DOI:10.32604/biocell.2022.022100

| BIOCELL DOI:10.32604/biocell.2022.022100 |  |
| Viewpoint |
How does FtsZ’s treadmilling help bacterial cells divide?
The Chinese Academy of Sciences Key Laboratory of Innate Immunity and Chronic Disease, School of Basic Medical Sciences, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, 230026, China
*Address correspondence to: Xinxing Yang, xinxingyang@ustc.edu.cn
Received: 21 February 2022; Accepted: 22 April 2022
Abstract: Most bacteria assemble a ring-like macromolecular machinery scaffolded by the essential cytoskeletal protein FtsZ for cell division. Studies have broadly explored how FtsZ could polymerize at the correct place and time. Recently, the FtsZ-ring was found to exhibit dynamic treadmilling along the circumference of the division site, driven by GTP hydrolysis. This apparently directional motion of FtsZ seems to drive the movement of septal cell wall synthesis enzymes and to play an important role in modulating cell envelope constriction and septum morphogenesis. However, the relationship between FtsZ’s treadmilling dynamics and cell wall synthesis varies in different bacteria. More importantly, the biophysical and molecular mechanisms governing these dynamic processes are unclear. In this viewpoint, we will focus on some new and exciting studies surrounding this topic and discuss potential mechanisms that underlie how FtsZ’s treadmilling dynamics might regulate septal cell wall synthesis and cell division.
Keywords: FtsZ; Bacterial cell division; sPG synthesis
FtsZ is a prokaryotic tubulin homolog (Nogales et al., 1998) that plays a central role in regulating bacterial cell division. Thirty years ago, Bi and Lutkenhaus (1991) discovered that FtsZ molecules can form a ring-like structure (termed the “Z-ring”) at the future division site and determined this ring to be the first known prokaryotic cytoskeleton. Similar to tubulin, FtsZ self-polymerizes upon GTP binding (Bramhill and Thompson, 1994) and subsequently hydrolyzes GTP molecules, inducing its depolymerization (Chen and Erickson, 2005; de Boer et al., 1992). The Z-ring recruits more than thirty cell division proteins, many of them essential and involved in cell wall synthesis, to assemble a macromolecular machinery collectively called the divisome (Du and Lutkenhaus, 2017).
Given that FtsZ is broadly conserved among eubacteria and archaea, we wondered whether there exists a universal mechanism for prokaryotes to control and regulate the cell division process via the Z-ring. One well-accepted function of the Z-ring is to act as a scaffold for the recruitment of other divisome proteins, especially cell wall synthases and remodeling enzymes (Egan et al., 2020; McQuillen and Xiao, 2020). Another potential function of the Z-ring was proposed to generate a mechanical constricting force based on the homology to eukaryotic microtubes. In this model, FtsZ-ring utilizes the energy from GTP hydrolysis by its constitutive monomers to pull the cell envelope inward and lead to cell constriction (Erickson et al., 2010). This idea was strongly supported by the impressive experiments by Osawa et al. (2008) showing that FtsZ can polymerize to rings that bend and constrict liposomes in vitro. While this mechanism is intriguing, it raises questions such as: What is the exact physical model in which the Z-ring generates a homogeneous contractile force? Is the force strong enough to deform both the inner membrane and peptidoglycan cell wall against turgor? For insight into these questions, I refer the readers to the extensive discussion in the review by McQuillen and Xiao (2020).
The FtsZ force-generation model was challenged by Coltharp et al. (2016) who found that the cell constriction rate of E. coli cells does not depend on FtsZ’s intrinsic GTPase activity (originally proposed to govern the force-generation). Instead, the rate-limiting step of E. coli cell division was identified as the cell wall synthesis rate. This result was later confirmed by labeling the nascent sPG using fluorescent d-amino acids (FDAAs) (Kuru et al., 2012): the amount of newly synthesized (E. coli) cell wall was not correlated with FtsZ’s GTPase activity (Yang et al., 2017). These results are consistent with previous studies in which FtsZ GTPase mutants can still complete cell division with deformed or abnormal septa (Bi and Lutkenhaus, 1992; Lutkenhaus et al., 2012). Considering the fact that the Z-ring constantly hydrolyzes GTP, it is important to understand why bacterial cells continuously consume GTP if the potential mechanical force is not essential.
FtsZ Dynamically Treadmills During Cell Division
Later, FtsZ’s GTP hydrolysis was found to drive an unseen type of dynamic behavior of FtsZ: treadmilling (Bisson-Filho et al., 2017; Loose and Mitchison, 2014; Yang et al., 2017). FtsZ subunits in the Z-ring are known to exchange constantly with monomers in the cytoplasmic pool (lifetime ~10 sec) (Stricker et al., 2002), where the exchange rate depends on the GTPase activity. This behavior and the corresponding kinetic mechanism were thoroughly discussed in a classic review article by Erickson et al. (2010). They also proposed that one possible scheme of FtsZ dynamics could be treadmilling, a behavior that widely exists in eukaryotic cytoskeletal filaments.
Margolin’s group was the first to observe rapid dynamics and oscillations of the Z-ring back in 2004 (Thanedar and Margolin, 2004), but the treadmilling behavior of FtsZ was not determined until total internal reflection fluorescence microscopy (TIRFm) was introduced to image reconstituted FtsZ filaments in vitro and the Z-ring in vivo. FtsZ was found to form chunks or clusters in the Z-ring using super-resolution fluorescence microscopy (Buss et al., 2015; Fu et al., 2010; Strauss et al., 2012). With much less phototoxicity, TIRFm allowed researchers to monitor FtsZ clusters for a long period of time with a high spatial-temporal resolution. Interestingly, those FtsZ clusters move directionally around the ring while individual FtsZ monomers stay immobile. This behavior is a hallmark of treadmilling (Bisson-Filho et al., 2017; Buss et al., 2013; Loose and Mitchison, 2014; Niu and Yu, 2008; Yang et al., 2017): FtsZ monomers are added on one end (polymerization) and lost on the other (depolymerization), thus the filament appeared to be moving forward (Fig. 1).
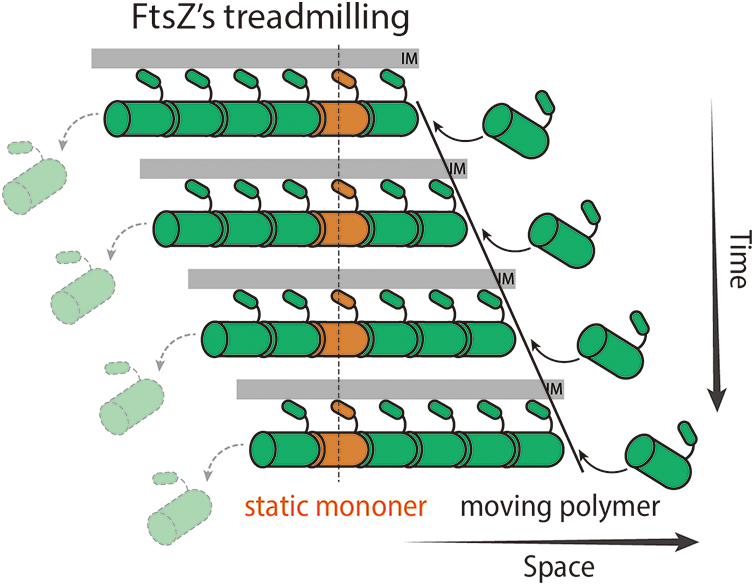
Figure 1: Diagram demonstrating FtsZ’s treadmilling. FtsZ monomers polymerize on the right end of the filament while the last subunit depolymerizes from the left end. The filament thus moves to the right with immobile FtsZ subunits (orange).
The apparent directional movement of FtsZ clusters implies an energy input to the system. Unlike the bacterial sidewall building system (the elongasome) that utilizes the peptidoglycan (PG) synthesis reaction to power its directional motion (Dominguez-Escobar et al., 2011; Garner et al., 2011; van Teeffelen et al., 2011), the FtsZ’s treadmilling speed is only correlated with its intrinsic GTPase activity but not with the FtsZ regulators or sPG synthesis (Bisson-Filho et al., 2017; Caldas et al., 2019; Perez et al., 2019; Ramirez-Diaz et al., 2018; Squyres et al., 2021; Whitley et al., 2021; Yang et al., 2017). Additionally, in vitro fluorescence and atomic force microscopic studies suggested that treadmilling dynamics also rely on FtsZ’s densities and the type of surface tethers (Gonzalez de Prado Salas et al., 2014; Loose and Mitchison, 2014; Marquez et al., 2019; Mateos-Gil et al., 2012; Ramirez-Diaz et al., 2018).
The treadmilling dynamics were later confirmed in other bacterial species such as S. mutans (Li et al., 2018), S. aureus (Monteiro et al., 2018), and S. pneumonia (Perez et al., 2019). Given the robustness of the treadmilling dynamics in living cells, it is natural to reason that the energy from GTP molecules is harvested to maintain FtsZ’s treadmilling which further assists the sPG synthesis and cell constriction. Indeed, the treadmilling was showed to be essential for successful and efficient cell division: In B. subtilis, abolishing FtsZ’s treadmilling by a small molecule inhibitor PC190723 stops cell division (Bisson-Filho et al., 2017). In E. coli, the ftsZD212G mutation with a slow treadmilling speed causes abnormal division and filamentous cells (Bi and Lutkenhaus, 1990; Stricker and Erickson, 2003; Yang et al., 2017).
FtsZ’s Treadmilling Drives the Directional Motion of sPG Synthesis
Back when Bi and Lutkenhaus discovered the Z-ring, they speculated that “formation of the ring would be the key point at which temporal and spatial control over division are exerted…perhaps by interacting directly with septal-specific peptidoglycan biosynthetic machinery at the leading edge of the invagination” (Bi and Lutkenhaus, 1991). This speculation turns out to be visionary even though it was not possible to monitor Z-ring dynamics or sPG synthesis in living cells at that time.
With novel single-molecule imaging and labeling techniques, we are now able to track single protein molecules in living cells and measure the spatial distribution of PG synthesis (Cho et al., 2016; Grimm et al., 2015; Kuru et al., 2012; Lee et al., 2016; Yu et al., 2006). The first tracked essential sPG synthase was the monofunctional transpeptidase (TPase) FtsI in E. coli (PBP2B in B. subtilis). These enzymes were observed to move processively along with the Z-ring (Bisson-Filho et al., 2017; Yang et al., 2017). Unlike their counterparts PBP2 (in E. coli) and PBP2A (in B. subtilis) in the elongasome, whose movement is driven by PG synthesis (Dominguez-Escobar et al., 2011; Garner et al., 2011; van Teeffelen et al., 2011), the average speeds of FtsI and PBP2b are highly correlated with FtsZ’s treadmilling speed. This exciting result indicated that FtsZ may convert the chemical energy in GTP into kinetic energy by treadmilling to drive the directed motion of these sPG enzymes. In other words, FtsZ could use its polymer dynamics to function as a linear motor to deliver cargos (sPG synthases) at target sites to initiate local septum synthesis.
This hypothesis is supported from pulse-labeling experiments showing that the newly synthesized septum is uneven and clustered, akin to the discontinuous, clustered Z-ring structure in E. coli, B. subtilis, and S. pneumoniae (Bisson-Filho et al., 2017; Perez et al., 2021; Yang et al., 2017). Such a discrete sPG pattern suggests that sPG synthases are not efficient in building the whole septum all at once, but that they, instead, work ‘locally’ producing ‘patches’ of new sPG around the ring. They therefore require regulators to modulate their spatial-temporal distribution to evenly construct the whole septum over time. As FtsZ clusters treadmill around the future septum, they likely guide enzymes to circle around the cell. Indeed, experiments have shown that the new septum becomes uneven or abnormal in FtsZ mutation strains with slow treadmilling speeds (Bisson-Filho et al., 2017; Perez et al., 2021; Yang et al., 2017). Furthermore, the cell division rate in B. subtilis was shown positively correlated with FtsZ’s treadmilling speed, indicating FtsZ’s treadmilling may guide both the distribution and activity of sPG synthases (Bisson-Filho et al., 2017).
However, the results from the gram-negative bacterium E. coli speaks to the contrary: FtsZ mutants with decreased GTPase activity and slower treadmilling only impact the septum morphology but do NOT alter the overall sPG synthesis or the constriction rate in most cases (Coltharp and Xiao, 2017; Yang et al., 2017). These results suggested that FtsZ only regulates the spatial-temporal distribution but not the enzymatic activity in E. coli. This discrepancy grew more puzzling after several other findings revealed that FtsZ may be dispensable for sPG synthesis under specific conditions.
sPG Synthesis Can be Independent of FtsZ’s Treadmilling
Soon after the discovery of FtsZ’s treadmilling dynamics, Pinho’s group monitored the septum closure process in S. aureus when FtsZ’s treadmilling was inhibited. Surprisingly, they found that FtsZ’s treadmilling is no longer required for sPG synthesis and cell constriction at a later stage of cell division, once the entire divisome is assembled (after the recruitment of the Lipid II lipase MurJ) (Monteiro et al., 2018).
In S. pneumonia, the Winkler lab then found that bPBP2x, the essential sPG TPase, moved directionally yet in an FtsZ independent manner (Perez et al., 2019). GTPase-defective FtsZ mutants did not decrease the moving speed of bPBP2x as that in E. coli or B. subtilis. Rather, inhibiting sPG synthesis slowed bPBP2x. Moreover, the sPG synthesis level of FtsZ mutant strains remained the same as that of wild-type cells, while the pattern of nascent sPG became irregular (Perez et al., 2021; Perez et al., 2019), very similar to the case of E. coli (Yang et al., 2017).
More recently, Kevin et al. from the Holden lab re-examined how inhibition of FtsZ’s treadmilling would disrupt cell division at different stages in B. subtilis. Similar to S. aureus, they found that a fraction of cells that have already proceeded to a later cell division stage continued to divide even when FtsZ’s treadmilling was inhibited (Whitley et al., 2021). Other early-stage cells could not constrict or build septal cell walls, as previous results (Bisson-Filho et al., 2017). The authors also carefully measured the septum closure rate under different cell growth rates, finding that they are highly correlated. Given the insignificant change in the FtsZ’s treadmilling speed under those conditions, there might be other regulatory factors of sPG synthesis besides FtsZ in B. subtilis.
In fact, FtsZ-independent cell constriction has been found years ahead. Soderstrom et al. (2014) determined the sequence of division proteins leaving the septum and found that the FtsZ-ring disassembled before the end of cytokinesis in E. coli (Soderstrom et al., 2014). This result showed that FtsZ (and its treadmilling) is also dispensable in the last part of division in E. coli. All the above observations complicated the understanding of how FtsZ’s treadmilling regulates sPG synthesis and cell division.
Rethink and Unify the Function of FtsZ in Cell Division
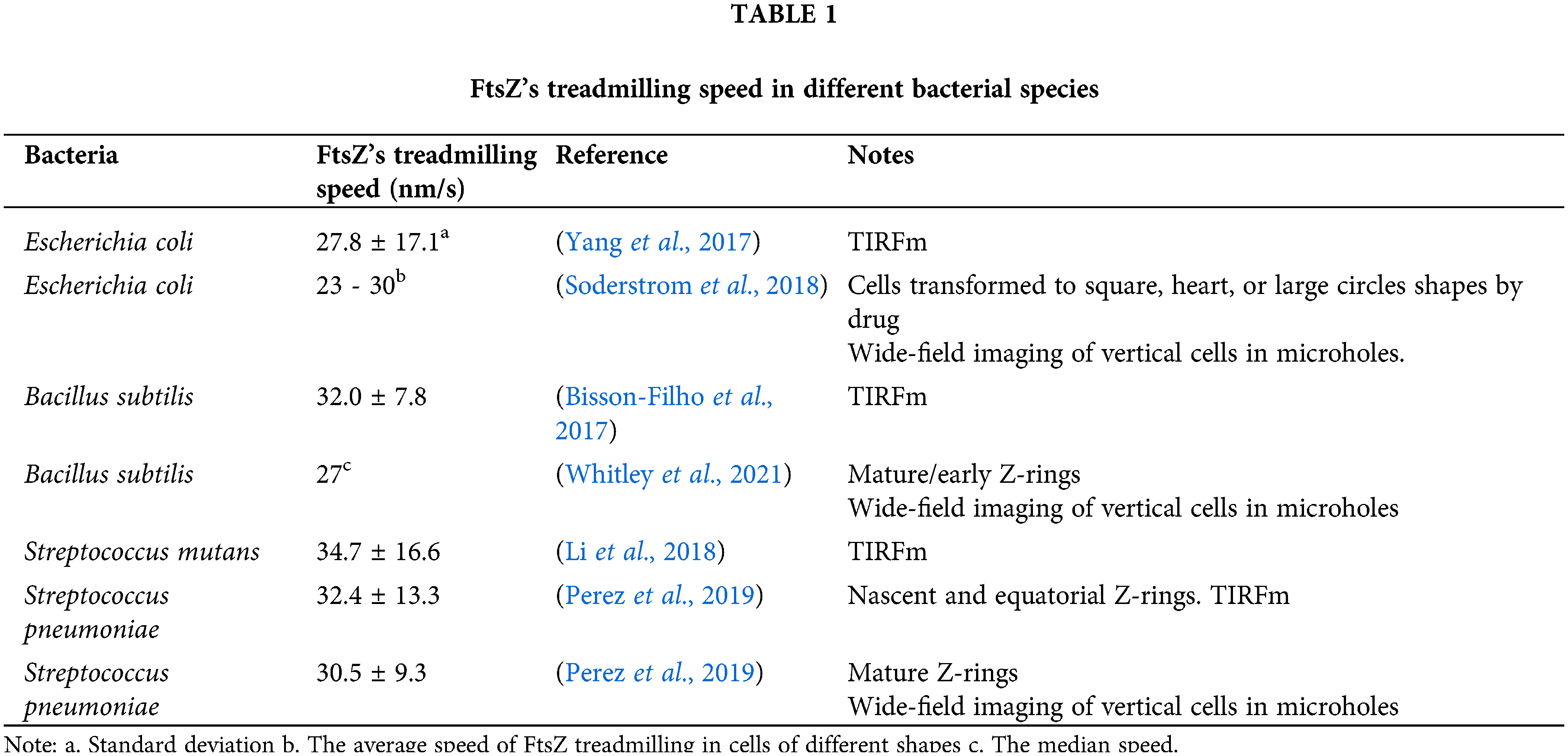
On one hand, in all bacteria tested thus far, FtsZ treadmills at approximately 30 nm/s regardless of gram-positive or gram-negative, rod or ovoid cell shapes, and even in artificial rectangular or heart-shaped cells (Table 1). Considering the significant differences in divisome composition and cell physiology among those bacteria, FtsZ’s treadmilling seems to be highly conserved. It is possible that this dynamic property of FtsZ (or the Z-ring) serves as a basic and robust mechanism in regulating the cell division process.
On the other hand, bacteria live in drastically different environments and have evolved for billion years, developing different cell shapes, sizes, cell wall thicknesses and PG synthesis/hydrolysis enzymes. It is not unreasonable to assume that they also have very different ways to build the septum and constrict the cell envelope.
1. FtsZ’s treadmilling condenses the divisome to the mid-cell
As the master regulator, FtsZ must first recruit and scaffold for other proteins to the future cell division site (i.e., the middle of the long axis in most rod shape bacteria). Recently, Squyres et al. (2021) from the Garner lab found that the Z-ring is not only simply positioned to mid-cell with sPG synthases. The FtsZ ring condenses along the progression of division, which facilitates the assembly of cell wall synthases such as PBP2b and FtsW in B. subtilis and enables them to function correctly. The reason might be that the enzymes have low concentrations inside the cell. Their functions often rely on other partners or activators (such as FtsQLB and FtsN in E. coli (Liu et al., 2015; Tsang and Bernhardt, 2015)). A condensed Z-ring can generate a confined volume trapping downstream divisome components and thus raise their local concentration.
At the same time, Whitley et al. (2021) showed that FtsZ’s treadmilling facilitates the aggregation of a narrow and matured Z-ring in the early division stage. After the tight ring formed, treadmilling becomes less important. It is worth mentioning that treadmilling was shown to facilitate transient FtsZ assemblies to localize to the correct cell division site (Walker et al., 2020). These studies suggested that the first regulatory mechanism of FtsZ’s treadmilling is to enable FtsZ filaments to dynamically encounter each other, locate at the mid-cell and condense to a “narrow ring” by lateral interactions. The condensed Z-ring subsequently helps other components assemble and trigger sPG synthesis complex formation (Fig. 2A).
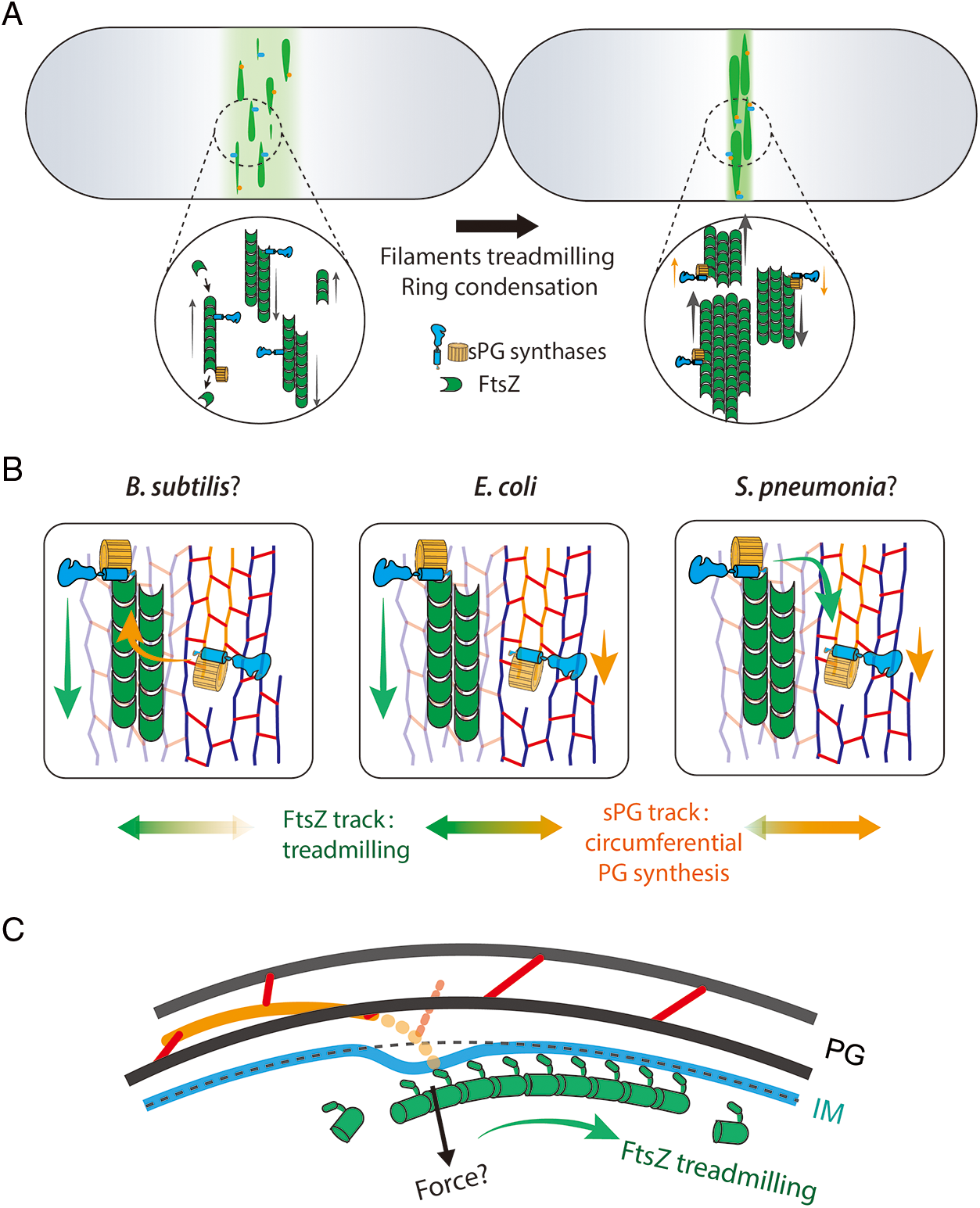
Figure 2: Potential models of FtsZ’s treadmilling regulates the cell division in three different dimensions. A. FtsZ filaments (green) travel around the cell circumference via treadmilling (left). The treadmilling dynamics facilitate the formation of a narrow and mature Z-ring along the long axis (right) that concentrate and activate the sPG synthases (blue and orange parts) (Squyres et al., 2021; Whitley et al., 2021). B. The treadmilling direction is approximately along the glycan strands in septum. sPG synthases can be transported by FtsZ’ treadmilling to different positions (green arrow) or synthesize new sPG processively in a different speed (orange arrow). In B. subtilis, synthases tend to associate more on the Z-track (Bisson-Filho et al., 2017) (left) while enzymes in S. pneumoniae were found to move with PG synthesis (Perez et al., 2019) (right). E. coli synthases move on both tracks (Yang et al., 2021) (middle). C. FtsZ may generate contractile force radially toward the cell center. The deformation of the inner membrane could displace the synthesis complexes thus build the new septal cell wall inside the old one (Nguyen et al., 2019).
2. FtsZ’s treadmilling regulates the spatial distribution of sPG synthase complex along the ring
The next step of cell division is to synthesize new sPG material along the circumference of the cell. In rod-shaped bacteria, such as E. coli and B. subtilis, the glycan strands of PG are arranged circumferentially, while the peptide stems are along the long axis of the cell (Holtje, 1998). If the glycan strands are perfectly aligned and spaced, sPG synthesis could use the old PG strand as a template, at least along the circumferential direction. Unfortunately, the glycan chains are not perfectly organized, as visualized by atomic force microscopy (AFM) (Pasquina-Lemonche et al., 2020; Turner et al., 2018). Since FtsZ filaments constantly treadmill around the cell’s circumference and scaffold sPG synthases, it is natural to reason that FtsZ keeps the enzymes moving (perhaps synthesis too) along the ring. Three supporting observations were obtained: 1) FtsI and PBP2b molecules move along the ring at a similar speed as FtsZ’s treadmilling; 2) the PG synthesis pattern is clustered like that of the Z-ring; and 3) new septa in some ftsZ mutation strains exhibit helical shapes similar to the Z-rings (Bisson-Filho et al., 2017; Yang et al., 2017).
However, it is difficult to comprehend how enzymes or enzymatic complexes can travel directionally along the Z-ring while every single FtsZ protein is static. The Liu and Xiao labs proposed a Brownian ratchet model in which minus end-attaching enzymes (i.e., FtsI) on a treadmilling filament will fall off while the last FtsZ subunit dissociates (Fig. 3). Although falling FtsZ monomers likely diffuses away in the cytoplasm, the sPG synthase is confined to the membrane and cannot escape the surrounding zone in a short time window. It could catch up and associate with the FtsZ subunit on the new ‘minus-end’ of the filament again. As such, the enzyme follows the FtsZ filament and moves forward (McCausland et al., 2021). In this way, FtsZ transports sPG synthases to different septal sites and generates new clustered sPG. Even in S. pneumoniae, whose PG synthases move slower than FtsZ’s treadmilling, the nascent sPG still exhibits a clear clustered pattern along the ring (Perez et al., 2021; Perez et al., 2019), indicating that FtsZ still determines the spatial distribution of PG synthases (perhaps in a passive way). Using a reconstituted lipid bilayer system, Baranova et al. (2020) observed the collective co-immigration of the cytoplasmic portion of FtsQ and FtsN with FtsZ. Even though the truncated proteins did not display a processive motion, the results demonstrated that the late divisome proteins could be redistributed by FtsZ’s treadmilling.
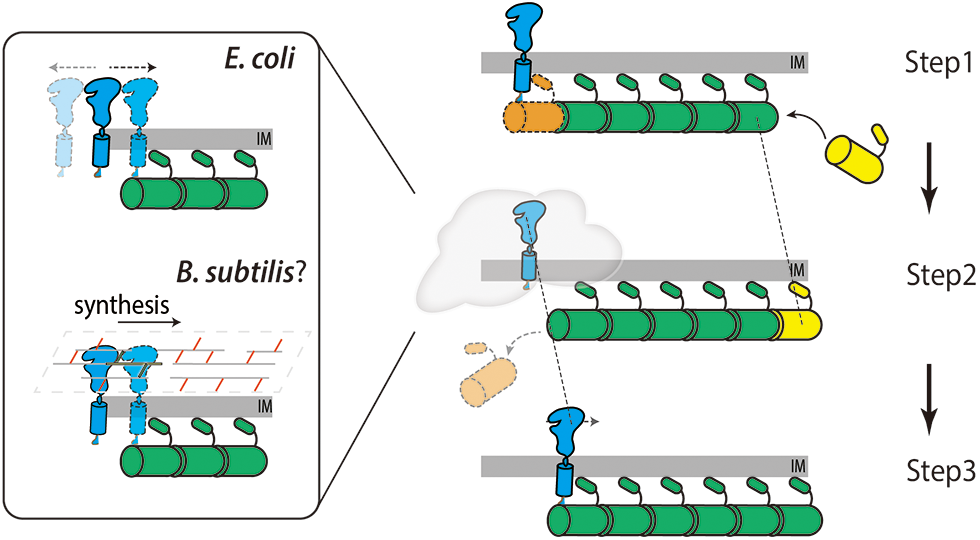
Figure 3: Diagram showing the Brownian ratchet model that FtsZ’s treadmilling guides sPG synthases’ directional movement. Step 1. The sPG synthase (blue) binds with the FtsZ subunit on the minus end of an FtsZ filament (orange). Step 2. The last FtsZ subunit dissociates from the filament leaving the synthase diffuses on the membrane (in E. coli, top left) or synthesizes sPG shortly (in B. subtilis, bottom left). Step 3. The synthase rebinds to the new end of FtsZ filament thus moves forward to the right at the same speed as that of the FtsZ filament (dashed lines).
3. FtsZ’s treadmilling-associated synthase complexes may or may not be active
In addition to the spatial distribution, it is also important to understand whether FtsZ’s treadmilling determines the sPG synthesis rate. On this aspect, the results seem to differ among bacteria. Between the completely dependent case of B. subtilis and the independent case of S. pneumoniae, we found that the motion of E. coli sPG synthases partially depends on FtsZ’s treadmilling. These enzymes split into two populations: one goes with FtsZ, while the other was driven by the active sPG synthesis (Yang et al., 2021). The results suggested a two-track model in which FtsZ filaments (clusters) carry enzymes (or synthase complexes) from one place to another along the ring until these enzyme molecules stochastically dissociation and/or encounter activators such as FtsN (Lyu et al., 2021). The enzymes could then synthesize new sPG for ~200 nm in a highly processive manner. After the complexes terminate on the PG (stochastically or being inactivated), they are able to catch on a treadmilling FtsZ filament again (Fig. 2B middle, Fig. 3). Thus, treadmilling FtsZ filaments act as trains to collect free synthases and keep them on track to synthesize hot spots along the ring. This “two-track model” might explain the different behaviors in bacteria. Enzymes in S. pneumoniae might have low binding affinities to FtsZ and therefore cannot follow treadmilling for a sufficient time to be detected in experiments; only molecules processively synthesizing sPG were observed (Fig. 2B, right). In contrast, B. subtilis may have enzymes with fast sPG synthesis rates but not very processive, showing Z-track related motion only (Fig. 2B, left). Mechanistically, B. subtilis sPG synthases may rapidly proceed a couple of synthesis cycles (a few nanometers) after leaving the Z-track and departing from PG. FtsZ filaments then carry them until the next synthesis ‘hot spot’ (Fig. 3). This hypothesis might be able to explain the correlation of the sPG synthesis rate and FtsZ’s treadmilling speed in B. subtilis (Bisson-Filho et al., 2017). More simulations and experiments are required to reveal the true lying mechanism. For example, it would be interesting to examine the motion of the synthases and sPG synthesis pattern when FtsZ’s treadmilling is abolished in B. subtilis and S. aureus.
4. Does FtsZ’s treadmilling generate a constriction force to reduce the radius of the new sPG?
The synthesis of sPG is intrinsically different from that of the lateral cell wall. The new sPG material must be inserted inside of the old PG template to reduce the septal radius gradually, which means that the synthesis complex must break the symmetry and add new glycan strands biased to the cell’s center. However, as introduced above and discussed in a detailed review article (McQuillen and Xiao, 2020), the FtsZ-ring might not be able to generate enough force to counter the turgor pressure and bend the cell wall mechanically. A possible mechanism is that the sPG synthases (transmembrane proteins) are pulled inside together with the inner membrane, which is deformed inward by FtsZ (Fig. 2C). Subsequently, the newly crossed linked sPG could lie under the old PG and gradually constrict the cell envelope. In C. crescentus cells, sPG can grow as bulges instead of invagination at division sites with FtsZ’s C-terminal linker (CTL) truncated variants, which might affect the transduction of the force to the inner membrane. (Sundararajan et al., 2015),
Although it remains unclear how much force FtsZ produces in living cells, Schwille’s group found that treadmilling FtsZ filaments could deform lipid tubes in vitro and wall-less E. coli cells (Ramirez-Diaz et al., 2021). The authors were able to measure the force (1–2 pN/μm) that is partially coupled with GTP hydrolysis (or treadmilling). However, FtsZ GTPase defective mutation could still bend the liposome. This finding is consistent with the results that treadmilling-inhibited B. subtilis cells can still divide yet at a slower rate (the static ring generating less force) (Whitley et al., 2021). The force is orders of magnitude smaller than the expected value (~5 nN/μm) to synthesize the new sPG on the inner side of the old PG against the turgor pressure from inside (Nguyen et al., 2019). We speculate that either FtsZ filaments can generate greater force in vivo or there are unknown factors supporting the new sPG in the inward position, relieving the tension from the turgor. Undoubtedly more theoretical and experimental works need to be done to predict and measure the true mechanical force generated by FtsZ filaments or the treadmilling dynamics.
Future: Figure Out the Unknowns of FtsZ’s Treadmilling in Bacterial Cell Division
From the limited number of organisms tested, the master cytoskeleton protein FtsZ presents almost identical properties in organization, GTPase activity, and treadmilling dynamics. A general model starts to emerge: the treadmilling Z-ring acts as a dynamic scaffold to assemble sPG synthases and keep them ‘on track’ on both the long axis and along the circumference. At least in some periods (aka, the late division stage after the whole divisome is established), FtsZ and its treadmilling become less important. In the gram-negative bacterium E. coli, for instance, FtsN was shown to form a separate ring structure and might serve as a scaffold in the later cell division stage (Lyu et al., 2021; Soderstrom et al., 2018). It is likely that different types of bacteria evolved their PG synthesis strategies by altering the affinity of PG synthases to FtsZ or sPG, enzymatic activity and processivity, lipid II substrate concentration, and the types of activators. Thus, cells can divide robustly with different cell sizes, growth rates, and environmental conditions. Current studies are primarily based on model bacteria that have similar cell dimensions and division rates. To further explore the dynamics and functions of FtsZ’s treadmilling, more studies on other bacteria with unconventional shapes and different division rates are needed. Considering the complexity of sPG synthesis and consequently septal cell wall remodeling during growth and division, careful studies of how PG enzymes are regulated and work upon different cell wall structures, lipid II concentrations, and different activation or inhibition pathways are required to understand the full picture of FtsZ and sPG synthases and the septal cell wall.
We still do not know how FtsZ filaments arrange and treadmill on a molecular level in living cells yet. FtsZ protofilaments may organize into diverse structures in vitro, such as bundles, sheets, and other high-order structures (Gonzalez de Prado Salas et al., 2014; Lu et al., 2000; Mateos-Gil et al., 2012; Sundararajan and Goley, 2017). In vivo super-resolution microscope revealed heterogenous cluster-like structures (Buss et al., 2015; Coltharp et al., 2016; Fu et al., 2010; Holden et al., 2014; Rowlett and Margolin, 2014) not in agree with the well-aligned continuous or short filaments from Cryo-EM images (Li et al., 2007; Szwedziak et al., 2014; Yao et al., 2017). Resolving the organization of FtsZ inside divisome is critical to understand the fundamental physical mechanism of the treadmilling dynamics. New imaging techniques such as MinFLUX combined with functional FtsZ labeling methods might be a promising method to depict the molecular organization of FtsZ in cells (Balzarotti et al., 2017; Moore et al., 2017; Schmidt et al., 2021).
Another amazing fact not explored in this article is that the cells could manage to maintain FtsZ in a steady and robust treadmilling state while reconstituted FtsZ requires a proper protein density and certain membrane tethers to trigger the treadmilling (Loose and Mitchison, 2014; Ramirez-Diaz et al., 2018). Recent studies suggested that FtsA and ZipA may be with complicated structures and functions to regulate FtsZ filaments rather than simple membrane-attachments (Conti et al., 2018; Krupka et al., 2017). The cooperative treadmilling of multiple FtsZ filaments was shown enhanced by FtsZ binding protein ZapA in vitro (Caldas et al., 2019). This FtsZ “crosslinker” has been found important to keep a narrow Z-ring in vivo (Buss et al., 2013), indicating treadmilling itself might not be enough to create the tight ring mentioned in the previous section. To understand the molecule level mechanism of treadmilling regulation, computational modeling could reinforce our toolbox to explore a broad range of parameters that are difficult to test by experiments. In fact, simulations based on structural information and kinetic measurements have already provided valuable insights (Mateos-Gil et al., 2019).
How much force can be generated by FtsZ’s treadmilling in living cells is the other major question difficult to answer. New techniques or methods such as genetically encoded force sensors (Wang et al., 2011) are needed to be carefully designed to measure the amount of constriction force generated by FtsZ during cytokinesis in real time. The force measurement companying with structural studies and computational simulations, we imagine, could complete one important piece of the puzzle.
As FtsZ’s treadmilling was shown to regulate the sPG synthesis, it may also participate to coordinate the remodeling and reconstruction the outer membrane in gram-negative bacteria. A recent study had shown that FtsZ or some early division proteins can recruit the essential outer membrane protein folding complex, BAM (Consoli et al., 2021). Whether FtsZ’ treadmilling directly regulates the outer membrane remodeling in time and space like the sPG should be further studied.
Last but not the least, given its essential role in cell division, FtsZ has attracted a lot of attention as a target for new antibiotic development. Many potent molecules were discovered or designed to inhibit the GTPase activity, polymerization, or depolymerization property of FtsZ (Pradhan et al., 2021; Ur Rahman et al., 2020). We hope this viewpoint article could provide some new angles for antimicrobial developers to think about targeting FtsZ’s conserved treadmilling behavior or the regulatory pathways. In fact, some small molecules such as PC190723 have been shown to abolish the treadmilling and become a powerfully tool in the studies mentioned above (Bisson-Filho et al., 2017; Monteiro et al., 2018; Whitley et al., 2021).
Acknowledgement: We thank Dr. Jie Xiao, Dr. Ryan McQuillen and Joshua McCausland for their critical reading and valuable advices on the manuscript.
Availability of Data and Materials: Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
Author Contribution: The authors confirm contribution to the paper as follows: draft manuscript and prepare figures: X. Y. and R. L. All authors reviewed the final version of the manuscript.
Ethics Approval: Not applicable.
Funding Statement: This work is supported by the start-up funding by the University of Science and Technology of China KJ2070000083 (X.Y) and KY9100000035 (X.Y).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Balzarotti F, Eilers Y, Gwosch KC, Gynna AH, Westphal V et al. (2017). Nanometer resolution imaging and tracking of fluorescent molecules with minimal photon fluxes. Science 355: 606–612. DOI 10.1126/science.aak9913. [Google Scholar] [CrossRef]
Baranova N, Radler P, Hernandez-Rocamora VM, Alfonso C, Lopez-Pelegrin M et al. (2020). Diffusion and capture permits dynamic coupling between treadmilling FtsZ filaments and cell division proteins. Nature Microbiology 5: 407–417. DOI 10.1038/s41564-019-0657-5. [Google Scholar] [CrossRef]
Bi E, Lutkenhaus J (1990). Analysis of ftsZ mutations that confer resistance to the cell division inhibitor SulA (SfiA). Journal of Bacteriology 172: 5602–5609. DOI 10.1128/jb.172.10.5602-5609.1990. [Google Scholar] [CrossRef]
Bi E, Lutkenhaus J (1992). Isolation and characterization of FtsZ alleles that affect septal morphology. Journal of Bacteriology 174: 5414–5423. [Google Scholar]
Bi EF, Lutkenhaus J (1991). FtsZ ring structure associated with division in Escherichia coli. Nature 354: 161–164. DOI 10.1038/354161a0. [Google Scholar] [CrossRef]
Bisson-Filho AW, Hsu YP, Squyres GR, Kuru E, Wu F et al. (2017). Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science 355: 739–743. DOI 10.1126/science.aak9973. [Google Scholar] [CrossRef]
Bramhill D, Thompson CM (1994). GTP-dependent polymerization of Escherichia coli FtsZ protein to form tubules. Proceedings of the National Academy of Sciences 91: 5813–5817. [Google Scholar]
Buss J, Coltharp C, Huang T, Pohlmeyer C, Wang SC et al. (2013). In vivo organization of the FtsZ-ring by ZapA and ZapB revealed by quantitative super-resolution microscopy. Molecular Microbiology 89: 1099–1120. DOI 10.1111/mmi.12331. [Google Scholar] [CrossRef]
Buss J, Coltharp C, Shtengel G, Yang X, Hess H et al. (2015). A multi-layered protein network stabilizes the Escherichia coli FtsZ-ring and modulates constriction dynamics. PLoS Genetics 11: e1005128. DOI 10.1371/journal.pgen.1005128. [Google Scholar] [CrossRef]
Caldas P, Lopez-Pelegrin M, Pearce DJG, Budanur NB, Brugues J et al. (2019). Cooperative ordering of treadmilling filaments in cytoskeletal networks of FtsZ and its crosslinker ZapA. Nature Communications 10: 5744. DOI 10.1038/s41467-019-13702-4. [Google Scholar] [CrossRef]
Chen Y, Erickson HP (2005). Rapid in vitro assembly dynamics and subunit turnover of FtsZ demonstrated by fluorescence resonance energy transfer. Journal of Biological Chemistry 280: 22549–22554. [Google Scholar]
Cho H, Wivagg CN, Kapoor M, Barry Z, Rohs PDA et al. (2016). Bacterial cell wall biogenesis is mediated by SEDS and PBP polymerase families functioning semi-autonomously. Nature Microbiology 1: 16172. DOI 10.1038/nmicrobiol.2016.172. [Google Scholar] [CrossRef]
Coltharp C, Buss J, Plumer TM, Xiao J (2016). Defining the rate-limiting processes of bacterial cytokinesis. Proceedings of the National Academy of Sciences 113: E1044–1053. [Google Scholar]
Coltharp C, Xiao J (2017). Beyond force generation: Why is a dynamic ring of FtsZ polymers essential for bacterial cytokinesis? Bioessays 39: 1–11. DOI 10.1002/bies.201600179. [Google Scholar] [CrossRef]
Consoli E, Luirink J, den Blaauwen T (2021). The Escherichia coli outer membrane beta-barrel assembly machinery (BAM) crosstalks with the divisome. International Journal of Molecular Sciences 22: 1853. DOI 10.3390/ijms22041853. [Google Scholar] [CrossRef]
Conti J, Viola MG, Camberg JL (2018). FtsA reshapes membrane architecture and remodels the Z-ring in Escherichia coli. Molecular Microbiology 107: 558–576. DOI 10.1111/mmi.13902. [Google Scholar] [CrossRef]
de Boer P, Crossley R, Rothfield L (1992). The essential bacterial cell-division protein FtsZ is a GTPase. Nature 359: 254–256. [Google Scholar]
Dominguez-Escobar J, Chastanet A, Crevenna AH, Fromion V, Wedlich-Soldner R et al. (2011). Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science 333: 225–228. DOI 10.1126/science.1203466. [Google Scholar] [CrossRef]
Du S, Lutkenhaus J (2017). Assembly and activation of the Escherichia coli divisome. Molecular Microbiology 105: 177–187. [Google Scholar]
Egan AJF, Errington J, Vollmer W (2020). Regulation of peptidoglycan synthesis and remodelling. Nature Reviews Microbiology 18: 446–460. [Google Scholar]
Erickson HP, Anderson DE, Osawa M (2010). FtsZ in bacterial cytokinesis: Cytoskeleton and force generator all in one. Microbiology and Molecular Biology Reviews 74: 504–528. DOI 10.1128/MMBR.00021-10. [Google Scholar] [CrossRef]
Fu G, Huang T, Buss J, Coltharp C, Hensel Z et al. (2010). In vivo structure of the E. coli FtsZ-ring revealed by photoactivated localization microscopy (PALM). PLoS One 5: e12682. [Google Scholar]
Garner EC, Bernard R, Wang W, Zhuang X, Rudner DZ et al. (2011). Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science 333: 222–225. DOI 10.1126/science.1203285. [Google Scholar] [CrossRef]
Gonzalez de Prado Salas P, Horger I, Martin-Garcia F, Mendieta J, Alonso A et al. (2014). Torsion and curvature of FtsZ filaments. Soft Matter 10: 1977–1986. DOI 10.1039/c3sm52516c. [Google Scholar] [CrossRef]
Grimm JB, English BP, Chen J, Slaughter JP, Zhang Z et al. (2015). A general method to improve fluorophores for live-cell and single-molecule microscopy. Nature Methods 12: 244–250. DOI 10.1038/nmeth.3256. [Google Scholar] [CrossRef]
Holden SJ, Pengo T, Meibom KL, Fernandez Fernandez C, Collier J et al. (2014). High throughput 3D super-resolution microscopy reveals Caulobacter crescentus in vivo Z-ring organization. Proceedings of the National Academy of Sciences of the United States of America 111: 4566–4571. DOI 10.1073/pnas.1313368111. [Google Scholar] [CrossRef]
Holtje JV (1998). Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiology and Molecular Biology Reviews 62: 181–203. DOI 10.1128/MMBR.62.1.181-203.1998. [Google Scholar] [CrossRef]
Krupka M, Rowlett VW, Morado D, Vitrac H, Schoenemann K et al. (2017). Escherichia coli FtsA forms lipid-bound minirings that antagonize lateral interactions between FtsZ protofilaments. Nature Communications 8: 15957. DOI 10.1038/ncomms15957. [Google Scholar] [CrossRef]
Kuru E, Hughes HV, Brown PJ, Hall E, Tekkam S et al. (2012). In situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent D-amino acids. Angewandte Chemie International Edition 51: 12519–12523. DOI 10.1002/anie.201206749. [Google Scholar] [CrossRef]
Lee TK, Meng K, Shi H, Huang KC (2016). Single-molecule imaging reveals modulation of cell wall synthesis dynamics in live bacterial cells. Nature Communications 7: 13170. DOI 10.1038/ncomms13170. [Google Scholar] [CrossRef]
Li Y, Shao S, Xu X, Su X, Sun Y et al. (2018). MapZ forms a stable ring structure that acts as a nanotrack for FtsZ treadmilling in streptococcus mutans. ACS Nano 12: 6137–6146. DOI 10.1021/acsnano.8b02469. [Google Scholar] [CrossRef]
Li Z, Trimble MJ, Brun YV, Jensen GJ (2007). The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO Journal 26: 4694–4708. DOI 10.1038/sj.emboj.7601895. [Google Scholar] [CrossRef]
Liu B, Persons L, Lee L, de Boer PA (2015). Roles for both FtsA and the FtsBLQ subcomplex in FtsN-stimulated cell constriction in Escherichia coli. Molecular Microbiology 95: 945–970. DOI 10.1111/mmi.12906. [Google Scholar] [CrossRef]
Loose M, Mitchison TJ (2014). The bacterial cell division proteins FtsA and FtsZ self-organize into dynamic cytoskeletal patterns. Nature Cell Biology 16: 38–46. DOI 10.1038/ncb2885. [Google Scholar] [CrossRef]
Lu C, Reedy M, Erickson HP (2000). Straight and curved conformations of FtsZ are regulated by GTP hydrolysis. Journal of Bacteriology 182: 164–170. DOI 10.1128/JB.182.1.164-170.2000. [Google Scholar] [CrossRef]
Lutkenhaus J, Pichoff S, Du S (2012). Bacterial cytokinesis: From Z ring to divisome. Cytoskeleton 69: 778–790. [Google Scholar]
Lyu Z, Yahashiri A, Yang X, McCausland JW, Kaus GM et al. (2021). FtsN activates septal cell wall synthesis by forming a processive complex with the septum-specific peptidoglycan synthase in E. coli. BioRxiv 1: 16172. DOI 10.1101/2021.08.23.457437. [Google Scholar] [CrossRef]
Marquez I, Diaz-Haro G, Velez M (2019). Surface Orientation and binding strength modulate shape of FtsZ on lipid surfaces. International Journal of Molecular Sciences 20: 2545. DOI 10.3390/ijms20102545. [Google Scholar] [CrossRef]
Mateos-Gil P, Paez A, Horger I, Rivas G, Vicente M et al. (2012). Depolymerization dynamics of individual filaments of bacterial cytoskeletal protein FtsZ. Proceedings of the National Academy of Sciences of the United States of America 109: 8133–8138. DOI 10.1073/pnas.1204844109. [Google Scholar] [CrossRef]
Mateos-Gil P, Tarazona P, Velez M (2019). Bacterial cell division: Modeling FtsZ assembly and force generation from single filament experimental data. FEMS Microbiology Reviews 43: 73–87. DOI 10.1093/femsre/fuy039. [Google Scholar] [CrossRef]
McCausland JW, Yang X, Squyres GR, Lyu Z, Bruce KE et al. (2021). Treadmilling FtsZ polymers drive the directional movement of sPG-synthesis enzymes via a Brownian ratchet mechanism. Nature Communications 12: 609. DOI 10.1038/s41467-020-20873-y. [Google Scholar] [CrossRef]
McQuillen R, Xiao J (2020). Insights into the structure, function, and dynamics of the bacterial cytokinetic FtsZ-ring. Annual Review of Biophysics 49: 309–341. DOI 10.1146/annurev-biophys-121219-081703. [Google Scholar] [CrossRef]
Monteiro JM, Pereira AR, Reichmann NT, Saraiva BM, Fernandes PB et al. (2018). Peptidoglycan synthesis drives an FtsZ-treadmilling-independent step of cytokinesis. Nature 554: 528–532. DOI 10.1038/nature25506. [Google Scholar] [CrossRef]
Moore DA, Whatley ZN, Joshi CP, Osawa M, Erickson HP (2017). Probing for binding regions of the FtsZ protein surface through site-directed insertions: Discovery of fully functional FtsZ-fluorescent proteins. Journal of Bacteriology 199: e00553-16. DOI 10.1128/JB.00553-16. [Google Scholar] [CrossRef]
Nguyen LT, Oikonomou CM, Ding HJ, Kaplan M, Yao Q et al. (2019). Simulations suggest a constrictive force is required for Gram-negative bacterial cell division. Nature Communications 10: 1259. DOI 10.1038/s41467-019-09264-0. [Google Scholar] [CrossRef]
Niu L, Yu J (2008). Investigating intracellular dynamics of FtsZ cytoskeleton with photoactivation single-molecule tracking. Biophysical Journal 95: 2009–2016. DOI 10.1529/biophysj.108.128751. [Google Scholar] [CrossRef]
Nogales E, Downing KH, Amos LA, Lowe J (1998). Tubulin and FtsZ form a distinct family of GTPases. Nature Structural & Molecular Biology 5: 451–458. [Google Scholar]
Osawa M, Anderson DE, Erickson HP (2008). Reconstitution of contractile FtsZ rings in liposomes. Science 320: 792–794 [Google Scholar]
Pasquina-Lemonche L, Burns J, Turner RD, Kumar S, Tank R et al. (2020). The architecture of the Gram-positive bacterial cell wall. Nature 582: 294–297. DOI 10.1038/s41586-020-2236-6. [Google Scholar] [CrossRef]
Perez AJ, Boersma MJ, Bruce KE, Lamanna MM, Shaw SL et al. (2021). Organization of peptidoglycan synthesis in nodes and separate rings at different stages of cell division of Streptococcus pneumoniae. Molecular Microbiology 115: 1152–1169. DOI 10.1111/mmi.14659. [Google Scholar] [CrossRef]
Perez AJ, Cesbron Y, Shaw SL, Bazan Villicana J, Tsui HT et al. (2019). Movement dynamics of divisome proteins and PBP2x: FtsW in cells of Streptococcus pneumoniae. Proceedings of the National Academy of Sciences 116: 3211–3220. DOI 10.1073/pnas.1816018116. [Google Scholar] [CrossRef]
Pradhan P, Margolin W, Beuria TK (2021). Targeting the achilles heel of FtsZ: The interdomain cleft. Frontiers in Microbiology 12: 732796. DOI 10.3389/fmicb.2021.732796. [Google Scholar] [CrossRef]
Ramirez-Diaz DA, Garcia-Soriano DA, Raso A, Mucksch J, Feingold M et al. (2018). Treadmilling analysis reveals new insights into dynamic FtsZ ring architecture. PLoS Biology 16: e2004845. DOI 10.1371/journal.pbio.2004845. [Google Scholar] [CrossRef]
Ramirez-Diaz DA, Merino-Salomon A, Meyer F, Heymann M, Rivas G et al. (2021). FtsZ induces membrane deformations via torsional stress upon GTP hydrolysis. Nature Communications 12: 3310. DOI 10.1038/s41467-021-23387-3. [Google Scholar] [CrossRef]
Rowlett VW, Margolin W (2014). 3D-SIM super-resolution of FtsZ and its membrane tethers in Escherichia coli cells. Biophysical Journal 107: L17–L20. DOI 10.1016/j.bpj.2014.08.024. [Google Scholar] [CrossRef]
Schmidt R, Weihs T, Wurm CA, Jansen I, Rehman J et al. (2021). MINFLUX nanometer-scale 3D imaging and microsecond-range tracking on a common fluorescence microscope. Nature Communications 12: 1478. DOI 10.1038/s41467-021-21652-z. [Google Scholar] [CrossRef]
Soderstrom B, Badrutdinov A, Chan H, Skoglund U (2018). Cell shape-independent FtsZ dynamics in synthetically remodeled bacterial cells. Nature Communications 9: 4323. DOI 10.1038/s41467-018-06887-7. [Google Scholar] [CrossRef]
Soderstrom B, Skoog K, Blom H, Weiss DS, von Heijne G et al. (2014). Disassembly of the divisome in Escherichia coli: evidence that FtsZ dissociates before compartmentalization. Molecular Microbiology 92: 1–9. DOI 10.1111/mmi.12534. [Google Scholar] [CrossRef]
Squyres GR, Holmes MJ, Barger SR, Pennycook BR, Ryan J et al. (2021). Single-molecule imaging reveals that Z-ring condensation is essential for cell division in Bacillus subtilis. Nature Microbiology 6: 553–562. DOI 10.1038/s41564-021-00878-z. [Google Scholar] [CrossRef]
Strauss MP, Liew AT, Turnbull L, Whitchurch CB, Monahan LG et al. (2012). 3D-SIM super resolution microscopy reveals a bead-like arrangement for FtsZ and the division machinery: implications for triggering cytokinesis. PLoS Biology 10: e1001389. DOI 10.1371/journal.pbio.1001389. [Google Scholar] [CrossRef]
Stricker J, Erickson HP (2003). In vivo characterization of Escherichia coli ftsZ mutants: Effects on Z-ring structure and function. Journal of Bacteriology 185: 4796–4805. DOI 10.1128/JB.185.16.4796-4805.2003. [Google Scholar] [CrossRef]
Stricker J, Maddox P, Salmon ED, Erickson HP (2002). Rapid assembly dynamics of the Escherichia coli FtsZ-ring demonstrated by fluorescence recovery after photobleaching. Proceedings of the National Academy of Sciences 99: 3171–3175. DOI 10.1073/pnas.052595099. [Google Scholar] [CrossRef]
Sundararajan K, Goley ED (2017). The intrinsically disordered C-terminal linker of FtsZ regulates protofilament dynamics and superstructure in vitro. Journal of Biological Chemistry 292: 20509–20527. DOI 10.1074/jbc.M117.809939. [Google Scholar] [CrossRef]
Sundararajan K, Miguel A, Desmarais SM, Meier EL, Casey Huang K et al. (2015). The bacterial tubulin FtsZ requires its intrinsically disordered linker to direct robust cell wall construction. Nature Communications 6: 7281. DOI 10.1038/ncomms8281. [Google Scholar] [CrossRef]
Szwedziak P, Wang Q, Bharat TA, Tsim M, Lowe J (2014). Architecture of the ring formed by the tubulin homologue FtsZ in bacterial cell division. Elife 3: e04601. DOI 10.7554/eLife.04601. [Google Scholar] [CrossRef]
Thanedar S, Margolin W (2004). FtsZ exhibits rapid movement and oscillation waves in helix-like patterns in Escherichia coli. Current Biology 14: 1167–1173. DOI 10.1016/j.cub.2004.06.048. [Google Scholar] [CrossRef]
Tsang MJ, Bernhardt TG (2015). A role for the FtsQLB complex in cytokinetic ring activation revealed by an FtsL allele that accelerates division. Molecular Microbiology 95: 925–944. DOI 10.1111/mmi.12905. [Google Scholar] [CrossRef]
Turner RD, Mesnage S, Hobbs JK, Foster SJ (2018). Molecular imaging of glycan chains couples cell-wall polysaccharide architecture to bacterial cell morphology. Nature Communications 9: 1263. DOI 10.1038/s41467-018-03551-y. [Google Scholar] [CrossRef]
Ur Rahman M, Wang P, Wang N, Chen Y (2020). A key bacterial cytoskeletal cell division protein FtsZ as a novel therapeutic antibacterial drug target. Bosnian Journal of Basic Medical Sciences 20: 310–318. [Google Scholar]
van Teeffelen S, Wang S, Furchtgott L, Huang KC, Wingreen NS et al. (2011). The bacterial actin MreB rotates, and rotation depends on cell-wall assembly. Proceedings of the National Academy of Sciences 108: 15822–15827. DOI 10.1073/pnas.1108999108. [Google Scholar] [CrossRef]
Walker BE, Mannik J, Mannik J (2020). Transient membrane-linked FtsZ assemblies precede Z-Ring formation in Escherichia coli. Current Biology 30: 499–508 e496. DOI 10.1016/j.cub.2019.12.023. [Google Scholar] [CrossRef]
Wang Y, Meng F, Sachs F (2011). Genetically encoded force sensors for measuring mechanical forces in proteins. Communicative & Integrative Biology 4: 385–390. DOI 10.4161/cib.15505. [Google Scholar] [CrossRef]
Whitley KD, Jukes C, Tregidgo N, Karinou E, Almada P et al. (2021). FtsZ treadmilling is essential for Z-ring condensation and septal constriction initiation in Bacillus subtilis cell division. Nature Communications 12: 2448. DOI 10.1038/s41467-021-22526-0. [Google Scholar] [CrossRef]
Yang X, Lyu Z, Miguel A, McQuillen R, Huang KC et al. (2017). GTPase activity-coupled treadmilling of the bacterial tubulin FtsZ organizes septal cell wall synthesis. Science 355: 744–747. DOI 10.1126/science.aak9995. [Google Scholar] [CrossRef]
Yang X, McQuillen R, Lyu Z, Phillips-Mason P, de La Cruz A et al. (2021). A two-track model for the spatiotemporal coordination of bacterial septal cell wall synthesis revealed by single-molecule imaging of FtsW. Nature Microbiology 6: 584–593. DOI 10.1038/s41564-020-00853-0. [Google Scholar] [CrossRef]
Yao Q, Jewett AI, Chang YW, Oikonomou CM, Beeby M et al. (2017). Short FtsZ filaments can drive asymmetric cell envelope constriction at the onset of bacterial cytokinesis. EMBO Journal 36: 1577–1589. DOI 10.15252/embj.201696235. [Google Scholar] [CrossRef]
Yu J, Xiao J, Ren X, Lao K, Xie XS (2006). Probing gene expression in live cells, one protein molecule at a time. Science 311: 1600–1603. DOI 10.1126/science.1119623. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |