DOI:10.32604/biocell.2021.015652

www.techscience.com/journal/biocell
| BIOCELL DOI:10.32604/biocell.2021.015652 |  www.techscience.com/journal/biocell |
| Review |
Advances in molecular regulation of goat lipid metabolism and FAS structure and function regulation
1School of Nursing, Yangzhou University, Yangzhou, 225009, China.
2College of Animal Science and Technology, Yangzhou University, Yangzhou, 225009, China.
3Hubei Blue Valley Microbial Technology Co., Ltd., Yichang, 443100, China
*Address correspondence to: Yuan Yuan, 006707@yzu.edu.cn
Received: 02 January 2021; Accepted: 24 February 2021
#These authors contributed equally to this work.
Abstract: Goat milk is widely recognized for its nutritional value. Fatty acid synthase (FAS) is the crucial enzyme of fatty acid de novo synthesis. It plays an important role in the formation of goat milk fat. In this paper, we first introduced the molecular regulation process of goat milk fat metabolism based on the structure research of FAS. Secondly, we reviewed some key factors in FAS transcription and post-transcriptional regulation of the goat mammary gland and preliminarily constructed the expression network of the goat mammary gland FAS gene. The purpose of this paper is to systematically introduce the role of FAS in goat milk fat metabolism and to provide a reference for future studies on the mechanism of goat milk fat metabolism.
Keywords: Fatty acid synthase; Molecular regulation; FAS; Goat’s milk
List of abbreviations
| SCFA = | short chain fatty acid |
| ACC = | Acetyl-CoA carboxylase |
| FAS = | fatty acid synthase |
| KS = | β-ketoacyl synthase |
| MAT = | malonyl-CoA transacylases |
| DH = | Dehydratase |
| ER = | Enoyl reductase |
| KR = | Ketoacyl reductase |
| ACP = | Acyl carrier protein |
| TE = | Thioesterase |
| ACSL1 = | acyl-CoA synthetase long-chain family member 1 |
| ACSS2 = | acetyl-CoA synthetase 2 |
| FABP3 = | Fat Acid Binding Protein3 |
| SCD = | stearoyl-ACP desaturase |
| CLA = | Conjugated Linoleic Acid |
| sn = | Stereos- pecifically Numbering, |
| GPAT = | Glyceroltriphosphate acyl transferase |
| DGAT = | Diacylglycerol acyltransferase |
| ADFP = | Adipose Differentiation-Related Protein |
| TIP47 = | Tail-interacting protein 47 |
| PLIN1 = | Perilipin1 |
| XDH = | xanthine dehydrogenase |
| BTN1A1 = | Recombinant Butyrophilin Sub family1, Member A1 |
| PMSF = | Phenylmethanesulfonyl fluoride |
| SREBP1 = | Sterol-regulatory element binding proteins1 |
| PPARG = | Peroxisome Proliferator Activated Receptor Gamma |
| THRSP = | Thyroid hormone responsive spot |
| INSIG1 = | insulin induced gene |
| PANK = | Panthenic acid kinase |
Goat’s milk is rich in fatty acids, proteins, and 8 essential amino acids needed by the human body. The volume of fat particles in goat’s milk is only one-third that of cow’s milk, making it easy to digest and absorb. The fatty acids in goat’s milk, namely, dodecylic acid, myristic acid, stearic acid, and palmitic acid, are the main energy substances in the human body, accounting for approximately 5, 10, 8, and 27% of the total fatty acids, respectively, while the unsaturated fatty acids are mainly oleic acid and linoleic acid, accounting for 23% and 4% of the total fatty acids, respectively (Garard, 1939). The total dry matter content of goat’s milk is 11.4% higher than that of milk and contains 3.28% milk fat and 8.13% nonfat milk solid (4.29% lactose, 3.20% milk protein and 0.64% ash). The fat in goat’s milk is in the form of fat globules, the average diameter of which is approximately 3 µm, which is smaller than that of milk (approximately 6 µm). Goat’s milk is rich in short- and medium-chain fatty acids and unsaturated fatty acids (Luo et al., 2005). Goat’s milk contains more caproic acid (C6:0), caprylic acid (C8:0), and decanoic acid (C10:0) than milk. The hydrolysis of triglycerides by lipase releases fatty acids, which enhance the flavor (and odor) of dairy products (Gelais et al., 2005).
Goat’s milk is rich in short- and medium-chain fatty acids, which can be used to prevent and treat some metabolic diseases in humans (Williams, 2000).
These fatty acids play important roles in improving nutritional absorption disorder syndrome and intestinal dysfunction, inhibiting and reducing blood cholesterol content (Dostalova, 1992; Haenlein, 2001; Kusunoki et al., 2007), lowering blood pressure, preventing and treating atherosclerosis (Luna et al., 2008), improving hyperlipidemia (Galina et al., 1999; Bhattacharya et al., 2006) and inhibiting tumor growth (Beppu et al., 2006). Short-chain fatty acids (SCFAs) are important energy substrates in the body, as well as the most basic functional substances in the body of ruminants, and they are involved in the absorption of water and sodium. In addition, SCFAs play important roles in maintaining normal intestinal blood circulation and gastrointestinal hormone secretion. Since SCFAs can improve intestinal microcirculation and promote intestinal cell proliferation and mucosal growth, they are widely used in intestinal surgery (Miller, 2004; Jing et al., 2019). In conclusion, the difference between goat’s milk and other milk, especially milk, is largely due to the difference in fatty acid composition, which directly reflects the unique nutritional value of goat’s milk. The study of the mechanism of fatty acid metabolism at the molecular level to increase the composition of beneficial fatty acids in goat’s milk is an important aspect of goat breeding and thus of great significance to the development of the dairy goat industry.
Fatty acid synthase (FAS) is the crucial enzyme of fatty acid de novo synthesis. It plays an important role in the formation of goat milk fat. It is of great significance to study the structure, function, and gene regulation network of fatty acid synthase.
Molecular regulation of milk fat formation
Goat fatty acids are synthesized in the liver, kidney, brain, lung, mammary gland, fat, and other tissues and cells, but the synthesis of fatty acids in different parts of goats in different ages and nutritional states will have certain differences.
No matter where in the body, the main expressions of FAS in tissues were 16:0, 18:0, and C9-18:1. PUFA levels of n-6 and n-3 are low in AT but higher in the secretory tissues of the mammary gland. PUFA concentrations of 10:01, 12:01, 14:0, and C9, T11-18:2 were the highest in mammary gland tissue, but all ultra-long chain PUFAs were much more abundant in the liver (Toral et al., 2013). The following content will focus on the synthesis of fatty acids in the goat mammary gland.
De novo synthesis of goat fatty acids
Goat mammary gland fatty acids have two sources, short- chain, medium-chain fatty acids and one-half of the total palmitic acid, which is obtained from the de novo synthesis of mammary epithelial cells, and long-chain fatty acids and the other half of the total palmitic acid pool are obtained from blood (Bernard et al., 2005b). SCFAs, also known as volatile fatty acids (VFAs), are organic fatty acids with 1–6 carbon atoms mainly including acetic acid, propionic acid, butyric acid, isobutyric acid, pentanoic acid, caproic acid or hexanoic acid (Miller, 2004). The acetic acid, propionic acid, and butyric acid levels are the highest, accounting for approximately 90–95% of the short-chain fatty acids, and acetic acid is the main fatty acid (Schmitt Jr. et al., 1977). The synthesis of fats in goat is accomplished by a series of enzymatic reactions that require two substrates as precursors: α-glycerophosphate and aliphatic CoA (or fatty acids). Alpha-phosphate glycerol is mainly derived from dihydroxyacetone phosphate and glycerol phosphate. Acyl-CoA (or fatty acid) is synthesized by acetyl-CoA carboxylase (ACACA) and FAS. Since α-glycerophosphate is mainly derived from glucose metabolism, ACAC A and FAS in goat play rate-limiting and decisive roles in fat synthesis (Zhu et al., 2014).
Lactation has been an important process throughout biological evolution, characterized by breast milk, which provides complete nutrition for offspring. Lactation is a microcosm of the basic biological process of an organism, including all the processes of cell proliferation, differentiation, growth, and apoptosis, which determine the nutritional composition of the milk (Lemay et al., 2007; Bionaz and Loor, 2008). In goat (including their milk), there are two sources of fatty acids: exogenous intake from the diet and de novo synthesis. The process of biosynthesis in vivo is driven mainly via (FAS, which acts as a catalyst (Menendez et al., 2009). As a key enzyme for the de novo synthesis of fatty acids in cells, fatty acid synthase plays a core regulatory role in cell lipid metabolism (Chirala et al., 1997). Currently, it is known that FAS mainly produces long-chain fatty acids in cells, and long-chain fatty acids are the synthetic substrates of ultralong-chain fatty acids. FAS can catalyze all the steps of de novo synthesis of a long-chain stearic acid (such as palmitate) with acetyl-CoA and malonyl-CoA as substrates (Jenni et al., 2007; Livore et al., 2007) (Fig. 1). The decrease of FAS expression also resulted in a significant decrease of relative contents of decanoic acid (C10:0) and lauric acid (C12:0) in GMEC (Zhu et al., 2014). Sun et al. (2016) treat goat mammary gland epithelial cells (GMECs) with acetate, propionate, or butyrate and find that messenger RNA (mRNA) expression of FAS and LXRα was not affected by propionate but reduced by butyrate.
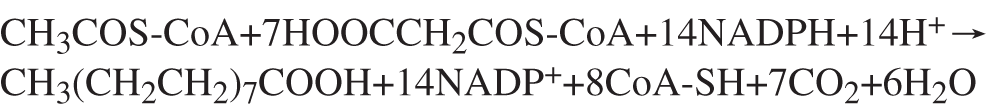
Figure 1: Formula of palmitic synthesis.
Goat mammary gland fatty acid synthesis occurs in the cytoplasm of mammary epithelial cells and consists of two connected phases (Glass et al., 1969). In the first step, acetyl-CoA is carboxylated to malonyl-CoA under the action of ACACA. The second step is catalyzed by fatty acid synthase complexes. The whole process of synthesis begins with the deacylation of malonyl-CoA transacylases (MAT). First, MAT transfers acetyl groups (i.e., initial substrates) to acyl carrier protein (ACP), and they are transferred from ACP to β-ketoacyl synthase (KS). At the same time, MAT catalyzes the deacylation of malonyl-CoA to transfer malonic acid monoacyl (prolonging substrates) to the pan-acyl-triethylamine arm of ACP. Then, by KS catalysis, a carbon unit is integrated into acetyl-ACP compounds and by Ketoacyl reductase (KR) catalytic Nicotinamide Adenine Dinucleotide Phosphate (NADPH)-dependent beta-carbon reduction. Upon the hydrolysis of Dehydratase (DH), the acyl compounds are transferred to the alpha- or beta-olefin fatty acyl intermediate. With the participation of NADPH, Enoyl reductase (ER) catalyzes the decrease in the olefin fatty acyl intermediate and the generation of four-carbon acyl chains, which enter into the next cycle. When the acyl chain is extended to 16–18 carbons, Thioesterase (TE) catalyzes the release of the acyl chain from ACP to form the final long-chain fatty acid (Asturias, 2006) (Fig. 2).
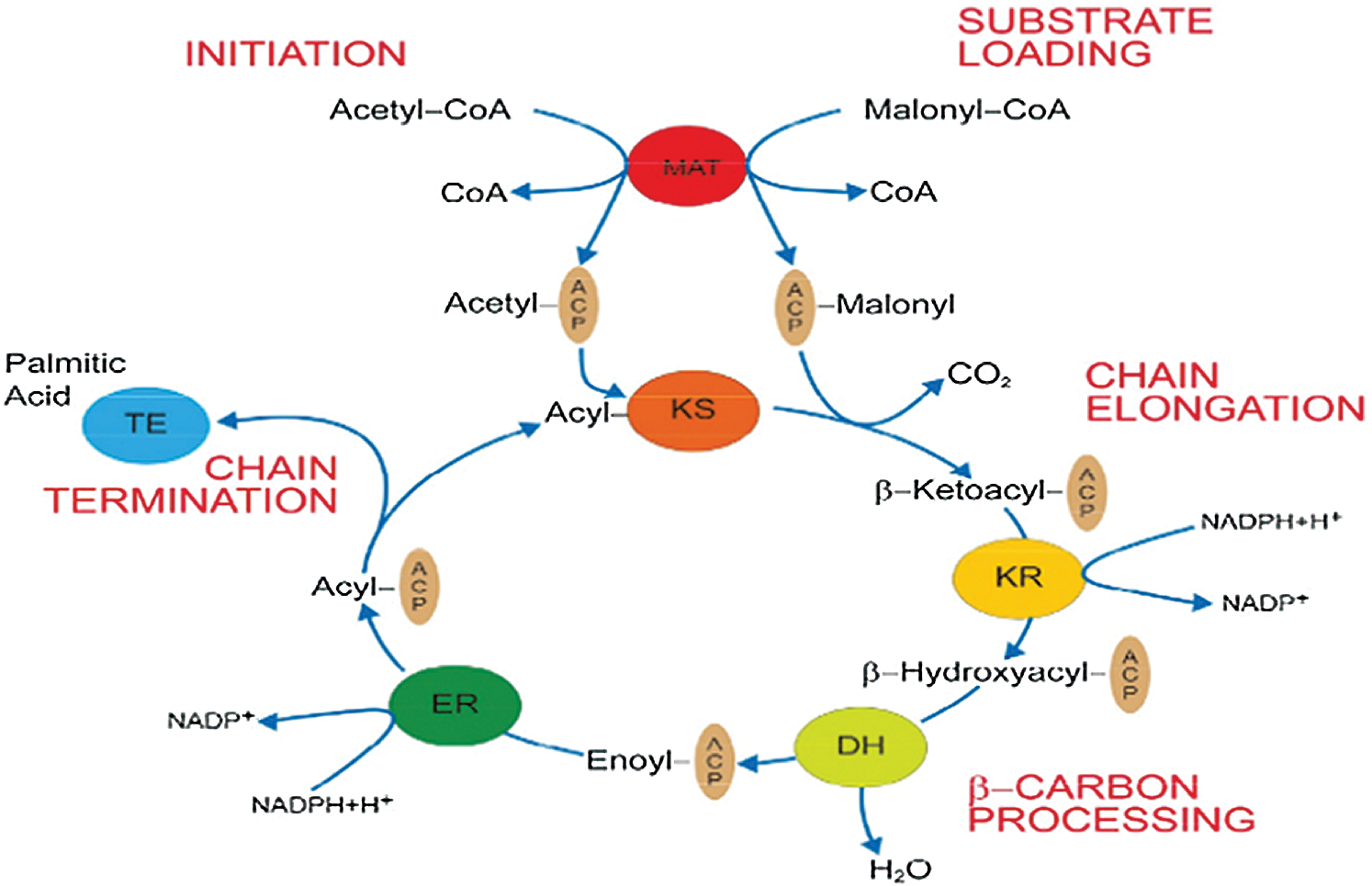
Figure 2: De novo fatty acid synthesis catalyzed by FAS (Asturias, 2006).
Studies have shown that, in ruminants, acetyl-CoA and malonyl-CoA are tra nsferred into the fatty acid synthesis cycle through the activity of acetyl/malonyl transferases. Knudsen et al. (Knudsen et al., 1976; Libertini and Smith, 1978; Mikkelsen et al., 1985) found that short-chain fatty acids are formed by transferase activity. Both butyryl-CoA and caproyl-CoA are sensitive substrates of fatty acid synthase, and they complete the loading process of the fatty acid synthesis cycle through the activity of acetyl/malonic acid monoacyl transferases. However, FAS of nonruminants can only effectively transfer acetyl-CoA and butyryl-CoA (Bloch and Vance, 1977; Knudsen and Grunnet, 1980), and ruminants can also transfer C12 fatty acids (Smith, 2009). Competitive inhibition experiments showed that acetyl/malonic acid monoacyl transferases had not only acetyl/malonic acid monoacyl transferase activity but also butyryl and acyltransferase activity, and the activity of goat mammary acid synthase was higher than that of acetyl, butyryl, or diacyl transfer (Mikkelsen et al., 1985). This result is consistent with that of Carey and Dils (1970; 1972). Sequencing results and active site analysis showed that the sequence and site of the transferase region binding to the acyl group were consistent with those of the acetyl/malonic acid monoacyl transferase region. This finding indicated that acetyl/malonic acid monoacyl transferase acted to transfer acyl groups. In addition, only one active site of an acyltransferase was found on goat fatty acid synthase. These results also illustrate that short-chain fatty acid synthesis in ruminants and nonruminant animals is dependent on b/single acyltransferase activity and malonic acid. The formation of chain fatty acids in nonruminant animals is undertaken by sulfur esterase II. Ruminant acetyltransferase/propylene acid in the basic form can also form a chain of fatty acids.
In 1998, Fielding and Frayn (1999) reported that the ruminant mammary gland is capable of absorbing free and bound fatty acids from the blood. Triglycerides in the blood are mainly in the form of high-density lipoprotein, low-density lipoprotein, and chylomicron. Under the catalyzed action of lipoprotein lipase (LPL), low-density lipoprotein and chylomicron can be hydrolyzed to release free long-chain fatty acids that are absorbed by mammary epithelial cells. CD36 is an important protein in the body that is involved in the uptake of Long Chain Fatty Acids (LCFAs) by fat and muscle cells. In 1997, Sfeir et al. (1997) conducted adipocyte transport studies and found CD36 gene expression and fatty acid uptake during adipocyte induction and differentiation. In 1998, Watanabe et al. (1998) reported that the transfer of the CD36 gene into cells without endogenous CD36 expression enhanced the binding ability of LCFAs, and this binding effect was reduced after the application of relevant protein inhibitors. CD36 in tissue lipoprotein and lipid metabolism. However, mammary cells absorb foreign fatty acids in a complicated process for which the exact mechanism is still unclear.
Transportation and desaturation of fatty acids
Massimo and Loor (2008) reported that long-chain fatty acids play a role in mammary epithelial cells in mammary gland tissue. The initial step is the transfer of the long-chain fatty acids from plasma into cells and their activation. Previous studies have suggested that CD36 plays an extremely important role in the uptake of long-chain fatty acids into mammary epithelial cells. Mashek et al. (2006) reported that acyl-CoA synthetase long-chain family member 1 (ACSL1) and acetyl-CoA synthetase 2 (ACSS2) catalyze the intracellular long-chain and short-chain fatty acid conversion into their active forms, respectively. McArthur et al. (1999) reported that Fat Acid Binding Protein3 (FABP3) and FABP4 are fatty acid-binding proteins critical mainly for the transport of fatty acids in cells. Yonezawa et al. (2004) found that oleic acid and linoleic acid can upregulate the expression of CD36, FABP4, and ACSL1. Kadegowda et al. (2009b) found that stearic acid has a greater up-regulatory effect on CD36 and FABP4 than do other unsaturated fatty acids. Massimo and Loor (2008) found that the expression of FABP3 was inhibited by the addition of long-chain fatty acids. They believed that FABP3 mainly provided fatty acids for SCD and catalyzed the de novo synthesis of fatty acids. After adding linolenic acid, the expression of CD36 was upregulated, while the expression of ACSL1 and FABP3 was downregulated. It has been reported that long-chain fatty acids can inhibit the de novo synthesis of short-chain fatty acids (Bauman et al., 2008). In the study of Kadegowda et al. (2009a), unsaturated fatty acids inhibited the expression ofACSS2 to a greater extent than did saturated fatty acids.
The desaturation of fatty acid chains is catalyzed by the product of stearoyl-ACP desaturase (SCD (in vivo)) genes, which introduces a cis double bond at delta 9 to generate a delta 9 unsaturated fatty acid. Active fatty acids mainly target C14–C19. In contrast to monogastric animals, ruminants have only one SCD gene, which encodes a 5 kb transcript in goats (Bernard et al., 2005a). In goats, the SCD gene is highly expressed in mammary gland tissue and subcutaneous adipose tissue (Corl et al., 2001). In cow mammary glands, most cis-9, trans-11 (Griinari et al., 2000; Loor et al., 2005; Shingfield et al., 2016) and trans-7, cis-9 (Corl et al., 2002). Conjugated Linoleic Acid (CLAs) are catalyzed by SCD. The transcription enhancer element (STE) in the promoter region of SCD plays an important role in the desaturation effect of SCD. STE can block the generation of trans-10 and cis-12 fatty acids by SCD and inhibit the production of cis-9 and trans-11 CLAs (Corl et al., 2001; Bernard et al., 2008).
Synthesis of triglycerides and the secretion of lipid droplets
Fat is an important part of milk and plays an important role in energy supply and improving milk quality. Milk fat is composed of 98% triglycerides, a small amount of cholesterol, diglycerides, glycerides, and free fatty acids (Dils, 1986). Triglycerides constitute the main part of milk fat, which is mainly composed of 95% fatty acids and 1% phospholipids combined with glycerol (Bernard et al., 2008). Triglycerides are selective in the utilization of fatty acids, which determines the function and nutritional value of different triglycerides (German et al., 1997). Of the fatty acids, 56% to 62% are mainly bound at the sn-1 and sn-2 position of triglyceride, respectively, and are mainly composed of medium- and long-chain saturated fatty acids (C10–C18), of which C16:0 is evenly distributed at sn-1 and sn-2, while C8:0, C10:0, C12:0, and C14:0 are mainly bound to sn-2 and C18:0 is mainly bound to sn-1. In addition, C18:1 accounts for 24% of sn-1 fatty acid sources. Short-chain fatty acids (44%) C4:0, C6:0, C8:0, and oleic acid (27%) bind to Stereo-specifically Numbering (sn)-3. When hydrolyzed, the sn-1 and sn-3 fatty acids are specifically released under the action of an esterase, while the sn-2 fatty acids are preferentially absorbed and utilized in the form of glycerol monoester. The synthesis of triglycerides begins with the preferential esterification at the sn-1 site of glycerol 3-phosphate. This process is catalyzed by Glyceroltriphosphate acyltransferase (GPAT). GPAT in mammals can be divided into two subtypes according to its different location; namely, GPAT1 is located in the endoplasmic reticulum, and GPAT2 is located in mitochondria. GPAT1 is sensitive to ethyl maleimide (NEM), while GPAT2 is resistant to NEM. Both subtypes of GPAT catalyze the cooperation of mouse triglycerides (Coleman, 2004). AGPAT/LPAAT catalyzes the esterification of sn-2 fatty acids. AGPAT has a higher affinity for saturated medium- and long-chain fatty acids, and the esterification sequence is C16 > C14 > C12 > C10 > C8 (Marshall and Knudsen, 1977; Mistry and Medrano, 2002). C16:0 accounts for 43% of the sn-2 chains. However, the concentration of the substrate can also change the fatty acid composition at sn-2. The third step of triglyceride synthesis is catalyzed by the Diacylglycerol acyltransferase (DGAT) gene. The DGAT protein is located on the endoplasmic reticulum and is the only triglyceride synthesis function-specific protein (Mayorek et al., 1989). DGAT plays an important role in the synthesis of milk fat (Harris et al., 2011). There are two forms of DGAT: DGAT1 and DGAT2. These two forms can catalyze the synthesis of triglycerides. DGAT1 is highly expressed in the small intestine, where it plays an important role in the absorption of TG. DGAT1 also accounting for more than 90% of the DGAT activity in the small intestine (Cases et al., 2001; Buhman et al., 2002). The triglyceride content was significantly reduced in dgat1-deficient mice (Smith et al., 2000). When the activity of DGAT1 decreases, DGAT2 can compensate and exert the activity of the DGAT1 enzyme, thus ensuring the normal synthesis of triglycerides in the small intestine. However, DGAT2 is mainly found in the liver and white adipose tissue (Smith et al., 2000). Loss of DGAT2 in adipose tissue can significantly reduce triglyceride content (Meegalla et al., 2002) (Fig. 3). In addition, there are functional differences between the two DGAT subtypes. Studies have shown that DGAT1 is mainly expressed in response to exogenous fatty acids, while DGAT2 is expressed in response to endogenous fatty acid synthesis (Smith et al., 2000). From lactating mammary gland tissue, triglycerides are mainly secreted in the form of milk fat globules. The formation of lipid globules begins with the formation of lipid droplets. The formation of fat droplets is mainly controlled by PAT family genes, including the Adipose Differentiation-Related Protein (ADFP), Tail-interacting protein 47(TIP47), and Perilipin1 (PLIN1) genes.
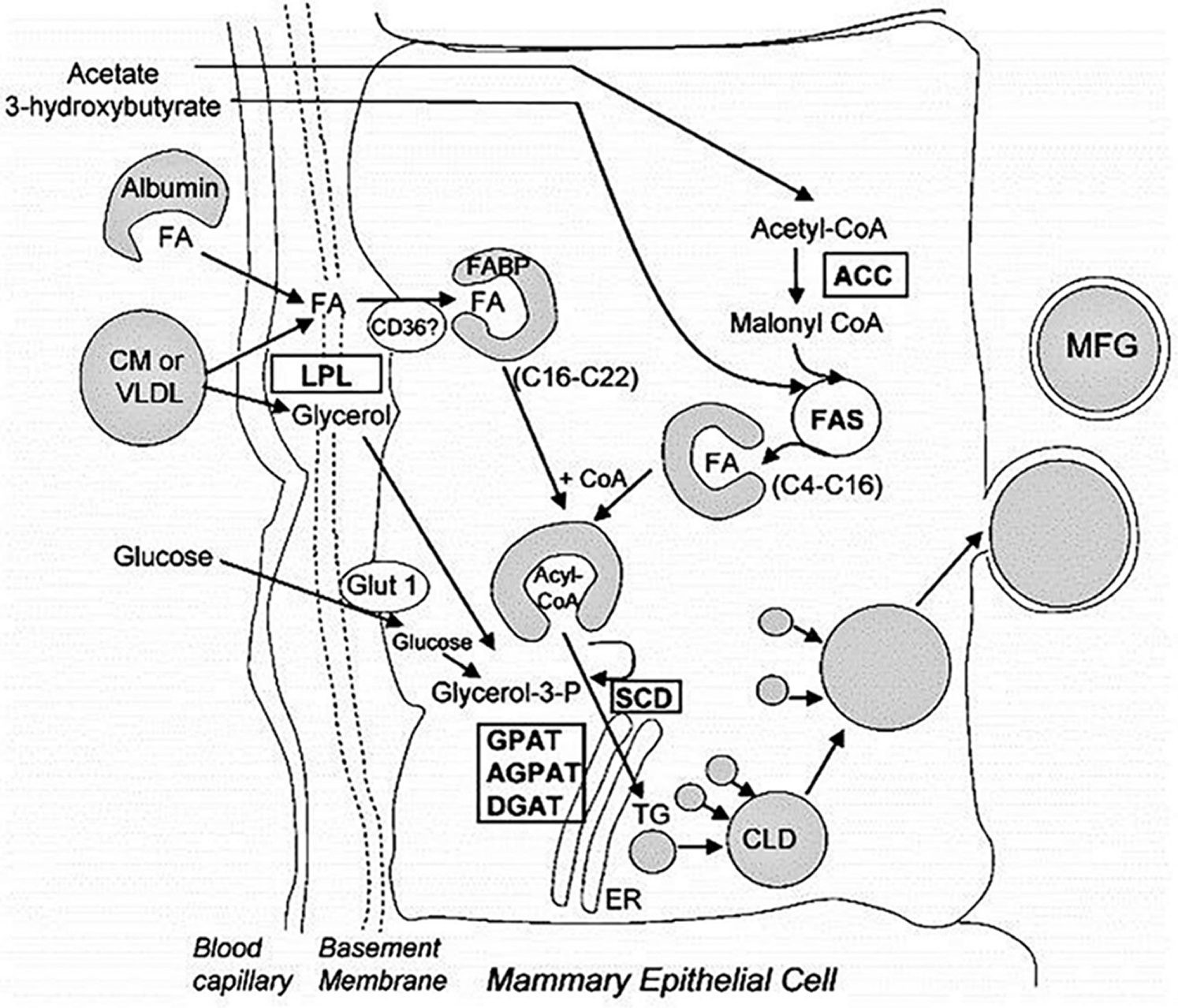
Figure 3: Triglyceride synthesis and milk fat secretion (Meegalla et al., 2002).
Lipid droplets (less than 0.5 μm in diameter) released from the endoplasmic reticulum are released and then coated with proteins and polar lipids. Some of these droplets undergo a series of enzymatic fusion events that lead to the formation of large droplets in the cytoplasm that are transported to the cell membrane for secretion. However, many lipid droplets do not fuse and are secreted at their initial size (Bauman et al., 2006). While the average droplet diameter is close to 4 μm, more than 80% have a diameter of less than 1 μm.
The secretion of lipid droplets is mainly facilitated by three proteins. First, the newly formed lipid droplets are aggregated upon binding to ADFP and fused with other lipid droplets and then transported to the cell membrane. Under the action of Recombinant Butyrophilin Sub family1, Member A1 (BTN1A1) and xanthine dehydrogenase (XDH) are released through the cell membrane by exocytosis. Among these proteins, ADFP plays the role of coordinator in the whole process by binding to BTN1A1, which mediates the binding of ADFP to lipid droplets. The recruitment of XDH under the action of BTN1A1 promotes the formation of a three-step process, which leads to the release of lipid droplets (Chong et al., 2011) (Fig. 4).
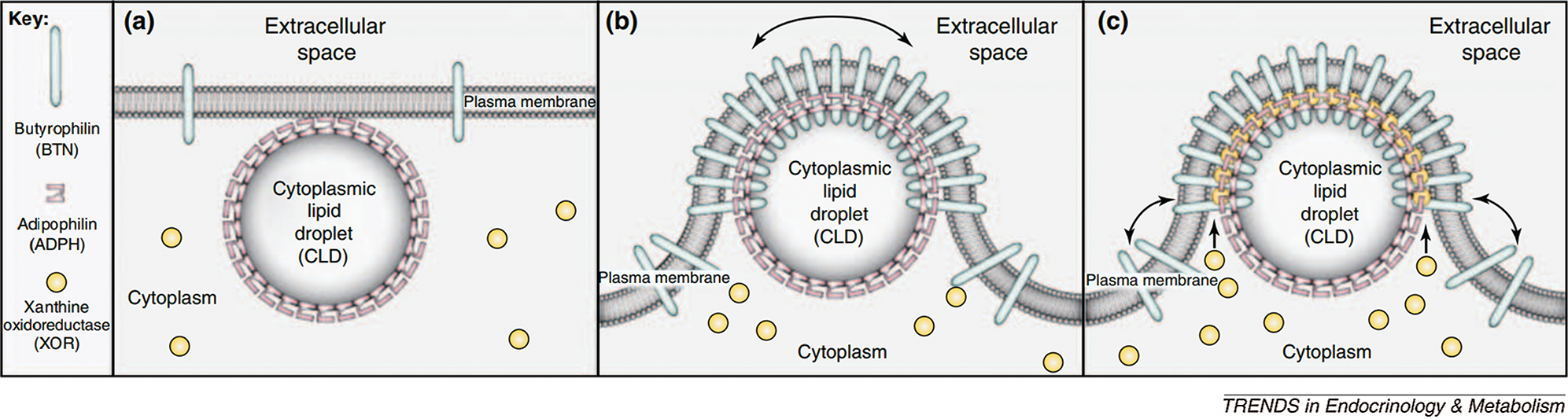
Figure 4: Milk droplet model of milk lipid secretion (Chong et al., 2011).
FAS structure and function regulation
The distribution of fatty acid synthase gene expression in goat organs was significantly different. The mRNA expression of FAS was the lowest in the liver, which was significantly lower than that in mesenteric fat and muscle fat (P < 0.05). The mRNA expression level of FAS was the highest in mammary gland tissues and was significantly higher than that in mesenteric adipose tissues (P < 0.05). This phenomenon indicates that endogenous fatty acid synthesis is less in the liver and more in mammary gland tissues. The following content will mainly introduce the FAS structure and function regulation of goats.
Naturally occurring FAS is categorized into two types: FASI and FASII. In animals, FASI is the main form (Smith et al., 2003). Mammalian fatty acid synthase must be in the form of a dimer to render its catalytic activity for fatty acid synthesis. The monomer of fatty acid synthase does not have full catalytic ability; but, studies have shown that some of the functional domains in the monomer have catalytic activity, primarily to transfer the substrate in advance to the ACP functional areas and interact with ACP functional domains (Pappenberger et al., 2010).
Two models have been proposed to modify the structure of FAS in mammals (Asturias et al., 2005; Pappenberger et al., 2010): the conventional model and the revised model. In the conventional model, the two monomers functionally interact in inversely parallel forms, with clear boundaries between the monomers (Fig. 5). The revised model is based on the interaction between two monomers connected at the N-terminus and C-terminus (Asturias et al., 2005). Cryo-electron microscopy also revealed that the FAS monomers are arranged in a coiled manner (Fig. 6c). The crystallographic density map of FAS reveals the structural interface of a central region formed by the combination of the two monomers, with DH and ER, and MAT located on both sides of KS (Smith, 2009) (Fig. 6b). Grunnet and Knudsen (1979) confirmed for the first time in goats that MAT has not only transferase activity but also thioesterase activity. The systematic study on the overall spatial structure of goat FAS has not been reported yet, but based on cDNA sequence-related studies, the FAS structure of goat is different from that of other mammals.
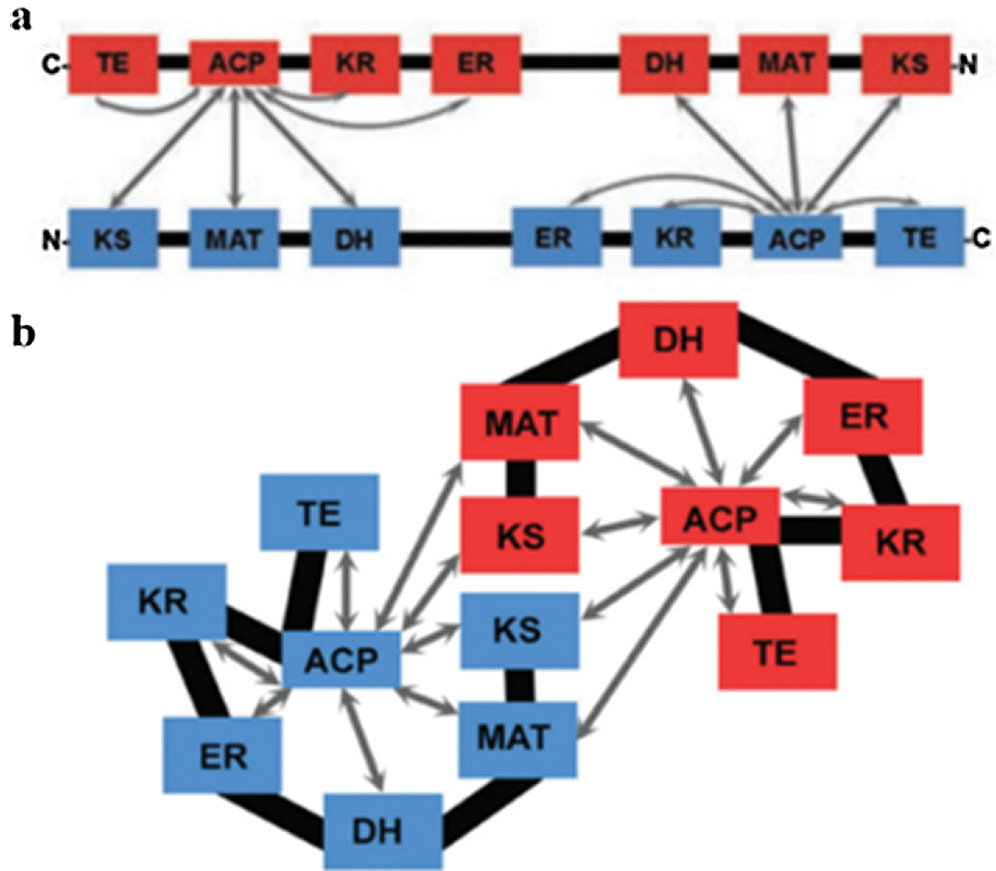
Figure 5: Structure model of fatty acid synthase. (a) Conventional model. (b) Revised model (Asturias et al., 2005).
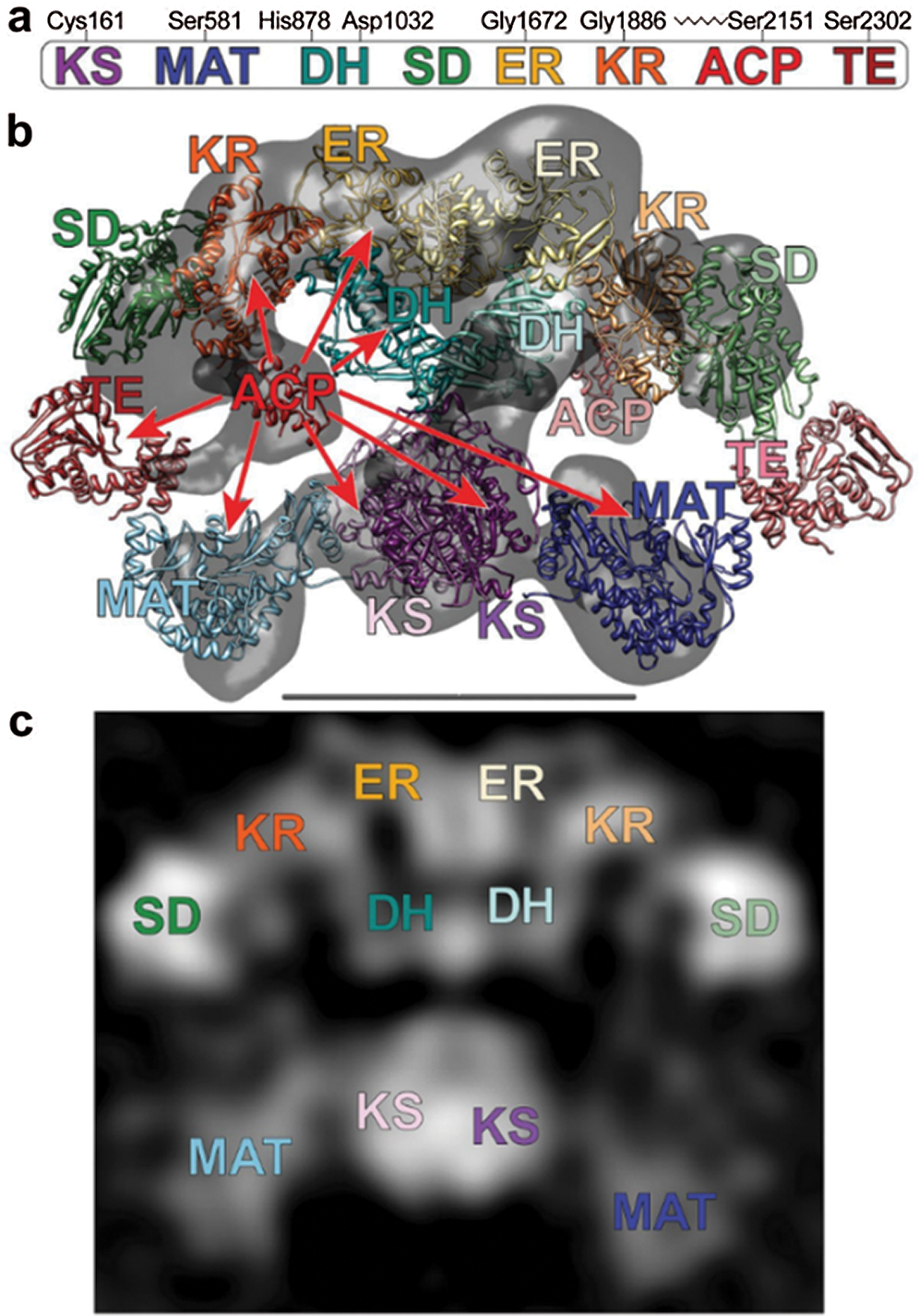
Figure 6: (a) The domains of FAS. (b) Crystal and cryo-EM structures capture different conformations of FAS. (c) Images of FAS molecules (Smith, 2009).
In Xinong dairy goats, the FAS gene is located in chromosome 19 and encodes the homodimeric multifunctional enzyme. It has 8217 bp comprised of 42 exons, an ORF of 7545 bp, and 5’- and 3’-UTR regions of 88 bp and 584 bp, respectively, with the start codon located in the 2nd exon, and the stop codon is in the 42nd exon. It encompasses seven active functional domains, which participate in all the processes of fatty acid synthesis. It encodes 3514 amino acids with an approximate molecular weight (MW) of 273.8 kDa (Zhu et al., 2014). The similarity of the FAS gene exon 9–15 in the cells from the mammary gland of Saanen goats was 95, 85.7, 82.7, 73.2, 92.9, 77.7, 82.3, and 64.7%, respectively, with that of cow (NM_001012669), human (NM_004104), rat (NM_017332) and chicken (NM_205155). Its catalytic structure, starting at the N-terminus, has the following order (Fig. 7): β-ketoacyl synthase (KS), acetyl-CoA and malonyl-CoA transacylase (MAT), dehydratase (DH), enoyl reductase (ER), and thioesterase (TE). FAS can be divided into three regions: regional I contains KS, MAT, and DH, regional II contains ER, KR, and ACP, and regional III consists of TE (Chirala and Wakil, 2004). The active sites in the AT/MT region were highly conserved serine residues among all species, but exon 10 in the Saanen goat had one less amino acid than the other species, which may have an important impact on the spatial conformation and physiological functions of the AT/MT region.

Figure 7: The linear map of fatty acid synthase domains of Saanen goat (Zhu et al., 2014).
Regulation of Milk fat metabolism by goat FAS
Wang et al. (2010) used RNA interference technology to interfere with the expression of the FAS gene in goats and found that, after FAS gene silencing, the content of C10:0, C12:0, C16:0, and C18:0 in the cells decreased by 42, 42.37, 18.51, and 29.3%, respectively. The C14:0 content increased by 29.82%. Quantitative real-time PCR results showed that the expression levels of the leptin receptor (LEPR) gene and liver X receptor (LXRα) gene were 1.71- and 1.36-fold that of the control group, respectively. The expression levels of the fatty acid-binding protein (AFABP) gene and lipoprotein lipase (LPL) gene were decreased by 32 and 25%, respectively. These results indicate that the FAS gene plays an important role in regulating fatty acids, according to chain length, in mammary gland epithelial cells.
The termination mechanism of medium-chain fatty acids in ruminants is quite different from that in nonruminants. Medium-chain fatty acids in ruminants are terminated by the activity of a transferase. Knudsen and Grunnet (1982) found that purified mammary fatty acid synthase from lactating swine and rat cannot produce fatty acid when incubated with goat microsomes, and only goat and cattle fatty acid synthase can prompt the synthesis of C10 fatty acids. Furthermore, research shows that these fatty acids are not present in the ruminant mammary gland independent of TE II. These findings indicate that the activity of the medium-chain fatty acid synthase is terminated in goat (Grunnet and Knudsen, 1978). Goat FAS can terminate the synthesis of long-chain fatty acids and may terminate fatty acid chain synthesis when treated with the proteasome inhibitor Phenylmethanesulfonyl fluoride (PMSF), for which the synthase of long-chain fatty acid termination activity was significantly affected but not sensitive. The authors speculated that goat fatty acid synthase might contain two hydrolase sites or that two hydrolysis activities are undertaken. Kundsen and Grunnet (1982) found that the ratio of C10 to C12 was significantly higher in the presence of 5.2 mg/mL albumin, and the content of C10 was 4-fold higher when 10.4 mg/mL albumin was used. The addition of monoacyl-CoA of malonate inhibits the effect of albumin and promotes the formation of C12. However, changing the concentration of monoacyl-CoA of malonate cannot fundamentally change the chain length of fatty acids. The addition of globulin also had the same effect, but only at a higher concentration and only in ruminants.
Shi et al. (2015) used GO and KOGG enrichment analysis to obtain genes related to milk fat metabolism in goat mammary tissue and established a regulatory network model of these genes. They found that genes encoding FABP3, FAS, SCD, PLIN2, whey proteins, and caseins at 100 and 310 days postpartum increased significantly compared with the non-lactating period (Fig. 8).
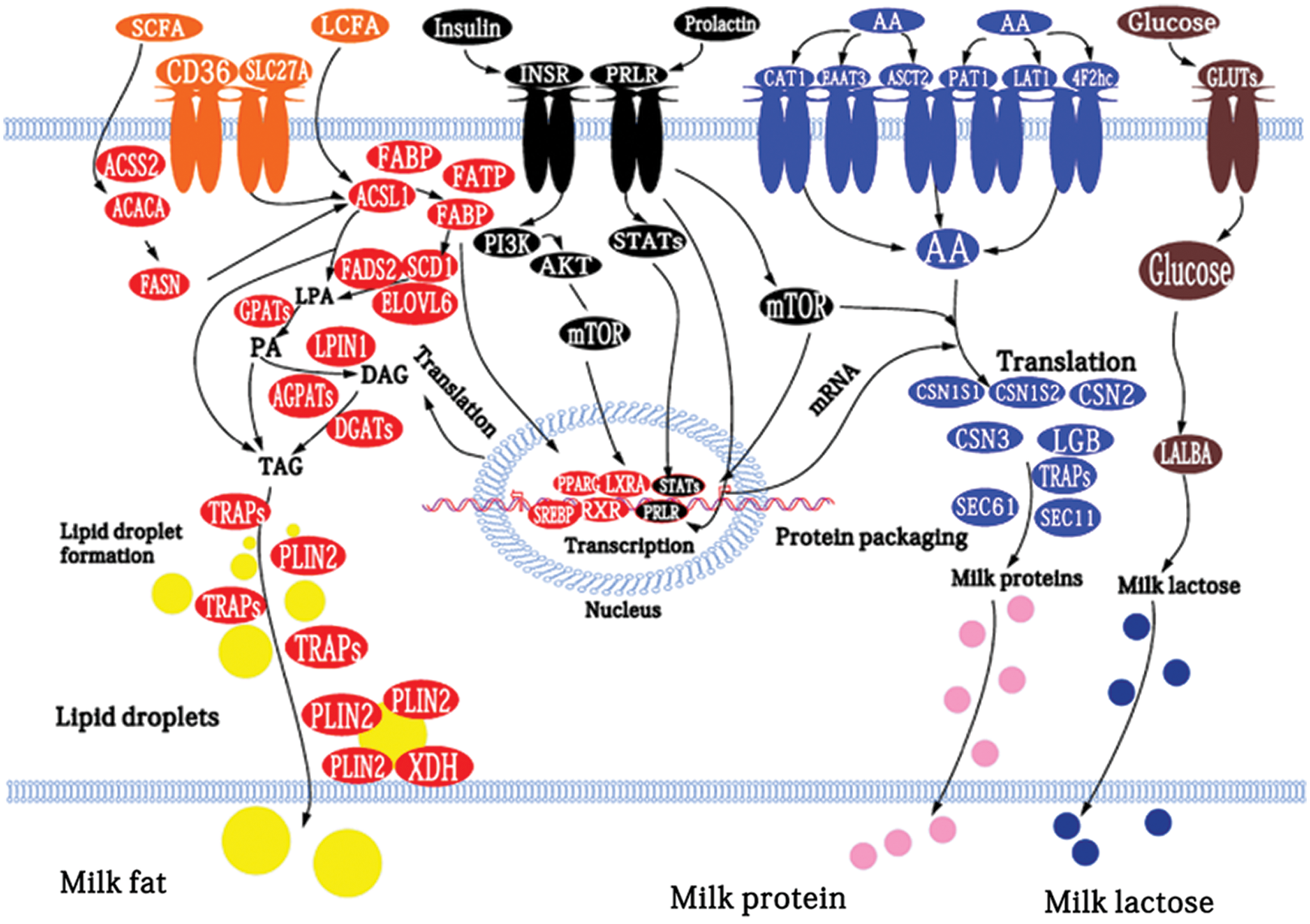
Figure 8: Model of the networks of factors (proteins, hormones, and enzymes) potentially involved in the regulation of milk fat, protein, and lactose biosynthesis in goat mammary tissue (Shi et al., 2015).
In 2014, Zhao et al. (2014), using goat mammary epithelial cells (GMEC), knockdown of LPL reduced the expression of mRNA sterol-regulatory element-binding proteins-1 (SREBP-1), FAS, and Peroxisome Proliferator-Activated Receptor Gamma (PPARG), results show that LPL is the key gene for FAS regulation. The prolactin receptor (PRLR) gene is the same as LPL, both in the upstream of FAS gene regulation. PRLR, as a transmembrane receptor, results in a large variety of physiological processes (Bole-Feysot et al., 1998). PRLR signaling during lactation triggers the copious synthesis of triglycerides meanwhile controls these substance’s secretions (Rudolph et al., 2011). Treatment of goat mammary epithelial cells (GMECs) with prolactin decreased the expression of PPARG, SREBP1, FAS, and ACACA, which relative to the control group averaged 76, 55, 52, and 68%. Some scholars have also studied the relationship between SREBP-1, PPARG, and FAS (Shi et al., 2016). Cholesterol regulatory element-binding protein 1 (SREBP1) is a membrane-bound transcription factor involved in many roles of lipid homeostasis. SREBP1 is central in the transcription regulation of many genes related to milk fat synthesis and secretion in dairy cattle (Li et al., 2014). Li et al. (2015) found that SREBP-1 overexpression and knockdown by small interference RNA in goat mammary epithelial cells influenced the abundance of endogenous FAS. SREBP-1 regulates FAS expression at the transcriptional level. Peroxisome proliferator-activated receptors (PPAR) are ligand-activated transcription factors that belong to the nuclear receptor superfamily and include 3 closely related members: PPAR alpha (PPARA), gamma (PPARG), and delta (PPARD). ALL of them have been proved to affect the expression of the FAS gene in goats.
Goat PPARG has two isoforms which are PPARG1 and PPARG2. PPARG1 upregulates the transcription regulators SREBF1 and FAS, ACACA, and SCD; these data suggest that PPARG1 is the isoform controlling lipogenesis in goat mammary cells (Shi et al., 2014). PPARD is associated primarily with the catabolism of fatty acids in goats. Previous studies have found that PPARD expresses higher levels of PPARG in the bovine mammary gland (Bionaz et al., 2013). Shi et al. (2017b) found that the expression of fatty acid synthesis (FAS) gene downregulated significantly after knockdown of PPARD in goat mammary gland cells incubated with GW. The involvement of PPARA in fatty acid metabolism was also confirmed this year. Activation of PPARA up-regulated FAS, SCD1, ACSL1, DGAT1, FABP4, and CD36 in goat mammary gland epithelial cells, but down-regulated DGAT2 and PGC1A abundance (Tian et al., 2020).
The researchers found that knockdown of THRSP reduced the incorporation of breast cancer cytoplasmic acetate into lipids (Martel et al., 2006). Yao et al. (2016) found overexpression of THRSP upregulated the expression of fatty acid synthase (FAS) in goat mammary epithelial cells. In contrast, overexpression of THRSP led to downregulation of thrombospondin receptor CD36. Thyroid hormone responsive (THRSP) is a crucial protein for cellular de novo lipogenesis in goat. Tian et al. (2018) used CRISPR/Cas9 knockout SCD1gene in GMEC. The deletion of SCD1 decreased the expression of other genes involved in de novo fatty acid synthesis, including SREBF1 and FAS. SCD1 is regulated by Peroxisome proliferator-activated receptor-(PPARG). Shi et al. (2013) observed a positive correlation between PPARG and SCD expression in the goat mammary gland at peak lactation. Overexpression of PPARG leads to increased SCD gene expression.
LXR is an important nuclear receptor that regulates lipid and cholesterol homeostasis (Ulven et al., 2005). Overexpression of LXRB dramatically upregulated SREBP1c and FAS to levels higher than overexpression of LXRA. These results highlight an important role for LXRB in the transcriptional regulation of SREBP1c and FAS in goat mammary epithelial cells (Shi et al., 2017a).
Posttranscriptional regulation of goat FAS gene
Post-transcriptional control refers to the regulation of gene expression at the post-transcriptional level (RNA), including a series of modifications and processing of the transcription products of eukaryotic genes (Liu et al., 2020; Yang and Lu, 2020; Zheng et al., 2020). MicroRNA (miRNA) is a small non-coding RNA molecule that regulates mRNA expression at the post-transcriptional level by binding to the 3’-untranslated region (3’-UTR) of the target mRNA (Bartel, 2009). In recent years, several microRNAs have been reported to modify cellular lipid metabolism by regulating the expression of lipid-related genes. For instance, miR-221 regulates lipid metabolism in mammary epithelial cells (MECs) and is expressed differentially at various stages in mice (Chu et al., 2018a).
Synergistic regulation among microRNAs (miRNAs) is important to understand the mechanisms underlying the complex molecular regulatory networks in goats. Goat milk fat synthesis is driven by a gene network that involves many biological processes in the mammary gland. These biological processes are affected by several miRNAs rather than a single miRNA. Lin et al. (2013b) found that the expression of 11 miRNAs that have the potential to regulate milk fat synthesis in the goat mammary gland. They also found that prolactin promotes the expression of four miRNAs (miR-23a, miR-27b, miR-103, and miR-200a).
Functional verification of these microRNAs has also been widely reported. Downregulation of miR-26a/b and their host genes in goat mammary gland cells decreased the expression of genes relate to fatty acid synthesis (PPARG, LXRA, SREBF1, FAS, ACACA, GPAM, LPIN1, DGAT1, and SCD1 [in vitro]). Luciferase reporter assays confirmed INSIG1 as a direct target of miR-26a/b. This suggests that FAS and other lactate metabolism genes regulate insulin-induced gene1 (INSIG1 [in vitro]) in the goat nucleus (Wang et al., 2016).
It was found that miR-103 expression level was significantly higher in the mid-lactation period than in the dry period. The miR-103 family has three members, miR-103-1, miR-103-2, and miR-107, which reside in the sense oriented intron 5 of three members of the pantothenic acid kinase (PANK [in vitro]) gene family members across species: PANK3, PANK2, and PANK1, respectively (Rock et al., 2000). Overexpression of miR-103 in goat mammary gland epithelial cells resulted in increased expression of FAS, ACACA, and other fat metabolic-related genes (Lin et al., 2013a) (Fig. 9). miR-103 plays an important role in the molecular regulation of milk fat metabolism.
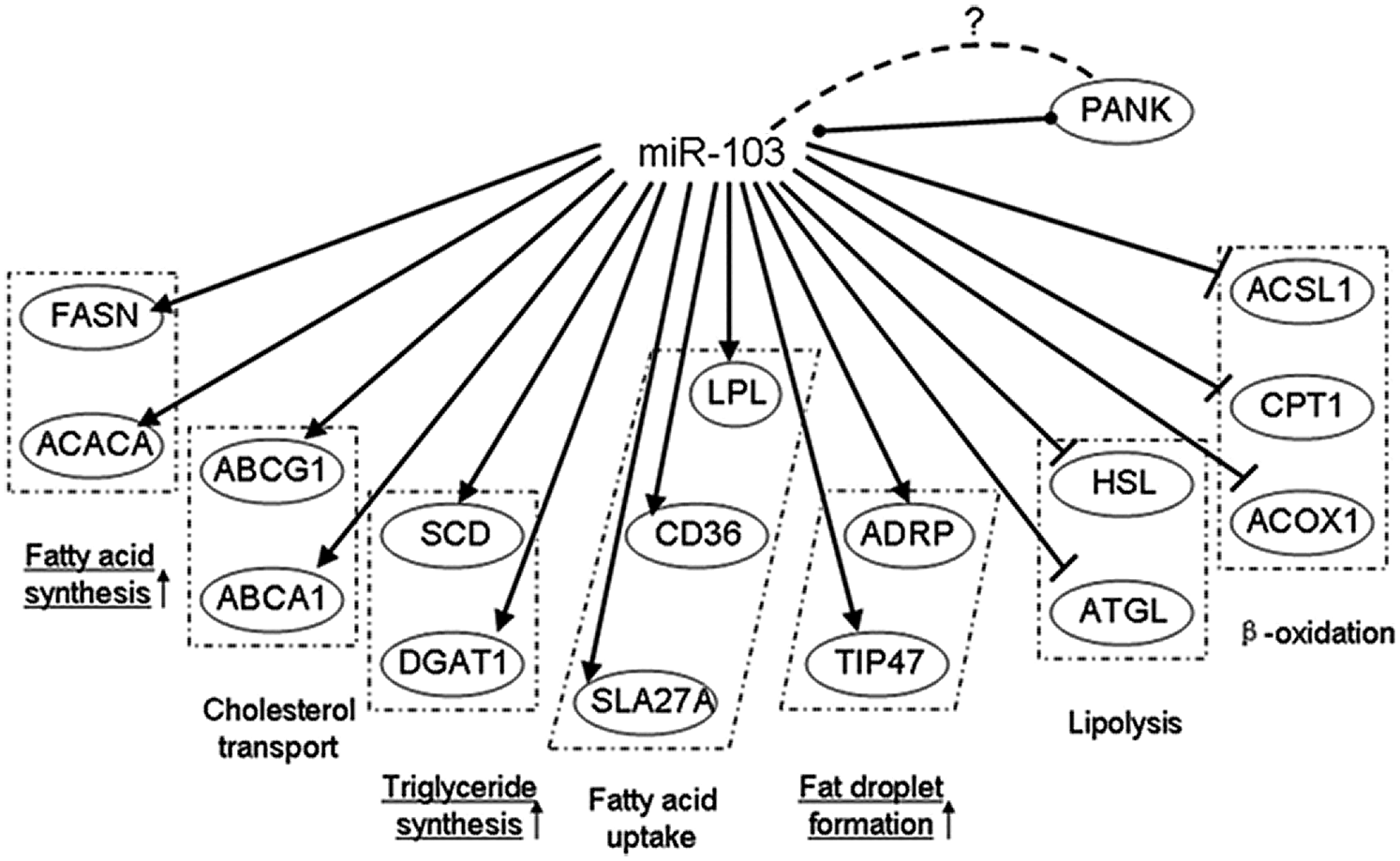
Figure 9: Gene networks modulated by miR-103 (Lin et al., 2013a).
MiR-145 expression level in goat mammary gland was significantly higher in mid-lactation period than in dry period. Overexpression of miR-145 increased transcription of genes associated with milk fat synthesis. In contrast, the silencing of miR-145 impaired fatty acid synthesis. Inhibition of miR-145 increased methylation levels of fatty acid synthase (FAS), stearoyl-CoA desaturase 1 (SCD1), peroxisome proliferator-activated receptor gamma (PPARG) (Wang et al., 2017). The role of Mir-15b in mammary fatty acid metabolism in goats has been studied. Mir-15b down-regulates lipid metabolism in goat mammary gland epithelial cells and its expression was lower during lactation. Overexpression of miR-15b reduced lipid content in mammary epithelial cells with decreasing levels of FAS (Chu et al., 2018b). Some miRNAs do not directly regulate FAS expression but can indirectly affect FAS gene expression by affecting the expression levels of other milk fat metabolism genes like miR-148a and miR-17-5p. They regulate milk TAG synthesis via PPARGC1A and PPARA in goat mammary epithelial cells, which affect the whole milk fat metabolism network (Chen et al., 2017). In summary, miRNA plays a very important role in the post-transcriptional regulation of FAS genes in goats.
FAS is a key enzyme in the de novo synthesis of fatty acids and is considered to play an important role in the formation of goat milk fat. Genes that regulate FAS in goat mammary epithelial cells are LPL, CD36, ACACA, SCD, SERBP-1, PPARG, PPARD, PPARA, PRLR, LXRB, THRSP. Some genes regulate FAS directly, and some regulate FAS indirectly. MicroRNA is an important factor in FAS post-transcriptional regulation which contained miR-145, miR-26a/b, miR-103, Mir-15b.
Currently, there are few studies on the gene expression profile of milk goats at different lactation stages and that address how FAS regulates milk fat formation. The systematic study of the role of FAS in milk fat formation in milk goats is of great significance for discovering the molecular regulatory mechanism of milk fat formation.
Author Contributions: Conceptualization, Jiahe Guo and Zhi Chen and Yuan Yuan; software, Yongjiang Mao; validation, Yongjiang Mao, Zhangping Yang and Zhi Chen; formal analysis, Yongjiang Mao; resources, Yongjiang Mao; writing—original draft preparation, Jiahe Guo and Zhi Chen; writing—review and editing, Jiahe Guo; visualization, Zhangping Yang; supervision, Zhangping Yang and Zhi Chen; project administration, Zhi Chen; funding acquisition, Zhi Chen. All authors have read and agreed to the published version of the manuscript.
Image Copyright Description: All images quoted in this article have been authorized by the authors.
Funding Statement: This research was supported by the National Natural Science Foundation of China (Grant Nos. 31802035, 31872324 and 31601915), the China Postdoctoral Science Foundation (Grant Nos. 2017M621841 and 2019T120472).
Conflicts of Interest: The authors declare no conflict of interest, financial or otherwise
Asturias FJ (2006). Mammalian fatty acid synthase: X-ray structure of a molecular assembly line. ACS Chemical Biology 1: 135–138. DOI 10.1021/cb6001448. [Google Scholar] [CrossRef]
Asturias FJ, Chadick JZ, Cheung IK, Stark H, Witkowski A, Joshi AK, Smith S (2005). Structure and molecular organization of mammalian fatty acid synthase. Nature Structural & Molecular Biology 12: 225–232. DOI 10.1038/nsmb899. [Google Scholar] [CrossRef]
Bartel DP (2009). MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233. DOI 10.1016/j.cell.2009.01.002. [Google Scholar] [CrossRef]
Bauman DE, Mather IH, Wall RJ, Lock AL (2006). Major advances associated with the biosynthesis of milk. Journal of Dairy Science 89: 1235–1243. DOI 10.3168/jds.S0022-0302(06)72192-0. [Google Scholar] [CrossRef]
Bauman DE, Perfield 2nd J W, Harvatine KJ, Baumgard LH (2008). Regulation of fat synthesis by conjugated linoleic acid: lactation and the ruminant model. Journal of Nutrition 138: 403–409. DOI 10.1093/jn/138.2.403. [Google Scholar] [CrossRef]
Beppu F, Hosokawa M, Tanaka L, Kohno H, Tanaka T, Miyashita K (2006). Potent inhibitory effect of trans9, trans11 isomer of conjugated linoleic acid on the growth of human colon cancer cells. Journal of Nutritional Biochemistry 17: 830–836. DOI 10.1016/j.jnutbio.2006.01.007. [Google Scholar] [CrossRef]
Bernard L, Leroux C, Bonnet M, Rouel J, Martin P, Chilliard Y (2005a). Expression and nutritional regulation of lipogenic genes in mammary gland and adipose tissues of lactating goats. Journal of Dairy Research 72: 250–255. DOI 10.1017/S0022029905000786. [Google Scholar] [CrossRef]
Bernard L, Leroux C, Chilliard Y 2008. Expression and nutritional regulation of lipogenic genes in the ruminant lactating mammary gland. In: Bösze Z (eds.Bioactive Components of Milk. Advances in Experimental Medicine and Biology. vol. 606, pp. 67–108. New York, NY: Springer. [Google Scholar]
Bernard L, Rouel J, Leroux C, Ferlay A, Faulconnier Y, Legrand P, Chilliard Y (2005b). Mammary lipid metabolism and milk fatty acid secretion in alpine goats fed vegetable lipids. Journal of Dairy Science 88: 1478–1489. DOI 10.3168/jds.S0022-0302(05)72816-2. [Google Scholar] [CrossRef]
Bhattacharya A, Banu J, Rahman M, Causey J, Fernandes G (2006). Biological effects of conjugated linoleic acids in health and disease. Journal of Nutritional Biochemistry 17: 789–810. DOI 10.1016/j.jnutbio.2006.02.009. [Google Scholar] [CrossRef]
Bionaz M, Chen S, Khan MJ, Loor JJ (2013). Functional role of PPARs in ruminants: potential targets for fine-tuning metabolism during growth and lactation. PPAR Research 2013: 1–28. DOI 10.1155/2013/684159. [Google Scholar] [CrossRef]
Bionaz M, Loor JJ (2008). Gene networks driving bovine milk fat synthesis during the lactation cycle. BMC Genomics 9: 366. DOI 10.1186/1471-2164-9-366. [Google Scholar] [CrossRef]
Bloch K, Vance D (1977). Control mechanisms in the synthesis of saturated fatty acids. Annual Review of Biochemistry 46: 263–298. DOI 10.1146/annurev.bi.46.070177.001403. [Google Scholar] [CrossRef]
Bole-Feysot C, Goffin V, Edery M, Binart N, Kelly PA (1998). Prolactin (PRL) and its receptor: actions, signal transduction pathways and phenotypes observed in PRL receptor knockout mice. Endocrine Reviews 19: 225–268. DOI 10.1210/edrv.19.3.0334. [Google Scholar] [CrossRef]
Buhman KK, Smith SJ, Stone SJ, Repa JJ, Wong JS, Knapp FF, Jr, Burri BJ, Hamilton RL, Abumrad NA, Farese RV, Jr (2002). DGAT1 is not essential for intestinal triacylglycerol absorption or chylomicron synthesis. Journal of Biological Chemistry 277: 25474–25479. DOI 10.1074/jbc.M202013200. [Google Scholar] [CrossRef]
Carey EM, Dils R (1970). Fatty acid biosynthesis. V. Purification and characterisation of fatty acid synthetase from lactating-rabbit mammary gland. Biochimica et Biophysica Acta 210: 371–387. DOI 10.1016/0005-2760(70)90033-0. [Google Scholar] [CrossRef]
Carey EM, Dils R (1972). The pattern of fatty acid synthesis in lactating rabbit mammary gland studied in vivo. Biochemical Journal 126: 1005–1007. DOI 10.1042/bj1261005. [Google Scholar] [CrossRef]
Cases S, Stone SJ, Zhou P, Yen E, Tow B, Lardizabal KD, Voelker T, Farese RV,Jr. (2001). Cloning of DGAT2, a second mammalian diacylglycerol acyltransferase, and related family members. Journal of Biological Chemistry 276: 38870–38876. DOI 10.1074/jbc.M106219200. [Google Scholar] [CrossRef]
Chen Z, Luo J, Sun S, Cao D, Shi H, Loor JJ (2017). miR-148a and miR-17-5p synergistically regulate milk TAG synthesis via PPARGC1A and PPARA in goat mammary epithelial cells. RNA Biology 14: 326–338. DOI 10.1080/15476286.2016.1276149. [Google Scholar] [CrossRef]
Chirala SS, Huang WY, Jayakumar A, Sakai K, Wakil SJ (1997). Animal fatty acid synthase: functional mapping and cloning and expression of the domain I constituent activities. Proceedings of the National Academy of Sciences of the United States of America 94: 5588–5593. DOI 10.1073/pnas.94.11.5588. [Google Scholar] [CrossRef]
Chirala SS, Wakil SJ (2004). Structure and function of animal fatty acid synthase. Lipids 39: 1045–1053. DOI 10.1007/s11745-004-1329-9. [Google Scholar] [CrossRef]
Chong BM, Reigan P, Mayle-Combs KD, Orlicky DJ, McManaman JL (2011). Determinants of adipophilin function in milk lipid formation and secretion. Trends in Endocrinology & Metabolism 22: 211–217. DOI 10.1016/j.tem.2011.04.003. [Google Scholar] [CrossRef]
Chu M, Zhao Y, Yu S, Hao Y, Zhang P, Feng Y, Zhang H, Ma D, Liu J, Cheng M, Li L, Shen W, Cao H, Li Q, Min L (2018a). MicroRNA-221 may be involved in lipid metabolism in mammary epithelial cells. The International Journal of Biochemistry & Cell Biology 97: 118–127. DOI 10.1016/j.biocel.2018.02.014. [Google Scholar] [CrossRef]
Chu M, Zhao Y, Yu S, Hao Y, Zhang P, Feng Y, Zhang H, Ma D, Liu J, Cheng M, Li L, Shen W, Cao H, Li Q, Min L (2018b). miR-15b negatively correlates with lipid metabolism in mammary epithelial cells. American Journal of Physiology—Cell Physiology 314: C43–C52. DOI 10.1152/ajpcell.00115.2017. [Google Scholar] [CrossRef]
Coleman R (2004). Enzymes of triacylglycerol synthesis and their regulation. Progress in Lipid Research 43: 134–176. DOI 10.1016/S0163-7827(03)00051-1. [Google Scholar] [CrossRef]
Corl BA, Baumgard LH, Dwyer DA, Griinari JM, Phillips BS, Bauman DE (2001). The role of Δ9-desaturase in the production of cis-9, trans-11 CLA. The Journal of Nutritional Biochemistry 12: 622–630. DOI 10.1016/S0955-2863(01)00180-2. [Google Scholar] [CrossRef]
Corl BA, Baumgard LH, Griinari JM, Delmonte P, Morehouse KM, Yurawecz MP, Bauman DE (2002). Trans-7,cis-9 CLA is synthesized endogenously by Δ9-desaturase in dairy cowsin dairy cows. Lipids 37: 681–688. DOI 10.1007/s11745-002-0949-4. [Google Scholar] [CrossRef]
Dils RR (1986). Comparative aspects of milk fat synthesis. Journal of Dairy Science 69: 904–910. DOI 10.3168/jds.S0022-0302(86)80480-5. [Google Scholar] [CrossRef]
Dostalova J (1992). Keeping of goats and the use of goat milk and meat in human nutrition, Studijni Informace Zivocisna Vyroba (CSFR) 4: 55P. [Google Scholar]
Fielding BA, Frayn KN (1998). Lipoprotein lipase and the disposition of dietary fatty acids. British Journal of Nutrition 80: 495–502. DOI 10.1017/S0007114598001585. [Google Scholar] [CrossRef]
Galina MA, Hummel JD, Sánchez M (2004). Rambouillet lambs with corn stubble or alfalfa, slow intake urea supplementation or balanced concentrate. Small Ruminant Research 53: 89–98. DOI 10.1016/j.smallrumres.2003.08.008. [Google Scholar] [CrossRef]
Garard ID (1939). Report on difference between dairy products made from cow’s milk and those made from the milk of other animals. Journal of the Association of Official Agricultural Chemists 3: 3. [Google Scholar]
German JB, Morand L, Dillard CJ, Xu R (1997). Milk fat composition: targets for alteration of function and nutrition. In: Welch RAS, Burns DJW, Davis SR, Popay AI, Prosser CG (eds.) Milk Composition, Production and Biotechnology, pp. 39–72. New York, NY: CAB International. [Google Scholar]
Glass RL, Jenness R, Lohse LW (1969). Comparative biochemical studies of milks—V. The triglyceride composition of milk fats. Comparative Biochemistry and Physiology 28: 783–786. DOI 10.1016/0010-406X(69)92113-6. [Google Scholar] [CrossRef]
Griinari JM, Corl BA, Lacy SH, Chouinard PY, Nurmela KVV, Bauman DE (2000). Conjugated linoleic acid is synthesized endogenously in lactating dairy cows by Δ9-desaturase. The Journal of Nutrition 130: 2285–2291. DOI 10.1093/jn/130.9.2285. [Google Scholar] [CrossRef]
Grunnet I, Knudsen J (1978). Medium chain acyl-thioester hydrolase activity in goat and rabbit mammary gland fatty acid synthetase complexes. Biochemical and Biophysical Research Communications 80: 745–749. DOI 10.1016/0006-291X(78)91308-6. [Google Scholar] [CrossRef]
Grunnet I, Knudsen J (1979). Fatty-acid synthesis in lactating-goat mammary gland. 1. Medium-chain fatty-acid synthesis. European Journal of Biochemistry 95: 497–502. DOI 10.1111/j.1432-1033.1979.tb12989.x. [Google Scholar] [CrossRef]
Haenlein GF (2001). Past, present, and future perspectives of small ruminant dairy research. Journal of Dairy Science 84: 2097–2115. DOI 10.3168/jds.S0022-0302(01)74655-3. [Google Scholar] [CrossRef]
Harris CA, Haas JT, Streeper RS, Stone SJ, Kumari M, Yang K, Han X, Brownell N, Gross RW, Zechner R, Farese JRV (2011). DGAT enzymes are required for triacylglycerol synthesis and lipid droplets in adipocytes. Journal of Lipid Research 52: 657–667. DOI 10.1194/jlr.M013003. [Google Scholar] [CrossRef]
Jenni S, Leibundgut M, Boehringer D, Frick C, Mikolasek B, Ban N (2007). Structure of fungal fatty acid synthase and implications for iterative substrate shuttling. Science 316: 254–261. DOI 10.1126/science.1138248. [Google Scholar] [CrossRef]
Jing W, Li C, Lu Y, Feng L (2019). MicroRNA expression profile and lipid metabolism characteristics in liver of rat undergoing high-fat diet. Biocell 43:129–138. DOI 10.32604/biocell.2019.06087. [Google Scholar] [CrossRef]
Kadegowda AK, Bionaz M, Thering B, Piperova LS, Erdman RA, Loor JJ (2009a). Identification of internal control genes for quantitative polymerase chain reaction in mammary tissue of lactating cows receiving lipid supplements. Journal of Dairy Science 92: 2007–2019. DOI 10.3168/jds.2008-1655. [Google Scholar] [CrossRef]
Kadegowda AKG, Bionaz M, Piperova LS, Erdman RA, Loor JJ (2009b). Peroxisome proliferator-activated receptor-γ activation and long-chain fatty acids alter lipogenic gene networks in bovine mammary epithelial cells to various extents. Journal of Dairy Science 92: 4276–4289. DOI 10.3168/jds.2008-1932. [Google Scholar] [CrossRef]
Knudsen J, Clark S, Dils R (1976). Purification and some properties of a medium-chain acyl-thioester hydrolase from lactating-rabbit mammary gland which terminates chain elongation in fatty acid synthesis. Biochemical Journal 160: 683–691. DOI 10.1042/bj1600683. [Google Scholar] [CrossRef]
Knudsen J, Grunnet I (1980). Primer specificity of mammalian mammary gland fatty acid synthetases. Biochemical and Biophysical Research Communications 95: 1808–1814. DOI 10.1016/S0006-291X(80)80109-4. [Google Scholar] [CrossRef]
Knudsen J, Grunnet I (1982). Transacylation as a chain-termination mechanism in fatty acid synthesis by mammalian fatty acid synthetase. Synthesis of medium-chain-length (C8-C12) acyl-CoA esters by goat mammary-gland fatty acid synthetase. Biochemical Journal 202: 139–143. DOI 10.1042/bj2020139. [Google Scholar] [CrossRef]
Kusunoki M, Tsutsumi K, Nakayama M, Kurokawa T, Nakamura T, Ogawa H, Fukuzawa Y, Morishita M, Koide T, Miyata T (2007). Relationship between serum concentrations of saturated fatty acids and unsaturated fatty acids and the homeostasis model insulin resistance index in Japanese patients with type 2 diabetes mellitus. The Journal of Medical Investigation 54: 243–247. DOI 10.2152/jmi.54.243. [Google Scholar] [CrossRef]
Lemay DG, Neville MC, Rudolph MC, Pollard KS, German JB (2007). Gene regulatory networks in lactation: identification of global principles using bioinformatics. BMC Systems Biology 1: 56. DOI 10.1186/1752-0509-1-56. [Google Scholar] [CrossRef]
Li J, Luo J, Xu H, Wang M, Zhu J, Shi H, Haile AB, Wang H, Sun Y (2015). Fatty acid synthase promoter: characterization, and transcriptional regulation by sterol regulatory element binding protein-1 in goat mammary epithelial cells. Gene 561: 157–164. DOI 10.1016/j.gene.2015.02.034. [Google Scholar] [CrossRef]
Li N, Zhao F, Wei C, Liang M, Zhang N, Wang C, Li QZ, Gao XJ (2014). Function of SREBP1 in the milk fat synthesis of dairy cow mammary epithelial cells. International Journal of Molecular Sciences 15: 16998–17013. DOI 10.3390/ijms150916998. [Google Scholar] [CrossRef]
Libertini LJ, Smith S (1978). Purification and properties of a thioesterase from lactating rat mammary gland which modifies the product specificity of fatty acid synthetase. Journal of Biological Chemistry 253: 1393–1401. DOI 10.1016/S0021-9258(17)34879-2. [Google Scholar] [CrossRef]
Lin X, Luo J, Zhang L, Wang W, Gou D (2013a). MiR-103 controls milk fat accumulation in goat (Capra hircus) mammary gland during lactation. PLoS One 8: e79258. DOI 10.1371/journal.pone.0079258. [Google Scholar] [CrossRef]
Lin X, Luo J, Zhang L, Zhu J (2013b). MicroRNAs synergistically regulate milk fat synthesis in mammary gland epithelial cells of dairy goats. Gene Expression the Journal of Liver Research 16: 1–13. DOI 10.3727/105221613X13776146743262. [Google Scholar] [CrossRef]
Livore VI, Tripodi KEJ, Uttaro AD (2007). Elongation of polyunsaturated fatty acids in trypanosomatids. FEBS Journal 274: 264–274. DOI 10.1111/j.1742-4658.2006.05581.x. [Google Scholar] [CrossRef]
Loor JJ, Ferlay A, Ollier A, Ueda K, Doreau M, Chilliard Y (2005). High-concentrate diets and polyunsaturated oils alter trans and conjugated isomers in bovine rumen, blood, and milk. Journal of Dairy Science 88: 3986–3999. DOI 10.3168/jds.S0022-0302(05)73085-X. [Google Scholar] [CrossRef]
Liu W, Lu Y, Zhang D, Shi L, Zu G et al. (2020). MicroRNA-708 inhibits the proliferation and chemoresistance of pancreatic cancer cells. Biocell 44:73–80. DOI 10.32604/biocell.2020.08613. [Google Scholar] [CrossRef]
Luna P, Bach A, Juarez M, de la Fuente MA (2008). Effect of a diet enriched in whole linseed and sunflower oil on goat milk fatty acid composition and conjugated linoleic acid isomer profile. Journal of Dairy Science 91: 20–28. DOI 10.3168/jds.2007-0447. [Google Scholar] [CrossRef]
Luo J, Shan CY, Liu LP, Wang HB, Sun XQ, Wang XQ, Wu HJ (2005). Preliminary study on effect of parity on short- and medium-chain fatty acids composition in milk of Xinong Saanen goat. Journal of Northwest Sci-Tech University of Agriculture and Forestry 33: 22–28. [Google Scholar]
Marshall MO, Knudsen J (1977). The specificity of 1-acyl-sn-glycerol 3-phosphate acyltransferase in microsomal fractions from lactating cow mammary gland towards short, medium and long chain acyl-CoA esters. Biochimica et Biophysica Acta (BBA)—Lipids and Lipid Metabolism 489: 236–241. DOI 10.1016/0005-2760(77)90142-4. [Google Scholar] [CrossRef]
Martel PM, Bingham CM, McGraw CJ, Baker CL, Morganelli PM (2006). S14 protein in breast cancer cells: direct evidence of regulation by SREBP-1c, superinduction with progestin, and effects on cell growth. Experimental Cell Research 312: 278–288. [Google Scholar]
Mashek DG, Li LO, Coleman RA (2006). Rat long-chain acyl-CoA synthetase mRNA, protein, and activity vary in tissue distribution and in response to diet. Journal of Lipid Research 47: 2004–2010. DOI 10.1194/jlr.M600150-JLR200. [Google Scholar] [CrossRef]
Massimo B, Loor JJ (2008). ACSL1, AGPAT6, FABP3, LPIN1, and SLC27A6 are the most abundant isoforms in bovine mammary tissue and their expression is affected by stage of lactation. Journal of Nutrition 138: 1019–1024. DOI 10.1093/jn/138.6.1019. [Google Scholar] [CrossRef]
Mayorek N, Grinstein I, Bar-Tana J (1989). Triacylglycerol synthesis in cultured rat hepatocytes. The rate-limiting role of diacylglycerol acyltransferase. European Journal of Biochemistry 182: 395–400. DOI 10.1111/j.1432-1033.1989.tb14844.x. [Google Scholar] [CrossRef]
McArthur MJ, Atshaves BP, Frolov A, Foxworth WD, Kier AB, Schroeder F (1999). Cellular uptake and intracellular trafficking of long chain fatty acids. Journal of Lipid Research 40: 1371–1383. DOI 10.1016/S0022-2275(20)33379-4. [Google Scholar] [CrossRef]
Meegalla RL, Billheimer JT, Cheng D (2002). Concerted elevation of acyl-coenzyme A: diacylglycerol acyltransferase (DGAT) activity through independent stimulation of mRNA expression of DGAT1 and DGAT2 by carbohydrate and insulin. Biochemical and Biophysical Research Communications 298: 317–323. DOI 10.1016/S0006-291X(02)02466-X. [Google Scholar] [CrossRef]
Menendez JA, Vazquez-Martin A, Ortega FJ, Fernandez-Real JM (2009). Fatty acid synthase: association with insulin resistance, type 2 diabetes, and cancer. Clinical Chemistry 55: 425–438. DOI 10.1373/clinchem.2008.115352. [Google Scholar] [CrossRef]
Mikkelsen J, Hojrup P, Hansen HF, Hansen JK, Knudsen J (1985). Evidence that the medium-chain acyltransferase of lactating-goat mammary-gland fatty acid synthetase is identical with the acetyl/malonyltransferase. Biochemical Journal 227: 981–985. DOI 10.1042/bj2270981. [Google Scholar] [CrossRef]
Miller SJ (2004). Cellular and physiological effects of short-chain fatty acids. Mini Reviews in Medicinal Chemistry 4: 839–845. DOI 10.2174/1389557043403288. [Google Scholar] [CrossRef]
Toral PG, Bernard L, Delavaud C, Gruffat D, Leroux C, Chilliard Y (2013). Effects of fish oil and additional starch on tissue fatty acid profile and lipogenic gene mRNA abundance in lactating goats fed a diet containing sunflower-seed oil. Animal 7: 948–956. DOI 10.1017/S1751731113000049. [Google Scholar] [CrossRef]
Mistry DH, Medrano JF (2002). Cloning and localization of the bovine and ovine lysophosphatidic acid acyltransferase (LPAAT) genes that codes for an enzyme involved in triglyceride biosynthesis. Journal of Dairy Science 85: 28–35. DOI 10.3168/jds.S0022-0302(02)74049-6. [Google Scholar] [CrossRef]
Pappenberger G, Benz J, Gsell B, Hennig M, Ruf A et al. (2010). Structure of the human fatty acid synthase KS–MAT didomain as a framework for inhibitor design. Journal of Molecular Biology 397: 508–519. DOI 10.1016/j.jmb.2010.01.066. [Google Scholar] [CrossRef]
Rock CO, Calder RB, Karim MA, Jackowski S (2000). Pantothenate kinase regulation of the intracellular concentration of coenzyme A. Journal of Biological Chemistry 275: 1377–1383. DOI 10.1074/jbc.275.2.1377. [Google Scholar] [CrossRef]
Rudolph MC, Russell TD, Webb P, Neville MC, Anderson SM (2011). Prolactin-mediated regulation of lipid biosynthesis genes in vivo in the lactating mammary epithelial cell. American Journal of Physiology—Endocrinology & Metabolism 300: E1059–E1068. DOI 10.1152/ajpendo.00083.2011. [Google Scholar] [CrossRef]
Schmitt MGJr., Soergel KH, Wood CM, Steff J (1977). Absorption of short-chain fatty acids from the human ileum. American Journal of Digestive Diseases 22: 340–347. DOI 10.1007/BF01072192. [Google Scholar] [CrossRef]
Sfeir Z, Ibrahimi A, Amri E, Grimaldi P, Abumrad N (1997). Regulation of FAT/CD36 gene expression: further evidence in support of a role of the protein in fatty acid binding/transport. Prostaglandins, Leukotrienes and Essential Fatty Acids 57: 17–21. DOI 10.1016/S0952-3278(97)90487-7. [Google Scholar] [CrossRef]
Shi H, Zhang T, Yi Y, Wang H, Luo J (2016). Long form PRLR (lPRLR) regulates genes involved in the triacylglycerol synthesis in goat mammary gland epithelial cells. Small Ruminant Research 139: 7–14. DOI 10.1016/j.smallrumres.2016.04.008. [Google Scholar] [CrossRef]
Shi H, Zhu J, Luo J, Cao W, Shi H, Yao D, Li J, Sun Y, Xu H, Yu K, Loor JJ (2015). Genes regulating lipid and protein metabolism are highly expressed in mammary gland of lactating dairy goats. Functional & Integrative Genomics 15: 309–321. DOI 10.1007/s10142-014-0420-1. [Google Scholar] [CrossRef]
Shi HB, Luo J, Yao DW, Zhu JJ, Xu HF, Shi HP, Loor JJ (2013). Peroxisome proliferator-activated receptor-gamma stimulates the synthesis of monounsaturated fatty acids in dairy goat mammary epithelial cells via the control of stearoyl-coenzyme A desaturase. Journal of Dairy Science 96: 7844–7853. DOI 10.3168/jds.2013-7105. [Google Scholar] [CrossRef]
Shi HB, Zhang CH, Xu ZA, Xu XF, Lv ZB et al. (2017a). Nuclear receptor subfamily 1 group H member 2 (LXRB) is the predominant liver X receptor subtype regulating transcription of 2 major lipogenic genes in goat primary mammary epithelial cells. Journal of Dairy Science 100: 6743–6752. DOI 10.3168/jds.2016-12510. [Google Scholar] [CrossRef]
Shi HB, Zhang CH, Zhao W, Luo J, Loor JJ (2017). Peroxisome proliferator-activated receptor delta facilitates lipid secretion and catabolism of fatty acids in dairy goat mammary epithelial cells. Journal of Dairy Science 100: 797–806. DOI 10.3168/jds.2016-11647. [Google Scholar] [CrossRef]
Shi HB, Zhao WS, Luo J, Yao DW, Sun YT, Li J, Shi HP, Loor JJ (2014). Peroxisome proliferator-activated receptor gamma1 and gamma2 isoforms alter lipogenic gene networks in goat mammary epithelial cells to different extents. Journal of Dairy Science 97: 5437–5447. DOI 10.3168/jds.2013-7863. [Google Scholar] [CrossRef]
Shingfield KJ, Ahvenjärvi S, Toivonen V, Ärölä A, Nurmela KVV, Huhtanen P, Griinari JM (2003). Effect of dietary fish oil on biohydrogenation of fatty acids and milk fatty acid content in cows. Animal Science 77: 165–179. DOI 10.1017/S1357729800053765. [Google Scholar] [CrossRef]
Smith S (2009). Conformational flexibility of metazoan fatty acid synthase enables catalysis. Nature Structural & Molecular Biology 16: 1218–1223. DOI 10.1038/nsmb.1702. [Google Scholar] [CrossRef]
Smith S, Witkowski A, Joshi AK (2003). Structural and functional organization of the animal fatty acid synthase. Progress in Lipid Research 42: 289–317. DOI 10.1016/S0163-7827(02)00067-X. [Google Scholar] [CrossRef]
Smith SJ, Cases S, Jensen DR, Chen HC, Sande E, Tow B, Sanan DA, Raber J, Eckel RH, Farese RVJr (2000). Obesity resistance and multiple mechanisms of triglyceride synthesis in mice lacking Dgat. Nature Genetics 25: 87–90. DOI 10.1038/75651. [Google Scholar] [CrossRef]
St-Gelais D, Turcot S, Ould Baba Ali A (2005). Chemical composition and properties of milk from five goat breeds. Milchwissenschaft-Milk Science International 60: 140–143. [Google Scholar]
Sun Y, Luo J, Zhu J, Shi H, Li J, Qiu S, Wang P, Loor JJ (2015). Effect of short-chain fatty acids on triacylglycerol accumulation, lipid droplet formation and lipogenic gene expression in goat mammary epithelial cells. Animal Science Journal 87: 242–249. DOI 10.1111/asj.12420. [Google Scholar] [CrossRef]
Tian H, Luo J, Shi H, Chen X, Wu J, Liang Y, Li C, Loor JJ (2020). Role of peroxisome proliferator-activated receptor-alpha on the synthesis of monounsaturated fatty acids in goat mammary epithelial cells. Journal of Animal Science 98: 433. DOI 10.1093/jas/skaa062. [Google Scholar] [CrossRef]
Tian H, Luo J, Zhang Z, Wu J, Zhang T, Busato S, Huang L, Song N, Bionaz M (2018). CRISPR/Cas9-mediated stearoyl-CoA desaturase 1 (SCD1) deficiency affects fatty acid metabolism in goat mammary epithelial cells. Journal of Agricultural and Food Chemistry 66: 10041–10052. DOI 10.1021/acs.jafc.8b03545. [Google Scholar] [CrossRef]
Ulven SM, Dalen KT, Gustafsson JA, Nebb HI (2005). LXR is crucial in lipid metabolism. Prostaglandins, Leukotrienes and Essential Fatty Acids 73: 59–63. DOI 10.1016/j.plefa.2005.04.009. [Google Scholar] [CrossRef]
Wang H, Luo J, Zhang T, Tian H, Ma Y, Xu H, Yao D, Loor JJ (2015). MicroRNA-26a/b and their host genes synergistically regulate triacylglycerol synthesis by targeting the INSIG1 gene. RNA Biology 13: 500–510. DOI 10.1080/15476286.2016.1164365. [Google Scholar] [CrossRef]
Wang H, Shi H, Luo J, Yi Y, Yao D, Zhang X, Ma G, Loor JJ (2017). MiR-145 regulates lipogenesis in goat mammary cells via targeting INSIG1 and epigenetic regulation of lipid-related genes. Journal of Cellular Physiology 232: 1030–1040. DOI 10.1002/jcp.25499. [Google Scholar] [CrossRef]
Watanabe K, Ohta Y, Toba K, Ogawa Y, Hanawa H, Hirokawa Y, Kodama M, Tanabe N, Hirono S, Ohkura Y, Nakamura Y, Kato K, Aizawa Y, Fuse I, Miyajima S, Kusano Y, Nagamoto T, Hasegawa G, Naito M (1998). Myocardial CD36 expression and fatty acid accumulation in patients with type I and II CD36 deficiency. Annals of Nuclear Medicine 12: 261–266. DOI 10.1007/BF03164911. [Google Scholar] [CrossRef]
Williams CM (2000). Dietary fatty acids and human health. Annales de Zootechnie 49: 165–180. DOI 10.1051/animres:2000116. [Google Scholar] [CrossRef]
Yang X, LU L (2020). MicroRNA-145-3p suppresses the malignant behaviors of T-cell acute lymphoblastic leukemia Jurkat cells via inhibiting the NFkappaB signaling pathway. Biocell 44:101–110. DOI 10.32604/biocell.2020.08324. [Google Scholar] [CrossRef]
Yao DW, Luo J, He QY, Wu M, Shi HB, Wang H, Wang M, Xu HF, Loor JJ (2016). Thyroid hormone responsive (THRSP) promotes the synthesis of medium-chain fatty acids in goat mammary epithelial cells. Journal of Dairy Science 99: 3124–3133. DOI 10.3168/jds.2015-10632. [Google Scholar] [CrossRef]
Yonezawa T, Yonekura S, Sanosaka M, Hagino A, Katoh K, Obara Y (2004). Octanoate stimulates cytosolic triacylglycerol accumulation and CD36 mRNA expression but inhibits Acetyl coenzyme A carboxylase activity in primary cultured bovine mammary epithelial cells. Journal of Dairy Research 71: 398–404. DOI 10.1017/S0022029904000408. [Google Scholar] [CrossRef]
Zhao WS, Hu SL, Yu K, Wang H, Wang W, Loor J, Luo J (2014). Lipoprotein lipase, tissue expression and effects on genes related to fatty acid synthesis in goat mammary epithelial cells. International Journal of Molecular Sciences 15: 22757–22771. DOI 10.3390/ijms151222757. [Google Scholar] [CrossRef]
Zhang Y, Zhang P, Zhu J, Zhang L,Zeng W (2020). MicroRNA-382 inhibits the proliferation of mouse spermatogonia by targeting Kmt5a. Biocell 44:201–207. DOI 10.32604/biocell.2020.08770. [Google Scholar] [CrossRef]
Zhu JJ, Luo J, Wang W Yu, Wang K, Shi HB, Sun YT, Lin XZ, Li J (2014). Inhibition of FAS reduces the synthesis of medium-chain fatty acids in goat mammary gland. Animal 8: 1469–1478. DOI 10.1017/S1751731114001323. [Google Scholar] [CrossRef]
Wang W, Luo J, Zhao W, Li J, Zhang X, Wang L (2010). Sequence screening of shRNA of Fas gene in Xinongsaneng sheep and construction of its adenovirus vector, Acta Agriculturae Boreali-occidentalis Sinica 10–16. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |