DOI:10.32604/biocell.2022.021008

| BIOCELL DOI:10.32604/biocell.2022.021008 |  |
| Article |
Human cytomegalovirus autophagy is related to the interferon synthesis and mTOR signal pathway
1Department of Clinical Laboratory, Third Affiliated Hospital of Anhui Medical University, Hefei, 230032, China
2Department of Gastroenterology, Sanmen People’s Hospital, Taizhou, 317100, China
3Department of Microbiology, Anhui Medical University, Hefei, 230032, China
*Address correspondence to: Hongzhang Li, smyylhz@163.com; Jun Zhao, anhuiimmune@aliyun.com
#Dongmei Gao and Jiaoe Chen contributed equally to this work and should be considered as co-first authors
Received: 23 December 2021; Accepted: 18 February 2022
Abstract: Introduction: Human cytomegalovirus (HCMV) is reported to be involved in the occurrence of many human diseases. To further investigate the biological changes of HCMV, we analyzed the relevant factors that affect the autophagy caused by HCMV infection. Methods: Firstly, we cultured human embryonic lung fibroblasts (HELF) cells with HCMV infection, and evaluated the effects of HELF cells infected with different viruses through Enzyme-linked immunoabsorbent assay (ELISA), Real-time quantitative Polymerase Chain Reaction (RT-qPCR), Acridine orange (AO) staining and Western blotting (WB) experiments. Results: Through the above experiments, we found that the combined treatment of HCMV infection and carbamazepine, rapamycin and si-mTOR promoted the increase of interferon (IFN)-α/β expression and protein level, and caused the increase of LC3B protein level in HELF cells. In addition, HCMV infection could also affect the biological activities of HELF cells by regulating signal pathways like the JAK/STAT. Conclusion: Autophagy induced by HCMV is affected by the changes in the biological behavior of HELF cells, especially IFN-α/β synthesis and mTOR signal pathway. These findings might shed new light on HCMV-related disease treatment.
Keywords: HCMV; HELF cell; mTOR; IFN-α; IFN-β
Viruses must depend on host cell systems to conduct activities required for viral reproduction since they are intracellular parasites with limited genetic resources (König et al., 2010). Although host cell defense mechanism can inactivate most hijacked processes, viruses have devised strategies to keep certain cellular processes functioning, and can also destroy them for their objectives. Human cytomegalovirus (HCMV) is a herpes virus, widely existing among individuals with low immunity, such as bone marrow transplant recipients and hematological malignancy patients (Gilbert and Boivin, 2005). It has been clinically proven that HCMV can be infected through breastfeeding, kissing, sexual contact, blood transfusion, etc., causing symptoms like cytomegalovirus hepatitis, cytomegalovirus encephalitis, cytomegalovirus retinitis (Ray and Bala, 2013). The prevalence of HCMV ranges from 40% to 100%, resulting in lifelong asymptomatic infection in most people. HCMV has become a major public health burden in the world.
Macroautophagy (alias autophagy) refers to a homeostatic process, in which cytoplasmic components are digested and removed by the lysosomal pathway (Lee, 2009). This mechanism seems to be constitutively active in animal cells, and it is blocked or increased in response to a range of factors, nutritional status, specific hormones and intracellular signaling pathways included (He and Klionsky, 2009). Current studies have pointed out that there is a certain correlation between autophagy and viral infection. Mao et al. (2019) proposed that autophagy control of viral infection was a multi-faceted process, which could not only destroy the virus and regulate the inflammatory response, but also enhance its biological activity. The study of Abdoli et al. (2018) exemplified the interaction between influenza virus and autophagy, and found NS1 and M2 viral proteins were related to autophagy signals, which could maintain autophagy in a steady state. Moreover, autophagy also has a protective effect on hepatitis C virus, Huntington’s disease and Parkinson’s disease, etc. Based on these, autophagy might bring a fresh perspective for the therapies of viral infection and autoimmune diseases. Studies have shown that HCMV infection can promote autophagy in THP-1 cells, and the level of autophagy may be reduced during the latent period (Liu et al., 2017). However, the effect of HCMV on autophagy in HELF cells remains to be further explored.
Mammalian target of rapamycin (mTOR) is a serine/threonine kinase containing two complexes (mTORC1 and mTORC2) which are important regulators of cell growth and proliferation. For example, some studies mention that mTORC1 is the main regulator of cell metabolism and autophagy. The mutual regulation of mTORC1 and AMPK can trigger the autophagy of phosphorylated complexes, thereby changing changes in cell energy levels. Besides, the combination of mTOR and AMPK can damage lysosomes and induce autophagy in cells (Yang et al., 2020). Altman et al. (2019) studied the specific role of the PI3K/Akt/mTOR axis in different stages of HCMV, and found it not only met the metabolic requirements during cell lysis and infection, but also changed cell transcription and maintained monocyte survival. A large number of studies have demonstrated the mTOR signaling pathway acts a non-negligible role in material metabolism, cell apoptosis, autophagy and disease development (Yu et al., 2021).
IFN-α and IFN/β are members of the interferons (IFNs) family, and also a set of signal proteins synthesized and released by host cells in response to pathogens. Among them, IFN-α protein is produced by white blood cells and is mainly involved in innate immunity in response to viral infections (Heim, 2013). IFN-β plays an important role in inducing non-specific antiviral infection, and is related to cell proliferation and immune response regulation (Fujita et al., 1989). There is research showing IFN-α/β may be related to the cell process of HCMV infection. For example, Sainz et al. (2005) through viral replication, RT-PCR, immunofluorescence and other experiments proved that the combined action of IFN-α/β and IFN-γ could effectively inhibit the replication of HCMV. Pautasso et al. (2018) mentioned that the apolipoprotein B editing enzyme catalytic subunit 3 (APOBEC3) protein family was related to the defense mechanism against viral infections through deamination. DeFilippis et al. (2010) pointed out that Z-DNA binding protein 1 (ZBP1) was essential in the process of HCMV activation of interferon regulatory factor 3 (IRF3) and transcription of INF-β, and this process also depended on JAK/STAT signal transduction. Moreover, the report by Miller et al. (1998) explained that the JAK/STAT pathway stimulated by IFN-γ was related to the up-regulation of class II transactivator and major histocompatibility complex class II (MHC II) transcriptional activation, thereby affecting the cellular immune detection of HCMV. In this experiment, we explored the effects of carbamazepine, rapamycin and si-mTOR on HCMV-infected cells, the level of interferon (IFN)-α/β, and the expression of signal pathways in the human embryonic lung fibroblast (HELF) autophagy.
Cell culture and virus infection
For this experiment, we purchased HELF from Wuhan Procell Life Technology Co., Ltd. After that, the cells were cultured in Dulbecco’s Modified Eagle Media (DMEM, Life Technologies, Grand Island, NY) medium containing 10% fetal bovine serum (FBS), and maintained at 37°C. Finally, the HCMV laboratory standard strain AD169 (Wuhan Research Institute, Chinese Academy of Sciences) was used for virus infection.
The 3-Methyladenine (3-MA) (autophagy inhibitor), carbamazepine (autophagy inducer), rapamycin (RAPA, mTOR inhibitor) and 3BDO (mTOR activator) used in this experiment were all from Sigma-Aldrich Corporation (St. Louis, MO, USA). Antibodies against IFN-α/β, STAT1/STAT2/JAK1/Tyk2, p-STAT1/p-STAT2/p-JAK1/p-Tyk2 and LC3B (autophagy marker protein) were collected from Thermo Fisher Scientific, TRIzol reagent from Invitrogen and SYBR-Green Master Mix from Roche Company. Three siRNAs targeting mTOR were purchased from Genechem Co., Ltd. (Shanghai). According to the manufacturer’s instructions, we use Lipofectamine® 3000 reagent (Invitrogen, Carlsbad, CA, USA) to transfect HELF cells with si-mTOR. si-RNA sequences are as follow: mTOR si1, 5’-CCACCCGAAUUGGCAGAUUTT-3’; mTOR si2, 5’- GCAUCCAGCAGGAUAUCAATT-3’; mTOR si3, 5’- CCAAGAUACCAUGAACCAUTT-3’; si-NC, 5’-UUCUCCGAACGUGUCACGUTT-3’.
ELISA is an immunoassay method for detecting and quantifying biomolecules, such as antibodies, proteins, hormones or peptides, as well as characterizing protein-protein and protein-nucleic acid interactions with a microplate reader (Borges et al., 2022). Herein, following the manufacturer’s instructions, we used an ELISA kit (Wuhan Genemei Technology Co., Ltd.) to detect the concentration of IFN-α and IFN-β in HCMV-treated HELF (including 6 types in total), and then put them in each well plate for observation.
The total RNA was separated and removed from HELF cells using TRIzol reagent (Invitrogen, USA). Then, RNA was converted into complementary DNA (cDNA). RT-PCR amplification was carried out with SYBR Green master mix (BioRed, USA) with the following primers. The specific primer sequences used were mTOR: Forward, 5’-GGCCAATGACCCAACATCTC-3’ and Reverse, 5’-CATGATGCGATGCTCGATGT-3’; IFN-α: Forward, 5’-CTGCAAGTCAAGCTGCTCTC-3’ and Reverse, 5’-CATTTGTGCCAGGAGCATCA-3’; IFN-β: Forward, 5’-GCTTGGATTCCTACAAAGAAGCA-3’ and Reverse, 5’-ATAGATGGTCAATGCGGCGTC-3’; β-actin: Forward, 5’-CCCTGGAGAAGAGCTACGAG-3’ and Reverse, 5’-GGAAGGAAGGCTGGAAGAGT-3’. This method could detect the relative levels of IFN-α, IFN-β and mTOR, and set the conditions for the 30 s at 95°C (one time), then 95°C for 15 s, 60°C for 30 s, 40 cycles in total. β-actin was the control gene to normalize target gene expression. The final result was analyzed using the 2–ΔΔCT method.
Acridine orange (AO) staining assay
AO is a commonly used fluorescent dye, which can display different colors of fluorescence after combining with DNA and RNA in primary cells. It is often used in experiments as fluorescent indicators, tumor cells, nucleic acid stains, and cell autophagy detection (Lin et al., 2017). In 6-well plates, HELF cells were kept the whole night. Being washed three times with PBS later, cells were treated with 2 mg/ml of AO for 15 minutes at 37°C. Then, cell was rinsed by PBS three times and immediately inspected under a fluorescence microscope (excitation, 488 nm).
HELF cells after treatment were resolved in RIPA buffer supplemented with protease and phosphatase inhibitors. To separate all the protein, sodium dodecyl sulfate-polyacrylamide gel electrophoresis was utilized, and the separated protein was subsequently filtered on nitrocellulose membrane. Iry antibodies were then used to incubate the membrane.
This time, we followed the WB experimental procedure to detect the protein expression levels of IFN-α/β, and then detected the levels of LC3B protein in the 6 groups of infected HELF cells. Finally, the IFNAR1, JAK1, Tyk2, STAT1, STAT2, p-JAK1, p-IFNAR1, p-Tyk2, p-STAT1 and p-STAT2 signaling pathways were detected and analyzed. All of the above operations used ImageJ software to analyze the protein expression level by density and normalize it to β-actin.
The data obtained in all the above experiments were expressed by the mean ± standard deviation. Each experiment was repeated at least three times. The results were processed by one-way analysis of variance, and difference analysis by two-tailed Student’s t-test through the GraphPad Prism 8 software (GraphPad Software, Inc., San Diego, CA, USA). When P < 0.05, the outcomes of this experiment were statistically significant.
mTOR inhibits the expression of IFN-α/β in HCMV-infected HELF cells
As shown in Fig. 1A, after HCMV-infected HELF cells were transfected with siRNA targeting mTOR, the expression of mTOR was significantly reduced, and mTOR si3 was the most significant one. Later, we used HELF cells transfected with HCMV as a control, and found that IFN-α/β expression was higher in HELF cells treated with HCMV + carbamazepine, HCMV + RAPA and HCMV + si-mTOR (Figs. 1B and 1C). Not only that, the WB experiment also showed the copy number and expression of IFN-α/β mRNA (Fig. 1D). From the results of the ELISA experiment, we could see that compared with HELF cells infected with HCMV, the concentrations of IFN-α/β in HELF cells infected with HCMV + carbamazepine, HCMV + RAPA, HCMV + si-mTOR were obviously higher. On the contrary, the contents of IFN-α/β in HCMV+3-MA and HCMV+3BDO cells were relatively lower (Figs. 2A and 2B). IFN-α/β increased significantly after knocking down m-TOR in HELF cells infected by HCMV.

Figure 1: Autophagy and infection of HCMV cells. (A) The relative expression of mTOR si-RNA in HELF. (B) The relative expression of IFN-α in six different groups of HELF cells. (C) The relative expression of IFN-β in six different groups of HELF cells. (D) IFN-α/β expression detected by WB. **P < 0.01, ***P < 0.001, ****P < 0.0001.

Figure 2: ELISA test. The effects of six groups of different HCMV on the expression level of IFN-α/β in HELF cells under the treatment of autophagy regulators. (A) IFN-α. (B) IFN-β. ****P < 0.0001.
mTOR inhibits autophagy in HCMV-infected HELF cells
By AO staining on 6 different groups of HELF cells, we could find that HELF cells of HCMV + carbamazepine, HCMV + RAPA, HCMV + si-mTOR had autophagy, HELF autophagy in HCMV + 3-MA, HCMV + 3BDO and HCMV infection groups was not obvious (Fig. 3A). WB analysis found that HELF cells with HCMV + carbamazepine, HCMV + RAPA, HCMV + si-mTOR contained more LC3B protein, which promoted the level of autophagy (Fig. 3B). The above results indicated that mTOR played an important role in inhibiting autophagy in HELF cells infected by HCMV.

Figure 3: AO staining experiment. (A) Fluorescence dot-like aggregation of 6 groups of cells. (B) The expression level of LC3B protein in 6 groups of cells based on WB detection.
3-MA and 3BDO inhibit phosphorylation of JAK/STAT in HELF cell line after HCMV infection
WB analysis of different treatment groups in the HELF cells after HCMV infection showed that the total protein expressions of TYK2, JAK1, STAT1, STAT2 and IFNAR1 were not significantly different (Fig. 4). However, the phosphorylation status of these proteins found that HCMV + 3-MA and HCMV + 3BDO were significantly down-regulated, while other treatment groups and control groups were up-regulated (Fig. 4).
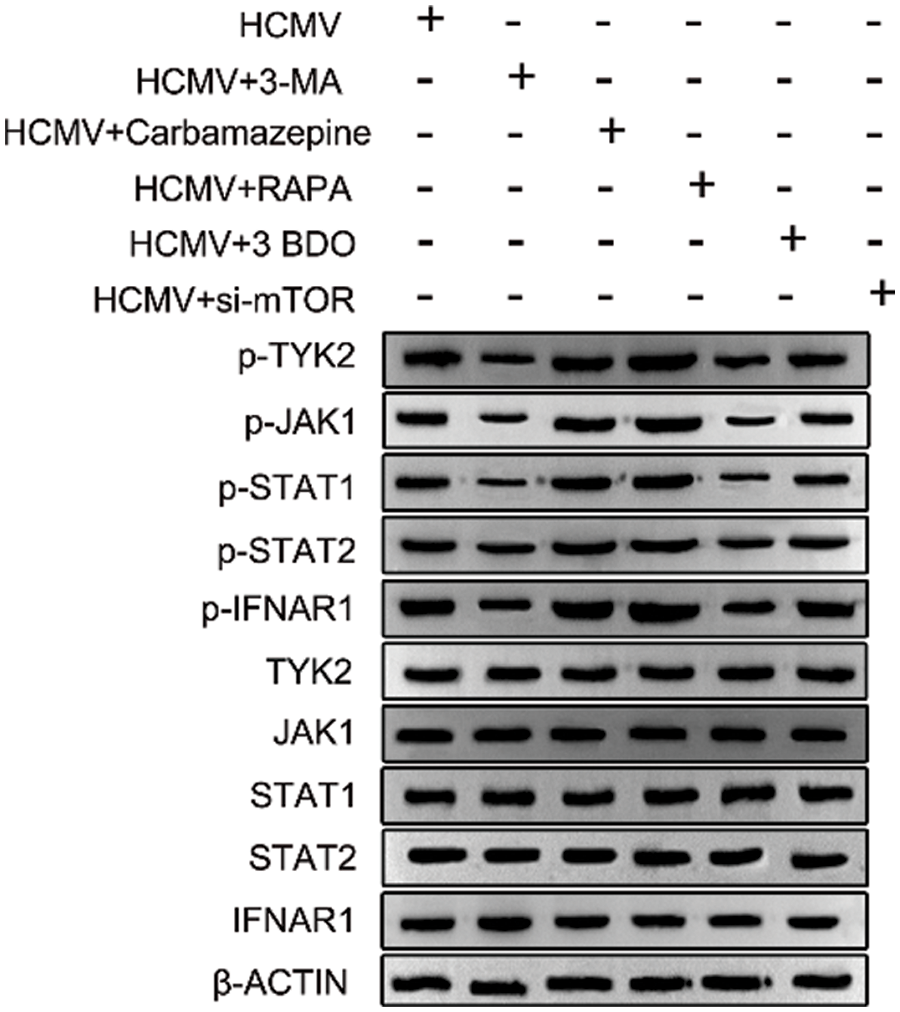
Figure 4: WB analysis. The expression levels of key signal pathways (IFNAR1, p-IFNAR1, JAK1, p-JAK1, TYK2, p-TYK2, STAT1, p-STAT1, STAT1, STAT2, p-STAT2) in 6 groups of cells were detected by the WB method.
HCMV is a virus that is widely infected among humans, especially the immunodeficiency group (Cappuyns et al., 2005). Surveys show that in developed countries, the positive rate of HCMV antibodies in adults is about 50%, and 10% to 15% of children are infected with HCMV before the age of 5, and after 5 years of age, it drops significantly. HCMV can spread through saliva, cervical secretions, urine and so on (Cannon et al., 2014; Ekema et al., 2006). For example, pregnant women infected with the HCMV virus can cause miscarriage, intrauterine growth retardation and congenital malformations of the fetus. In addition, studies have shown that HCMV can be combined with IFITM3rs12252 to be highly expressed in the brain of Rasmussen’s encephalitis (RE) patients, and IFITM3rs12252-C can promote RE development by promoting the continuous infection of HCMV in the brain tissues (Wang et al., 2021). At present, the infection of HCMV to the host cell will cause a variety of diseases, and the signal pathways regulating cell behavior after HCMV infection remain to be discovered.
Herein, we analyzed the effects of HELF cell autophagy and mTOR signaling pathway on the synthesis of IFN-α/β interferon and the expressions of JAK/STAT signaling pathway during HCMV infection. Specifically, after knocking down the expression of mTOR in HELF cells, we treated HELF cells with autophagy inhibitor (3-MA), autophagy inducer (carbamazepine), mTOR inhibitor (rapamycin), mTOR inducer (3BDO) and HCMV combination. The PCR and ELISA results demonstrated the levels of IFN-α/β interferon in the carbamazepine, RAPA and si-mTOR groups were markedly higher, compared to those of HCMV + 3-MA and HCMV + 3BDO groups. In addition, the results of AO staining and WB also showed that in the autophagy of HELF cells infected with HCMV, the level of LC3B protein elevated, and the protein level of P-TYK2/p-JAK1/p-STAT1/p-STAT2/p-IFNAR1 signaling pathway was also up-regulated. Interestingly, we detected the total protein expression of TYK2/JAK1/STAT1/STAT2/IFNAR1 by WB, and found there was no significant difference. These data do not exclude the role of TYK2/JAK1/STAT1/STAT2/IFNAR1 in regulating the autophagy in HELF cells infected by HCMV, but these proteins need to be activated after phosphorylation to participate in regulating autophagy.
At present, various studies have shown that there is a certain correlation between mTOR and HCMV, that is, the activation and replication of HCMV virus depend on the mTOR pathway. For example, Clippinger et al. (2011) pointed out that mTORC1 and mTORC2 were activated during HCMV infection, and mTOR kinase could stably synthesize protein during this process. Poglitsch et al. (2012) used the HCMV infection model of human macrophages and found that mTOR activation could be observed in the late stage of infection, and the late viral proteins Pul-44 and pp 65 were produced, indicating that mTOR activation was necessary for virus replication.
Besides, our research also showed that a variety of signaling pathways were also involved in the expression of HELF cells in the autophagy process, and combined with HCMV infection to promote cell autophagy, which was confirmed by many studies. Hu et al. (2021) found that HCMV infection triggers a stress response in glioma cells and activates the survival factor ATF5 in tumor cells by downregulating specific miRNAs. Huang et al. (2021) pointed out that HCMV targets Wnt and Notch signaling pathways to cause hearing loss. In order to study the effects of HCMV on primary human hepatocytes (PHH) and HepG2 cells, Lepiller et al. (2013) conducted ELISA, WB and colony formation experiments, and found that HCMV infected HepG2 and PHH cells to produce IL-6. The 6R-JAK-STAT3 pathway triggered cell proliferation, induced PHH transformation and HepG2 tumor formation. Baron and Davignon (2008) pointed out that the JAK/STAT pathway was regulated by the phosphorylation step, which led to the nuclear translocation of tyrosine phosphorylated STAT1 (STAT1-P-Tyr). In addition, SHP2 as a phosphatase is also involved in the regulation of IFN-γ-mediated tyrosine phosphorylation. The pertinent data show that SHP2 activation caused by HCMV infection is associated with IFN-γ-induced down-regulation of STAT1 tyrosine phosphorylation. On the basis of these current studies, we can determine that HCMV infection can affect autophagy of diseased cells by activating signal pathways like mTOR/TAK/STAT, participating in the biological process of cells, and enabling subsequent disease target analysis.
Several questions need to be addressed in future studies. First, the experimental results of this study need to be further verified in vivo; secondly, the regulatory mechanism of autophagy on apoptosis needs to be further explored. In future research, we will conduct the above exploration.
All in all, we have studied the regulatory mechanism of autophagy in HELF cells during HCMV infection, and found that the mTOR autophagy signaling pathway was closely related to the infection of HCMV. During HCMV infection, autophagy inducer (carbamazepine), mTOR inhibitor (rapamycin) and si-mTOR may promote HELF cell autophagy through JAK/STAT signaling pathway. These findings might shed new light on the HCMV-related disease treatment.
Availability of Data and Materials: The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Author Contribution: The authors confirm contribution to the paper as follows: study conception and design Hongzhang Li, Jun Zhao; data collection: Dongmei Gao; analysis and interpretation of results: Jiaoe Chen; draft manuscript preparation: Dongmei Gao, Jiaoe Chen. Dongmei Gao and Jiaoe Chen contributed equally to this work and should be considered as co-first authors. All authors reviewed the results and approved the final version of the manuscript.
Funding Statement: This study was supported by Basic Public Welfare Research Project of Zhejiang Province (LY19C010006), and Research Programs of Zhejiang Provincial Medicine and Health Science and Technology Plan Project (2020382731), and Natural Science Fund of Anhui Province (Grant No. 1808085MC75), and Project of Sanmen County Science and Technology Planning (No. 16310).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Abdoli A, Alirezaei M, Mehrbod P, Forouzanfar F (2018). Autophagy: The multi-purpose bridge in viral infections and host cells. Reviews in Medical Virology 28: e1973. DOI 10.1002/rmv.1973. [Google Scholar] [CrossRef]
Altman AM, Mahmud J, Nikolovska-Coleska Z, Chan G (2019). HCMV modulation of cellular PI3K/AKT/mTOR signaling: New opportunities for therapeutic intervention? Antiviral Research 163: 82–90. DOI 10.1016/j.antiviral.2019.01.009. [Google Scholar] [CrossRef]
Baron M, Davignon JL (2008). Inhibition of IFN-γ-induced STAT1 tyrosine phosphorylation by human CMV is mediated by SHP2. Journal of Immunology 181: 5530–5536. DOI 10.4049/jimmunol.181.8.5530. [Google Scholar] [CrossRef]
Borges A, Onasenko I, Nag A (2022). Binding characterization of cyclic peptide ligands to target proteins and chemical epitopes using ELISA and Fluorescence Polarization Assays. In: Peptide Macrocycles, pp. 335–354. New York: Springer. [Google Scholar]
Cannon MJ, Stowell JD, Clark R, Dollard PR, Johnson D, Mask K, Stover C, Wu K, Amin M, Hendley W (2014). Repeated measures study of weekly and daily cytomegalovirus shedding patterns in saliva and urine of healthy cytomegalovirus-seropositive children. BMC Infectious Diseases 14: 1–10. DOI 10.1186/s12879-014-0569-1. [Google Scholar] [CrossRef]
Cappuyns I, Gugerli P, Mombelli A (2005). Viruses in periodontal disease—A review. Oral Diseases 11: 219–229. DOI 10.1111/j.1601-0825.2005.01123.x. [Google Scholar] [CrossRef]
Clippinger AJ, Maguire TG, Alwine JC (2011). The changing role of mTOR kinase in the maintenance of protein synthesis during human cytomegalovirus infection. Journal of Virology 85: 3930–3939. DOI 10.1128/JVI.01913-10. [Google Scholar] [CrossRef]
DeFilippis VR, Alvarado D, Sali T, Rothenburg S, Früh K (2010). Human cytomegalovirus induces the interferon response via the DNA sensor ZBP1. Journal of Virology 84: 585–598. DOI 10.1128/JVI.01748-09. [Google Scholar] [CrossRef]
Ekema G, Pedersini P, Milianti S, Ubertazzi M, Minoli D, Manciana A (2006). Colonic stricture mimicking Hirschsprung’s disease: A localized cytomegalovirus infection. Journal of Pediatric Surgery 41: 850–852. DOI 10.1016/j.jpedsurg.2005.12.029. [Google Scholar] [CrossRef]
Fujita T, Kimura Y, Miyamoto M, Barsoumian EL, Taniguchi T (1989). Induction of endogenous IFN-α and IFN-β genes by a regulatory transcription factor, IRF-1. Nature 337: 270–272. DOI 10.1038/337270a0. [Google Scholar] [CrossRef]
Gilbert C, Boivin G (2005). Human cytomegalovirus resistance to antiviral drugs. Antimicrobial Agents and Chemotherapy 49: 873–883. DOI 10.1128/AAC.49.3.873-883.2005. [Google Scholar] [CrossRef]
He C, Klionsky DJ (2009). Regulation mechanisms and signaling pathways of autophagy. Annual Review of Genetics 43: 67–93. DOI 10.1146/annurev-genet-102808-114910. [Google Scholar] [CrossRef]
Heim MH (2013). Innate immunity and HCV. Journal of Hepatology 58: 564–574. DOI 10.1016/j.jhep.2012.10.005. [Google Scholar] [CrossRef]
Hu M, Yu B, Zhang B, Wang B, Qian D, Li H, Ma J, Liu DX (2021). Human cytomegalovirus infection activates glioma activating transcription factor 5 via microRNA in a stress-induced manner. ACS Chemical Neuroscience 12: 3947–3956. DOI 10.1021/acschemneuro.1c00576. [Google Scholar] [CrossRef]
Huang SN, Zhou YP, Jiang X, Yang B, Cheng H, Luo MH (2021). Hearing loss caused by HCMV infection through regulating the Wnt and notch signaling pathways. Viruses 13: 623. DOI 10.3390/v13040623. [Google Scholar] [CrossRef]
König R, Stertz S, Zhou Y, Inoue A, Hoffmann HH, Bhattacharyya S, Alamares JG, Tscherne DM, Ortigoza MB, Liang Y (2010). Human host factors required for influenza virus replication. Nature 463: 813–817. DOI 10.1038/nature08699. [Google Scholar] [CrossRef]
Lee JA (2009). Autophagy in neurodegeneration: Two sides of the same coin. BMB Reports 42: 324–330. DOI 10.5483/BMBRep.2009.42.6.324. [Google Scholar] [CrossRef]
Lepiller Q, Abbas W, Kumar A, Tripathy MK, Herbein G (2013). HCMV activates the IL-6-JAK-STAT3 axis in HepG2 cells and primary human hepatocytes. PLoS One 8: e59591. DOI 10.1371/journal.pone.0059591. [Google Scholar] [CrossRef]
Lin YC, Lin JF, Tsai TF, Chen HE, Chou KY, Yang SC, Tang YM, Thomas I, Hwang S (2017). Acridine orange exhibits photodamage in human bladder cancer cells under blue light exposure. Scientific Reports 7: 1–11. DOI 10.1038/s41598-017-13904-0. [Google Scholar] [CrossRef]
Liu Y, Pan J, Liu L, Li W, Tao R et al. (2017). The influence of HCMV infection on autophagy in THP-1 cells. Medicine 96: e8298. [Google Scholar]
Mao J, Lin E, He L, Yu J, Tan P, Zhou Y (2019). Autophagy and viral infection. Autophagy Regulation of Innate Immunity 1209: 55–78. DOI 10.1007/978-981-15-0606-2. [Google Scholar] [CrossRef]
Miller DM, Rahill BM, Boss JM, Lairmore MD, Durbin JE, Waldman JW, Sedmak DD (1998). Human cytomegalovirus inhibits major histocompatibility complex class II expression by disruption of the Jak/Stat pathway. Journal of Experimental Medicine 187: 675–683. DOI 10.1084/jem.187.5.675. [Google Scholar] [CrossRef]
Pautasso S, Galitska G, Dell’Oste V, Biolatti M, Cagliani R, Forni D, de Andrea M, Gariglio M, Sironi M, Landolfo S (2018). Strategy of human cytomegalovirus to escape interferon beta-induced APOBEC3G editing activity. Journal of Virology 92: e01224–01218. DOI 10.1128/JVI.01224-18. [Google Scholar] [CrossRef]
Poglitsch M, Weichhart T, Hecking M, Werzowa J, Katholnig K, Antlanger M, Krmpotic A, Jonjic S, Hörl W, Zlabinger G (2012). CMV late phase-induced mTOR activation is essential for efficient virus replication in polarized human macrophages. American Journal of Transplantation 12: 1458–1468. DOI 10.1111/j.1600-6143.2012.04002.x. [Google Scholar] [CrossRef]
Ray K, Bala M (2013). Human cytomegalovirus infection. In: Sexually Transmitted Infections-E-Book. [Google Scholar]
Sainz B, LaMarca HL, Garry RF, Morris CA (2005). Synergistic inhibition of human cytomegalovirus replication by interferon-alpha/beta and interferon-gamma. Virology Journal 2: 1–13. DOI 10.1186/1743-422X-2-1. [Google Scholar] [CrossRef]
Wang YS, Luo QL, Guan YG, Fan DY, Luan GM, Jing A (2021). HCMV infection and IFITM3 rs12252 are associated with Rasmussen’s encephalitis disease progression. Annals of Clinical and Translational Neurology 8: 558–570. DOI 10.1002/acn3.51289. [Google Scholar] [CrossRef]
Yang Y, Wang Q, Song D, Zen R, Zhang L, Wang Y, Yang H, Zhang D, Jia J, Zhang J (2020). Lysosomal dysfunction and autophagy blockade contribute to autophagy-related cancer suppressing peptide-induced cytotoxic death of cervical cancer cells through the AMPK/mTOR pathway. Journal of Experimental & Clinical Cancer Research 39: 1–18. DOI 10.1186/s13046-020-01701-z. [Google Scholar] [CrossRef]
Yu F, Ma R, Liu C, Zhang L, Feng K, Wang M, Yin D (2021). SQSTM1/p62 promotes cell growth and triggers autophagy in papillary thyroid cancer by regulating the AKT/AMPK/mTOR signaling pathway. Frontiers in Oncology 11: 1015. DOI 10.3389/fonc.2021.638701. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |