DOI:10.32604/biocell.2021.015178

www.techscience.com/journal/biocell
| Biocell DOI:10.32604/biocell.2021.015178 |  www.techscience.com/journal/biocell |
| Article |
Es-ATG7 is required for spermatogenesis of Eriocheir sinensis and modulates p53-dependent apoptosis in germ cells
The Sperm Laboratory, College of Life Sciences, Zhejiang University, Hangzhou, 310058, China
*Address correspondence to: Wan-xi Yang, wxyang@zju.edu.cn
Received: 27 November 2020; Accepted: 24 February 2021
Abstract: Spermatogenesis is a complicated and highly regulated male gamete differentiation process that begins with the proliferation and differentiation of spermatogonia to the release of the mature spermatozoa. The autophagy-related gene atg7 has been reported as closely related to spermatogenesis and communication of Sertoli cell-germ cells in mice, including acrosome biogenesis, sperm flagellum development, and ectoplasmic specialization assembly. However, the function of es-ATG7 and its molecular regulatory mechanism during spermatogenesis in Crustacea remain largely unknown. Here, we cloned and identified es-atg7 from the testes of the Chinese mitten crabs Eriocheir sinensis and found that the expression of es-atg7 was relatively high in testes through semi-quantitative RT-PCR. The dynamic localization of es-ATG7 detected by immunofluorescence may convey information about its role in the spermatogenesis of E. sinensis. Furthermore, a knockdown of es-atg7 revealed that the malformed sperm with irregular sperm shape or loose nuclear cup and germ cell apoptosis were increased significantly. Accompanying this, we found an up-regulated expression of es-p53 during spermatogenesis in es-atg7 knockdown groups. Altogether, our results indicate that es-ATG7 plays an essential role during spermatogenesis of E. sinensis, and we demonstrated that es-ATG7 acts as an antagonist for p53-dependent apoptosis induction in this process.
Keywords: Es-ATG7; Spermatogenesis; Es-p53; Apoptosis; Eriocheir sinensis
Spermatogenesis is an important and remarkable event for all sexually reproducing animals during their reproductive period. Spermatogenesis, as a process, is divided into three main phases: the mitotic phase, the meiotic phase, and the spermiogenesis phase. In the mitotic phase, spermatogonia proliferate to maintain their amount or differentiate to produce spermatocytes; and then enter the meiotic phase. Haploid spermatids were generated from the meiotic divisions of spermatocytes. Finally, the spermiogenesis phase where the haploid spermatids undergo complicated morphological changes and transform to mature sperm (Hermo et al., 2010; Turek, 2016). Crustaceans are unique among the invertebrates. In terms of sperm morphology, crustaceans are not only atypical in shape but also aflagellate in structure, which needs special transport mechanisms to complete their fertilization (Koch and Lambert, 1990). In Crustacea, it is similar to most animals in the process of spermatogenesis. There is the proliferation of spermatogonia, the formation of spermatocytes, where spermatids are produced through meiosis in the testis. These then enter into the process of spermiogenesis and emerge as mature sperm. Mature sperm are transported from the vasa deferentia into the spermatophores (Subramoniam, 2017). However, spermiogenesis is extensively different in various crustacean taxa because their mature sperm show species-specific morphology and structure (Tudge, 2009; Poljaroen et al., 2010). The mature sperm of the Chinese mitten crab, Eriocheir sinensis, shows the typical structure of the brachyuran sperm. They are cup-shaped with a globular acrosome cupped by the nucleus (Medina and Rodríguez, 1992).
Autophagy-related-7 (ATG7) is homologous to the ubiquitin-activating enzyme E1 (Uba1), which facilitates post-translational modifications in the ubiquitination-like process and is essential for the formation of autophagosomes (Xie and Klionsky, 2007; Walls et al., 2010). ATG7 is one of the most important members among a series of ATG proteins and is essential to autophagy regulation. Autophagy, with a rather conserved molecular machinery, is triggered by LC3, which is activated by ATG7 (Yang and Klionsky, 2009). In mammals, the acrosome is also identified as a novel lysosome-related organelle (LRO) (Berruti and Paiardi, 2011; Kang-Decker et al., 2001). Wang et al. (2014) assumed that autophagy is likely involved in acrosome biogenesis based on the possibility of the acrosome originating from a modified autolysosome. A new function of Atg7 in acrosome biogenesis was uncovered, because of almost complete infertility with aberrant acrosome formation in Atg7-mutant male mice, which also supports that the acrosome originates from the autolysosome (Wang et al., 2014). In addition, they also found a malformation in the “9+2” sperm flagellum and an inefficient cytoplasm removal in Atg7-null spermatozoa, except for the deficiency of acrosome biogenesis. Atg7 knockout in spermatids induced the accumulation of PDLIM1, a negative regulator of the organization of the cytoskeleton, which resulted in the disruption of cytoskeleton organization and the inefficient removal of cytoplasm during spermiogenesis (Shang et al., 2016). Therefore, ATG7 plays an important role in mammal spermatogenesis.
In addition, ATG7 is not only a key factor for autophagy induction but also an upstream mediator of lysosome dysfunction-induced cell death (Walls et al., 2010). During starvation, ATG7-induced autophagy plays a cytoprotective role, and ATG7 has a unique pro-apoptotic function in lysosome dysfunction-induced death. Once atg7 is knocked down, the phosphorylation of p53 declines. This causes a deficiency of ATG7 stimulating p53-dependent apoptosis (Walls et al., 2010). Mice with cartilage-specific ablation of atg7 (atg7cKO) show growth retardation associated with decreased cell proliferation and enhanced chondrocyte cell death (Vuppalapati et al., 2015). Autophagy promotes chondrocyte survival, and autophagy inhibition leads to growth retardation in mice due to caspase-dependent apoptosis of chondrocytes (Vuppalapati et al., 2015). However, previous studies reported that the crosstalk between autophagy and apoptosis is quite complicated in the regulation of cell death. Autophagy can work as a partner or an antagonist with apoptosis during cell death (Eisenberg-Lerner et al., 2009; Ullman et al., 2008). In high-fat diet (HFD)-induced obesity mice, autophagy is overactivated in the testis, and the process of spermatogenesis is impaired. When autophagy is inhibited by chloroquine (CQ), the deficiency of spermatogenesis by HFD is improved, which suggests that autophagy is closely involved in the impairment of HFD-induced spermatogenesis (Mu et al., 2017). In addition, heat stress was reported to reduce the expression of ATG7 protein in germ cells with a notable decrease of the autophagic level. Lower cell apoptosis and impaired spermatogenesis were detected (Zhang et al., 2012). Therefore, the accurate functions of ATG7 during cell death and spermatogenesis are still not fully understood, particularly in Crustacea, research in this field is almost non-existing.
The Chinese mitten crab is a suitable model for studying the molecular mechanism during spermatogenesis in Crustacea. The purpose of this study was to explore whether es-ATG7 plays a role during spermatogenesis of E. sinensis and whether it is related to apoptosis of the germ cells in the testis. To solve this problem, we obtained the full-length cDNA of the es-atg7 gene from the testis of E. sinensis. The expression pattern of es-ATG7 protein during spermatogenesis indicated that es-ATG7 may affect the process of spermatogenesis in E. sinensis. More than that, we conducted RNAi to knock down the expression of es-ATG7 by dsRNA injection and dsRNA transfection in vivo and in vitro, respectively.
Animals and sample preparation
The adult male Chinese mitten crabs were bought from Chongming Island Research Base of Shanghai Ocean University, Shanghai, China. All crabs were randomly cultured in the breeding troughs in a manner of five crabs in a group and kept the temperature about 22°C for 1 week to ensure that they are in a stable, healthy state. After the adaptation period, we selected 6 crabs from the breeding troughs and then quickly sacrificed them to collect various tissues, including heart, hepatopancreas, muscle, gill, intestine, seminal vesicle, and testis on ice. Among them, various tissues from three crabs were quickly frozen in liquid nitrogen and preserved at −80°C for RNA analysis. Testes from the remaining three crabs were immediately fixed in 4% paraformaldehyde (PFA) dissolved in phosphate-buffered saline (PBS, 0.1 M, pH 7.4) at 4°C overnight, then dehydrated orderly in 15% (for 8 h) and 25% (4 h) sucrose solution in PBS and then embedded them in O.C.T. for sectioning in the frozen state. Those samples were stored at −80°C for future experiments.
Total RNA extraction and cDNA reverse transcription
Total RNA of various tissues was extracted according to the manufacturer’s instruction of RNAiso Plus (Takara, Dalian, China). Each sample was dissolved using the RNAiso Plus through a homogenizer. The homogenate was ordinally dealt with chloroform, isopropanol, and 75% ethanol to get the RNA precipitate. Finally, we dissolved the obtained precipitate from each tissue in 100 μL RNase-free H2O. After we finished the extraction, we measured the concentration using the micro-spectrophotometer (Nano-100, Allsheng, Hangzhou, China). Some total RNA was used to prepare cDNA, and the remaining cDNA was stored at −80°C.
The PrimeScript® RT reagent Kit (Takara, Dalian, China) was used for RNA reverse transcription. The testis cDNA was used for es-atg7 gene cloning. The cDNA from different tissues was used for semi-quantitative PCR analysis of es-atg7 mRNA expression. The 5’ and 3’ end cDNA were reverse transcribed by the Smart RACE cDNA Amplification Kit (CloneTech, Dalian, China) and the 3’-Full RACE Core Set with PrimeScript™ RTase (Takara, Dalian, China) following the manufacturer’s instructions, respectively. All cDNA samples were stored at −20°C.
Cloning of es-atg7 full-length and its sequence analysis
A partial sequence of es-atg7 mRNA was obtained from our transcriptome data, and primers for es-atg7 cloning were designed by the software Primer Premier 5.0 (Tab. 1). The 2×Flash Hot Start MasterMix (CoWin Biosciences, Beijing, China) was used for PCR, and the program was run as follows: 35 cycles at 98°C for 5 s, 60°C for 10 s, and 72°C for 15 s. SanPrep column DNA gel extraction kit (Sangon Biotech, Shanghai, China) was used to purify the PCR production. Finally, the purified fragment was linked into PMD19-T-vector (Takara, Dalian, China), propagated in competent cells of Escherichia coli DH5α (Takara, Dalian, China), and then sent to Healthy Creatures Biological Technology Company for sequencing. For 3’ RACE and 5’ RACE, two forward primers (atg7-F’1311 and atg 7-F’1859) and two reverse primers (atg7-R’292 and atg7-R’228) were designed by Primer Premier 5 (Tab. 1). The Nested Touchdown PCR was performed with primers in each kit. The amplification procedures were as follows: 98°C for 5 s, 10 cycles of the touchdown program (98°C for 5 s, 65°C for 10 s, 72°C for 15 s, followed by a 0.5°C decrease of the annealing temperature per cycle), and then followed by 25 cycles (98°C for 5 s, 60°C for 10 s, and 72°C for 15 s). The same operation was conducted with the PCR product like before, and the sequencing results were acquired from the company.
The full-length sequence of the es-atg7 gene was assembled and checked by a software named Seqman (DNASTAR, Inc.). The amino acid sequence of es-ATG7 was translated through the online ExPASy translate tool (http://web.expasy.org/translate/). Multiple sequence alignment was completed in the Vector NTI10 software (Invitrogen). The secondary structure and 3-D structure of es-ATG7 were predicted with an online tool SMART (http://smart.embl-heidelberg.de/) and I-TASSER (http://zhanglab.ccmb.med.umich.edu/I-TASSER), respectively. A neighbor-joining (NJ) phylogenetic tree was constructed according to a deduced amino acid sequence using MEGA 6 software. The sequences of ATG7 used in this study were downloaded from NCBI. Their Genbank accession numbers were as follows: Homo sapiens (BC000091.2), Bos taurus (BC134445.1), Sus scrofa (HM046511.1), Mus musculus (BC058597.1), Rattus norvegicus (BC082059.1), Xenopus tropicalis (BC167363.1), Macrobrachium nipponense (MH587643.1), Locusta migratoria (KY053223.1), Penicillium chrysogenum (EF107740.1), Pichia angusta (EF102887.1), Brachionus koreanus (MH231881.1), Dugesia japonicas (KX268473.2).
Es-atg7 and es-p53 mRNA expression detected through semi-quantitative RT-PCR
The primer of es-atg7 (atg7-RT-F2 and atg7-RT-R2), es-p53 (p53-RT-F1 and p53-RT-R1), and β-actin (actin-RT-F1 and actin-RT-R1) used for semi-quantitative RT-PCT were designed by Primer 5.0 software (Tab. 1). The 2×Flash Hot Start MasterMix (CoWin Biosciences, Beijing, China) was used for PCR, and the program was run as follows: 30 cycles of 98°C for 5 s, 55°C for 10 s, and 72°C for 15 s. The Image J and GraphPad Prism 7 were used to analyze the expression of es-atg7 and es-p53 mRNA in different samples.
Antibody preparation for es-ATG7 protein
A partial sequence of es-ATG7 was selected and amplified with primers atg7-KT-F3 and atg7-KT-R3 (Tab. 1) into the expression vector pET28a. The prokaryotic expression of this recombinant plasmid was performed in BL21 strain with 0.5 mM isopropyl-β-d-thiogalactoside (IPTG). Recombinant es-ATG7 protein was purified with His-Tagged Protein Purification Kit (Inclusion Body Protein) (CoWin Biosciences, Beijing, China), and then the purified protein was dispensed into mice by intraperitoneal injection four times in a month (one time/week). In the first time of intraperitoneal injection, the purified protein (100 μL) was well blended with Complete Freunds Adjuvant (100 μL) (Sigma-Aldrich, Missouri, USA). In the remaining three times, the purified protein (100 μL) was intensively mixed with incomplete Freund adjuvant (100 μL) (Sigma-Aldrich). Five days after the fourth injection, the antibody of es-ATG7 protein was obtained from the blood serum of immunized mice.
Table 1: Primers used in the present study for es-atg7 cloning, semi-quantitative RT-PCR, and the construction of recombinant plasmids for prokaryotic expression and dsRNA synthesis

Total protein extraction and western blot
Various tissues from the Chinese crabs were ground in RIPA Lysis Buffer (Beyotime, Shanghai, China), and a 1% protease inhibitor was added. The total protein was separated by 10% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) gels and transferred to polyvinylidene fluoride (PVDF) membranes (Millipore) with 200 mA, 80 min. The membranes were firstly blocked in 5% non-fat milk in PBST buffer (0.1% Tween-20 in PBS, pH 7.2–7.4) for 2 h, and then incubated overnight in primary antibodies at 4°C (anti-ATG7 mouse polyclonal antibody (1:500), anti-p53 rabbit polyclonal antibody (1:1000, made by our lab) (Xu and Yang, 2018), or anti-ACTB rabbit polyclonal antibody (1:2000, Sangon Biotech, Shanghai, China)). Then the membranes were incubated with secondary goat-anti-rabbit HRP-conjugated antibody (1:4000, Sangon Biotech) or secondary goat-anti-mouse HRP-conjugated antibody (1:4000, Sangon Biotech) for 1 h after being washed for three times (15 min each) in PBST. After washing for three times like before, an enhanced chemiluminescent kit (Beyotime, Shanghai, China) was used to examine the protein blots through chemiluminescence imaging. β-actin was used as a control.
The prepared samples were taken out from the −80°C freezer and cut into 8 μm sections by a Cryostat microtome (Thermo Scientific, HM525 NX, Waltham, USA). The sections were permeabilized with 0.25% Triton X-100 in PBS for 15 min. Then, the sections were blocked in 5% BSA for 1 h. After that, they were incubated with es-ATG7 mouse polyclonal antibody (1:100) or es-p53 rabbit polyclonal antibody (1:100) overnight at 4°C. The negative control sections were incubated with a non-immune primary antibody (IgG, 1:100). They were washed in PBST three times and each time 15 min. Subsequently, the sections were incubated with secondary Alexa Fluor 555-conjugated donkey-anti-rabbit antibody (1:500, Beyotime) and secondary Alexa Fluor 488-conjugated donkey-anti-mouse antibody (1:500, Beyotime) for 1 h at room temperature. The sections were then incubated with DAPI (Beyotime) to stain the nucleus for 5 min after being washed in PBST three times. Finally, the sections were mounted with Antifade Mounting Medium (Beyotime), observed, and documented by taking photos with a laser scanning confocal microscope (Carl Zeiss, FV3000, Jena, Germany).
DsRNA synthesis and RNA interference (RNAi)
Two pairs of specific primers of es-atg7 and one pair of egfp negative control primers (Tab. 1) with restriction sites XbaI/KpnI were designed by Primer Premier 5 software. We cloned the target sequences into L4440 plasmid with dual T7 promoters. The recombinant plasmid was then transferred into HT115 competent cells (Weidi Biotechnology, Shanghai, China) to synthesize the dsRNA with 0.5 mM IPTG. The quality and concentration of synthetic dsRNA were examined by 1% agarose gel electrophoresis and a micro-spectrophotometer (Nano-100, Allsheng, Hangzhou, China), respectively.
Healthy adult male crabs, selected for an RNA interference experiment, were divided into four groups (PBS group, EGFP group, RNAi-1 group, and RNAi-2 group, 5 individuals in each group). In vivo injection of dsRNA on the crabs was conducted at the base of the fifth pleopod (Liu et al., 2016b). In the EGFP, RNAi-1, and RNAi-2 groups, each crab was injected with 120 μg dsRNA diluted in 120 μL RNase-free PBS every three days. In the PBS group, each crab was only injected with 120 μL RNase-free PBS every three days. And the in vivo injection experiment was continued for two weeks.
Primary germ cell culture and transfection
After thoroughly disinfecting the crabs with 1% potassium permanganate solution steeped for about 1 h, we dissected them, took out their testes, and washed them in PBS three times. Then we transferred the testes into PBS with 100 U/mL penicillin/streptomycin (Genom, Shanghai, China) for 5 min. We cultured them using 6-well cell culture plates after cutting them into small pieces of about 1 mm3. The culture medium we used was Dulbecco’s modified Eagle’s medium (DMEM; Genom, Shanghai, China) with 10% fetal bovine serum (Evergreen, Hangzhou, China) and 1% penicillin/streptomycin. The plates were placed in a 28°C stationary incubator without carbon dioxide. After 24 h, when most germ cells were released from the small tissue, we removed all tissue pieces and conducted primary germ cell cultures. Two days later, we renewed the culture medium and continued the primary germ cell culture in fresh medium for another two days. Subsequently, the primary germ cells were transfected with the dsRNA (100 pmol for each well) using Lipofectamine 8000 (Byotime, Shanghai, China). We collected all cells for the total RNA extraction, for total protein extraction, and cell smear preparation 48 h later (Wei et al., 2019).
Hematoxylin-eosin (HE) staining and the malformation rate calculation
The smeared sperm slides were fixed in 4% paraformaldehyde in PBS for 15 min. Followed by staining in hematoxylin for 15 min, washing with tap water for 5 min, color separation with 10% hydrochloric acid and 1% ammonium hydroxide for 15 s, washing with tap water for 5 min again, dehydrating with graded ethanol (70%, 80%, 90%, 95%, 100%, 100%) for 2 min each step, dyeing in eosin for 3 min and dehydrating with graded ethanol (95%, 95%, 100%, 100%) again, and finally in xylene for 5 min. Neutral balsam medium was used to mount the slides, and the pictures were captured by a light microscope (Olympus BX 40, Tokyo, Japan).
We randomly selected four pictures from the es-atg7 RNAi group and the control group, counted the total sperm and the abnormal sperm in each picture, and then calculated the malformation rate, respectively. The total number of sperms, counted in the es-atg7 RNAi group and the control group, was 661 and 1258, respectively. We indicated the results as mean ± SD.
The One Step TUNEL Apoptosis Assay Kit (Beyotime, Shanghai, China) was used for a TUNEL assay. The prepared sections (tissue sections or smeared sperm slides) were fixed in 4% PFA for 30 min and then transferred to 0.5% Triton X-100 in PBS for 5 min for permeabilized treatment. After that, the sections were washed with PBS two times, 10 min each time. Subsequently, these sections were incubated with a TUNEL reaction mixture for 1 h at 37°C in the dark, washed three times (10 min each time) in PBS, and then stained with DAPI (Beyotime, Shanghai, China) for 5 min. Finally, the sections were mounted with Antifade Mounting Medium (Beyotime, Shanghai, China), examined, and documented by taking photos with a laser scanning confocal microscope (Carl Zeiss, FV3000, Jena, Germany).
Statistical analyses were performed by the GraphPad Prism 7.0 software. A t-test was used to determine the significance between the two groups. Statistical significance was presented as *P < 0.05; **P < 0.01; ***P < 0.001, and no significance (ns). We indicated the results as means ± SEM.
Identification and characterization of the es-atg7 gene from the testis of E. sinensis
Based on the partial sequence of es-atg7 from the transcriptome data, we obtained its full-length through 3’/5’ RACE from the testis of E. sinensis. The full-length of the es-atg7 gene we obtained was 2298 bp (GenBank accession number: MT543027), which consisted of 63 bp 5’ untranslated region (UTR), 2076 bp coding sequence (CDS), and 159 bp 3’ UTR (Suppl. Fig. S1). It can be translated into the es-ATG7 protein with 691 amino acids (aa) (Suppl. Fig. S1), whose molecular weight and isoelectric point were predicted by an online ExPASy-ProtParam tool (https://web.expasy.org/protparam/) to be 76.15 kDa and 6.41, respectively. There were two important domains in this protein predicted by an online tool named SMART: the ATG7_N domain at the N-terminal from 7 aa to 319 aa and the ThiF domain at the C-terminal from 333 aa to 630 aa (Suppl. Fig. S2a). Phylogenetic analysis indicated that the es-ATG7 protein of E. sinensis formed a sister clade with its homologs of Macrobrachium nipponense (Palaemonidae, Decapoda, Crustacea), and this was in accordance with its evolutionary position (Suppl. Fig. S2b). The analysis of multiple alignments with ATG7 homologs showed that the ThiF domain at the C-terminal was relatively conserved among different species (Suppl. Fig. S3). The similarity of es-ATG7 was highest with that of M. nipponense, and their similarity was 65.28%.
Es-atg7 mRNA is highly expressed in testis of E. sinensis
In order to determine the expression of the es-atg7 gene in different tissues of this crab, we conducted a semi-quantitative RT-PCR to examine it. The results of the semi-quantitative RT-PCR showed that es-atg7 mRNA expressed in all selected tissues had the highest expression in the testis (Figs. 1a–1b).
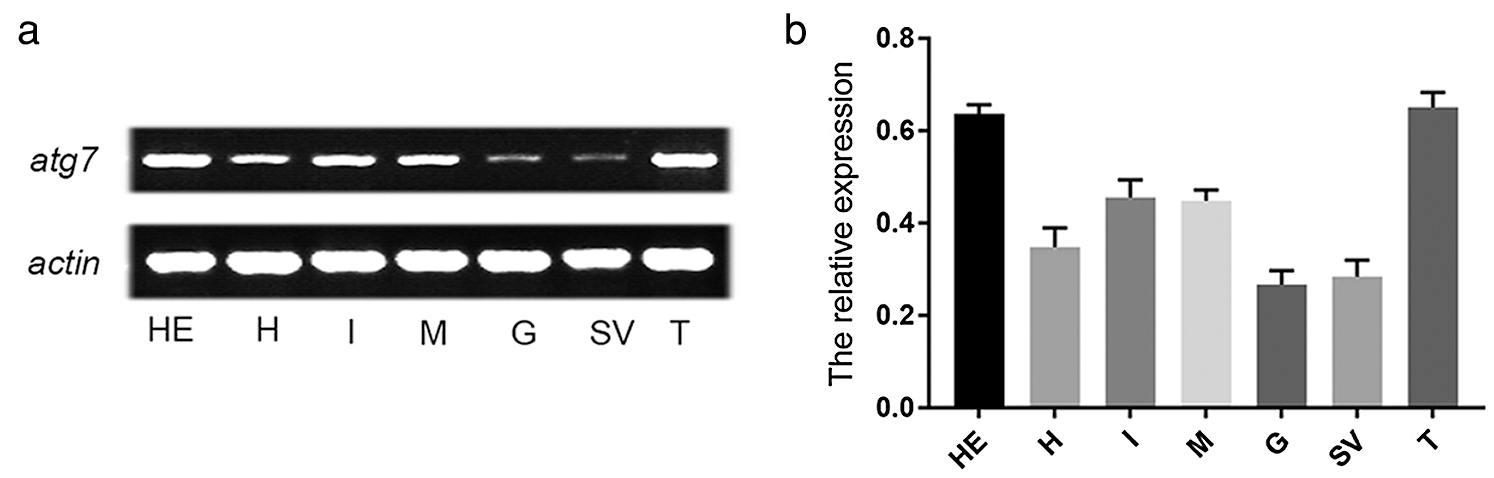
Figure 1: The expression of es-atg7 mRNA and es-ATG7 protein in different tissues of E. sinensis. (a) Semi-quantitative RT-PCR of es-atg7 mRNA in different tissues. (b) The analyses of es-atg7 expression. The expression of es-atg7 mRNA was highest in testis. H: heart, HE: hepatopancreas, M: muscle, G: gill, I: intestine, SV: seminal vesicle, and T: testis.
Distribution of es-ATG7 protein at various phases during spermatogenesis of E. sinensis
To investigate the functions of es-ATG7 during spermatogenesis of E. sinensis, we performed IF to determine its localization in different phases of the developing germ cells. In the early stage of spermatogenesis from the spermatogonial phase to the early primary spermatocyte phase, the signal of es-ATG7 was dispersed in the whole cytoplasm (Figs. 2a–2b). In the metaphase primary spermatocytes of the first meiosis, es-ATG7 signals were gathering around the nucleus (Fig. 2c). At the beginning of spermiogenesis, the es-ATG7 signal was not only detected in the cytoplasm of the early spermatids, but also in the cavity of the nucleus, where the proacrosomal vesicle was developing (Fig. 2d). With the developing spermatids, they began to further reshape. At this moment, es-ATG7 was mainly detected at the dorsal sides of the nuclei and acrosome complex (Fig. 2e). Finally, in the mature sperm, the es-ATG7 signal was just found around the acrosome complex (Fig. 2f). In the control group, no signal of es-ATG7 was detected from the spermatogonia until the mature sperm was developing (Suppl. Figs. S4a–S4e). In addition, we determined the specificity of our es-ATG7 antibody. The results showed that specific bands at 76.0 kDa and 30 kDa were obtained from the total protein of E. sinensis testis and the es-ATG7 fusion protein, respectively (Suppl. Fig. S4f). When we blocked the es-ATG7 antibody by the es-ATG7 fusion protein, the specific bands disappeared (Suppl. Fig. S4g). So, our es-ATG7 antibody was specific and effective.

Figure 2: Immunofluorescent localization of es-ATG7 during spermatogenesis in the testis of E. sinensis. (a) Spermatogonia. The signal of es-ATG7 was dispersed in the whole cytoplasm (white arrows). (b) Early primary spermatocyte. The distribution of the es-ATG7 signal is the same as that in spermatogonia (white arrows). (c) The metaphase primary spermatocytes of the first meiosis. At this time, the es-ATG7 signal was gathered around the nucleus (white arrows). (d) Early spermatid. Es-ATG7 signal was detected in the cytoplasm and in the cavity of the nucleus (white arrows). (e) Middle spermatid. Es-ATG7 was mainly detected on the dorsal sides of the nuclei and acrosome complex (white arrows). (f) Mature sperm. Es-ATG7 signal was just found around the acrosome complex (white arrows). AC: acrosomal cap, AT: acrosomal tubule, AV: acrosomal vesicle, FL: fibrous layer, LS: lamellar structure, MC: membrane complex, ML: intermediate layer, N: nucleus, NC: nuclear cup, PV: proacrosomal vesicle.
Deficiency of es-atg7 in vivo results in sperm malformation in the testes of E. sinensis
To further illustrate the role of es-ATG7 during spermatogenesis of the Chinese mitten crab, the models of es-atg7 RNAi crabs were constructed by dsRNA injection in vivo. The PBS injection group and EGFP injection group were used as control. The efficiency of es-atg7 knockdown was examined by semi-quantitative RT-PCR and western blot. Both the band results and gray analysis results revealed that the expression of es-atg7 mRNA and es-ATG7 protein were significantly declined in the RNAi groups compared to the two control groups (Figs. 3a–3d). Therefore, these two groups of es-atg7 RNAi were effective, and both were used in follow-up studies.
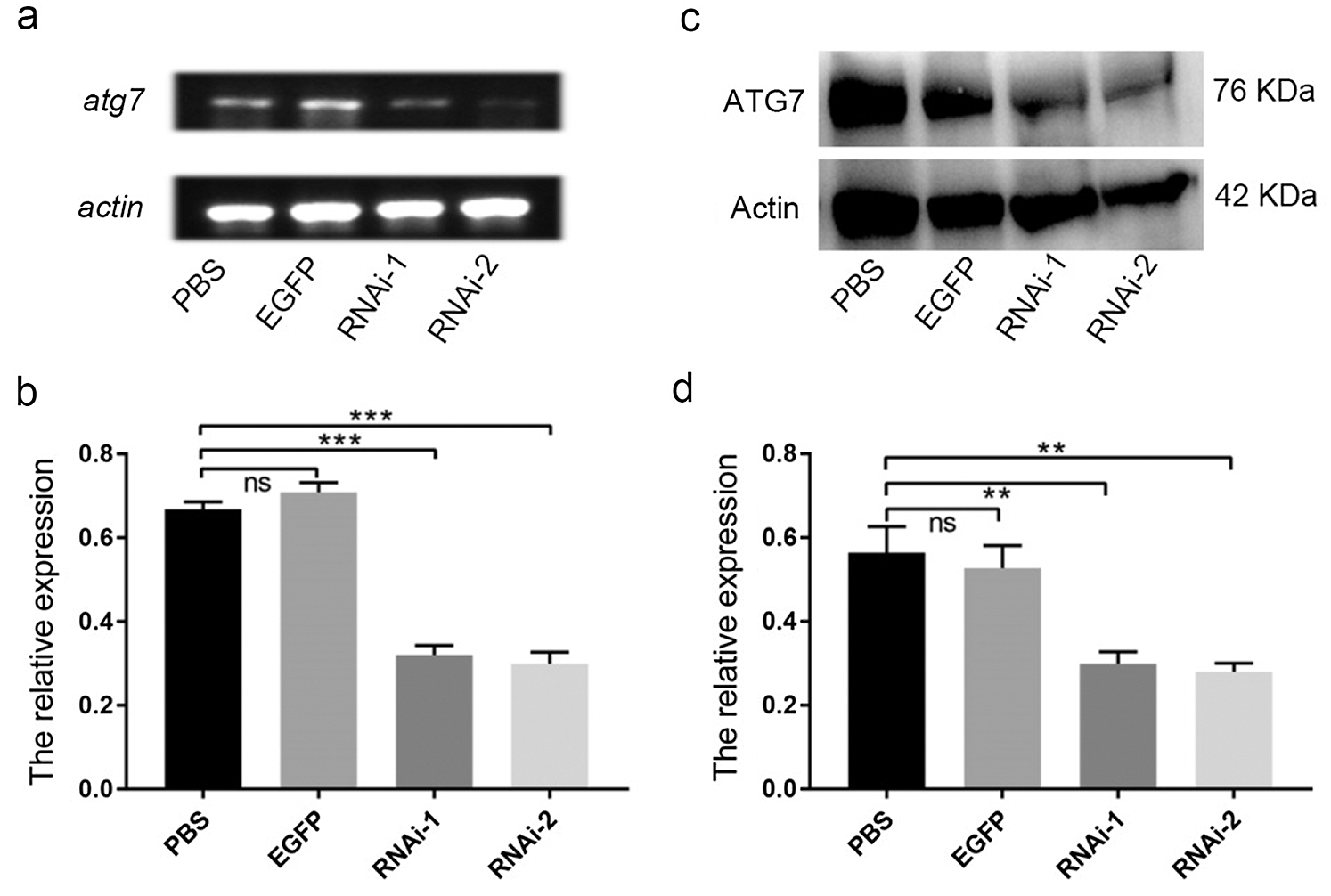
Figure 3: Knockdown efficiency of es-ATG7 by dsRNA injection in vivo. (a) The expression of es-atg7 mRNA was examined by semi-quantitative RT-PCR analysis in the testis. (b) A quantitative analysis of es-atg7 mRNA expression in the testis was determined by Image J. (c) The expression of the es-ATG7 protein was examined by Western blot in the testis. (d) A quantitative analysis of es-ATG7 protein was determined by Image J. PBS and EGFP were used as control groups, which were injected with PBS and dsRNA of egfp, respectively. RNAi-1 and RNAi-2 were injected with two dsRNA of es-atg7, respectively. β-actin was used as an internal control.
After confirming the low es-ATG7 expression in testes, we conducted morphological detection of the mature sperm through sperm smear by HE-staining. In the control group of EGFP, the sperm was perfectly circular with nuclear rings, and clear acrosomal tubes were observed from the upside and oval-shaped with nuclear cups, and clear acrosomal tubes were observed in profile (Figs. 4b, 4b1 and 4b2). However, in the es-atg7 RNAi group, some abnormal sperm were detected, most with loose nuclei and irregular sperm morphology (Figs. 4a, 4a1 and 4a2). In addition, we counted the teratosperm ratios. We found that the teratosperm ratio in the es-atg7 RNAi group was as high as 17.89%, being significantly increased when compared to the EGFP group (1.38%) (Fig. 4c). These results demonstrated, therefore, that es-ATG7 was essential for the spermatogenesis of E. sinensis, and its deficiency resulted in sperm malformation.

Figure 4: Knockdown of es-atg7 induces sperm malformation and germ cell apoptosis. (a) The histomorphological changes of E. sinensis spermatid in the RNAi group by HE-staining. (a1, a2) The enlargements of abnormal sperm (white arrows) from the RNAi group. (b) The histomorphological changes of E. sinensis spermatid in the control group by HE-staining. (b1, b2) The enlargements of normal sperm (white arrows) from the control group. Scale bar: 20 μm. (c) Quantification of the abnormal spermatozoa in a and b. (d, e) No apoptosis signal was detected in spermatogonia (d) and spermatocytes (e) in the RNAi group. (f-h) Es-atg7 knockdown resulted in apoptosis (white arrows) of the metaphase primary spermatocytes of the first meiosis (f), early spermatid (g) and middle spermatid (h). (i) No apoptosis signal was detected in mature. (j-o) No apoptosis signal was detected in each stage of spermatogenesis in the control group. Scale bar: 10 μm.
Es-atg7 knockdown in vivo induced germ cell apoptosis in the testes of E. sinensis
The lower expression of es-ATG7 could produce abnormal sperm. But did it affect germ cell apoptosis in E. sinensis? To solve this problem, the TUNEL assay was performed. The results showed that the signal of apoptosis was detected in primary spermatocytes experiencing the first meiotic division, the early spermatids and reshaping the middle spermatids after es-ATG7 was knocked down (Figs. 4f–4h), while almost no apoptosis signal was found in the spermatogonial phase, the early primary spermatocyte phase, and the mature sperm phase (Figs. 4d, 4e, and 4i). We also examined apoptosis in the control group, and no apoptosis signal was detected throughout the whole spermatogenesis process (Figs. 4j–4o). These results indicated that es-ATG7 played a key function during spermatogenesis of E. sinensis, and its deficiency led to germ cell apoptosis.
Depletion of es-atg7 in vivo up-regulates the expression of es-p53 in testis of E. sinensis
In order to further expound the mechanism on germ cell apoptosis induction related to es-atg7 knockdown, we examined the expression of the tumor-suppressor gene es-p53 by semi-RT PCR and western blot. Surprisingly, we found an obviously higher expression of both es-p53 mRNA and es-p53 protein detected in es-atg7 RNAi groups than in the control groups (Fig. 5), which suggested es-ATG7 was likely to regulate the expression of es-p53 to participate in the germ cell apoptosis process.

Figure 5: Depletion of es-ATG7 in vivo up-regulates the expression of es-p53 in the testis of E. sinensis. (a) The expression of es-p53 mRNA was examined by semi-quantitative RT-PCR analysis in the testis. (b) A quantitative analysis of es-p53 mRNA expression in the testis. (c) The expression of the es-p53 protein was examined by Western blot in the testis. (d) A quantitative analysis of es-p53 protein. PBS and EGFP were used as control groups, which were injected with PBS and dsRNA of egfp, respectively. RNAi-1 and RNAi-2 were injected with two dsRNA of es-atg7, respectively. β-actin was used as an internal control.
Es-atg7 knockdown increased the expression of es-p53 and spermatid apoptosis in vitro
To further verify our results, we carried out the primary germ cell culture from the testes of E. sinensis. We checked the cultured primary germ cells by microscopy and found that most cells were spermatids. Then we performed dsRNA transfection on primary cultured germ cells in vitro. The knockdown efficiency of es-atg7 in cultured germ cells was examined with the same methods as above, and the results testified a successful knockdown of es-atg7 in cultured germ cells (Figs. 6a–6d). The expression level of es-p53 was increased in the dsRNA transfected groups at both the transcriptional level and the translational level, which was agreed with the in vivo results (Figs. 6e–6h). Finally, we also verified germ cell apoptosis or spermatid apoptosis in the cultured cells from the testes of the crabs. Plenty of TUNEL-positive signals were presented in the dsRNA transfected groups, while they rarely appeared in the EGFP group (Figs. 6i–6l).

Figure 6: Depletion of es-ATG7 resulted in up-regulation of es-p53 and apoptosis in the primary cultured germ cells of E. sinensis. (a–b) The expression of es-atg7 mRNA was examined in the primary cultured germ cells. (c–d) The expression of the es-ATG7 protein was examined by Western blot in primary cultured germ cells. (e–f) The expression of es-p53 mRNA was determined in the primary cultured germ cells. (g–h) The expression of the es-p53 protein was determined by Western blot in the primary cultured germ cells. (i) Plenty of TUNEL-positive signals (white arrows) were detected in the primary cultured germ cells of the RNAi group. (j) The enlargements of i. (k) TUNEL-positive signals (white arrows) were rarely found in the primary cultured germ cells of the EGFP group. PBS and EGFP were used as control groups, which were injected with PBS and dsRNA of egfp, respectively. RNAi-1 and RNAi-2 were injected with two dsRNA of es-atg7, respectively. β-actin was used as an internal control. Scale bar: 10 μm.
Atg7 is a key member of a series of autophagy-related genes and indispensable for initiating the formation of autophagic vacuoles (Ravikumar et al., 2010). Although several studies focused on ATG7 participating in the regulation of autophagy, the functions of ATG7 during spermatogenesis were just reported from mice. Therefore, to fill in gaps among Crustacea, we identified and characterized es-ATG7 from the testes of E. sinensis, and our results suggest that es-ATG7 is likely to play a critical role in the spermatogenesis of Crustacea. Besides, es-atg7 knockdown up-regulates the expression of es-p53 and induces the p53-dependent apoptosis of the germ cells in E. sinensis.
Spermatogenesis is the male gamete production process and accompanied by successive cellular renewal and differentiation throughout the mitosis of spermatogonia, meiosis of spermatocyte, and spermiogenesis of spermatids (Hess and Renato de Franca, 2008, Ma et al., 2017). In atg7-null mice, the spermatozoan motility declined sharply with the defect of flagellar and cytoskeleton organization (Shang et al., 2016). It was reported that ATG7 can modulate cytoskeleton organization by the regulation of negative regulators of cytoskeleton organization in spermatids, Sertoli cells, and even cardiomyocytes (Li et al., 2016; Liu et al., 2016a). In addition, several studies also reported that the expression change of ATG7 in porcine oocytes is involved in abnormal spindle formation and rescue (Lee et al., 2014; Lin et al., 2018). In the present study, we obtained the sequence of the es-atg7 gene from the testes of the Chinese mitten crabs, and its expression in the testes is also quite high among all selected tissues suggesting an important function for es-ATG7 in the spermatogenesis of E. sinensis. Subsequently, from the results of es-ATG7 localization in different germ cell types, we found that the es-ATG7 signal gathered around the nucleus of the metaphase primary spermatocytes during the first meiosis, but not being dispersed in the whole cytoplasm like in the earlier stages. These dynamics of es-ATG7 may convey information about its role in the process of meiosis.
The acrosome is a specialized organelle that is surrounded tightly by the nuclear cup with a number of radial arms in E. sinensis (Du et al., 1988). Although the acrosomal morphology in crustaceans is quite different from that of mammals, their origination is evolutionarily conserved (Berruti and Paiardi, 2011; Du et al., 1988). In mammals, ATG7 is essential to acrosome biogenesis during spermiogenesis. A serious defect in acrosome biogenesis was observed in atg7 deficient mice. Meanwhile, ATG7 and even all autophagy molecular machinery were validated to be involved in the fusion and transportation of proacrosomal vesicles, similar to their function in the induction of autophagy (Wang et al., 2014). We found that es-ATG7 localization is related to the formation of the acrosome with the developing spermatids, suggesting its function in acrosome biogenesis.
The malformation of the nucleus is always accompanied by defected acrosomes in several studies (Kanemori et al., 2016; Liu et al., 2017). In atg7 deficient mice, a defect nucleus and acrosome were also observed together (Wang et al., 2014). There is no exception during spermiogenesis in es-atg7 knockdown of E. sinensis. We not only detected that the es-ATG7 protein localization changed with the progress of nuclear architecture but also observed abnormal shape and chromatinic state of the nucleus. Therefore, our data revealed that es-ATG7 is critical for the development of the nuclear cup during spermiogenesis of E. sinensis.
Some crosstalk mechanisms between autophagy and apoptosis have been identified through the models with ATG7 deficiency since ATG7 can trigger autophagy directly (Kroemer et al., 2010; Maiuri et al., 2007; Ravikumar et al., 2010). In other words, ATG7 is closely related to cell apoptosis to a certain degree. However, crosstalk between autophagy and apoptosis is quite complicated and remains controversial. Two kinds of opposite interplay between them were shown in different studies: partnership or antagonist (Eisenberg-Lerner et al., 2009; Zhang et al., 2012). Therefore, ATG7 is likely to perform dual functions in apoptosis induction and regulation (Han et al., 2014, Walls et al., 2010). Targeted deletion of atg7 gene in different cell lines (Gomez-Puerto et al., 2016; Liu et al., 2015; Vuppalapati et al., 2015; Yang et al., 2012), tissues (Ghavami et al., 2012; Ni et al., 2012), and diseases (Folkerts et al., 2017) has been evidenced that ATG7 induces and promotes cell apoptosis in a caspase-dependent manner or p53-dependent manner. In mouse embryonic fibroblasts with prolonged metabolic stress, ATG7 deficiency led to enhanced DNA damage with increased p53-dependent apoptosis (Lee et al., 2012). Knockdown of atg7 triggered apoptosis and accompanied by increased p53 expression in leukemic cell lines and primary human CD34+ acute myeloid leukemia cells coincided with the results in human hematopoietic stem/progenitor cells (Folkerts et al., 2017; Ghavami et al., 2012). However, some other studies show that a decreased ATG7 expression inhibits cell apoptosis and promotes cell survival (Walls et al., 2010; Wang et al., 2018). Moreover, ATG7 plays a key function in the impairment of spermatogenesis after heat treatment and in obesity-induced spermatogenesis deficiency (Mu et al., 2017; Zhang et al., 2012). We explored the role of es-ATG7 during the spermatogenesis of crustaceans for the first time. Our results indicate that es-atg7 knockdown in E. sinensis can increase germ cell apoptosis with the up-regulation of es-p53, which suggests es-ATG7 works as an antagonist in germ cell apoptosis induction during spermatogenesis of E. sinensis.
In conclusion, we identified and characterized es-ATG7 from the testes of E. sinensis for the first time. The spatial and temporal distribution of es-ATG7 during spermatogenesis of E. sinensis was examined by IF, suggesting that the dynamics of es-ATG7 may convey information about its role in the spermatogenesis of E. sinensis. Knockdown of es-atg7 augmented spermatogenesis impairment and significantly increased the production of abnormal sperm. Our study also confirmed that es-ATG7 modulated germ cell apoptosis as an antagonist with an inhibited expression of es-p53. In addition, combined with the reports in other species that we discussed above, our results showed that the functions of atg7 were likely to conserve among species in spermatogenesis and apoptosis.
Acknowledgement: The authors greatly appreciate all members of the Sperm Laboratory in Zhejiang University for their kind assistance and support.
Funding Statement: This project was supported by the National Natural Science Foundation of China (No. 32072954 and No. 41776144) and Zhejiang Province Public Welfare Technology Application Research Project (including Natural Science Foundation) (No. LGF20C120001).
Conflicts of Interest: The authors declare they have no conflict of interest.
Berruti G, Paiardi C (2014). Acrosome biogenesis: Revisiting old questions to yield new insights. Spermatogenesis 1: 95–98. DOI 10.4161/spmg.1.2.16820. [Google Scholar] [CrossRef]
Du NS, Xue LZ, Lai W (1988). Studies on the sperm of Chinese mitten-handed crab, Eriocheir sinensis (Crustacea, Decapoda) II. Spermatogenesis. Oceanologia Et Limnologia Sinica 19: 71–75. [Google Scholar]
Eisenberg-Lerner A, Bialik S, Simon HU, Kimchi A (2009). Life and death partners: Apoptosis, autophagy and the cross-talk between them. Cell Death & Differentiation 16: 966–975. DOI 10.1038/cdd.2009.33. [Google Scholar] [CrossRef]
Folkerts H, Hilgendorf S, Wierenga ATJ, Jaques J, Mulder AB, Coffer PJ, Schuringa JJ, Vellenga E (2017). Inhibition of autophagy as a treatment strategy for p53 wild-type acute myeloid leukemia. Cell Death & Disease 8: e2927. DOI 10.1038/cddis.2017.317. [Google Scholar] [CrossRef]
Ghavami S, Mutawe MM, Schaafsma D, Yeganeh B, Unruh H, Klonisch T, Halayko AJ (2012). Geranylgeranyl transferase 1 modulates autophagy and apoptosis in human airway smooth muscle. American Journal of Physiology-Lung Cellular and Molecular Physiology 302: L420–L428. DOI 10.1152/ajplung.00312.2011. [Google Scholar] [CrossRef]
Gomez-Puerto MC, Folkerts H, Wierenga AT, Schepers K, Schuringa JJ, Coffer PJ, Vellenga E (2016). Autophagy proteins ATG5 and ATG7 are essential for the maintenance of human CD34+ hematopoietic stem-progenitor cells. Stem Cells 34: 1651–1663. DOI 10.1002/stem.2347. [Google Scholar] [CrossRef]
Han J, Hou W, Goldstein LA, Stolz DB, Watkins SC, Rabinowich H (2014). A complex between Atg7 and Caspase-9: A novel mechanism of cross-regulation between autophagy and apoptosis. Journal of Biological Chemistry 289: 6485–6497. DOI 10.1074/jbc.M113.536854. [Google Scholar] [CrossRef]
Hermo L, Pelletier RM, Cyr DG, Smith CE (2010). Surfing the wave, cycle, life history, and genes/proteins expressed by testicular germ cells. Part 1: background to spermatogenesis, spermatogonia, and spermatocytes. Microscopy Research and Technique 73: 241–278. DOI 10.1002/jemt.20783. [Google Scholar] [CrossRef]
Hess RA, Renato de Franca L 2008. Spermatogenesis and cycle of the seminiferous epithelium. In: Cheng CY (eds.Molecular Mechanisms in Spermatogenesis. Advances in Experimental Medicine and Biology, vol. 636. New York, NY: Springer. [Google Scholar]
Kanemori Y, Koga Y, Sudo M, Kang W, Kashiwabara S, Ikawa M, Hasuwa H, Nagashima K, Ishikawa Y, Ogonuki N, Ogura A, Baba T (2016). Biogenesis of sperm acrosome is regulated by pre-mRNA alternative splicing of Acrbp in the mouse. Proceedings of the National Academy of Sciences of the United States of America 113: E3696–E3705. DOI 10.1073/pnas.1522333113. [Google Scholar] [CrossRef]
Kang-Decker N, Mantchev GT, Juneja SC, McNiven MA, van Deursen JM (2001). Lack of acrosome formation in Hrb-deficient mice. Science 294: 1531–1533. DOI 10.1126/science.1063665. [Google Scholar] [CrossRef]
Koch RA, Lambert CC (1990). Ultrastructure of sperm, spermiogenesis, and sperm-egg interactions in selected invertebrates and lower vertebrates which use external fertilization. Journal of Electron Microscopy Technique 16: 115–154. DOI 10.1002/jemt.1060160204. [Google Scholar] [CrossRef]
Kroemer G, Mariño G, Levine B (2010). Autophagy and the integrated stress response. Molecular Cell 40: 280–293. DOI 10.1016/j.molcel.2010.09.023. [Google Scholar] [CrossRef]
Lee IH, Kawai Y, Fergusson MM, Rovira II, Bishop AJ, Motoyama N, Cao L, Finkel T (2012). Atg7 modulates p53 activity to regulate cell cycle and survival during metabolic stress. Science 336: 225–228. DOI 10.1126/science.1218395. [Google Scholar] [CrossRef]
Lee SE, Kim EY, Choi HY, Moon JJ, Park MJ, Lee JB, Jeong CJ, Park SP (2014). Rapamycin rescues the poor developmental capacity of aged porcine oocytes. Asian-Australasian Journal of Animal Sciences 27: 635–647. DOI 10.5713/ajas.2013.13816. [Google Scholar] [CrossRef]
Li S, Liu C, Gu L, Wang L, Shang Y, Liu Q, Wan J, Shi J, Wang F, Xu Z, Ji G, Li W (2016). Autophagy protects cardiomyocytes from the myocardial ischaemia-reperfusion injury through the clearance of CLP36. Open Biology 6: 160177. DOI 10.1098/rsob.160177. [Google Scholar] [CrossRef]
Lin T, Oqani RK, Lee JE, Kang JW, Kim SY, Cho ES, Jeong YD, Baek JJ, Jin DI (2018). α-Solanine impairs oocyte maturation and quality by inducing autophagy and apoptosis and changing histone modifications in a pig model. Reproductive Toxicology 75: 96–109. DOI 10.1016/j.reprotox.2017.12.005. [Google Scholar] [CrossRef]
Liu C, DeRoo EP, Stecyk C, Wolsey M, Szuchnicki M, Hagos EG (2015). Impaired autophagy in mouse embryonic fibroblasts null for Krüppel-like Factor 4 promotes DNA damage and increases apoptosis upon serum starvation. Molecular Cancer 14: 20009. DOI 10.1186/s12943-015-0373-6. [Google Scholar] [CrossRef]
Liu C, Song Z, Wang L, Yu H, Liu W, Shang Y, Xu Z, Zhao H, Gao F, Wen J, Zhao L, Gui Y, Jiao J, Gao F, Li W (2017). Sirt1 regulates acrosome biogenesis by modulating autophagic flux during spermiogenesis in mice. Development 144: 441–451. DOI 10.1242/dev.147074. [Google Scholar] [CrossRef]
Liu C, Wang H, Shang Y, Liu W, Song Z, Zhao H, Wang L, Jia P, Gao F, Xu Z, Yang L, Gao F, Li W (2016a). Autophagy is required for ectoplasmic specialization assembly in sertoli cells. Autophagy 12: 814–832. DOI 10.1080/15548627.2016.1159377. [Google Scholar] [CrossRef]
Liu ZQ, Jiang XH, Qi HY, Xiong LW, Qiu GF (2016b). A novel SoxB2 gene is required for maturation of sperm nucleus during spermiogenesis in the Chinese mitten crab, Eriocheir sinensis. Scientific Reports 6: 240. DOI 10.1038/srep32139. [Google Scholar] [CrossRef]
Ma DD, Wang DH, Yang WX (2017). Kinesins in spermatogenesis. Reproductive Biology 96: 267–276. DOI 10.1095/biolreprod.116.144113. [Google Scholar] [CrossRef]
Maiuri MC, Zalckvar E, Kimchi A, Kroemer G (2007). Self-eating and self-killing: Crosstalk between autophagy and apoptosis. Nature Reviews Molecular Cell Biology 8: 741–752. DOI 10.1038/nrm2239. [Google Scholar] [CrossRef]
Medina A, Rodríguez A (1992). Structural changes in sperm from the fiddler crab, Uca tangeri (Crustacea, Brachyuraduring the acrosome reaction. Molecular Reproduction and Development 33: 195–201. DOI 10.1002/mrd.1080330212. [Google Scholar] [CrossRef]
Mu Y, Yan WJ, Yin TL, Zhang Y, Li J, Yang J (2017). Diet-induced obesity impairs spermatogenesis: A potential role for autophagy. Scientific Reports 7: 1. DOI 10.1038/srep43475. [Google Scholar] [CrossRef]
Ni HM, Boggess N, McGill MR, Lebofsky M, Borude P, Apte U, Jaeschke H, Ding WX (2012). Liver-specific loss of Atg5 causes persistent activation of Nrf2 and protects against acetaminophen induced liver injury. Toxicological Sciences 127: 438–450. DOI 10.1093/toxsci/kfs133. [Google Scholar] [CrossRef]
Poljaroen J, Vanichviriyakit R, Tinikul Y, Phoungpetchara I, Linthong V, Weerachatyanukul W, Sobhon P (2010). Spermatogenesis and distinctive mature sperm in the giant freshwater prawn, Macrobrachium rosenbergii (De Man, 1879). Zoologischer Anzeiger 249: 81–94. DOI 10.1016/j.jcz.2010.03.002. [Google Scholar] [CrossRef]
Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia-Arencibia M, Green-Thompson ZW, Jimenez-Sanchez M, Korolchuk VI, Lichtenberg M, Luo S, Massey DC, Menzies FM, Moreau K, Narayanan U, Renna M, Siddiqi FH, Underwood BR, Winslow AR, Rubinsztein DC (2010). Regulation of mammalian autophagy in physiology and pathophysiology. Physiological Reviews 90: 1383–1435. DOI 10.1152/physrev.00030.2009. [Google Scholar] [CrossRef]
Shang Y, Wang H, Jia P, Zhao H, Liu C, Liu W, Song Z, Xu Z, Yang L, Wang Y, Li W (2016). Autophagy regulates spermatid differentiation via degradation of PDLIM1. Autophagy 12: 1575–1592. DOI 10.1080/15548627.2016.1192750. [Google Scholar] [CrossRef]
Subramoniam T 2017. Sexual biology and reproduction in crustaceans. Chapter 11–Spermatogenesis, pp. 293–324. Academic Press. [Google Scholar]
Tudge C (2009). Spermatozoal morphology and its bearing on decapod phylogeny. Decapod crustacean phylogenetics. Crustacean 18: 101–119. [Google Scholar]
Turek P 2016. Male reproductive physiology. In: Wein AJ, Kavoussi LR, Partin AW, Peters CA (eds.Campbell-Walsh Urology (11th ed.), pp. 516–537. Philadelphia, PA: Elsevier Saunders. [Google Scholar]
Ullman E, Fan Y, Stawowczyk M, Chen HM, Yue Z, Zong WX (2008). Autophagy promotes necrosis in apoptosis-deficient cells in response to ER stress. Cell Death & Differentiation 15: 422–425. DOI 10.1038/sj.cdd.4402234. [Google Scholar] [CrossRef]
Vuppalapati KK, Bouderlique T, Newton PT, Kaminskyy VO, Wehtje H, Ohlsson C, Zhivotovsky B, Chagin AS (2015). Targeted deletion of autophagy genes Atg5 or Atg7 in the chondrocytes promotes caspase-dependent cell death and leads to mild growth retardation. Journal of Bone and Mineral Research 30: 2249–2261. DOI 10.1002/jbmr.2575. [Google Scholar] [CrossRef]
Walls KC, Ghosh AP, Franklin AV, Klocke BJ, Ballestas M, Shacka JJ, Zhang J, Roth KA (2010). Lysosome dysfunction triggers Atg7-dependent neural apoptosis. Journal of Biological Chemistry 285: 10497–10507. DOI 10.1074/jbc.M110.103747. [Google Scholar] [CrossRef]
Wang H, Wan H, Li X, Liu W, Chen Q, Wang Y, Yang L, Tang H, Zhang X, Duan E, Zhao X, Gao F, Li W (2014). Atg7 is required for acrosome biogenesis during spermatogenesis in mice. Cell Research 24: 852–869. DOI 10.1038/cr.2014.70. [Google Scholar] [CrossRef]
Wang JL, Wang JJ, Cai ZN, Xu CJ (2018). The effect of curcumin on the differentiation, apoptosis and cell cycle of neural stem cells is mediated through inhibiting autophagy by the modulation of Atg7 and p62. International Journal of Molecular Medicine 42: 2481–2488. [Google Scholar]
Wei YL, Yang T, Kovacs T, Yang WX (2019). C-terminal kinesin motor es-KIFC1 regulates nuclear formation during spermiogenesis in Chinese mitten crab Eriocheir sinensis. Gene 719: 144074. DOI 10.1016/j.gene.2019.144074. [Google Scholar] [CrossRef]
Xie Z, Klionsky DJ (2007). Autophagosome formation: core machinery and adaptations. Nature Cell Biology 9: 1102–1109. DOI 10.1038/ncb1007-1102. [Google Scholar] [CrossRef]
Xu YR, Yang WX (2018). Roles of three Es-Caspases during spermatogenesis and Cadmium-induced apoptosis in Eriocheir sinensis. Sedentary Life and Nutrition 10: 1146–1165. [Google Scholar]
Yang F, Chen WD, Deng R, Li DD, Wu KW, Feng GK, Li HJ, Zhu XF (2012). Hirsutanol A induces apoptosis and autophagy via reactive oxygen species accumulation in breast cancer MCF-7 cells. Journal of Pharmacological Sciences 119: 214–220. DOI 10.1254/jphs.11235FP. [Google Scholar] [CrossRef]
Yang Z, Klionsky DJ (2009). An overview of the molecular mechanism of autophagy. In: Levine B, Yoshimori T, Deretic V (eds.Autophagy in Infection and Immunity. Current Topics in Microbiology and Immunology, vol. 335. Berlin, Heidelberg: Springer. [Google Scholar]
Zhang M, Jiang M, Bi Y, Zhu H, Zhou Z, Sha J (2012). Autophagy and apoptosis act as partners to induce germ cell death after heat stress in mice. PLoS One 7: e41412. DOI 10.1371/journal.pone.0041412. [Google Scholar] [CrossRef]
Supplementary Data

Supplementary Figure S1: The full length of es-atg7 cDNA from Eriocheir sinensis testis. The full length of es-atg7 cDNA consists of a 63 bp 5’ untranslated region, a 159 bp 3’ untranslated region, and a 2076 bp coding sequence encoding 691 amino acids. The amino acid sequence is shown below the nucleotide sequence

Supplementary Figure S2: The prediction of major structural features of es-ATG7 and phylogenetic tree of its homologs. (a) Two main domains are shown in this figure. The N-terminus from 7 aa to 319 aa is the ATG_N domain, and the C-terminus from 333 aa to 630 aa is the ThiF. (b) The phylogenetic tree of es-ATG7 and its homologues were constructed by the neighbor-joining method through the software Mega 6. The number of bootstrap replications we used here was 1000. Es-ATG7 of E. sinensis is most closely related to that of Macrobrachium nipponense

Supplementary Figure S3: Multiple sequence alignment of es-ATG7 protein homologs

Supplementary Figure S4: The negative control for es-ATG7 during spermatogenesis in the testis of E. sinensis (a–e) and the specificity of the es-ATG7 antibody (f–g). (a) Spermatogonia. (b) Spermatocyte. (c) Early spermatid. (d) Middle spermatid. (e) Mature sperm. No red signal was detected at each stage during spermatogenesis. Scale bar: 10 μm. (f) The western blot results of the total protein of E. sinensis testis (Testis) and the fusion protein of es-ATG7 (ATG7-FP) using our es-ATG7 antibody. (g) The western blot results of the total protein of E. sinensis testis (Testis) and the fusion protein of es-ATG7 (ATG7-FP) using the blocked es-ATG7 antibody by the fusion es-ATG7 protein
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |