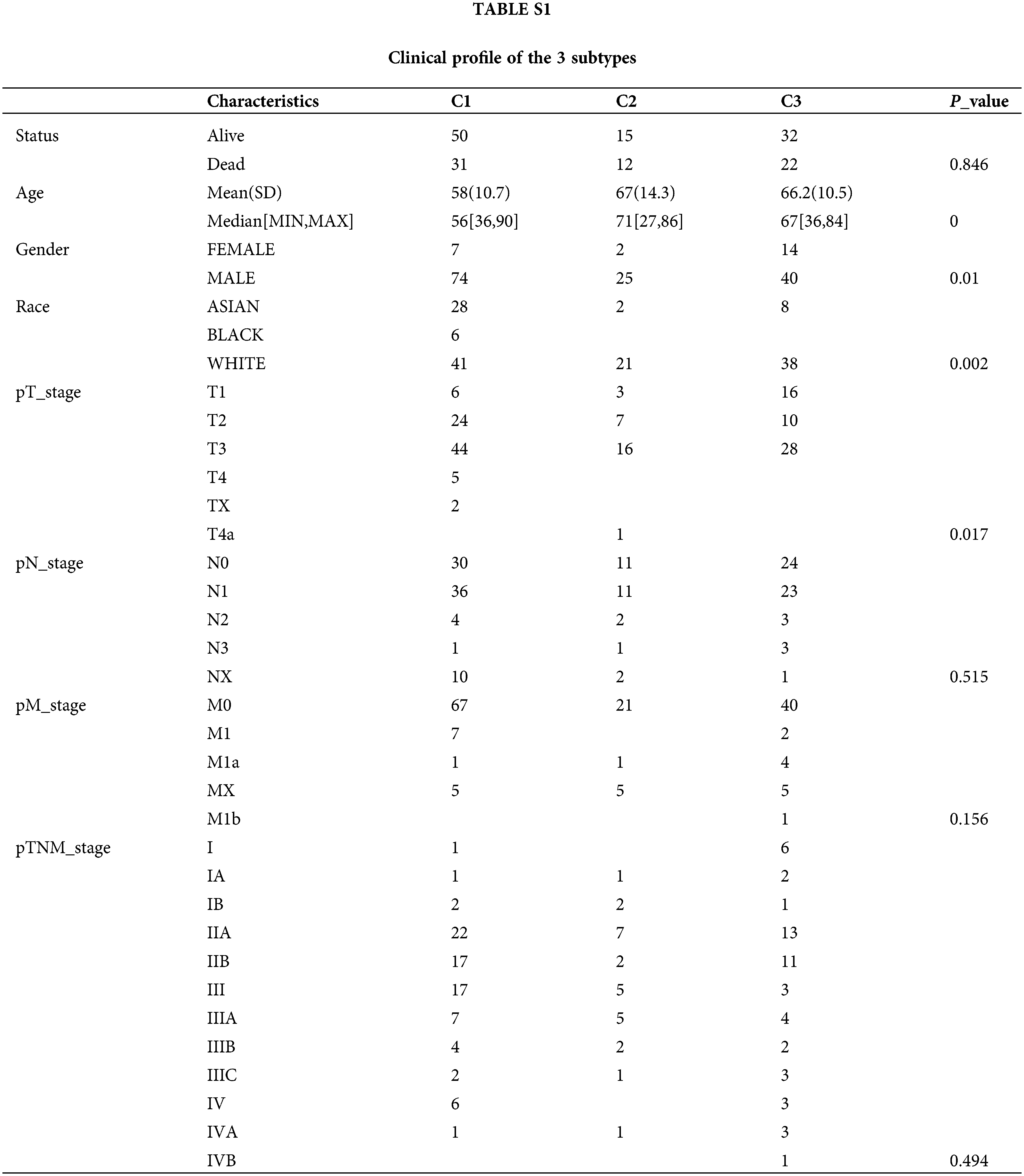
Clinical profile of the 3 subtypes
To reveal the clinical relevance of DDR based subtypes, gender, race, T stage, N stage, M stage and grade were compared between patients with the 3 DDR subtypes (Table 1). Clinically, we observed cluster 1 and cluster 2 have a markedly higher proportion of male patients, while cluster 3 have a markedly higher proportion of female patients (Fig. 5A). Cluster 1 has a markedly higher proportion of Asian patients and cluster 2 and cluster 3 have a markedly higher proportion of white patients (Fig. 5B). Clusters C1 and C2 showed a similar frequency of T2, which was remarked higher than that in cluster 3 (Fig. 5C). However, N stage, M stage and was not different among the 3 groups (Figs. 5D–5F).
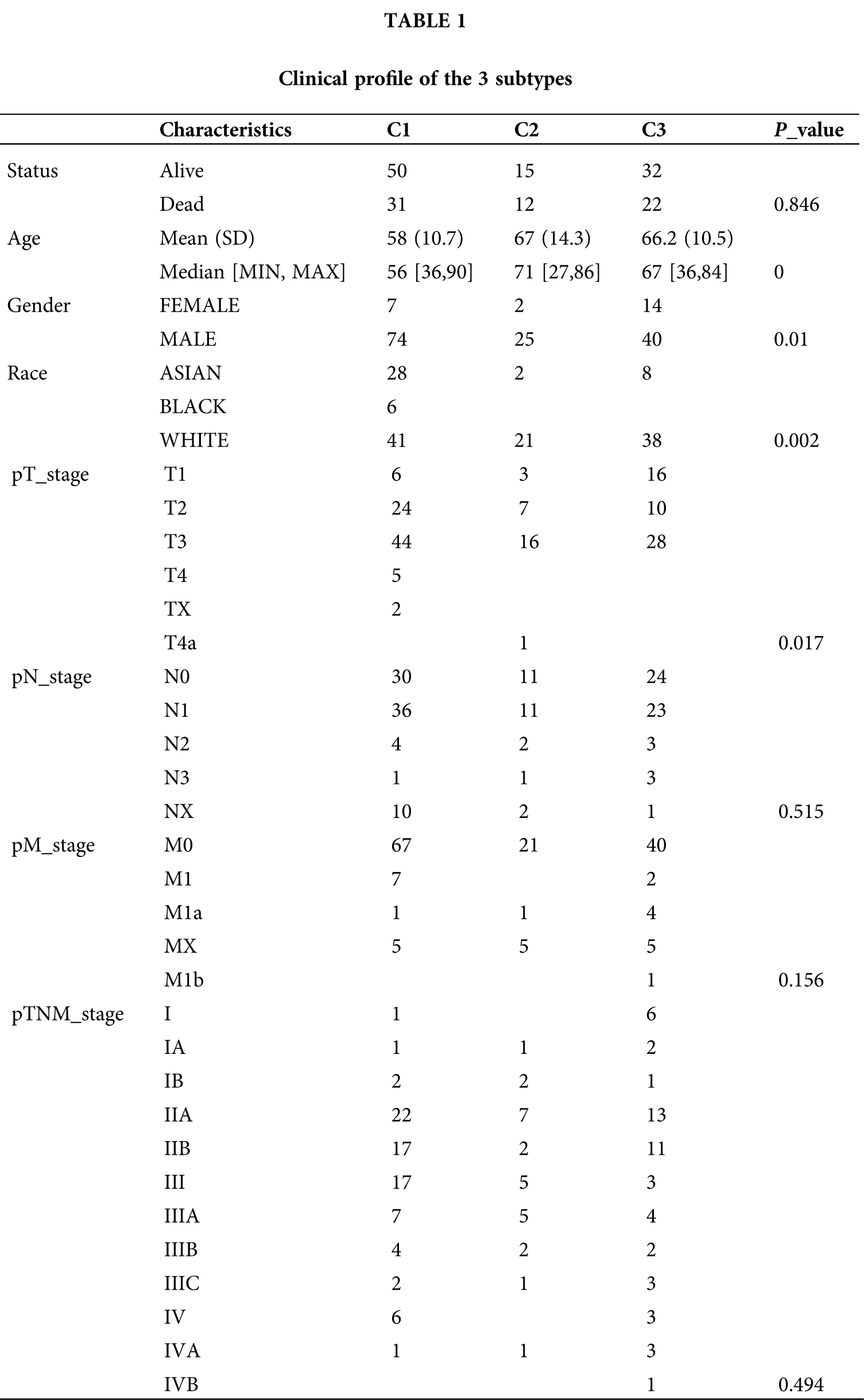
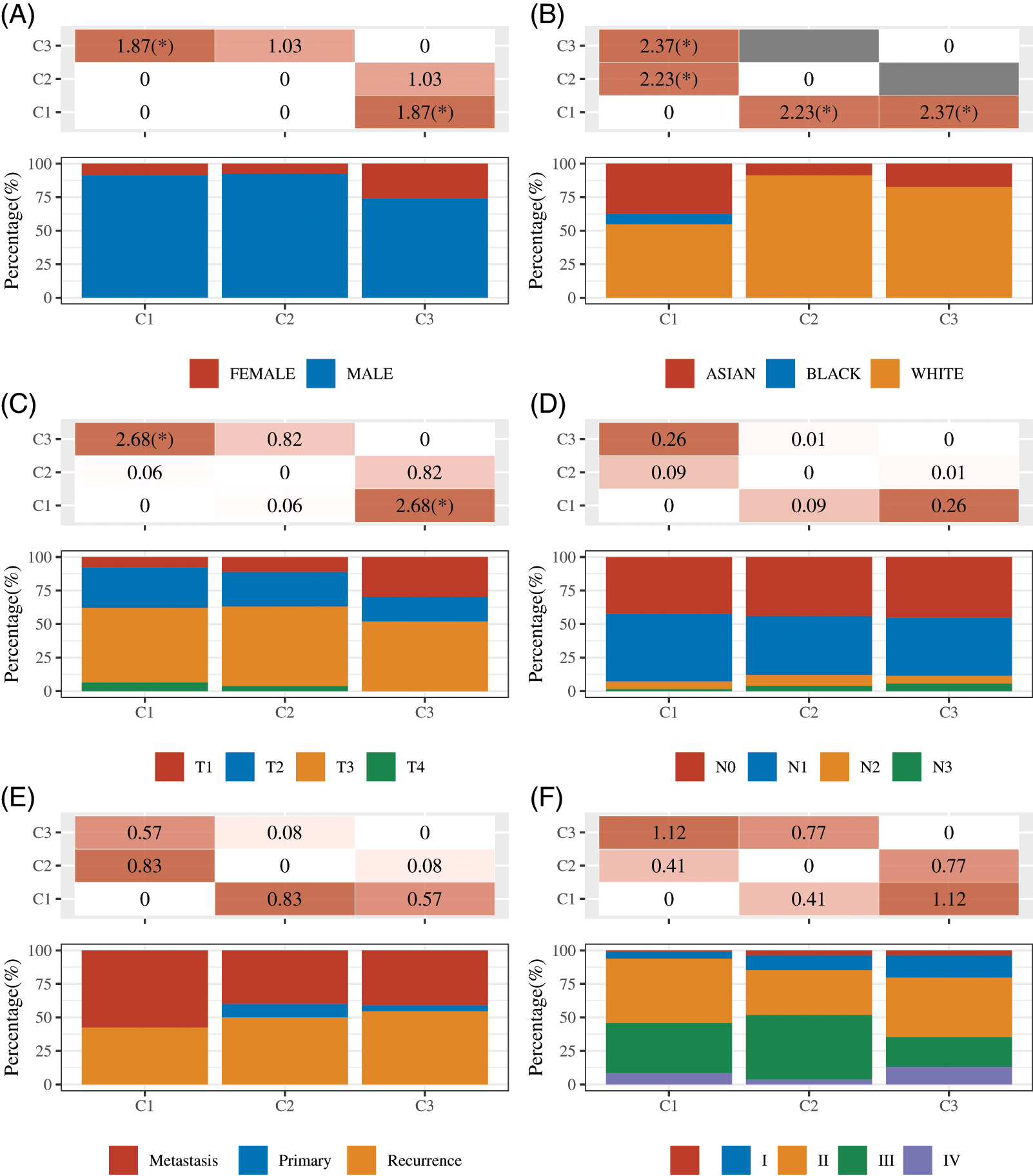
Figure 5: Clinical profile of the 3 subtypes were analyzed. (A–F) The status of Gender (A), race (B), T stage (C), N stage (D), M stage (E) and grade (F) were compared between patients with the 3 DDR subtypes. *P < 0.05.
Landscape of immune infiltration in 3 subtypes of esophageal cancer
To further analyze the differences in immune cell inflation of 3 subtypes of esophageal cancer, the TIMER algorithm was used to investigate the differences in the proportion of 6 immune cell subsets. The results showed infiltration levels of T cell CD8+, B cell and NK cells were markedly higher in cluster 2 than that in cluster 1 and cluster 3. The results showed T cell CD4+ infiltration levels were markedly higher in cluster 1 than that in cluster 2 and cluster 3 (Figs. 6A and 6B). The infiltration levels of Endothelial cell were markedly higher in cluster 2 and 3 than that in cluster 1 (Figs. 6A and 6B). Fig. 6 presents the immune cell distribution in the EC samples.
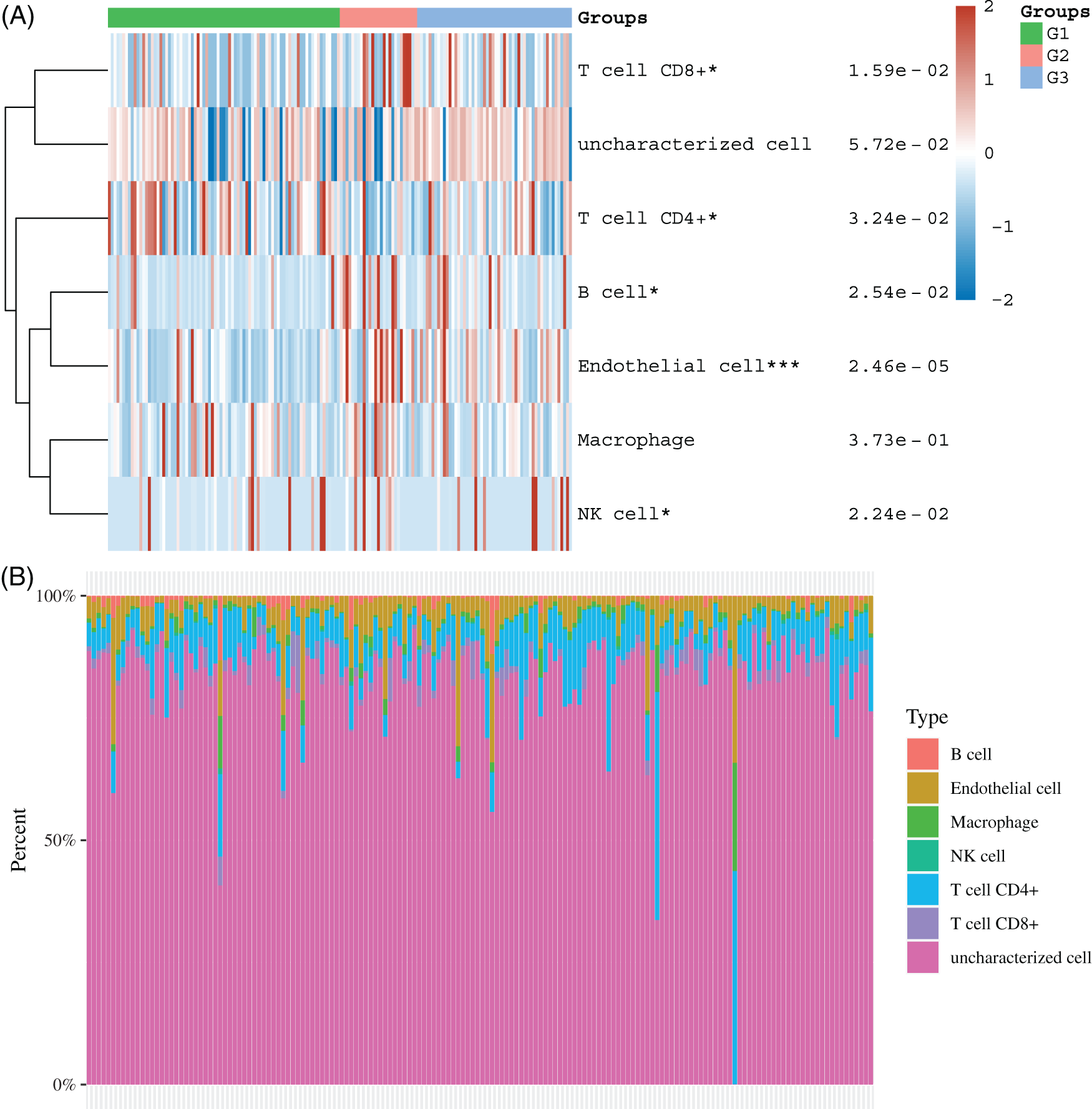
Figure 6: Landscape of immune infiltration in 3 subtypes of esophageal cancer. (A–B) The immune cell distribution in the different clusters of EC samples were shown. *P < 0.05; ***P < 0.001.
Identification of differently expressed genes among 3 subtypes of esophageal cancer
Then, we identified differently expressed genes among 3 subtypes. As present in Fig. 6, 1071 overexpressed and 215 suppressed genes were found in cluster 1 in comparison with cluster 2, 419 overexpressed and 378 suppressed genes were identified in cluster 1 in comparison with cluster 3, and 48 overexpressed and 142 suppressed genes were identified in cluster 2 in comparison with 3 (Figs. 7A, 7D and 7G). The KEGG analysis showed these DEGs were related to multiple pathways regulation.
Figure 7: Identification of differently expressed genes among 3 subtypes of esophageal cancer. (A) Heatmap analysis of DEGs between cluster 1 and cluster 2. (B–C) KEGG analysis of DEGs between subtype 1 and subtype 2. (D) Heatmap analysis of DEGs between subtype 1 and subtype 3. (E–F) KEGG analysis of DEGs between subtype 1 and subtype 3. (G) Heatmap analysis of DEGs between subtype 2 and subtype 3. (H–I) KEGG analysis of DEGs between subtype 2 and subtype 3.
For example, the DEGs between cluster 1 and cluster 2 were significantly related to Base excision repair, Cell cycle, DNA replication, Cellular senescence, Fanconi anemia pathway, Homologous recombination, Mismatch repair, Nucleotide excision repair, Wnt pathway, p53 pathway, IL−17 pathway, Insulin resistance, Fat digestion and absorption, Fatty acid degradation (Figs. 7B and 7C). The DEGs between cluster 1 and cluster 3 were significantly related to Hedgehog, Hippo, PI3K−Akt, and Wnt signaling, Glycolysis/Gluconeogenesis, Glycosphingolipid biosynthesis, IL−17 signaling pathway (Figs. 7E and 7F). the DEGs between cluster 2 and cluster 3 were significantly related to Fatty acid degradation, Gastric acid secretion, Glycolysis/Gluconeogenesis, PPAR signaling pathway, Cell cycle, Cellular senescence, IL−17 pathway, NF−kappa B signaling (Figs. 7H and 7I).
EC is an aggressive malignant neoplasm characterized by high morbidity and poor prognosis. Alcohol along with its metabolite, acetaldehyde, and ALDH2 have been reported to exhibit an associated with esophageal carcinogenesis. In recent years, much advance has been made in EC’s diagnosis and treatment, but EC’s prognosis is yet poor (Wang et al., 2020a). The mechanism underlying in EC’s tumorigenesis and development stays unclear. In this study, by comparing EC samples and normal samples, we found a total of 132 DDR expression with a significant difference. Moreover, we revealed higher expression of POLN, PALB2, ATM, PER1, TOP3B and lower expression of HMGB1, UBE2B were correlated to longer OS in EC. In addition, a prognostic risk score based on 7 DDR gene expression was constructed for the prognosis of EC. Meanwhile, EC cancer samples were divided into 3 subtypes based on 132 DDR genes expressions. Finally, we perform bioinformatics analysis of DEGs among 3 subtypes of EC to understand the mechanisms underlying the regulation of DDR in EC progression.
Previous studies had demonstrated that DDR signaling played important regulatory role in EC. For example, in ESCC, polymorphisms of base excision repair (BER) genes were probably related to ESCC’s susceptibility (Yang et al., 2015). Genetic variants of the nucleotide excision repair (NER) gene had a connection with Chinese ESCC patients’ survival outcome (Zhang et al., 2018). In addition, the genetic polymorphisms of XRCC6 displayed a link to a high risk of ESCC (Li et al., 2015). Besides, hypermethylation of the promoter of mismatch repair (MMR) gene MLH1 is of importance for maintaining genome stability, indicating a potential indicator of male ESCC patients’ prognosis (Wu et al., 2017). The present study, we revealed 119 genes with up-regulation and 13 genes with down-regulation in EC, such as DEGS, HAP1, RDM1, RAD54L and POLI, XPA, PER1, further demonstrating the crucial roles of DDR in EC.
Till now, many genes, such as VEGF, and miR-21 have been confirmed to have important clinical significance in the prediction of EC prognosis. Even so, more clinical studies are needed to obtain more biomarkers related to the prognosis of EC, thus providing a preliminary basis for clinical management of EC. Of note, in this study, we revealed higher expression of POLN, PALB2, ATM, PER1, TOP3B and lower expression of HMGB1, UBE2B were correlated to longer OS in EC. These results showed that dysregulation of DDR genes may serve as potential prognostic markers for EC. DNA topoisomerases was considered to be an important anti-cancer drug target (Bai et al., 2016). Type IA (TOP3A and TOP3B) enzymes are the only ones relaxing negative supercoiling. Convincing evidence shown that they interact with SGS1 helicase (Bai et al., 2016) and other members of RecQ family (Staker et al., 2002), indicating that they have the function of maintaining genomic stability. UBE2B have been reported to be related to survival in esophageal adenocarcinoma (EAC). It also took part in worse OS for EAC patients. UBE2B was greatly higher in EAC patients of stage II than stage I, suggesting it was probably increased with the progression of EAC at early stage (Wang et al., 2020b). Li et al. (2017b) showed that HMGB1 had an obvious link to OS in the ESCC cohort. And HMGB1 was adversely prognostic genes in ESCC. PALB2 exhibits importantly in the maintenance of genomic integrity. PALB2 has become a crucial tumor suppressor related to genetic susceptibility to breast and pancreatic carcinomas, since being identified as a BRCA2 interaction partner (Nepomuceno et al., 2017). Li et al. (2016) indicated that circadian protein PER2 level was largely raised, while E-cadherin was greatly reduced in the tissues of human metastatic EC when compared with non-metastatic EC. PER2 overexpression in EC cells markedly inhibited the expression of E-cadherin. Compared with non-metastatic EC tissues, pHDAC1 was more common in human metastatic EC tissues. After PER2 overexpression, the repressors of pHDAC1 and E-cadherin at the E-cadherin promoter locus were increased. Overexpressing PER2 significantly heightened the migration ability of EC cells, while inhibiting HDAC1 eliminated this migration ability, all data suggested that PER2 played importantly in the metastasis of EC cells. Based on above, we presumed that PER1 might function the same as PER2 in EC. More researches needs to be further verified. In addition, a prognostic risk score based on 7 DDR gene expression was constructed for the prognosis of EC. Kaplan–Meier (K–M) plots shown that patients with high risk scores had poor OS. It is worth noting that EC patient were classified into three subtypes in the light of DDR genes expression. Clinically, we observed these subtypes were significantly enriched in different stages of EC.
Recently, researches on immune checkpoint inhibitors in ESCC (e.g., PD-1/PD-L1 inhibitors, CTLA-4 inhibitors, etc.) presented satisfactory results. Additionally, combining immune checkpoint inhibitors and traditional strategies against neoplasms to treat ESCC has aroused widespread interest, and the results are encouraging. As previously described, PD-L1 expression, TMB, MSI-H, as well as other biomarkers displayed a relation to the immunotherapy’s efficacy (Jiao et al., 2019). More and more evidence showed that TMB was an important factor in carcinogenesis with regard to EC (Yuan et al., 2020). TMB displayed an association with abundant neoantigens and enhanced immunogenicity (Yuan et al., 2020), along with high immunogenicity (Champiat et al., 2014). High TMB was a marker of associated antigen mutations, indicating an increase in lymphocyte infiltration in the tumor microenvironment. High TMB group exhibited higher level of Treg cell infiltration than low TMB group in EC patients free of radiotherapy. Nevertheless, no difference of Treg cell infiltration was observed in EC patients with radiotherapy. Some researchers indicated that, however, high TMB generated a plenty of neoantigens that motivated immune responses against neoplasms (Rizvi et al., 2015), and that high TMB displayed a relation to genomic instability, bringing about induced immune responses to neoplasms (Rizvi et al., 2015). The significance of PD-L1 expression levels in tumor cells remained controversial. A majority of researches suggested that PD-L1 expression in neoplastic cells was the most expected biomarker for immune therapy. Nonetheless, immune therapies have shown good efficacy in some PD-L1 negative patients, but ineffective in some PD-L1 positive patients. The inconsistency of these results was mainly related to the heterogeneity of PD-L1 expression, samples variation and inconsistent detection standards, etc. (Yuan et al., 2020). Besides, several clinical researches of PD-1/PD-L1 inhibitors along with CTLA-4 inhibitors for treating EC are also in progress (Champiat et al., 2014). As previous study shown, MSI-H has a positive correlation with high TMB in the immunotherapy for colorectal carcinoma (Fabrizio et al., 2018). Le et al. (2017) (Asaoka et al., 2015) discovered that mismatch repair status could be utilized as a predictor of the effect of pembrolizumab in clinical, and found that deficiencies mismatch repair (dMMR) neoplasms had an association with prolonged progression-free survival (PFS) relative to mismatch repair-proficient neoplasms, regardless of the carcinomas’ origin tissues. Even though the incidence of MSI-H in ESCC stayed low, with estimated 8%, the function of this biomarker is extremely important since it perhaps exert an effect on the efficiency of immune checkpoint inhibitors (Le et al., 2017). Immune cell infiltration has been linked to EC, and it has been shown to have a significant impact on tumor progression, treatment, and patient outcomes. More recent studies have shown that immune cell infiltration is a potential predictive marker for immune therapy response in human malignancies, including EC (Liu et al., 2021; Zhang et al., 2020). A few regulators have been linked to immune cell infiltration; for example, Liu et al. (2021) found that COL9A3, GFRA2, and VSIG4 were linked to levels of infiltration of CD4 memory T cells, M1 and M2 macrophages. Moreover, we found the immune cell inflation levels were significantly changed in different subtypes of EC. The results showed infiltration levels of T cell CD8+, B cell and NK cells were greatly higher in cluster 2. The results showed T cell CD4+ infiltration levels were dramatically higher in cluster 1. These results showed DDR based subtypes could predict the potential response of immune therapy in EC.
In order to reveal the underlying mechanism in different subtypes of EC. We identified differently expressed genes and performed functional analysis based on these DEGs. Our results showed the DEGs between cluster 1 and cluster 2 were significantly related to Base excision repair, Cell cycle, Cellular senescence, DNA replication, DNA damage repair related signaling. The DEGs between cluster 1 vs. 3 were significantly related to Hedgehog, Hippo, PI3K−Akt signaling. The DEGs between cluster 2 vs. 3 were significantly related to Glycolysis/Gluconeogenesis, PPAR signaling pathway. Previous research has found that the HH pathway is highly activated in esophageal cancer (Wang et al., 2019). In esophageal cancer, several key members of the HH pathway, such as GLI1 and PTCH1, are significantly overexpressed (Wang et al., 2019). The Hedgehog (HH) pathway is linked to cancer stemness and therapy resistance (Wang et al., 2019). Inhibiting the HH pathway was linked to fewer cells with a cancer stem cell-associated phenotype (CD44+/CD24-), decreased spheroid formation, and increased radiosensitivity, according to a recent study (Wang et al., 2019). The glycolytic pathway is important in the occurrence and progression of esophageal cancer (Zhu et al., 2021). Previous research has shown that glycolysis can affect EAC immunotherapy by altering the microenvironment caused by genetic mutations (Zhu et al., 2021). CLDN9, GFPT1, HMMR, RARS, and STMN1 are glycolysis-related gene signatures that predict prognosis in patients with esophageal adenocarcinoma. DNA damage repair pathways are essential for genome integrity (Kang et al., 2020). A large number of DNA damage repair genes, such as RAD54B, and PRKDC, were significantly amplified and overexpressed in esophageal cancer. Amplification of DSB repair pathway genes are associated with poorer outcome in EC. Inhibiting the DSB repair pathway can improve ESCC radiosensitivity. APE1-mediated DNA damage repair promoted EC survival in response to acidic bile salts, according to a recent study (Hong et al., 2016).
Several limitations of this study should be noted. Firstly, only TCGA was analyzed in this study. In the future study, more validation will be performed using clinical samples. Secondly, several hub DDR genes were identified, such as RAD54B, NBS1, RAD51B. However, the molecular functions of these DDR genes were not confirmed. The experimental validation of these genes will be performed in the future study. Thirdly, the findings of this article are still a long way from clinical practice. More translational medicine work will be done in the future study.
In summary, our findings showed DDR genes were significantly differently expressed in EC and correlated to the OS time in EC. Moreover, a prognostic risk score based on 7 DDR gene expression was constructed to predict OS of patients with EC. In addition, EC cancer samples were divided into 3 subtypes based on 132 DDR genes expressions. We found the immune cell inflation levels were significantly changed in different subtypes of EC. These results showed DDR genes may serve as new target for the treatment of EC.
Acknowledgement: None to declare.
Availability of Data and Materials: All the data were extracted from TCGA dataset.
Author Contribution: The authors confirm contribution to the paper as follows: study conception and design: Daowen Jiang; data collection: Jiaming Zhan; analysis and interpretation of results: Jiaming Zhan, Weihua Wang, Yanlei Tang and Ning Zhou; draft manuscript preparation: Jiaming Zhan, Weihua Wang, Yanlei Tang and Ning Zhou. All authors reviewed the results and approved the final version of the manuscript.
Ethics Approval: None to declare.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
Asaoka Y, Ijichi H, Koike K (2015). PD-1 blockade in tumors with mismatch-repair deficiency. New England Journal of Medicine 373: 1979. DOI 10.1056/NEJMc1510353. [Google Scholar] [CrossRef]
Bai Y, Li LD, Li J, Lu X (2016). Targeting of topoisomerases for prognosis and drug resistance in ovarian cancer. Journal of Ovarian Research 9: 35. DOI 10.1186/s13048-016-0244-9. [Google Scholar] [CrossRef]
Broustas CG, Lieberman HB (2014). DNA damage response genes and the development of cancer metastasis. Radiation Research 181: 111–130. [Google Scholar]
Cancer Genome Atlas Research N, Analysis Working Group: Asan U, Agency BCC, Brigham, Women’s H et al. (2017). Integrated genomic characterization of oesophageal carcinoma. Nature 541: 169–175. [Google Scholar]
Champiat S, Ferte C, Lebel-Binay S, Eggermont A, Soria JC (2014). Exomics and immunogenics: Bridging mutational load and immune checkpoints efficacy. Oncoimmunology 3: e27817. DOI 10.4161/onci.27817. [Google Scholar] [CrossRef]
Curtin NJ (2012). DNA repair dysregulation from cancer driver to therapeutic target. Nature Reviews Cancer 12: 801–817. DOI 10.1038/nrc3399. [Google Scholar] [CrossRef]
da Silva MS (2021). DNA double-strand breaks: A double-edged sword for trypanosomatids. Frontiers in Cell and Developmental Biology 9: 669041. DOI 10.3389/fcell.2021.669041. [Google Scholar] [CrossRef]
Fabrizio DA, George TJJr, Dunne RF, Frampton G, Sun J et al. (2018). Beyond microsatellite testing: Assessment of tumor mutational burden identifies subsets of colorectal cancer who may respond to immune checkpoint inhibition. Journal of Gastrointestinal Oncology 9: 610–617. DOI 10.21037/jgo.2018.05.06. [Google Scholar] [CrossRef]
Freedman ND, Abnet CC, Leitzmann MF, Mouw T, Subar AF et al. (2007). A prospective study of tobacco, alcohol, and the risk of esophageal and gastric cancer subtypes. American Journal of Epidemiology 165: 1424–1433. DOI 10.1093/aje/kwm051. [Google Scholar] [CrossRef]
Global Burden of Disease Cancer C, Fitzmaurice C, Abate D, Abbasi N, Abbastabar H et al. (2019). Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 29 cancer groups, 1990 to 2017: A systematic analysis for the global burden of disease study. JAMA Oncology 5: 1749–1768. DOI 10.1001/jamaoncol.2019.2996. [Google Scholar] [CrossRef]
Gu C, Shi X, Dai C, Shen F, Rocco G et al. (2020). RNA m6A modification in cancers: Molecular mechanisms and potential clinical applications. Innovation 1: 100066. DOI 10.1016/j.xinn.2020.100066. [Google Scholar] [CrossRef]
Gu C, Chen J, Dang X, Chen C, Huang Z et al. (2021). Hippo pathway core genes based prognostic signature and immune infiltration patterns in lung squamous cell carcinoma. Frontiers in Oncology 11: 680918. DOI 10.3389/fonc.2021.680918. [Google Scholar] [CrossRef]
Hong J, Chen Z, Peng D, Zaika A, Revetta F et al. (2016). APE1-mediated DNA damage repair provides survival advantage for esophageal adenocarcinoma cells in response to acidic bile salts. Oncotarget 7: 16688–16702. DOI 10.18632/oncotarget.7696. [Google Scholar] [CrossRef]
Huang FL, Yu SJ (2018). Esophageal cancer: Risk factors, genetic association, and treatment. Asian Journal of Surgery 41: 210–215. DOI 10.1016/j.asjsur.2016.10.005. [Google Scholar] [CrossRef]
Jiao R, Luo H, Xu W, Ge H (2019). Immune checkpoint inhibitors in esophageal squamous cell carcinoma: progress and opportunities. OncoTargets and Therapy 12: 6023–6032. DOI 10.2147/OTT. [Google Scholar] [CrossRef]
Kang H, Wang N, Wang X, Zhang Y, Lin S et al. (2020). A glycolysis-related gene signature predicts prognosis of patients with esophageal adenocarcinoma. Aging 12: 25828–25844. DOI 10.18632/aging.104206. [Google Scholar] [CrossRef]
Le DT, Durham JN, Smith KN, Wang H, Bartlett BR et al. (2017). Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357: 409–413. DOI 10.1126/science.aan6733. [Google Scholar] [CrossRef]
Lewis SJ, Smith GD (2005). Alcohol, ALDH2, and esophageal cancer: A meta-analysis which illustrates the potentials and limitations of a Mendelian randomization approach. Cancer Epidemiology, Biomarkers & Prevention 14: 1967–1971. DOI 10.1158/1055-9965.EPI-05-0196. [Google Scholar] [CrossRef]
Li K, Yin X, Yang H, Yang J, Zhao J et al. (2015). Association of the genetic polymorphisms in XRCC6 and XRCC5 with the risk of ESCC in a high-incidence region of North China. Tumori Journal 101: 24–29. DOI 10.5301/tj.5000206. [Google Scholar] [CrossRef]
Li S, Qin X, Chai S, Qu C, Wang X et al. (2016). Modulation of E-cadherin expression promotes migration ability of esophageal cancer cells. Scientific Reports 6: 21713. DOI 10.1038/srep21713. [Google Scholar] [CrossRef]
Li T, Fan J, Wang B, Traugh N, Chen Q et al. (2017a). TIMER: A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Research 77: e108–e110. DOI 10.1158/0008-5472.CAN-17-0307. [Google Scholar] [CrossRef]
Li Y, Lu Z, Che Y, Wang J, Sun S et al. (2017b). Immune signature profiling identified predictive and prognostic factors for esophageal squamous cell carcinoma. Oncoimmunology 6: e1356147. DOI 10.1080/2162402X.2017.1356147. [Google Scholar] [CrossRef]
Liu M, Li F, Liu B, Jian Y, Zhang D et al. (2021). Profiles of immune cell infiltration and immune-related genes in the tumor microenvironment of esophageal squamous cell carcinoma. BMC Medical Genomics 14: 75. DOI 10.1186/s12920-021-00928-9. [Google Scholar] [CrossRef]
Nepomuceno TC, de Gregoriis G, de Oliveira FMB, Suarez-Kurtz G, Monteiro AN et al. (2017). The role of PALB2 in the DNA damage response and cancer predisposition. International Journal of Molecular Sciences 18: 1886. DOI 10.3390/ijms18091886. [Google Scholar] [CrossRef]
Ohashi S, Miyamoto S, Kikuchi O, Goto T, Amanuma Y et al. (2015). Recent advances from basic and clinical studies of esophageal squamous cell carcinoma. Gastroenterology 149: 1700–1715. DOI 10.1053/j.gastro.2015.08.054. [Google Scholar] [CrossRef]
Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V et al. (2015). Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348: 124–128. DOI 10.1126/science.aaa1348. [Google Scholar] [CrossRef]
Ru B, Wong CN, Tong Y, Zhong JY, Zhong SSW et al. (2019). TISIDB: An integrated repository portal for tumor-immune system interactions. Bioinformatics 35: 4200–4202. DOI 10.1093/bioinformatics/btz210. [Google Scholar] [CrossRef]
Staker BL, Hjerrild K, Feese MD, Behnke CA, Burgin ABJr et al. (2002). The mechanism of topoisomerase I poisoning by a camptothecin analog. PNAS 99: 15387–15392. DOI 10.1073/pnas.242259599. [Google Scholar] [CrossRef]
Turati F, Tramacere I, La Vecchia C, Negri E (2013). A meta-analysis of body mass index and esophageal and gastric cardia adenocarcinoma. Annals of Oncology 24: 609–617. DOI 10.1093/annonc/mds244. [Google Scholar] [CrossRef]
Turgeon MO, Perry NJS, Poulogiannis G (2018). DNA damage, repair, and cancer metabolism. Frontiers in Oncology 8: 15. DOI 10.3389/fonc.2018.00015. [Google Scholar] [CrossRef]
Vidotto T, Nersesian S, Graham C, Siemens DR, Koti M (2019). DNA damage repair gene mutations and their association with tumor immune regulatory gene expression in muscle invasive bladder cancer subtypes. Journal for ImmunoTherapy of Cancer 7: 148. DOI 10.1186/s40425-019-0619-8. [Google Scholar] [CrossRef]
Wang D, Nagle P, Wang H, Smit J, Faber H et al. (2019). Hedgehog pathway as a potential intervention target in esophageal cancer. Cancers 11: 821. DOI 10.3390/cancers11060821. [Google Scholar] [CrossRef]
Wang G, Guo S, Zhang W, Li Z, Xu J et al. (2020a). A comprehensive analysis of alterations in DNA damage repair pathways reveals a potential way to enhance the radio-sensitivity of esophageal squamous cell cancer. Frontiers in Oncology 10: 575711. DOI 10.3389/fonc.2020.575711. [Google Scholar] [CrossRef]
Wang Y, Liang N, Xue Z, Xue X (2020b). Identifying an eight-gene signature to optimize overall survival prediction of esophageal adenocarcinoma using bioinformatics analysis of ceRNA network. OncoTargets and Therapy 13: 13041–13054. DOI 10.2147/OTT.S287084. [Google Scholar] [CrossRef]
Wu D, Chen X, Xu Y, Wang H, Yu G et al. (2017). Prognostic value of MLH1 promoter methylation in male patients with esophageal squamous cell carcinoma. Oncology Letters 13: 2745–2750. DOI 10.3892/ol.2017.5759. [Google Scholar] [CrossRef]
Yang X, Zhu H, Qin Q, Yang Y, Yang Y et al. (2015). Genetic variants and risk of esophageal squamous cell carcinoma: A GWAS-based pathway analysis. Gene 556: 149–152. DOI 10.1016/j.gene.2014.11.049. [Google Scholar] [CrossRef]
Yuan C, Xiang L, Cao K, Zhang J, Luo Y et al. (2020). The prognostic value of tumor mutational burden and immune cell infiltration in esophageal cancer patients with or without radiotherapy. Aging 12: 4603–4616. DOI 10.18632/aging.102917. [Google Scholar] [CrossRef]
Zhang D, Qian C, Wei H, Qian X (2020). Identification of the prognostic value of tumor microenvironment-related genes in esophageal squamous cell carcinoma. Frontiers in Molecular Biosciences 7: 599475. DOI 10.3389/fmolb.2020.599475. [Google Scholar] [CrossRef]
Zhang R, Zhou F, Cheng L, Yu A, Zhu M et al. (2018). Genetic variants in nucleotide excision repair pathway predict survival of esophageal squamous cell cancer patients receiving platinum-based chemotherapy. Molecular Carcinogenesis 57: 1553–1565. DOI 10.1002/mc.22877. [Google Scholar] [CrossRef]
Zhu L, Yang F, Li X, Li Q, Zhong C (2021). Glycolysis changes the microenvironment and therapeutic response under the driver of gene mutation in esophageal adenocarcinoma. Frontiers in Genetics 12: 743133. DOI 10.3389/fgene.2021.743133. [Google Scholar] [CrossRef]
Supplementary Materials
Cite This Article
 Copyright © 2022 The Author(s). Published by Tech Science Press.
Copyright © 2022 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools